Introduction
Cholangiocarcinoma (CCA), or bile duct cancer,
arises from the epithelium in the biliary tract. CCA development
has been associated with infection by carcinogenic liver flukes,
Opisthorchis viverrini, therefore, this type of cancer
exhibits the highest incidence and mortality rates in Southeast
Asia, particularly in Thailand (1). Diagnosis and treatment of CCA is
difficult in a majority of cases, as the cancer is often detected
when the patients are at an advanced stage, with metastases to the
liver, lungs, lymph nodes or other secondary organs (2,3). A
previous model was established for studying the in vitro
metastasis of CCA using a pair of human CCA cell lines without and
with high metastatic activity, namely KKU-213 and KKU-213L5,
respectively. KKU-213 is the parental cell line and KKU-213L5 was
selected in vivo through the fifth serial passage of tissues
from pulmonary metastases formed following the tail vein injection
in NOD/scid/Janus kinase 3 mice. KKU-213L5 cells were revealed to
exhibit the most prominent metastatic phenotype compared with the
parental KKU-213 cells. The mRNA expression profiles of 77
metastasis associated genes were determined using a quantitative
(q)PCR array, which revealed that anterior gradient-2 (AGR2) was
the most highly upregulated among 77 genes that were predominantly
upregulated in KKU-213L5 cells compared with parental KKU-213 cells
(4).
AGR2 is a protein that localizes in the anterior
border of the embryonic ectoderm and is crucial in cementing gland
development in the early embryonic development in Xenopus
laevis (5). Human AGR2 is
classified as an enzyme in the protein disulfide isomerases (PDIs)
family. The 13,304 base pair (bp) of the AGR2 gene on chromosome 7
encodes for a 996-bp 8-exon mRNA, which translates into a 175-amino
acid protein (6). AGR2 is
typically localized in the endoplasmic reticulum (ER) and is
involved in the production of cysteine-rich proteins, including the
mucin family in mucus-secreting cells/tissues, including the
respiratory tract, stomach, colon, prostate gland and female
reproductive system, and is highly expressed in various types of
cancer tissues (7).
With regards to the functional involvement of AGR2
in ER, it directly functions as the isomerase enzyme for the
folding of proteins and corrects protein misfolding by catalyzing
the cysteine disulfide bond to produce productive functional
proteins (8). Under abnormal
conditions in humans, including cancer, the upregulation of AGR2 is
associated with disease development and progression, and it has
been revealed to promote pancreatic cancer cell proliferation and
survival (9). In addition, the
dimerization of monomeric AGR2 is required, particularly when the
cells are under ER stress. For example, cancer cells exhibit a
marked increase in their protein synthesis ability to support cell
proliferation, resulting in the accumulation of proteins in the ER
for post-translational modification into functional proteins
(10). ER stress is caused by an
accumulation of unfolded proteins or the presence of mutated
proteins, which cannot fold correctly, and AGR2 is a key enzyme
that serves an important function in protein-folding homeostasis
under these ER conditions (8). A
previous study demonstrated that an AGR2 homodimer is required to
interact with binding immunoglobulin protein
(BiP)/glucose-regulated protein 78 kDa (GRP78) to activate the
unfolded protein response (UPR) pathway, a cellular stress response
mechanism that is directly associated with ER stress (10). The UPR pathway is initiated by
three ER transmembrane-resident proteins, including
inositol-requiring enzyme 1 (IRE1), activating transcription factor
6 (ATF6) and protein kinase RNA-like endoplasmic reticulum kinase
(PERK). Under non-stress conditions, the three
ER-transmembrane-resident proteins bind with BiP or GRP78 to remain
inactive. Under ER stress conditions, BiP dissociates from these
ER-transmembrane sensors, resulting in their activation (11). Activated IRE1 induces the splicing
of X-box binding protein 1 (XBP1) mRNA to XBP1s, which translocates
to the nucleus and functions as a transcription factor for the
upregulation of UPR target genes (12). Activated ATF6 translocates to
nucleus to function as a transcription factor, which modulates the
expression of the chaperones and enzymes required for ER function
(13). One of the downstream
targets of ATF6 is GRP94, which is upregulated for the folding of
newly synthesized proteins and prevents the accumulation of
unfolded or misfolded proteins (14). Activated PERK phosphorylates a
downstream target, eukaryotic initiation factor 2 (eIF2), and
phosphorylated (p-)eIF2α promotes the expression of transcription
factor ATF4, which regulates numerous UPR pathway target genes
involved in ER stress-mediated apoptosis, including C/EBP
homologous protein (CHOP) (15).
In 2014, the first evidence of AGR2 splicing was
reported in prostate cancer, including 6 spliced variant
transcripts, including AGR2vB, AGR2vC, AGR2vE, AGR2vF, AGR2vG and
AGR2vH (16). A previous study
reported the aberrant splicing of AGR2 in CCA cells, which
characterized the highly upregulated AGR2vH transcript and its
function in promoting the metastasis-associated phenotypes of CCA
cells, including migration, invasion and adhesion capacities. Of
note, only AGR2vH was predictably translatable into a protein
isoform that consists of 67 amino acids, which were truncated from
175 amino acids in AGR2 (17,18). The oncogenic properties of AGR2vH,
which enhance the metastatic ability of CCA cells, were recently
demonstrated (17). It was
previously reported that the suppression of AGR2vH in highly
metastatic KKU-213L5 cells reduced their migration and invasion
abilities, whereas the overexpression of AGR2vH in parental KKU-213
cells promoted cancer cell migration, invasion, adhesion and
proliferation (17).
Regarding the characterization of AGR2, it is a gene
that is highly and specifically upregulated in the metastatic CCA
cell line (4) and the
upregulation of this gene coincides with the aberrant splicing of
AGR2 mRNA, and AGR2vH are specific to metastatic CCA cells
(17). Previous results have
demonstrated that the AGR2vH isoform enables metastatic-associated
phenotypes in CCA cells. Additionally, another important function
of AGR2vH was proposed (17).
Taking into account experiments in other cancer types that proved
that the AGR2 wild-type is required for interaction with BiP/GRP78,
which is the UPR pathway activator as mentioned above (10), the present study attempted to
investigate the functional ability of AGR2vH in the activation of
the UPR pathway in a model of CCA.
Prospectively, AGR2vH may serve as an alternative
partner molecule contributing to the survival of CCA cells. The aim
of the present study was to determine the effect of AGR2vH on UPR
pathway response and cell viability/apoptosis following the
overexpression and knockdown of AGR2vH in CCA cells, particularly
when experimentally inducing ER stress in the cancer cells. The
activation of the UPR pathway was investigated by the expression of
UPR-sensitive markers and UPR pathway activation proteins. In
addition, the number of dead cells and the activity of caspase
enzymes in the apoptosis pathway and the survival of CCA cells were
investigated.
Materials and methods
Cell lines and cell culture
The two CCA cell lines used in the present study
included KKU-213, which was obtained from the Japanese Collection
of Research Bioresources Cell Bank, and KKU-213L5, a highly
metastatic CCA cell line derived from the parental KKU-213 cell,
which was established in a previous study (4). Cells were provided from the
Cholangiocarcinoma Research Institute, Faculty of Medicine, Khon
Kaen University (Khon Kaen, Thailand). The cell lines were cultured
in Dulbecco's modified Eagle's medium supplemented with 10% v/v
fetal bovine serum with 100 U/ml penicillin and 100 µg/ml
streptomycin (Gibco; Thermo Fisher Scientific, Inc.) and maintained
at 37°C in a humidified incubator in a 5%
CO2atmosphere.
Transfection and overexpression of AGR2vH
in CCA cells
AGR2vH-overexpressing KKU-213 cells were established
as previous described (17).
Briefly, AGR2vH mRNA was amplified by specific primers, as listed
in Table I, with the relevant
restriction sites to clone into the pCR®
2.1-TOPO® cloning vector (Invitrogen; Thermo Fisher
Scientific, Inc.). The AGR2vH nucleotide sequences were analyzed
and confirmed before being sub-cloned into the p3XFLAG-CMV-14
expression vector (Sigma-Aldrich; Merck KGaA). Either pCMV14-AGR2vH
or pCMV14-Empty vector (5 µg/µl) were transfected
into KKU-213 cells by Lipofectamine 2000 (Thermo Fisher Scientific,
Inc.) and cultured for 48 h at 37°C, and the single clones were
selected using 2 mg/ml Geneticin G418 (Thermo Fisher Scientific,
Inc.) and subjected to expansion and culture.
 | Table IPrimer sequences for reverse
transcription-PCR and quantitative PCR. |
Table I
Primer sequences for reverse
transcription-PCR and quantitative PCR.
| Name | Forward primer
5′-3′ | Reserve primer
5′-3′ |
|---|
| AGR2vH |
CAGACATATGAAGAAAGCTCTCAAGT |
TCCACACTAGCCAGTCTTCTCA |
| AGR2vH (with
restriction sites) | AAGCTTATGGAGAAAATTCCAGTGTC | GAATTCGTCTTCAGCAACTTG |
| ATF6 |
GAACCATTGCTTTACATTCCTCCAC |
CTGCTTGACTTGGTCCTTTCTACTTC |
| BiP/GRP78 |
GTTCTTCAATGGCAAGGAACCATCTC |
CCATCCTTTCGATTTCTTCAGGTGGAA |
| CHOP |
TGAACGGCTCAAGCAGGAAATCG |
GGATTGAGGGTCACATCATTGGCACT |
| eIF2 |
GCCAAATTGCCCTATCTCAA |
CAGAAAAATGGGCAAAGGAA |
| GRP94 |
TGGGAAGAGGTTCCAGAATG |
GTTGCCAGACCATCCGTACT |
| XBP1 |
TTACGAGAGAAAACTCATGGCC |
GGGTCCAAGTTGTCCAGAATGC |
| XBP1s |
TGCTGAGTCCGCAGCAGGT |
GCTGGAGGCTCTGGGGAA |
| β-actin |
GTGCGTGACATTAAGGAG |
GGAAGGCTGGAAGAGTG |
Experimental induction of ER stress
Tunicamycin was used to block the activity of
glycosylase, which resulted in the accumulation of
unglycosylated-proteins in the ER. Tunicamycin (Sigma-Aldrich;
Merck KGaA) was dissolved in dimethyl sulf-oxide. The optimal
concentration was determined by testing tunicamycin at 0.5, 1, 2, 4
and 8 µg/µl in culture media for 24 h, in order to
examine cytotoxicity using an MTT assay (Bio Basic, Inc.), as
previously described (19) (data
not shown). The resulting formazan crystals were dissolved in DMSO
and the absorbance at 540 nm was measured by using a Synergy HT
Multi-Detection Microplate Reader (BioTek Instruments, Inc.)
Subsequently, the expression of ER stress-sensitive markers,
including XBP1s and BiP/GRP78, was determined by reverse
transcription (RT)-PCR and qPCR.
Depletion of AGR2vH by small interfering
RNA (siRNA)
AGR2vH-overexpressing cells were transfected with
siAGR2vH (antisense: 5′-UUG AGA GCU UUC UUC AUA UGU CUG-3′) as
previously described (17).
Briefly, AGR2vH-overexpressing cells were plated in a 6-well plate
at 2.5×104 cells per well for 24 h. Then, the cells were
transfected with 75 nmol siAGR2vH or negative control siRNA
(Ambion; Thermo Fisher Scientific, Inc.) using Lipofectamine™ 2000
(Thermo Fisher Scientific, Inc.) in Opti-MEM I reduced-serum medium
(Gibco; Thermo Fisher Scientific, Inc.) and incubated for 6 h at
37°C. After that, the media was removed and replaced with fresh
complete media. At 48 h after transfection, cells were harvested
for used in further experiments.
Preparation of RNA and RT-PCR
Total RNA was isolated from the cells using an
E.Z.N.A® Total RNA kit I (Omega Bio-Tek, Inc.). The
concentrations of RNA samples were measured, and 1 µg total
RNA was used to synthesize the complementary DNA using the
HisenScriptTM RH [-] RT PreMix kit (Intron Biotechnology, Inc.)
according to the manufacturer's protocol. All cDNA samples were
stored at -80°C until use. For the determination of gene expression
by the amplification of synthesized cDNA, PCR was performed under
optimized conditions. The reaction mixture contained 0.2 µg
cDNA template, 0.4 µM each the forward and reverse primers
with a total volume of 20 µl of 1X MyTaqTM HS Red Mix
(Bioline Reagents Limited). The house-keeping gene β-actin was used
as an internal control for semi-quantitative normalization. The
primers for the target genes were based on previous studies
including AGR2vH (17), XBP1
(20), BiP/GRP78 (11), ATF6, CHOP (21), eIF2a and GRP94 (22) and are listed in Table I. The thermocycling conditions
were as follows: 95°C for 5 min for a pre-denature, followed by 30
cycles of 95°C for 30 sec, 55°C for 30 sec and 72°C for 30 sec, and
final extension at 72°C for 5 min. The PCR products were mixed with
6X non-mutagenic fluorescent (SYBR-Green) DNA staining reagent
(Novel Juice, Gene DireX, Inc.) and analyzed by 2% agarose gel
electrophoresis at 90 volts for 30 min and visualized under
ultraviolet illumination of agarose gels detected by ImageQuant™
LAS 500 (GE Healthcare Life Sciences) and quantitated using
ImageQuant TL 7.0 software (GE Healthcare Life Sciences).
qPCR
qPCR was performed for the relative quantification
of gene expression, including AGR2vH expression in
AGR2vH-overexpressing cells and the expression of ER
stress-sensitive markers. The reaction mixture (10 µl)
contained a cDNA template, forward and reverse primers for each
target gene (Table I) and 1X
LightCycler® 480 SYBR Green I Master (Roche Applied
Science). Primers for the target genes were based on previous
studies including XBP1s (23) and
BiP/GRP78. The thermocycling conditions were as follows: 95°C for
10 min, followed by 45 cycles of 95°C for 10 sec, 60°C for 10 sec
and 72°C for 10 sec. All reactions were experimentally performed in
biological triplicate and analyzed using the
LightCycler® 480 systems (Roche Applied Science). The
expression levels of the target genes were normalized with β-actin
using the relative quantification formula of 2-ΔΔCq
(24).
Protein extraction and western blot
analysis
Cells were lysed with lysis buffer containing 7M
urea, 2M thiourea, 4%
3-[3-cholamidopropyl)dimethylammonio]-1-propane-sulfonate, and
protease and phosphatase inhibitors (Roche Diagnostics). The
protein concentration was determined using the Bradford assay
(Bio-Rad Laboratories, Inc.). Equal amounts of protein (30
µg/lane) were separated by 12% SDS-PAGE and western blot
analysis were performed as previously described (17). Briefly, membranes were blocked
with 5% non-fat milk in TBST for 2 h at room temperature.
Subsequently, membranes were incubated with primary antibodies
overnight at 4°C and incubated with secondary antibodies for 1 h at
room temperature. Antibodies against BiP/GRP78 (cat no. E-AB-31742;
1:2,000), p-eIF2a (cat no. E-AB-20864; 1:2,000), B-cell lymphoma-2
(Bcl-2; cat no. E-AB-15522; 1:650) and Bcl-2-associated X (BAX; cat
no. E-AB-30629; 1:1,000) were purchased from Elabscience (Houston,
TX, USA) whereas antibodies for β-actin (cat no. A5441; 1:1,000),
horseradish peroxidase (HRP)-conjugated anti-rabbit immunoglobulin
G (IgG) (cat no. AP182P; 1:5,000) and HRP-conjugated anti-mouse IgG
(cat no. 12-349; 1:5,000) were purchased from Sigma-Aldrich; Merck
KGaA. Protein bands were detected with an enhanced
chemiluminescence system (Bio-Rad Laboratories, Inc.) and
visualised with ImageQuant™ LAS 500 (GE Healthcare Life
Sciences).
Flow cytometry
Apoptosis was assessed by Annexin V-Phy
coerythrin/7-Amino-Actinomycin staining using a Muse™ Annexin V and
Dead Cell assay kit (Merck KGaA). Cells were plated in a 6-well
plate at 2.5×105 cells per well for 24 h prior to
tunicamycin treatment. After 24 h treatment at 37°C, 100 µl
Muse™ Annexin V & Dead Cell reagent and an equal volume of
4×105 cells from each of the groups were mixed.
Subsequent to incubating for 20 min at room temperature, the
numbers of live, dead and apoptotic cells were analyzed using
Muse® Cell Analyzer and Muse 1.5.0.0 Analysis software
(Merck KGaA) (25).
Caspase 3/7 activity assay
Cells were plated in a 96-well black plate at
2×104 cells per well for 24 h prior to tunicamycin
treatment. After 24 h treatment, caspase 3/7 activities were
analyzed using an Apo-ONE® Homogeneous caspase 3/7 assay
(Promega Corporation), based on the cleavage of the non-fluorescent
caspase substrate Z-DEVD-R110 by caspase-3/7 to create fluorescent
Rhodamine 110 (26), according to
the manufacturer's protocol. The fluorescence signal of each well
was measured by a fluorescence microplate reader (EnSpire Multimode
Plate reader; PerkinElmer, Inc.). With regards to the measurement
of the fluorescence intensities, the assay suggested that the
excitation wavelength be set at 499 nm, and the emission wavelength
at 521 nm (27,28).
Cell viability assay
Cells were plated in a 96-well plate at
3×103 cells per well for 24 h prior to tunicamycin
treatment. After 24 h treatment, 10 µl Cell Counting Kit-8
(CCK-8) reagent (Sigma-Aldrich; Merck KGaA) was added to each well.
Cells were incubated for 4 h at 37°C, and the absorbance at 450 nm
was measured using a Synergy HT Multi-Detection Microplate Reader
(BioTek Instruments, Inc.).
Statistical analysis
Experiments were performed in biological triplicate.
Data were calculated and presented as the mean ± standard
deviation. Statistical significance between two groups was
determined using a Student's t-test (two tailed). One-way analysis
of variance with a Least Significant Difference post-hoc test was
performed to compare multiple groups using SPSS 17 (SPSS, Inc.).
P<0.05 was considered to indicate a statistically significant
difference.
Results
Expression of AGR2vH on
AGR2vH-overexpressing cells
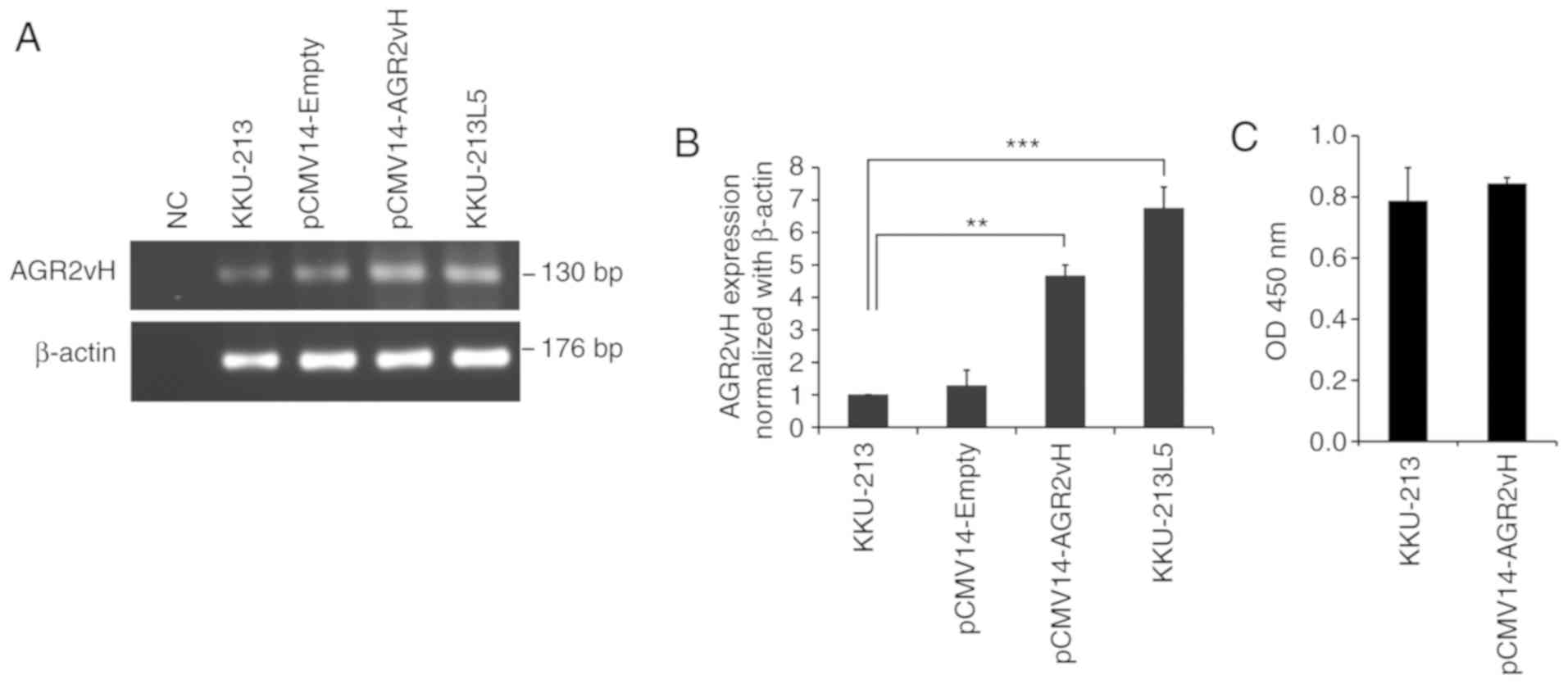
Semi-quantitative RT-PCR was performed to evaluate
the expression of AGR2vH once the cells were transfected with
pCMV14-Empty and pCMV14-AGR2vH vector. The expression of AGR2vH was
substantially increased in AGR2vH-overexpressing cells when
compared with the untransfected control cells, but the expression
level of AGR2vH in AGR2vH-overexpressing cells was still lower
compared with KKU-213L5 cells (Fig.
1A). Furthermore, RT-qPCR confirmed the results of RT-PCR
(P<0.01; Fig. 1B). In
addition, the cell viability following AGR2vH transfection into
KKU-213 cells was not significantly altered at 48 h
post-transfection compared with KKU-213 cells (untransfected
control) (Fig. 1C).
Experimentally induced ER stress
To optimize the different concentrations of
tunicamycin, the expression of ER stress markers (XBP1s and
BiP/GRP78) was determined after 24 h treatment using <4
µg/ml tunicamycin. XBP1s was significantly upregulated at 2
µg/ml when compared with untreated cells, as demonstrated
using RT-PCR and confirmed by qPCR (P<0.05; Fig. 2A). Similar results were obtained
for BiP/GRP78 by RT-PCR and qPCR (P<0.05; Fig. 2B). Therefore, 2 µg/ml
tunicamycin was selected for ER stress induction.
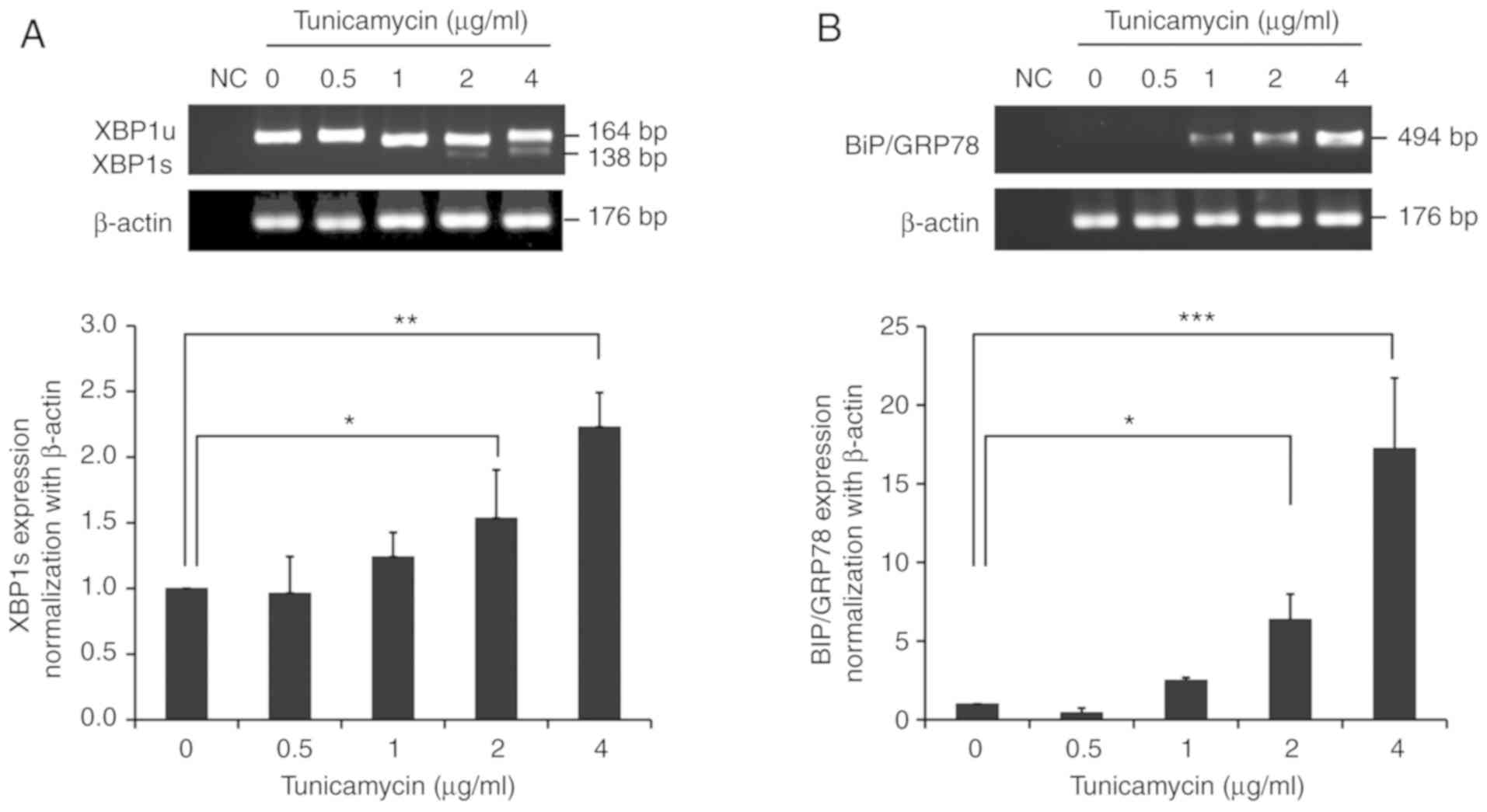 | Figure 2Effects of tunicamycin on the
expression of endoplasmic reticulum stress markers. (A) Expression
of XBP1 (XBP1u and XBP1s) using RT-PCR and of XBP1s using qPCR. (B)
Expression of BiP/GRP78 after 24 h tunicamycin treatment at 0.5, 1,
2 and 4 µg/ml using RT-PCR and qPCR. All data are presented
as the mean ± standard deviation. *P<0.05,
**P<0.01 and ***P<0.001 with
comparisons shown by lines. XBP1u, unspliced X-box binding protein
1; XBP1s, spliced X-box binding protein 1; BiP, Binding
immunoglobulin protein; GRP, Glucose-regulated protein; RT-PCR,
reverse transcription-PCR; qPCR, quantitative PCR; bp, base pairs;
NC, negative control. |
Activation of the UPR pathway and UPR
downstream
The present study sought to investigate UPR response
following AGR2vH overexpression and knockdown in CCA cells and UPR
response under ER stress-inducing conditions. On determination of
the UPR marker gene expression, the notable upregulation of XBP1s,
ATF6 and eIF2a was observed when the AGR2vH-overexpressing cells
were under ER stress conditions (Fig.
3A). Similar results were observed in AGR2vH-depleted cells,
with the downregulation of these markers under ER stress
conditions, particularly ATF6 and eIF2a (Fig. 3B). The expression of GRP94 mRNA,
an ER chaperone downstream of the UPR pathway, was upregulated in
AGR2vH-overexpressing cells (Fig.
3C) and downregulated in AGR2vH-depleted cells, particularly
under ER stress conditions (Fig.
3D). In addition, the activation of the UPR pathway was
confirmed by determination of BiP/GRP78 and p-eIF2a protein
expression under ER stress-inducing conditions. It was revealed
that BiP/GRP78 and p-eIF2a protein expression was upregulated in
AGR2vH-overexpressing cells (Fig.
3E), whereas they were downregulated in AGR2vH-depleted cells
(Fig. 3F).
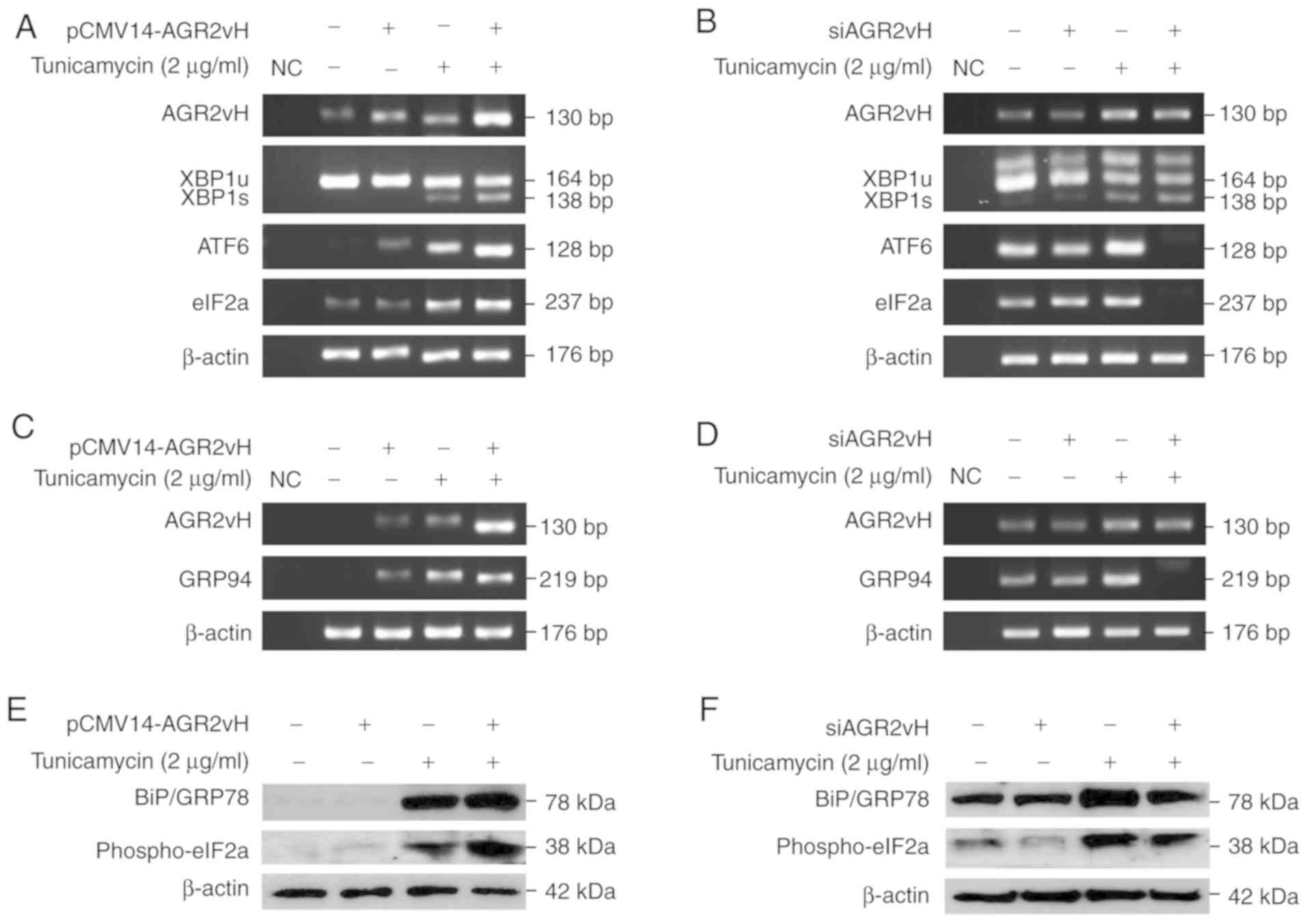 | Figure 3Status of UPR pathway activation and
expression of UPR downstream markers in AGR2vH-overexpressing and
AGR2vH-depleted cells. (A) mRNA expression of XBP1s, ATF6 and eIF2a
in AGR2vH-overexpressing cells. (B) mRNA expression of XBP1s, ATF6
and eIF2a in AGR2vH-depleted cells. (C) mRNA expression of GRP94 in
AGR2vH-overexpressing cells. (D) mRNA expression of GRP94 in
AGR2vH-depleted cells. (E) Expression of BiP/GRP78 and p-eIF2a
proteins in AGR2vH-overexpressing cells. (F) Expression of
BiP/GRP78 and p-eIF2a proteins in AGR2vH-depleted cells. UPR,
unfolded protein response; AGR2vH, Anterior gradient-2 spliced
variant H; XBP1s, spliced X-box binding protein 1; BiP, Binding
immunoglobulin protein; GRP, Glucose-regulated protein; ATF6,
Activating transcription factor 6; eIF2, Eukaryotic initiation
factor 2; BiP, Binding immunoglobulin protein; GRP,
Glucose-regulated protein; NC, negative control. |
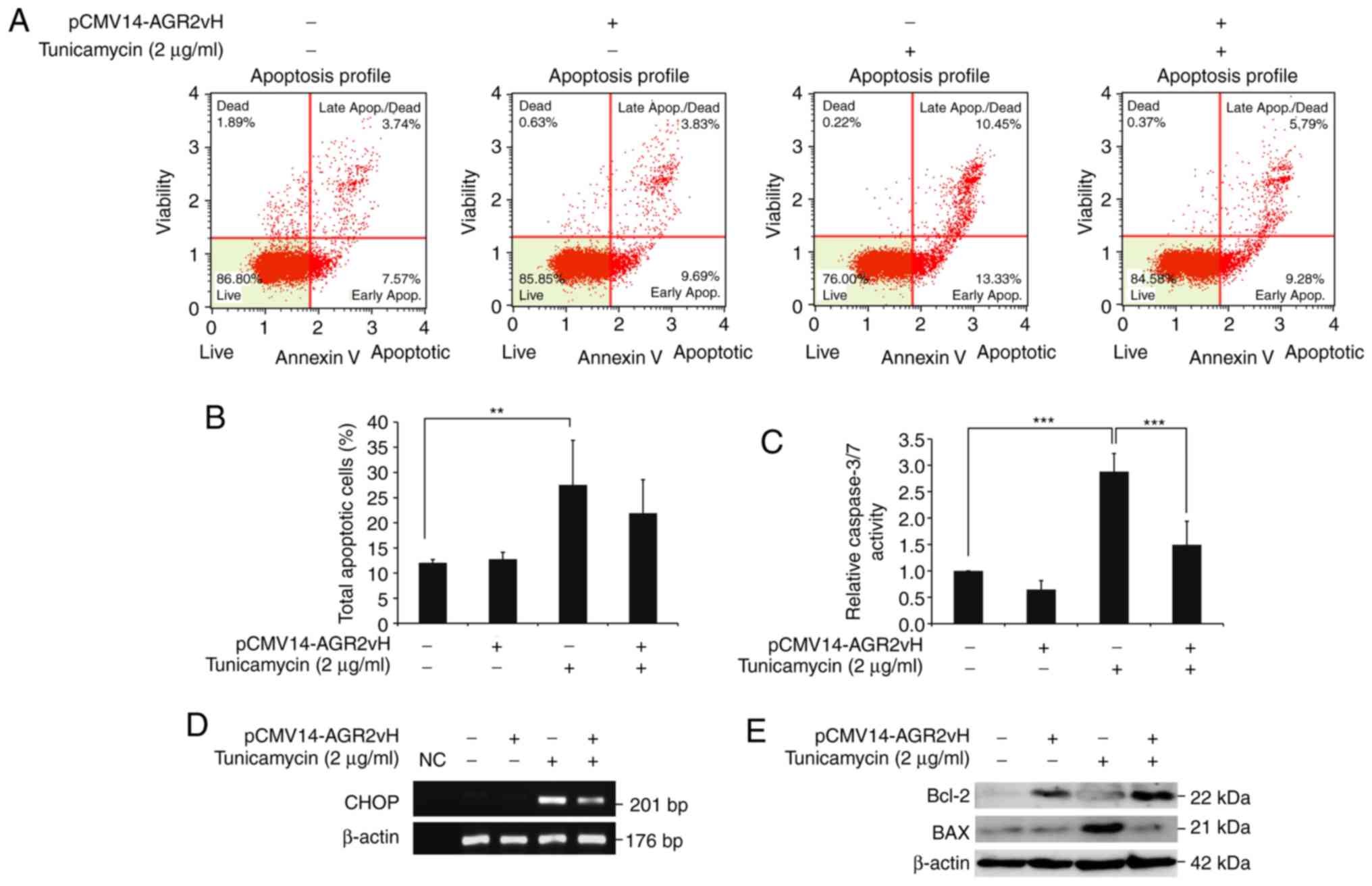
Effects of AGR2vH on cell apoptosis
The apoptosis of CCA cells was determined using flow
cytometry. Reduction of the apoptotic cell population, particularly
cells in late apoptosis, was observed in AGR2vH-overexpressing
cells under ER stress-inducing conditions when compared with empty
vector-transfected cells (Fig. 4A
and B). The results were also confirmed by caspase 3/7 activities,
which were significantly decreased in AGR2vH-overexpressing cells
under ER stress conditions when compared with empty
vector-transfected cells (P<0.001; Fig. 4C). In addition, the present study
observed the downregulation of CHOP mRNA, which is the ER
stress-induced apoptosis gene (Fig.
4D), and upregulation of the anti-apoptotic Bcl-2 protein and
downregulation of the apoptotic BAX protein (Fig. 4E), particularly when the
AGR2vH-overexpressing cell were under ER stress.
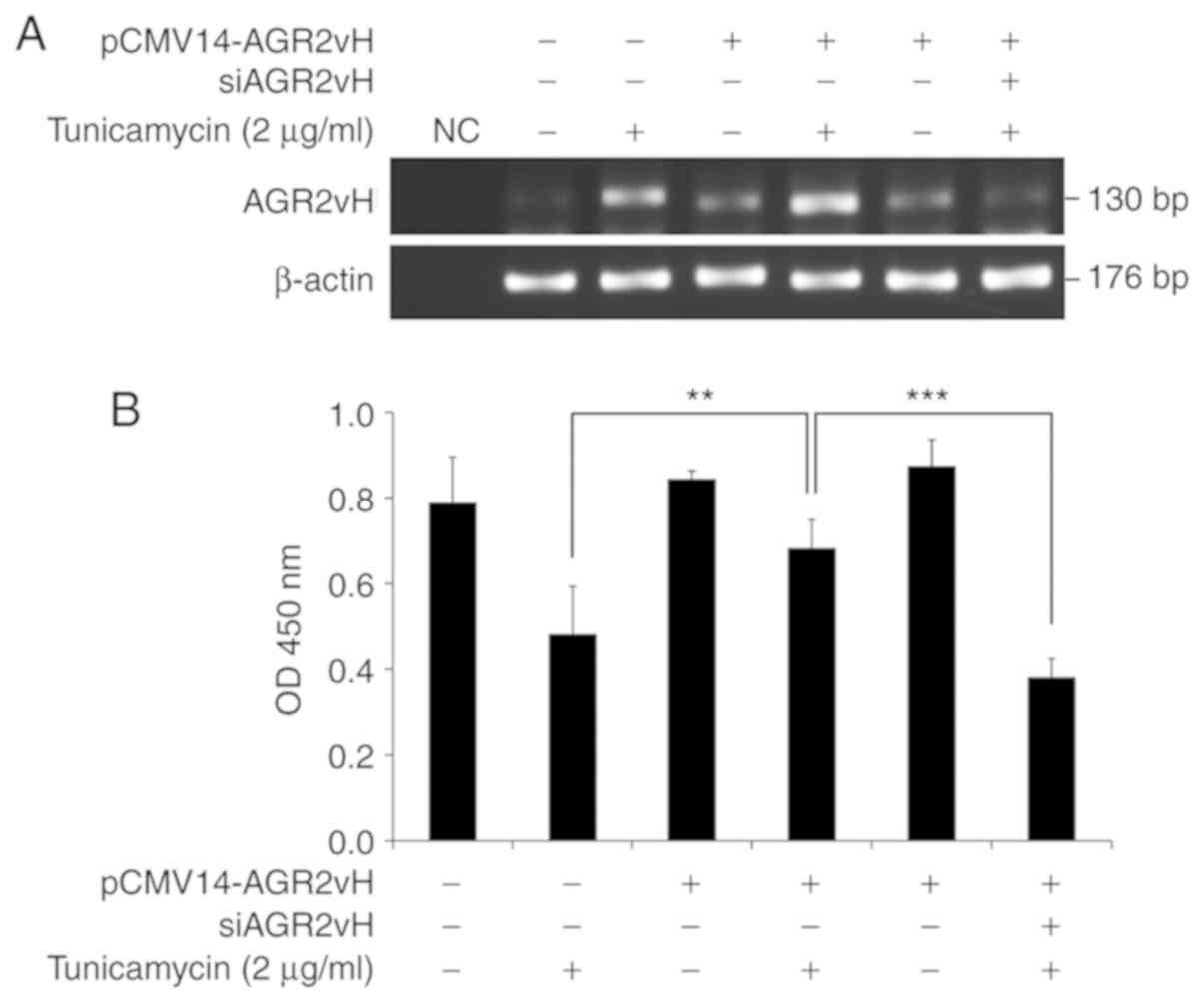
Effects of AGR2vH on cell survival
The mRNA expression of AGR2vH was upregulated in
AGR2vH-overexpressing cells under ER stress-inducing conditions,
and downregulated in AGR2vH-overexpressing cells with AGR2vH
depletion (Fig. 5A). The
expression of AGR2vH was directly associated with the survival of
CCA cells, which was determined using the CCK-8 assay. Cell
survival was significantly increased with AGR2vH overexpression in
cells under ER stress-inducing condition when compared with empty
vector-transfected cells (P<0.01), while the cell viability of
AGR2vH-overexpressing cells with AGR2vH depletion under ER
stress-inducing conditions was significantly decreased when
compared with AGR2vH-overexpressing cells (P<0.001; Fig. 5B).
Discussion
AGR2vH, produced from the aberrant splicing of AGR2,
promotes the metastatic phenotype of CCA cells. Of note, only
AGR2vH is predicted to be translatable into a 67-amino acid protein
isoform (17). In addition,
AGR2vH was revealed to contribute to the migration and invasion of
CCA cells (17,18). The dimerization of AGR2 is
required to activate the UPR pathway by interaction with BiP/GRP78
for the recovery of cellular ER stress and the increase of cancer
cell survival (10).
Prospectively, AGR2vH may serve as an alternative partner molecule,
which may readily interact with BiP/GRP78 to activate the UPR
pathway when ER stress occurs in cancer cells.
For the verification of ER stress, the BiP/GRP78
protein chaperone, which activates the UPR pathway, was
upregulated. In addition, XBP1 was spliced, which removes a
26-nucleotide intron from XBP1 mRNA, into XBP1s, resulting in the
increased expression of the chaperon proteins (29). In the present study, BiP/GRP78 and
XBP1s were upregulated. In addition, AGR2 was demonstrated to be
upregulated under ER stress conditions to facilitate protein
folding in the cells (30). In
the present study, AGR2vH expression was induced.
The activation of the UPR pathway under ER stress
conditions is based on three ER transmembrane receptors, namely
IRE1, ATF6 and PERK (31). The
present study investigated the expression of unspliced
(XBP1u)/XBP1s downstream of IRE1. AGR2vH-overexpressing cells
exhibited XBP1u downregulation and XBP1s upregulation, which
induces the expression of genes involved in restoring protein
folding, including BiP/GRP78 and PDIs (12). A previous study demonstrated that
the expression of eIF2a, a downstream target of PERK, was
upregulated following tunicamycin treatment (22). The present study reported that
eIF2a mRNA and activated p-eIF2a protein were upregulated under
conditions of ER stress by 2 µg/ml tunicamycin, and were
upregulated in AGR2vH-overexpressing cells. Furthermore, in the
present study, the expression of GPR94, a downstream target of ATF6
(32), was upregulated in
association with the increased expression of ATF6.
In addition, the expression of CHOP, a molecule
involved in ER stress-induced apoptosis, is low under non-stress
conditions but increases under ER stress conditions (33). In the present study, the
expression of CHOP in CCA cells was upregulated under ER
stress-inducing conditions, but was downregulated in
AGR2vH-overexpressing cells. Of note, the present study also
demonstrated changes in the expression of anti-apoptotic and
apoptotic proteins, and revealed that the expression of Bcl-2 and
BAX were associated with the survivability of CCA cells with AGR2vH
overexpression and knockdown.
In conclusion, the upregulation of AGR2vH activated
the UPR pathway and the expression of UPR downstream markers,
decreasing cell apoptosis via decreased caspase-3/7 activity and
contributing to the survival of CCA cells, particularly under ER
stress-inducing conditions. These results support the potential
application of this molecule as an alternative therapeutic target
for CCA.
Funding
The present study was supported by The Naresuan
University Research Scholarship for Graduate Students, the Thailand
Research Fund and Medical Research Council-UK (Newton Fund; grant
no. DBG5980004) and the Thailand Research Fund and Office of the
Higher Education Commission (grant no. TRF-MRG6080014).
Availability of data and materials
The datasets used and/or analyze during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
WK and SP designed the experiments. GS and JY
performed the experiments. GS and WK contributed to data
acquisition and wrote the manuscript. SW contributed to project
design and edited the manuscript. SJ made substantial contributions
to conception and design, and revised the manuscript for important
intellectual content. All authors have read and approved the
manuscript, and agree to be accountable for all aspects of the
research in ensuring that the accuracy and integrity of any part of
the work are appropriately investigated and resolved.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Abbreviations:
|
AGR2
|
anterior gradient-2
|
|
AGR2vH
|
anterior gradient-2 spliced variant
H
|
|
ATF6
|
activating transcription factor 6
|
|
BAX
|
Bcl-2-associated X protein
|
|
Bcl-2
|
B-cell leukemia/lymphoma 2
|
|
BiP/GRP78
|
binding immunoglobulin
protein/glucose-regulated protein 78
|
|
CCA
|
cholangiocarcinoma
|
|
CHOP
|
C/EBP homologous protein
|
|
eIF2
|
Eukaryotic initiation factor 2
|
|
GPR94
|
glucose-regulated protein 94
|
|
IRE1
|
inositol-requiring enzyme 1
|
|
PERK
|
protein kinase RNA-like endoplasmic
reticulum kinase
|
|
XBP1
|
X-box binding protein
|
Acknowledgments
Not applicable.
References
|
1
|
Shin HR, Oh JK, Masuyer E, Curado MP,
Bouvard V, Fang YY, Wiangnon S, Sripa B and Hong ST: Epidemiology
of cholangio-carcinoma: An update focusing on risk factors. Cancer
Sci. 101:579–585. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Sripa B and Pairojkul C:
Cholangiocarcinoma: Lessons from thailand. Curr Opin Gastroenterol.
24:349–356. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Sripa B, Bethony JM, Sithithaworn P,
Kaewkes S, Mairiang E, Loukas A, Mulvenna J, Laha T, Hotez PJ and
Brindley PJ: Opisthorchiasis and opisthorchis-associated
cholangiocarcinoma in Thailand and Laos. Acta Trop. 120(Suppl 1):
S158–S168. 2011. View Article : Google Scholar
|
|
4
|
Uthaisar K, Vaeteewoottacharn K, Seubwai
W, Talabnin C, Sawanyawisuth K, Obchoei S, Kraiklang R, Okada S and
Wongkham S: Establishment and characterization of a novel human
cholangiocarcinoma cell line with high metastatic activity. Oncol
Rep. 36:1435–1446. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Aberger F, Weidinger G, Grunz H and
Richter K: Anterior specification of embryonic ectoderm: The role
of the Xenopus cement gland-specific gene XAG-2. Mech Dev.
72:115–130. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Petek E, Windpassinger C, Egger H, Kroisel
PM and Wagner K: Localization of the human anterior gradient-2 gene
(AGR2) to chromosome band 7p21.3 by radiation hybrid mapping and
fluorescence in situ hybridisation. Cytogenet Cell Genet.
89:141–142. 2000. View Article : Google Scholar
|
|
7
|
Obacz J, Takacova M, Brychtova V, Dobes P,
Pastorekova S, Vojtesek B and Hrstka R: The role of AGR2 and AGR3
in cancer: Similar but not identical. Eur J Cell Biol. 94:139–147.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Higa A, Mulot A, Delom F, Bouchecareilh M,
Nguyên DT, Boismenu D, Wise MJ and Chevet E: Role of pro-oncogenic
protein disulfide isomerase (PDI) family member anterior gradient 2
(AGR2) in the control of endoplasmic reticulum homeostasis. J Biol
Chem. 286:44855–44868. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ramachandran V, Arumugam T, Wang H and
Logsdon CD: Anterior gradient 2 is expressed and secreted during
the development of pancreatic cancer and promotes cancer cell
survival. Cancer Res. 68:7811–7818. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ryu J, Park SG, Lee PY, Cho S, Lee DH, Kim
GH, Kim JH and Park BC: Dimerization of pro-oncogenic protein
anterior gradient 2 is required for the interaction with BiP/GRP78.
Biochem Biophys Res Commun. 430:610–615. 2013. View Article : Google Scholar
|
|
11
|
Oslowski CM and Urano F: Measuring ER
stress and the unfolded protein response using mammalian tissue
culture system. Methods Enzymol. 490:71–92. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Suh DH, Kim MK, Kim HS, Chung HH and Song
YS: Unfolded protein response to autophagy as a promising druggable
target for anticancer therapy. Ann N Y Acad Sci. 1271:20–32. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Eizirik DL, Miani M and Cardozo AK:
Signalling danger: Endoplasmic reticulum stress and the unfolded
protein response in pancreatic islet inflammation. Diabetologia.
56:234–241. 2013. View Article : Google Scholar
|
|
14
|
Zhu G and Lee AS: Role of the unfolded
protein response, GRP78 and GRP94 in organ homeostasis. J Cell
Physiol. 230:1413–1420. 2015. View Article : Google Scholar
|
|
15
|
Harding HP, Novoa I, Zhang Y, Zeng H, Wek
R, Schapira M and Ron D: Regulated translation initiation controls
stress-induced gene expression in mammalian cells. Mol Cell.
6:1099–1108. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Neeb A, Hefele S, Bormann S, Parson W,
Adams F, Wolf P, Miernik A, Schoenthaler M, Kroenig M, Wilhelm K,
et al: Splice variant transcripts of the anterior gradient 2 gene
as a marker of prostate cancer. Oncotarget. 5:8681–8689. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Yosudjai J, Inpad C, Chomwong S, Dana P,
Sawanyawisuth K, Phimsen S, Wongkham S, Jirawatnotai S and Kaewkong
W: An aberrantly spliced isoform of anterior gradient-2, AGR2vH
promotes migration and invasion of cholangiocarcinoma cell. Biomed
Pharmacother. 107:109–116. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Yosudjai J, Wongkham S, Jirawatnotai S and
Kaewkong W: Aberrant mRNA splicing generates oncogenic RNA isoforms
and contributes to the development and progression of
cholan-giocarcinoma. Biomed Rep. 10:147–155. 2019.PubMed/NCBI
|
|
19
|
Thamrongwaranggoon U, Seubwai W, Phoomak
C, Sangkhamanonz S, Cha'on U, Boonmars T and Wongkham S: Targeting
hexokinase II as a possible therapy for cholangiocar-cinoma.
Biochem Biophys Res Commun. 484:409–415. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Nami B, Donmez H and Kocak N:
Tunicamycin-induced endoplasmic reticulum stress reduces in vitro
subpopulation and invasion of CD44+/CD24- phenotype breast cancer
stem cells. Exp Toxicol Pathol. 68:419–426. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Li Q, Liu Z, Guo J, Chen J, Yang P, Tian
J, Sun J, Zong Y and Qu S: Cholesterol overloading leads to hepatic
L02 cell damage through activation of the unfolded protein
response. Int J Mol Med. 24:459–464. 2009.PubMed/NCBI
|
|
22
|
Dioufa N, Kassi E, Papavassiliou AG and
Kiaris H: Atypical induction of the unfolded protein response by
mifepristone. Endocrine. 38:167–173. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Van Schadewijk A, van't Wout EF, Stolk J
and Hiemstra PS: A quantitative method for detection of spliced
X-box binding protein-1 (XBP1) mRNA as a measure of endoplasmic
reticulum (ER) stress. Cell Stress Chaperones. 17:275–279. 2012.
View Article : Google Scholar :
|
|
24
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(Delta Delta C (T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
25
|
Khan A, Gillis K, Clor J and Tyagarajan K:
Simplified evaluation of apoptosis using the muse cell analyzer.
Postepy Biochem. 58:492–496. 2012.PubMed/NCBI
|
|
26
|
Sačková V, Kuliková L, Kello M, Uhrinová I
and Fedoročko P: Enhanced antiproliferative and apoptotic response
of HT-29 adenocarcinoma cells to combination of photoactivated
hypericin and farnesyltransferase inhibitor manumycin A. Int J Mol
Sci. 12:8388–8405. 2011. View Article : Google Scholar
|
|
27
|
Yip NK and Ho WS: Berberine induces
apoptosis via the mitochondrial pathway in liver cancer cells.
Oncol Rep. 30:1107–1112. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Zheng YM, Shen JZ, Wang Y, Lu AX and Ho
WS: Anti-oxidant and anti-cancer activities of Angelica dahurica
extract via induction of apoptosis in colon cancer cells.
Phytomedicine. 23:1267–1274. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Wang M and Kaufman RJ: The impact of the
endoplasmic reticulum protein-folding environment on cancer
development. Nat Rev Cancer. 14:581–597. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Dumartin L, Alrawashdeh W, Trabulo SM,
Radon TP, Steiger K, Feakins RM, di Magliano MP, Heeschen C,
Esposito I, Lemoine NR, et al: ER stress protein AGR2 precedes and
is involved in the regulation of pancreatic cancer initiation.
Oncogene. 36:3094–3103. 2017. View Article : Google Scholar :
|
|
31
|
Ron D and Walter P: Signal integration in
the endoplasmic reticulum unfolded protein response. Nat Rev Mol
Cell Biol. 8:519–529. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Yamamoto K, Sato T, Matsui T, Sato M,
Okada T, Yoshida H, Harada A and Mori K: Transcriptional induction
of mammalian ER quality control proteins is mediated by single or
combined action of ATF6alpha and XBP1. Dev Cell. 13:365–376. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Nishitoh H: CHOP is a multifunctional
transcription factor in the ER stress response. J Biochem.
151:217–219. 2012. View Article : Google Scholar : PubMed/NCBI
|