Introduction
Patients with type 2 diabetes mellitus have a high
risk of developing atherosclerosis (1), and a higher likelihood of restenosis
after percutaneous coronary intervention (2). However, injecting insulin into a
patient with diabetes does not affect restenosis (3). High glucose causes vascular smooth
muscle cell (VSMC) alteration (4-6),
which contributes to diabetic macrovasculopathy.
Atherosclerosis is a chronic disease and could lead
to sudden death (7). As clinical
consequences of atherosclerosis, myocardial infarction or stroke is
not a consequence of gradual narrowing in the lumen but the
thrombotic disease is related to acute rupture or erosion of an
unstable plaque (8). A previous
clinical imaging study identified that plaque instability led to
ruptures, such as a thin fibrous cap-formed by extracellular matrix
molecules from VSMCs, releasing lipids that accumulated
extracellularly to form the necrotic core of the plaque (9). In addition, VSMCs in advanced
lesions are generally regarded as having athero-protective
properties, and a previous study demonstrated that VSMC
proliferation even helped relieve atherogenesis (10). However, a previous study
demonstrated that formation of atherosclerotic plaques can be
inhibited by suppressing VSMC migration and proliferation (11). Hence, VSMC proliferation,
migration and invasion are related to atherosclerosis; however, the
specific mechanism of action still requires further
elucidation.
MicroRNA (miRNA/miR), which consist of 17-25
nucleotides, can lead to target mRNA degradation or translational
suppression by interacting with the 3′ untranslated region (3′UTR)
of its target gene (12). Hence,
miRNAs have been characterized throughout the creature genome
(http://www.mirbase.org/), with potentially more
to be found. Each miRNA could repress the translation of hundreds
of mRNA (13,14), producing complex changes in the
protein expression field. A previous study has shown that miRNA
expression took part in regulation networks in cell processes,
including cell proliferation, apoptosis, invasion and migration
(15), and miRNA levels were also
correlated with patient survial and prognosis in cancer (16,17). miRNAs have been increasingly
reported to be involved in dysfunction of VSMCs, including VSMC
calcification, proliferation and migration (18-20). Therefore, studying the association
between miRNAs and VSMCs may comfirm whether miRNAs are biomarkers
for VSMCs. miR-19a, which belongs to the miR-17-92 cluster, is an
miRNA with pleiotropic functions in cancer cell survival, apoptosis
and migration (21-23). However, whether miR-19a plays an
important role in VSMC proliferation, migration and invasion
remains unknown.
As a small GTPase, ras homolog family member B
(RHOB) is the only member of the Rho family and can be modified by
palmitoylation (24). RHOB has
different functions, which are realized depending on its exact
locations, for example, RHOB protects keratinocytes from UVB injury
and RHOB determines tumor aggressiveness (25-27). RHOB is regarded as a tumor
suppressor protein (28), and
more importantly, a target of miRNAs in cancer (29,30). In previous years, studies have
increasingly suggested that activation of RHOB plays a critical
role in the migration of glucose-stimulated VSMCs (31-33). Therefore, the present study
investigated the effect of miR-19a on VSMC proliferation, migration
and invasion, and whether RHOB is a regulatory factor of miR-19a in
affecting the expression of other genes or proteins.
Materials and methods
Cell culture and transfection
A-10 cells are derived from the thoracic aorta of an
embryonic rat and possess many properties of smooth muscle cells.
The A-10 cell line was purchased from The American Type Culture
Collection. The cells were cultured in complete growth medium with
high-glucose DMEM (Gibco; Thermo Fisher Scientific, Inc.), 10% FBS
(Gibco; Thermo Fisher Scientific, Inc.), 1% 5,000 units/ml
penicillin and 5,000 µg/ml streptomycin (Gibco; Thermo
Fisher Scientific, Inc.). miR-19a mimics (5′-UGU GCA AAU CCA UGC
AAA ACU GA-3′), miR-19a inhibitor (5′-ACA CGU UUA GGU ACG UUU UGA
CU-3′) and small interfering (si)RHOB (sense, 5′-CCG UCU UCG AGA
ACU AUG U-3′; antisense, 5′-ACA UAG UUC UCG AAG ACG G-3′) were
obtained from OBiO Technology (Shanghai) Corp., Ltd.
Lipofectamine® (Gibco; Thermo Fisher Scientific, Inc.),
50 nM miR-19a mimics, 100 nM miR-19a inhibitor and 25 mM siRNA were
diluted with FBS-free high-glucose DMEM. Lipofectamine®
solution mixed with miR-19a mimics solution or miR-19a inhibitor
solution or siRNA was added to the cells and incubated for 3-4 h,
and the mixed solution was then replaced by complete growth medium
and cultured for 24 or 48 h.
Cell proliferation
Cell Counting Kit-8 (CCK-8) was used to measure the
cell proliferation. The cells were seeded in a 96-well plate
(Corning, Inc.) at a density of 5×103 cells/well and
treated with mimics, inhibitor or siRNA for 24 or 48 h. CCK-8
(Sigma-Aldrich; Merck KGaA) solution was diluted with FBS-free
high-glucose DMEM at a ratio of 1:9. CCK-8 solution was added the
cells for 2 h after the culture medium had been removed. The
96-well plate was put into a microplate reader (Thermo Fisher
Scientific, Inc.), which was used to detect the optical density
value at a wavelength of 490 nm.
Cell cycle
The cells were resuspended and collected by trypsin
(Gibco; Thermo Fisher Scientific, Inc.) using cold PBS (Gibco;
Thermo Fisher Scientific, Inc.) and added to ethyl alcohol
(Sigma-Aldrich; Merck KGaA) at 4°C for 12 h. Next, the cells were
centrifuged (Cence Company) at 1,000 × g for 5 min at 4°C, PBS was
added and the cells were centrifuged again at 1,000 × g for 5 min
at 4°C. Propidium iodide (PI; Sigma-Aldrich; Merck KGaA) solution
was added to the cells, and the cell cycle was determined using a
BD FACS Calibur flow cytometer (BD Biosciences), and data were
analyzed using BD CellQuestTM Pro Software version 5.1
(BD Biosciences). In total, 50 µg/ml PI was diluted in 0.25
mg/ml RNaseA (Sigma-Aldrich; Merck KGaA) and mixed with PBS.
Cell scratch
Cell scratching is used to assess cell migration.
When the serum-starved cells filled the plate (Corning, Inc.), a
200-µl pipette tip (Sigma-Aldrich; Merck KGaA) was used to
scratch the cells, which were then washed with PBS twice, and the
location and images were recorded. The images were taken using a
light microscope (Olympus Corporation; magnification, ×200). The
wound width was calculated using the following formula: Wound
width=(width of 24 h or 0 h)/(width of 0 h in the Control group)
×100.
Transwell Matrigel assay
A Transwell Matrigel assay is performed to assess
cell invasive ability. Matrigel (Sigma-Aldrich; Merck KGaA) was
diluted with FBS-free high-glucose DMEM (Gibco; Thermo Fisher
Scientific, Inc.) at a ratio of 1:8. The Matrigel solution was
placed in the top of the Transwell plate (Corning, Inc.) in an
incubator (Thermo Fisher Scientific, Inc.) at 37°C and incubated
for 6 h. The top of the Transwell plate was then washed with
FBS-free high-glucose DMEM three times. The cells were resuspended
by centrifugation (Cence Company; 1,000 × g) at 4°C for 10 min with
FBS-free high-glucose DMEM after the treatment with mimics,
inhibitor or siRNA. Next, the cells (1×105) were added
to the upper chamber of the Transwell plate, while high-glucose
DMEM with 20% FBS was added to the lower chamber. The cells in the
Transwell plate were incubated in an incubator at 37°C for 24 h.
The cells in the upper chamber of the Transwell plate were removed,
fixed with 10% methanol (Thermo Fisher Scientific, Inc.) for 30 min
at 4°C, whereas those in the lower chamber of the Transwell plate
were stained using crystal violet (Beijing Solarbio Science &
Technology Co., Ltd.) for 30 min at 37°C. The images of the invaded
cells were taken using a light microscope (Olympus Corporation;
magnification, ×200). The invasion rate was calculated using the
following formula: Invasion rate, %=(the amount of invasive
cells)/(the amount of invasive cells in the Control group)
×100.
Dual luciferase assay
TargetScan 7.2 (www.targetscan.org) was used to identify three
potential targets of miR-19a. The wild-type RHOB 3′UTR or mutant
RHOB 3′UTR were cloned into psi-CHECK-2 (Promega Corporation). The
cells were transfected by carrier material with the target gene
using Lipofectamine® (Gibco; Thermo Fisher Scientific,
Inc.) for 6 h. The fluorescence was detected using a Dual
Luciferase Reporter Gene Detection kit (Beijing Solarbio Science
& Technology Co., Ltd.) 24 h after transfection, according to
the manufacturer's protocol. Firefly luciferase activity was
normalized to Renilla luciferase activity.
Western blotting
Cell lysis buffer (Thermo Fisher Scientific, Inc.)
was used to extract total protein, the concentration of which was
detected using a bicinchoninic assay kit (Thermo Fisher Scientific,
Inc.) according to the manufacturer's protocol. An equal amount of
protein (20 µg) was loaded and added to an 12% SDS-PAGE
machine (Bio-Rad Laboratories, Inc.) to be separated. Next, the
protein was transferred to PVDF membranes (Sigma-Aldrich; Merck
KGaA) by cataphoresis. 5% bovine serum albumin (Sigma-Aldrich;
Merck KGaA) was used to block the blank site of the membranes at
37°C for 30 min, which were subsequently incubated with antibodies
against dual specificity phosphatase Cdc25A (CDC25A; cat. no.
ab2357; Abcam; 1:1,000), cyclinD1 (cat. no. ab134175; Abcam;
1:1,000), matrix metal-loproteinase (MMP)-2 (cat. no. ab37150;
Abcam; 1:1,000), MMP-9 (cat. no. ab73734; Abcam; 1:1,000), α-smooth
muscle actin (α-SMA; cat. no. ab5694; Abcam; 1:1,000), smooth
muscle 22α (SM22α; cat. no. ab14106; Abcam; 1:1,000), RHOB (cat.
no. ab155149; Abcam; 1:1,000) and GAPDH (cat. no. ab8245; Abcam;
1:5,000) overnight at 4°C. TBS with Tween-20 (Beijing Solarbio
Science & Technology Co., Ltd.) was used to wash the membranes
three times. The membranes were incubated with a secondary antibody
(cat. no. ab7090; Abcam; 1:5,000) at room temperature for 2-3 h and
stained using an ECL kit (Sigma-Aldrich; Merck KGaA). The western
blots were analyzed using Bio-Rad ChemiDoc™ XRS+ System with Image
Lab™ Software version 4.1 (Bio-Rad Laboratories, Inc.).
Reverse transcription-quantitative PCR
(RT-qPCR)
Total RNA was extracted with TRIzol®
(Sigma-Aldrich; Merck KGaA) by centrifuging the cells at 6,500 × g
for 10 min at 4°C. The Sensiscript RT kit (Promega Corporation) was
used to synthesize cDNA at 45°C for 15 min and at 95°C for 3 min.
The amplification reactants were cDNA, ddH2O, forward
primer, reverse primer and Fast SYBR-Green Master mix (Thermo
Fisher Scientific, Inc.).
The primer sequences used were as follows: miR-19a
(forward: 5′-GTT TTG CAT AGT TGC ACT A-3′; reverse: 5′-GAA CAT GTC
TGC GTA TCT C-3′) MMP-2 primer (forward: 5′-TTC CCC CGC AAG CCC AAG
TG-3′; reverse: 5′-GAG AAA AGC GCA GCG GAG TGA CG-3′); MMP-9 primer
(forward: 5′-CAC CAC CAC AAC TGA ACC-3′; reverse: 5′-GCC TAG ACC
CAA CTT ATC C-3′); α-SMA primer (forward: 5′-AGC CAG TCG CCA TCG
GAA C-3′; reverse: 5′-CCG GAG CCA TTG TCA CAC AC-3′); SM22α primer
(forward: 5′-TTC TGC CTC AAC ATG GCC AAC-3′; reverse: 5′-CAC CTT
CAC TGG CTT GGA TC-3′); RHOB primer (forward: 5′-TGC TGATCG TGT TCA
GTA AG-3′; reverse: 5′-AGC ACA TGA GAA TGA CGT CG-3′); suppressor
of cytokine signaling 3 (SOCS3) primer (forward: 5′-CCC AAG CTT ATG
GTC ACC CAC AGC-3′; reverse: 5′-CGC GGA TCC TAC TGG TCC AGG-3′);
T-cell intracellular antigen-1 (TIA1) primer (forward: 5′-CAG ATG
GGT GGC CAG TGG CT-3′; reverse: 5′-TGA CCT TCA ATG GTA GTA CCA-3′);
cyclinD1 primer (forward: 5′-GTA GCA GCG AGC AGC AGA GT-3′;
reverse: 5′-CTC CTC GCA CTT CTG TTC CTC-3′); CDC25A primer
(forward: 5′-CCA AAG GAA CCA TTG AGA AC-3′; reverse: 5′-CAG ATG CCA
TAA TTT CTG GAG-3′); and U6 primer as the loading control (forward:
5′-TGA GAA CTG AAT TCC ATG GGT T-3′; reverse: 5′-ACG CTT CAC GAA
TTT GCG T-3′). The conditions of the PCR amplification were 95°C
for 30 sec, followed by 40 cycles of 95°C for 30 sec and 75°C for
15 sec. The relative level of mRNA was determined using the
2−ΔΔCq method (34).
Statistical analysis
Each experiment was repeated three time. Data are
presented as the mean ± SD and were analyzed by SPSS 16.0 software
(SPSS, Inc.) using ANOVA with Turkey's multiple comparisons test.
P<0.05 was considered to indicate a statistically significant
difference.
Results
miR-19a promotes VSMC proliferation and
cell cycle
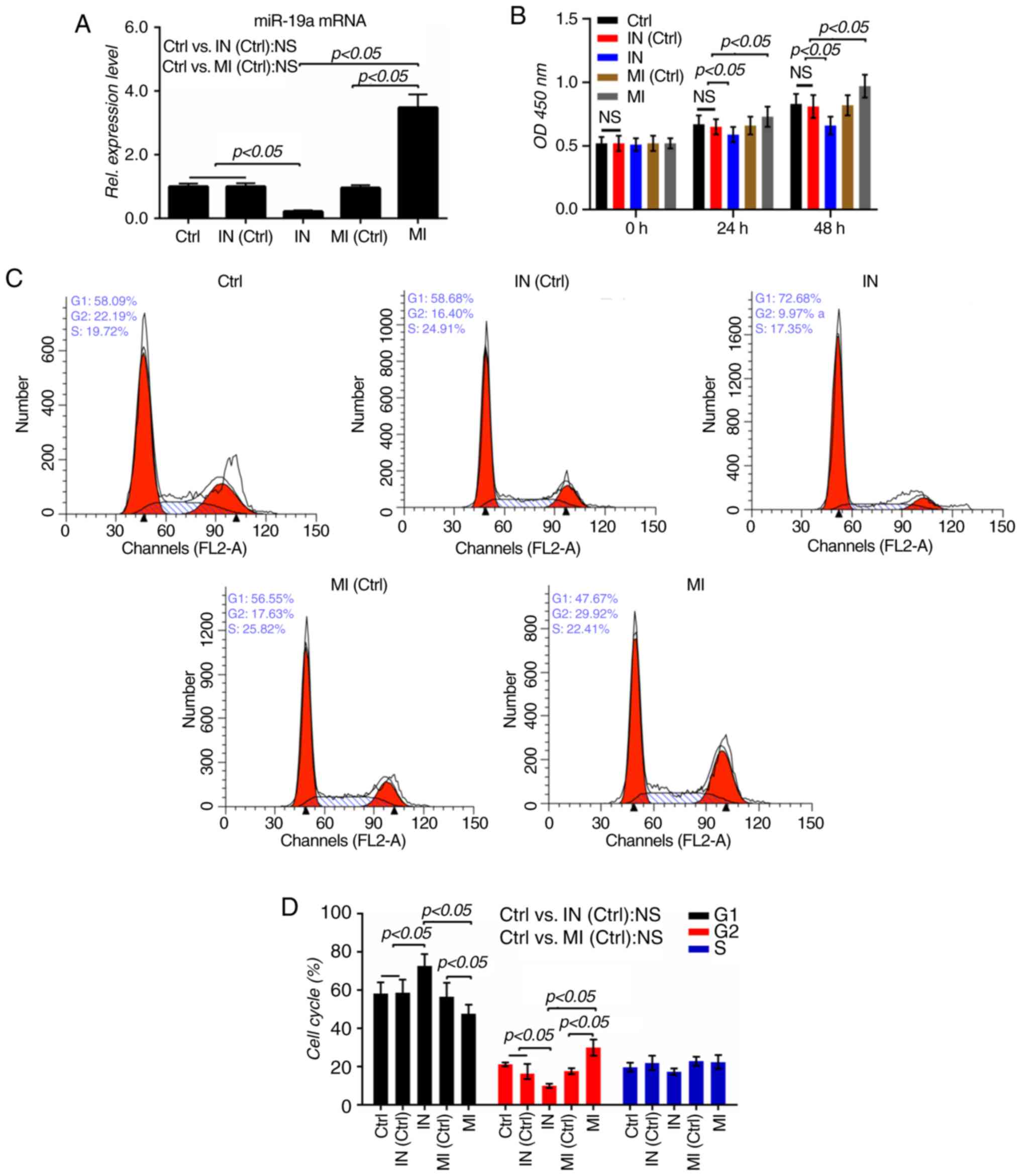
To validate the roles of miR-19a in VSMCs, miR-19a
mimics and inhibitor were transfected into VSMCs to increase or
decrease miR-19a levels, respectively (P<0.05; Fig. 1A). The effects of miR-19a on the
proliferation of VSMCs were subsequently determined. It was
observed that elevated miR-19a significantly increased the
proliferation of VSMCs, while downregulated miR-19a noticeably
repressed the proliferation (P<0.05; Fig. 1B). The cell cycle of the VSMCs was
analyzed by flow cytometry, as shown in Fig. 1C and D, the proportion of cells in
the G1 phase was notably reduced from 56.55% in the
mimic control group to 47.67% in the mimic group, while the number
of cells in the G2 phase increased from 17.63% in the
mimic control group to 29.92% in the mimic group (P<0.05). By
contrast, the numbers of cells in the G1 phase was
notably increased from 58.68% in the inhibitor control group to
72.68% in the inhibitor group, while those in the G2
phase decreased from 16.4% in the inhibitor control group to 9.97%
in the inhibitor group (P<0.05). The present results suggested
that overexpression of miR-19a could promote the cell cycle, while
inhibition of miR-19a could induce G1 cell cycle arrest.
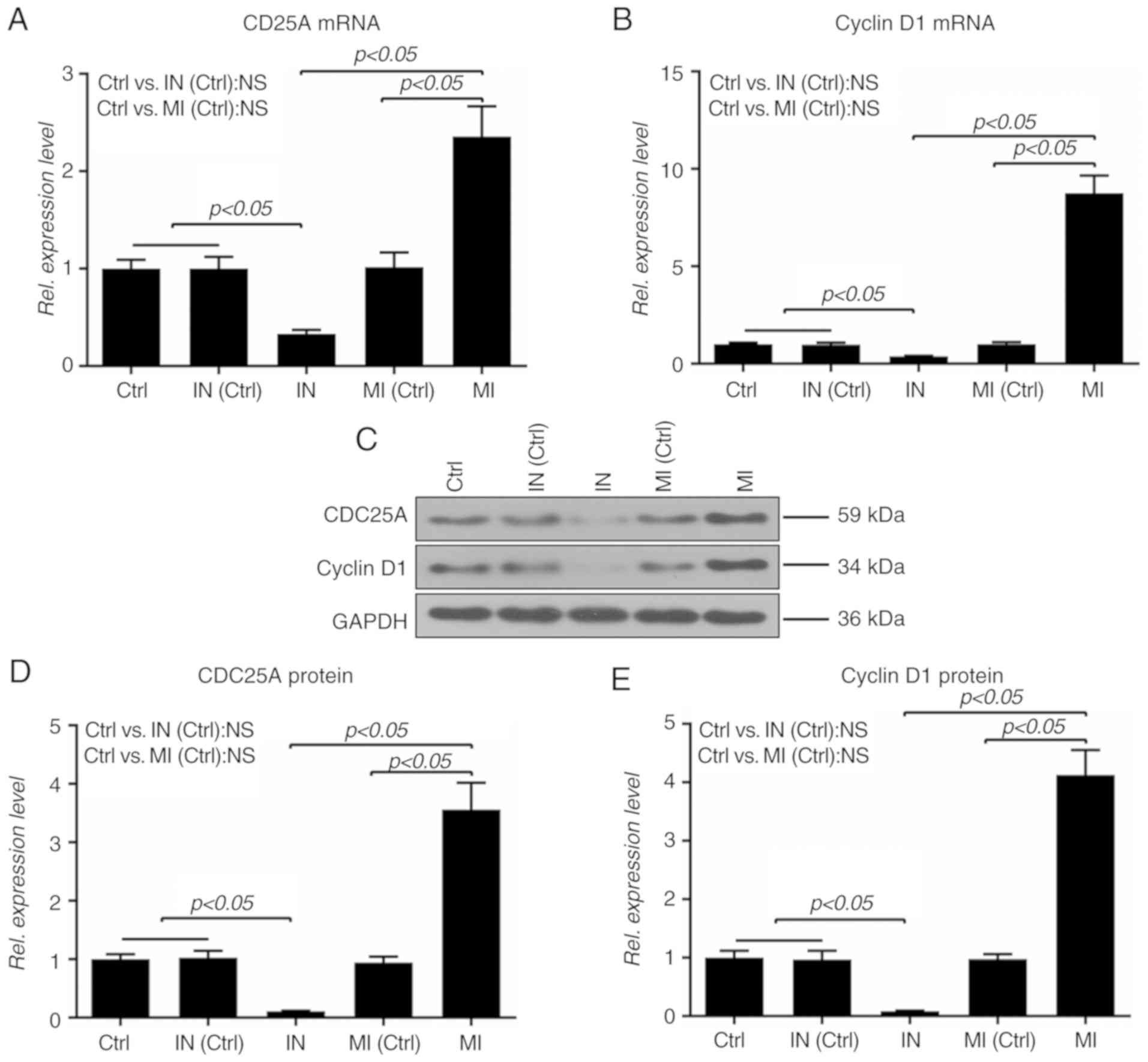
To further explore the effect of miR-19a on the cell cycle, the
expressions of CDC25A and cyclinD1 at the mRNA and protein levels
were determined; it was demonstrated that upregulation of miR-19a
increased the mRNA and protein expressions of CDC25A and cyclinD1,
which were repressed by the inhibitor (P<0.05; Fig. 2).
miR-19a promotes the migration and
invasion of VSMCs
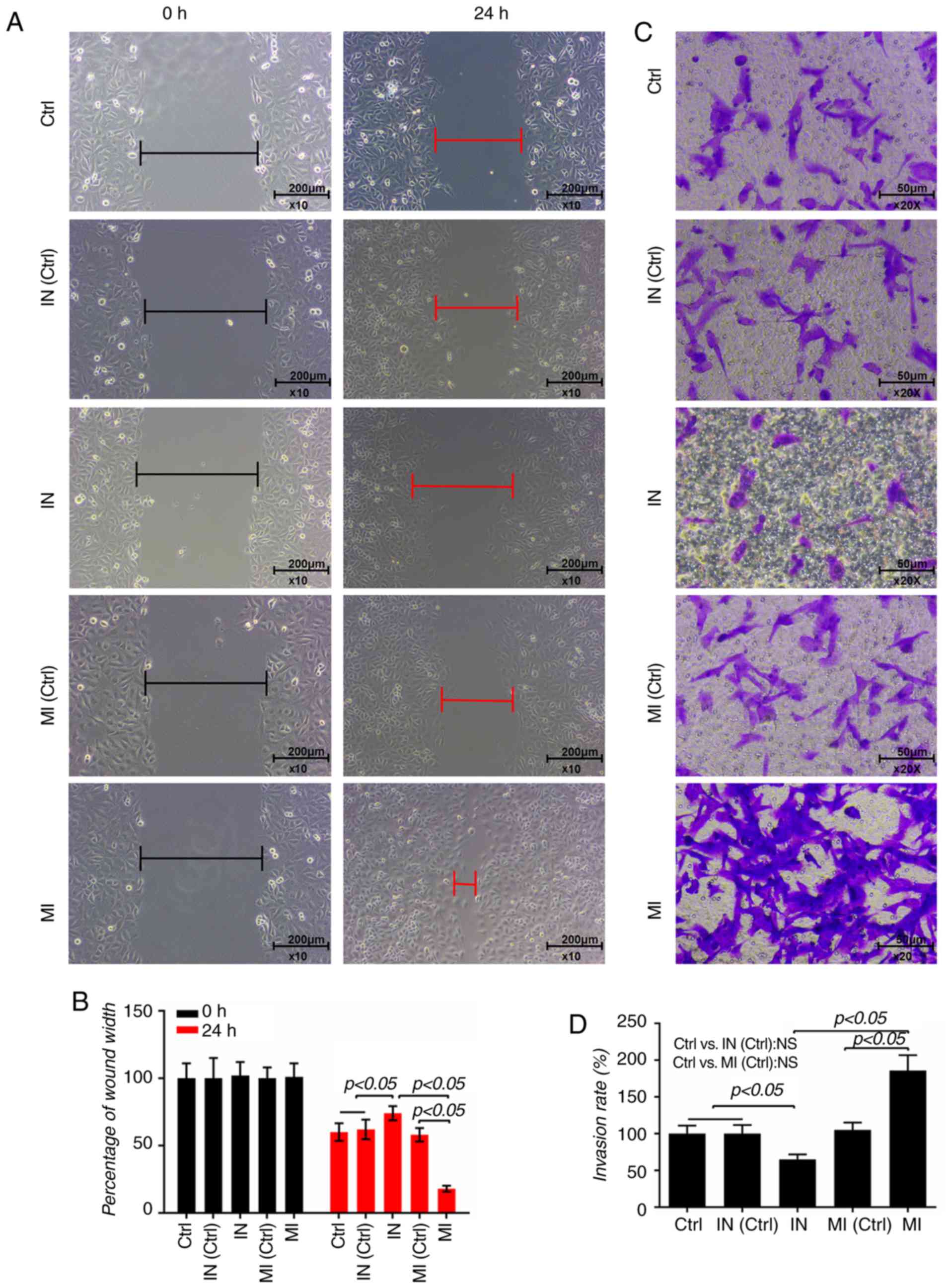
The shortest distance of a scratch was observed in
the cells transfected with miR-19a mimics for 24 h (P<0.05;
Fig. 3A and B). Moreover, the
proportion of invasive cells in the miR-19a mimics group was higher
than the control group, and the cells treated with miR-19a
inhibitor had a lower rate of invasion (P<0.05; Fig. 3C and D), suggesting that miR-19a
promoted the invasion of VSMCs.
To verify the role of miR-19a in the migration and
invasion of VSMCs, the expressions of several related molecules
were additionally detected. It was observed that the miR-19a mimics
significantly upregulated the mRNA levels of MMP-2, MMP-9, α-SMA
and SM22α, and that the miR-19a inhibitor exerted an opposite
effect on these mRNAs (P<0.05; Fig. 4A-D). Also, the protein levels of
MMP-2, MMP-9, α-SMA and SM22α were similar to the mRNA expressions
of those molecules (P<0.05; Fig.
4E and F).
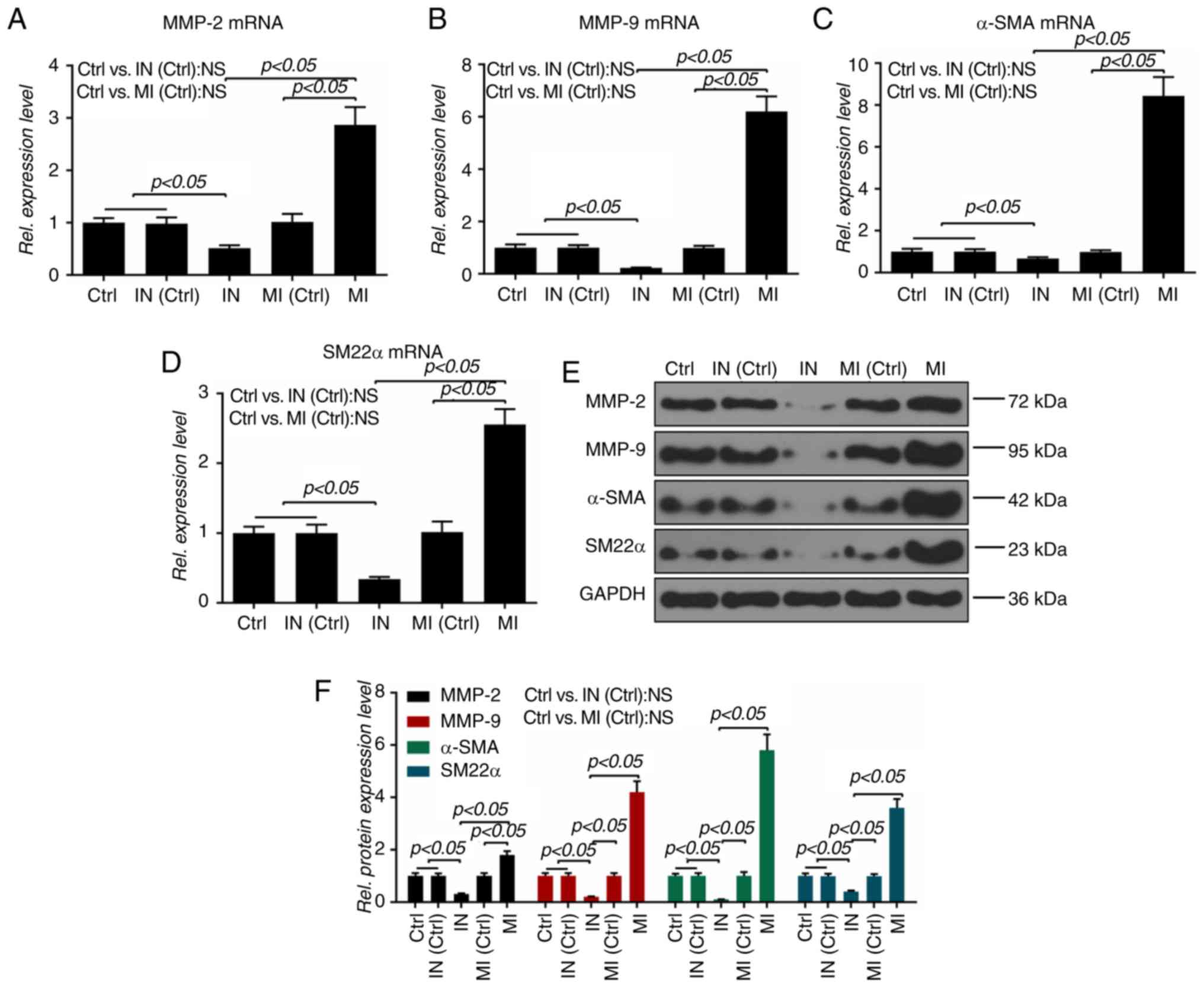 | Figure 4Effect of microRNA-19a on
migration-related molecules. The mRNA levels of (A) MMP-2, (B)
MMP-9, (C) α-SMA and (D) SM22α were measured by reverse
transcription-quantitative PCR. Protein levels of MMP-2, MMP-9,
α-SMA and SM22α were determined by (E) western blotting and (F)
densitometry. All values are presented as the mean ± SD. The data
were analyzed by ANOVA with Turkey's multiple comparisons test.
MMP, matrix metalloproteinase; α-SMA, α-smooth muscle actin; SM22α,
smooth muscle 22α; MI, microRNA-19a mimics; IN, microRNA-19a
inhibitor; Ctrl, control; Rel. relative; NS, not significant. |
RHOB is a direct target of miR-19a
TIA1, RHOB and SOCS3 were identified as three
potential targets of miR-19a. Although miR-19a did not affect the
level of TIA1, overexpression of miR-19a inhibited the expressions
of RHOB and SOCS3, and downregulation of miR-19a increased the
levels of RHOB and SOCS3, and the effect of miR-19a on RHOB was
more marked compared with SOCS3 (P<0.05; Fig. 5A). Therefore, it was predicted
that RHOB is a potential target of miR-19a, and the potential
binding site of miR-19a was identified in the 3′-UTR of RHOB mRNA
(Fig. 5B). Additionally, the
luciferase activity of the wild-type RHOB-3′-UTR was significantly
lower in the miR-19a group than in the control group (P<0.05);
however, it was significantly increased by the inhibitor
(P<0.05; Fig. 5C).
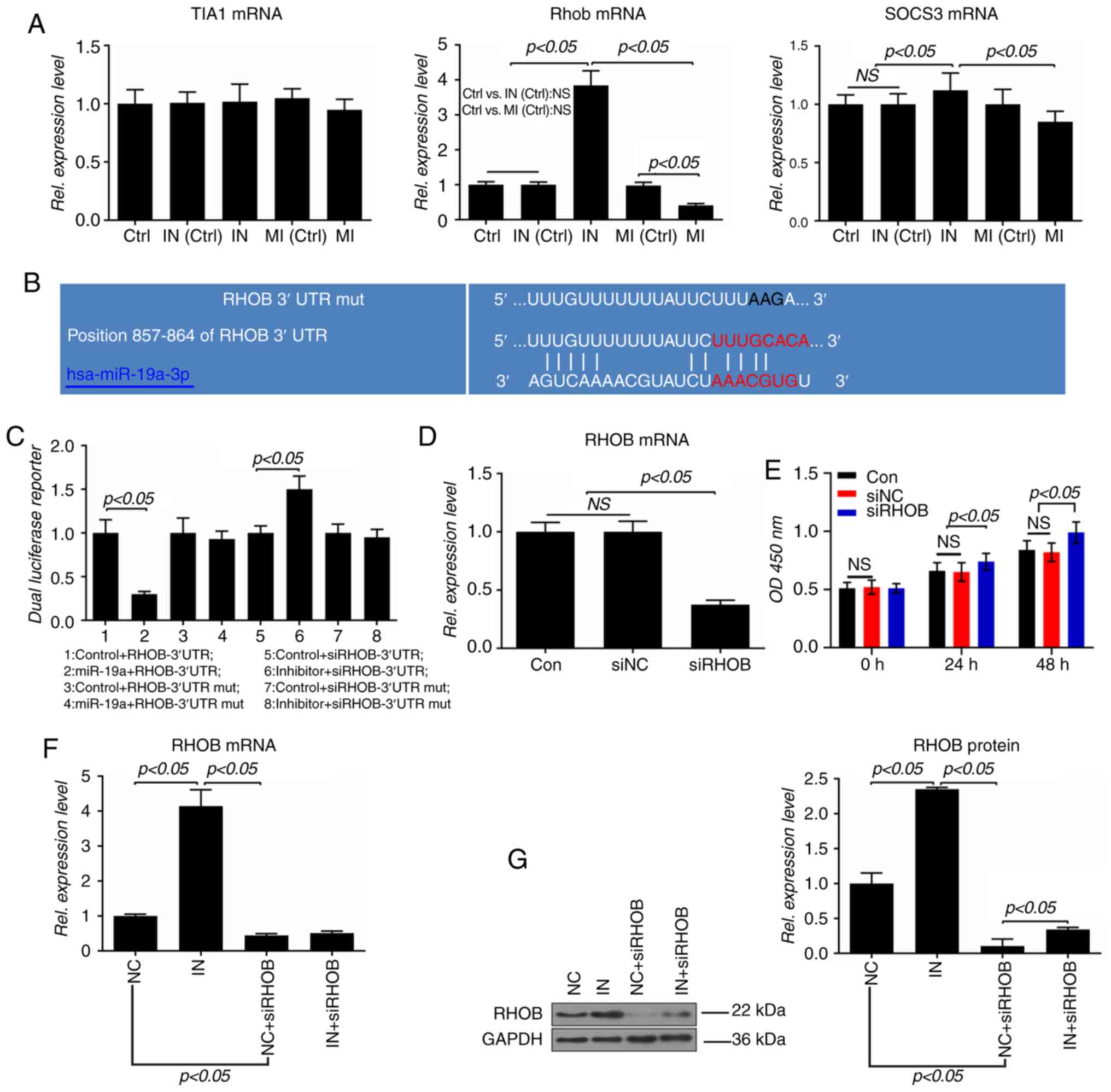 | Figure 5RHOB is a direct target gene of
miR-19a in VSMCs. (A) mRNA levels of TIA1, RHOB and SOCS3 were
measured by RT-qPCR. (B) Possible miR-19a binding site in the
3′-UTR of RHOB mRNA was predicted using TargetScan7.2. (C) A
dual-luciferase reporter assay was performed to analyze the
relative luciferase activity in VSMCs. VSMCs were transfected in
24-well plates with the indicated luciferase reporter plasmid or
its mutant. VSMCs were also co-transfected with miR-19a mimics,
inhibitor or corresponding control. At 48 h post-transfection, the
luciferase activities were measured using a microplate reader with
a dual luciferase kit. (D) mRNA expression of RHOB in VSMCs
transfected with siRHOB or negative control was analyzed by
RT-qPCR. (E) Proliferation of VSMCs was detected by a Cell Counting
Kit-8 assay using a microplate reader at 490 nm. (F) mRNA and (G)
protein expressions of RHOB in VSMCs transfected with inhibitor and
siRNAs were determined by RT-qPCR and western blotting,
respectively. All values are presented as the mean ± SD. The data
were analyzed by ANOVA with Turkey's multiple comparisons test.
RHOB, Ras homolog family member B; miR, microRNA; VSMC, vascular
smooth muscle cell; TIA1, T-cell intracellular antigen-1; SOCS3,
suppressor of cytokine signaling 3; RT-qPCR, reverse
transcription-quantitative PCR; 3′UTR, 3′ untranslated region; si,
small interfering; Ctrl/Con, control; MI, miR-19a mimics; IN,
miR-19a inhibitor; NS/ns, not significant; Rel., relative; NC,
negative control; OD, optical density; mut, mutant. |
To further explore the role of RHOB in VSMCs, RHOB
expression was silenced using siRNA. RHOB expression was
significantly suppressed by siRNA, and the proliferation of VSMCs
was significantly increased by the siRHOB (P<0.05; Fig. 5D and E). To further confirm that
RHOB was a direct target gene of miR-19a, VSMCs were co-transfected
with inhibitor and siRHOB. The results showed that the mRNA
expression of RHOB was increased by the inhibitor (P<0.05);
however, the inhibitor showed no effect on the expression in the
siRHOB group (Fig. 5F).
Furthermore, the present data demonstrated that the protein levels
of RHOB in the inhibitor group were significantly upregulated, and
that the inhibitor could increase the low expression of RHOB caused
by siRNA (P<0.05; Fig.
5G).
RHOB suppression restores the inhibitory
effects of miR-19a inhibitor in VSMCs
The results of the CCK-8 assay demonstrated that
co-transfection of miR-19a inhibitor and siRHOB could increase the
proliferation of VSMCs, compared with the inhibitor group
(P<0.05; Fig. 6A). As RHOB
serves a direct role in the migration of VSMCs (31), the role of RHOB in the regulatory
effect of miR-19a inhibitor in VSMCs was investigated. The scratch
width in the co-transfection group was longer than that in the
siRHOB group (P<0.05) but slightly shorter than the inhibitor
group (P>0.05; Fig. 6B and C).
Furthermore, the invasive cells in the co-transfection group
significantly increased compared with the inhibitor group
(P<0.05; Fig. 6D and E).
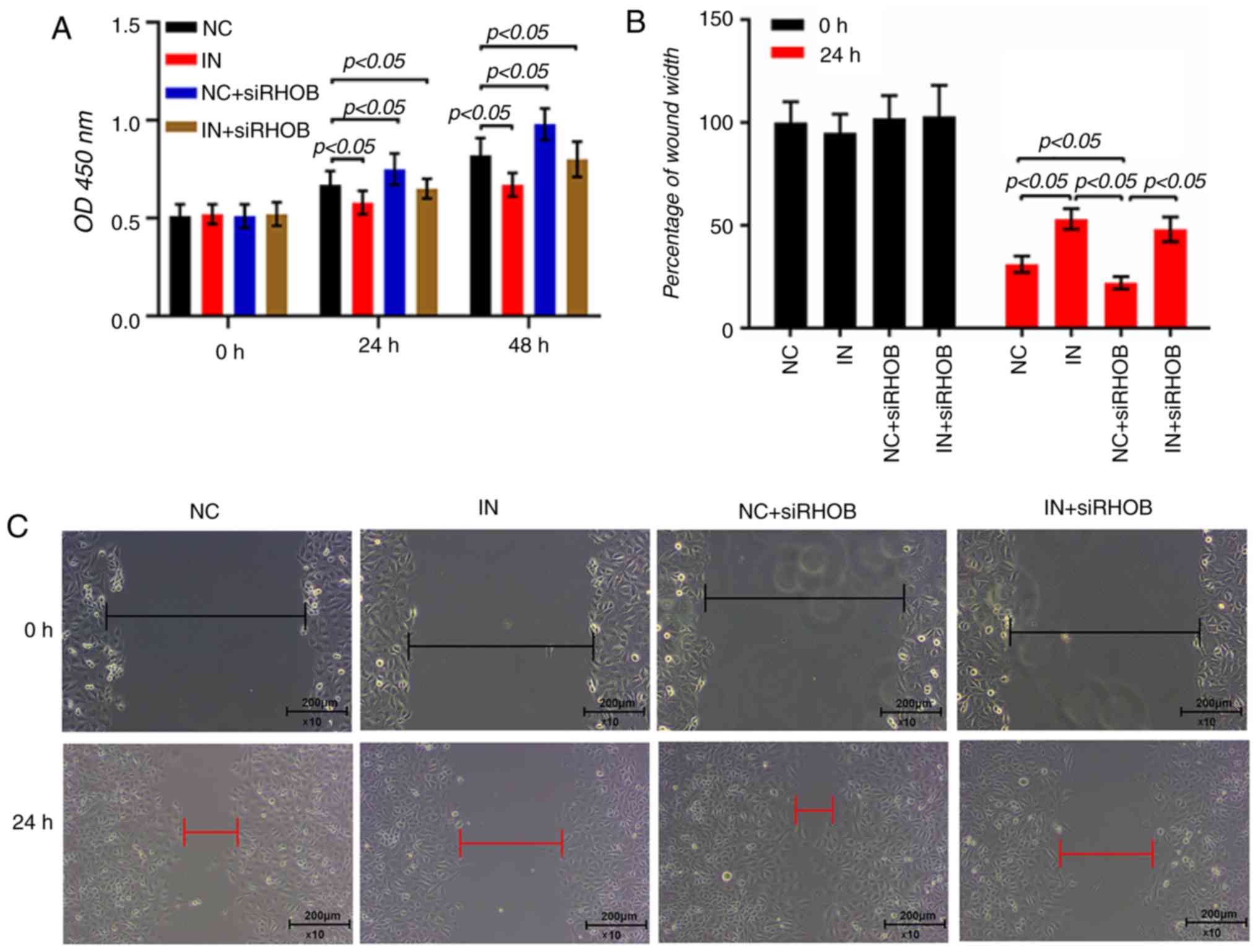 | Figure 6Inhibition of RHOB restores miR-19a
inhibitor-induced inhibitory effects on VSMC proliferation,
migration and invasion. (A) miR-19a inhibitor and siRHOB alone or
in combination were transfected into the VSMCs and cultured for 48
h. Cell proliferation was measured by a Cell Counting Kit-8 assay.
A wound-healing assay was performed in VSMCs that had been
transfected with inhibitor and siRHOB, and the relative wound width
was measured. (B) Percentage of wound width and (C) representative
images are presented. A transwell Matrigel assay was used in VSMCs
that had been treated with inhibitor and siRHOB, and the invasion
rate was calculated. (D) Invasion rate and (E) representative
images of cells stained with crystal violet are presented. All
values are presented as the mean ± SD. The data were analyzed by
ANOVA with Turkey's multiple comparisons test. RHOB, Ras homolog
family member B; miR, microRNA; VSMC, vascular smooth muscle cell;
si, small interfering; MI, miR-19a mimics; IN, miR-19a inhibitor;
NC, negative control; OD, optical density. |
To further explore the effect of RHOB on the
migration and invasion of VSMCs, the expressions of molecules
associated with migration and invasion were detected. Compared with
the inhibitor group, the co-transfection group showed significantly
increased mRNA expressions of MMP-2, MMP-9, α-SMA and SM22α
(P<0.05; Fig. 7A-D). Moreover,
the changes at the protein level in each group were similar to
those of mRNA, except SM22α, as the protein level of SM22α in the
co-transfection group exhibited a slight increase in comparison
with the inhibitor group (Fig.
7E-I).
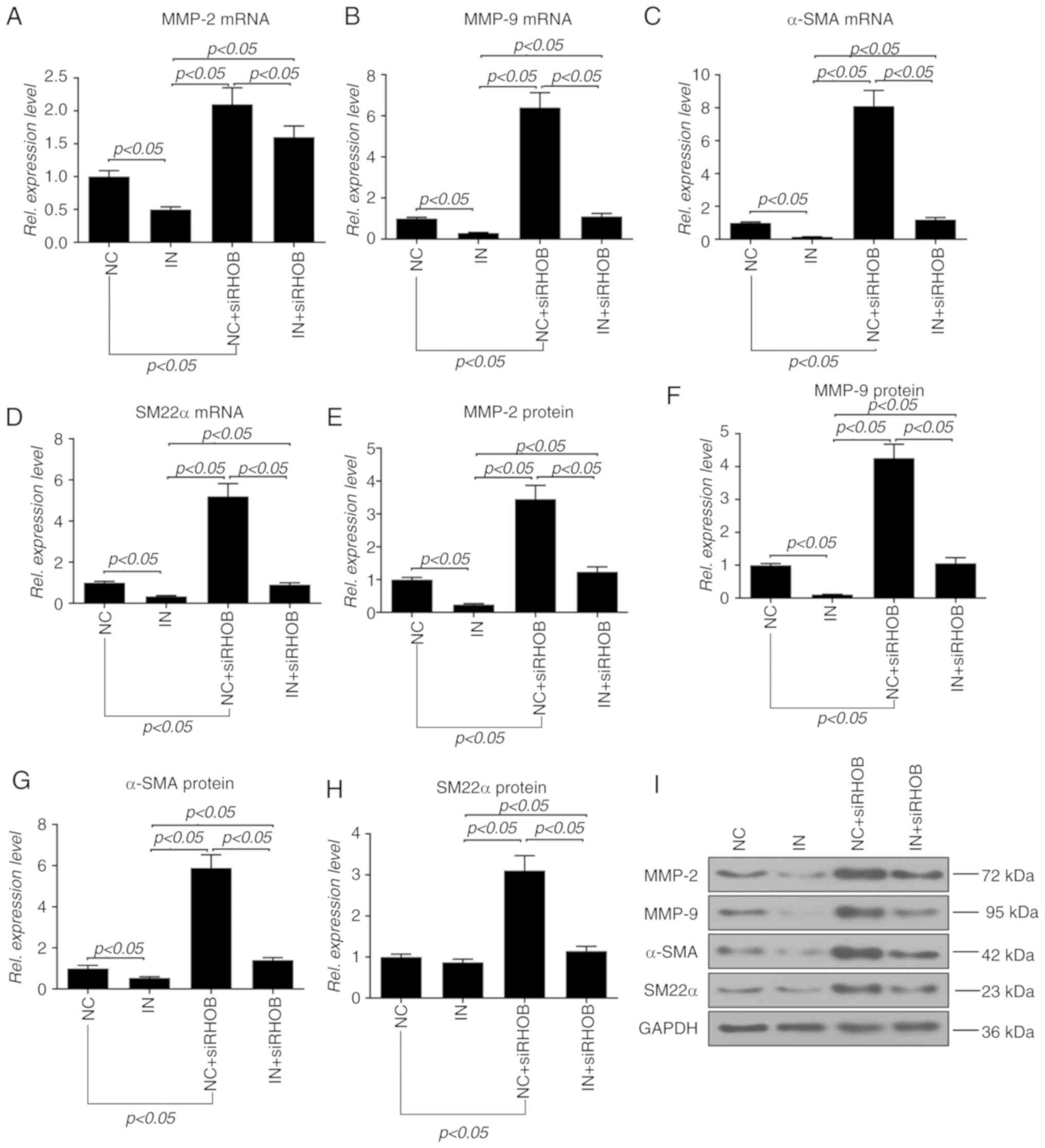 | Figure 7Inhibition of RHOB restores miR-19a
inhibitor-induced inhibitory effects on molecules related to
migration and invasion in VSMCs. miR-19a inhibitor and siRHOB alone
or in combination were transfected into the VSMCs and cultured for
48 h. The mRNA levels of (A) MMP-2, (B) MMP-9, (C) α-SMA and (D)
SM22α were measured by reverse transcription-quantitative PCR. The
protein levels of (E) MMP-2, (F) MMP-9, (G) α-SMA and (H) SM22α
were detected and determined by (I) western blotting followed by
densitometry analysis. All values are presented as the mean ± SD.
The data were analyzed by ANOVA with Turkey's multiple comparisons
test. RHOB, Ras homolog family member B; miR, microRNA; VSMC,
vascular smooth muscle cell; si, small interfering; MMP, matrix
metalloproteinase; α-SMA, α-smooth muscle actin; SM22α, smooth
muscle 22α; IN, miR-19a inhibitor; NC, negative control; Rel.,
relative. |
Discussion
The present study is the first, to the best of the
authors' knowledge, to demonstrate that miR-19a promoted VSMC
proliferation, migration and invasion, although VSMC proliferation
is implicated in atherogenesis. Moreover, miR-19a contributed to
the growth of VSMCs by promoting G2 cell generation. The
effect of miR-19a on the expressions of related proteins or genes
was also investigated.
CDK activation and inactivation play a key role in
cell cycle progression (35). CDK
activation, a critical step in the cell cycle, is realized by
inhibiting CDK phosphorylation through the CDC25 family
dual-specificity phosphatases. CDC25 isoforms include CDC25A,
CDC25B and CDC25C in mammals (36), and CDC25A has the most important
function of the CDC25 isoforms, and a defect in CDC25A is lethal in
early stages of embryogenesis, indicating that CDC25A plays an
indispensable role in cell division (37). CDC25A activates Cyclin A/E to
accelerate G1/S transition (38). Moreover, CDC25A also contributes
to Cyclin B contribution in G2/M transition (39). Cell mitosis requires cells to
leave the resting state (G0/G1) and proceed
to the phase of DNA synthesis (S) and mitosis (G2/M).
Noticeably, there is a critical moment between
G0/G1 and S, during which, cyclinD1, an
important cyclin, allows cells to get through the critical moment
between S and G2/M (40). A higher cyclinD1 expression can
promote cell proliferation (41).
The present data showed that miR-19a altered cell cycle progression
by upregulating cyclinD1 and CDC25A expressions.
MMPs are a large family, in which zinc-dependent and
calcium-dependent endopeptidases are responsible for degrading
multiple extracellular matrix proteins (42). MMP-2 and MMP-9, two gelatin enzyme
subgroups of MMP, have the ability to hydrolyze the basement
membrane and have been regarded as key molecules involved in cell
metastasis, migration and invasion (43-46). Encoded by the actin α 2, smooth
muscle, α-SMA is an isoform of vascular smooth muscle actin, which
is typically expressed in VSMCs and contributes to vascular cell
motility and contraction (47).
SM22α, an actin-binding protein, is abundant in smooth muscle cells
of vertebrates (48). Yuan
(47) demonstrated that SM22α
accelerated actin filament assembly into bundles, which possibly
enhanced VSMC contractility and mobility, and SM22α activation
helped balance the VSMC differentiated phenotype. In the present
study, inhibition of RHOB promoted VSMC proliferation, invasion and
migration, and partly reversed the inhibitory effect of the miR-19a
inhibitor on VSMC proliferation, invasion and migration. miR-19a
increased VSMC migration and invasion by promoting the
MMP/α-SMA/SM22α signaling pathway. Moreover, inhibited RHOB also
promoted the MMP/α-SMA/SM22α signaling pathway, suggesting that the
signaling pathway could be affected through RHOB.
SOCS3 inactivation was shown to enhance the cell
survival signaling pathway (49).
A previous study demonstrated that SOCS3 was a target of miR-19a,
in which miR-19a could promote rheumatoid arthritis fibroblast-like
synoviocytes by targeting SOCS3 (50). miR-19a can also enhance the
proliferation and insulin secretion of pancreatic β cells, and
inhibit the apoptosis of pancreatic β cells by targeting SOCS3
(51). In the present study,
overexpression of miR-19a suppressed the SOCS3 level, while
inhibition of miR-19a increased SOCS3 expression. However, the
effect of miR-19a on SOCS3 was weaker than that on RHOB, suggesting
that SOCS3 may play a less crucial role in the proliferation,
migration and invasion of VSMCs. As a DNA/RNA binding protein, TIA1
is associated with granules of cytolytic lymphocytes and apoptosis
by DNA fragmentation (52).
Transfection of miR-19a mimics and miR-19a inhibitor did not change
the RNA metabolism in VSMCs, suggesting that TIA1 may not be
involved in the regulation of miR-19a. However, the present study
only conducted in vivo experiments and the role of miR-19a
in animal models with cardiovascular diseases will be explored in
the future.
In conclusion, the present study demonstrated that
miR-19a promotes VSMC proliferation, migration and invasion via the
MMP/α-SMA/SM22α signaling pathway and the cyclinD1/CDC25A signaling
pathway. Moreover, miR-19a promotes VSMC proliferation, migration
and invasion via the MMP/α-SMA/SM22α signaling pathway by
inhibiting RHOB. This may provide understanding for myocardial
infarction or stroke, or even atherosclerosis.
Abbreviations:
|
VSMCs
|
vascular smooth muscle cells
|
|
RHOB
|
Ras homolog family member B
|
|
CCK-8
|
Cell Counting Kit-8
|
|
miRNA/miR
|
microRNA
|
|
miR-19
|
microRNA-19a
|
|
PI
|
propidium iodide
|
|
RT-qPCR
|
reverse transcription-quantitative
PCR
|
|
MMP
|
matrix metalloproteinase
|
|
α-SMA
|
α-smooth muscle actin
|
|
SM22α
|
smooth muscle 22α
|
|
SOCS3
|
suppressor of cytokine signaling 3
|
|
TIA1
|
T-cell intracellular antigen-1
|
Acknowledgments
Not applicable.
Funding
No funding was received.
Availability of data and materials
The analyzed datasets generated during the study are
available from the corresponding author on reasonable request.
Authors' contributions
GS and SW made substantial contributions to the
conception and design of the study. SW, HS and GS were involved in
data acquisition, analysis and interpretation. SW and HS drafted
the article or critically revised it for important intellectual
content. HS and GS agreed to be accountable for all aspects of the
work in ensuring that questions related to the accuracy or
integrity of the work are appropriately investigated and resolved.
All authors read and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Beckman JA, Creager MA and Libby P:
Diabetes and atherosclerosis: Epidemiology, pathophysiology, and
management. JAMA. 287:2570–2581. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Barsness GW, Peterson ED, Ohman EM, Nelson
CL, DeLong ER, Reves JG, Smith PK, Anderson RD, Jones RH, Mark DB
and Califf RM: Relationship between diabetes mellitus and long-term
survival after coronary bypass and angioplasty. Circulation.
96:2551–2556. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Natarajan MK, Strauss BH, Rokoss M, Buller
CE, Mancini GB, Xie C, Sheth TN, Goodhart D, Cohen EA, Seidelin P,
et al: Randomized trial of insulin versus usual care in reducing
reste-nosis after coronary intervention in patients with diabetes.
The STent restenosis and metabolism (STREAM) study. Cardiovasc
Revasc Med. 13:95–100. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
El Akoum S, Cloutier I and Tanguay JF:
Vascular smooth muscle cell alterations triggered by mice
adipocytes: Role of high-fat diet. J Atheroscler Thromb.
19:1128–1141. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Hattori Y, Hattori S, Sato N and Kasai K:
High-glucose-induced nuclear factor kappaB activation in vascular
smooth muscle cells. Cardiovasc Res. 46:188–197. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wu WY, Yan H, Wang XB, Gui YZ, Gao F, Tang
XL, Qin YL, Su M, Chen T and Wang YP: Sodium tanshinone IIA silate
inhibits high glucose-induced vascular smooth muscle cell
proliferation and migration through activation of AMP-activated
protein kinase. PLoS One. 9:e949572014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Virmani R, Kolodgie FD, Burke AP, Farb A
and Schwartz SM: Lessons from sudden coronary death: A
comprehensive morphological classification scheme for
atherosclerotic lesions. Arterioscler Thromb Vasc Biol.
20:1262–1275. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Bajaj A and Sethi A, Rathor P, Suppogu N
and Sethi A: Acute complications of myocardial infarction in the
current Era: Diagnosis and management. J Investig Med. 63:844–855.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Libby P, Ridker PM and Hansson GK:
Progress and challenges in translating the biology of
atherosclerosis. Nature. 473:317–325. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Bennett MR, Sinha S and Owens GK: Vascular
smooth muscle cells in atherosclerosis. Circ Res. 118:692–702.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Kim J, Jang SW, Park E, Oh M, Park S and
Ko J: The role of heat shock protein 90 in migration and
proliferation of vascular smooth muscle cells in the development of
atherosclerosis. J Mol Cell Cardiol. 72:157–167. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Pan HW, Li SC and Tsai KW: MicroRNA
dysregulation in gastric cancer. Curr Pharm Des. 19:1273–1284.
2013.
|
|
13
|
Lim LP, Lau NC, Garrett-Engele P, Grimson
A, Schelter JM, Castle J, Bartel DP, Linsley PS and Johnson JM:
Microarray analysis shows that some microRNAs downregulate large
numbers of target mRNAs. Nature. 433:769–773. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Lewis BP, Burge CB and Bartel DP:
Conserved seed pairing, often flanked by adenosines, indicates that
thousands of human genes are microRNA targets. Cell. 120:15–20.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhou Y, Li S, Li J, Wang D and Li Q:
Effect of microRNA-135a on Cell Proliferation, migration, invasion,
apoptosis and tumor angio-genesis through the IGF-1/PI3K/Akt
signaling pathway in non-small cell lung cancer. Cell Physiol
Biochem. 42:1431–1446. 2017. View Article : Google Scholar
|
|
16
|
Wu P, Agnelli L, Walker BA, Todoerti K,
Lionetti M, Johnson DC, Kaiser M, Mirabella F, Wardell C, Gregory
WM, et al: Improved risk stratification in myeloma using a
microRNA-based classifier. Br J Haematol. 162:348–359. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Tian Z, Zhao JJ, Tai YT, Amin SB, Hu Y,
Berger AJ, Richardson P, Chauhan D and Anderson KC: Investigational
agent MLN9708/2238 targets tumor-suppressor miR33b in MM cells.
Blood. 120:3958–3967. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Li T, Yang GM, Zhu Y, Wu Y, Chen XY, Lan
D, Tian KL and Liu LM: Diabetes and hyperlipidemia induce
dysfunction of VSMCs: Contribution of the metabolic
inflammation/miRNA pathway. Am J Physiol Endocrinol Metab.
308:E257–E269. 2015. View Article : Google Scholar
|
|
19
|
Mackenzie NC, Staines KA, Zhu D, Genever P
and Macrae VE: miRNA-221 and miRNA-222 synergistically function to
promote vascular calcification. Cell Biochem Funct. 32:209–216.
2014. View
Article : Google Scholar : PubMed/NCBI
|
|
20
|
Chen Q, Yang F, Guo M, Wen G, Zhang C,
Luong le A, Zhu J, Xiao Q and Zhang L: miRNA-34a reduces neointima
formation through inhibiting smooth muscle cell proliferation and
migration. J Mol Cell Cardiol. 89:75–86. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Hayashita Y, Osada H, Tatematsu Y, Yamada
H, Yanagisawa K, Tomida S, Yatabe Y, Kawahara K, Sekido Y and
Takahashi T: A polycistronic microRNA cluster, miR-17-92 is
overexpressed in human lung cancers and enhances cell
proliferation. Cancer Res. 65:9628–9632. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zhao D, Chen Y, Chen S, Zheng C, Hu J and
Luo S: MiR-19a regulates the cell growth and apoptosis of
osteosarcoma stem cells by targeting PTEN. Tumour Biol.
39:10104283177053412017. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhong B, Guo S, Zhang W and Zhang C, Wang
Y and Zhang C: Bioinformatics prediction of miR-30a targets and its
inhibition of cell proliferation of osteosarcoma by up-regulating
the expression of PTEN. BMC Med Genomics. 10:642017. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Roberts PJ, Mitin N, Keller PJ, Chenette
EJ, Madigan JP, Currin RO, Cox AD, Wilson O, Kirschmeier P and Der
CJ: Rho Family GTPase modification and dependence on CAAX
motif-signaled posttranslational modification. J Biol Chem.
283:25150–25163. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Etienne-Manneville S and Hall A: Rho
GTPases in cell biology. Nature. 420:629–635. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Meyer N, Peyret-Lacombe A, Canguilhem B,
Médale-Giamarchi C, Mamouni K, Cristini A, Monferran S, Lamant L,
Filleron T, Pradines A, et al: RhoB promotes cancer initiation by
protecting keratinocytes from UVB-induced apoptosis but limits
tumor aggressiveness. J Invest Dermatol. 134:203–212. 2014.
View Article : Google Scholar
|
|
27
|
Calvayrac O, Pradines A, Raymond-Letron I,
Rouquette I, Bousquet E, Lauwers-Cances V, Filleron T, Cadranel J,
Beau-Faller M, Casanova A, et al: RhoB determines tumor
aggressiveness in a murine EGFRL858R-induced adenocarcinoma model
and is a potential prognostic biomarker for lepidic lung cancer.
Clin Cancer Res. 20:6541–6550. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Huang M and Prendergast GC: RhoB in cancer
suppression. Histol Histopathol. 21:213–218. 2006.
|
|
29
|
Niu S, Ma X, Zhang Y, Liu YN, Chen X, Gong
H, Yao Y, Liu K and Zhang X: MicroRNA-19a and microRNA-19b promote
the malignancy of clear cell renal cell carcinoma through targeting
the tumor suppressor RhoB. PLoS One. 13:e01927902018. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Chen Q, Guo W, Zhang Y, Wu Y and Xiang J:
MiR-19a promotes cell proliferation and invasion by targeting RhoB
in human glioma cells. Neurosci Lett. 628:161–166. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Chan KC, Wu CH, Huang CN, Lan KP, Chang WC
and Wang CJ: Simvastatin inhibits glucose-stimulated vascular
smooth muscle cell migration involving increased expression of RhoB
and a block of Ras/Akt signal. Cardiovasc Ther. 30:75–84. 2012.
View Article : Google Scholar
|
|
32
|
Chan KC, Lin MC, Huang CN, Chang WC and
Wang CJ: Mulberry 1-deoxynojirimycin pleiotropically inhibits
glucose-stimulated vascular smooth muscle cell migration by
activation of AMPK/RhoB and down-regulation of FAK. J Agric Food
Chem. 61:9867–9875. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Zhang N, Fu L, Bu Y, Yao Y and Wang Y:
Downregulated expression of miR-223 promotes Toll-like
receptor-activated inflammatory responses in macrophages by
targeting RhoB. Mol Immunol. 91:42–48. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
35
|
Malumbres M and Barbacid M: Mammalian
cyclin-dependent kinases. Trends Biochem Sci. 30:630–641. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Boutros R, Lobjois V and Ducommun B: CDC25
phosphatases in cancer cells: Key players? Good targets? Nat Rev
Cancer. 7:495–507. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Ray D, Terao Y, Nimbalkar D, Hirai H,
Osmundson EC, Zou X, Franks R, Christov K and Kiyokawa H:
Hemizygous disruption of Cdc25A inhibits cellular transformation
and mammary tumorigenesis in mice. Cancer Res. 67:6605–6611. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Blomberg I and Hoffmann I: Ectopic
expression of Cdc25A accelerates the G(1)/S transition and leads to
premature activation of cyclin E- and cyclin A-dependent kinases.
Mol Cell Biol. 19:6183–6194. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Timofeev O, Cizmecioglu O, Settele F,
Kempf T and Hoffmann I: Cdc25 phosphatases are required for timely
assembly of CDK1-cyclin B at the G2/M transition. J Biol Chem.
285:16978–16990. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Cai FG, Xiao JS and Ye QF: Effects of
ischemic preconditioning on cyclinD1 expression during early
ischemic reperfusion in rats. World J Gastroenterol. 12:2936–2940.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Gopalakrishnan N, Saravanakumar M,
Madankumar P, Thiyagu M and Devaraj H: Colocalization of β-catenin
with Notch intracellular domain in colon cancer: A possible role of
Notch1 signaling in activation of CyclinD1-mediated cell
proliferation. Mol Cell Biochem. 396:281–293. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Visse R and Nagase H: Matrix
metalloproteinases and tissue inhibitors of metalloproteinases:
Structure, function, and biochemistry. Circ Res. 92:827–839. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Chambers AF and Matrisian LM: Changing
views of the role of matrix metalloproteinases in metastasis. J
Natl Cancer Inst. 89:1260–1270. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Polette M, Nawrocki-Raby B, Gilles C,
Clavel C and Birembaut P: Tumour invasion and matrix
metalloproteinases. Crit Rev Oncol Hematol. 49:179–186. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Li WD, Hu N, Lei FR, Wei S, Rong JJ,
Zhuang H and Li XQ: Autophagy inhibits endothelial progenitor cells
migration via the regulation of MMP2, MMP9 and uPA under normoxia
condition. Biochem Biophys Res Commun. 466:376–380. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Fatar M, Stroick M, Griebe M and Hennerici
M: Matrix metal-loproteinases in cerebrovascular diseases.
Cerebrovasc Dis. 20:141–151. 2005. View Article : Google Scholar
|
|
47
|
Yuan SM: α-Smooth muscle actin and ACTA2
gene expressions in vasculopathies. Braz J Cardiovasc Surg.
30:644–649. 2015.
|
|
48
|
Zhang JC, Kim S, Helmke BP, Yu WW, Du KL,
Lu MM, Strobeck M, Yu Q and Parmacek MS: Analysis of
SM22alpha-deficient mice reveals unanticipated insights into smooth
muscle cell differentiation and function. Mol Cell Biol.
21:1336–1344. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Wang J, Zhou H, Han Y, Liu X, Wang M, Wang
X, Yin G, Li X and Xiang M: SOCS3 methylation in synergy with Reg3A
over-expression promotes cell growth in pancreatic cancer. J Mol
Med (Berl). 92:1257–1269. 2014. View Article : Google Scholar
|
|
50
|
Chen Y, Wang W, Chen Y, Tang Q, Zhu W, Li
D and Liao L: MicroRNA-19a-3p promotes rheumatoid arthritis
fibroblast-like synoviocytes via targeting SOCS3. J Cell Biochem.
2019.Epub ahead of print.
|
|
51
|
Li Y, Luo T, Wang L, Wu J and Guo S:
MicroRNA-19a-3p enhances the proliferation and insulin secretion,
while it inhibits the apoptosis of pancreatic β cells via the
inhibition of SOCS3. Int J Mol Med. 38:1515–1524. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Tian Q, Streuli M, Saito H, Schlossman SF
and Anderson P: A polyadenylate binding protein localized to the
granules of cytolytic lymphocytes induces DNA fragmentation in
target cells. Cell. 67:629–639. 1991. View Article : Google Scholar : PubMed/NCBI
|