Introduction
Lung cancer is one of the most aggressive types of
cancer and the primary cause of cancer-related mortality, with
>2.0 million new cases diagnosed and 1.7 million deaths annually
worldwide (1). Non-small-cell
lung cancer (NSCLC) cases comprise 80–85% of total lung cancer
cases. NSCLC includes adenocarcinoma, squamous cell carcinoma,
large-cell carcinoma and other poorly differentiated subtypes
(2). Despite significant advances
in lung cancer diagnosis and treatment, the 5-year survival rate is
currently <15% (3). One of the
most common causes of treatment failure is metastasis, but the
exact underlying mechanism remains unclear (4).
Recent studies have demonstrated that
post-translational histone modifications are implicated in various
diseases. These histone modifications include acetylation,
methylation and phosphorylation on N-terminal histone tails.
Histone tail methylation is key to the regulation of gene
transcription and protein function (5). [Su(var)3–9, enhancer of zeste,
Trithorax] domain-containing protein 7 (SETD7), also known as SET7,
SET9, SET7/9 or KMT7, belongs to the protein lysine
methyltransferase family (6).
SETD7 was initially shown to catalyze histone monomethylation of
H3K4 (7). However, recent studies
have demonstrated that SETD7 also post-translationally modifies
non-histone proteins, including p53, Msx2-interacting protein, and
signal transducer and activator of transcription 3 (STAT3)
(7–9).
It was previously indicated that SETD7 plays an
important role in several types of cancer. SETD7 expression was
found to be lost or reduced in gastric cancer, and low SETD7
expression was associated with poor prognosis (10). The expression of SETD7 was also
decreased in breast cancer, and knockdown of SETD7 enhanced cell
proliferation, migration, invasion and tumor growth in vivo
(11). Liu et al
demonstrated that resveratrol inhibited colorectal cancer cell
growth via upregulating the expression of SETD7, suggesting that
SETD7 is implicated in colorectal cancer cell growth (12). However, the role of SETD7 in lung
cancer metastasis has not been fully elucidated.
The aim of the present study was to assess the
expression of SETD7 in lung cancer tissues and cell lines,
investigate its effect on lung cancer metastatic potential, and
determine whether the Janus kinase 2 (JAK2)/signal transducer and
activator of transcription 3 (STAT3) signaling pathway is involved
in this process.
Materials and methods
Reagents and antibodies
The STAT3 inhibitor Stattic was purchased from
Sigma-Aldrich; Merck KGaA. Antibodies against SETD7 (cat. no.
ab14820) and H3K4me2 (cat. no. ab7766) were purchased from Abcam;
antibodies against p-JAK2 (cat. no. 3771), p-STAT3 (cat. no. 9145),
STAT3 (cat. no. 9132), N-cadherin (cat. no. 13116) and vimentin
(cat. no. 5741) were purchased from Cell Signaling Technology,
Inc.; antibody against H3 (cat. no. BMS-33042m) was purchased from
Bioss; antibody against E-cadherin (cat. no. AF748) was purchased
from R&D Systems; and antibody against β-actin was purchased
from Sigma-Aldrich: Merck KGaA (cat. no. F3022). All antibodies
were diluted at 1:1,000, except β-actin, which was diluted at
1:5,000.
Cell culture
The human lung cancer cell lines A549, H1299 and
H661, and the human lung bronchial epithelial cell line BEAS-2B,
were purchased from American Type Culture Collection. A549 and
BEAS-2B cells were grown in DMEM, and H1299 and H661 cells were
grown in RPMI-1640 medium (Gibco; Thermo Fisher Scientific, Inc.),
at 37°C with 5% CO2. The medium was supplemented with
10% fetal bovine serum (FBS), 1% penicillin and streptomycin
(Gibco; Thermo Fisher Scientific, Inc.).
The lung cancer specimens were obtained from
patients treated at the Tianjin Medical University General Hospital
(TMUGH; Tianjin, China). None of the patients underwent
chemotherapy or targeted therapy prior to tissue collection
(Table I). The study protocol was
approved by the Institutional Review Board of TMUGH.
 | Table IPatient characteristics. |
Table I
Patient characteristics.
| Number of
patients | Sex | Age, years | Histology | Tumor
differentiation |
|---|
| 1 | Male | 59 | Squamous cell
carcinoma | I |
| 2 | Male | 76 | Squamous cell
carcinoma | I |
| 3 | Female | 66 | Adenocarcinoma | I |
| 4 | Male | 64 | Adenocarcinoma | I |
| 5 | Male | 80 | Squamous cell
carcinoma | I |
| 6 | Female | 65 | Mixed (65% squamous
cell carcinoma and 35% adenocarcinoma) | I |
| 7 | Male | 42 | Mediastinal
tumor | I |
| 8 | Male | 70 | Adenocarcinoma | I |
| 9 | Male | 82 | Adenocarcinoma | I |
| 10 | Male | 50 | Squamous cell
carcinoma | I |
Cell viability
Cells (5x103) were placed in 96-well
plates. After treatment, cell viability was assessed using the Cell
Counting Kit-8 assay (Dojindo Molecular Technologies, Inc.),
according to the manufacturer’s instructions.
Transfection and RNA interference
SETD7 was cloned into the pCMV-tag2B vector (Agilent
Technologies, Inc.), according to the manufacturer’s instructions.
The primers were as follows: 5′-CGCGGATCCATGGATAGCGACGACGAGA-3′
(forward); 5′-CCGCTCGAGTCACTTTTGCTGGGTGGCC-3′
(reverse). The forward primer included a BamHI restriction
site (underlined), and the reverse primer included a XhoI
restriction site (underlined) (Takara Biotechnology Co., Ltd.).
The siRNA duplex for SETD7 was purchased from
Genepharma. The sequences of the siRNA duplex were as follows:
Sense, 5′-CACUCCUUCACUCCAAACUTT-3′, and antisense,
5′-AGUUUGGAGUGAAGGAGUGTT-3′. The sequences of the NC-siRNA duplex
were as follows: Sense 5′-UUCUCCGAACGUGUCACGUTT-3′, and antisense
5′-ACGUGACACGUUCGGAGAATT-3′.
Lung cancer cells were plated in 6-well plates at a
density of 2.5x105 cells/well, and pSETD7 or si-SEDT7
were transfected into the cells using Lipofectamine 2000
(Invitrogen; Thermo Fisher Scientific, Inc.).
Wound healing assay
Cell migration was assessed with the wound healing
assay as previously described (13). Cells were cultured in 6-well
plates at 37°C for 24 h until they reached 80% confluence. A
sterile 200-μl pipette tip was used to scratch the cell monolayer,
and PBS was used to remove cell debris. Cells were then cultured at
37°C in DMEM or RPMI-1640 medium in the absence of FBS for 48 h,
and cell migration was analyzed by evaluating wound closure over
time using an inverted microscope (Nikon Corporation) at a
magnification of x40. The closure rate was calculated as follows:
Closure rate=(1-final wound area/initial wound area) x100%.
Cell invasion assay
Cell invasion ability was examined using the
Transwell assay as previously described (14). Briefly, the upper chamber of the
Transwell insert (Costar; Corning, Inc.) was coated with Matrigel
(BD Biosciences, Inc.). SETD7-overexpressing or -silenced cells
(1x105) were added to the upper chamber in serum-free
medium. Complete medium with 10% FBS was added to the lower
chamber. After 48 h, non-invading cells in the upper chamber were
removed with a cotton swab. Invading cells in the lower chamber
were stained with 0.1% crystal violet solution (Beyotime Institute
of Biotechnology), and quantified by counting the cell number from
5 random fields under a light microscope (Nikon Corporation) at a
magnification of x200.
Reverse transcription-quantitative PCR
(RT-qPCR) analysis
RT-qPCR was performed as previously described
(15). Briefly, RNA was extracted
from cells or tumor tissues using TRIzol (Invitrogen; Thermo Fisher
Scientific, Inc.). RT was performed using a Takara kit following
the manufacturer’s instructions. The reaction protocol was 37°C for
15 min, and then 85°C for 5 sec. Gene expression was analyzed by
qPCR using the Power SYBR-Green Master Mix (Applied Biosystems;
Thermo Fisher Scientific, Inc.), on an ABI Prism 7500 Sequence
Detector System. The reaction protocol was as follows: Stage 1 at
94°C for 3 min; stage 2 at 94°C for 30 sec and 60°C for 30 sec, for
40 cycles; and stage 3 at 95°C for 15 sec and 60°C for 1 min. GAPDH
was used as an internal control. Primers were designed using Primer
5.0 (PREMIER Biosoft International) and synthesized by the Beijing
Genomics Institute. The primer sequences are listed in Table II. The gene expression levels
were calculated using the 2−ΔΔCq method (16), by comparing the Cq values of the
target genes to the Cq values of GAPDH.
 | Table IIPCR primer sequences. |
Table II
PCR primer sequences.
| Primers | Sequence
(5′-3′) | Length of amplicons
(bp) |
|---|
| MMP2 |
| Forward |
GCGGCGGTCACAGCTACTT | 76 |
| Reverse |
CACGCTCTTCAGACTTTGGTTCT | |
| Twist |
| Forward |
GCCAATCAGCCACTGAAAGG | 83 |
| Reverse |
TGTTCTTATAGTTCCTCTGATTGTTACCA | |
| VEGF |
| Forward |
AGGAGGAGGGCAGAATCATCA | 76 |
| Reverse |
CTCGATTGGATGGCAGTAGCT | |
| SETD7 |
| Forward |
CACGGAGAAAAGAACGGACG | 141 |
| Reverse |
GTCTACATACGTGCCCTGGA | |
| GAPDH |
| Forward |
TGCACCACCAACTGCTTAGC | 87 |
| Reverse |
GGCATGGACTGTGGTCATGAG | |
Western blot analysis
Western blotting was conducted as previously
described (17). Proteins were
isolated from cells using RIPA buffer (Beyotime Institute of
Biotechnology) containing protease inhibitor (Sigma-Aldrich; Merck
KGaA). Protein was quantified using Pierce™ BCA Protein Assay kit
(Thermo Fisher Scientific, Inc.). Protein (20 μg) was loaded and
12% SDS-PAGE was performed. The proteins were transferred to
nitrocellulose membranes (EMD Millipore). The membranes were
blocked with 5% milk in 0.1% TBS-Tween for 1 h at room temperature.
The membranes were then incubated with primary antibodies at 4°C
overnight. After washing, the membranes were probed with the
corresponding HRP-conjugated secondary antibodies. The protein
bands were visualized using the enhanced chemiluminescence
technique (EMD Millipore).
Statistical analysis
The data are presented as mean ± standard deviation,
and they were obtained from at least three independent experiments.
Statistical analysis was performed using SPSS version 13.0 (SPSS,
Inc.). The differences between two groups were analyzed using
Student’s t-test, and among multiple groups using one-way analysis
of variance followed by Tukey’s post hoc test. P<0.05 was
considered to indicate statistically significant differences.
Results
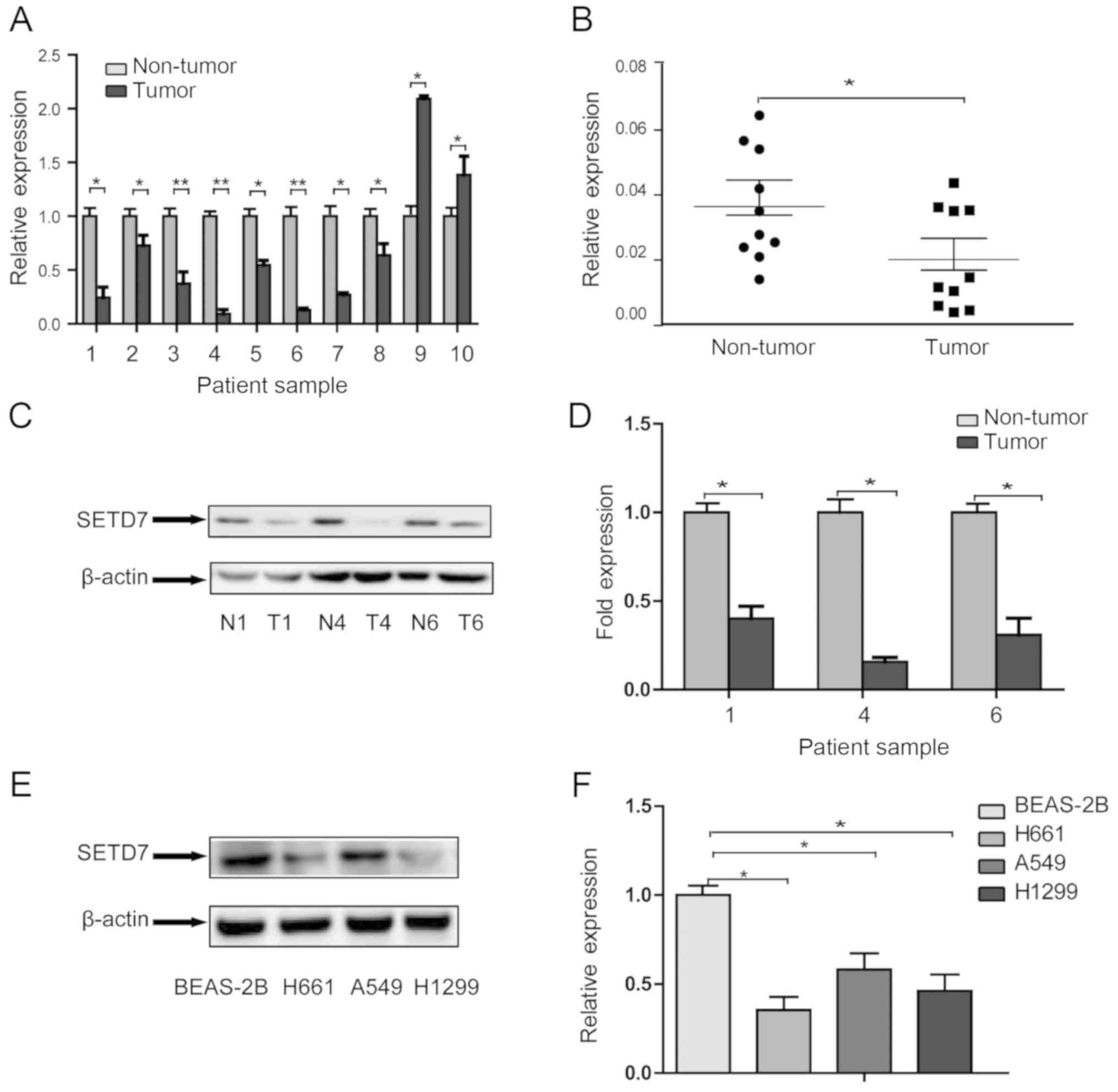
SETD7 is downregulated in human lung
cancer tissues and cell lines
Ten pairs of lung cancer specimens, including tumor
tissues and matched non-tumor tissues, were collected to study the
role of SETD7 in human lung tumorigenesis. The mRNA levels were
first compared by qPCR. As shown in Fig. 1A and B, in the majority of the
samples (8/10), SETD7 expression was lower in tumor tissues
compared with that in non-tumor tissues, although SETD7 was
expressed at higher levels in the tumors in two pairs of tissues
(2/10). The characteristics of the two patients in whom the
expression of SETD7 was higher in tumor tissues were further
reviewed; however, no specific characteristics were observed. This
may due to individual differences, such as genetic background.
Protein expression was then detected in three pairs of tissues
representing lung squamous cell carcinoma, adenocarcinoma, and
mixed lung squamous cell carcinoma and adenocarcinoma. Western blot
analysis also confirmed that the protein level of SETD7 in tumor
tissues was reduced compared with that in non-tumor tissues
(Fig. 1C and D).
We next compared the expression level of SETD7 in
three human NSCLC cell lines, A549 cells (adenocarcinoma), H1299
cells (NSCLC) and H661 cells (large-cell carcinoma), to that in
BEAS-2B cells (human normal bronchial epithelial cells) by western
blotting (Fig. 1E and F). The
expression of SETD7 was reduced by 35.3, 58.1 and 46.1% in H661,
A549 and H1299 cells, respectively, compared with that in BEAS-2B
cells. These findings indicated that SETD7 was markedly
downregulated in lung cancer cells.
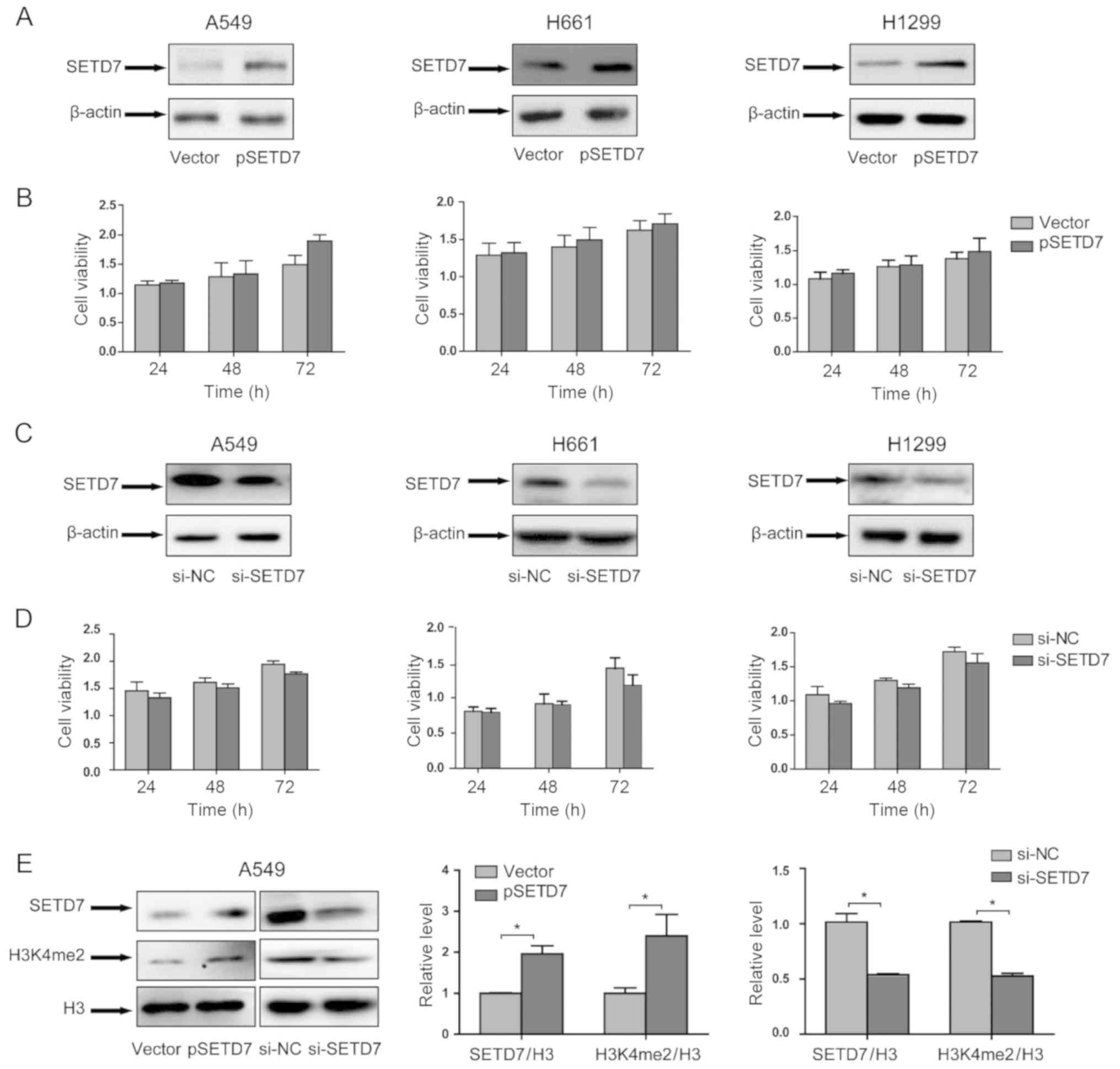
SETD7 exerts no effect on the viability
of lung cancer cells
The role of SEDT7 in lung cancer cell viability was
investigated. As SEDT7 is downregulated in lung cancer cells, SEDT7
expression was evaluated in lung cancer cell lines. Lung cancer
cells were transfected with pSETD7, and the overexpression of SEDT7
was determined by western blotting (Fig. 2A). After transfection, cell
viability was assessed by the Cell Counting Kit-8 assay. As shown
in Fig. 2B, SEDT7 overexpression
did not affect lung cancer cell viability in any of the three lung
cancer cell lines.
To further confirm the role of SEDT7, the expression
of SEDT7 was knocked down by RNAi technology. As shown in Fig. 2C, SEDT7 expression was
significantly reduced; however, SEDT7 knockdown did not affect cell
viability (Fig. 2D). These
results demonstrated that SEDT7 exerted no effect on lung cancer
cell viability.
As SEDT7 is a protein lysine methyltransferase that
methylates histone H3K4, we also investigated the effect of SEDT7
on histone methylation status. The histone methylation status can
be assessed by western blotting (18,19). In particular, Chen et al
reported that the expression of SEDT7 is associated with the
expression of H3K4me2 (18);
therefore, the H3K4me2 level was measured in SETD7 overexpression
and knockdown samples. The results demonstrated that SETD7
overexpression increased H3K4me2 expression, whereas SETD7
knockdown reduced H3K4me2 expression (Fig. 2E).
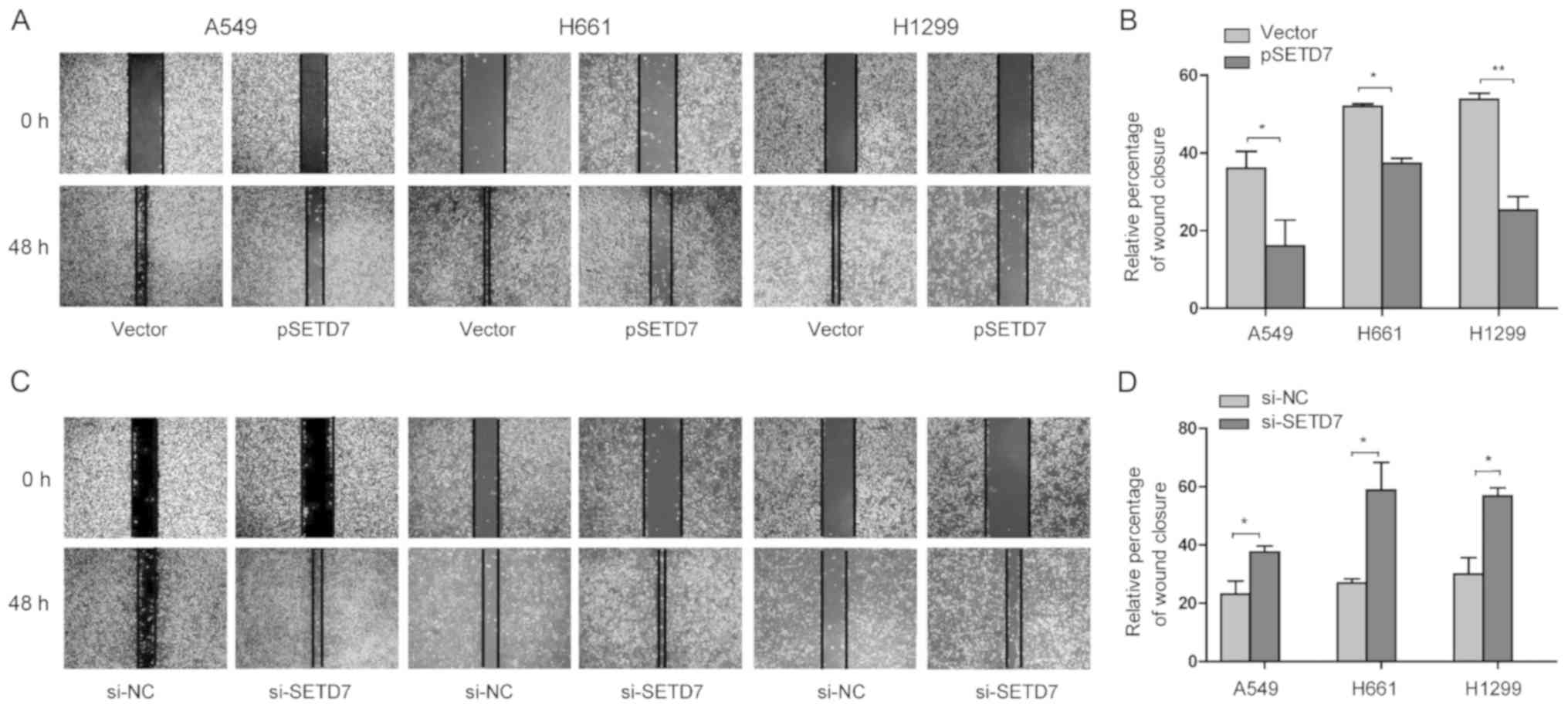
SETD7 downregulation enhances the
migration of lung cancer cells
The migration ability of cancer cells is responsible
for tumor metastasis (20).
Therefore, the effect of SETD7 on the migration of lung cancer
cells was investigated by the wound healing assay. A549, H1299 and
H661 cells were transfected with pSETD7, and cell migration was
assessed at 48 h after transfection. As shown in Fig. 3A and B, the relative rates of
wound closure at 48 h for A549, H661 and H1299 cells were 42.56,
55.09 and 59.12%, respectively. Interestingly, when SETD7 was
overexpressed in lung cancer cells, the relative percentages of
wound closure were reduced to 14.27% in A549 cells, 40.43% in H661
cells, and 31.72% in H1299 cells. The strongest inhibitory effect
on migration was observed in A549 cells.
By contrast, when SEDT7 expression was reduced by
RNAi knockdown, the relative rates of wound closure were increased
in all three cell lines (Fig. 3C and
D). These data indicated that SEDT7 downregulation enhances
lung cancer cell migration.
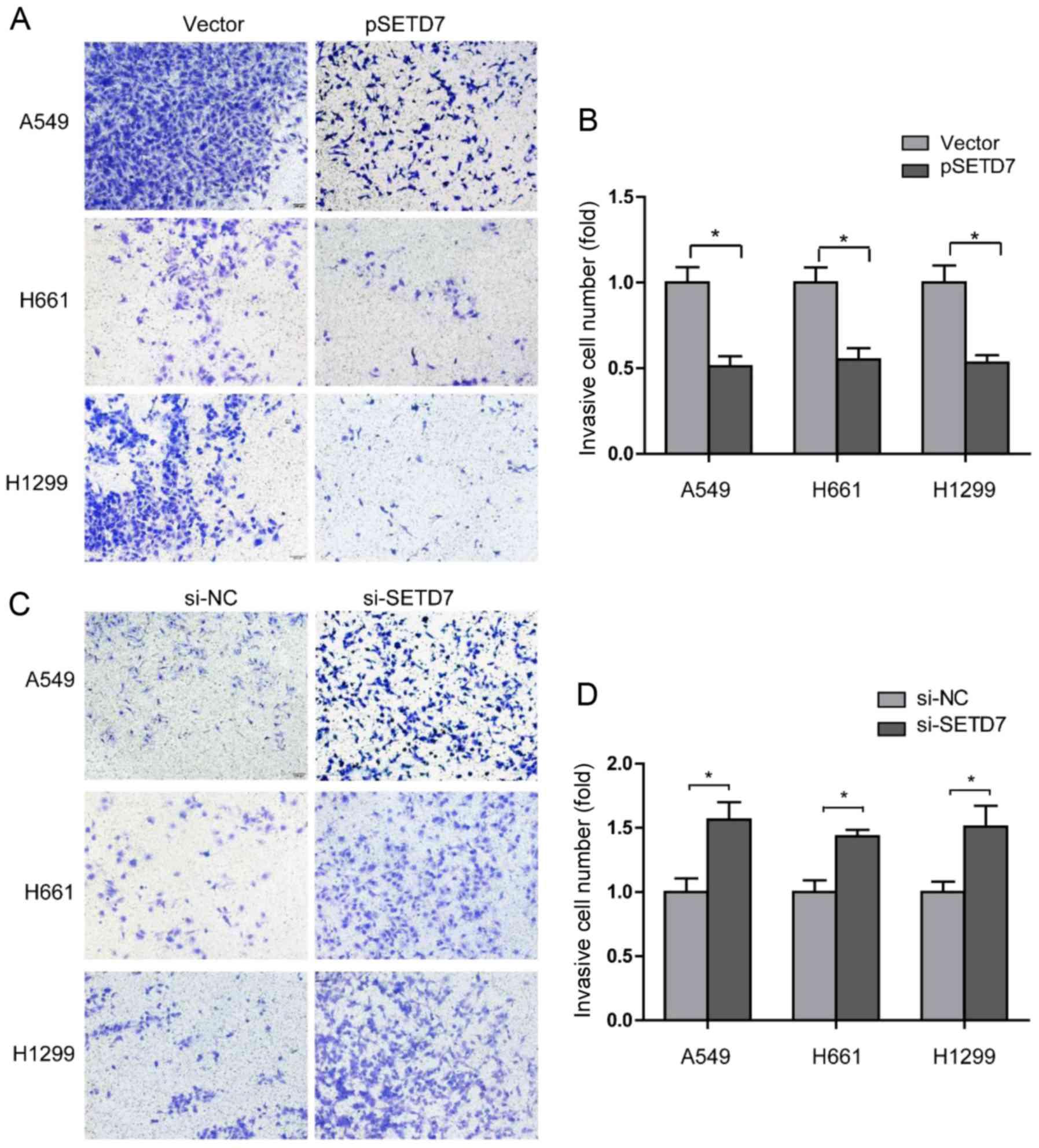
SETD7 downregulation enhances the
invasion of lung cancer cells
Cell invasion is also an important step in tumor
metastasis (20). The effect of
SETD7 on cell invasion was examined by the Transwell assay. Lung
cancer cells were transfected with pSETD7, and the invading lung
cancer cells were stained and counted at 48 h after transfection. A
marked suppression of cell invasion ability was observed, as the
invading cell numbers were decreased to 0.53-fold in A549 cells,
0.60-fold in H661 cells, and 0.57-fold in H1299 cells (Fig. 4A and B).
Next, the effect of the reduction of SETD7 on cell
invasion was investigated. As shown in Fig. 4C and D, suppression of SETD7
expression enhanced lung cancer cell invasion. Taken together,
these results demonstrated that SETD7 downregulation enhances the
metastatic potential of lung cancer cells.
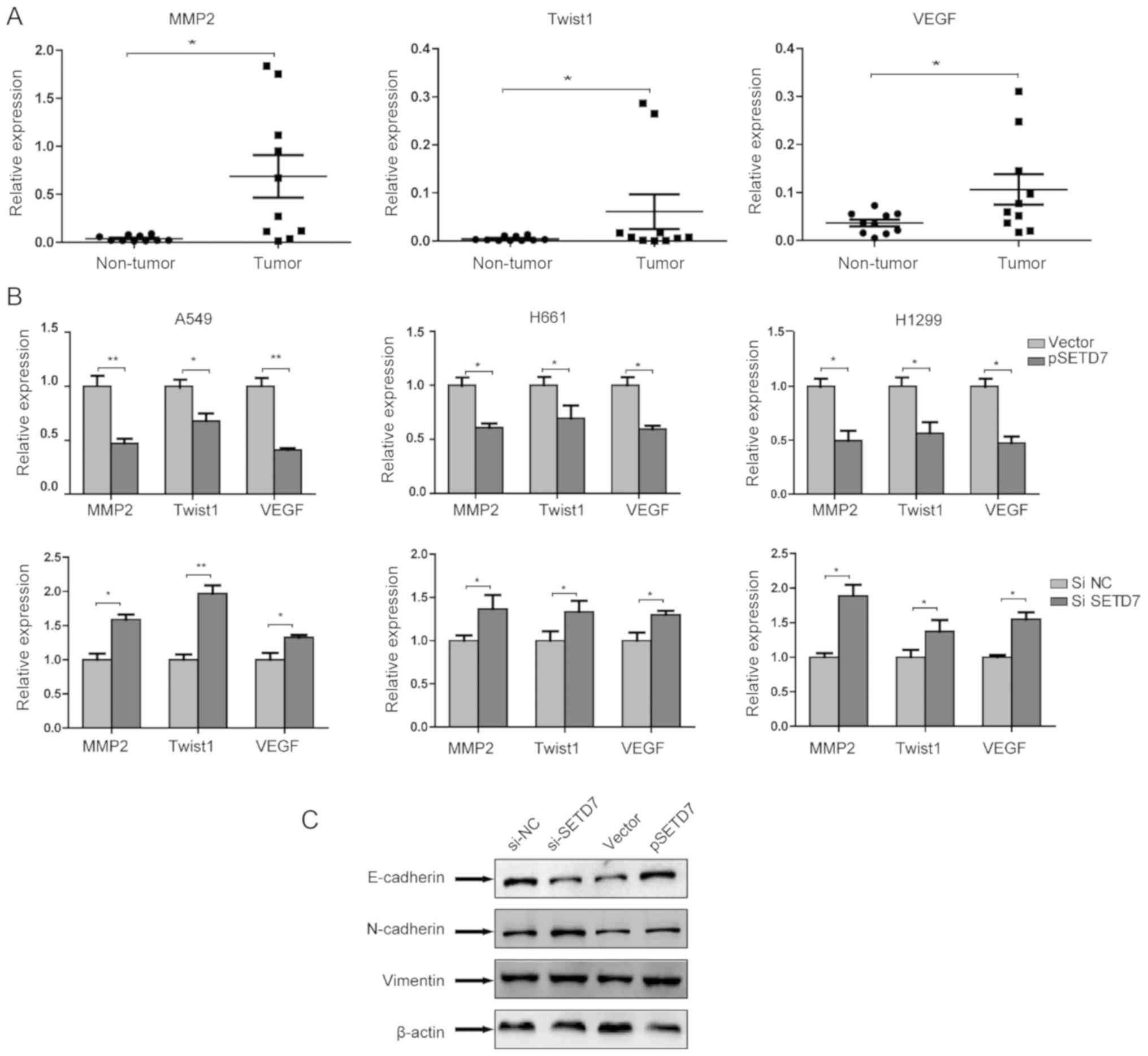
SETD7 regulates metastasis-related gene
expression
Given the observed inhibitory effect of SETD7 on the
migration and invasion of lung cancer cells, its effect on the
expression of metastasis-related genes was further investigated.
Numerous studies have demonstrated that matrix metallopeptidase
(MMP)2, Twist and vascular endothelial growth factor (VEGF) play
important roles in tumor metastasis.
qPCR was used to compare the mRNA expressions of
these three genes in tumor and non-tumor tissues. As shown in
Fig. 5A, tumor tissues expressed
higher levels of MMP2, Twist and VEGF compared with non-tumor
tissues.
In order to determine the role of SETD7 in the
expression of metastasis-related genes, SETD7 was first
overexpressed in lung cancer cells. It was observed that the
expression of these genes was markedly downregulated by SETD7. By
contrast, when SETD7 expression was knocked down, the expression of
these genes was upregulated (Fig.
5B). These results suggested that SETD7 suppresses the
metastatic potential of lung cancer cells through modulating
metastasis-related genes.
As epithelial-to-mesenchymal transition (EMT) is
crucial for cancer cell invasion and metastasis, the role of SETD7
in EMT was further assessed. The expression of EMT markers was
examined following overexpression or knockdown of SETD7 in A549
cells. It was observed that, after SETD7 knockdown, the expression
of the epithelial phenotype marker E-cadherin was decreased,
whereas the expression of the mesenchymal phenotype markers
N-cadherin and vimentin was increased. These results indicate that
SETD7 knockdown promotes EMT. SETD7 overexpression exerted no
effect on EMT (Fig. 5C).
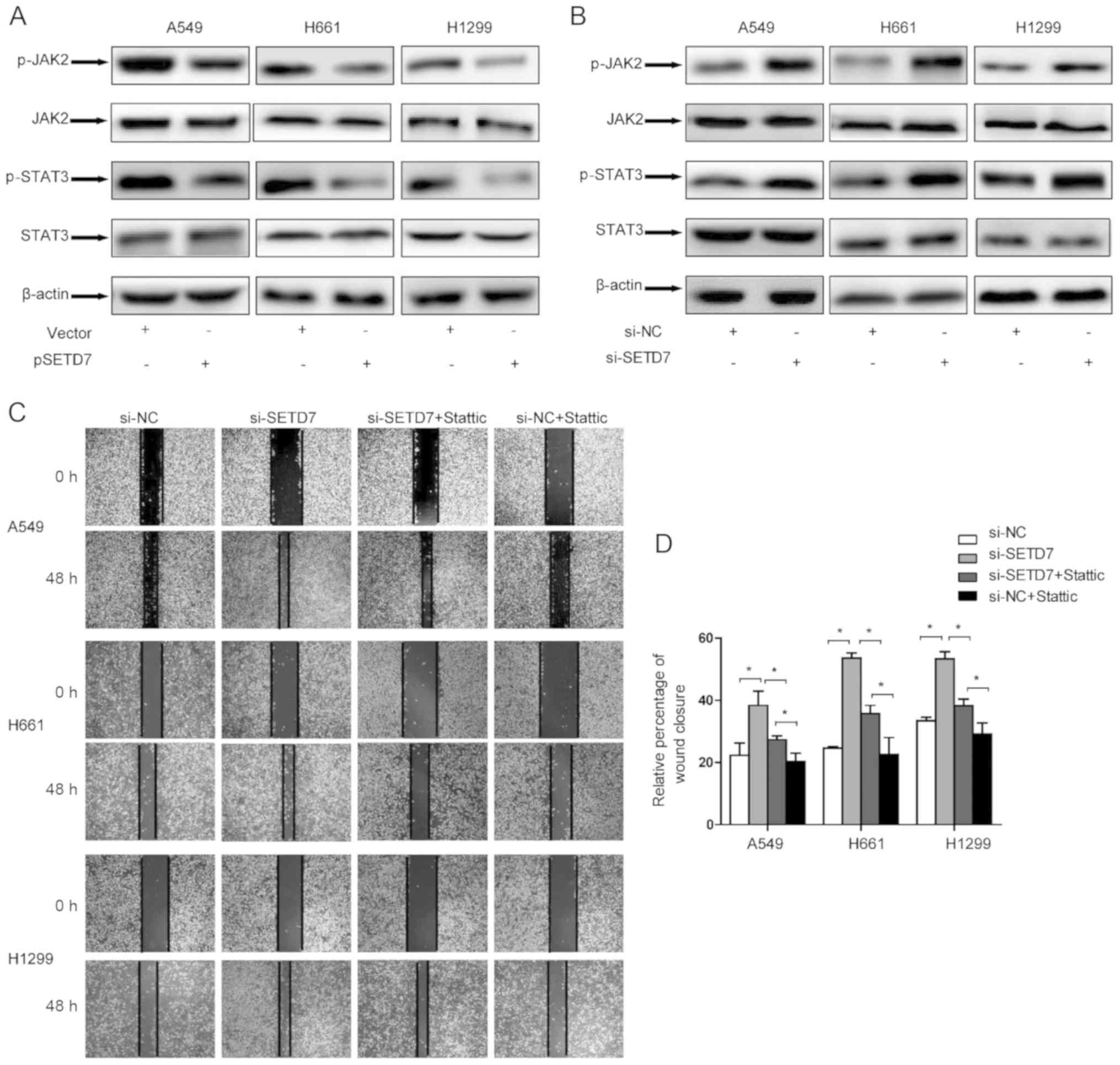
The JAK2/STAT3 signaling pathway mediates
the effects of SETD7
To further elucidate the mechanism underlying the
inhibitory role of SETD7 in lung cancer metastasis,
metastasis-related signaling pathways were investigated. It was
previously demonstrated that the JAK/STAT3 pathway plays a crucial
role in cell proliferation, survival, differentiation and, in
particular, tumor invasion and metastasis (21). This prompted us to explore the
effect of SETD7 on the JAK/STAT3 pathway.
By transfecting pSETD7 in lung cancer cells, it was
observed that SETD7 effectively suppressed the phosphorylation
level of JAK2 and STAT3, while the total STAT3 level remained
unchanged. As expected, when SETD7 was downregulated, the
phosphorylation levels of both JAK2 and STAT3 were increased
(Fig. 6A and B).
Furthermore, the role of the JAK/STAT3 pathway on
cell migration was determined by introducing the STAT3-specific
inhibitor Stattic. SETD7 expression was knocked down in lung cancer
cells and the cells were treated with Stattic. The wound healing
assay demonstrated that Stattic effectively attenuated the effect
of SETD7 on cell migration (Fig. 6C
and D). Taken together, these results indicate that the
JAK/STAT3 pathway may be responsible for the effect of SEDT7 on
metastatic potential.
As it was demonstrated that SETD7 overexpression
increased H3K4me2 expression, whereas SETD7 knockdown reduced
H3K4me2 expression (Fig. 2E),
these results suggest that histone methylation may be implicated in
the effects of SEDT7 on metastatic potential.
Discussion
SETD7 is a protein lysine monomethylase that
methylates not only histone H3K4, but also non-histone proteins
(22,23). The methylation on proteins alters
their activity and affects a series of biological processes. The
role of SETD7 in cancer development has recently attracted
attention. In the present study, SETD7 was found to be
downregulated in both lung cancer tissues and lung cancer cell
lines. The expression of SETD7 was assessed by western blotting and
qPCR analysis. Due to the limitations regarding the amount of
tissue available, we were unable to perform immunohistochemistry
assay, which can reveal the distribution of SETD7 expression across
the tumor macrostructure. As the number of tissue samples was
relatively small, more lung tumor tissues must be assessed in order
to establish the correlation of SETD7 expression with tumor size
and pathological stage.
The effect of SETD7 on cancer cell growth is
controversial. SETD7 inhibits cell growth in colorectal cancer
(12), acute myeloid leukemia
(24), gastric cancer (10), cervical cancer and colon cancer
(25). On the contrary, SETD7
promotes hepatocellular carcinoma cell growth (26). Interestingly, in breast cancer,
Song et al demonstrated that SETD7 inhibited cell growth
through regulation of Gli-1 expression (11), whereas Zhang et al reported
that SETD7 promoted tumor growth by interacting with the
transcription factor GATA1 (27).
The opposite roles of SETD7 in the same cancer may be due to the
physiological subtype and stage of the tumor. In the present study,
SETD7 did not affect lung cancer cell viability.
Cancer metastasis is the primary cause of treatment
failure, and 90% of lung cancer patients eventually succumb to
metastatic disease (20). Tumor
metastasis is a complex process. Cell migration and invasion are
crucial steps during metastasis. Therefore, the role of SETD7 in
cell migration and invasion was investigated. The results
demonstrated that SETD7 downregulation enhanced both cell migration
and invasion. The role of SETD7 in migration and invasion is also
supported by other studies. Akiyama et al demonstrated that
SETD7 downregulation promoted gastric cancer cell migration and
invasion (10), and Gu et
al reported that SETD7 downregulation significantly decreased
cell migration and invasion in hepatocellular carcinoma (26).
As our results demonstrated that SETD7 exerted no
effect on cell viability, it was inferred that the role of SETD7 in
metastasis relies on its regulation of metastasis-related genes,
rather than the stimulation of cell proliferation. A number of
genes have been demonstrated to be involved in metastasis. MMP2
mediates the promoting effect of chorionic gonadotropin on
epithelial ovarian cancer metastasis (28). lncRNA TP73-AS1 stimulates ovarian
cancer cell metastasis via regulation of MMP2 and MMP9 (29). In NSCLC, miR-149 inhibits cell
growth and metastasis by targeting MMP2 (30). Furthermore, the transcription
factor Twist1 is also involved in metastasis by regulating gene
transcription.
Twist1 induces EMT and promotes metastasis (31); Twist1 is also responsible for
hypoxia-induced angiogenesis and metastasis (32). In NSCLC, Li et al reported
that promoting protein family member 3 enhances cell invasion and
tumor metastasis via the STAT3/Twist1 pathway (33). VEGF is another common
metastasis-related gene. VEGF binds to the VEGF receptor, induces
angiogenesis and promotes metastasis and tumor progression
(34). Zhang et al
demonstrated that a novel oncogene, KIF26B, promotes gastric cancer
metastasis via activating the VEGF pathway (35). Zhou et al demonstrated that
oxymatrine inhibits lung cancer cell migration by regulating
miR-520/VEGF (36). In the
present study, it was observed that lung cancer tissues expressed
higher levels of MMP2, Twist1 and VEGF, and lower levels of SETD7,
compared with non-cancerous tissue. Overexpression of SETD7 in lung
cancer cells downregulated MMP2, Twist1 and VEGF, indicating that
SETD7 modulates the expression of metastasis-related genes.
To further elucidate the mechanism underling the
role of SETD7 in metastasis, we investigated signaling pathways
involved in cell invasion and metastasis, among which the JAK/STAT3
pathway attracted our attention. Indeed, it was demonstrated that
the JAK2/STAT3 signaling pathway mediated the effect of SETD7. An
et al reported that STAT3 induces long non-coding RNA
LINC00668 expression to promote migration and invasion of NSCLC
cells (37). Jin et al
demonstrated that TRIM14 enhances colorectal cancer cell invasion
through the STAT3 pathway (38).
He et al reported that mesenchymal stem cells inhibit breast
cancer progression by suppressing the STAT3 signaling pathway
(39). Therefore, STAT has
emerged as a promising target for cancer treatment (40,41).
To the best of our knowledge, the direct regulation
of SETD7 expression has not been reported to date, and the
mechanism of SETD7 regulation remains unclear. miRNAs regulate
their target genes, thereby regulating tumor development and
progression. SETD7 may also be regulated by miRNAs, particularly
those that have been found to be dysregulated in tumors. Further
investigation on these miRNAs may help elucidate the SETD7 network
in tumors.
In summary, the present study demonstrated that
SETD7 is downregulated in lung cancer, and SETD7 downregulation
enhanced the migration and invasion of lung cancer cells. It was
further demonstrated that downregulation of SETD7 promoted the
metastatic potential of lung cancer cells via the JAK2/STAT3
pathway. These findings may provide useful information regarding
the potential of SETD7 as a novel therapeutic candidate for
metastatic lung cancer treatment.
Acknowledgements
Not applicable.
Funding
The present study was supported by the National
Natural Science Foundation of China (grant no. 81372519), the
Specialized Research Fund for the Doctoral Program of Higher
Education of China (grant no. 20131202110005), the Key Project of
Natural Science Foundation of Tianjin (grant no. 18JCZDJC98500) and
the start-up fund of Tianjin Medical University General Hospital
(grant no. 303079401501).
Availability of data and materials
All data generated or analyzed during the present
study are included in this published article.
Authors’ contributions
LC, YR, XG, LW, QZ and XL performed the experiments.
XW contributed to the data analysis. ZM contributed to the study
design and data analysis. KX contributed to the study design, data
analysis and wrote the manuscript. All the authors have read and
approved the final manuscript.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of Tianjin Medical University General Hospital. Written
informed consent was obtained from all patients.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that there are no competing
interests.
References
|
1
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Goldstraw P, Ball D, Jett JR, Le Chevalier
T, Lim E, Nicholson AG and Shepherd FA: Non-small-cell lung cancer.
Lancet. 378:1727–1740. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2017. CA Cancer J Clin. 67:7–30. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Turajlic S and Swanton C: Metastasis as an
evolutionary process. Science. 352:169–175. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Keating ST and El-Osta A: Transcriptional
regulation by the Set7 lysine methyltransferase. Epigenetics.
8:361–372. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Xiao B, Jing C, Wilson JR, Walker PA,
Vasisht N, Kelly G, Howell S, Taylor IA, Blackburn GM and Gamblin
SJ: Structure and catalytic mechanism of the human histone
methyltransferase SET7/9. Nature. 421:652–656. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Li Y, Reddy MA, Miao F, Shanmugam N, Yee
JK, Hawkins D, Ren B and Natarajan R: Role of the histone H3 lysine
4 methyltransferase, SET7/9, in the regulation of
NF-kappaB-dependent inflammatory genes. Relevance to diabetes and
inflammation. J Biol Chem. 283:26771–26781. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Dhayalan A, Kudithipudi S, Rathert P and
Jeltsch A: Specificity analysis-based identification of new
methylation targets of the SET7/9 protein lysine methyltransferase.
Chem Biol. 18:111–120. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Yang J, Huang J, Dasgupta M, Sears N,
Miyagi M, Wang B, Chance MR, Chen X, Du Y, Wang Y, et al:
Reversible methylation of promoter-bound STAT3 by histone-modifying
enzymes. Proc Natl Acad Sci USA. 107:21499–21504. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Akiyama Y, Koda Y, Byeon SJ, Shimada S,
Nishikawaji T, Sakamoto A, Chen Y, Kojima K, Kawano T, Eishi Y, et
al: Reduced expression of SET7/9, a histone mono-methyltransferase,
is associated with gastric cancer progression. Oncotarget.
7:3966–3983. 2016. View Article : Google Scholar :
|
|
11
|
Song Y, Zhang J, Tian T, Fu X, Wang W, Li
S, Shi T, Suo A, Ruan Z, Guo H and Yao Y: SET7/9 inhibits oncogenic
activities through regulation of Gli-1 expression in breast cancer.
Tumour Biol. 37:9311–9322. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Liu Z, Wu X, Lv J, Sun H and Zhou F:
Resveratrol induces p53 in colorectal cancer through SET7/9. Oncol
Lett. 17:3783–3789. 2019.PubMed/NCBI
|
|
13
|
Lin G, Liu B, Meng Z, Liu Y, Li X, Wu X,
Zhou Q and Xu K: MiR-26a enhances invasive capacity by suppressing
GSK3β in human lung cancer cells. Exp Cell Res. 352:364–374. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Song Q, Liu B, Li X, Zhang Q, Cao L, Xu M,
Meng Z, Wu X and Xu K: MiR-26a-5p potentiates metastasis of human
lung cancer cells by regulating ITGβ8-JAK2/STAT3 axis. Biochem
Biophys Res Commun. 501:494–500. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wang L, Li X, Ren Y, Geng H, Zhang Q, Cao
L, Meng Z, Wu X, Xu M and Xu K: Cancer-associated fibroblasts
contribute to cisplatin resistance by modulating ANXA3 in lung
cancer cells. Cancer Sci. 110:1609–1620. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
17
|
Wang H, Wang L, Cao L, Zhang Q, Song Q,
Meng Z, Wu X and Xu K: Inhibition of autophagy potentiates the
anti-metastasis effect of phenethyl isothiocyanate through
JAK2/STAT3 pathway in lung cancer cells. Mol Carcinog. 57:522–535.
2018. View
Article : Google Scholar
|
|
18
|
Chen Y, Yang S, Hu J, Yu C, He M and Cai
Z: Increased expression of SETD7 promotes cell proliferation by
regulating cell cycle and indicates poor prognosis in
hepatocellular carcinoma. PLoS One. 11:e01549392016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Salz T, Deng C, Pampo C, Siemann D, Qiu Y,
Brown K and Huang S: Histone methyltransferase hSETD1A is a novel
regulator of metastasis in breast cancer. Mol Cancer Res.
13:461–469. 2015. View Article : Google Scholar
|
|
20
|
Welch DR and Hurst DR: Defining the
hallmarks of metastasis. Cancer Res. 79:3011–3027. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Xin P, Xu X, Deng C, Liu S, Wang Y, Zhou
X, Ma H, Wei D and Sun S: The role of JAK/STAT signaling pathway
and its inhibitors in diseases. Int Immunopharmacol. 80:1062102020.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Fu L, Wu H, Cheng SY, Gao D, Zhang L and
Zhao Y: Set7 mediated Gli3 methylation plays a positive role in the
activation of Sonic Hedgehog pathway in mammals. Elife. 5:pii:
e15690. 2016. View Article : Google Scholar
|
|
23
|
Lezina L, Aksenova V, Ivanova T, Purmessur
N, Antonov AV, Tentler D, Fedorova O, Garabadgiu AV, Talianidis I,
Melino G and Barlev NA: KMTase Set7/9 is a critical regulator of
E2F1 activity upon genotoxic stress. Cell Death Differ.
21:1889–1899. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Gu Y, Wang Y, Wang X, Gao L, Yu W and Dong
WF: Opposite effects of SET7/9 on apoptosis of human acute myeloid
leukemia cells and lung cancer cells. J Cancer. 8:2069–2078. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Shen C, Wang D, Liu X, Gu B, Du Y, Wei FZ,
Cao LL, Song B, Lu X, Yang Q, et al: SET7/9 regulates cancer cell
proliferation by influencing β-catenin stability. FASEB J.
29:4313–4323. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Gu Y, Wang X, Liu H, Li G, Yu W and Ma Q:
SET7/9 promotes hepatocellular carcinoma progression through
regulation of E2F1. Oncol Rep. 40:1863–1874. 2018.PubMed/NCBI
|
|
27
|
Zhang Y, Liu J, Lin J, Zhou L, Song Y, Wei
B, Luo X, Chen Z, Chen Y, Xiong J, et al: The transcription factor
GATA1 and the histone methyltransferase SET7 interact to promote
VEGF-mediated angiogenesis and tumor growth and predict clinical
outcome of breast cancer. Oncotarget. 7:9859–9875. 2016.PubMed/NCBI
|
|
28
|
Wu W, Gao H, Li X, Peng S, Yu J, Liu N,
Zhan G, Zhu Y, Wang K and Guo X: β-hCG promotes epithelial ovarian
cancer metastasis through ERK/MMP2 signaling pathway. Cell Cycle.
18:46–59. 2019. View Article : Google Scholar
|
|
29
|
Wang X, Yang B, She Y and Ye Y: The lncRNA
TP73-AS1 promotes ovarian cancer cell proliferation and metastasis
via modulation of MMP2 and MMP9. J Cell Biochem. 119:7790–7799.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zhao L, Liu L, Dong Z and Xiong J: miR-149
suppresses human non-small cell lung cancer growth and metastasis
by inhibiting the FOXM1/cyclin D1/MMP2 axis. Oncol Rep.
38:3522–3530. 2017.PubMed/NCBI
|
|
31
|
Lai YJ, Yu WN, Kuo SC, Ho CT, Hung CM, Way
TD and Chen CT: CSC-3436 inhibits TWIST-induced
epithelial-mesenchymal transition via the suppression of
Twist/Bmi1/Akt pathway in head and neck squamous cell carcinoma. J
Cell Physiol. 234:9118–9129. 2019. View Article : Google Scholar
|
|
32
|
Patra K, Jana S, Sarkar A, Mandal DP and
Bhattacharjee S: The inhibition of hypoxia-induced angiogenesis and
metastasis by cinnamaldehyde is mediated by decreasing HIF-1α
protein synthesis via PI3K/Akt pathway. Biofactors. 45:401–415.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Li Y, Bai M, Xu Y, Zhao W, Liu N and Yu J:
TPPP3 promotes cell proliferation, invasion and tumor metastasis
via STAT3/Twist1 pathway in non-small-cell lung carcinoma. Cell
Physiol Biochem. 50:2004–2016. 2018. View Article : Google Scholar
|
|
34
|
Hsu MC, Pan MR and Hung WC: Two birds, one
stone: Double hits on tumor growth and lymphangiogenesis by
targeting vascular endothelial growth factor receptor 3. Cells.
8:E2702019. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Zhang H, Ma RR, Wang XJ, Su ZX, Chen X,
Shi DB, Guo XY, Liu HT and Gao P: KIF26B, a novel oncogene,
promotes proliferation and metastasis by activating the VEGF
pathway in gastric cancer. Oncogene. 36:5609–5619. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Zhou W, Wu Y, Pan M, Liu D and Liu B:
Proliferation and migration of lung cancer could be inhibited by
oxymatrine through the regulation for miR-520/VEGF. Am J Chin Med.
47:865–878. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
An YX, Shang YJ, Xu ZW, Zhang QC, Wang Z,
Xuan WX and Zhang XJ: STAT3-induced long noncoding RNA LINC00668
promotes migration and invasion of non-small cell lung cancer via
the miR-193a/KLF7 axis. Biomed Pharmacother. 116:1090232019.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Jin Z, Li H, Hong X, Ying G, Lu X, Zhuang
L and Wu S: TRIM14 promotes colorectal cancer cell migration and
invasion through the SPHK1/STAT3 pathway. Cancer Cell Int.
18:2022018. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
He N, Kong Y, Lei X, Liu Y, Wang J, Xu C,
Wang Y, Du L, Ji K, Wang Q, et al: MSCs inhibit tumor progression
and enhance radiosensitivity of breast cancer cells by
down-regulating Stat3 signaling pathway. Cell Death Dis.
9:10262018. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Chong PSY, Chng WJ and de Mel S: STAT3: A
promising therapeutic target in multiple myeloma. Cancers (Basel).
11:E7312019. View Article : Google Scholar
|
|
41
|
Qin JJ, Yan L, Zhang J and Zhang WD: STAT3
as a potential therapeutic target in triple negative breast cancer:
A systematic review. J Exp Clin Cancer Res. 38:1952019. View Article : Google Scholar : PubMed/NCBI
|