Introduction
Leukemia is recognized as a fatal malignant clonal
disorder of hematopoietic multi-potent stem cells and immature
progenitors with increased leucocytes in human blood and bone
marrow (1,2). Benzene is an industrial chemical and
component of gasoline and it is commonly recognized that benzene
expo-sure in either occupational or environmental conditions is an
affirmative contributor to leukemia (3). Clonal hematopoiesis is confirmed as
an important risk factor for hematological cancers, and acquired
mutations may lead to the age distribution of acute myeloid
leukemia, while physical abnormalities have a greater influence on
acute lymphoblastic leukemia (1).
It has also been determined that a younger age, the female sex and
greater physical activities are associated with an evidently
reduced risk of developing chronic myeloid leukemia; however,
smoking and obesity are risk factors (4). Approximately 25% of patients
suffering from chronic lymphocytic leukemia will undergo secondary
autoimmune disorders, complicating the symptoms and treatment of
leukemia, decreasing their quality of life, and even endangering
their lives (5). Adriamycin (ADM)
is an antibiotic and is a widely used chemotherapeutic drug for the
treatment of solid tumors and hematological disorders (6). In particular, it is the most
effective drug for the treatment of childhood acute lymphoblastic
leukemia (7). Leukemic stem cells
have the ability of limitless self-renewal and possess potent
functions to maintain leukemia (8). Leukemic stem cells are being studied
as potential therapeutic targets, and the signaling pathways that
control their development and survival are of particular interest
(9).
MicroRNAs (miRNAs or miRs) are a family of small
non-coding RNAs, functioning as key post-transcriptional mediators
of genes involved in multiple fundamental processes, such as
differentiation, proliferation, apoptosis and cancer drug
resistance (10). Several miRNAs
participate in the differentiation of various hematopoietic cells,
and their dysregulation is clearly associated with cancer
development and particularly, with leukemia (11). The expression of miR-145 has been
to be markedly increased in normal hematopoietic progenitor cells
(12). A lower expression of
miR-145 was identified as an independent risk factor by Xia et
al and miR-145 overexpression was shown to suppress tumor cell
growth in adult T-cell leukemia/lymphoma cell lines (13). In the present study, through
bioinformatics prediction and dual-luciferase reporter gene assay,
it was found that miR-145 targeted adenosine triphosphate
(ATP)-binding cassette (ABC) transporter E1 (ABCE1) to inhibit its
expression. ABCE1 is a less extensively studied member of the ABC
multigene family and plays key roles in diverse biological events,
such as viral infection, cell proliferation and anti-apoptosis
(14). ABC transporters play
important roles in numerous disorders, particularly in acute
myeloid leukemia, while the overexpression of certain ABC members
in leukemic cells has a strong link with the poor outcome of
patients afflicted with acute myeloid leukemia (15). Based on the above-mentioned
information, it was hypothesized that miR-145 and ABCE1 may play a
role in the biological processes of leukemia and in cell
sensitivity to ADM.
Materials and methods
Cells and cell culture
The human leukemia cell line, K562, and
corresponding ADM-resistant cells, K562/ADM cells, were obtained
from the Kunming Cell Bank of Chinese Academy of Sciences and
cultured in Roswell Park Memorial Institute (RPMI)-1640 medium
(Gibco; Thermo Fisher Scientific, Inc.) with 10% fetal bovine serum
(FBS) (HyClone; GE Healthcare Life Sciences) in an incubator (37°C,
5% CO2). Cells were passaged once for 2-3 days with a
total of 3 passages. The corresponding K562/ADM cells were
continuously cultured in the above-mentioned medium containing 1.0
µg/ml ADM, and then cultured in normal medium 2 weeks before
the experiment.
Cell transfection and treatment
K562/ADM cells in good growing conditions (i.e.,
cells have good light transmittance, less granular matter in the
cytoplasm, smaller volume of the cytoplasm, compact shape of the
whole cell, and even cell growth without hypertrophy) were assigned
into the K562/ADM group, mimic-negative control (NC) group
(transfected with miR-145 mimic NC), miR-145 mimic group
(transfected with miR-145 mimic) (from Shanghai GenePharma Co.,
Ltd.), miR-145 mimic + overexpression (EO)-NC group (transfected
with miR-145 mimic and ABCE1 empty plasmid (Shanghai GenePharma
Co., Ltd.) and miR-145 mimic + ABCE1 group (transfected with
miR-145 mimic and ABCE1 overexpression plasmid). K562 cells in good
growing conditions (i.e., cells have good light transmittance, less
granular matter in the cytoplasm, smaller volume of the cytoplasm,
compact shape of the whole cell, and even cell growth without
hypertrophy) were assigned into the K562 group, inhibitor-NC group
(transfected with miR-145 inhibitor NC), and miR-145 inhibitor
group (transfected with miR-145 inhibitor). The inhibitor NC and
miR-145 inhibitor were provided by Shanghai GenePharma Co., Ltd.
Transfection was conducted using Lipofectamine® 2000
according to the instructions of the manufacturer (Invitrogen;
Thermo Fisher Scientific, Inc.). The final concentration of the
plasmid was 50 nM. miR-145 expression was measured by reverse
transcription-quantitative polymerase chain reaction (RT-qPCR) at
24 h following transfection to verify the transfection
efficiency.
RT-qPCR
Total RNA was extracted from tissues and cells using
TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.), and
ultraviolet analysis combined with formaldehyde denaturation
electrophoresis then confirmed the high quality of the extracted
RNA. RNA (1 µg) was reverse transcribed into cDNA using
avian myeloblastosis virus reverse transcriptase (Beijing Solarbio
Science & Technology Co., Ltd.). qPCR was conducted according
to SYBR-Green method (Thermo Fisher Scientific, Inc.) with U6 as
the internal reference of miR-145 and β-actin as the internal
reference of ABCE1. A NC with PCR products, but no primers was set.
PCR primers were devised and synthesized by Shanghai Sangon
Biotechnology Co., Ltd. (Table
I). The PCR system included cDNA 1.0 µl, 2X SYBR-Green
Mix 10 µl, Forward Primer (10 µM) 0.5 µl,
Reverse Primer (10 µM) 0.5 µl, and was supplemented
into 20 µl with the addition of RNase-free water. The
reaction conditions were as follows: 5 min pre-denaturation at
94°C, 40 cycles of 40 sec denaturation at 94°C, 40 sec annealing at
60°C, 60 sec extension at 72°C, and finally 10 min extension at
72°C. The products were tested by agarose gel electrophoresis. The
data were analyzed using the 2−ΔΔCq method (16), which indicated the multiple
association between the experimental group and the control group.
ΔΔCq=[Cq (target gene)-Cq (reference gene)]experimental
group-[Cq (target gene)-Cq (reference gene)]control
group. The normally cultured K562 and K562/ADM cells served
as the controls.
 | Table IPrimer sequences of RT-qPCR. |
Table I
Primer sequences of RT-qPCR.
| Gene | Sequences |
|---|
| miR-145 | F:
5′-GGCACTGCTGAAGGCATCTC-3′ |
| R:
5′-CTGTTAAGCCATGACCTCAAGAAC-3′ |
| U6 | F:
5′-CTCGCTTCGGCAGCACA-3′ |
| R:
5′-AACGCTTCACGAATTTGCGT-3′ |
| ABCE1 | F:
5′-CCAGGTGAAGTTTTGGGATTAG-3′ |
| R:
5′-AGGTTTGATGATGGCTTTTAGG-3′ |
| MRP1 | F:
5′-CATTCAGCTCGTCTTGTCCTG-3′ |
| R:
5′-GGATTAGGGTCGTGGATGGTT-3′ |
| β-actin | F:
5′-GGCATCACACTTTCTACAACG-3′ |
| R:
5′-GGCAGGAACATTAAAGGTTTC-3′ |
Western blot analysis
The RIPA lysis buffer (Beyotime Institute of
Biotechnology) was used to extract the proteins, and the extracted
proteins was determined based on the instructions of bicinchoninic
acid (BCA) kit (Wuhan Boster Biological Technology Co., Ltd.). The
extracted proteins were placed in loading buffer, boiled at 95°C
for 10 min, and loaded on each well (each for 30 µg), and
then separated by 6% sodium dodecyl sulfate (SDS) polyacrylamide
gel electrophoresis (PAGE) with the voltage changing from 80 to 120
v. The proteins were transferred onto a polyvinylidene fluoride
(PVDF) membrane by semi-dry transfer at 100 mv for 30-45 min. The
membrane was then sealed for 1 h in 5% bovine serum albumin (BSA)
at room temperature, followed by incubation with the following
primary antibodies (all from Abcam): ABCE1 (1 µg/ml,
ab32270), multidrug resistance protein 1 (MRP1) (1:500, ab32574),
P-glycoprotein (P-gp) (1:500, ab216656), and β-actin (1:5,000,
ab8227) at 4°C overnight. Subsequently, the membrane was rinsed
with Tris-buffered saline Tween (TBST) 3 times (5 min/time), and
then incubated for 1 h with the secondary antibody [horseradish
peroxidase labeled goat anti-rabbit IgG (H+L), ZB-2301, 1:2,500,
unconjugated; or horseradish peroxidase labeled goat anti-mouse IgG
(H+L), ZB-2305, 1:2,500, unconjugated; ZSGB-Bio Co., Ltd.] at room
temperature. Finally, the membrane was washed 3 times (5 min/time)
and developed by chemiluminescence reagent and bands were
visualized using the Bio-Rad Gel Dol EZ imager (Bio-Rad
Laboratories). The target band was analyzed using Image J software
(National Institutes of Health) for gray value analysis.
3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT)
colorimetric assay
After counting the number, cells were adjusted into
5×104 cells/l and 180 µl/well in a 96-well plate
and cultured in an incubator (37°C, 5% CO2) for 6 h
until cell adherence. K562 cells (concentrations of ADM used were
0.1, 0.2, 0.4, 0.6 and 0.8 µmol/ml) and K562/ADM cells
(concentrations of ADM used were 1, 2, 4, 6 and 8 µmol/ml)
were placed in RPMI-1640 medium (HyClone; GE Healthcare Life
Sciences), respectively for incubation in a 37°C incubator (Heal
Force Bio-Meditech Holdings Group) with 5% CO2 for 48 h,
with 3 duplicated wells set for each group. After discarding the
supernatant, 90 µl fresh medium and 20 µl of 5 g/l
MTT solution were added to the cells for 4 h of culture. Following
the removal of the supernatant, 150 µl dimethyl sulphoxide
(DMSO) solution (Beijing Solarbio Science & Technology Co.,
Ltd.) was added to each well, followed by shaking for 10 min at a
low speed on a shaking table. The optical density (OD) value was
detected at 490 nm wavelength using a microplate reader (Shenzhen
Rayto Life Science Co., Ltd.). The cell survival rate was
calculated as follows: Cell survival rate=(OD value in the
experiment group/mean OD value in the control group) ×100%
(17). The half maximal
inhibitory concentration (IC50) was calculated by
mid-efficacy analysis (Logit method) with SPSS 11.0 software (SPSS
Inc.) (18). The detection of the
proliferation of K562 cells overexpressing miR-145 and of the
K562/ADM cells with a low expression of miR-145 was performed as
described above.
Colony formation assay
Following treatment of the K562 cells
(concentrations of ADM used were 0.1, 0.2, 0.4, 0.6 and 0.8
µmol/ml) and K562/ADM cells (concentrations of ADM used were
1, 2, 4, 6 and 8 µmol/ml), and culture medium removal, the
adherent cells were then rinsed with phosphate-buffered saline
(PBS) once, and detached with 0.25% trypsin [without ethylene
diamine tetraacetic acid (EDTA)]. The cells were then triturated
into a single cell suspension and suspended in complete medium, and
200 cells/well were inoculated into a 6-well plate. The plate was
shaken gently, so that the cells were evenly dispersed and were
cultured for 1-2 weeks. When cell clones (>50) were visible to
the naked eye, following cultivation termination and culture medium
removal, the cells were washed 3 times with PBS and fixed 10 min
with anhydrous methanol. Subsequently, the cells were stained with
Giemsa solution (Sigma-Aldrich; Merck KGaA) for 15 min, and the
dying solution was washed away slowly with running water and the
cells were dried and photographed under a microscope (Leica,
AF6000, Leica Microsystems GmbH). The experiment was repeated 3
times. Under the microscope (low power microscope) (AF6000, Leica
Microsystems GmbH), the colonies with >10 cells were counted.
The data are expressed as the means ± standard deviation.
Annexin V-fluorescein isothiocyanate
(FITC)/propidium iodide (PI) assay
The ADM-treated K562 cells and K562/ADM cells, and
the K562 cells in which miR-145 was intervened with, and the
K562/ADM cells were detached with trypsin and collected to form a
single cell suspension. Following centrifugation (4°C, 300 × g, 5
min) and supernatant removal, cells were washed by pre-cooled PBS
twice, resuspended in the binding buffer and reacted with 5
µl Annexin V working fluid and 1 µl PI working fluid
(Sigma-Aldrich; Merck KGaA) at room temperature for 15 min.
Subsequently, the cells were mixed with 300 µl of binding
buffer gently. Approximately 10,000 cells were measured using a
flow cytometer (MoFloAstrios EQ, Beckman Coulter, Inc.), and the
percentage of AV+ (apoptotic cells) was then calculated.
The experiment was repeated 3 times, The apoptotic rate was
calculated by flow cytometry (FACS420, BD Biosciences). The data
are expressed as the mean ± standard deviation.
To investigate the effects of the overexpression of
miR-145 on the apoptosis of K562/ADM stem cells, approximately
2×108 K562/ADM cells were resuspended in 600 µl
magnetically activated cell separation (MACS) solution according to
the requirements of magnetic bead sorting. The cells were then
incubated with 200 µl Fc receptor (FcR) blocker to block FcR
on the cell surface, and 200 µl CD34+ Multisort
MicroBeads (Miltenyi Biotec) and 10 µl CD38-FITC antibodies
were incubated with the cells in a dark room at 4-8°C for 10 min.
Following centrifugation and supernatant removal, the cells were
resuspended in 500 µl MACS solution, and stem cells were
sorted and collected by magnetic bead separation column in a
magnetic field. Subsequently, the cells were resuspended with 1 ml
MACS solution, and cultured with 20 µl Multisort releasing
agent at 4°C for 10 min. Following centrifugation (300 × g, 4°C for
5 min) and supernatant removal, the cells were suspended again in
40 µl MACS solution, and cultivated with 60 µl MACS
Multisort terminating agent and 100 µl anti-FITC MicroBeads
at 4°C for 30 min. CD34+CD38 stem cells were obtained by
sorting with magnetic bead separation column in magnetic field. The
obtained CD34+CD38 stem cells were transfected with
miR-145 mimic for 24 h as described above, followed by apoptosis
detection. Following centrifugation removal (4°C, 400 × g, 5 min)
and supernatant, the cells were incubated with 100 µl
binding buffer, 1 µl Annexin V-FITC, and 2 µl PI at
4°C for 30 min. The cell apoptotic rate was measured using a flow
cytometer (FACS420, BD Biosciences). The experiment was repeated 3
times. The apoptotic rate was calculated by flow cytometry
(FACS420, BD Biosciences). The data are expressed as the means ±
standard deviation.
Acridine orange/ethidium bromide (AO/EB)
double fluorescence staining
K562 cells and K562/ADM cells in each group were
mixed with 2 µl AO/EB working fluid (Beijing Leagene
Biotechnology Co., Ltd.) gently in each group. Following
centrifugation at 4°C, 500 × g for 3 min with the supernatant
removed, the cells were resuspended with AO/EB buffer, and adjusted
to (0.5-5) ×106 cells/ml. The cells were then fully
mixed with 1 µl AO/EB working fluid. Clean slides were taken
and dripped with 5 µl cell suspension. The slides were then
lightly covered and cells were examined under a fluorescence
microscope [TS100 Nikon Instruments (Shanghai) Co., Ltd.]. The
experiment was repeated 3 times.
Dual luciferase reporter gene assay
Bioinformatics software and the http://www.microrna.org/ website were utilized to
predict the binding site of miR-145 to ABCE1, and the binding site
of miR-145 to MRP1 was examined through the website, http://www.targetscan.org/cgi-bin/targetscan/vert_71/view_gene.cgi?rs.
The 3′UTR sequences of ABCE1 and MRP1 containing the binding site
of miR-145 were synthesized and the wild-type (WT) plasmid of ABCE1
and MRP1 3′UTR (ABCE1-WT and MRP1-WT) was constructed. On the basis
of ABCE1-WT and MRP1-WT, the mutant type (MUT) plasmid of ABCE1
3′UTR (ABCE1-MUT) and MRP1 3′UTR (MRP1-MUT) was constructed by the
mutation binding site. The constructed plasmids were then mixed
with miR-145 mimic NC and miR-145 mimic plasmids, respectively, and
transfected into 293T cells (Cell Bank, Shanghai Institute of Life
Sciences, Chinese Academy of Sciences), cultured in DMEM containing
10% FBS, 1% Glutamax and 1% penicillin-streptomycin at 37°C with 3%
CO2 and 95% relative humidity. The cells were collected
and lysed using Passive lysis buffer (E1941, Promega Corp.) at 48 h
following transfection. Luciferase activity was measured using a
luciferase detection kit (BioVision, Inc.) and Glomax 20/20
luminometer fluorescence detector (Promega Corp.). The experiment
was repeated 3 times. The relative fluorescence expression was
calculated by taking the dual luciferase value of the control group
as 1 and the other groups as multiple of the control group.
Statistical analysis
SPSS 21.0 (IBM Corp.) was used for data analysis.
The Kolmogorov-Smirnov test was used to deter-mine whether the data
were normally distributed. The results are presented as the means ±
standard deviation. Comparisons between 2 groups were analyzed
using a t-test, and comparisons among multiple groups were analyzed
by one- or two-way analysis of variance (ANOVA); pairwise
comparisons after ANOVA were conducted with Tukey's multiple
comparisons test. The P-values obtained were two-tailed and a value
of P<0.05 was considered to indicate a statistically significant
difference.
Results
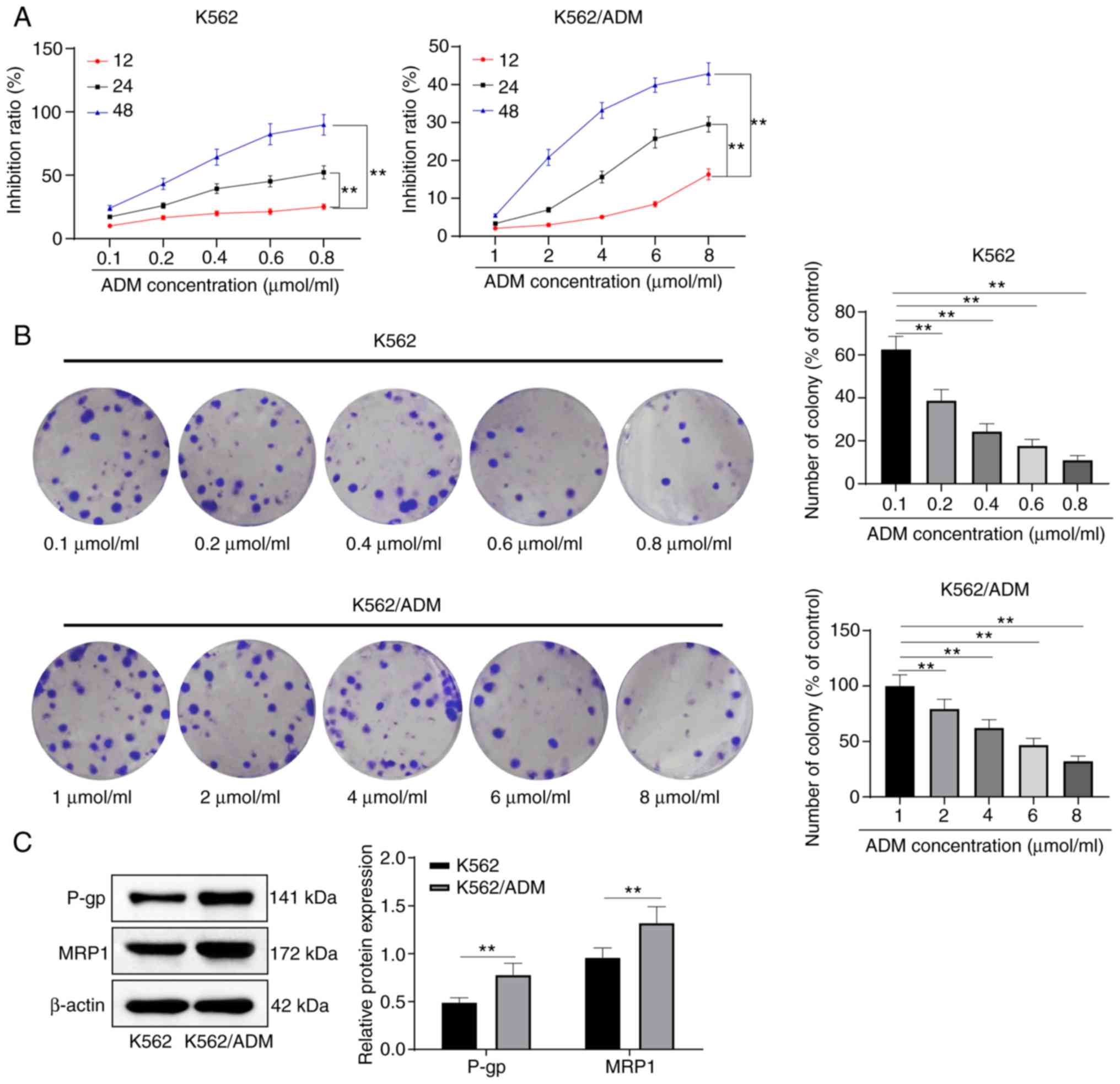
K562/ADM cells exhibit potent drug
resistance
The drug resistance of K562/ADM cells was examined
with K562 cells as the reference. K562 and K562/ADM cells were
treated with increasing concentrations of ADM for different periods
of time. The results revealed that ADM suppressed the growth of
K562/ADM and K562 cells in varying degrees. The cytotoxicity
induced by ADM was dose- and time-dependent in both cell lines.
Compared with the K562 cells, the K562/ADM cells were more
resistant to ADM (P<0.05) (Fig.
1A and Table II).
 | Table IIIC50 values of ADM in K562
cells and K562/ADM cells. |
Table II
IC50 values of ADM in K562
cells and K562/ADM cells.
| Time (h) | K562 cells
(µmol/l) | K562/ADM cells
(µmol/l) |
|---|
| 12 | 2.48±0.34 | 56.34±0.51 |
| 24 | 1.13±0.29 | 48.92±4.79 |
| 48 | 0.21±0.03 | 14.98±1.22 |
The results revealed that the number of cell
colonies of K562/ADM cells significantly decreased with the
increasing ADM concentration (Fig.
1B), and the levels of MRP1 and P-gp in the K562/ADM cells were
notably higher than those in the parental drug-sensitive K562 cells
(Fig. 1C) (all P<0.05).
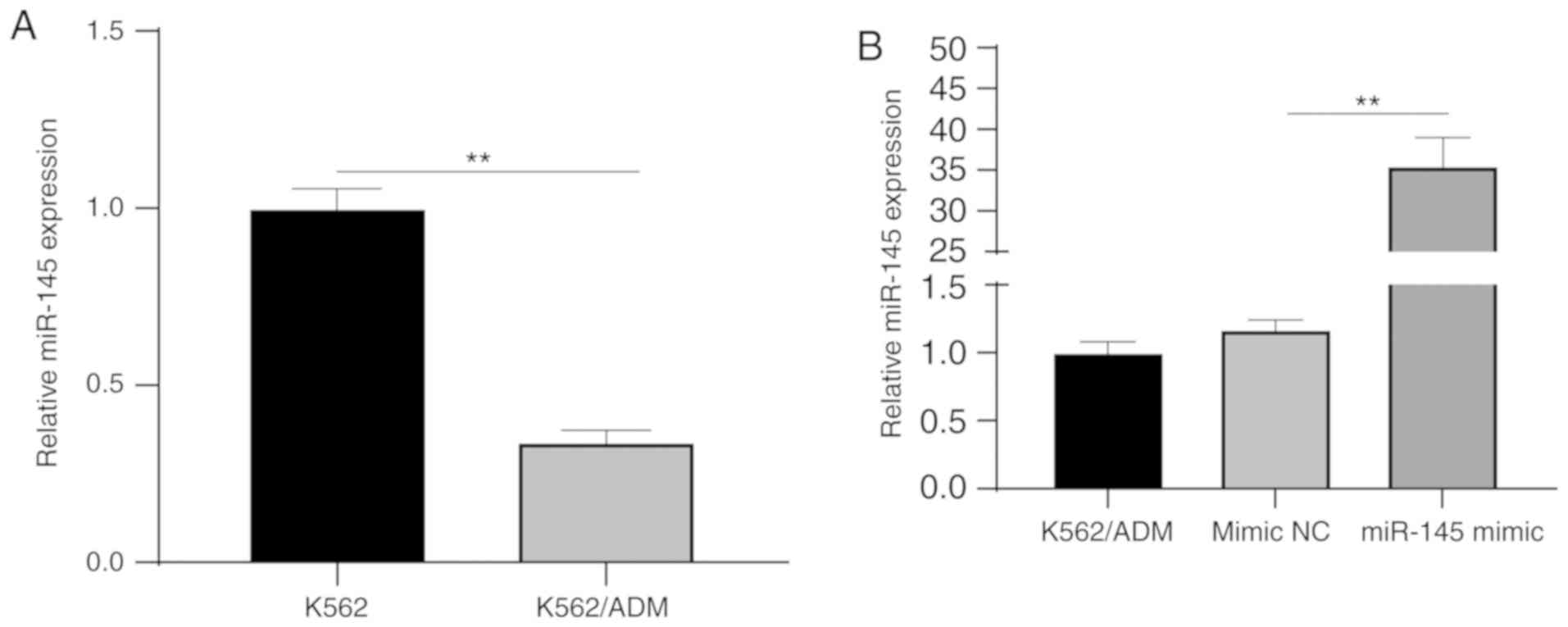
miR-145 is downregulated in K562/ADM
cells
RT-qPCR was used to detect miR-145 expression in
K562/ADM and K562 cells, and it was demonstrated that miR-145
expression was at a lower level in the K562/ADM cells than in the
K562 cells (P<0.05) (Fig. 2A).
To further examine the effect of miR-145 on K562/ADM cells, mimic
NC and miR-145 mimic were transfected into the K562/ADM cells,
respectively. miR-145 expression was detected following
transfection, and the results revealed a significant increase in
miR-145 expression in the miR-145 mimic group as compared with the
NC group (P<0.05) (Fig. 2B),
indicating successful transfection.
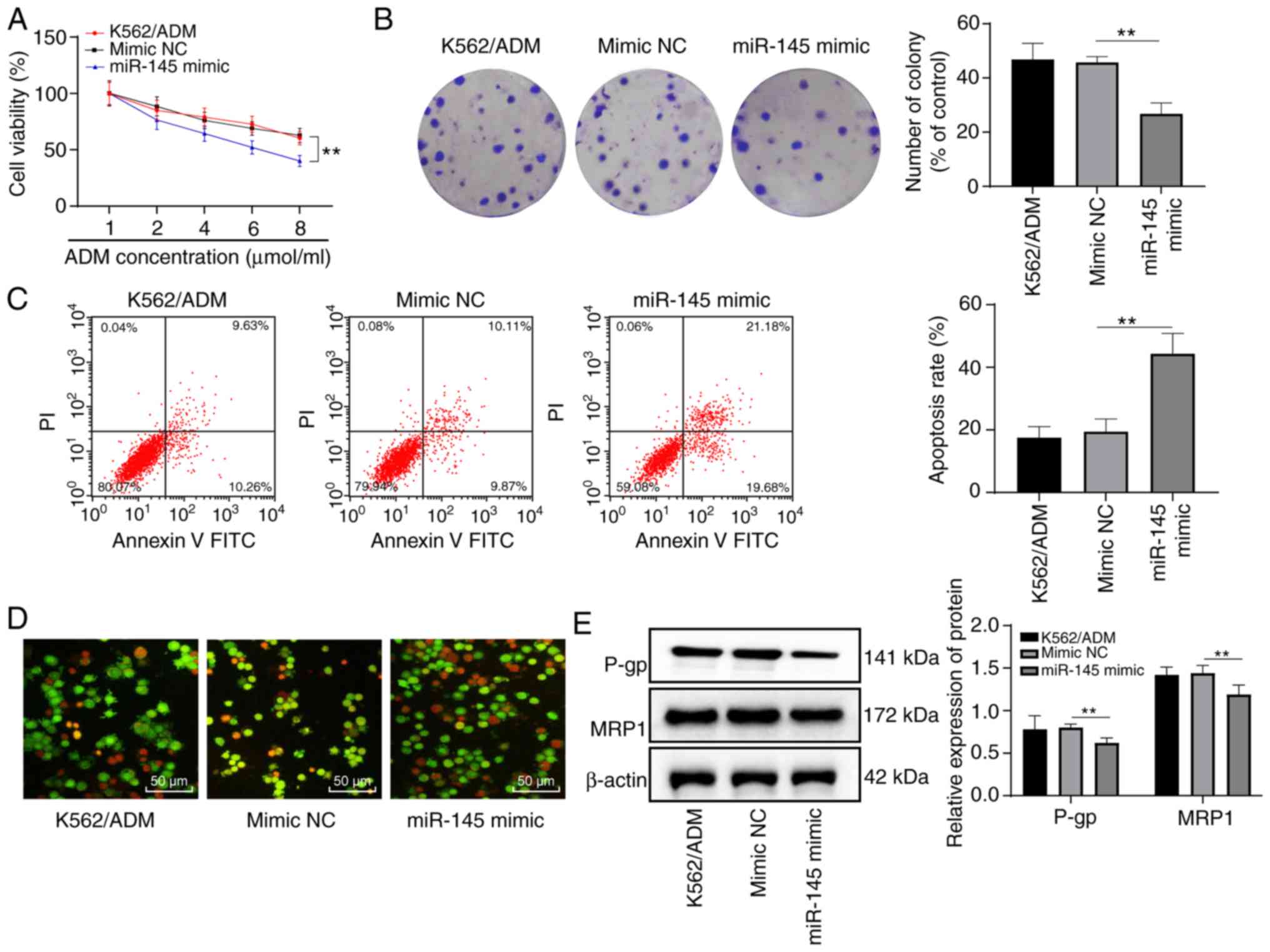
Overexpression of miR-145 enhances the
sensitivity of K562/ADM cells to ADM
Following transfection, the K562/ADM cells further
treated with various concentrations of ADM. The results
demonstrated that the cells in each group exhibited a dose
dependence on ADM. Compared with the K562/ADM group, there was no
notable difference in cell proliferation in the mimic NC group
(P>0.05); however, the miR-145 mimic group exhibited a decreased
cell proliferation (P<0.05). In addition, when the ADM
concentration was 6 µmol/l, cell viability was >50%, and
cell viability was <50% when the ADM concentration was 8
µmol/l (Fig. 3A).
Therefore, the ADM concentration of 6 µmol/l was selected
for data determination in subsequent experiments.
Following treatment with ADM at a concentration of 6
µmol/l, no significant difference was observed between the
K562/ADM group and the NC group as regards the number of cell
colonies, apoptosis rate and protein levels of MRP1 and P-gp (all
P>0.05). Compared with the NC group, the number of cell colonies
and the levels of MRP1 and P-gp in the miR-145 mimic group were
decreased, while the apoptotic rate increased significantly (all
P<0.05), and the majority of nuclear chromatins of the cells
were orange-red in color with a solid or bead-like shape (Fig. 3B-E). This suggested that the
overexpression of miR-145 enhanced the sensitivity of
drug-resistant K562/ADM cells to ADM.
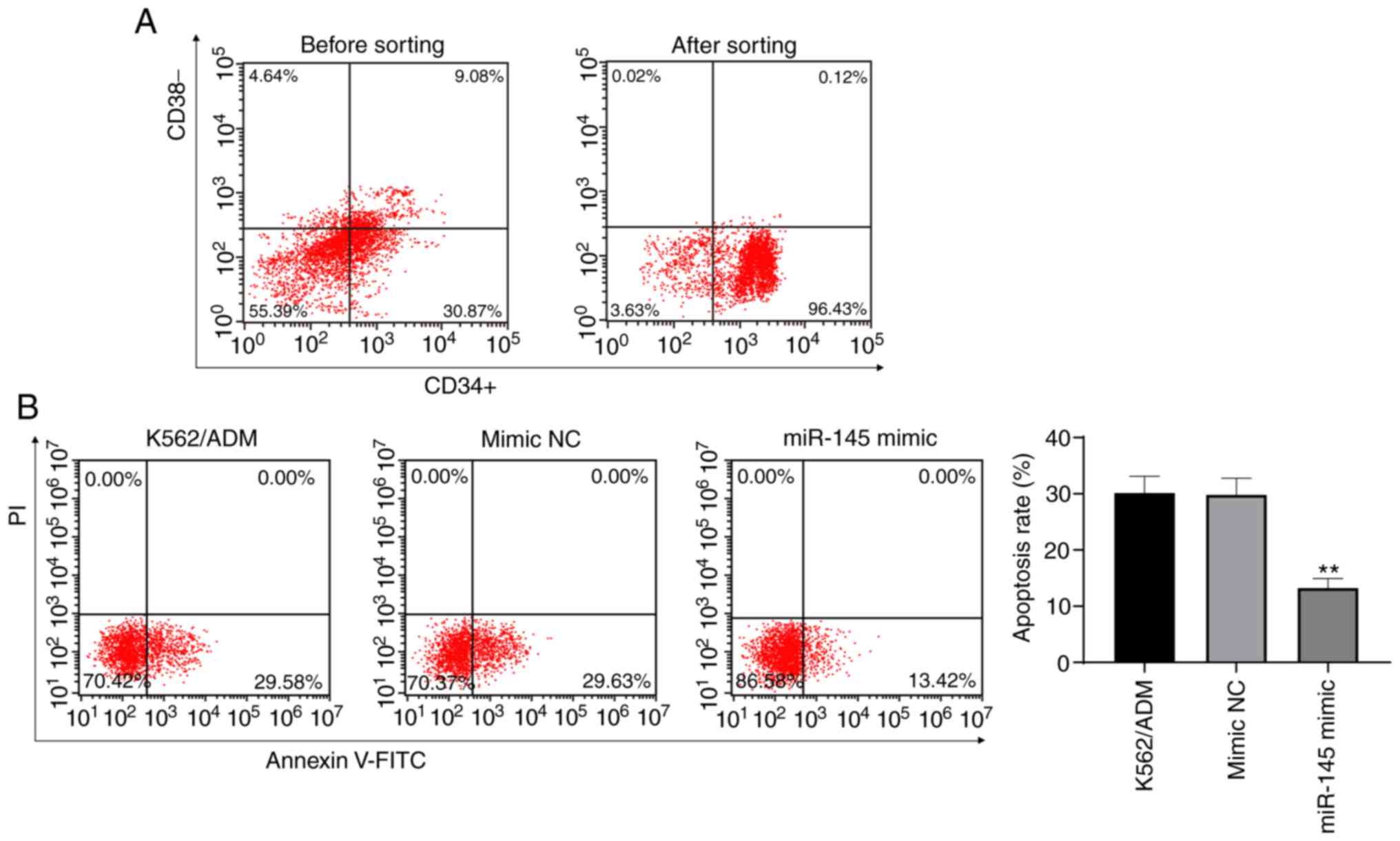
Overexpression of miR-145 promotes the
apoptosis of K562/ADM stem cells
CD34+CD38−K562/ADM stem cells
were sorted from K562/ADM stem cells using magnetic bead sorting
methods. Flow cytometry verified that the purity of the
CD34+CD38−K562/ADM stem cells was 94.69±1.93%
(this can be seen from CD34+ CD38− in the
lower right quadrant of the flow chart after sorting) (Fig. 4A). To examine the effects of the
overexpression of miR-145 on K562/ADM stem cells, flow cytometry
was utilized to detect the apoptotic rate of the K562/ADM stem
cells. It was revealed that the overexpression of miR-145 decreased
the apoptotic rate of the K562/ADM stem cells (P<0.05) (Fig. 4B). The results demonstrated that
the overexpression of miR-145 promoted the apoptosis of stem
cells.
Low miR-145 expression reduces the
sensitivity of K562 cells to ADM
To examine the effects of miR-145 on K562 cells,
miR-145 inhibitor and inhibitor NC were transfected into the K562
cells, respectively. RT-qPCR was performed to detect miR-145
expression in each group and it was found miR-145 expression in the
miR-145 inhibitor group was notably decreased as compared to the
inhibitor NC group (P<0.05) (Fig.
5A). To verify the effects of a low miR-145 expression on the
sensitivity of K562 cells to ADM, the cells in each group were
treated with various concentrations of ADM. It was revealed that
with the increase in the ADM concentration, cell proliferation in
each group was inhibited (P<0.05), although no significant
difference was observed in cell viability at each concentration
between the K562 group and the NC group (P>0.05). Compared with
the NC group, K562 cell proliferation was significantly increased
with the low expression of miR-145 (P<0.05). In addition, when
the ADM concentration was 0.6 µmol/l, cell viability was
>50% (Fig. 5B). Therefore, the
ADM concentration of 0.6 µmol/l was selected for use in
subsequent experiments.
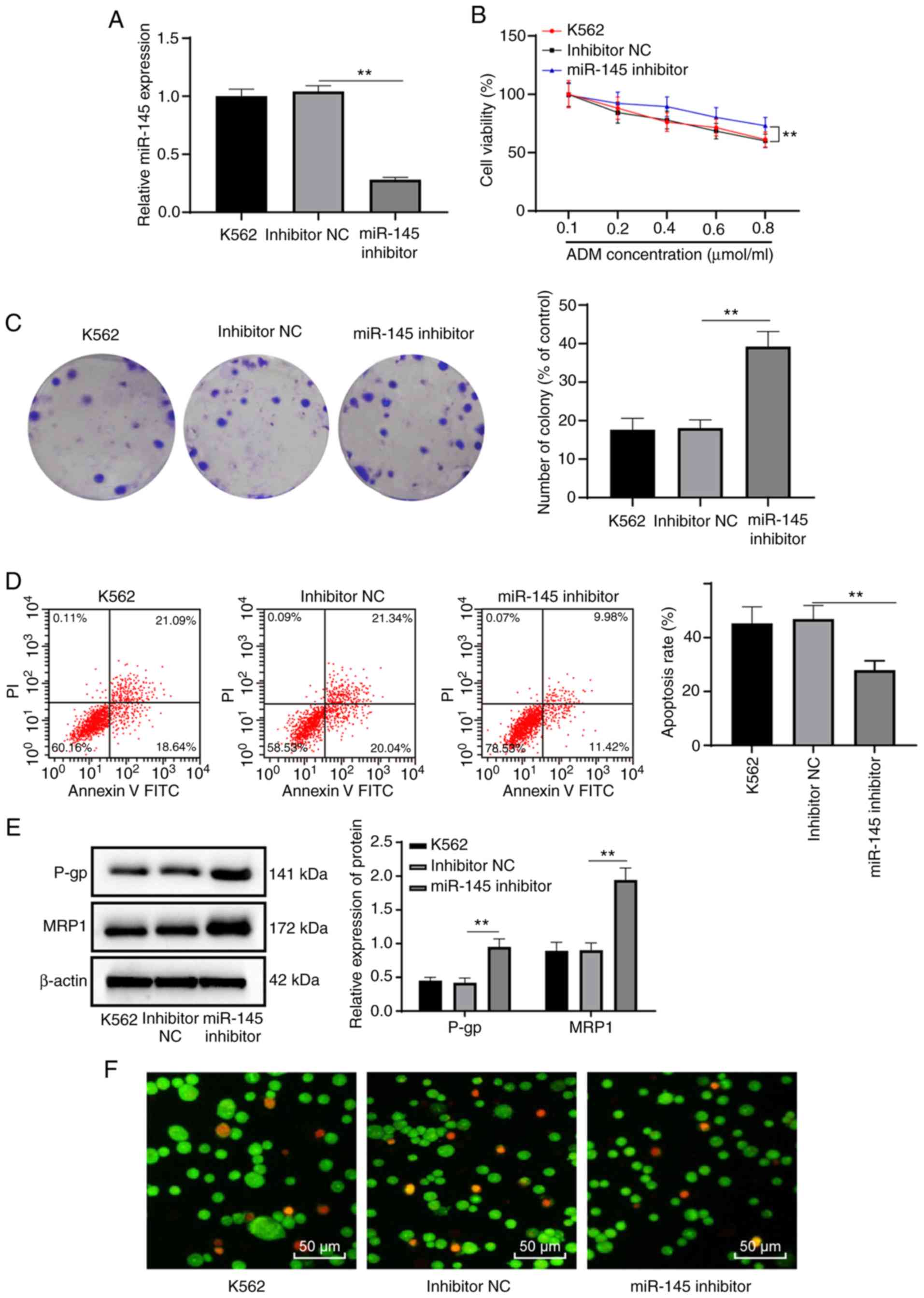 | Figure 5Low expression of miR-145 reduces the
sensitivity of K562 cells to ADM. (A) Relative miR-145 expression
in K562 cells detected by RT-qPCR. (B) Relative viability of K562
cells treated with various concentrations of ADM measured by MTT
assay. (C) Relative cell colonies of K562 cells treated with 0.6
µmol/l ADM measured by colony formation assay. (D) Relative
cell apoptosis of K562 cells treated with 0.6 µmol/l ADM
measured by flow cytometry. (E) Levels of MRP1 and P-gp of K562
cells treated with 0.6 µmol/l ADM measured by western blot
analysis. (F) The morphology and apoptosis of K562 cells treated
with 0.6 µmol/l ADM were observed under a fluorescence
microscope. **P<0.01; n=3. Data in (A, C and D) were
analyzed by one-way ANOVA, while data in (B and E) were analyzed by
two-way ANOVA. Tukey's multiple comparisons test was used as a post
hoc test. ADM, adriamycin; miR-145, microRNA-145; RT-qPCR, reverse
transcription quantitative polymerase chain reaction; MTT,
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide; MRP1,
multidrug resistance protein 1; P-gp, P-glycoprotein; ANOVA,
analysis of variance. |
Following treatment with ADM at a concentration of
0.6 µmol/l, no significant difference was observed between
the K562 group and the NC group as regards the number of cell
colonies, apoptotic rate, and the protein levels of MRP1 and P-gp
(all P>0.05). Compared with the NC group, the number of cell
colonies, and the levels of MRP1 and P-gp in the miR-145 inhibitor
group were increased, while the apoptotic rate decreased
significantly (all P<0.05), and cell morphology was normal
(Fig. 5C-E). These results
suggested that the low expression of miR-145 reduced the
sensitivity of K562 cells to ADM.
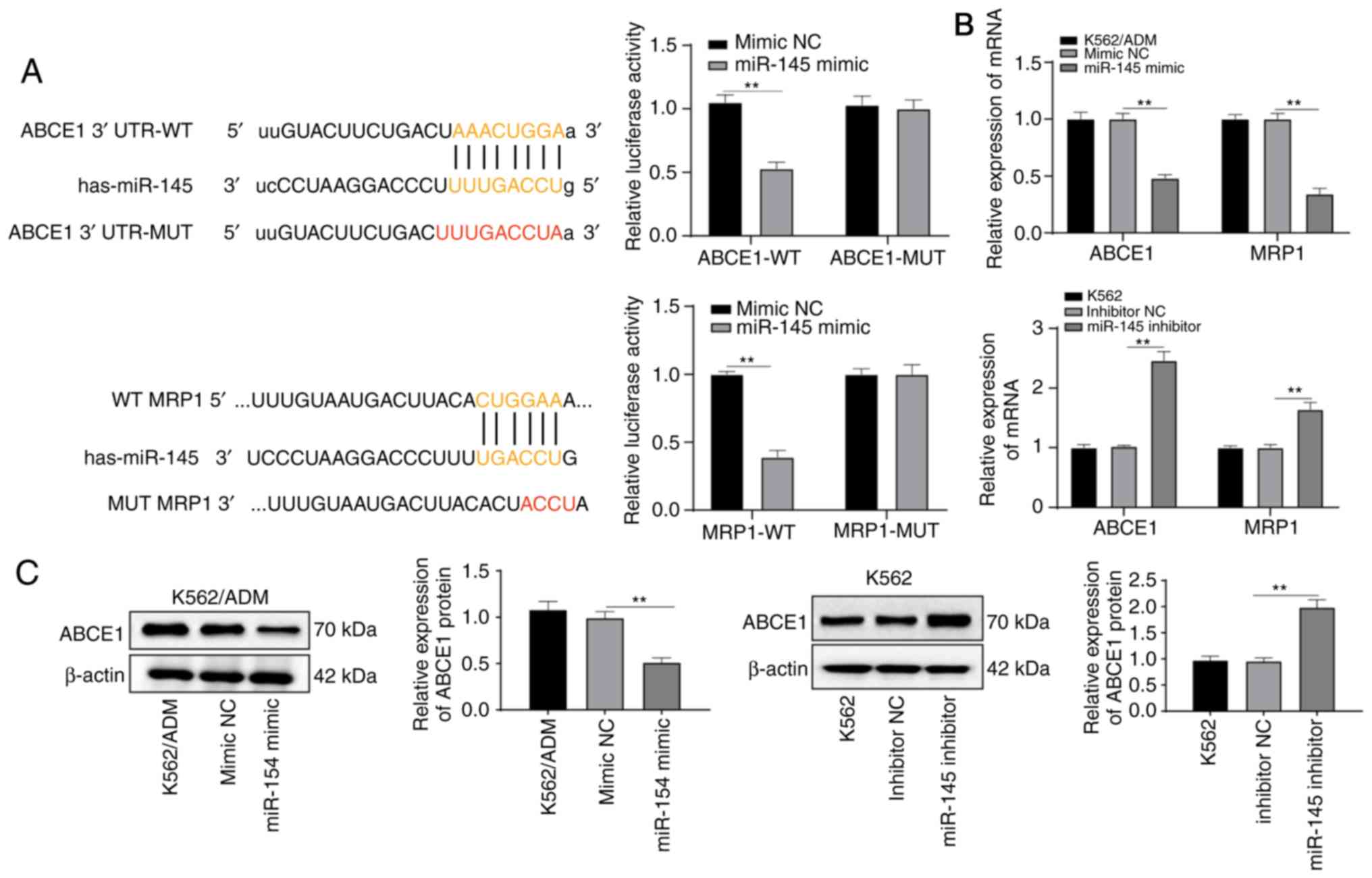
miR-145 targets and inhibits the
expression of ABCE1
As demonstrated above, the levels of the
drug-resistant gene, MRP1, were altered following the intervention
of miR-145 expression. Through website prediction, it was found
that there was a binding sequence between miR-145 and MRP1, and
their targeting association was verified by a dual luciferase
reporter gene assay (Fig. 6A). In
addition, the results of RT-qPCR revealed that the overexpression
of miR-145 inhibited the mRNA expression of MRP1 (P<0.05)
(Fig. 6B).
The binding of miR-145 to ABCE1 was predicted
through http://www.microrna.org/. It was noted
that there was a binding site between the 3′ UTR of miR-145 and
ABCE1. The results of dual-luciferase reporter gene assay verified
the targeting association between miR-145 and ABCE1 (P<0.05)
(Fig. 6A). The ABCE1 mRNA and
protein levels were significantly decreased in the K562/ADM cells
overexpressing miR-145, but were substantially increased following
transfection with miR-145 inhibitor (all P<0.05) (Fig. 6B and C), indicating that miR-145
targeted ABCE1.
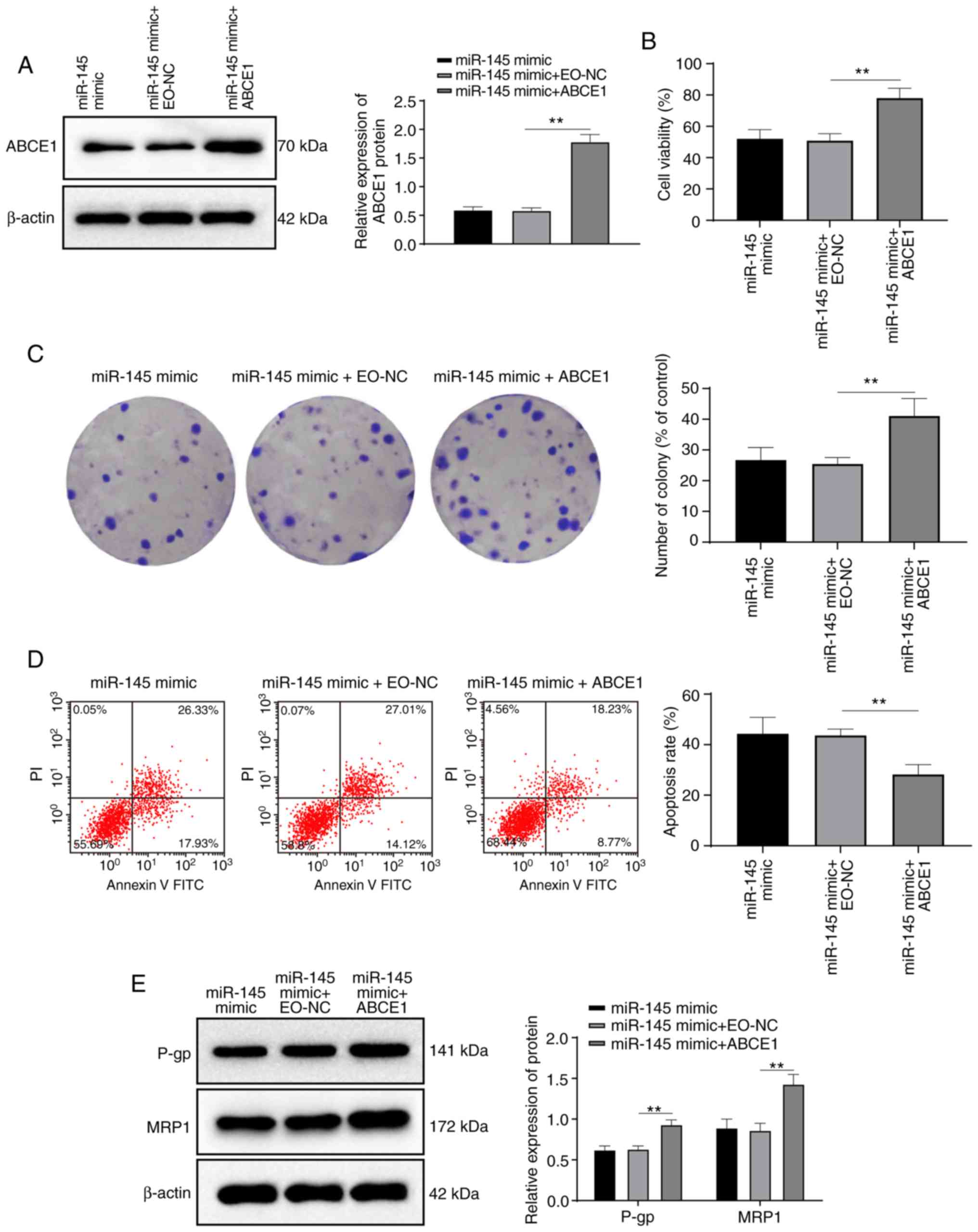
Activation of ABCE1 reduces the promoting
effect of miR-145 on the sensitivity of K562/ADM cells to ADM
Compared with miR-145 overexpression alone, the
combination of ABCE1 activation and miR-145 overexpression
significantly increased K56/ADM cell viability, the number of
colonies, the levels of MRP1 and P-gp, and reduced the apoptotic
rate (all P<0.05) (Fig. 7),
suggesting that ABCE1 activation reversed the promoting effects of
miR-145 overexpression on K562/ADM cell sensitivity to ADM.
Discussion
There is only one feasible option, namely allogeneic
hematopoietic stem cell transplant for patients afflicted with
recurrent or refractory leukemia whose cure rates are almost 10%
with current standard chemotherapeutic regimens (19). Recently, it has been shown that a
majority of therapeutic failures are due to cell resistance to
leukemic therapies (20). miRNAs
have been confirmed to be involved in normal hematopoiesis,
suggesting that the dysregulation of miRNAs may be a contributing
factor to leukemogenesis (21).
In the present study, the mechanisms of miR-145 and ABCE1 in the
biological processes of drug resistance to leukemia were examined.
Consequently, it was found that the overexpression of miR-145
promoted leukemic stem cell apoptosis and K562/ADM cell sensitivity
to ADM by inhibiting ABCE1.
To select an appropriate ADM concentration, a
pre-experiment was performed. According to a previous study
(22), following ADM treatment
(0.1, 0.2, 0.4, 0.6 and 0.8 µmol/ml), the activity of K562
cells was significantly inhibited (Fig. 1A, left panel). The K562/ADM cells
were also treated with ADM (0.1, 0.2, 0.4, 0.6, and 0.8
µmol/ml). It was found that ADM at this concentration
gradient exerted minimal inhibitory effects on the K562/ADM cells
(Fig. S1). Thus, according to a
previous study (23), it was
found that the difference between the ADM treatment concentration
of the K562 and K562/ADM cells was approximately 10-fold.
Therefore, the present study used (1, 2, 4, 6, 8 µmol/ml)
ADM to treat the K562/ADM cells, and it was found that the activity
of the K562/ADM cells was significantly inhibited (Fig. 1A, right panel). Finally, ADM was
used at 0.1, 0.2, 0.4, 0.6 and 0.8 µmol/ml to treat the K562
cells, and ADM was at 1, 2, 4, 6 and 8 µmol/ml to treat the
K562/ADM cells.
ADM, also known as doxorubicin, is considered a
potent drug for the treatment of childhood with acute lymphoblastic
leukemia (7). P-gp (also known as
ABCB1) and MRP1 (also known as ABCC1), two major ABC transportation
proteins grant resistance to a number of anticancer agents, lead to
multidrug resistance (24). The
first major result if the present study was that K562/ADM cells
were more resistant to ADM, presenting with less cell colonies and
cell growth inhibition with the increasing ADM concentrations, and
markedly higher levels of MRP1 and P-gp. As an important
chemotherapeutic drug with a variety of cellular targets, ADM is
frequently used in combination therapies for leukemia, multiple
myeloma and solid tumors, and it can also stimulate the
differentiation of K562 cells (23). A previous study revealed that high
levels of MDR1 and P-gp were associated with chemotherapy
sensitivity, high-grade tumors, lymph node metastasis, a poor
response and a shorter survival (25). Ge et al stated that ADM
induced the overexpression of P-gp in breast cancer cells, which,
in turn, increased the intracellular efflux of ADM (26). The present study further
highlighted that K562/ADM cells were more resistant to ADM, which
may provide new insight into leukemic therapies.
A previous study found that miRNAs are important for
the drug resistance of leukemia cells (K562/ADM) (18). It was then found miR-145 was
downregulated in K562/ADM cells. miR-145 was identified as a
tumor-suppressor and to be downregulated in several types of
cancer, such as glioma, lung cancer, colon cancer, breast cancer
and gastric cancer (27).
Similarly, miR-145 expression has been shown to be significantly
decreased in A549/cisplatin cells when compared with A549 cells
(28). The decreased expression
of miR-145 in hematopoietic stem cells contributes to an increased
platelet count in blood and the abnormal development of
megakaryocytes (12).
Additionally, the present study indicated that the overexpression
of miR-145 suppressed proliferation and accelerated the apoptosis
of K562/ADM cells, markedly decreasing the levels of MRP1 and P-gp,
and enhancing the sensitivity of K562/ADM cells to ADM. miR-145
overexpression has also been shown to suppress cell proliferation
and facilitate the apoptosis of human esophageal carcinomas cells
(29). Xia et al found
that the overexpression of miR-145 inhibited adult T-cell
leukemia/lymphoma cell proliferation and growth (13). Similarly, a high expression of
miR-145 has been shown to enhance breast cancer cell sensitivity to
ADM via intracellular ADM accumulation and MRP1 inhibition
(30). CD38, an antigen present
on the surface of human cells, is a type II multifunctional
transmembrane glycoprotein broadly distributed in hematopoietic
cells, and its expression is used as a phenotypic marker for the
proliferation and activation of T and B lymphocytes (31). Furthermore, non-thorough
chemotherapeutic obliteration of CD34+CD38−
stem cells is prone to leukemia relapse (32). In the present study, the number of
CD34+CD38− subsets decreased markedly and the
apoptosis of leukemic stem cells was promoted following the
overexpression of miR-145. Yalçintepe et al considered that
CD38 may play an essential role in the process of drug resistance
to ADM in K562 cells (33).
Moreover, in the present study, experiments using
the K562 cells revealed that a low expression of miR-145 increased
cell proliferation, decreased cell apoptosis, and increased the
levels of MRP1 and P-gp, thus reducing the sensitivity of K562
cells to ADM. A previous study conducted by Ikemura et al
found that miR-145 negatively regulated the expression and
functions of P-gp by direct interaction with MRP1, and the
downregulated miR-145 elevated P-gp expression following liver
ischemia-reperfusion injury (34). miR-145 has also been shown to
increase the sensitivity of esophageal squamous cell carcinoma to
cisplatin, and to facilitate ccisplatin-induced apoptosis to
decrease MRP1 and P-gp expression (35). MRP1 protein expression in leukemic
stem cells is a negative prognostic marker in acute myeloid
leukemia patients (36). Through
bioinformatics prediction and dual-luciferase reporter gene assay,
in the present study, it was verified that miR-145 targeted ABCE1
and MRP1. miR-145 suppressed MRP1 expression by targeting MRP1
3′-UTR, and miR-145 overexpression sensitized breast cancer cells
to ADM by inducing intracellular ADM accumulation via MRP1
inhibition (30). ABC
transportation genes exhibit a high expression in hematopoietic
stem cells; thus, they may play roles in the pathogenesis of stem
cell-derived leukemia (37). The
overexpression of ABC family members may lead to chemotherapy
failure (33). Additionally, as
observed in the present study, the activation of ABCE1 reduced the
promoting effects of miR-145 on the sensitivity of drug-resistant
K562/ADM cells to ADM. ABCE1 has been found to be overexpressed in
drug-resistant cancer cells, and ABCE1 silencing enhances the
sensitivity of lung cancer A549 cells to 5-Fluorouracil (38). Thus, the effects of a high miR-145
expression on ADM sensitivity in leukemia cells may be achieved via
the inhibition of ABCE1 and MRP1.
In conclusion, the present study demonstrated that
the overexpression of miR-145 promoted leukemic stem cell apoptosis
and enhanced the sensitivity of K562/ADM cells to ADM by inhibiting
ABCE1. These results reveal a potential target for cell resistance
to leukemic therapies. Due to the limitations of the experimental
conditions and costs, the present study did not explore the other
effects of miR-145 on leukemic stem cells, apart from apoptosis.
Another limitation of the present study is that normal,
non-cancerous cells were not used as a negative control. If the
experimental conditions permit in the future, the authors aim to
explore the effects of miR-145 on other characteristics of leukemic
stem cells. Although the present findings provide insight into the
treatment of leukemia, the experimental results and effective
application into clinic practice warrant further in-depth
validation.
Supplementary Data
Funding
The present was supported by the National Natural
Science Foundation of China (grant no. 81460035).
Availability of data and materials
All the data generated or analyzed during this study
are included in this published article.
Authors' contributions
ZW and HW are the guarantees of the integrity of the
entire study and all authors (ZW, HW, QS, HZ, MH, ST, LX, YC and
XH) contributed to the study concept and the design and definition
of the intellectual content of this study. HZ, LX, YC and MH
contributed to the experimental studies, data acquisition and
statistical analysis. ZW contributed to the preparation of the
manuscript. XH contributed to the manuscript review. All authors
read and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgments
Not applicable.
References
|
1
|
Juliusson G and Hough R: Leukemia. Prog
Tumor Res. 43:87–100. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Yamashita Y, Yuan J, Suetake I, Suzuki H,
Ishikawa Y, Choi YL, Ueno T, Soda M, Hamada T, Haruta H, et al:
Array-based genomic resequencing of human leukemia. Oncogene.
29:3723–3731. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Steinmaus C and Smith MT: Steinmaus and
Smith respond to 'proximity to gasoline stations and childhood
leukemia'. Am J Epidemiol. 185:5–7. 2017. View Article : Google Scholar
|
|
4
|
Kabat GC, Wu JW, Moore SC, Morton LM, Park
Y, Hollenbeck AR and Rohan TE: Lifestyle and dietary factors in
relation to risk of chronic myeloid leukemia in the NIH-AARP Diet
and Health Study. Cancer Epidemiol Biomarkers Prev. 22:848–854.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Kosmider O, Gelsi-Boyer V, Slama L,
Dreyfus F, Beyne-Rauzy O, Quesnel B, Hunault-Berger M, Slama B, Vey
N, Lacombe C, et al: Mutations of IDH1 and IDH2 genes in early and
accelerated phases of myelodysplastic syndromes and
MDS/myeloproliferative neoplasms. Leukemia. 24:1094–1096. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Papeta N, Zheng Z, Schon EA, Brosel S,
Altintas MM, Nasr SH, Reiser J, D'Agati VD and Gharavi AG: Prkdc
participates in mitochondrial genome maintenance and prevents
adriamycin-induced nephropathy in mice. J Clin Invest.
120:4055–4064. 2010. View
Article : Google Scholar : PubMed/NCBI
|
|
7
|
Escherich G, Zimmermann M and Janka-Schaub
G; CoALL study group: Doxorubicin or daunorubicin given upfront in
a therapeutic window are equally effective in children with newly
diagnosed acute lymphoblastic leukemia. A randomized comparison in
trial CoALL 07-03. Pediatr Blood Cancer. 60:254–257. 2013.
View Article : Google Scholar
|
|
8
|
Wang Y, Krivtsov AV, Sinha AU, North TE,
Goessling W, Feng Z, Zon LI and Armstrong SA: The Wnt/beta-catenin
pathway is required for the development of leukemia stem cells in
AML. Science. 327:1650–1653. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Dick JE: Stem cell concepts renew cancer
research. Blood. 112:4793–4807. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Gomes BC, Rueff J and Rodrigues AS:
MicroRNAs and cancer drug resistance. Methods Mol Biol.
1395:137–162. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Bousquet M, Harris MH, Zhou B and Lodish
HF: MicroRNA miR-125b causes leukemia. Proc Natl Acad Sci USA.
107:21558–21563. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Lindsley RC and Ebert BL: Molecular
pathophysiology of myelodysplastic syndromes. Annu Rev Pathol.
8:21–47. 2013. View Article : Google Scholar
|
|
13
|
Xia H, Yamada S, Aoyama M, Sato F, Masaki
A, Ge Y, Ri M, Ishida T, Ueda R, Utsunomiya A, et al: Prognostic
impact of microRNA-145 down-regulation in adult T-cell
leukemia/lymphoma. Hum Pathol. 45:1192–1198. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Tian Y, Tian X, Han X, Chen Y, Song CY,
Jiang WJ and Tian DL: ABCE1 plays an essential role in lung cancer
progression and metastasis. Tumour Biol. 37:8375–8382. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
de Jonge-Peeters SD, Kuipers F, de Vries
EG and Vellenga E: ABC transporter expression in hematopoietic stem
cells and the role in AML drug resistance. Crit Rev Oncol Hematol.
62:214–226. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
17
|
Li W, Jiang Y, Wang Y, Yang S, Bi X, Pan
X, Ma A and Li W: MiR-181b regulates autophagy in a model of
Parkinson's disease by targeting the PTEN/Akt/mTOR signaling
pathway. Neurosci Lett. 675:83–88. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Liu W, He J, Yang Y, Guo Q and Gao F:
Upregulating miR-146a by physcion reverses multidrug resistance in
human chronic myelogenous leukemia K562/ADM cells. Am J Cancer Res.
6:2547–2560. 2016.PubMed/NCBI
|
|
19
|
Bose P, Vachhani P and Cortes JE:
Treatment of relapsed/refractory acute myeloid leukemia. Curr Treat
Options Oncol. 18:172017. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhang X, Ai Z, Chen J, Yi J, Liu Z, Zhao H
and Wei H: Glycometabolic adaptation mediates the insensitivity of
drug-resistant K562/ADM leukaemia cells to adriamycin via the
AKT-mTOR/c-Myc signalling pathway. Mol Med Rep. 15:1869–1876. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Lv M, Zhang X, Jia H, Li D, Zhang B, Zhang
H, Hong M, Jiang T, Jiang Q, Lu J, et al: An oncogenic role of
miR-142-3p-in human T-cell acute lymphoblastic leukemia (T-ALL) by
targeting gluco-corticoid receptor-α and cAMP/PKA pathways.
Leukemia. 26:769–777. 2012. View Article : Google Scholar
|
|
22
|
Wang F, Chen J, Zhang Z, Yi J, Yuan M,
Wang M, Zhang N, Qiu X, Wei H and Wang L: Differences of basic and
induced autophagic activity between K562 and K562/ADM cells.
Intractable Rare Dis Res. 6:281–290. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Yang MY, Lin PM, Liu YC, Hsiao HH, Yang
WC, Hsu JF, Hsu CM and Lin SF: Induction of cellular senescence by
doxorubicin is associated with upregulated miR-375 and induction of
autophagy in K562 cells. PLoS One. 7:e372052012. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Stefan K, Schmitt SM and Wiese M:
9-Deazapurines as broad-spectrum inhibitors of the ABC transport
proteins P-glycoprotein, multidrug resistance-associated protein 1,
and breast cancer resistance protein. J Med Chem. 60:8758–8780.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Bao L, Haque A, Jackson K, Hazari S, Moroz
K, Jetly R and Dash S: Increased expression of P-glycoprotein is
associated with doxorubicin chemoresistance in the metastatic 4T1
breast cancer model. Am J Pathol. 178:838–852. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Ge C, Cao B, Feng D, Zhou F, Zhang J, Yang
N, Feng S, Wang G and Aa J: The down-regulation of SLC7A11 enhances
ROS induced P-gp over-expression and drug resistance in MCF-7
breast cancer cells. Sci Rep. 7:37912017. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Tang M, Lin L, Cai H, Tang J and Zhou Z:
MicroRNA-145 downregulation associates with advanced tumor
progression and poor prognosis in patients suffering osteosarcoma.
Onco Targets Ther. 6:833–838. 2013.PubMed/NCBI
|
|
28
|
Zhang H, Luo Y, Xu W, Li K and Liao C:
Silencing long inter-genic non-coding RNA 00707 enhances cisplatin
sensitivity in cisplatin-resistant non-small-cell lung cancer cells
by sponging miR-145. Oncol Lett. 18:6261–6268. 2019.PubMed/NCBI
|
|
29
|
Zhang JH, Du AL, Wang L, Wang XY, Gao JH
and Wang TY: Episomal lentiviral vector-mediated miR-145
overexpression inhibits proliferation and induces apoptosis of
human esopha-geal carcinomas cells. Recent Pat Anticancer Drug
Discov. 11:453–460. 2016. View Article : Google Scholar
|
|
30
|
Gao M, Miao L, Liu M, Li C, Yu C, Yan H,
Yin Y, Wang Y, Qi X and Ren J: miR-145 sensitizes breast cancer to
doxorubicin by targeting multidrug resistance-associated protein-1.
Oncotarget. 7:59714–59726. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Mantei K and Wood BL: Flow cytometric
evaluation of CD38 expression assists in distinguishing follicular
hyperplasia from follicular lymphoma. Cytometry B Clin Cytom.
76:315–320. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Wang F, Wang XK, Shi CJ, Zhang H, Hu YP,
Chen YF and Fu LW: Nilotinib enhances the efficacy of conventional
chemo-therapeutic drugs in CD34+CD38− stem
cells and ABC transporter overexpressing leukemia cells. Molecules.
19:3356–3375. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Yalçintepe L, Halis E and Ulku S: Effect
of CD38 on the multi-drug resistance of human chronic myelogenous
leukemia K562 cells to doxorubicin. Oncol Lett. 11:2290–2296. 2016.
View Article : Google Scholar
|
|
34
|
Ikemura K, Yamamoto M, Miyazaki S,
Mizutani H, Iwamoto T and Okuda M: MicroRNA-145
post-transcriptionally regulates the expression and function of
P-glycoprotein in intestinal epithelial cells. Mol Pharmacol.
83:399–405. 2013. View Article : Google Scholar
|
|
35
|
Zheng TL, Li DP, He ZF and Zhao S: miR-145
sensitizes esophageal squamous cell carcinoma to cisplatin through
directly inhibiting PI3K/AKT signaling pathway. Cancer Cell Int.
19:2502019. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Paprocka M, Bielawska-Pohl A, Rossowska J,
Krawczenko A, Duś D, Kiełbiński M, Haus O, Podolak-Dawidziak M and
Kuliczkowski K: MRP1 protein expression in leukemic stem cells as a
negative prognostic marker in acute myeloid leukemia patients. Eur
J Haematol. 99:415–422. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Raaijmakers MH: ATP-binding-cassette
transporters in hematopoietic stem cells and their utility as
therapeutical targets in acute and chronic myeloid leukemia.
Leukemia. 21:2094–2102. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Kara G, Tuncer S, Türk M and Denkbas EB:
Downregulation of ABCE1 via siRNA affects the sensitivity of A549
cells against chemotherapeutic agents. Med Oncol. 32:1032015.
View Article : Google Scholar : PubMed/NCBI
|