Introduction
Bladder cancer (BC) is one of the leading causes of
cancer-associated mortality worldwide, with a high incidence rate
(1). Alterations in DNA repair
pathwaya and signaling pathways, angiogenesis, genetic mutations
and hypoxia all contribute to the development of BC (2). Surgical resection, immunotherapy,
intravesical chemotherapy, and radical cystectomy with neoadjuvant
chemotherapy have been used in the treatment of non-muscle-invasive
or muscle-invasive BC (3).
However, minimal improvements have been made in the cure rates and
long-term survival of patients with BC over the past several
decades (4).
Sulforaphane (SFN) is a natural isothiocyanate
extracted from cruciferous plants. SFN has been used in the
treatment of diabetic cardiomyopathy (5), angiogenesis (6) and cancer (7). The main pathway regulated by SFN is
the Keap-1/nuclear factor erythroid 2-related factor 2 (Nrf2)
pathway (8). Research has found
that exposure to SF enables Nrf2 to escape from Keap1-dependent
degradation, leading to the stabilization of Nrf2, the increased
nuclear localization of Nrf2 and the activation of Nrf2-dependent
cancer-protective genes (9,10).
SFN also regulates cell apoptosis. For instance, it has been shown
that mouse embryonic fibroblasts (MEFs) treated with 40 mM SF
exhibit increased levels of Bax and Bak proteins at treatment for
4-8 h, although such effects are abolished in SF-treated MEFs
derived from Bax/Bak double-knockout mutant mice (11). It has been found that JNK/MAPK is
involved in SFN-mediated apoptosis (12,13). Of note, SFN also inhibits
apoptosis and cell cycle progression (14,15). Thus, the complex function and
mechanisms of SFN warrant further investigation. A previous study
demonstrated that SFN suppressed the development and decreased the
risk of developing BC. For instance, SFN combined with carbonic
anhydrase acetazolamide inhibits tumor growth (16). SFN has also been shown to induce
the apoptosis and cell cycle arrest of BC cells via the Nrf2
pathway and ROS-dependent pathway (17,18). However, the mechanisms of action
of SFN in BC are not yet fully understood.
FAT atypical cadherin 1 (FAT1) is an atypical
cadherin, and plays a role in a variety of human cancers. More
specifically, FAT1 suppresses epithelial-mesenchymal transition
(EMT), which plays an important role in tumor metastasis (19). FAT1 knockdown in hypoxic
glioblastoma cells has been shown to significantly decrease the
expression levels of EMT/stemness markers (20). FAT1 however, has also been shown
to inhibits the migration and invasion of esophageal squamous cell
carcinoma cells (21). It has
also been demonstrated that FAT1 significantly enhances the
migratory and invasive properties of glioma cells (22). Moreover, protein-inactivating
mutations in FAT1 have been detected in BC by whole-genome
sequencing (23). However, the
function of FAT1 in BC has not been identified to date.
Therefore, the present study aimed to investigate
the roles of SFN and FAT1 in BC cells, in order to provide insight
into the mechanisms underlying the role of SFN in BC.
Materials and methods
Sample collection
In the present study, 85 bladder cancer tissues and
27 adjacent normal tissues were collected from patients with BC who
attended the Shenzhen Hospital of Southern Medical University from
2010 to 2014 for treatment. All the tissues were stored at −80°C.
The present study was reviewed and approved by the Committee for
Ethical Review of Research at Shenzhen Hospital of Southern Medical
University, and all the patients signed an informed consent. The
relative expression of FAT1 (high or low) was determined according
to the median expression of FAT1 in BC tissues.
Cell culture and treatment
The normal bladder cell line, SV-HUC-1 (CRL-9520),
and the BC cell lines, 5637 (HTB-9), T24 (HTB-4), J82 (HTB-1),
SW780 (CRL-2169) and UM-UC-3 (CRL-1749), were purchased from the
American Type Culture Collection (ATCC). The BC cell lines, 5637,
J82 and UM-UC-3, were cultured in MEM (12492013, Gibco; Thermo
Fisher Scientific, Inc.); SV-HUC-1 cells were cultured in Ham's
F-12K medium (21127030, Gibco; Thermo Fisher Scientific, Inc.); T24
cells were grown in McCoy's medium (16600082, Gibco; Thermo Fisher
Scientific, Inc.); SW780 cells were grown in RPMI-1640 medium
(31870082, Gibco; Thermo Fisher Scientific, Inc.). All the media
were supplemented with 10% FBS (10099141, Gibco; Thermo Fisher
Scientific, Inc.), 100 units/ml penicillin and 100 µg/ml
streptomycin (15140163, Gibco; Thermo Fisher Scientific, Inc.). The
cells were grown at 37°C with 5% CO2.
The T24 and SW780 cells were separately cultured in
a 24-well plates at a density of 3×105 (cell/ml) for 24
h. For the silencing FAT1 siRNA targeting FAT1 were used. the
sequences of the siRNAs were obtained from GenePharma and
demonstrated as follows: siFAT1 sense, 5′-GGG CCA GUC AAG UUU GAA
A-3′ and antisense, 5′-CCC GGU CAG UUC AAA CUU U-3′); and siNC as
the siRNA control sense, 5′-UUCUCC GAA CGU GUC ACG UTT-3′ and
antisense, 5′-ACG UGA CAC GUU CGG AGA ATT-3′. The empty pcDNA3.1
plasmid (NC) and FAT1-pcDNA3.1 plasmid (FAT1) were purchased from
Sigma-Aldrich; Merck KGaA. The medium in the 24-well plates was
then replaced after 24 h, and the T24 and SW780 cells were
separately transfected with 10 pmol siFAT1, siNC, NC, or FAT1 using
Lipofectamine 2000 transfection reagent (Thermo Fisher Scientific,
Inc.). The cells were harvested 24 h following transfection and
used in subsequent experiments. To examine the effects of SFN
(Sigma-Aldrich; Merck KGaA) on the BC cells, the T24 and SW780
cells were incubated with SFN (0, 10, 20, 40 and 80 µmol/l)
for 24 h at 37°C. The T24 and SW780 cells transfected with siNC,
siFAT1, NC or FAT1 overexpression plasmid were incubated with SFN
(20 µmol/l) for 24 h at 37°C.
Western blot analysis
The tissues were homogenized by bead milling for
approximately 5 min at 4°C, and the cells were treated with lysis
buffer for 30 min on ice. Proteins were then collected by
centrifugation at 1,000 × g, at 4°C for 30 min. Subsequently,
proteins (approximately 50 µg) were denatured, separated on
12% SDS/PAGE gels, and then transfected onto a PVDF membrane
(LC2002, Invitrogen; Thermo Fisher Scientific, Inc.). This was
followed by blocking of the membranes with 5% fat-free milk.
Primary antibodies to FAT-1 (1:2,000, ab190242, Abcam) and GAPDH
(1:2,000, ab8245, Abcam) were then separately incubated with the
membrane for >8 h at 4°C. Subsequently, anti-rabbit IgG antibody
(1:5,000, 7074, Cell Signaling Technology, Inc.) and anti-mouse IgG
antibody (31430, Thermo Fisher Scientific, Inc.) were incubated
with the membrane at room temperature for 2 h. Finally, the protein
signal of the membrane was detected using ECL Chemiluminescent
Substrate (WP20005, Thermo Fisher Scientific, Inc.) and analyzed
using an ImageQuant ECL Imager (28-9605-63, GE Healthcare).
Reverse transcription-quantitative PCR
(RT-qPCR)
Tissues were homogenized by bead milling with TRIzol
reagent for approximately 5 min at 4°C, and the cells were treated
with lysis buffer for 30 sec on ice. RNA was then isolated from the
tissues and cells at 4°C using chloroform and isopropanol.
Subsequently, 1 µg RNA was used for reverse transcription
with the PrimeScript™ II 1st Strand cDNA Synthesis kit (6210B,
Takara Bio, Inc.). SYBR®-Green PCR Master Mix (4312704,
ABI) and Bio-Rad CFX 96 Touch Real-Time PCR Detection System
(1855196, Bio-Rad Laboratories, Inc.) were used for qPCR analysis.
The parameters were set up as follows: 95°C for 5 min, 40 cycles of
95°C for 15 sec, 60°C for 30 sec, and 70°C for 10 sec. GAPDH served
as an internal control, and the relative gene expression levels
were calculated using the 2−ΔΔCq method (24). The sequences of the primers used
were as follows: FAT-1 forward, 5′-CAT CCT GTC AAG ATG GGT GTT T-3′
and reverse, 5′-TCC GAG AAT GTA CTC TTC AGC TT-3′; and GAPDH
forward, 5′-GGA GCG AGA TCC CTC CAA AAT-3′ and reverse, 5′-GGC TGT
TGT CAT ACT TCT CAT GG-3′.
MTT assay
Briefly, cells (4,000 cells/well) were seeded in a
96-well plate. Following culture for 48 h, the medium was removed
and 10 µl MTT solution mixed with 110 µl fresh medium
were added to each well. Following incubation at 37°C for 4 h, the
medium was removed, and 150 µl DMSO was added to each well.
The absorbance value at 570 nm was then detected using a microplate
reader (PLUS 384, Molecular Devices, LLC).
Wound-healing assay
A scratch was generated using a 10 µl pipette
tip when cells in a 6-well plate reached 80% confluence.
Subsequently, the cells were cultured in serum-free medium for 48
h. The scratch area was observed under a microscope (TS100, Nikon
Corp.) at 0 and 48 h. In total, 10 fields of the scratch area of
each group were selected to be observed under a microscope (TS100,
Nikon Corp.) for statistical analysis conducted by SPSS 19.0
software (SPSS, Inc.).
Transwell assay
The upper chamber of a Transwell plate (3428,
Corning, Inc.) was coated with Matrigel. The cells
(2×103 cells/well) were added to the upper chamber with
serum-free medium. Moreover, 600 µl normal medium with 20%
FBS was added to the lower chamber. Following incubation at 37°C
for 24 h, the invaded cells were fixed with methanol and stained
with 0.1% crystal violet (R40052, Thermo Fisher Scientific, Inc.)
at room temperature for approximately 30 min. Subsequently, 10
fields were randomly selected and the numbers of invaded cells were
counted under a microscope (TS100, Nikon). The assay was
independently repeated in triplicate, and the statistical analysis
was conducted using SPSS 19.0 software (SPSS, Inc.).
Flow cytometry
The Annexin-V kit (70-AP101-100-AVF, MultiSciences)
was used to detect cell apoptosis according to the instructions
provided with the kit. In brief, the cells (2×105 cells)
were cultured in a 6-well plate for 24 h. The cells were then
collected by trypsinization at 4°C for 2 min and centrifugation at
450 × g, 4°C for 5 min, and then washed with pre-cold PBS.
Subsequently, the cells were incubated with 300 µl binding
buffer, supplemented with 5 µl Annexin V-FITC and incubated
at room temperature for 20 min. PI (5 µl) was then incubated
with the cells at 4°C for 15 min to stain the nuclei. Finally, 200
µl binding buffer were incubated with the cells at room
temperature for 5 min. Cell apoptosis was detected using a
FACSCalibur flow cytometer (342973, BD Biosciences). The BD
FACSCanto™ system software v2.4 (646602, BD Biosciences) was used
for further analysis.
Statistical analysis
Data are presented as the means ± SD. One-way
analysis of variance followed by Tukey's multiple-comparison test
was used to analyze the statistical differences between groups
using SPSS 19.0 software (SPSS, Inc.). Kaplan-Meier analysis was
applied for survival analysis in the study, and the log-rank test
was used to calculate the P-values. P<0.05 was considered to
indicate a statistically significant difference.
Results
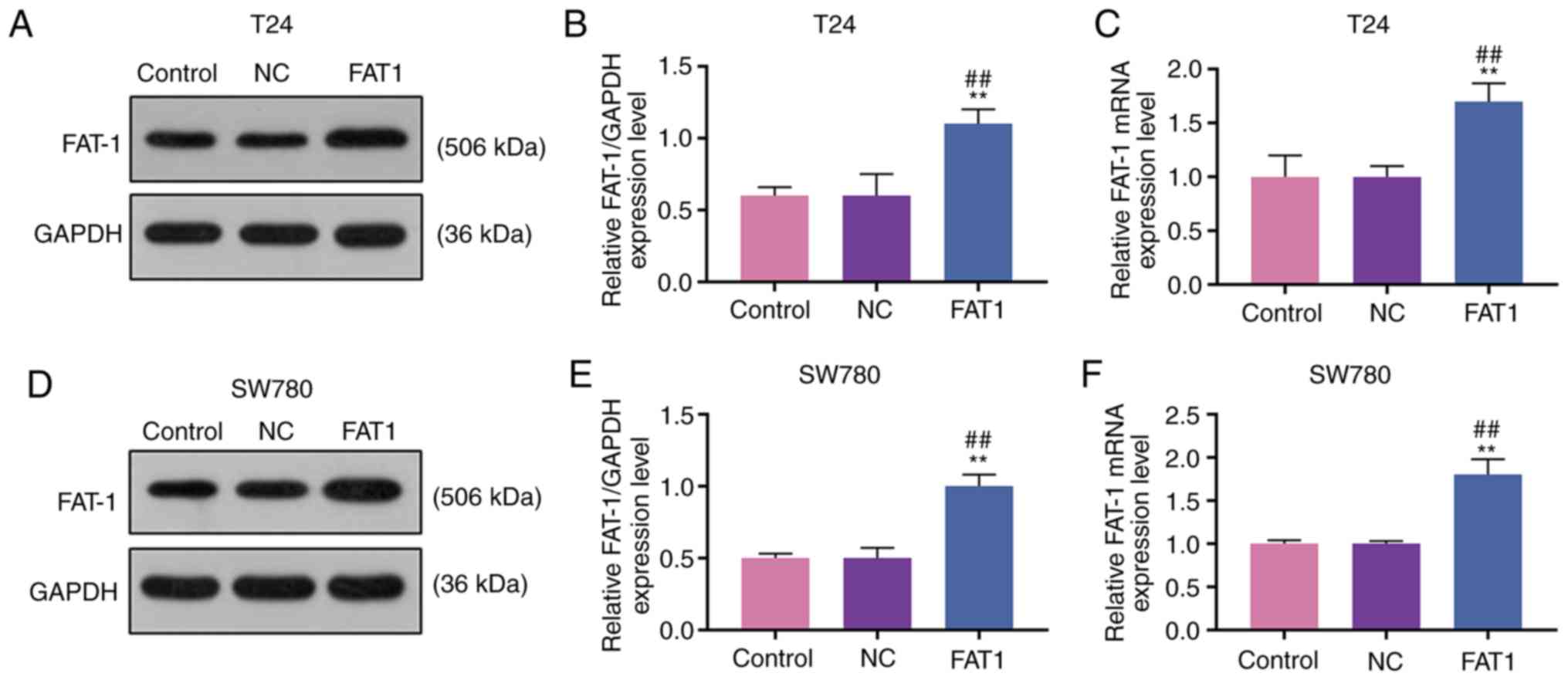
FAT-1 expression is upregulated in BC and
is associated with a lower survival rate of patients with BC
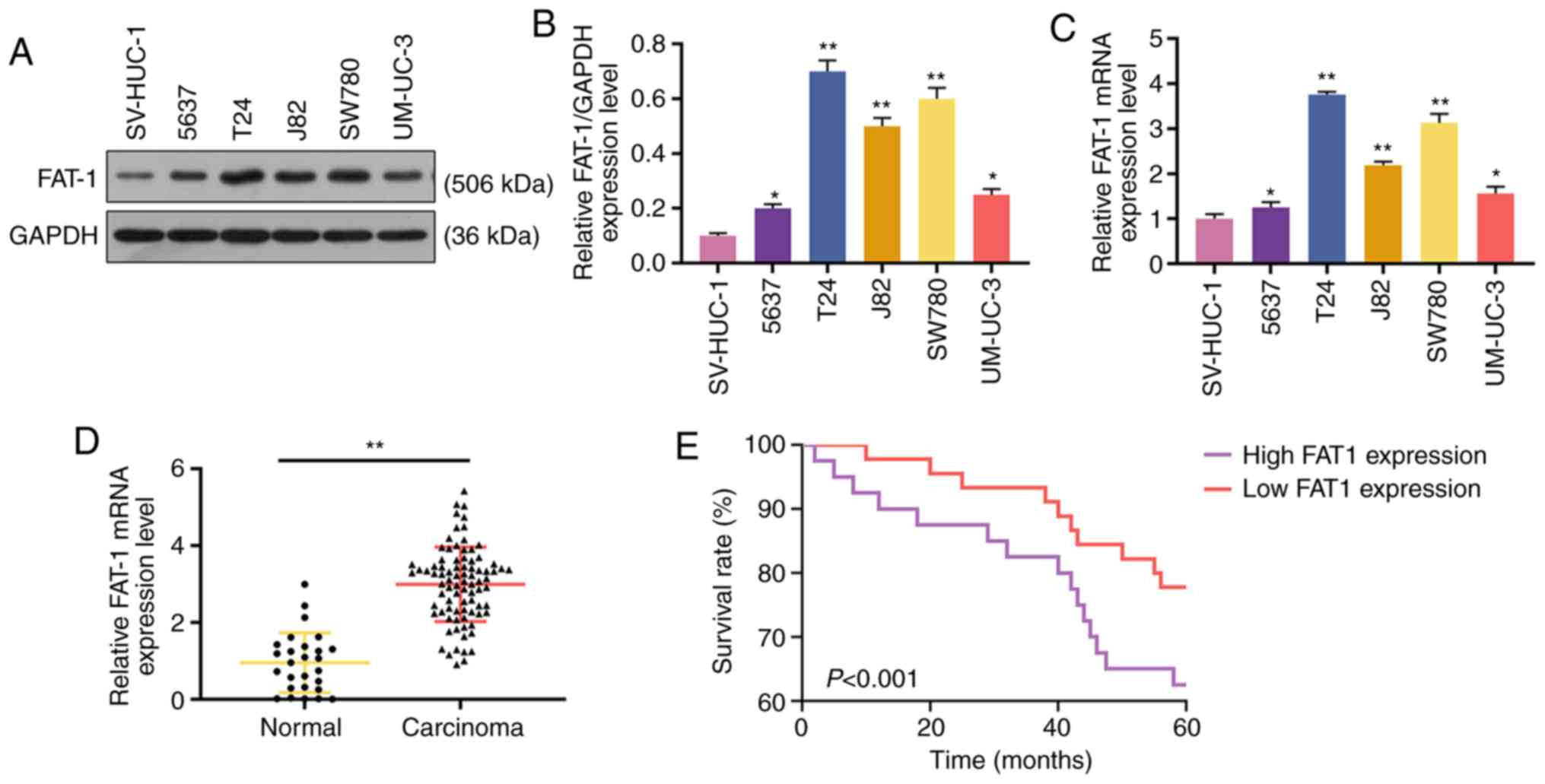
Western blot analysis and RT-qPCR were performed to
analyze the expression of FAT-1 in BC cells and tissues to explore
the role of FAT-1 in BC. The results revealed that FAT-1 was
significantly upregulated in BC cells and tissues (P<0.05 or
P<0.001, Fig. 1A-D). In
addition, patients with a relatively high expression (compared with
median expression) of FAT-1 exhibited a shorter 5-year survival
than those with a relatively low expression (compared with median
expression) of FAT-1 (P<0.001, Fig. 1E). As the levels of FAT-1 were
relatively higher in the T24 and SW780 BC cells, these two cell
lines were therefore used for further analysis.
SFN inhibits the viability and the
expression of FAT-1 in BC cells
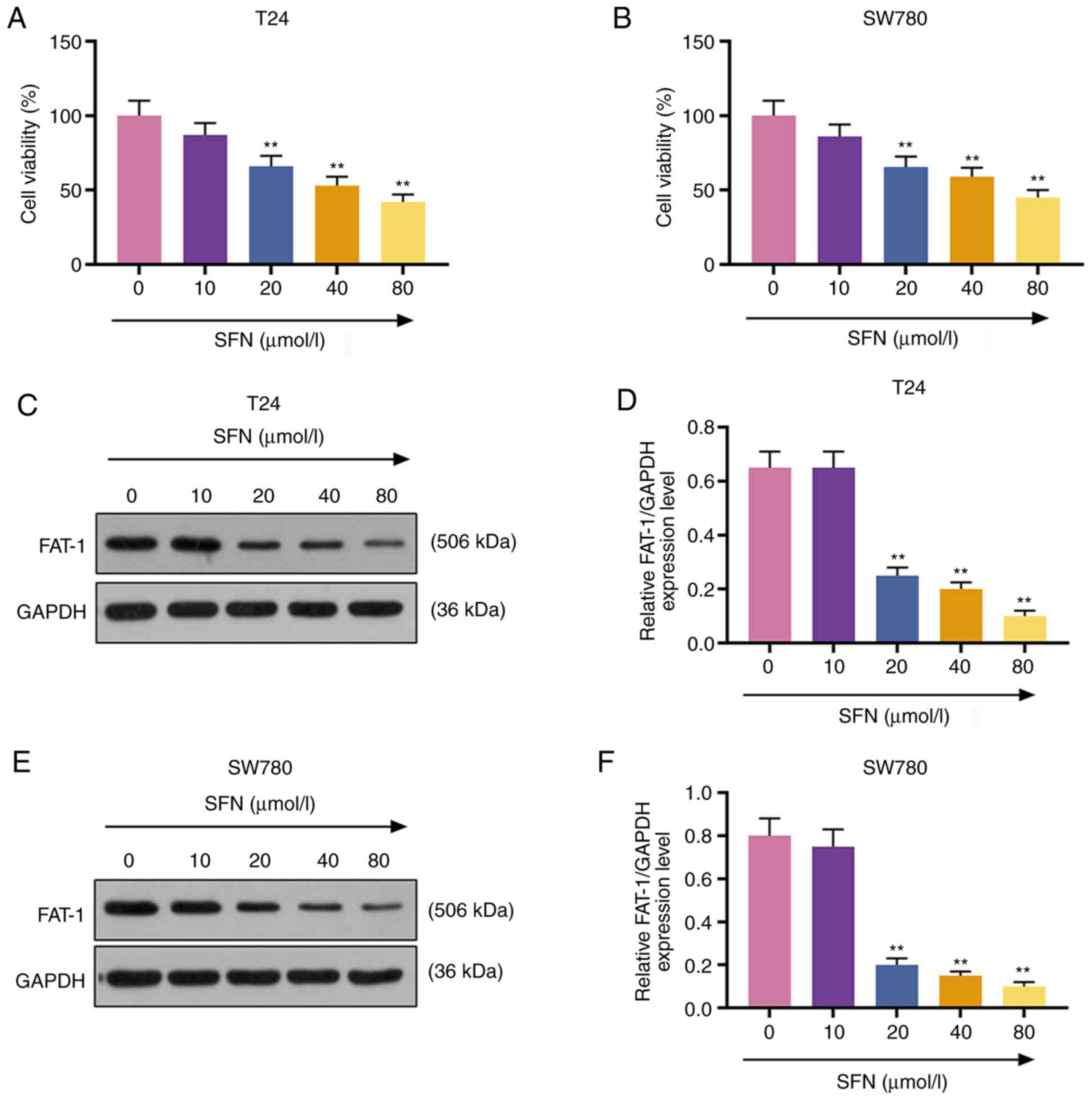
To examine the effects of SFN on the development of
BC cells, T24 and SW780 BC cells were exposed to SFN at
concentrations of 0, 10, 20, 40 and 80 µmol/l for 24 h. Cell
viability was measured by MTT assay. The results demonstrated that
SFN at 20, 40 and 80 µmol/l exerted a suppressive effect on
the viability of the T24 and SW780 cells (P<0.001, Fig. 2A and B). It was also found that
FAT-1 expression in the T24 and SW780 cells exposed to SFN at 20,
40 and 80 µmol/l (P<0.001, Fig. 2C-F) was markedly decreased. SFN at
20 µmol/l was used in subsequent experiments.
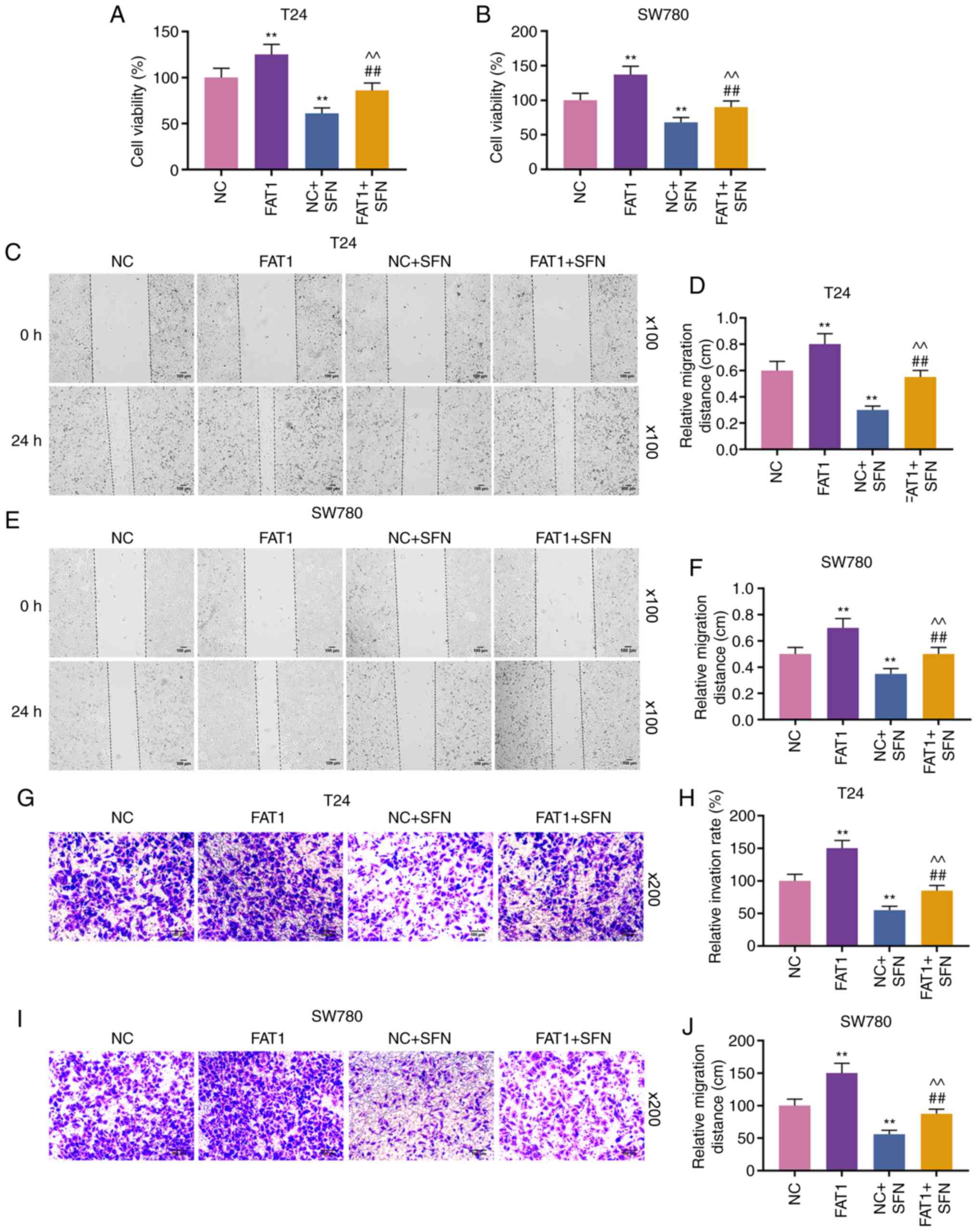
SFN inhibits the viability, migration and
invasion of BC cells by decreasing the expression of FAT-1
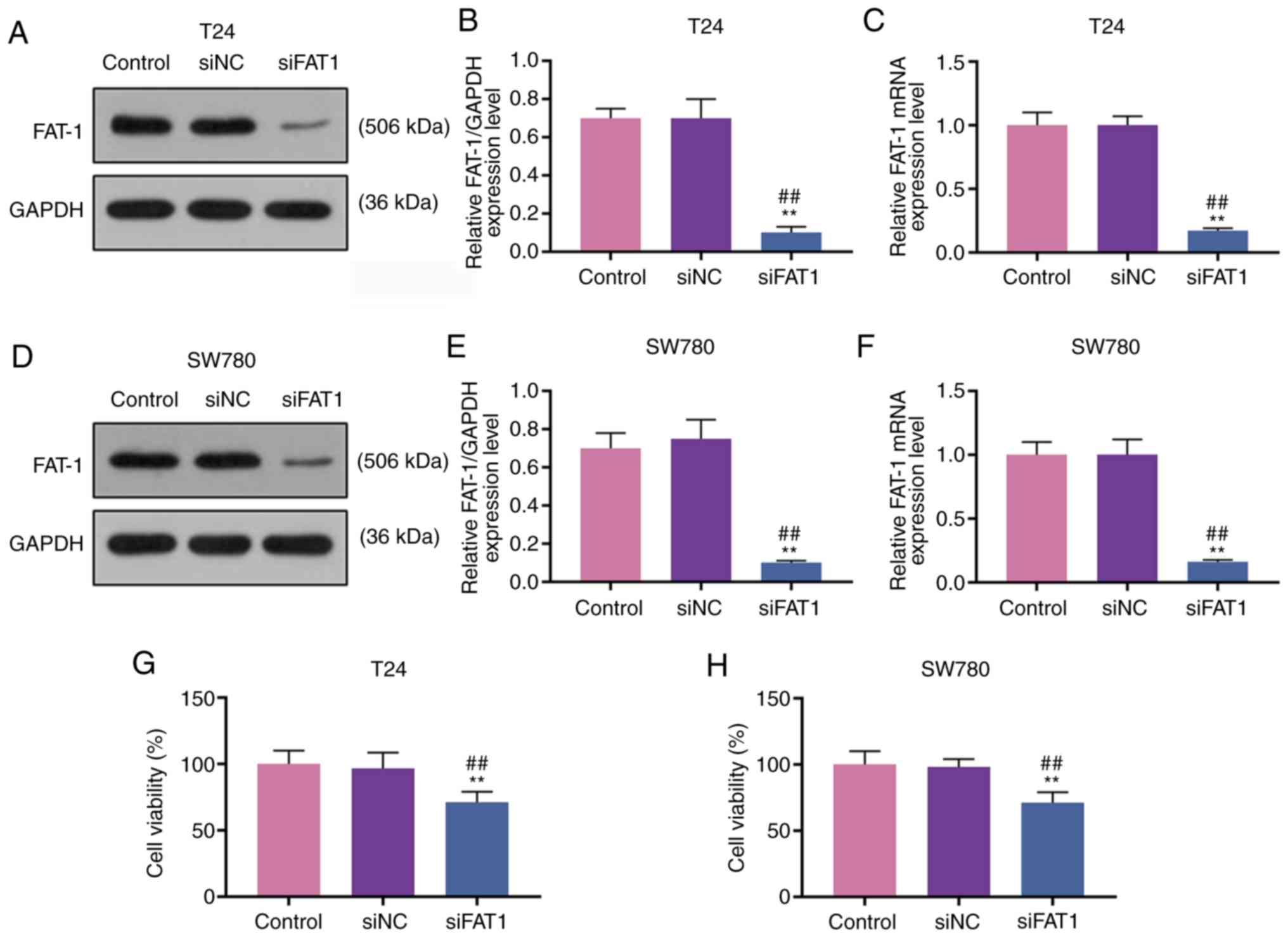
To explore the function of FAT-1 and SFN in BC, T24
and SW780 cells were transfected with siFAT-1 (P<0.001, Fig. 3A-F). MTT assay revealed that the
knockdown of FAT-1 inhibited the viability of the T24 and SW780
cells (P<0.001, Fig. 3G and
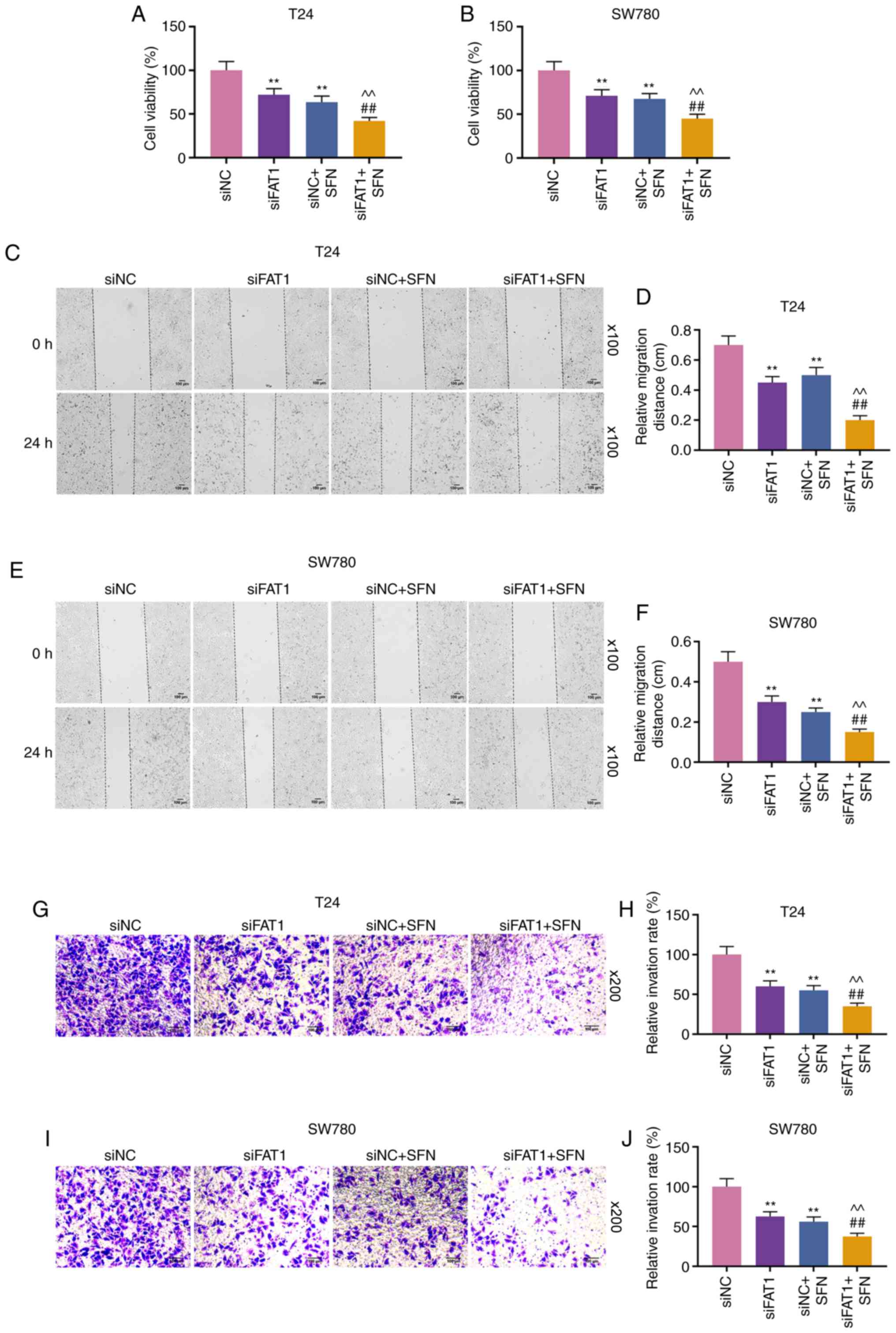
H). Compared with the results shown in Fig. 2, it could be observed that SFN
suppressed the viability, and the migratory and invasive abilities
of the two cell lines, while SFN combined with siFAT-1 enhanced
these effects on the above-mentioned cell behaviors (P<0.001,
Fig. 4). Although siFAT-1 only
slightly decreased viability, the difference was statistically
significant.
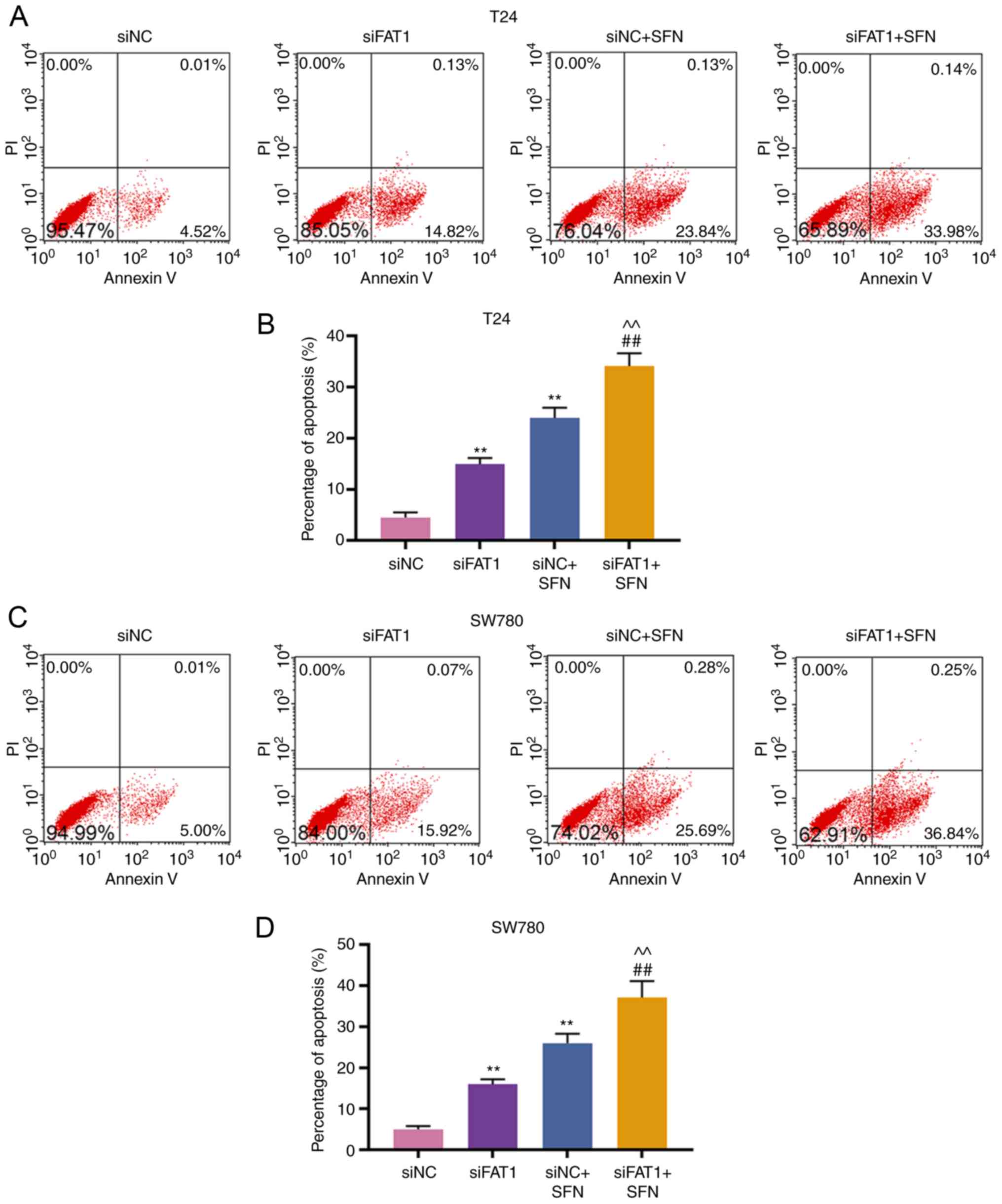
SFN promotes BC cell apoptosis by
inhibiting the expression of FAT-1
Subsequently, the apoptosis and FAT-1 expression in
T24 and SW780 cells transfected with siNC, siFAT-1, treated with
SFN, or co-treated with siFAT-1 and SFN were detected. The results
revealed that SFN alone or siFAT-1 induced cell apoptosis, and
inhibited the expression of FAT-1, as compared with the siNC group.
siFAT-1 combined with SFN treatment enhanced cell apoptosis and
markedly inhibited the expression of FAT-1 (P<0.001, Fig. 5).
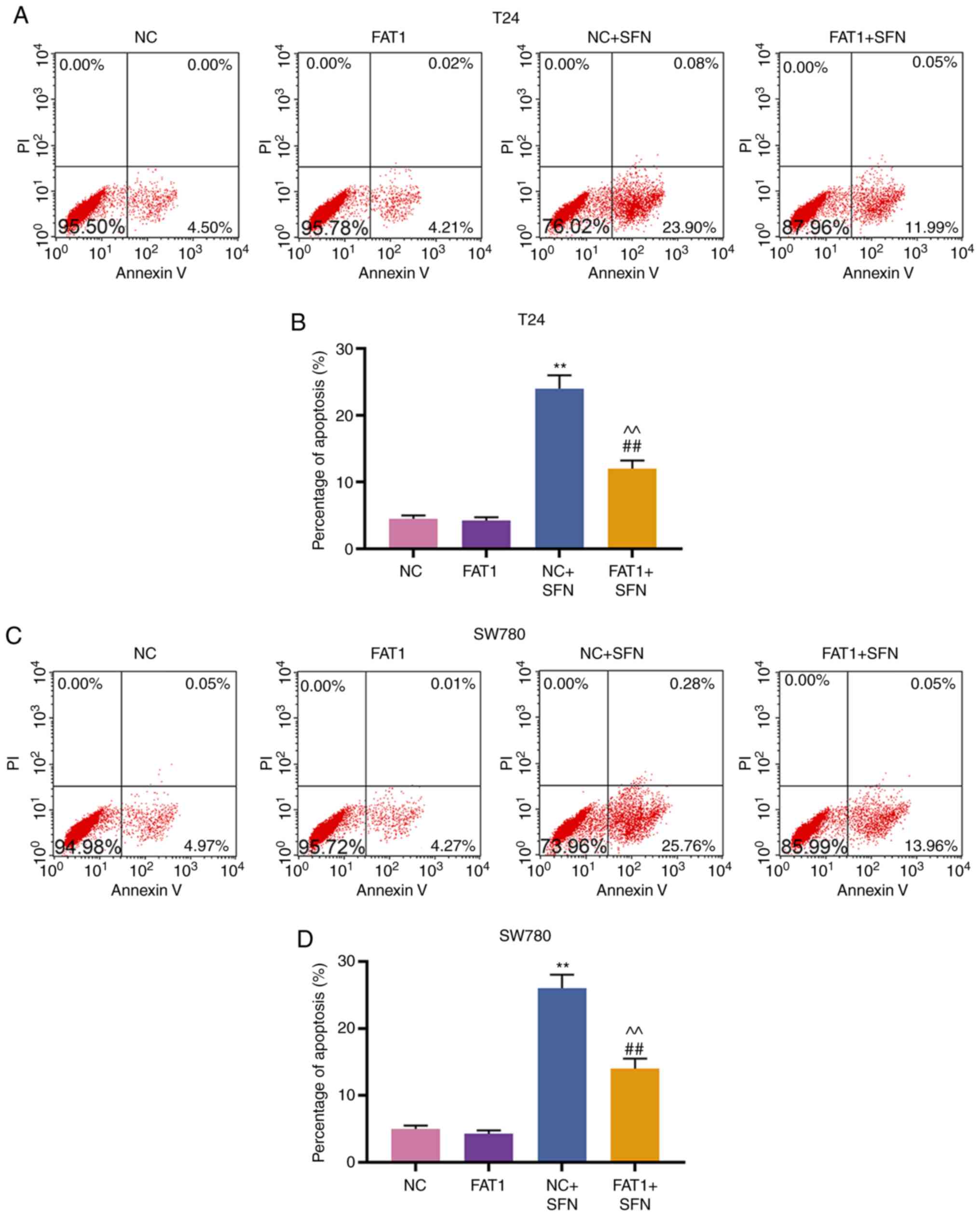
FAT-1 overexpression reverses the effects
of SFN on viability, metastasis and apoptosis of T24 and SW780
cells
To further confirm the association between SFN and
FAT-1, the FAT-1 overexpression plasmid, pc-FAT1, was transfected
into T24 and SW780 cells treated with or without SFN. As shown in
Fig. 6, FAT-1 was successfully
overexpressed in the T24 and SW780 cells (P<0.001).
Subsequently, cell behaviors were examined, and the results
revealed that FAT-1 overexpression enhanced the viability,
migration and invasion, and decreased the cell apoptotic rate of
the T24 and SW780 cells. However, SFN counteracted the promoting
effects of FAT-1 overexpression on the proliferation, migration and
invasion, and the suppressive effects on the apoptosis of the two
cell lines (P<0.001, Figs. 7
and 8A-D). The change in FAT-1
expression was then detected by western blot analysis and RT-qPCR.
It was observed that SFN blocked the expression of FAT-1 to a
modest, yet significant degree, even in comparison with pc-FAT-1
overexpression in T24 and SW780 cells (P<0.001, Fig. 8E-J).
Discussion
The overgrowth and metastasis of tumor cells are the
major cause of cancer recurrence and associated mortality (25). Therefore, effective antitumor
therapy should not only focus only on the inhibition of tumor cell
growth, but also on the prevention of metastasis. It has been
demonstrated that SFN exerts anti-carcinogenic effects on various
types of cancer, such as ovarian cancer (26), gastric cancer (27), lung cancer (28) and bladder cancer (18). However, the molecular mechanisms
of action of SFN in BC remain unknown. The present study found that
SFN exerted antitumor effects through the suppression of the
growth, migration and invasion, and the induction of apoptosis of
BC cells. Moreover, these effects of SFN were largely mediated by
the inhibition of the expression of FAT1.
FAT1 is considered a tumor suppressor in various
types of cancer. For instance, a low expression of FAT1 is
frequently observed in head and neck squamous cell carcinoma and
oral squamous cell carcinoma (29,30); antibody targeting FAT1 could
possibly be a novel therapeutic strategy for the treatment of
colorectal cancer (31); however,
FAT1 is upregulated in grade IV glioma cells, and is an upstream
regulator of oncogenic and inflammatory pathways (22). Moreover, glioblastoma cells in
which FAT1 is knocked down are more susceptible to death
receptor-mediated apoptosis (32). In BC, protein-inactivating
mutations in FAT1 have been previously identified (23). However, the present study found
that FAT1 was upregulated in BC and was associated with a low
5-year survival rate, suggesting that FAT1 may be involved in the
progression and prognosis of BC. Further experiments indicated that
the silencing of FAT1 suppressed the viability and metastasis of
T24 and SW780 cells, whereas the overexpression of FAT1 produced
opposite effects on the two cell lines. Rescue assays also
demonstrated that the apoptosis of the two cell lines was enhanced
by the knockdown of FAT1 in combination with SFN treatment, whereas
it was inhibited by the overexpression of FAT1 combined with SFN
stimulation. Thus, the current findings indicate that FAT1 may
function as an oncogenic driver in BC.
SFN is a natural product, and previous studies have
demonstrated that SFN exerts an antitumor effect on BC. For
instance, SFN has been shown to inhibit cell viability and induce
cell apoptosis in a dose-dependent manner, and such an effect is
involved in mitochondrial dysfunction (17). Moreover, SFN has been found to
regulate the metastasis and recurrence of BC by modulating EMT
(33). Consistent with the
findings of previous studies, the present study found that SFN
inhibited the viability, and suppressed the migration and invasion
of T24 and SW780 cells. Notably, it was also found that SFN
decreased the expression of FAT1 in a dose-dependent manner.
Further experiments found that BC cells exposed to SFN exhibited a
reduced viability, migration and invasion, and these effects were
enhanced by the silencing of FAT1 and were suppressed by FAT1
overexpression.
However, the molecular mechanisms of FAT1 in
development of bladder cancer were not investigated in the present
study. It has previously been demonstrated that FAT1 is involved in
regulating the Hippo pathway (34,35), the Wnt/β-catenin pathway (36) and the MAPK/ERK pathway (37); however, whether these pathways are
regulated in BC via FAT1 warrants further investigation. Moreover,
the function of FAT1 in BC was not examined by an in vivo
assay in the present study. It has been reported that FAT1-specific
monoclonal antibody mAb198.3 can suppress the growth of colon
cancer xenograft models; therefore, it can be developed as a
potential drug for colorectal cancer (38). However, whether the FAT1-specific
antibody can be applied to the treatment of BC remains unknown.
Additionally, the mechanisms through which SFN regulates FAT1
remain unknown and warrant further investigation. A previous study
demonstrated that SFN inhibits the invasion of glioblastoma cells
by increasing the expression levels of E-cadherin, and decreasing
the expression levels of MMP-2, MMP-9 and galectin-3 (39). Recently, a study found that SFN
plays an anticancer role by targeting multiple molecules and
pathways, such as Nrf2, histone deacetylases (HDACs),
poly(ADP-ribose) polymerase (PARP), Bcl-2 family, MAPKs,
hypoxia-inducible factor (HIF)1α and NF-κB (8). However, the mechanisms underlying
the regulation of FAT1 by SFN remain unclear. A limitation of the
present study is that clinical data from a TCGA BC dataset were not
compared. In addition, the association of other BC cadherins
related to cell interactions or organization, and apoptosis
related-factors were not investigated. Although the highest doses
of SFN may not be achievable in vivo, drugs or approaches to
tackle FAT-1 expression warrant further investigation.
In conclusion, the findings of the present study
demonstrate that FAT1 is upregulated in BC tissues and cells. A
high expression of FAT1 is predictive of a low 5-year survival rate
of patients with BC. Moreover, SFN exerts a suppressive effect on
the growth and metastasis of BC. Furthermore, the therapeutic
effect of SFN on BC is possibly mediated by FAT1. Thus, the current
findings provide a novel understanding of the role of SFN in
BC.
Abbreviations:
|
FAT1
|
FAT atypical cadherin 1
|
|
SFN
|
sulforaphane
|
|
BC
|
bladder cancer
|
|
MEFs
|
mouse embryonic fibroblasts
|
|
RT-qPCR
|
reverse transcription-quantitative
PCR
|
Acknowledgments
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
FW and PL made substantial contributions to the
conception and design of the study. PL and HA were involved in data
acquisition, data analysis and interpretation, as well as in
experiments including MTT, wound healing, and flow cytometry. FW
and YZ were involved in the drafting of the manuscript or
critically revising it for important intellectual content, as well
as in experiments including RT-qPCR and western blot analysis. All
authors read and approved the final manuscript. Agreement to be
accountable for all aspects of the work in ensuring that questions
related to the accuracy or integrity of the work are appropriately
investigated and resolved.
Ethics approval and consent to
participate
All procedures performed in studies involving human
participants were in accordance with the ethical standards of the
institutional and/or national research committee and with the 1964
Helsinki declaration and its later amendments or comparable ethical
standards. The present study was reviewed and approved by the
Committee For Ethical Review Of Research Involving Human Subjects
at Shenzhen Hospital of Southern Medical University and all
patients associated with the present study signed an informed
consent.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Antoni S, Ferlay J, Soerjomataram I, Znaor
A, Jemal A and Bray F: Bladder cancer incidence and mortality: A
global overview and recent trends. Eur Urol. 71:96–108. 2017.
View Article : Google Scholar
|
|
2
|
Robertson AG, Kim J, Al-Ahmadie H,
Bellmunt J, Guo G, Cherniack AD, Hinoue T, Laird PW, Hoadley KA,
Akbani R, et al: comprehensive molecular characterization of
muscle-invasive bladder cancer. Cell. 171:540–556.e25. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Kamat AM, Hahn NM, Efstathiou JA, Lerner
SP, Malmström PU, Choi W, Guo CC, Lotan Y and Kassouf W: Bladder
cancer. Lancet. 388:2796–2810. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Ebrahimi H, Amini E, Pishgar F, Moghaddam
SS, Nabavizadeh B, Rostamabadi Y, Aminorroaya A, Fitzmaurice C,
Farzadfar F, Nowroozi MR, et al: Global, regional and national
burden of bladder cancer, 1990 to 2016: Results from the GBD study
2016. J Urol. 201:893–901. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Bai Y, Cui W, Xin Y, Miao X, Barati MT,
Zhang C, Chen Q, Tan Y, Cui T, Zheng Y and Cai L: Prevention by
sulforaphane of diabetic cardiomyopathy is associated with
up-regulation of Nrf2 expression and transcription activation. J
Mol Cell Cardiol. 57:82–95. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wang G, Nie JH, Bao Y and Yang X:
Sulforaphane rescues ethanol-suppressed angiogenesis through
oxidative and endoplasmic reticulum stress in chick embryos. J
Agric Food Chem. 66:9522–9533. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Liu P, Atkinson SJ, Akbareian SE, Zhou Z,
Munsterberg A, Robinson SD and Bao Y: Sulforaphane exerts
anti-angiogenesis effects against hepatocellular carcinoma through
inhibition of STAT3/HIF-1α/VEGF signalling. Sci Rep. 7:126512017.
View Article : Google Scholar
|
|
8
|
Russo M, Spagnuolo C, Russo GL,
Skalicka-Woźniak K, Daglia M, Sobarzo-Sánchez E, Nabavi SF and
Nabavi SM: Nrf2 targeting by sulforaphane: A potential therapy for
cancer treatment. Crit Rev Food Sci Nutr. 58:1391–1405. 2018.
View Article : Google Scholar
|
|
9
|
McMahon M, Itoh K, Yamamoto M and Hayes
JD: Keap1-dependent proteasomal degradation of transcription factor
Nrf2 contributes to the negative regulation of antioxidant response
element-driven gene expression. J Biol Chem. 278:21592–21600. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Jeong WS, Keum YS, Chen C, Jain MR, Shen
G, Kim JH, Li W and Kong AN: Differential expression and stability
of endogenous nuclear factor E2-related factor 2 (Nrf2) by natural
chemopreventive compounds in HepG2 human hepatoma cells. J Biochem
Mol Biol. 38:167–176. 2005.PubMed/NCBI
|
|
11
|
Choi S and Singh SV: Bax and Bak are
required for apoptosis induction by sulforaphane, a cruciferous
vegetable-derived cancer chemopreventive agent. Cancer Res.
65:2035–2043. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Yeh CT and Yen GC: Effect of sulforaphane
on metallothionein expression and induction of apoptosis in human
hepatoma HepG2 cells. Carcinogenesis. 26:2138–2148. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Hu R, Kim BR, Chen C, Hebbar V and Kong
AN: The roles of JNK and apoptotic signaling pathways in
PEITC-mediated responses in human HT-29 colon adenocarcinoma cells.
Carcinogenesis. 24:1361–1367. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Liu ZM, Chen GG, Ng EK, Leung WK, Sung JJ
and Chung SC: Upregulation of heme oxygenase-1 and p21 confers
resistance to apoptosis in human gastric cancer cells. Oncogene.
23:503–513. 2004. View Article : Google Scholar
|
|
15
|
Gamet-Payrastre L, Lumeau S, Gasc N,
Cassar G, Rollin P and Tulliez J: Selective cytostatic and
cytotoxic effects of glucosinolates hydrolysis products on human
colon cancer cells in vitro. Anticancer Drugs. 9:141–148. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Islam SS, Mokhtari RB, Akbari P, Hatina J,
Yeger H and Farhat WA: Simultaneous targeting of bladder tumor
growth, survival, and epithelial-to-mesenchymal transition with a
novel therapeutic combination of acetazolamide (AZ) and
sulforaphane (SFN). Target Onco. 11:209–227. 2016. View Article : Google Scholar
|
|
17
|
Jo GH, Kim GY, Kim WJ, Park KY and Choi
YH: Sulforaphane induces apoptosis in T24 human urinary bladder
cancer cells through a reactive oxygen species-mediated
mitochondrial pathway: the involvement of endoplasmic reticulum
stress and the Nrf2 signaling pathway. Int J Oncol. 45:1497–1506.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Park HS, Han MH, Kim GY, Moon SK, Kim WJ,
Hwang HJ, Park KY and Choi YH: Sulforaphane induces reactive oxygen
species-mediated mitotic arrest and subsequent apoptosis in human
bladder cancer 5637 cells. Food Chem Toxicol. 64:157–165. 2014.
View Article : Google Scholar
|
|
19
|
Wang S, Zhao X, Yang S, Chen B and Shi J:
Salidroside alleviates high glucose-induced oxidative stress and
extracellular matrix accumulation in rat glomerular mesangial cells
by the TXNIP-NLRP3 inflammasome pathway. Chem Biol Interact.
278:48–53. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Srivastava C, Irshad K, Dikshit B,
Chattopadhyay P, Sarkar C, Gupta DK, Sinha S and Chosdol K: FAT1
modulates EMT and stemness genes expression in hypoxic
glioblastoma. Int J Cancer. 142:805–812. 2018. View Article : Google Scholar
|
|
21
|
Hu X, Zhai Y, Shi R, Qian Y, Cui H, Yang
J, Bi Y, Yan T, Yang J, Ma Y, et al: FAT1 inhibits cell migration
and invasion by affecting cellular mechanical properties in
esophageal squamous cell carcinoma. Oncol Rep. 39:2136–2146.
2018.PubMed/NCBI
|
|
22
|
Dikshit B, Irshad K, Madan E, Aggarwal N,
Sarkar C, Chandra PS, Gupta DK, Chattopadhyay P, Sinha S and
Chosdol K: FAT1 acts as an upstream regulator of oncogenic and
inflammatory pathways, via PDCD4, in glioma cells. Oncogene.
32:3798–3808. 2013. View Article : Google Scholar
|
|
23
|
Cazier JB, Rao SR, McLean CM, Walker AK,
Wright BJ, Jaeger EE, Kartsonaki C, Marsden L, Yau C, Camps C, et
al: Whole-genome sequencing of bladder cancers reveals somatic
CDKN1A mutations and clinicopathological associations with mutation
burden. Nat Commun. 5:37562014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
25
|
Ranjit M, Motomura K, Ohka F, Wakabayashi
T and Natsume A: Applicable advances in the molecular pathology of
glioblastoma. Brain Tumor Pathol. 32:153–162. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Kan SF, Wang J and Sun GX: Sulforaphane
regulates apoptosis- and proliferation-related signaling pathways
and synergizes with cisplatin to suppress human ovarian cancer. Int
J Mol Med. 42:2447–2458. 2018.PubMed/NCBI
|
|
27
|
Choi YH: ROS-mediated activation of AMPK
plays a critical role in sulforaphane-induced apoptosis and mitotic
arrest in AGS human gastric cancer cells. Gen Physiol Biophys.
37:129–140. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Wang DX, Zou YJ, Zhuang XB, Chen SX, Lin
Y, Li WL, Lin JJ and In ZQ: Sulforaphane suppresses EMT and
metastasis in human lung cancer through miR-616-5p-mediated
GSK3β/β-catenin signaling pathways. Acta Pharmacol Sin. 38:241–251.
2017. View Article : Google Scholar
|
|
29
|
Lin SC, Lin LH, Yu SY, Kao SY, Chang KW,
Cheng HW and Liu CJ: FAT1 somatic mutations in head and neck
carcinoma are associated with tumor progression and survival.
Carcinogenesis. 39:1320–1330. 2018.PubMed/NCBI
|
|
30
|
Hayes TF, Benaich N, Goldie SJ, Sipilä K,
Ames-Draycott A, Cai W, Yin G and Watt FM: Integrative genomic and
functional analysis of human oral squamous cell carcinoma cell
lines reveals synergistic effects of FAT1 and CASP8 inactivation.
Cancer Lett. 383:106–114. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Pileri P, Campagnoli S, Grandi A, Parri M,
De Camilli E, Song C, Ganfini L, Lacombe A, Naldi I, Sarmientos P,
et al: FAT1: A potential target for monoclonal antibody therapy in
colon cancer. Br J Cancer. 115:40–51. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Kranz D and Boutros M: A synthetic lethal
screen identifies FAT1 as an antagonist of caspase-8 in extrinsic
apoptosis. EMBO J. 33:181–197. 2014.PubMed/NCBI
|
|
33
|
Shan Y, Zhang L, Bao Y, Li B, He C, Gao M,
Feng X, Xu W, Zhang X and Wang S: Epithelial-mesenchymal
transition, a novel target of sulforaphane via COX-2/MMP2, 9/Snail,
ZEB1 and miR-200c/ZEB1 pathways in human bladder cancer cells. J
Nutr Biochem. 24:1062–1069. 2013. View Article : Google Scholar
|
|
34
|
Ahmed AF, de Bock CE, Sontag E,
Hondermarck H, Lincz LF and Thorne RF: FAT1 cadherin controls
neuritogenesis during NTera2 cell differentiation. Biochem Biophys
Res Commun. 514:625–631. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Martin D, Degese MS, Vitale-Cross L,
Iglesias-Bartolome R, Valera JLC, Wang Z, Feng X, Yeerna H, Vadmal
V, Moroishi T, et al: Assembly and activation of the Hippo
signalome by FAT1 tumor suppressor. Nat Commun. 9:23722018.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Alamoud KA and Kukuruzinska MA: Emerging
insights into Wnt/β-catenin signaling in head and neck cancer. J
Dent Res. 97:665–673. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Hu X, Zhai Y, Kong P, Cui H, Yan T, Yang
J, Qian Y, Ma Y, Wang F, Li H, et al: FAT1 prevents epithelial
mesenchymal transition (EMT) via MAPK/ERK signaling pathway in
esophageal squamous cell cancer. Cancer Lett. 397:83–93. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Grifantini R, Taranta M, Gherardini L,
Naldi I, Parri M, Grandi A, Giannetti A, Tombelli S, Lucarini G,
Ricotti L, et al: Magnetically driven drug delivery systems
improving targeted immunotherapy for colon-rectal cancer. J Control
Release. 280:76–86. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Zhang Z, Li C, Shang L, Zhang Y, Zou R,
Zhan Y and Bi B: Sulforaphane induces apoptosis and inhibits
invasion in U251MG glioblastoma cells. Springerplus. 5:2352016.
View Article : Google Scholar : PubMed/NCBI
|