Introduction
Non-alcoholic fatty liver disease (NAFLD) is a
progressive chronic liver disorder that is a global public health
problem. The disease has exhibited increased prevalence due to the
increasing rates of obesity and diabetes. NAFLD increases the risk
of developing nonalcoholic steatohepatitis, liver fibrosis and
liver cancer, due to the accelerated rates of inflammation and
hepatocyte injury (1,2). In China, sedentary lifestyles and
excessive dietary fat intake are becoming the leading causes of
obesity, metabolic syndrome and NAFLD (3). Oxidative stress has been proposed to
be the primary mechanism of hepatocellular damage (4). Intracellular oxidative damage to
cellular macromolecules is based on the disruption of the
homeostasis between oxidative and antioxidative chemical species
(5). This condition results from
the release of toxic substances and inflammatory cytokines and
contributes to the induction of hepatocyte apoptosis (4).
CYP4A is a member of the CYP4 enzyme family and is
involved in the metabolism of medium- and long-chain fatty acids,
such as arachidonic acid, palmitate, laurate and compounds with
highly regionally selective hydroxylated terminal ω-carbon
(6). CYP4A is considered to serve
a crucial role in the pathogenesis of NAFLD (7). CYP4A exhibits different pattern of
metabolism across different species. Human CYP4A11 and CYP4A22, and
mouse CYP4A10, CYP4A12 and CYP4A14 are responsible for the
ω-hydroxylation of fatty acids (8). CYP4A11 primarily metabolizes
endobiotics and catalyzes the ω-hydroxylation of fatty acids in
human liver and kidney tissues. CYP4A11 is a functionally active
member of the human CYP4A family of enzymes (9).
In patients with NAFLD, triglyceride (TG) synthesis
was identified to be increased compared with that of the TG export
from the liver cells (10). Liver
free fatty acids (FFAs) that are derived from circulating
non-esterified fatty acids (NEFAs) are stored as TGs. NEFAs are
released from adipose tissues, hepatic de novo lipogenesis
and the remnants of the TG rich lipoproteins, namely
very-low-density lipoproteins (VLDL) and chylomicrons (11). These FFAs enter the mitochondria
and undergo β-oxidation in order to produce ATP equivalents.
Alternatively, they are esterified to triglycerides (TGs) and
subsequently expelled from the hepatocytes as VLDLs (12). During the mitochondrial
dysfunction caused by NAFLD, the CYP4-mediated ω-hydroxylation of
fatty acids is markedly increased (13). This process results in the
excessive production of reactive oxygen species (ROS) (13). Cytochrome P450 (CYP) enzymes
primarily depend on NADPH to produce superoxide and hydrogen
peroxide (14). ROS serve a
central role in activating NF-κB. In addition, oxidative stress
increases the release of pro-inflammatory cytokines and is
associated with activation of the NF-κB signaling pathway (15,16). Therefore, ROS production by
CYP4A11 metabolism of fatty acids may cause inflammatory
reactions.
In the present study, the CYP4A11 protein content
was determined in the plasma of patients with NAFLD. In addition,
the study aimed to investigate the regulation of oxidative stress
and lipid peroxidation (LPO) by CYP4A11 in a cell line mode of
NAFLD. These interactions were examined by inducing and inhibiting
CYP4A expression in a cell line model, in order to affect the
production of ROS.
Materials and methods
Patients and study design
In the present study, 59 patients with NAFLD and 30
normal healthy subjects were enrolled between March 2019 and April
2019. The study protocol was approved by The First Affiliated
Hospital of the Anhui Medical University Institutional Review
Board. Blood samples were collected at The First Affiliated
Hospital of the Anhui Medical University following provision of
informed consent for scientific research from the patients on March
4, 2019. The criteria for patient selection were as described
previously (17): i) No history
of alcohol consumption, or alcohol intake per week was <140 g in
men and <70 g in women; ii) no presence of specific diseases
that may result in fatty liver, such as viral hepatitis,
drug-induced liver disease, total parenteral nutrition and Wilson's
disease; iii) besides clinical manifestations of the primary
disease, other non-specific symptoms and signs, such as fatigue,
dyspepsia, dull liver pain and hepatosplenomegaly occur; iv)
symptoms of metabolic syndromes, such as overweight and visceral
obesity, hyperglycemia, blood lipid disorder and hypertension
occur; v) mild to moderate increases in serum levels of
transaminase and γ-glutamyl transpeptidase (<5-fold the upper
normal limit), usually presenting as an increase of alanine
aminotransferase (ALT); vi) the results of the liver imaging
studies met the imaging diagnostic criteria of diffuse fatty liver.
In addition, laboratory tests were performed, including Color
Doppler and blood biochemistry. The patients with liver disease
selected did not suffer from kidney disease.
Biochemical analysis
After blood samples were centrifuged at 1,006 × g at
4°C for 15 min, the serum concentrations of TG, alanine
aminotransferase (ALT) and aspartate amino-transferase (AST) were
measured separately using TG (cat. no. A110-1-1), ALT (cat. no.
C009-1-1), and AST commercial analysis kits (cat. no. C010-1-1; all
Nanjing Jiancheng Bioengineering Institute Co., Ltd.).
Analysis of plasma levels of lipid
peroxidation products (LPO) and of CYP4A11 expression
Laboratory investigations were performed using
plasma samples. Following centrifugation at 1,341 × g at 4°C for 20
min, the plasma supernatant was collected and stored at -20°C. LPO
and CYP4A11 levels were quantified in plasma by the human LPO ELISA
kit (cat. no. JL12392; Shanghai Jianglai Biological Technology Co.,
Ltd.) and the human CYP4A11 ELISA kit (cat. no. YX-032516H;
Shanghai Preferred Biotechnology Co., Ltd.), respectively following
a 5-fold dilution, according to the manufacturer's protocol.
FFA-induced steatosis
The liver cancer HepG2 cell line was purchased from
Shanghai GeneChem Co., Ltd., and was seeded in a 6-well plate. A
total of 2.5×105 cells were plated and maintained in
normal growth conditions for 24 h. The cells were then treated with
1 mM FFA for an additional 24 h. The FFA solution used in the
present study was a mixture of oleic acid (Sigma-Aldrich; Merck
KGaA) and palmitic acid (Sigma-Aldrich; Merck KGaA) at a specific
concentration ratio (2:1) as previously described (18). The expression of CYP4A11 was
regulated by simultaneous treatment of the cells with 100 mM
clofibrate (Dalian Meilun Biotechnology Co., Ltd.) and FFA
solution. HepG2 cells were incubated with 1 µM HET0016
(Cayman Chemical Company) for 1 h at 37°C prior to the addition of
FFA.
Oil Red O staining
After incubation of HepG2 cells with FFA solution
for 24 h, the cells were washed gently with PBS and fixed with 10%
neutral formaldehyde for 30 min at room temperature. The cells were
then stained with freshly diluted Oil Red storage solution (3:2
ratio of Oil Red:deionized water) at room temperature for 10 min.
After the Oil Red O working solution was removed, the cells were
washed with 60% isopropanol for 5 sec. Finally, the cells were
observed under a fluorescent inverted microscope (Olympus
Corporation).
Cell transfection
The small interfering RNA (siRNA) oligo-nucleotides
against the CYP4A11 gene and the plasmids for CYP4A11
overexpression were both designed and synthesized by Shanghai
Sangon Biotech Corporation. The siRNA sequences were as follows:
Human CYP4A11-siRNA sense, 5′-CCG UGU GAG GAA UGC CUU UTT-3′; and
antisense: 5′-AAA GGC AUU CCU CAC ACG GTT-3′; negative control (NC)
sense: 5′-UUC UCC GAA CGU GUC ACG UTT-3′; and anti-sense: 5′-ACG
UGA CAC GUU CGG AGA ATT-3′. HepG2 cells were seeded at a density of
80-90% and were transfected with 20 µM CYP4A11 small
interfering RNA (siRNA) or 20 µM CYP4A11 plasmid using
Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific, Inc.)
according to the manufacturer's protocol. Following 6 h, the
culture medium was changed and the FFA (1 mM) solution was added to
the cells for 24 h.
Western blot analysis
HepG2 cells were lysed with RIPA lysis buffer
(Beyotime Institute of Biotechnology) containing 1 mM
phenylmethylsulfonyl fluoride. The upper supernatant was collected
following centrifugation at 4,024 × g at 4°C for 30 min. The
supernatant was quantitatively administered using a BCA protein
kit. The protein (10 µl/lane) was separated by 10% SDS-PAGE.
The proteins were electrotransferred to PVDF membranes (EMD
Millipore). The membranes were washed in TBS-0.5% Tween-20 buffer
following blocking with 5% fat free milk powder for 2 h at room
temperature and incubated with the specific primary antibodies at
4°C overnight. Horseradish peroxidase-conjugated goat anti-rabbit
IgG or anti-mouse IgG antibodies(1:5,000, cat. nos. ZB-2301 or
ZB-2305; Beijing Zhongshan Golden Bridge Biotechnology Co., Ltd.;
OriGene Technologies, Inc.). were used for 1 h at room temperature
for signal detection. The membrane was developed with enhanced
chemiluminescence (Thermo Fisher Scientific, Inc.). Quantitative
densitometric analyses of western blot gel images were performed
using Image J 1.45s software (National Institutes of Health). The
following primary antibodies were used: CYP4A11 (1:1,000; cat. no.
DF4749; Affinity Biosciences), p65 (1:500; cat. no. 21014;
Signalway Antibody LLC), phosphorylated (p)-p65 (1:500, 11014;
Signalway Antibody LLC), and β-actin (1:1,000; cat. no. TA-09;
Beijing Zhongshan Golden Bridge Biotechnology Co., Ltd.; OriGene
Technologies, Inc.).
Measurement of oxidative stress
activity
ROS levels were measured using a ROS kit (Shanghai
Beibo Biotechnology Co., Ltd.) according to the manufacturer's
protocol. The fluorescent probe 2′,7′-dichlorofluorescein (DCF)
diacetate was added to HepG2 cells at 10 µM and the cells
were collected following incubation for 20 min at 37°C in the dark.
The fluorescence intensity of DCF was excited at 488 nm and the
emission was detected at 525 nm using an inverted fluorescence
microscope (magnification ×10). Fluorescence intensity was
quantified using Image J.
Measurement of malondialdehyde (MDA) and
superoxide dismutase (SOD) levels
Following cell collection, the HepG2 cells were
lysed and centrifuged at 4,024 × g for 30 min at 4°C to obtain the
supernatant samples. The levels of MDA were determined using an MDA
assay kit (Beyotime Institute of Biotechnology) and SOD was
measured using a SOD assay kit (Beyotime Institute of
Biotechnology) according to the manufacturer's protocol. The
protein concentration levels in the lysates were determined using a
BCA protein assay kit (Beyotime Institute of Biotechnology). The
results from the MDA and SOD assays are presented as nmol/mg and
U/mg protein, respectively.
Determination of mRNA levels using
reverse transcription-quantitative polymerase chain reaction
(RT-qPCR)
Total RNA was extracted from cultured HepG2 cells
using the TRIzol reagent (Invitrogen; Thermo Fisher Scientific,
Inc.). The RNA was eluted with RNasefree water and the
concentration was measured by UV detection at 260 nm. Subsequently,
cDNA was synthesized using the Transcriptor First-Strand cDNA
Synthesis kit (Takara Bio, Inc.). qPCR was performed with
SYBR-Green Master Mix (Takara Bio, Inc.). The thermocycling
conditions were as follows: Initial denaturation at 95°C for 30
sec, followed by 40 cycles of 95°C for 10 sec and 60°C for 30 sec.
The RT-qPCR primers were purchased from Sangon Biotech Co., Ltd.
The fold-change for mRNA relative to β-actin was determined using
the 2-∆∆Cq method (19). The specific primer sequences were
as follows: CYP4A11 (human) forward, 5′-ACG GCT TGC TCC TGT
TGA AT GG-3′; CYP4A11 reverse, 5′-AGA GGT CAG GCT GTA GAT
GGT GTC-3′; IL-6 (human) forward, 5′-CAC ACA GAC AGC CAC TCA
CC-3′; IL-6 reverse, 5′-AGT GCC TCT TTG CTG CTT TC-3′;
TNF-α (human) forward, 5′-AAC CTC CTC TCT GCC ATC AA-3′;
TNF-α reverse, 5′-CTG AGT CGG TCA CCC TTC TC-3′;
IL-1β forward, 5′-GGA CAA GCT GAG GAA GAT GC-3′;
IL-1β reverse, 5′-TCG TTA TCC CAT GTG TCG AA-3′;
β-actin forward, 5′-TTG CTG ACA GGA TGC AGA A-3′; and
β-actin reverse, 5′-ACC AAT CCA CAC AGA GTA CTT-3′.
Statistical analysis
The data are expressed as the mean ± standard
deviation. Pearson correlation analysis was used to analyze
correlation between variables. A Student's t-test, or one-way
analysis of variance followed by Tukey post hoc test were used to
compare the differences among the various groups. P<0.05 was
considered to indicate a statistically significant difference. All
statistical analyses were performed with the SPSS v.16.0 software
(SPSS, Inc.).
Results
Detection of LPO and CYP4A11 levels in
plasma samples
The clinical and biochemical characteristics of the
patients with NAFLD and of the control subjects involved in the
present study are presented in Table
I. A total of 77.5% of the participants were male, with an age
range of 24-91 years. No significant differences were noted in the
ALT levels between the patients with NAFLD and the control
subjects, whereas the AST levels were slightly increased in the
patients with NAFLD. However, this increase was not significant,
and the levels were within the normal range of reference values. In
contrast to these results, the mean BMI value of the patients with
NAFLD was significantly increased compared with that noted for the
healthy subjects, and this value exceeded the threshold of obesity
according to the body-mass index for Asian populations (20). Plasma VLDL and TG levels in the
patients with NAFLD were significantly increased compared with
those of the control subjects.
 | Table IClinical and biochemical features of
the patient cohort. |
Table I
Clinical and biochemical features of
the patient cohort.
| Parameters | Control group | NAFLD group | P-value |
|---|
| Sex
(male/female) | 25/5 | 44/15 | - |
| Age, years (range,
24-91) | 54.57±17.30 | 52.53±15.79 | 0.58 |
| ALT, U/l
(9-50)a | 25.37±13.81 | 30.56±16.47 | 0.09 |
| AST, U/l (15-40)a | 20.47±6.98 | 22.76±6.38 | 0.03b |
| BMI,
kg/m2 | 21.43±1.83 | 25.53±2.37 | 0.06 |
| TG, mmol/l
(0.56-1.70)a | 1.52±1.69 | 2.47±1.60 | 0.04b |
| VLDL, mmol/l
(0.21-0.60)a | 0.56±0.26 | 0.92±0.56 | 0.04b |
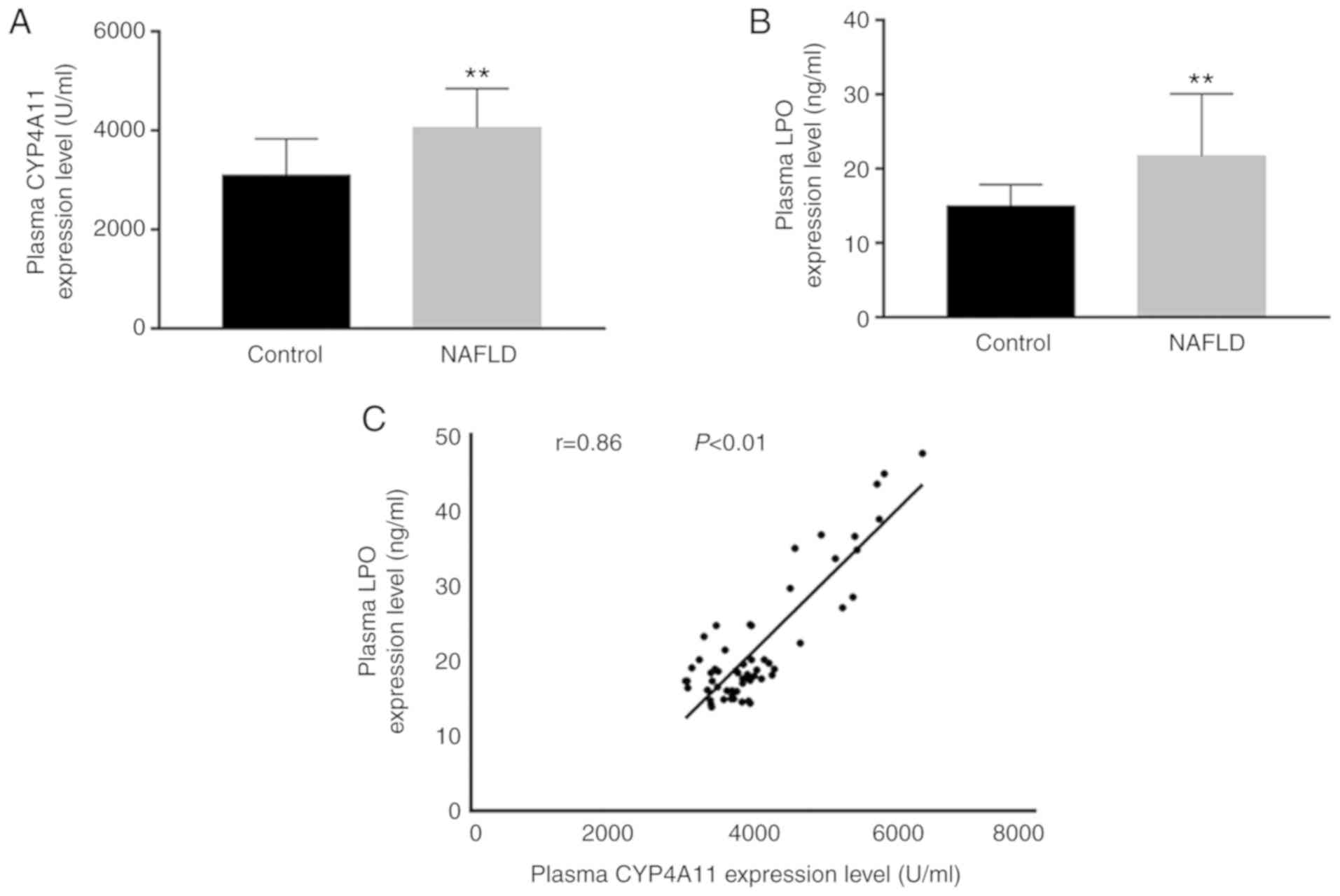
Plasma LPO levels were apparently elevated in the
patients with NAFLD compared with those observed in the control
subjects (Fig. 1). The plasma
levels of CYP4A11 in the patients with NAFLD were significantly
increased compared with those noted in control subjects. The levels
of LPO were increased in a similar trend as that observed in the
CYP4A11 levels in the patients with NAFLD. Further linear
regression mapping indicated that these two parameters were closely
associated (r=0.86).
Lipid accumulation in FFA-treated HepG2
cells
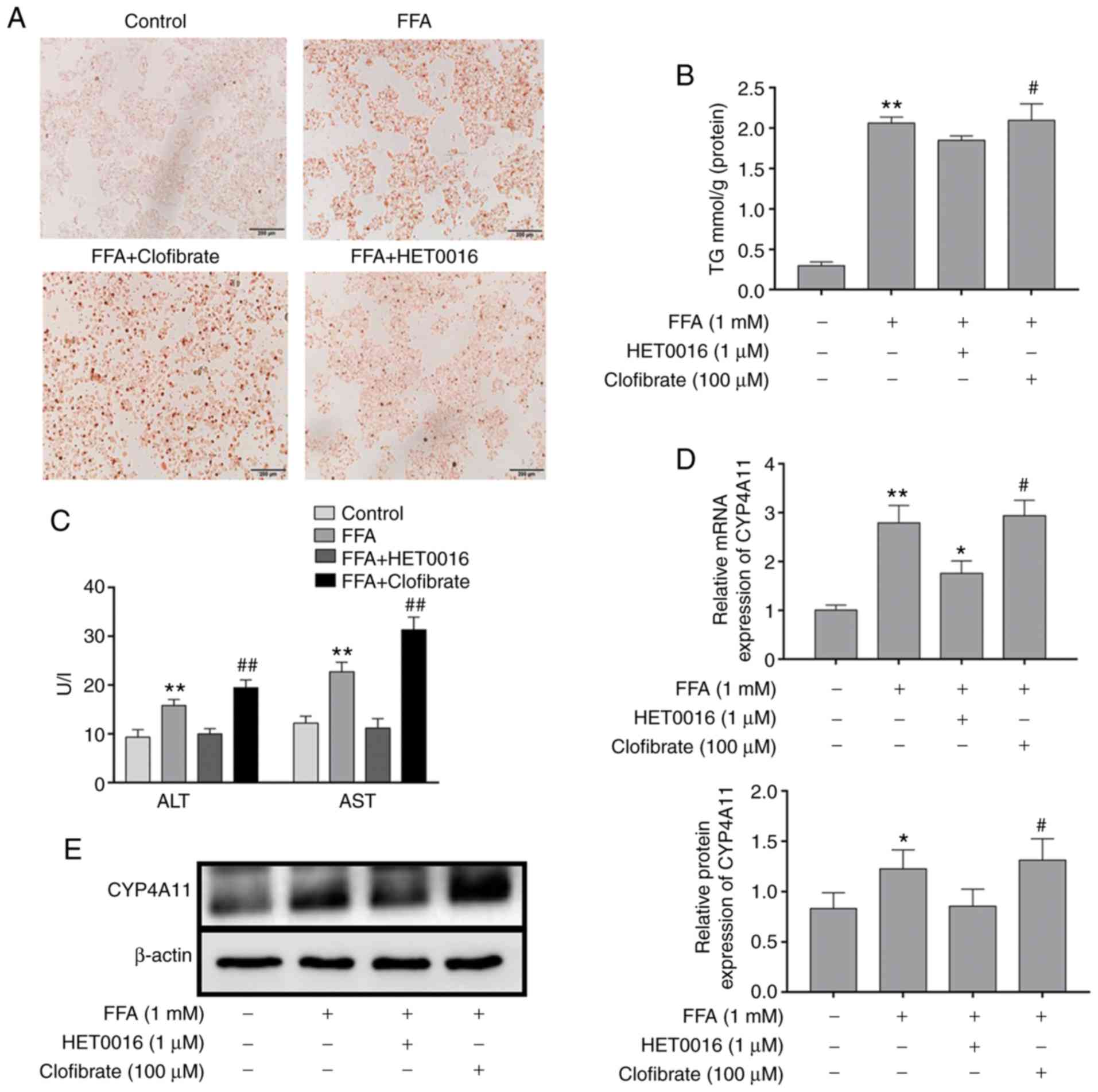
The effects of FFA treatment on lipid accumulation
were confirmed by Oil Red O staining (Fig. 2A). The results indicated that the
intracellular lipid deposition in the FFA-treated group was
considerably increased compared with that of the control group.
However, following the addition of HET0016 to the FFA-treated HepG2
cells, the level of lipid deposition was significantly decreased.
The highest degree of lipid deposition was observed in the presence
of clofibrate + FFA solution. The levels of the intracellular TGs
followed the same trend (Fig.
2B).
Effect of FFA on the transaminase levels
of HepG2 cells
Following FFA stimulation of the cells for 24 h, the
cell supernatant was collected for ALT and AST determination and
the results are shown in Fig. 2C.
The results demonstrated that the levels of ALT and AST in the
supernatant were increased in the FFA-treated HepG2 cells compared
with those noted in the normal control group. HET0016 treatment
decreased FFA-induced ALT and AST activity, whereas clofibrate
promoted the FFA-induced activity of ALT and AST.
Detection of CYP4A11 expression in
FFA-treated HepG2 cells
CYP4A11 levels were detected following FFA treatment
of HepG2 cells. The protein and mRNA expression levels of CYP4A11
were elevated in FFA-treated HepG2 cells compared with those of the
normal control group (Fig. 2D and
E). In addition, western blot analysis and RT-qPCR analysis
demonstrated that clofibrate treatment resulted in a significant
upregulation in the expression level of CYP4A11. In addition,
FFA-induced CYP4A11 expression in HepG2 cells was inhibited by
HET0016 treatment.
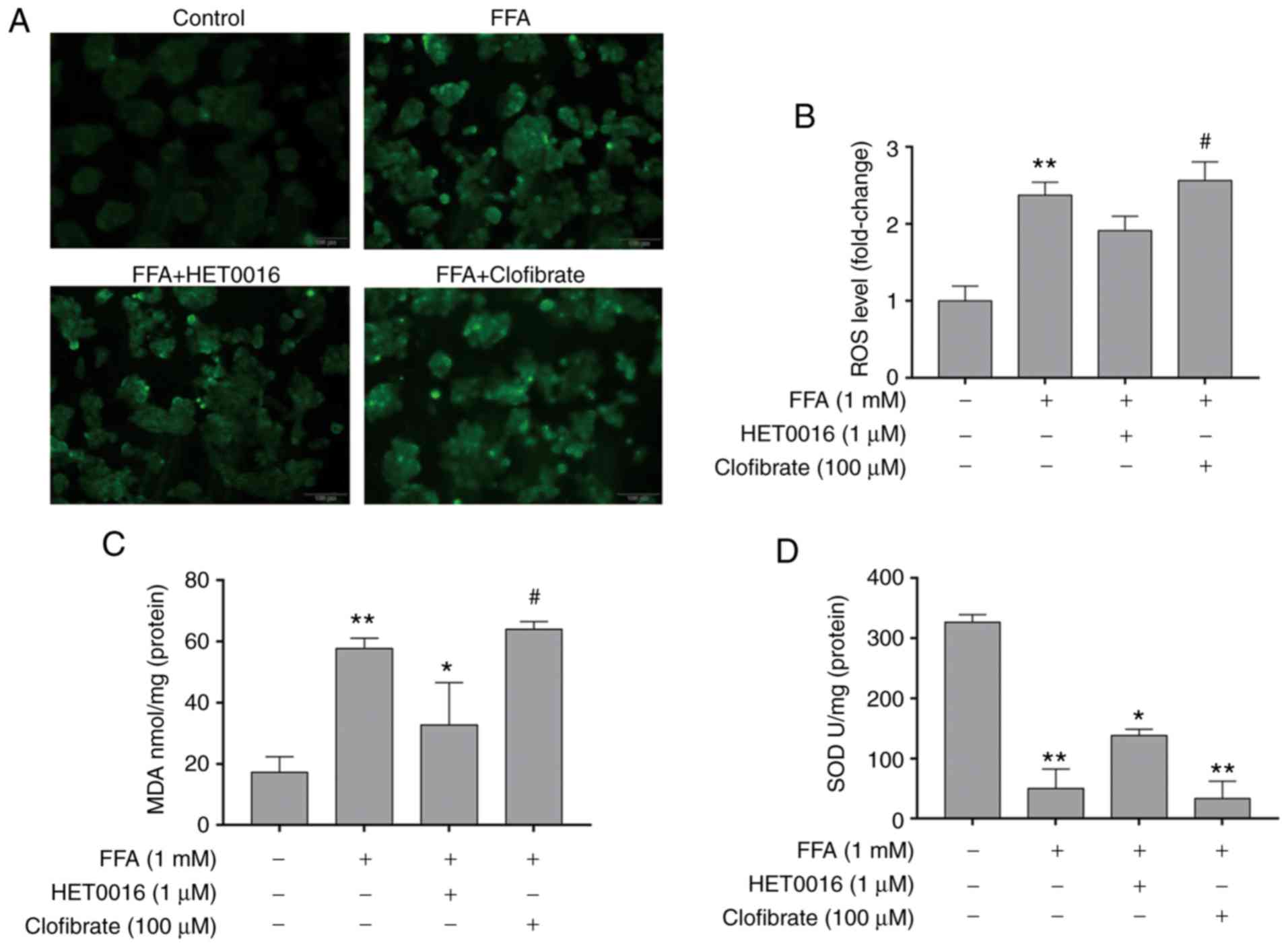
Association between CYP4A11 and oxidative
stress
Pre-treatment of the HepG2 cells with FFA for 24 h
increased the intracellular ROS content compared with that observed
in the control group. In addition, the levels of ROS were
signifi-cantly increased in the clofibrate group, as demonstrated
by fluorescence detection. Conversely, HET0016 significantly
inhibited the intracellular ROS production compared with that of
the clofibrate group (Fig. 3A and
B). The changes noted in the MDA levels were in accordance with
the trend noted for the ROS levels among the different treatment
groups. In contrast to the ROS and MDA levels, SOD levels were
decreased following an increase in ROS levels (Fig. 3C and D).
Effects of CYP4A11 on inflammatory
cytokine production in FFA-treated HepG2 cells
The expression levels of CYP4A11 were elevated
following the transfection of pcDNA3.1-CYP4A11 into the HepG2
cells, as determined by western blot analysis and RT-qPCR. The
levels of CYP4A11 in the pCDNA3.1-CYP4A11 group were significantly
increased compared with those of the model group (Fig. 4A and B). Moreover, the expression
levels of the inflammatory cytokines TNF-α, IL-6 and IL-1β were
examined by RT-qPCR. The expression levels of TNF-α, IL-6 and IL-1β
were significantly increased in the pCDNA3.1-CYP4A11 group
(Fig. 4E). The role of CYP4A11 on
the secretion of inflammatory cytokines was further confirmed by
transfection of siRNA-CYP4A11. The results demonstrated that the
mRNA and protein expression levels of CYP4A11 in the siRNA-CYP4A11
group were significantly decreased (Fig. 4C and D). With regard to
inflammatory cytokine secretion, RT-qPCR analysis demonstrated that
CYP4A11 gene silencing inhibited the expression levels of
TNF-α, IL-6 and IL-1β (Fig. 4F).
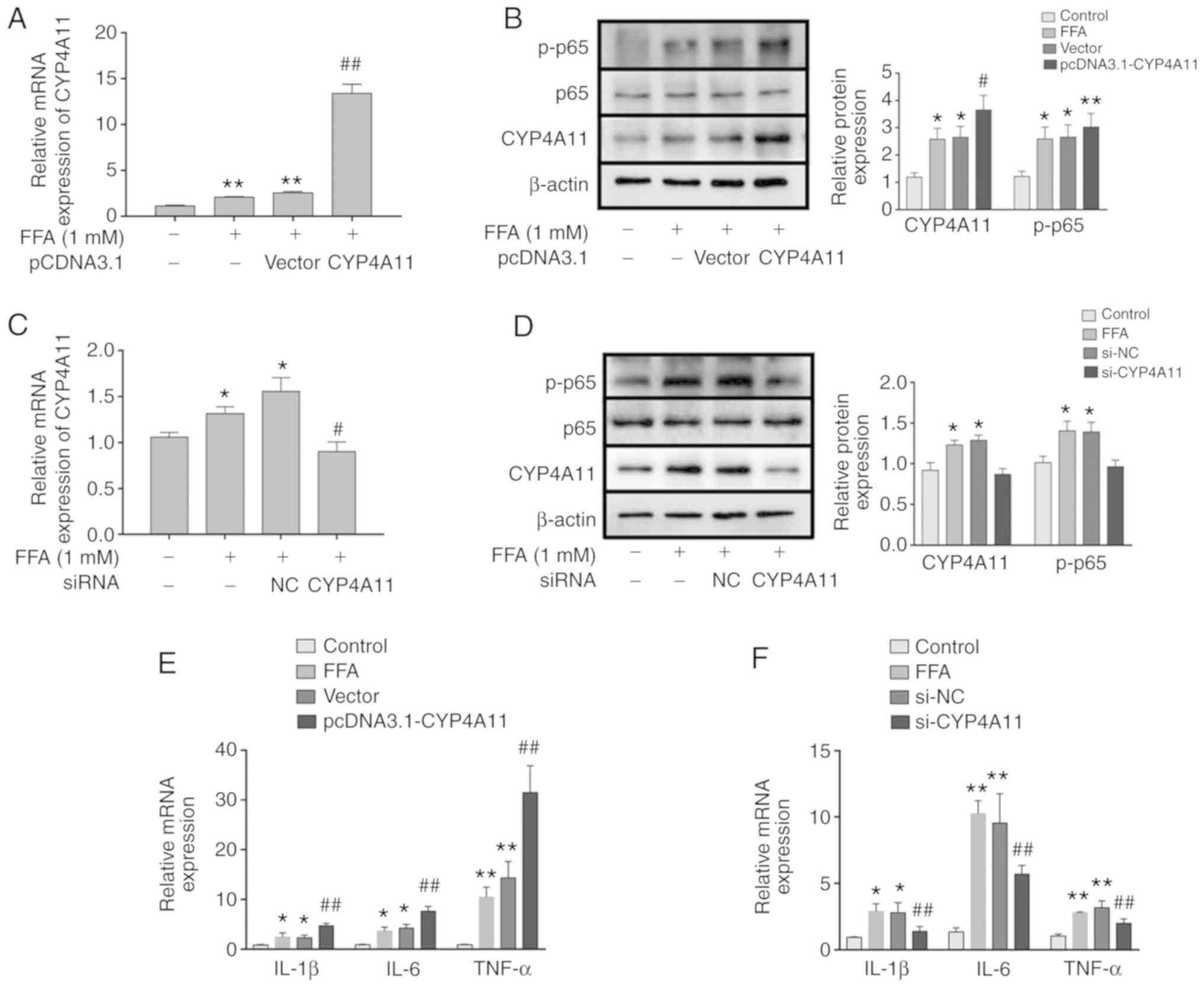 | Figure 4Effect of CYP4A11 gene on
phosphorylation of p65 protein and synthesis of TNF-α, IL-6 and
IL-1β. (A and C) The mRNA level of CYP4A11 was examined by RT-qPCR
in the (A) pCDNA3.1-transfected and (C) siRNA-transfected groups.
(B and D) The protein levels of CYP4A11 and p-p65 in (B)
pCDNA3.1-transfected and (D) siRNA-transfected groups were assessed
by western blot analysis. (E and F) IL-1β, IL-6 and TNF-α
expression levels were determined by RT-qPCR in the (E)
pCDNA3.1-transfected and (F) siRNA-transfected groups. The results
are presented as the mean ± standard deviation.
*P<0.05 and **P<0.01 vs. control,
#P<0.05 and ##P<0.01 vs. vector or NC
group. CYP4A11, cytochrome P450 4A11; p65, transcription factor
p65; TNF-α, tumor necrosis factor α; IL, interleukin; RT-qPCR,
reverse transcription-quantitative polymerase chain reaction;
siRNA, small interfering RNA; p-, phosphorylated; NC, negative
control. |
Effect of CYP4A11 on the NF-κB signaling
pathway in vitro
The expression levels of the primary protein
associated with the NF-κB signaling pathway were detected by
western blot analysis. The levels of p-p65 were apparently
increased following the overexpression of CYP4A11 compared with
those of the vector group (Fig.
4B). CYP4A11-siRNA inhibited the expression of p-p65 compared
with that of the siRNA-NC group (Fig.
4D).
Discussion
It has been demonstrated that CYP4A activity is
mediated via peroxisome proliferator-activated receptor α (PPARα)
activation and that it is involved in hepatic fatty acid disposal,
leading to an increase in the levels of ROS and LPO (21). A previous study considered CYP4A
as a potential target for the treatment of fatty liver disease
(22). CYP4A induces oxidative
stress by enhancing NADPH-dependent superoxide anion generation.
The primary sources of ROS production that can cause oxidative
stress damage to the cells are H2O2, singlet
oxygen and lipid hydroperoxides (23,24). Excessive levels of ROS are the
result of a disruption of the homeostasis between ROS production
and the antioxidant system (25).
Mitochondrial CYP2E1 is a direct source of ROS and it has also been
demonstrated to have a greater ability to generate ROS compared to
CYP3A4 and CYP1A1 (26,27). CYP2E1 and CYP4A are vital hepatic
microsomal oxidases associated with fatty acid oxidation and the
formation of ROS and NOS (24).
In addition, similar to CYP2E1, CYP4A is an alternative initiator
of oxidative stress and promotes LPO in non-alcoholic
steatohepatitis (NASH) (22).
Although CYP4A serves an important role in the regulation of
oxidative stress and LPO during a high-fat diet, it is not clear
whether it promotes the development of NAFLD. CYP4A11 is considered
to be an active ω-hydroxylase in the human liver and is a major
member of the human CYP4A family of enzymes. Another member of the
human CYP4A family is the enzyme CYP4A22, which is not considered
to serve a major role in the metabolism of fatty acids, as its
expression level is extremely low in the human liver (28). In the present study, the
functional role of CYP4A11 was primarily investigated.
It was initially demonstrated that the expression
levels of CYP4A11 were increased in the plasma of patients with
NAFLD, as determined by ELISA. Although this method did not
directly determine the amount of protein per liver tissue, it was
considered to be a rough estimation of the changes that occurred in
the expression of CYP4A11 in the liver of patients with NAFLD. This
is due to the limited applications of liver biopsy methods, which
are not commonly used to detect the presence of NAFLD. Therefore,
it is particularly difficult to obtain liver tissue. Plasma LPO
levels were measured by ELISA. The present study sought to identify
a potential association between CYP4A11 expression and LPO levels
in the plasma of patients with NAFLD. CYP4A is associated with
hepatic microsomal LPO and previous studies have reported that this
enzyme may be responsible for the increase of in vitro
microsomal LPO (14,21). The present data indicated that
hepatic CYP4A11 was significantly increased and that the plasma LPO
levels were upregulated in patients with NAFLD. The linear
regression between CYP4A11 expression and LPO levels indicated that
they were highly correlated. The present study suggested that
CYP4A11 was involved in LPO in patients with NAFLD.
The majority of the aforementioned studies involved
animal experiments. In order to further understand the mechanism of
the association between CYP4A11 expression and LPO levels in
vitro, cell-based assays were conducted in the present study.
An in vitro NAFLD model was established by pre-treating
HepG2 cells with FFAs, which was similar to certain previously
described in vivo NAFLD models (29). FFA is a suitable stimulus for
HepG2 cells as it is more steatogenic and causes less damage
compared with PA, thereby increasing intracellular TG accumulation
(18). In the present study, the
results demonstrated that the expression levels of CYP4A11 in HepG2
cells treated with FFA were significantly increased, corresponding
to the high CYP4A expression induced by high-fat diet or methionine
and choline-deficient diet in animal models (30,31). This is attributed to a large
amount of FFA entering the cell that can maximize β-oxidation in
the mitochondria, leading to the activation of the peroxisome
β-oxidation and the microsomal ω-oxidation pathways, which in turn
increases CYP4A11 expression (32). Clofibrate, a PPARα agonist,
increases the expression of CYP4A (33). The addition of clofibrate to FFA
will further increase the expression of CYP4A11. HET0016 is a known
CYP4 family inhibitor that selectively inhibits the formation of
20-HETE (34). In the present
study, the highest expression level of CYP4A11 protein was observed
following addition of clofibrate, whereas HET0016 treatment
decreased the expression levels of CYP4A11 to approximately the
same levels as those noted for the normal group. CYP4A enzymes have
been demonstrated to induce liver fat accumulation by increasing
the expression of platelet glycoprotein 4 (22). The present study demonstrated that
the increase in the expression levels of CYP4A11 increased the
synthesis of TG, while the inhibition of CYP4A11 expression
decreased the synthesis of TG. Therefore, CYP4A11 was closely
associated with the production of TG, affecting the lipid toxicity
of HepG2 cells and the increase in the levels of ALT and AST.
Elevated plasma FFA concentration leads to high
intracellular ROS production due to the metabolism of FFAs in the
mitochondria, peroxisomes and microsomes, which are considered the
primary sources of ROS that cause oxidative stress (15,35). The present study demonstrated that
clofibrate-induced CYP4A11 expression may increase ROS levels by
increasing FFA, while HET0016 attenuated ROS production by
inhibiting CYP4A11 expression. These data indicated that CYP4A11
may promote the production of ROS. ROS induces LPO of membranes
leading to cell necrosis or apop-tosis. If ROS scavengers are used,
we hypothesize that ROS scavengers will greatly decrease cytotoxic
damage, but they may have little effect on the CYP4A11 effect. MDA
is released as a result of LPO, which triggers a harmful
immunological response to liver cells (36). In the present study, the severity
of the antioxidant system disorders was reflected by the decrease
of SOD concentration and the increased deposition of MDA, which
were closely associated with the increase in ROS levels.
FFAs may be considered important mediators of
lipo-toxicity due to their potential cytotoxicity. In addition,
they can lead to excessive accumulation of lipids and further
stimulate TNF-α expression (37).
ROS production and the products of LPO have been identified as a
trigger for the production of proinflammatory cytokines, which in
turn leads to neutrophil chemotaxis and development of diverse
lesions in NASH subjects (38).
The present study demonstrated that the mRNA expression levels of
TNF-α, IL-6 and IL-1β were significantly
increased in the FFA-treated HepG2 cells. The expression levels of
CYP4A11 were controlled at the genetic level, in order to cause an
amplification of its effect on the expression of pro-inflammatory
cytokines. The expression levels of TNF-α, IL-6 and IL-1β were
increased following pCDNA3.1-CYP4A11 transfection into the cells.
In contrast to this model, knockdown of CYP4A11 was achieved by
siRNA transfection. The mechanism by which CYP4A11 regulates the
expression of pro-inflammatory cytokines may be due to ROS and
lipid peroxide production induced by CYP4A11. Excess ROS production
and release of pro-inflammatory cytokines can activate NF-Kb
(39,40). ROS levels can activate and inhibit
NF-κB signaling. The interaction between ROS and the NF-κB pathway
is complex (41). The present
study further investigated whether ROS induction by CYP4A11
affected the activation of the NF-κB signaling pathway. The data
confirmed that upregulation of CYP4A11 by pCDNA3.1-CYP4A11
increased the expression levels of the NF-κB signaling
pathway-associated protein p-p65. In addition, a reverse trend was
noted in the presence of si-CYP4A11. This result suggested that
CYP4A11 may affect the activation of the NF-κB signaling pathway,
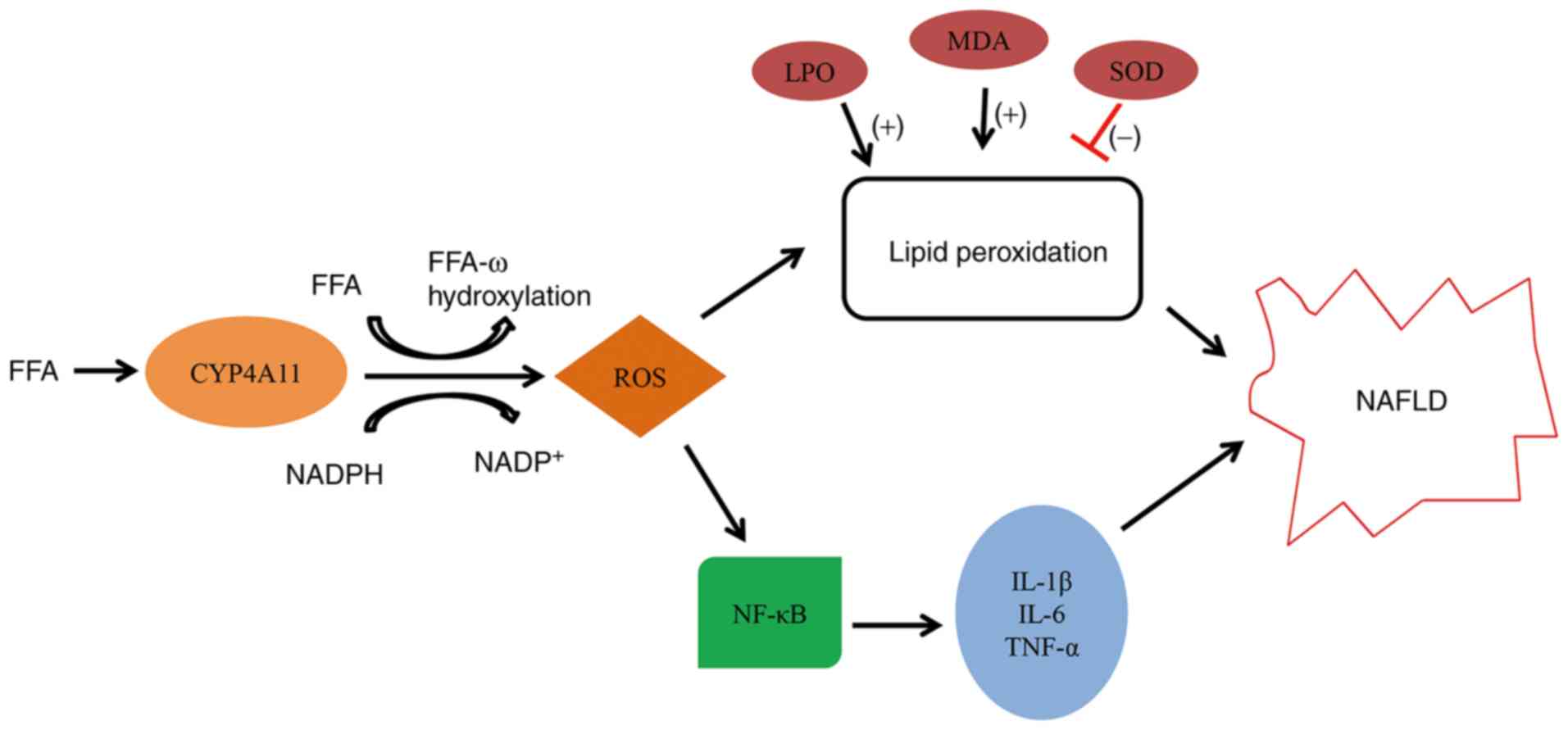
which may be mediated by ROS production (Fig. 5).
Taken collectively, the data of the present study
demonstrated the significant role of CYP4A11 in the metabolism of
fatty acids. CYP4A11 may serve as a mediator of oxidative stress
and LPO. However, additional experimental studies are required to
fully investigate the mechanisms of fatty acid metabolism by
CYP4A11, in order to aid the development of novel therapeutic
strategies for the treatment of NAFLD.
Funding
The present study was supported by the Anhui
Province Natural Science Foundation (grant no. 1808085MH235) and
the Scientific Research Foundation of the Institute of Anhui
Province Transformational Medicine (grant no. 2017zhyx32).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
HG and YJ conceived the study and analyzed the
relevant literature. YC, HX and XZ performed the literature review
and were involved in conducting experiments. HG and YJ performed
experiments, and were involved in writing, reviewing and editing
the manuscript. All authors read and approved the final version of
the manuscript.
Ethics approval and consent to
participate
All experiments were authorized by the Biomedical
Ethics Committee of Anhui Medical University. All procedures
involving human participants were performed in accordance with the
ethical standards of the institutional, and with the 1964 Helsinki
declaration and its later amendments or comparable ethical
standards. Written informed consent was provided by each
patient.
Patient consent for publication
Written informed consent was provided by each
patient.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgments
The authors would like to thank Mr Yuan Hong Xu of
the First Affiliated Hospital of Anhui Medical University for
assisting with the collection of patient blood samples.
References
|
1
|
Bedossa P: Pathology of non-alcoholic
fatty liver disease. Liver Int. 37:85–89. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Brunt EM: Pathology of nonalcoholic fatty
liver disease. Nat Rev Gastroenterol Hepatol. 7:195–203. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Loomba R and Sanyal AJ: The global NAFLD
epidemic. Nat Rev Gastroenterol Hepatol. 10:686–690. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Basaranoglu M, Basaranoglu G and Senturk
H: From fatty liver to fibrosis: A tale of 'second hit'. World J
Gastroenterol. 19:1158–1165. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Robertson G, Leclercq I and Farrel GC:
Nonalcoholic steatosis and steatohepatitis II. Cytochrome P-450
enzymes and oxidative stress. Am J Physiol Gastrointest Liver
Physiol. 281:G1135–G1139. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Sharma RK, Doig MV, Lewis DF and Gibso GG:
Role of hepatic and renal cytochrome P-450 IVA1 in the metabolism
of lipid substrates. Biochem Pharmacol. 38:3621–3629. 1989.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Leclercq IA, Farrell GC, Field J, Bell DR,
Gonzalez FJ and Robertson GR: CYP2E1 and CYP4A as microsomal
catalysts of lipid peroxides in murine nonalcoholic
steatohepatitis. J Clin Invest. 105:1067–1075. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Roman RJ: P-450 metabolites of arachidonic
acid in the control of cardiovascular function. Physiol Rev.
82:131–185. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Kim D, Cha GS, Nagy LD, Yun CH and
Guengerich FP: Kinetic analysis of lauric acid hydroxylation by
human cytochrome P450 4A11. Biochemistry. 53:6161–6172. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zhang J, Hu J, Zhang C, Jiao Y, Kong X and
Wang W: Analyses of risk factors for polycystic ovary syndrome
complicated with non-alcoholic fatty liver disease. Exp Ther Med.
15:4259–4264. 2018.PubMed/NCBI
|
|
11
|
Shojaee-Moradie F, Cuthbertson DJ, Barrett
M, Jackson NC, Herring R, Thomas EL, Bell J, Kemp GJ, Wright J and
Umpleby AM: Exercise training reduces liver fat and increases rates
of VLDL clearance but not VLDL production in NAFLD. J Clin
Endocrinol Metab. 101:4219–4228. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Mansouri A, Gattolliat CH and Asselah T:
Mitochondrial dysfunction and signaling in chronic liver diseases.
Gastroenterology. 155:629–647. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Hardwick JP: Cytochrome P450 omega
hydroxylase (CYP4) function in fatty acid metabolism and metabolic
diseases. Biochem Pharmacol. 75:2263–2275. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Kuthan H, Tsuji H, Graf H and Ullrich V:
Generation of superoxide anion as a source of hydrogen peroxide in
a reconstituted monooxygenase system. FEBS Lett. 91:343–345. 1978.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Tripathy D, Mohanty P, Dhindsa S, Syed T,
Ghanim H, Aljada A and Dandona P: Elevation of free fatty acids
induces inflammation and impairs vascular reactivity in healthy
subjects. Diabetes. 52:2882–2887. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Lugrin J, Rosenblatt-Velin N, Parapanov R
and Liaudet L: The role of oxidative stress during inflammatory
processes. Biol Chem. 395:203–230. 2014. View Article : Google Scholar
|
|
17
|
Zeng MD, Fan JG, Lu LG, Li YM, Chen CW,
Wang BY and Mao YM: Chinese National Consensus Workshop on
Nonalcoholic Fatty Liver Disease: Guidelines for the diagnosis and
treatment of nonalcoholic fatty liver diseases. J Dig Dis.
9:108–112. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Ricchi M, Odoardi MR, Carulli L, Anzivino
C, Ballestri S, Pinetti A, Fantoni LI, Marra F, Bertolotti M, Banni
S, et al: Differential effect of oleic and palmitic acid on lipid
accumulation and apoptosis in cultured hepatocytes. J Gastroenterol
Hepatol. 24:830–840. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
20
|
WHO Expert Consultation: Appropriate
body-mass index for Asian populations and its implications for
policy and intervention strategies. Lancet. 363:157–163. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Ip E, Farrell GC, Robertson G, Hall P,
Kirsch R and Leclercq I: Central role of PPARα-dependent hepatic
lipid turnover in dietary steatohepatitis in mice. Hepatology.
38:123–132. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zhang X, Li S, Zhou Y, Su W, Ruan X, Wang
B, Zheng F, Warner M, Gustafsson JA and Guan Y: Ablation of
cytochrome P450 omega-hydroxylase 4A14 gene attenuates hepatic
steatosis and fibrosis. Proc Natl Acad Sci USA. 114:3181–3185.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Garcia MC, Amankwa-Sakyi M and Flynn TJ:
Cellular glutathione in fatty liver in vitro models. Toxicol In
Vitro. 25:1501–1506. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Eid AA, Gorin Y, Fagg BM, Maalouf R,
Barnes JL, Block K and Abboud HE: Mechanisms of podocyte injury in
diabetes: Role of cytochrome P450 and NADPH oxidases. Diabetes.
58:1201–1211. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Sozen E and Ozer NK: Impact of high
cholesterol and endoplasmic reticulum stress on metabolic diseases:
An updated mini-review. Redox Biol. 12:456–461. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Duwaerts CC and Maher JJ: Mechanisms of
liver injury in non-alcoholic steatohepatitis. Curr Hepatol Rep.
13:119–129. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zangar RC, Davydov DR and Verma S:
Mechanisms that regulate production of reactive oxygen species by
cytochrome P450. Toxicol Appl Pharmacol. 199:316–331. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Hsu MH, Savas U, Griffin KJ and Johnson
EF: Human cytochrome p450 family 4 enzymes: Function, genetic
variation and regulation. Drug Metab Rev. 39:515–538. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Chavez-Tapia NC, Rosso N and Tiribelli C:
In vitro models for the study of nonalcoholic fatty liver disease.
Curr Med Chem. 18:1079–1084. 2011. View Article : Google Scholar
|
|
30
|
Kang H and Koppula S: Houttuynia cordata
alleviates high-fat diet-induced non-alcoholic fatty liver in
experimental rats. Pharm Biol. 53:414–422. 2015. View Article : Google Scholar
|
|
31
|
Vornoli A, Pozzo L, Della Croce CM,
Gervasi PG and Longo V: Drug metabolism enzymes in a steatotic
model of rat treated with a high fat diet and a low dose of
streptozotocin. Food Chem Toxicol. 70:54–60. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Kohjima M, Enjoji M, Higuchi N, Kato M,
Kotoh K, Yoshimoto T, Fujino T, Yada M, Yada R, Harada N, et al:
Re-Evaluation of fatty acid metabolism-related gene expression in
nonalcoholic fatty liver disease. Int J Mol Med. 20:351–358.
2007.PubMed/NCBI
|
|
33
|
Zhou Y, Luo P, Chang HH, Huang H, Yang T,
Dong Z, Wang CY and Wang MH: Colfibrate attenuates blood pressure
and sodium retention in DOCA-salt hypertension. Kidney Int.
74:1040–1048. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Miyata N, Taniguchi K, Seki T, Ishimoto T,
Sato-Watanabe M, Yasuda Y, Doi M, Kametani S, Tomishima Y, Ueki T,
et al: HET0016, a potent and selective inhibitor of 20-HETE
synthe-sizing enzyme. Br J Pharmacol. 133:325–329. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Pessayre D: Role of mitochondria in
non-alcoholic fatty liver disease. J Gastroenterol Hepatol.
22(Suppl 1): S20–S27. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Byrne CD: Fatty liver: Role of
inflammation and fatty acid nutrition. Prostaglandins Leukot Essent
Fatty Acids. 82:265–271. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Feldstein AE, Werneburg NW, Canbay A,
Guicciardi ME, Bronk SF, Rydzewski R, Burgart LJ and Gores GJ: Free
fatty acids promote hepatic lipotoxicity by stimulating TNF-alpha
expression via a lysosomal pathway. Hepatology. 40:185–194. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Rolo AP, Teodoro JS and Palmeira CM: Role
of oxidative stress in the pathogenesis of nonalcoholic
steatohepatitis. Free Radic Biol Med. 52:59–69. 2012. View Article : Google Scholar
|
|
39
|
Cnop M, Foufelle F and Velloso LA:
Endoplasmic reticulum stress, obesity and diabetes. Trends Mol Med.
18:59–68. 2012. View Article : Google Scholar
|
|
40
|
Hui JM, Hodge A, Farrell GC, Kench JG,
Kriketos A and George J: Beyond insulin resistance in NASH:
TNF-alpha or adiponectin? Hepatology. 40:46–54. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Morgan MJ and Liu ZG: Crosstalk of
reactive oxygen species and NF-κB signaling. Cell Res. 21:103–115.
2011. View Article : Google Scholar
|