Introduction
Ankylosing spondylitis (AS) is the prototypic and
debilitating type of spondyloarthritis, which is an autoimmune
disorder (1). AS often affects
the axial joints, including the sacroiliac joint and spine, and
induces new bone formation and ultimately ankylosis (2,3).
AS is associated with the interplay of genetic risks
and environmental triggers, and its etiology and pathogenesis
remain largely unknown. A number of studies have reported a strong
association between the major histocompatibility complex class I
allele human leukocyte antigen B27 (HLA-B27) and AS (4-6).
However, the mechanism by which HLA-B27 causes a predisposition to
AS remains unclear. T cells and a number of immune pathways,
including the interleukin (IL)-17/IL-23 pathway and control of
nuclear factor-κB (NF-κB) activation, have been reported to be
closely associated with AS (5,6).
Currently, it is difficult to diagnose AS during the early stages
due to the lack of accurate diagnostic biomarkers.
Long non-coding RNA (lncRNA) is a type of
non-protein-coding transcript with a length of >200 nucleotides;
they are emerging as key regulators in various biological processes
(7). Recent studies have
indicated that lncRNAs, including lncRNA-AK001085, lnc-zinc finger
protein 354A (ZNF354A)-1, lnc-Lin-54 DREAM MuvB core complex
component (LIN54)-1, lnc-Facioscapulohumeral muscular dystrophy
region gene 2 family member C (FRG2C)-3 and lnc-ubiquitin specific
peptidase 50 (USP50)-2, serve roles in AS (8,9).
Some researchers have also revealed that mRNAs, such as programmed
cell death 1 (PDCD1) (1), caspase
recruitment domain-containing protein 11 (CARD11) (10), phospholipase Cγ1 (PLCG1) (11,12) and DNA methyltransferase 1 (DNMT1)
(13), may be involved in the
pathogenesis of AS.
In the present study, the key differentially
expressed (DE) mRNAs (DEmRNAs) and DElncRNAs in AS were identified
using RNA sequencing and bioinformatics analysis. DElncRNA-DEmRNA
co-expression network construction, identification of nearby target
DEmRNAs of DElncRNAs and functional annotation of DEmRNAs were
performed in order to understand the biological functions of the
key DEmRNAs and DElncRNAs in AS.
Materials and methods
Patients and samples
Three patients with AS and three normal controls
were enrolled in the present study from 2nd Affiliated Hospital,
School of Medicine, Zhejiang University (Zhejiang, China). The
patients with AS were aged 37, 36 and 40 years old, and all were
HLA-B27+ and male. These patients were diagnosed with AS
based on kyphotic deformity, bilateral damage of the sacroiliac
joint observed in the computed tomography results, and spinal
fusion and sacroiliac arthrodesis observed in the X-ray results.
All of the patients had not received treatment with non-steroidal
anti-inflammatory drugs or biologics and they had not exhibited
complications. The normal controls were aged 35, 36 and 37 years
old and were male. None of the six participants had any other type
of autoimmune disease. Blood samples were obtained from all six
participants. All of the participants submitted written informed
consent and the present study was approved by the Ethics Committee
of 2nd Affiliated Hospital, School of Medicine, Zhejiang
University.
Library preparation and high-throughput
sequencing
Total RNA was isolated from the blood samples with
TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.,
Waltham, MA, USA) according to the manufacturer's instructions. A
NanoDrop ND-2000 spectrophotometer (Thermo Fisher Scientific, Inc.)
was used to check the concentration and purity of the RNA. The
integrity of the RNA was confirmed using 2% agarose gel and the RNA
integrity number (RIN) was obtained using an Agilent 2100
Bioanalyzer instrument (Agilent Technologies, Inc., Santa Clara,
CA, USA). The thresholds of total RNA for cDNA library construction
were as follows: i) Amount of RNA, >5 μg; ii)
concentration of RNA; ≥200 ng/ml; iii) 1.8<optical density
(OD)260/280<2.2; iv) 2.0<OD260/280; and
v) RIN value, >8.0.
Firstly, the ribosomal RNA was removed from the
total RNA using a Ribo-Zero Magnetic kit (Epicentre; Illumina,
Inc., San Diego, CA, USA). In addition, 'Soap' software (version
1.03; soap.genomics.org.cn/SOAPdenovo-Trans.html#intro2) was
used to compare the reads of the rRNA, and then to write a perl
script to remove it from the FastQ file. Subsequently, the cDNA
library for RNA sequencing was contrasted using the TruSeq RNA
Sample Prep kit (Illumina, Inc.). Briefly, the retrieved RNA was
fragmented by adding First Strand Master Mix and then first-strand
cDNA was generated using the First Strand Master mix and Super
Script II reverse transcription (Invitrogen; Thermo Fisher
Scientific, Inc.) with the following reaction conditions: 25°C for
10 min, 42°C for 50 min and 70°C for 15 min. Following purification
of the product with Agencourt RNA Clean XP beads (Beckman Coulter,
Inc., Brea, CA, USA), Second Strand Master mix and dATP, dGTP, dCTP
and dUTP mix (Beckman Coulter, Inc.) were added to synthesize the
second-strand cDNA (16°C for 1 h). Subsequently, the purified cDNA
was combined with End Repair Mix (Thermo Fisher Scientific, Inc.)
and incubated at 30°C for 30 min. Following purification with beads
(Qiagen, Inc., Valencia, CA, USA), A-Tailing mix (Qiagen, Inc.) was
added into the reaction system, which was incubated at 37°C for 30
min. Adenylate 3' Ends DNA, Index Adapter and Ligation mix (Qiagen,
Inc.) were combined and incubated at 30°C for 10 min. Subsequently,
the Uracil-N-Glycosylase enzyme was added into the purified
ligation product and incubated at 37°C for 10 min. A total of 15
rounds of polymerase chain reaction (PCR) amplification were
conducted with PCR Primer Cocktail (Illumina, Inc.) and PCR Master
Mix (Illumina, Inc.) to enrich the cDNA fragments. The PCR products
were then purified with AMPure XP beads (Qiagen, Inc.). The
qualified libraries were amplified on cBot (Illumina, Inc.) to
generate a cluster on the flow cell (TruSeq PE Cluster kit
v3-cBot-HS; Illumina, Inc.). The amplified flow cell was sequenced
on an Illumina HiSeq X Ten platform (Illumina, Inc.).
Quality control of raw sequence
Quality control of the raw reads derived from the
RNA sequencing was performed to obtain clean reads of high quality.
Briefly, quality control involved trimming low-quality reads,
including adaptor sequences, sequences with a quality score <20
and sequences with an N-base rate of raw reads >10%, using
SeqPrep (version 1.2; github.com/jstjohn/SeqPrep) and Sickle (version
V3.4.0; github.com/najoshi/sickle).
Clean reads mapping
Sequences were aligned to the human reference genome
GRCh38.p7 (Ensembl v84; www.ensembl.org/index.html) using hierarchical
indexing for the spliced alignment of the transcripts programme
HISAT2 (version 2.1.0; ccb.jhu.edu/software/hisat2/index.shtml). Then,
StringTie (version v1.3.4; ccb.jhu.edu/software/stringtie/) was used to assemble
and quantify the transcripts in each sample. Ultimately,
differential gene expression analysis was performed with Ballgown
(version 3.7; www.bioconductor.org/packages/release/bioc/html/ballgown.html)
in R environment.
Identification of DEmRNAs and
DElncRNAs
Using Ballgown, the DEmRNAs and DElncRNAs between
the patients with AS and normal controls were identified with
P<0.05. The false discovering rate (FDR)-adjusted P-value of the
test statistic was used. Hierarchical clustering of the DElncRNAs
and DEmRNAs expression profile was performed using hcluster in R
language (version 3.3.3;
stat.ethz.ch/R-manual/R-devel/library/stats/html/hclust.html).
AS-specific protein-protein interaction
(PPI) network construction
PPI networks of the top 30 up- and down-regulated
DEmRNAs, respectively, were constructed using BioGRID (version 3.5;
thebiogrid.org).
AS-specific weighted DEmRNA-DElncRNA
co-expression network analyses
Weighted Gene Co-expression Network Analysis
(WGCNA_1.64-1; horvath.genetics.ucla.edu/html/CoexpressionNetwork/Rpackages/WGCNA/)
(14) is an R package for
weighted correlation network analysis, which is also known as
weighted gene co-expression network analysis. Using WGCNA,
AS-specific weighted DEmRNA-DElncRNA co-expression network analysis
was performed. The pairwise Pearson's correlation coefficients
(PCCs) between the DElncRNAs and DEmRNAs in patients with AS were
calculated. DElncRNA-DEmRNA pairs with |PCC value ≥0.90| and
P<0.001 were used to construct an AS-specific weighted
DEmRNA-DElncRNA co-expression network, which was deciphered using
the online-based software GeneCodis3
(genecodis.cnb.csic.es/analysis).
Functional analyses of DEmRNAs
co-expressed with DElncRNAs in AS
The DEmRNAs of these DElncRNA-DEmRNA pairs with |PCC
value ≥0.90| and P<0.001 were used to conduct Gene Ontology (GO;
www.geneontology.org/) and Kyoto
Encyclopedia of Genes and Genomes (KEGG; www.genome.jp/kegg/) molecular pathway enrichment
analysis using GeneCoDis3.
DEmRNAs close to DElncRNAs with
cis-regulatory effects
Previous studies have reported that lncRNAs can
regulate genes that are transcribed near to them, consistent with
activity in cis-regulatory elements (15,16). Therefore, in the present study
DEmRNAs that were close cis targets of DElncRNAs were identified by
searching for DEmRNAs that were transcribed within a 100-kb window
up- or downstream of DElncRNAs between the patients with AS and the
normal controls (17).
Validation of the expression of DEmRNAs
and DElncRNAs using the GSE73754 and GSE25101 datasets
The mRNA expression profile of 52 patients with AS
and 20 normal controls (Canadian cohort) in the GSE73754 dataset
(GPL10558 Illumina HumanHT-12 V4.0 expression beadchip), which was
downloaded from the Gene Expression Omnibus (GEO; www.ncbi.nlm.nih.gov/gds). The mRNA and lncRNA
expression profile of 16 patients with AS and 16 normal controls
(Australian cohort) in the GSE25101 dataset (GPL6947 Illumina
HumanHT-12 V3.0 expression beadchip) was also downloaded from the
GEO database. The expression levels of the selected DEmRNAs and
DElncRNAs between the patients with AS and normal controls in the
present study were validated using the GSE73754 and GSE25101
datasets, and the difference in the expression levels was
visualized using box plots.
Receiver operating characteristic (ROC)
analyses
To assess the diagnostic value of the DEmRNAs in AS,
ROC analyses were conducted using pROC package (version 1.13.0) in
R language (cran.r-project.org/web/packages/pROC/index.html). The
area under the curve (AUC) under the binomial exact confidence
interval was calculated. ROC curves were then generated.
Statistical analysis
Values are displayed as the mean ± standard
deviation. Student's t-test was used to assess differences among
the groups, and P≤0.05 was considered to indicate a statistically
significant difference. Co-expression associations between the
lncRNAs and the protein-coding genes were estimated using pairwise
PCC analysis using R language (version 3.3.3; stat.
ethz.ch/R-manual/R-devel/library/stats/html/hclust.html).
Results
Identification of DEmRNAs and DElncRNAs
between patients with AS and normal controls
Compared with the normal controls, 1,072 DEmRNAs
(320 upregulated and 752 down-regulated) and 372 DElncRNAs (230
upregulated and 142 downregulated) in patients with AS were
identified. The top 10 up- and downregulated DEmRNAs and DElncRNAs
between the patients with AS and normal controls are shown in
Tables I and II, respectively.
 | Table ITop 10 up- and downregulated DEmRNAs
between ankylosing spondylitis and normal controls. |
Table I
Top 10 up- and downregulated DEmRNAs
between ankylosing spondylitis and normal controls.
| DEmRNAs | log2.Fold
change | P-value | SD_fpkm_C | SD_fpkm_T | Regulation |
|---|
| IGHG1 | −2.021979095 | 0.047255959 | 8.80361236 | 0.515381703 | Down |
| IGHG4 | −1.859252007 | 0.025210597 | 3.368882506 | 0.227307135 | Down |
| TRAJ18 | −1.643179443 | 0.035866398 | 10.07677638 | 4.09159482 | Down |
| TRAJ11 | −1.524711275 | 0.014505975 | 8.654159887 | 4.620202967 | Down |
| TRAJ5 | −1.49467071 | 0.000440647 | 6.737567008 | 2.426348758 | Down |
| TRAJ44 | −1.46271809 | 0.001143618 | 4.42364856 | 1.848651282 | Down |
| ETS1 | −1.335519855 | 0.031765089 | 32.5763664 | 9.312805914 | Down |
| LAIR2 | −1.302776176 | 0.044790098 | 0.834353066 | 1.050550336 | Down |
| ESYT1 | −1.259200819 | 0.002673524 | 15.14634661 | 6.658023234 | Down |
| NCR3 | −1.247053856 | 0.033379705 | 5.641796936 | 2.719067002 | Down |
| CLC | 1.194865831 | 0.043003696 | 21.60725157 | 83.0330712 | Up |
| S100A12 | 0.98788469 | 0.00518168 | 40.98384277 | 110.0364144 | Up |
| CMPK2 | 0.904925259 | 0.01474919 | 0.622597776 | 2.021786682 | Up |
| ZFP36 | 0.842014426 | 0.012071156 | 6.481954564 | 7.910723637 | Up |
| MME | 0.746984037 | 0.031312145 | 9.013396028 | 20.24882244 | Up |
| LIN7A | 0.711395439 | 0.049387957 | 3.456739121 | 4.043733587 | Up |
| CYP4F3 | 0.707298362 | 0.025967428 | 4.648357447 | 8.950444777 | Up |
| CXCL8 | 0.651965182 | 0.048148598 | 6.882829746 | 11.04384805 | Up |
| CLEC4D | 0.636486225 | 0.047074356 | 2.579585737 | 3.692774365 | Up |
| FAM118A | 0.634156346 | 0.010715671 | 0.992441488 | 2.574184644 | Up |
 | Table IITop 10 up- and downregulated
DElncRNAs between the patients with ankylosing spondylitis and the
normal controls. |
Table II
Top 10 up- and downregulated
DElncRNAs between the patients with ankylosing spondylitis and the
normal controls.
| DElncRNAs | log2.Fold
change | P-value | SD_fpkm_C | SD_fpkm_T | Regulation |
|---|
| MSTRG.9221 | −2.900321700 | 0.018550654 | 4.278074312 | 0.276811741 | Down |
| MSTRG.1368 | −2.619486415 | 0.003067593 | 1.673345961 | 0 | Down |
| AL928768.3 | −2.320417178 | 0.018613919 | 1.432426837 | 0.766958784 | Down |
| WDR86-AS1 | −1.749482818 | 0.045879379 | 5.820719606 | 0.430167793 | Down |
| GS1-393G12.12 | −1.595574045 | 0.025413087 | 2.892968281 | 0.573066828 | Down |
| RP11-75C10.7 | −1.578454562 | 0.028918409 | 1.681378986 | 0.27755832 | Down |
| MSTRG.22984 | −1.515487489 | 0.028151217 | 6.307372767 | 6.856887292 | Down |
| RP1-29C18.8 | −1.491219357 | 0.007182308 | 1.813751882 | 0.780203396 | Down |
| RP11-20I20.4 | −1.489870084 | 0.045895116 | 1.882568915 | 0.702213888 | Down |
| CTD-2531D15.5 | −1.452569255 | 0.044459184 | 1.198212257 | 0 | Down |
| AC010084.1 | 1.812545185 | 0.005654266 | 0 | 0.808113485 | Up |
| PSMD5-AS1 | 1.788651473 | 0.011076928 | 8.927660428 | 20.42027077 | Up |
| RP11-535M15.1 | 1.639437785 | 0.00820557 | 0 | 1.477784502 | Up |
| LLPH-AS1 | 1.498657545 | 0.005471274 | 0.230318651 | 2.298519226 | Up |
| RP4-811H24.9 | 1.452389476 | 0.004968106 | 0.053490925 | 0.499762058 | Up |
| MSTRG.30231 | 1.342076952 | 0.040559246 | 2.874551067 | 6.364192991 | Up |
| TMEM92-AS1 | 1.339293051 | 0.014889801 | 0 | 1.352052251 | Up |
| WI2-87327B8.2 | 1.289651543 | 0.017494757 | 1.241932483 | 1.714464098 | Up |
| AC099668.5 | 1.25464443 | 0.023121561 | 0 | 1.059053016 | Up |
| LINC01151 | 1.231390756 | 0.00790838 | 0.384761558 | 0.385097694 | Up |
MANSC domain containing 1 (MANSC1) and DNMT1 were
the most significantly up- and downregulated DEmRNAs between the
patients with AS and normal controls, respectively (data not
shown). NOTCH1 associated lncRNA in T-cell acute lymphoblastic
leukemia 1 (NALT1) and RP11-837J7.4 were the most significantly up-
and downregulated DElncRNAs between the patients with AS and normal
controls, respectively (data not shown).
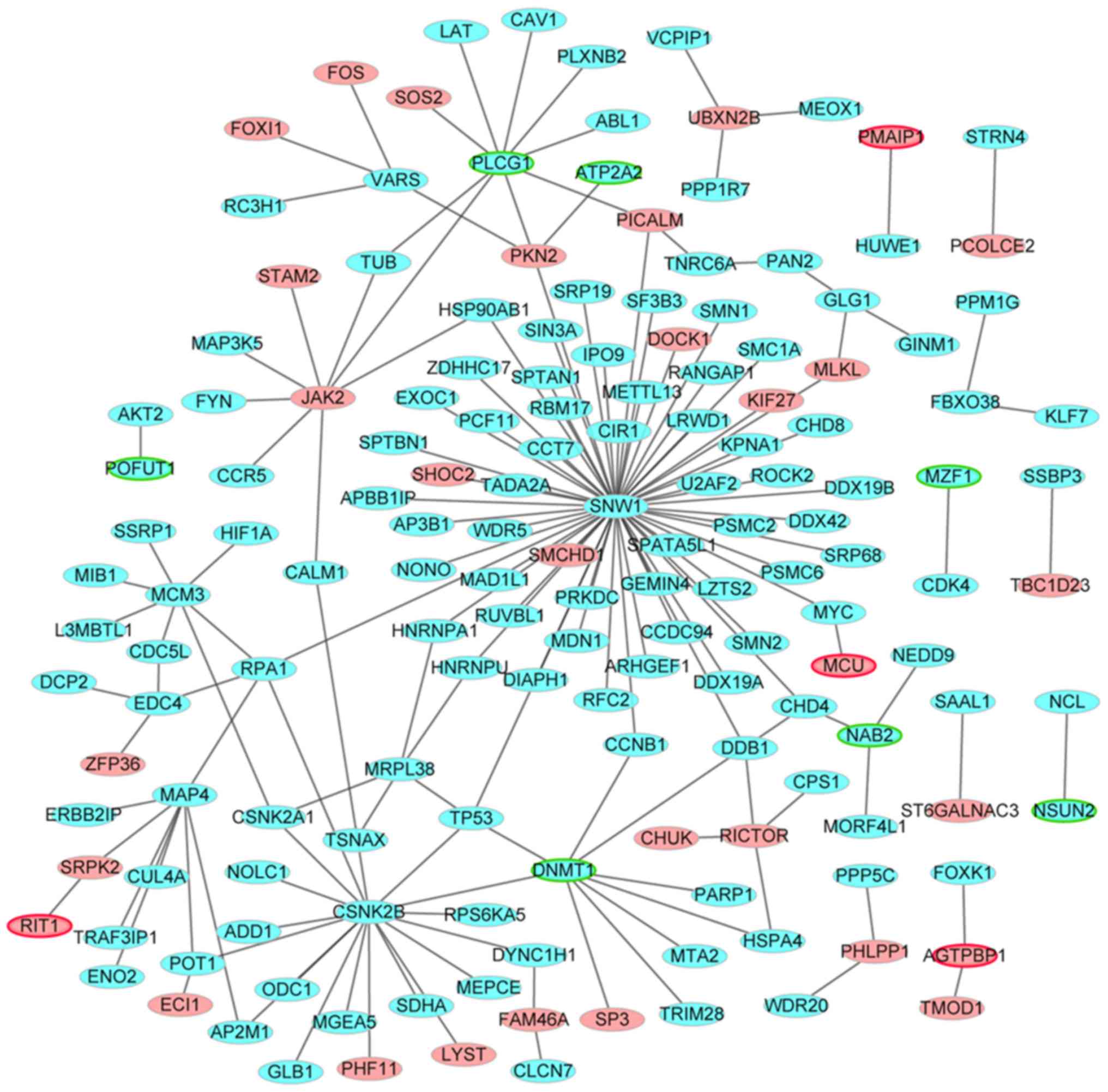
AS-specific PPI network construction
The AS-specific PPI network consisted of 159 nodes
and 164 edges. The top 10 mRNAs that had the highest degrees were
Myc proto-oncogene basic helix-loop-helix transcription factor
(MYC; degree=65), heterogeneous nuclear ribonucleoprotein A1
(HNRNPA1; degree= 40), spectrin-a non-erythrocytic 1 (SPTAN1;
degree=18), TATA-box binding protein associated factor 10 (TAF10;
degree=9), ETS proto-oncogene 1 transcription factor (degree=7),
NCK adaptor protein 1 (degree=6), eukaryotic translation initiation
factor 4B (degree=5), KIAA1033 (degree=5), extended synaptotagmin 1
(degree=5) and tumor protein 53 (degree=5); of these, MYC, HNRNPA1,
SPTAN1 and TAF10 were hub proteins of the AS-specific PPI network
(Fig. 1).
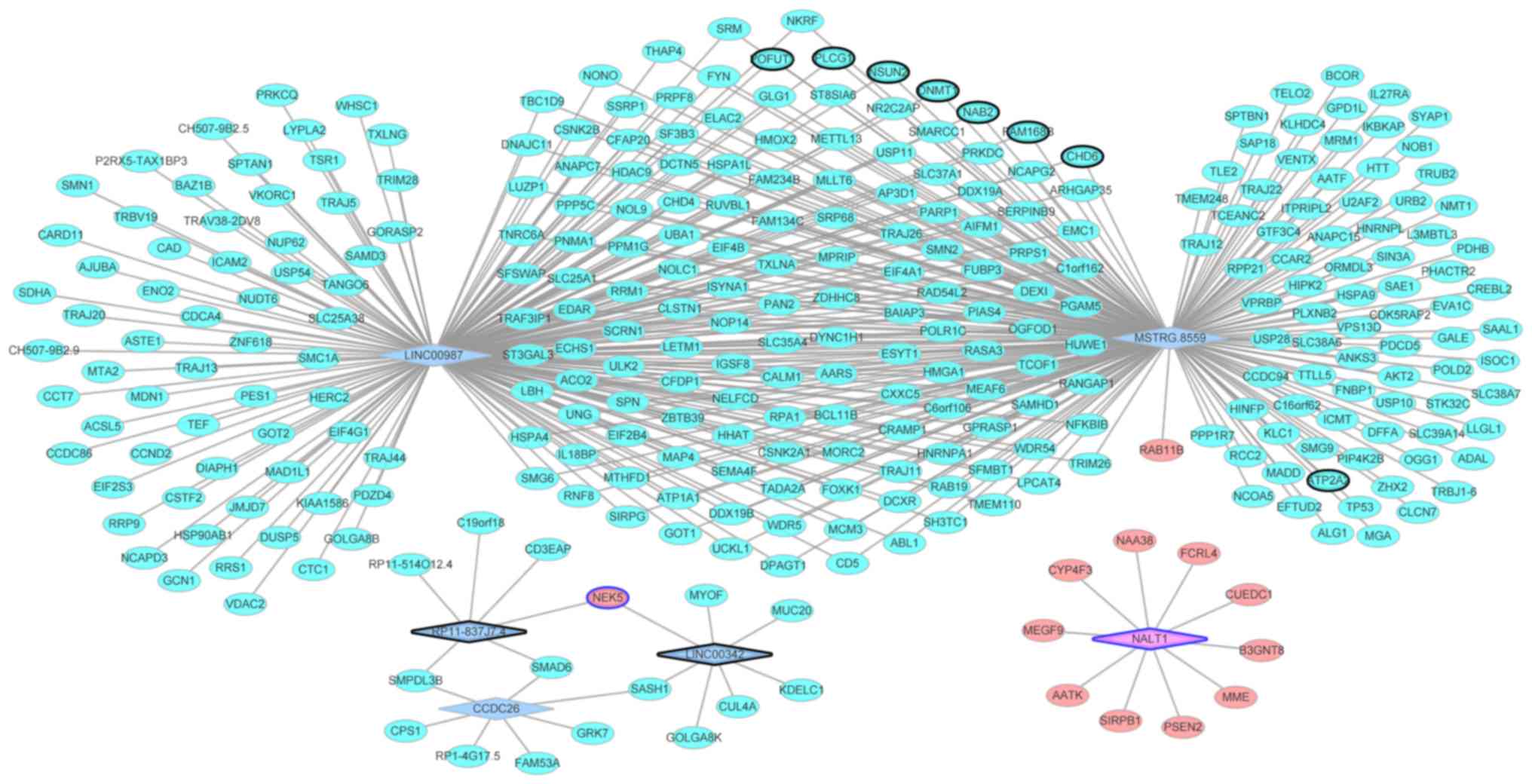
AS-specific weighted DEmRNA-DElncRNA
co-expression network analysis
A total of 3,505 lncRNA-mRNA pairs were identified,
which included 302 DElncRNAs and 602 DEmRNAs with |PCC value ≥0.90|
and P<0.001. Based on these lncRNA-mRNA pairs, the negatively
and positively co-expressed DEmRNA-DElncRNA interaction networks
were constructed, respectively. The top 100 co-expressed
DEmRNA-DElncRNA interaction network is presented in Table III. Based on the positively
co-expressed DEmRNA-DElncRNA interaction network, MSTRG.8559
(degree=226) and long intergenic non-protein coding RNA 987
(LINC00987; degree=209) were the top 2 DElncRNAs that were
co-expressed with the greatest number of DEmRNAs (Fig. 2). The selected co-expression
DEmRNA-DElncRNA interaction network is presented in Fig. 2.
 | Table IIIThe top 100 co-expressed
differentially expressed mRNA-differentially expressed long
non-coding RNA interaction network. |
Table III
The top 100 co-expressed
differentially expressed mRNA-differentially expressed long
non-coding RNA interaction network.
| mRNAsig | lncRNAsig | corsig | pvalsig |
|---|
| OR5B2 | RP11-524O1.4 | 0.999999784 |
7.02×10−14 |
| ADAMTS14 | RP5-906A24.2 | 0.999998855 |
1.97×10−12 |
| ADAMTS14 | KCNMB2-AS1 | 0.99999619 |
2.18×10−11 |
| DMRTC2 | RP11-756H6.1 | 0.9999959 |
2.52×10−11 |
| PRY | RP11-355N15.3 | 0.999995538 |
2.99×10−11 |
| CAMSAP3 | LINC01337 | 0.999993952 |
5.49×10−11 |
| GPR62 | LINC01337 | 0.999993931 |
5.52×10−11 |
| ACTN3 | AC004471.9 | 0.999985033 |
3.36×10−10 |
| COL21A1 | LINC01337 | 0.999984742 |
3.49×10−10 |
| OR5B2 | RP11-91I20.2 | 0.9999843 |
3.70×10−10 |
| SFTA2 | AC004471.9 | 0.999975676 |
8.88×10−10 |
| ADAMTS14 | TSPEAR-AS1 | 0.999948189 |
4.03×10−9 |
| DMRTC2 | TSPEAR-AS1 | 0.999936548 |
6.04×10−9 |
| FOXI1 | RP5-906A24.2 | 0.999888814 |
1.85×10−8 |
| FOXI1 | AC022431.3 | 0.999855298 |
3.14×10−8 |
| ERICH6 | RP1-72A23.4 | 0.999853976 |
3.20×10−8 |
| DMRTC2 | KCNMB2-AS1 | 0.99982545 |
4.57×10−8 |
| FOXI1 | KCNMB2-AS1 | 0.999815958 |
5.08×10−8 |
| OR2C3 | RP11-355N15.3 | 0.999806865 |
5.59×10−8 |
| FOXI1 | RP11-320N21.1 | 0.999759427 |
8.68×10−8 |
| PAK6 | PSMD5-AS1 | 0.999743004 |
9.91×10−8 |
| DMRTC2 | RP5-906A24.2 | 0.999736476 |
1.04×10−7 |
| CAMSAP3 | RP11-254I22.2 | 0.999726506 |
1.12×10−7 |
| GPR62 | RP11-254I22.2 | 0.99972637 |
1.12×10−7 |
| SFTA2 | RP11-320N21.1 | 0.99970509 |
1.30×10−7 |
| ADAMTS14 | RP11-756H6.1 | 0.999704563 |
1.31×10−7 |
| ERICH6 | RP11-254I22.2 | 0.999685951 |
1.48×10−7 |
| COL21A1 | RP11-254I22.2 | 0.999676565 |
1.57×10−7 |
| OR2C3 | RP11-524O1.4 | 0.999662773 |
1.71×10−7 |
| FOXI1 | TSPEAR-AS1 | 0.999646107 |
1.88×10−7 |
| SFTA2 | AC022431.3 | 0.999573412 |
2.73×10−7 |
| PRY | RP11-203E8.1 | 0.999549564 |
3.04×10−7 |
| PRY | RP11-104N10.1 | 0.999534887 |
3.24×10−7 |
| OR2C3 | RP11-91I20.2 | 0.999522111 |
3.43×10−7 |
| PRY | RP4-736L20.3 | 0.999509557 |
3.61×10−7 |
| FCRL4 | CTD-2534I21.8 | 0.999489932 |
3.90×10−7 |
| ADAMTS14 | AC022431.3 | 0.999441016 |
4.69×10−7 |
| PRY | AC008781.7 | 0.999421699 |
5.02×10−7 |
| ACTN3 | RP11-320N21.1 | 0.999325423 |
6.82×10−7 |
| FZD9 | AC008781.7 | 0.999315145 |
7.03×10−7 |
| RP11-505K9.4 | RP11-756H6.1 | 0.999273631 |
7.91×10−7 |
| ADAMTS14 | RP11-320N21.1 | 0.999264308 |
8.12×10−7 |
| FZD9 | RP4-736L20.3 | 0.999211985 |
9.31×10−7 |
| FZD9 | RP11-104N10.1 | 0.99917912 |
1.01×10−6 |
| FOXI1 | RP11-756H6.1 | 0.999170485 |
1.03×10−6 |
| FZD9 | RP11-203E8.1 | 0.99915935 |
1.06×10−6 |
| PDZD4 | RP11-159N11.4 | 0.999141006 |
1.11×10−6 |
| ACTN3 | AC022431.3 | 0.999132502 |
1.13×10−6 |
| RP11-219A15.1 | RP11-457M11.7 | 0.999106184 |
1.20×10−6 |
| CAMSAP3 | RP1-72A23.4 | 0.999004264 |
1.49×10−6 |
| GPR62 | RP1-72A23.4 | 0.999004022 |
1.49×10−6 |
| OR5B2 | RP11-355N15.3 | 0.998989141 |
1.53×10−6 |
| ERICH6 | LINC01337 | 0.998988895 |
1.53×10−6 |
| CXXC5 | LINC01588 | 0.998981114 |
1.56×10−6 |
| FZD9 | RP1-72A23.4 | 0.998951532 |
1.65×10−6 |
| COL21A1 | RP1-72A23.4 | 0.99891675 |
1.76×10−6 |
| NDUFC2-KCTD14 | RP11-45A17.2 | -0.998877807 |
1.89×10−6 |
| MAP4 | MSTRG.8559 | 0.998862021 |
1.94×10−6 |
| PRY | RP11-524O1.4 | 0.998818651 |
2.09×10−6 |
| SRGN | CTD-2562G15.3 | 0.998661182 |
2.69×10−6 |
| RP11-505K9.4 | TSPEAR-AS1 | 0.998635484 |
2.79×10−6 |
| OR2C3 | RP11-203E8.1 | 0.998613936 |
2.88×10−6 |
| OR2C3 | RP11-104N10.1 | 0.998588286 |
2.99×10−6 |
| FOXI1 | AC004471.9 | 0.998585363 |
3.00×10−6 |
| PRY | RP11-91I20.2 | 0.998566098 |
3.08×10−6 |
| ZNF804A | TCEAL3-AS1 | 0.99856452 |
3.09×10−6 |
| FAM111A | MSTRG.6714 | 0.998552192 |
3.14×10−6 |
| OR2C3 | RP4-736L20.3 | 0.998544416 |
3.18×10−6 |
| DMRTC2 | AC022431.3 | 0.998494339 |
3.40×10−6 |
| OR2C3 | AC008781.7 | 0.99839571 |
3.86×10−6 |
| HHAT | AC012074.2 | 0.998323875 |
4.21×10−6 |
| IL18BP | MSTRG.8559 | 0.998273402 |
4.47×10−6 |
| RP5-862P8.2 | RP11-1H8.5 | 0.998269298 |
4.49×10−6 |
| SMAD6 | RP5-1092A3.5 | 0.998257122 |
4.55×10−6 |
| CFAP20 | MSTRG.8559 | 0.998252424 |
4.58×10−6 |
| RP11-505K9.4 | KCNMB2-AS1 | 0.998220553 |
4.75×10−6 |
| DMRTC2 | RP11-320N21.1 | 0.998212166 |
4.79×10−6 |
| PCOLCE2 | RP11-105C19.2 | -0.998176846 |
4.98×10−6 |
| SFTA2 | RP5-906A24.2 | 0.998131862 |
5.23×10−6 |
| FZD9 | RP11-254I22.2 | 0.998116114 |
5.32×10−6 |
| ARL11 | DNAJC3-AS1 | 0.998052713 |
5.68×10−6 |
| MYOF | LINC01151 | -0.998035735 |
5.78×10−6 |
| NT5C2 | RP11-139H15.6 | 0.998022562 |
5.86×10−6 |
| MME | RP11-295I5.4 | 0.998001494 |
5.99×10−6 |
| RP11-505K9.4 | RP5-906A24.2 | 0.99795661 |
6.26×10−6 |
| IL18BP | MYCBP2-AS2 | 0.997935664 |
6.39×10−6 |
| HSPA1L | LINC01588 | 0.997924492 |
6.46×10−6 |
| SAMHD1 | MYCBP2-AS2 | 0.997902735 |
6.59×10−6 |
| SFTA2 | KCNMB2-AS1 | 0.997861652 |
6.85×10−6 |
| CSNK2B | RP11-214K3.23 | 0.997833473 |
7.04×10−6 |
| NOLC1 | MSTRG.8559 | 0.997817608 |
7.14×10−6 |
| TNXB | RP11-441F2.2 | 0.997791211 |
7.31×10−6 |
| RHOB | LINC00671 | 0.997708778 |
7.87×10−6 |
| DDB1 | RP11-429J17.2 | 0.997652855 |
8.26×10−6 |
| ORMDL3 | MYCBP2-AS2 | 0.997626959 |
8.44×10−6 |
| OR5B2 | RP11-927P21.2 | 0.99758326 |
8.75×10−6 |
| ADAMTS14 | AC004471.9 | 0.997577319 |
8.80×10−6 |
| TRIM73 | RP11-565F19.2 | 0.997494288 |
9.41×10−6 |
| PEG3 | RP11-17E13.2 | 0.997467797 |
9.61×10−6 |
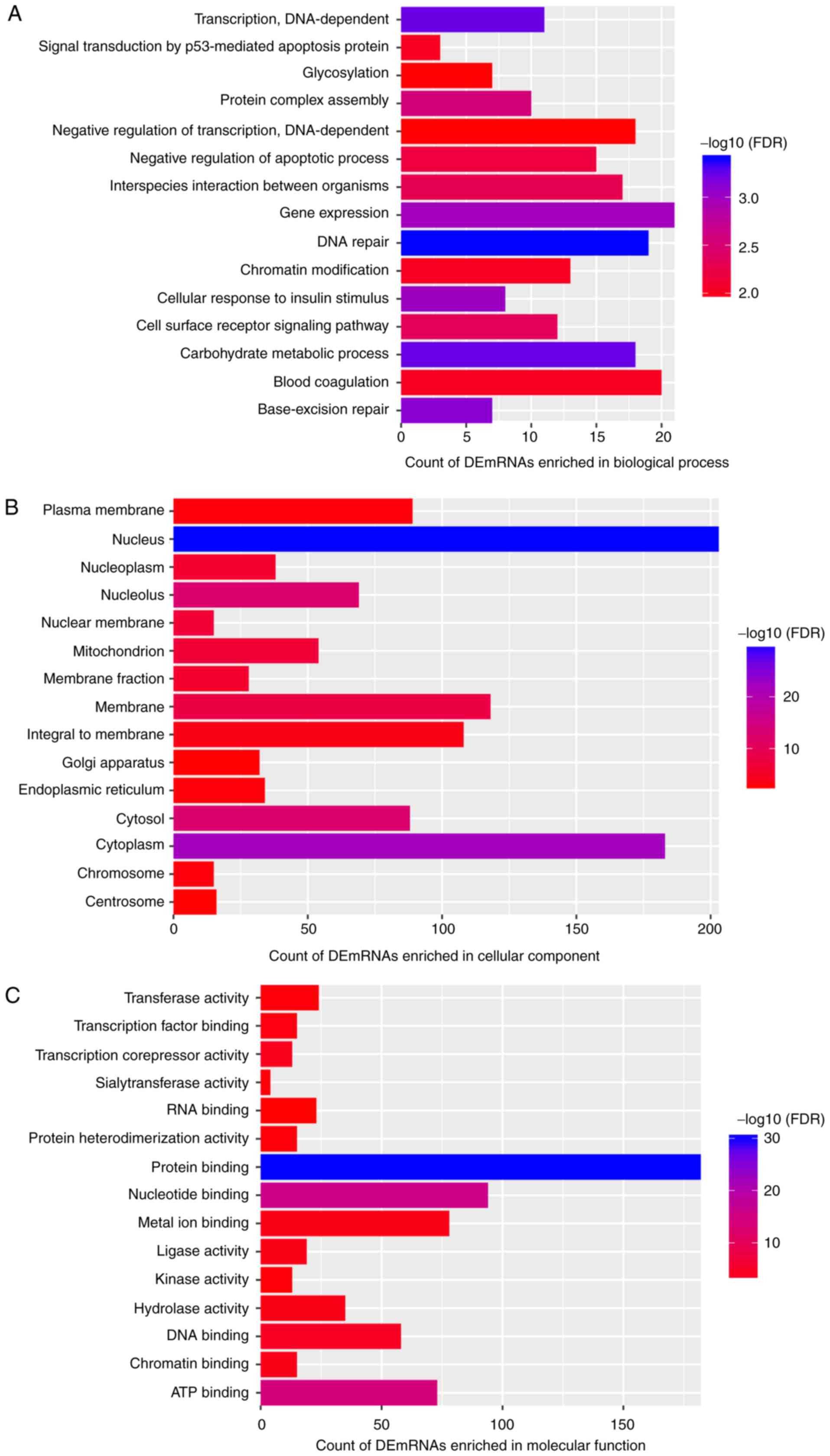
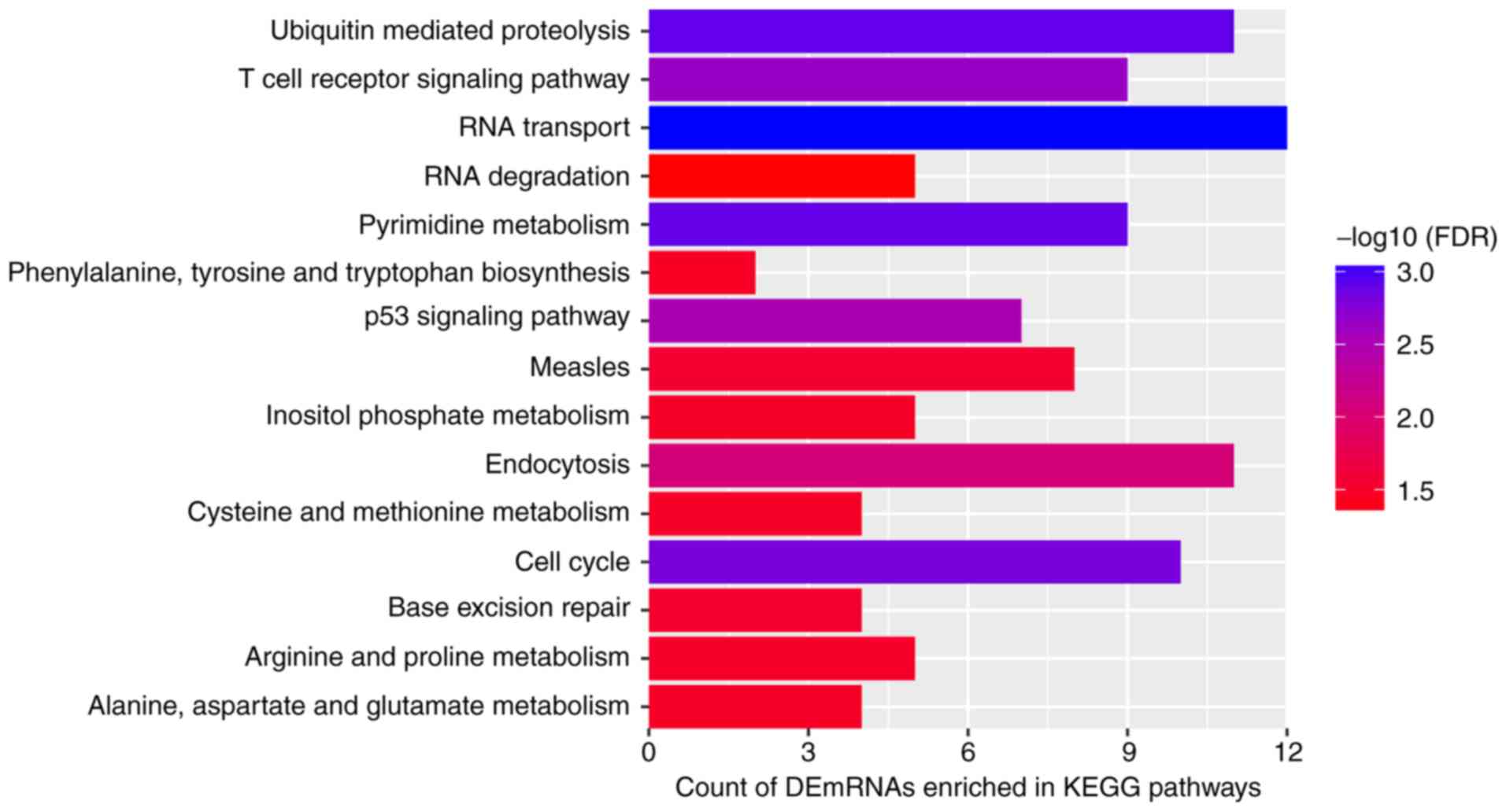
Functional analysis of DEmRNAs
co-expressing DElncRNAs between patients with AS and normal
controls
Based on the GO enrichment analysis (Fig. 3) of the 602 DEmRNAs that were
co-expressed with DElncRNAs in patients with AS and normal
controls, the significantly enriched GO terms were as follows: 'DNA
repair' (FDR=3.94×10−4; Fig. 3A), 'carbohydrate metabolic
process' (FDR=5.66×10−4; Fig. 3A), 'nucleus'
(FDR=3.34×10−30; Fig.
3B) and 'protein binding' (FDR=7.54×10−31; Fig. 3C). 'T-cell receptor signaling
pathway' (FDR=2.29×10−3; Fig. 4) and 'cell cycle'
(FDR=1.52×10−3; Fig.
4) were the significantly enriched KEGG pathways.
DEmRNAs close to DElncRNAs with
cis-regulatory effects
A total of 84 DElncRNAs and nearby cis target DEmRNA
pairs, which included 73 DElncRNAs and 82 DEmRNAs, were obtained
(Table IV).
 | Table IVNearby targeted DEmRNAs of DElncRNAs
between ankylosing spondylitis and normal controls. |
Table IV
Nearby targeted DEmRNAs of DElncRNAs
between ankylosing spondylitis and normal controls.
| Chr | DElncRNA
| Nearby DEmRNA
|
|---|
| Symbol | Start -100 kb | End +100 kb | Symbol | Start | End |
|---|
| chr1 | PIK3CD-AS1 | 9,552,610 | 9,754,586 | CLSTN1 | 9,729,026 | 9824526 |
| chr14 | LINC01588 | 49,827,571 | 50,192,643 | SOS2 | 50,117,120 | 50,231,558 |
| chr22 | AC004471.9 | 19,021,529 | 19,224,503 | SLC25A1 | 19,175,575 | 19,178,830 |
| chr2 | AC007879.5 | 207,139,864 | 207,629,795 | KLF7 | 207,074,137 | 207,167,267 |
| ChrX | INE1 | 47,104,921 | 47,305,865 | UBA1 | 47,190,861 | 472,151,28 |
| ChrX | INE1 | 47,104,921 | 47,305,865 | USP11 | 47,232,690 | 47,248,328 |
| chr1 | TAF1A-AS1 | 222,489,825 | 222,693,032 | MIA3 | 222,618,086 | 222,668,012 |
| chr7 | AC007285.6 | 29,888,600 | 30,125,660 | SCRN1 | 29,920,103 | 29,990,289 |
| chr1 | RP11-318C24.2 | 175,804,762 | 176,020,513 | RFWD2 | 175,944,831 | 176,207,493 |
| chr5 | AC008781.7 | 141,518,414 | 141,726,481 | DIAPH1 | 141,515,016 | 141,619,055 |
| chr3 | U73166.2 | 50,160,303 | 50,363,358 | HYAL3 | 50,292,831 | 50,299,468 |
| ChrX | RP13-314C10.5 | 23,672,992 | 23,882,956 | PRDX4 | 23,664,262 | 23,686,399 |
| chr1 | RP11-195C7.1 | 176,107,648 | 176,329,330 | RFWD2 | 175,944,831 | 176,207,493 |
| chr21 | AP001056.1 | 44,075,489 | 44,276,453 | PWP2 | 44,107,290 | 44,131,181 |
| chr21 | AP001056.1 | 44,075,489 | 44,276,453 | TRAPPC10 | 44,012,319 | 44,106,552 |
| chr1 | RP11-7O11.3 | 43,844,370 | 44,046,551 | ST3GAL3 | 43,705,824 | 43,931,165 |
| chr3 | RP11-379K17.4 | 169,839,353 | 170,066,734 | GPR160 | 170,037,929 | 170,085,403 |
| chr2 | AC092620.2 | 138469090 | 138,674,458 | SPOPL | 138,501,801 | 138,573,547 |
| chr3 | LINC00969 | 195,558,062 | 195,839,964 | MUC20 | 195,720,882 | 195,741,123 |
| chr4 | RP11-15B17.1 | 99,850,006 | 100,295,099 | LAMTOR3 | 99,878,336 | 99,894,490 |
| chr5 | SCAMP1-AS1 | 78,242,365 | 78,460,507 | AP3B1 | 78,000,525 | 78,294,755 |
| chr15 | RP11-30K9.6 | 58,668,072 | 58,870,974 | ADAM10 | 58,588,807 | 58,749,978 |
| chr8 | BAALC-AS1 | 103,056,990 | 103,398,772 | SLC25A32 | 103,39,8635 | 103,415,189 |
| chr5 | RP11-159F24.5 | 43,415,274 | 43,625,310 | NNT | 43,602,692 | 43,707,405 |
| chr5 | LINC01187 | 170,091,579 | 170,299,141 | FOXI1 | 17,010,5897 | 170,109,725 |
| chr8 | LINC01151 | 122,570,385 | 122,794,106 | ZHX2 | 122,781,394 | 122,974,512 |
| chr11 | RP11-727A23.4 | 83,080,144 | 83,284,520 | PCF11 | 83,156,988 | 83,187,451 |
| chr8 | GS1-393G12.12 | 144,214,590 | 144,415,138 | BOP1 | 144,262,102 | 144,291,370 |
| chr12 | RP11-561P12.5 | 8,448,361 | 8,667,613 | CLEC4E | 8,533,305 | 8,540,963 |
| chr12 | RP11-561P12.5 | 8,448,361 | 8,667,613 | CLEC4D | 8,509,475 | 8,522,366 |
| chr12 | RP11-996F15.2 | 29,180,418 | 29,417,848 | ERGIC2 | 29,337,352 | 29,381,189 |
| chr14 | RP11-44N21.1 | 104,993,609 | 105,199,004 | CDCA4 | 105,009,573 | 105,021,148 |
| chr14 | RP11-44N21.1 | 104,993,609 | 105,199,004 | C14 or f79 | 104,985,775 | 105,010,482 |
| chr12 | RP11-753H16.3 | 54,253,661 | 54,597,688 | HNRNPA1 | 54,280,193 | 54,287,088 |
| chr14 | RP11-193F5.1 | 60,779,714 | 61,082,585 | SLC38A6 | 60,981,114 | 61,083,733 |
| chr14 | RP5-1021I20.1 | 73,687,360 | 73,903,270 | PNMA1 | 73,711,783 | 73,714,372 |
| chr3 | AC099668.5 | 49,584,480 | 49,784,983 | MST1 | 49,683,947 | 49,689,501 |
| chr6 | RP11-425D10.10 | 109,282,795 | 109,483,666 | SMPD2 | 109,440,763 | 109,443,919 |
| chr14 C | TD-2547L24.4 | 91,158,299 | 91,359,003 | CCDC88C | 91,271,323 | 91,417,844 |
| chr14 | RP11-524O1.4 | 21,284,292 | 21,484,920 | CHD8 | 21,385,194 | 21,456,126 |
| chr14 | RP11-524O1.4 | 21,284,292 | 21,484,920 | SUPT16H | 21,351,472 | 21,384,266 |
| chr16 | RP11-459F6.3 | 58,029,529 | 58,259,133 | CFAP20 | 58,113,588 | 58,129,450 |
| chr16 | RP11-264B17.3 | 28,874,804 | 29,090,775 | LAT | 28,984,826 | 28,990,783 |
| chr1 | RP11-196G18.22 | 149,744,498 | 149,949,024 | HIST2H2BF | 149,782,689 | 149,812,373 |
| chr16 | LA16c-306E5.3 | 3,358,071 | 3,615,564 | NLRC3 | 3,539,033 | 3,577,400 |
| chr16 | RP11-461A8.4 | 3,550,636 | 3,751,703 | NLRC3 | 3,539,033 | 3,577,400 |
| chr17 | RP1-59D14.5 | 2,275,061 | 2,479,306 | TSR1 | 2,322,503 | 2,337,507 |
| chr17 | RP1-59D14.5 | 2,275,061 | 2,479,306 | METTL16 | 2,405,562 | 2,511,891 |
| chr17 | RP1-59D14.5 | 2,275,061 | 2,479,306 | SMG6 | 2,059,839 | 2,303,771 |
| chr17 | RP11-927P21.2 | 64,799,766 | 65,000,716 | LRRC37A3 | 64,854,312 | 64,919,480 |
| chr17 |
RP11-1094M14.11 | 35,468,109 | 35,674,843 | SLFN14 | 35,548,125 | 35,558,098 |
| chr17 C | TD-2534I21.8 | 44,847,912 | 45,048,939 | EFTUD2 | 44,849,943 | 44,899,662 |
| chr19 | PTOV1-AS1 | 49,738,639 | 49,951,676 | NUP62 | 49,906,825 | 49,929,763 |
| chr20 | RP4-591C20.9 | 63,761,212 | 63,964,293 | UCKL1 | 63,939,829 | 63,956,415 |
| chr19 | AC007292.3 | 4,256,637 | 4,458,448 | CCDC94 | 4,247,079 | 4,269,090 |
| chr19 | RAB11B-AS1 | 8,274,373 | 8,490,685 | RAB11B | 8,389,981 | 8,404,434 |
| chr5 | CTD-2033C11.1 | 65,824,629 | 66,025,135 | ERBIN | 65,926,475 | 66,082,549 |
| chr4 | RP11-572O17.1 | 1,612,821 | 1,813,622 | FAM53A | 1,617,915 | 1,684,302 |
| chr4 | RP11-572O17.1 | 1,612,821 | 1,813,622 | LETM1 | 1,811,479 | 1,856,247 |
| chr1 | RP4-736L20.3 | 10,329,881 | 10,530,677 | DFFA | 10,456,522 | 10,472,526 |
| chr10 | RP11-574K11.29 | 73,603,735 | 73,813,581 | USP54 | 73,497,538 | 73,625,953 |
| chr1 | RP11-156E8.1 | 244,869,350 | 245,071,088 | EFCAB2 | 244,969,705 | 245,127,164 |
| chr3 | RP11-767C1.2 | 12,732,219 | 12,932,728 | IQSEC1 | 12,897,220 | 13,073,117 |
| chr1 | RP11-11N7.4 | 244,764,738 | 244,965,272 | HNRNPU | 244,840,638 | 244,864,560 |
| chr3 | RP11-778D9.13 | 184,032,942 | 184,233,561 | AP2M1 | 184,174,689 | 184,184,091 |
| chr12 | RP1-197B17.4 | 47,631,908 | 47,832,351 | HDAC7 | 47,782,722 | 47,833,132 |
| chr17 | AC142472.6 | 45,046,730 | 45,248,470 | NMT1 | 45,051,610 | 45,109,016 |
| chr15 C | TD-2562G15.3 | 75,352,964 | 75,553,947 | SIN3A | 75,369,379 | 75,455,842 |
| chr17 | RP11-333J10.3 | 36,898,598 | 37,100,034 | AATF | 36,948,875 | 37,056,871 |
| chr15 C | TD-2382E5.6 | 41,808,204 | 42,008,714 | JMJD7 | 41,828,085 | 41,837,581 |
| chr18 | RP11-405M12.3 | 74,970,197 | 75,171,091 | ZNF407 | 74,597,870 | 75,065,671 |
| chr17 | HEXDC-IT1 | 82,325,498 | 82,527,310 | NARF | 82,458,180 | 82,490,537 |
| chr17 | RP11-159D12.6 | 57,906,674 | 58,108,187 | CUEDC1 | 57,861,243 | 57,955,323 |
| chr15 | RP11-1H8.5 | 34315,450 | 34,520,273 | LPCAT4 | 34,358,618 | 34,367,278 |
| chr15 | RP11-1H8.5 | 34,315,450 | 34,520,273 | SLC12A6 | 34,229,996 | 34,338,060 |
| chr5 | CTC-487M23.6 | 112,794,933 | 112,996,531 | SRP19 | 112,861,222 | 112,869,788 |
| chr5 | CTC-487M23.6 | 112,794,933 | 112,996,531 | DCP2 | 112,976,702 | 113,020,970 |
| chr5 | AC005593.2 | 131,697,415 | 131,897,929 | FNIP1 | 131,641,714 | 131,797,063 |
| chr17 | RP13-638C3.5 | 82,548,849 | 82,750,657 | FN3KRP | 82,716,683 | 82,730,328 |
| chr14 | RP11-298I3.6 | 22,923,083 | 23,124,217 | AJUBA | 22,971,174 | 22,982,642 |
| chr16 | RP11-451N19.3 | 58,605,799 | 58,806,297 | SLC38A7 | 58,665,109 | 58,685,104 |
| chr16 | RP11-451N19.3 | 58,605,799 | 58,806,297 | GOT2 | 58,707,131 | 58,734,357 |
| chr19 | CTD-2233K9.1 | 46,846,535 | 47,049,156 | ARHGAP35 | 46,918,676 | 47,005,077 |
| chr6 |
XXbac-BPG283O16.9 | 30,182,349 | 30,386,054 | RPP21 | 30,345,131 | 30,346,884 |
| chr6 |
XXbac-BPG283O16.9 | 30,182,349 | 30,386,054 | TRIM26 | 30,184,455 | 30,213,427 |
Validation of the expression of DEmRNAs
and DElncRNAs using the GSE73754 and GSE25101 datasets
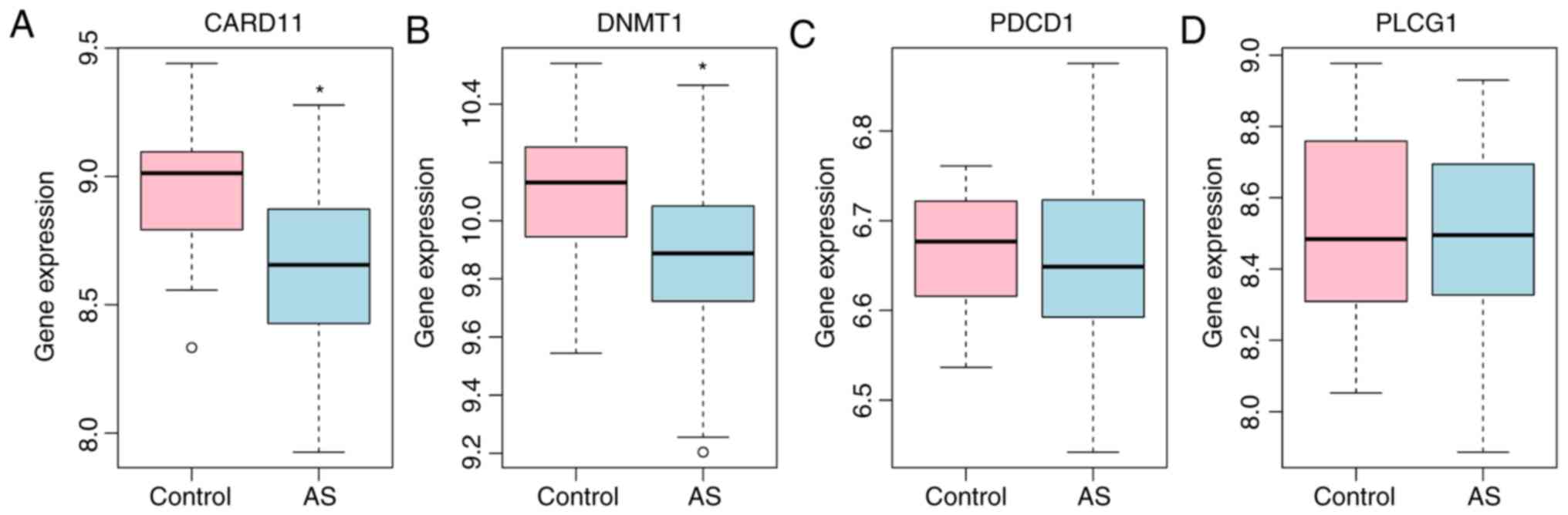
A total of 4 DEmRNAs (DNMT1, PDCD1, CARD11 and
PLCG1) were selected to perform expression validation using the
GSE73754 dataset (Fig. 5). The
expression of all four DElncRNAs was downregulated in patients with
AS when compared with the normal controls, which was generally
consistent with the RNA sequencing data (DNMT1, P<0.05; CARD11,
P<0.05; PDCD1, P>0.05; PLCG1, P>0.05). Three lncRNAs (cat
eye syndrome chromosome region candidate 7, hydatidiform mole
associated and imprinted and KIAA0125) and seven DEmRNAs [MANSC1,
adenosine triphosphatase sarcoplasmic/endoplasmic reticulum
Ca2+ transporting 2 (ATP2A2), myeloid zinc finger 1
(MZF1), PDCD1, DNMT1, CARD11 and PLCG1] were selected to perform
expression validation using the GSE25101 dataset (Fig. 6). The expression of KIAA0125
(P>0.05), MANSC1 (P>0.05), ATP2A2 (P>0.05), MZF1
(P>0.05), PDCD1 (P>0.05), DNMT1 (P<0.0001), CARD11
(P<0.05) and PLCG1 (P>0.05) was generally consistent with the
RNA sequencing data.
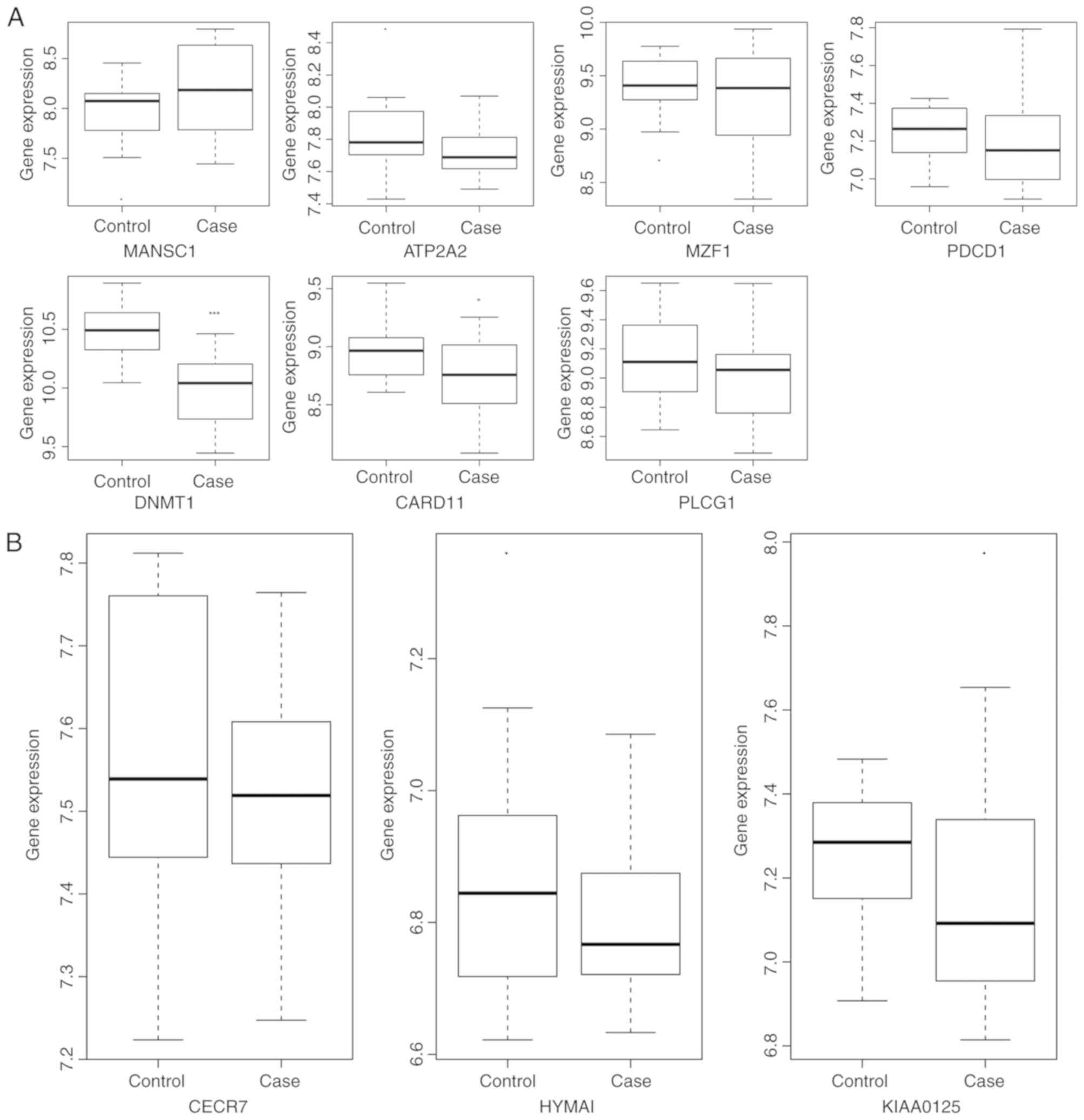 | Figure 6Validation of the expression of
selected DEmRNAs and DElncRNAs between patients with AS and the
normal controls in the GSE25101 dataset. The x-axis presents the AS
and normal control groups and the y-axis presents the expression
levels. (A) The selected DEmRNAs and (B) The selected DElncRNAs.
AS, ankylosing spondylitis; DEmRNA, differentially expressed mRNA;
DElncRNA, differentially expressed long non-coding RNA; CARD11,
caspase recruitment domain-containing protein 11; DNMT1, DNA
methyltransferase 1; PDCD1, programmed cell death 1; PLCG1,
phospholipase Cγ1; MANSC1, MANSC domain containing 1; ATP2A2,
adenosine triphosphatase sarcoplasmic/endoplasmic reticulum
Ca2+ transporting 2; MZF1, myeloid zinc finger 1; CECR7,
cat eye syndrome chromosome region candidate 7; HYMAI, hydatidiform
mole associated and imprinted. |
ROC curve analysis
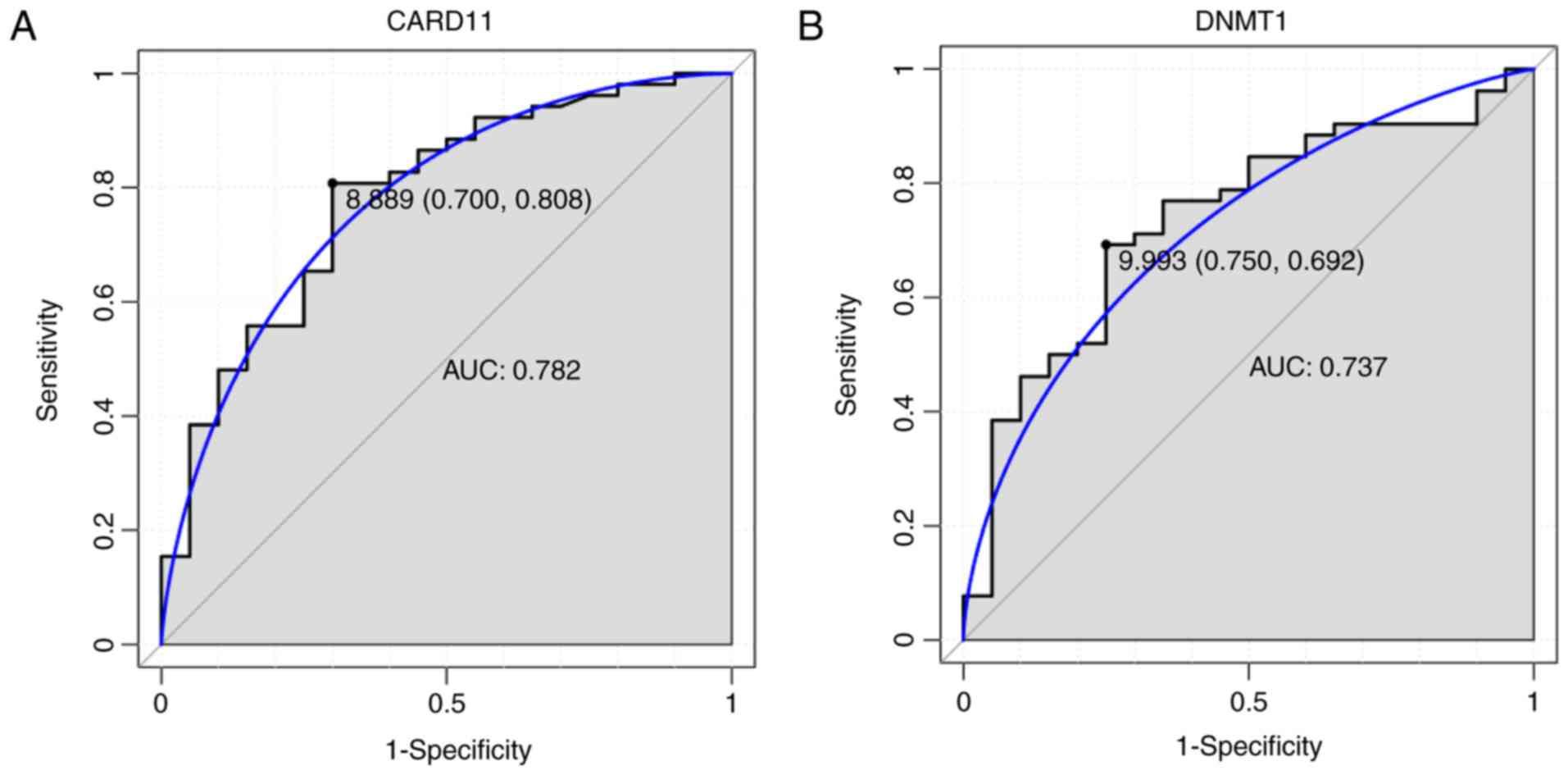
ROC curve analyses and the AUC were used to assess
the discriminatory ability of the four DEmRNAs (DNMT1, PDCD1,
CARD11 and PLCG1) among the 52 patients with AS and 20 normal
controls of the GSE73754 dataset. The AUCs of PDCD1 and PLCG1 were
<0.7 (data not shown). The AUCs of CARD11 and DNMT1 were 0.782
and 0.737, respectively (Fig. 7).
For AS diagnosis, the sensitivity (proportion of true positive) and
1-specificity (proportion of false positive) of CARD11 were 70.0
and 80.8%, respectively, and for DNMT1 were 75.0 and 69.2%,
respectively (Fig. 7).
Discussion
AS is a type of autoimmune disorder that is
associated with HLB-27 and T-cells; however, its etiology and
pathogenesis remain unclear (18). The delays in the diagnosis of AS
and the insufficient responses to the currently available
therapeutics supports the requirement for a greater understanding
of its pathogenesis.
A previous microarray study identified four lncRNAs,
lnc-ZNF354A-1, lnc-LIN54-1, lnc-FRG2C-3 and lnc-USP50-2, that are
involved in the abnormal osteogenic differentiation of mesenchymal
stem cells (MSCs) in patients with AS. The expression of these four
lncRNAs was positively correlated with that of bone morphogenetic
protein 2 and Noggin in MSCs from healthy donors (8). A recent study reported that
lncRNA-AK001085 was downregulated in patients with AS and served as
a potential diagnostic indicator, thus, lncRNA-AK001085 was
considered to be a potential suppressor of AS (9). Due to the lack of research, the
regulatory mechanism of the majority of lncRNAs in AS remains
unknown. In the present study, the key DEmRNAs and DElncRNAs
associated with AS were identified and their functions in AS were
investigated using RNA sequencing and bioinformatics analysis.
LINC00342 was demonstrated to be upregulated in
patients with non-small cell lung cancer and its expression was
positively correlated with lymph node metastasis and the
Tumor-Node-Metastasis stage (19). Coiled-coil domain containing 26
(CCDC26) is also a tumor-associated lncRNA that regulates the
growth of glioma, pancreatic cancer and myeloid leukemia cells
(20-22). In the present study, LINC00342 and
CCDC26 were downregulated in patients with AS, which suggests that
they may serve roles in AS. Further studies are required in order
to identify their precise function in AS.
Furthermore, the results of the present study
indicated that many novel DElncRNAs may be involved in AS. In order
to investigate their functions in AS, a weighted DElncRNA-DEmRNA
co-expression network was constructed and functional annotation of
the DElncRNAs co-expressed with DElncRNAs was performed. A total of
3,505 DElncRNA-DEmRNA co-expression pairs, which included 302
DElncRNAs and 602 DEmRNAs, were obtained. Based on the DEmRNAs
co-expressed with DElncRNAs, 'T-cell receptor signaling pathway'
was a significantly enriched pathway. T-cells have been
demonstrated by previous studies to serve important roles in the
pathology of AS (23-25). HLA-B27-reactive cluster of
differentiation-4+ T-cells have been reported to be
involved in the pathogenesis of spondyloarthropathies (26). The number of peripheral T-helper
(Th)-2 and Th17 lymphocytes has been demonstrated to be increased
in AS, which is suggestive of their potential roles in AS (27,28). Therefore, the results of the
present study support those of previous studies as well as the
importance of the T-cell receptor signaling pathway in AS.
Furthermore, the DEmRNAs that were enriched in the T-cell receptor
signaling pathway, including NF-κB inhibitor β, CARD11, P21
(Ras-related C3 botulinum toxin substrate 1) activated kinase 6,
protein kinase Cθ, PLCG1, PDCD1, FYN proto-oncogene Src family
tyrosine kinase, protein kinase B2 and Vav guanine nucleotide
exchange factor 3, may be involved with AS by regulating the T-cell
receptor signaling pathway.
Among these DEmRNAs, PDCD1 is a known AS-associated
gene. PDCD1 is a member of the immunoglobulin superfamily that is
expressed on the surface of peripheral T-cells, and regulates
T-cell responses and the maintenance of peripheral tolerance
(29,30). A previous study demonstrated that
the expression levels of PDCD1 on activated T-cells were decreased
in patients with AS (1).
Downregulation of PDCD1 may be involved in AS by stimulating the
activity of T-cells and elevating the production of cytokines,
which promotes spinal inflammation and destruction in patients with
AS (1). Downregulated PDCD1 was
also identified in patients with AS in the present study, which
provides evidence to support the results of the previous study.
CARD11 is a shared member of the CARD and
CARD-containing membrane-associated guanylate kinase protein 1
families that has been reported to serve a vital role in regulation
of inflammation and the immune response (31). Although to the best of our
knowledge, there have been no studies that have reported on the
association between CARD11 and AS, downregulated CARD11 has been
implicated in another type of autoimmune disease, rheumatoid
arthritis, via NF-κB activation, reduced Th17 responses and the
decreased production of proinflammatory cytokines, including IL-1β,
IL-6 and IL-17 (10,32,33). Joint inflammation and destruction
were demonstrated to be attenuated by CARD11 small interfering RNA
treatment in mice (10).
Considering the crucial roles of NF-κB activation, Th17 cells and
proinflammatory cytokines in AS, it was hypothesized that CARD11
may also be a key regulator of AS.
Similar to CARD11, PLCG1 has been reported to be
associated with other types of autoimmune disease, including
multiple sclerosis and lymphoproliferative syndrome (11,12). Furthermore, the interaction
between PLCG1 and linker for activation of T-cells (LAT) was
revealed to be involved with the activation and proliferation of
T-cells (11,12). The production of IL-6 by T-cells
is also regulated by PLCG1-LAT (11,12). Therefore, downregulated PLCG1 in
the patients with AS in the present study may serve important roles
in the progression of AS by regulating T-cells and the production
of IL-6.
DNMT1 encodes an enzyme that establishes and
regulates patterns of methylated cytosine residues (13). A previous study observed
downregulated and hypermethylated DNMT1 in the peripheral blood
mononuclear cells of patients with AS when compared with normal
controls, which suggests that DNMT1 may be a potential biomarker of
AS (13). Thus, the
downregulation of DNMT1 observed in patients with AS in the present
study is consistent with this previous study.
Based on the ROC analysis in the present study,
CARD11 and DNMT1 may have great diagnostic value for AS, and
therefore may be potential biomarkers.
LINC00987 was a downregulated lncRNA in the patients
with AS in the present study, and was co-expressed with DNMT1,
CARD11 and PLCG1. In addition, DNMT1 and PLCG1 were co-expressed
with another downregulated lncRNA, MSTRG.8559, in the patients with
AS. Furthermore, LINC00987 and MSTRG.8559 were two hub lncRNAs of
the positively co-expressed DElncRNA-DEmRNA network, which
regulates the majority of the DEmRNAs in AS. It was hypoth-esized
that these two DElncRNAs may serve crucial roles by regulating the
expression of DNMT1, the T-cell receptor signaling pathway and its
associated genes. Further studies are required to further
investigate the biological functions of LINC00987 and MSTRG.8559,
particularly those in AS.
RP11-837J7.4 and NALT1 were the most significantly
down- and upregulated, respectively, DElncRNAs in patients with AS
in the present study; however, their biological functions remain
known. Further studies are required in order to identify whether
these two DElncRNAs could serve as diagnostic biomarkers for
AS.
Previous studies have revealed the prevalence of
lncRNA-mediated cis regulation on nearby transcription (34-36). The 84 AS-specific DElncRNA and
nearby cis target DEmRNA pairs obtained in the present study
provide novel information for understanding the biological
functions of lncRNAs in AS.
In conclusion, the present study obtained lncRNA and
mRNA expression profiles from patients with AS and normal controls
using RNA sequencing. A number of key genes, including PDCD1,
DNMT1, CARD11 and PLCG1, that are involved in AS were identified.
In addition, the results indicated that numerous novel DElncRNAs
may be involved in AS. Furthermore, the functions of DElncRNAs in
AS were investigated using functional annotation of DEmRNAs
co-expressed with DElncRNAs and through the identification of
nearby target DEmRNAs of DElncRNAs. These results may support
further studies on the potential roles of lncRNAs in AS. However,
the sample size for RNA-seq was small, which is a limitation of the
present study, therefore, studies with larger sample sizes are
required in order to confirm this conclusion.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
ZX and GC conceived and designed the experiments.
ZX, XZ and HL performed the experiments. ZX, XZ and QC analyzed the
data. HL and QC were significant contributors in the manuscript.
All authors revised the manuscript and have agreed to the
publication of this manuscript.
Ethics approval and consent to
participate
All of the participants submitted written informed
consent and the present study was approved by the Ethics Committee
of the 2nd Affiliated Hospital, School of Medicine, Zhejiang
University (Zhejiang, China).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Abbreviations:
|
AS
|
ankylosing spondylitis
|
|
DEmRNAs
|
differentially expressed mRNAs
|
|
DElncRNAs
|
differentially expressed long
non-coding RNAs
|
|
GO
|
Gene Ontology
|
|
KEGG
|
Kyoto Encyclopedia of Genes and
Genomes
|
|
PPI
|
protein-protein interaction
|
Acknowledgments
The authors would like to thank Beijing Yangshen
Bioinformatic Technology (Beijing, China) for their assistance
during high-throughput sequencing and data analysis.
References
|
1
|
Zhou L, Zhang Y, Xu H, Hu L, Zhang C, Sun
L, Xie Y, Lu H, Zhang Z, Hu W and Lin X: Decreased programmed
death-1 expression on the T cells of patients with ankylosing
spondylitis. Am J Med Sci. 349:488–492. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Assassi S, Reveille JD, Arnett FC, Weisman
MH, Ward MM, Agarwal SK, Gourh P, Bhula J, Sharif R, Sampat K, et
al: Whole-blood gene expression profiling in ankylosing spondylitis
shows upregulation of toll-like receptor 4–5. J Rheumatol.
38:87–98. 2011. View Article : Google Scholar
|
|
3
|
El Maghraoui A: Extra-articular
manifestations of ankylosing spondylitis: Prevalence,
characteristics and therapeutic implications. Eur J Intern Med.
22:554–560. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Smith JA: Update on ankylosing
spondylitis: Current concepts in pathogenesis. Curr Allergy Asthma
Rep. 15:4892015. View Article : Google Scholar
|
|
5
|
Evans DM, Spencer CC, Pointon JJ, Su Z,
Harvey D, Kochan G, Oppermann U, Dilthey A, Pirinen M, Stone MA, et
al: Interaction between ERAP1 and HLA-B27 in ankylosing spondylitis
implicates peptide handling in the mechanism for HLA-B27 in disease
susceptibility. Nat Genet. 43:761–767. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Davidson SI, Liu Y, Danoy PA, Wu X, Thomas
GP, Jiang L, Sun L, Wang N, Han J, Han H, et al: Association of
STAT3 and TNFRSF1A with ankylosing spondylitis in han chinese. Ann
Rheum Dis. 70:289–292. 2011. View Article : Google Scholar
|
|
7
|
Wright MW and Bruford EA: Naming 'junk':
Human non-protein coding RNA (ncRNA) gene nomenclature. Hum
Genomics. 5:90–98. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Xie Z, Li J, Wang P, Li Y, Wu X, Wang S,
Su H, Deng W, Liu Z, Cen S, et al: Differential expression profiles
of long noncoding RNA and mRNA of osteogenically differentiated
mesenchymal stem cells in ankylosing spondylitis. J Rheumatol.
43:1523–1531. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Li X, Chai W, Zhang G, Ni M, Chen J, Dong
J, Zhou Y, Hao L, Bai Y and Wang Y: Down-regulation of
lncRNA-AK001085 and its influences on the diagnosis of ankylosing
spondylitis. Med Sci Monit. 23:11–16. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wang H, Zhao J, Zhang H, Huang Y, Wang S,
Tu Q and Yang N: CARD11 blockade suppresses murine collagen-induced
arthritis via inhibiting CARD11/Bcl10 assembly and T helper type 17
response. Clin Exp Immunol. 176:238–245. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Mahurkar S, Moldovan M, Suppiah V and
O'Doherty C: Identification of shared genes and pathways: A
comparative study of multiple sclerosis susceptibility, severity
and response to interferon beta treatment. PLoS One. 8:e576552013.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
O'Brien SA, Zhu M and Zhang W: The
importance of IL-6 in the development of LAT-mediated autoimmunity.
J Immunol. 195:695–705. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Aslani S, Mahmoudi M, Garshasbi M,
Jamshidi AR, Karami J and Nicknam MH: Evaluation of DNMT1 gene
expression profile and methylation of its promoter region in
patients with ankylosing spondylitis. Clin Rheumatol. 35:2723–2731.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Langfelder P and Horvath S: WGCNA: An R
package for weighted correlation network analysis. BMC
Bioinformatics. 9:5592008. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Jiang W, Liu Y, Liu R, Zhang K and Zhang
Y: The lncRNA DEANR1 facilitates human endoderm differentiation by
activating FOXA2 expression. Cell Rep. 11:137–148. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wang P, Ning S, Zhang Y, Li R, Ye J, Zhao
Z, Zhi H, Wang T, Guo Z and Li X: Identification of
lncRNA-associated competing triplets reveals global patterns and
prognostic markers for cancer. Nucleic Acids Res. 43:3478–3489.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Fei Q, Bai X, Lin J, Meng H, Yang Y and
Guo A: Identification of aberrantly expressed long non-coding RNAs
in postmenopausal osteoporosis. Int J Mol Med. 41:3537–3550.
2018.PubMed/NCBI
|
|
18
|
Chen WC, Wei CC, Lu HF, Wong HS, Woon PY,
Hsu YW, Huang JD and Chang WC: rs657075 (CSF2) is associated with
the disease phenotype (BAS-G) of ankylosing spondylitis. Int J Mol
Sci. 18:E832017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wang L, Chen Z, An L, Wang Y, Zhang Z, Guo
Y and Liu C: Analysis of long non-coding RNA expression profiles in
non-small cell lung cancer. Cell Physiol Biochem. 38:2389–2400.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Peng W and Jiang A: Long noncoding RNA
CCDC26 as a potential predictor biomarker contributes to
tumorigenesis in pancreatic cancer. Biomed Pharmacother.
83:712–717. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Hirano T, Yoshikawa R, Harada H, Harada Y,
Ishida A and Yamazaki T: Long noncoding RNA, CCDC26, controls
myeloid leukemia cell growth through regulation of KIT expression.
Mol Cancer. 14:902015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wang S, Hui Y, Li X and Jia Q: Silencing
of lncRNA-CCDC26 restrains the growth and migration of glioma cells
in vitro and in vivo via targeting miR-203. Oncol Res.
26:1143–1154. 2018. View Article : Google Scholar
|
|
23
|
Xu H, Liew LN, Kuo IC, Huang CH, Goh DL
and Chua KY: The modulatory effects of
lipopolysaccharide-stimulated B cells on differential T-cell
polarization. Immunology. 125:218–228. 2010. View Article : Google Scholar
|
|
24
|
Huan J, Kaler LJ, Mooney JL, Subramanian
S, Hopke C, Vandenbark AA, Rosloniec EF, Burrows GG and Offner H:
MHC class II derived recombinant T Cell receptor ligands protect
DBA/1LacJ mice from collagen-induced arthritis. J Immunol.
180:1249–1257. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Landers-Ramos RQ, Sapp RM, Jenkins NT,
Murphy AE, Cancre L, Chin ER, Spangenburg EE and Hagberg JM:
Chronic endurance exercise affects paracrine action of
CD31+ and CD34+ cells on endothelial tube formation. Am
J Physiol Heart Circ Physiol. 309:H407–H420. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Azizi E, Massoud A, Amirzargar AA,
Mahmoudi M, Soleimanifar N, Rezaei N, Jamshidi AR, Nikbin B and
Nicknam MH: Association of CTLA4 gene polymorphism in Iranian
patients with ankylosing spondylitis. J Clin Immunol. 30:268–271.
2010. View Article : Google Scholar
|
|
27
|
Yang PT, Kasai H, Zhao LJ, Xiao WG, Tanabe
F and Ito M: Increased CCR4 expression on circulating CD4(+) T
cells in ankylosing spondylitis, rheumatoid arthritis and systemic
lupus erythematosus. Clin Exp Immunol. 138:342–347. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Jandus C, Bioley G, Rivals JP, Dudler J,
Speiser D and Romero P: Increased numbers of circulating
polyfunctional Th17 memory cells in patients with seronegative
spondylarthritides. Arthritis Rheum. 58:2307–2317. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Keir ME, Butte MJ, Freeman GJ and Sharpe
AH: PD-1 and its ligands in tolerance and immunity. Annu Rev
Immunol. 26:677–704. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Shinohara T, Taniwaki M, Ishida Y,
Kawaichi M and Honjo T: Structure and chromosomal localization of
the human PD-1 gene (PDCD1). Genomics. 23:704–706. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Bertin J, Wang L, Guo Y, Jacobson MD,
Poyet JL, Srinivasula SM, Merriam S, DiStefano PS and Alnemri ES:
CARD11 and CARD14 are novel caspase recruitment domain
(CARD)/membrane-associated guanylate kinase (MAGUK) family members
that interact with BCL10 and activate NF-kappa B. J Biol Chem.
276:11877–11882. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Chabaud M, Fossiez F, Taupin JL and
Miossec P: Enhancing effect of IL-17 on IL-1-induced IL-6 and
leukemia inhibitory factor production by rheumatoid arthritis
synoviocytes and its regulation by Th2 cytokines. J Immunol.
161:409–414. 1998.PubMed/NCBI
|
|
33
|
Maddur MS, Miossec P, Kaveri SV and Bayry
J: Th17 cells: Biology, pathogenesis of autoimmune and inflammatory
diseases, and therapeutic strategies. Am J Pathol. 181:8–18. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Yan P, Luo S, Lu JY and Shen X: Cis- and
trans-acting lncRNAs in pluripotency and reprogramming. Curr Opin
Genet Dev. 46:170–178. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Pian L, Wen X, Kang L, Li Z, Nie Y, Du Z,
Yu D, Zhou L, Jia L, Chen N, et al: Targeting the IGF1R pathway in
breast cancer using antisense lncRNA-mediated promoter cis
competition. Mol Ther Nucleic Acids. 12:105–117. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Lipovich L: Abstract IA3: Regulatory
networks in onco-lncRNAomics: Cis-regulation and non-conservation.
Indian J Microbiol. 52:400–405. 2012.
|