Introduction
Enterovirus 71, a single-stranded RNA virus, is one
of the major causative pathogens of contagious hand, foot and mouth
disease (HFMD) that mainly affects children under the age of 5
(1,2). HFMD is an emerging public health
issue worldwide, especially in Asia-Pacific countries (1,3,4).
Although HFMD is considered as a self-limited disease characterized
by ulcerating vesicles in the mouth and viral rashes on hands and
feet (5-7), a small proportion of cases are
severe and even fatal due to cardiopulmonary or neurological
complications (8,9). EV71 infection accounts for ≥80% of
severe cases of HFMD and 90% of HFMD-related deaths in China
(10). Increasing evidence
indicates that EV71 may target neurons in the central nervous
system (CNS), leading to neuronal degeneration, severe neurological
disorders and even death (11-13). A previous study by our group
demonstrated that neuronal necrosis and neuronophagia were present
in the brainstem in fatal EV71-infected cases (14). Despite the neurotropic
characteristics of EV71, the pathogenesis and molecular mechanisms
of EV71-induced neuronal damage remain largely unknown. EV71
possesses four structural proteins including structural viral
protein (VP) 1, VP2, VP3 and VP4. VP1 homodimers are the main
component of the characteristic icosahedral capsid contributing to
the pathogenicity and stability of the EV71 virus to survive in the
environment of the gastrointestinal tract (15,16).
Autophagy is a cellular process mediated by a unique
organelle named autophagosome that transports cytoplasmic
components to the lysosomes for degradation (17,18). Changes in autophagy in the nervous
system is associated with various neurodegenerative and
neurometabolic disorders, such as Alzheimer's disease and
Niemann-Pick disease (19-21).
Autophagy can be observed by transmission electron microscopy (TEM)
and assessed by measuring the conversion of the
microtubule-associated protein 1 light chain 3 α (LC3) to
phosphatidylethanolamine-conjugated LC3 (LC3II) localized in
autophagosome membranes, which reflects the number of
autophagosomes or the degree of autophagy (22-24). EV71 has been demonstrated to
induce autophagy in infected human rhabdomyosarcoma and
neuroblastoma cells (25,26). A previous study by our group
demonstrated that EV71 VP1 also induces autophagy in cultured
primary EV71-infected brain-stem neurons, which can be inhibited by
salubrinal (SAL), an inhibitor of endoplasmic reticulum (ER) stress
(27), suggesting an essential
role of ER stress in VP1-induced autophagy.
ER stress is triggered by the accumulation of
unfolded or misfolded proteins in the ER (28,29). Although the relationship between
ER stress and autophagy is not yet fully understood, it is well
established that there is a dynamic crosstalk between these two
systems, and ER stress either stimulates or inhibits autophagy
(28,30,31). Since ER stress and autophagy are
commonly concurrent in some human pathologies, such as
cardiovascular diseases, cancers and neurodegenerative disorders
(31-33), it is of great importance to
identify ER stress-associated factors as positive or negative
regulators of autophagy. Peripheral myelin protein 22 (PMP22) is a
trans-membrane glycoprotein highly expressed in the myelinating
Schwann cells of peripheral neurons, and contributes to synthesis
and function of myelin sheaths (34). In Schwann cells, newly synthesized
PMP22 is transiently retained in ER and Golgi before being
transported to the plasma membrane (35,36). Under pathological conditions,
excessive mature or premature (unfolded or misfolded) PMP22
accumulates in the ER and interacts with calnexin, a
Ca2+-binding chaperone, leading to ER retention and
activation of ER stress (37,38). Nevertheless, it remains unknown
whether the relationship between PMP22 and ER stress is associated
with autophagy.
The present study hypothesized that PMP22 may be a
downstream effector of ER stress and may trigger the activation of
autophagy in response to the EV71 capsid protein VP1. To examine
this hypothesis, mouse Schwann cells were transfected with
VP1-expressing vectors, and the effect of VP1 overexpression in
autophagy and PMP22 expression was explored. The results suggested
that ER stress induced the expression of PMP22 and that it was
essential for autophagy of mouse Schwann cells, suggesting an
involvement of the VP1/ER stress/PMP22 axis in the autophagy
regulation of mouse Schwann cells. Targeting the VP1/ER
stress/PMP22 axis may serve in the future as a novel therapeutic
strategy against EV71 infection-induced neuronal damage.
Materials and methods
Cell line and culture
Mouse Schwann cells were purchased from ScienCell
Research Laboratories, Inc. and maintained in Schwann cell medium
(ScienCell Research Laboratories, Inc.) containing penicillin (100
U/ml) and streptomycin (100 µg/ml; Hyclone; GE Healthcare
Life Sciences) in poly-L-lysine-coated (2 µg/cm2)
flasks at 37°C in a humidified atmosphere of 5% CO2.
EV71 was isolated from clinical specimens including
throat, anal swabs and stools of a patient with HFMD due to EV71
infection, provided by the Center for Disease Control and
Prevention of Guangdong Province (Guangzhou, China). The patient
was diagnosed by Guangxi Medical University (Nanning, China) based
on pathological analysis by the Forensic Identification Center,
Zhongshan School of Medicine, Sun Yat-sen University (Guangzhou,
China).
The study was approved by the Ethics Committee of
the Center for Disease Control and Prevention of Guangdong Province
(no. 2017020812). All procedures performed involving human
participants were in accordance with the ethical standards of the
institutional and/or national research committee and with the 1964
Helsinki declaration and its later amendments or comparable ethical
standards. Written informed consent was obtained from the family of
the patient from whom EV71 was isolated for the use of their
clinical samples for research purposes.
Gene cloning and transfection
Total RNA was extracted from EV71 using TRIzol
(Thermo Fisher Scientific, Inc.) and reverse transcribed to obtain
cDNA (cat. no. A3800; Promega Corporation). The 894-bp VP1 cDNA was
synthesized by reverse transcription-polymerase chain reaction
(RT-PCR; cat. no. 2011A; Takara Bio, Inc.) using the primers:
Forward 5′-CCGCTCGAGGCCACCATGGGTGATGGAATTGCAGACATGA-3′ and reverse
5′-CGCGGATCCTAGTGTTGTTATTTTGTCCCTACTTGTGC-3′ (Genewiz, Inc.). The
thermocycling conditions were: 95°C for 2 min, followed by 30
cycles at 95°C for 10 sec and 58°C for 30 sec, then 60°C for 5 min.
The PCR products were subcloned into the pEGFP-C3 expression vector
(Clontech Laboratories, Inc.) and sequencing was performed by
Sangon Biotech for confirmation of successful cloning. The results
were compared with the VP1 cDNA sequence reported in the GenBank
database (GenBank accession number: U55763). Cells were transiently
transfected with 2 mg plasmids using Lipofectamine 2000
(Invitrogen; Thermo Fisher Scientific, Inc.) for 24 h, according to
the manufacturer's instructions.
Small interfering RNA (siRNA)
The siRNA against PMP22 (siPMP22; 100 nmol) was from
Santa Cruz Biotechnology, Inc. (sc-42037) and transfected using the
siRNA transfection reagent (Santa Cruz Biotechnology, Inc.;
sc-37007). Scramble negative control siRNA (NCsiRNA; 100 nmol) was
used as a control. The cells were transfected for 24 h prior to
experiments.
Reverse transcription-quantitative PCR
(RT-qPCR)
Total RNA was extracted from cells using TRIzol
(Thermo Fisher Scientific, Inc.), according to the manufacturer's
instructions, and was reverse transcribed into cDNA using reverse
transcriptase (cat. no. A3800; Promega Corporation). qPCR was
performed using SYBR Green qPCR SuperMix (cat. no. 11733046; Thermo
Fisher Scientific, Inc.), with the following conditions: 95°C for 2
min; 40 cycles at 95°C for 10 sec and 55°C for 30 sec; and final
step 55°C for 10 min. Primers sequences are listed in Table I. GAPDH was used as an internal
control. Relative fold changes in mRNA expression were calculated
using the formula 2−ΔΔCq (39).
 | Table IPrimers used for quantitative
PCR. |
Table I
Primers used for quantitative
PCR.
| Gene | Forward primer
(5′-3′) | Reverse primer
(5′-3′) |
|---|
| PMP22 |
CTGCCAGCTCTTCACTCTCA |
GTTGACATGCCACTCACTGT |
| GAPDH |
GGCCTCCAAGGAGTAAGAAA |
GCCCCTCCTGTTATTATGG |
Western blot analysis
Mouse Schwann cells were lysed using a RIPA lysis
and extraction buffer (cat. no. 89900; Thermo Fisher Scientific,
Inc). Protein concentration in the lysates was determined using the
bicinchoninic acid protein assay reagent (Beyotime Institute of
Biotechnology). Then, 50 ng of proteins were separated by 10%
SDS-PAGE and transferred to polyvinylidene fluoride membranes. The
membranes were blocked with 5% nonfat milk powder in Tris-buffered
saline/Tween 20 (TBST) at room temperature and incubated with
anti-GAPDH: (1:1,000; Abcam; ca. no. ab181602) or anti-LC3II
(1:1,000; Abcam; cat. no. ab51520) for 1-2 h at room temperature.
Following three washes with cold TBST, the membranes were incubated
with peroxidase-conjugated secondary antibody (1:4,000; Thermo
Fisher Scientific, Inc.; cat. no. 31460) for 1 h at room
temperature. After three washes with TBST, the protein expression
was detected using an enhanced chemiluminescent development reagent
(GE Healthcare) and densitometry analysis was performed using
Quantity One (Bio-Rad Laboratories, Inc.).
Immunofluorescence
Mouse Schwann cells were seeded on sterile
coverslips 48 h after transfection and incubated overnight at 37°C.
Cells were fixed with 4% paraformaldehyde for 30 min, followed by
incubation with 0.2% Triton X-100 at 4°C for 5 min. After PBS
washes, the cells were blocked with 10% normal goat serum (Jackson
ImmunoResearch Laboratories, Inc.) for 30 min and incubated with
anti-PMP22 antibody (1:200; Abcam; cat. no. ab211052) overnight at
4°C. The cells were incubated with fluorescence-conjugated
secondary antibodies (1:1,000; Thermo Fisher Scientific, Inc.; cat.
no. 35510) for 1 h at room temperature. In addition, Fluo-8 AM
(1:100; cat. no. 21080; AAT Bioquest, Inc.), a green fluorescent
indicator that monitors Ca2+ conncetration and flux in
cells, was used as a positive control for ER stress inhibition by
SAL. The images were captured with a Leica confocal microscope
(Leica Microsystems GmbH).
Transmission electron microscopy (TEM)
analysis
Mouse Schwann cells were prefixed with 2.5%
glutaraldehyde for 2 h and post-fixed with 1% osmic acid for 2 h at
4°C, followed by gradient dehydration in 30, 50 and 70% ethanol (10
min each), then 80, 90 and 95% acetone (10 min each), and 100%
acetone (10 min twice). The cells were embedded in resin at 35°C,
45°C, 60°C for 24 h respectively, then sectioned to 60 nm thick,
and stained with lead citrate for 15 min at room temperature. The
stained cells were observed and imaged under a Hitachi H-7500
transmission electron microscope (Hitachi, Ltd.).
Statistical analysis
All experiments were repeated at least three times.
Data are expressed as mean ± standard error (SE). Statistical
significance was assessed by the Student's t test or one-way
ANOVA with the Least Significance Difference post hoc test, using
the SPSS 16.0 statistical software (SPSS, Inc.). P<0.05 was
considered to indicate a statistically significant difference.
Results
Cloning and identification of VP1
cDNA
To determine if the VP1 cDNA was successfully cloned
into the pEGFP-C3 vector, plasmids from transformed bacteria were
prepared and digested with BamHI and XhoI. The
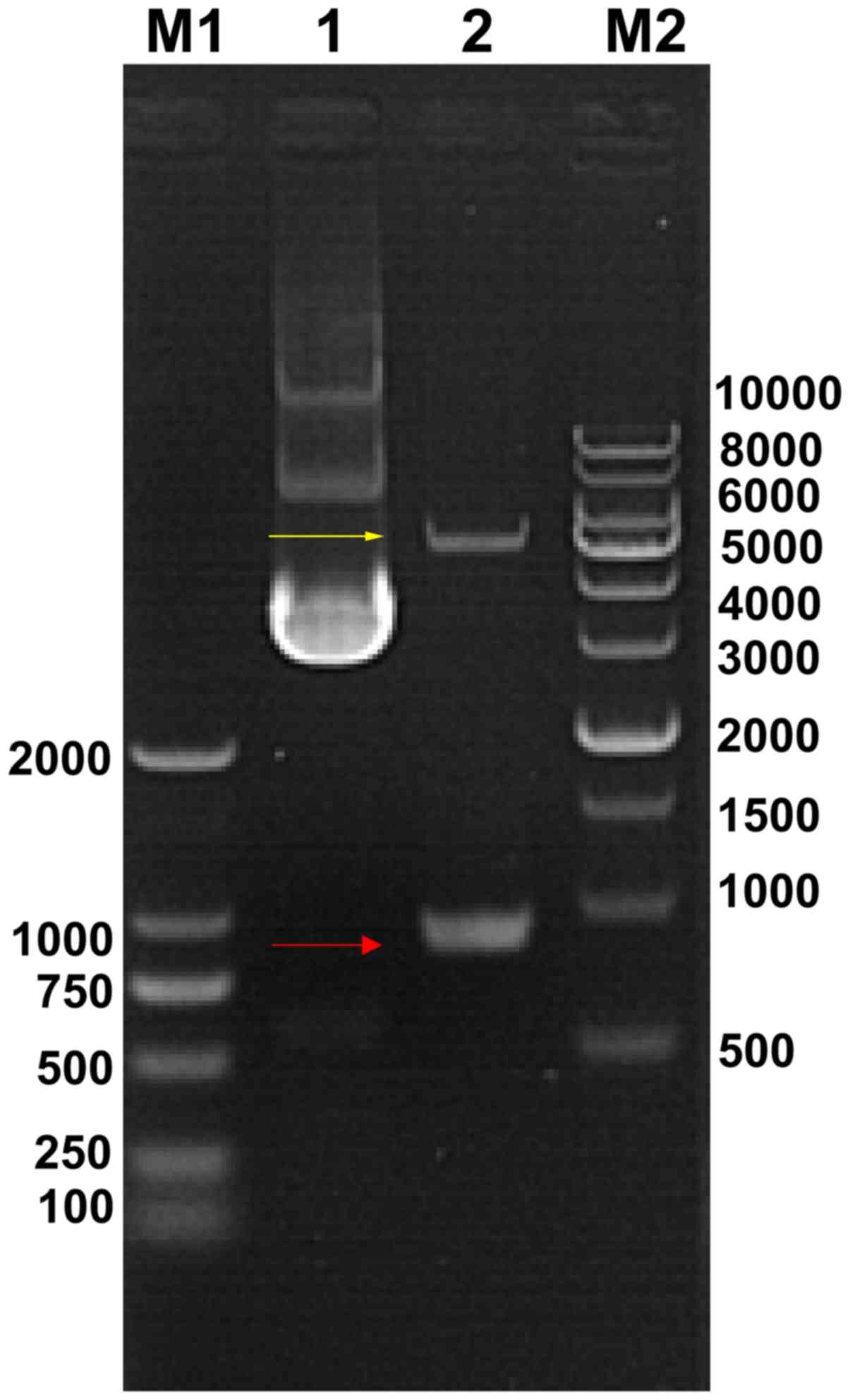
results of agarose electrophoresis demonstrated that a band was
located between 750 and 1,000 bp (Fig. 1), which is consistent with the
size of the VP1 cDNA (894 bp), according to the GenBank database.
The sequencing results also indicated that the cloned fragment was
identical to the VP1 cDNA sequence (data not shown), suggesting
that VP1 cDNA was successfully cloned into the vector, without any
mutation.
Overexpression of VP1 activates mouse
Schwann cell autophagy
To examine whether VP1 has an effect on mouse
Schwann cell autophagy, the cellular and subcellular morphology of
VP1-overexpressing mouse Schwann cells was analyzed using TEM. As
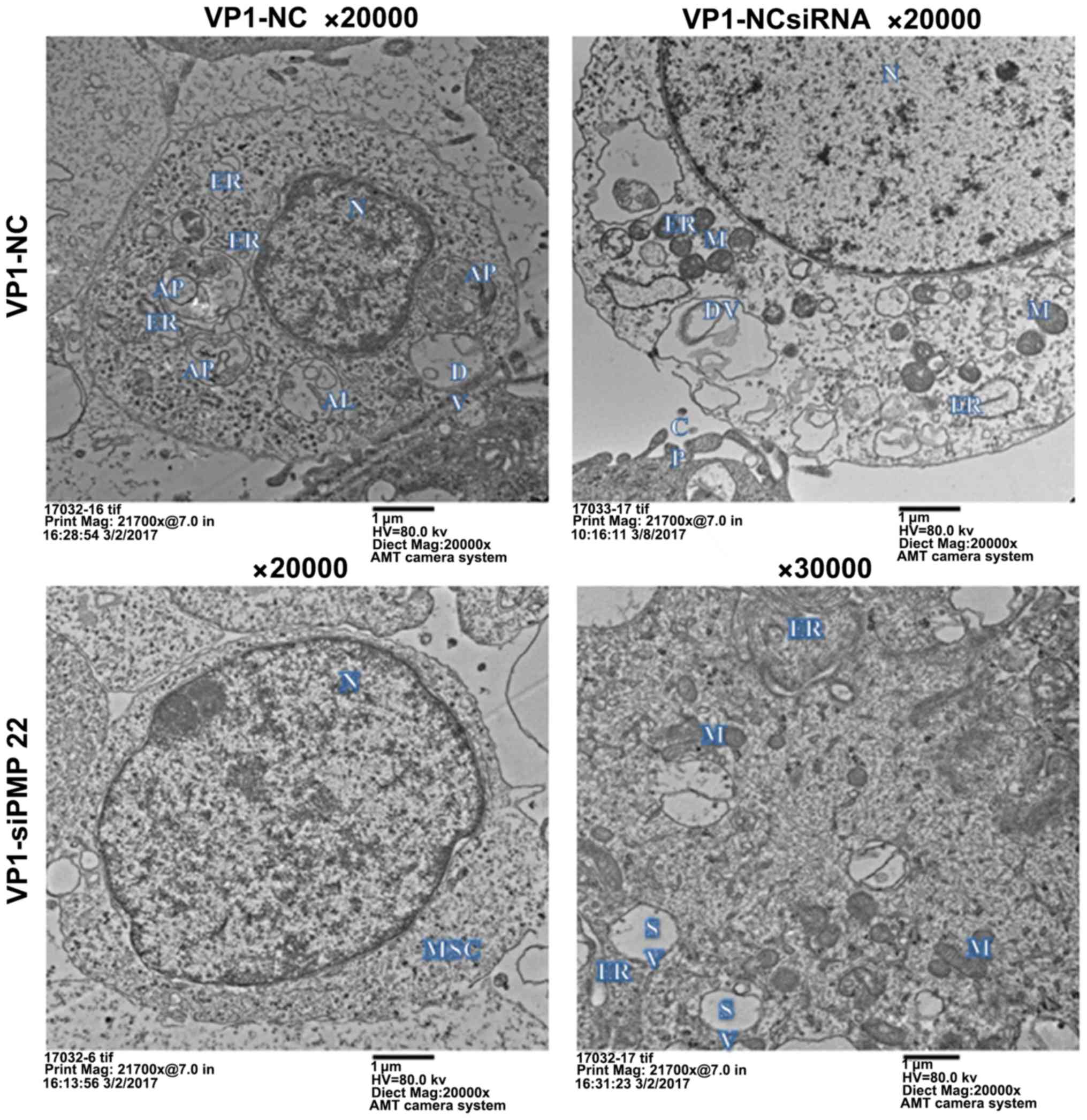
presented in Fig. 2,
VP1-overexpressing mouse Schwann cells exhibited characteristic
features of autophagy, including swelling mitochondria, dilation
and degranulation of rough ER and vesicle-like dilation of Golgi
(40). By contrast, the
organelles in untransfected and empty vector-transfected control
mouse Schwann cells were morphologically normal (Fig. 2). These results suggested that VP1
may activate autophagy in mouse Schwann cells.
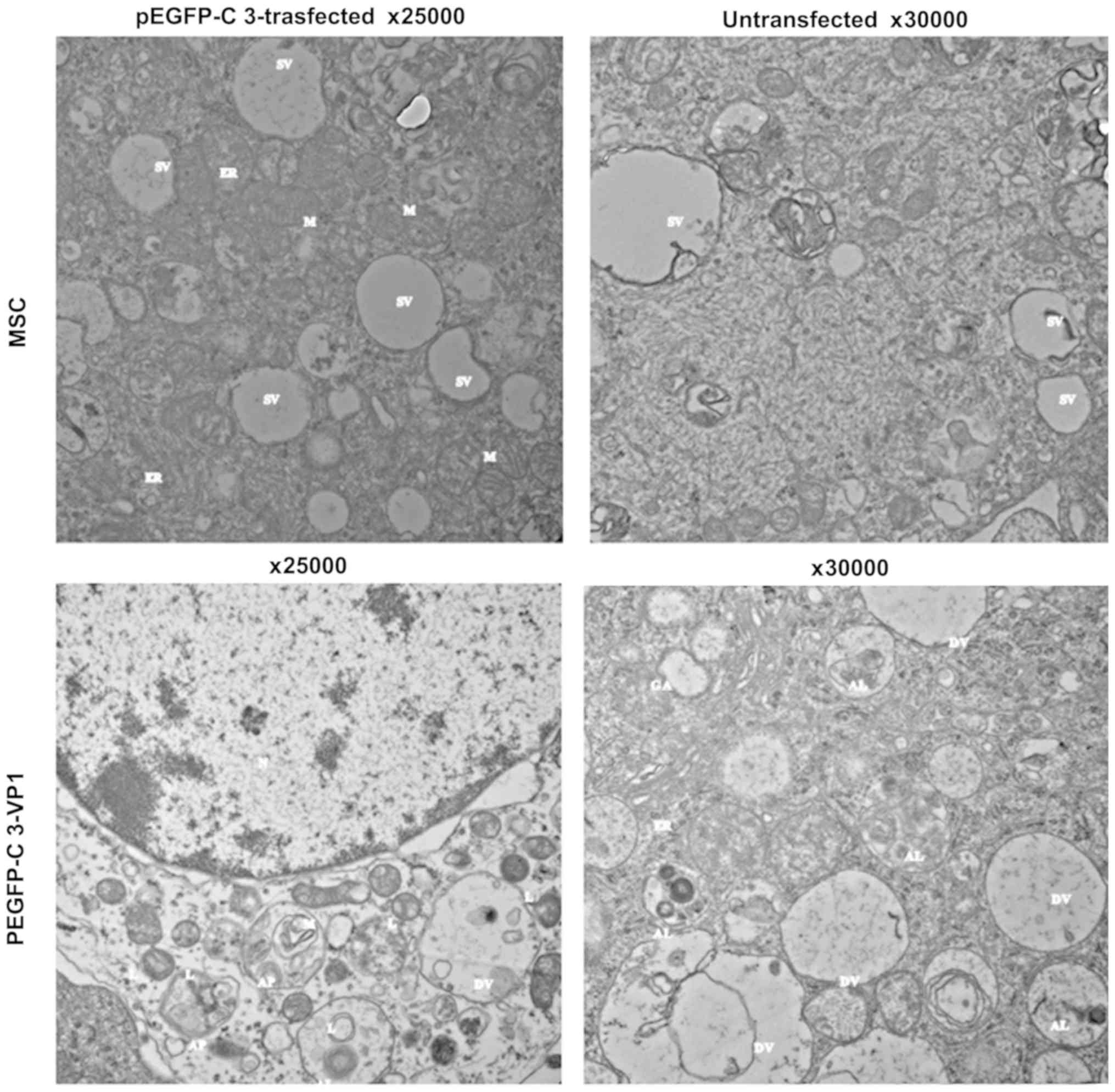 | Figure 2Effect of VP1 overexpression on mouse
Schwann cell autophagy. Mouse Schwann cells were transfected with
pEGFP-C3-VP1 for 48 h. Untransfected and pEGFP-C3-trasfected cells
were used as blank and negative controls, respectively.
Representative transmission electron microscopic images depicting
subcellular structures of mouse Schwann cells are shown.
VP1-overexpressing mouse Schwann cells exhibited the features of
autophagy such as swelling mitochondria, dilation and degranulation
of rough ER, and vesicle-like dilation of Golgi, whereas the
organelles in untransfected and pEGFP-C3-transfected control mouse
Schwann cells were still morphologically normal. VP1, structural
viral protein 1; N, nucleus; M, mitochondrion; L, lysosome; AP,
autophago-some; AL, autolysosome; DV, degradation vesicles; GA,
Golgi apparatus; ER, endoplasmic reticulum; SV, secretory vesicles;
MSC, mouse Schwann cell. |
Overexpression of VP1 upregulates PMP22
expression in mouse Schwann cells
A previous study from our group has indicated an
essential role of ER stress in VP1-induced autophagy in primary
cultured EV71-infected brainstem neurons (14). Because PMP22 is abundant in
Schwann cells and associated with ER stress activation (34,37,38), the present study hypothesized that
PMP22 might correlate with VP1 and serve an important role in
VP1-induced autophagy. To examine this hypothesis, the mRNA and
protein expression levels of PMP22 were detected in
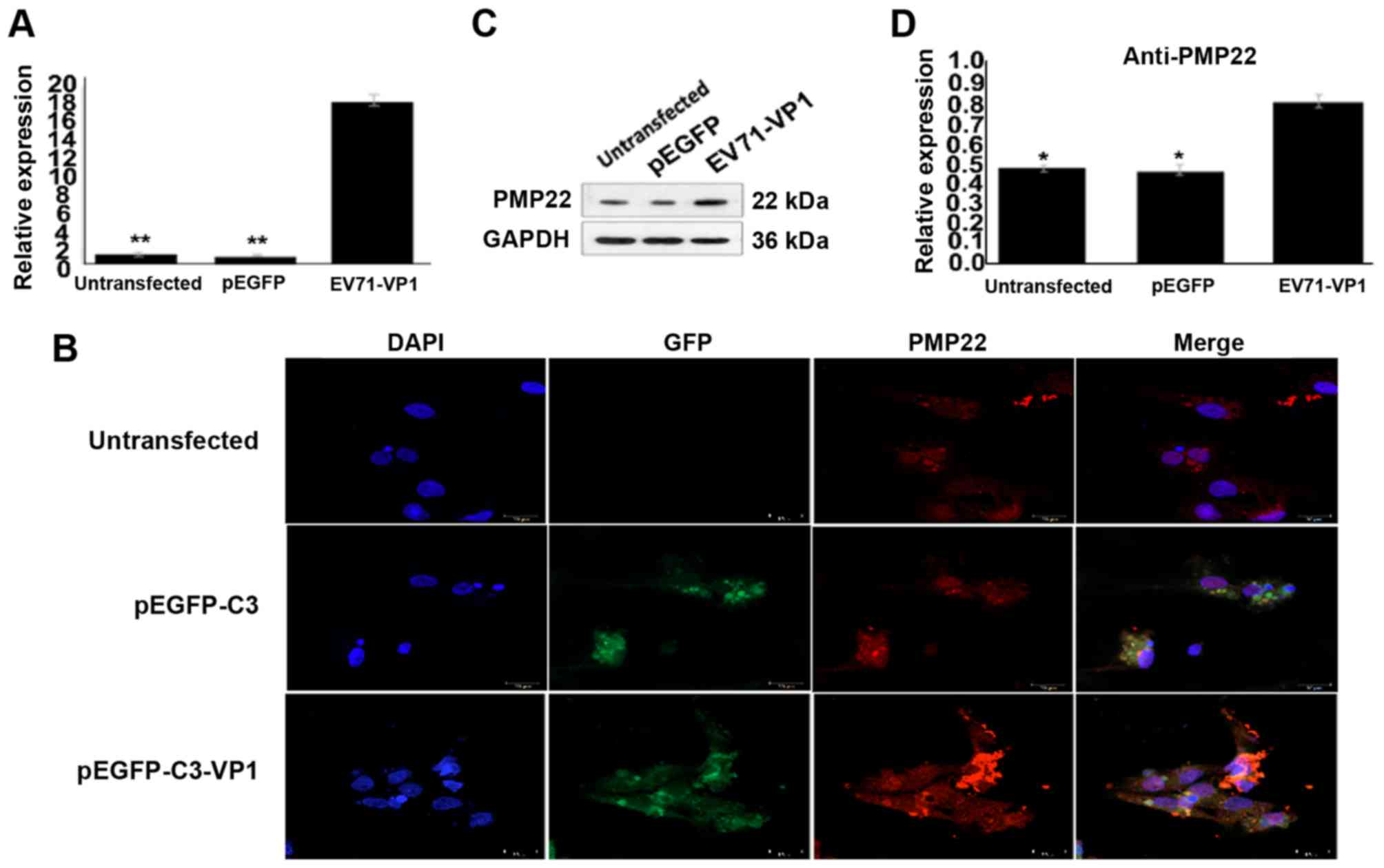
VP1-overexpressing mouse Schwann cells. As presented in Fig. 3A, the mRNA expression levels of
PMP22 were significantly increased in VP1-overexpressing mouse
Schwann cells compared with empty vector-transfected cells.
Immunofluorescence analysis demonstrated similar results (Fig. 3B). Finally, western blot analysis
confirmed that PMP22 protein expression levels were significantly
increased in VP1-overexpressing mouse Schwann cells compared with
empty vector-transfected cells (Fig.
3C and D). These data indicated that VP1 was an upstream
regulator of PMP22, suggesting a possible involvement of PMP22 in
VP1-mediated activation of mouse Schwann cell autophagy.
PMP22 is essential for mouse Schwann
cells autophagy
Next, the present study sought to investigate
whether PMP22 may be involved in mouse Schwann cell autophagy.
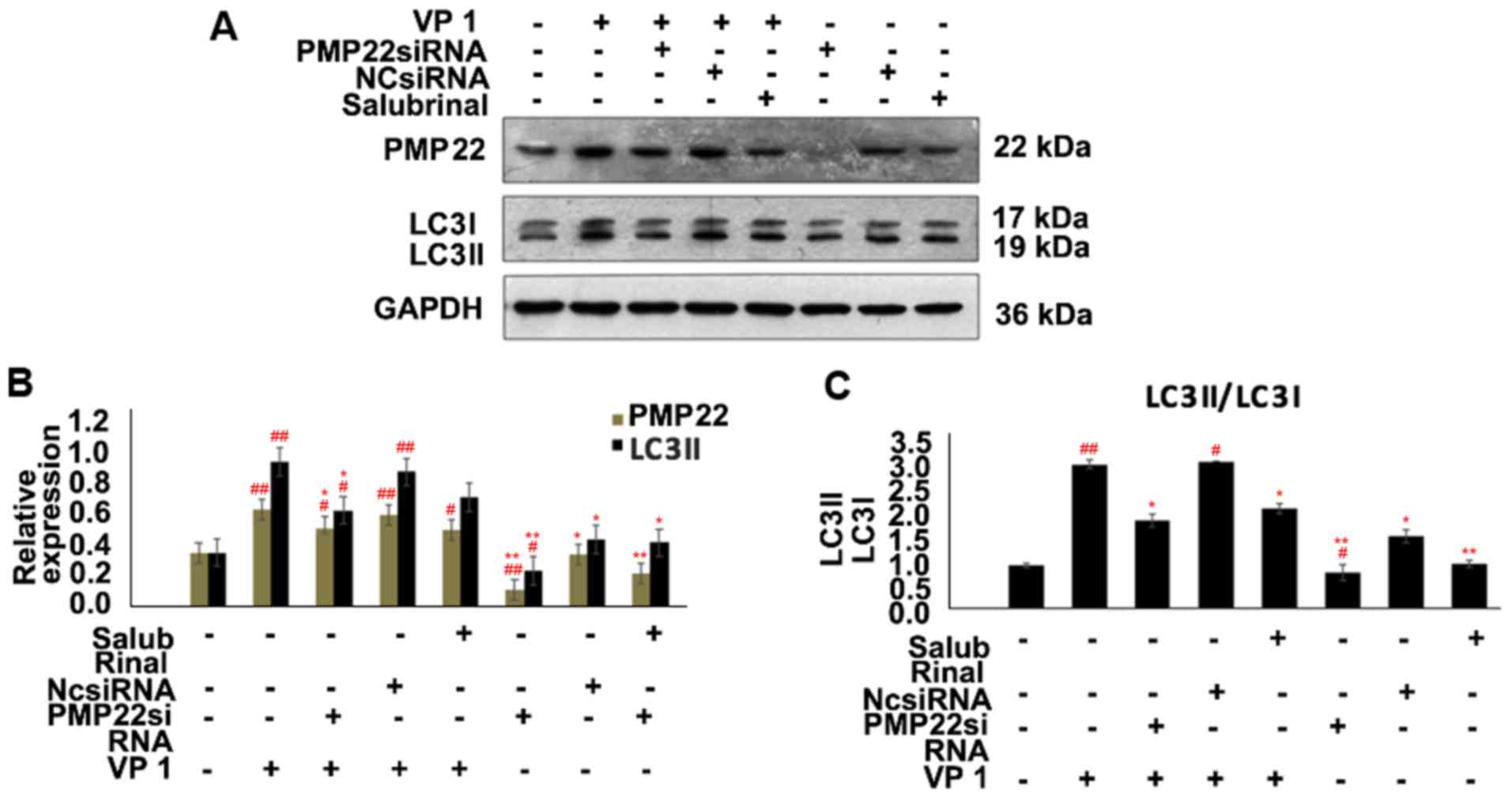
PMP22 was silenced by siRNA, as confirmed by significantly
decreased expression of PMP22 in siPMP22-transfected mouse Schwann
cells (Fig. 4A and B). Notably,
compared with NCsiRNA-transfected groups, PMP22 silencing
significantly downregulated the expression of LC3 isoform LC3II, an
established autophagy marker (41), while VP1 overexpression reversed
this phenomenon (Fig. 4A and B).
Consistently, the ratio of LC3II to LC3I in PMP22-deficient mouse
Schwann cells was also significantly lower compared with the
NCsiRNA-transfected groups (Fig.
4C). Compared with the negative control group, SAL treatment
alone decreased PMP22 and increased LC3II expression, while
siPMP22-transfected cells exhibited decreased PMP22 and LC3II
expression (Fig. 4). Furthermore,
TEM images revealed that there was no observable autophagic
structure in siPMP22-transfected mouse Schwann cells compared with
NCsiRNA-transfected cells (Fig.
5). Taken together, these data suggested that PMP22 was
required for the VP1-mediated activation of autophagy in mouse
Schwann cells.
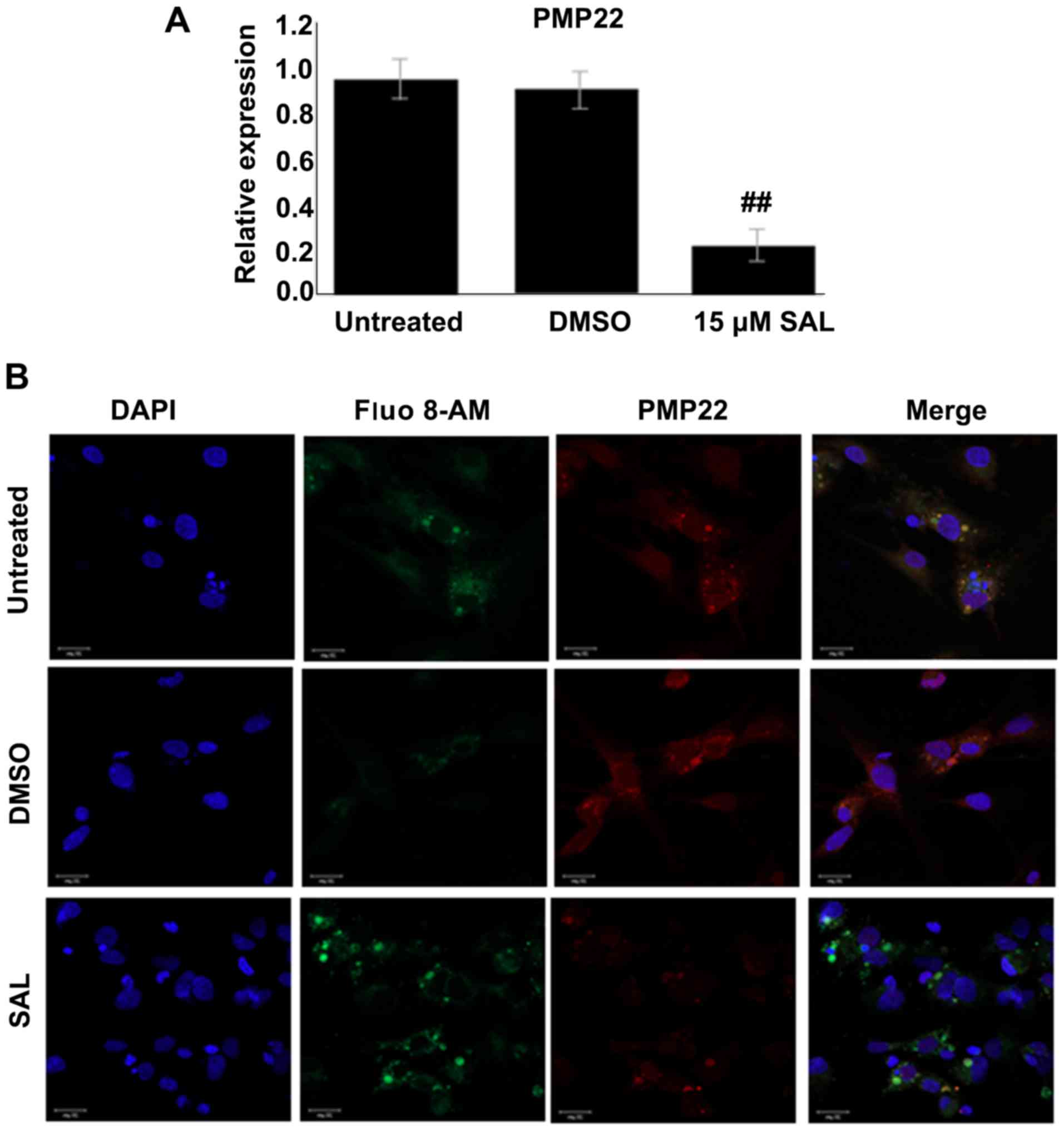
ER stress induces PMP22 expression in
mouse Schwann cells
Since PMP22 is closely associated with ER stress and
since both PMP22 and ER stress are essential for activation of
autophagy, the present study next sought to examine the
relationship between PMP22 and ER stress in mouse Schwann cells
using the selective ER stress inhibitor SAL. As presented in
Fig. 6A, compared with the
control groups, mRNA expression of PMP22 was significantly
downregulated following SAL treatment. Consistently, markedly weak
fluorescent staining of PMP22 was observed in SAL-treated mouse
Schwann cells (Fig. 6B),
suggesting that PMP22 expression in mouse Schwann cells is induced
by ER stress. These results indicate that the VP1/ER stress/PMP22
signaling axis was an important component in mouse Schwann cells
autophagy.
Discussion
The present study investigated the role and
mechanism of the EV71 capsid protein VP1 in mouse Schwann cell
autophagy, and demonstrated for the first time that VP1 promoted
mouse Schwann cell autophagy through ER stress-mediated PMP22
upregulation, suggesting that the VP1/ER stress/PMP22 axis may
serve as a novel potential target against EV71-induced neuronal
disorder in severe HFMD cases.
The current results demonstrated that VP1 promoted
mouse Schwann cell autophagy (Fig.
2), which is consistent with our previous findings (27). Nevertheless, the effect of
VP1-induced autophagy on mouse Schwann cell survival remains
unclear, because autophagy has dual roles in the nervous system.
Excessive autophagy may be protective in chronic neurodegenerative
diseases but detrimental in acute neural damages (20,42). It has been reported that the
inhibition of EV71-induced autophagy in human rhabdomyosarcoma
cells inhibits cell apoptosis at the autophagosome formation and
execution stages, but promotes apoptosis at the
autophagosome-lysosome fusion stage. Furthermore, the inhibition of
autophagy in the autophagosome formation stage decreases the
release of EV71 viral particles, which is an effective strategy
against virus infection (43). On
the other hand, EV71-induced autophagy promotes viral replication
in human rhabdomyosarcoma and neuroblastoma cells, and aggravates
physiopathological parameters, including weight loss, disease
symptoms, and mortality in mouse models (25,26). Further in vitro and in
vivo studies are required to clarify the exact role of
VP1-induced autophagy in neuron cells.
The present results also demonstrated that VP1
overexpression upregulated the ER stress-associated protein PMP22
in mouse Schwann cells, suggesting an involvement of ER stress
activation in VP1-induced autophagy. It is well-established that
the presence of excessive or premature PMP22 in the ER induces ER
stress (37,38). Nevertheless, the effect of ER
stress activation on PMP22 expression has not been investigated to
date. The current data revealed for the first time that inhibition
of ER stress significantly downregulated the expression of PMP22 in
mouse Schwann cells (Fig. 6),
suggesting that PMP22 is a downstream effector of ER stress. It
seems that there is a positive feedback loop between ER stress and
PMP22 in mouse Schwann cells. Furthermore, the present results
demonstrated that in PMP22-deficient mouse Schwann cells there was
no morphological signs of autophagy and the autophagy marker LC3II
was downregulated (Figs. 4 and
5), suggesting that PMP22 was
essential for mouse Schwann cell autophagy. Of note, in an
EV71-infected mouse model, VP1 was found to co-localize with LC3
and/or autophagosome-like vesicles in neurons, and VP1 expression
was positively correlated with LC3II expression, aggregation and
autophagosome formation (26).
Upregulation of LC3II expression was also observed in
VP1-transfected 293 cells (27).
Considering the regulatory role of VP1 in both mouse Schwann cell
autophagy and PMP22 expression (Fig.
3), it can be concluded that the VP1/ER stress/PMP22 axis may
have an important role in the activation of autophagy in mouse
Schwann cells. Nevertheless, whether ER stress may contribute to
autophagy independently of PMP22 still requires additional studies.
In addition, EV71 could increase PMP22 expression through
ER-dependent and independent mechanisms, which also requires
additional studies. Finally, the present study was performed in
mouse Schwann cells, while the mechanistic studies of EV71 have
been mainly performed in tumor cells such as rhabdomyosar-coma.
Caution is advised when comparing the results of the present study
with the literature.
In summary, the present study demonstrated that
mouse Schwann cell autophagy was activated by the EV71 capsid
protein VP1. Mechanistically, the expression of the ER
stress-associated protein PMP22 was significantly upregulated by
VP1 overexpression, suggesting that ER stress-mediated PMP22
upregulation was at least partly responsible for VP1-induced
autophagy activation. The VP1/ER stress/PMP22 axis may eventually
serve as a target for novel therapeutic strategies against the
neuronal damage induced by EV71 infection. Additional studies are
necessary prior to clinical applications.
Abbreviations:
|
CNS
|
central nervous system
|
|
ER
|
endoplasmic reticulum
|
|
EV71
|
enterovirus 71
|
|
HFMD
|
hand, foot and mouth disease
|
|
PMP22
|
peripheral myelin protein 22
|
|
SAL
|
salubrinal
|
|
siRNA
|
small interfering RNA
|
|
TEM
|
transmission electron microscopy;
|
|
VP1
|
viral protein 1
|
Acknowledgments
Not applicable.
Funding
No funding was received.
Availability of data and material
The data analyzed during the present study are
available from the corresponding author on reasonable request.
Authors' contributions
SY and PL conceived and supervised the study. PL and
DH designed experiments. HZ, JL and DW performed experiments. GL
and JL provided new tools and reagents. SN developed new software
and performed simulation studies. HY analyzed data. PL wrote the
manuscript. SY and PL made manuscript revisions. All authors
reviewed the results and approved the final version of the
manuscript.
Ethics approval and consent to
participate
The study was approved by the Ethics Committee of
the Center for Disease Control and Prevention of Guangdong
Province. All procedures performed involving human participants
were in accordance with the ethical standards of the institutional
and/or National Research Committee and with the 1964 Helsinki
declaration and its later amendments or comparable ethical
standards. Written informed consent was obtained from the family of
the patient from whom EV71 was isolated for use of the clinical
samples in research.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Liu MY, Liu J, Lai W, Luo J, Liu Y, Vu GP,
Yang Z, Trang P, Li H and Wu J: Characterization of enterovirus 71
infection and associated outbreak of Hand, Foot, and Mouth disease
in shawo of China in 2012. Sci Rep. 6:384512016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Wang Y, Zou G, Xia A, Wang X, Cai J, Gao
Q, Yuan S, He G, Zhang S, Zeng M and Altmeyer R: Enterovirus 71
infection in children with hand, foot, and mouth disease in
Shanghai, China: Epidemiology, clinical feature and diagnosis.
Virol J. 12:832015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Aswathyraj S, Arunkumar G, Alidjinou EK
and Hober D: Hand, foot and mouth disease (HFMD): Emerging
epidemiology and the need for a vaccine strategy. Med Microbiol
Immunol. 205:397–407. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Shimizu H and Nakashima K: Surveillance of
hand, foot, and mouth disease for a vaccine. Lancet Infect Dis.
14:262–263. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Zhang Y, Tan XJ, Wang HY, Yan DM, Zhu SL,
Wang DY, Ji F, Wang XJ, Gao YJ, Chen L, et al: An outbreak of hand,
foot, and mouth disease associated with subgenotype C4 of human
entero-virus 71 in Shandong, China. J Clin Virol. 44:262–267. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Chong CY, Chan KP, Shah VA, Ng WY, Lau G,
Teo TE, Lai SH and Ling AE: Hand, foot and mouth disease in
Singapore: A comparison of fatal and non-fatal cases. Acta
Paediatr. 92:1163–1169. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Sabanathan S, Tan le V, Thwaites L, Wills
B, Qui PT and Rogier van Doorn H: Enterovirus 71 related severe
hand, foot and mouth disease outbreaks in South-East Asia: Current
situation and ongoing challenges. J Epidemiol Community Health.
68:500–502. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Solomon T, Lewthwaite P, Perera D, Cardosa
MJ, McMinn P and Ooi MH: Virology, epidemiology, pathogenesis, and
control of enterovirus 71. Lancet Infect Dis. 10:778–790. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Gui J, Liu Z, Zhang T, Hua Q, Jiang Z,
Chen B, Gu H, Lv H and Dong C: Epidemiological characteristics and
spatial-temporal clusters of hand, foot, and mouth disease in
zhejiang province, China, 2008-2012. PLoS One. 10:e01391092015.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Xing W, Liao Q, Viboud C, Zhang J, Sun J,
Wu JT, Chang Z, Liu F, Fang VJ, Zheng Y, et al: Hand, foot, and
mouth disease in China, 2008-12: An epidemiological study. Lancet
Infect Dis. 14:308–318. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Hsueh C, Jung SM, Shih SR, Kuo TT, Shieh
WJ, Zaki S, Lin TY, Chang LY, Ning HC and Yen DC: Acute
encephalomyelitis during an outbreak of enterovirus type 71
infection in Taiwan: Report of an autopsy case with pathologic,
immunofluorescence, and molecular studies. Mod Pathol.
13:1200–1205. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kuo RL, Kung SH, Hsu YY and Liu WT:
Infection with enterovirus 71 or expression of its 2A protease
induces apoptotic cell death. J Gen Virol. 83:1367–1376. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Li ML, Hsu TA, Chen TC, Chang SC, Lee JC,
Chen CC, Stollar V and Shih SR: The 3C protease activity of
enterovirus 71 induces human neural cell apoptosis. Virology.
293:386–395. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Yang SD, Li PQ, Li YM, Li W, Lai WY, Zhu
CP, Tao JP, Deng L, Liu HS, Ma WC, et al: Clinical manifestations
of severe enterovirus 71 infection and early assessment in a
Southern China population. BMC Infect Dis. 17:1532017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Klionsky DJ: Autophagy: From phenomenology
to molecular understanding in less than a decade. Nat Rev Mol Cell
Biol. 8:931–937. 2007. View
Article : Google Scholar : PubMed/NCBI
|
|
16
|
Mizushima N and Klionsky DJ: Protein
turnover via autophagy: Implications for metabolism. Annu Rev Nutr.
27:19–40. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Ebrahimi-Fakhari D, Wahlster L, Hoffmann
GF and Kolker S: Emerging role of autophagy in pediatric
neurodegenerative and neurometabolic diseases. Pediatr Res.
75:217–226. 2014. View Article : Google Scholar
|
|
18
|
Ginet V, Spiehlmann A, Rummel C, Rudinskiy
N, Grishchuk Y, Luthi-Carter R, Clarke PG, Truttmann AC and Puyal
J: Involvement of autophagy in hypoxic-excitotoxic neuronal death.
Autophagy. 10:846–860. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Lee JH, Yu WH, Kumar A, Lee S, Mohan PS,
Peterhoff CM, Wolfe DM, Martinez-Vicente M, Massey AC, Sovak G, et
al: Lysosomal proteolysis and autophagy require presenilin 1 and
are disrupted by Alzheimer-related PS1 mutations. Cell.
141:1146–1158. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Klionsky DJ, Abdelmohsen K, Abe A, Abedin
MJ, Abeliovich H, Acevedo Arozena A, Adachi H, Adams CM, Adams PD,
Adeli K, et al: Guidelines for the use and interpretation of assays
for monitoring autophagy (3rd edition). Autophagy. 12:1–222. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Tanida I, Ueno T and Kominami E: LC3 and
autophagy. Methods Mol Biol. 445:77–88. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Benito-Cuesta I, Diez H, Ordonez L and
Wandosell F: Assessment of autophagy in neurons and brain tissue.
Cells. 6:E252017. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Huang SC, Chang CL, Wang PS, Tsai Y and
Liu HS: Enterovirus 71-induced autophagy detected in vitro and in
vivo promotes viral replication. J Med Virol. 81:1241–1252. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Lee YR, Wang PS, Wang JR and Liu HS:
Enterovirus 71-induced autophagy increases viral replication and
pathogenesis in a suckling mouse model. J Biomed Sci. 21:802014.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Hu DD, Mai JN, He LY, Li PQ, Chen WX, Yan
JJ, Zhu WD, Deng L, Wei D, Liu DH, et al: Glucocorticoids prevent
enterovirus 71 capsid protein VP1 induced calreticulin surface
exposure by alleviating neuronal ER stress. Neurotox Res.
31:204–217. 2017. View Article : Google Scholar
|
|
26
|
Rashid HO, Yadav RK, Kim HR and Chae HJ:
ER stress: Autophagy induction, inhibition and selection.
Autophagy. 11:1956–1977. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Lin W and Popko B: Endoplasmic reticulum
stress in disorders of myelinating cells. Nat Neurosci. 12:379–385.
2009. View
Article : Google Scholar : PubMed/NCBI
|
|
28
|
Lee WS, Yoo WH and Chae HJ: ER stress and
autophagy. Curr Mol Med. 15:735–745. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Cai Y, Arikkath J, Yang L, Guo ML,
Periyasamy P and Buch S: Interplay of endoplasmic reticulum stress
and autophagy in neurodegenerative disorders. Autophagy.
12:225–244. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zou X, Xu J, Yao S, Li J, Yang Y and Yang
L: Endoplasmic reticulum stress-mediated autophagy protects against
lipopoly-saccharide-induced apoptosis in HL-1 cardiomyocytes. Exp
Physiol. 99:1348–1358. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Cheng X, Liu H, Jiang CC, Fang L, Chen C,
Zhang XD and Jiang ZW: Connecting endoplasmic reticulum stress to
autophagy through IRE1/JNK/beclin-1 in breast cancer cells. Int J
Mol Med. 34:772–781. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Snipes GJ, Suter U, Welcher AA and Shooter
EM: Characterization of a novel peripheral nervous system myelin
protein (PMP-22/SR13). J Cell Biol. 117:225–238. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Pareek S, Suter U, Snipes GJ, Welcher AA,
Shooter EM and Murphy RA: Detection and processing of peripheral
myelin protein PMP22 in cultured Schwann cells. J Biol Chem.
268:10372–10379. 1993.PubMed/NCBI
|
|
34
|
Ryan MC, Notterpek L, Tobler AR, Liu N and
Shooter EM: Role of the peripheral myelin protein 22 N-linked
glycan in oligomer stability. J Neurochem. 75:1465–1474. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Dickson KM, Bergeron JJ, Shames I, Colby
J, Nguyen DT, Chevet E, Thomas DY and Snipes GJ: Association of
calnexin with mutant peripheral myelin protein-22 ex vivo: A basis
for 'gain-of-function' ER diseases. Proc Natl Acad Sci USA.
99:9852–9857. 2002. View Article : Google Scholar
|
|
36
|
Hara T, Hashimoto Y, Akuzawa T, Hirai R,
Kobayashi H and Sato K: Rer1 and calnexin regulate endoplasmic
reticulum retention of a peripheral myelin protein 22 mutant that
causes type 1A Charcot-Marie-Tooth disease. Sci Rep. 4:69922014.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Shibutani ST and Yoshimori T: A current
perspective of autophagosome biogenesis. Cell Res. 24:58–68. 2014.
View Article : Google Scholar :
|
|
38
|
Barth S, Glick D and Macleod KF:
Autophagy: Assays and artifacts. J Pathol. 221:117–124. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
40
|
Shi M, Zhou Y, Cao L, Ding C, Ji Y, Jiang
Q, Liu X, Li X, Hou X, Peng H and Shi W: Expression of enterovirus
71 capsid protein VP1 in Escherichia coli and its clinical
application. Braz J Microbiol. 44:1215–1222. 2013. View Article : Google Scholar
|
|
41
|
Lal SK, Kumar P, Yeo WM, Kar-Roy A and
Chow VT: The VP1 protein of human enterovirus 71 self-associates
via an interaction domain spanning amino acids 66-297. J Med Virol.
78:582–590. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Puyal J, Ginet V, Grishchuk Y, Truttmann
AC and Clarke PG: Neuronal autophagy as a mediator of life and
death: Contrasting roles in chronic neurodegenerative and acute
neural disorders. Neuroscientist. 18:224–236. 2012. View Article : Google Scholar
|
|
43
|
Xi X, Zhang X, Wang B, Wang T, Wang J,
Huang H, Wang J, Jin Q and Zhao Z: The Interplays between autophagy
and apoptosis induced by enterovirus 71. PLoS One. 8:e569662013.
View Article : Google Scholar : PubMed/NCBI
|