Introduction
Anillin, an actin binding protein, serves crucial
roles in cytokinesis. Anillin forms scaffolds upon which the
actomyosin contractile ring forms and therefore functions as a
central factor of the ingression and activity of the cleavage
furrow (1–3). The transcription of the anillin
(ANLN) gene locus and the resultant protein expression are
cell cycle regulated (4).
Furthermore, there are marked changes in localization during the
cell cycle (5). In interphase
cells, the anillin protein is restricted to the nucleus. However
with the dissolution of the nuclear membrane at the onset of
mitosis, anillin becomes phosphorylated (2) and relocates to the cell cortex,
associating with the cell membrane prior to being accumulated at
the developing cleavage furrow (5). Here, it serves as a scaffold,
associating with actin (6),
septins (7) myosin II (8) and also Rho like signaling complexes,
including racGAP components (9)
and a range of other proteins (10). The ubiquitin-mediated degradation
of anillin, occurring at the end of mitosis, is facilitated by
anaphase-promoting complex (11).
The importance of anillin and anillin-associated proteins in
cytokinesis has been demonstrated in several studies, including
examining the effect of anillin mutations on cytokinesis (2,3,12).
However, other functions of anillin in the nucleus or in
association with other actomyosin activities including migration,
cannot be excluded (13).
The coordinated and efficient execution of mitosis
and cell division are important events that militate against
oncogenic events associated with inaccurate or incorrect chromosome
segregation and the mechanics of cell division (14). Therefore, it is not surprising
that alterations in anillin expression have been described in
neoplasia. Hall et al (15) performed global analyses of anillin
in diverse samples of normal and diseased tissue, the results of
which revealed that anillin overexpression occurred in neoplasia.
Anillin mRNA is expressed at increased levels in human tumors.
Furthermore, in many different types of tumor, a progressive
increase in anillin mRNA levels has been associated with tumor
spread and stage: For example, anillin is overexpressed in breast
tumors, notably at the transition from in situ to invasive
disease (16). Similarly, anillin
has been demonstrated to be overexpressed in gastric carcinoma
(17), pancreatic carcinomas
(18), hepatocellular melanoma
(19) and in head and neck
squamous carcinoma (20). Further
data from Suzuki et al (21) demonstrated anillin overexpression
in lung cancer, which was associated with anillin dysfunction and
perturbations of Rho and AKT signaling.
The highly prevalent overexpression of anillin in
multiple different types of tumors and its association with tumor
progression is unusual and requires investigation. The increased
expression of anillin is not a consequence of increased tumor
growth fraction and it should be noted that the highest levels of
anillin mRNA occur within the adult central nervous system, a
tissue characterized by a non-proliferative phenotype (15). Furthermore, Mirza et al
(22) demonstrated that a number
of genes are repressed by p53, including anillin. Given the
prevalence of p53 pathway defects in human cancer and previous
evidence demonstrating that putative p53 binding sites are located
within the 5′-untranslated region of the ANLN gene, the
present study hypothesized that the tumor-associated loss of p53
function may negate its normally repressive effects on anillin
expression. This would be consistent with the prevalence of anillin
deregulation and with the association of increased anillin
expression with tumor progression. The present study assessed the
role of p53 in the regulation of anillin and demonstrated that
anillin mRNA and protein may be regulated in a p53-dependent
manner.
Materials and methods
The cancer genome Atlas (TCGA) database
analysis
Anillin overexpression in patients with colorectal
cancer was analyzed using data obtained from TCGA database
(https://portal.gdc.cancer.gov/genes/ENSG00000011426).
Anillin protein expression were analyzed in colorectal (n=239) or
breast carcinomas (n=539) cases; normal colon (n=19) or breast
tissues (n=61) were included as controls.
Bioinformatics
The upstream 3 kb region of the initiating ATG
sequence was evaluated using UCSC Genome Browser software
(http://www.genome.ucsc.edu) (23). Additionally, putative p53
responsive elements (REs) were identified using the online
transcription factor prediction tool, Genomatix (http://www.genomatix.de/; Interxon Bioinformatics
Germany GmbH).
Cell culture and transfection
p53 wild-type and isogenic p53 null HCT116 cells
were donated by Dr B. Vogelstein (Oncology Center, Johns Hopkins
University School of Medicine) and authentication was performed
using the GenePrint10 system (Promega Corporation; cat. no. B9510).
The MCF7 cell line was purchased from the American Type Culture
Collection (cat. no. ATCC® HTB-22™). Cells were
maintained at 37°C in McCoy’s 5A medium (Thermo Fisher Scientific,
Inc.) under standard conditions and routinely tested for mycoplasma
contamination using the MycAway-Color One-Step Mycoplasma detection
kit [Yeasen Biotechnology (Shanghai) Co., Ltd.; cat. no.
40611ES60]. The full-length (−1 to −3,000), mutant A (−1,721 to
−3,000) and mutant B (−2,105 to −3,000) constructs were cloned into
pGL3 luciferase reporter vectors (cat. no. E1751; Promega
Corporation). The transient transfection of these plasmids was then
conducted for 48 h prior to western blotting to determine the
transfection efficiency. HCT116 cells was subsequently performed
using 24-well plates. Cells were grown at a density of
~1x105 cells/well in a 24-well plate 1 day prior to
transfection. For each well, HCT116 p53+/+ cells were
co-transfected with 100 ng luciferase reporter plasmid and 20 ng
β-galactosidase plasmid. HCT116 p53−/− cells were then
co-transfected with 100 ng luciferase plasmids, 20 ng
β-galactosidase plasmids and wild-type or mutant p53 (R175H or
R248W) plasmids using GeneJuice® (Merck KGaA) according
to the manufacturer’s protocol.
Western blot analysis
HCT116 p53+/+ or HCT116 p53−/−
cells were treated with 0.5 μM doxorubicin for 0, 4, 12, 18, 24, 36
and 48 h. Cells were then lysed in RIPA buffer (50 mM Tris-base,
150 mM NaCl, 1% Triton X-100, 0.5% sodium deoxycholate, 0.5% SDS, 2
mM EDTA) and total protein was collected at 12, 18. 24 and 48 h
time points. A total of 50 μg protein was electrophoresed on 8%
SDS-PAGE gels, and semi-dry transferred to a PVDF membrane. Blots
were then blocked in 2.5% milk for 30 min at room temperature,
followed by incubation with either anillin S4 or mouse monoclonal
p53 DO-1 antibodies (Santa Cruz Biotechnology, Inc.; cat. no.
sc-126; 1:1,000) at 4°C overnight. A goat anti-mouse horseradish
peroxidase-conjugated IgG secondary antibody (Dako; Agilent
Technologies, Inc; cat. no. P044701-2; 1:2,000) was then incubated
with the PVDF membranes for 1 h at room temperature. ECL™ Western
Blotting Detection Reagent (GE Healthcare Life Sciences) was then
used to visualize the bands. Expression was quantified using
ImageJ2 software (National Institutes of Health).
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA (2 μg) extracted from HCT116
p53+/+ and HCT116 p53−/− cells was reverse
transcribed using M-MLV reverse transcriptase and random primers
(Thermo Fisher Scientific, Inc.). qPCR was performed using
SYBR-Green (cat. no. 04707516001; Roche Diagnostics) and the
following gene-specific primers: Anillin, forward
5′-GAAAAGGTGACCGAAAACCA-3′, reverse 5′-TTTCGTCATTTCGCATTCAG-3′.
Anillin mRNA level was normalized to β-actin (forward
5′-AGGCACCAGGGCGTGAT-3′, reverse 5′-GCCCACATAGGAATCCTTCTGAC-3′).
Amplification was performed in a LightCycler® 480 (Roche
Diagnostics) using the following protocol: Initial denaturation at
95°C for 5 min, followed by amplification at 95°C for 10 sec and
72°C for 30 sec for 40 cycles, and finally melting curve analysis
at 95°C for 5 sec, 65°C for 1 min followed by cooling at 4°C. An
Opticon® 2 continuous fluorescence detection system
equipped with Opticon® Monitor v2.02 software was also
employed. The relative quantification cycle (Cq) method
(24) was applied to analyze the
relative expression of anillin.
Plasmid construction
Genomic DNA was prepared from HCT116
p53+/+ cells for the construction of ANLN
promoter-driven luciferase reporter plasmids. Reporter plasmids
containing −1,318 (encoding the promoter region from −1,318 to −1,
which included potential binding sites A and B), A (encoding the
promoter region from −1,310 to −1, with a deletion from −901 to
−561, excluding the binding site of B) and B (encoding the promoter
region from −921 to −1, with a deletion from −661 to −561,
excluding binding site A) regions were constructed into the
XhoI/HindIII restriction sites of the pGL3-basic
vector (Promega Corporation).
Chromatin immunoprecipitation (ChIP)
MCF7 cells were cultured and treated as
aforementioned. Chromatin was sheared into 300–600 bp fragments
using a bioruptor (Diagenode, Inc.) for 4 rounds of 10 cycles of 30
sec ON/30 sec OFF at 4°C. Fragmented chromatin was centrifuged at
500 x g for 10 min at 4°C to pellet the remaining insoluble
material and the supernatant was pre-cleared overnight at 4°C with
600 μl magnetic protein-G Dynal beads (Thermo Fisher Scientific,
Inc.). The fragmented chromatin (50 μl) was reserved as an input
control. The remaining chromatin fragments were precipitated
overnight at 4°C with magnetic protein-G beads bound with
antibodies. For each ChIP reaction, either 10 μg anti-p53 DO1
antibody (Santa Cruz Biotechnology, Inc.; cat. no. sc-126;
1:1,000), 10 μg mouse IgG immunoglobulin (1:2,000; cat. no. R0480;
Dako; Agilent Technologies, Inc.) or no antibody was added and
incubated with 600 μl sheep anti-mouse IgG protein G (Dako; Agilent
Technologies, Inc.) beads at 4°C overnight. Beads were washed 8
times with RIPA buffer (50 mM HEPES at pH 8.0, 500 mM EDTA at pH
8.0, 10% NP-40, 10% Deoxycholate, 8 M LiCl, protease inhibitor
cocktail) followed by 1 wash in 1 ml 1X TE buffer. Each reaction
was transferred into a fresh 1.5 ml Eppendorf tube, and centrifuged
at 1,000 x g for 3 min at 4°C. Immune complexes were eluted with 50
μl elution buffer [10 mmol/l Tris-Cl (pH 8.0), 1% SDS, 5 mmol/l
EDTA] at 65°C for 10 min and pelleted at 14,000 x g for 30 sec.
Supernatant was then transferred into a fresh 1.5 ml Eppendorf tube
prior to elution with 120 μl elution buffer. Cross-links were
reversed by incubating at 65°C overnight. Eluted material was
purified using a PCR clean up kit (Qiagen, Inc.). Promoter regions
containing the potential p53 binding sites of A and B were
amplified with GoTaq® DNA polymerase (cat. no. M3001;
Promega Corporation) using the following site specific primers:
p53a forward, 5′-GGAGGAATAGTTCTGTTTTG-3′; p53a reverse,
5′-TCTCCTGCTTATTCTTTGTA-3′; p53b forward,
5′-AAATTGTGCATGAACGCTT-3′; p53b reverse, 5′-TTGGCCTTCAGTAGCTTTG-3′;
p53cd forward, 5′-GGGTCCCAGTTCAAGCAAT-3′. Amplification was
performed in an Eppendorf Master cycler thermal cycler using the
following temperature protocol: 95°C for 5 min, followed by 95°C
for 30 sec, 55°C for 1 min, 72°C for 3 min 10 sec for 25 cycles,
and then 72°C for 5 min. PCR products were analyzed via
electrophoresis on 1.5% agarose gels and visualized using ethidium
bromide.
Luciferase reporter assay
HCT116 p53+/+ and HCT116
p53−/− cells were cultured, treated and transfected as
aforementioned. At 24 h post-transfection, cells were lysed in 1X
passive lysis buffer on a shaker for 15 min at room temperature.
Cells were pelleted via centrifugation at 14,000 x g for 5 min at
4°C. Supernatants were subsequently transferred into corresponding
tubes. Luciferase substrate stock solution was thawed to room
temperature (22–26°C) and the required quantity was diluted 1:2 in
ddH2O, and placed in foil-wrapped Sterilin™ (Thermo
Fisher Scientific, Inc.) prior to use. A Luciferase Assay System
(Promega Corporation; cat. no. E1500) was used and β-galactosidase
(Roche Diagnostics; cat. no. 11291963103) activity was detected
according to the manufacturers’ protocols. The reaction was
incubated at 37°C for ~30 min until the color turned dark red. The
activity was then assayed at 570 nm using a plate reader (Bio-Rad
Laboratories, Inc.). Luciferase activity was normalized to
Renilla luciferase activity (cat. no. E2231; Promega
Corporation).
Statistical analysis
An unpaired Student’s t-test was performed to
compare the differences between two groups. A two-way analysis of
variance followed by Tukey’s multiple comparisons post hoc test was
performed to compare the differences between the fold changes of
anillin promoter luciferase activities with respect to p53
responsiveness. Statistical analysis was performed and graphs were
generated by GraphPad prism v6.01 software (GraphPad, Software,
Inc.). Data are presented as mean ± standard deviation of
triplicate cultures and are representative of 3 independent
experiments. P<0.05 was considered to indicate a statistically
significant difference.
Results
Anillin serves as a biomarker of
colorectal or breast cancer
Previous studies have demonstrated the
overexpression of anillin in various types of tumors and its
association with disease progression (16–19). To confirm its role as a biomarker,
the present study assessed the expression of anillin using the TCGA
database. The results revealed that anillin was highly expressed in
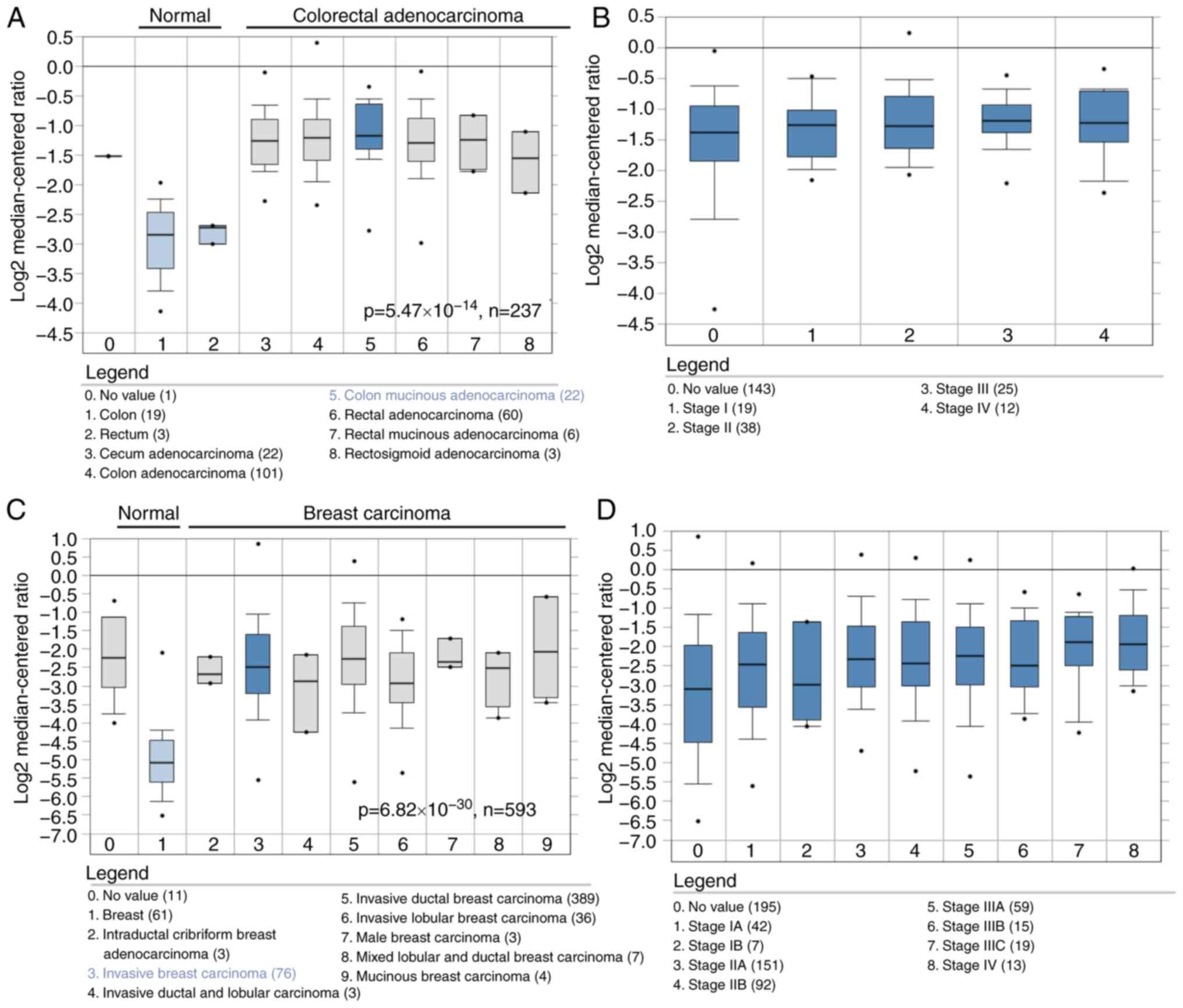
the colorectal adenocarcinoma cohort (Fig. 1A; n=237) and the breast carcinoma
cohort (Fig. 1B; n=593) when
compared with the respective normal colon, rectum or breast
tissues. Furthermore, its expression was increased in colorectal
(Fig. 1C) or breast (Fig. 1D) tumors where tumor progression
had been observed.
Bioinformatics analysis and ChIP
identification of the putative p53 REs in the ANLN upstream 3 kb
promoter
The classic p53 consensus DNA binding sequence
contained the following: A half site of 5′-RRRCWWGYYY, a spacer
consisting of 0–25 bases and a second half site of RRRCWWGYYY-3′,
where R represents purine, W represents adenine or thymidine, Y
represents pyrimidine, G represents guanine and C represents
cytosine. The 3 kb sequence located upstream of the ANLN promoter
sequence was analyzed using Genomatix software, an online
transcriptional factor prediction system. According to the classic
p53 consensus DNA binding sequence, 4 putative p53 binding sites
were identified and located (Fig.
2A): p53 RE-a, p53 RE-b, p53 RE-c and p53 RE-d. The sequences,
the strand on which they were located and the start and ending
positions of each p53 binding site are presented in Fig. 2B. By performing ChIP, the present
study elucidated that sites p53 RE-a and -b were capable of binding
p53 (Fig. 2C). However, other two
putative binding sites, p53 RE-c/d, were not identified by the ChIP
assay (Fig. S1). Mirza et
al (22) previously
demonstrated via a ChIP assay that RE-b was capable of binding p53
and repressed anillin expression. To further define the
functionality of the anillin promoter with respect to p53
responsiveness, the present study constructed a series of
luciferase reporter plasmids containing activated or mutated RE-a
and/or RE-b (Figs. 2D and
S2). Wild-type or p53 null
counterparts containing either activated or mutated binding sites
were subsequently transfected into the HCT116 cells. The results
revealed that each site acted in cis to inhibit luciferase
activity following doxorubicin treatment in a p53-dependent manner
(Fig. 2E). By contrast, the
inhibition of anillin promoter luciferase activities was negated by
mutating the binding sites in the reporter plasmids (Fig. S2). In subsequent experiments, the
constructs were co-transfected into p53-null cells with mutant or
wild-type p53 constructs (Figs.
2F and S3). It was
demonstrated that exogenous wild-type p53 repressed
RE-a/b-containing luciferase constructs, while the mutants
activated luciferase activity (Fig.
2F).
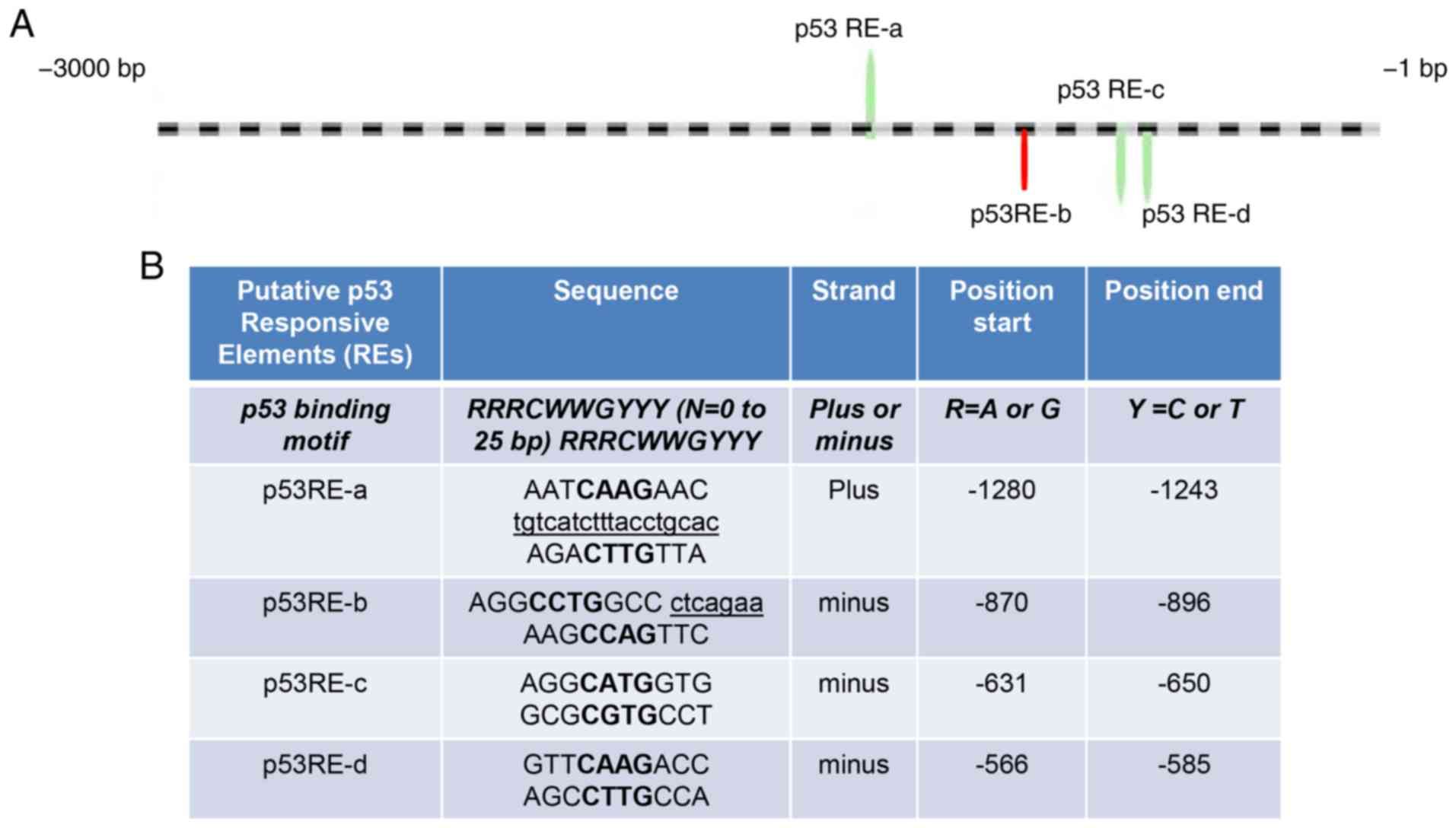 | Figure 2p53 directly binds to ANLN
promoter to inhibit its transcription. (A) Schematic representation
of the 4 putative p53 binding sites on ANLN 3 kb upstream
promoter region. The striped line represents the ANLN
upstream 3 kb promoter region. The 4 putative p53 REs were
identified in this region. p53 RE-a started at −1,721 bp, with
region a located at (+) strand of ANLN 3 kb region. In
addition, p53 RE-c and p53 RE-d began at −2,351 bp and d started at
−2,415 bp were closely located to each other on (−) strand.
Therefore, they were grouped together as one region for the
chromatin immunoprecipitation (ChIP) assay. The p53 RE-b of
ANLN, represented by the red bar, started at −2,105 bp and
was also included to confirm the results. (B) The p53 binding motif
was included for comparison. The uppercase letters in bold
represent the 4 core nucleotides of each half-site. The underlined
and lowercase letters sequences represent mismatches to the
consensus sequence. The half-site sequences separated from one
another and from spacer nucleotides are indicated by a space. (C)
MCF7 cells were treated with 0.5 μM doxorubicin. The chromatin
immunoprecipitates were obtained using p53 antibody or mouse normal
IgG, or without antibody, and analyzed using PCR. Chromatin inputs
from doxorubicin-treated or non-treated MCF7 cells were also used
as positive controls for PCR, while double-distilled H2O
was used as a PCR template for negative control. The PCR products
were resolved on a 2% agarose gel. (D) Schematic representation of
the ANLN promoter luciferase constructs. (E) β-galactosidase
expression plasmids were co-transfected with the pGL3 vector or
reporter plasmids FL, A or B into both HCT116/p53+/+ and
HCT116/p53−/− cells for 24 h. Cells were treated with
0.5 μM doxorubicin for an additional 24 h. Luciferase reporter
activity was measured and normalized to β-galactosidase activity in
the same sample, and presented as the fold decrease. (F)
β-galactosidase expression plasmids were co-transfected with pGL3
vector or reporter plasmids FL, A or B, and pcDNA3, wild-type p53,
R175H or R248W into HCT116/p53−/− cells. Luciferase
reporter activity was measured after 24 h and normalized to
β-galactosidase activity in the same sample, and presented as the
fold decrease. The data are presented as the mean ± standard
deviation of triplicate transfections, and are representative of 3
independent experiments. *P<0.05,
**P<0.01 and ***P<0.001. ANLN, anillin;
FL, full-length. |
Repression of anillin mRNA and protein
levels by the doxorubicin-induced accumulation of p53 in
HCT116/p53+/+ cells
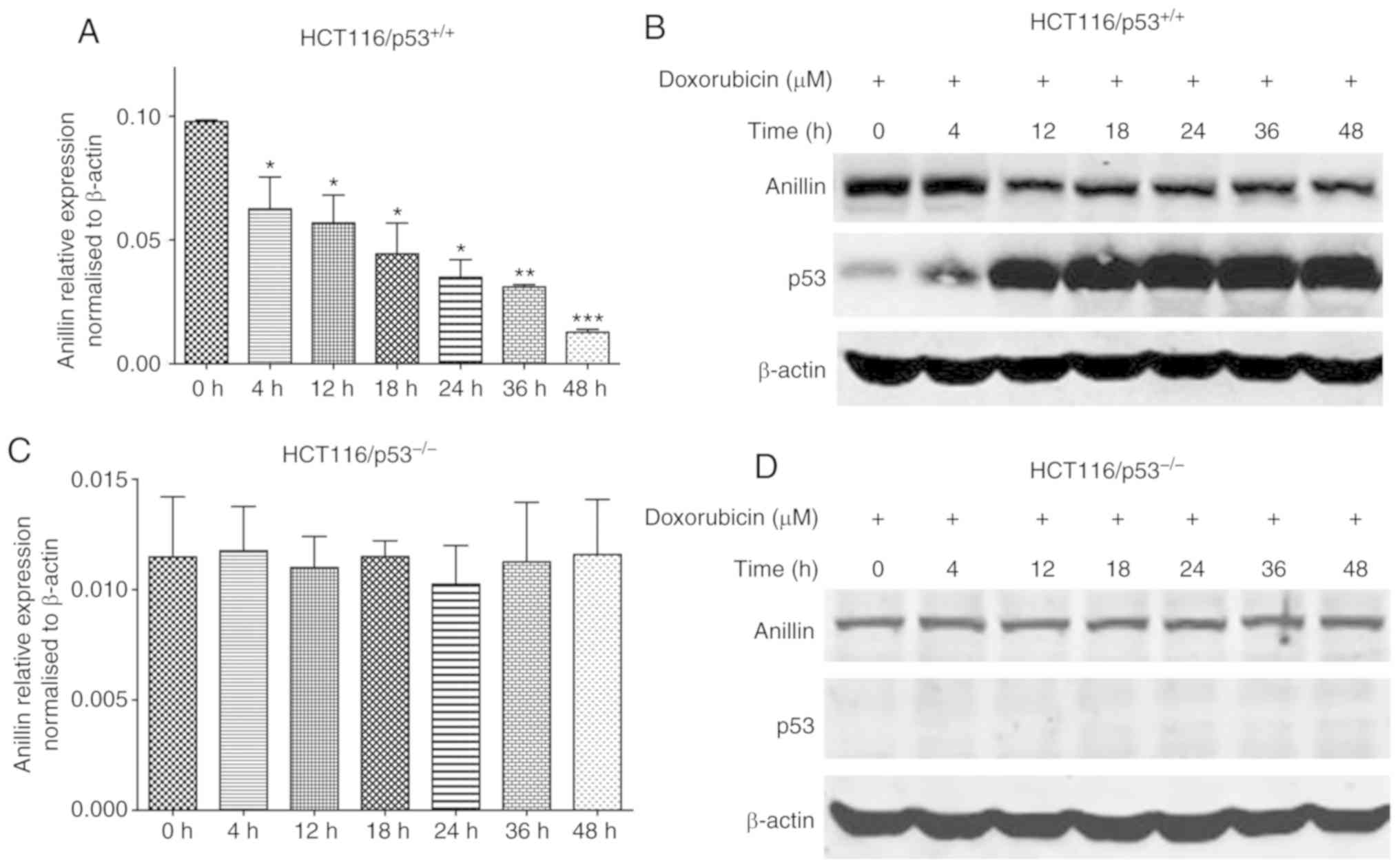
To investigate whether anillin was regulated during
apoptosis, HCT116/p53+/+ cells were treated with
doxorubicin to induce DNA damage. The results of the qPCR analysis
revealed that anillin mRNA expression in HCT116/p53+/+
cells was inhibited at 4 h after doxorubicin treatment (Fig. 3A). To confirm this result, cell
lysates were collected at different time points following
doxorubicin treatment and subjected to western blot analysis. The
results revealed that the expression of anillin was downregulated
in a time-dependent manner following doxorubicin treatment
(Fig. 3B; top row). In addition,
p53 expression was significantly increased following doxorubicin
treatment (Fig. 3B; middle row),
indicating that p53 may be involved in the regulation of anillin
expression. Therefore, HCT116/p53−/− cells were treated
with doxorubicin, following which qPCR and anillin western blot
analysis were performed. Neither the qPCR nor western blot analysis
assays revealed any alterations in anillin expression following
doxorubicin treatment in the HCT116/p53−/− cells
(Fig. 3C and D). These data
indicated that p53 was a key transcription factor that regulated
anillin expression during apoptosis.
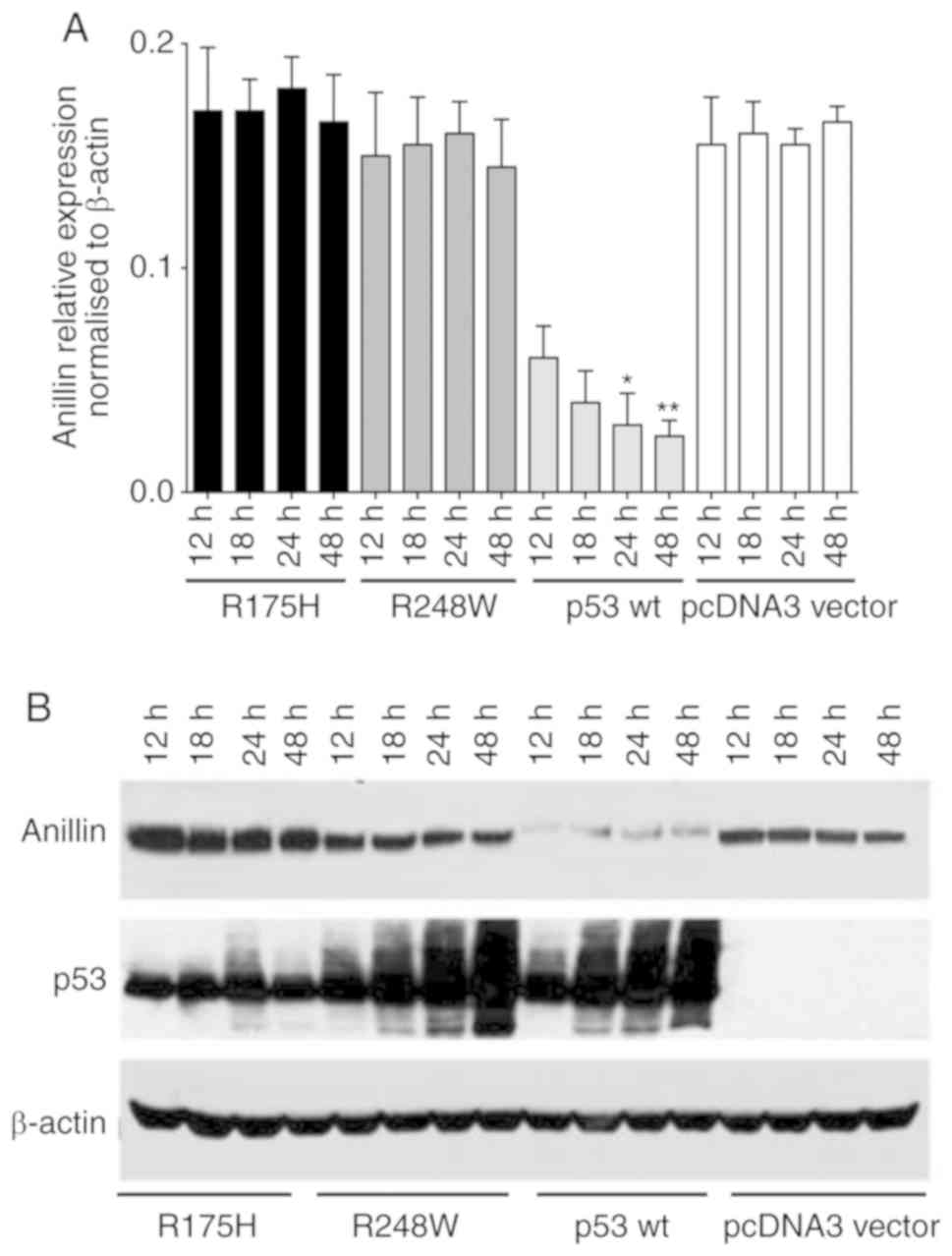
Anillin expression is inhibited in
p53-transfected HCT116/p53−/− cells
To additionally determine whether anillin functions
a transcription factor to p53, 2 p53 mutants that were deficient in
DNA-binding were employed. HCT116/p53−/− cells were
transfected with wild-type or mutant p53 plasmids. The results of
the qPCR analysis demonstrated that anillin mRNA overexpression was
inhibited using the p53 wild-type construct. However, neither of
the mutant constructs used in the present study (R175H or R248W)
exerted an effect in HCT116/p53−/− cells (Fig. 4A). Additionally, anillin protein
levels were suppressed by overexpressing p53 using the wild-type
construct in HCT116 p53 null cells. However, no effect was observed
in cells transfected with R175H or R248W p53 mutant constructs
(Fig. 4B). These data indicated
that p53 may regulate anillin transcription by directly binding to
the ANLN promoter.
Discussion
By fulfilling the criteria set out by Riley et
al (25), in which state that
the p53 binding motif is composed of a half-site RRRCWWGYYY
followed by a spacer (0–21 bp) and subsequently a second half-site
RRRCWWGYYY sequence, the present study demonstrated that anillin
may be a p53 responsive gene. This has relevance in 2 contexts:
Firstly, the frequent overexpression of anillin in neoplasia and
its association with tumor progression may be a consequence of p53
tumor suppressor gene loss of function, which itself is highly
prevalent in human neoplasia and associated with tumor progression.
This model was based on the suggestion that anillin was
transcriptionally repressed by p53 and that p53 mutations may
reverse this effect. The present data is consistent with the
previously described model (24),
since the potent transcriptional repression of the anillin promoter
was induced by wild-type p53. This may explain why anillin
overexpression is common in neoplasia and frequently associated
with tumor progression.
The second inference made from the data of the
present study is associated with the roles of p53 in cytokinesis.
Cell cycle regulation of p53 expression has been well documented,
despite the physiological role of this process being unclear
(26). Previous studies have
indicated the role of p53 in interphase and mitosis (27,28). It has been established that
cytokinesis failure initiates the p53 pathway (29). However, emerging reports have
demonstrated that certain p53 regulated genes have roles in
cytokinesis, including protein regulator cytokinesis 1 (30,31). In addition, the p53-mediated
regulation of the LIM domain kinase (LIMK2) splice variant
LIMK2b links cell cycle checkpoint control with actin
dynamics (32). Changes in cell
shape are key processes in the cell cycle, and this is disrupted in
incidences of neoplasia. Given the role of anillin as an actin
interactor, it is hypothesized that the p53 dependent regulation of
ANLN may modulate cytokinetic processes under conditions of
cellular stress. Other studies have proposed an association between
the p53 response and Septins, which are proteins that interact with
anillin and have roles in cytokinesis (33,34).
Recent data have suggested associations between p53
and stem cell function and, in particular, between p53 and
polarized asymmetric cell division (35,36). Anillin also serves roles across
phylogeny in asymmetric cell divisions. In the development of
Caenorhabditis elegans, protease-activated receptor 4
protein regulates polarity by altering the anillin scaffold and
hence, actin and myosin dynamics (37). Similarly, anillin is crucial for
the asymmetric divisions that give rise to polar bodies in nematode
oocyte development (38).
Asymmetric divisions are central to all eukaryote cell development
(39), particularly in stem cell
function. Evolutionary conservation of the structural and
regulatory elements is also crucial for the determination of
polarity (40).
Finally, the role of p53-regulated anillin
expression may indicate the activities of anillin that are not
involved with cytokinesis. Although the focus of the studies
investigating anillin has been its significant roles in
cytokinesis, substantial anillin protein levels have been observed
in the interphase nucleus. While this may be a storage form of the
protein (1,41), it may also indicate that anillin
serves roles in regulating aspects of nuclear actin function
(42). It is also notable that
other cytokinesis proteins, including Pav and Tum, have important
nuclear roles in the regulation of Wnt signaling (43). In addition to anillin, these
proteins also interact with Rac GTPase-activating protein 1
(44). These observations, and
the results of the present study, indicate that the scope of
anillin function may be greater than previously considered.
Supplementary Information
Acknowledgements
The authors would like to thank Dr B. Vogelstein at
Oncology Center, Johns Hopkins University School of Medicine for
providing the p53 wild-type and isogenic p53 null HCT116 cells. The
results shown in this manuscript are in part based upon data
generated by the TCGA Research Network: https://www.cancer.gov/tcga.
Funding
This project was supported in part by Shanghai
Pujiang Program (grant no. 17PJ1405500 to JM), Shanghai Natural
Science Foundation (grant no. 17ZR1415600 to JM), and National
Natural Science Foundation of China (grant no. 81700134 to JM).
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors’ contributions
JM designed and performed the experiments, analyzed
the data and wrote the manuscript. XL, RM, WL, XS and PL drafted
parts of the manuscript. HZ conceived and supervised the project,
designed the experiments and wrote the manuscript. All authors read
and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Field CM and Alberts BM: Anillin, a
contractile ring protein that cycles from the nucleus to the cell
cortex. J Cell Biol. 131:165–178. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Kim H, Johnson JM, Lera RF, Brahma S and
Burkard ME: Anillin phosphorylation controls timely membrane
association and successful cytokinesis. PLoS Genet.
13:e10065112017. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Pollard TD and O’Shaughnessy B: Molecular
mechanism of cytokinesis. Annu Rev Biochem. 88:661–689. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Zeng S, Yu X, Ma C, Song R, Zhang Z, Zi X,
Chen X, Wang Y, Yu Y, Zhao J, et al: Transcriptome sequencing
identifies ANLN as a promising prognostic biomarker in bladder
urothelial carcinoma. Sci Rep. 7:31512017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Beaudet D, Akhshi T, Phillipp J, Law C and
Piekny A: Active Ran regulates anillin function during cytokinesis.
Mol Biol Cell. 28:3517–3531. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zhang S, Nguyen LH, Zhou K, Tu HC, Sehgal
A, Nassour I, Li L, Gopal P, Goodman J, Singal AG, et al: Knockdown
of anillin actin binding protein blocks cytokinesis in hepatocytes
and reduces liver tumor development in mice without affecting
regeneration. Gastroenterology. 154:1421–1434. 2018. View Article : Google Scholar :
|
|
7
|
Erwig MS, Patzig J, Steyer AM, Dibaj P,
Heilmann M, Heilmann I, Jung RB, Kusch K, Möbius W, Jahn O, et al:
Anillin facilitates septin assembly to prevent pathological
outfoldings of central nervous system myelin. Elife. 8:pii: e43888.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Yasuda T, Takaine M, Numata O and Nakano
K: Anillin-related protein Mid1 regulates timely formation of the
contractile ring in the fission yeast Schizosaccharomyces
japonicus. Genes Cells. 21:594–607. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Gregory SL, Ebrahimi S, Milverton J, Jones
WM, Bejsovec A and Saint R: Cell division requires a direct link
between microtubule-bound RacGAP and anillin in the contractile
ring. Curr Biol. 18:25–29. 2008. View Article : Google Scholar
|
|
10
|
Glotzer M: Cytokinesis in metazoa and
fungi. Cold Spring Harb Perspect Biol. 9:pii: a022343. 2017.
View Article : Google Scholar
|
|
11
|
Zhao WM and Fang G: Anillin is a substrate
of anaphase-promoting complex/cyclosome (APC/C) that controls
spatial contractility of myosin during late cytokinesis. J Biol
Chem. 280:33516–33524. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Perez AM and Thorner J: Septin-associated
proteins Aim44 and Nis1 traffic between the bud neck and the
nucleus in the yeast Saccharomyces cerevisiae. Cytoskeleton
(Hoboken). 76:15–32. 2019. View
Article : Google Scholar
|
|
13
|
Wang D, Chadha GK, Feygin A and Ivanov AI:
F-actin binding protein, anillin, regulates integrity of
intercellular junctions in human epithelial cells. Cell Mol Life
Sci. 72:3185–3200. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Stauffer S, Zeng Y, Zhou J, Chen X, Chen Y
and Dong J: CDK1-mediated mitotic phosphorylation of PBK is
involved in cytokinesis and inhibits its oncogenic activity. Cell
Signal. 39:74–83. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Hall PA, Todd CB, Hyland PL, McDade SS,
Grabsch H, Dattani M, Hillan KJ and Russell SE: The septin-binding
protein anillin is overexpressed in diverse human tumors. Clin
Cancer Res. 11:6780–6786. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Magnusson K, Gremel G, Rydén L, Pontén V,
Uhlén M, Dimberg A, Jirström K and Pontén F: ANLN is a prognostic
biomarker independent of Ki-67 and essential for cell cycle
progression in primary breast cancer. BMC Cancer. 16:9042016.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Pandi NS, Manimuthu M, Harunipriya P,
Murugesan M, Asha GV and Rajendran S: In silico analysis of
expression pattern of a Wnt/β-catenin responsive gene ANLN in
gastric cancer. Gene. 545:23–29. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Idichi T, Seki N, Kurahara H, Yonemori K,
Osako Y, Arai T, Okato A, Kita Y, Arigami T, Mataki Y, et al:
Regulation of actin-binding protein ANLN by antitumor miR-217
inhibits cancer cell aggressiveness in pancreatic ductal
adenocarcinoma. Oncotarget. 8:53180–53193. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Lian YF, Huang YL, Wang JL, Deng MH, Xia
TL, Zeng MS, Chen MS, Wang HB and Huang YH: Anillin is required for
tumor growth and regulated by miR-15a/miR-16-1 in HBV-related
hepatocellular carcinoma. Aging (Albany NY). 10:1884–1901. 2018.
View Article : Google Scholar
|
|
20
|
Shimizu S, Seki N, Sugimoto T, Horiguchi
S, Tanzawa H, Hanazawa T and Okamoto Y: Identification of molecular
targets in head and neck squamous cell carcinomas based on
genome-wide gene expression profiling. Oncol Rep. 18:1489–1497.
2007.PubMed/NCBI
|
|
21
|
Suzuki C, Daigo Y, Ishikawa N, Kato T,
Hayama S, Ito T, Tsuchiya E and Nakamura Y: ANLN plays a critical
role in human lung carcinogenesis through the activation of RHOA
and by involvement in the phosphoinositide 3-kinase/AKT pathway.
Cancer Res. 65:11314–11325. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Mirza A, Wu Q, Wang L, McClanahan T,
Bishop WR, Gheyas F, Ding W, Hutchins B, Hockenberry T, Kirschmeier
P, et al: Global transcriptional program of p53 target genes during
the process of apoptosis and cell cycle progression. Oncogene.
22:3645–3654. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Kent WJ, Sugnet CW, Furey TS, Roskin KM,
Pringle TH, Zahler AM and Haussler D: The human genome browser at
UCSC. Genome Res. 12:996–1006. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
25
|
Riley T, Sontag E, Chen P and Levine A:
Transcriptional control of human p53-regulated genes. Nat Rev Mol
Cell Biol. 9:402–412. 2008. View
Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zhong B, Shingyoji M, Hanazono M, Nguyễn
TTT, Morinaga T, Tada Y, Hiroshima K, Shimada H and Tagawa M: A
p53-stabilizing agent, CP-31398, induces p21 expression with
increased G2/M phase through the YY1 transcription factor in
esophageal carcinoma defective of the p53 pathway. Am J Cancer Res.
9:79–93. 2019.PubMed/NCBI
|
|
27
|
Johmura Y and Nakanishi M: Multiple facets
of p53 in senescence induction and maintenance. Cancer Sci.
107:1550–1555. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Luo Q, Guo H, Kuang P, Cui H, Deng H, Liu
H, Lu Y, Wei Q, Chen L, Fang J, et al: Sodium fluoride arrests
renal G2/M phase cell-cycle progression by activating
ATM-Chk2-P53/Cdc25C signaling pathway in mice. Cell Physiol
Biochem. 51:2421–2433. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Meitinger F, Anzola JV, Kaulich M,
Richardson A, Stender JD, Benner C, Glass CK, Dowdy SF, Desai A,
Shiau AK and Oegema K: 53BP1 and USP28 mediate p53 activation and
G1 arrest after centrosome loss or extended mitotic duration. J
Cell Biol. 214:155–166. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zhang B, Shi X, Xu G, Kang W, Zhang W,
Zhang S, Cao Y, Qian L, Zhan P, Yan H, et al: Elevated PRC1 in
gastric carcinoma exerts oncogenic function and is targeted by
piperlongumine in a p53-dependent manner. J Cell Mol Med.
21:1329–1341. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Tsuchihara K, Lapin V, Bakal C, Okada H,
Brown L, Hirota-Tsuchihara M, Zaugg K, Ho A, Itie-Youten A,
Harris-Brandts M, et al: Ckap2 regulates aneuploidy, cell cycling,
and cell death in a p53-dependent manner. Cancer Res. 65:6685–6691.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Hsu FF, Lin TY, Chen JY and Shieh SY:
p53-mediated transactivation of LIMK2b links actin dynamics to cell
cycle checkpoint control. Oncogene. 29:2864–2876. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Patzig J, Erwig MS, Tenzer S, Kusch K,
Dibaj P, Möbius W, Goebbels S, Schaeren-Wiemers N, Nave KA and
Werner HB: Septin/anillin filaments scaffold central nervous system
myelin to accelerate nerve conduction. Elife. 5:pii: e17119. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Kremer BE, Adang LA and Macara IG: Septins
regulate actin organization and cell-cycle arrest through nuclear
accumulation of NCK mediated by SOCS7. Cell. 130:837–850. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Levine AJ, Puzio-Kuter AM, Chan CS and
Hainaut P: The role of the p53 protein in stem-cell biology and
epigenetic regulation. Cold Spring Harb Perspect Med. 6:pii:
a026153. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Itahana Y, Zhang J, Göke J, Vardy LA, Han
R, Iwamoto K, Cukuroglu E, Robson P, Pouladi MA, Colman A and
Itahana K: Histone modifications and p53 binding poise the p21
promoter for activation in human embryonic stem cells. Sci Rep.
6:281122016. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Chartier NT, Salazar Ospina DP, Benkemoun
L, Mayer M, Grill SW, Maddox AS and Labbé JC: PAR-4/LKB1 mobilizes
nonmuscle myosin through anillin to regulate C. elegans embryonic
polarization and cytokinesis. Curr Biol. 21:259–269. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Sharif B, Fadero T and Maddox AS: Anillin
localization suggests distinct mechanisms of division plane
specification in mouse oogenic meiosis I and II. Gene Expr
Patterns. 17:98–106. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Fox S, Southam P, Pantin F, Kennaway R,
Robinson S, Castorina G, Sánchez-Corrales YE, Sablowski R, Chan J,
Grieneisen V, et al: Spatiotemporal coordination of cell division
and growth during organ morphogenesis. PLoS Biol. 16:e20059522018.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Pillitteri LJ, Guo X and Dong J:
Asymmetric cell division in plants: Mechanisms of symmetry breaking
and cell fate determination. Cell Mol Life Sci. 73:4213–4229. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Oegema K, Savoian MS, Mitchison TJ and
Field CM: Functional analysis of a human homologue of the
Drosophila actin binding protein anillin suggests a role in
cytokinesis. J Cell Biol. 150:539–552. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Gieni RS and Hendzel MJ: Actin dynamics
and functions in the interphase nucleus: Moving toward an
understanding of nuclear polymeric actin. Biochem Cell Biol.
87:283–306. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Jones WM, Chao AT, Zavortink M, Saint R
and Bejsovec A: Cytokinesis proteins Tum and Pav have a nuclear
role in Wnt regulation. J Cell Sci. 123:2179–2189. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Guillemot L, Guerrera D, Spadaro D, Tapia
R, Jond L and Citi S: MgcRacGAP interacts with cingulin and
paracingulin to regulate Rac1 activation and development of the
tight junction barrier during epithelial junction assembly. Mol
Biol Cell. 25:1995–2005. 2014. View Article : Google Scholar : PubMed/NCBI
|