Introduction
Dendritic cells (DCs) are commonly derived from the
bone marrow, spleen, lymph glands and peripheral blood. The
conventional and stable method for obtaining DCs is the cultivation
and differentiation of DC precursors (pre-DCs), which are derived
from the primary cells of the bone marrow (1). DCs serve pivotal roles in the immune
response. Immature DCs exhibit potent antigen-uptake and weak
antigen-presenting activities, whereas mature DCs possess weak
antigen-uptake and potent antigen-presenting activities (2).
B lymphocyte-induced maturation protein-1 (Blimp1)
is a transcriptional inhibitor with five zinc finger motifs that is
known to act as a master regulator of B-cell terminal
differentiation. Blimp1 modulates plasmocyte differentiation
(3,4), and regulates the development and
stability of T cells, including T helper cells and regulatory T
cells (Tregs) (5,6). In addition, Blimp1 modulates
lipopolysaccharide (LPS)-induced differentiation of primary B
lymphocytes (7), induces the
differentiation of macrophages (8) and affects the maturation of DCs by
regulating various signaling cascades (9). Furthermore, it affects stem cell
development and tissue formation by regulating sagittal axis
development and head structure formation (10,11). Although the basal expression of
Blimp1 is low in bone marrow-derived DCs, Blimp1 serves a critical
role in the tolerogenic function of DCs; diminished Blimp1
expression in DCs can result in aberrant activation of the adaptive
immune system (12-14), which may be associated with DC
differentiation and maturation.
Genetic manipulation is a notable method for
modulating the function of target cells. Each vector, non-viral or
viral, has distinct features. Although the liposome method is
conventional and cheap, it has low efficiency in primary and/or
non-dividing cells, and can alter the expression of certain genes
by affecting the physiological function of cells (15). Although electroporation has high
transfection efficiency, it causes severe cell damage. Adenoviral
vectors have high efficiency during cell infection, but can lead to
immune responses due to their immunogenicity (16,17). Lentiviral vectors have the ability
to efficiently transduce non-dividing and dividing cells by
inserting a large genetic segment in the host chromatin, in order
to sustain stable long-term transgene expression. Furthermore,
lentiviral vectors can be used to generate the induction of
pluripotent stem cells (18,19). Lentiviruses have recently been
accepted as promising transduction vectors for stem cells (20). Their applications have been
examined in bone marrow cells, which contain numerous stem cells
and precursor cells, which possess the potential to differentiate
into a wide range of cells (21).
Based on previous studies, it was hypothesized that
lentiviral-mediated Blimp1 short hairpin RNA (shRNA)
(lenti-shRNA-Blimp1) gene transduction may affect the
differentiation and maturation of DCs, and consequently regulate
subsequent antigen presentation to T cells. In the present study,
primary bone marrow cells were harvested from mice and the cells
were transduced with lenti-shRNA-Blimp1. During the differentiation
of pre-DCs to DCs, transduction efficiency, and the mRNA and
protein expression levels of Blimp1 were assessed. The effects of
Blimp1 downregulation on differentiation, maturation and
alloreactive T cell stimulation of DCs were also elucidated. In
addition, the disadvantages of this method were identified. The
findings of the present study may aid the establishment of a novel
model for generating immature DCs from marrow-derived cells, in
order to regulate immune reactions.
Materials and methods
Animals
Male C57BL/6 and BALB/C mice (weight, ~20 g; age,
6-8 weeks) were purchased from Beijing HFK Bioscience Co. Ltd.
(Beijing, China) and were maintained under specific pathogen-free
conditions (humidity, 50-60%; temperature, 18-22°C; 12-h light/dark
cycle; free access to food and water). Six C57BL/6 mice were used
for reverse transcription-quantitative polymerase chain reaction
(RT-qPCR). Six C57BL/6 mice were used for western blotting. Six
C57BL/6 mice and two BALB/C mice were used for mixed lymphocyte
reaction (MLR). Six C57BL/6 mice were used for ELISA and flow
cytometry. All experiments were approved by the Institutional
Animal Care and Use Committee at Tongji Medical College, Huazhong
University of Science and Technology (Wuhan, China; ethical
approval no. TJ-A201111211).
Culture of DCs
The culture of DCs from mouse bone marrow cells has
been reported previously (22-25). Briefly, the femurs and tibiae of
male C57BL/6 mice were removed, purified and maintained in RPMI
1640 (cat. no. SH30809.01B; HyClone; GE Healthcare, Logan, UT, USA)
supplemented with 10% inactivated fetal bovine serum (FBS; cat. no.
16000044; Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA).
Both ends were then cut and the bone marrow was flushed. The
clusters within the marrow suspension were disintegrated by
vigorous pipetting and the red blood cells were lysed using red
blood cell lysis buffer (cat. no. R1010; Beijing Solarbio Science
& Technology Co., Ltd., Beijing, China). Approximately
1-1.5×107 bone marrow-derived mononuclear cells (BMDMCs)
were obtained per femur and/or tibia. On day 0, 1×106
BMDMCs were added to each well (6-well plate) in 2.5 ml medium
consisting of complete RPMI 1640, 20 ng/ml recombinant mouse (rm)
granulocyte-macrophage colony-stimulating factor (GM-CSF; cat. no.
315-03) and 10 ng/ml rm interleukin (IL)-4 (cat. no. cat. no.
214-14; both PeproTech, Inc., Rocky Hill, NJ, USA). On day 3, the
medium containing non-adherent cells was discarded and 2.5 ml fresh
medium with supplemented GM-CSF and IL-4 was added for further
culture. On day 6, the medium containing non-adherent cells was
collected and centrifuged at 300 × g for 5 min at 25°C. The cell
pellet was resuspended and incubated in 2.5 ml RPMI 1640
supplemented with 10% inactivated FBS (25-28). On day 7, 500 ng/ml LPS (cat. no.
L2630; Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) was added to
the culture for 24 h, in order to induce DC maturation. On day 8,
cell culture was terminated and the cells were harvested for
further investigation. The morphology and proliferation of cultured
cells were observed and recorded under a microscope (Nikon Eclipse
TE2000-E; Nikon Corporation, Tokyo, Japan).
Design and synthesis of shRNA
The shRNA sequence that was designed for Blimp1
interference was 5′-GGAAAGGACCUCUACCGUUTT-3′ (29). The negative control shRNA sequence
was 5′-TTCTCCGAACGTGTCACGT-3′. The loop structure in the shRNA
template was selected as TTC AAG AGA, in order to avoid the
termination signal, and the shRNA sequence of transcriptional
termination was selected as the T6 structure. The CACC sequence was
added at the 5′-end in the sense strand, which could combine with
the sticky end of BbsI, which was produced by enzyme
digestion. The GATC sequence was added at the 5′-end in the
antisense strand, which could combine with the sticky end of
BamHI, which was produced by enzyme digestion. DNA oligos
(shRNA sequences) were dissolved in TE (pH 8.0) solution at a
concentration of 100 µM. PCR amplification was used to
complete the annealing process and the products were preserved at
4°C (30). The shRNA samples were
obtained following the annealing process. Their concentration was
10 µM and they were diluted 500 times to a final
concentration of 20 nM, which was used for ligation.
Construction of lentiviral vectors
The shRNA was ligated into the pGPU6/GFP/Neo vector
to establish the pGPU6/GFP/Neo-shRNA vector. The green fluorescent
protein (GFP) gene uses a separate CMV promoter, which is different
from the U6 promoter. For transformation, shRNA vectors (100 ng)
were mixed with 50 µl Escherichia coli (TOP10;
Invitrogen; Thermo Fisher Scientific, Inc.) and incubated in LB
culture medium containing 10 g tryptone, 5 g yeast extract and 5 g
sodium chloride per 1 l (cat. nos. LP0042B, LP0021T and LP0005B;
Thermo Fisher Scientific, Inc.) at 37°C for 90 min with constant
agitation at 200 rpm. Five positive transformant colonies were
selected on LB agar plates (cat. no. 22700041; Invitrogen; Thermo
Fisher Scientific, Inc.) containing 50 µg/ml kanamycin (cat.
no. E004000; Sigma-Aldrich; Merck KGaA) following incubation at
37°C for 14 h. The pGPU6/GFP/Neo-shRNA vector plasmids were
extracted using the alkaline lysis method and were identified by
BamHI and PstI enzyme digestion. Subsequently, 9
µg lentiviral packaging plasmid (Virapower™ Packaging Mix;
cat. no. K497500; Thermo Fisher Scientific, Inc.) and 3 µg
lentiviral vector were each diluted in 1.5 ml Opti-minimum
essential medium (MEM) culture medium (cat. no. SH30024.01;
HyClone; GE Healthcare). The two components were then mixed.
Typically, 36 µl Lipofectamine® 2000 (Invitrogen;
Thermo Fisher Scientific, Inc.) was diluted in 1.5 ml Opti-MEM
culture medium and was then mixed with the viral vector dilution
after 5 min. The mixture was incubated for 20 min at room
temperature and was then transfected into 293T cells (CRL-3216;
American Type Culture Collection, Manassas, VA, USA), which were
cultured in high-glucose Dulbecco’s Modified Eagle’s Medium (cat.
no. SH30022.01B; HyClone, GE Healthcare) supplemented with 10%
inactivated FBS (cat. no. 16000044; Gibco; Thermo Fisher
Scientific, Inc.) for 24 h at 37°C in an atmosphere containing 5%
CO2 and 95% humidity. The supernatant containing the
virus was collected between 48 and 72 h post-transfection and was
centrifuged at 300 × g for 5 min at 4°C. The cellular debris was
removed using Lenti-X™ Maxi Purification kit (cat. no. 631233;
Takara Bio Inc., Otsu, Japan) and viruses were preserved at −80°C.
The viral titer was detected in the 293T cell line. Positive
colonies were selected, and the lentiviral titer was calculated by
crystal violet staining.
Lentiviral transduction of bone marrow
cells
On culture day 1, the lentiviral solution
(1×109 TU/ml) was thawed at 37°C and added to the
cultures (80 µl/well, 8×107 TU) at a multiplicity
of infection of 100. Furthermore, polybrene was added at a final
concentration of 5 µg/ml. On day 2, the medium was replaced
to remove the residual virus, and cells were incubated with 20
ng/ml GM-CSF and 20 ng/ml IL-4.
Grouping
According to lentiviral transduction, cultured cells
were divided into the following three groups: Empty-control group,
in which the cells received no intervention; lenti-control group,
in which the cells received blank lentiviral transduction; and
lenti-shRNA-Blimp1 group, in which cells received
lenti-shRNA-Blimp1-shRNA transduction.
Detection of mRNA expression by
RT-qPCR
A total of 72 h post-transduction, total RNA was
extracted from the immature cells in each group using
TRIzol® (cat. no. 15596018; Invitrogen; Thermo Fisher
Scientific, Inc.) and RNA concentration was adjusted to 1 g/l.
β-actin, which is a housekeeping gene for normalization, served as
an internal reference. The primers used for detection of the
corresponding gene transcripts were as follows: β-actin (accession
no. NC_000071) forward, 5′-AGTGTGACGTTGACATCCGTA-3′ and reverse,
5′-GCCAGAGCAGTAATCTCCTTCT-3′; Blimp1 (accession no. NC_000076)
forward, 5′-GTATTGTCGGGACTTTGCGGAG-3′ and reverse,
5′-TCACTACTGTATTGCTTTGGGTTGC-3′; class II major histocompatibility
complex (MHC) transactivator (CIITA) (accession no. NC_000082)
forward, 5′-TGCAGGCGACCAGGAGAGACA-3′ reverse,
5′-GAAGCTGGGCACCTCAAAGAT-3′; and c-myc (accession no. NC_000081)
forward, 5′-CACCAGCAGCGACTCTGAA-3′ and reverse,
5′-GCCCGACCTCTTG-3′. Total RNA was reverse transcribed into cDNA
using the PrimeScript™ RT reagent kit (Perfect Real Time; cat. no.
RR036A; Takara Bio Inc.), according to manufacturer’s protocol,
which was then used as a template for PCR amplification. qPCR was
conducted using TransStart Top Green qPCR SuperMix (+Dye I) (cat.
no. AQ132-11; Beijing TransGen Biotech Co., Ltd., Beijing, China),
with the following reaction conditions: Pre-denaturation at 95°C
for 1 min, followed by 40 cycles of denaturation at 95°C for 15
sec, annealing at 58°C for 20 sec and extension at 72°C for 20 sec,
and a final extension step at 72°C for 5 min. Relative quantity
(RQ) was used to determine the mRNA expression levels, according to
melting curve analysis. The detection was performed in triplicate
(31).
Detection of protein expression by
western blotting
Blimp1, and its direct target proteins CIITA and
c-myc, were examined by western blotting. A total of 96 h
post-transduction, the immature cells in each group were lysed in
lysis buffer (cat. no. P0013; Beyotime Institute of Biotechnology,
Shanghai, China) and the total protein was quantified as previously
described (32,33). The protein concentration was
adjusted to 1.5 g/l and the proteins were stored at −80°C.
Subsequently, 30 µg proteins were loaded per lane, separated
by 10% SDS-PAGE, subjected to electrophoresis (80 V for 30 min and
110 V for 30 min) and electrotransferred onto a polyvinylidene
fluoride membrane at a constant current of 200 mA for 150 min. The
membrane was blocked with nonfat milk for 2 h at 37°C in order to
reduce nonspecific binding, and was then incubated with the
following primary antibodies (1:500) at 4°C overnight: Blimp1 (cat.
no. 9115; Cell Signaling Technology, Inc., Danvers, MA, USA), CIITA
(cat. no. sc-13556; Santa Cruz Biotechnology, Inc., Dallas, TX,
USA) and c-myc (cat. no. 5605; Cell Signaling Technology, Inc.).
Subsequently, the membrane was washed three times in Tris-buffered
saline-0.1% Tween-20 (15 min/wash) and incubated with secondary
antibodies (1:6,000; cat. nos. A0216 and A0208; Beyotime Institute
of Biotechnology) at 37°C for 1.5 h. The membrane was then treated
with enhanced chemiluminescence reagents (cat. no. P0018AM;
Beyotime Institute of Biotechnology), in order to visualize the
bands, which were scanned and densitometrically analyzed by ImageJ
1.6.0_20 (National Institutes of Health, Bethesda, MD, USA).
Densitometric values were normalized to β-actin (1:1,000; cat. no.
3700; Cell Signaling Technology, Inc.) immunoreactivity, in order
to correct for loading and transfer differences among the samples.
The detection procedure was repeated in triplicate.
ELISA
The concentration levels of a direct target of
Blimp1, IL-6, were detected in the culture supernatant on culture
day 7 by ELISA. The IL-6 ELISA kit (cat. no. DKW12-2060-096; Dakewe
Biotech Co., Ltd., Shenzhen, China) was used according to the
manufacturer’s protocol. The detection was repeated in
triplicate.
MLR
Lymph nodes from BALB/C mice were harvested from the
armpits, groin and mesenteric lymph nodes. Lymph nodes were minced
and filtered through a 70-µm cell strainer (cat. no. 352350;
BD Biosciences, Franklin Lakes, NJ, USA) to obtain alloreactive T
cells (22,34-36). Total T cells were stained with 2
µM PKH26 (cat. no. MINI26; Sigma-Aldrich; Merck KGaA) at
room temperature for 5 min. On day 8, the transduced C57BL/6 DCs
were harvested and co-cultured with 105 alloreactive T
cells at a ratio of 1:5 in 96-well round-bottom plates in
triplicate; non-transduced C57BL/6 DCs were used as a control.
After 96 h incubation at 37°C, 5% CO2 and 95% humidity,
T cell proliferation was measured by flow cytometry. T cells that
were cultured with non-stimulated DCs were used as a negative
control. As a positive control, at the beginning of MLR, T cells
were stimulated by cluster of differentiation (CD)3 and CD28
antibodies (1:2,000; cat. nos. 16-0032-85 and 16-0281-85;
eBiosciences; Thermo Fisher Scientific, Inc.) at 37°C for 96 h.
Flow cytometry
At the end of DC culture, the cells in each group
were centrifuged at 300 × g for 5 min. The supernatant was removed
and the cells were washed with PBS (cat. no. SH30256.01B; HyClone;
GE Healthcare) containing 0.5% bovine serum albumin (cat. no.
B2064; Sigma-Aldrich; Merck KGaA) at room temperature for 5 min,
after which they were incubated with anti-mouse CD11c, CD80, CD86,
MHC-I and MHC-II antibodies (1:100; cat. nos. 17-0114-82,
17-0801-82, 12-0860-83, 17-5958-82 and 12-0920-82; eBiosciences;
Thermo Fisher Scientific, Inc.) in the dark at 4°C for 30 min.
Subsequently, the cells were centrifuged at 300 × g for 5 min at
4°C, resuspended in 1 ml PBS, and analyzed by flow cytometry and
FlowJo 7.6 (FlowJo LLC, Ashland, OR, USA). Alloreactive T cells
were collected at the end of MLR and were stained with a CD4
antibody (cat. no. 45-0042-82; eBiosciences; Thermo Fisher
Scientific, Inc.) in 1 ml PBS in the dark at 4°C for 30 min. With
regards to intracellular staining, the cells were fixed and
permeabilized with Fixation & Permeabilization Buffer (cat. no.
88-8824-00; eBiosciences; Thermo Fisher Scientific, Inc.) at 4°C
for 30 min, stained with forkhead box P3 (Foxp3) antibodies (1:50;
cat. no. 17-5773-82; eBiosciences; Thermo Fisher Scientific, Inc.)
at 4°C for 30 min, and were then analyzed by flow cytometry. All
antibodies were diluted in PBS. Forward scatter and side scatter
were employed to gate the lymphocytes and exclude other types of
cells. Green fluorescence was detected in cells by flow cytometry
using the 530/40 filter (GFP); these cells were defined as
GFP+ cells. The proportion of CD11c+,
CD80+, CD86+, MHC-I+,
MHC-II+, CD4+, Foxp3+ and
GFP+ cells was estimated. Mean fluorescence intensity
(MFI) of MHC-II+ cells, DC transduction efficiency,
proportion of DCs, cell surface marker expression and T cell MLR
were analyzed by FlowJo 7.6 (FlowJo LLC). The procedure was
repeated in triplicate.
Statistical analysis
Data were analyzed with one-way analysis of variance
followed by a Student Newman Keuls-q post hoc test, or with a
Pearson correlation analysis using SPSS 22.0 (IBM Corporation,
Armonk, NY, USA). Data are presented as the means ± standard
deviation. P<0.05 was considered to indicate a statistically
significant difference.
Results
Lenti-shRNA-Blimp1 exhibits high
transduction efficiency in pre-DCs
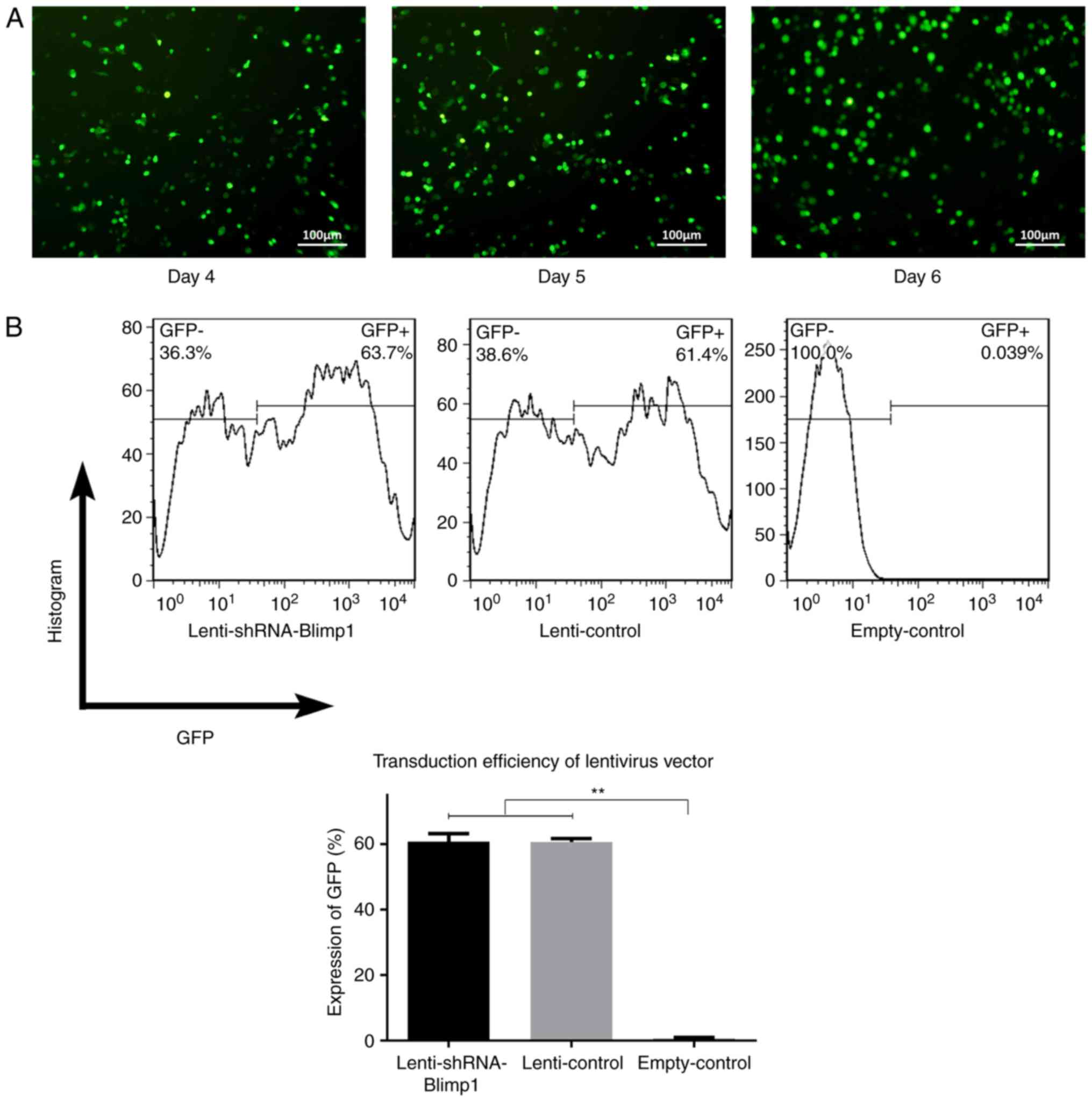
The cells demonstrated green fluorescence on day 3
following lenti-shRNA-Blimp1 gene transduction (day 4 of culture),
and its intensity gradually increased, exhibiting continuous GFP
protein expression (Fig. 1A). The
transduction efficiency was calculated as the proportion of
GFP+ cells by flow cytometry. The values were 60.83±1.39
and 60.64±0.65% in the lenti-Blimp1 and lenti-control groups
(n=3/group), respectively. No significant differences were observed
with regards to the transduction efficiency between the two groups
(Fig. 1B). No green fluorescence
was detected in the empty-control group.
Lenti-shRNA-Blimp1 gene transduction
downregulates Blimp1 expression
A total of 72 h post-transduction, the cells were
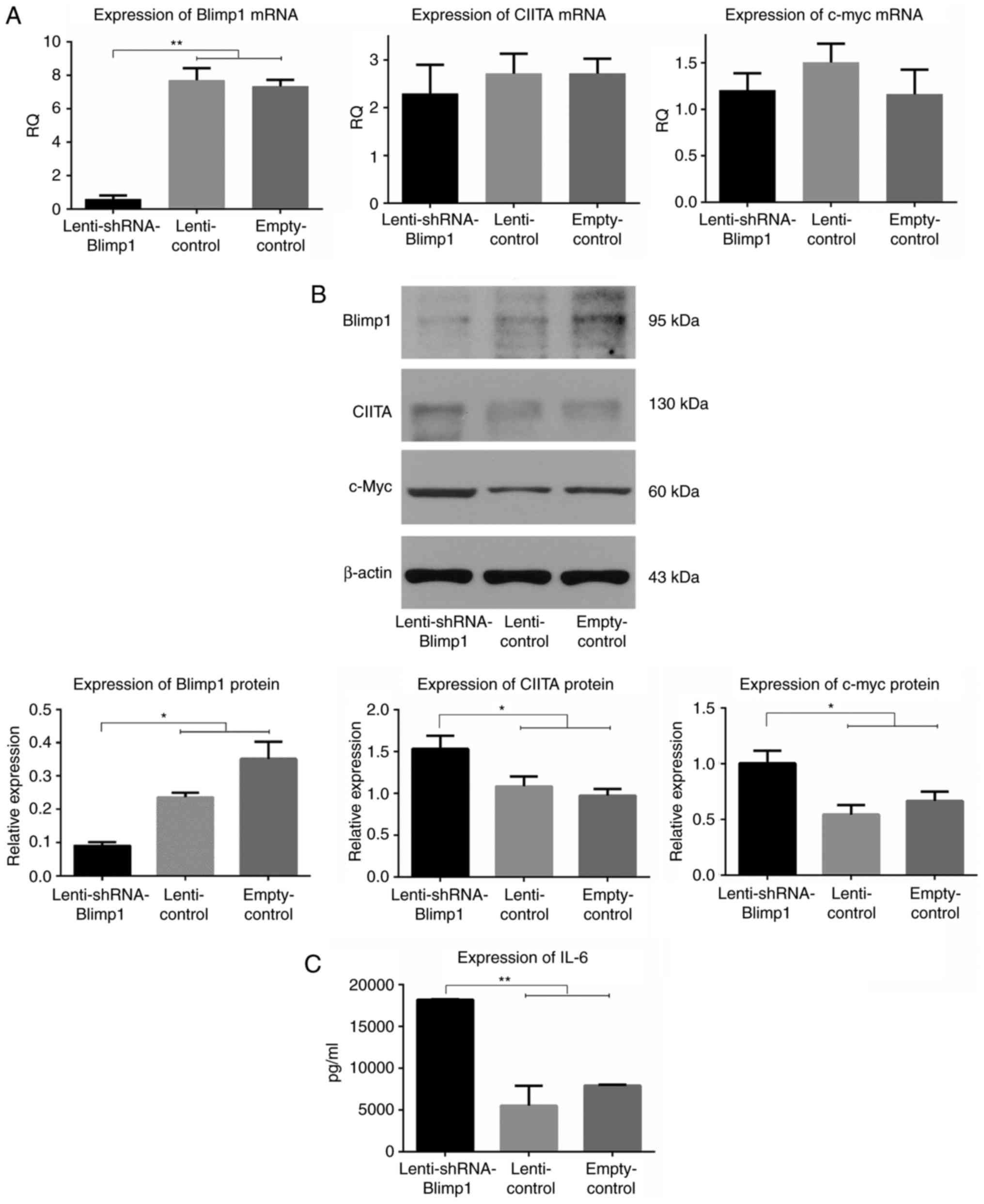
harvested in order to assess the mRNA expression levels of Blimp1,
CIITA and c-myc (Fig. 2A). The RQ
values of Blimp1 mRNA were 0.59±0.16, 7.69±0.02 and 7.33±0.28 in
the lenti-shRNA-Blimp1, lenti-control and empty-control groups
(n=3/group), respectively (P<0.01). The RQ values of CIITA mRNA
were 2.29±0.35, 2.71±0.03 and 2.71±0.02, respectively (P>0.05).
The RQ values of c-myc mRNA were 1.20±0.11, 1.50±0.01 and
1.16±0.06, respectively (P>0.05).
A total of 96 h post-transduction, the cells were
harvested in order to examine the protein expression levels of
Blimp1, and its direct target proteins CIITA and c-myc
(3, 5.52±2.38×103 and
7.92±1.13×103 pg/ml, respectively (P<0.01; Fig. 2C). Furthermore, a significant
correlation was identified between Blimp1 and CIITA, c-myc and IL-6
expression, with the following r-values: −0.819, −0.622 and −0.745,
respectively.
Lenti-shRNA-Blimp1 gene transduction has
no effect on the differentiation of bone marrow cells to DCs
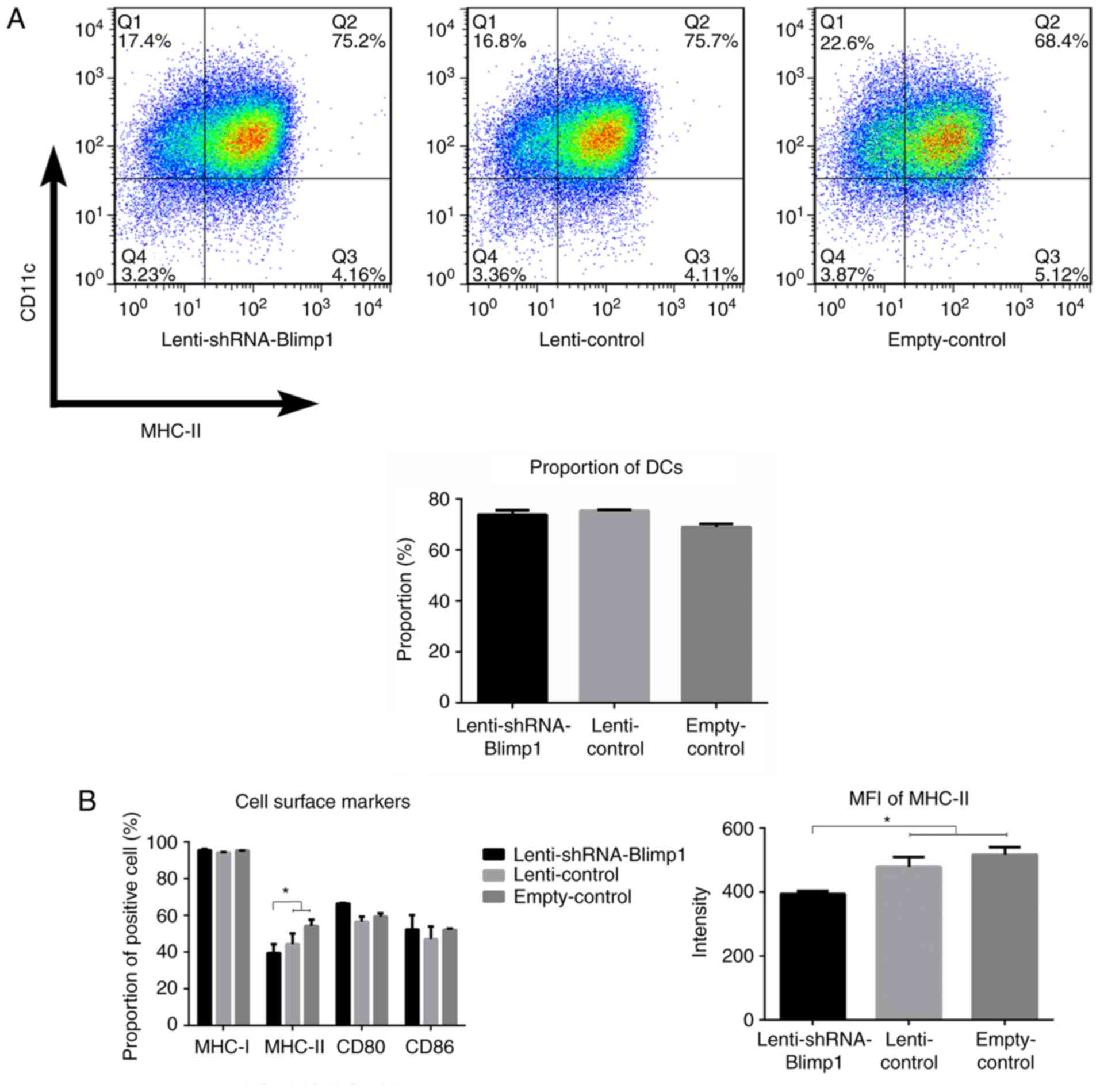
On day 8, the percentage of
CD11c+MHC-II+ cells was 73.80±1.69,
75.35±0.42 and 68.97±1.23% in the lenti-shRNA-Blimp1, lenti-control
and empty-control groups (n=3/group), respectively (Fig. 3A). No significant differences
(P>0.05) were observed between the lenti-shRNA-Blimp1 group and
the two control groups.
Lenti-shRNA-Blimp1 gene transduction
induces a downregulation in MHC-II expression on DCs, whereas it
has no effect on the expression levels of CD80/CD86 and MHC-I
With regards to antigen presentation, DCs transfer
signals to T cells through signal 1 molecules (MHC-I/MHC-II) and
signal 2 molecules (CD80/CD86). On day 8, CD11c+ cells
were harvested and were examined with regards to their maturation.
The percentage of MHC-I+ cells was 96.78±1.30,
95.38±0.51 and 96.46±0.75% in the lenti-shRNA-Blimp1, lenti-control
and empty-control groups (n=3/group), respectively (P>0.05). The
percentage of MHC-II+ cells was 38.46±4.71, 42.58±5.52
and 54.48±3.67%, respectively (P<0.05). MFI of
MHC-II+ DCs was 393.50±9.19, 478.00±31.11 and
516.36±23.34, respectively (P<0.05; Fig. 3B). Furthermore, a significant
correlation was identified between Blimp1 expression and the MFI of
MHC-II, with an r-value of 0.966 (data not shown). The percentage
of CD80+ cells was 69.17±0.3, 58.00±3.95 and
60.30±1.01%, and of CD86+ cells was 51.17±4.82,
44.00±4.95 and 51.30±0.91% in the lenti-shRNA-Blimp1, lenti-control
and empty-control groups, respectively (P>0.05).
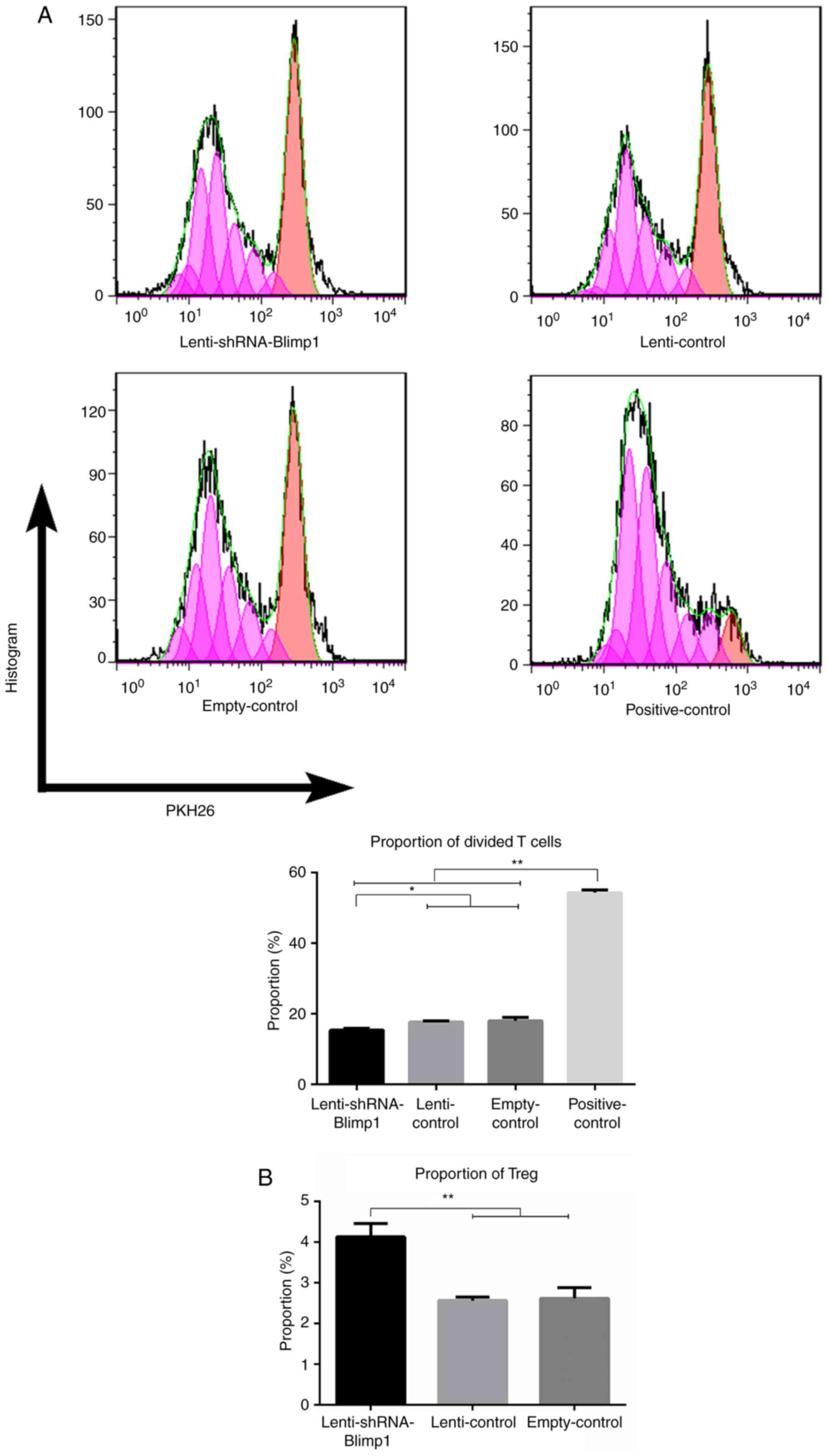
Stimulation by
lenti-shRNA-Blimp1-transduced DCs alleviates alloreactive T cell
proliferation and induces Treg expansion
To clarify the effects of DCs on alloantigen
presentation, alloreactive T cells were stimulated by DCs. At the
end of the MLR, the proportion of divided alloreactive T cells in
the empty-control group was 6.46±3.59 under a DC:alloreactive T
cell ratio of 1:10, compared with 18.05±0.92 under a ratio of 1:5
(data not shown). Therefore, 2×104 DCs were co-cultured
with 1×105 alloreactive T cells in a 96-well
round-bottom plate. The proportion of divided T cells was
15.35±0.50, 17.67±0.35, 18.05±0.92 and 54.32±0.82 in the
lenti-shRNA-Blimp1, lenti-control, empty-control and positive
control groups (n=3/group), respectively (Fig. 4A). Significant differences
(P<0.05) were observed between the lenti-shRNA-Blimp1 group and
all of the control groups.
The yield of Tregs was detected following
stimulation of alloreactive T cells by DCs transduced with
lenti-shRNA-Blimp1. The percentage of Tregs was 4.13±0.33,
2.56±0.09 and 2.62±0.26% in the lenti-shRNA-Blimp1, lenti-control
and empty-control groups (n=3/group), respectively. Significant
differences (P<0.01) were observed between the
lenti-shRNA-Blimp1 group and the control groups (Fig. 4B). Furthermore, a significant
correlation was identified between Blimp1 expression and Tregs with
an r-value of −0.719, whereas the r-value between Blimp1 and
divided T cells was 0.937.
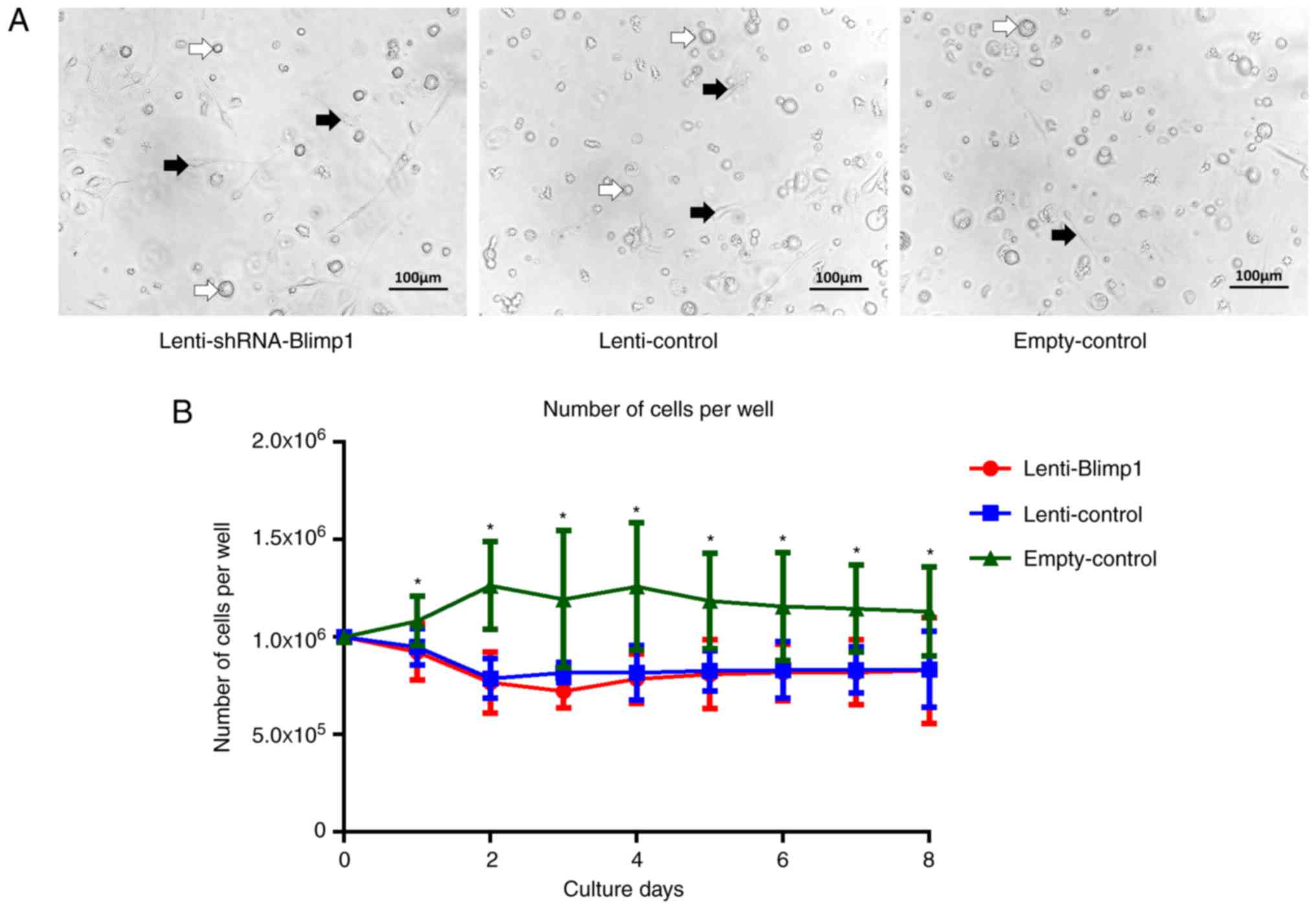
Lentiviral vectors cause mild damage to
DC morphology
On culture day 2, cells in the empty-control group
were round, granular and suspended in medium, whereas the number of
adherent cells was increased. From day 2, the size of cells
increased. On day 5, the suspended cells were round, whereas the
adherent cells were irregular in shape or spindle-shaped. On day 8,
the adherent cells reached ~90% confluence, and were similar in
size and morphology, and the majority of the cells had dendritic
features. In the lenti-control and lenti-shRNA-Blimp1 groups,
certain fragmented cells appeared on day 2 and cell density was
lower than that in the empty-control group. Subsequently, the
suspended cells varied in size, and the adherent cells were
irregular in shape and/or spindle-shaped. The morphology on day 6
is shown in Fig. 5A. Cells in the
lenti-control and lenti-shRNA-Blimp1 groups exhibited similar
morphological features.
Lentiviral vector, but not Blimp1, mildly
suppresses the proliferation of DCs
Untransduced cells exhibited logarithmic growth
within 3 days, and the highest number of cells
(1.26±0.33×106/well) was evident on day 4. This number
was decreased to 1.16±0.28×106/well on day 6, and was
maintained at 1.13±0.23×106/well on day 8. Cells in both
the lenti-control and lenti-shRNA-Blimp1 groups did not proliferate
further on day 2 and all remained at a level
<0.83±0.19×106/well (Fig. 5B). A significant difference
(P<0.05) was obtained following comparison of the lenti-control
and lenti-shRNA-Blimp1 groups with the empty-control group. The
detection procedure was repeated six times. No significant
difference (P>0.05) was noted between the lenti-control and
lenti-shRNA-Blimp1 groups.
Discussion
DCs have been recognized as pivotal elements in the
determination of either an immunogenic or tolerogenic effect,
according to their mature states (37,38). The establishment of a reliable
culture system for the generation and manipulation of DCs is
required to serve as a cornerstone for immunological studies
(23-26,34,38). It is well known that large numbers
of myeloid-derived DCs can be generated from bone marrow cultures.
The present study used an 8-day DC culture system derived from bone
marrow cells (22). On day 3, all
of the non-adherent cells were discarded, since the suspended cells
consisted of heterogeneous clusters including granulocytes, red
blood cells and DCs, whereas the adherent cells consisted mainly of
pre-DCs. Removing the suspended cells from the culture decreased
the yield but increased the final purity of the DCs; this is a key
procedure performed at various time points (from hour 3 to day 4)
in different protocols (22-26). On day 6, both non-adherent and
adherent cells predominantly consisted of DCs and therefore were
restored for further culture under LPS stimulation for maturation.
The present strategy is reasonable and operable with a superior
final DC yield of 1-1.5×108 DCs raised from a single
mouse, a number 20-30 folds higher than that reached in previous
studies (22,25,34). Based on this procedure, bone
marrow cells were induced to differentiate into pre-DCs and DCs
with a large yield.
Lentiviral-mediated Blimp1-shRNA gene transduction
was used to effectively downregulate Blimp1 expression in DCs.
Firstly, it was verified that the lentiviral vector had a high
efficiency to transduce bone marrow cells. GFP was detected 3 days
post-transduction and was noted in ~60.83±1.39% cells 8 days
post-transduction in both the lenti-control and lenti-shRNA-Blimp1
groups. Furthermore, Blimp1 expression levels were detected in
pre-DCs that were transduced with lenti-shRNA-Blimp1. The RQ of
Blimp1 mRNA was reduced to 8.0% (0.59/7.33) 72 h post-transduction,
which suggested that RNA interference was successful. Following 24
h of incubation, the relative protein expression levels of Blimp1
were downregulated to 22.5% (0.09/0.40) of the normal level. To
date, Blimp1 has been identified as an essential factor for cell
growth, and it has been reported that, in mammals, complete Blimp1
knockout leads to mortality (5).
Therefore, lenti-shRNA-Blimp1 transduction was used for the
immunological modulation of DCs as an appropriate strategy to
partially downregulate Blimp1 expression. The present study further
detected the expression levels of downstream target proteins of
Blimp1, namely CIITA, c-myc and IL-6 (39-41), demonstrating that the expression
of these proteins was regulated accordingly. Notably, this
modulation may be a post-transcriptional effect, since the mRNA
expression levels of these target proteins were stable in the
presence and/or absence of lenti-shRNA-Blimp1. Collectively, these
findings confirmed the efficiency and safety of lenti-shRNA-Blimp1
transduction.
The present study demonstrated that the decrease in
Blimp1 expression had no effect on the committed differentiation of
DCs. At the end of the culture procedure, the proportion of
CD11c+ cells in the lenti-control and lenti-shRNA-Blimp1
groups indicated no difference compared with in the empty-control
group. However, downregulation of Blimp1 during the differentiation
process from bone marrow cells to DCs exhibited an impact on the
maturation of DCs. The expression of MHC-II, but not MHC-I,
molecules on the surface membrane of DCs was significantly
decreased by lenti-shRNA-Blimp1 gene transduction, thus indicating
the immature status of the cultured DCs. The process of antigen
presentation to T-cell receptors (signal 1) may be hampered by the
decreased amount of MHC-II molecules on the surface membrane of
immature DCs, which may result in inactivation of antigen-specific
alloreactive T cells. Notably, the expression of the co-stimulatory
molecules CD80 and CD86 were not affected, thus suggesting that the
inactivation of alloreactive T cells was not mediated via a
co-stimulatory (signal 2) mechanism. Subsequently, MLR was employed
to examine the effects of the induced immature DCs; the results
revealed that alloreactive T cell proliferation was alleviated
following stimulation of cells with lenti-shRNA-Blimp1-transduced
DCs compared with non-transduced DCs. Furthermore, CD4+
FoxP3+ Tregs were expanded following stimulation by
immature DCs, which may partly contribute to the alteration in
alloreactive T cell proliferation. Taken together, these data
indicated that lenti-shRNA-Blimp1 gene transduction inhibited DC
maturation via MHC-II molecule suppression, which in turn inhibited
the subsequent alloreactive T cell reaction.
In the present study, lentiviral vectors exhibited
mild toxicity on DCs during culture. Without intervention, the
cultured cells exhibited a marked increase in burr-like
protrusions, changed from a circular to dendritic morphology, and
reached 90% confluence by the end of cell culture. Lentiviral
transduction, either with lenti-vector or lenti-shRNA-Blimp1,
induced variations in size and irregular shapes, with a decreased
density in certain types of cells. Furthermore, the proliferation
curves demonstrated that the cell number was slightly lower
following lentiviral transduction compared with in cultures without
intervention. Lenti-vector and lenti-shRNA-Blimp1 transduction
arrested proliferation of DCs at similar levels, thus suggesting
that the toxicity was derived from the lentiviral vector itself,
and not Blimp1 shRNA. The toxicity of the lentiviral vector on DCs,
although mild, requires further technical improvement. Furthermore,
additional caution should be exercised with regards to the
biosecurity of the lentivirus (42-44).
In conclusion, lenti-shRNA-Blimp1 transduction was
able to modulate the maturation of DCs via the suppression of
MHC-II molecules, and inhibited the subsequent alloreactive T cell
activation. The findings of the present study support a
Blimp1-based intervention as a novel approach and mechanism for
inducing immature DCs, which may be adopted for certain aims, such
as modulating the immunological alloreaction of T cells.
Funding
The present study was supported by grants to NQ Gong
from the National Natural Science Foundation of China (grant no.
81570678 and 81072441), the Major State Basic Research Development
Program of China (grant no. 2013CB530803) and the Clinical Research
Physician Program of Tongji Medi cal College, HUST.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors’ contributions
NG and JS designed the experiments. JX, JZ, XL, XD,
JW, LW and YH performed the experiments and analyzed the data. NG
and JX wrote the paper. JX and JZ contributed equally to the
present study. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
All experiments were approved by the Institutional
Animal Care and Use Committee at Tongji Medical College, Huazhong
University of Science and Technology (ethical approval no.
TJ-A201111211).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgments
Not applicable.
References
|
1
|
Mossink MH, de Groot J, Van Zon A,
Fränzel-Luiten E, Schoester M, Scheffer GL, Sonneveld P, Scheper RJ
and Wiemer EA: Unimpaired dendritic cell functions in MVP/LRP
knockout mice. Immunology. 110:58–65. 2003. View Article : Google Scholar
|
|
2
|
Steinman RM, Inaba K, Turley S, Pierre P
and Mellman I: Antigen capture, processing, and presentation by
dendritic cells: Recent cell biological studies. Hum Immunol.
60:562–567. 1999. View Article : Google Scholar
|
|
3
|
Ying W, Fei H, Jun D, Xi-chuan Y, Bai-yu Z
and Qing-yi Y: Reversible transfection of human melanocytes
mediated by Cre/loxP site-specific recombination system and SV40
large T antigen. Exp Dermatol. 16:437–444. 2007. View Article : Google Scholar
|
|
4
|
Martins G and Calame K: Regulation and
functions of Blimp-1 in T and B lymphocytes. Annu Rev Immunol.
26:133–169. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Minnich M, Tagoh H, Bönelt P, Axelsson E,
Fischer M, Cebolla B, Tarakhovsky A, Nutt SL, Jaritz M and
Busslinger M: Multifunctional role of the transcription factor
Blimp-1 in coordinating plasma cell differentiation. Nat Immunol.
17:331–343. 2016. View
Article : Google Scholar
|
|
6
|
Martins GA, Cimmino L, Shapiro-Shelef M,
Szabolcs M, Herron A, Magnusdottir E and Calame K: Transcriptional
repressor Blimp-1 regulates T cell homeostasis and function. Nat
Immunol. 7:457–465. 2006. View
Article : Google Scholar
|
|
7
|
Magnúsdóttir E, Kalachikov S, Mizukoshi K,
Savitsky D, Ishida-Yamamoto A, Panteleyev AA and Calame K:
Epidermal terminal differentiation depends on B lymphocyte-induced
maturation protein-1. Proc Natl Acad Sci USA. 104:14988–14993.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Lin FR, Kuo HK, Ying HY, Yang FH and Lin
KI: Induction of apoptosis in plasma cells by B lymphocyte-induced
maturation protein-1 knockdown. Cancer Res. 67:11914–11923. 2007.
View Article : Google Scholar
|
|
9
|
Chan YH, Chiang MF, Tsai YC, Su ST, Chen
MH, Hou MS and Lin KI: Absence of the transcriptional repressor
Blimp-1 in hematopoietic lineages reveals its role in dendritic
cell homeostatic development and function. J Immunol.
183:7039–7046. 2009. View Article : Google Scholar
|
|
10
|
Kurimoto K, Yamaji M, Seki Y and Saitou M:
Specification of the germ cell lineage in mice: A process
orchestrated by the PR-domain proteins, Blimp1 and Prdm14. Cell
Cycle. 7:3514–3518. 2008. View Article : Google Scholar
|
|
11
|
Takahashi K and Yamanaka S: Induction of
pluripotent stem cells from mouse embryonic and adult fibroblast
cultures by defined factors. Cell. 126:663–676. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kim SJ, Zou YR, Goldstein J, Reizis B and
Diamond B: Tolerogenic function of Blimp-1 in dendritic cells. J
Exp Med. 208:2193–2199. 2011. View Article : Google Scholar
|
|
13
|
Yang H, Qiu Q, Gao B, Kong S, Lin Z and
Fang D: Hrd1-mediated BLIMP-1 ubiquitination promotes dendritic
cell MHCII expression for CD4 T cell priming during inflammation. J
Exp Med. 211:2467–2479. 2014. View Article : Google Scholar
|
|
14
|
Kim SJ, Gregersen PK and Diamond B:
Regulation of dendritic cell activation by microRNA let-7c and
BLIMP1. J Clin Invest. 123:823–833. 2013.
|
|
15
|
Datiles MJ, Johnson EA and Mccarty RE:
Inhibition of the ATPase activity of the catalytic portion of ATP
synthases by cationic amphiphiles. Biochim Biophys Acta.
1777:362–368. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Min Kim J, Young Choi J, Sun Kim M and
Chang Kim S: In vivo excision and amplification of large human
genomic segments using the Cre/loxP -and large T antigen/SV40 ori
-mediated machinery. J Biotechnol. 110:227–233. 2004. View Article : Google Scholar
|
|
17
|
Ashihara E, Kawata E and Maekawa T: Future
prospect of RNA interference for cancer therapies. Curr Drug
Targets. 11:345–360. 2010. View Article : Google Scholar
|
|
18
|
Rubinson DA, Dillon CP, Kwiatkowski AV,
Sievers C, Yang L, Kopinja J, Rooney DL, Zhang M, Ihrig MM, McManus
MT, et al: A lentivirus-based system to functionally silence genes
in primary mammalian cells, stem cells and transgenic mice by RNA
interference. Nat Genet. 33:401–406. 2006. View Article : Google Scholar
|
|
19
|
Picanco-Castro V, de Sousa
Russo-Carbolante EM and Tadeu Covas D: Advances in lentiviral
vectors: A patent review. Recent Pat DNA Gene Seq. 6:82–90. 2012.
View Article : Google Scholar
|
|
20
|
Escors D and Breckpot K: Lentiviral
vectors in gene therapy: Their current status and future potential.
Arch Immunol Ther Exp (Warsz). 58:107–119. 2010. View Article : Google Scholar
|
|
21
|
Watt FM and Hogan BL: Out of Eden: Stem
cells and their niches. Science. 287:1427–1430. 2000. View Article : Google Scholar
|
|
22
|
Wang J, Zhao LB, Chang S, Ming CS, Yang J
and Gong NQ: Dynamic changes of phenotypes and secretory functions
during the differentiation of pre-DCs to mature DCs. J Huazhong
Univ Sci Technolog Med Sci. 37:191–196. 2017. View Article : Google Scholar
|
|
23
|
Wheat WH, Pauken KE, Morris RV and Titus
RG: Lutzomyia longipalpis salivary peptide maxadilan alters murine
dendritic cell expression of CD80/86, CCR7 and cytokine secretion
and reprograms dendritic cell-mediated cytokine release from
cultures containing allogeneic T cells. J Immunol. 180:8286–8298.
2008. View Article : Google Scholar
|
|
24
|
Machen J, Harnaha J, Lakomy R, Styche A,
Trucco M and Giannoukakis N: Antisense oligonucleotides
down-regulating costimulation confer diabetes-preventive properties
to nonobese diabetic mouse dendritic cells. J Immunol.
173:4331–4341. 2004. View Article : Google Scholar
|
|
25
|
Inaba K, Inaba M, Romani N, Aya H, Deguchi
M, Ikehara S, Muramatsu S and Steinman RM: Generation of large
numbers of dendritic cells from mouse bone marrow cultures
supplemented with granulocyte/macrophage colony-stimulating factor.
J Exp Med. 176:1693–1702. 1992. View Article : Google Scholar
|
|
26
|
Lutz MB, Kukutsch N, Ogilvie AL, Rössner
S, Koch F, Romani N and Schuler G: An advanced culture method for
generating large quantities of highly pure dendritic cells from
mouse bone marrow. J Immunol Methods. 223:77–92. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Brasel K, De Smedt T, Smith JL and
Maliszewski CR: Generation of murine dendritic cells from
flt3-ligand-supplemented bone marrow cultures. Blood. 96:3029–3039.
2000.
|
|
28
|
Gallucci S, Lolkema M and Matzinger P:
Natural adjuvants: Endogenous activators of dendritic cells. Nat
Med. 5:1249–1255. 1999. View
Article : Google Scholar
|
|
29
|
Tooze RM, Stephenson S and Doody GM:
Repression of IFN-gamma induction of class II transactivator: A
role for PRDM1/Blimp-1 in regulation of cytokine signaling. J
Immunol. 177:4584–4593. 2006. View Article : Google Scholar
|
|
30
|
Islam MF, Watanabe A, Wong L, Lazarou C,
Vizeacoumar FS, Abuhussein O, Hill W, Uppalapati M, Geyer CR and
Vizeacoumar FJ: Enhancing the throughput and multiplexing
capabilities of next generation sequencing for efficient
implementation of pooled shRNA and CRISPR screens. Sci Rep.
7:10402007. View Article : Google Scholar
|
|
31
|
Galluzzi L, Ceccarelli M, Diotallevi A,
Menotta M and Magnani M: Real-time PCR applications for diagnosis
of leishmaniasis. Parasit Vectors. 11:2732018. View Article : Google Scholar
|
|
32
|
Wu RC, Chen DF, Liu MJ and Wang Z: Dual
effects of cycloheximide on U937 apoptosis induced by its
combination with VP-16. Biol Pharm Bull. 27:1075–1080. 2004.
View Article : Google Scholar
|
|
33
|
O’Sullivan AM, O’Callaghan YC, O’Grady MN,
Queguineur B, Hanniffy D, Troy DJ, Kerry JP and O’Brien NM: In
vitro and cellular antioxidant activities of seaweed extracts
prepared from five brown seaweeds harvested in spring from the west
coast of Ireland. Food Chem. 126:1064–1070. 2011. View Article : Google Scholar
|
|
34
|
Wang J, Dai X, Hsu C, Ming C, He Y, Zhang
J, Wei L, Zhou P, Wang CY, Yang J and Gong N: Discrimination of the
heterogeneity of bone marrow-derived dendritic cells. Mol Med Rep.
16:6787–6793. 2017. View Article : Google Scholar
|
|
35
|
Marelli-Berg FM, Hargreaves RE, Carmichael
P, Dorling A, Lombardi G and Lechler RI: Major histocompatibility
complex class II-expressing endothelial cells induce allospecific
nonresponsiveness in naive T cells. J Exp Med. 183:1603–1612. 1996.
View Article : Google Scholar
|
|
36
|
Wallace PK and Muirhead KA: Cell tracking
2007: A proliferation of probes and applications. Immunol Invest.
36:527–561. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Banchereau J and Steinman RM: Dendritic
cells and the control of immunity. Nature. 392:245–252. 1998.
View Article : Google Scholar
|
|
38
|
Beriou G, Moreau A and Cuturi MC:
Tolerogenic dendritic cells: Applications for solid organ
transplantation. Curr Opin Organ Transplant. 17:42–47. 2012.
View Article : Google Scholar
|
|
39
|
Wang P, Xue Y, Han Y, Lin L, Wu C, Xu S,
Jiang Z, Xu J, Liu Q and Cao X: The STAT3-binding long noncoding
RNA lnc-DC controls human dendritic cell differentiation. Science.
344:310–313. 2014. View Article : Google Scholar
|
|
40
|
Yoon HS, Scharer CD, Majumder P, Davis CW,
Butler R, Zinzow-Kramer W, Skountzou I, Koutsonanos DG, Ahmed R and
Boss JM: ZBTB32 is an early repressor of the CIITA and MHC class II
gene expression during B cell differentiation to plasma cells. J
Immunol. 189:2393–2403. 2012. View Article : Google Scholar :
|
|
41
|
Xia Y, Xu-Monette ZY, Tzankov A, Li X,
Manyam GC, Murty V, Bhagat G, Zhang S, Pasqualucci L, Visco C, et
al: Loss of PRDM1/BLIMP-1 function contributes to poor prognosis of
activated B-cell-like diffuse large B-cell lymphoma. Leukemia.
31:625–636. 2017. View Article : Google Scholar
|
|
42
|
Kim SJ, Goldstein J, Dorso K, Merad M,
Mayer L, Crawford JM, Gregersen PK and Diamond B: Expression of
Blimp-1 in dendritic cells modulates the innate inflammatory
response in dextran sodium sulfate-induced colitis. Mol Med.
20:707–719. 2015.
|
|
43
|
Muruve DA: The innate immune response to
adenovirus vectors. Hum Gene Ther. 15:1157–1166. 2004. View Article : Google Scholar
|
|
44
|
Scanlon KJ: Cancer gene therapy:
Challenges and opportunities. Anticancer Res. 24:501–504.
2004.PubMed/NCBI
|