Introduction
Osteosarcoma is a primary bone malignancy that
predominantly occurs in children and adolescents, and accounts for
60% of bone tumors cases (1,2).
In the past decade, great progress has been made in the treatment
of osteosarcoma, and current therapeutic strategies include
surgical resection, chemotherapy and radiotherapy. However, due to
a high degree of metastasis and recurrence, the outcome of patients
with osteosarcoma remains unsatisfactory (2). Although the molecular mechanisms of
osteosarcoma have attracted widespread attention, those involved in
disease progression have not been fully elucidated (3). Therefore, investigating the
molecular mechanism underlying the development of osteosarcoma may
aid the identification of novel biomarkers and potential targets
for the diagnosis and treatment of the disease (4).
MicroRNAs (miRNA/miR) are non-coding RNAs that have
been widely reported to inhibit the expression of their target
genes, and play key roles in cellular processes including
proliferation, differentiation and metastasis (5). Various studies have demonstrated
that miRNA dysregulation is one of the main factors leading to the
development of osteosarcoma (6).
Wang et al (7) revealed
that miR-193a is involved in the chemoresistance of osteosarcoma
cells and that upregulating the expression of miR-193a promotes
chemosensitivity. Liu et al (8) demonstrated that miR-377 may serve as
a tumor-suppressive miRNA by inducing apoptosis in osteosarcoma
cells. Hu et al (9)
revealed that miR-1285-3p exerts a tumor suppressor effect in
osteosarcoma and may serve as a novel biomarker for the diagnosis
of the disease. In addition, Chu et al (10) found that miR-136 expression was
low in a collection of osteosarcoma tissues compared with adjacent
normal tissues, and that this was negatively associated with
prognosis. Furthermore, Wang et al (11) revealed that miR-628-5p increased
the proliferation and migration of osteosarcoma cells by decreasing
the expression of interferon induced protein 44 like. Therefore, a
clear understanding of the effects and relative molecular
mechanisms of miRNAs may facilitate the identification of novel
biomarkers for the diagnosis of osteosarcoma, as well as potential
therapeutic targets (12).
The present study investigated the biological
functions and the latent mechanism of miR-145-5p in osteosarcoma
using a series of molecular biological experiments including CCK-8,
colony formation, cell cycle distribution analysis, western blot,
RT-qPCR and luciferase assays. The results obtained in the present
study suggest that miR-145-5p may serve as a diagnostic biomarker
and therapeutic target in patients with osteosarcoma, which has
potential to inhibit the proliferation of osteosarcoma via
targeting E2F transcription factor 3 (E2F3).
Materials and methods
Patient samples
A total of 20 patients were enrolled in the present
study, who were pathologically diagnosed with osteosarcoma and had
received surgical resection at Guizhou Orthopedics Hospital
(Guizhou, China) between March 2014 and May 2019. The patients
included 14 males and 6 females with a mean age of 17.15±15.17
years (range 7–45 years); 11 patients were diagnosed in the early
stage, while 9 patients were diagnosed in the advanced stage of
disease. The inclusion criteria were as follows: i) The tissues
were obtained during surgery and osteosarcoma was diagnosed by two
pathologists; ii) the patients were diagnosed and treated for the
first time; and iii) the patients were willing to participate. The
exclusion criteria were as follows: i) Patients with other
malignancies; ii) patients with other systemic diseases; iii)
patients who had received treatment prior to admission; and iv)
patients (and/or their families) who refused to participate. All 20
patients provided osteosarcoma tissue samples, while only 10
patients also provided adjacent normal tissues. The present study
was approved by the Committee of Guizhou Orthopedics Hospital and
was performed in accordance with the Declaration of Helsinki. All
patients provided written informed consent.
Cell culture and lentivirus
transfection
In total, five osteosarcoma cell lines (U2OS, Saos2,
MG63, SJSA-1 and 143B) and a normal osteoblast line (hFOB) were
purchased from The Cell Bank of Type Culture Collection of the
Chinese Academy of Sciences. Well5 cells (5) were purchased from the Chinese
National Infrastructure of Cell Line Resource (cat. no.
3142C0001000000722; http://www.cellresource.cn/index.aspx). All cell lines
were previously authenticated by short tandem repeat typing. The
cells were cultured in Dulbecco’s Modified Eagle Medium (DMEM;
Gibco; Thermo Fisher Scientific, Inc.) supplemented with 10% fetal
bovine serum (FBS; Gibco; Thermo Fisher Scientific, Inc.) and
maintained in a humidified incubator at 37°C (5% CO2).
Lentiviruses containing miR-145-5p- and non-targeting miRNA DNA
encoding plasmids (LV-miR-145-5p and LV-miR-NC, respectively) were
purchased from Shanghai GeneChem Co., Ltd. Transfection was
performed using Lipo2000 (Shanghai GeneChem Co., Ltd) according to
the manufacturer’s protocol (MOI=20 for 143B and Well5 cells). The
miR-145-5p sequence used for lentiviral construction was
5′-GUCCAGUUUUCCCAGGAAUCCCU-3′, and the LV-miR-NC sequence was
5′-UUCUCCGAACGUGUCACGUTT-3′. E2F3 (NM_001949.5) was used as a
backbone and sub-cloned into the pcDNA3.1 vector to construct an
E2F3 overexpression plasmid (Shanghai GeneChem Co., Ltd.), and an
empty pcDNA3.1 vector was used as a negative control (NC).
Reverse transcription-quantitative PCR
(RT-qPCR)
Total RNA was extracted from the osteosarcoma
tissues and cell lines using TRIzol® reagent, and
subsequently reverse transcribed into cDNA using an mRNA First
Strand cDNA Synthesis kit (both Yeasen Biotech Co., Ltd.) according
to the manufacturer’s protocols. miRNAs were reverse transcribed
using the miRNA First Strand cDNA Synthesis kit, and qPCR was
performed using the qPCR SYBR® Green Master Mix (both
Yeasen Biotech Co., Ltd.). The following primer pairs were used:
miR-145-5p forward, 5′-GTCCAGTTTTCCCAGGAATC-3′ and reverse,
5′-AGAACAGTATTTCCAGGAAT-3′; E2F3 forward,
5′-AGAAAGCGGTCATCAGTACCT-3′ and reverse,
5′-TGGACTTCGTAGTGCAGCTCT-3′; GAPDH forward,
5′-GGAGCGAGATCCCTCCAAAAT-3′ and reverse,
5′-GGCTGTTGTCATACTTCTCATGG-3′; and U6 forward,
5′-GCTTCGGCAGCACATATACTAAAAT-3′ and reverse,
5′-CGCTTCACGAATTTGCGTGTCAT-3′. Relative expression levels were
quantified using the 2−ΔΔCq method (13). GAPDH was used as the reference
gene for E2F3, and U6 was used as the reference for miR-145-5p
expression. The qPCR thermocycling conditions were as follows: 95°C
for 5 min, 40 cycles of 95°C for 25 sec, annealing at 60°C for 40
sec, and a final elongation step at 72°C for 30 sec.
Cell Counting Kit-8 (CCK-8) assay
Following transfection with the miR-145-5p and NC
lentiviruses (and subsequent incubation for 48 h), 143B and Well5
cells were seeded into 96-well plates at a density of
3x103 cells/well, and cultured for 24, 48, 72 and 96 h.
Subsequently, 10 μl CCK-8 solution (Wuhan Boster Biological
Technology, Ltd.) was added to each well according to the
manufacturers protocol, and the cells were cultured for an
additional 2 h. The absorbance was measured at a wavelength of 450
nm.
Colony formation assay
At 48 h post-transfection with the miR-145-5p and NC
lentiviruses, 143B and Well5 cells were seeded into a 6-well plate
at a density of 1×103 cells/well and cultured for 2
weeks. The cells were subsequently fixed with 4% polyoxymethylene
(Wuhan Boster Biological Technology, Ltd.) for 30 min and stained
with 1% crystal violet (Wuhan Boster Biological Technology, Ltd.)
for 20 min, both at room temperature. The cells were photographed
and the colonies were counted by eye.
Cell cycle distribution analysis
Osteosarcoma 143B and Well5 cells were harvested and
fixed using 70% cold ethanol for 24 h at 4°C. The cells were then
stained with propidium iodide (Invitrogen; Thermo Fisher
Scientific, Inc.) for 30 min at room temperature in the dark. The
cell cycle distribution was analyzed using a NovoCyte flow
cytometer (Agilent Technologies, Inc.,) and FlowJo software (FlowJo
LLC, Version: 7.6.1).
Western blotting
Total protein was extracted from osteosarcoma
tissues and 143B and Well5 cells using RIPA lysis buffer
supplemented with protease inhibitor cocktail (1:100) and
phenylmethylsulfonyl fluoride (1:100) (all Wuhan Boster Biological
Technology, Ltd.). The protein concentration was determined using a
bicinchoninic acid assay, and the protein samples (30 μg
each) were separated via SDS-PAGE using a 10% gel. The separated
proteins were transferred onto a PVDF membrane and blocked using 5%
skim milk for 2 h at room temperature. The membrane was
subsequently incubated with primary antibodies against cyclin D1
(cat. no. 19532-1-AP; 1:1,000), cyclin E1 (cat. no. 11554-1-AP;
1:1,000), cyclin dependent kinase (CDK) 2 (cat. no. 10122-1-AP;
1:1,000), CDK4 (cat. no. 11026-1-AP; 1:1,000), CDK6 (cat. no.
14052-1-AP; 1:1,000), E2F3 (cat. no. 27615-1-AP; 1:1,000) and GAPDH
(cat. no. 60004-1-Ig; 1:1,000) (all ProteinTech, Inc.) for 14 h at
4°C. The membrane was then washed using TBS-T and incubated with
horseradish peroxidase (HRP)-conjugated secondary goat anti-mouse
(cat. no. BA1050; 1:3,000) or goat anti-rabbit antibodies (cat. no.
BA1054; 1:3,000) (both Wuhan Boster Biological Technology, Ltd.)
for 2 h at room temperature. Finally, the protein bands were
visualized using ECL reagent (Wuhan Boster Biological Technology,
Ltd) and quantified using Image Lab v2 (Bio-Rad Laboratories Inc,).
GAPDH served as the loading control.
Animal experiments
Female BALB/c nude mice (n=10; age, 4–6 weeks;
weight, 15–18 g) were obtained from the Laboratory Animal Center of
Guizhou Medical University (Guizhou, China). Mice were housed under
specific pathogen-free conditions at 25°C with a 12-h light/dark
cycle and free access to food and water. A total of
1×107 143B cells overexpressing miR-145-5p or NC were
injected into the subcutaneous tissues of the upper-right flank.
The tumor volume was measured weekly as follows: Volume
(mm3) = (length × width2)/2. The mice were
sacrificed after 5 weeks and the tumor weights were measured. The
animal experiments were approved by the Ethics Committee of Guizhou
Medical University (Guizhou, China).
Immunohistochemical (IHC) staining
The tumor tissues acquired from the nude mice were
dehydrated and embedded in paraffin (Wuhan Servicebio Technology
Co., Ltd.) at room temperature. After being cut into 4-μm
slices, the tissues were deparaffinized using xylene and rehydrated
in a descending alcohol series at room temperature. After
restoration with sodium citrate the samples were treated with 3%
H2O2 to block endogenous peroxidase activity,
and then blocked using 5% bovine serum albumin (BSA; Wuhan
Servicebio Biotechnology Co., Ltd.) for 30 min at room temperature.
The specimens were subsequently incubated with a primary anti-KI67
antibody (cat. no. 27309-1-AP; 1:400; ProteinTech) or anti-PCNA
antibody (cat. no. 10205-2-AP; 1:400; ProteinTech) for 12 h at 4°C,
followed by a second incubation with HRP-conjugated secondary
antibodies (cat. no. G1210-2-A-100; 1:200; Wuhan Servicebio
Biotechnology Co., Ltd.) for 2 h at room temperature. After
subsequent development using the Cell and Tissue Staining HRP-DAB
kit (Beyotime Institute of Biotechnology) according to the
manufacturer’s protocol, images were captured with an orthophoto
microscope (magnification, ×400).
Luciferase reporter assay
TargetScan software (www.targetscan.org; Version 7.2) was used to predict
the target genes of miR-145-5p. A dual-luciferase reporter assay
was conducted to verify whether miR-145-5p binds to and regulates
the expression of E2F3. The wild type (Wt) and mutant (Mut) 3′
untranslated regions (UTR) of E2F3 were synthesized and sub-cloned
into the psiCHECK-2 luciferase reporter vector (Promega
Corporation). A total of 5×103 143B and Well5 cells were
cultured for 24 h and co-transfected with the Wt/Mut E2F3
luciferase reporter vector, and the miR-145-5p mimic or NC, using
LipofectamineTM 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.). Finally, a dual-luciferase reporter assay system
(Promega Corporation) was used to quantify luciferase activity 24 h
post-transfection. Luciferase activity was normalized to that of
Renilla luciferase.
Statistical analysis
Statistical analyses were performed using SPSS
software (version 21.0; IBM Corp.). The paired t-test was used to
compare two groups while one-way analysis of variance and Dunnett’s
post hoc test were used for the comparison of multiple groups.
P<0.05 was considered to indicate a statistically significant
difference.
Results
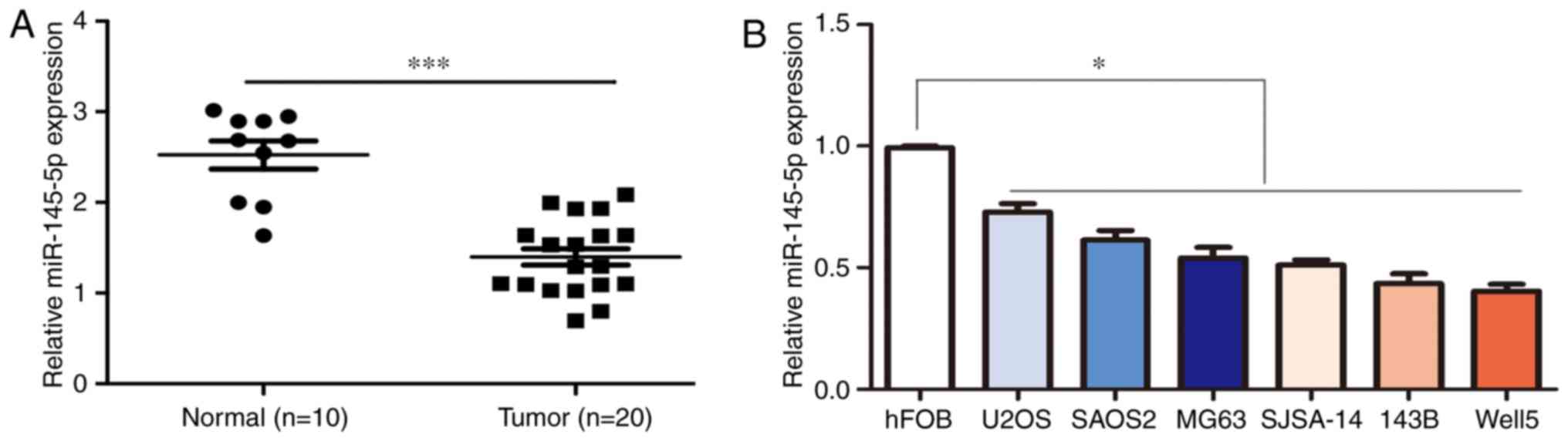
miR-145-5p expression is decreased in
osteosarcoma tissues
The expression levels of miR-145-5p in osteosarcoma
and normal adjacent tissues were quantified using RT-qPCR.
miR-145-5p expression was significantly decreased in osteosarcoma
tissues (n=20) compared with adjacent normal tissues (n=10;
Fig. 1A). The expression level of
miR-145-5p was also decreased in six osteosarcoma cell lines (U2OS,
Saos2, MG63, SJSA-1, 143B and Well5) compared with the normal hFOB
osteoblasts (Fig. 1B). Due to the
evidence that the expression of miR-145-5p was decreased most
significantly in 143B and Well5 cells, we then used these two cell
lines to detect biology function of miR-145-5p in osteosarcoma.
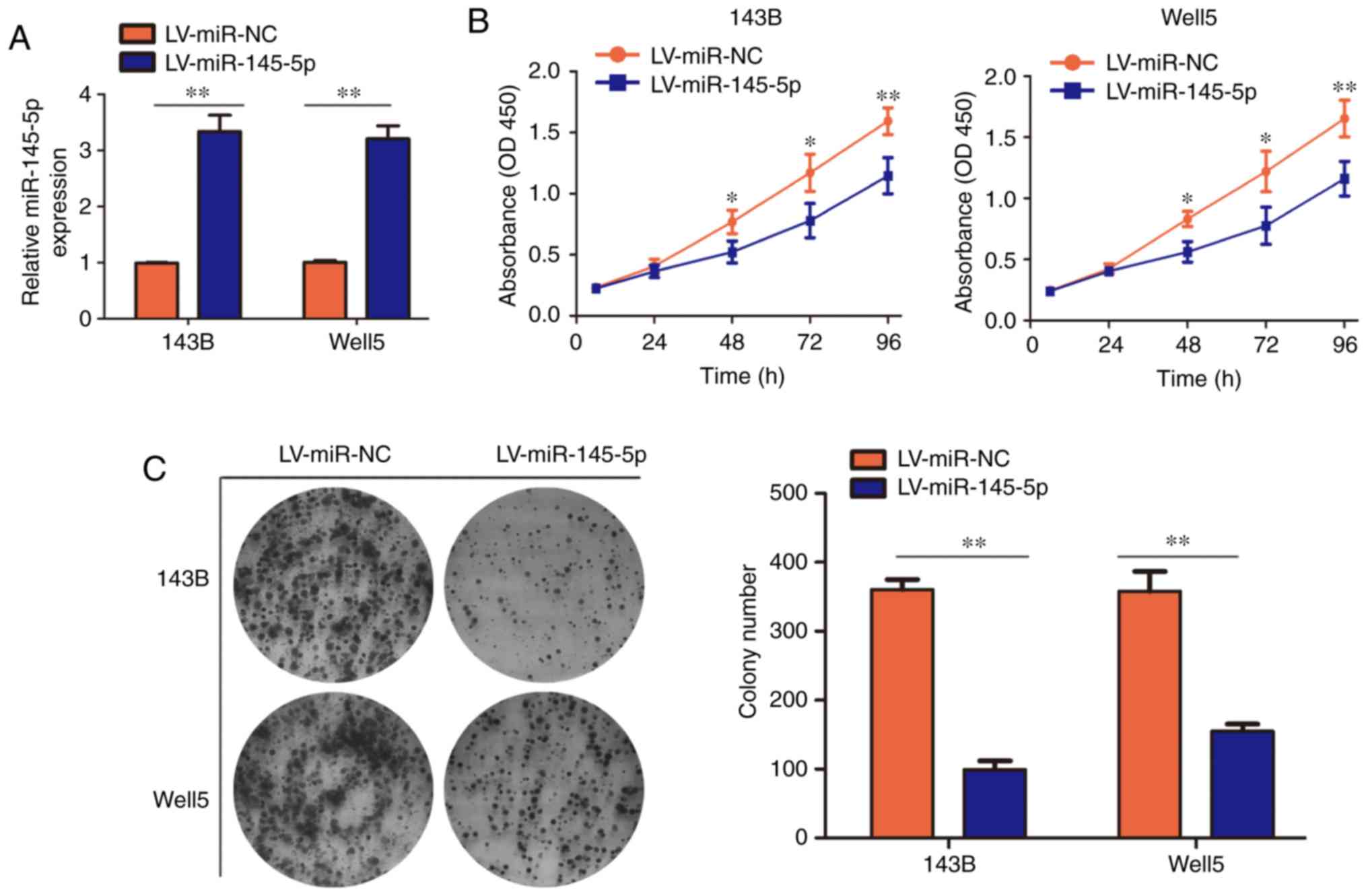
miR-145-5p inhibits proliferation in
vitro
In order to investigate the role of miR-145-5p in
osteosarcoma, an osteosarcoma cell line that overexpressed
miR-145-5p (LV-miR-145-5p) and a corresponding normal control cell
line (LV-miR-NC) were successfully constructed (Fig. 2A). The results of the CCK-8 assay
revealed that miR-145-5p overexpression significantly inhibited the
proliferation of 143B and Well5 cells at 48, 72 and 96 h, compared
with LV-miR-NC cells (Fig. 2B).
Similarly, the results of the colony formation assay demonstrated
that miR-145-5p overexpression significantly reduced colony
formation in osteosarcoma cells compared with the LV-miR-NC cell
line (Fig. 2C).
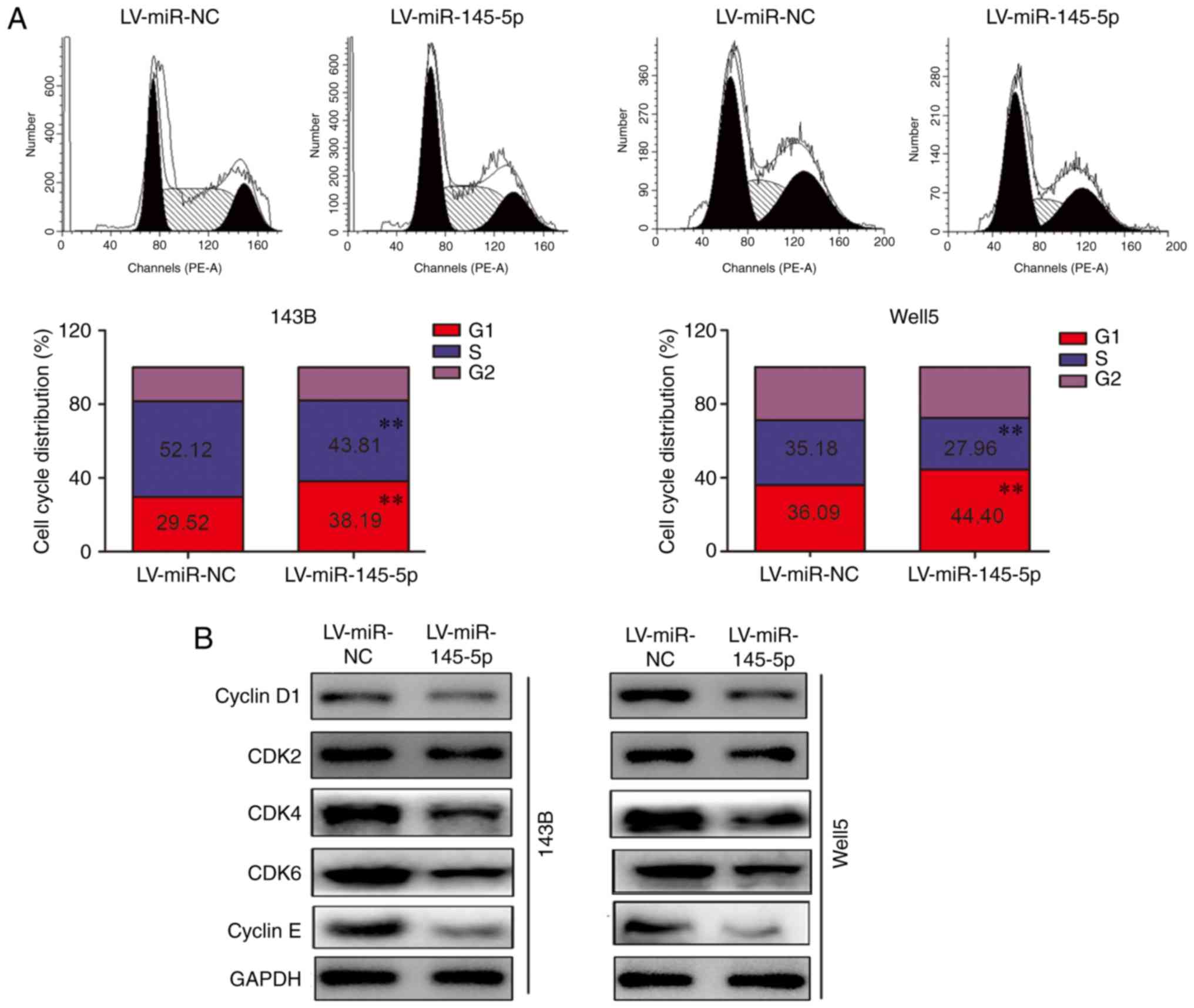
miR-145-5p induces G1 phase
arrest
The results of cell cycle distribution analysis
revealed that miR-145-5p overexpression significantly increased the
proportion of 143B and Well5 cells in the G1 phase of
the cell cycle, and decreased the proportion of cells in the S
phase (Fig. 3A). Previous studies
have revealed that cyclin D-CDK4/CDK6 and cyclin E/CDK2 are
involved in cell cycle regulation at the G1 phase, and
that increased expression of these proteins is required for cells
in G1 phase to pass the G1/S check point (14,15). In the present study, western
blotting revealed that the protein expression levels of cyclin D1,
cyclin E, CDK2, CDK4 and CDK6 were decreased in cells
overexpressing miR-145-5p compared with the LV-miR-NC cells
(Fig. 3B).
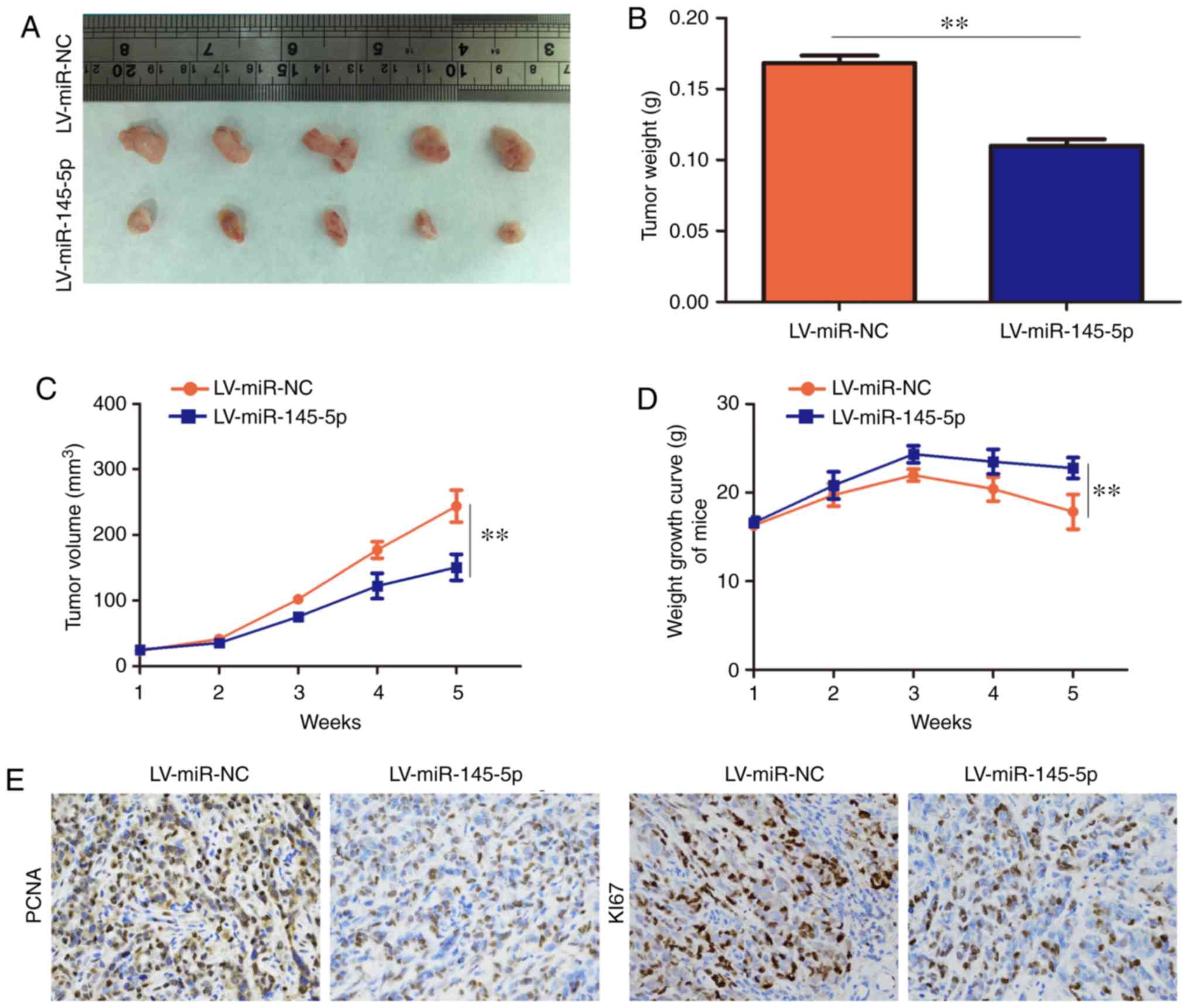
miR-145-5p inhibits osteosarcoma tumor
growth in vivo
miR-145-5p overexpression inhibited the
proliferation of osteosarcoma cells in vitro. Therefore, the
effect of miR-145-5p overexpression in vivo was also
investigated. The results revealed that mice injected with 143B
cells overexpressing miR-145-5p exhibited slower tumor growth and
lower tumor volumes compared with the LV-miR-NC group (Fig. 4A–C). Furthermore, the weight-loss
rate of the mice injected with 143B cells overexpressing miR-145-5p
was slower than that of the LV-miR-NC group (Fig. 4D). Moreover, the expression levels
of KI67 and PCNA were decreased in the miR-145-5p-over-expression
group compared with the LV-miR-NC group (Fig. 4E). Collectively, these results
suggest that increasing the expression level of miR-145-5p inhibits
tumor growth in vivo.
E2F3 is directly regulated by
miR-145-5p
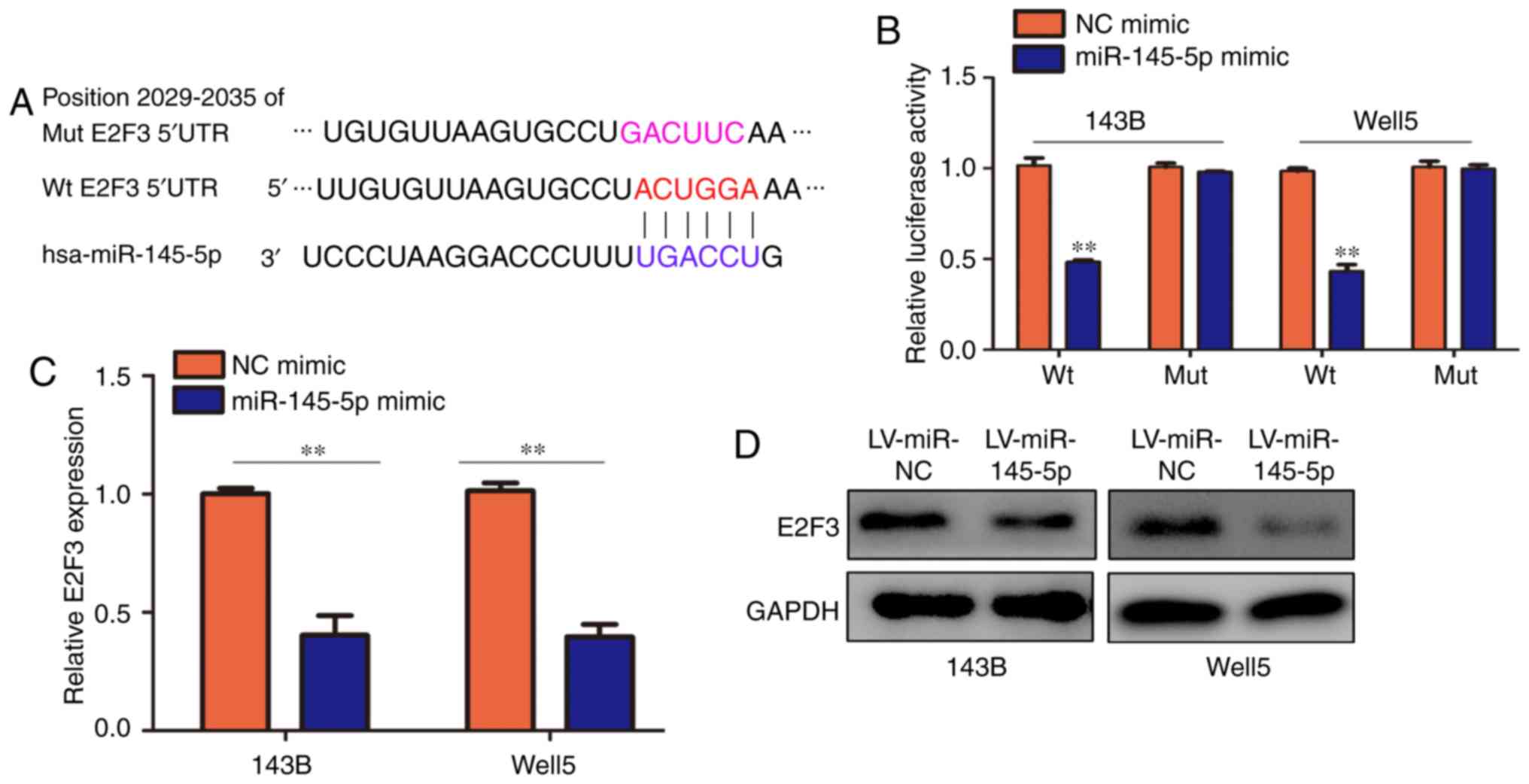
Using TargetScan, E2F3 (which has conservative
binding sites for miR-145-5p; Fig.
5A) was identified as one of the downstream targets of
miR-145-5p. Previous studies showed that miR-145-5p was indicated
as a novel biomarker in osteosarcoma (16), and therefore, E2F3 was
hypothesized to be a key target of miR-145-5p in osteosarcoma. A
luciferase reporter assay was subsequently used to validate this
result. Co-transfection of miR-145-5p mimics in 143B and Well5
cells significantly inhibited the luciferase activity of the WT
E2F3 3′-UTR sequence, but failed to inhibit that of the MUT E2F3
3′-UTR (Fig. 5B). The mRNA and
protein expression levels of E2F3 were also significantly decreased
in cells overexpressing miR-145-5p compared with the LV-miR-NC
cells (Fig. 5C and D).
Furthermore, the mRNA expression levels of E2F3 were negatively
associated with miR-145-5p in osteosarcoma tissues (Fig. 6A), and E2F3 expression was
increased in osteosarcoma tissues (Fig. 6B) and cell lines (Fig. 6C and D).
 | Figure 6E2F3 is highly expressed in
osteosarcoma tissues and cell lines. (A) miR-145-5p expression was
inversely associated with that of E2F3 in osteosarcoma tissues. (B)
The mRNA expression levels of E2F3 were detected in 20 osteosarcoma
tissues and 10 normal adjacent tissues using RT-qPCR. (C) The mRNA
expression levels of E2F3 in the U2OS, Saos2, MG63, SJSA-1, 143B
and Well5 osteosarcoma cell lines, and in human normal osteoblast
hFOB cells were detected using RT-qPCR. (D) The protein expression
levels of E2F3 in U2OS, Saos2, MG63, SJSA-1, 143B and Well5 cells,
and in the human normal osteoblasts (hFOB) were detected using
RT-qPCR. *P<0.05 and **P<0.01. E2F3,
E2F transcription factor 3; miR, microRNA; RT-qPCR, reverse
transcription-quantitative PCR. |
Restoration of E2F3 decreases the
inhibitory effect induced by miR-145-5p
To investigate whether E2F3 was involved in the
miR-145-5p-induced inhibitory effect on proliferation, the
expression of E2F3 in miR-145-5p-overexpressing cells was restored
(Fig. 7A). CCK-8 and colony
formation assays revealed that restoring E2F3 expression decreased
the inhibitory effect of miR-145-5p on the proliferation of
osteosarcoma cells (Fig. 7B and
C). Furthermore, cell cycle distribution analysis revealed that
co-transfection with lentiviruses delivering miR-145-5p and E2F3
significantly decreased the proportion of osteosarcoma cells in the
G1 phase (Fig. 7D).
Additionally, co-transfection increased the expression of cyclin
D1, cyclin E, CDK2, CDK4 and CDK6 (Fig. 7E).
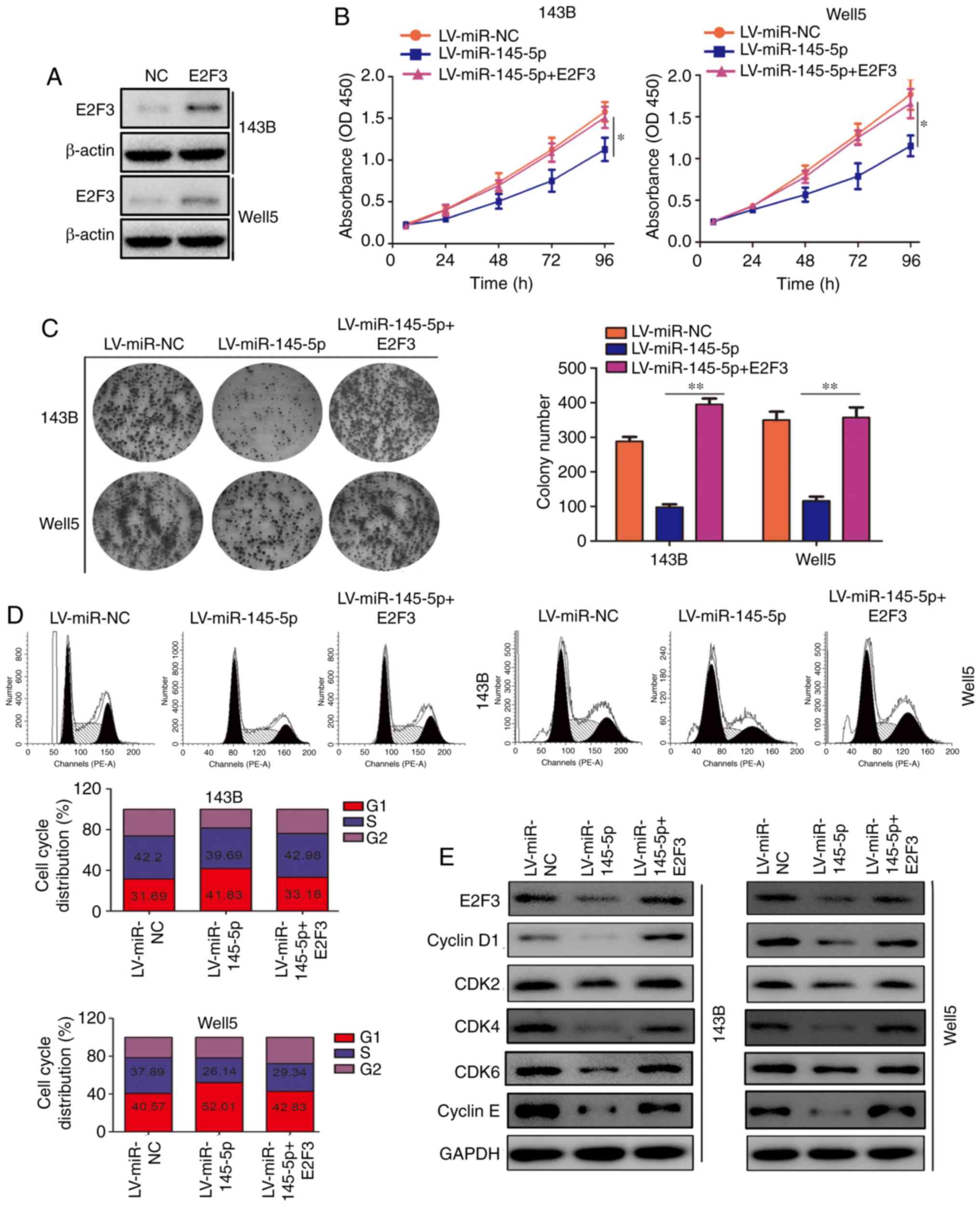 | Figure 7Restoration of E2F3 reverses the
inhibitory effects of miR-145-5p. (A) The efficiency of the E2F3
overexpression plasmid was detected using western blotting. 143B
and Well5 cells were divided into 3 groups and transfected with LV,
LV-miR-145-5p or LV-miR-145-5p + E2F3 plasmids. (B) Cell Counting
Kit-8 and (C) colony formation assays were used to detect
proliferation in each group. (D) Cell distribution analysis was
also performed. (E) Western blotting was used to detect the
expression levels of E2F3, CDK2, CDK4, CDK6, cyclin E and cyclin D1
in each group. *P<0.05 and **P<0.01.
E2F3, E2F transcription factor 3; LV, lentivirus; miR, microRNA;
NC, negative control; LV, lentivirus; CDK, cyclin depen dent
kinase. |
Discussion
The biological function of miR-145-5p has been
demonstrated in various cancer types, including bladder cancer
(17), laryngocarcinoma (18) and gall bladder carcinoma (19). Wang et al (20) indicated that miR-145-5p was
downregulated in hepatocellular carcinoma and served as a tumor
suppressor. Tang et al (21) demonstrated that the expression
level of miR-145-5p was decreased in breast cancer tissues compared
with adjacent tissues, and was negatively associated with overall
survival rate. Jian et al (22) revealed that miR-145-5p decreased
the drug resistance of prolactinoma and increased sensitivity to
chemotherapy. In addition, Zhou et al (23) showed that miR-145-5p affected the
differentiation of gastric cancer cells by inhibiting the
expression of Kruppel like factor 5, and Wu et al (24) demonstrated that downregulation of
miR-145-5p increased the proliferation and migration of bladder
cancer cells. In the present study, miR-145-5p was found to be
downregulated in osteosarcoma tissues and cell lines compared with
the corresponding normal controls. Overexpression of miR-145-5p
decreased the proliferation of osteosarcoma cells, inhibited colony
formation and induced G1 phase arrest. Similarly,
overexpression of miR-145-5p significantly decreased the expression
of cyclin D, CDK4, CDK6, cyclin E and CDK2 in osteosarcoma,
preventing the cells from passing the G1 phase check
point. To the best of our knowledge, the present study is the first
to suggest that miR-145-5p serves as a tumor-suppressive miRNA in
osteosarcoma.
It is widely reported that miRNAs regulate
transcription by binding to their target genes (11). Bioinformatics analysis has
revealed a number of potential target genes of miR-145-5p,
including E2F3, an important member of the E2F family that
participates in the cell cycle, metastasis and proliferation
(25). The role of E2F3 in
several types of cancer, including osteosarcoma, has been reported
(26–28). E2F3 was indicated to be involved
in the progression of melanoma, and to promote cellular
proliferation (29).
Downregulation of E2F3 also decreased the proliferation and
migration of hepatocellular carcinoma cells (30) and inhibited the progression of
pancreatic cancer (31,32). Furthermore, various studies have
indicated that the downregulation of E2F3 may suppress the
proliferation and metastasis of osteosarcoma (26), and that E2F3 may serve as a
biomarker for this disease (16).
The present study revealed that E2F3 is directly and
negatively regulated by miR-145-5p. In addition, overexpression of
miR-145-5p inhibited the expression of E2F3. E2F3 was highly
expressed in osteosarcoma tissues and cell lines, and its
expression was negatively associated with that of miR-145-5p.
Restoration of E2F3 reversed the inhibitory effects on
proliferation induced by miR-145-5p. To the best of our knowledge,
the present study is the first to demonstrate the potential link
between miR-145-5p and E2F3 in osteosarcoma.
In conclusion, the present study demonstrated that
miR-145-5p may act as a tumor-suppressive miRNA in osteosarcoma.
miR-145-5p suppressed osteosarcoma cell proliferation by targeting
E2F3; therefore, miR-145-5p and E2F3 may serve as novel diagnostic
biomarkers and therapeutic targets for osteosarcoma.
Acknowledgements
Not applicable.
Funding
The present study was supported by Houping Chen
[Qiankehe; grant. no. SY(2013)3042].
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors’ contributions
HL, RP, QL and CR performed the experiments; CR also
collected the samples. JS and HW contributed to the data analysis
and presentation. JW and HC designed the experiment and wrote the
manuscript. All authors read and approved the final version of the
manuscript.
Ethics approval and consent to
participate
The present study was approved by the ethics
Committee of Guizhou Orthopedics Hospital approved. Written
informed consent was obtained from the patients who provided the
specimens. The present study was performed in accordance with the
principles outlined in the Declaration of Helsinki.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Fox E, Levin K, Zhu Y, Segers B, Balamuth
N, Womer R, Bagatell R and Balis F: Pantoprazole, an inhibitor of
the organic cation transporter 2, does not ameliorate
cisplatin-related ototoxicity or nephrotoxicity in children and
adolescents with newly diagnosed osteosarcoma treated with
methotrexate, doxorubicin, and cisplatin. Oncologist. 23:e762–e779.
2018. View Article : Google Scholar
|
|
2
|
Endicott AA, Morimoto LM, Kline CN,
Wiemels JL, Metayer C and Walsh KM: Perinatal factors associated
with clinical presentation of osteosarcoma in children and
adolescents. Pediatr Blood Cancer. 64:e263492017. View Article : Google Scholar
|
|
3
|
Kebudi R, Ozger H, Kizilocak H, Bay SB and
Bilgic B: Osteosarcoma after hematopoietic stem cell
transplantation in children and adolescents: Case report and review
of the literature. Pediatr Blood Cancer. 63:1664–1666. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Lee JA, Paik EK, Seo J, Kim DH, Lim JS,
Yoo JY and Kim MS: Radiotherapy and gemcitabine-docetaxel
chemotherapy in children and adolescents with unresectable
recurrent or refractory osteosarcoma. Jpn J Clin Oncol. 46:138–143.
2016.
|
|
5
|
Han Q, Li C, Cao Y, Bao J, Li K, Song R,
Chen X, Li J and Wu X: CBX2 is a functional target of miRNA let-7a
and acts as a tumor promoter in osteosarcoma. Cancer Med.
8:3981–3991. 2019.PubMed/NCBI
|
|
6
|
Huang C, Wang Q, Ma S, Sun Y, Vadamootoo
AS and Jin C: A four serum-miRNA panel serves as a potential
diagnostic biomarker of osteosarcoma. Int J Clin Oncol. 24:976–982.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Wang H, Zhao F, Cai S and Pu Y: MiR-193a
regulates chemoresistance of human osteosarcoma cells via
repression of IRS2. J Bone Oncol. 17:1002412019. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Liu XG, Xu J, Li F, Li MJ and Hu T:
Down-regulation of miR-377 contributes to cisplatin resistance by
targeting XIAP in osteosarcoma. Eur Rev Med Pharmacol Sci.
22:1249–1257. 2018.PubMed/NCBI
|
|
9
|
Hu XH, Dai J, Shang HL, Zhao ZX and Hao
YD: MiR-1285-3p is a potential prognostic marker in human
osteosarcoma and functions as a tumor suppressor by targeting YAP1.
Cancer Biomark. 25:1–10. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Chu Y, Hu X, Wang G, Wang Z and Wang Y:
Downregulation of miR-136 promotes the progression of osteosarcoma
and is associated with the prognosis of patients with osteosarcoma.
Oncol Lett. 17:5210–5218. 2019.PubMed/NCBI
|
|
11
|
Wang JY, Lu SB and Wang JQ: MiR-628-5p
promotes the growth and migration of osteosarcoma by targeting
IFI44L. Biochem Cell Biol. Apr 24–2019.(Epub ahead of print).
|
|
12
|
Ouyang L, Liu P, Yang S, Ye S, Xu W and
Liu X: A three-plasma miRNA signature serves as novel biomarkers
for osteosarcoma. Med Oncol. 30:3402013. View Article : Google Scholar
|
|
13
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
14
|
Bertoli C, Skotheim JM and de Bruin RA:
Control of cell cycle transcription during G1 and S phases. Nat Rev
Mol Cell Biol. 14:518–528. 2013. View
Article : Google Scholar : PubMed/NCBI
|
|
15
|
Crncec A and Hochegger H: Triggering
mitosis. FEBS Lett. 593:2868–2888. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Schwentner R, Papamarkou T, Kauer MO,
Stathopoulos V, Yang F, Bilke S, Meltzer PS, Girolami M and Kovar
H: EWS-FLI1 employs an E2F switch to drive target gene expression.
Nucleic Acids Res. 43:2780–2789. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zhang H, Jiang M, Liu Q, Han Z, Zhao Y and
Ji S: MiR-145-5p inhibits the proliferation and migration of
bladder cancer cells by targeting TAGLN2. Oncol Lett. 16:6355–6360.
2018.PubMed/NCBI
|
|
18
|
Zhuang S, Liu F and Wu P: Upregulation of
long noncoding RNA TUG1 contributes to the development of
laryngocarcinoma by targeting miR-145-5p/ROCK1 axis. J Cell
Biochem. 120:13392–13402. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Goeppert B, Truckenmueller F, Ori A, Fritz
V, Albrecht T, Fraas A, Scherer D, Silos RG, Sticht C, Gretz N, et
al: Profiling of gallbladder carcinoma reveals distinct miRNA
profiles and activation of STAT1 by the tumor suppressive
miRNA-145-5p. Sci Rep. 9:47962019. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wang B, Dong W and Li X: MiR-145-5p acts
as a novel tumor suppressor in hepatocellular carcinoma through
targeting RAB18. Technol Cancer Res Treat. 18:1533033819850189.
2019. View Article : Google Scholar
|
|
21
|
Tang W, Zhang X, Tan W, Gao J, Pan L, Ye
X, Chen L and Zheng W: MiR-145-5p suppresses breast cancer
progression by inhibiting SOX2. J Surg Res. 236:278–287. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Jian M, Du Q, Zhu D, Mao Z, Wang X, Feng
Y, Xiao Z, Wang H and Zhu Y: Tumor suppressor miR-145-5p sensitizes
prolactinoma to bromocriptine by downregulating TPT1. J Endocrinol
Invest. 42:639–652. 2019. View Article : Google Scholar
|
|
23
|
Zhou T, Chen S and Mao X: MiR-145-5p
affects the differentiation of gastric cancer by targeting KLF5
directly. J Cell Physiol. 234:7634–7644. 2019. View Article : Google Scholar
|
|
24
|
Wu Z, Huang W, Wang X, Wang T, Chen Y,
Chen B, Liu R, Bai P and Xing J: Circular RNA CEP128 acts as a
sponge of miR-145-5p in promoting the bladder cancer progression
via regulating SOX11. Mol Med. 24:402018. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Shen ZG, Liu XZ, Chen CX and Lu JM:
Knockdown of E2F3 inhibits proliferation, migration, and invasion
and increases apoptosis in glioma cells. Oncol Res. 25:1555–1566.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Dong D, Gong Y, Zhang D, Bao H and Gu G:
MiR-874 suppresses the proliferation and metastasis of osteosarcoma
by targeting E2F3. Tumour Biol. 37:6447–6455. 2016. View Article : Google Scholar
|
|
27
|
Jin Y, Wei J, Xu S, Guan F, Yin L and Zhu
H: MiR210-3p regulates cell growth and affects cisplatin
sensitivity in human ovarian cancer cells via targeting E2F3. Mol
Med Rep. 19:4946–4954. 2019.PubMed/NCBI
|
|
28
|
Liu J, Si L and Tian H: MicroRNA-148a
inhibits cell proliferation and cell cycle progression in lung
adenocarcinoma via directly targeting transcription factor E2F3.
Exp Ther Med. 16:5400–5409. 2018.PubMed/NCBI
|
|
29
|
Feng Z, Peng C, Li D, Zhang D, Li X, Cui
F, Chen Y and He Q: E2F3 promotes cancer growth and is
overexpressed through copy number variation in human melanoma. Onco
Targets Ther. 11:5303–5313. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Han R, Chen X, Li Y, Zhang S, Li R and Lu
L: MicroRNA-34a suppresses aggressiveness of hepatocellular
carcinoma by modulating E2F1, E2F3, and Caspase-3. Cancer Manag
Res. 11:2963–2976. 2019. View Article : Google Scholar :
|
|
31
|
Sun FB, Lin Y, Li SJ, Gao J, Han B and
Zhang CS: MiR-210 knockdown promotes the development of pancreatic
cancer via upregulating E2F3 expression. Eur Rev Med Pharmacol Sci.
22:8640–8648. 2018.PubMed/NCBI
|
|
32
|
Yang J, Zhang HF and Qin CF: MicroRNA-217
functions as a prognosis predictor and inhibits pancreatic cancer
cell proliferation and invasion via targeting E2F3. Eur Rev Med
Pharmacol Sci. 21:4050–4057. 2017.PubMed/NCBI
|