Introduction
Breast cancer (BC) is one of the most common types
of cancer and main causes of cancer-associated mortality in women
worldwide (1). The onset of BC
typically occurs between 20 and 60 years of age, and the incidence
of this disease has markedly increased in China in recent years
(2). Due to improvements in
diagnostic and therapeutic methods, including surgery,
radiotherapy, chemotherapy, hormone therapy, immunotherapy, stem
cell therapy and gene silencing, the mortality rate of BC has
markedly decreased (3-5). However, the molecular mechanisms of
BC remain to be fully elucidated. It is well known that an
accumulation of hereditary and epigenetic changes, including
oncogene activation, tumor suppressor gene inactivation (6), changes in intercellular complex
signal networks, microenvironment (7,8)
and epigenetic changes (9,10)
contribute to the formation of malignant tumors. An increasing
number of studies have indicated that epigenetic changes may
represent important events in the pathogenesis of BC (11-13), and that gene silencing at the
post-transcriptional level may be an important epigenetic change
(14,15).
Although prognostic microRNAs (miRNAs), including
miRNA (miR)-21, miR-489 and miR125b, have been identified in BC
(16), the underlying pathways
that regulate the invasiveness of BC remain to be elucidated.
miRNAs are able to bind to their target mRNAs to induce degradation
or suppress translation (17).
miRNAs are involved in cellular processes, including cell
differentiation, cell proliferation, apoptosis and tumor inhibition
(18-20). Previous studies have shown that
miRNAs are critical in the development and progression of BC, for
example, by functioning as tumor suppressors or oncogenes.
miR-421 is a tumor suppressor that is aberrantly
expressed in several types of cancer. For example, miR-421 inhibits
the proliferation and metabolism of prostate cancer cells by
targeting Cullin 4B (21) and
suppresses BC metastasis by targeting metastasis associated 1
(22). Furthermore, the higher
positive detection rate of miR-421 compared with serum
carcino-embryonic antigen in gastric cancer indicates that miR-421
may serve as a superior diagnostic marker (23). A number of studies have reported
that miR-421 acts as an oncogene, however, its underlying
mechanisms in BC remain to be fully elucidated.
The aim of the present study was to investigate the
regulatory role of miR-421 in BC and the underlying molecular
mechanism responsible.
Materials and methods
Patients
A total of 52 BC tissue samples were collected from
52 patients (52 females; 43-68 years old; mean age 55.6±12.5 years)
histologically diagnosed with BC between 2015 and 2017 at the
Second Affiliated Hospital of Xi’an Medical University (Xi’an,
China). In addition, 52 normal tissue samples (52 females; 44-74
years old; mean age 50.2±10.4 years) were obtained from the same
hospital. All protocols were approved by the Ethics Committee of
the Second Affiliated Hospital of Xi’an Medical University and
written informed consent was provided by all participants. The
clinical characteristics of the patients with BC are summarized in
Table I.
 | Table IAssociation between
clinicopathological factors and miR-421 levels. |
Table I
Association between
clinicopathological factors and miR-421 levels.
| Clinicopathological
factor | No. of patients
(n=52) | Level of miR-421
| P-value |
|---|
| High, n (%)
(n=28) | Low, n (%)
(n=24) |
|---|
| Age (years) | | | | <0.001 |
| <55 | 20 | 11 (39.28) | 9 (37.50) | |
| ≥55 | 32 | 17 (60.71) | 15 (62.50) | |
| Pathological
stage | | | | <0.001 |
| I+II | 38 | 20 (71.4) | 18 (75.00) | |
| III+IV | 14 | 8 (28.57) | 6 (25.00) | |
| Histological
grade | | | | <0.001 |
| I+II | 34 | 18 (64.28) | 16 (66.66) | |
| III+IV | 18 | 10 (35.71) | 8 (33.33) | |
| Lymph node
status | | | | <0.001 |
| Negative | 32 | 16 (57.14) | 16 (66.66) | |
| Positive | 20 | 12 (42.85) | 8 (33.33) | |
| ER status | | | | 0.069 |
| Negative | 26 | 15 (53.57) | 11 (45.83) | |
| Positive | 26 | 13 (46.42) | 13 (54.16) | |
| PR status | | | | 0.053 |
| Negative | 30 | 17 (60.71) | 13 (54.16) | |
| Positive | 22 | 11 (39.28) | 11 (45.83) | |
| HER-2 status | | | | 0.074 |
| Negative | 40 | 21 (75.00) | 19 (79.16) | |
| Positive | 12 | 7 (25.00) | 5 (20.83) | |
| Ki67 | | | | 0.081 |
| <15% | 29 | 17 (60.71) | 12 (50.00) | |
| ≥15% | 23 | 11 (39.28) | 12 (50.00) | |
Cell culture
The MCF-7 and MDA-MB-231 BC cells, in addition to
the normal Hs578bst cell line, were purchased from the Cell
Resource Center of the Shanghai Academy of Sciences (Shanghai,
China). The cells were cultured in an atmosphere containing 5%
CO2 at 37°C in DMEM supplemented with 10% FBS (both from
Hyclone; GE Healthcare Life Sciences, Logan, UT, USA), 100 U/ml
penicillin and 100 mg/ml streptomycin.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR) analysis
Total RNA was extracted from the BC tissues, MCF-7
cells and MDA-MB-231 cells using TRIzol (Invitrogen; Thermo Fisher
Scientific, Inc., Waltham, MA, USA) and was quantified using a
photometer. cDNA was synthesized using M-MLV reverse transcriptase
(Invitrogen; Thermo Fisher Scientific, Inc.) according to the
manufacturer’s protocol. All PCR primers were designed and
synthesized by Invitrogen; Thermo Fisher Scientific, Inc. qPCR
analysis was performed according to the manufacturer’s protocol.
The reaction mixture comprised the following: cDNA (0.5 µl),
forward primer (0.5 µl), reverse primer (0.5 µl), 2.5
mM dNTPs (2 µl), 10 U/µl DNA polymerase (0.5
µl), 5X buffer (4 µl) and ddH2O (12
µl). The thermocycling conditions were as follows: 35 cycles
at 95°C for 5 min, 95°C for 15 sec and 56°C for 40 sec. U6 was used
as an internal reference. The primers used were as follows:
miR-421, forward, 5′-ACA CTC CAG CTG GGA TCA ACA GAC ATT AAT TG-3′
and reverse, 5′-TGG TGT CGT GGA GTC G-3′; U6, forward, 5′-CTC GCT
TCG GCA GGA CA-3′ and reverse, 5′-AAC GCT TCA CGA ATT TGC GT-3′;
PDCD4, forward, 5′-TCT GGG AAA GGA AGG GGA CTA C-3′ and reverse,
5′-TTC ATA AAC ACA GTT CTC CTG GTC AT-3′; U6 forward, 5′-CTC GCT
TCG GCA GCA CA-3′ and reverse, 5′-AAC GCT TCA CGA ATT TGC GT-3′.
The results were quantified using the 2−ΔΔCq method
(24).
Immunohistochemistry
Tissues slides (1.2×1.2 cm) were incubated with
primary PDCD4 antibody (dilution 1:600; cat. no. PA5-20309;
Invitrogen; Thermo Fisher Scientific, Inc.) at 37°C for 30 min and
were washed twice with cold PBS. Subsequently, the slides were
incubated with horseradish peroxidase-conjugated goat anti-rabbit
IgG (H+L) secondary antibody (dilution 1:100; cat. no. 31460;
Invitrogen; Thermo Fisher Scientific, Inc.) for 30 min at room
temperature. The slides were developed with 3,3′-diaminobenzidine
and the nuclei were lightly stained with hematoxylin. The
expression of PDCD4 was evaluated by observing at least five fields
of the slides under an inverted non-confocal microscope (CKX53
type, 4000K LED light; Olympus Corporation, Tokyo, Japan), with at
least 100 cells per field assessed.
Cell transfection
miR-421 mimics, miR-421 inhibitors, mock miRNAs and
the luciferase reporter plasmid were designed and synthesized by
Invitrogen; Thermo Fisher Scientific, Inc. The MCF-7 and MDA-MB-231
cells were transfected with miR-421 mimics, miR-421 inhibitors or
mock miRNAs using Lipofectamine® 2000 (Invitrogen;
Thermo Fisher Scientific, Inc.) according to the manufacturer’s
protocol.
Western blot analysis
The cells were lysed and proteins were quantified
using a BCA assay. The proteins (25 µg/lane) were separated
by 10% SDS-PAGE and transferred onto PVDF membranes (Bio-Rad
Laboratories, Inc., Hercules, CA, USA). The membranes were
incubated with primary antibodies against PDCD4 (dilution 1:600;
cat. no. PA5-20309; Invitrogen; Thermo Fisher Scientific, Inc.) and
GAPDH (dilution 1:800; cat. no. MA5-15738; Invitrogen; Thermo
Fisher Scientific, Inc.) at 37°C for 30 min. The membranes were
subsequently incubated with horseradish peroxidase-conjugated goat
anti-rabbit IgG (H+L) secondary antibody. The band intensities were
evaluated using ImageJ 1.47 software (National Institutes of
Health, Bethesda, MD, USA).
Luciferase reporter assay
A search for putative targets of miR-421 was
performed with TargetScan Human7.2 software. Target genes of
miR-421 were predicted by TargetScan (http://www.targetscan.org/vert_71/). To determine
whether PDCD4 is a direct target gene of miR-421, luciferase
reported assays were performed. The luciferase reporter plasmids
pGL3-PDCD4 3′UTR wild-type (WT) and pGL3-PDCD4 3′UTR mutant type
(Mut) were designed and synthesized by Invitrogen; Thermo Fisher
Scientific, Inc. The MCF-7 and MDA-MB-231 cells were seeded in
24-well plates and cultured until they reached 40% confluence. The
cells were then transfected with miR-421 mimics, inhibitors or mock
miRNA using Lipofectamine® 2000 according to the
manufacturer’s protocol. After 24 h incubation at 37°C, the cells
were harvested and luciferase activity was assessed using the
Dual-Luciferase Reporter Assay system (Promega Corporation,
Madison, WI, USA). Each experiment was performed in triplicate.
Cell proliferation assay
Following transfection, the proliferation of the
MCF-7, MDA-MB-231 and Hs578bst cells was determined using Cell
Counting Kit-8 (Beijing Solarbio Science & Technology Co.,
Ltd., Beijing, China) according to the manufacturer’s protocol. For
each experimental group, six duplicate wells were assessed.
Cell migration and invasion assays
The MCF-7 and MDA-MB-231 cells were cultured and
treated as described above, following which cell migration and
invasion were assessed using Transwell assays. The filters (Corning
Incorporated, Corning, NY, USA) were washed in serum-free DMEM and
placed into 24-well plates. The lower Transwell chamber contained
DMEM supplemented with 10% FBS. A total of 3×104 BC
cells were seeded in the upper chamber with 200 µl DMEM
supplemented with 0.1% bovine serum albumin (BD Biosciences,
Franklin Lakes, NJ, USA), with a 2 mg/ml Matrigel-coated membrane
containing 8-m pores. The Transwell chamber was subsequently
incubated at 37°C in an atmosphere containing 5% CO2 for
24 h. Following incubation, any cells remaining on the upper
membrane surface were removed with a cotton swab, whereas cells on
the lower surface of the membrane were fixed in 10% formalin at
room temperature for 30 min and stained with 0.5% crystal violet.
Images were captured of six randomly selected fields of view using
an inverted microscope (Nikon Corporation, Otsu, Japan) at x200
magnification. For the migration assay, transfected cells
(2×104 cells/chamber) were seeded in the top chamber, as
above, without Matrigel. After 24 h, the migrated cells were lysed
in glacial acetic acid and the solution was transferred to a
96-well culture plate. Colorimetry was performed at 560 nm to
determine the optical density. Each experiment was performed in
triplicate.
Statistical analysis
All results were analyzed using SPSS 16.0 (SPSS,
Inc., Chicago, IL, USA) and GraphPad 5.0 (GraphPad Software, Inc.,
La Jolla, CA, USA). χ2 or Fisher’s exact test was used
to compare categorical variables as appropriate. Student’s t-test,
Mann-Whitney U test and one-way analysis of variance with Tukey’s
post hoc test were used to compare continuous data. P<0.05 was
considered to indicate a statistically significant difference.
Results
Patient characteristics
To investigate the expression of miR-421 in patients
with BC with different clinicopathological features, 52 patients
with BC were divided into two groups (28 in the miR-421 high
expression group and 24 in the low expression group). Patients’
characteristics are summarized in Table I. No significant differences were
observed in clinicopathological factors, including estrogen
receptor status, progesterone receptor status, human epidermal
growth factor receptor-2 status, and Ki-67 status, between the
miR-421 high expression group and miR-421 low expression group.
However, an older age at diagnosis was associated with a high
expression of miR-421 in patients with BC (P<0.001). The
pathological stage, histological grade and lymph node status were
also closely associated with the expression of miR-421
(P<0.001).
Protein expression of PDCD4 in BC
tissues
The protein expression of PDCD4 in tissues of
patients with BC were analyzed by immunohistochemistry, although
only 15 pairs of immunohistochemistry results are presented. PDCD4
protein expression was detected in three sets of five normal and
five BC tissues in each set, using immunohistochemistry (Fig. 1A-C), and was decreased
substantially in BC tissues compared with normal control
tissues.
Expression of miR-421 in BC tissues and
cell lines
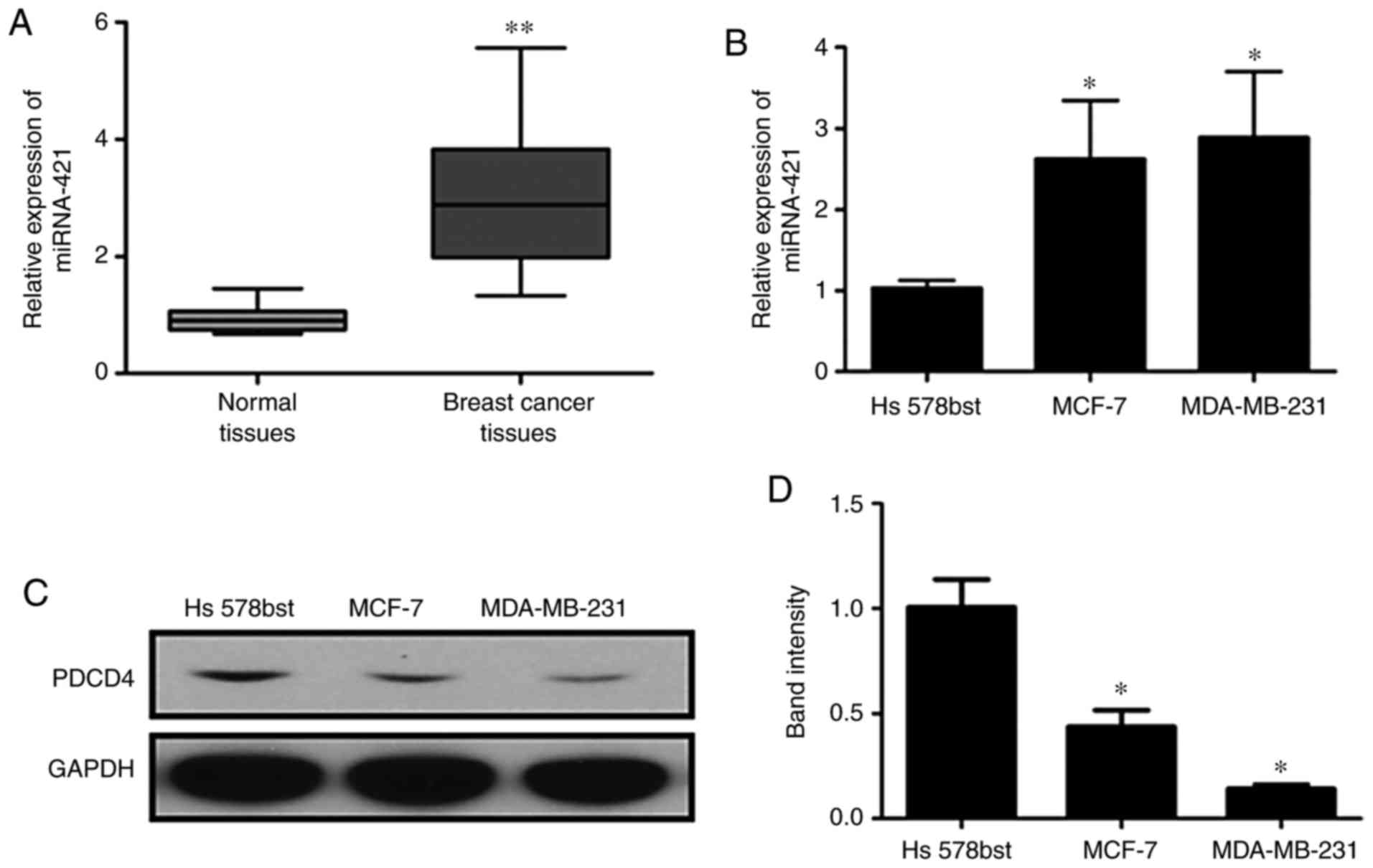
RT-qPCR analysis was performed to assess the
expression of miR-421 in BC tissues, Hs578bst cells, MCF-7 cells
and MDA-MB-231 cells (Fig. 2A and
B). The expression of miR-421 was significantly increased in
the BC tissues and cells compared with the normal controls. By
contrast, the protein expression of PDCD4 was markedly
downregulated in BC cells compared with the controls (Fig. 2C and D). These results suggested
that the over-expression of miR-421 was associated with the
downregulation of PDCD4 in BC.
PDCD4 is a direct target gene of
miR-421
PDCD4 was found as a potential target gene of
miR-421. Luciferase reporter assays were performed to further
confirm whether PDCD4 is a direct target of miR-421. The 3′-UTR of
PDCD4 mRNAs, including putative binding sites of miR-421 together
with their corresponding mutated sequences, were cloned into
vectors and co-transfected into BC cells with miR-421 mimics,
inhibitors or mock miRNA (Fig.
3A). Transfection with miR-421 mimics suppressed the luciferase
activity significantly in MCF-7 cells (Fig. 3B) and MDA-MB-231 cells (Fig. 3C); by contrast, miR-421 inhibitors
significantly enhanced the luciferase activity in MCF-7 cells
(Fig. 3B) and MDA-MB-231 cells
(Fig. 3C). The functions of the
miR-421 mimics and inhibitors were abrogated when mutated 3′-UTR
PDCD4-vector constructs were used, suggesting that miR-421 directly
targets the 3′-UTRs of PDCD4.
miR-421 promotes BC cell proliferation in
vitro
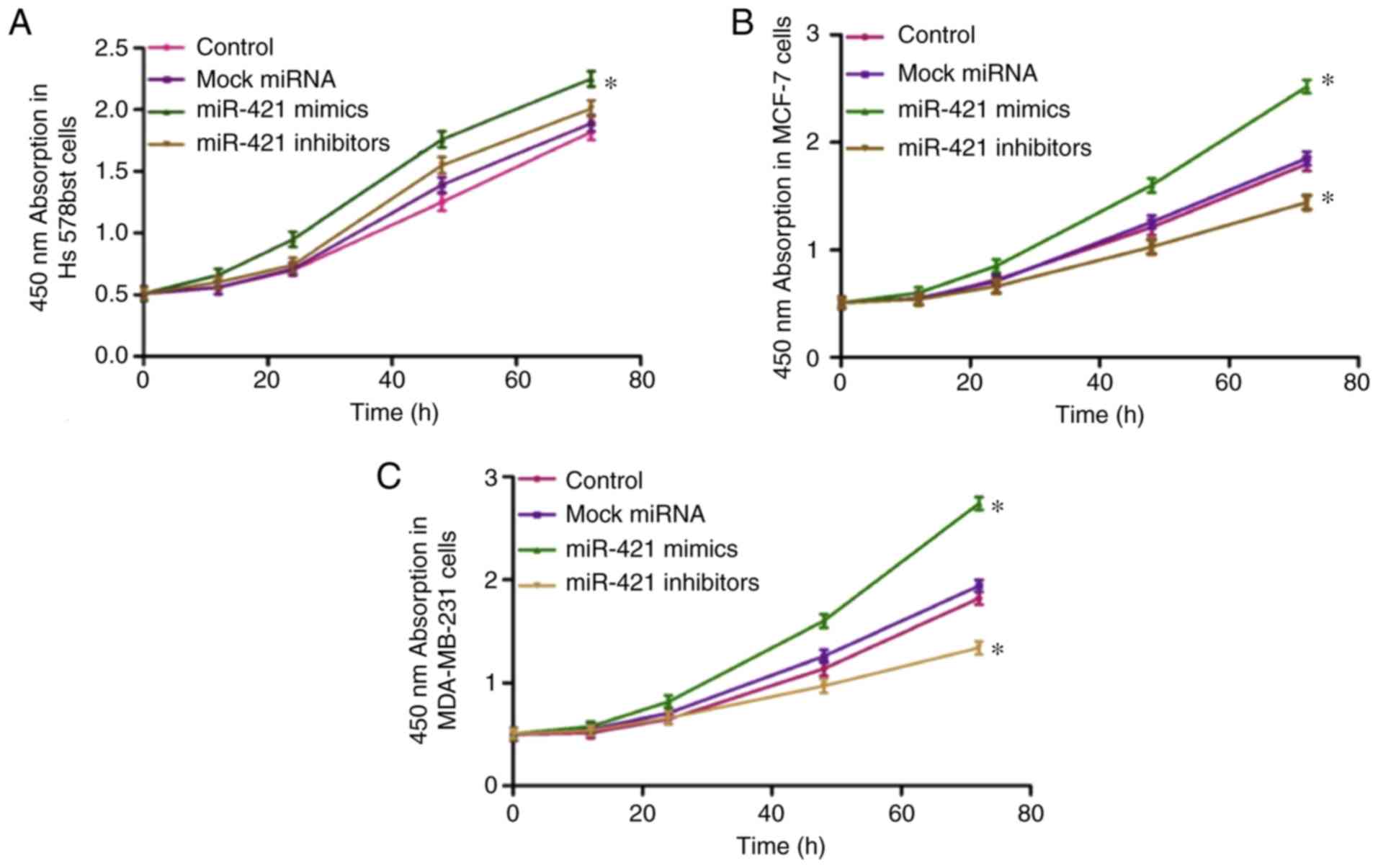
The results revealed that transfection with miR-421
mimics enhanced the proliferation of Hs578bst (Fig. 4A), MCF-7 (Fig. 4B) and MDA-MB-231 (Fig. 4C) cells; however, transfection
with miR-421 inhibitors had the opposite effect. These results
suggested that the overexpression of miR-421 accelerates BC cell
proliferation in vitro.
miR-421 promotes the migration and
invasion of BC cells
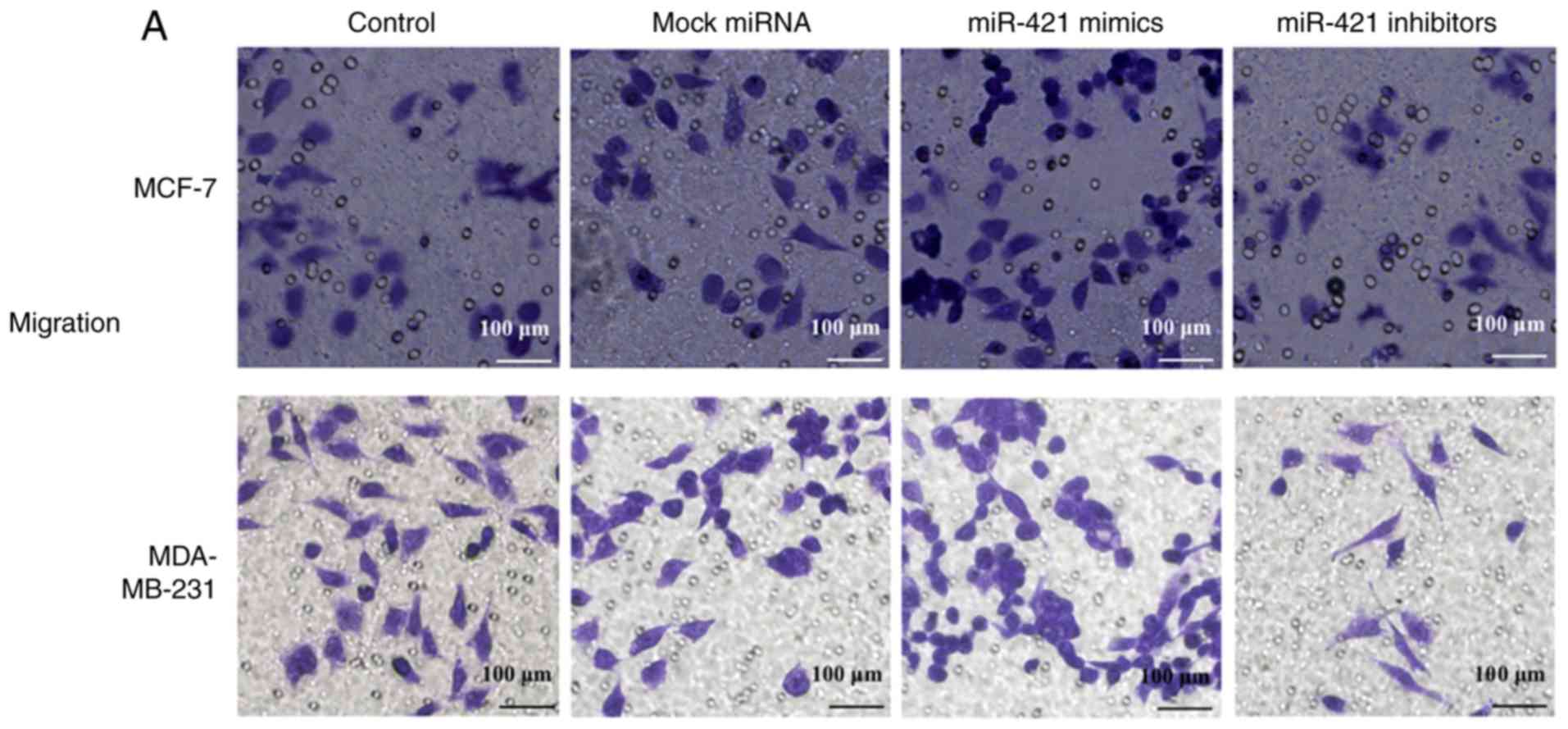
The Transwell results revealed that the
overexpression of miR-421 significantly promoted the migration of
MCF-7 and MDA-MB-231 cells, whereas miR-421 knockdown inhibited
cell migration (Fig. 5A). The
Matrigel assay revealed that the invasive abilities of the MCF-7
and MDA-MB-231 cells were enhanced following transfection with
miR-421 mimics and downregulated by miR-421 inhibitors (Fig. 5B). These results suggested that
the expression of miR-421 increases the motility of MCF-7 and
MDA-MB-231 BC cells.
miR-421 influences BC cell apoptosis
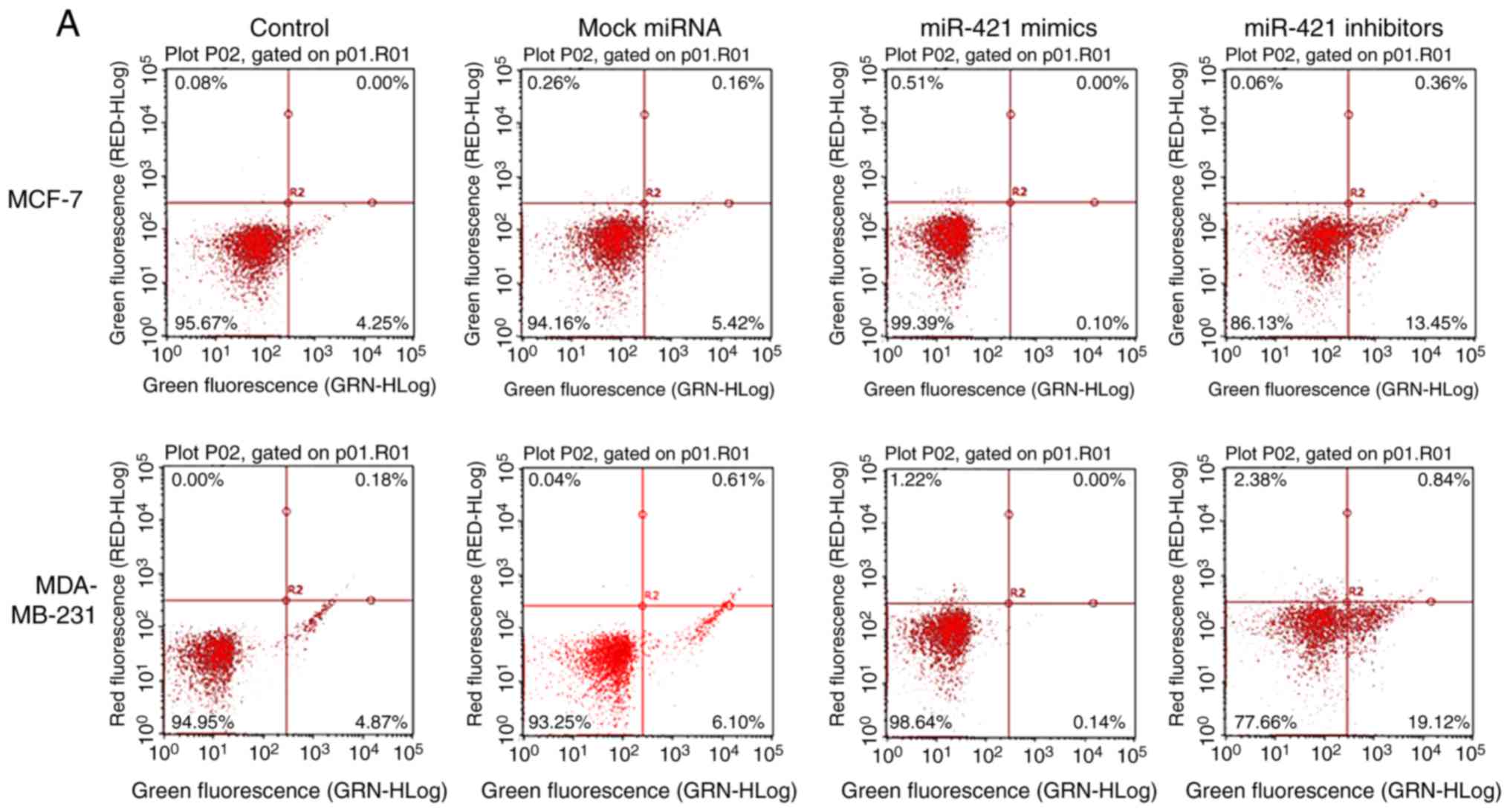
The flow cytometry results suggested that the number
of apoptotic MCF-7 and MDA-MB-231 cells (Fig. 6A) was significantly increased
following transfection with miR-421 inhibitors compared with the
control group (Fig. 6B),
suggesting that miR-421 affects the apoptosis of MCF-7, MDA-MB-231
cells via regulating PDCD4.
miR-421 regulates the protein expression
of PDCD4
The RT-qPCR and western blot analyses revealed that
the expression of miR-421 was increased in MCF-7 and MDA-MB-231
cells transfected with miR-421 mimics, but decreased in cells
transfected with miR-421 inhibitors (Fig. 7A). Furthermore, the overexpression
of miRNA-421 suppressed the expression of PDCD4 at the mRNA
(Fig. 7B) and protein (Fig. 7C and D) levels in MCF-7 and
MDA-MB-231 cells. By contrast, miR-421 knockdown upregulated the
mRNA and protein levels of PDCD4, suggesting that PDCD4 is directly
regulated by miR-421 and may function as an anticancer gene.
Discussion
miRNAs have previously been reported to be
associated with key developmental pathways, and various miRNA
disorders contribute to a number of human ailments, including
inflammatory diseases, infection, developmental disabilities and
cancer (25). Abnormal miRNA
expression has been reported in a number of types of cancer, with
miRNAs serving tumor-suppressive or oncogenic roles to modulate
tumor cell growth, cell cycle progression, migration and metastasis
(26). miR-421, which is located
in Xq13.2, has been reported to be involved in the
post-transcriptional regulation of gene expression by affecting the
stability and translation of genes in multicellular organisms,
while regulating the expression of multiple tumor-promoting genes
(27). Previous studies have
shown that miR-421 is significantly overexpressed in gastric cancer
tissues and promotes gastric cancer cell proliferation by
downregulating the expression of caspase-3 (28). Furthermore, miR-421 may act as a
tumor promoter in pancreatic cancer by targeting DPC4/Smad4
(29).
In the present study, it was found that miR-421 was
significantly upregulated in BC tissues and cells compared with
normal controls. However, miR-421 knockdown inhibited the
proliferation, migration and invasion abilities of BC cells, and
promoted apoptosis. PDCD4 was predicted as a target gene of miR-421
using TargetScan software. PDCD4 is a tumor suppressor protein,
which is able to induce apoptosis in tumor cells (30,32) by binding to the translation
initiation factor eukaryotic initiation factor-4A, inhibiting
RNA-helicase activity and in turn suppressing protein translation
(33). Furthermore, PDCD4 is able
to inhibit activator protein-1-mediated transactivation (34) and induce the expression of
cyclin-dependent kinase inhibitor p21 (35). PDCD4 is involved in tumorigenesis
and tumor progression (36,37) and is downregulated in BC (38). These results suggest that
miRNA-421 may function as an onco-miRNA in BC cells by regulating
PDCD4.
The results of the present study indicated that
miR-421 regulates the proliferation, migration, invasion and
apoptosis of BC cells by targeting PDCD4. PDCD4 is important in a
number of cellular physiological activities. In conclusion, PDCD4
may represent a novel therapeutic target for BC, and miR-421 may be
used as a potential treatment for BC.
Funding
No funding was received.
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors’ contributions
YW and ZL completed all experiments and wrote the
manuscript. JS designed and guided all experiments in the paper. In
addition, all authors revised the manuscript. All authors read and
approved the final manuscript.
Ethics approval and consent to
participate
All protocols were approved by the Ethics Committee
of the Second Affiliated Hospital of Xi’an Medical University
(Xi’an, China) and written informed consent was provided by all
participants.
Patient consent for publication
All patients involved in the present study consented
to the publication of their data.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgments
Not applicable.
References
|
1
|
Siegel R, Naishadham D and Jemal A: Cancer
statistics, 2013. CA Cancer J Clin. 63:11–30. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Breast carcinoma: Updates in molecular
profiling 2018. Clin Lab Med. 38:401–420. 2018.
|
|
3
|
Cheng CW, Yu JC, Hsieh YH, Liao WL, Shieh
JC, Yao CC, Lee HJ, Chen PM, Wu PE and Shen CY: Increased cellular
levels of microRNA-9 and microRNA-221 correlate with cancer
stemness and predict poor outcome in human breast Cancer. Cell
Physiol Biochem. 48:2205–2218. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Mansoori B, Mohammadi A, Shirjang S and
Baradaran B: HMGI-C suppressing induces P53/caspase9 axis to
regulate apoptosis in breast adenocarcinoma cells. Cell Cycle.
15:2585–2592. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Mansoori B, Mohammadi A, Goldar S,
Shanehbandi D, Mohammadnejad L, Baghbani E, Kazemi T, Kachalaki S
and Baradaran B: Silencing of high mobility group isoform IC
(HMGI-C) enhances paclitaxel chemosensitivity in breast
adenocarcinoma cells (MDA-MB-468. Adv Pharm Bull. 6:171–177. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Raval GN, Bharadwaj S, Levine EA,
Willingham MC, Geary RL, Kute T and Prasad GL: Loss of expression
of tropomyosin-1, a novel class II tumor suppressor that induces
anoikis, in primary breast tumors. Oncogene. 22:6194–203. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Volinsky N, McCarthy CJ, von Kriegsheim A,
Saban N, Okada-Hatakeyama M, Kolch W and Kholodenko BN: Signalling
mechanisms regulating phenotypic changes in breast cancer cells.
Biosci Rep. 35:e001782015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Sauter ER: Breast cancer prevention:
Current approaches and future Directions. Eur J Breast Health.
14:64–71. 2018.PubMed/NCBI
|
|
9
|
Zhuo D and Li X: Biological roles of
aberrantly expressed glycosphingolipids and related enzymes in
human cancer development and progression. Front Physiol. 9:4662018.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Bronchud MH, Tresserra F and Zantop BS:
Epigenetic changes found in uterine decidual and placental tissues
can also be found in the breast cancer microenvironment of the same
unique patient: description and potential interpretations.
Oncotarget. 9:6028–6041. 2017.
|
|
11
|
Jovanovic J, Rønneberg JA, Tost J and
Kristensen V: The epigenetics of breast cancer. Mol Oncol.
4:242–254. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Weidle UH, Dickopf S, Hintermair C,
Kollmorgen G, Birzele F and Brinkmann U: The role of micro RNAs in
breast cancer metastasis: Preclinical validation and potential
therapeutic targets. Cancer Genomics Proteomics. 15:17–39.
2018.
|
|
13
|
Byler S, Goldgar S, Heerboth S, Leary M,
Housman G, Moulton K and Sarkar S: Genetic and epigenetic aspects
of breast cancer progression and therapy. Anticancer Res.
34:1071–1077. 2014.PubMed/NCBI
|
|
14
|
Dworkin AM, Huang THM and Toland AE:
Epigenetic alterations in the breast: Implications for breast
cancer detection, prognosis and treatment. Semin Cancer Biol.
19:165–171. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Hosseinahli N, Aghapour M, Duijf PHG and
Baradaran B: Treating cancer with microRNA replacement therapy: A
literature review. J Cell Physiol. 233:5574–5588. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Sieuwerts AM, Mostert B, Bolt-de Vries J,
Peeters D, de Jongh FE, Stouthard JM, Dirix LY, van Dam PA, Van
Galen A, de Weerd V, et al: mRNA and microRNA expression profiles
in circulating tumor cells and primary tumors of metastatic breast
cancer patients. Clin Cancer Res. 17:3600–3618. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Ranji N, Sadeghizadeh M, Karimipoor M,
Shokrgozar MA and Ebrahimzadeh-Vesal R: AKT family and miRNAs
expression in IL-2 induced CD4(+) T cells. Iran J Basic Med Sci.
17:886–894. 2014.
|
|
18
|
Blondal T, Jensby Nielsen S, Baker A,
Andreasen D, Mouritzen P, Wrang Teilum M and Dahlsveen IK:
Assessing sample and miRNA profile quality in serum and plasma or
other biofluids. Methods. 59:S1–S6. 2013. View Article : Google Scholar
|
|
19
|
Ranji N, Sadeghizadeh M, Shokrgozar MA,
Bakhshandeh B, Karimipour M, Amanzadeh A and Azadmanesh K:
MiR-17-92 cluster: An apoptosis inducer or proliferation enhancer.
Mol Cell Biochem. 380:229–238. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Sarkar FH, Li Y, Wang Z, Kong D and Ali S:
Implication of microRNAs in drug resistance for designing novel
cancer therapy. Drug Resist Updat. 13:57–66. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Meng D, Yang S, Wan X, Zhang Y, Huang W,
Zhao P, Li T, Wang L, Huang Y, Li T and Li Y: A transcriptional
target of androgen receptor, miR-421 regulates proliferation and
metabolism of prostate cancer cells. Int J Biochem Cell Biol.
73:30–40. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Pan Y, Jiao G, Wang C, Yang J and Yang W:
MicroRNA-421 inhibits breast cancer metastasis by targeting
metastasis associated 1. Biomed Pharmacother. 83:1398–1406. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Jiang Z, Guo J, Xiao B, Miao Y, Huang R,
Li D and Zhang Y: Increased expression of miR-421 in human gastric
carcinoma and its clinical association. J Gastroenterol. 45:17–23.
2010. View Article : Google Scholar
|
|
24
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2-ΔΔCT method. Methods. 25:402–408. 2001. View Article : Google Scholar
|
|
25
|
Mehrgou A and Akouchekian M: Therapeutic
impacts of microRNAs in breast cancer by their roles in regulating
processes involved in this disease. J Res Med Sci. 22:1302017.
View Article : Google Scholar
|
|
26
|
Petrovic N and Ergun S: miRNAs as
potential treatment targets and treatment options in cancer. Mol
Diagn Ther. 22:157–168. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Wu J, Li G, Yao Y, Wang Z, Sun W and Wang
J: MicroRNA-421 is a new potential diagnosis biomarker with higher
sensitivity and specificity than carcinoembryonic antigen and
cancer antigen 125 in gastric cancer. Biomarker. 20:58–63. 2015.
View Article : Google Scholar
|
|
28
|
Wu JH, Yao YL, Gu T, Wang ZY, Pu XY, Sun
WW, Zhang X, Jiang YB and Wang JJ: MiR-421 regulates apoptosis of
BGC-823 gastric cancer cells by targeting caspase-3. Asian Pac J
Cancer Prev. 15:5463–5468. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Hao J, Zhang S, Zhou Y, Liu C, Hu X and
Shao C: MicroRNA-421 suppresses DPC4/Smad4 in pancreatic cancer.
Biochem Biophys Res Commun. 406:552–557. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Lankat-Buttgereit B and Göke R: The tumour
suppressor Pdcd4: recent advances in the elucidation of function
and regulation. Biol Cell. 101:309–317. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Göke R, Gregel C, Göke A, Arnold R,
Schmidt H and Lankat-Buttgereit B: Programmed cell death protein 4
(PDCD4) acts as a tumor suppressor in neuroendocrine tumor cells.
Ann N Y Acad Sci. 1014:220–221. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Yang HS, Jansen AP, Komar AA, Zheng X,
Merrick WC, Costes S, Lockett SJ, Sonenberg N and Colburn NH: The
transformation suppressor Pdcd4 is a novel eukaryotic translation
initiation factor 4A binding protein that inhibits translation. Mol
Cell Biol. 23:26–37. 2003. View Article : Google Scholar :
|
|
33
|
Yang HS, Cho MH, Zakowicz H, Hegamyer G,
Sonenberg N and Colburn NH: A novel function of the MA-3 domains in
transformation and translation suppressor Pdcd4 is essential for
its binding to eukaryotic translation initiation factor 4A. Mol
Cell Biol. 24:3894–3906. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Yang HS, Jansen AP, Nair R, Shibahara K,
Verma AK, Cmarik JL and Colburn NH: A novel transformation
suppressor, Pdcd4, inhibits AP-1 transactivation but not NF-kappaB
or ODC transactivation. Oncogene. 20:669–676. 2011. View Article : Google Scholar
|
|
35
|
Goke R, Barth P, Schmidt A, Samans B and
Lankat-Buttgereit B: Programmed cell death protein 4 suppresses
CDK1/cdc2 via induction of p21(Waf1/Cip1). Am J Physiol Cell
Physiol. 287:C1541–C1546. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Zhang Z, Wang J, Li J, Wang X and Song W:
MicroRNA-150 promotes cell proliferation, migration, and invasion
of cervical cancer through targeting PDCD4. Biomed Pharmacother.
97:511–517. 2018. View Article : Google Scholar
|
|
37
|
Schmid T, Jansen AP, Baker AR, Hegamyer G,
Hagan JP and Colburn NH: Translation inhibitor Pdcd4 is targeted
for degradation during tumor promotion. Cancer Res. 68:1254–1260.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Wen YH, Shi X, Chiriboga L, Matsahashi S,
Yee H and Afonja O: Alterations in the expression of PDCD4 in
ductal carcinoma of the breast. Oncol Rep. 18:1387–1393.
2007.PubMed/NCBI
|