Introduction
In total, >80% of lung carcinomas are non-small
cell lung carcinoma (NSCLC), which includes large cell carcinoma,
squamous cell carcinoma, adenocarcinoma, and unclassifiable NSCLC.
The prognosis of patients with lung cancer is significantly
affected by the cell type (for example, small cell vs. NSCLC),
clinical staging, and the overall condition of the patients.
Unfortunately, NSCLC is frequently diagnosed at a late stage, thus
limiting the number of available treatment options. Since pulmonary
nodules may be observed in specific NSCLC cases, the difference
between malignant and benign pulmonary nodules has been used as a
diagnostic criterion of NSCLC (1).
As a family member of immunoglobulin (Ig) proteins,
cluster of differentiation 200 (CD200; also known as OX-2) carries
two extracellular immunoglobulin domains and one small 19 amino
acid intracellular domain with no known signaling motifs (2). CD200 is expressed in a wide range of
human cancer types, including malignant B carcinoma, myeloid
leukemia, ovarian cancer and human melanoma (3–6). A
previous study has demonstrated that intravenous injection of
CD200+ B16 cells into CD200R-deficient mice induced
significant tumor growth in multiple organs, including kidney,
lung, liver and the peritoneal cavity (7).
As small non-coding RNAs containing 19–25
nucleotides, microRNAs (miRNAs) are able to mediate the expression
of numerous protein-coding genes in animals and plants. In
addition, miRNAs have important roles in a number of cellular
processes, including stem cell renewal, cell metabolism,
proliferation, apoptosis and differentiation (8). Due to the important functions of
miRNAs, their abnormal expression has been implicated in many human
diseases, including obesity, cardiovascular diseases, cancer,
psoriasis, schizophrenia, chronic hepatitis and diabetes (9-14).
For example, a previous study (15) demonstrated that miR-499a-5p serves
as a tumor suppressor to regulate the expression of vav guanine
nucleotide exchange factor 3 (VAV3), and that low expression levels
of miR-499a-5p are correlated with a poor prognosis in NSCLC.
Experimental data has additionally demonstrated that overexpression
of miR-499a-5p inhibits the proliferation and metastasis of NSCLC
cells by inducing their apoptosis (15). As a type of genetic variation
discovered in precursor miRNAs (pre-miRNAs), single nucleotide
polymorphisms (SNPs) may influence the expression of their host
miRNAs. As a result, SNPs have been implicated in altered
biological functions and gene expression. For example, the
miR-499A>G (rs3746444) SNP is closely associated with the
pathogenesis of diabetes, cardiovascular disease, hypertension and
cancer (16).
It has been reported that the presence of SNP
rs3746444 in miR-499a affects miR-499a expression (17). In the present study, by searching
an online miRNA database, CD200 was identified as a possible target
of miR-499a. Notably, CD200 has been reported to function as a
tumor suppressor in the development of lung cancer (18,19). Therefore, the association between
SNP rs3746444 and the risk of lung cancer was evaluated in a
Chinese population. In addition, the effect of SNP rs3746444 on the
expression of miR-499a and CD200 was investigated.
Patients and methods
Sample collection
All participants underwent imaging examinations to
confirm the presence of pulmonary nodules. Peripheral blood and
tissue samples were collected from the participants at the
Affiliated Hospital of Qingdao University (Qingdao, China) between
January 2017 and August 2017 to determine the malignance of their
pulmonary nodules. In total, 476 patients with benign pulmonary
nodules and 425 patients with malignant pulmonary nodules were
enrolled into the study. The tissue samples were additionally
examined by genotyping analyses, and 30 samples were determined as
benign while 32 were determined as malignant.
The Human Research Ethics Committee of The
Affiliated Hospital of Qingdao University approved the present
study. Written informed consent was obtained from all patients or
their first-degree relatives prior to the start of the research
project. The research protocols additionally complied with the
latest version of the Declaration of Helsinki.
TaqMan assay
A QuantStudio 12K Flex instrument (Thermo Fisher
Scientific, Inc., Waltham, MA, USA) was used to perform TaqMan
assays and to identify the genotypes of SNP rs3746444 in all
samples.
RNA isolation and reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
TRIzol® reagent (Thermo Fisher Scientific, Inc.) was
used to extract total RNA from tissue samples and A549 and A427
cells. The extracted RNA samples were transcribed to cDNA using a
ThermoScript RT-PCR system (Invitrogen; Thermo Fisher Scientific,
Inc.) at 50°C for 15 min. Subsequently, the expression of CD200
mRNA [CD200 mRNA-forward (F), 5′-CAGAGGCATAGTGGTAACACCT-3′ and
reverse (R): 5′-GTGCCATTGCCCCAGTATTCT-3′] and miR-499a (miR-499a-F,
5′-GGCCAACATCACAGCAAGTCTG-3′ and R, 5′-AGTGCAGGGTCCGAGGTAT-3′) was
measured using a MyiQ RT-PCR Detection System with Bio-Rad iQTM
SYBR-Green Supermix (both Bio-Rad Laboratories, Inc., Hercules, CA,
USA). The thermocycling protocol for RT-PCR was 10 min at 95°C,
followed by 40 cycles of 30 sec at 95°C, 30 sec at 60°C and 30 sec
at 72°C. The 2−ΔΔCq method (20) was used to determine the relative
expression of CD200 mRNA and miR-499a. The expression of U6 (U6-F,
5′-CTCGCTTCGGCAGCACA-3′ and R, 5′-AACGCTTCACGAATTTGCGT-3′) and
GAPDH (GAPDH-F, 5′-TGCACCACCAACTGCTTA-3′ and R,
5′-GGATGCAGGGATGATGTT-3′) were used as internal controls.
Cell culture and transfection
Dulbecco's modified Eagle's medium (DMEM) containing
10% fetal bovine serum (FBS; both Gibco; Thermo Fisher Scientific,
Inc.), 100 mg/ml streptomycin and 100 U/ml penicillin was used to
culture A549 and A427 cells, at 37°C and 5% CO2. A549
cells were purchased from the Type Culture Collection of the
Chinese Academy of Sciences (Shanghai, China). A427 cells were
purchased from the American Type Culture Collection (Manassas, VA,
USA). The possibility of contamination was excluded using a
mycoplasma stain assay kit (Beyotime Institute of Biotechnology,
Haimen, China) according to the manufacturer's protocol. When the
cell confluence reached 80%, the cells were transfected with
miR-499a mimics (sequence, 5′-UUAAGACUUGCAGUGAUGUUU-3′; 50 nM),
miR-499a inhibitors (sequence, 5′-AAACAUCACUGCAAGUCUUAA-3′; 50 nM),
CD200 short hairpin RNA 1 (shRNA1; sequence, 5′-CGTACGTAGTCGTAGTCA
GTA-3′; 50 nM) or CD200 shRNA2 (sequence,
5′-CGACGTACGTATCGTAACGA-3′; 50 nM; all Shanghai GenePharma Co.,
Ltd., Shanghai, China) for 24 h using Lipofectamine® 2000
(Invitrogen; Thermo Fisher Scientific, Inc.) according to the
manufacturer's protocol. For the shRNA, an empty vector was used as
the control, and for the anti-miR, a scramble sequence
(5′-GCCGACATACCAAGAAGCTTGC-3′) was used as the control.
Luciferase assay
A fragment of the 3′-untranslated region (UTR) of
CD200 mRNA containing the miR-499a binding site was amplified by
PCR. At the same time, a site-directed mutagenesis kit was used to
generate a mutation in the miR-499a binding site of CD200. The two
different PCR products were respectively inserted into pGL3-Basic
vectors (Promega Corporation, Madison, WI, USA) to generate
luciferase reporter constructs for wild-type and mutant CD200.
Subsequently, A549 and A427 cells were seeded into 48-well plates
and cotransfected with wild-type or mutant CD200 luciferase
reporter vectors and miR-499a mimics using Lipofectamine® 2000
(Thermo Fisher Scientific, Inc.) according to the manufacturer's
protocol. After 24 h of trans-fection, a 96-well plate luminometer
(Berthold Detection System, Pforzheim, Germany) was used to analyze
the luciferase activity (normalized to Renilla luciferase
activity) of transfected cells with a Dual-Luciferase Reporter
Assay System (Promega Corporation).
Western blot analysis
A urea buffer (containing 200 mM
ammoniumbicarbonate, 6 M urea, and 2% SDS) containing a protease
inhibitor cocktail (Complete mini) or a complete lysis-M reagent
(both Roche Diagnostics GmbH, Mannheim, Germany) was used to
extract proteins from A549 and A427 cells, according to the
manufacturer's protocol. A bicin-choninic acid protein assay
(Pierce; Thermo Fisher Scientific, Inc.) was used to measure the
protein concentration. The extracted proteins (35 μg/lane)
were subsequently denatured in hot water and resolved by 15%
SDS-PAGE, followed by semi-dry transfer onto a nitrocellulose
membrane (Trans-Blot; both Bio-Rad Laboratories, Inc.). TBS with
Tween-20 containing 5% bovine serum albumin (Thermo Fisher
Scientific, Inc.) or 5% non-fat dry milk was used to block the
membrane for 60 min at room temperature. Subsequently, the membrane
was incubated at 4°C for 12 h with anti-CD200 primary antibodies
(cat. no. ab173498; 1:2,000; Abcam, Cambridge, UK), followed by
another 2 h of incubation with horseradish peroxidase-conjugated
anti-mouse secondary antibodies (cat. no. 7076s; 1:10,000; Cell
Signaling Technology, Inc., Danvers, MA, USA). The membrane was
subsequently developed with an enhanced chemiluminescence kit (GE
Healthcare, Little Chalfont, Buckinghamshire, UK) and exposed to
X-rays. ImageJ software (v1.8.0; National Institutes of Health,
Bethesda, MD, USA) was used to perform protein densitometric
analyses.
Statistical analysis
All data are presented as the mean ± standard
deviation. The bioinformatics tool TargetScan (21) was used for the sequence analysis
between CD200 mRNA and miR-499a. Student's t-test was used to
evaluate the difference between two different groups, while
Student-Newman-Keuls multiple comparison test and one-way analysis
of variance with Bonferroni's test was used to examine the
difference among multiple groups. Count data were compared using
χ2 tests. Logistic regression analysis was used to
analyze the correlation between SNP rs3746444 and the risk of
NSCLC, in addition to the correlation between miR-499a and CD200
expression. P<0.05 was considered to indicate a statistically
significant difference. SPSS 15.0 statistical package (SPSS, Inc.,
Chicago, IL, USA) was used to perform the statistical analysis.
Each experiment was repeated three times.
Results
Patient characteristics
The demographic and clinicopathological
characteristics of the 425 patients with malignant pulmonary
nodules and 476 patients with benign pulmonary nodules are listed
in Table I. No obvious difference
was found between the two groups of subjects in terms of sex and
age (P>0.05). However, a significant difference in smoking
history was observed between the two groups. The 476 patients with
benign pulmonary nodules were further divided into four groups
according to the genotypes of their SNP rs3746444: AA (n=346), AG
(n=118), GG (n=12) and AG+GG (n=120). At the same time, the 425
patients with malignant pulmonary nodules were also divided into
four groups: AA (n=260), AG (n=138), GG (n=27) and AG+GG (n=165). A
logistic regression test was used to analyze the correlation
between the genotypes of SNP rs3746444 and the risk of NSCLC. The
results revealed that the AG, GG and AG+GG genotypes of SNP
rs3746444 were significantly associated with an increased risk of
NSCLC (95% CI=1.16-2.08, OR=1.55, P=0.032; 95% CI=1.70-7.55,
OR=3.59, P=0.0007; and 95% CI=1.29-2.27, OR=1.71, P=0.0002,
respectively). In addition, the 476 patients with benign pulmonary
nodules were divided into two groups: A (n=810) and G (n=138).
Similarly, the 425 patients with malignant pulmonary nodules were
also divided into two groups: A (n=658) and G (n=192). A logistic
regression test was used to analyze the correlation between the A/G
allele of SNP rs3746444 and the risk of NSCLC. The results
demonstrated that the A/G allele of SNP was significantly
associated with the risk of NSCLC (95% CI=1.34-2.18; OR=1.71;
P<0.0001).
 | Table IDemographic and clinicopathological
characteristics of the participants enrolled in the study. |
Table I
Demographic and clinicopathological
characteristics of the participants enrolled in the study.
| Characteristic | Benign PNs, n=476
(%) | Malignant PNs,
n=425 (%) | OR (95% CI) | P-value |
|---|
| Sex | | | | 0.5468 |
| Male | 292 (61.3) | 269 (63.3) | | |
| Female | 184 (38.7) | 156 (36.7) | | |
| Age, years | 55.6±17.7 | 57.6±12.4 | | 0.4128 |
| Smoke | | | | <0.0001 |
| Yes | 121 (25.4) | 202 (47.5) | | |
| Never | 355 (74.6) | 223 (52.5) | | |
| Rs3746444
polymorphism in miR-499a | | | | |
| AA | 346 | 260 | | |
| AG | 118 | 138 | 1.55
(1.16-2.08) | 0.0032 |
| GG | 10 | 27 | 3.59
(1.70-7.55) | 0.0007 |
| AG+GG | 128 | 165 | 1.71
(1.29-2.27) | 0.0002 |
| A | 810 | 658 | | |
| G | 138 | 192 | 1.71
(1.34-2.18) | <0.0001 |
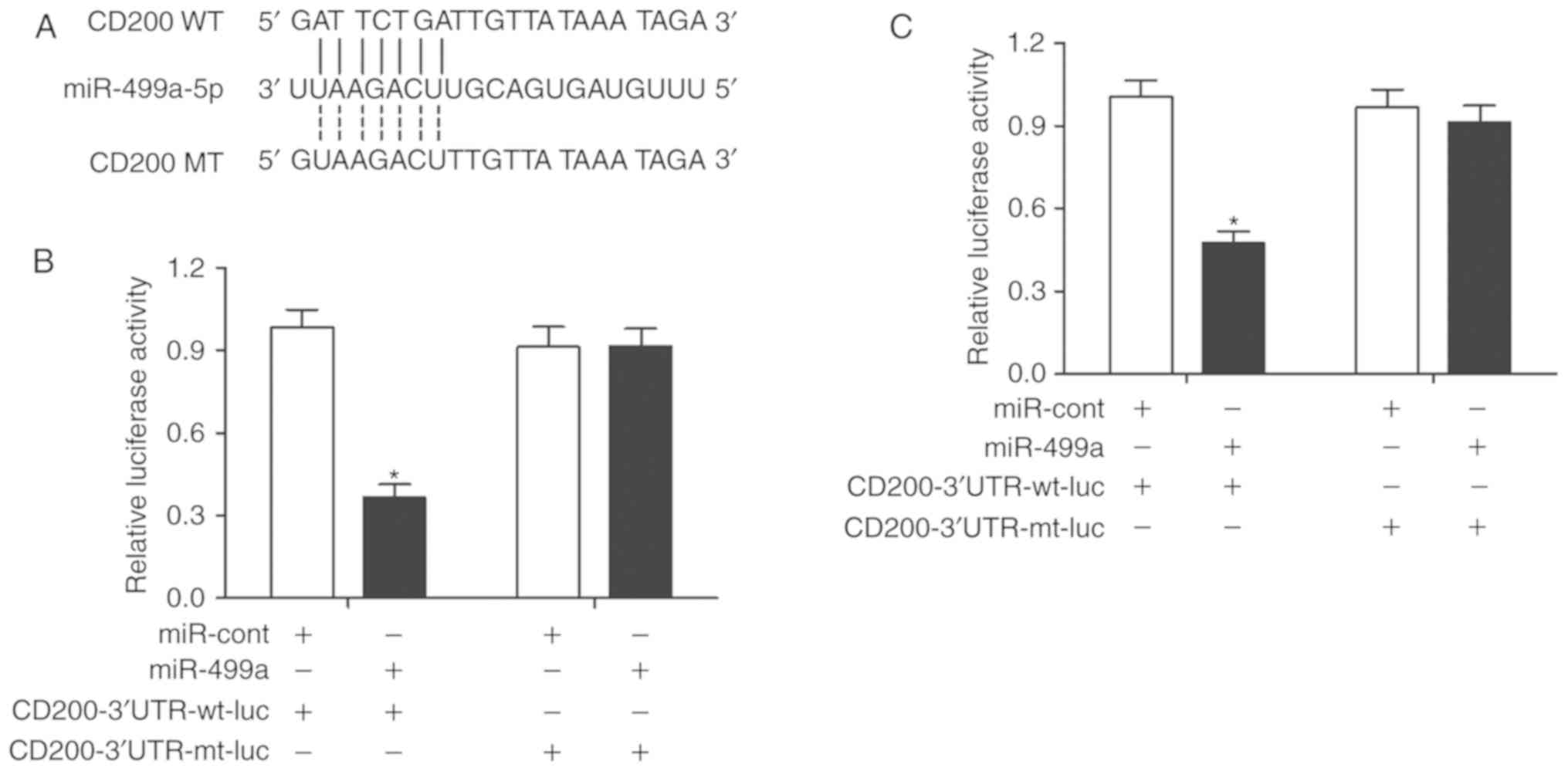
CD200 is a direct target of miR-499a
As illustrated in Fig.
1A, CD200 mRNA was identified as a potential target of miR-499a
by the online bioinformatics tool TargetScan (21). To determine if CD200 was a direct
target of miR-499a, a luciferase reporter assay was conducted in
A549 and A427 cells. As presented in Fig. 1, the luciferase activity in A549
(Fig. 1B) and A427 (Fig. 1C) cells was significantly reduced
when cotransfected with miR-499a mimics and wild-type 3′UTR of
CD200, suggesting that CD200 is a direct target of miR-499a.
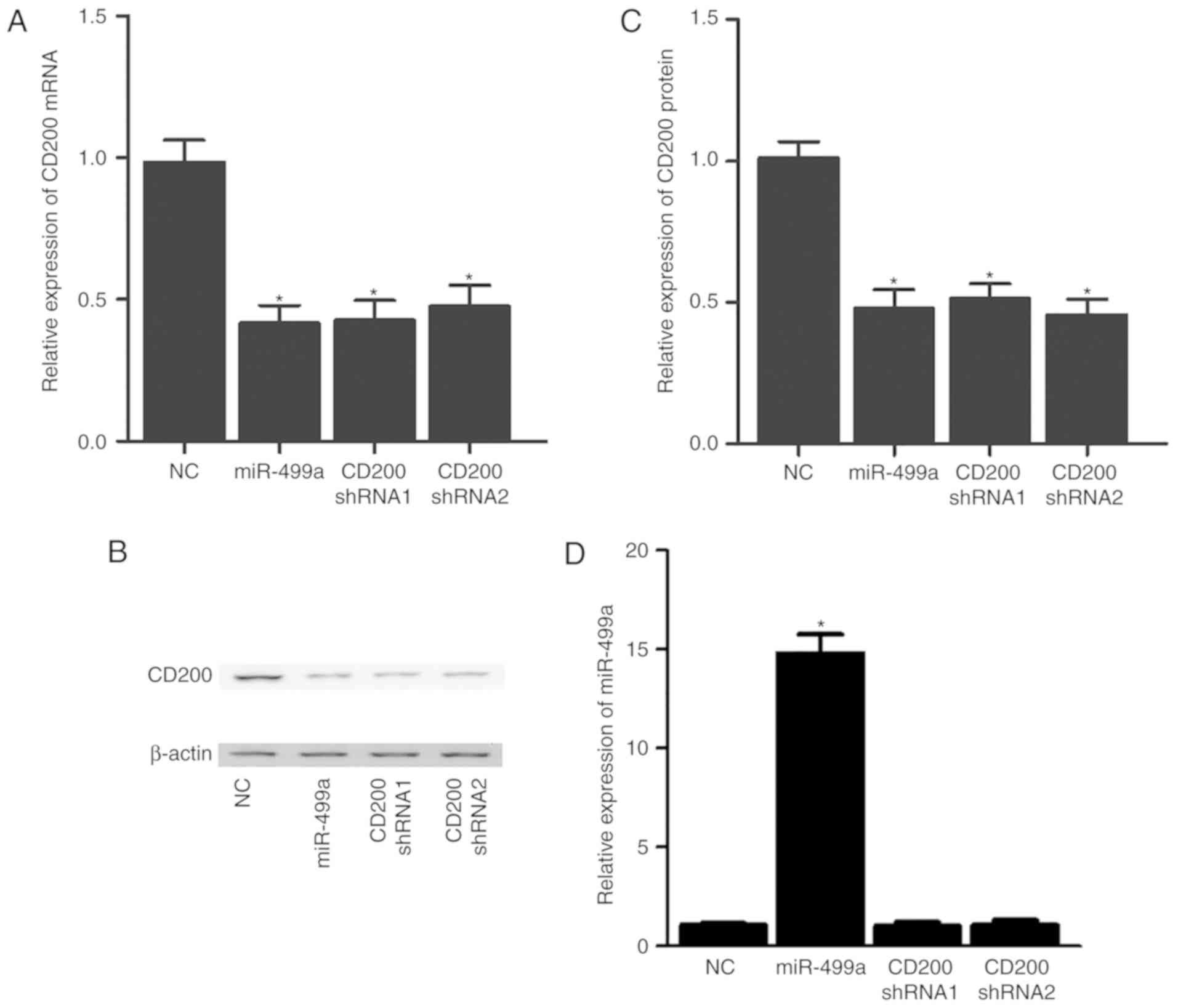
miR-499a inhibits CD200 expression
To confirm the effect of miR-499a on CD200
expression, A549 and A427 cells were transfected with miR-499a
mimics, miR-499a inhibitors, CD200 shRNA1 or CD200 shRNA2.
Subsequently, the mRNA and protein expression of CD200 was measured
using RT-qPCR and western blot analyses. As presented in Fig. 2, the expression of CD200 mRNA
(Fig. 2A) and protein (Fig. 2B and C) in A549 cells was
significantly reduced upon successful transfection with miR-499a
mimics (Fig. 2D), CD200 shRNA1 or
CD200 shRNA2. As presented Fig.
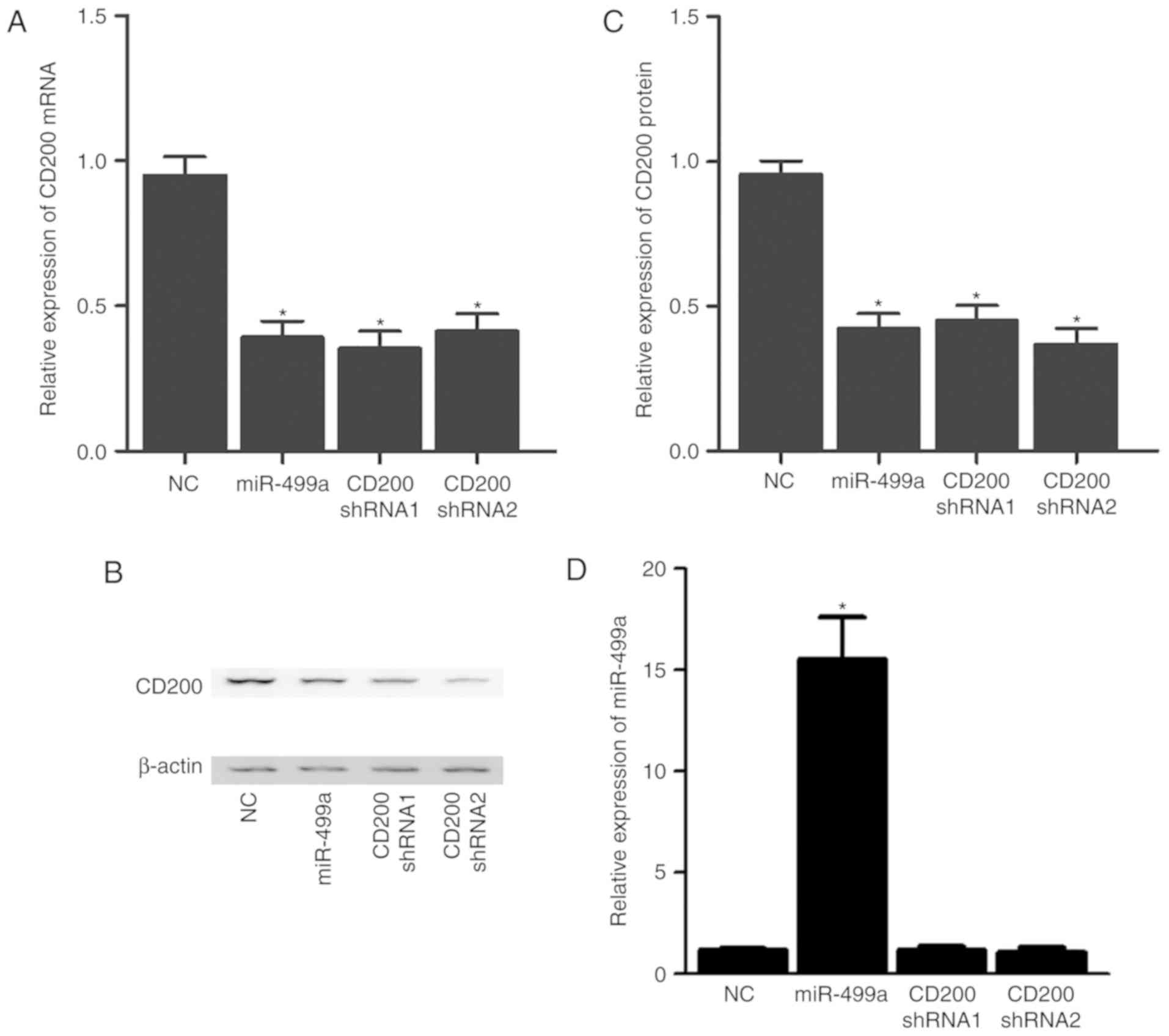
3, the expression of CD200 mRNA (Fig. 3A) and protein (Fig. 3B and C) in A427 cells was
significantly reduced upon successful transfection with miR-499a
mimics (Fig. 3D), CD200 shRNA1 or
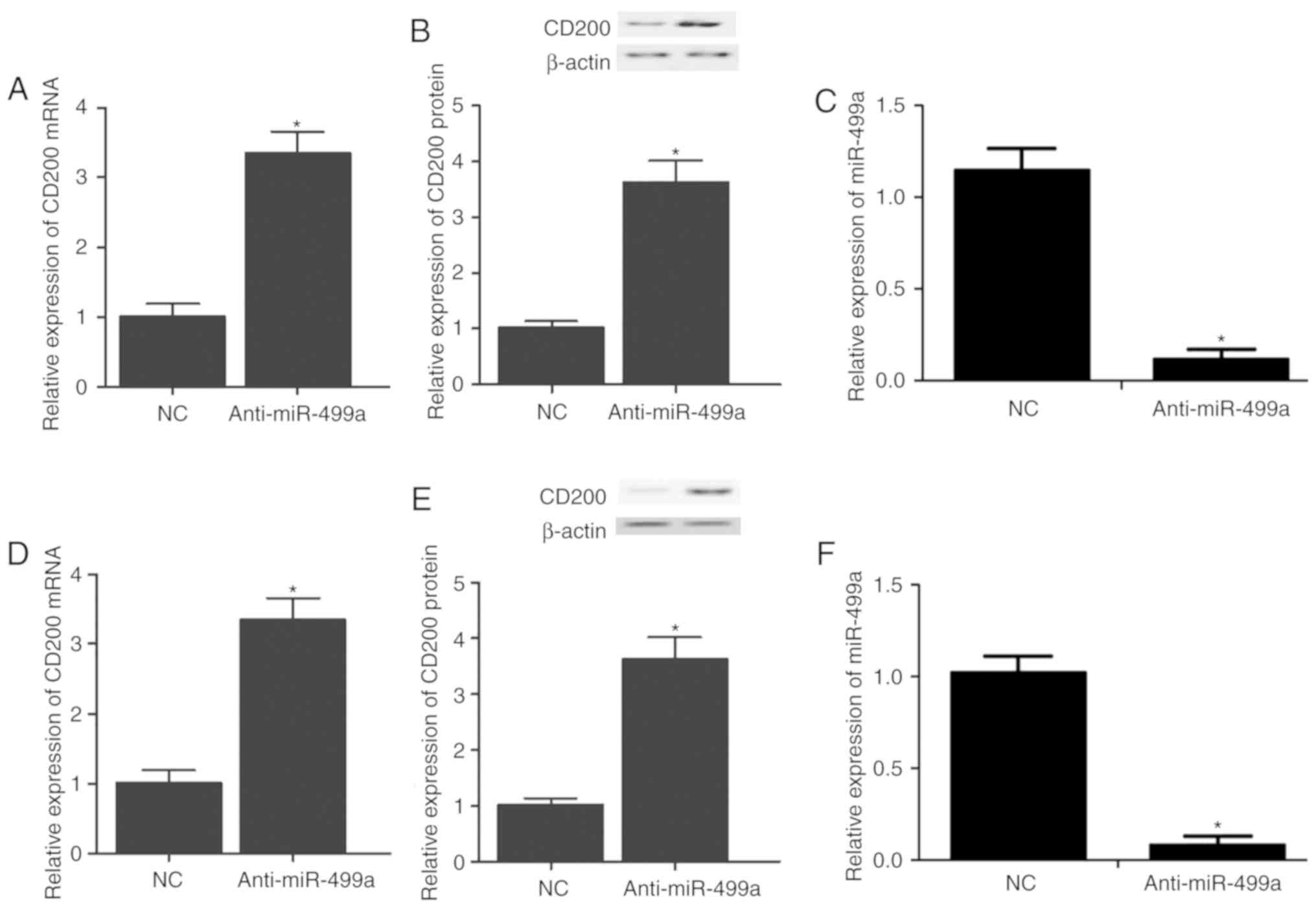
CD200 shRNA2. By contrast, the expression of CD200 mRNA (Fig. 4A and D) and protein (Fig. 4B, C, E and F) was significantly
increased in A549 (Fig. 4A–C) and
A427 (Fig. 4D–F) cells
transfected with miR-499a inhibitors.
Determination of miR-499a and CD200
expression in different groups of patients
A total of 32 malignant pulmonary nodule samples and
30 benign pulmonary nodule samples were collected in the present
study. These 62 tissue samples were divided into three groups
according to their genotypes of SNP rs3746444: AA (n=32), AG
(n=14), and GG (n=6). RT-qPCR and western blot analyses were then
performed to compare the expression of miR-499a and CD200 among
different samples. Furthermore, the expression of miR-499a and
CD200 was compared among the AA, AG and GG groups. As illustrated
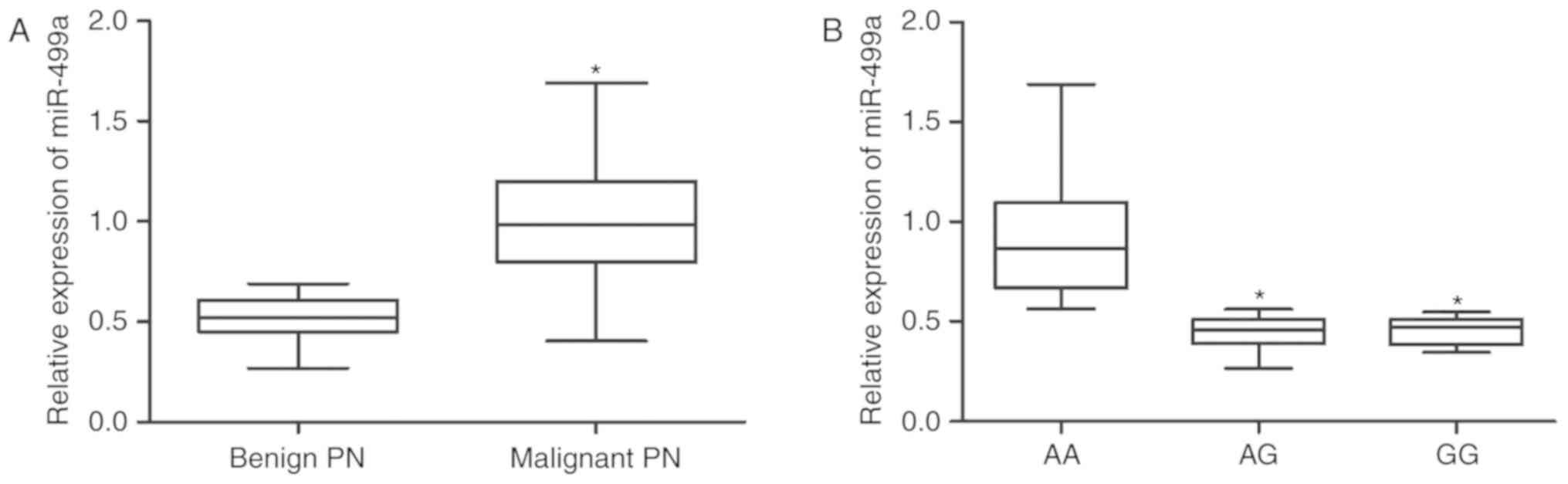
in Fig. 5A, the levels of
miR-499a in malignant pulmonary nodule samples were significantly
higher compared with benign pulmonary nodule samples. In addition,
miR-499a expression in the AA group was significantly higher
compared with the AG and GG groups, while miR-499a expression in
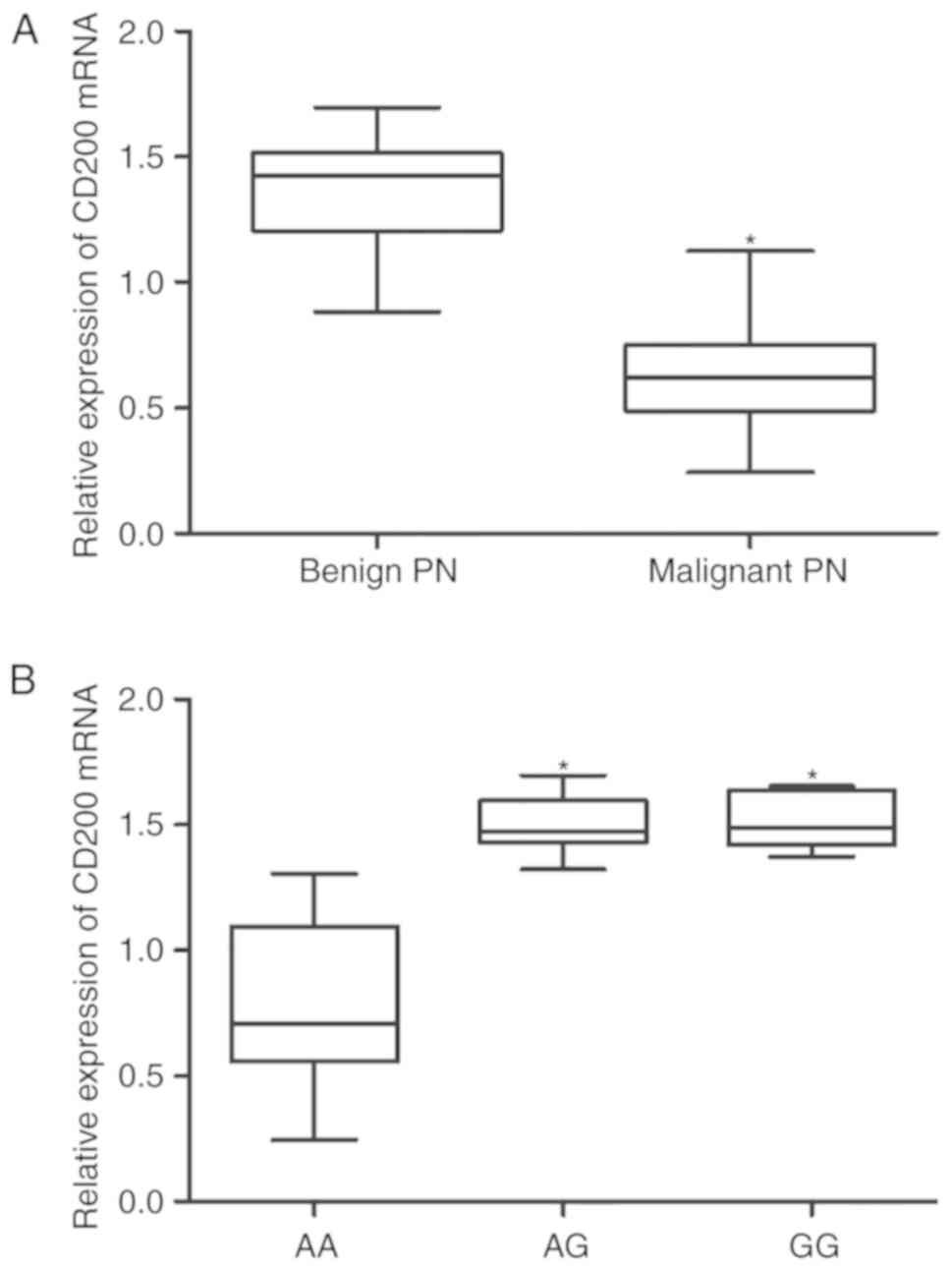
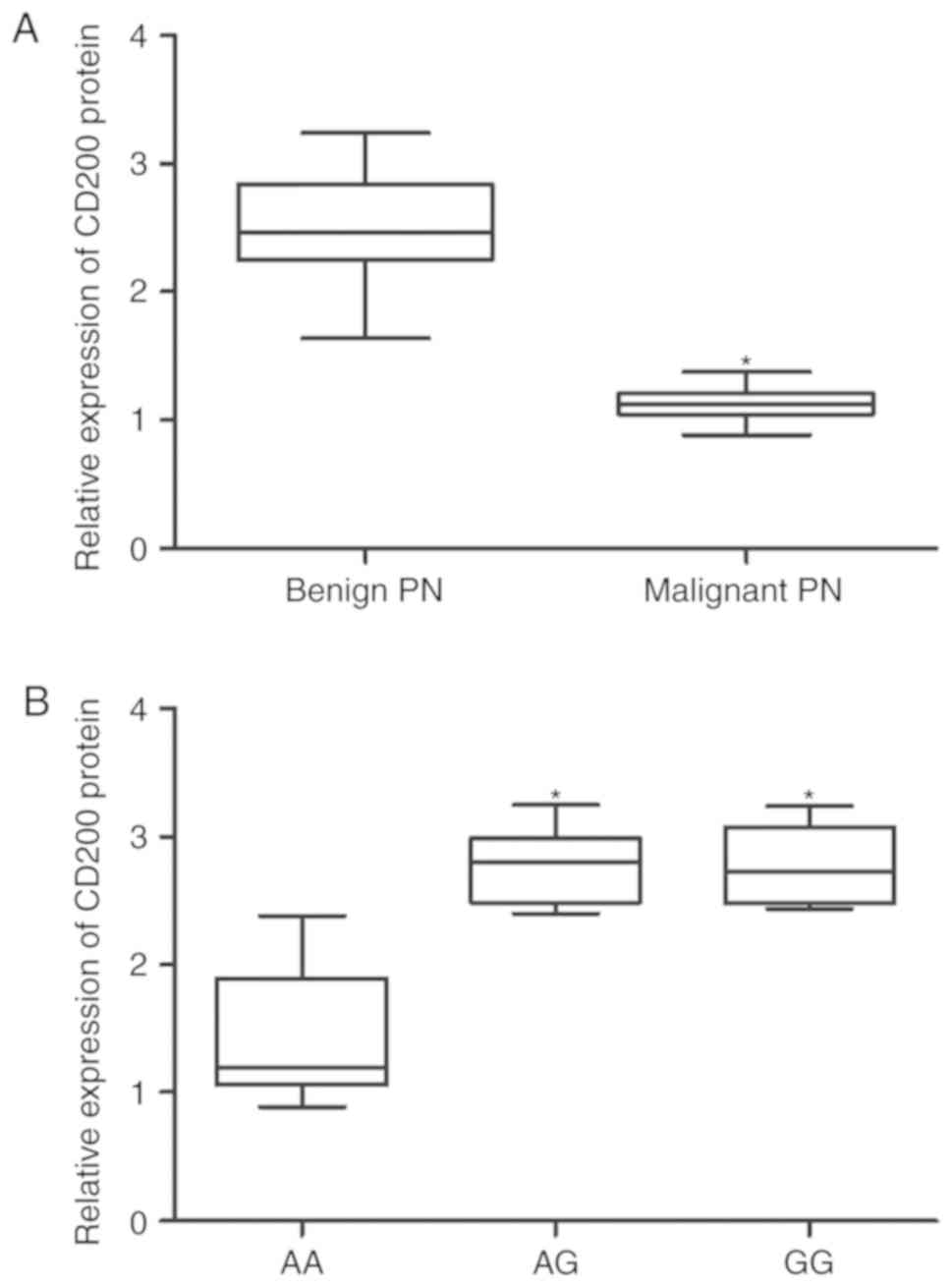
the AG and GG groups was comparable (Fig. 5B). By contrast, the levels of
CD200 mRNA (Fig. 6A) and protein
(Fig. 7A) were significantly
lower in malignant pulmonary nodule samples compared with benign
pulmonary nodule samples. Furthermore, the levels of CD200 mRNA
(Fig. 6B) and protein (Fig. 7B) in the AA group were lower
compared with the AG and GG groups, whereas, the expression levels
of CD200 mRNA (Fig. 6B) and
protein (Fig. 7B) in the AG and
GG groups were comparable.
Discussion
miRNAs have been reported to serve essential roles
in the pathogenesis of many diseases (22). In addition, SNPs located in miRNAs
may affect their expression, thus inducing diseases, including
colorectal cancer (23).
Furthermore, it is hypothesized that such SNPs may additionally
affect the binding between the host miRNAs and the target mRNAs,
thus altering the expression of target mRNAs (24).
SNP rs3746444 (A>G) is located in the seed
sequence of miR-499a-3p, a region crucial to the process of
miRNA-mediated gene silencing. For example, the SNP rs3746444 has
been demonstrated to increase the risk of ankylosing spondylitis,
Behcet's disease, coronary artery disease and rheumatoid arthritis
(25-28). The SNP rs3746444 additionally
increases the risk of hepatocellular carcinoma among the Chinese
population, and is used as a biomarker to predict the
susceptibility to coronary artery disease (29). In the present study, 476 samples
of malignant pulmonary nodules and 425 samples of benign pulmonary
nodules were collected, and it was demonstrated that the SNP
rs3746444 was significantly associated with the risk of NSCLC.
Furthermore, the expression of miR-499a was demonstrated to be
higher, whereas, the expression of CD200 was lower in malignant
nodules. Lung tissue samples were additionally divided based on
their genotypes of SNP rs3746444 and the results identified that
the expression levels of miR-499a in the AA group were much higher,
whereas, the CD200 expression in the AA group was significantly
reduced.
To validate the regulatory relationship between
miR-499a and CD200, in silico analyses and luciferase assays
were performed to confirm the role of CD200 as a direct target of
miR-499a. In addition, the effect of miR-499a on CD200 expression
was investigated using RT-qPCR and western blot analyses. The
results demonstrated that transfection of miR-499a mimics, CD200
shRNA1 and CD200 shRNA2 into A549 and A427 cells significantly
reduced the mRNA and protein expression levels of CD200. By
contrast, transfection of miR-499a inhibitors into the cells
increased the mRNA and protein expression of CD200.
As a family member of Ig proteins, CD200 carries two
extracellular immunoglobulin domains and one small 19 amino acid
intracellular domain with no known signaling motifs (30). CD200 is expressed in a wide range
of normal tissues and lymphoid cells, such as activated T cells and
B lymphocytes (31,32). CD200R, a receptor of CD200, is
mainly expressed in mast cells, neutrophils and macrophages, and
the expression levels of CD200R are similar in human and mouse
(4). The activation of CD200R
inhibits the in vitro activity of myeloid cells, while the
binding between CD200R and CD200 suppresses the activation of
myeloid cells (33). Unlike other
Ig receptors, CD200R does not contain immunoreceptor tyrosine-based
inhibition motif (ITIM) domains (34). In mast cells and macrophages, the
signaling cascade of CD200 suppresses the phosphorylation of P38,
c-Jun N-terminal kinase (JNK) and extracellular signal-regulated
kinase (ERK) (35). The
expression of CD200 can also be detected in most cases of plasma
cell myeloma. Nevertheless, the results regarding the levels of
CD200 in polytypic plasma cells are inconsistent (36-39). In addition, the post-treatment
levels of CD200 are stable in myeloma, suggesting that CD200 can be
used as a diagnostic biomarker for minimal residual disease of
myeloma (36). Despite this, the
effort to evaluate the prognostic value of CD200 in multiple
myeloma produced conflicting results (36,39). For example, CD200 is expressed in
normal hematogones, while the abnormal expression of CD200 was
detected in ~95% of B-cell acute lymphocytic leukemia (B-ALL)
(40). Consequently, the levels
of CD200 may be used in the evaluation of minimal residual disease
of B-ALL (41). In addition,
significantly upregulated CD200 expression has been observed in the
TEL-AML1 subtype of B-ALL, although the relationship between CD200
expression and the prognosis of B-ALL (TEL-AML1) remains elusive
(41). Nevertheless, the abnormal
level of CD200 may suggest a poor prognosis for ALL (5). Additionally, CD200 can increase the
efficacy of gefitinib in the treatment of lung cancer by decreasing
the number of drug-resistant cancer cells (21). Similarly, a microarray analysis
study revealed that the levels of CD200 were significantly
increased in cancer associated fibroblasts (CAFs), whereas the
silencing of CD200 decreased the sensitizing potential of CAFs. An
immunohistochemical analysis of samples collected from patients
undergoing gefitinib treatments revealed that the patients
harboring CD200+ CAFs were associated with longer
progression-free survival (PFS) (42). Expression of CD200 has been
observed in melanoma, neuroblastoma, ovarian carcinoma, and renal
carcinoma, but not in glioblastoma, astrocytoma, breast carcinoma,
lung carcinoma, and prostatic carcinoma (43). In addition, a sub-group of
basal-cell carcinoma cells have been found to express CD200 during
the initiation of tumor growth (44). CD200 has also been demonstrated to
induce the metastasis of cutaneous squamous cell carcinoma
(45). However, the expression
profile of CD200 has not been studied in pulmonary small cell
carcinoma.
In summary, using an online miRNA database and
luciferase reporter assays, CD200 was confirmed to be a target of
miR-499a. Furthermore, the SNP rs3746444 located in miR-499a was
demonstrated to affect the risk of NSCLC by regulating the
expression of CD200. Given that CD200 functions as a tumor
suppressor in lung cancer development, the genotypes of SNP
rs3746444 may be used as the biomarker to differentiate malignant
and benign pulmonary nodules.
Funding
No funding was received.
Availability of data and materials
The analyzed datasets generated during the study are
available from the corresponding author on reasonable request.
Authors' contributions
WJ and CL contributed to the conception of the
study, guaranteed the integrity of the entire study and reviewed
the manuscript. NG defined the intellectual content, designed the
study, was involved in data collection and reviewed the manuscript.
CM and QU collected and analyzed the data, and prepared the draft
of the manuscript. BH and YW visualized the data, and prepared the
draft of the manuscript. LX and XY collected the literature, and
prepared the draft of the manuscript. All authors contributed to
the data collection and analysis, and read and approved the final
manuscript.
Ethics approval and consent to
participate
The Human Research Ethics Committee of The
Affiliated Hospital of Qingdao University approved the present
study. Written informed consent was obtained from all patients or
their first-degree relatives prior to the start of the research
project.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgments
Not applicable.
References
|
1
|
Vilmar AC and Sorensen JB: Customising
chemotherapy in advanced nonsmall cell lung cancer: Daily practice
and perspectives. Eur Respir Rev. 20:45–52. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Barclay AN, Clark MJ and McCaughan GW:
Neuronal/lymphoid membrane glycoprotein MRC OX-2 is a member of the
immunoglobulin superfamily with a light-chain-like structure.
Biochem Soc Symp. 51:149–157. 1986.PubMed/NCBI
|
|
3
|
Moreaux J, Hose D, Reme T, Jourdan E,
Hundemer M, Legouffe E, Moine P, Bourin P, Moos M, Corre J, et al:
CD200 is a new prognostic factor in multiple myeloma. Blood.
108:4194–4197. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Wright GJ, Cherwinski H, Foster-Cuevas M,
Brooke G, Puklavec MJ, Bigler M, Song Y, Jenmalm M, Gorman D,
McClanahan T, et al: Characterization of the CD200 receptor family
in mice and humans and their interactions with CD200. J Immunol.
171:3034–3046. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Tonks A, Hills R, White P, Rosie B, Mills
KI, Burnett AK and Darley RL: CD200 as a prognostic factor in acute
myeloid leukaemia. Leukemia. 21:566–568. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Moreaux J, Veyrune JL, Reme T, De Vos J
and Klein B: CD200: A putative therapeutic target in cancer.
Biochem Biophys Res Commun. 366:117–122. 2008. View Article : Google Scholar
|
|
7
|
Liu JQ, Talebian F, Wu L, Liu Z, Li MS,
Zhu J, Markowitz J, Carson WE III, Basu S and Bai XF: A critical
role for CD200R signaling in limiting the growth and metastasis of
CD200+ melanoma. J Immunol. 197:1489–1497. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Karp X and Ambros V: Developmental
biology. Encountering microRNAs in cell fate signaling. Science.
310:1288–1289. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Weiler J, Hunziker J and Hall J:
Anti-miRNA oligonucleotides (AMOs): Ammunition to target miRNAs
implicated in human disease? Gene Ther. 13:496–502. 2006.
View Article : Google Scholar
|
|
10
|
Latronico MV, Catalucci D and Condorelli
G: Emerging role of microRNAs in cardiovascular biology. Circ Res.
101:1225–1236. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Chen Y, Fu LL, Wen X, Liu B, Huang J, Wang
JH and Wei YQ: Oncogenic and tumor suppressive roles of microRNAs
in apoptosis and autophagy. Apoptosis. 19:1177–1189. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Sonkoly E, Wei T, Janson PC, Sääf A,
Lundeberg L, Tengvall-Linder M, Norstedt G, Alenius H, Homey B,
Scheynius A, et al: MicroRNAs: Novel regulators involved in the
pathogenesis of psoriasis? PLoS One. 2:e6102007. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Cheng L, Gao S, Song X, Dong W, Zhou H,
Zhao L and Jia L: Comprehensive N-glycan profiles of hepatocellular
carcinoma reveal association of fucosylation with tumor progression
and regulation of FUT8 by microRNAs. Oncotarget. 7:61199–61214.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Hansen T, Olsen L, Lindow M, Jakobsen KD,
Ullum H, Jonsson E, Andreassen OA, Djurovic S, Melle I, Agartz I,
et al: Brain expressed microRNAs implicated in schizophrenia
etiology. PLoS One. 2:e8732007. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Li M, Zhang S, Wu N, Wu L, Wang C and Lin
Y: Overexpression of miR-499-5p inhibits non-small cell lung cancer
proliferation and metastasis by targeting VAV3. Sci Rep.
6:231002016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wang XH, Wang FR, Tang YF, Zou HZ and Zhao
YQ: Association of miR-149C>T and miR-499A>G polymorphisms
with the risk of hepatocellular carcinoma in the Chinese
population. Genet Mol Res. 13:5048–5054. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Bohling SD, Davis E, Thompson K, Kussick
SJ and Love J: Flow cytometric analysis of CD200 expression by
pulmonary small cell carcinoma. Cytometry B Clin Cytom. 90:493–498.
2016. View Article : Google Scholar
|
|
19
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
20
|
Lewis BP, Burge CB and Bartel DP:
Conserved seed pairing, often flanked by adenosines, indicates that
thousands of human genes are microRNA targets. Cell. 120:15–20.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Ishibashi M, Neri S, Hashimoto H,
Miyashita T, Yoshida T, Nakamura Y, Udagawa H, Kirita K, Matsumoto
S, Umemura S, et al: CD200-positive cancer associated fibroblasts
augment the sensitivity of epidermal growth factor receptor
mutation-positive lung adenocarcinomas to EGFR Tyrosine kinase
inhibitors. Sci Rep. 7:466622017. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Schetter AJ, Leung SY, Sohn JJ, Zanetti
KA, Bowman ED, Yanaihara N, Yuen ST, Chan TL, Kwong DL, Au GK, et
al: MicroRNA expression profiles associated with prognosis and
therapeutic outcome in colon adenocarcinoma. JAMA. 299:425–436.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Li L, Pan X, Li Z, Bai P, Jin H, Wang T,
Song C, Zhang L and Gao L: Association between polymorphisms in the
promoter region of miR-143/145 and risk of colorectal cancer. Hum
Immunol. 74:993–997. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Saunders MA, Liang H and Li WH: Human
polymorphism at microRNAs and microRNA target sites. Proc Natl Acad
Sci USA. 104:3300–3305. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Xu HY, Wang ZY, Chen JF, Wang TY, Wang LL,
Tang LL, Lin XY, Zhang CW and Chen BC: Association between
ankylosing spondylitis and the miR-146a and miR-499 polymorphisms.
PLoS One. 10:e01220552015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Oner T, Yenmis G, Tombulturk K, Cam C,
Kucuk OS, Yakicier MC, Dizman D and Sultuybek GK: Association of
Pre-miRNA-499 rs3746444 and Pre-miRNA-146a rs2910164 polymorphisms
and susceptibility to behcet's disease. Genet Test Mol Biomarkers.
19:424–430. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Chen W, Shao D, Gu H, Gong J and Zhang J:
Hsa-mir-499 rs3746444 T/C polymorphism is associated with increased
risk of coronary artery disease in a Chinese population. Acta
Cardiol Sin. 33:34–40. 2017.PubMed/NCBI
|
|
28
|
Toraih EA, Ismail NM, Toraih AA, Hussein
MH and Fawzy MS: Precursor miR-499a variant but not miR-196a2 is
associated with rheumatoid arthritis susceptibility in an Egyptian
population. Mol Diagn Ther. 20:279–295. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Zou HZ and Zhao YQ: Positive association
between miR-499A>G and hepatocellular carcinoma risk in a
Chinese population. Asian Pac J Cancer Prev. 14:1769–1772. 2013.
View Article : Google Scholar
|
|
30
|
Mantovani A, Allavena P, Sica A and
Balkwill F: Cancer-related inflammation. Nature. 454:436–444. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Wright GJ, Jones M, Puklavec MJ, Brown MH
and Barclay AN: The unusual distribution of the neuronal/lymphoid
cell surface CD200 (OX2) glycoprotein is conserved in humans.
Immunology. 102:173–179. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Koning N, Swaab DF, Hoek RM and Huitinga
I: Distribution of the immune inhibitory molecules CD200 and CD200R
in the normal central nervous system and multiple sclerosis lesions
suggests neuron-glia and glia-glia interactions. J Neuropathol Exp
Neurol. 68:159–167. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Jenmalm MC, Cherwinski H, Bowman EP,
Phillips JH and Sedgwick JD: Regulation of myeloid cell function
through the CD200 receptor. J Immunol. 176:191–199. 2006.
View Article : Google Scholar
|
|
34
|
Zhang S, Cherwinski H, Sedgwick JD and
Phillips JH: Molecular mechanisms of CD200 inhibition of mast cell
activation. J Immunol. 173:6786–6793. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Minas K and Liversidge J: Is the
CD200/CD200 receptor interaction more than just a myeloid cell
inhibitory signal? Crit Rev Immunol. 26:213–230. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Olteanu H, Harrington AM, Hari P and Kroft
SH: CD200 expression in plasma cell myeloma. Br J Haematol.
153:408–411. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Pojero F, Casuccio A, Parrino MF,
Cardinale G, Colonna Romano G, Caruso C and Gervasi F: Old and new
immunophenotypic markers in multiple myeloma for discrimination of
responding and relapsing patients: The importance of 'normal'
residual plasma cell analysis. Cytometry B Clin Cytom. 88:165–182.
2015. View Article : Google Scholar
|
|
38
|
Conticello C, Giuffrida R, Parrinello N,
Buccheri S, Adamo L, Sciuto MR, Colarossi C, Aiello E, Chiarenza A,
Romano A, et al: CD200 expression in patients with multiple
Myeloma: Another piece of the puzzle. Leuk Res. 37:1616–1621. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Cannizzo E, Bellio E, Sohani AR,
Hasserjian RP, Ferry JA, Dorn ME, Sadowski C, Bucci JJ, Carulli G
and Preffer F: Multiparameter immunophenotyping by flow cytometry
in multiple myeloma: The diagnostic utility of defining ranges of
normal antigenic expression in comparison to histology. Cytometry B
Clin Cytom. 78:231–238. 2010.PubMed/NCBI
|
|
40
|
Alapat D, Coviello-Malle J, Owens R, Qu P,
Barlogie B, Shaughnessy JD and Lorsbach RB: Diagnostic usefulness
and prognostic impact of CD200 expression in lymphoid malignancies
and plasma cell myeloma. Am J Clin Pathol. 137:93–100. 2012.
View Article : Google Scholar
|
|
41
|
Coustan-Smith E, Song G, Clark C, Key L,
Liu P, Mehrpooya M, Stow P, Su X, Shurtleff S, Pui CH, et al: New
markers for minimal residual disease detection in acute
lymphoblastic leukemia. Blood. 117:6267–6276. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Kapoor A, Kumar N, Narayan S, Nirban RK,
Maharia S, Beniwal SK, Jakhar SL and Kumar HS: 101pda prospective
randomized open label phase III study of geftinib versus docetaxel
as second or third line therapy in patients with advanced non small
cell lung cancer in asian indians. Ann Oncol. 26(Suppl 1): S29–S44.
2015.
|
|
43
|
Siva A, Xin H, Qin F, Oltean D, Bowdish KS
and Kretz-Rommel A: Immune modulation by melanoma and ovarian tumor
cells through expression of the immunosuppressive molecule CD200.
Cancer Immunol Immunother. 57:987–996. 2008. View Article : Google Scholar
|
|
44
|
Colmont CS, Benketah A, Reed SH, Hawk NV,
Telford WG, Ohyama M, Udey MC, Yee CL, Vogel JC and Patel GK:
CD200-expressing human basal cell carcinoma cells initiate tumor
growth. Proc Natl Acad Sci USA. 110:1434–1439. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Stumpfova M, Ratner D, Desciak EB, Eliezri
YD and Owens DM: The immunosuppressive surface ligand CD200
augments the metastatic capacity of squamous cell carcinoma. Cancer
Res. 70:2962–2972. 2010. View Article : Google Scholar : PubMed/NCBI
|