Introduction
Thyroid cancer is the most commonly diagnosed
malignant endocrine tumor worldwide (1). Globally, there are ~300,000 new
cases of thyroid cancer and ~40,000 mortalities are reported every
year, and these numbers are set to increase (2,3).
Thyroid cancer includes four subtypes: Papillary thyroid carcinoma,
follicular thyroid carcinoma, poorly differentiated thyroid
carcinoma and anaplastic thyroid carcinoma (4). Papillary thyroid carcinoma, the most
common form of thyroid cancer, accounts for >80% of thyroid
cancer cases (5). Multimodal
therapeutic approaches, including surgical resection, radioiodine
ablation, and long-term thyrotropin inhibitory therapy, have led to
marked reductions in mortality in recent years; however, the
development of recurrence and metastasis remains the most common
cause of death for patients with thyroid cancer (6). Although numerous genes have been
demonstrated to be closely associated with the malignant
development of thyroid cancer, the underlying molecular mechanisms
have not been fully investigated (7). Therefore, a complete understanding
of the molecular events that occur during thyroid cancer formation
and progression will aid in the development of effective
therapeutic techniques.
MicroRNAs (miRNAs/miRs) are 17-23-nucleotide-long,
single-stranded, non-coding RNAs (8). miRNAs post-transcriptionally silence
a gene or inhibit protein expression by base-pairing with the
3′-untranslated regions (UTRs) of their target genes (9). Over half of the human miRNAs are
located in cancer-related genomic regions or in fragile sites,
suggesting that miRNAs may serve critical roles in carcinogenesis
and cancer progression (10). The
aberrant expression of miRNAs in thyroid cancer has been widely
reported in recent years. For example, miR-129 (11) miR-539 (12), and miR-791 (13) are downregulated in thyroid cancer,
and inhibit the malignant phenotype. On the contrary, miR-1270
(14), miR-221 (15), miR-222 (16), and miR-625-3p (17) are upregulated and promote thyroid
cancer aggressiveness. Hence, further investigation into the
detailed functions of cancer-associated miRNAs in thyroid cancer
may aid the identification of novel targets for preventing the
development of thyroid cancer.
Long noncoding RNA (lncRNA) is a type of noncoding
RNA >200 nucleotides in length (18). Several studies have reported the
aberrant expression of lncRNAs in thyroid cancer and revealed that
the dysregulation of lncRNAs is involved in various biological
processes (19-21). At present, the lncRNA-miRNA-mRNA
network is considered the most widespread mechanism underlying the
action of lncRNAs in tumorigenesis and tumor development (22). Therefore, further investigation
into the interaction between lncRNAs and miRNAs may facilitate the
identification of effective approaches for managing patients with
thyroid cancer.
Downregulation of miR-592 has frequently been
observed in multiple human cancer types, including non-small cell
lung cancer (23), breast cancer
(24), glioma (25,26), hepatocellular carcinoma (27-29), prostate cancer (30), and colorectal cancer (31,32). However, its expression profile and
functions in thyroid cancer are yet to be determined. In this
study, we examined miR-592 expression in thyroid cancer and
assessed its regulatory roles in thyroid cancer progression. In
addition, the mechanisms underlying miR-592-mediated restriction of
the aggressive phenotypes of thyroid cancer cells were explored in
detail.
Materials and methods
Human tissue samples
The Ethics Committee of Jilin Central General
Hospital approved the use of human tissue samples. Written informed
consent was obtained from all patients prior to surgical resection.
Primary thyroid cancer tissues and paired adjacent normal tissues
(ANTs) were collected from 51 patients (24 males, 27 females; age
range, 46-69 years) with thyroid cancer who underwent surgical
resection in the Jilin Central General Hospital. None of these
patients had been treated with chemotherapy, radioiodine ablation,
or long-term thyrotropin inhibitory therapy before surgery. All
patients were graded according to TNM staging system (33), and were divided into either the
miR-592-low or miR-592-high expression group based on the median
value of miR-592 expression of thyroid cancer tissues. Freshly
resected tissues were snap-frozen in liquid nitrogen and stored at
−80°C.
Cell culture
The thyroid anaplastic carcinoma cell line HTH83,
papillary thyroid carcinoma cell line TPC-1, thyroid cancer cell
line BCPAP, as well as a normal human thyroid cell line (HT-ori3)
were purchased from the American Type Culture Collection. All cell
lines were maintained in Dulbecco's Modified Eagle's medium (DMEM;
Gibco; Thermo Fisher Scientific, Inc.) containing 10% fetal bovine
serum (FBS; Gibco; Thermo Fisher Scientific, Inc.) and 1%
penicillin-streptomycin (Sigma-Aldrich; Merck KGaA) and incubated
at 37°C in an incubator supplied with 5% CO2.
Transfection assay
The agomir-592, agomir-NC, NEAT1 small
interfering RNA [siRNA; si-nuclear paraspeckle assemble transcript
1 (NEAT1)], and nontargeting control siRNA (si-NC) were designed
and synthesized by Shanghai GenePharma Co., Ltd. The agomir-592
sequence was 5′-UUG UGU CAA UAU GCG AUG AUG U-3′ and the agomir-NC
sequence was 5′-UUC UCC GAA CGU GUC ACG UTT-3′. The si-NEAT1
sequence was 5′-GUG AGA AGU UGC UUA GAAACU UUC C-3′ and the si-NC
sequence was 5′-UAC UGU CUA GUC GCC GUA C-3′. The coding sequences
of neuro-oncological ventral antigen 1 (NOVA1) were
amplified by GeneRay and cloned into the pcDNA3.1 plasmid (GeneRay;
restriction sites, HindIII + XhoI) to produce the
pcDNA3.1-NOVA1 (pc-NOVA1) plasmid. The NEAT1 overexpression
plasmid pcDNA3.1-NEAT1 (pc-NEAT1) was also chemically synthe-sized
by GeneRay. Empty pcDNA3.1 plasmid was utilized as a negative
control. Cells were plated in 6-well plates 24 h prior to
transfection, and were transfected with agomir-592 (50 nM),
agomir-NC (50 nM), siRNA (100 pmol), or plasmid (4 µg) using
Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.) following the manufacturer's protocol. After
incubation at 37°C for different periods, transfected cells were
harvested and used for subsequent analysis. Reverse
transcription-quantitative polymerase chain reaction (RT-qPCR),
Cell Counting Kit-8 (CCK-8) assay and xenograft tumor model was
carried out at 24 h post-transfection. Migration and invasion
assays and western blotting were performed at 48 and 72 h after
transfection, respectively.
RT-qPCR
Total RNA was extracted using TRIzol®
(Thermo Fisher Scientific, Inc.) from tissues and cultured cells.
To quantify miR-592 expression, an miRcute miRNA First-Strand cDNA
Synthesis kit (Tiangen Biotech, Co., Ltd.) was used to prepare cDNA
from total RNA, according to the manufacturer's protocols. The
synthesized cDNA was then subjected to qPCR using a miRcute miRNA
qPCR Detection kit SYBR-Green (Tiangen Biotech, Co., Ltd.). U6
small nuclear RNA was used as an internal control for miR-592
expression. The cycling conditions for qPCR were as follows: 15 min
at 95°C, followed by 45 cycles of 94°C for 20 sec and 60°C for 34
sec. To measure the mRNA levels of NOVA1 and NEAT1,
total RNA was reverse transcribed into cDNA with a
PrimeScript® RT reagent kit (Takara Bio, Inc.),
according to the manufacturer's protocols. Thereafter, qPCR was
performed on an ABI 7500 PCR System (Applied Biosystems; Thermo
Fisher Scientific, Inc.) using a SYBR® Premix Ex Taq™ II
kit (Takara Bio, Inc.). The cycling conditions for qPCR were as
follows: 5 min at 95°C, followed by 40 cycles of 95°C for 30 sec
and 65°C for 45 sec. The relative mRNA expression of NOVA1
and NEAT1 was normalized against that of glyceraldehyde
3-phosphate dehydrogenase (GAPDH). Relative gene expression
was analyzed using the 2-ΔΔCq method (34). The primers were designed as
follows: miR-592, 5′-CCA TGA CAT TGT GTC AAT ATG CGA-3′ (forward)
and 5′-CGT CAT GAT GTT GCG TCA CC-3′ (reverse); U6, 5′-GCT TCG GCA
GCA CAT ATA CTA AAA T-3′ (forward) and 5′-CGC TTC ACG AAT TTG CGT
GTC AT-3′ (reverse); NOVA1, 5′-GGT CTC AGC CAA GCA GCA GCA
A-3′ (forward) and 5′-TTG CAG CAG TAG CAG CAG CCA G-3′ (reverse);
NEAT1, 5′-TGG CTA GGC TCA GGC TT C AG-3′ (forward) and
5′-TCT CCT TGC CAA GCT TCC TTC-3′ (reverse); and GAPDH,
5′-TGA ACG GGA AGC TCC TGG-3′ (forward) and 5′-TCC ACC CCT GTT GCT
GTA-3′ (reverse).
Cell Counting Kit-8 (CCK-8) assay
Transfected cells were seeded in 96-well plates at a
density of 2×103 cells/well. The CCK-8 assay was
conducted to measure cellular proliferation every 24 h for a period
of 72 h. Briefly, 10 µl of CCK-8 solution (Dojindo Molecular
Technologies, Inc.) was added to each well prior to incubation at
37°C for another 2 h. Finally, the absorbance at 450 nm was
detected using a microplate reader (BioTek Instruments, Inc.).
Migration and invasion assays
Transfected cells were detached using 0.25% trypsin
(Gibco; Thermo Fisher Scientific, Inc.) and then resuspended in
FBS-free DMEM at a concentration of 1×105 cells/ml. For
the migration assay, 200 µl cell suspension was added to the
upper compartments of Transwell inserts (8 µm pore size,
Costar; Corning, Inc.). For the invasion assay, an equivalent
number of cells were added to the upper compartments of Transwell
inserts that were coated with Matrigel (BD Biosciences). In both
assays, the bottom compartments were covered with 600 µl
DMEM containing 20% FBS (Gibco; Thermo Fisher Scientific, Inc.).
After 24 h of routine culture, non-migrated and non-invaded cells
were gently removed with a cotton swab. Cells that migrated/invaded
to the lower surface of the Transwell inserts were fixed with 4%
paraformaldehyde at room temperature for 30 min and stained with
0.5% crystal violet at room temperature for 30 min. The number of
migrated/invaded cells in five randomly selected visual fields was
counted under an IX83 inverted microscope (×200 magnification;
Olympus Corporation).
Xenograft tumor model
TPC-1 cells transfected with agomir-592 or agomir-NC
were harvested after 24 h of incubation and subcutaneously injected
into the flanks of 4-6-week-old BALB/c nude mice (n=4 for each
group) purchased from Beijing Vital River Laboratory Animal
Technology Co., Ltd. All nude mice were maintained under specific
pathogen-free conditions (25°C; 50% humidity; 10-h light/14-h dark
cycle; free access to food and water). After 14 days following
inoculation, tumor size was measured every 2 days for 14 days using
a caliper, and tumor volumes were determined based on the following
formula: Tumor volume = length × (width)2/2. All nude
mice were sacrificed 4 weeks post-injection, and tumor weights were
measured. The tumor xenografts were resected and stored for western
blotting and RT-qPCR analysis. Animal experiments were approved by
the Animal Care and Use Committee of Jilin Central General Hospital
and were performed in accordance with the Animal Protection Law of
the People's Republic of China-2009 for experimental animals.
Bioinformatics analysis
miRDB (last modified: January 22, 2019; http://mirdb.org/) and TargetScan (Release 7.2: March
2018; http://www.targetscan.org) were utilized
for predicting the potential target gene of miR-592. The binding
site between miR-592 and NEAT1 was predicted using DIANA
tools -LncBase Experimental v2 (http://carolina.imis.athena-innovation.gr/diana_tools/web/index.php?r=lncbasev2%2findex-experimental).
Luciferase reporter assay
The wild-type (wt) NEAT1 containing the
predicted miR-592 binding site and mutant (mut) NEAT1 were
amplified by Shanghai GenePharma Co., Ltd. and inserted into the
pmiR-REPORT reporter plasmid (Promega Corporation) to generate
NEAT1-wt and NEAT1-mut, respectively. The NOVA1-wt and NOVA1-mut
reporter plasmids were synthesized in a similar manner. Cells were
plated in 24-well plates with a density of 1.0×105 cells
per well and transfected with a synthetic luciferase reporter
plasmid, along with agomir-592 or agomir-NC, using
Lipofectamine® 2000. Then, 48 h after transfection,
luciferase activity was quantified using a Dual Luciferase Assay
(Promega Corporation). Firefly luciferase activity was normalized
to that of Renilla luciferase activity.
Western blot analysis
Total protein was isolated from tissue specimens,
cells, or tumor xenografts was conducted using
radioimmunoprecipitation assay lysis buffer (Beyotime Institute of
Biotechnology. The total protein concentration was measured with a
BCA Protein Assay kit (Pierce; Thermo Fisher Scientific, Inc.).
Equal amounts of protein (30 µg) were loaded, separated by
electrophoresis using 10% SDS-PAGE, and then transferred onto
polyvinylidene fluoride membranes (Beyotime Institute of
Biotechnology). Prior to overnight incubation at 4°C with primary
antibodies against NOVA1 (ab183024; 1:1,000 dilution; Abcam) or
GAPDH (ab128915; 1:1,000 dilution; Abcam), the membranes were
blocked at room temperature with 5% skim milk diluted in
Tris-buffered saline buffer containing 0.1% Tween-20 (TBST) for 2
h. Thereafter, the membranes were incubated with a goat anti-rabbit
horseradish peroxidase-linked IgG secondary antibody (ab97051;
1:5,000 dilution; Abcam) at room temperature for 1 h. Finally,
protein signals were visualized using an Enhanced Chemiluminescence
Detection System (Pierce; Thermo Fisher Scientific, Inc.). Quantity
One software version 4.62 (Bio-Rad Laboratories, Inc.) was utilized
for quantify protein expression.
Statistical analysis
All results are presented the mean ± standard
deviation from at least three separate experiments, and data
processing was performed with SPSS 21.0 statistical software (IBM
Corp.). The χ2 test was used to examine the association
between miR-592 and clinicopathological parameters in patients with
thyroid cancer. One-way analysis of variance followed by the
Bonferroni post-hoc test was used to analyze the data among
multiple groups; the difference between two groups was assessed
using a two-sided Student's t-test. The correlation between genes
was examined via Spearman's correlation analysis. A log-rank test
was applied to determine the overall survival rates, following
Kaplan-Meier analysis. P<0.05 was considered to indicate a
statistically significant difference.
Results
miR-592 is downregulated in thyroid
cancer, and its downregulation is inversely associated with the
prognosis of patients with thyroid cancer
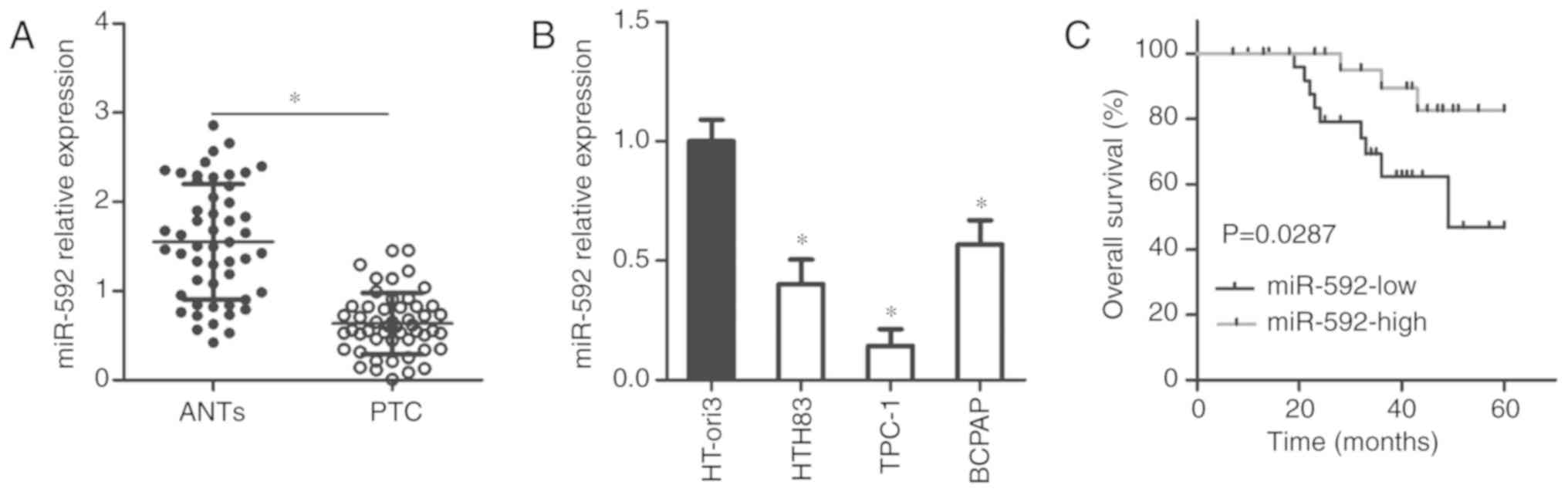
To explore the roles of miR-592 during the
development of thyroid cancer, we first measured miR-592 expression
in primary thyroid cancer tissues and ANTs obtained from 51
patients with thyroid cancer. The expression of miR-592 was
significantly downregulated in thyroid cancer tissues compared with
that in ANTs, as demonstrated by RT-qPCR (P<0.05; Fig. 1A). miR-592 expression in three
thyroid cancer cell lines and a normal human thyroid cell line
(HT-ori3) was also detected via RT-qPCR. The results indicated that
the miR-592 levels were significantly downregulated in thyroid
cancer cell lines than in HT-ori3 cells (P<0.05; Fig. 1B).
All patients were divided into either the
miR-592-low or miR-592-high expression group based on the median
value of miR-592 expression in thyroid cancer tissues. To examine
the clinical value of miR-592 in thyroid cancer, we investigated
the association between miR-592 expression and clinicopathological
parameters in patients with thyroid cancer. As presented in
Table I, decreased miR-592
expression was associated with a higher incidence of increased
lymph node metastasis (P=0.002) and advanced tumor-node-metastasis
(TNM) stage (P=0.009). In addition, expression levels of miR-592
exhibited an inverse association with overall survival in patients
with thyroid cancer (P=0.0287; Fig.
1C). Thus, miR-592 was determined to be downregulated in
thyroid cancer, and its decreased expression could predict poor
prognosis for patients with thyroid cancer.
 | Table IAssociation between the
clinicopathological features and miR-592 expression in patients
with papillary thyroid cancer. |
Table I
Association between the
clinicopathological features and miR-592 expression in patients
with papillary thyroid cancer.
| Features | miR-592 expression
| P-value |
|---|
| Low | High |
|---|
| Age (years) | | | 0.579 |
| <60 | 15 | 12 | |
| ≥60 | 11 | 13 | |
| Sex | | | 0.095 |
| Male | 9 | 15 | |
| Female | 17 | 10 | |
| Tumor size
(cm) | | | 0.264 |
| <1 cm | 12 | 16 | |
| ≥1 cm | 14 | 9 | |
| Lymph node
metastasis | | | 0.002a |
| Negative | 13 | 23 | |
| Positive | 13 | 2 | |
| TNM stage | | | 0.009a |
| I-II | 15 | 23 | |
| III-IV | 11 | 2 | |
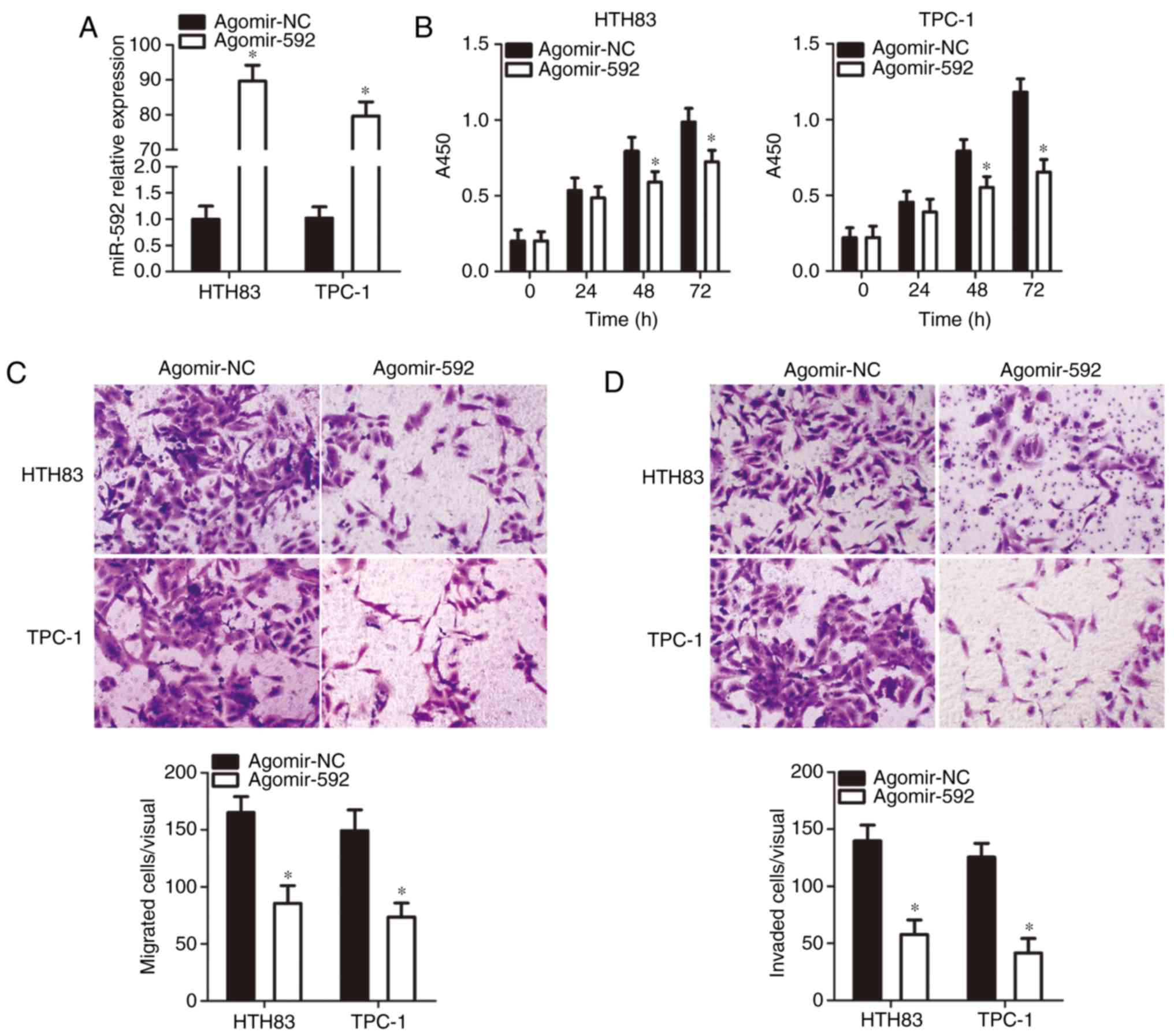
Restoring miR-592 expression suppresses
the proliferation, migration, and invasion of thyroid cancer
cells
To restore miR-592 expression, the HTH83 and TPC-1
cell lines, which had the lowest miR-592 expression among the three
thyroid cancer cell lines examined, were transfected with
agomir-592. A significant increase in miR-592 expression in HTH83
and TPC-1 cells after agomir-592 transfection was confirmed via
RT-qPCR analysis (P<0.05; Fig.
2A). Subsequently, the influence of miR-592 upregulation on
thyroid cancer cell proliferation, migration, and invasion was
determined by CCK-8, and migration and invasion assays,
respectively. The proliferation (P<0.05; Fig. 2B), migration (P<0.05; Fig. 2C), and invasion (Fig. 2D, P<0.05) of HTH83 and TPC-1
cells were significantly suppressed by miR-592 restoration. These
results suggested that the upregulation of miR-592 can impair the
malignancy of thyroid cancer cells in vitro.
NOVA1 is a novel target of miR-592 in
thyroid cancer cells
To identify the molecular mechanism of miR-592 in
regulating the malignant phenotypes of thyroid cancer cells,
NOVA1 was validated as a putative target of miR-592 using
bioinformatics analysis (Fig.
3A). To determine whether miR-592 could recognize and interact
with the 3′-UTR of NOVA1, a luciferase reporter assay was
conducted in HTH83 and TPC-1 cells after co-transfection with
agomir-592 or agomir-NC, and NOVA1-wt or NOVA1-mut. Restoring
miR-592 expression significantly reduced the luciferase activity of
the NOVA1-wt (P<0.05; Fig.
3B), but not the NOVA1-mut in HTH83 and TPC-1 cells. In
addition, RT-qPCR and western blot analysis were conducted to
determine whether endogenous NOVA1 expression could be
directly regulated by miR-592 in thyroid cancer cells. The results
indicated that expression levels of NOVA1 mRNA (P<0.05;
Fig. 3C) and protein (P<0.05;
Fig. 3D) were significantly
downregulated in HTH83 and TPC-1 cells transfected with agomir-592
compared with the control. Furthermore, the expression of
NOVA1 mRNA in thyroid cancer tissues and ANTs was detected
using RT-qPCR, and the results showed that NOVA1 mRNA was
markedly upregulated in thyroid cancer tissues in comparison with
that in ANTs (P<0.05; Fig.
3E). Spearman's correlation analysis also demonstrated that the
expression of miR-592 was inversely correlated with NOVA1
mRNA expression in the same thyroid cancer tissues
(R2=0.3204, P<0.0001; Fig. 3F). These results suggest that
NOVA1 is a novel direct target of miR-592 in thyroid cancer
cells.
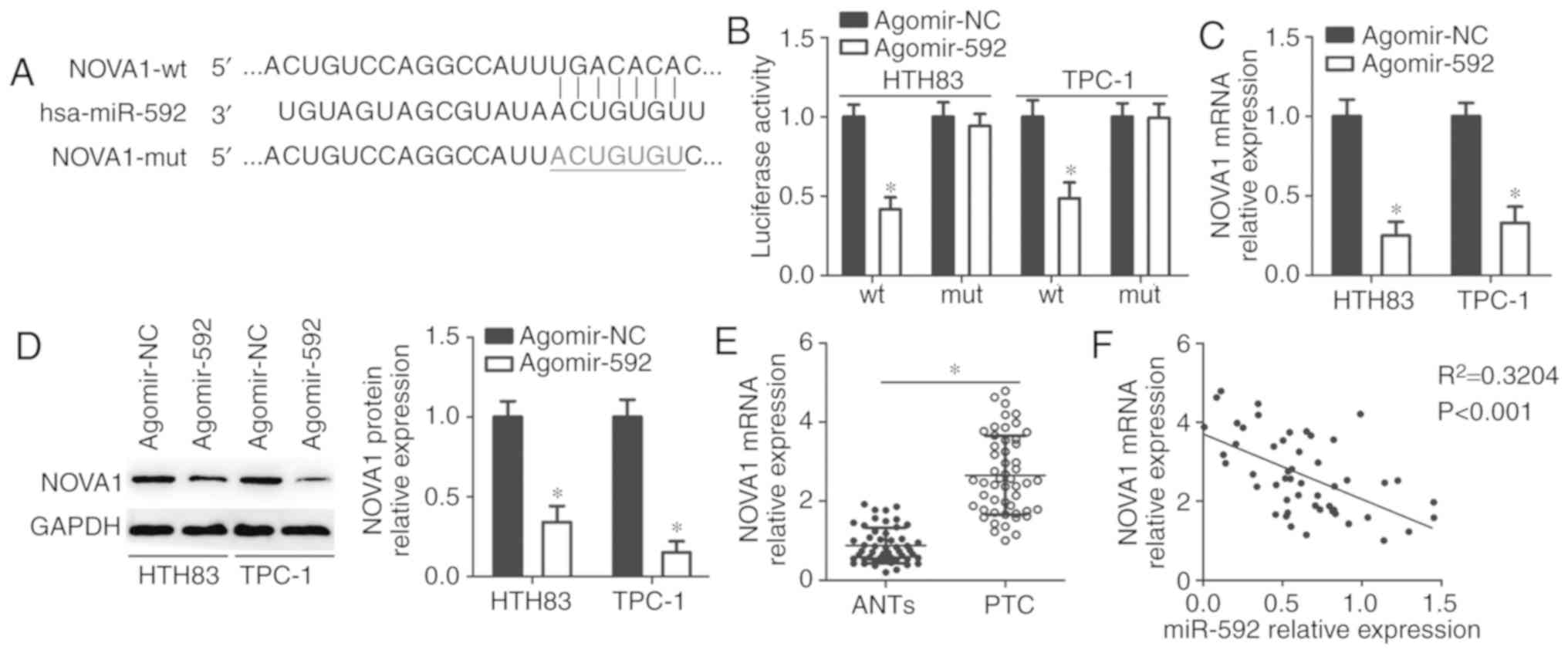 | Figure 3NOVA1 is a novel direct target
of miR-592 in thyroid cancer cells. (A) Sequence of the wt and mut
miR-592 binding sequences within the 3′-untranslated region of the
NOVA1 gene. (B) HTH83 and TPC-1 cells were co-transfected
with agomir-592 or agomir-NC and NOVA1-wt or NOVA1-mut reporter
plasmids. Luciferase activities were measured 48 h after
transfection. *P<0.05 vs. agomir-NC. (C and D) The
mRNA and protein expression levels of NOVA1 in
miR-592-overexpressing HTH83 and TPC-1 cells were analyzed by
RT-qPCR and western blot analysis, respectively.
*P<0.05 vs. agomir-NC. (E) RT-qPCR analysis revealed
the increased expression level of NOVA1 in thyroid cancer
tissues compared with that in ANTs. *P<0.05 vs. ANTs.
(F) Spearman's correlation analysis was used to determine the
correlation between miR-592 and NOVA1 mRNA expression in
thyroid cancer tissues. R2=0.3204, P<0.0001. ANTs,
adjacent normal tissues; hsa, homo sapiens; NOVA1,
neuro-oncological ventral antigen 1; miR, microRNA; Mut, mutant;
NC, negative control; PTC, papillary thyroid cancer; RT-qPCR,
reverse transcription-quantitative polymerase chain reaction; wt,
wild-type. |
Ectopic NOVA1 expression mitigates the
effects of miR-592 upregulation in thyroid cancer cells
Rescue experiments were further used to determine
whether the tumor-suppressing effects of miR-592 in thyroid cancer
cells were achieved through decreasing NOVA1 expression.
Agomir-592 was co-transfected with pc-NOVA1 or pcDNA3.1 into HTH83
and TPC-1 cells. First, RT-qPCR analysis indicated that the
transfection of pc-NOVA1 significantly increased NOVA1 expression
in HTH83 and TPC-1 cells compared with the control (P<0.05;
Fig. 4A). Downregulation of
NOVA1 caused by miR-592 over-expression was recovered in
HTH83 and TPC-1 cells through co-transfection with pc-NOVA1
(P<0.05; Fig. 4B). Of note,
the suppressive effects of miR-592 overexpression on HTH83 and
TPC-1 cell proliferation (P<0.05; Fig. 4C), migration (P<0.05; Fig. 4D), and invasion (P<0.05;
Fig. 4E) were nearly abolished by
restoring NOVA1 expression. Therefore, these results
indicated that NOVA1 is a functionally relevant target of
miR-592 in thyroid cancer cells.
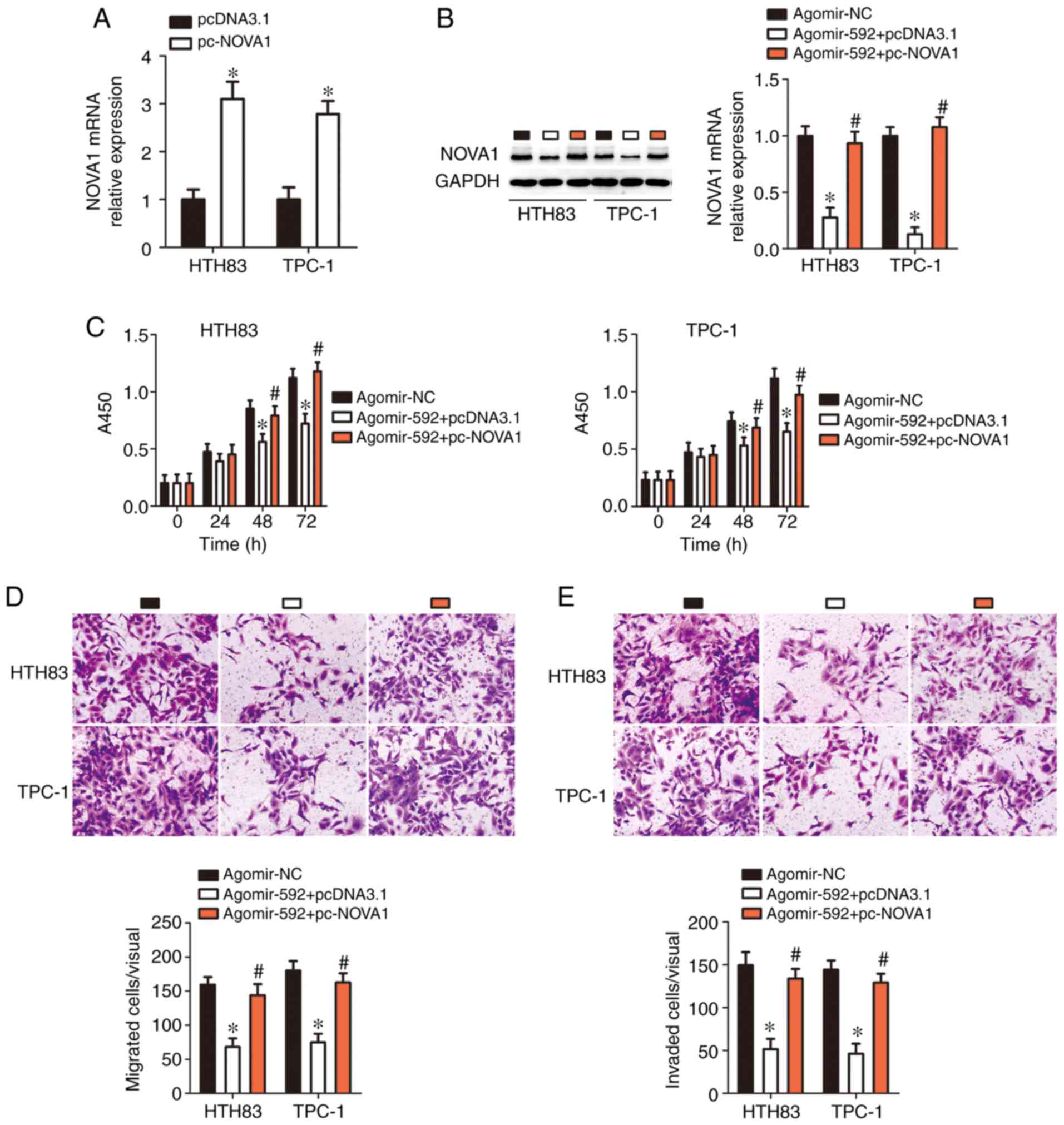 | Figure 4Restoring NOVA1 expression
abolishes the tumor-suppressing effects of microRNA-592 on thyroid
cancer cell proliferation, migration, and invasion in vitro.
(A) pc-NOVA1 or pcDNA3.1 was introduced into HTH83 and TPC-1 cells.
After transfection, reverse transcription-quantitative polymerase
chain reaction analysis was used for determination of NOVA1
expression. *P<0.05 vs. pcDNA3.1. (B) pc-NOVA1 or
pcDNA3.1 in combination with agomir-592 was transfected into HTH83
and TPC-1 cells. Decreased NOVA1 expression, induced by
miR-592 overexpression, was successfully restored in HTH83 and
TPC-1 cells after co-transfection with pc-NOVA1.
*P<0.05 vs. agomir-NC. #P<0.05 vs.
agomir-NC. (C-E) The proliferation, migration (×200 magnification),
and invasion (×200 magnification) of HTH83 and TPC-1 cells treated
as aforementioned were assessed using Cell Counting Kit-8, and
migration and invasion assays, respectively. *P<0.05
vs. agomir-NC. #P<0.05 vs. agomir-592 + pcDNA3.1.
NOVA1, neuro-oncological ventral antigen 1; NC, negative
control. |
NEAT1 acts as a competing endogenous RNA
(ceRNA) for miR-592 in thyroid cancer cells
lncRNAs may act as molecular sponges for miRNAs and
play an important role in regulating miRNAs (22). Bioinformatics analysis revealed a
putative binding site for miR-592 in the lncRNA NEAT1
(Fig. 5A), which has been well
studied and confirmed to play an oncogenic role in thyroid cancer
progression (20,35-37). A luciferase reporter assay was
used to confirm this prediction. The results showed that compared
with the agomir-NC group, recovery of miR-592 expression
significantly decreased the luciferase activity of NEAT1-wt
compared with the control (P<0.05; Fig. 5B); however, the luciferase
activity of the NEAT1-mut was markedly unchanged after agomir-592
transfection. In addition, silencing and increasing NEAT1
expression via the transfection of si-NEAT1 (P<0.05; Fig. 5C) and pc-NEAT1 (P<0.05;
Fig. 5D), significantly increased
(P<0.05; Fig. 5E) and
decreased (P<0.05; Fig. 5F)
miR-592 expression in HTH83 and TPC-1 cells, respectively compared
with the controls. Furthermore, the expression levels of
NEAT1 in thyroid cancer tissues was were upregulated than
that in ANTs (P<0.05; Fig.
5G). An inverse correlation was identified between the
expression levels of NEAT1 and miR-592 in thyroid cancer
tissues (R2=0.4555, P<0.0001; Fig. 5H). Collectively, these results
suggest that miR-592 may be sponged by NEAT1 in thyroid
cancer cells.
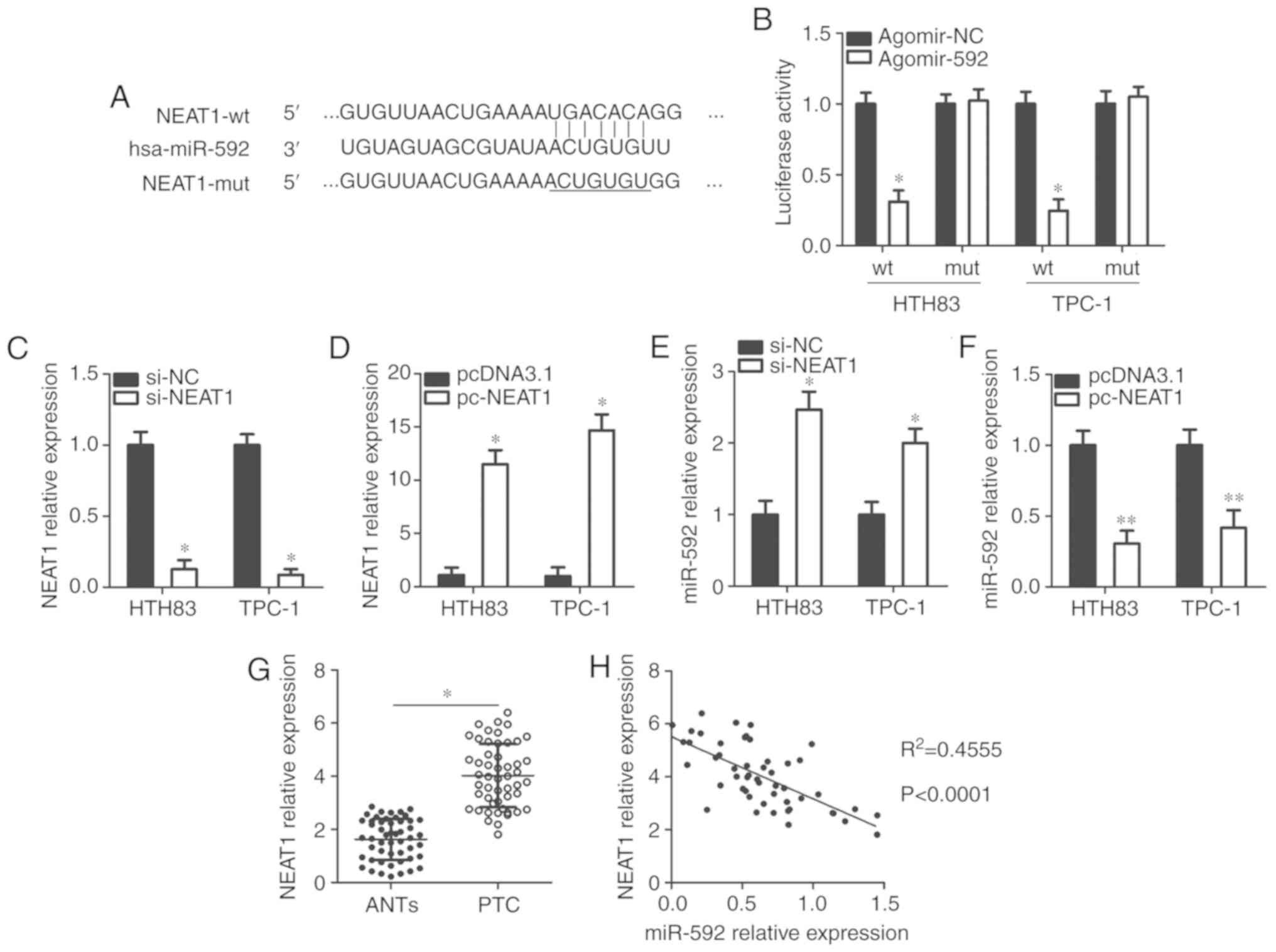 | Figure 5miR-592 is sponged by NEAT1 in
thyroid cancer cells. (A) The wt and mut miR-592 binding sites in
NEAT1, as predicted by bioinformatics analysis. (B)
Luciferase reporter assays were carried out using HTH83 and TPC-1
cells that were co-transfected with agomir-592 or agomir-NC, and
NEAT1-wt or NEAT1-mut reporter plasmids. *P<0.05 vs.
agomir-NC. (C) NEAT1 expression in HTH83 and TPC-1 cells
after transfection with si-NEAT1 or si-NC was examined using
RT-qPCR. *P<0.05 vs. si-NC. (D) HTH83 and TPC-1 cells
were transfected with pcDNA3.1 or pc-NEAT1. After transfection 48
h, RT-qPCR was conducted to measure NEAT1 expression.
*P<0.05 vs. pcDNA3.1. (E and F) Expression of miR-592
in NEAT1-silenced or NEAT1-overexpressing HTH83 and
TPC-1 cells was analyzed using RT-qPCR. *P<0.05 vs.
si-NC. **P<0.05 vs. pcDNA3.1. (G) The expression
levels of NEAT1 in 51 pairs of thyroid cancer tissues and
ANTs was examined through RT-qPCR. *P<0.05 vs. ANTs.
(H) The correlation between the expression of NEAT1 and
miR-592 in thyroid cancer tissues was explored via Spearman's
correlation analysis. R2=0.4555, P<0.0001. ANTs,
adjacent normal tissues; hsa, homo sapiens; miR, microRNA;
NEAT1, nuclear paraspeckle assemble transcript 1; mut, mutant; NC,
negative control; RT-qPCR, reverse transcription-quantitative
polymerase chain reaction; si, small interfering RNA; wt,
wild-type. |
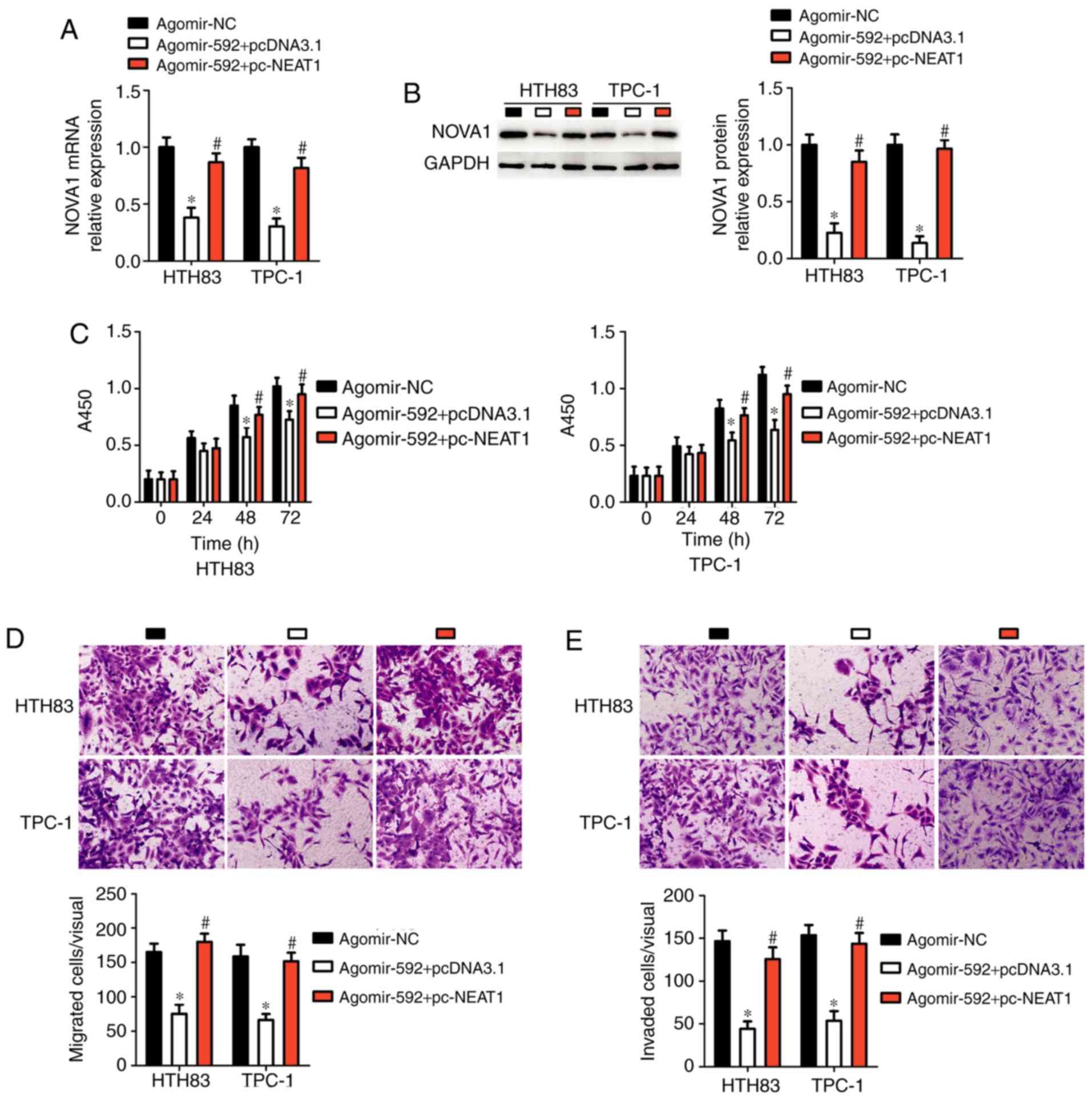
Ectopic NEAT1 expression reverses the
miR-592-mediated inhibitory effects in thyroid cancer cells
Since we proposed that NEAT1 acts as a ceRNA
for miR-592 in thyroid cancer cells, we then determined whether the
effects of miR-592 on thyroid cancer cell proliferation, migration,
and invasion were modulated by NEAT1. Thus, pc-NEAT1 or
pcDNA3.1 was co-transfected with agomir-592 into HTH83 and TPC-1
cells. The mRNA (P<0.05; Fig.
6A) and protein (P<0.05; Fig.
6B) levels of NOVA1 in HTH83 and TPC-1 cells, which were
decreased by agomir-592 transfection, were restored by
co-transfection with the NEAT1 overexpression plasmid
pc-NEAT1. Functionally, the miR-592-mediated decreases in HTH83 and
TPC-1 cell proliferation (P<0.05; Fig. 6C), migration (P<0.05; Fig. 6D), and invasion (P<0.05;
Fig. 6E) were substantially
reversed after co-transfection with pc-NEAT1. Thus, these results
indicated that miR-592 acts as the downstream RNA of NEAT1
in thyroid cancer cells.
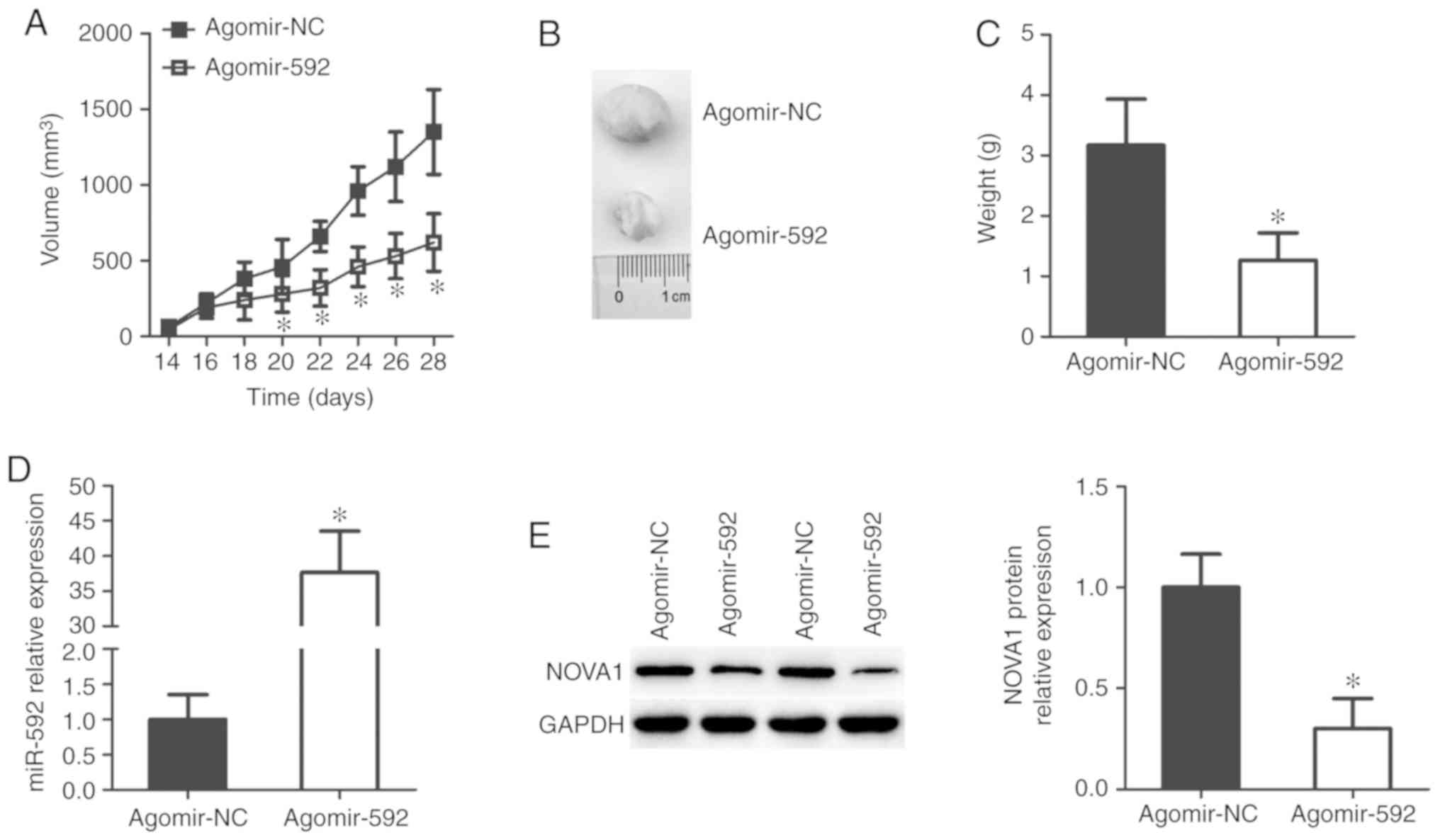
miR-592 impairs tumor growth in vivo
Finally, a xenograft tumor model was established to
assess the effect of miR-592 on tumor growth in vivo. The
tumor xenografts derived from agomir-592-transfected TPC-1 cells
exhibited significantly slower growth patterns than those obtained
from the agomir-NC-transfected TPC-1 cells (P<0.05; Fig. 7A and B). At the end point of the
experiment, the tumor xenografts were resected and weighed.
Xenograft tumors in the agomir-592 group weighed significantly less
than tumors in the agomir-NC group (P<0.05; Fig. 7C). Furthermore, the expression of
miR-592 was significantly upregulated (P<0.05; Fig. 7D), whereas NOVA1 protein
expression was decreased (P<0.05; Fig. 7E), in the tumor xenografts derived
from agomir-592-transfected TPC-1 cells, compared with the control.
These results suggest that miR-592 overexpression impairs the tumor
growth of thyroid cancer cells in vivo.
Discussion
Numerous studies have demonstrated that various
miRNAs are aberrantly expressed in thyroid cancer and play
indispensable roles in the malignancy of thyroid cancer (38-40). Almost all aggressive
characteristics of thyroid cancer have been demonstrated to be
modulated by miRNAs via regulating the expression of their target
genes (41). In the past few
decades, multiple miRNAs have been extensively studied in thyroid
cancer (13,14,42); however, many miRNAs remain to be
investigated. Further exploration of the specific functions of
miRNAs in the development of thyroid cancer may aid in identifying
potential targets for anticancer therapy. In this study, for the
first time, we determined the expression profile of miR-592 in
thyroid cancer and examined the prognostic significance of miR-592
in patients with thyroid cancer. Notably, the biological roles of
miR-592 with regards to the malignant phenotypes of thyroid cancer
and the related molecular mechanisms were explored in detail.
miR-592 is downregulated in non-small cell lung
cancer, and its downregulation is significantly associated with TNM
stage and lymph node metastasis (23). The expression of miR-592 is
decreased in breast cancer and is associated with malignant
clinicopathological characteristics (24). miR-592 expression was also
reported to be reduced in glioma (25,26) and hepatocellular carcinoma
(27-29). On the contrary, miR-592 is
upregulated in prostate (30) and
colorectal (31,32) cancers. These inconsistent findings
led us to explore the expression profile of miR-592 in thyroid
cancer. In the present study, we found that miR-592 was
downregulated in thyroid cancer tissues and cell lines. Low miR-592
expression was associated with lymph node metastasis and TNM stage.
Patients with thyroid cancer samples exhibiting low miR-592
expression exhibited poorer overall survival than patients with
high miR-592 expression. These findings suggest that miR-592 could
be developed as a novel prognostic marker for patients with thyroid
cancer.
miR-592 exerts an inhibitory role in breast cancer
through regulating cell growth and metastasis in vitro, as
well as tumor growth in vivo (24). miR-592 was also identified as a
tumor-suppressing miRNA in non-small cell lung cancer (23), glioma (25,26), and hepatocellular carcinoma
(27-29), serving dispensable roles in
regulating a wide range of biological behaviors. On the contrary,
miR-592 was found to play tumor-promoting roles in prostate
(30) and colorectal (31,32) cancers. These observations indicate
that the functional roles of miR-592 in cancer genesis and
development exhibit tissue specificity. However, the detailed roles
of miR-592 in regulating the malignant development of thyroid
cancer had not been studied yet. In the present study, ectopic
miR-592 expression suppressed the proliferation, migration, and
invasion of thyroid cancer cells in vitro. In addition,
miR-592 overexpression suppressed the tumor growth of thyroid
cancer cells in vivo. Accordingly, miR-592 might be an
effective target for treating patients with thyroid cancer.
Understanding the molecular mechanisms underlying
the tumor-suppressing roles of miR-592 in thyroid cancer is
important for identifying effective therapeutic techniques. In the
present study, NOVA1 was validated as a direct target of
miR-592 in thyroid cancer cells, and NEAT1 was proposed to
function as a ceRNA to modulate NOVA1 expression by sponging
miR-592. Multiple studies reported that NEAT1 is expressed
at high levels and exerts oncogenic roles in a variety of human
cancers, including cervical cancer (43), nasopharyngeal carcinoma (44), breast cancer (45), non-small cell lung cancer
(46), colorectal cancer
(47,48), and oral squamous cell carcinoma
(49). NEAT1 is also
upregulated in thyroid cancer (35-37), and upregulation of NEAT1 is
positively correlated with TNM stage and tumor size (35). NEAT1 is closely involved in
the aggressiveness of thyroid cancer by promoting cell
proliferation, migration, invasion, and radioactive iodine
resistance in vitro; inhibiting cell survival and apoptosis
in vitro; and impairing tumor growth in vivo
(20,35-37). NOVA1, a member of the NOVA
family of neuron-specific RNA-binding proteins, is aberrantly
downregulated in multiple human cancers and affects a variety of
pathophysiological processes associated with carcinogenesis and
cancer progression (50-52). The expression and roles of NEAT1
and roles have been studied in other human cancers (43-49). Herein, we identified a novel
NEAT1-miR-592-NOVA1 pathway, and this pathway could serve crucial
roles in the aggressiveness of thyroid cancer. Our results suggest
that silencing NEAT1 expression and restoring miR-592
expression, thereby decreasing NOVA1 expression, represents
a promising therapeutic approach for preventing thyroid cancer.
This study could not use the TCGA database to
validate the relevance of the NEAT1/miR-592/NOVA1 pathway in
thyroid cancer, which poses as a limitation of our study, and we
aim to resolve this issue in our future studies.
In conclusion, miR-592 was downregulated in thyroid
cancer tissues and cell lines. Low miR-592 expression was
determined to be associated with the poor prognosis of patients
with thyroid cancer. Silencing NEAT1 expression was proposed
to decrease the expression of NOVA1 by sponging miR-592,
thereby suppressing the progression of thyroid cancer.
Acknowledgments
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
This study was designed by LW and YL. YL performed
RT-qPCR and the luciferase reporter assays. Migration and invasion
assays, and the xenograft tumor model were conducted by TH, JZ, and
MZ. PS carried out western blot analysis and the CCK-8 assay. LW
and YL wrote the manuscript. All authors read and approved the
final manuscript.
Ethics approval and consent to
participate
The Ethics Committee of Jilin Central General
Hospital approved the use of human tissue samples. Written informed
consent was obtained from all patients before they underwent
surgical resection.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2015. CA Cancer J Clin. 65:5–29. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ferlay J, Soerjomataram I, Dikshit R, Eser
S, Mathers C, Rebelo M, Parkin DM, Forman D and Bray F: Cancer
incidence and mortality worldwide: Sources, methods and major
patterns in GLOBOCAN 2012. Int J Cancer. 136:E359–E386. 2015.
View Article : Google Scholar
|
|
3
|
La Vecchia C, Malvezzi M, Bosetti C,
Garavello W, Bertuccio P, Levi F and Negri E: Thyroid cancer
mortality and incidence: A global overview. Int J Cancer.
136:2187–2195. 2015. View Article : Google Scholar
|
|
4
|
Xing M: Molecular pathogenesis and
mechanisms of thyroid cancer. Nat Rev Cancer. 13:184–199. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Fröhlich E and Wahl R: The current role of
targeted therapies to induce radioiodine uptake in thyroid cancer.
Cancer Treat Rev. 40:665–674. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Shi X, Liu R, Basolo F, Giannini R, Shen
X, Teng D, Guan H, Shan Z, Teng W, Musholt TJ, et al: Differential
clinicopathological risk and prognosis of major papillary thyroid
cancer variants. J Clin Endocrinol Metab. 101:264–274. 2016.
View Article : Google Scholar :
|
|
7
|
Kim WW, Ha TK and Bae SK: Clinical
implications of the BRAF mutation in papillary thyroid carcinoma
and chronic lymphocytic thyroiditis. J Otolaryngol Head Neck Surg.
47:42018. View Article : Google Scholar
|
|
8
|
Kim VN, Han J and Siomi MC: Biogenesis of
small RNAs in animals. Nat Rev Mol Cell Biol. 10:126–139. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Huang W: MicroRNAs: Biomarkers,
diagnostics, and therapeutics. Methods Mol Biol. 1617:57–67. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Calin GA, Sevignani C, Dumitru CD, Hyslop
T, Noch E, Yendamuri S, Shimizu M, Rattan S, Bullrich F, Negrini M
and Croce CM: Human microRNA genes are frequently located at
fragile sites and genomic regions involved in cancers. Proc Natl
Acad Sci USA. 101:2999–3004. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Gao X, Chen Z, Li A, Zhang X and Cai X:
miR-129 regulates growth and invasion by targeting MAL2 in
papillary thyroid carcinoma. Biomed Pharmacother. 105:1072–1078.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Xu CB, Liu XS, Li JQ, Zhao X, Xin D and Yu
D: MicroRNA-539 functions as a tumor suppressor in papillary
thyroid carcinoma via the transforming growth factor β1/Smads
signaling pathway by targeting secretory leukocyte protease
inhibitor. J Cell Biochem. 120:10830–10846. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Gao XB, Chen CL, Tian ZL, Yuan FK and Jia
GL: MicroRNA-791 is an independent prognostic factor of papillary
thyroid carcinoma and inhibits the proliferation of PTC cells. Eur
Rev Med Pharmacol Sci. 22:5562–5568. 2018.PubMed/NCBI
|
|
14
|
Yi T, Zhou X, Sang K, Zhou J and Ge L:
MicroRNA-1270 modulates papillary thyroid cancer cell development
by regulating SCAI. Biomed Pharmacother. 109:2357–2364. 2019.
View Article : Google Scholar
|
|
15
|
Diao Y, Fu H and Wang Q: miR-221
exacerbate cell proliferation and invasion by targeting TIMP3 in
papillary thyroid carcinoma. Am J Ther. 24:e317–e328. 2017.
View Article : Google Scholar :
|
|
16
|
Huang Y, Yu S, Cao S, Yin Y, Hong S, Guan
H, Li Y and Xiao H: MicroRNA-222 promotes invasion and metastasis
of papillary thyroid cancer through targeting protein phosphatase 2
regulatory subunit B alpha expression. Thyroid. 28:1162–1173. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Fang L, Kong D and Xu W: MicroRNA-625-3p
promotes the proliferation, migration and invasion of thyroid
cancer cells by up-regulating astrocyte elevated gene 1. Biomed
Pharmacother. 102:203–211. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Ponting CP, Oliver PL and Reik W:
Evolution and functions of long noncoding RNAs. Cell. 136:629–641.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Huang F, Zhang Q, Chen W, Zhang H, Lu G,
Chen J and Qiu C: Long noncoding RNA cancer susceptibility
candidate 2 suppresses papillary thyroid carcinoma growth by
inactivating the AKT/ERK1/2 signaling pathway. J Cell Biochem.
120:10380–10390. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Liu C, Feng Z, Chen T, Lv J, Liu P, Jia L,
Zhu J, Chen F, Yang C and Deng Z: Downregulation of NEAT1 reverses
the radioactive iodine resistance of papillary thyroid carcinoma
cell via miR-101-3p/FN1/PI3K-AKT signaling pathway. Cell Cycle.
18:167–203. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Li X, Zhong W, Xu Y, Yu B and Liu H:
Silencing of lncRNA LINC00514 inhibits the malignant behaviors of
papillary thyroid cancer through miR-204-3p/CDC23 axis. Biochem
Biophys Res Commun. 508:1145–1148. 2019. View Article : Google Scholar
|
|
22
|
Chan JJ and Tay Y: Noncoding RNA:RNA
regulatory networks in cancer. Int J Mol Sci. 19:2018. View Article : Google Scholar
|
|
23
|
Li Z, Li B, Niu L and Ge L: miR-592
functions as a tumor suppressor in human non-small cell lung cancer
by targeting SOX9. Oncol Rep. 37:297–304. 2017. View Article : Google Scholar
|
|
24
|
Hou W, Zhang H, Bai X, Liu X, Yu Y, Song L
and Du Y: Suppressive role of miR-592 in breast cancer by
repressing TGF-β2. Oncol Rep. 38:3447–3454. 2017.PubMed/NCBI
|
|
25
|
Gao S, Chen J, Wang Y, Zhong Y, Dai Q,
Wang Q and Tu J: miR-592 suppresses the development of glioma by
regulating Rho-associated protein kinase. Neuroreport.
29:1391–1399. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Peng T, Zhou L, Qi H, Wang G, Luan Y and
Zuo L: miR-592 functions as a tumor suppressor in glioma by
targeting IGFBP2. Tumour Biol. 39:10104283177192732017. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Wang W, Zhang H, Tang M, Liu L, Zhou Z,
Zhang S and Wang L: MicroRNA-592 targets IGF-1R to suppress
cellular proliferation, migration and invasion in hepatocellular
carcinoma. Oncol Lett. 13:3522–3528. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Jia YY, Zhao JY, Li BL, Gao K, Song Y, Liu
MY, Yang XJ, Xue Y, Wen AD and Shi L: miR-592/WSB1/HIF-1α axis
inhibits glycolytic metabolism to decrease hepatocellular carcinoma
growth. Oncotarget. 7:35257–35269. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Li X, Zhang W, Zhou L, Yue D and Su X:
MicroRNA-592 targets DEK oncogene and suppresses cell growth in the
hepatocellular carcinoma cell line HepG2. Int J Clin Exp Pathol.
8:12455–12463. 2015.
|
|
30
|
Lv Z, Rao P and Li W: miR-592 represses
FOXO3 expression and promotes the proliferation of prostate cancer
cells. Int J Clin Exp Med. 8:15246–15253. 2015.PubMed/NCBI
|
|
31
|
Liu M, Zhi Q, Wang W, Zhang Q, Fang T and
Ma Q: Up-regulation of miR-592 correlates with tumor progression
and poor prognosis in patients with colorectal cancer. Biomed
Pharmacother. 69:214–220. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Fu Q, Du Y, Yang C, Zhang D, Zhang N, Liu
X, Cho WC and Yang Y: An oncogenic role of miR-592 in tumorigenesis
of human colorectal cancer by targeting Forkhead Box O3A (FoxO3A).
Expert Opin Ther Targets. 20:771–782. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Schmid KW, Synoracki S, Dralle H and
Wittekind C: Proposal for an extended pTNM classification of
thyroid carcinoma: Commentary on deficits of the 8th edition of the
TNM classification. Pathologe. 40:18–24. 2019. View Article : Google Scholar
|
|
34
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
35
|
Sun W, Lan X, Zhang H, Wang Z, Dong W, He
L, Zhang T, Zhang P, Liu J and Qin Y: NEAT1_2 functions as a
competing endogenous RNA to regulate ATAD2 expression by sponging
microRNA-106b-5p in papillary thyroid cancer. Cell Death Dis.
9:3802018. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Li JH, Zhang SQ, Qiu XG, Zhang SJ, Zheng
SH and Zhang DH: Long non-coding RNA NEAT1 promotes malignant
progression of thyroid carcinoma by regulating miRNA-214. Int J
Oncol. 50:708–716. 2017. View Article : Google Scholar
|
|
37
|
Zhang H, Cai Y, Zheng L, Zhang Z, Lin X
and Jiang N: Long noncoding RNA NEAT1 regulate papillary thyroid
cancer progression by modulating miR-129-5p/KLK7 expression. J Cell
Physiol. 233:6638–6648. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Liu Y, Li J, Li M, Li F, Shao Y and Wu L:
MicroRNA-510-5p promotes thyroid cancer cell proliferation,
migration, and invasion through suppressing SNHG15. J Cell Biochem.
2019.
|
|
39
|
Chen J, Yin J, Liu J, Zhu RX, Zheng Y and
Wang XL: miR-202-3p-functions as a tumor suppressor and reduces
cell migration and invasion in papillary thyroid carcinoma. Eur Rev
Med Pharmacol Sci. 23:1145–1150. 2019.PubMed/NCBI
|
|
40
|
Ma Y and Sun Y: miR-29a-3p inhibits
growth, proliferation, and invasion of papillary thyroid carcinoma
by suppressing NF-κB signaling via direct targeting of OTUB2.
Cancer Manag Res. 11:13–23. 2018. View Article : Google Scholar :
|
|
41
|
Zembska A, Jawiarczyk-Przybyłowska A,
Wojtczak B and Bolanowski M: MicroRNA expression in the progression
and aggressiveness of papillary thyroid carcinoma. Anticancer Res.
39:33–40. 2019. View Article : Google Scholar
|
|
42
|
Jia M, Shi Y, Li Z, Lu X and Wang J:
MicroRNA-146b-5p as an oncomiR promotes papillary thyroid carcinoma
development by targeting CCDC6. Cancer Lett. 443:145–156. 2019.
View Article : Google Scholar
|
|
43
|
Yuan LY, Zhou M, Lv H, Qin X, Zhou J, Mao
X, Li X, Xu Y, Liu Y and Xing H: Involvement of NEAT1/miR-133a axis
in promoting cervical cancer progression via targeting SOX4. J Cell
Physiol. 2019. View Article : Google Scholar
|
|
44
|
Ji Y, Wang M, Li X and Cui F: The long
noncoding RNA NEAT1 targets miR-34a-5p and drives nasopharyngeal
carcinoma progression via Wnt/β-catenin signaling. Yonsei Med J.
60:336–345. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Shin VY, Chen J, Cheuk IW, Siu MT, Ho CW,
Wang X, Jin H and Kwong A: Long non-coding RNA NEAT1 confers
oncogenic role in triple-negative breast cancer through modulating
chemo-resistance and cancer stemness. Cell Death Dis. 10:2702019.
View Article : Google Scholar
|
|
46
|
Li B, Gu W and Zhu X: NEAT1 mediates
paclitaxel-resistance of non-small cell of lung cancer through
activation of Akt/mTOR signalling pathway. J Drug Target. 1–7.
2019.
|
|
47
|
Zhu Z, Du S, Yin K, Ai S, Yu M, Liu Y,
Shen Y, Liu M, Jiao R, Chen X and Guan W: Knockdown long noncoding
RNA nuclear paraspeckle assembly transcript 1 suppresses colorectal
cancer through modulating miR-193a-3p/KRAS. Cancer Med. 8:261–275.
2019. View Article : Google Scholar
|
|
48
|
Yu HM, Wang C, Yuan Z, Chen GL, Ye T and
Yang BW: LncRNA NEAT1 promotes the tumorigenesis of colorectal
cancer by sponging miR-193a-3p. Cell Prolif. 52:e125262019.
View Article : Google Scholar
|
|
49
|
Liu X, Shang W and Zheng F: Long
non-coding RNA NEAT1 promotes migration and invasion of oral
squamous cell carcinoma cells by sponging microRNA-365. Exp Ther
Med. 16:2243–2250. 2018.PubMed/NCBI
|
|
50
|
Jin L, Li H, Wang J, Lin D, Yin K, Lin L,
Lin Z, Lin G, Wang H, Ying X, et al: MicroRNA-193a-5p exerts a
tumor suppressor role in glioblastoma via modulating NOVA1. J Cell
Biochem. 120:6188–6197. 2019. View Article : Google Scholar
|
|
51
|
Yu X, Zheng H, Chan MTV and Wu WKK: NOVA1
acts as an oncogene in melanoma via regulating FOXO3a expression. J
Cell Mol Med. 22:2622–2630. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Shen B, Zhang Y, Yu S, Yuan Y, Zhong Y, Lu
J and Feng J: MicroRNA-339, an epigenetic modulating target is
involved in human gastric carcinogenesis through targeting NOVA1.
FEBS Lett. 589:3205–3211. 2015. View Article : Google Scholar : PubMed/NCBI
|