Introduction
Alcoholic liver disease is a chronic liver disease
caused by long-term alcohol intake. Early manifestations of the
disease are fatty liver, alcoholic hepatitis, liver fibrosis and
liver cirrhosis, which also occurs during the disease progression,
causing extensive necrosis of liver cells and even liver failure
when the disease is severe (1,2).
Increasing levels of alcohol consumption has led to an increase in
the incidence of alcoholic liver disease. It has been reported that
alcohol-associated liver deaths accounted for up to 48% of
cirrhosis-associated deaths in the United States in 2007 and are
also major contributors to liver disease-related mortality in other
countries (3). Data showed that
up to 90% of patients with heavy alcohol intake have fatty liver,
which could be rapidly reversible with abstinence (4). If the alcohol abuse persists, liver
fibrosis progressively develops, ultimately resulting in cirrhosis
(4,5). Evidence indicates that multiple
pathways are involved in the progression of alcoholic liver
disease. For example, ethanol induces binding of
lipopoly-saccharides to monocyte differentiation antigen CD14,
which then combines with Toll-like receptor 4 to activate multiple
cytokine genes involved in inflammatory cytokine production and to
decrease the expression of STATs in Kupffer cells, thereby causing
alcoholic liver disease (6).
Besides, long-term alcohol consumption also induces mitochondrial
damage, reactive oxygen species production and lipid peroxidation,
which cause liver injury (7,8).
Previous reports have revealed that the activation
of hepatic stellate cells serves a critical role in hepatic
fibrosis development (9,10). Activated hepatic stellate cells
exhibit a proliferative and myofibroblastic phenotype with
increased production of type I collagen and other extracellular
matrix proteins. A recent report showed that suppression of YAP can
alleviate the development of hepatic fibrosis through enhancing
apoptosis and reversion of hepatic stellate cells (11). Another study indicated that
silencing of NADPH oxidase 5 can inhibit the proliferation and
collagen type I levels in hepatic stellate cells (12). Nevertheless, the factors that
regulate the proliferation and apoptosis of hepatic stellate cells
have not been comprehensively identified yet.
Previous studies have identified that that microRNAs
(miRNAs) are associated with the pathogenesis of alcoholic liver
fibrosis (ALF) (13-15). miRNAs measure 20-25 nucleotides in
length, and serve important roles in regulating the expression
levels of cellular genes and proteins (16,17). It was also revealed that genes
from the miR-148 family function critically in the liver by
promoting a liver-specific phenotype and inhibiting invasion in
transformed cells (18,19). miR-148a-3p has been demonstrated
to act as a tumor suppressor by participating in the occurrence and
development of various tumors (20-22). An integrated analysis of miRNAs
from Gene Expression Omnibus datasets showed that miR-148a-3p is
one of a number of functional regulatory modules in the development
of liver fibrosis (23). Notably,
some studies indicate that miR-148a-3p had a negative effect on
hepatic stellate cell activation by targeting transforming growth
factor β receptor type (TGFBR)1 and TGFBR2 (24) or ubiquitin carboxyl-terminal
hydrolase 4 (25). Overexpression
of miR-148a-3p also promotes autophagic and apoptotic activity of
hepatic stellate cells (26).
However, the role and molecular mechanisms of miR-148a-3p in liver
fibrosis and activation of hepatic stellate cells remain largely
unknown.
Receptor tyrosine-protein kinase erbB-3 (ERBB3) is a
membrane bound protein that belongs to the epidermal growth factor
receptor (EGFR/ERBB) family of receptor tyrosine kinases. Several
studies have indicated that ERBB3 participates in various liver
diseases including liver fibrosis (27-29). A recent study indicated that ERBB3
can be regulated by miR-125a-5p in hepatitis B virus-associated
hepatocellular carcinoma (30).
Whether ERBB3 can be regulated by miR-148a-3p and participated in
the viability of hepatic stellate cells requires further
exploration.
miRNAs can regulate disease processes by identifying
target genes and degrading themselves or inhibiting the expression
of target genes according to different degrees of complementarity
(31). TargetScan7.2 (32) is a well-known online analysis tool
by which the potential interaction between miRNAs and mRNAs can be
predicted. Rat hepatic stel-late cells HSC-T6 have been commonly
used in the study of hepatic stellate cells in ALF (33,34). In the present study, TargetScan7.2
predicted that ERBB3 was the direct target gene for miR-148a-3p,
and based on this, the association between miR-148a-3p and ERBB3
was studied, and their roles in the pathogenesis of alcoholic liver
fibrosis were further investigated in HSC-T6 cells.
Materials and methods
Establishing ALF model in rats
Male Sprague-Dawley rats (8 weeks old; weight,
278-289 g) were purchased from Tongji Hospital, Tongji University
School of Medicine. Rats were kept in cages at room temperature
(23±3°C) with a constant humidity (50±10%) with free access to
food/water and a 12-h/12-h light/dark cycle. The animal
experimental protocol was approved by the Tongji Hospital of Tongji
University and performed in accordance with the Guidance and Ethics
Committee of Tongji Hospital, Tongji University School of Medicine
(approval no. THTU20160810A). Lentiviral vectors with overexpressed
miR-148a-3p and negative controls were synthesized by Sangon
Biotech Co., Ltd. The rats were randomly divided into 5 groups:
Control; miR-148-3p; Model; Model + NC; and Model + miR-148-3p,
with 16 rats in each group. Rats in the miR-148-3p and Model +
miR-148-3p groups were selected and injected with 200 µl
overexpressed miR-148a-3p lentivirus vectors, while the rats in the
Model + NC group were injected with 200 µl negative control
(NC) vectors. For the surgical procedure, the rats were held in
place, and their blood vessels were gently rubbed with an
incandescent lamp and repeatedly swabbed by 75% alcohol (cat. no.
10009218, Sinopharm Chemical Reagent Beijing Co., Ltd.) to fully
dilate the vessels and soften the cuticle of the skin to facilitate
puncture. The lentivirus and NC were slowly injected into one-third
of the tail vein. The control and Model groups were administered
distilled water by gavage. The next day, the rats from the model
group received irrigation with 56% alcohol (9.2 mg/kg) once a day.
Rats from the control group received irrigation with
double-distilled water (10 ml/kg) once a day. During the protocol,
all rats were fed with standard food and water.
The Model, Model + NC and Model + miR-148-3p groups
were used to establish the ALF model. Briefly, the rats were
treated by alcohol gavage for 9 weeks. At the end of 9 weeks, all
rats were anesthetized by intramuscular injections of 60 mg/kg
ketamine (cat. no. ZTR-K165295; Shanghai ZZBIO Co., Ltd.) and 10
mg/kg of xylazine (cat. no. B27154, Shanghai Yuanye Bio-Technology
Co., Ltd.) mixed in a 1:1 ratio. The blood was then collected via
the posterior orbital venous plexus of the rats. All rats were then
injected with an overdose of ketamine (180 mg/kg) + xylazine (30
mg/kg) immediately for euthanasia. The weight of the animals at the
time of sacrifice was at 300.06-319.69 g. Then, the complete livers
were removed from the abdominal cavity immediately. The liver
tissues were weighed and the liver index was calculated using the
following formula: Liver index=wet weight of the liver/body weight.
The blood was allowed to coagulate at room temperature for 2 h, and
then centrifuged at 1,000 x g at 4°C for 20 min. The serum was
removed and cryopreserved at -20°C for serum detection. Half of the
liver was immersed in 10% neutral formalin (cat. no. 181101;
Nanchang Yulu Experimental Equipment Co., Ltd.) for histological
analysis, while the other half was placed in liquid nitrogen and
maintained at -80°C for western blotting and reverse
transcription-quantitative (RT-q)PCR test.
Hematoxylin and eosin (H&E)
staining
The tissues (1x1 cm) were fixed using 10% neutral
formalin, dehydrated by a graded ethanol series (80, 90, 95 and
100%) for 2 h and then treated with 50% xylene for 30 min. Next,
the tissues were immersed in wax, embedded and sectioned. The
sections were examined using H&E staining. The samples were
dewaxed by xylene (cat. no. 10023418; Sinopharm Chemical Reagent
Co., Ltd.) and then dephenylated using a graded ethanol series
(100, 95, 80 and 70%) for 2 min. Next, the tissues were rehydrated
by rinsed in distilled water twice to, stained using 0.5%
hematoxylin (Beijing Zhongshan Jinqiao Biotechnology Co., Ltd.) for
20 min at room temperature and then washed under running water. The
slices were then rinsed in acidification solution comprised of
hydrochloric acid (cat. no. 10011008; Sinopharm Chemical Reagent
Co., Ltd.) and 75% ethanol at a ratio of 1:99 for 1 min to remove
the blue-stained cytoplasm and then washed with tap water for 10
min. Then the tissues were stained using 0.5% eosin (Beijing
Zhongshan Jinqiao Biotechnology Co., Ltd.) for 15 min at room
temperature, dehydrated in 100% ethanol for 15 min and treated with
xylene for 15 min. Finally, neutral gum was used to seal the film,
and pathological changes were observed using a CKX31 light optical
microscope (magnification, x100 and x200; Olympus Corporation).
Treated culture
The rat hepatic stellate HSC-T6 cell line was
purchased from Shanghai Institutes for Biological Sciences, Chinese
Academy of Sciences. The cells were cultured in RPMI medium 1640
(Gibco; Thermo Fisher Scientific, Inc.) containing 10% fetal bovine
serum (FBS; Gibco; Thermo Fisher Scientific, Inc.) at 37°C in a 5%
CO2 atmosphere.
Detection of lactate dehydrogenase (LDH),
aspartate aminotransferase (AST), alanine transaminase (ALT) and
alkaline phosphatase (ALP) activity
The activities of LDH, AST, ALT and ALP were
detected by LDH (cat. no. MAK066; Sigma-Aldrich; Merck KGaA), AST
(cat. no. MAK055; Sigma-Aldrich; Merck KGaA), ALT (cat. no. MAK052;
Sigma-Aldrich; Merck KGaA) and ALP (cat. no. MAK262; Sigma-Aldrich;
Merck KGaA) assay kits. The cells were inoculated in a 96-well
plate and allowed to adhere to the walls of the wells. In each
group, blank and standard wells were generated, and
double-distilled water (25 µl) and matrix buffer (25
µl) were added into the blank wells, while double distilled
water (5 µl), 0.2 mmol/l standard solution (20 µl)
and matrix buffer (25 µl) were added into standard wells. In
each group, testing and control wells were generated, and the cell
suspension (20 µl), matrix buffer (25 µl) and
coenzyme I (5 µl) were added into the testing wells, while
the cell suspension (20 µl), matrix buffer (25 µl)
and double distilled water (5 µl) were added into the
control well. The 96-well plate was held in a water bath at 37°C
for 15 min, then 2, 4-dinitrobenzene hydrazine (25 µl) was
added into each group. Next, the cells were mixed with 0.4 mol/l
sodium hydroxide solution (250 µl) and left to stand at room
temperature for 5 min. LDH absorbance was measured at 450 nm, ALT
and AST absorbance values were measured at 510 nm, and ALP
absorbance was measured at 405 nm using a Multiskan™ GO microplate
reader (Thermo Fisher Scientific, Inc.).
RT-qPCR
Total RNA of cells were extracted using
TRIzol® reagent (cat. no. 15596018; Thermo Fisher
Scientific, Inc.), and RNA concentration was measured using a
UV1700PC Nanodrop spectrophotometer (Nanodrop Technologies; Thermo
Fisher Scientific, Inc.) and diluted to 500 ng/µl. Reverse
transcription was performed using Superscript II first-strand cDNA
synthesis System (Invitrogen; Thermo Fisher Scientific, Inc.). The
mRNA expression levels were determined by RT-qPCR using SYBR Green
Real Time PCR kit (cat. no. 204057; Qiagen China, Co., Ltd.) in a
7500 Real-Time PCR system (Applied Biosystems, Inc.). A volume of
0.5 µl forward primer (10 µM), 0.5 µl reverse
primer (10 µM), 4 µl cDNA, 5 µl SYBR Green I
nucleic acid gel stain (cat. no. S9430; Sigma-Aldrich; Merck KGaA)
were mixed into 10 µl solution. The thermocycler conditions
for the qPCR assay was set as follows: Pretreatment at 95°C for 1
min, followed by 40 cycles of 95°C for 30 sec, 58°C for 20 sec and
70°C for 20 sec. The final extension was performed at 72°C for 7
min and then held at 4°C. The sequences of primers are listed in
Table I. The 2−ΔΔCq
method was used to determine the expression levels of RT-PCR
products (35).
 | Table IPrimers used in reverse transcription
quantitative PCR analysis. |
Table I
Primers used in reverse transcription
quantitative PCR analysis.
| Gene | Primer
sequence | Species |
|---|
| miR-148a-3p | Forward:
5′-AGCAGTTCAGTGCACTACAG-3′ | Mouse |
| Reverse:
5′-GCAGGGTCCGAGGTATTC-3′ | |
| α-SMA | Forward:
5′-ATCCACGAAACCACCTATAACA-3′ | Mouse |
| Reverse:
5′-CCACCGATCCAGACAGACTA-3′ | |
| Collagen I | Forward:
5′-TCACCTACTGCACGCTTGTGG-3′ | Mouse |
| Reverse:
5′-TTGGCTTTTGGGGAAATTGA-3′ | |
| Bcl-2 | Forward:
′5′-TGCACCTGAGCGCCTTCAC-3′ | Mouse |
| Reverse:
5′-TAGCTGATTCGACCATTTGCCTGA-3′ | |
| Bax | Forward:
′5′-CCACCAGCTCTGAACAGTT-3′ | Mouse |
| Reverse:
5′-TCAGCCCATCTTCTTCCAG-3′ | |
| ERBB3 | Forward:
′5′-AGATCTGCACCATTGACGTC-3′ | Mouse |
| Reverse:
5′-TAGGTCTAGGTCCAGTTCTG-3′ | |
| GAPDH | Forward:
5′-CCAACCGCGAGAAGATGA-3′ | Mouse |
| Reverse:
5′-CCAGAGGCGTACAGGGATAG-3′ | |
Transfection
For the transfection assay, miR-148a-3p mimic
(5′-UCAGUGCACUACAGAACUUUG-3′), ERBB3 overexpression plasmid
(ERBB3-pcDNA3.1) and negative control (NC) were synthesized by
Sangon Biotech Co., Ltd. The cells were digested, thoroughly mixed
and seeded at 1x106/ml into the 6-well plate. The next
day, the cells were transfected after reaching 80-90% confluence.
Briefly, 20 pmol mimic or 1 µg plasmid DNA was dissolved
into 50 µl Dulbecco′s modified Eagle medium (DMEM; Hyclone;
GE Healthcare) and mixed together with the transfection solution A,
respectively. A total of 50 µl DMEM was dissolved in 1
µl Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.), set aside for 5 min at room temperature and then
mixed with the transfection solution A as the transfection solution
B. Finally, the trans-fection solution B was added into the
corresponding wells of the 6-well plate, and the cell culture plate
was placed in a cell culture box at 37°C with 5% CO2 for
further culture. The culture medium was refreshed 24 h after the
transfection, and then the cells were collected after 72 h.
Luciferase activity assay
For dual-luciferase reporter assays, sequence
information of ERBB 3′ untranslated region (UTR) region was
extracted and constructed into the pMIR-report Luciferase vector
(Promega Corporation) to construct ERBB3 wild-type (WT) vector (3′
UTR-WT). The target-binding region of miR-148a-3p and ERBB3 was
predicted using TargetScan version 7.2 (http://www.targetscan.org/vert_72/). As there were 2
binding sites identified between miR-148a-3p and ERBB3, the 2 sites
were mutated simultaneously to construct an ERBB3 mutant (MUT)
plasmid (3′ UTR-MUT). The pmirGLO vectors containing WT and MUT
ERBB3 3′ UTR were co-transfected with miR-148a-3p mimic into HSC-T6
cells using Lipofectamine® 2000 (Invitrogen; Thermo
Fisher Scientific, Inc.). A total of 48 h after transfection, the
relative luciferase activities of the cells were measured by
Dual-Luciferase Reporter Assay (Promega Corporation).
Renilla luciferase activity was used for normalization.
Cell viability detection
The cells were inoculated into the 96-well plate at
1x105 per well, and cultured with 10% fetal bovine serum
(FBS; Gibco; Thermo Fisher Scientific, Inc.) at 37°C and 5%
CO2. Next, 20 µl MTT solution (cat. no. C0009;
Beyotime Institute of Biotechnology) was added into each well and
incubated for 4 h. Then, 150 µl 0.5% DMSO were added and
gently shaken for 10 min to fully dissolve the purple crystals.
Absorbance at 492 nm was read using a microplate reader (cat. no.
E0225; Beyotime Institute of Biotechnology). All experiments were
repeated 3 times and the average value was calculated.
Cell apoptosis
Following transfection for 24 h, the cells
(1x106/ml) were resuspended in the solution composed of
500 µl 1X Annexin binding buffer (V13246; Thermo Fisher
Scientific, Inc.), 5 µl fluorescein isothiocyanate (FITC)
Annexin V (C1062S; Beyotime Institute of Biotechnology) and 1
µl 100 µg/ml propidium iodide (P1304MP Thermo Fisher
Scientific, Inc.). Then, after the reaction, 300 µl 1X
Annexin Binding Buffer was added to the cell suspension for 15 min
at room temperature. Finally, the stained cells were analyzed using
a FACSCalibur™ flow cytometer (BD Bioscience) and FlowJo software
(v.10.0 (FlowJo LLC).
Western blotting
The total proteins of cells were lysed using RIPA
buffer (Tianjin Yitailong Science & Technology Co., Ltd.).
Next, the proteins (30 µg/lane) were boiled for 5 min at
100°C for denaturation, separated using 15% SDS-PAGE and then
transferred to polyvinylidene fluoride membranes (EMD Millipore).
The membranes were blocked using 5% milk at room temperature for 1
h. The blots were then probed with the primary antibodies: Rabbit
anti-alpha-smooth muscle actin (anti-α-SMA; 1:1,000; cat. no.
ab32575; Abcam); rabbit anti-collagen I (1:1,000; cat. no. ab34710;
Abcam); rabbit anti-Bcl-2 (1:1,000; cat. no. ab59348; Abcam);
rabbit anti-Bax (1:1,000; cat. no. ab32503; Abcam); rabbit
anti-ERBB3 (1:1,000; cat. no. ab5470; Abcam); rabbit anti-cleaved
Caspase-3 (1:1,000; cat. no. ab2302; Abcam); and rabbit anti-GAPDH
(1:2,000; cat. no. ab181602; Abcam) at 4°C overnight. The membranes
were washed using PBS 3 times, and then incubated with goat
anti-rabbit IgG (H+L) horseradish peroxidase-conjugated secondary
antibody (1:2,000; cat. no. SA00001-2; ProteinTech Group, Inc.) for
2 h at room temperature. ECL (Thermo Scientific, Thermo Fisher
Scientific, Inc.) was used to detect the protein bands, which were
then scanned by a multifunctional imager with Image J software
v.4.7 (National Institutes of Health).
Statistical analysis
GraphPad Prism 6 (v.6.01; GraphPad Software, Inc.)
was used for data analysis. The data were shown as mean ± standard
deviation of at least 3 independent experiments, and the
differences between continuous variables from two groups was
compared by t-test. A one-way analysis of variance (ANOVA) followed
by Tukey's post hoc test was performed to analyze the differences
among multiple groups. P<0.05 was considered to indicate a
statistically significant difference.
Results
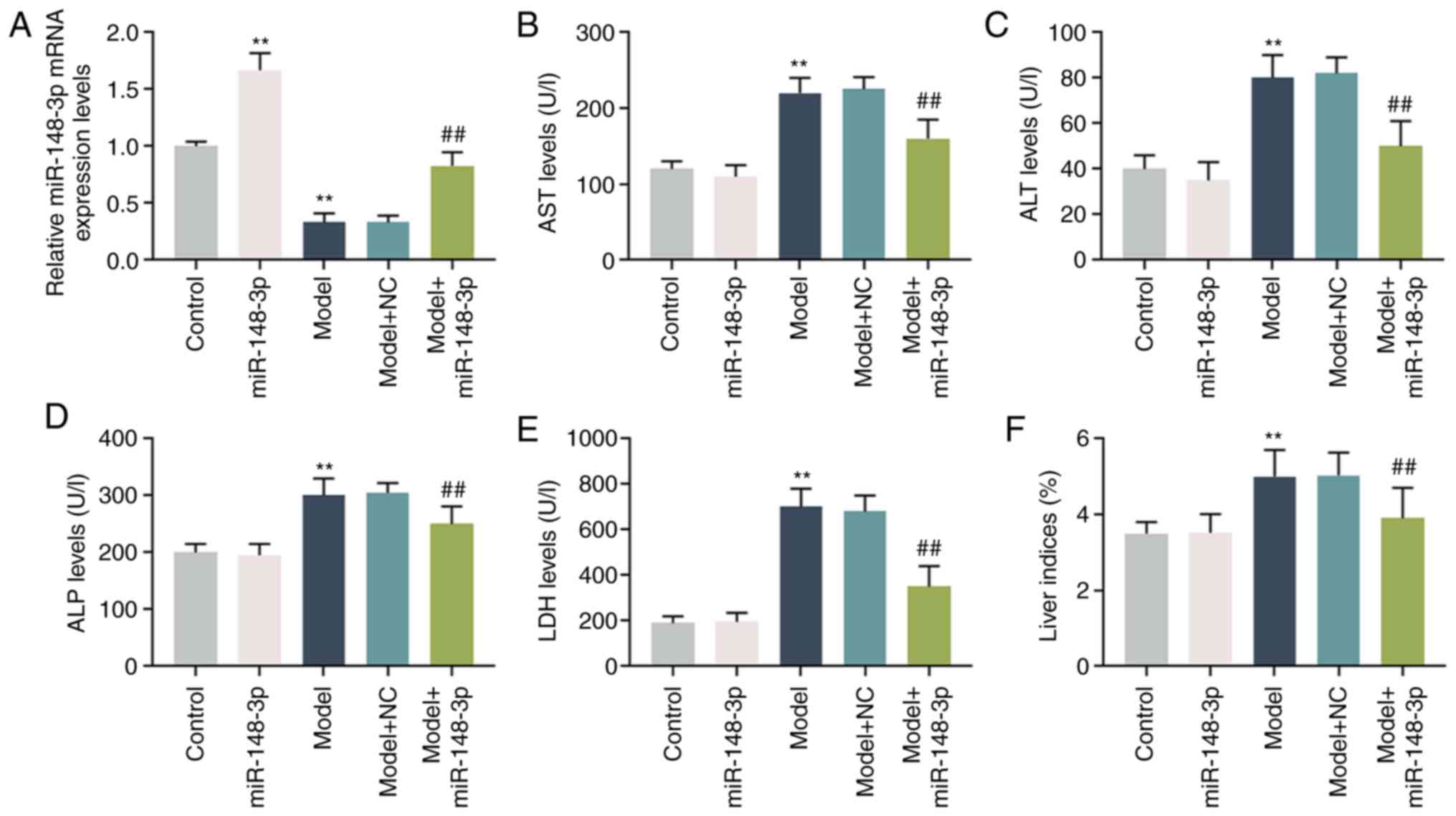
miR-148a-3p expression in ALF model in
rats and the degree of liver injury of model
The expression of miR-148a-3p decreased
significantly in the ALF model of rats; however, miR-148a-3p
expression in the group treated by miR-148a-3p was significantly
increased compared with that of the model group (P<0.001;
Fig. 1A). The expression levels
of AST, ALT, ALP and LDH were markedly upregulated in the model,
but significantly downregulated by miR-148a-3p (P<0.001;
Fig. 1B-E). Moreover, the liver
indices increased significantly when the rats were treated with
alcohol, but the indices decreased noticeably following miR-148a-3p
treatment (P<0.001; Fig.
1F).
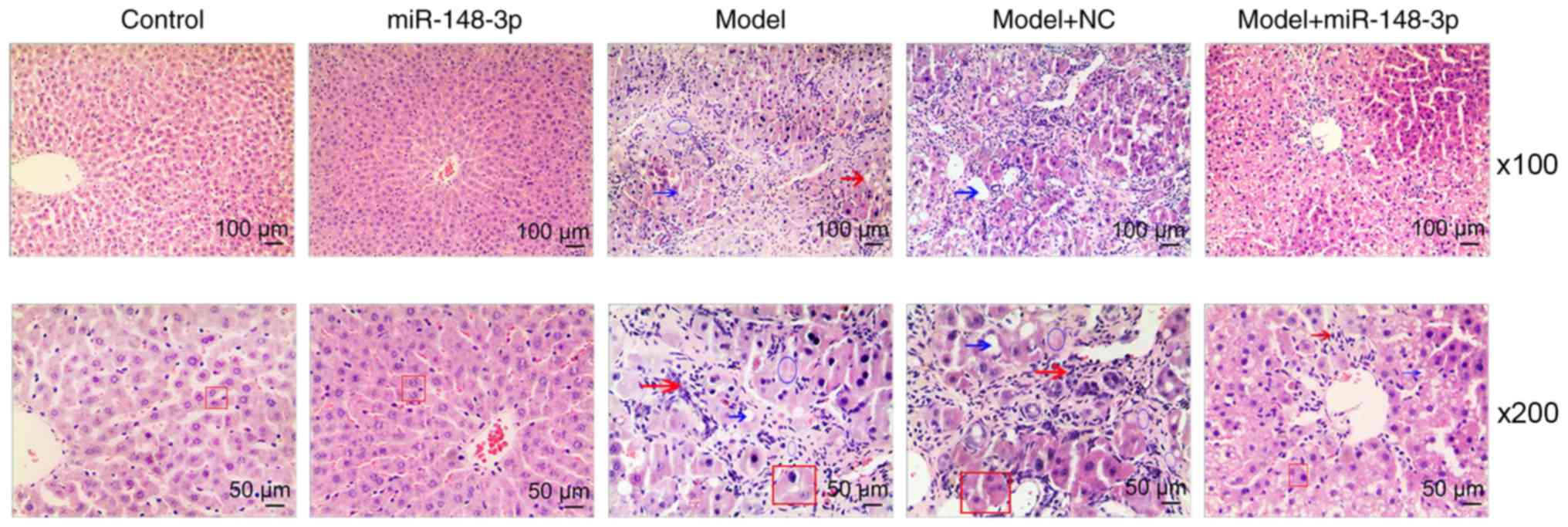
Pathological changes of ALF model of
rats
As shown in Fig.
2, the H&E staining results after 9 weeks indicated that
the hepatocytes in the liver tissues of rats in the normal control
group were normal in size, clear in structure and abundant in
cytoplasm. However, in the ALF model group, a large amount of
steatosis in the liver, massive inflammatory cell infiltration,
obvious interlobular connective tissue hyperplasia and formation of
fibrous septum were observed. Moreover, treatment with miR-148a-3p
mimics partially inhibited the liver fibrosis-like changes in rats
in the alcohol model group. Compared with the model group, the
model group treated with miR-148a-3p exhibited less infiltration of
inflammatory cells in the liver and less fatty degeneration
(Fig. 2).
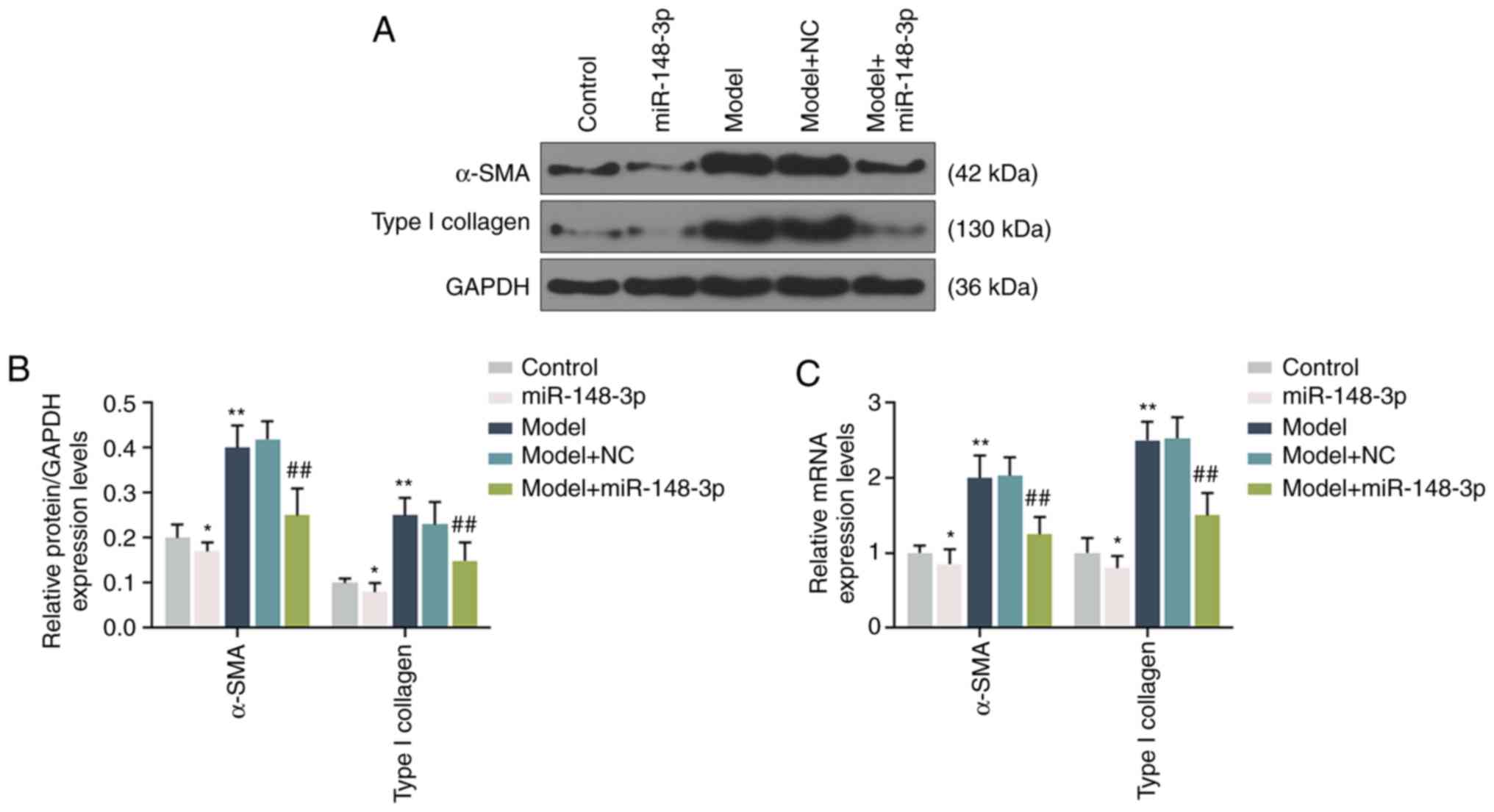
Fibrosis in the ALF model
RT-qPCR and western blotting were performed to
detect the expression levels of α-SMA and type I collagen, and the
results indicated that the expression levels of α-SMA and type I
collagen in the model group were significantly increased compared
with those in the normal control group. Compared with the model
group, following miR-148a-3p intervention, the protein expression
levels of α-SMA and type I collagen in the liver tissues were
significantly downregulated, but still increased compared with
those in the normal control group (P<0.001; Fig. 3A-C).
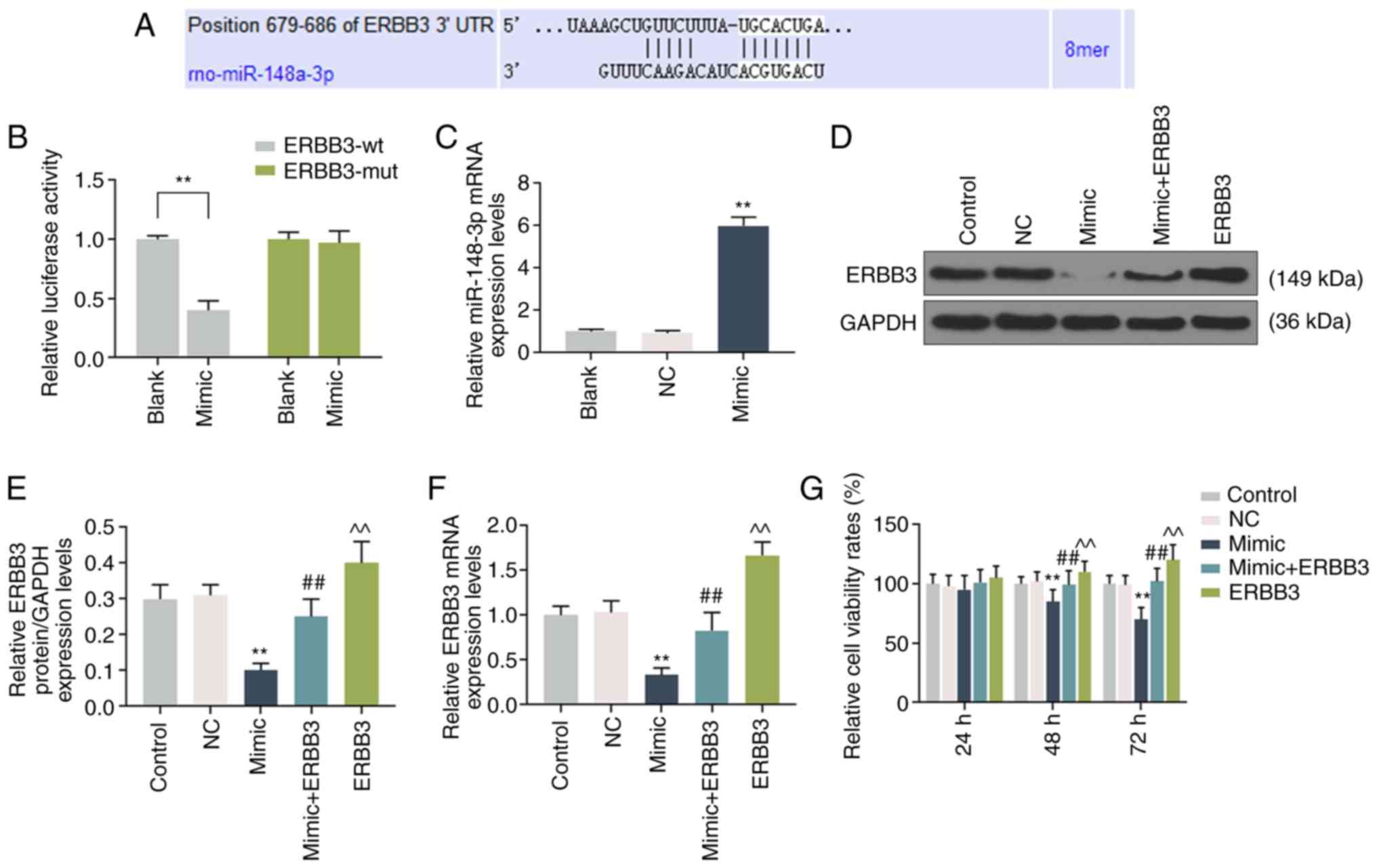
ERBB3 directly targets miR-148a-3p in
liver cells
TargetScan7.2 software (http://www.targetscan.org/vert_72/) predicted that
miR-148a-3p and ERBB3 shared binding sites (Fig. 4A). PmirGLO dual-luciferase
reporter vectors were constructed by respectively transfecting
ERBB3-WT and ERBB3-MUT with miR-148a-3p mimic into HSC-T6 cells,
and it was identified that the levels of luciferase activity in the
ERBB3-WT cells were significantly suppressed (P<0.001; Fig. 4B). In addition, the association
between ERBB3 and miR-148a-3p was explored. Firstly, the
miR-148a-3p mimic was transfected into HSC-T6 cells and the
transfection efficiency was assessed using RT-qPCR (Fig. 4C). Next, cells were transfected
with miR-148a-3p mimics, ERBB3 overex-pression plasmids or
co-transfected with miR-148a-3p mimics and ERBB3 overexpression
plasmids. It was identified that the expression of ERBB3 decreased
noticeably in HSC-T6 cells transfected with miR-148a-3p mimic, but
was significantly increased when the mimic + ERBB3 was transfected
into the cells (P<0.001; Fig.
4D-F).
Effects of miR-148a-3p on the viability
and apoptosis of liver cells by targeting ERBB3
The cells were transfected with miR-148a-3p mimic
and ERBB3 overexpression plasmids for 48 h. The data demonstrated
that the cell viability was decreased significantly in cells
transfected with the mimic, but markedly increased following
transfection with the ERBB3 plasmid. However, the mimic inhibited
the effect of the ERBB3 plasmids on the promotion of cell viability
(P<0.001; Fig. 4G). Similarly,
transfection with the mimic reversed the effect of ERBB3 on
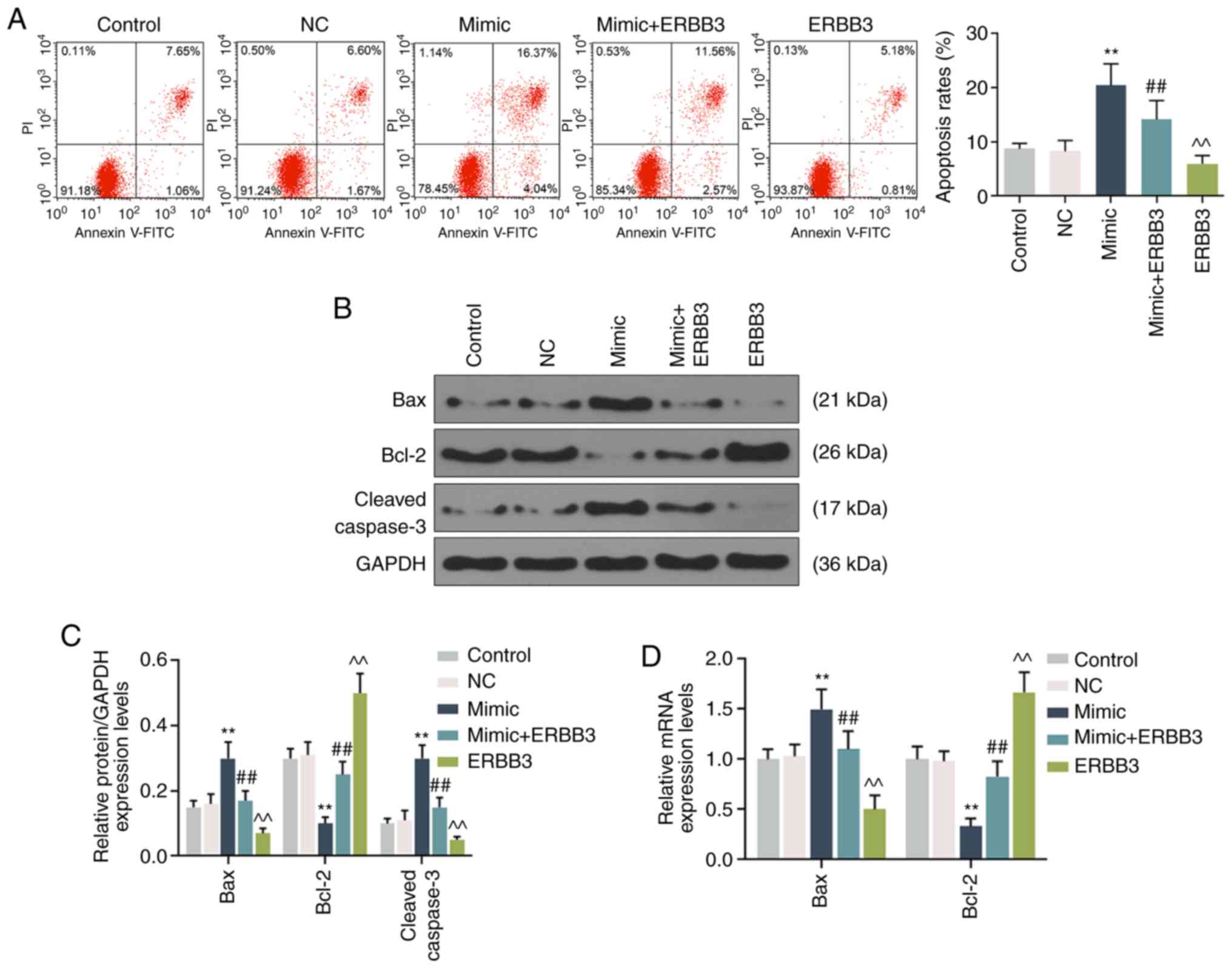
inhibiting cell apoptosis (P<0.001; Fig. 5A). Concomitantly, the Bcl-2
expression level was observed to be significantly decreased, but
the expression levels of Bax and c-cleaved-3 were markedly
increased in the cells treated by miR-148a-3p. Moreover, following
ERBB3 transfection, the expression level of these
apoptosis-associated proteins showed the opposite results, however,
mimic transfection reversed the effect of ERBB3 on the expression
levels of these proteins (P<0.001; Fig. 5B-D).
Discussion
Since the majority of miRNAs involved in various
physiological and pathological processes are highly conserved, and
miRNA expression is tissue-specific, studying the characteristics
of miRNAs can provide novel insights into the detection and
treatment of certain associated diseases (36,37). Alcoholic liver disease, which is a
common condition affecting the digestive system, is characterized
by impaired liver function (38).
In previous studies, miRNAs have been identified to serve an
important role in cell differentiation and function of the liver;
therefore, the comprehensively study of miRNAs may also provide
novel insights into the pathogenesis of alcoholic liver disease in
humans (39,40). It has been demonstrated that genes
from the miR-148 family, for example, miR-148a (miR-148a-3p), serve
important roles in physiological liver function through promoting
the hepatospecific phenotype and inhibiting the invasiveness of
transformed cells (18,19). miR-148a-3p is also considered to
act as a tumor suppressor gene of hepatocel-lular carcinoma, and
has potential value in evaluating the prognosis of patients with
hepatocellular carcinoma (41).
In addition, miR-148a knockout promoted liver lipid metabolism in
mice (42). However, the role of
the miR-148 family in the regulation of alcoholic liver disease has
not been reported in detail, and therefore became the focus of the
present study.
Previous data revealed that the expression levels of
miR-132 and miR-155 in the liver and brain were significantly
increased following long-term consumption of alcohol in mice
(43). However, a study conducted
on the association between liver aging and miRNA expression
profiles in rats identified that miR-148a-3p expression was greatly
downregulated in the livers of aged rats (44). Therefore, the present study
further explored the association between miR-148a-3p and alcoholic
liver disease in rats. The rats were treated with miR-148-3p and
received alcohol gavage for 9 weeks. The results demonstrated that
the expression of miR-148a-3p in the livers of the alcohol-treated
rats was significantly decreased; moreover, the miR-148a-3p
expression levels in rats treated with miR-148a-3p mimics were
promoted. These results were consistent with those from the study
by Mimura et al (44).
In previous years, the mechanisms of the involvement
of miRNAs in the pathological process of liver diseases have been
widely studied. For example, Bala et al (14) identified that miR-155 has a
certain protective effect on alcohol-treated mouse liver, and may
decreased oxidative stress and steatosis. miR-122 was also observed
to protect liver function by lowering AST and ALT levels, which are
used as markers of liver injury (45). Therefore, the present study
investigated the effect of miR-148a-3p on the liver pathology of
the rats with alcoholic liver disease. LDH, ALT, AST and ALP are
used as indicators to evaluate the degree of liver injury, as they
have high levels in various liver diseases (46-48). The present study identified that
miR-148a-3p inhibited the expression levels of AST, ALT, ALP and
LDH in the model. In addition, a higher liver index indicates a
larger amount of lipid deposition in the liver. In the present
study, following treatment with miR-148a-3p mimics in the rats, it
was identified that the liver indices of the rats treated with
ethanol were significantly decreased compared with the blank
control group, indicating the protective effect of miR-148a-3p in
the rat model.
Overconsumption of alcohol can increase levels of
hepatic fibrosis, which involves the increase of activated
fibroblasts or myofibroblasts when necrosis occurs in hepatocytes.
It is a tissue repair process observed in various chronic liver
injuries, and can result in cirrhosis in severe cases (49,50). Increased α-SMA and type I collagen
expression levels are important markers of liver fibrosis (51), and according to the results of the
present study, miR-148a-3p treatment markedly inhibited α-SMA and
type I collagen expression levels in the ALF rats, indicating that
miR-148a-3p can improve liver fibrosis caused by alcohol.
A previous study showed that mir-148a-3p can bind to
other target genes to regulate the progress of various diseases,
and miR-148a can inhibit the proliferation and migration of
pancreatic cancer cells by downregulating the expression of ERBB3
(52). miR-148a-3p in combination
with ERBB3 inhibited the proliferation of bladder cancer and
epithelial mesenchymal transition (21). In addition, ERBB3 overexpression
can lead to cell degeneration and the occurrence and development of
tumors, and is currently considered as a proto-oncogene with
important research and clinical application potential (53,54). In the present study, TargetScan7.2
software predicted that ERBB3 was a possible target gene of
miR-148a-3p, which was further confirmed by a luciferase reporter
assay. Furthermore, ERBB3 overexpression plasmids were transfected
with miR-148a-3p mimic into hepatic stellate HSC-T6 cells, and the
experimental results confirmed that the overexpression of
miR-148a-3p could inhibit the expression of ERBB3 in the hepatic
stellate cells.
The level of ERBB3 is closely associated with the
risk of developing liver cancer (55,56). The absence of ERBB3 in hepatocytes
affects the occurrence of liver cancer and can delay the formation
of liver tumors and cell proliferation (29). Besides that, it has been reported
that EGFR-ERBB3 signaling in hepatocytes is also required for the
activation of hepatic stellate cells and liver fibrosis (27). The present study identified that
ERBB3 can promote cell viability, inhibit cell apoptosis of
stellate cells, and that the expression of miR-148a-3p partially
reversed the effects of ERBB3 on promoting cell viability and
suppressing cell apoptosis, suggesting that miR-148a-3p can be
targeted by ERBB3 to regulate the activation of hepatic stellate
cells.
The present study identified a significant effect of
miR-148-a-3p on rat liver fibrosis. It was also observed that ERBB3
may be the downstream gene of miR-148-a-3p in hepatic stellate
cells. Nevertheless, the present study had a number of limitations,
due to funding limitations. Firstly, the effect of
miR-148-a-3p/ERBB3 signaling axis on hepatic stellate cell fibrosis
was not detected. Secondly, whether the miR-148-a-3p/ERBB3
signaling axis and associated function identified in HSC-T6 cells
are consistent with those in rats primary hepatic stellate cells
requires further exploration. Besides, whether the
miR-148-a-3p/ERBB3 signaling axis and its associated function
contribute to the liver fibrosis in vivo also requires
confirmation.
In conclusion, the present study revealed that
miR-148-a-3p regulated ALF, and the viability and apoptosis of
hepatic stellate cells through targeting ERBB3. However, since
in vivo experiments were only performed in rats in the
present study, further human sample collection is required to
verify the results.
Abbreviations:
|
LDH
|
lactate dehydrogenase
|
|
AST
|
aspartate aminotransferase
|
|
ALT
|
alanine transaminase
|
|
ALP
|
alkaline phosphatase
|
|
miRNA
|
microRNAs
|
|
RT-qPCR
|
reverse transcription-quantitative
PCR
|
|
NC
|
negative control
|
Acknowledgments
Not applicable.
Funding
The present study was supported by the National
Natural Science Foundation of China (grant nos. 81700509 and
81400663) and the Fundamental Research Funds for the Central
Universities (grant no. 22120180619).
Availability of data and materials
The analyzed data sets generated during the study
are available from the corresponding author on reasonable
request.
Authors' contributions
JX was responsible for substantial contributions to
the conception and design of the study, and agreed to be
accountable for all aspects of the work in ensuring that questions
related to the accuracy or integrity of the work are appropriately
investigated and resolved. JN was involved in data acquisition,
data analysis and interpretation. CC and KW were involved in
drafting the article or critically revising it for important
intellectual content. All authors provided final approval of the
version to be published.
Ethics approval and consent to
participate
The animal experimental protocol was approved by the
Tongji Hospital of Tongji University and performed in accordance
with the Guidance and Ethics Committee of Tongji Hospital, Tongji
University School of Medicine (approval no. THTU20160810A).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Ding L, Wo L, Du Z, Tang L, Song Z and Dou
X: Danshen protects against early-stage alcoholic liver disease in
mice via inducing PPARα activation and subsequent 4-HNE
degradation. PLoS One. 12:e01863572017. View Article : Google Scholar
|
|
2
|
Singal AK, Kamath PS, Gores GJ and Shah
VH: Alcoholic hepatitis: Current challenges and future directions.
Clin Gastroenterol Hepatol. 12:555–564; quiz e31-e32. 2014.
View Article : Google Scholar :
|
|
3
|
Gao B and Bataller R: Alcoholic liver
disease: Pathogenesis and new therapeutic targets.
Gastroenterology. 141:1572–1585. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Bataller R and Gao B: Liver fibrosis in
alcoholic liver disease. Semin Liver Dis. 35:146–156. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Voican CS, Louvet A, Trabut JB,
Njiké-Nakseu M, Dharancy S, Sanchez A, Corouge M, Lamouri K, Lebrun
A, Balian A, et al: Transient elastography alone and in combination
with FibroTest® for the diagnosis of hepatic fibrosis in
alcoholic liver disease. Liver Int. 37:1697–1705. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Roh YS and Seki E: Toll-like receptors in
alcoholic liver disease, non-alcoholic steatohepatitis and
carcinogenesis. J Gastroenterol Hepatol. 28(Suppl 1): S38–S42.
2013. View Article : Google Scholar
|
|
7
|
Devi BG, Henderson GI, Frosto TA and
Schenker S: Effect of ethanol on rat fetal hepatocytes: Studies on
cell replication, lipid peroxidation and glutathione. Hepatology.
18:648–659. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Olguín-Martínez M, Hernández-Espinosa DR
and Hernández- Muñoz R: High α-tocopherol dosing increases lipid
metabolism by changing redox state in damaged rat gastric mucosa
and liver after ethanol treatment. Clin Sci (Lond). 132:1257–1272.
2018. View Article : Google Scholar
|
|
9
|
Li YL, Wu J, Wei D, Zhang DW, Feng H, Chen
ZN and Bian H: Newcastle disease virus represses the activation of
human hepatic stellate cells and reverses the development of
hepatic fibrosis in mice. Liver Int. 29:593–602. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Higashi T, Friedman SL and Hoshida Y:
Hepatic stellate cells as key target in liver fibrosis. Adv Drug
Deliv Rev. 121:27–42. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Yu HX, Yao Y, Bu FT, Chen Y, Wu YT, Yang
Y, Chen X, Zhu Y, Wang Q, Pan XY, et al: Blockade of YAP alleviates
hepatic fibrosis through accelerating apoptosis and reversion of
activated hepatic stellate cells. Mol Immunol. 107:29–40. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Andueza A, Garde N, García-Garzón A,
Ansorena E, López-Zabalza MJ, Iraburu MJ, Zalba G and
Martínez-Irujo JJ: NADPH oxidase 5 promotes proliferation and
fibrosis in human hepatic stellate cells. Free Radic Biol Med.
126:15–26. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Hartmann P and Tacke F: Tiny RNA with
great effects: miR-155 in alcoholic liver disease. J Hepatol.
64:1214–1216. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Bala S, Csak T, Saha B, Zatsiorsky J,
Kodys K, Catalano D, Satishchandran A and Szabo G: The
pro-inflammatory effects of miR-155 promote liver fibrosis and
alcohol-induced steatohepa-titis. J Hepatol. 64:1378–1387. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Leti F, Malenica I, Doshi M, Courtright A,
Van Keuren-Jensen K, Legendre C, Still CD, Gerhard GS and DiStefano
JK: High-throughput sequencing reveals altered expression of
hepatic microRNAs in nonalcoholic fatty liver disease-related
fibrosis. Transl Res. 166:304–314. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Hayes J, Peruzzi PP and Lawler S:
MicroRNAs in cancer: Biomarkers, functions and therapy. Trends Mol
Med. 20:460–469. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Trionfini P and Benigni A: MicroRNAs as
master regulators of glomerular function in health and disease. J
Am Soc Nephrol. 28:1686–1696. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Gailhouste L, Gomez-Santos L, Hagiwara K,
Hatada I, Kitagawa N, Kawaharada K, Thirion M, Kosaka N, Takahashi
RU, Shibata T, et al: miR-148a plays a pivotal role in the liver by
promoting the hepatospecific phenotype and suppressing the
invasiveness of transformed cells. Hepatology. 58:1153–1165. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Jung KH, Zhang J, Zhou C, Shen H, Gagea M,
Rodriguez-Aguayo C, Lopez-Berestein G, Sood AK and Beretta L:
Differentiation therapy for hepatocellular carcinoma: Multifaceted
effects of miR-148a on tumor growth and phenotype and liver
fibrosis. Hepatology. 63:864–879. 2016. View Article : Google Scholar :
|
|
20
|
Bhattacharya S, Chalk AM, Ng AJ, Martin
TJ, Zannettino AC, Purton LE, Lu J, Baker EK and Walkley CR:
Increased miR-155-5p and reduced miR-148a-3p contribute to the
suppression of osteosarcoma cell death. Oncogene. 35:5282–5294.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Wang X, Liang Z, Xu X, Li J, Zhu Y, Meng
S, Li S, Wang S, Xie B, Ji A, et al: miR-148a-3p represses
proliferation and EMT by establishing regulatory circuits between
ERBB3/AKT2/ c-myc and DNMT1 in bladder cancer. Cell Death Dis.
7:e25032016. View Article : Google Scholar
|
|
22
|
Bellissimo T, Russo E, Ganci F, Vico C,
Sacconi A, Longo F, Vitolo D, Anile M, Disio D, Marino M, et al:
Circulating miR-21-5p and miR-148a-3p as emerging non-invasive
biomarkers in thymic epithelial tumors. Cancer Biol Ther. 17:79–82.
2016. View Article : Google Scholar :
|
|
23
|
Chen W, Zhao W, Yang A, Xu A, Wang H, Cong
M, Liu T, Wang P and You H: Integrated analysis of microRNA and
gene expression profiles reveals a functional regulatory module
associated with liver fibrosis. Gene. 636:87–95. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Li Z, Wang J, Zeng Q, Hu C, Zhang J, Wang
H, Yan J, Li H and Yu Z: Long noncoding RNA HOTTIP promotes mouse
hepatic stellate cell activation via downregulating miR-148a. Cell
Physiol Biochem. 51:2814–2828. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zhu J, Luo Z, Pan Y, Zheng W, Li W, Zhang
Z, Xiong P, Xu D, Du M, Wang B, et al: H19/miR-148a/USP4 axis
facilitates liver fibrosis by enhancing TGF-β signaling in both
hepatic stellate cells and hepatocytes. J Cell Physiol.
234:9698–9710. 2019. View Article : Google Scholar
|
|
26
|
Liu XY, He YJ, Yang QH, Huang W, Liu ZH,
Ye GR, Tang SH and Shu JC: Induction of autophagy and apoptosis by
miR-148a through the sonic hedgehog signaling pathway in hepatic
stellate cells. Am J Cancer Res. 5:2569–2589. 2015.PubMed/NCBI
|
|
27
|
Scheving LA, Zhang X, Threadgill DW and
Russell WE: Hepatocyte ERBB3 and EGFR are required for maximal
CCl4-induced liver fibrosis. Am J Physiol Gastrointest Liver
Physiol. 311:G807–G816. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Cao K, Gong H, Qiu Z, Wen Q, Zhang B, Tang
T, Zhou X, Cao T, Wang B, Shi H and Wang R: Hepatitis B virus X
protein reduces the stability of Nrdp1 to up-regulate ErbB3 in
hepatocellular carcinoma cells. Tumour Biol. 37:10375–10382. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Scheving LA, Zhang X, Stevenson MC,
Weintraub MA, Abbasi A, Clarke AM, Threadgill DW and Russell WE:
Loss of hepatocyte ERBB3 but not EGFR impairs
hepatocarcinogen-esis. Am J Physiol Gastrointest Liver Physiol.
309:G942–G954. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Li G, Zhang W, Gong L and Huang X:
MicroRNA 125a-5p inhibits cell proliferation and induces apoptosis
in hepatitis B virus-related hepatocellular carcinoma by
downregulation of ErbB3. Oncol Res. 27:449–458. 2019. View Article : Google Scholar
|
|
31
|
Shi J: Regulatory networks between
neurotrophins and miRNAs in brain diseases and cancers. Acta
Pharmacol Sin. 36:149–157. 2015. View Article : Google Scholar :
|
|
32
|
Agarwal V, Bell GW, Nam JW and Bartel DP:
Predicting effective microRNA target sites in mammalian mRNAs.
Elife. 4:e050052015. View Article : Google Scholar :
|
|
33
|
Ni YH, Huo LJ and Li TT: Antioxidant axis
Nrf2-keap1-ARE in inhibition of alcoholic liver fibrosis by IL-22.
World J Gastroenterol. 23:2002–2011. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Ma X, Luo Q, Zhu H, Liu X, Dong Z, Zhang
K, Zou Y, Wu J, Ge J and Sun A: Aldehyde dehydrogenase 2 activation
ameliorates CCl4-induced chronic liver fibrosis in mice
by up-regulating Nrf2/HO-1 antioxidant pathway. J Cell Mol Med.
22:3965–3978. 2018. View Article : Google Scholar :
|
|
35
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
36
|
Yuan DZ, Yu LL, Qu T, Zhang SM, Zhao YB,
Pan JL, Xu Q, He YP, Zhang JH and Yue LM: Identification and
characterization of progesterone- and estrogen-regulated MicroRNAs
in mouse endometrial epithelial cells. Reprod Sci. 22:223–234.
2015. View Article : Google Scholar :
|
|
37
|
Zhao F, Xu G, Zhou Y, Wang L, Xie J, Ren
S, Liu S and Zhu Y: MicroRNA-26b inhibits hepatitis B virus
transcription and replication by targeting the host factor CHORDC1
protein. J Biol Chem. 289:35029–35041. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Osaki K, Shimizu Y, Yamamoto T, Miyake F,
Kondo S and Yamaguchi H: Improvement of liver function by the
administration of oyster extract as a dietary supplement to
habitual alcohol drinkers: A pilot study. Exp Ther Med. 10:705–710.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Zhang Y, Huang F, Wang J, Peng L and Luo
H: MiR-15b mediates liver cancer cells proliferation through
targeting BCL-2. Int J Clin Exp Pathol. 8:15677–15683. 2015.
|
|
40
|
Roderburg C and Trautwein C: Cell-specific
functions of miRNA in the liver. J Hepatol. 66:655–656. 2017.
View Article : Google Scholar
|
|
41
|
Pan L, Huang S, He R, Rong M, Dang Y and
Chen G: Decreased expression and clinical significance of miR-148a
in hepatocellular carcinoma tissues. Eur J Med Res. 19:682014.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Cheng L, Zhu Y, Han H, Zhang Q, Cui K,
Shen H, Zhang J, Yan J, Prochownik E and Li Y: MicroRNA-148a
deficiency promotes hepatic lipid metabolism and
hepatocarcinogenesis in mice. Cell Death Dis. 8:e29162017.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Bala S and Szabo G: MicroRNA signature in
alcoholic liver disease. Int J Hepatol. 2012:4982322012. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Mimura S, Iwama H, Kato K, Nomura K,
Kobayashi M, Yoneyama H, Miyoshi H, Tani J, Morishita A, Himoto T,
et al: Profile of microRNAs associated with aging in rat liver. Int
J Mol Med. 34:1065–1072. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Roderburg C, Benz F, Vargas Cardenas D,
Koch A, Janssen J, Vucur M, Gautheron J, Schneider AT, Koppe C,
Kreggenwinkel K, et al: Elevated miR-122 serum levels are an
independent marker of liver injury in inflammatory diseases. Liver
Int. 35:1172–1184. 2015. View Article : Google Scholar
|
|
46
|
Lv Y, Zhang B, Xing G, Wang F and Hu Z:
Protective effect of naringenin against acetaminophen-induced acute
liver injury in metallothionein (MT)-null mice. Food Funct.
4:297–302. 2013. View Article : Google Scholar
|
|
47
|
Rahim SM, Taha EM, Al-janabi MS, Al-douri
BI, Simon KD and Mazlan AG: Hepatoprotective effect of cymbopogon
citratus aqueous extract against hydrogen peroxide-induced liver
injury in male rats. Afr J Tradit Complement Altern Med.
11:447–451. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Ferriero R, Nusco E, De Cegli R, Carissimo
A, Manco G and Brunetti-Pierri N: Pyruvate dehydrogenase complex
and lactate dehydrogenase are targets for therapy of acute liver
failure. J Hepatol. 69:325–335. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Oh Y, Park O, Swierczewska M, Hamilton JP,
Park JS, Kim TH, Lim SM, Eom H, Jo DG, Lee CE, et al: Systemic
PEGylated TRAIL treatment ameliorates liver cirrhosis in rats by
eliminating activated hepatic stellate cells. Hepatology.
64:209–223. 2016. View Article : Google Scholar :
|
|
50
|
Yang JA, Kong WH, Sung DK, Kim H, Kim TH,
Lee KC and Hahn SK: Hyaluronic acid-tumor necrosis factor-related
apoptosis-inducing ligand conjugate for targeted treatment of liver
fibrosis. Acta Biomater. 12:174–182. 2015. View Article : Google Scholar
|
|
51
|
Wang T, Wu D, Li P, Zhang K, Tao S, Li Z
and Li J: Effects of Taohongsiwu decoction on the expression of
α-SMA and TGF-β1 mRNA in the liver tissues of a rat model of
hepatic cirrhosis. Exp Ther Med. 14:1074–1080. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Feng H, Wang Y, Su J, Liang H, Zhang CY,
Chen X and Yao W: MicroRNA-148a suppresses the proliferation and
migration of pancreatic cancer cells by down-regulating ErbB3.
Pancreas. 45:1263–1271. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Zhang K, Wong P, Salvaggio C, Salhi A,
Osman I and Bedogni B: Synchronized targeting of Notch and ERBB
signaling suppresses melanoma tumor growth through inhibition of
Notch1 and ERBB3. J Invest Dermatol. 136:464–472. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Humtsoe JO, Pham E, Louie RJ, Chan DA and
Kramer RH: ErbB3 upregulation by the HNSCC 3D microenvironment
modulates cell survival and growth. Oncogene. 35:1554–1564. 2016.
View Article : Google Scholar
|
|
55
|
Sayagués JM, Corchete LA, Gutiérrez ML,
Sarasquete ME, Del Mar Abad M, Bengoechea O, Fermiñán E, Anduaga
MF, Del Carmen S, Iglesias M, et al: Genomic characterization of
liver metastases from colorectal cancer patients. Oncotarget.
7:72908–72922. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Shi DM, Li LX, Bian XY, Shi XJ, Lu LL,
Zhou HX, Pan TJ, Zhou J, Fan J and Wu WZ: miR-296-5p suppresses EMT
of hepatocellular carcinoma via attenuating NRG1/ERBB2/ERBB3
signaling. J Exp Clin Cancer Res. 37:2942018. View Article : Google Scholar : PubMed/NCBI
|