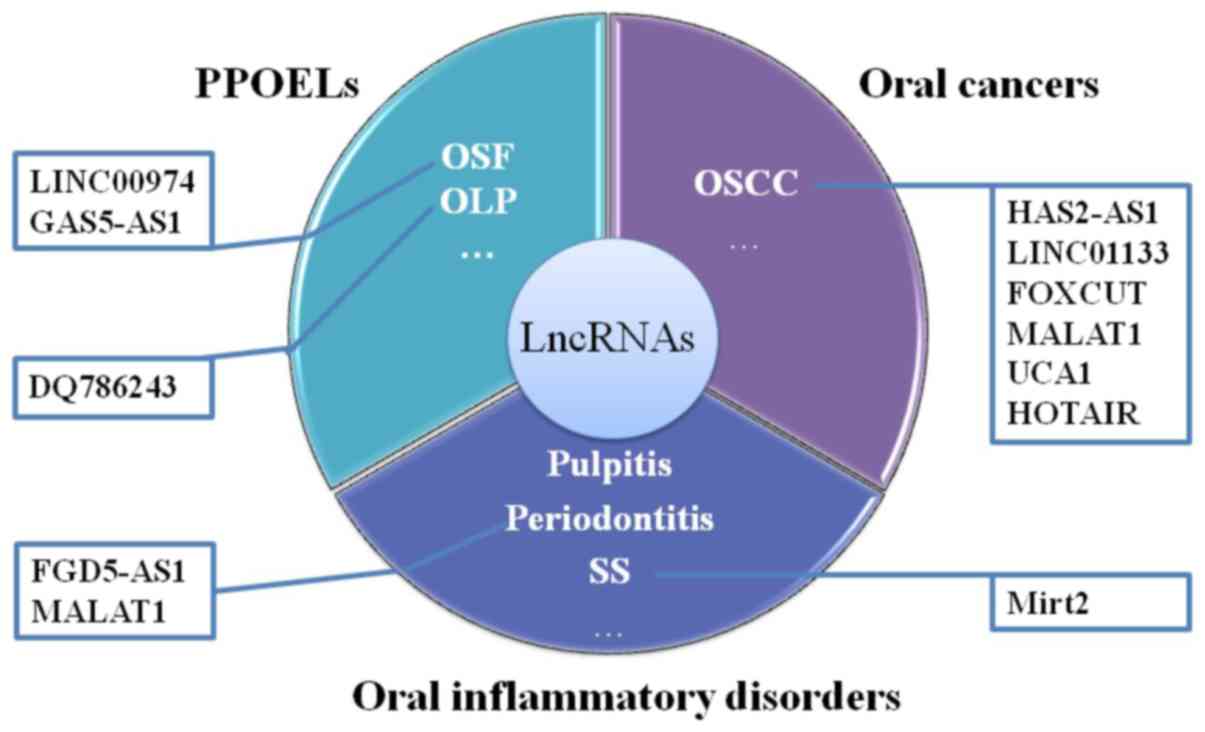
1. Introduction
Oral inflammatory disorder is a series of processes
associated with microbial infections (such as periodontal,
endodontic and periapical diseases) and immune-mediated damage [for
example Sjögren's syndrome (SS)] (1). Numerous factors, including
non-coding (nc)RNAs, leukocytes, cytokines and complement
components, are involved in this process (2,3).
Long non-coding (lnc)RNAs affect oral inflammation by sponging
microRNAs (miRNA/miR) (4) or
activating downstream target miRNAs (5). Research on competing endogenous
(ce)RNA mechanisms, in which lncRNAs sponge specific miRNAs to
suppress their target genes, has increased in numbers in over the
last 2 years (6). For example,
lncRNA metastasis associated lung adenocarcinoma transcript 1
(MALAT1) acts as a sponge of miR-20a to induce Toll-like receptor 4
(TLR4) signaling and results in an inflammatory reaction from human
gingival fibroblasts (HGFs) (7).
Several potentially premalignant oral epithelial
lesions (PPOELs) are associated with the disease process of chronic
inflammatory disorders (8,9).
PPOEL is a broad term to define both histological and clinical oral
lesions that have malignant potential, including oral lichen planus
(OLP), oral submucous fibrosis (OSF), and oral dysplasia (10). In addition, other vital epigenetic
and subcellular regulatory non-coding transcripts, such as lncRNAs
and miRNAs are also known to regulate the mRNA expression of
inflammation-related cytokines, and disturbance of the
miRNA-mRNA-cytokine regulatory network is one of the common
pathological features of PPOELs (11,12). For example, the significant
upregulation of miR-31 and downregulation of its target gene, C-X-C
motif chemokine ligand 12 (CXCL12) contributed to progression of
PPOELs (13).
As the 7th hallmark of cancer, chronic inflammation
has been linked to various stages of tumorigenesis (14). Several studies have reported that
numerous regulators, including inflammatory cytokines and ncRNAs,
facilitate tumor development (15,16). LncRNAs participate in the
transformation of chronic inflammation into cancer by altering the
expression of various inflammatory signaling pathways such as NF-κB
and STAT3 and proinflammatory cytokines [such as tumor necrosis
factor (TNF) family]. Interleukin (IL)-6-dependent STAT3 signaling
activation contributes to the occurrence of colorectal cancer
(17). The major histological
type of oral cancer is oral squamous cell carcinoma (OSCC)
(18). Jia et al (19), identified that differentially
expressed (DE) lncRNAs and genes between OSCC, oral dysplasia, and
normal oral tissues may control the initiation and development of
OSCC through phosphatidylinositol-3-kinases (PI3K)/Akt signaling
and mast cell NF-κB functional pathways.
LncRNAs play an essential role in the occurrence and
development of inflammation and cancer (20,21). The oral diseases associated with
the function of lncRNAs are shown in Fig. 1. According to previous reports,
the DE lncRNAs between healthy and pathological oral tissues may
affect the occurrence and process of oral diseases (Table I). The present review outlines the
current understanding of the established functions and underlying
mechanisms of lncRNAs in various oral inflammatory disorders,
PPOELs and OSCC.
 | Table IExpression profile of lncRNAs in
diseased oral tissues. |
Table I
Expression profile of lncRNAs in
diseased oral tissues.
| Disease | No. of
sample/subjects | Method | Confirmation | Findings | Comments | (Refs.) |
|---|
| Pulpitis | 7 inflamed pulp
tissues and 5 healthy pulp tissues | Microarray | RT-qPCR; 10 normal
human pulp tissue and 10 inflamed samples | 338 upregulated and
414 downregulated lncRNAs; FC>2; P<0.05 | Inflamed pulp
samples were collected from patients with pulpitis, to reflect the
real pathological state | (31) |
| Periodontitis | Inflamed and
healthy adjacent gingival tissue from two patients with chronic
periodontitis | Microarray | RT-qPCR; 15 normal
tissue samples and 30 chronic periodontitis tissue samples | 4313 upregulated
and 4612 downregulated lncRNAs; FC≥2.0; P≤0.05 | First time to
analyze DE lncRNAs between chronic periodontitis tissues samples
and adjacent normal tissues | (36) |
| SS | PBMCs from 8
patients with pSS and 8 healthy subjects | Microarray | qRT-PCR; No sample
size reported | 199 dysregulated
lncRNAs; FC≥1.5; P≤0.01a | No validation;
first time to analyze DE lncRNAs in PBMCs of pSS | (43) |
| SS | LSGs from 4 pSS
patients and 4 healthy individuals | Microarray | RT-qPCR; 30 pSS
patients and 16 controls | 890 upregulated
lncRNAs and 353 downregulated lncRNAs; FC>2; P<0.05 | First time to
analyze DE lncRNAs in LSGs of pSS | (44) |
| OSF | 20 OSCC samples, 10
OSF samples and 13 normal mucous samples | RNA-sequencing | RT-qPCR; 20 OSCC
samples, 10 OSF samples and 13 normal mucous samples | 231 upregulated
lncRNAs and 456 downregulated lncRNAs; FC>2; P<0.05 | LncRNAs involved in
the malignant transformation process were firstly analyzed and
assessed | (56) |
| OLP | 1
papillomavirus-related OSCC tissue, one OLP tissue and 1 normal
oral mucosa tissue samples | RNA-sequencing | - | 76.2% intergenic
lncRNAs and 16.5% sense lncRNAs; A coding potential calculator
score <0, and CPAT probability ≤0.364 | 3 types of tissues
samples but sample numbers were too small; lncRNAs involved in the
malignant transformation process were firstly analyzed and
assessed | (70) |
| OSCC | 72 OSCC tissues and
adjacent normal tissues | Microarray | RT-qPCR; 72 OSCC
tissues and adjacent normal tissues | 933 upregulated and
1361 downregulated lncRNAs; FC>2.0; P<0.05 | 3 types of OSCC
tissues (tongue cancer, gingival carcinoma and carcinoma of the
buccal mucosa) were analyzed and compared, and 4 critical lncRNA
nodes were identified | (62) |
2. LncRNA biogenesis
LncRNAs are a class of RNA molecules whose
transcript length is >200 nucleotides (22). Different lncRNA classifications
have been established based on different criteria. The first
criteria was presented by Jarroux et al (23), and lncRNAs which are >10 kb
belong to the groups of very long intergenic RNAs and macro
lncRNAs. Examples of macro lncRNAs include antisense of IGF2R
non-protein coding RNA, KCNQ1 opposite strand/antisense transcript
1 and GNAS antisense RNA1, which act as cis-silencers in mouse
genomic imprinting (24). The
second criterion, location with respect to protein coding genes, is
commonly used. In this classification, 5 types (sense, antisense,
bidirectional, intronic and intergenic) of lncRNAs are included
(23). An example of nature
antisense transcripts, which is one type of antisense lncRNAs, is
anti-sense non-coding RNA in the INK4 locus, which is
encoded by the NK4b-ARF-INK4a locus on chromosome 9p21 (25). According to the classification
based on association with other DNA regulatory elements and loci,
lncRNAs are divided into pseudogenes, enhancer lncRNAs and
promoter-associated lncRNAs and 3′-untranslated region-associated
RNAs (23). Long intergenic
non-coding (LINC)RNA-p21 is an enhancer RNA, which originates from
a p53 binding site associated with regulation of cyclin-dependent
kinase inhibitor 1A (26). The
associated biochemical pathways or stability of lncRNAs serve as
characteristics for their classification, as demonstrated by stable
unannotated transcripts (27),
Xrn1 sensitive unstable transcripts (XUTs) (such as 5′-long
terminal repeat antisense TY1 RNA and XUT1678) (28), Nrd1-unterminated transcripts
(29) and cryptic unstable
transcripts (for example promoter upstream transcript) (30). Furthermore, several subgroups of
lncRNAs with a precise subcellular localization have been defined.
Long non-coding mitochondrial RNAs (ncmtRNAs) are cytoplasmic
lncRNAs while GAA repeat-containing RNAs and chromatin-enriched
RNAs (cheRNAs) locate in the nucleus (23). For example, antisense
mitochondrial non-coding RNA-2 and hemin-induced chromatin-enriched
RNA down-stream of fetal hemoglobin are ncmtRNAs and cheRNAs,
respectively (31,32). Lastly, hypoxia-induced non-coding
ultra-conserved transcripts (HINCUTs), stress-induced lncRNA
(si-lncRNA), senescence-associated lncRNA (SAL), non-annotated stem
transcript, prostate cancer-associated transcripts serve as
subgroups of another attribute used for lncRNA classification:
Association with specific biological processes (23). Hypoxia-inducible factor (HIF)
induces HINCUTs to elevate transcription of nearby genes, which are
involved in cellular signaling pathways and processes, such as
glucose metabolism (33).
Oxidative stress causes rapid and transient dynamics of si-lncRNAs
in the nucleus and the cytosol, leading to their accumulation at
polysomes, which subsequently induces transcription (34). Antisense very long intergenic
ncRNA (VAD), one type of SAL, modulates chromatin structure
in cis and increases gene expression in trans at the
INK4 locus, which encodes cell cycle inhibitors, that are important
to senescence-associated cell proliferation arrest (35). The schematic diagram illustrating
various classes of lncRNAs are presented in Fig. 2.
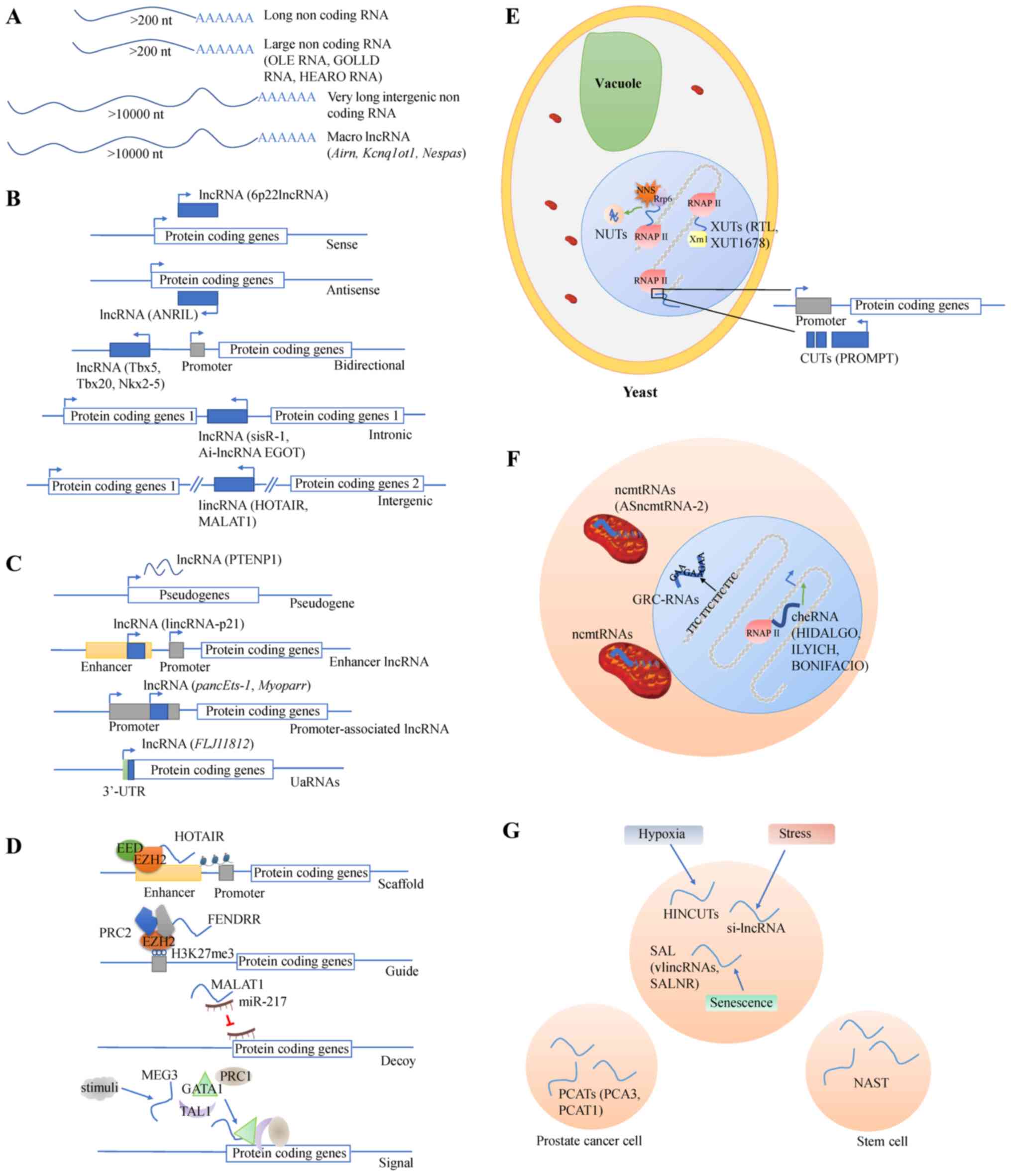 | Figure 2Schematic diagram of the various
classes of lncRNAs. Classification according to lncRNAs (A) length,
(B) location with respect to protein coding genes, (C) location
with specific DNA regulatory elements and loci, (D) lncRNAs
function, (E) biogenesis pathway and stability, (F) subcellular
localization or origin. (G) association with specific biological
processes. The green arrow indicates induction, and the red T
indicates inhibition. HOTAIR, HOX transcript antisense RNA; MALAT1,
metastasis associated lung adenocarcinoma transcript 1; UTR,
untranslated region; uaRNAs, 3′-untranslated region-associated
RNAs; EED, embryonic ectoderm development; EZH2, enhancer of zeste
homolog 2; PRC2, polycomb repressive complex 2; RNAPII, RNA
polymerase II; NUTs, Nrd1-unterminated transcripts; CUTs, cryptic
unstable transcripts; XUTs, Xrn1-sensitive unannotated transcripts;
PROMPT, promoter upstream transcript; ncmtRNAs, long noncoding
mitochondrial RNAs; GRC-RNAs, GAA repeat-containing RNAs; cheRNA,
chromatin-enriched RNA; si-lncRNA, stress-induced lncRNA; SAL,
senescence-associated lncRNA; vlincRNAs, very long intergenic
non-coding RNAs; SALNR, senescence-associated long non coding RNA;
PCATs, prostate cancer-associated transcripts; PCA3, prostate
cancer antigen 3; NAST, non-annotated stem transcript. |
LncRNA can also be classified according to their
function. LncRNAs exert or execute their functions in four main
ways: Signal, decoy, guide, and scaffold. They have been shown to
impact cell macromolecular (protein, RNA and DNA) stability
(36). The regulatory mechanism
of lncRNAs vary based on their locations within the cells. They
participate in chromatin modification and transcription in the
nucleus, while they interact with RNA-binding proteins or modulate
mRNA translation in the cytoplasm (37,38). There are a variety of mutual
regulatory mechanisms for lncRNAs and miRNAs. LncRNAs can not only
be mediated by miRNAs but also act as miRNA precursors. It is
noteworthy that the ceRNA network is one of the common sites of
posttranscriptional regulation (12). In the last 5 years, it has been
demonstrated that lncRNAs are important in the regulation of a
healthy immune system, which in turn is important for healthy oral
tissue (39,40,41). An increasing number of lncRNAs
have been reported to regulate the differentiation and activation
of immune cells. The differentiation of granulocytes is partly
mediated by HOX antisense intergenic RNA myeloid 1 (HOTAIRM1), an
antisense lncRNA within the HOXA gene locus. Hallmark myeloid
maturation-associated genes such as HOXA1/A2 would be inhibited by
silencing HOTAIRM1 (42). The
potential importance of lncRNAs in the immune response,
inflammation and even cancers is emerging (43-45).
3. Role of lncRNAs in oral inflammatory
disorders
Pulpitis
Pulpitis is a state of inflammation of the dental
pulp. Most cases are due to penetration of a carious lesion into
the pulp chamber (46). Pulpitis
is classified as either reversible or irreversible (47). Huang and Chen (48) conducted a microarray analysis to
establish lncRNA profiles of inflamed (n=7) and normal (n=5) pulp
tissues. A total of 752 lncRNAs (338 upregulated and 414
downregulated) were significantly expressed. A total of 460
significantly upregulated genes were enriched in biological
processes, such as immune system processes, immune and defense
response, the response to stress, and cell activation. The results
indicated that most lncRNAs might play roles in the immune system
and inflammatory responses of dental pulp. Lei et al
(49), performed a comprehensive
analysis of a lncRNA-miRNA-mRNA ceRNA network by integrating the
lncRNA profile from Huang and Chen (48), the miRNA profile from Zhong et
al (50) and the gene
expression profile from Galicia et al (51). A ceRNA regulatory network was
created, which was composed of the lncRNA plasmacytoma variant
translocation 1, miR-455-5p, and the mRNAs, suppressor of cytokine
signaling 3 and Plexin C1. To the best of our knowledge, only one
lncRNA microarray has been performed with a comprehensive analysis
(49). There has been no
mechanistic study focusing on specific lncRNAs in pulpitis. Thus,
more research is required to explore the regulatory role of
specific lncRNAs in pulpitis.
Periodontitis
Periodontitis is an inflammatory disease, which is
primarily caused by bacterial infection (52). It can cause inflammation of the
gingivae, loss of alveolar bone and loss of attachment (53). Tooth loss, which occurs in adults
is largely due to periodontitis (54). In 2015, Zou et al (55) revealed for the first time that
lncRNAs have critical roles in the pathogenesis of periodontitis. A
total of 2 pairs of chronic periodontitis gingival samples and
adjacent healthy samples were collected for lncRNA analysis and
8,925 DE lncRNAs were detected, of which 4,313 were upregulated and
4,612 were downregulated. Functional analysis of the nearby
protein-coding genes revealed that different lncRNAs can regulate a
common gene, and a single lncRNA can be regulated by different
genes. Thus, lncRNAs might play crucial and dual roles in
periodontitis.
Chen et al (4) found that lncRNA FGD5 antisense RNA1
(FGD5-AS1) was downregulated in the gingival samples from patients
with chronic periodontal compared with that in healthy samples.
FGD5-AS1 inhibited NF-κB signaling via the
FGD5-AS1/miR-142-3p/suppressor of cytokine signaling 6 (SOCS6)
ceRNA network and subsequently reduced the secretion of TNF-α,
IL-6, IL-1β and IL-8. Thus, the axis may provide a promising
strategy for the treatment of periodontitis.
Several lncRNAs inhibit periodontitis; however,
results from recent research have revealed that lncRNAs could
promote the inflammatory process of periodontal-derived cells,
including MALAT1 (7) and
papillary thyroid carcinoma susceptibility candidate 3 (56). Li et al (7), explored the role of MALAT1 in
inflammatory cytokine production in HGFs. The study indicated that
MALAT1 bound to miR-20a, as a ceRNA and consequently led to
increased mRNA levels of TLR4, which contributed to the activation
of inflammation. Therefore, the effect and mechanism of MALAT1 in
periodontal inflammation have been characterized. SS. SS is
a chronic systemic autoimmune disease characterized by reduced
secretions of the salivary and lacrimal glands and associated
neuroendocrine disturbances (57). The disturbances of neuroendocrine
include release of hormones (i.e., glucocorticoids) via the
hypothalamic-pituitary-adrenal axis stimulation, production of
mediators within the sympathetic innervation of immune organs
(i.e., thymus) and production of proinflammatory cytokines (i.e.,
IL-2 and TNF-α) during the inflammatory response (58). The primary hallmark of SS is the
infiltration of inflammatory mediators and cells, particularly T
and B cells into the salivary and lacrimal glands (59). In addition, gland tissues have
damaged acinar cells, fibrosis and increased adiposity with severe
inflammatory lesions (60,61).
Therefore, exploration into the relevant molecular mechanisms
underlying SS is required.
In 2019, Dolcino et al (62) performed a high-throughput gene and
lncRNA expression profiling in peripheral blood mononuclear cell
samples from 8 patients with primary SS (pSS) and 8 healthy
subjects. Among the 199 lncRNAs that were identified,
CTD-2020K17.1, LINC00511 and LINC00657 and their target genes were
found to be involved in apoptosis, immune response, cell
proliferation, and several proinflammatory pathways. Shi et
al (63), compared the
expression profiles of lncRNAs from labial salivary glands between
patients with pSS and healthy individuals. The gene ontology and
pathway analysis results found 28 DE mRNAs associated with 8 DE
lncRNAs were involved in chemokine signaling pathways, the NF-κB
signaling pathway, and the TNF signaling pathway. Taken together,
the results suggest that samples from multiple tissues could be
utilized for investigating the same oral autoimmune disease. A
comparison of DE lncRNA sets from different sample types of the
same disease could provide valuable clues to the discovery of novel
therapeutic targets to treat oral autoimmune diseases.
Subsequently, Xin et al (5) demonstrated that lncRNA myocardial
infarction associated transcript 2 (Mirt2) reduces apoptosis and
inflammatory levels in interferon (IFN)-γ-induced inflammation in
salivary gland epithelial cells. It is hypothesized that Mirt2
might block NF-κB, and Janus kinase (JAK)/STAT3 signaling by
increasing miR-377 expression levels.
Research investigating lncRNAs typically involves
the fields of medicine and biology; however, research regarding
lncRNAs in oral inflammatory disorders is still in the early
stages. To date, the collection of clinical oral tissue samples and
subsequent microarray or sequencing analysis has been the primary
method of investigation in this field. In periodontitis, the
majority of lncRNAs serve as ceRNAs (4,6,7).
There are a high number of research models investigating oral
tissue-derived cells treated with related inflammatory stimuli
(i.e., lipopolysaccharide and IFN-γ) (5,7).
Periodontitis animal models and lncRNAs have been well-established
(64-66), while animal models involving
lncRNAs in other oral inflammatory disorders requires further
investigation. Thus, additional research is required to determine
the mechanism of lncRNAs in oral inflammatory disorders.
4. Role of lncRNAs in PPOELs
OSF
OSF is a chronic, occult oral mucosal disease
associated with chewing betel nut, characterized by a
juxta-epithelial inflammatory response followed by generalized
submucosal fibrosis (67,68). As a result, OSF typically leads to
difficulty in opening the mouth and an increased malignant
transformation rate (69,70). Therefore, the identification of
molecules associated with OSF pathological progression is urgent.
In the course of OSF, endothelial dysfunction may be accompanied by
the dysregulation of multiple lncRNAs (71). For example, lncRNA growth arrest
specific 5 antisense 1 (GAS5-AS1) was inhibited, while lncRNA
hypoxia-inducible factor 1-α antisense RNA 1 (HIF1A-AS1) was
upregulated during the development of OSF (72,73). Research into lncRNAs has increased
in the last 5 years; however, the function of numerous lncRNAs in
OSF remains unclear. To date, only four articles have illustrated
the relative issues (72-75).
The lncRNA sequencing conducted in 2019 by Zhou
et al (75) included 13
normal mucous samples, 10 OSF samples, and 20 OSCC samples. A total
of 5 DE candidate lncRNAs were found to participate in the
inflammatory signaling pathway and contributed to inflammatory and
fibro-elastic pathogenetic changes by deregulating their cis-target
and trans-target genes in OSF malignant development. Further
functional analysis of these lncRNAs is required to provide
conclusive evidence supporting an underlying regulatory mechanism
during OSF.
Fang et al (74), determined that arecoline-induced
myofibroblast trans-differentiation occurred via LINC00974-mediated
activation of the transforming growth factor-β (TGF-β)/Smad
signaling pathway. According to their study, collagen gel
contractility and myofibroblast migration ability was increased in
fibrotic buccal mucosal fibroblasts (fBMFs) overexpressing
LINC00974. Increased expression of another lncRNAHIF1A-AS1 also
positively modulates the TGF-β/Smad signaling pathway, similar to
LINC00974 (72). However, the
lncRNA GAS5-AS1 presented contrary results in arecoline-treated
BMFs and fBMFs (73).
OLP
OLP is a chronic inflammatory disease affecting the
oral mucosa with characteristic relapses and remissions (76-78). Emerging evidence shows that OLP
may be premalignant (79).
Unstable molecular changes can induce the production of several
inflammatory cytokines and subsequently contribute to the course of
OLP (77,80). For example, pathogen associated
molecular patterns and adaptor molecules (i.e., myeloid
differentiation factor 88) activation leads to nuclear
translocation of NF-κB and augments the transcription of
inflammatory genes (i.e., IL-6 and IL-8) (81). As with OSF, there have only been
two studies on the role of lncRNAs in OLP to date.
Yang et al (82), examined DE genes and lncRNA
targets in human papillomavirus-related OSCC (n=1), normal (n=1),
and OLP (n=1) samples. Of the identified lncRNAs, most (697; 76.2%)
were intergenic lncRNAs, followed by 151 sense lncRNAs (16.5%).
Keratinization and MHC class I antigen processing and presentation
were significantly enriched by OSCC-associated DE genes and lncRNA
targets, and the olfactory transduction pathway was enriched by
OLP- and OSCC-related DE genes. To the best of our knowledge, this
has been the only study investigating the lncRNA profile of OLP so
far; however, the number of tissue samples was too small to
guarantee the validity of these findings. In addition, there is
evidence suggesting that lncRNA DQ786243 significantly enhances the
expression levels of miR-146a by inducing Forkhead box P3 (Foxp3),
which subsequently blocks NF-κB signaling during OLP. Moreover,
Foxp3+ regulatory T cells significantly suppressed the
function of other CD4+ T cells, such as
CD4+IL-1+ helper T cells and
CD4+IL-17+ helper T cells, by inhibiting the
mRNA expression levels of IFN-γ and IL-17 (83).
LncRNA microarray analysis is considered a
reasonable option for comparing and determining DE lncRNAs in
normal, PPOEL and OSCC tissues, which are difficult to
simultaneously acquire in one patient. Animal models which
investigate both lncRNAs and PPOELs are expected to be explored
extensively, and to primarily include fBMFs treated with related
stimuli (i.e., arecoline). Mechanistically, the classical signaling
pathways in PPOELs focus on the TGF-β/Smad and NF-κB signaling
pathways. Therefore, it is worth exploring more regulatory
mechanisms [i.e., p38/MAPK and JAK/STAT3 signaling pathways] in
subsequent studies on PPOELs.
5. Role of lncRNAs in OSCC
Oral cancers are cancers that exist in the oral
cavity, such as the mucosal surfaces of the lips, floor of the
mouth, tongue, buccal mucosa, lower and upper gingival surfaces,
hard palate, and retromolar trigone (84). The histology of oral cancers
varies widely; the majority of them are OSCCs (84). An increasing number of reports
have revealed that lncRNAs play a broad role in the oncogenesis and
progression of OSCC through transcriptional regulation,
posttranscriptional modulation and epigenetic modifications
(85). LncRNAs, functioning in
oral cancer migration, epithelial-mesenchymal transition (EMT),
metastasis, progression and invasion (86), could serve as biomarkers or
therapeutic targets for OSCC diagnosis, prognosis and treatment
(87).
With the development of whole transcriptome
analyses, including serial analyses of gene expression, RNA
sequencing and microarray data, several oral cancer-associated
lncRNAs have been identified. In 2019, Qiu et al (88) screened 2,294 DE lncRNAs (933
upregulated and 1,361 downregulated) in OSCC tissues (n=72)
compared with paired adjacent normal tissues. A total of four
lncRNA-mRNA coexpression networks were constructed, and low
expression levels of the four lncRNA nodes contributed to poor
median progression-free survival and overall survival. This study
provided novel insights into the role of lncRNAs in OSCC.
LncRNAs could transcriptionally regulate the
progression of OSCC via interactions with proteins or interactions
with RNA and DNA molecules (85).
Zhu et al (89), verified
that hypoxia induces the overexpression of hyaluronan synthase 2
antisense 1 (HAS2-AS1) in a HIF-1α- and NF-κB-dependent manner.
HAS2-AS1 mediates hypoxia-induced EMT and invasiveness of OSCC
cells by binding and stabilizing the HAS2 gene. Additional research
found that LINC01133 inhibited OSCC cell migration and invasion by
inhibiting growth and differentiation factor 15 (GDF15) protein
expression and formed a feedback regulatory loop with GDF15
(90). Kong et al
(91), found that expression of
lncRNA FOXC1 upstream transcript, which is the adjacent promoter
upstream of the Fork head box C1 (FOXC1) gene, was positively
correlated with FOXC1 mRNA expression and promoted OSCC
proliferation and migration. Thus, a vast amount of research has
elucidated transcriptional regulatory mechanisms to address the
role of lncRNAs in OSCC.
Posttranscriptional regulation through pre-mRNA
alternative splicing, mRNA decay acceleration, mRNA protection, or
translational activation or repression is also one of the primary
mechanisms of lncRNAs (92).
Chang and Hu (93), demonstrated
that MALAT1 could function as a ceRNA to mediate STAT3 expression
by sponging miR-125b in OSCC. OSCC cell viability and growth were
enhanced by increasing expression levels of MALAT1, and a role of
MALAT1/miR-125b/STAT3 axis was confirmed in vivo using a
nude mouse xenograft model with OSCC Tca8113 cells. Fang et
al (94), found that lncRNA
urothelial cancer associated 1 (UCA1) facilitated proliferation,
enhanced cisplatin chemoresistance, and suppressed apoptosis in
OSCC cells by suppressing miR-184 expression to increase the mRNA
expression levels of splicing factor 1. Thus, ceRNA appears to be a
promising posttranscriptional regulatory mechanism in OSCC.
In addition, lncRNAs could affect the
characteristics of OSCC through epigenetic modifications, including
DNA methylation, chromatin modification and imprinting (95). LncRNA HOX transcript antisense RNA
(HOTAIR) repressed the expression of E-cadherin by binding to
enhancer of zeste homolog 2, the enzymatic component of polycomb
repressive complex 2 (PRC2) and H3K27me3 at the E-cadherin promoter
(96). HOTAIR silenced
transcription factors by interacting with PRC2, lysine-specific
histone demethylase-1 and RE-1 elements, leading to chromatin
remodelling, thus trans-inhibiting the expression of the homeobox D
cluster gene and promoting the occurrence, invasion and metastasis
of tumors (96). Additional
lncRNAs, participating in the progression of OSCC through
epigenetic modifications, are expected to be detected, as there is
an increasing number of the research being performed (95,97).
Accumulating evidence has shown that lncRNAs
modulate the metastasis, proliferation, invasiveness and migration
of oral cancer, especially OSCC, in cellular physiological
processes (86,98,99). As aforementioned, lncRNAs interact
with different cellular macromolecules, including chromatin,
protein and RNA (36). According
to the aforementioned studies, unstimulated OSCC cell lines are
typically used for external models. Animal models are also
well-established to provide more convincing evidence to confirm the
role of lncRNAs in OSCC (89,93,94). This suggests that lncRNAs have the
potential to be prognostic and therapeutic markers, providing valid
approaches for clinical treatment.
6. Conclusion
With the advent of genomic technologies, including
micro-arrays and RNA sequencing, investigations of lncRNA genomic
profiles have been widely performed in the last 5 years (6,100,101). A large number of studies include
micro-array analyses of clinical tissue samples, which enhances the
clinical value and significance of the findings (48,55,88). The present review not only
summarized the lncRNA profiles but also elucidated the potential
underlying mechanisms of lncRNAs in oral diseases (Table II and Fig. 3). These mechanisms may be
significant to the clinical diagnosis and therapy of oral diseases.
Moreover, the present review focused on the identification and
associations of oral inflammatory disorders, PPOELs and OSCC and
discussed the functions of lncRNAs in the pathological process. By
summarizing the studies involving the mechanisms of lncRNAs in oral
diseases, ceRNA regulation was found to be the most common, while
lncRNAs interacting with proteins is relatively rare. Thus,
additional research is required to expand the early findings and
characterize the mechanisms of DE lncRNAs. Overall, lncRNAs, the
novel candidates in oral inflammatory disorders, PPOELs and oral
cancer, should be investigated further, and their diagnostic and
therapeutic functions may have significant value.
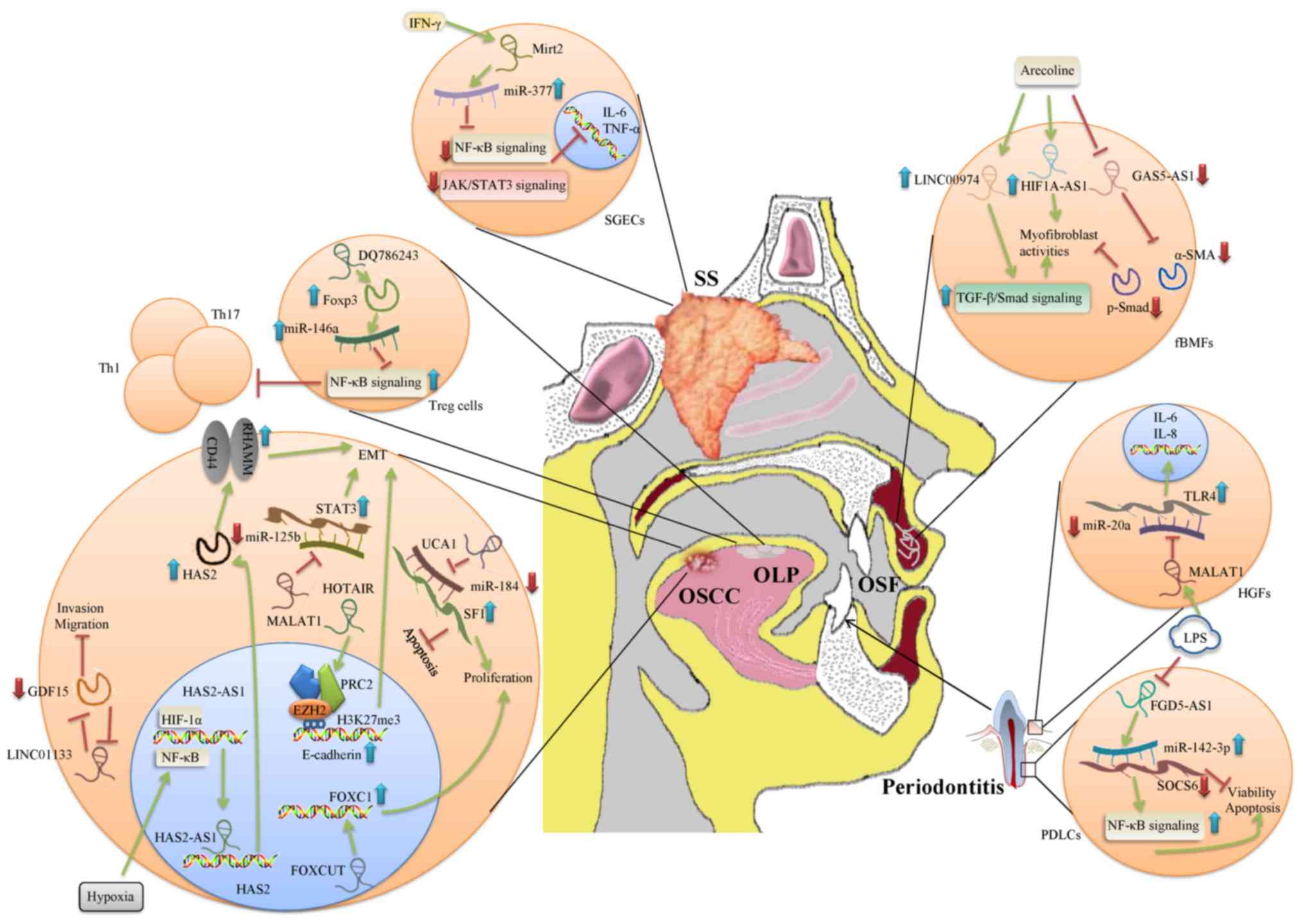 | Figure 3Schematic view of the biological
processes involving lncRNAs in multiple oral diseases. The green
arrow indicates induction, and the red T indicates inhibition. The
blue arrow indicates an increase in expression levels of molecules
or activation of pathways. The red arrow indicates a decrease in
expression levels of molecules or inhibition of pathways. Lnc, long
non-coding; HGFs, human gingival fibroblasts; SGECs, salivary gland
epithelial cells; fBMFs, fibrotic buccal mucosal fibroblasts; SS,
Sjögren's syndrome; OSCC, oral squamous cell carcinoma; OLP, oral
lichen planus; OSF, oral submucous fibrosis; IFN, interferon;
HIF1-α, hypoxia-inducible factor 1-α; Mirt2, lncRNA Myocardial
infarction associated transcript 2; FGD5-AS1, FGD5 antisense RNA 1;
MALAT1, lncRNA Metastasis Associated Lung Adenocarcinoma Transcript
1; HOTAIR, lncRNA HOX transcript antisense RNA; α-SMA,
myofibroblasts α-smooth muscle actin; Foxp3, Forkhead box P3; HAS2,
Hyaluronan Synthase 2; GDF15, growth and differentiation factor 15;
FOXC1, Fork head box C1; SF1, splicing factor 1; TNF-α, tumor
necrosis factor-α; RHAMM, receptor for hyaluronan-mediated
motility; IL, interleukin; NF-κB, nuclear factor-κ B; TGF-β,
transforming growth factor-β; EMT, epithelial mesenchymal
transition. |
 | Table IIRegulatory effect of lncRNAs in oral
inflammatory disorders, PPOELs and OSCC. |
Table II
Regulatory effect of lncRNAs in oral
inflammatory disorders, PPOELs and OSCC.
| lncRNA | Cell category | Cell stimulation
(bacteria) | Disease | Effects | Modes of
action | Associated targets
or pathways | (Refs.) |
|---|
| FGD5-AS1 | PDLCs | LPS (P.
g) | Periodontitis | Inhibit apoptosis
and reduce inflammatory cytokine production | Sponging
miR-142-3p | SOCS6 and
NF-κB | (37) |
| MALAT1 | HGFs | LPS (P. g or
E. coli) | Periodontitis
inflammatory | Enhance cytokine
production | Sponging
miR-20a | TLR4 | (7) |
| Mirt2 | SGECs | IFN-γ | SS | Repress apoptosis
inflammatory cytokine production | Facilitate miR-377
and enhance | JAK/STAT3 and
NF-κB | (5) |
| LINC00974 | fBMFs | - | OSF | Increase
myofibroblasts activation | - | TGF-β/Smad | (55) |
| HIF1A-AS1 | fBMFs | - | OSF | Increase
myofibroblasts activation | - | - | (53) |
| GAS5-AS1 | fBMFs | Arecoline | OSF | Inhibit
myofibroblasts activities in OSF | Inhibiting p-Smad
and α-SMA | TGF-β/Smad | (54) |
| DQ786243 | CD4+ Treg
cells | - | OLP | Suppress the
function of CD4+ T cells such as Th1 and Th17 | Elevating
Foxp3 | miR-146a/NF-κB | (63) |
| HAS2-AS1 | Human OSCC cell
lines | Hypoxia | OSCC | Mediate
hypoxia-induced EMT and invasiveness of OSCC | Stabilizing
HAS2 | HF-1α and
NF-κB | (69) |
| LINC01133 | Human OSCC cell
lines | - | OSCC | Inhibit OSCC cell
migration and invasion | - | GDF15 | (70) |
| FOXCUT | Human OSCC cell
lines | - | OSCC | Promote OSCC
proliferation and migration ability | - | FOXC1 | (71) |
| MALAT1 | Human OSCC cell
lines | - | OSCC | Accelerates EMT and
development of OSCC | Sponging
miR-125b | STAT3 | (73) |
| UCA1 | Human OSCC cell
lines | - | OSCC | Facilitate
proliferation, enhance CDDP chemoresistance, and suppresses
apoptosis | Sponging
miR-184 | SF1 | (74) |
| HOTAIR | Human OSCC cell
lines | - | OSCC | Promote occurrence,
invasion and metastasis | EZH2 and
H3K27me3 | E-cadherin | (76) |
Funding
This study was supported by the National Natural
Science Foundation of China (grant nos. 81600882 and 81870755), a
China Postdoctoral Science Foundation funded project (grant no.
2019M663009) and the President Foundation of Nanfang Hospital,
Southern Medical University (grant no. 2019B002).
Availability of data and materials
Not applicable.
Authors' contributions
FF and BW contributed to the conception and design
of the review. KZ contributed to the writing and drafting of the
manuscript. FF and WQ contributed to the critical revision of the
manuscript for important intellectual content. All the authors have
given approval of the final version to be published and agree to be
accountable for all aspects of the work.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgments
Not applicable.
References
|
1
|
Brogden KA, Johnson GK, Vincent SD, Abbasi
T and Vali S: Oral inflammation, a role for antimicrobial peptide
modulation of cytokine and chemokine responses. Expert Rev Anti
Infect Ther. 11:1097–1113. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Grivennikov SI, Greten FR and Karin M:
Immunity, inflammation, and cancer. Cell. 140:883–899. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Kulkarni V, Uttamani JR, Naqvi AR and
Nares S: microRNAs: Emerging players in oral cancers and
inflammatory disorders. Tumour Biol. 39:10104283176983792017.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Chen H, Lan Z, Li Q and Li Y: Abnormal
expression of long noncoding RNA FGD5-AS1 affects the development
of periodontitis through regulating miR-142-3p/SOCS6/NF-kB pathway.
Artif Cells Nanomed Biotechnol. 47:2098–2106. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Xin M, Liang H, Wang H, Wen D, Wang L,
Zhao L, Sun M and Wang J: Mirt2 functions in synergy with miR-377
to participate in inflammatory pathophysiology of sjogren's
syndrome. Artif Cells Nanomed Biotechnol. 47:2473–2480. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Li S, Liu X, Li H, Pan H, Acharya A, Deng
Y, Yu Y, Haak R, Schmidt J, Schmalz G and Ziebolz D: Integrated
analysis of long noncoding RNA-associated competing endogenous RNA
network in periodontitis. J Periodontal Res. 53:495–505. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Li J, Wang M, Song L, Wang X, Lai W and
Jiang S: LncRNA MALAT1 regulates inflammatory cytokine production
in lipopolysaccharide-stimulated human gingival fibroblasts through
sponging miR-20a and activating TLR4 pathway. J Periodontal Res.
55:182–190. 2020. View Article : Google Scholar
|
|
8
|
Lee PH, Chu PM, Hsieh PL, Yang HW, Chueh
PJ, Huang YF, Liao YW and Yu CC: Glabridin inhibits the activation
of myofi-broblasts in human fibrotic buccal mucosal fibroblasts
through TGF-β/smad signaling. Environ Toxicol. 33:248–255. 2018.
View Article : Google Scholar
|
|
9
|
Ganesh D, Sreenivasan P, Ohman J,
Wallström M, Braz-Silva PH, Giglio D, Kjeller G and Hasséus B:
Potentially malignant oral disorders and cancer transformation.
Anticancer Res. 38:3223–3229. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Awadallah M, Idle M, Patel K and Kademani
D: Management update of potentially premalignant oral epithelial
lesions. Oral Surg Oral Med Oral Pathol Oral Radiol. 125:628–636.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Han X, Wei YB, Tian G, Tang Z, Gao JY and
Xu XG: Screening of crucial long non-coding RNAs in oral epithelial
dysplasia by serial analysis of gene expression. Genet Mol Res.
14:11729–11738. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Chen L, Zhou Y and Li H: LncRNA, miRNA and
lncRNA-miRNA interaction in viral infection. Virus Res. 257:25–32.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Chattopadhyay E, Singh R, Ray A, Roy R,
Sarkar ND, Paul RR, Pal M, Aich R and Roy B: Expression
deregulation of mir31 and CXCL12 in two types of oral precancers
and cancer: Importance in progression of precancer and cancer. Sci
Rep. 6:327352016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Fiaschi T and Chiarugi P: Oxidative
stress, tumor microenvironment, and metabolic reprogramming: A
diabolic liaison. Expression deregulation of mir31 and CXCL12 in
two types of oral precancers and cancer: Importance in progression
of precancer and cancer. Int J Cell Biol. 2012:7628252012.
View Article : Google Scholar
|
|
15
|
Naylor MS, Stamp GW, Foulkes WD, Eccles D
and Balkwill FR: Tumor necrosis factor and its receptors in human
ovarian cancer. Potential role in disease progression. J Clin
Invest. 91:2194–2206. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Yang G, Lu X and Yuan L: LncRNA: A link
between RNA and cancer. Biochim Biophys Acta. 1839:1097–1109. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Yang ZH, Dang YQ and Ji G: Role of
epigenetics in transformation of inflammation into colorectal
cancer. World J Gastroenterol. 25:2863–2877. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Katsanos KH, Roda G, Brygo A, Delaporte E
and Colombel JF: Oral cancer and oral precancerous lesions in
inflammatory bowel diseases: A systematic review. J Crohns Colitis.
9:1043–1052. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Jia H, Wang X and Sun Z: Exploring the
long noncoding RNAs-based biomarkers and pathogenesis of malignant
trans-formation from dysplasia to oral squamous cell carcinoma by
bioinformatics method. Eur J Cancer Prev. 29:174–181. 2020.
View Article : Google Scholar
|
|
20
|
Camacho CV, Choudhari R and Gadad SS: Long
noncoding RNAs and cancer, an overview. Steroids. 133:93–95. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhang J and Chu M: Targeting of
IL-6-relevant long noncoding RNA profiles in inflammatory and
tumorous disease. Inflammation. 42:1139–1146. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
St LG, Wahlestedt C and Kapranov P: The
landscape of long noncoding RNA classification. Trends Genet.
31:239–251. 2015. View Article : Google Scholar
|
|
23
|
Jarroux J, Morillon A and Pinskaya M:
History, discovery, and classification of lncRNAs. Adv Exp Med
Biol. 1008:1–46. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Guenzl PM and Barlow DP: Macro lncRNAs: A
new layer of cis-regulatory information in the mammalian genome.
RNA Biol. 9:731–741. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Beltrami C, Angelini TG and Emanueli C:
Noncoding RNAs in diabetes vascular complications. J Mol Cell
Cardiol. 89:42–50. 2015. View Article : Google Scholar
|
|
26
|
Allen MA, Andrysik Z, Dengler VL, Mellert
HS, Guarnieri A, Freeman JA, Sullivan KD, Galbraith MD, Luo X,
Kraus WL, et al: Global analysis of p53-regulated transcription
identifies its direct targets and unexpected regulatory mechanisms.
Elife. 3:e22002014. View Article : Google Scholar
|
|
27
|
Xu Z, Wei W, Gagneur J, Perocchi F,
Clauder-Münster S, Camblong J, Guffanti E, Stutz F, Huber W and
Steinmetz LM: Bidirectional promoters generate pervasive
transcription in yeast. Nature. 457:1033–1037. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Neil H, Malabat C, D'Aubenton-Carafa Y, Xu
Z, Steinmetz LM and Jacquier A: Widespread bidirectional promoters
are the major source of cryptic transcripts in yeast. Nature.
457:1038–1042. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Fox MJ, Gao H, Smith-Kinnaman WR, Liu Y
and Mosley AL: The exosome component Rrp6 is required for RNA
polymerase II termination at specific targets of the Nrd1-Nab3
pathway. PLoS Genet. 11:e10049992015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Yu D, Ma X, Zuo Z, Wang H and Meng Y:
Classification of tran-scription boundary-associated RNAs (TBARs)
in animals and plants. Front Genet. 9:1682018. View Article : Google Scholar
|
|
31
|
Bianchessi V, Badi I, Bertolotti M, Nigro
P, D'Alessandra Y, Capogrossi MC, Zanobini M, Pompilio G, Raucci A
and Laur A: The mitochondrial lncRNA ASncmtRNA-2 is induced in
aging and replicative senescence in endothelial cells. J Mol Cell
Cardiol. 81:62–70. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Werner MS, Sullivan MA, Shah RN, Nadadur
RD, Grzybowski AT, Galat V, Moskowitz IP and Ruthenburg AJ:
Chromatin-enriched lncRNAs can act as cell-type specific activators
of proximal gene transcription. Nat Struct Mol Biol. 24:596–603.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Ferdin J, Nishida N, Wu X, Nicoloso MS,
Shah MY, Devlin C, Ling H, Shimizu M, Kumar K, Cortez MA, et al:
HINCUTs in cancer: Hypoxia-induced noncoding ultraconserved
transcripts. Cell Death Differ. 20:1675–1687. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Giannakakis A, Zhang J, Jenjaroenpun P,
Nama S, Zainolabidin N, Aau MY, Yarmishyn AA, Vaz C, Ivshina AV,
Grinchuk OV, et al: Contrasting expression patterns of coding and
noncoding parts of the human genome upon oxidative stress. Sci Rep.
5:97372015. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Lazorthes S, Vallot C, Briois S,
Aguirrebengoa M, Thuret JY, St Laurent G, Rougeulle C, Kapranov P,
Mann C, Trouche D and Nicolas E: A vlincRNA participates in
senescence maintenance by relieving H2AZ-mediated repression at the
INK4 locus. Nat Commun. 6:59712015. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Dahariya S, Paddibhatla I, Kumar S,
Raghuwanshi S, Pallepati A and Gutti RK: Long non-coding RNA:
Classification, biogenesis and functions in blood cells. Mol
Immunol. 112:82–92. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Chen LL and Carmichael GG: Decoding the
function of nuclear long non-coding RNAs. Curr Opin Cell Biol.
22:357–364. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Fang F, Zhang K, Chen Z and Wu B:
Noncoding RNAs: New insights into the odontogenic differentiation
of dental tissue-derived mesenchymal stem cells. Stem Cell Res
Ther. 10:2972019. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Geng F, Liu J, Guo Y, Li C, Wang H, Wang
H, Zhao H and Pan Y: Persistent exposure to porphyromonas
gingivalis promotes proliferative and invasion capabilities, and
tumorigenic proper-ties of human immortalized oral epithelial
cells. Front Cell Infect Microbiol. 7:572017. View Article : Google Scholar
|
|
40
|
Song Y, Pan Y and Liu J: Functional
analysis of lncRNAs based on competitive endogenous RNA in tongue
squamous cell carcinoma. PeerJ. 7:e69912019. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Ranzani V, Rossetti G, Panzeri I, Arrigoni
A, Bonnal RJ, Curti S, Gruarin P, Provasi E, Sugliano E, Marconi M,
et al: The long intergenic noncoding RNA landscape of human
lymphocytes highlights the regulation of T cell differentiation by
linc-MAF-4. Nat Immunol. 16:318–325. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Zhang X, Lian Z, Padden C, Gerstein MB,
Rozowsky J, Snyder M, Gingeras TR, Kapranov P, Weissman SM and
Newburger PE: A myelopoiesis-associated regulatory intergenic
noncoding RNA transcript within the human HOXA cluster. Blood.
113:2526–2534. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Elling R, Chan J and Fitzgerald KA:
Emerging role of long noncoding RNAs as regulators of innate immune
cell development and inflammatory gene expression. Eur J Immunol.
46:504–512. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Heward JA and Lindsay MA: Long non-coding
RNAs in the regulation of the immune response. Trends Immunol.
35:408–419. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Qu Q, Fang F, Wu B, Hu Y, Chen M, Deng Z,
Ma D, Chen T, Hao Y and Ge Y: Potential role of long non-coding RNA
in osteogenic differentiation of human periodontal ligament stem
cells. J Periodontol. 8:e127–e137. 2016. View Article : Google Scholar
|
|
46
|
Bjørndal L, Simon S, Tomson PL and Duncan
HF: Management of deep caries and the exposed pulp. Int Endod J.
52:949–973. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Hui T, Wang C, Chen D, Zheng L, Huang D
and Ye L: Epigenetic regulation in dental pulp inflammation. Oral
Dis. 23:22–28. 2017. View Article : Google Scholar
|
|
48
|
Huang X and Chen K: Differential
expression of long noncoding RNAs in normal and inflamed human
dental pulp. J Endod. 44:62–72. 2018. View Article : Google Scholar
|
|
49
|
Lei F, Zhang H and Xie X: Comprehensive
analysis of an lncRNA-miRNA-mRNA competing endogenous RNA network
in pulpitis. PeerJ. 7:e71352019. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Zhong S, Zhang S, Bair E, Nares S and Khan
AA: Differential expression of microRNAs in normal and inflamed
human pulps. J Endod. 38:746–752. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Galicia JC, Henson BR, Parker JS and Khan
AA: Gene expression profile of pulpitis. Genes Immun. 17:239–243.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Mombelli A: Microbial colonization of the
periodontal pocket and its significance for periodontal therapy.
Periodontol 2000. 76:85–96. 2018. View Article : Google Scholar
|
|
53
|
Singhrao SK, Harding A, Poole S, Kesavalu
L and Crean S: Porphyromonas gingivalis periodontal infection and
its putative links with Alzheimer's disease. Mediators Inflamm.
2015:1373572015. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Michaud DS, Fu Z, Shi J and Chung M:
Periodontal disease, tooth loss, and cancer risk. Epidemiol Rev.
39:49–58. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Zou Y, Li C, Shu F, Tian Z, Xu W, Xu H,
Tian H, Shi R and Mao X: lncRNA expression signatures in
periodontitis revealed by microarray: The potential role of lncRNAs
in periodontitis pathogenesis. J Cell Biochem. 116:640–647. 2015.
View Article : Google Scholar
|
|
56
|
Liu W, Zheng Y, Chen B, Ke T and Shi Z:
LncRNA papillary thyroid carcinoma susceptibility candidate 3
(PTCSC3) regulates the proliferation of human periodontal ligament
stem cells and toll-like receptor 4 (TLR4) expression to improve
periodontitis. BMC Oral Health. 19:1082019. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Malathi N, Mythili S and Vasanthi HR:
Salivary diagnostics: A brief review. ISRN Dent.
2014:1587862014.PubMed/NCBI
|
|
58
|
Tzioufas AG, Tsonis J and Moutsopoulos HM:
Neuroendocrine dysfunction in Sjogren's syndrome.
Neuroimmunomodulation. 15:37–45. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Zoukhri D: Effect of inflammation on
lacrimal gland function. Exp Eye Res. 82:885–898. 2006. View Article : Google Scholar :
|
|
60
|
Gliozzi M, Greenwell-Wild T, Jin W,
Moutsopoulos NM, Kapsogeorgou E, Moutsopoulos HM and Wahl SM: A
link between interferon and augmented plasmin generation in
exocrine gland damage in Sjögren's syndrome. J Autoimmun.
40:122–133. 2013. View Article : Google Scholar
|
|
61
|
Reksten TR, Jonsson MV, Szyszko EA, Brun
JG, Jonsson R and Brokstad KA: Cytokine and autoantibody profiling
related to histopathological features in primary Sjogren's
syndrome. Rheumatology (Oxford). 48:1102–1106. 2009. View Article : Google Scholar
|
|
62
|
Dolcino M, Tinazzi E, Vitali C, Del PN,
Puccetti A and Lunardi C: Long non-coding RNAs modulate Sjögren's
syndrome associated gene expression and are involved in the
pathogenesis of the disease. J Clin Med. 8:13492019. View Article : Google Scholar
|
|
63
|
Shi H, Cao N, Pu Y, Xie L, Zheng L and Yu
C: Long non-coding RNA expression profile in minor salivary gland
of primary Sjögren's syndrome. Arthritis Res Ther. 18:1092016.
View Article : Google Scholar
|
|
64
|
Jia B, Qiu X, Chen J, Sun X, Zheng X, Zhao
J, Li Q and Wang Z: A feed-forward regulatory network
lncPCAT1/miR-106a-5p/E2F5 regulates the osteogenic differentiation
of periodontal ligament stem cells. J Cell Physiol.
234:19523–19538. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Huang Y, Han Y, Guo R, Liu H, Li X, Jia L,
Zheng Y and Li W: Long non-coding RNA FER1L4 promotes osteogenic
differentiation of human periodontal ligament stromal cells via
miR-874-3p and vascular endothelial growth factor A. Stem Cell Res
Ther. 11:52020. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Wang L, Wu F, Song Y, Li X, Wu Q, Duan Y
and Jin Z: Long noncoding RNA related to periodontitis interacts
with miR-182 to upregulate osteogenic differentiation in
periodontal mesenchymal stem cells of periodontitis patients. Cell
Death Dis. 7:e23272016. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Canniff JP, Harvey W and Harris M: Oral
submucous fibrosis: Its pathogenesis and management. Br Dent J.
160:429–434. 1986. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Tilakaratne WM, Klinikowski MF, Saku T,
Peters TJ and Warnakulasuriya S: Oral submucous fibrosis: Review on
aetiology and pathogenesis. Oral Oncol. 42:561–568. 2006.
View Article : Google Scholar
|
|
69
|
Sharma M and Radhakrishnan R: Limited
mouth opening in oral submucous fibrosis: Reasons, ramifications,
and remedies. J Oral Pathol Med. 46:424–430. 2017. View Article : Google Scholar
|
|
70
|
Arakeri G, Patil SG, Aljabab AS, Lin KC,
Merkx MAW, Gao S and Brennan PA: Oral submucous fibrosis: An update
on pathophysiology of malignant transformation. J Oral Pathol Med.
46:413–417. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Sharma M, Shetty SS and Radhakrishnan R:
Oral submucous fibrosis as an overhealing wound: Implications in
malignant transformation. Recent Pat Anticancer Drug Discov.
13:272–291. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Wang YK, Liu CM, Lin T, Fang CY, Yu CC and
Yu CH: Inhibition of HIF1A-AS1 impedes the arecoline-induced
migration activity of human oral mucosal fibroblasts. J Formos Med
Assoc. 119:879–883. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Lin CY, Liao YW, Hsieh PL, Lu MY, Peng CY,
Chu PM, Yang HW, Huang YF, Yu CC and Yu CH: LncRNA GAS5-AS1
inhibits myofibroblasts activities in oral submucous fibrosis. J
Formos Med Assoc. 117:727–733. 2018. View Article : Google Scholar
|
|
74
|
Fang CY, Yu CC, Liao YW, Hsieh PL, Lu MY,
Lin KC, Wu CZ and Tsai LL: LncRNA LINC00974 activates TGF-β/Smad
signaling to promote oral fibrogenesis. J Oral Pathol Med.
48:151–158. 2019.
|
|
75
|
Zhou S, Zhu Y, He Z, Zhang D, Guo F, Jian
X and Zhang C: Long non-coding RNA expression profile associated
with malignant progression of oral submucous fibrosis. J Oncol.
2019:68351762019. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Lodi G, Scully C, Carrozzo M, Griffiths M,
Sugerman PB and Thongprasom K: Current controversies in oral lichen
planus: Report of an international consensus meeting. Part 1. Viral
infections and etiopathogenesis. Oral Surg Oral Med Oral Pathol
Oral Radiol Endod. 100:40–51. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Eisen D, Carrozzo M, Bagan SJ and
Thongprasom K: Number V oral lichen planus: Clinical features and
management. Oral Dis. 11:338–349. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Scully C, Beyli M, Ferreiro MC, Ficarra G,
Gill Y, Griffiths M, Holmstrup P, Mutlu S, Porter S and Wray D:
Update on oral lichen planus: Etiopathogenesis and management. Crit
Rev Oral Biol Med. 9:86–122. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Lončar-Brzak B, Klobučar M, Veliki-Dalic
I, Sabol I, Pavelić SK, Krušlin B and Mravak-Stipetić M: Expression
of small leucine-rich extracellular matrix proteoglycans biglycan
and lumican reveals oral lichen planus malignant potential. Clin
Oral Investig. 22:1071–1082. 2018. View Article : Google Scholar
|
|
80
|
Santoro A, Majorana A, Bardellini E, Festa
S, Sapelli P and Facchetti F: NF-kappaB expression in oral and
cutaneous lichen planus. J Pathol. 201:466–472. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Groeger S and Meyle J: Oral mucosal
epithelial cells. Feonr Immunol. 10:2082019.
|
|
82
|
Yang Q, Xu B, Sun H, Wang X, Zhang J, Yu X
and Ma X: A genome-wide association scan of biological processes
involved in oral lichen planus and oral squamous cell carcinoma.
Medicine (Baltimore). 96:e70122017. View Article : Google Scholar
|
|
83
|
Wang J, Zhai X, Guo J, Li Y, Yang Y, Wang
L, Yang L and Liu F: Long non-coding RNA DQ786243 modulates the
induction and function of CD4(+) Treg cells through
Foxp3-miR-146a-NF-kB axis: Implications for alleviating oral lichen
planus. Int Immunopharmacol. 75:1057612019. View Article : Google Scholar
|
|
84
|
Huang SH and O'Sullivan B: Oral cancer:
Current role of radio-therapy and chemotherapy. Med Oral Patol Oral
Cir Bucal. 18:e233–e240. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Gomes CC, de Sousa SF, Calin GA and Gomez
RS: The emerging role of long noncoding RNAs in oral cancer. Oral
Surg Oral Med Oral Pathol Oral Radiol. 123:235–241. 2017.
View Article : Google Scholar
|
|
86
|
Luo X, Qiu Y, Jiang Y, Chen F, Jiang L,
Zhou Y, Dan H, Zeng X, Lei YL and Chen Q: Long non-coding RNA
implicated in the invasion and metastasis of head and neck cancer:
Possible function and mechanisms. Mol Cancer. 17:142018. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Zhang L, Meng X, Zhu XW, Yang DC, Chen R,
Jiang Y and Xu T: Long non-coding RNAs in Oral squamous cell
carcinoma: Biologic function, mechanisms and clinical implications.
Mol Cancer. 18:1022019. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Qiu YL, Liu YH, Ban JD, Wang WJ, Han M,
Kong P and Li BH: Pathway analysis of a genomewide association
study on a long noncoding RNA expression profile in oral squamous
cell carcinoma. Oncol Rep. 41:895–907. 2019.
|
|
89
|
Zhu G, Wang S, Chen J, Wang Z, Liang X,
Wang X, Jiang J, Lang J and Li L: Long noncoding RNA HAS2-AS1
mediates hypoxia-induced invasiveness of oral squamous cell
carcinoma. Mol Carcinog. 56:2210–2222. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Kong J, Sun W, Zhu W, Liu C, Zhang H and
Wang H: Long noncoding RNA LINC01133 inhibits oral squamous cell
carcinoma metastasis through a feedback regulation loop with GDF15.
J Surg Oncol. 118:1326–1334. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Kong XP, Yao J, Luo W, Feng FK, Ma JT, Ren
YP, Wang DL and Bu RF: The expression and functional role of a
FOXC1 related mRNA-lncRNA pair in oral squamous cell carcinoma. Mol
Cell Biochem. 394:177–186. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Li Y, Zhang J, Pan J, Feng X, Duan P, Yin
X, Xu Y, Wang X and Zou S: Insights into the roles of lncRNAs in
skeletal and dental diseases. Cell Biosci. 8:82018. View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Chang SM and Hu WW: Long non-coding RNA
MALAT1 promotes oral squamous cell carcinoma development via
microRNA-125b/STAT3 axis. J Cell Physiol. 233:3384–3396. 2018.
View Article : Google Scholar
|
|
94
|
Fang Z, Zhao J, Xie W, Sun Q, Wang H and
Qiao B: LncRNA UCA1 promotes proliferation and cisplatin resistance
of oral squamous cell carcinoma by sunppressing miR-184 expression.
Cancer Med. 6:2897–2908. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Gonzalez-Ramirez I, Soto-Reyes E,
Sanchez-Perez Y, Herrera LA and Garcia-Cuellar C: Histones and long
non-coding RNAs: The new insights of epigenetic deregulation
involved in oral cancer. Oral Oncol. 50:691–695. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Wu Y, Zhang L, Zhang L, Wang Y, Li H, Ren
X, Wei F, Yu W, Liu T, Wang X, et al: Long non-coding RNA HOTAIR
promotes tumor cell invasion and metastasis by recruiting EZH2 and
repressing E-cadherin in oral squamous cell carcinoma. Int J Oncol.
46:2586–2594. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Yang CM, Wang TH, Chen HC, Li SC, Lee MC,
Liou HH, Liu PF, Tseng YK, Shiue YL, Ger LP and Tsai KW: Aberrant
DNA hypermethylation-silenced SOX21-AS1 gene expression and its
clinical importance in oral cancer. Clin Epigenetics. 8:1292016.
View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Shao TR, Zheng ZN, Chen YC, Wu QQ, Huang
GZ, Li F, Zeng WS and Lv XZ: LncRNA AC007271.3 promotes cell
proliferation, invasion, migration and inhibits cell apoptosis of
OSCC via the Wnt/β-catenin signaling pathway. Life Sci.
239:1170872019. View Article : Google Scholar
|
|
99
|
Chen F, Qi S, Zhang X, Wu J, Yang X and
Wang R: lncRNA PLAC2 activated by H3K27 acetylation promotes cell
proliferation and invasion via the activation of Wnt/β-catenin
pathway in oral squamous cell carcinoma. Int J Oncol. 54:1183–1194.
2019.PubMed/NCBI
|
|
100
|
Meseure D, Drak AK, Nicolas A, Bieche I
and Morillon A: Long noncoding RNAs as new architects in cancer
epigenetics, prognostic biomarkers, and potential therapeutic
targets. Biomed Res Int. 2015:3202142015. View Article : Google Scholar : PubMed/NCBI
|
|
101
|
Ju H, Zhang L, Mao L, Wu Y, Liu S, Ruan M,
Hu J and Ren G: A comprehensive genome-wide analysis of the long
noncoding RNA expression profile in metastatic lymph nodes of oral
mucosal melanoma. Gene. 675:44–53. 2018. View Article : Google Scholar : PubMed/NCBI
|