Introduction
Osteoblasts are responsible osteogenesis and bone
regeneration. The balance between osteoblasts and osteoclasts is
crucial for the maintenance of normal morphology and bone tissue
strength, in which osteogenic differentiation from mesenchymal stem
cells serves an important role (1,2). A
previous study demonstrated that disturbances in osteogenic
differentiation leads to bone loss (3). This may result in health problems
such as osteoporosis (OP) and osteoarthritis (4). Furthermore, bone defects following
trauma due to the inhibition of osteogenic differentiation severely
compromise patient quality of life (5). Therefore, understanding the
molecular mechanisms associated with osteogenic differentiation is
essential for the treatment of osteogenic disorders.
Non-protein coding RNAs encoded by the human genome
are known to be important regulatory transcripts associated with
multiple biological processes and pathologies (6). Among them, microRNAs (miRNAs/miRs)
are small non-coding RNAs composed of 1-25 nucleotides. miRNAs
regulate the expression of multiple genes by disrupting mRNA
stability and/or inhibiting translation in conjunction with the
RNA-induced silencing complex (6). An increasing number of miRNAs have
emerged as pivotal regulators of osteoblast differentiation.
miR-34a has been identified as an inhibitor of human stromal cell
differentiation through targeting Jagged 1 and downregulating Notch
signaling, resulting in bone formation abnormalities (7). miR-628-3p has been shown to regulate
osteoblast differentiation by targeting runt-related transcription
factor 2 (RUNX2), the master osteoblast transcription factor
(8). miR-375 serves as a
regulator of osteogenic differentiation in human adipose-derived
stem cells, and overexpression of miR-375 promotes osteogenic
differentiation via the Yes-associated protein 1/DEP domain
containing mTOR-interacting protein/protein kinase B regulatory
network (9). These findings
highlight the essential role of miRNAs in the process of osteoblast
differentiation. However, the number of miRNAs regulating
osteoblast differentiation is predicted to be large, and further
research focusing on the role of miRNAs in the regulatory
mechanisms of osteogenesis is required.
miR-223 participates in multiple biological
processes, including myocardial infarction, inflammation and cancer
development (10-14). miR-223 may protect myocardial
cells from hypoxia-induced apoptosis and excessive autophagy by
targeting poly(ADP-ribose) polymerase 1 (10). In addition, the miR-223-5p/-3p
duplex has been verified to cooperatively inhibit
ischemia/reperfusion-induced cardiac necroptosis (11). miR-223 is involved in the
regulation of neutrophil wound response and nuclear factor-κB
activation by directly targeting Cullin1a/b, tumor necrosis factor
receptor-associated factor 6, and transforming growth
factor-β-activated kinase 1 (MAP3K7)-binding protein 1 (12). miR-223-5p inhibits the migration
and invasion of bladder cancer cells by regulating anillin
actin-binding protein (13).
Decreased miR-223 levels abate the osteogenic differentiation
potential of bone marrow mesenchymal stromal cells derived from
patients with multiple myeloma (14), suggesting that that miR-223 may
serve as a regulator of osteogenic differentiation.
The present study demonstrated that miR-223-5p has
critical function in osteoblast differentiation. miR-223-5p was
upregulated during consecutive osteogenic induction, and promoted
osteogenic differentiation. Furthermore, histone deacetylase 2
(HDAC2) was identified as a target of miR-223-5p, and
downregulation of HDAC2 by miR-223-5p induced osteoblast
differentiation. Therefore, miR-223-5p may represent a potential
therapeutic target for bone regeneration-associated diseases.
Materials and methods
Cell culture and osteogenic
differentiation
MC3T3-E1 cells were obtained from the American Type
Culture Collection (Manassas, VA, USA) and routinely maintained in
growth medium (GM) consisting of α-modified Eagle's medium (α-MEM)
supplemented with 10% fetal bovine serum (both Gibco; Thermo Fisher
Scientific, Inc., Waltham, MA, USA), penicillin (50 U/ml) and
streptomycin (50 µg/ml) at 37°C in a humidified atmosphere
with 5% CO2. To induce osteogenic differentiation, the
cells were incubated with osteogenic medium (OM), containing 10
ng/ml β-glycerophosphate (Sigma-Aldrich; Merck KGaA, Darmstadt,
Germany), 10−7 mmol/l dexamethasone (Sigma-Aldrich;
Merck KGaA), and 50 µg/ml vitamin C (Sigma-Aldrich; Merck
KGaA) for 7 or 14 days. Next, the induced cells were digested for
further detection. For the CAY10683 treatment, CAY10683 (Selleck
Chemicals, Houston, TX USA) was added to the OM at concentrations
of 0.01, 0.1 and 1 µM for 7 days. CAY10683 was dissolved in
dimethyl sulfoxide (Sigma-Aldrich; Merck KGaA).
Cell transfection
miR-223-5p mimics, inhibitor and their corresponding
negative controls (NCs) were purchased from Guangzhou RiboBio Co.,
Ltd. (Guangzhou, China). The sequences were as follows: miR-223-5p
mimic, 5′-CGUGUAUUUGACAAGCUGAGUUG-3′; miR-223-5p inhibitor,
5′-CAACUCAGCUUGUCAAAUACACG-3′; mimics-NC,
5′-UUUGUACUACACAAAAGUACUG-3′; and inhibitor-NC,
5′-UUUGUACUACACAAAAGUACU G-3′. Small interfering (si)RNAs against
HDAC2 (5′-AAGCCUCAUAGAAUCCGCAUG-3′) and plasmid overexpressing
HDAC2 (HDAC2-pcDNA3.1) were purchased from Shanghai GenePharma Co.,
Ltd. (Shanghai, China). MC3T3-E1 cells were cultured in six-well
plates at a concentration of 2×105 cells/well. When
cells reached 70-80% confluence, mimics (50 nM), inhibitor (25 nM),
siRNAs (25 nM) or plasmids (2.5 µg/ml) were transfected into
cells using Lipofectamine® 3000 (Invitrogen; Thermo
Fisher Scientific, Inc.) according to the manufacturer's protocol.
After 48 h incubation, cells were subjected to further
experiments.
RNA isolation and reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was extracted with TRIzol®
reagent (Invitrogen; Thermo Fisher Scientific, Inc.) and the
reverse transcription reaction was performed with 1 µg total
RNA using a PrimeScript™ RT reagent kit according to the
manufacturer's protocol. RT-qPCR was conducted with SYBR-Green I
(both Takara Bio, Inc., Otsu, Japan) on the LightCycler 480 II
system (Roche Diagnostics, Basel, Switzerland). The thermocycling
conditions were as follows: 95°C for 30 sec, followed by 36 cycles
of denaturation at 95°C for 15 sec, annealing at 60°C for 15 sec
and extension at 72°C for 15 sec. Primer sequences were as follows:
miR-223-5p, 5′-CGCGCGTGTATTTGACAAGC-3′, and
5′-AGTGCAGGGTCCGAGGTATT-3′; U6, 5′-CTCGCTTCGGCAGCACA-3′, and
5′-AACGCTTCACGAATTTGCGT-3′; Alkaline phosphatase (ALP),
5′-TGGCTCTGCCTTTATTCCCTAGT-3′, and 5′-AAATAAGGTGCTTTGGGAATCTGT-3′;
Osteocalcin (OCN), 5′-GCCATCACCCTGTCTCCTAA-3′, and
5′-GCTGTGGAGAAGACACACGA-3′; RUNX2, 5′-GCCGGGAATGATGAGAACTA-3′, and
5′-GGTGAAACTCTTGCCTCGTC-3′; HDAC2, and
5′-GCTATTCCAGAAGATGCTGTTC-3′, 5′-GTTGCTGAGCTGTTCTGATTTG-3′;
β-actin, 5′-TCACCCACACTGTGCCCAT-3′, and 5′-CTCTTGCTCGAAGTCCAGGG-3′.
The 2−ΔΔCq method was used to quantify mRNA and miRNA
expression. Data were normalized to β-actin (mRNA) or U6
(miRNA).
Measurement of alkaline phosphatase (ALP)
activity
Cultured cells were rinsed in PBS three times, and
total protein was extracted using radioimmunoprecipitation assay
(RIPA) lysis buffer (Sigma-Aldrich; Merck KGaA) and quantified with
a bicinchoninic acid (BCA) protein assay. ALP activity was measured
with the Alkaline Phosphatase, Diethanolamine Detection kit (cat.
no. AP0100; Sigma-Aldrich; Merck KGaA). Equal volumes of cell
lysate (50 µl) were added to each well of the 96-well
plates, and incubated with an ALP staining solution at 37°C for 1
h. Following the addition of stop solution, ALP activity was
measured spectrophotometrically at 405 nm, and normalized to total
protein concentration.
Alizarin Red S staining
Mineralization was determined by Alizarin red S
staining. Cells were plated in six-well plates (5×105
cells/well), fixed with 4% paraformaldehyde for 10 min at room
temperature and stained with 0.1% Alizarin red staining solution
(pH 4.2; Sigma-Aldrich; Merck KGaA) at room temperature for 20 min.
The cells were washed with PBS, and images were captured under a
light microscope (×200).
Western blotting
The cells were harvested and lysed with RIPA lysis
buffer containing phenylmethane sulfonyl fluoride and protease
inhibitors (both Sigma-Aldrich; Merck KGaA). The concentration of
each sample was determined by a BCA protein assay. Equal amounts of
protein (30 µg/lane) were loaded and separated by 10%
SDS-PAGE, followed by transfer onto 0.4 µm polyvinylidene
difluoride membranes (EMD Millipore, Billerica, MA, USA). The
membranes were blocked with 5% non-fat milk at room temperature for
1 h, and incubated with different primary antibodies overnight at
4°C. The primary antibodies included anti-ALP rabbit polyclonal
antibody (1:1,000; cat. no. 11187-1-AP; ProteinTech Group, Inc.,
Chicago, IL, USA), anti-OCN rabbit polyclonal antibody (1:500; cat.
no. 23418-1-AP; ProteinTech Group, Inc.), anti-RUNX2 rabbit
polyclonal antibody (1:800; cat. no. 12556; Cell Signaling
Technology, Inc., Danvers, MA, USA), anti-HDAC2 rabbit polyclonal
antibody (1:1,000; cat. no. 12922-3-AP; ProteinTech Group, Inc.),
and anti-β-actin rabbit polyclonal antibody (1:2,000; cat. no.
20536-1-AP; ProteinTech Group, Inc.). Membranes were next probed
with horseradish peroxidase-conjugated anti-rabbit secondary
antibody (1:5,000; cat. no. 7074; Cell Signaling Technology, Inc.),
and protein signals were obtained with enhanced chemiluminescence
plus substrate (EMD Millipore).
In vivo transplantation
All animal procedures were approved by the Animal
Care Committee of Southern Medical University. Healthy female
NOD/SCID mice (5 weeks old; ~20 g) were purchased from Guangdong
Medical Laboratory Animal Center (Guangdong, China) were randomly
divided into two groups, with six mice per group. Transfected cells
(5×106) were loaded onto 20 mg hydroxyapatite-tricalcium
phosphate scaffold (HA-TCP; Sigma-Aldrich; Merck KGaA) and
subcutaneously implanted into the dorsal region of NOD/SCID mice
under anesthesia. After 4 weeks, xenografts were removed, fixed
with 4% paraformaldehyde for 2 days at room temperature and
decalcified in 10% EDTA (pH 6.0) for another 7 days at room
temperature. Xenografts were then embedded in paraffin, sectioned
at 4 µm thickness and stained with hematoxylin and eosin
(H&E; Beyotime Institute of Biotechnology, Shanghai, China), or
Masson's Trichrome stain (Sigma-Aldrich; Merck KGaA), according to
the manufacturer's protocols.
Statistical analysis
All data were presented as the mean ± standard
deviation. Statistical analyses were performed with SPSS software,
version 19.0 (IBM Corp., Armonk, NY, USA). The significance of mean
values between two groups was analyzed using a two-tailed unpaired
Student's t-test. Differences in multiple groups were determined by
one-way analysis of variance with subsequent Bonferroni correction.
P<0.05 was considered to indicate a statistically significant
difference.
Results
miR-223-5p is upregulated in osteogenic
differentiation
To investigate the role of miR-223-5p in osteogenic
differentiation, the present study detected its dynamic expression
profiles in MC3E3T1 cells incubated with osteogenic inducers. The
expression levels of osteogenesis-associated genes, including ALP,
OCN and RUNX2 were significantly upregulated under
differentiation-inducing conditions. On day 14, they had increased
by 3 (ALP), 5.1 (OCN) and 3.9 (RUNX2) fold, compared with day 0
(Fig. 1A). The upregulation of
RUNX2, ALP and OCN protein was also confirmed by western blotting
(Fig. 1B). Additionally, the
activity of ALP, an indicator of mineralization induced by
osteogenic differentiation, was promoted following induction
(Fig. 1C). Furthermore,
miR-223-5p expression in MC3T3-E1 cells gradually increased during
osteogenic differentiation, reaching a >4 fold increase at day
14, compared with day 0 (Fig.
1D), indicating the potential involvement of miR-223-5p in
osteogenic induction.
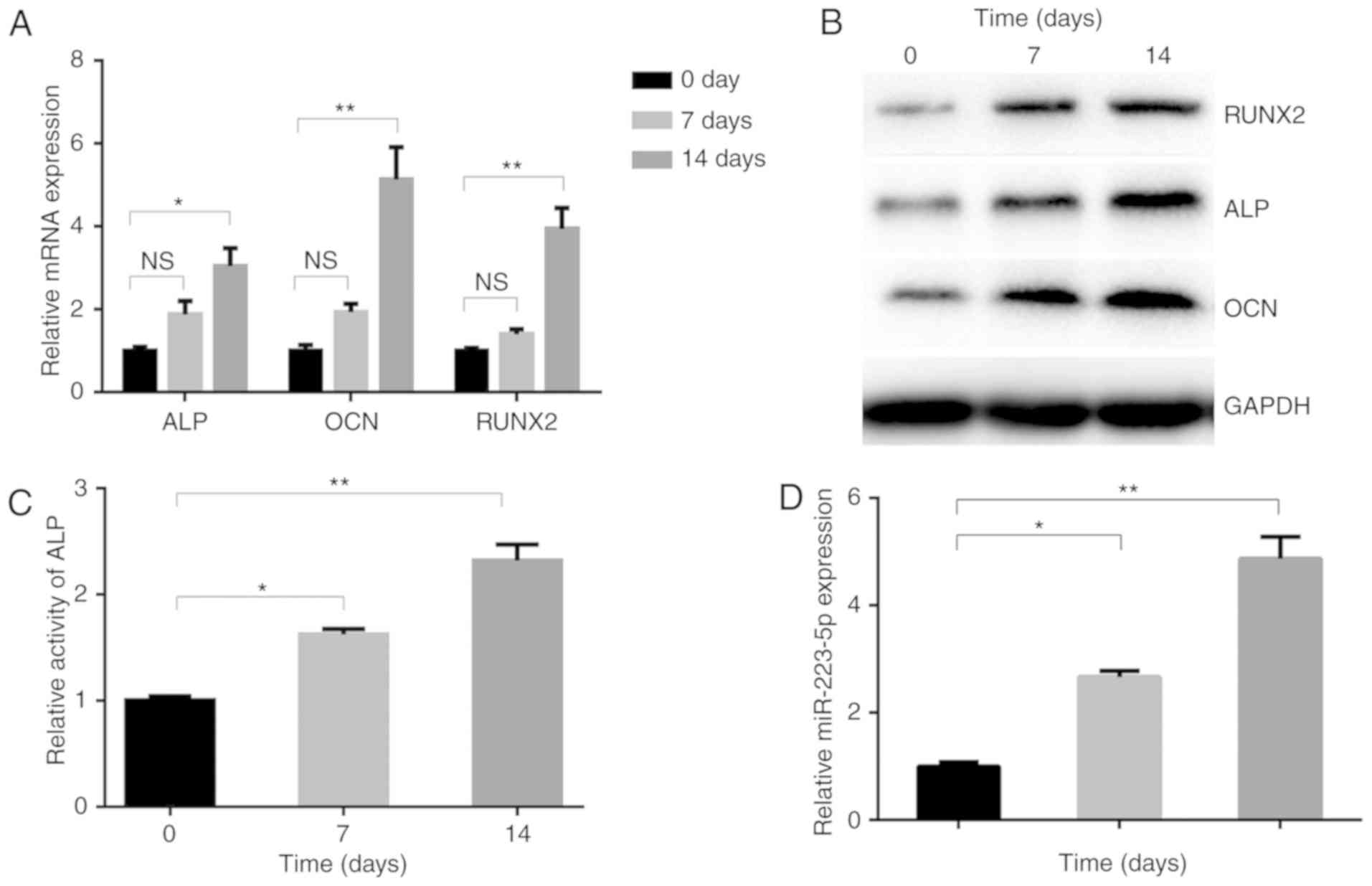 | Figure 1miR-223-5p is upregulated in
osteogenic differentiation. (A) Osteogenesis-associated gene and
(B) protein expression, including ALP, OCN and RUNX2, in MC3T3-E1
cells following treatment with OM for 0, 7 and 14 days. (C) ALP
activity and (D) the expression of miR-223-5p was assayed by
RT-qPCR at 0, 7 and 14 days after OM treatment.
*P<0.05, **P<0.01. OM, osteogenic
medium; miR, microRNA; RT-qPCR, reverse transcription-quantitative
polymerase chain reaction; ALP, alkaline phosphatase; OCN,
osteocalcin; RUNX2, runt-related transcription factor 2; NS, not
significant. |
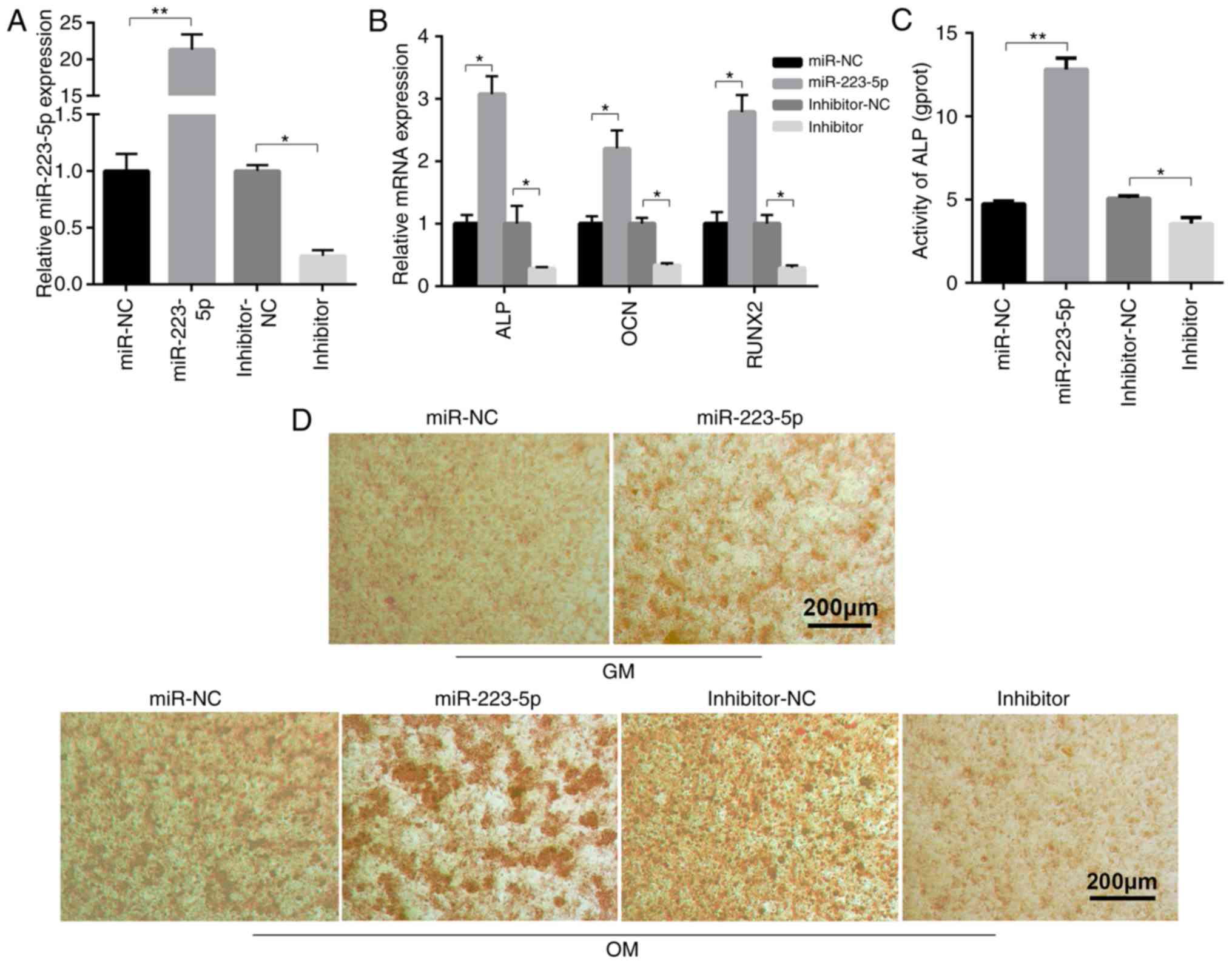
miR-223-5p contributes to osteogenic
differentiation and mineralization
To clarify the biological roles of miR-223-5p in
osteogenic differentiation, miR-233-5p overexpression and
inhibition was induced in MC3T3-E1 cells. The effect of miR-223-5p
and HDAC2 on osteogenic differentiation can be detected in 7 days
OM treatment. The expression of ALP, OCN and RUNX2 increased in 7
days OM treatment although they had no statistical significance.
While miR-223-5p treatment promoted the expression of ALP, OCN and
RUNX2 after 7 days of OM treatment, which was confirmed by RT-qPCR.
miR-223-5p overexpression and silencing was shown to be successful
by RT-qPCR (Fig. 2A). miR-223-5p
mimic transfection markedly increased the expression of
ossification-associated genes, compared to transfection with
negative control (NC) miRNA mimics. In contrast, inhibiting
miR-223-5p expression significantly reduced ossification-associated
gene expression (Fig. 2B).
Further results revealed that ALP activity in the differentiated
MC3T3-E1 cells was inhibited by transfection with miR-223-5p
mimics, but enhanced by the miR-223-5p inhibitor, compared with the
corresponding NC group (Fig. 2C).
The degree of cell mineralization, as determined by Alizarin red S
staining, increased following miR-223-5p mimic transfection
compared with the control group in MC3T3-E1 cells cultured with
either GM or OM medium. miR-223-5p inhibitor reduced osteogenic
differentiation, as shown by the notable reduction in
mineralization nodules (Fig. 2D).
Together, these results indicated that miR-223-5p served an
important role in osteogenic differentiation and
mineralization.
HDAC2 is a negative target of miR-223-5p
and is involved in osteogenic differentiation
It has previously been reported that HDAC2 is a
downstream target of miR-223-5p in chronic obstructive pulmonary
disease (15), but their
relationship in osteogenic differentiation remains to be
elucidated. In the present study, it was investigated whether
miR-223-5p regulated HDAC2 expression. RT-qPCR and western blotting
demonstrated that HDAC2 gene expression was reduced by 70% of the
NC group in the presence of miR-223-5p mimics, and increased >3
fold when transfected with miR-223-5p inhibitor, compared with the
inhibitor NC group (Fig. 3A).
HDAC2 protein expression was also decreased in
miR-223-5p-overex-pressing MC3T3-E1 cells, and increased in
MC3T3-E1 cells with downregulated miR-223-5p (Fig. 3B). RT-qPCR revealed that HDAC2
mRNA expression markedly decreased during osteoblast
differentiation (Fig. 3C). To
evaluate the role of HDAC2 in osteoblast differentiation, the
present study trans-fected MC3T3-E1 cells with si-HDAC2 (Fig. 3D). ALP, OCN and RUNX2 mRNA
(Fig. 3E) and protein (Fig. 3F) expression was increased in
cells transfected with si-HDAC2. Consistently, CAY10683, a HDAC2
inhibitor, induced a dose-dependent decrease in HDAC2 expression,
and a consequent increase in osteogenic-associated gene and protein
expression (Fig. 3G and H).
Furthermore, si-HDAC2 transfection increased ALP activity, compared
with the NC group (Fig. 3I).
Alizarin red S staining also revealed that HDAC2 silencing
increased mineralized bone matrix formation (Fig. 3J). Next, MC3E3T1 from control
groups and si-HDAC2 groups were loaded onto HA-TCP and then
implanted into NOD/SCID mice. The results showed that the MC3E3T1
in the si-HDAC2 groups group formed more osteoids than those in the
negative control groups (Fig.
3K).
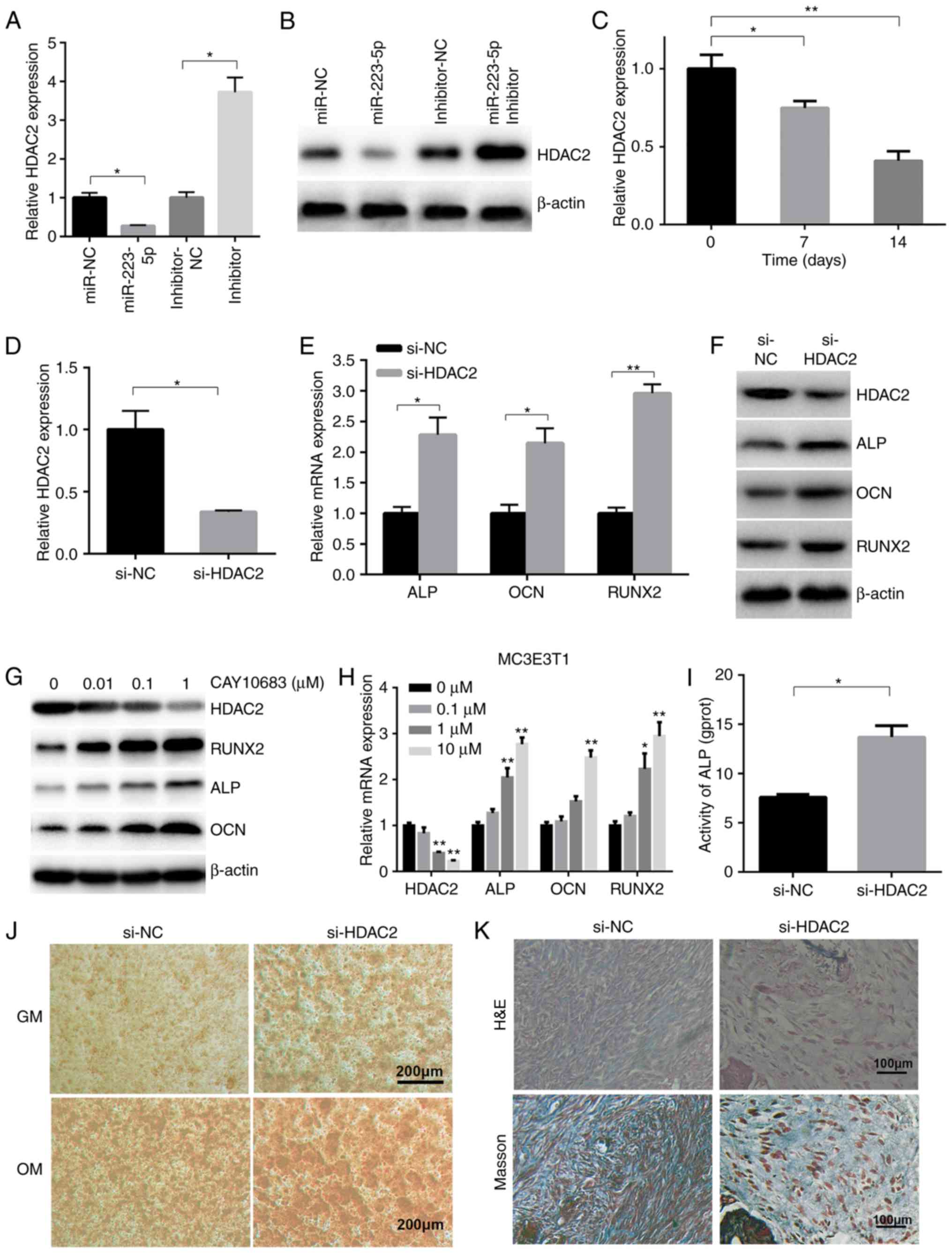 | Figure 3HDAC2 is a target of miR-223-5p and
is involved in osteogenic differentiation. (A) The gene and (B)
protein expression of HDAC2 was detected in transfected MC3T3-E1
cells. (C) After 0, 7 and 14 days of OM treatment, HDAC2 expression
was determined by RT-qPCR. (D) Silencing of HDAC2 expression was
validated in MC3T3-E1 cells transfected with si-HDAC2 or NC by
RT-qPCR. (E) ALP, OCN and RUNX2 mRNA, as well as (F) ALP, OCN,
RUNX2 and HDAC2 protein expression, was detected in MC3T3-E1 cells
transfected with si-HDAC2, 7 days after osteogenic induction.
*P<0.05, **P<0.01. (G) ALP, OCN, HDAC2
and RUNX2 mRNA and (H) protein expression was also detected in
MC3T3-E1 cells treated with OM and CAY10683 at the indicated
concentrations for 7 days. **P<0.01 vs. 0 µM
group. (I) ALP activity in si-HDAC2-transfected MC3T3-E1 cells
after 7 days of OM treatment. (J) The osteogenic differentiation of
MC3T3-E1 cells transfected with si-HDAC2 was determined by Alizarin
Red S staining. (K) Osteogenic differentiation of xenografts was
determined by H&E and Masson's Trichrome staining. miR,
microRNA; RT-qPCR, reverse transcription-quantitative polymerase
chain reaction; HDAC2, histone deacety-lase 2; ALP, alkaline
phosphatase; OCN, osteocalcin; RUNX2, runt-related transcription
factor 2; OM, osteogenic medium; H&E, hematoxylin and
eosin. |
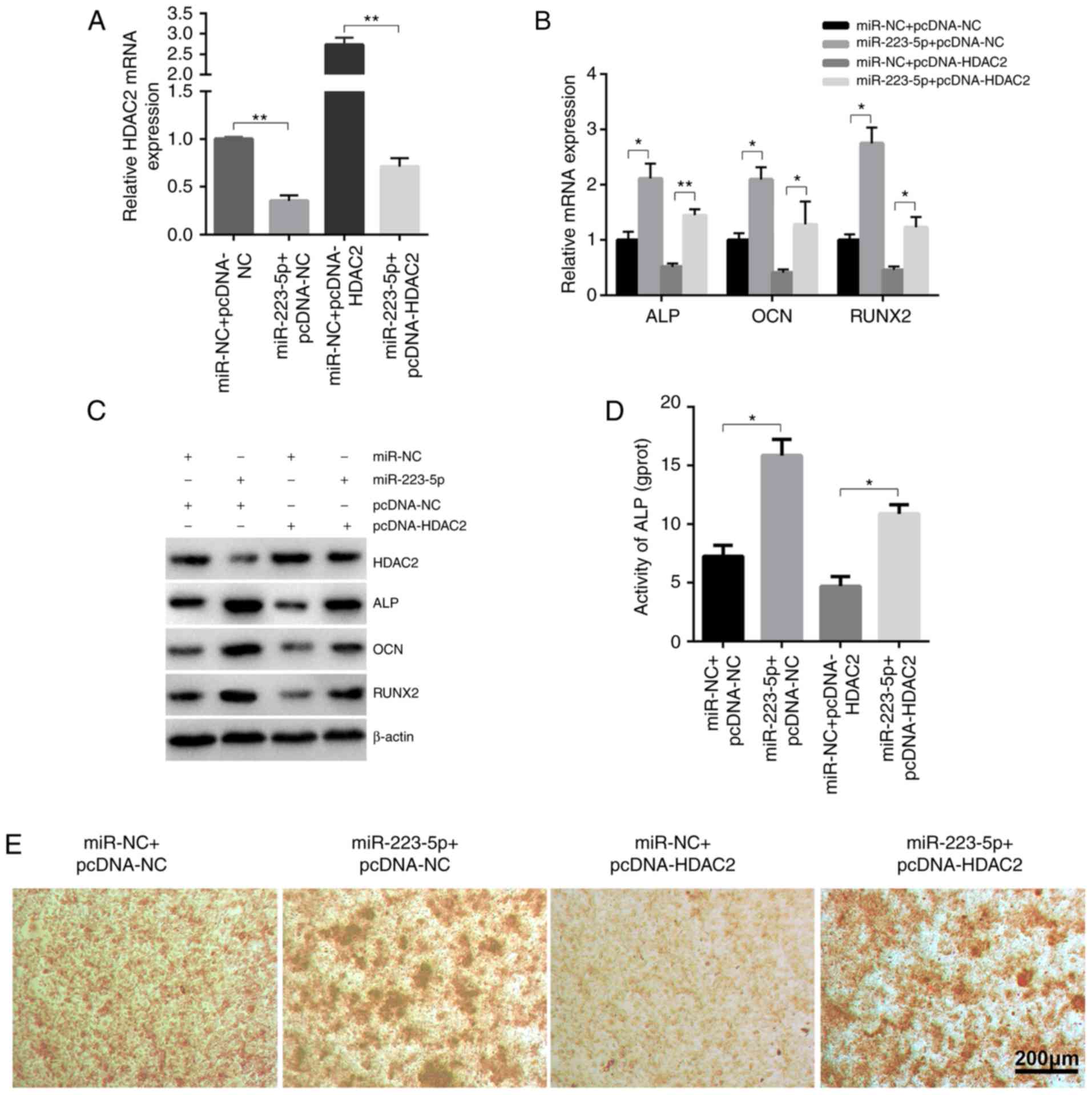
To further determine the role of HDAC2 in
miR-223-5p-mediated osteogenic differentiation,
HDAC2-overexpressing and control groups were established by
transfecting pcDNA3.1-HDAC2 plasmid and pcDNA3.1-NC, which were
then co-transfected with miR-223-5p mimics or mimics-NC (Fig. 4A). The upregulating effects of
miR-223-5p on the expression of ossification-associated genes were
markedly inhibited by HDAC2, as confirmed by RT-qPCR (Fig. 4B), western blot analysis (Fig. 4C), ALP activity (Fig. 4D) and Alizarin red S staining
(Fig. 4E). Collectively, these
findings suggested that HDAC2 may be a negative regulator of
osteogenic differentiation.
Discussion
OP is a common disease, with an incidence of 13.2%
in China (16). The serious
complications of OP, including osteoporotic fractures, pain and
misshapen bones, are associated with a high cost of treatment and
prevention, and pose a major socioeconomic burden (17). Abnormal stimulation of osteoclasts
generally results in bone loss, which leads to OP, and this disease
is primarily characterized by decreased bone mass and disorders of
the bone microstructure (18-20). Current therapies for OP, such as
bone resorption inhibitors and bisphosphonate, are mainly focused
on the balance of bone remodeling, which is critical during
maintenance and regeneration of bone tissue. However, effective
methods for promoting bone formation are still under development
(21,22). Therefore, novel effective methods
for treating OP and promoting bone synthesis are urgently
needed.
miRNAs belong to a small RNA family and have
important regulatory properties (23). miRNAs play key roles in the
physiological processes and in the pathogenic mechanism of numerous
diseases (24,25). An increasing number of studies
have confirmed the involvement of several miRNAs in the regulation
of bone biology; their potential role in osteogenesis has been
reported, but the relative functional significance is not yet
completely understood (5,7,26-28). A recent study using miRNA
microarray chip technology demonstrated that miR-223 is abnormally
expressed in patients with diffuse idiopathic skeletal hyperostosis
(DISH), which is associated with sclerostin metabolism (29). DISH is characterized by new bone
formation, constitutional abnormalities and metabolic
abnormalities, leading to the biomechanical alterations in the
musculoskeletal system and/or the formation of obstructive cervical
masses (30,31). However, the function of miR-223 in
osteoblasts remains unknown.
To the best of our knowledge, the present study is
the first to reveal that miR-223-5p was upregulated during
osteoblast differentiation. miR-223-5p expression increased
gradually in MC3T3-E1 cells over a 14 day period. Accordingly, the
expression levels of ALP, OCN and RUNX2 were all significantly
upregulated in a time-dependent manner. ALP (32), OCN (33) and RUNX2 (27) are classic biomarkers, that reflect
the functional status of osteoblast cells. The capacity of each
factor in regulating osteogenic differentiation is highly
associated with the bone formation process due to of the dynamic
feedback system in the human body. The expression of miR-223-5p
exhibited a similar tendency in the present study, suggesting that
miR-223-5p may be used as a biomarker for diagnostic purposes for
osteogenic differentiation. However, the mechanisms underlying the
role of miR-223-5p in this process has not yet been fully
elucidated.
The present study demonstrated that miR-223-5p was a
positive regulator of osteogenic differentiation, as its
overexpression led to the enhancement of osteogenic
differentiation, and its silencing had to the opposite effect.
Following transfection of miR-233 mimics, biomarkers of osteogenic
differentiation (ALP, OCN and RUNX2) were upregulated at 7 days,
and the Alizarin S red staining results revealed a higher number of
mineralized nodules compared with the control group. The
transfection of miR-233 inhibitor exerted the opposite effects. In
the animal experiment, the newly formed bone tissue stained red
with H&E and stained blue with Masson's Trichrome. The area of
new bone tissue was larger in implants containing si-HDAC2 cells,
compared with negative controls, indicating that HDAC2 silencing
increased mineralized bone matrix formation. These results
indicated that miR-223-5p may be a target for the treatment of bone
loss and the optimization of fracture healing. Several miRNAs
including miR-5100 (34), miR-192
(22) and miR-10a (5), have been proven to act as regulatory
factors, as their expression markedly affect osteogenic
differentiation. Accumulating evidence highlights the crucial role
of miRNAs during osteogenic differentiation, and these may exert
their effects through targeting their downstream genes. miR-590,
miR-9 and miR-5100 control osteogenic differentiation by targeting
mothers against decapentaplegic homolog 7, Dickkopf WNT Signaling
Pathway Inhibitor 1 and various other genes (27,35,36). However, the target genes of
miR-223-5p in osteogenic differentiation remain unknown.
The present study demonstrated that miR-223-5p
promoted osteogenic differentiation, at least in part by targeting
HDAC2. The experimental data demonstrated that miR-223-5p
downregulated HDAC2 gene expression, which has been described as an
anti-proliferative gene involved in cell cycle regulation (37,38). A study of chronic obstructive
pulmonary disease (COPD) demonstrated that miR-223-5p
overexpression decreased HDAC2 expression in human pulmonary artery
endothelial cells, whereas HDAC2 expression was preserved when
miR-223-5p was silenced (15).
These findings suggest that miR-223-5p controls the expression of
HDAC2 in COPD (15). Thus, it is
probable that miR-223-5p controlled the expression of HDAC2 in
osteoblasts, which is a novel regulatory axis.
In conclusion, HDAC2 expression at the mRNA and
protein level was altered following transfection with miR-223-5p
mimics or inhibitor in the present study. Therefore, it was
demonstrated that miR-223-5p regulated HDAC2 expression to promote
osteogenesis. Taken together, the results of the present study
indicated a novel potential therapeutic approach to the treatment
of osteogenic conditions.
Funding
No funding was received.
Availability of data and materials
All data generated or analyzed during this study are
available from the corresponding author upon reasonable
request.
Authors' contributions
JC contributed the central idea, analyzed the
majority of the data and wrote the initial draft of the manuscript.
GH and YW performed the in vivo experiments. DC contributed
to study design, revised and finalized the manuscript. All authors
read and approved the final manuscript.
Ethics approval and consent to
participate
The research protocols associated with the
experimental mice were approved by the Experimental Animal Ethics
Committee of Southern Medical University.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgments
Not applicable.
References
|
1
|
Kular J, Tickner J, Chim SM and Xu J: An
overview of the regulation of bone remodelling at the cellular
level. Clin Biochem. 45:863–873. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Wei J, Shi Y, Zheng L, Zhou B, Inose H,
Wang J, Guo XE, Grosschedl R and Karsenty G: miR-34s inhibit
osteoblast proliferation and differentiation in the mouse by
targeting SATB2. J Cell Biol. 197:509–521. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Dirckx N, Van Hul M and Maes C: Osteoblast
recruitment to sites of bone formation in skeletal development,
homeostasis, and regeneration. Birth Defects Res C Embryo Today.
99:170–191. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Blagojevic M, Jinks C, Jeffery A and
Jordan KP: Risk factors for onset of osteoarthritis of the knee in
older adults: A systematic review and meta-analysis. Osteoarthritis
Cartilage. 18:24–33. 2010. View Article : Google Scholar
|
|
5
|
Li J, Zhang Y, Zhao Q, Wang J and He X:
MicroRNA-10a influences osteoblast differentiation and angiogenesis
by regulating β-catenin expression. Cell Physiol Biochem.
37:2194–2208. 2015. View Article : Google Scholar
|
|
6
|
Mattick JS: RNA regulation: A new
genetics? Nat Rev Genet. 5:316–323. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Chen L, Holmstrøm K, Qiu W, Ditzel N, Shi
K, Hokland L and Kassem M: MicroRNA-34a inhibits osteoblast
differentiation and in vivo bone formation of human stromal stem
cells. Stem Cells. 32:902–912. 2014. View Article : Google Scholar
|
|
8
|
Chen H, Ji X, She F, Gao Y and Tang P:
miR-628-3p regulates osteoblast differentiation by targeting RUNX2:
Possible role in atrophic non-union. Int J Mol Med. 39:279–286.
2017. View Article : Google Scholar :
|
|
9
|
Li Z, Hassan MQ, Volinia S, van Wijnen AJ,
Stein JL, Croce CM, Lian JB and Stein GS: A microRNA signature for
a BMP2-induced osteoblast lineage commitment program. Proc Natl
Acad Sci USA. 105:13906–13911. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Liu X, Deng Y, Xu Y, Jin W and Li H:
MicroRNA-223 protects neonatal rat cardiomyocytes and H9c2 cells
from hypoxia-induced apoptosis and excessive autophagy via the
Akt/mTOR pathway by targeting PARP-1. J Mol Cell Cardiol.
118:133–146. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Qin D, Wang X, Li Y, Yang L, Wang R, Peng
J, Essandoh K, Mu X, Peng T, Han Q, et al: MicroRNA-223-5p and -3p
cooperatively suppress necroptosis in ischemic/reperfused hearts. J
Biol Chem. 291:20247–20259. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zhou W, Pal AS, Hsu AY, Gurol T, Zhu X,
Wirbisky-Hershberger SE, Freeman JL, Kasinski AL and Deng Q:
MicroRNA-223 suppresses the canonical NF-κB pathway in basal
keratinocytes to dampen neutrophilic inflammation. Cell Rep.
22:1810–1823. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Sugawara S, Yamada Y, Arai T, Okato A,
Idichi T, Kato M, Koshizuka K, Ichikawa T and Seki N: Dual strands
of the miR-223 duplex (miR-223-5p and miR-223-3p) inhibit cancer
cell aggressiveness: Targeted genes are involved in bladder cancer
pathogenesis. J Hum Genet. 63:657–668. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Berenstein R, Nogai A, Waechter M, Blau O,
Kuehnel A, Schmidt-Hieber M, Kunitz A, Pezzutto A, Dörken B and
Blau IW: Multiple myeloma cells modify VEGF/IL-6 levels and
osteogenic potential of bone marrow stromal cells via
Notch/miR-223. Mol Carcinog. 55:1927–1939. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Leuenberger C, Schuoler C, Bye H, Mignan
C, Rechsteiner T, Hillinger S, Opitz I, Marsland B, Faiz A,
Hiemstra PS, et al: MicroRNA-223 controls the expression of histone
deacetylase 2: A novel axis in COPD. J Mol Med (Berl). 94:725–734.
2016. View Article : Google Scholar
|
|
16
|
Lin X, Xiong D, Peng YQ, Sheng ZF, Wu XY,
Wu XP, Wu F, Yuan LQ and Liao EY: Epidemiology and management of
osteoporosis in the People's Republic of China: Current
perspectives. Clin Interv Aging. 10:1017–1033. 2015.PubMed/NCBI
|
|
17
|
Rau CS, Wu SC, Kuo PJ, Chen YC, Chien PC,
Hsieh HY and Hsieh CH: Epidemiology of bone fracture in female
trauma patients based on risks of osteoporosis assessed using the
osteoporosis self-assessment tool for Asians score. Int J Environ
Res Public Health. 14:E13802017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Lin J, Yang Y, Zhang X, Ma Z, Wu H, Li Y,
Yang X, Fei Q and Guo A: BFH-OSTM, a new predictive screening tool
for identifying osteoporosis in elderly Han Chinese males. Clin
Interv Aging. 12:1167–1174. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Cherian KE, Kapoor N, Shetty S, Naik D,
Thomas N and Paul TV: Evaluation of different screening tools for
predicting femoral neck osteoporosis in rural South Indian
postmenopausal women. J Clin Densitom. 21:119–124. 2018. View Article : Google Scholar
|
|
20
|
Zhu X, Luo J, Chen X, Wang J, Wang G, Li
H, Xu Y, Feng J and Tu H: Expression characteristic and
significance of interleukin-6, nuclear factor kappa beta, and bone
formation markers in rat models of osteoporosis. Transl Res.
152:18–23. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Hiligsmann M, Dellaert BG, Watson V and
Boonen A: Comment on: Patients' preferences for anti-osteoporosis
drug treatment: A cross-European discrete choice experiment: Reply.
Rheumatology (Oxford). 57:584–585. 2018. View Article : Google Scholar
|
|
22
|
Nogués X and Martinez-Laguna D: Update on
osteoporosis treatment. Med Clin (Barc). 150:pp. 479–486. 2018, In
English, Spanish. View Article : Google Scholar
|
|
23
|
Bushati N and Cohen SM: microRNA
functions. Annu Rev Cell Dev Biol. 23:175–205. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Chua JH, Armugam A and Jeyaseelan K:
MicroRNAs: Biogenesis, function and applications. Curr Opin Mol
Ther. 11:189–199. 2009.PubMed/NCBI
|
|
25
|
Foshay KM and Gallicano GI: Small RNAs,
big potential: The role of MicroRNAs in stem cell function. Curr
Stem Cell Res Ther. 2:264–271. 2007. View Article : Google Scholar
|
|
26
|
Ma Y, Yao N, Liu G, Dong L, Liu Y, Zhang
M, Wang F, Wang B, Wei X, Dong H, et al: Functional screen reveals
essential roles of miR-27a/24 in differentiation of embryonic stem
cells. EMBO J. 34:361–378. 2015. View Article : Google Scholar
|
|
27
|
Zhang Y, Gao Y, Cai L, Li F, Lou Y, Xu N,
Kang Y and Yang H: MicroRNA-221 is involved in the regulation of
osteoporosis through regulates RUNX2 protein expression and
osteoblast differentiation. Am J Transl Res. 9:126–135.
2017.PubMed/NCBI
|
|
28
|
Chung AC, Huang XR, Meng X and Lan HY:
miR-192 mediates TGF-beta/Smad3-driven renal fibrosis. J Am Soc
Nephrol. 21:1317–1325. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Xie Y, Zhang L, Gao Y, Ge W and Tang P:
The multiple roles of Microrna-223 in regulating bone metabolism.
Molecules. 20:19433–19448. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Mader R, Sarzi-Puttini P, Atzeni F,
Olivieri I, Pappone N, Verlaan JJ and Buskila D: Extraspinal
manifestations of diffuse idiopathic skeletal hyperostosis.
Rheumatology (Oxford). 48:1478–1481. 2009. View Article : Google Scholar
|
|
31
|
Verlaan JJ, Boswijk PF, de Ru JA, Dhert WJ
and Oner FC: Diffuse idiopathic skeletal hyperostosis of the
cervical spine: An underestimated cause of dysphagia and airway
obstraction. Spine J. 11:1058–1067. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Kim SK, Lee MH and Rhee MH: Studies on the
effects of biomedicinal agents on serum concentration of Ca2+, P
and ALP activity in osteoporosis-induced rats. J Vet Sci.
4:151–154. 2003.PubMed/NCBI
|
|
33
|
Yang X, Tao XA, Liang JQ, Huang YJ and
Yang XP: The dynamic changes of circulating OCN+ cells versus
insulinlike growth factor-I during primary healing of orthognathic
surgeries. Oral Surg Oral Med Oral Pathol Oral Radiol. 113:734–740.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Wang H, Cui Y, Luan J, Zhou X, Li C, Li H,
Shi L and Han J: MiR-5100 promotes osteogenic differentiation by
targeting Tob2. J Bone Miner Metab. 35:608–615. 2017. View Article : Google Scholar
|
|
35
|
Liu X, Xu H, Kou J, Wang Q, Zheng X and Yu
T: MiR-9 promotes osteoblast differentiation of mesenchymal stem
cells by inhibiting DKK1 gene expression. Mol Biol Rep. 43:939–946.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Swetha RG, Ramaiah S and Anbarasu A:
Molecular dynamics studies on D835N mutation in FLT3-its impact on
FLT3 protein structure. J Cell Biochem. 117:1439–1445. 2016.
View Article : Google Scholar
|
|
37
|
Noh JH, Jung KH, Kim JK, Eun JW, Bae HJ,
Xie HJ, Chang YG, Kim MG, Park WS, Lee JY and Nam SW: Aberrant
regulation of HDAC2 mediates proliferation of hepatocellular
carcinoma cells by deregulating expression of G1/S cell cycle
proteins. PLoS One. 6:e281032011. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Jung KH, Noh JH, Kim JK, Eun JW, Bae HJ,
Xie HJ, Chang YG, Kim MG, Park H, Lee JY and Nam SW: HDAC2
overexpression confers oncogenic potential to human lung cancer
cells by deregulating expression of apoptosis and cell cycle
proteins. J Cell Biochem. 113:2167–2177. 2012. View Article : Google Scholar : PubMed/NCBI
|