Introduction
Breast cancer (BC) is the most prevalent malignant
tumor among females worldwide. It is the most common cancer in
China, as well as the United States (1). Fan et al (2) reported that Chinese patients account
for over 12% of all newly diagnosed cases as well as 9% of all
deaths from BC globally. Similarly, Waks and Winer (3) reported that >250,000 new patients
were diagnosed in the United States in 2017 and they estimated that
12% of women over the course of their lifetimes will be diagnosed
with BC. Depending on levels of estrogen or progesterone receptors
(ER and PR, respectively), human epidermal growth factor-2 (Her-2),
as well as Ki-67, BC is classified into four major molecular
subtypes: Luminal A, Luminal B, Her-2 positive and basal-like.
Although the diagnosis, as well as treatment of BC has achieved
significant advances and overall survival (OS) has improved during
recent decades, there are still challenges regarding the early
detection and prediction of survival in BC. Moreover, the mechanism
of tumorigenesis is still unclear. Thus, it is important to develop
novel biomarkers for diagnosis and prognosis, as well as treatment
targets.
The authors' previous study reported that Rab22a is
the target gene of microRNAs (miRNAs/miR)-193b in BC (4). Studies of miRNAs have reported that
miR-520b (5), miR-204 (6) and miR-203 (7) are downregulated in BC or other types
of tumors. In addition, one of the potential targeting genes of the
above-mentioned miRNAs is Rab22a, which was confirmed using a
luciferase assay and is upregulated in these tumor tissues or cell
lines. All these findings lead to the conclusion that Rab22a may
act as an oncogene and participate in carcinogenesis. Furthermore,
emerging evidence suggests that dysregulated expression of Rab22a
affects tumor growth. He et al (8) found that Rab22a is upregulated in
most of the 11 hepatocellular carcinomas, as well as 1 case of
cholangiohepatoma. Okamoto et al (9) identified that Rab22a is positively
expressed in malignant melanoma cell lines and situated at the
areas of chromosomal breakpoints.
The coding gene of Rab22a, which belongs to the Ras
superfamily, is located at chromosome 20q13.32. Rab22a is a part of
the Ras-related proteins in brain (Rabs) GTPase subfamily, which
are small GTP-binding proteins. Zhou et al (10) reported that Rab22a participates at
several levels of the endocytic pathway. In addition, the
composition of the plasma membrane receptors is regulated by
endocytic recycling and endocytic uptake. Stenmark concluded that
Rab22a mediates trafficking among trans-Golgi network and early
endosomes (11). Kauppi et
al (12) reported that the
overexpression of Rab22a in vitro leads to the enlargement
of early endosomes. Previous studies have also indicated that the
recycling of the transferrin receptor and major histo-compatibility
complex-I (MHC)-I by endocytosed cargos is blocked by small
interfering RNAs that inhibit the function of Rab22a (13,14). It is widely reported that
dysregulation of endocytosis is the hallmark of several cancers.
Therefore, the abnormal expression of Rab GTPases may be involved
in tumorigenesis.
Wang et al (15) reported that the upregulation of
Rab22a mRNA in the primary tumor is related to worsening of overall
as well as metastasis-free survival, by analyzing The Cancer Genome
Atlas (TCGA) database of patients with BC. An additional study
found that knockdown of Rab22a inhibits BC metastasis in
vivo (15). Kang et al
(16) designed and synthe-sized
O(2)-3-Aminopropyl
diazeniumdiolates 3a-f, which prevents the growth and metastasis of
triple negative BC in vivo as well as in vitro by
MHC-I impairing microvesicle formation by altering the expression
of miR-203/Rab22a in a nitric oxide-dependent manner.
Although much of the research indicates that the
Rab22a gene can function as an oncogene in several cancer models
and indicates its potential therapy target (15,16), whether Rab22a may be developed as
a novel prognostic biomarker in BC is not well understood. The
current study applied immunohistochemistry (IHC) staining and
analyzed the association between Rab22a expression and
clinicopathological features, as well as OS status. The impact of
Rab22a on the progression of BC cells as well as the probable
pathways involved was also studied.
Materials and methods
Patients and samples
A total of 258 samples from female patients with BC
aged from 27 to 78, who had accepted modified radical mastectomy or
breast-conserving surgery at the China-Japan Union Hospital of
Jilin University from January 2012 to December 2013 were included
in the present study. Patient characteristics are shown in Table I. The clinical stage was
distinguished using the guidelines of the American Joint Committee
on Cancer. The patients who accepted chemotherapy or radiotherapy
and hormone therapy prior to surgery were excluded. Recurrent or
metastasis cases at the time of diagnosis were eliminated as well.
The last date of follow-up was in April 2019. A total of 56
para-tumor or non-tumor tissues were acquired as the controls. The
current study was conducted after obtaining written informed
consent from all patients and was approved by the Research Ethics
Committee of the China-Japan Union Hospital of Jilin University
(approval no. 2019022603).
 | Table IClinicopathological characteristics
of patient samples and expression of Rab22a in breast cancer. |
Table I
Clinicopathological characteristics
of patient samples and expression of Rab22a in breast cancer.
|
Characteristics | Rab22a expression
| P-value |
|---|
| Low | Middle | High |
|---|
| Age, years | | | | |
| <50 | 20 | 58 | 45 | |
| ≥50 | 10 | 67 | 58 | 0.079 |
| Menopause
stage | | | | |
| No | 20 | 68 | 55 | |
| Yes | 10 | 57 | 48 | 0.415 |
| Age at menarche,
years | | | | |
| <15 | 19 | 71 | 39 | |
| ≥15 | 11 | 54 | 64 | 0.005 |
| Clinical stage | | | | |
| I | 17 | 49 | 32 | |
| II | 7 | 57 | 34 | |
| III | 6 | 19 | 37 | 0.001 |
| T
classification | | | | |
| T1 | 22 | 80 | 53 | |
| T2 | 8 | 40 | 40 | |
| T3 | 0 | 5 | 9 | |
| T4 | 0 | 0 | 1 | 0.148 |
| N
classification | | | | |
| N0 | 19 | 71 | 47 | |
| N1 | 5 | 35 | 22 | |
| N2 | 2 | 13 | 13 | |
| N3 | 4 | 6 | 21 | 0.013 |
| Nipple
invasion | | | | |
| No | 30 | 124 | 97 | |
| Yes | 0 | 1 | 6 | 0.042 |
| Venous
invasion | | | | |
| No | 23 | 90 | 60 | |
| Yes | 7 | 35 | 43 | 0.044 |
| Nerve invasion | | | | |
| No | 26 | 99 | 82 | |
| Yes | 4 | 26 | 21 | 0.640 |
| Basal invasion | | | | |
| No | 30 | 124 | 101 | |
| Yes | 0 | 1 | 2 | 0.595 |
| Pectoralis minor
muscle invasion | | | | |
| No | 30 | 124 | 101 | |
| Yes | 0 | 1 | 2 | 0.595 |
| Pathological
differentiation | | | | |
| G1 | 4 | 10 | 7 | |
| G2 | 23 | 85 | 73 | |
| G3 | 3 | 30 | 23 | 0.446 |
| ER status | | | | |
| Negative | 5 | 32 | 33 | |
| Positive | 25 | 93 | 70 | 0.216 |
| PR status | | | | |
| Negative | 6 | 41 | 40 | |
| Positive | 24 | 84 | 63 | 0.151 |
| Expression of
Her-2 | | | | |
| Negative | 22 | 91 | 63 | |
| Positive | 8 | 34 | 40 | 0.140 |
| Expression of
Ki67 | | | | |
| Low
expression | 24 | 88 | 64 | |
| High
expression | 6 | 37 | 39 | 0.139 |
| Molecular
classification | | | | |
| Luminal A | 15 | 48 | 24 | |
| Luminal B
(Her-2-) | 7 | 21 | 25 | |
| Luminal B
(Her-2+) | 3 | 25 | 23 | |
| Her-2+ | 3 | 13 | 16 | |
| Basal like | 2 | 18 | 15 | 0.131 |
| Vital status (at
follow-up) | | | | |
| Alive | 28 | 112 | 772 | |
| Dead | 2 | 13 | 26 | 0.003 |
Breast cell lines
The human mammary gland epithelium cell line,
MCF-10A was cultured in a DMED/HAM F12 1:1 mixed medium (Hyclone;
GE Healthcare), which contains 5% horse serum (Gibco; Thermo Fisher
Scientific, Inc.), 0.5 mg/ml hydrocortisone (Sigma-Aldrich; Merck
KGaA), 100 ng/ml cholera toxin (Sigma-Aldrich; Merck KGaA), 10
µg/ml insulin (Sigma-Aldrich; Merck KGaA) and 20 ng/ml EFG
(Peprotech, Inc.). The human BC cell lines, MCF-7 as well as
SK-BR-3, were cultured in DMEM H21 (Hyclone; GE Healthcare) and
RPMI-1640 (Hyclone; GE Healthcare), each supplemented with 10%
fetal bovine serum (Gibco; Thermo Fisher Scientific, Inc.), 100
U/ml penicillin and 100 mg/ml streptomycin. All cell lines were
bought from American Type Culture Collection and cultured at 37°C
with 5% CO2, in a humidified atmosphere.
IHC staining
In order to identify Rab22a expression levels in the
BC tissues, IHC staining was performed by following the
manufacturer's protocol (Metal enhanced DAB for IHC; Invitrogen;
Thermo Fisher Scientific, Inc.). In brief, after fixation with
formalin at room temperature for 48 h and embedded with paraffin, 3
µm thick paraffin sections adhered to slides were incubated
at 65°C for 30 min, deparaffinized using xylene and then rehydrated
using alcohol. The slides were submerged in EDTA for antigen
retrieval. Next, the sections were treated with 3% hydrogen, heated
in a microwave at 100°C for 5 min and incubated in 1% bovine serum
albumin (Gibco; Thermo Fisher Scientific, Inc.). Following this,
they were incubated with a Rab22a antibody (cat. no. 12125-1-AP;
Proteintech, Inc.) at dilution of 1:100 overnight at 4°C. Normal
goat serum served as the negative control. After washing with
PBS+1% Tween, the sections were incubated with the secondary
antibody with streptavidin-horseradish peroxidase complex (cat. no.
31460; Invitrogen; Thermo Fisher Scientific, Inc.) at a dilution of
1:2,000 at room temperature for 1 h, followed by colorimetric
detection using metal enhanced DAB (cat. no. 34065; Invitrogen;
Thermo Fisher Scientific, Inc.) at room temperature for 5 min. All
sections were submerged in 3-amino-9-ethyl carbazole,
counterstained using 10% Mayer's hematoxylin at room temperature
for 2 min, dehydrated and mounted.
The intensity of IHC staining of the sections was
evaluated using a light microscope at ×100 magnification by two
senior pathologists independently, who were not aware of the
patient information and histopathological details. The plasma
staining of Rab22a was positive. The staining intensity as well as
positive staining percentage of tumor cells were used for
evaluation. The intensity of staining was categorized as previously
described (17): 0 (stainless), 1
(light yellow), 2 (yellow brown) and 3 (brown). The proportion of
positive staining tumor cells were 0 (0%), 1 (1-10%), 2 (11-25%), 3
(25-50%) and 4 (>50%). The score was calculated as staining
intensity x proportion of positive cells and was used as the
staining index to calculate IHC intensity (range from 0 to 12). A
staining index score of 0 was defined as no expression of Rab22
(-), 1-3 signified low expression of Rab22 (1+), 4-6 signified mild
expression of Rab22a (2+), while >6 signified high expression of
Rab22a (3+).
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA extraction was done using TRIzol (Ambion;
Thermo Fisher Scientific, Inc.). The reverse transcription of total
RNA was done using 5X PrimeScript RT Master Mix for Real Time
(Takara Bio, Inc.). RT-qPCR was performed using SYBR®
Premix DimerEraser™ Perfect Real Time (Takara Bio, Inc.). The
operation process was conducted following the manufacturer's
protocol. RT-qPCR analyses were conducted by Mastercycler
(Eppendorf). The thermocycling procedure was as follows: 30 sec at
95°C, followed by 40 cycles of 95°C for 30 sec, 57°C for 30 sec and
a final step of 68°C for 30 min. The primer sequences used were as
follows: Rab22a F: 5′-GTC CCT TAG CAC CAA TGT ACT ATC-3′, R: 5′-AAT
GGC AAC TAC AAT ATT AGG TGG-3′; GAPDH F: 5′-GAA GGC TGG GGC TCA TTT
GCA GGG-3′, R: 5′-GGT GCA GGA GGC ATT GCT GAT GAT-3′. The
comparative fold-change at mRNA level was calculated using the
2−ΔΔCq method (18).
GAPDH served as a reference control for normalization.
Short hairpin (sh)RNA knockdown
The lenti-virus of Rab22a shRNA was purchased from
Hanheng Biotechnology to target human Rab22a. The SK-BR-3 cells
were infected with Rab22a shRNA or a scramble control (SC)
lenti-virus as per the supplier's protocol. In brief,
2×105 SK-BR-3 cells were incubated with 2×107
transducing units of lenti-virus and 8 µg polybrene (Hanheng
Biotechnology) in 1 ml serum-free medium at 37°C with 5%
CO2 for 8 h. Consequently, 1 ml fresh growth medium was
added for 24 h and was changed with fresh growth media. Stable cell
lines were generated in the selected medium with 0.5 µg/ml
puromycin after 48 h of infection.
Western blotting
The cells were harvested in Lysis Buffer for the
western blot analysis and IP (cat. no. P0013; Shanghai Biyuntian
Bio-Technology Co., Ltd.) and the proteins were extracted. The
Bio-Rad Laboratories, Inc., method was applied to quantify protein
concentration. Proteins (25 µg/well) were separated on a 10%
SDS-PAGE gel and then transferred onto a nitrocellulose membrane.
After blocking with 5% fat-free milk at room temperature for 1 h,
the membranes were incubated with Rab22a (cat. no. 12125-1-AP;
Proteintech, Inc.) or GAPDH antibodies (cat. no. 60004-1-Ig,
Proteintech, Inc.) at a dilution of 1:1,000, overnight at 4°C.
Anti-rabbit secondary antibodies were conjugated with horseradish
peroxidase (cat. no. SA00001; Proteintech, Inc.) at room
temperature for 1 h. Immunoreactivity was detected using enhanced
chemiluminescence (BeyoECL Plus; Shanghai Biyuntian Bio-Technology
Co., Ltd.) and visualized through autoradiography.
Cell proliferation assay
Cell proliferation abilities were tested using Cell
Counting Kit-8 assays (CCK-8; Dalian Meilunbio Biotechnology Co.,
Ltd.) according to the manufacturer's protocol. A total of 3,500
cells/well (SK-BR-3 SC and shRNA) were suspended in 96-well plates
in serum free medium, overnight. Once the cells were adhered to the
plate, serum free medium was discarded and 100 µl growth
medium was added into all wells. After incubation for 24, 48, 72
and 96 h, 10 µl CCK-8 was added into each well and cultured
for 4 h. The optical density value was read at 450 nm using a
microplate reader (BioTek China).
Migration and invasion assay
Cell migration and invasion assays were conducted
using BD BioCoat Matrigel Invasion Chambers and Control Inserts (BD
Biosciences; Becton, Dickinson and Company). A total of
5×104 SC or knockdown of SK-BR-3 cells were re-suspended
in 0.5 ml medium with 1% fetal bovine serum (FBS), then seeded on
either upper control inserts or trans-well chambers with Matrigel
(BD Biosciences; Becton, Dickinson and Company). A total of 2 ml
medium containing 15% FBS was added onto the lower chamber, which
served as a chemo-attractant. A total of 20 h later, migrating or
invading cells that had attached to the lower surface of the
membrane insert were fixed at room temperature for 2 min and
stained by Giemsa at room temperature for 10 min, then counted
under a light microscope (Olympus IX51; Olympus Corporation) at
×200 magnification.
Mice xenograft model
A total of six female nude mice (Beijing Huafukang
Bioscience, Co., Ltd.) that weighed 20-25 g, aged 4-5 weeks were
separated into two groups. Each mouse was injected with
4×106 SK-BR-3 cells (Rab22a SC or shRNA3) into its
fourth mammary fat pad. Tumor size was measured using a caliper
every 3 days after injection. Tumor volume was calculated as
follows: Volume = (length x width2)/2. The mice were
sacrificed by inhalation of CO2 in 10%/min, the criteria
for confirmation of death is that heart and breath rate were
completely stopped, and pupillary response to light was
disappeared. Once the xenograft models were established for 30
days, the xenograft tumors were excised and weighed after
euthanasia. The mice were housed in the SPF facility, at 22±2°C
with a 12 h light/dark cycle and a relative humidity of 40-60%, and
had free access to food and water, following the Laboratory Animal
Care and Use guideline of Jilin University. Animal protocol were
approved by the Laboratory Animal Care and Use Committee of Jilin
University (approval no. 201905005).
Bioinformatics analysis
For this study, Gene Expression Omnibus (GEO)
datasets (GSE25487, GSE22796 and GSE18672) were downloaded from the
GEO database (19-21) (https://www.ncbi.nlm.nih.gov/geo/). The expression
difference between discrete variables was visualized using the
ggplot2 package of R software (version 3.5.2; http://www.R-project.org/bioconductor.org/), by
constructing boxplots. Gene set enrichment analysis (GSEA) was done
using GSEA software 3.0. First, the mRNA files consisting of 1,104
BC tumors and 114 normal tissues were downloaded from TCGA
database. The high and low mRNA group were defined as the median
value of mRNA level of Rab22a. Then, GSEA was performed as
previously described (22-24).
In brief, the annotated gene sets of 'h.all.v6.2.symbols.gmt',
which summarize and represent specific, biological states or
processes, for use with GSEA software were acquired from the
Molecular Signatures Database (MSigDB). The normalized enrichment
score (NES) was determined by investigating permutations for 1,000
times. A P<0.05 as well as false discovery rate (FDR) <0.25
were considered to signify the statistical significance of an
enriched gene set.
Statistical analysis
SPSS software version 21.0 (IBM Corps.) was used for
analysis. Analysis among expression of Rab22a and
clinicopathological characteristics of patients were carried out
using the chi-square test or Fisher's exact test. Bivariate
correlations among study variables were assessed using Spearman's
correlation analysis. Survival curves were constructed with
Kaplan-Meier plots and compared using the Log-rank test.
Multivariate Cox-regressive survival analysis was estimated by
including all the parameters that were significant for the
univariate Cox-regressive analysis. Data among or between groups
were investigated by one-way analysis of variance (ANOVA) or
Student's t-test, respectively. LSD (3 groups) and Tukey's (4
groups) post hoc tests were used following ANOVA. P<0.05 was
considered to indicate a statistically significant difference. All
experiments were conducted in triplicate and the outcomes are
presented as the mean ± standard deviation.
Results
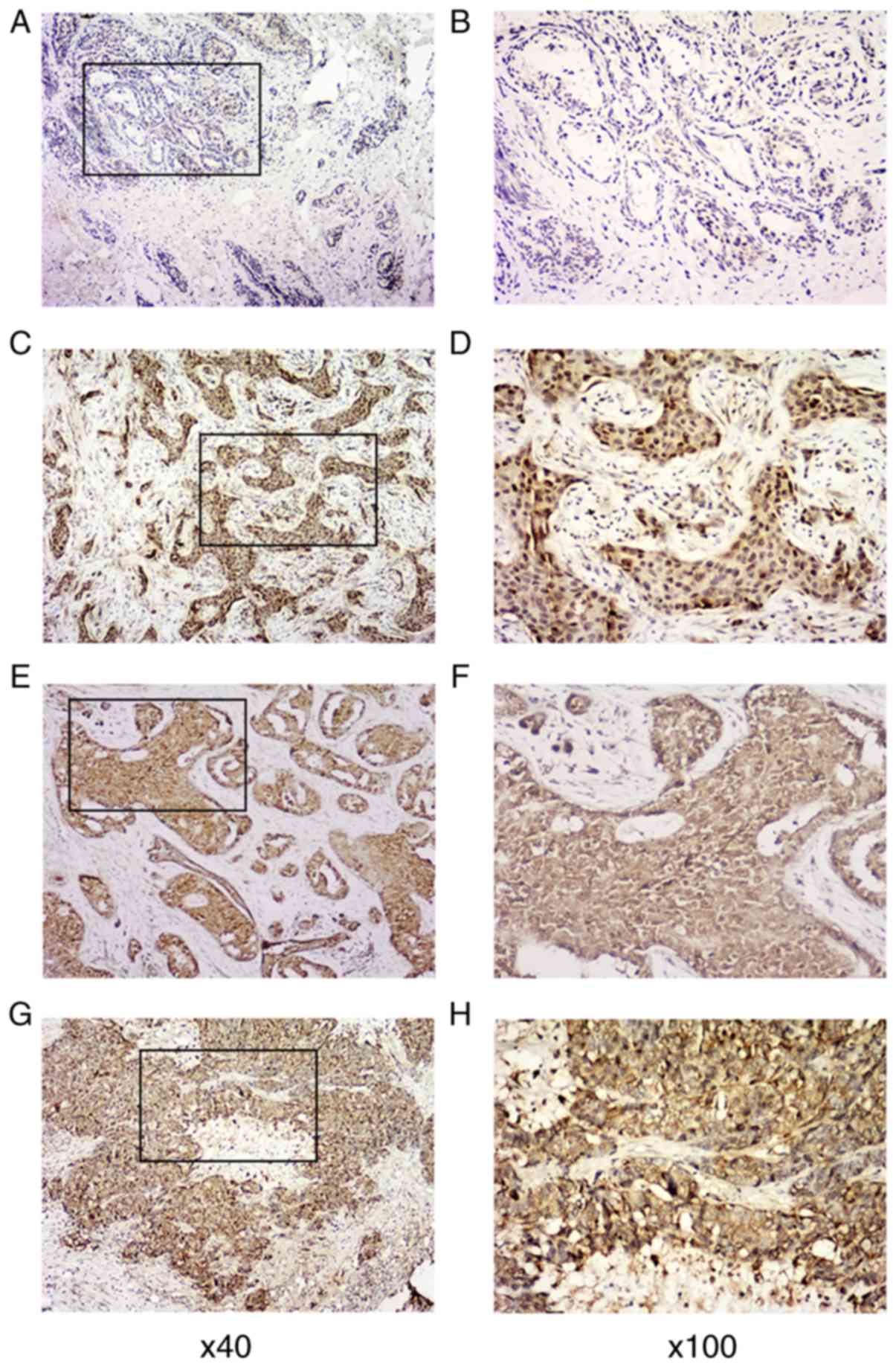
Rab22a is overexpressed in BC
tissues
In order to examine the status of Rab22a expression
in BC, its expression was inspected by IHC staining of 258
paraffin-embedded BC tissues, containing 98 cases of clinical stage
I BC, 98 cases of clinical stage II BC and 62 cases of clinical
stage III BC, and compared them with 56 non-tumor or para-tumor
samples. The clinical and pathological characteristics of all 258
BC patients are summarized in Table
I. Compared with the 56 cases of nearby non-tumor breast
tissues, in which Rab22a is untraceable or is expressed at low
levels, Rab22a was found to be overexpressed in BC tissues. Rab22a
expression in cancer tissues was successfully identified in 3
levels, including low expression (1+) in 30 cases, mild expression
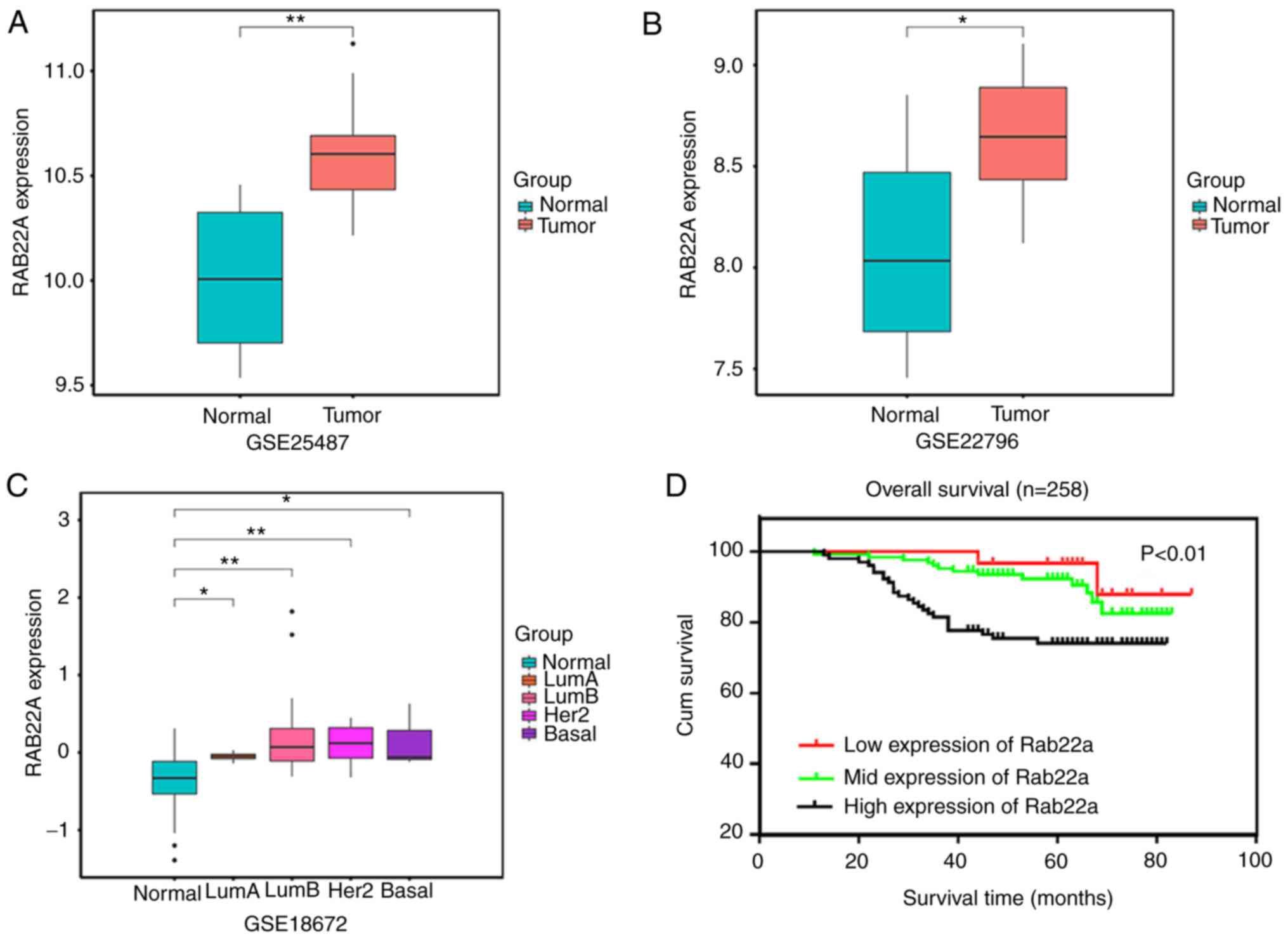
(2+) in 125 cases and high expression (3+) in 103 cases (Fig. 1). GEO datasets were employed to
verify Rab22a expression. Rab22a expression was significantly
increased in tumor samples, compared with normal samples (Fig. 2A and B). Furthermore, Rab22a
expression was significantly increased in all four molecular
subtypes (Luminal A and B, Her-2 and basal-like) compared with
normal tissues (Fig. 2C).
Rab22a overexpression is correlated with
the clinicopathological features of BC
The connection between expression of Rab22a and
clinicopathological characteristics of patients with BC was
analyzed (Table I). Rab22a
expression was found to be related with age at menarche (P=0.005),
clinical stage (P= 0.001), N classification (P= 0.013), nipple
invasion (P= 0.042), venous invasion (P=0.044) and vital states
(P=0.003). There was no difference based on age, menopause stage, T
classification, nerve invasion, basal invasion, pectoralis minor
muscle invasion, pathological differentiation, ER status, PR
status, expression of Her-2, expression of Ki67 and molecular
classification among the levels of Rab22a expression.
Spearman analysis of correlation among Rab22a and
clinicopathological features indicate that the expression of Rab22a
is significantly positively associated with age at menarche
(P=0.002), clinical stage (P=0.001), T classification (P=0.006), N
classification (P=0.008), nipple invasion (P=0.015), venous
invasion (P=0.014), expression of Her-2 (P=0.038) and molecular
classification (P=0.006), while negatively associated with vital
states (P=0.006) (Table II).
 | Table IISpearman analysis of correlation
between Rab22a and clinicopathological features. |
Table II
Spearman analysis of correlation
between Rab22a and clinicopathological features.
|
Characteristics | Rab22a expression
level
|
|---|
| Spearman
correlation | P-value |
|---|
| Age | 0.118 | 0.058 |
| Menopause
stage | 0.058 | 0.356 |
| Age at
menarche | 0.191 | 0.002 |
| Clinical stage | 0.199 | 0.001 |
| T
classification | 0.171 | 0.006 |
| N
classification | 0.165 | 0.008 |
| Nipple
invasion | 0.151 | 0.015 |
| Venous
invasion | 0.154 | 0.014 |
| Nerve invasion | 0.032 | 0.608 |
| Basal invasion | 0.063 | 0.310 |
| Pectoralis minor
muscle | 0.063 | 0.310 |
| invasion | | |
| Pathologic
differentiation | 0.066 | 0.288 |
| ER status | −0.016 | 0.090 |
| PR status | −0.112 | 0.073 |
| Expression of
Her-2 | 0.129 | 0.038 |
| Expression of
Ki67 | 0.104 | 0.095 |
| Molecular
classification | 0.170 | 0.006 |
| Vital states | −0.171 | 0.006 |
High Rab22a expression is associated with
the poor prognosis of patients with BC
In the Kaplan-Meier estimation, the median OS time
in patients with BC was 94.1 weeks for those with low expression of
Rab22a, 93.7 weeks for mild expression of Rab22a and 81.2 weeks for
high expression of Rab22a (P<0.01) (Fig. 2D). This indicates that patients
with high expression of Rab22a had poorer OS.
Univariate and multivariate Cox-regression analysis
were conducted to identify factors related with OS of patients with
BC (Table III). The univariate
Cox-regression analysis indicates that a high level of expression
of Rab22a (P=0.003) and Her-2 (P=0.044) were related to poor OS.
Then, a high level of expression of Rab22a (P=0.003) was identified
to be associated with short OS in the multivariate Cox-regression
analysis. These results infer that Rab22a could be an independent
predictor for BC.
 | Table IIIUnivariate and multivariate
Cox-regression analysis of various prognostic parameters in
patients with breast cancer. |
Table III
Univariate and multivariate
Cox-regression analysis of various prognostic parameters in
patients with breast cancer.
|
Characteristics | Univariate analysis
| Multivariate
analysis
|
|---|
| HR | 95% CI | P-value | HR | 95% CI | P-value |
|---|
| Age at
menarche | 0.958 | 0.804-1.142 | 0.634 | | | |
| Rab22a | 2.285 | 1.325-3.942 | 0.003 | 2.351 | 1.338-4.132 | 0.003 |
| Clinical stage | 1.300 | 0.844-2.004 | 0.234 | | | |
| T
classification | 0.997 | 0.609-1.630 | 0.989 | | | |
| N
classification | 1.320 | 0.986-1.768 | 0.062 | | | |
| Expression of
Her-2 | 1.326 | 1.008-1.745 | 0.044 | 1.282 | 0.958-1.714 | 0.094 |
| Molecular
classification | 1.647 | 0.925-2.930 | 0.090 | | | |
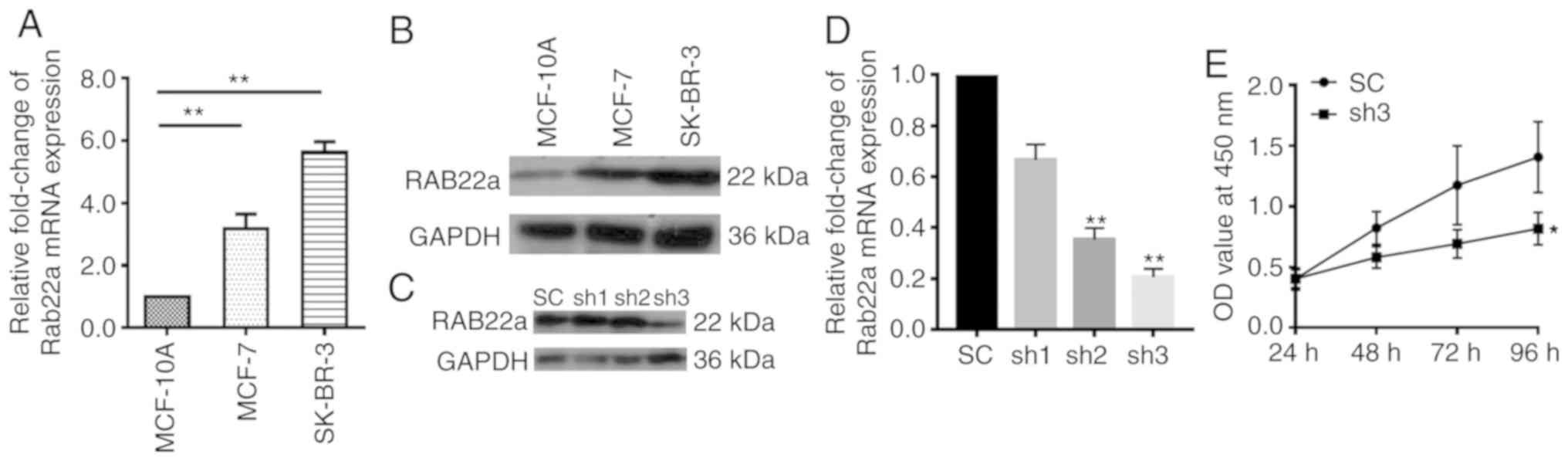
Rab22a is upregulated in BC cell lines
and required for cell progression
mRNAs were isolated from MCF-10A (control) and BC
cell lines MCF-7, as well as SK-BR-3, and examined using RT-qPCR.
The mRNA level of Rab22a in MCF-7 and SK-BR-3 was ~3 times greater
than that of MCF-10A (P<0.001; Fig. 3A). The results were confirmed at
the protein level using western blotting analysis (Fig. 3B). After a stable Rab22a shRNA
knockdown cell line was established in SK-BR-3 cells, mRNA and
protein levels of Rab22a were examined using RT-qPCR and western
blotting, respectively (Fig. 3C and
D). The mRNA level of Rab22a in SK-BR-3 shRNA2 and shRNA3 was
70% lower than that of the SK-BR-3 SC (P<0.001). The
proliferation of SK-BR-3 shRNA and SK-BR-3 SC cells were examined
continuously for 96 h. It was found that Rab22a-shRNA showed a
significantly lower proliferation rate than SK-BR-3 SC cells
(P<0.05; Fig. 3E). Moreover,
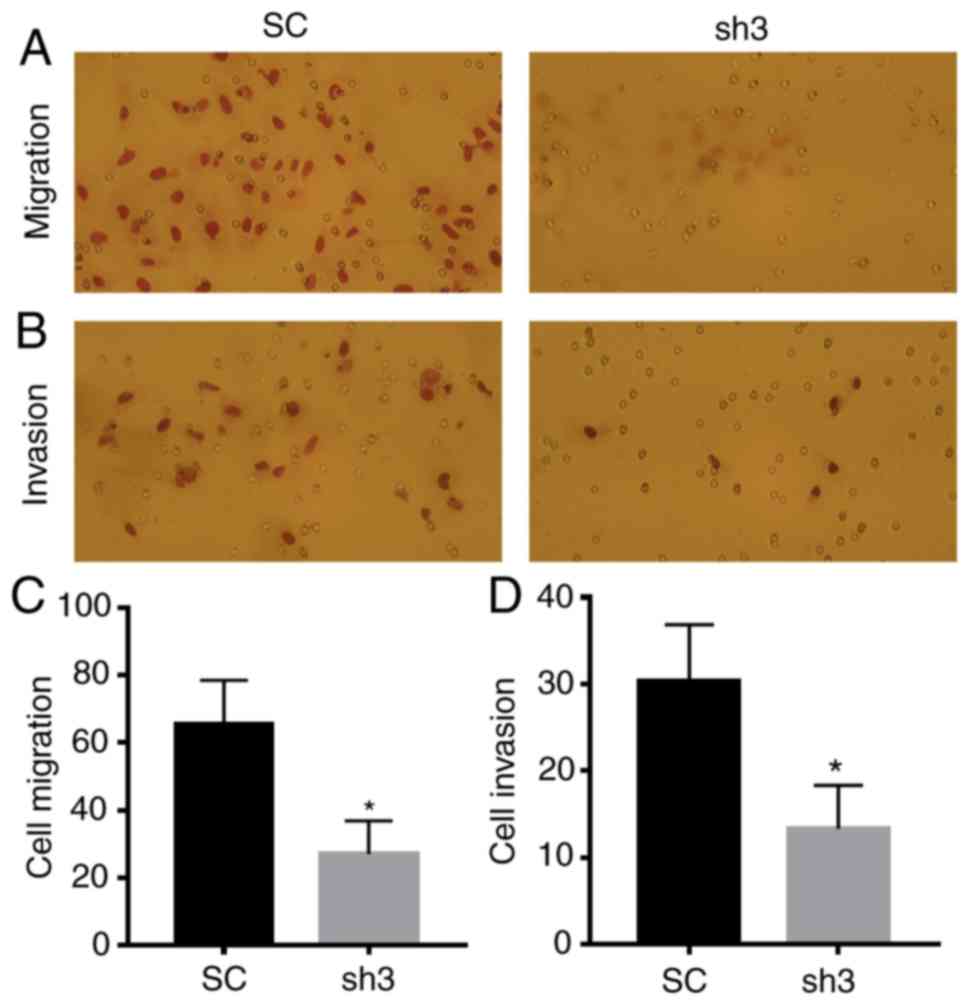
downregulation of Rab22a significantly inhibited the migration and
invasion of BC cells (P<0.05; Fig.
4).
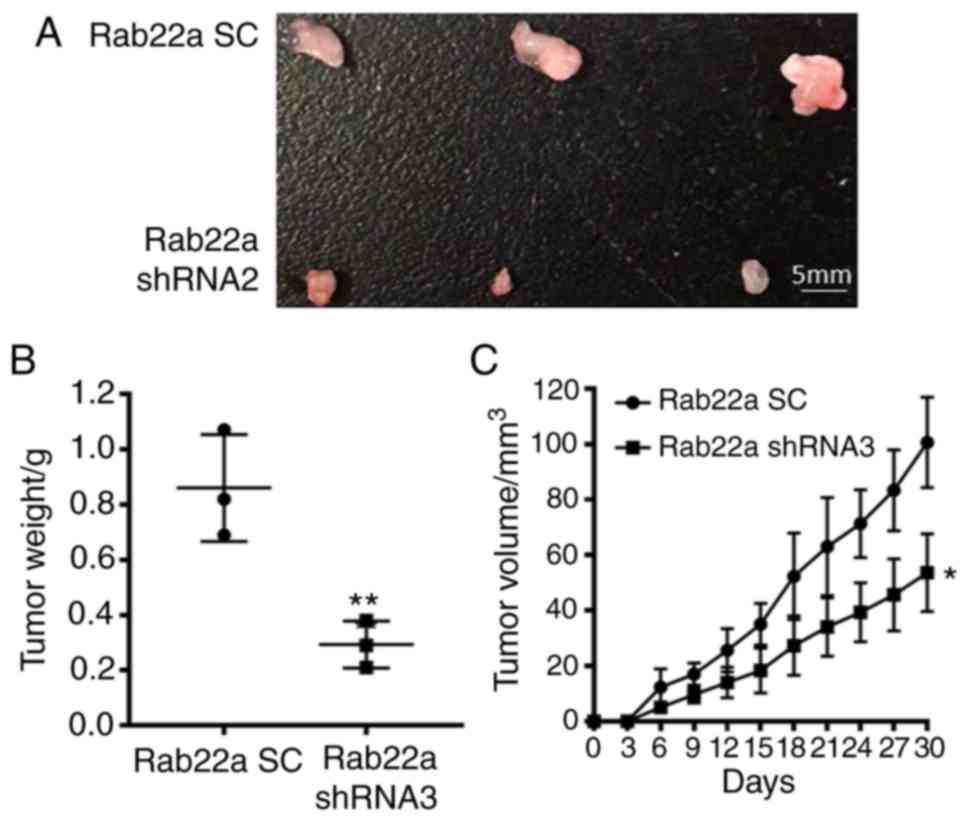
The xenograft mice model showed that the tumor
weight of Rab22a SC and knockdown group was 0.86±0.11 g vs.
0.29±0.05 g (P<0.01; Fig. 5A and
B) and tumor volume was 100.70±9.40 mm3 vs.
53.67±8.09 mm3 (P<0.05; Fig. 5C) at the endpoint date. The
results of the in vitro and in vivo experiments
indicate that the proliferation ability of BC is significantly
inhibited in Rab22a knockdown group, compared with the SC
group.
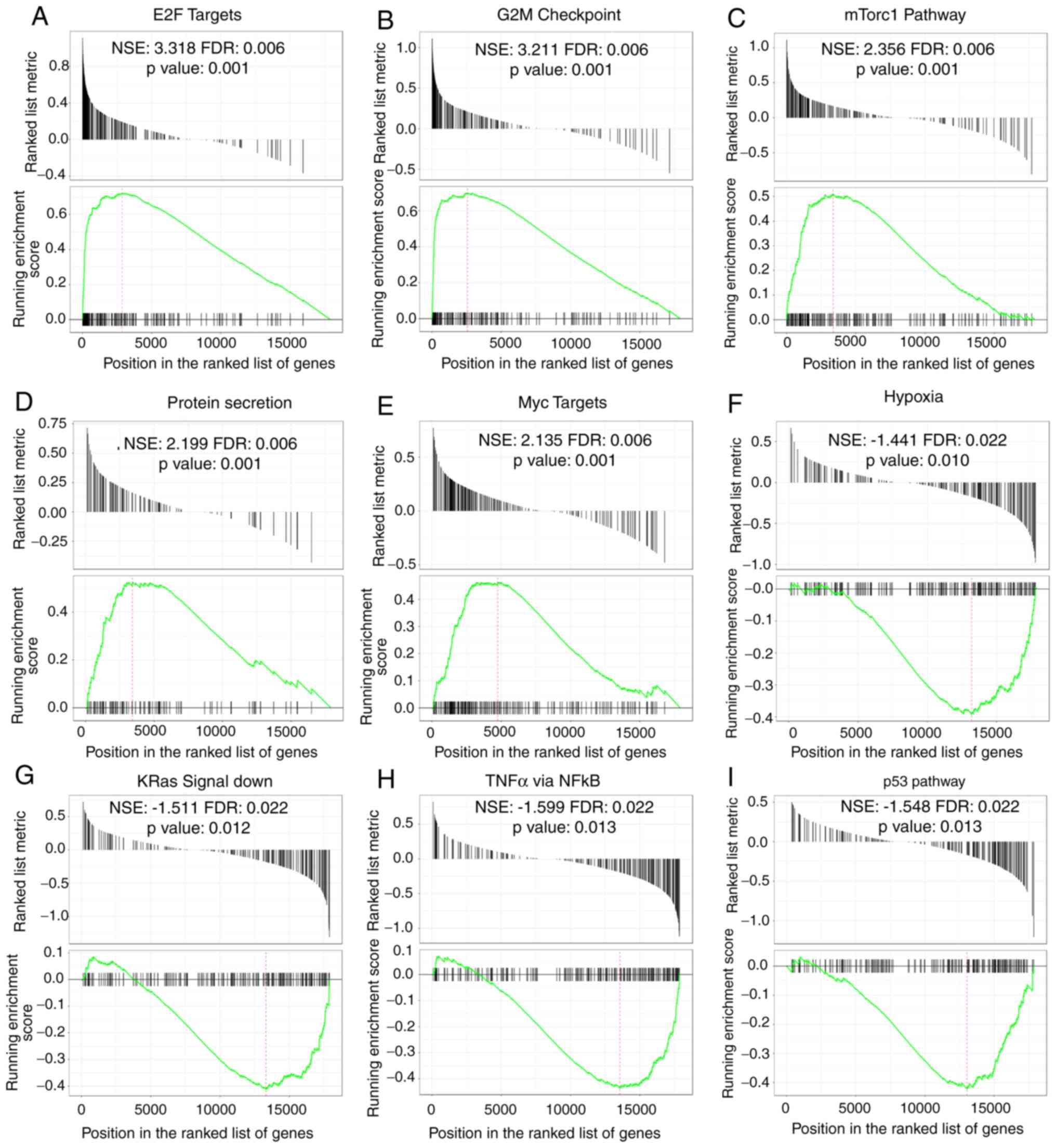
GSEA identifies the Rab22a-related
signaling pathways
In order to recognize signaling cascades related
with Rab22a in BC, GSEA was employed to analyze pathways
differentiated in the Rab22a high and low expressed data sets. GSEA
indicated noteworthy changes (FDR<0.25, P<0.05) in the
enrichment of the MSigDB Collection. The most significantly
enriched signaling cascades based on NES values are presented in
Fig. 6. GSEA found that in the
Rab22a overexpression group, 'E2F targets', 'G2M checkpoint',
'mammalian target of rapamycin complex 1 (mTorc1) pathway',
'protein secretion' and 'myc targets pathways' are upregulated
(Fig. 6A-E), while 'hypoxia',
'KRas signal-down', 'tumor necrosis factor (TNF)-α signal via
nuclear factor (NF)κB and p53 pathways' are downregulated (Fig. 6F-I).
Discussion
A number of studies on the possible function of
Rab22a have emerged during recent years (25,26). However, the role of Rab22a in BC
has not been discussed as much. In the present study, the
correlation between Rab22a expression and clinicopathological
characters and patient OS were examined. It was found that Rab22a
is overexpressed in BC tissues and that high expression of Rab22a
is an independent prognostic factor of BC. Moreover, Rab22a was
found to be required for the proliferation, migration and invasion
of BC cells.
Rab22a is a member of the RAS oncogene family and
the protein encoded gene is a member of the RAB family of small
GTPases. At present, >60 Rab members have been recognized
(27). The function of Rab
proteins is regulated by the cycling of the proteins between
GTP-bound active and GDP-bound inactive states, with the relative
amount of these two small GTPase states regulated by
GTPase-activating proteins and guanine nucleotide exchange factors
(28). Rab GTPases are considered
the main regulators of secretory and endocytic pathways (29). Endocytosis plays an imperative
role in cellular processes, including nutrient uptake and receptor
signaling (30). Some Rab GTPases
function as membrane trafficking regulators and determine protein
presentation for the neoplastic cell membrane (31). Defective vesicular trafficking of
growth factor receptors has been found to be a manifold hallmark of
cancerous cells (32). Rab22a has
been reported to control the trafficking of recycling endosomes
comprising clathrin-dependent factors, such as epidermal growth
factor receptor, transferrin receptor protein 2,
copper-transporting ATPase 1 and glucose transporter 4, as well as
clathrin-independent factors, such as MHC-I, β2-adrenergic receptor
and integrin b1 receptor are endocytosed cargoes to the cell
surface in non-melanocytes (26).
Rab22a is dysregulated in numerous cancer types.
However, the prognostic value of Rab22a has not been previously
investigated in BC. The present study found that Rab22a expression
is related to clinical stage, N classification, nipple invasion,
venous invasion and vital state. Increased expression of Rab22a in
the present BC cohorts is positively related to a poor prognosis in
patients. In the current study, it was shown that Rab22a is an
independent prognosis factor of BC. Therefore, Rab22a may be a
novel survival predictive biomarker for BC. These findings suggest
that the overexpression of Rab22a may act as a possible biomarker
to identify BC that may result in a poorer clinical outcome.
In order to investigate whether Rab22a affects BC
progression, Rab22a mRNA and protein levels were found to be
upregulated in BC cells compared with normal epithelium cells.
Knockdown of Rab22a inhibits BC cell proliferation in vitro
and in vivo. Moreover, loss function of Rab22a expression
decreases the migration and invasion capabilities of SK-BR-3 cells.
The present findings are consistent with other research reports.
Zhou et al (10) reported
that Rab22a is overexpressed in lung cancer and that knockdown of
Rab22a represses the migration and invasion of lung cancer cells.
Moreover, Rab22a improves recycling of cluster of differentiation
(CD)147 that is necessary for lung cancer cell migration and
invasion. A further study performed by Qi et al (33) identified that the transmembrane
protein, YIPY2, promotes CD147-medated malignant phenotypes by
recruiting and activating Rab5 and Rab22a in hepatocellular
carcinoma. Su et al (17)
found that the increased expression of Rab22a is related to an
advanced clinical stage in melanoma and shorter OS. The present
study suggests that Rab22a may act as a novel oncogene in BC and
may be a potential therapeutic target.
Atypical and uncontrolled cell proliferation is a
hallmark of malignancy and is triggered by the dysregulation of the
expression of numerous vital proteins. Although emerging evidence
shows the regulation between Rab22a and non-coding RNAs in
different types of tumors (7,34),
the regulatory mechanism of transcription or the signaling pathway
of Rab22a is not well studied. Therefore, a GSEA analysis was
performed to study the potential pathways related with Rab22a. GSEA
analyses indicated that in the Rab22a overexpression group, 'E2F
targets', 'G2M checkpoint', 'mTorc1 pathway', 'protein secretion'
and 'myc target pathways' are upregulated, while 'hypoxia', 'KRas
signal-down', 'TNF-α signal via NFκB and p53 pathways' are
downregulated. A set of Rab proteins have been reported to promote
tumor aggression by regulating intracellular signaling. The
signaling pathway identified by GSEA is consistent with that of
other published studies (35-37). Overexpression of Rab1A promotes
colorectal cancer progression and invasion by activating mTorc1
signaling. Moreover, it has been shown that BC stem cell
development is promoted by Rab2A through activation of
extracellular signaling (38).
While low Rab1B expression is reportedly correlated with poor
prognosis of BC, downregulation of Rab1B was found to promote
triple-negative breast cancer (TNBC) metastasis by activating
transforming growth factor-β/mothers against decapentaplegic
homolog signaling (39).
Hypoxia-inducible factors induces microvesicle (MV) generation by
upregulating Rab22a expression in human BC cells. However, Rab22a
knockdown abrogates MV generation under hypoxic conditions and
impairs lung metastasis (40).
However, the detailed underlying mechanism still requires
additional investigation. One limitation of the present research is
the loss-function study was only performed in one cell line;
further studies including gain-function will be conducted in
multiple BC cell lines.
In conclusion, this study has shown that Rab22a is
overexpressed in human BC tissues and cell lines, and is a
prognostic biomarker that is required for cell progression. The
present research suggests that Rab22a may be regarded as a possible
biomarker of prognosis and treatment of BC in the future.
Funding
The present study was supported by the Science and
Technology of Jilin Province Health and Family Planning Commission
Project 2017Q035(ZYY).
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
MH wrote the manuscript, and performed the data
analysis and interpretation. LS performed the statistical analysis
and followed-up the patients. CJ, ZK and GG performed the
immunochemistry staining estimation. KW collected the samples. YJ
performed the Gene Set Enrichment Analysis. LS, and YC performed
the in vivo experiments. ZK performed the in vitro
experiments. ZY designed the study. All authors read and
approved the final manuscript.
Ethics approval and consent to
participate
This study was approved by the Research Ethics
Committee of the China-Japan Union Hospital of Jilin University
(grant no. 2019022603). Animal protocol was approved by the
Laboratory Animal Care and Use Committee of Jilin University (grant
no. 201905005). This study was conducted with written informed
consent given by all patients.
Patient consent for publication
This study was conducted with written informed
consent from all patients.
Competing interest
The authors declare that there are no conflicts of
interest.
Abbreviations:
|
BC
|
breast cancer
|
|
Rab
|
Ras-related proteins in brain
|
|
ER
|
estrogen receptor
|
|
PR
|
progesterone receptor
|
|
Her-2
|
human epidermal growth factor-2
|
|
OS
|
overall survival
|
|
IHC
|
immunohistochemistry
|
|
SC
|
scramble control
|
|
GEO
|
Gene Expression Omnibus
|
|
TCGA
|
The Cancer Genome Atlas
|
|
GSEA
|
Gene Set Enrichment Analysis
|
|
NES
|
normalized enrichment score
|
|
FDR
|
false discovery rate
|
Acknowledgments
Not applicable.
References
|
1
|
DeSantis CE, Ma J, Gaudet MM, Newman LA,
Miller KD, Goding Sauer A, Goding Sauer A, Jemal A and Siegel RL:
Breast cancer statistics, 2019. CA Cancer J Clin. 69:438–451. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Fan L, Strasser-Weippl K, Li JJ, St Louis
J, Finkelstein DM, Yu KD, Chen WQ, Shao ZM and Goss PE: Breast
cancer in China. Lancet Oncol. 15:e279-e2892014. View Article : Google Scholar
|
|
3
|
Waks AG and Winer EP: Breast cancer
treatment: A review. JAMA. 321:288–300. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Sun L, He M, Xu N, Xu DH, Ben-David Y,
Yang ZY and Li YJ: Regulation of RAB22A by mir-193b inhibits breast
cancer growth and metastasis mediated by exosomes. Int J Oncol.
53:2705–2714. 2018.PubMed/NCBI
|
|
5
|
Zhang L and Yu S: Role of miR-520b in
non-small cell lung cancer. Exp Ther Med. 16:3987–3995.
2018.PubMed/NCBI
|
|
6
|
Xiong F, Liu K, Zhang F, Sha K, Wang X,
Guo X and Huang N: miR-204 inhibits the proliferation and invasion
of renal cell carcinoma by inhibiting RAB22A expression. Oncol Rep.
35:3000–3008. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Yang D, Liu G and Wang K: miR-203 acts as
a tumor suppressor gene in osteosarcoma by regulating RAB22A. PLoS
One. 10:e01322252015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
He H, Dai F, Yu L, She X, Zhao Y, Jiang J,
Chen X and Zhao S: Identification and characterization of nine
novel human small GTPases showing variable expressions in liver
cancer tissues. Gene Expr. 10:231–242. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Okamoto I, Pirker C, Bilban M, Berger W,
Losert D, Marosi C, Haas OA, Wolff K and Pehamberger H: Seven novel
and stable translocations associated with oncogenic gene expression
in malignant melanoma. Neoplasia. 7:303–311. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zhou Y, Wu B, Li JH, Nan G, Jiang JL and
Chen ZN: Rab22a enhances CD147 recycling and is required for lung
cancer cell migration and invasion. Exp Cell Res. 357:9–16. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Stenmark H: Rab GTPases as coordinators of
vesicle traffic. Nat Rev Mol Cell Biol. 10:513–525. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kauppi M, Simonsen A, Bremnes B, Vieira A,
Callaghan J, Stenmark H and Olkkonen VM: The small GTPase Rab22
interacts with EEA1 and controls endosomal membrane trafficking. J
Cell Sci. 115:899–911. 2002.PubMed/NCBI
|
|
13
|
Magadan JG, Barbieri MA, Mesa R, Stahl PD
and Mayorga LS: Rab22a regulates the sorting of transferrin to
recycling endosomes. Mol Cell Biol. 26:2595–2614. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Weigert R, Yeung AC, Li J and Donaldson
JG: Rab22a regulates the recycling of membrane proteins
internalized independently of clathrin. Mol Biol Cell.
15:3758–3770. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wang T, Gilkes DM, Takano N, Xiang L, Luo
W, Bishop CJ, Chaturvedi P, Green JJ and Semenza GL:
Hypoxia-inducible factors and RAB22A mediate formation of
microvesicles that stimulate breast cancer invasion and metastasis.
Proc Natl Acad Sci USA. 111:E3234–E3242. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Kang F, Zhu J, Wu J, Lv T, Xiang H, Tian
J, Zhang Y and Huang Z: O(2)-3-Aminopropyl diazeniumdiolates
suppress the progression of highly metastatic triple-negative
breast cancer by inhibition of microvesicle formation via nitric
oxide-based epigenetic regulation. Chem Sci. 9:6893–6898. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Su F, Chen Y, Zhu S, Li F, Zhao S, Wu L,
Chen X and Su J: RAB22A overexpression promotes the tumor growth of
melanoma. Oncotarget. 7:71744–71753. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
19
|
Bennett CN, Tomlinson CC, Michalowski AM,
Chu IM, Luger D, Mittereder LR, Aprelikova O, Shou J,
Piwinica-Worms H, Caplen NJ, et al: Cross-species genomic and
functional analyses identify a combination therapy using a CHK1
inhibitor and a ribonucleotide reductase inhibitor to treat
triple-negative breast cancer. Breast Cancer Res. 14:R1092012.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Tan MH, De S, Bebek G, Orloff MS,
Wesolowski R, Downs-Kelly E, Budd GT, Stark GR and Eng C: Specific
kinesin expression profiles associated with taxane resistance in
basal-like breast cancer. Breast Cancer Res Treat. 131:849–858.
2012. View Article : Google Scholar
|
|
21
|
Haakensen VD, Biong M, Lingjaerde OC,
Holmen MM, Frantzen JO, Chen Y, Navjord D, Romundstad L, Lüders T,
Bukholm IK, et al: Expression levels of uridine
5′-diphospho-gluc-uronosyltransferase genes in breast tissue from
healthy women are associated with mammographic density. Breast
Cancer Res. 12:R652010. View Article : Google Scholar
|
|
22
|
Mootha VK, Lindgren CM, Eriksson KF,
Subramanian A, Sihag S, Lehar J, Puigserver P, Carlsson E,
Ridderstråle M, Laurila E, et al: PGC-1alpha-responsive genes
involved in oxidative phosphorylation are coordinately
downregulated in human diabetes. Nat Genet. 34:267–273. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Subramanian A, Tamayo P, Mootha VK,
Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub
TR, Lander ES and Mesirov JP: Gene set enrichment analysis: A
knowledge-based approach for interpreting genome-wide expression
profiles. Proc Natl Acad Sci USA. 102:15545–15550. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Jiao Y, Fu Z, Li Y, Meng L and Liu Y: High
EIF2B5 mRNA expression and its prognostic significance in liver
cancer: A study based on the TCGA and GEO database. Cancer Manag
Res. 10:6003–6014. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Cebrian I, Croce C, Guerrero NA, Blanchard
N and Mayorga LS: Rab22a controls MHC-I intracellular trafficking
and antigen cross-presentation by dendritic cells. EMBO Rep.
17:1753–1765. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Shakya S, Sharma P, Bhatt AM, Jani RA,
Delevoye C and Setty SR: Rab22A recruits BLOC-1 and BLOC-2 to
promote the biogenesis of recycling endosomes. EMBO Rep.
19:e459182018. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Ishibashi K, Kanno E, Itoh T and Fukuda M:
Identification and characterization of a novel Tre-2/Bub2/Cdc16
(TBC) protein that possesses Rab3A-GAP activity. Genes Cells.
14:41–52. 2009. View Article : Google Scholar
|
|
28
|
Mesa R, Magadan J, Barbieri A, Lopez C,
Stahl PD and Mayorga LS: Overexpression of Rab22a hampers the
transport between endosomes and the Golgi apparatus. Exp Cell Res.
304:339–353. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Pfeffer SR: Rab GTPases: Master regulators
that establish the secretory and endocytic pathways. Mol Biol Cell.
28:712–715. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Maldonado-Baez L and Donaldson JG: Hook1,
microtubules, and Rab22: Mediators of selective sorting of
clathrin-independent endocytic cargo proteins on endosomes.
Bioarchitecture. 3:141–146. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Goldenring JR: A central role for vesicle
trafficking in epithelial neoplasia: Intracellular highways to
carcinogenesis. Nat Rev Cancer. 13:813–820. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Mosesson Y, Mills GB and Yarden Y:
Derailed endocytosis: An emerging feature of cancer. Nat Rev
Cancer. 8:835–850. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Qi S, Su L, Li J, Zhao P, Zhang Q, Niu X,
Liu J, Jia G, Wei X, Tavernier J, et al: YIPF2 is a novel Rab-GDF
that enhances HCC malignant phenotypes by facilitating CD147
endocytic recycle. Cell Death Dis. 10:4622019. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Zheng S, Jiang F, Ge D, Tang J, Chen H,
Yang J, Yao Y, Yan J, Qiu J, Yin Z, et al: LncRNA
SNHG3/miRNA-151a-3p/RAB22A axis regulates invasion and migration of
osteosarcoma. Biomed Pharmacother. 112:1086952019. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Mayorga LS and Cebrian I: Rab22a: A novel
regulator of immune functions. Mol Immunol. 11:87–92. 2018.
|
|
36
|
Rodriguez-Furlan C, Domozych D, Qian W,
Enquist PA, Li X, Zhang C, Schenk R, Winbigler HS, Jackson W,
Raikhel NV and Hicks GR: Interaction between VPS35 and RABG3f is
necessary as a checkpoint to control fusion of late compartments
with the vacuole. Proc Natl Acad Sci USA. 116:21291–21301. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Rajarajacholan UK, Thalappilly S and
Riabowol K: The ING1a tumor suppressor regulates endocytosis to
induce cellular senescence via the Rb-E2F pathway. PLoS Biol.
11:e10015022013. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Luo ML, Gong C, Chen CH, Hu H, Huang P,
Zheng M, Yao Y, Wei S, Wulf G, Lieberman J, et al: The Rab2A GTPase
promotes breast cancer stem cells and tumorigenesis via Erk
signaling activation. Cell Rep. 11:111–124. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Jiang HL, Sun HF, Gao SP, Li LD, Hu X, Wu
J and Jin W: Loss of RAB1B promotes triple-negative breast cancer
metastasis by activating TGF-β/SMAD signaling. Oncotarget.
6:16352–16365. 2015.PubMed/NCBI
|
|
40
|
Wu H, Carvalho P and Voeltz GK: Here,
there, and everywhere: The importance of ER membrane contact sites.
Science. 361:64012018. View Article : Google Scholar
|