Introduction
Osteosarcoma (OS) is a primary malignant tumor that
occurs in adolescents and children (1). The pathogenesis of OS is very
complex, with surgery and chemotherapy being the main treatment
methods (2,3). Moreover, the high recurrence and
metastasis of OS patients commonly occurs following standard
treatment. Therefore, it is necessary to explore the pathogenesis
of OS and to discover novel biomarkers for the targeted treatment
of OS.
Circular RNAs are non-coding RNAs with a closed
circular structure (4). Due to
their insensitivity to nucleases, circular RNAs are more stable and
more suitable as diagnostic markers (5,6).
Previous studies have demonstrated that circular RNAs play a
central regulatory role in the pathogenesis of a variety of cancer
types (7,8). Studies have also verified that
circular RNAs, as competitive endogenous RNAs, bind to miRNAs and
regulate the expression of downstream target genes (9-11).
For example, circular RNA circNASP has been shown to regulate OS
malignant behavior via the miR-1253/FOXF1 axis (12). CircSAMD4A, a novel circular RNA,
was derived from the SAMD4A gene and is located in
chr14:55168799-55169298. Yanbin and Jing identified that circSAMD4A
was involved in the cellular progression of OS by targeting
miR-1244 and regulating MDM2 expression (13). Therefore, the circular
RNA-mediated miRNA/mRNA regulatory network plays an essential role
in cancer.
MicroRNAs (miRNAs or miRs) are a class of non-coding
single-stranded RNAs, ranging from 18 to 22 nucleotides in length,
which degrade downstream target genes at the transcriptional and
post-transcriptional levels and affect gene expression (14-16). There is accumulating evidence to
suggest that miRNAs are widely involved in tumor progression,
including cell migration, invasion and proliferation, acting as
important regulators in the progression of human diseases, such as
hepatocellular carcinoma, osteoarthritis chondrocytes, colorectal
cancer and neuroblastoma (17-20). For example, miR-503 has been shown
to suppress cell proliferation and promote apoptosis by targeting
E2F3 in colorectal cancer cells (21). In addition, miRNAs have been
considered as biomarkers of cancer treatment and prognosis
(22-24). For example, serum miR-542-3p has
been verified as a prognostic biomarker in OS (25). miR-221-3p, miR-342-3p and
miR-491-3p have been shown to be associated with prognosis in colon
cancer (26). miR-21 and miR-29
could thus be used as biomarkers to improve diagnostic efficiency
of gastric cancer (27). These
data suggest that the regulatory function of miRNAs/mRNAs is
critical in the development of various tumors.
The present study demonstrated that CircSAMD4A was
highly expressed in OS tissues and cells, suggesting that it plays
an important role in the progression of OS. Therefore, biological
function assays were performed to further explore the function of
CircSAMD4A in OS and to provide novel regulatory mechanisms and
therapeutic targets for OS.
Materials and methods
Tumor specimens and control specimens
(pair-matched-adjacent normal tissues) were obtained from 30
patients diagnosed with OS (age, ≥25 years, n=8; age, <25 years,
n=22; male/female ratio, 16/14) at the Second Affiliated Hospital
of Guangzhou Medical University between August, 2016 and May, 2019.
All the patients underwent resection and signed written informed
consents. The experiment was approved by the Ethics Committee of
the Second Affiliated Hospital of Guangzhou Medical University. All
the samples were stored at -80°C until use in the experiments.
Cell culture and transfection
OS cell lines (HOS and U2OS) and the human
osteoblast cell line (hFOB 1.19) were purchased from the Chinese
Cell Bank of the Chinese Academy of Sciences (Shanghai, China) and
cultured in Dulbecco's modified Eagle's medium (DMEM; Gibco; Thermo
Fisher Scientific, Inc.) supplemented with 10% FBS (Gibco; Thermo
Fisher Scientific, Inc.), 100 U/ml of penicillin and 100 mg/ml of
streptomycin (Invitrogen; Thermo Fisher Scientific, Inc.) at 37°C
in a humidified incubator with 5% CO2.
Small interfering RNA against CircSAMD4A
(si-Circ-SAMD4A#1 and si-CircSAMD4A#2) and negative control si-NC,
sh-CircSAMD4A and sh-NC, pcDNA-CircSAMD4A and pcDNA-NC, miR-342-3p
mimics (miR-342-3p) and NC, anti-miR-342-3p and anti-NC, pcDNA-FZD7
(FZD7) and vector were purchased from GenePharma. The sequences of
si-CircSAMD4A were as follows: (5′-3′): si-CircSAMD4A#1, AGC ACA
AGT ACA AGA ATC ATT; si-CircSAMD4A#2, GCA CAA GTA CAA GAA TCA TTA;
miR-342-3p mimic sense, GGG TCT CAC ACA GAA ATC GC and antisense,
CAG TGC GTG TCG TGG AGT; miR-342-3p inhibitor, sense, GAG CAG GCT
GGA GAA; miRNA mimic negative control sense, UUU GUA CUA CAC AAA
AGU ACU G and antisense, CAG UAC UUU UGU GUA GUA CAA A; miRNA
inhibitor negative control sense, CAG UAC UUU UGU GUA GUA CAA A.
Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific, Inc.) was
used to transfect the vectors and oligonucleotides into OS cells
for 5 min at room temperature according to the manufacturer's
protocol.
CircRNA expression profile analysis
A gene expression profile (GSE96964) of OS was
downloaded from the Gene Expression Omnibus (GEO, https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE96964).
GSE96964 includes 8 samples (U2OS, U2OS/MTX300, HOS, MG63, 143B,
ZOS, ZOSM and hFOB1.19 cells). The differentially expressed genes
(DEGs) were compared by using GEO2R (https://www.ncbi.nlm.nih.gov/geo/geo2r/?acc=GSE96964).
RNase R treatment and RT-qPCR
Total RNA was isolated from tissues and cells using
TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.). The
PrimeScript® RT reagent kit (Takara) was applied to
reverse transcript RNA to cDNA. The Reverse Transcription Reagents
(Applied Biosystems) or TaqMan® MicroRNA Reverse
Transcription kit (Applied Biosystems) was used to detect
CircSAMD4A and Frizzled-7 (FZD7) or miR-342-3p expression,
respectively. Subsequently, qPCR assay was performed on an ABI 7500
Fast Real-Time PCR System (Applied Biosystems) with
SYBR® Premix Ex Taq™ reagent (Takara). Glyceraldehyde
3-phosphate dehydro-genase (GAPDH) was employed as the
normalization gene of CircSAMD4A and FZD7. U6 was employed as the
normalization gene of miR-342-3p. The mRNA expression levels of
CircSAMD4A and SAMD4A in both HOS and U2OS cell lines were detected
by RT-qPCR in the presence or absence of RNase R. The expression of
CircSAMD4A, FZD7 and miR-342-3p was calculated using
2−ΔΔCt method (28).
RT-qPCR was performed under the following thermocycling conditions:
60°C for 2 min, 95°C for 25 sec, and 40 circles of 95°C for 10 sec
and 60°C for 30 sec. The sequences or the primers were as follows:
U6 forward, 5′-CTC GCT TCG GCA GCA CA-3′ and reverse, 5′-AAC GCT
TCA CGA ATT TGC GT-3′; miR-342-3p forward, 5′-TGC GGT CTC ACA CAG
AAA TCG CAC-3′ and reverse, 5′-CCA GTG CAG GGT CCG AGG T-3′;
circSAMD4A forward, 5′-ACT GGC AGG ACA AAA GCA TG-3′ and reverse,
5′-CAG GAT TTT GGG CAG CAG TT-3′; FZD7 forward, 5′-TTC TCG GAC GAT
GGC TAC C-3′ and reverse, 5′-GAA CCA AGT GAG AGA CAG AAT GAC C-3′;
and GAPDH forward, 5′-GGA GCG AGA TCC CTC CAA AAT-3′ and reverse,
5′-GGC TGT TGT CAT ACT TCT CAT GG-3′.
Western blot analysis
Total protein was lysed and isolated from cells and
tissues using RIPA buffer (Beyotime Institute of Biotechnology) and
its quantification was measured using the BCA™ Protein Assay kit
(Pierce; Thermo Fisher Scientific, Inc.), and equal amount proteins
(20 µg) were then separated using sodium dodecyl
sulfate-polyacrylamide gel electrophoresis (10% SDS-PAGE). The
protein was transferred onto polyvinylidene fluoride (PVDF)
membranes (Millipore). The membranes were blocked with PBS
containing 5% non-fat milk for 1 h at room temperature and then
incubated at 4°C overnight with the following primary antibodies:
FZD7 (1:1,000 dilution, AV41251, Sigma-Aldrich; Merck KGaA),
C-caspase-3 (1:2,000 dilution, sc-70497, Santa Cruz Biotechnology,
Inc.), N-cadherin (1:2,000 dilution, sc-7939, Santa Cruz
Biotechnology, Inc.), proliferating cell nuclear antigen (PCNA;
1:1,000 dilution, MAB424R, Sigma-Aldrich; Merck KGaA), Vimentin
(1:2,000 dilution, sc-6260, Santa Cruz Biotechnology, Inc.),
E-cadherin (1:2,000 dilution, sc-21791, Santa Cruz Biotechnology,
Inc.), Ki-67 (1:1,000 dilution, ab15580, Abcam) and GAPDH (1:1,000
dilution, sc-25778, Santa Cruz Biotechnology, Inc.). The membranes
were then incubated with horseradish peroxidase (HRP)-conjugated
goat anti-rabbit secondary antibody (1:1,000 dilution, sc-2357,
Santa Cruz Biotechnology, Inc.) for 1 h at 37°C. Clarity Western
ECL substrate (Bio-Rad) and ChemiDocTM MP Imaging System (Bio-Rad)
were applied to detect the blots and for visualizion. In addition,
ImageJ software was used to quantify the blots.
Cell cytotoxicity and apoptosis
The cytotoxicity of the trans-fected cells was
measured using the Cell Growth Determination kit MTT based
(Sigma-Aldrich; Merck KGaA). The cells were cultured and seeded
into 96-well plates at a density of 2×103 cells per
well. Trypsin/EDTA 0.25% (Sigma-Aldrich; Merck KGaA) was then added
to each well to generate a single cell suspension. Following
incubation for 4 h at 37°C with 5% of CO2, 150 µl
MTT solvent (4 mM HCl, 0.1% NP40 in isopropanol) was added to each
well and incubated at 7°C for 3 h for conversion into formazan.
Finally, the optical density (OD) was determined at a wavelength of
450 nm with a spectrophotometric microplate reader (Beyotime
Institute of Biotechnology).
Cell apoptosis was tested using the Annexin
V-FITC/PI Apoptosis Detection kit (BD Biosciences) according to the
manufacturer's instructions. The transfected cells were collected
and Annexin V-FITC and propidium iodide (PI) were added to
incubation at 37°C in the dark for 15 min. Cell apoptosis was
performed and displayed using flow cytometry.
Cell invasion and migration
A Transwell assay was performed to detect the
invasion and migration of the transfected cells. Briefly, the
transfected cells (3×103) were harvested for 48 h in
serum-free DMEM and then seeded in the upper chambers with
8-µm pores coated with Matrigel (BD Biosciences) for
invasion assay. For Transwell migration assay, the upper chambers
were not coated with Matrigel. An amount of 10% FBS was added to
the medium as a chemoattractant and then the mixed medium was added
to the lower chamber. Following incubation for 48 h at 37°C, the
cells remaining in the upper chambers were removed. The cells
attached to the lower surface were stained for 20 min at 37°C using
crystal violet. The migrated and invasive cells were detected and
counted using a Countess Automatic Cell Counter (Invitrogen; Thermo
Fisher Scientific, Inc.).
Animal experiments
A total of 12 female nude mice (6 weeks old;
weighing 18-20 g) purchased from Beijing HFK Bioscience Co. Ltd.
were used in this study. The mice were housed in a standard animal
laboratory where the temperature was maintained at 25°C with a
humidity level of 40-70% and 12-h dark/light cycles under
pathogen-free conditions. The animals were given free access to
food and water. The animal experiments were approved by the Animal
Care and Welfare Committee of The Second Affiliated Hospital of
Guangzhou Medical University. The mice were divided into 2 groups
as follows: sh-CircSAMD4A (n=6) and sh-NC (n=6). Sh-CircSAMD4A and
sh-NC (20 µM) were transfected into the HOS and U2OS cells
and then injected subcutaneously into the right flanks of the
female nude mice. The tumor volume was measured every 5 days after
the tumors became visible. At 25 days after the injection, all the
12 mice were sacrificed by cervical dislocation that caused a sharp
section of the spinal cord followed by an instantaneous cardiac
arrest. Subsequently, the tumor weight was detected. The length (L)
and width (W) was measured using a vernier caliper. The tumor
volume (V) was calculated with the following formula:
V=(LxW2) ×0.5.
Dual-luciferase reporter assay
To validate the targeting association among
CircSAMD4A, FZD7 and miR-342-3p, a CircSAMD4A-wt, FZD7-wt vector
was constructed that contained the matching sequence of miR-342-3p
and CircSAMD4A-mut. A FZD7-mut vector was also constructed that
contained a mutant sequence. The vector is pmirGLO Dual-Luciferase
miRNA Target Expression Vector (Promega). CircSAMD4A-wt or
CircSAMD4A-mut and miR-342-3p or NC were then transfected into the
U2OS and HOS cells using Lipofectamine 2000 reagent (Invitrogen;
Thermo Fisher Scientific, Inc.) according to the manufacturer's
instructions. In addition, FZD7-wt or FZD7-mut and miR-342-3p or NC
were transfected into the U2OS and HOS cells using Lipofectamine
2000 reagent (Invitrogen; Thermo Fisher Scientific, Inc.).
Following transfection for 48 h, the Dual-Luciferase Reporter Assay
System (Promega) was used to determine the luciferase activity.
RNA immunoprecipitation (RIP)
RIP assay was applied using the EZ-Magna RIP kit
(Millipore) according to the manufacturer's protocol. The
transfected cells were collected and lysed using RIP buffer, and
then incubated with human anti-Argo-naute 2 (Ago2) antibody
(1:1,000 dilution, ab5072, Abcam) or normal mouse IgG (negative
control, 1:1,000 dilution, 12-349, Sigma-Aldrich; Merck KGaA).
Following incubation overnight at 4°C, proteinase K buffer (Abcam)
was added to remove the non-specific binding and the
immunoprecipitated RNA was extracted. The purified proteins were
detected by RT-qPCR.
Statistical analysis
All data are presented as the means ± standard
deviation (SD). The analysis of the data was performed using
GraphPad Prism 7.0 (GraphPad Software). The inter-action among
CircSAMD4A, miR-342-3p and FZD7 was analyzed by Pearson's
correlation analysis. All comparisons were analyzed using Student's
t-tests or one-way ANOVA followed by Tukey's test. P<0.05 was
considered to indicate a statistically significant difference.
Results
CircSAMD4A is highly expressed in OS
tissues and cell lines
The online data set (GSE96964) revealed that
CircSAMD4 (hsa_circRNA_0004846) was highly expressed in the MG-63,
U2OS, HOS and 143B cells compared with the hFOB 1.19 cells,
suggesting that CircSAMD4 may be associated with OS progression.
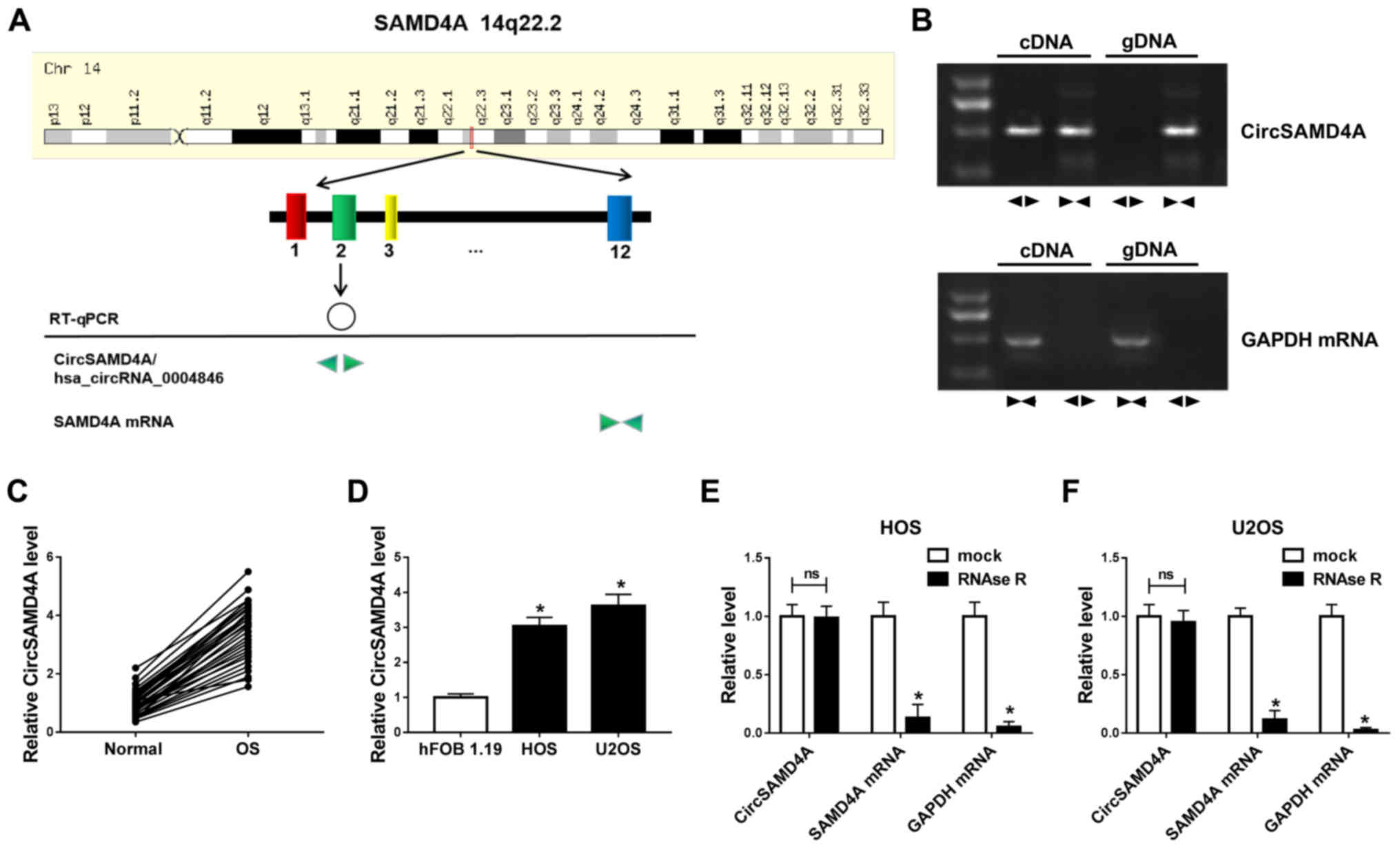
The present study demonstrated that CircSAMD4 was derived from exon
2 of the host gene SAMD4 according to circBase (http://www.circbase.org/) (Fig. 1A). Divergent primers were then
designed to amplify CircSAMD4 and convergent primers wer designed
to amplify SAMD4 mRNA. CircSAMD4 was only detectable in cDNA, but
not in genomic DNA (gDNA) from the U2OS cells, as shown by RT-qPCR
with divergent primers, while SAMD4 could be amplified in both cDNA
and gDNA using convergent primers (Fig. 1B). CircSAMD4A expression was
detected in OS tissues and cells. The data demonstrated that
CircSAMD4A expression was upregulated in OS tissues compared with
normal tissues (Fig. 1C).
Additionally, CircSAMD4A expression in the HOS and U2OS cells was
significantly higher than that in the hFOB 1.19 cells (Fig. 1D). Moreover, RNase R assay was
applied to confirm the stability of CircSAMD4A. The results
revealed that the expression of the linear forms of SAMD4A markedly
decreased following RNase R treatment; however, RNase R failed to
digest CircSAMD4A in the HOS and U2OS cells (Fig. 1E and F). Therefore, CircSAMD4A was
upregulated in OS cells and tissues, indicating that CircSAMD4A is
associated with OS progression.
Knockdown of CircSAMD4A promotes
cytotoxicity and apoptosis, and inhibits the migration, invasion
and epithelial-mesenchymal transition (EMT) of OS cells
To further examine the effects of CircSAMD4A on OS
progression, si-NC and si-CircSAMD4A were transfected into the HOS
and U2OS cells. The results indicated that CircSAMD4A was inhibited
in the HOS and U2OS cells, which were trans-fected with
si-CircSAMD4A, and 2 transfected cell lines (si-CircSAMD4A#1 and
si-CircSAMD4A#2) were selected for use in the following experiments
(Fig. 2A). As presented in
Fig. 2B, MTT assay suggested that
cell cytotoxicity was enhanced by CircSAMD4A downregulation.
Moreover, Ki-67 protein expression was decreased by si-CircSAMD4A
transfection (Fig. 2C). The
knockdown of CircSAMD4A suppressed OS cell growth. Flow cytometry
demonstrated that CircSAMD4A downregulation contributed to OS cell
apoptosis (Fig. 2D and E). The
protein level of C-caspase 3 was markedly induced by the knockdown
of CircSAMD4A (Fig. 2F).
Transwell assay confirmed that the migration and invasion of the
HOS and U2OS cells were decreased by the knockdown CircSAMD4A
(Fig. 2G and H). Furthermore, the
knockdown of CircSAMD4A inhibited N-cadherin and Vimentin protein
expression, whereas it increased E-cadherin protein expression in
the HOS and U2OS cells (Fig. 2I and
J). These data demonstrated that the knockdown of CircSAMD4A
suppressed the migration, invasion and EMT of OS cells, and
promoted the cytotoxicity and apoptosis of OS cells, suggesting
that CircSAMD4A plays a crucial role in OS progression.
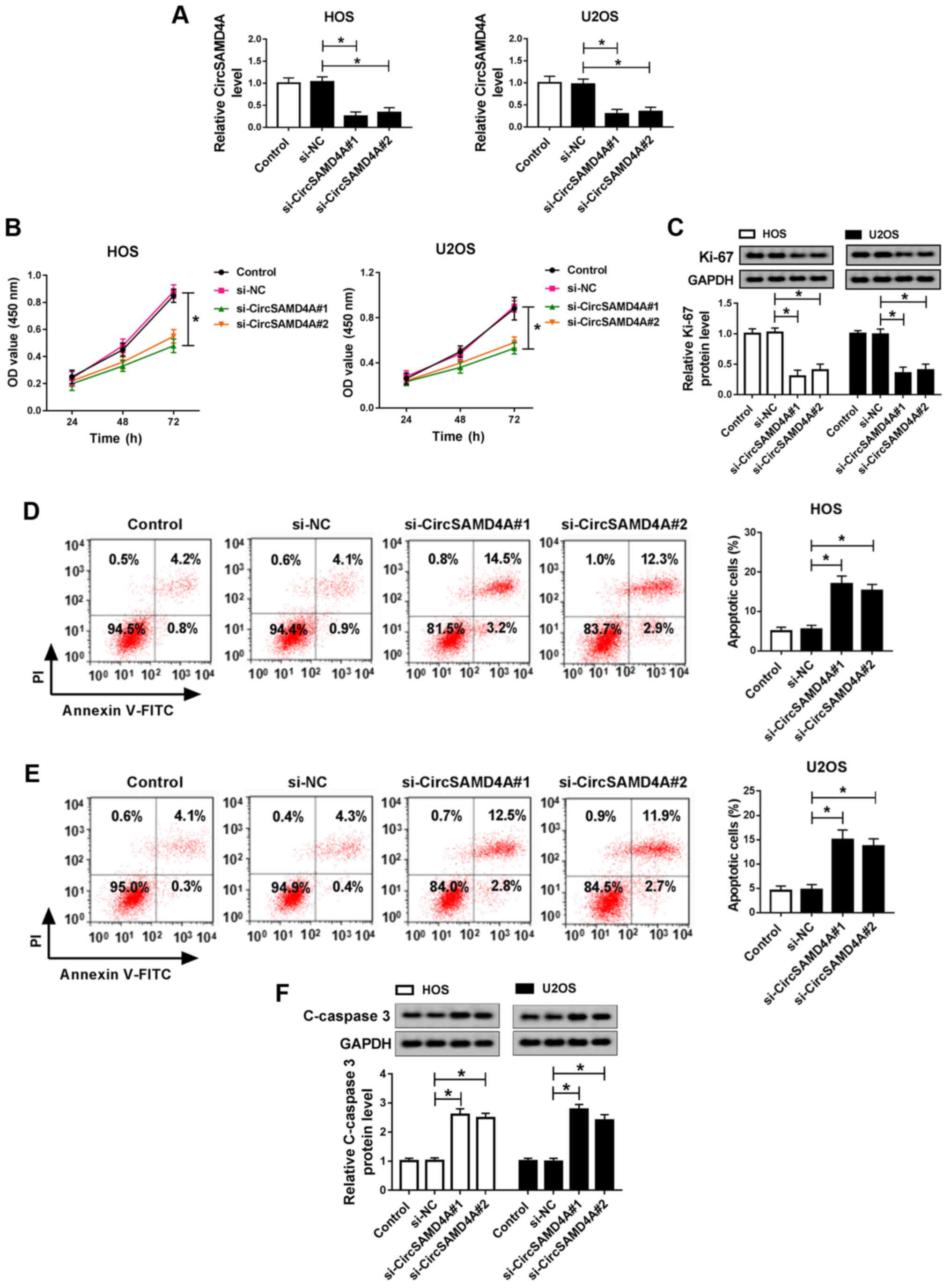 | Figure 2Knockdown of CircSAMD4A inhibits the
migration, invasion and EMT, and promotes the cytotoxicity and
apoptosis of OS cells. (A) CircSAMD4A expression of control, si-NC,
si-CircSAMD4A#1 or si-CircSAMD4A#2 groups was detected by RT-qPCR.
(B) MTT assay was applied to measure the cytotoxicity of HOS and
U2OS cells transfected with control, si-NC si-CircSAMD4A#1 and
si-CircSAMD4A#2. (C) The protein expression of Ki-67 was measured
in control, si-NC, si-CircSAMD4A#1 and si-CircSAMD4A#2 groups in
HOS and U2OS cells by western blot analysis. (D and E) Cell
apoptosis of control, si-NC si-CircSAMD4A#1 and si-CircSAMD4A#2
groups detected by flow cytometry in HOS and U2OS cells. (F) The
protein expression of C-caspase-3 was detected in control, si-NC
si-CircSAMD4A#1 and si-CircSAMD4A#2 groups in HOS and U2OS cells by
western blot analysis. (G and H) Cell migration and invasion of
control, si-NC si-CircSAMD4A#1 and si-CircSAMD4A#2 groups were
detected using Transwell assay in HOS and U2OS cells. (I and J) The
protein expression of N-cadherin, Vimentin and E-cadherin of
control, si-NC si-CircSAMD4A#1 and si-CircSAMD4A#2 groups was
measured by western blot analysis in HOS and U2OS cells.
*P<0.05, vs. respective control. OS, osteosarcoma;
EMT, epithelial-mesenchymal transition. |
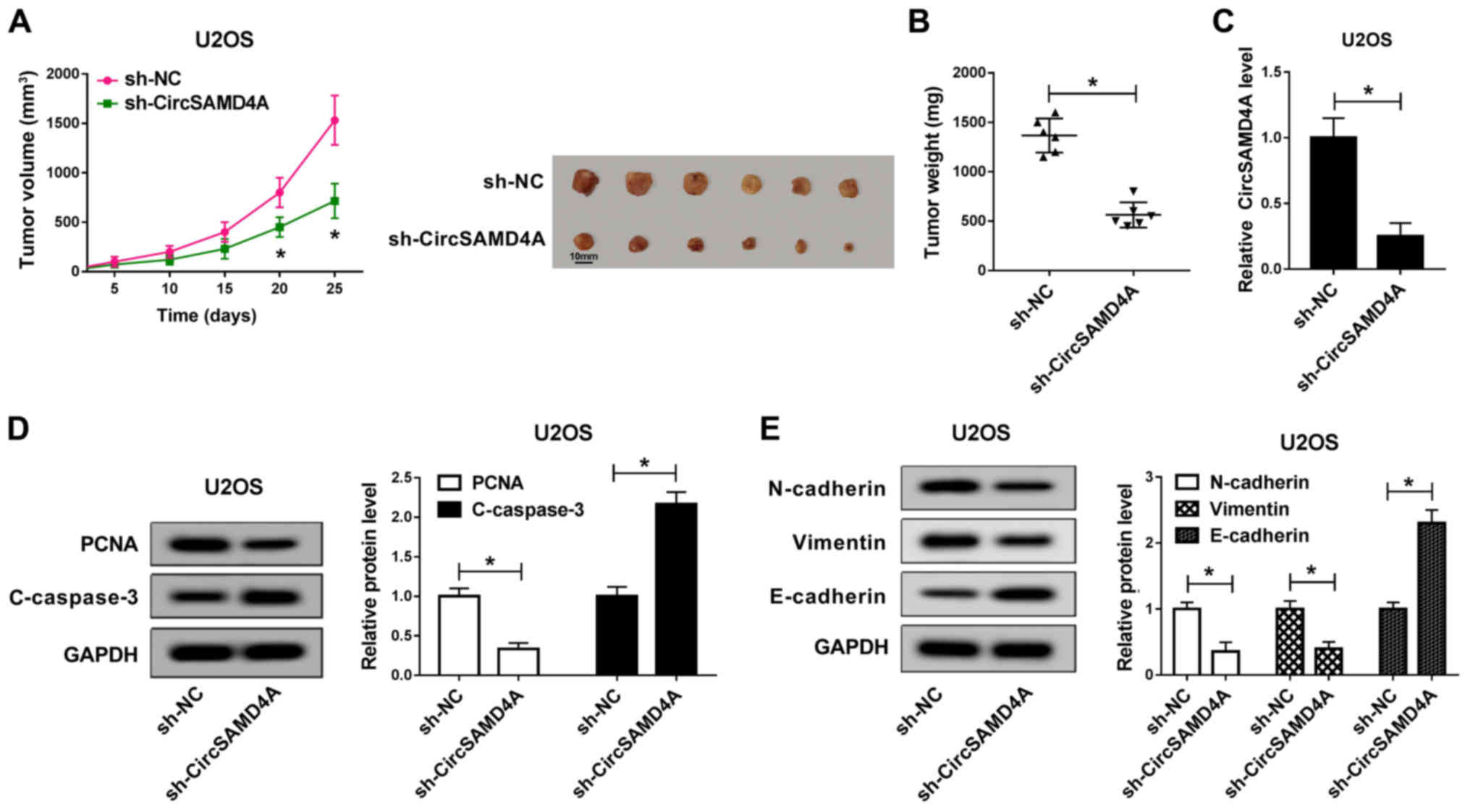
Knockdown of CircSAMD4A suppresses tumor
growth in vivo
Subsequently, the function of CircSAMD4A was
verified in a mouse experiment. sh-NC and sh-CircSAMD4A were
transfected into U2OS cells and the transfected cells were then
injected into mice. The results revealed that the tumor volume in
the CircSAMD4A knockdown group was significantly smaller than that
of the sh-NC group (Fig. 3A).
Similarly, CircSAMD4A knockdown significantly inhibited tumor
weight (Fig. 3B). In the tumor
tissues, the expression of CircSAMD4A was significantly reduced by
sh-CircSAMD4A (Fig. 3C).
Moreover, compared with the sh-NC group, PCNA protein expression
was markedly reduced by CircSAMD4A downregulation, while the
protein expression of C-caspase-3 was markedly increased (Fig. 3D). Moreover, the protein
expression of N-cadherin and Vimentin in the sh-Circ-SAMD4A group
was significantly lower than that in the sh-NC group, while the
protein expression of E-cadherin was markedly higher than that in
the sh-NC group (Fig. 3E). These
results revealed that the downregulation of CircSAMD4A
significantly inhibited the tumor growth of OS in vivo.
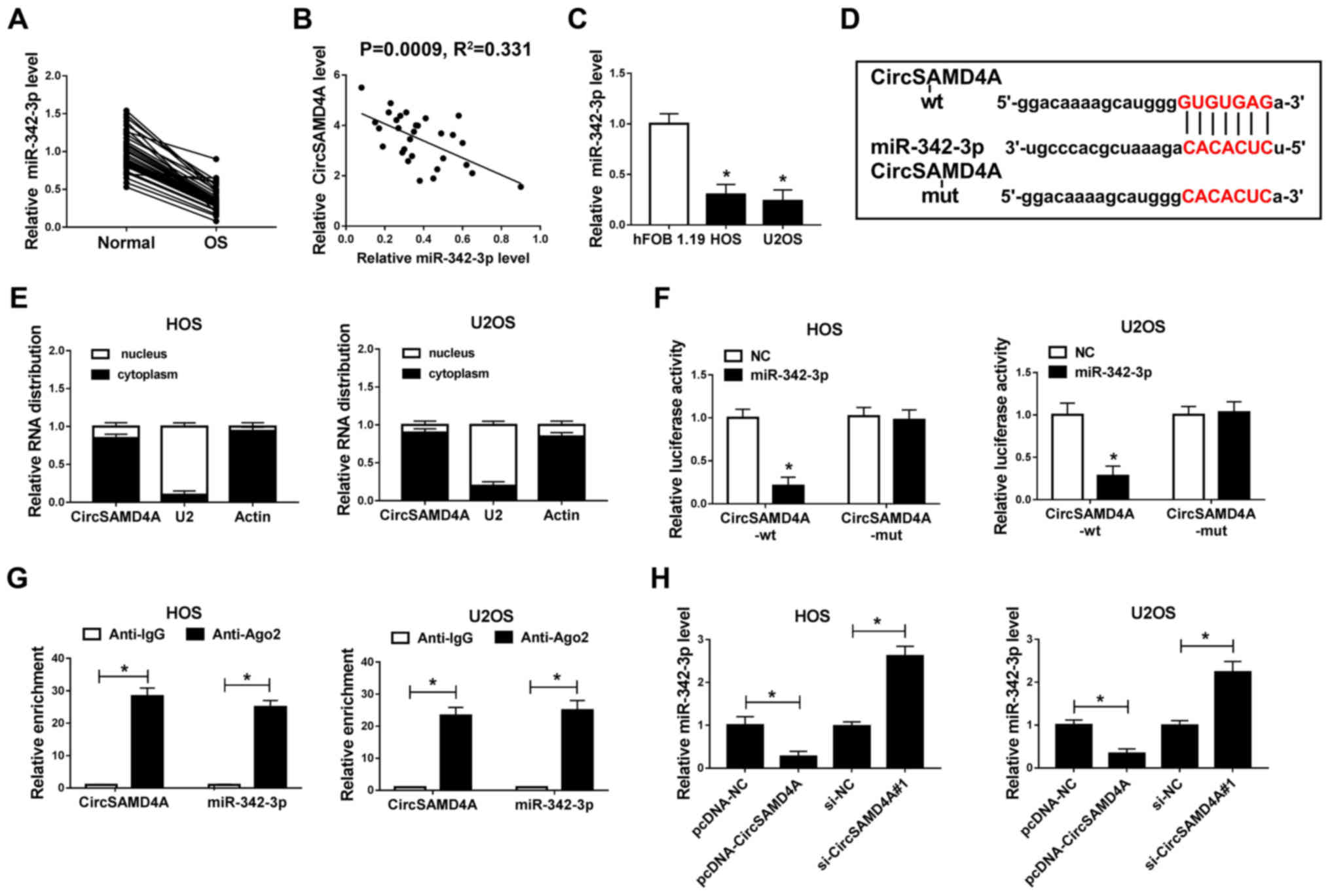
miR-342-3p expression is inhibited in OS
tissues and cells, and it is a target miRNA of CircSAMD4A in
OS
Based on bioinformatics analysis, the present study
demonstrated that miR-342-3p contained a binding site of
CircSAMD4A, predicting that miR-342-3p is a candidate target miRNA
of CircSAMD4A (Fig. 4D). Thus,
miR-342-3p expression was subsequently detected in OS tissues and
cells. The data revealed that miR-342-3p expression was
significantly decreased in OS tissues and cells, and that the
miR-342-3p level negatively correlated with CircSAMD4A expression
in OS (Fig. 4A-C). Subsequently,
nuclear and cytoplasmic separation experiments were performed using
actin as a marker in the cytoplasm and U2 as a marker to identify
the nuclear compartment, in order to determine the subcellular
localization of CircSAMD4A in the nucleus and cytoplasm. The
results indicated that the expression of CircSAMD4A in the
cytoplasm was significantly higher than that in the nucleus in the
HOS and U2OS cells (Fig. 4E). In
addition, the dual-luciferase reporter assay demonstrated that the
luciferase activity was significantly decreased when miR-342-3p was
bound to CircSAMD4A-wt; however, there was no significant change in
the luciferase activity when miR-342-3p was bound to CircSAMD4A-mut
in the HOS and U2OS cells (Fig.
4F). The experimental combination of RIP also demonstrated that
miR-342-3p is the target miRNA of CircSAMD4A in HOS and U2OS cells
(Fig. 4G). Moreover, miR-342-3p
expression in the pcDNA-NC, pcDNA-CircSAMD4A, si-NC and
si-Circ-SAMD4A groups was determined. The results revealed that
CircSAMD4A overexpression suppressed the expression level of
miR-342-3p, while CircSAMD4A knockdown promoted the miR-342-3p
level in the HOS and U2SO cells (Fig.
4H). These data indicated that miR-342-3p was the target miRNA
of CircSAMD4A in OS.
Inhibition of miR-342-3p reverses the
suppressive effects of si-CircSAMD4A on cell progression and EMT in
OS cells
To further clarify the function of CircSAMD4A and
miR-342-3p regulatory networks in OS cell progression, rescue
experiments were conducted. The HOS and U2OS cells were transfected
with si-NC, si-CircSAMD4A#1, si-CircSAMD4A#1+anti-NC or
si-CircSAMD4A#1+anti-miR -342-3p. miR-342-3p expression was
significantly induced by the knockdown of CircSAMD4A; however,
miR-342-3p inhibitor attenuated this effect (Fig. 5A). In addition, the results of MTT
assay and Transwell assay revealed that the knockdown of CircSAMD4A
significantly promoted cell cytotoxicity and inhibited cell
migration and invasion, while these effects were impaired by
miR-342-3p inhibition (Fig. 5B, F and
G). Moreover, reducing CircSAMD4A expression significantly
increased OS cells apoptosis, while this promotion effect was
significantly harbored by miR-342-3p downregulation (Fig. 5D). The results of western blot
analysis revealed that the knockdown of CircSAMD4A promoted the
expression of C-caspase-3 and E-cadherin, and inhibited the
expression of Ki-67, N-cadherin and Vimentin; these effects were
reversed by the inhibition of miR-342-3p (Fig. 5C, E, H and I). Overall, these
findings demonstrate that CircSAMD4A regulates OS cell progression
by targeting miR-342-3p.
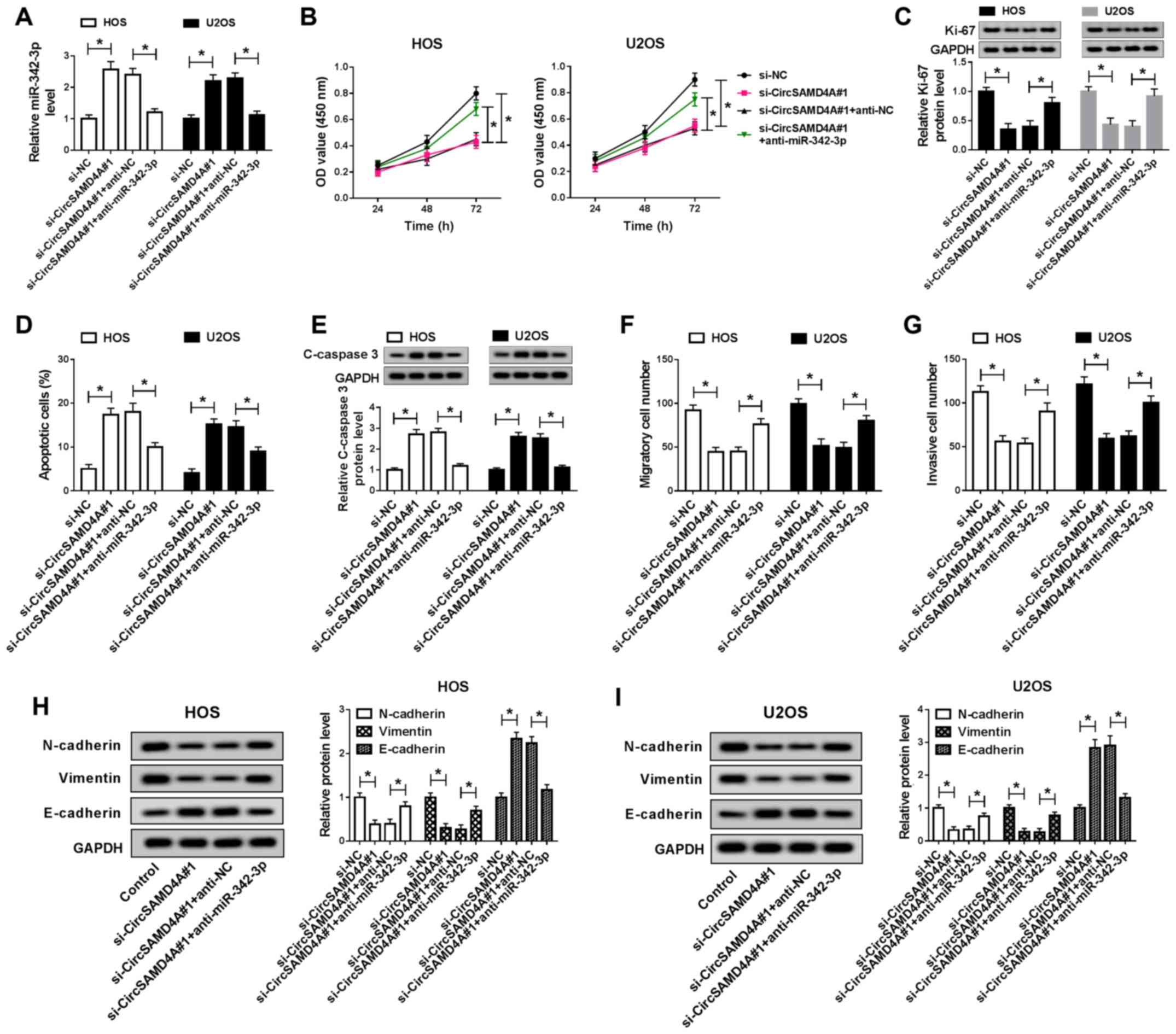 | Figure 5Inhibition of miR-342-3p reverses the
suppressive effects of si-CircSAMD4A on the proliferation and EMT
of OS cells. (A) CircSAMD4A expression of si-NC, si-CircSAMD4A#1,
si-CircSAMD4A#1+anti-NC and si-CircSAMD4A#1+anti-miR-342-3p groups
was detected by RT-qPCR in HOS and U2OS cells. (B) MTT assay was
applied to measure the cytotoxicity of HOS and U2OS cells
transfected with si-NC, si-CircSAMD4A, si-CircSAMD4A+anti-NC and
si-CircSAMD4A+anti-miR-342-3p. (C) The protein expression of Ki-67
was detected in si-NC, si-CircSAMD4A#1, si-CircSAMD4A#1+anti-NC and
si-CircSAMD4A#1+anti-miR-342-3p groups by western blot analysis.
(D) Cell apoptosis of si-NC, si-CircSAMD4A#1,
si-CircSAMD4A#1+anti-NC and si-CircSAMD4A#1+anti-miR-342-3p groups
was detected by flow cytometry in HOS and U2OS cells. (E) The
protein of C-caspase-3 was detected in si-NC, si-CircSAMD4A#1,
si-CircSAMD4A#1+anti-NC and si-CircSAMD4A#1+anti-miR-342-3p groups
in HOS and U2OS cells by western blot analysis. (F and G) Cell
migration and invasion of si-NC, si-CircSAMD4A#1,
si-CircSAMD4A#1+anti-NC and si-CircSAMD4A#1+anti-miR-342-3p groups
was detected using Transwell assay in HOS and U2OS cells. (H and I)
The protein expression of N-cadherin, Vimentin and E-cadherin of
si-NC, si-CircSAMD4A#1, si-CircSAMD4A#1+anti-NC and
si-CircSAMD4A#1+anti-miR-342-3p groups was measured by western blot
analysis in HOS and U2OS cells. *P<0.05, vs.
respective control. OS, osteosarcoma; EMT, epithelial-mesenchymal
transition. |
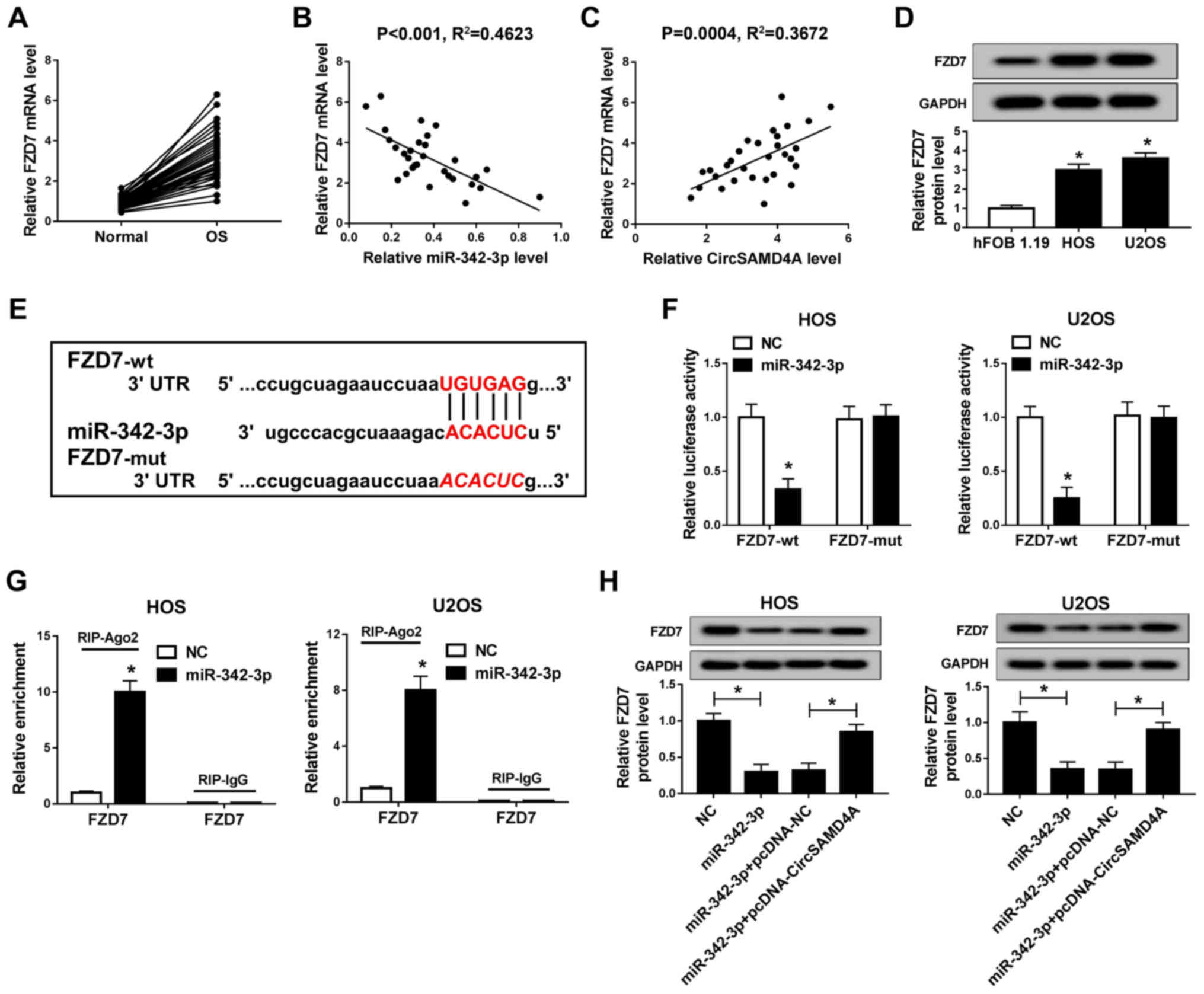
FZD7 is a direct target gene of
miR-342-3p in OS
It is well known that miRNAs can regulate the
downstream target genes to affect cell growth. Moreover, FZD7, a
Wnt signaling receptor, has been confirmed as a carcinogenic factor
in OS (29). The present study
demonstrated that FZD7 was a potential target gene of miR-342-3p,
with its 3′UTR containing the complement of miR-342-3p (Fig. 6E). In addition, it was found that
FZD7 was highly expressed in OS tissues and cells (Fig. 6A and D). As illustrated in
Fig. 6B and C, FZD7 expression
negatively correlated with the miR-342-3p level and positively
correlated with CircSAMD4A expression. Moreover, the luciferase
reporter assay demonstrated that when miR-342-3p was bound to
FZD7-wt 3′UTR, the luciferase activity was significantly decreased
(Fig. 6F). RIP assay revealed
that FZD7 was enriched in the miR-342-3p groups (Fig. 6G). These results indicated that
FZD7 was the target gene of miR-342-3p in the HOS and U2OS cells.
In addition, FZD7 expression in the HOS and U2OS was significantly
inhibited by miR-342-3p overexpression, while CircSAMD4A
overexpression reversed the inhibitory effect of miR-342-3p on FZD7
expression (Fig. 6H). In summary,
these results indicated that FZD7 was a target gene of
miR-342-3p.
The promotion of FZD7 attenuates the
suppressive effects of miR-342-3p overexpression on cell
progression and EMT in OS cells
To verify the effect of miR-342-3p and FDZ7
regulatory networks on OS cells, recovery experiments were
conducted. As illustrated in Fig.
7A, miR-342-3p overexpression inhibited FZD7 expression, and
this effect was impaired by increasing FZD7 expression in HOS and
U2OS cells. Furthermore, increasing miR-342-3p expression
significantly inhibited the migration and invasion, and increased
the cytotoxicity and apoptosis of HOS and U2OS cells, while these
effects were attenuated by FZD7 overexpression (Fig. 7B, D, F and G). Notably, miR-342-3p
overexpression induced an increase in C-caspase-3, E-cadherin
protein expression, and a decrease in Ki-67, N-cadherin and
Vimentin protein expression; however, these effects of miR-342-3p
overexpression were reversed by FZD7 upregulation (Fig. 7C, E, H and I). Taken together,
these findings demonstrate that miR-342-3p regulates OS cell growth
and EMT by targeting FZD7.
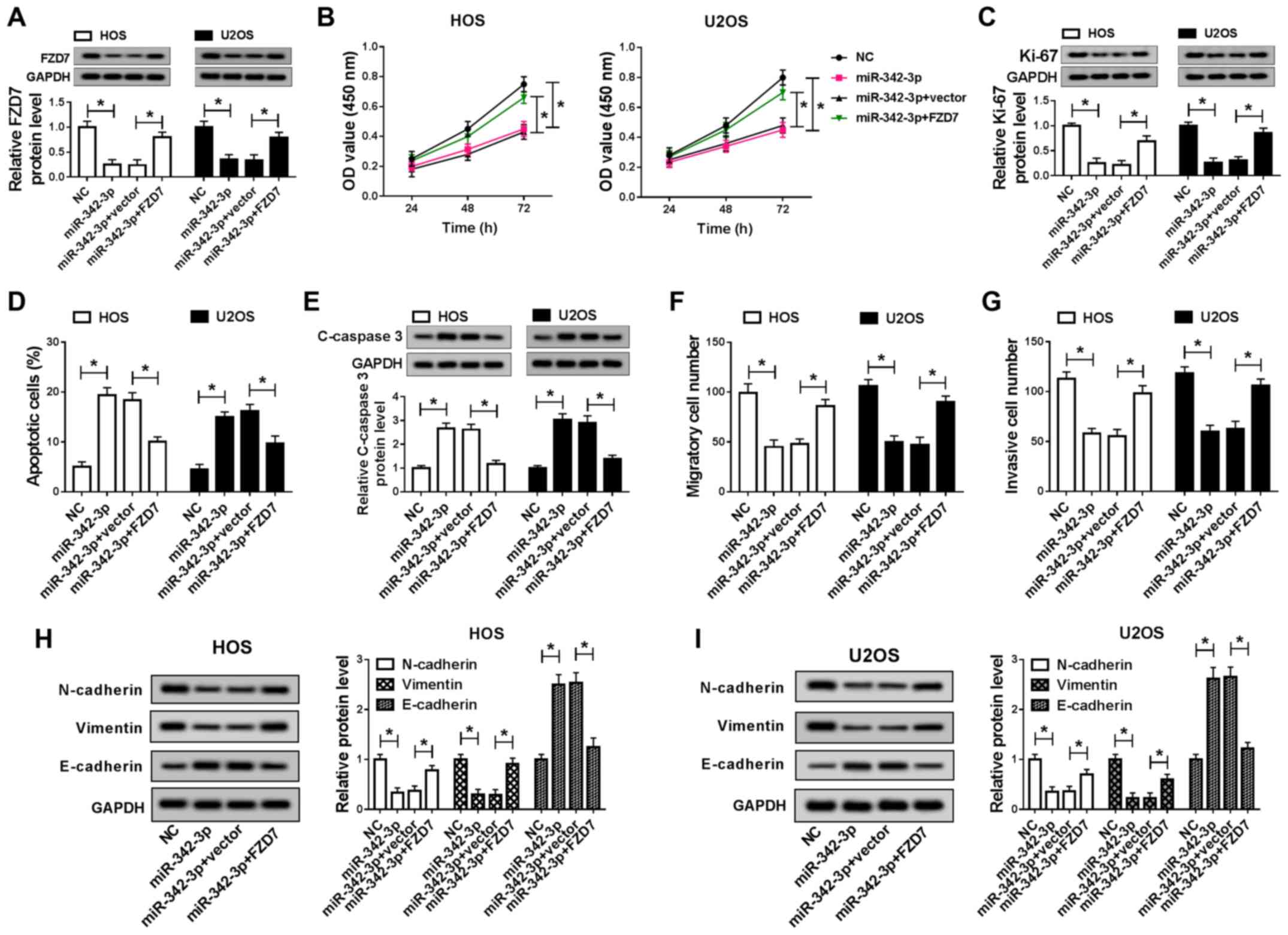 | Figure 7The promotion of FZD7 attenuates the
suppressive effects of miR-342-3p overexpression on the
proliferation and EMT of OS cells. (A) FZD7 expres-sion in NC,
miR-342-3p, miR-342-3p+vector and miR-342-3p+FZD7 groups in HOS and
U2OS cells was detected by RT-qPCR. (B) MTT assay was applied to
measure the cytotoxicity of HOS and U2OS cells transfected with NC,
miR-342-3p, miR-342-3p+vector and miR-342-3p+FZD7. (C) Ki-67
protein expression was detected in NC, miR-342-3p,
miR-342-3p+vector and miR-342-3p+FZD7 groups in HOS and U2OS cells
by western blot analysis. (D) Cell apoptosis was measured by flow
cytometry in NC, miR-342-3p, miR-342-3p+vector and miR-342-3p+FZD7
groups in HOS and U2OS cells. (E) The protein of C-caspase 3 was
detected in NC, miR-342-3p, miR-342-3p+vector and miR-342-3p+FZD7
groups in HOS and U2OS cells by western blot analysis. (F and G)
Cell migration and invasion of NC, miR-342-3p, miR-342-3p+vector
and miR-342-3p+FZD7groups was detected using Transwell assay in HOS
and U2OS cells. (H and I) The protein expression of N-cadherin,
Vimentin and E-cadherin of NC, miR-342-3p, miR-342-3p+vector and
miR-342-3p+FZD7 groups was measured by western blot analysis in HOS
and U2OS cells. *P<0.05, vs. respective control. OS,
osteosarcoma; EMT, epithelial-mesenchymal transition. |
Discussion
The diagnosis and treatment of OS have always been a
main concern for researchers. The advancement of biotechnology and
the development of next-generation sequencing technology are likely
to provide target therapy for OS. Cell proliferation, migration,
and invasion are essential phenomena with respect to cancer cell
metastasis and development. EMT is important in cancer metastasis
and is closely associated with cancer cell migration, invasion and
apoptosis. N-cadherin, E-cadherin and Vimentin proteins are
important markers in the cellular EMT mechanism, which can reflect
the degree of EMT in cells (30).
Cyclic RNAs have been found to be involved in tumor
progression in various cancer types. Circular RNAs have been
confirmed to exert various effects, including functioning as
biomarkers to improve the diagnostic efficiency and serving as a
regulatory factor in OS progression (12,31-33). In the study by Liu et al,
it was reported that circNT52 functioned as an oncogene to regulate
OS cell proliferation and metastasis by sponging miR-448 (34). CircSAMD4A has been reported to be
highly expressed in OS and to regulate cell proliferation by
targeting the miR-1244/MDM2 axis (27). In this study, CircSAMD4A was
identified to exhibit a significant high expression in OS tissues
and cells. CircSAMD4A deletion significantly attenuated the ability
of cell metastasis, and promoted cytotoxicity and apoptosis in OS
in vitro. In addition, the suppressive effect of CircSAMD4A
knockdown on OS tumor growth was confirmed in an experiment on nude
mice. These data proved that the downregulation of CircSAMD4A
suppressed OS progression in vitro and in vivo.
According to the results, CircSAMD4A expression was increased in OS
tissues and cells, and CircSAMD4A silencing inhibited proliferation
in OS (27). Moreover, the
present study revealed that miR-342-3p was a target miRNA of
CircSAMD4A. miR-342-3p has been demonstrated to be involved in
cellular growth in cancers, including pancreatic cancer,
hepatocellular carcinoma, prostate cancer and cervical cancer
(35-38). Zhang et al reported that
miR-342-3p was downregulated in OS tissues and inhibited cell
progression by targeting AEG-1 (39). However, the regulatory mechanism
of miR-342-3p in OS has not been fully elucidated. The present
study revealed that miR-342-3p expression was decreased in OS, a
finding which is in accordance with the previous study by Zhang
et al (39). miR-342-3p
overexpression inhibited OS cell growth and metastasis. Moreover,
the inhibition of miR-342-3p reversed the inhibitory effects of
CircSAMD4A deletion on OS cell progression. Thus, CircSAMD4A
affected OS cell growth and EMT by modulating miR-342-3p.
Finally, the present study also demonstrated that
FZD7 was a target gene of miR-342-3p. A number of studies have
indicated that FZD7 is an important regulator in the cellular
mechanism of cancers (40-42).
For example, FZD7 overexpression can induce cell proliferation in
glioma (43). Moreover, FZD7 has
been verified to be a novel prognostic marker and ti contribute to
the regulation of tumor metastasis (44). In addition, FZD7 has also been
shown to be associated with resistance and prognosis (45,46). This study revealed that FZD7 was
upregulated in OS tissues and cells. The accumulation of FDZ7
reversed the inhibitory effects of miR-342-3p overexpression on
cell growth and metastasis in OS, suggesting that CircSAMD4A
regulates OS progression by sponging miR-342-3p to modulate FDZ7
expression.
In conclusion, the present study demonstrated that
CircSAMD4A affected cell cytotoxicity, invasion, apoptosis,
migration and EMT by regulating the miR-342-3p/FDZ7 axis in OS,
thereby providing a novel regulatory mechanism of OS and a
potential therapeutic target for OS.
Funding
This study was supported by the National Natural
Science Foundation of China (81574002).
Availability of data and materials
All data generated or analyzed during this study are
included in this published article or are available from the
corresponding author on reasonable request.
Authors' contributions
CX was responsible for the conception and design of
the study. BC was involved in the development of the study
methodology. BW was involved in data acquisition. JG was involved
in the analysis and interpretation of the data. YS was involved in
the writing, reviewing and revision of the article and analyzed the
literature, and interpreted the data. YC was involved in providing
administrative, technical and material support, analyzed the
literature, interpreted the data and produced the figures. All
authors have reviewed and approved of the article prior to
submission and have read and approved the final article.
Ethics approval and consent to
participate
All patients underwent resection and signed written
informed consents. The use of patient samples was approved by the
Ethics Committee of the Second Affiliated Hospital of Guangzhou
Medical University. Animal experiments were approved by the Animal
Care and Welfare Committee of The Second Affiliated Hospital of
Guangzhou Medical University.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgments
Not applicable.
References
|
1
|
Meyers PA and Gorlick R: Osteosarcoma.
Pediatr Clin North Am. 44:973–989. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Longhi A, Errani C, De Paolis M, Mercuri M
and Bacci G: Primary bone osteosarcoma in the pediatric age: State
of the art. Cancer Treat Rev. 32:423–436. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Kempfbielack B, Bielack SS, Jürgens H,
Branscheid D, Berdel WE, Exner GU, Göbel U, Helmke K, Jundt G,
Kabisch H, et al: Osteosarcoma relapse after combined modality
therapy: An analysis of unselected patients in the cooperative
osteosarcoma study group (COSS). J Clin Oncol. 23:559–568. 2005.
View Article : Google Scholar
|
|
4
|
Sanger HL, Klotz G, Riesner D, Gross HJ
and Kleinschmidt AK: Viroids are single-stranded covalently closed
circular RNA molecules existing as highly base-paired rod-like
structures. Proc Natl Acad Sci USA. 73:3852–3856. 1976. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Guo JU, Agarwal V, Guo H and Bartel DP:
Expanded identification and characterization of mammalian circular
RNAs. Genome Biol. 15:4092014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Li Y, Zheng Q, Bao C, Li S, Guo W, Zhao J,
Chen D, Gu J, He X and Huang S: Circular RNA is enriched and stable
in exosomes: A promising biomarker for cancer diagnosis. Cell Res.
25:981–984. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Lux S and Bullinger L: Circular RNAs in
cancer. Adv Exp Mol Cancer. 1087:215–230. 2018.
|
|
8
|
Zhang M and Xin Y: Circular RNAs: A new
frontier for cancer diagnosis and therapy. J Hematol Oncol.
11:212018. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Sang M, Meng L, Liu S, Ding P, Chang S, Ju
Y, Liu F, Gu L, Lian Y and Geng C: Circular RNA ciRS-7 maintains
metastatic phenotypes as a ceRNA of miR-1299 to target MMPs. Mol
Cancer Res. 16:1665–1675. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wei H, Pan L, Tao D and Li R: Circular RNA
circZFR contributes to papillary thyroid cancer cell proliferation
and invasion by sponging miR-1261 and facilitating C8orf4
expression. Biochem Biophys Res Commun. 503:56–61. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zhong Y, Du Y, Yang X, Mo Y, Fan C, Xiong
F, Ren D, Ye X, Li C, Wang Y, et al: Circular RNAs function as
ceRNAs to regulate and control human cancer progression. Mol
Cancer. 17:792018. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Huang L, Chen M, Pan J and Yu W: Circular
RNA circNASP modulates the malignant behaviors in osteosarcoma via
miR-1253/FOXF1 pathway. Biochem Biophys Res Commun. 500:511–517.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Yanbin Z and Jing Z: CircSAMD4A
accelerates cell proliferation of osteosarcoma by sponging miR-1244
and regulating MDM2 mRNA expression. Biochem Biophys Res Commun.
516:102–111. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Qi W, Liang W, Jiang H and Miuyee Waye M:
The function of miRNA in hepatic cancer stem cell. Biomed Res Int.
2013:3589022013. View Article : Google Scholar
|
|
15
|
Chi SW, Zang JB, Mele A and Darnell RB:
Ago HITS-CLIP decodes miRNA-mRNA interaction maps. Nature.
460:479–486. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Allantaz F, Cheng DT, Bergauer T,
Ravindran P, Rossier MF, Ebeling M, Badi L, Reis B, Bitter H,
D'Asaro M, et al: Expression profiling of human immune cell subsets
identifies miRNA-mRNA regulatory relationships correlated with cell
type specific expression. PLoS One. 7:e299792012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Althoff K, Lindner S, Odersky A, Mestdagh
P, Beckers A, Karczewski S, Molenaar JJ, Bohrer A, Knauer S,
Speleman F, et al: miR-542-3p exerts tumor suppressive functions in
neuroblastoma by downregulating Survivin. Int J Cancer.
136:1308–1320. 2015. View Article : Google Scholar
|
|
18
|
Chen Y, Han X, Yin X, Zhou Y and Wu T:
Decreased expression of miR-132 in CRC tissues and its inhibitory
function on tumor progression. Open Life Sci. 11:130–135. 2016.
View Article : Google Scholar
|
|
19
|
Liu W, Kang L, Han J, Wang Y, Shen C, Yan
Z, Tai Y and Zhao C: miR-342-3p suppresses hepatocellular carcinoma
proliferation through inhibition of IGF-1R-mediated Warburg effect.
Onco Targets Ther. 11:1643–1653. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Chen H and Tian Y: MiR-15a-5p regulates
viability and matrix degradation of human osteoarthritis
chondrocytes via targeting VEGFA. Biosci Trends. 10:482–488. 2017.
View Article : Google Scholar
|
|
21
|
Chang SW, Yue J, Wang BC and Zhang XL:
miR-503 inhibits cell proliferation and induces apoptosis in
colorectal cancer cells by targeting E2F3. Int J Clin Exp Pathol.
8:12853–12860. 2015.
|
|
22
|
Hussein NA, Kholy ZA, Anwar MM, Ahmad MA
and Ahmad SM: Plasma miR-22-3p miR-642b-3p and miR-885-5p as
diagnostic biomarkers for pancreatic cancer. J Cancer Res Clin
Oncol. 143:83–93. 2017. View Article : Google Scholar
|
|
23
|
Kong XY, Du YQ Li L, Liu JQ, Wang GK, Zhu
JQ, Man XH, Gong YF, Xiao LN, Zheng YZ, et al: Plasma miR-216a as a
potential marker of pancreatic injury in a rat model of acute
pancreatitis. World J Gastroenterol. 16:4599–4604. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Liu BR and Xie L: microRNA: A new cancer
biomarker. Chin Clin Oncol. 15:1–5. 2010.In Chinese.
|
|
25
|
Li Q, Song S, Ni G, Li Y and Wang X: Serum
miR-542-3p as a prognostic biomarker in osteosarcoma. Cancer
Biomark. 21:521–526. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Tao K, Yang J, Guo Z, Hu Y, Sheng H, Gao H
and Yu H: Prognostic value of miR-221-3p miR-342-3p and miR-491-5p
expression in colon cancer. Am J Transl Res. 6:391–401. 2014.
|
|
27
|
Wang D, Fan Z, Liu F and Zuo J: Hsa-miR-21
and Hsa-miR-29 in tissue as potential diagnostic and prognostic
biomarkers for gastric cancer. Cell Physiol Biochem. 37:1454–1462.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
29
|
Li C, Wang F, Wei B, Wang L and Kong D:
LncRNA AWPPH promotes osteosarcoma progression via activation of
Wnt/β-catenin pathway through modulating miR-93-3p/FZD7 axis.
Biochem Biophys Res Commun. 514:1017–1022. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Yilmaz M and Christofori G: EMT, the
cytoskeleton, and cancer cell invasion. Cancer Metastasis Rev.
28:15–33. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Jin H, Jin X, Zhang H and Wang W: Circular
RNA hsacirc-0016347- promotes proliferation, invasion and
metastasis of osteosarcoma cells. Oncotarget. 8:25571–25581. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Jin J, Chen A, Qiu W, Chen Y, Li Q, Zhou X
and Jin D: Dysregulated circRNA_100876 suppresses proliferation of
osteosarcoma cancer cells by targeting microRNA-136. J Cell
Biochem. 120:15678–15687. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Pan Z, Sun X, Shan H, Wang N, Wang J, Ren
J, Feng S, Xie L, Lu C, Yuan Y, et al: MicroRNA-101 inhibited
postinfarct cardiac fibrosis and improved left ventricular
compliance via the FBJ osteosarcoma oncogene/transforming growth
factor-β1 pathway. Circulation. 126:840–850. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Liu X, Zhong Y, Li J and Shan A: Circular
RNA circ-NT5C2 acts as an oncogene in osteosarcoma proliferation
and metastasis through targeting miR-448. Oncotarget.
8:114829–114838. 2017. View Article : Google Scholar
|
|
35
|
Cheng D, Fan J, Ma Y, Zhou Y, Qin K, Shi M
and Yang J: LncRNA SNHG7 promotes pancreatic cancer proliferation
through ID4 by sponging miR-342-3p. Cell Biosci. 9:282019.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Gao Y, Zhang SG, Wang ZH and Liao JC:
Down-regulation of miR-342-3p in hepatocellular carcinoma tissues
and its prognostic significance. Eur Rev Med Pharmacol Sci.
21:2098–2102. 2017.PubMed/NCBI
|
|
37
|
Hu K, Mu X, Kolibaba H, Yin Q, Liu C,
Liang X and Lu J: Metadherin is an apoptotic modulator in prostate
cancer through miR-342-3p regulation. Saudi J Biol Sci. 25:975–981.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Jones D, Anene D, Aloway A, Anene P, Avila
D, Gobejishvili L, Barve S, Mcnally L and Kidd LC: Abstract 1844:
Reduced expression of miR-342-3p in prostate cancer. Cancer Res.
73:1844. 2013.
|
|
39
|
Zhang S, Liu L, Lv Z, Li Q, Gong W and Wu
H: MicroRNA-342-3p inhibits the proliferation, migration, and
invasion of osteosarcoma cells by targeting astrocyte-elevated
gene-1 (AEG-1). Oncol Res. 25:1505–1515. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Asad M, Wong MK, Tan TZ, Choolani M, Low
J, Mori S, Virshup D, Thiery JP and Huang RY: FZD7 drives in vitro
aggressiveness in Stem-A subtype of ovarian cancer via regulation
of non-canonical Wnt/PCP pathway. Cell Death Dis. 5:e13462014.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Deng B, Zhang Y, Zhang S, Wen F, Miao Y
and Guo K: MicroRNA-142-3p inhibits cell proliferation and invasion
of cervical cancer cells by targeting FZD7. Tumor Biol.
36:8065–8073. 2015. View Article : Google Scholar
|
|
42
|
Kirikoshi H, Sekihara H and Katoh M:
Up-regulation of Frizzled-7 (FZD7) in human gastric cancer. Int J
Oncol. 19:111–115. 2001.PubMed/NCBI
|
|
43
|
Qiu X, Jiao J, Li Y and Tian T:
Overexpression of FZD7 promotes glioma cell proliferation by
upregulating TAZ. Oncotarget. 7:85987–85999. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Cao TT, Xiang D, Liu BL, Huang TX, Tan BB,
Zeng CM, Wang ZY, Ming XY, Zhang LY, Jin G, et al: FZD7 is a novel
prognostic marker and promotes tumor metastasis via WNT and EMT
signaling pathways in esophageal squamous cell carcinoma.
Oncotarget. 8:65957–65968. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Cheng ZX, Song YX, Wang ZY, Wang Y and
Dong Y: miR-144-3p serves as a tumor suppressor by targeting FZD7
and predicts the prognosis of human glioblastoma. Eur Rev Med
Pharmacol Sci. 21:4079–4086. 2017.PubMed/NCBI
|
|
46
|
Chen Z, Huang C, Ma T, Jiang L, Tang L,
Shi T, Zhang S, Zhang L, Zhu P, Li J and Shen A: Reversal effect of
quercetin on multidrug resistance via FZD7/β-catenin pathway in
hepatocellular carcinoma cells. Phytomedicine. 43:37–45. 2018.
View Article : Google Scholar : PubMed/NCBI
|