Introduction
Contact lens (CL)-induced papillary conjunctivitis
(CLPC) is a chronic conjunctivitis that occurs in CL wearers.
Physical stimulation to the palpebral conjunctiva by the CL itself
and allergic reactions to substances adhering to the CL surface are
considered among the primary causes (1). Although the types of depositions
that induce an allergic reaction have not yet been confirmed,
bio-deposits of denatured protein are predicted to be one of the
etiologies (2,3). Bacteria adhering to the CL surface
are possible allergens for CLPC. However, this has not been
investigated in depth thus far. Tagliaferri et al cultivated
CLs collected from patients with CLPC and reported that there was
no association between CLPC and bacteria adhering to the CL surface
(4). However, the authors of that
study evaluated the association only by the use of conventional
cultivation methods, and they did not analyze the bacterial
population at the genus level. Since CL wear affects the ocular
surface through various waterborne bacteria of environmental origin
(5), and many of the
environmental bacteria are fastidious (6), the validation of CL and care
solutions in a culture-independent manner is desirable.
Recently, microbiome analysis employing
next-generation sequencing (NGS) has been applied to gut, skin,
oral, and vaginal samples (7-9).
Several studies have analyzed the ocular surface microbiome of
healthy subjects with NGS (10-12). These studies report that the
composition of the ocular surface microbiome inferred by NGS
greatly differs from that previously described using conventional
culture methods (10,12).
It is known that microbiomes are present in human
organs, where they contribute to maintaining the homeostasis of
each organ and/or human health. For example, in a study on the gut
microbiome, the alteration of the gut microbiome termed dysbiosis
was found to be involved in the development of non-infectious
diseases, such as obesity, diabetes mellitus, autoimmune disease
and inflammatory bowel disease (13,14). Similarly, the ocular surface
microbiome may play an important role in ocular surface
homeostasis, and its alteration may be related to the onset and
deterioration of some non-infectious eye diseases. Thus, micobiome
analysis using NGS of ocular surface and ophthalmic clinical
samples may help to elucidate the causal association between
microorganism and non-infectious ocular diseases.
In the present study, NGS analysis of the microbiome
in CL care solutions remaining in the lens storage cases and the
tear fluids of CL wearers with allergic symptoms (AS) was
performed. The findings of the present study suggest a possible
association between bacterial contamination in the lens storage
case and AS.
Materials and methods
Participant profiles
In total, 15 CL wearers (9 subjects with AS and 6
healthy controls, ranging in age from 19 to 24 years) were enrolled
in the present study (Table I).
The present study followed the tenets of the Declaration of
Helsinki and was approved by the Institutional Review Board of
Kindai University Faculty of Medicine (approval no. 28-137).
Informed consent was obtained from all subjects after explanation
of the nature and possible consequences of the study. They were all
volunteers who wore refractive soft hydrogel lenses, use
multi-purpose solution for their lens care, and answered the
questions on AS listed below (regarding the symptoms) after
providing the lens storage case. Although all the brand names of
lenses and care solutions could not be ascertained, the care
solutions enrolled in the present study contained polidronium
chloride or polyhexamethylene biguanide and all lenses were made of
silicone hydrogel. The diagnostic criteria of AS include soft CL
wearers who have symptoms of eye itching plus at least one of the
following symptoms during CL wear: Conjunctival hyperaemia, eye
discharge, foreign body sensation, or eye discomfort. Healthy
subjects were defined as soft CL wearers who did not have any of
the above-mentioned symptoms. Individuals who were administered any
topical medications of anti-allergic agents and/or antimicrobial
agents within 3 months prior to initiation of the present study
were excluded. Individuals diagnosed with dry eye or atopic
dermatitis were excluded.
 | Table ISample profiles and the results of
bacterial culture and histamine quantification. |
Table I
Sample profiles and the results of
bacterial culture and histamine quantification.
| Subject ID | Age, sex | Ocular status | Sample type | Bacterial culture
| Histamine
(µg/ml) |
|---|
| Total
counts/ml | Isolated
species |
|---|
| 1 | 24, Male | AS | Tear | ND | | <0.01 |
| CL | 2,000 | Streptococcus
tigurinus Chryseobacterium indologenes Microbacterium maritypicum
Comamonas koreensis Bacillus subtilis | |
| 2 | 21, Female | AS | Tear | ND | | 0.032 |
| CL | ND | | |
| 3 | 20, Female | AS | Tear | ND | | <0.01 |
| CL | 60 | Bacillus
subtilis Pseudomonas marginalis | |
| 4 | 21, Female | AS | Tear | ND | | <0.01 |
| CL | 60 | Pseudomonas
marginalis | |
| 5 | 20, Female | AS | Tear | ND | | <0.01 |
| CL | 841,000 | Delftia
tsuruhatensis Pseudomonas hibiscicola Microbacterium testaceum
Xenophilus aerolatus | |
| 6 | 20, Female | AS | Tear | ND | | <0.01 |
| CL | ND | | |
| 7 | 19, Female | AS | Tear | ND | | 0.223 |
| CL | ND | | |
| 8 | 21, Female | AS | Tear | ND | | <0.01 |
| CL | ND | | |
| 9 | 21, Male | AS | Tear | ND | | <0.01 |
| CL | 20 | Staphylococcus
epidermidis | |
| 10 | 24, Male | Healthy | Tear | ND | | <0.01 |
| CL | ND | | |
| 11 | 20, Female | Healthy | Tear | ND | | <0.01 |
| CL | ND | | |
| 12 | 21, Female | Healthy | Tear | ND | | <0.01 |
| CL | ND | | |
| 13 | 23, Female | Healthy | Tear | ND | | <0.01 |
| CL | 521,000 | Microbacterium
trichothecenolyticum Pseudomonas azotoformans Microbacterium
testaceum | |
| 14 | 21, Female | Healthy | Tear | ND | | <0.01 |
| CL | 14,900 | Pantoea
vagans | |
| 15 | 21, Female | Healthy | Tear | 200 | Staphylococcus
capitis | <0.01 |
| CL | ND | | |
Isolation of contaminated bacteria from
CL care solution
The CL care solutions remaining in the CL storage
cases were collected within 4 h of CL wear on the same day. The
solution was sampled (0.1 ml) and spread onto a brain heart
infusion (BHI; Eiken Chemical Co., Ltd.) agar plate. The plate was
incubated at 37°C for 48 h. The colonies formed on the plate were
individually inoculated into BHI broth. The culture was centrifuged
(10,000 × g, 5 min, 4°C), washed once with
Tris-ethylenediaminetetraacetic acid (TE) buffer [10 mM Tris-HCl
(pH 8.0), 1 mM ethylenediaminetetraacetic acid] and resuspended in
0.5 ml of TE buffer. Using a part of the suspension, Gram staining
was performed. After boiling the samples, they were centrifuged
(10,000 × g, 5 min, 4°C), and the supernatants were used as
templates for 16S rDNA amplification using universal primers 27F
(5′-AGA GTT TGA TCM TGG CTC AG-3′) and 1492R (5′-GGM TAC CTT GTT
ACG ACT T-3′). Species identification was performed by a BLAST
(https://blast.ncbi.nlm.nih.gov/Blast.cgi) search of
the obtained 16S rDNA sequences against the 16S ribosomal RNA
database in NCBI. The species of each colony was identified as the
best-hit microorganism (>97% identity) by a BLAST search.
Quantification of histamine by
high-performance liquid chromatography (HPLC)
Tear fluids were collected from the study subjects
during CL wearing at the same time when their CL care solutions
were collected by soaking in Schirmer test paper (Eagle Vision) and
distillation in phosphate-buffered saline (PBS). An additional
experiment using saline solution to measure the amount of tear
fluids per 1 mm of Schirmer test paper revealed that the paper was
soaked with approximately 0.5 µl of fluids per 1 mm
(Table SI). Therefore, the
amount of tear fluids collected from the subjects was approximately
5 µl, since they were collected when the scale of the
Schirmer test paper reached 10 mm. For 8 subjects with a
tear-soaked scale of ≥15 mm on the Schirmer test paper, 0.5
µl the tear fluids were used for western blot analysis as
described below. The tear fluids of the subjects with AS were
collected during the symptomatic period. Each diluted tear fluids
(0.1 ml) was mixed with 0.3 ml of 0.6 M HClO4. After
vortexing, the mixture was centrifuged for 15 min at 10,000 × g.
The supernatant was filtered through a 0.2-µm membrane. An
aliquot of 80 µl of the extract was used for derivatization.
The extract samples containing 0.8 µg/ml of
1,7-diaminoheptane as an internal standard were dansylated as
previously described by Chiacchierini et al (15). The concentration of dansylated
histamine was determined by HPLC. HPLC was performed with a Tosoh
8020 HPLC system equipped with a CCPM-II pump, a UV-8020
variable-wavelength detector, a CO-8020 column oven, and a Degasys
DG-1210 in-line degasser (Uniflows Co., Ltd.). The chromatographic
peak area was computed with a SIC chromatorecorder (System
Instruments Co., Ltd.). The separation was performed on a reverse
phase TSKgel ODS-80Tm column (4.6×150 mm, Tosoh Corporation). The
column temperature was maintained at 25°C, and absorbance was
monitored at 254 nm. The 50 µl each of the samples was
injected into the column and eluted at a flow rate of 1.2 ml/min.
The mobile phase was acetonitrile-H2O (60:40, v/v).
Histamine hydrochloride (Sigma-Aldrich; Merck KGaA) was used as a
standard.
Microbiome analysis
Tear fluids and CL care solutions were centrifuged
(10,000 × g, 5 min, 4°C), and the pellets were washed once with TE
buffer (pH 8.0). Following centrifugation (10,000 × g, 5 min, 4°C),
the pellets were frozen at -80°C until use. DNA extraction was
performed as per the method previously reported by Morita et
al (16). Sequencing
libraries were prepared by amplifying the V3-V4 region of the 16S
rDNA using the primers (5′-TCG TCG GCA GCG TCA GAT GTG TAT AAG AGA
CAG CCT ACG GGN GGC WGC AG-3′ and 5′-TCG TCG GCA GCG TCA GAT GTG
TAT AAG AGA CAG CCT ACG GGN GGC WGC AG-3′ and 5′-GTC TCG TGG GCT
CGG AGA TGT GTA TAA GAG ACA GGA CTA CHV GGG TAT CTA ATC C-3′)
previously described by Klindworth et al (17). Following initial amplification, a
second polymerase chain reaction was performed with Nextra XT Index
kit (cat. no. 15055294, Illumina, Inc.) to attach Illumina
adaptors, as well as barcodes that allow for multiplexing.
Amplification was performed in 25 µl reactions containing
2.5 µl of diluted template, 12.5 µl of 2X KAPA HiFi
HotStart Ready Mix (cat. no. KK2602, Kapa Biosystems), and 5
µl each of primers. Thermal cycling consisted of an initial
denaturation step (3 min at 95°C), followed by 25 cycles of
denaturation (30 sec at 95°C), annealing (30 sec at 55°C), and 30
sec extension at 72°C. The final extension was performed for 5 min
at 72°C. Amplicons were purified using AMPure XP beads (cat. no.
A63881, Beckman Coulter, Inc.). The integrity of the DNA amplicon
library was evaluated by an Agilent 2100 Bioanalyzer (Agilent
Technologies Japan, Ltd.) using an Agilent High Sensitivity DNA kit
(cat. no. 5067-4626, Agilent Technologies Japan, Ltd.). The loading
DNA concentration was adjusted to 6 pM each with Qubit Fluorometer
Q32857 (Thermo Fisher Scientific K.K.) using a Qubit dsDNA BR Assay
kit (cat. no. Q32853, Thermo Fisher Scientific K.K.). Sequencing
was performed on the Illumina MiSeq platform (cat. no. MS-102-3003,
MiSeq Reagent kit version 3,600 cycles, Illumina, Inc.) according
to the manufacturer's specifications to generate paired-end reads
of 300 bases in length in each direction. In total, 15,992,047
2×300 base pair reads with an average of 333,168 reads per sample
was obtained. Primer sequences were trimmed away, and the
paired-end reads were merged using fastqjoin (18) with default parameters and
processed with the QIIME 1.8.0 pipeline (19). Following a chimera check by
Usearch v6.1.544 (20), 20,000
Illumina reads per sample (average Phred >20) were randomly
selected for further analysis. Using the UCLUST (20) algorithm built into the QIIME
pipeline, sequences were clustered at 97% identity against the
Greengenes reference database (http://greengenes.lbl.gov), producing 744 operational
taxonomic units (OTUs). Using the QIIME pipeline, unweighted
UniFrac distances were produced and used for investigation of beta
diversity through plotting principal coordinate analysis (PCoA)
coordinates. QIIME analysis pipeline that include these software
was downloaded through QIIME (http://qiime.org/).
Western blot analysis against ocular
bacteria by tear IgE
Staphylococcus aureus 209P, Staphylococcus
epidermidis (isolated from the CL care solution of subject 9),
Staphylococcus capitis (isolated from the tear fluid of
subject 15), Streptococcus pneumoniae IID555 and
Streptococcus tigurinus (isolated from the CL care solution
of subject 1) were cultured under microaerophilic conditions (5%
CO2) in BHI at 37°C for 24 h. The bacterial cells were
centrifuged (10,000 × g, 5 min, 4°C), washed once with PBS, and
resuspended in 4% paraformaldehyde in PBS (PFA) to
OD600=1.0. The culture supernatants were mixed with PFA
at 1:1. The 2 µl each of the bacterial cell suspension,
culture supernatants, or purified human IgE (H+L; 1.0 µg/ml,
cat. no. ab65866, Abcam) were plotted onto a nitrocellulose
membrane and dried overnight at 25°C. Blocking was performed with
3% skim milk in PBS with Tween-20 (PBS-T) for 1 h at 25°C. The
membrane was washed 3 times with PBS-T. The membrane was incubated
with diluted tear fluids (10 µl each) in 3 ml of PBS-T at
30°C for 1 h. After washing 3 times with PBS-T, goat-anti human IgE
(H+L) HRP-conjugated (1:1,000) (cat. no. ab73901, Abcam) was added
and incubated at 30°C for 1 h with gentle agitation. The membrane
was washed 3 times with PBS-T, and the chemiluminescence signal
developed by ECL Prime Western Blotting Detection Reagent (GE
Healthcare UK Ltd.) was detected with LAS4000 (GE Healthcare UK
Ltd.). To examine the non-specific binding of IgE to the bacterial
cells, human IgE (1.0 µg/ml) was used instead of tear
fluids. Western dot-blot hybridization was applied to 6 subjects
with AS and 2 healthy subjects who were able to collect enough tear
fluids for the examination.
Statistical analysis
The minimum sample size required for the present
study was calculated using Easy R (EZR) version 1.41 (21). Quantitative microbiological
measurements were transformed to log10 prior to
conducting an analysis. All experiments were performed at least 3
times. The data were analysed by Chi-squared test, one-way analysis
of variance and Tukey's post hoc test (StatFlex ver.6, Artec Co.,
Ltd., Tokyo, Japan) to assess the significance. P-values <0.05
were considered to indicate statistically significant
differences.
Sequence data deposition
Sequence data have been deposited in the DDBJ
database (accession nos. DRA010383: https://ddbj.nig.ac.jp/DRASearch/submission?acc=DRA010383;
PRJDB10068: https://ddbj.nig.ac.jp/BPSearch/bioproject?acc=PRJDB10068;SAMD00232908-SAMD00232922:
https://ddbj.nig.ac.jp/DRASearch/experiment?acc=DRX224997,
https://ddbj.nig.ac.jp/DRASearch/experiment?acc=DRX224998,
https://ddbj.nig.ac.jp/DRASearch/experiment?acc=DRX224999,
https://ddbj.nig.ac.jp/DRASearch/experiment?acc=DRX225000,
https://ddbj.nig.ac.jp/DRASearch/experiment?acc=DRX225001,
https://ddbj.nig.ac.jp/DRASearch/experiment?acc=DRX225002,
https://ddbj.nig.ac.jp/DRASearch/experiment?acc=DRX225003,
https://ddbj.nig.ac.jp/DRASearch/experiment?acc=DRX225004,
https://ddbj.nig.ac.jp/DRASearch/experiment?acc=DRX225005,
https://ddbj.nig.ac.jp/DRASearch/experiment?acc=DRX225006,
https://ddbj.nig.ac.jp/DRASearch/experiment?acc=DRX225007,
https://ddbj.nig.ac.jp/DRASearch/experiment?acc=DRX225008,
https://ddbj.nig.ac.jp/DRASearch/experiment?acc=DRX225009,
https://ddbj.nig.ac.jp/DRASearch/experiment?acc=DRX225010,
https://ddbj.nig.ac.jp/DRASearch/experiment?acc=DRX225011).
Results
Sample collection, isolation of bacteria
and histamine level in tear fluids
Tear fluids and CL care solutions remaining in lens
storage cases were collected from 15 CL wearers (9 subjects with AS
and 6 healthy subjects) (Table
I). Bacterial cultures of CL care solutions were positive in 5
of the 9 subjects with AS and in 2 of the 6 healthy subjects, and
diverse bacteria were detected. The culture positive ratios between
CL care solutions from subjects with AS and healthy subjects did
not exhibit any statistically significant differences (P=0.3980,
data not shown). All the tear cultures were negative apart from 1
participant (subject 15) from whom the Staphylococcus
capitis was isolated. The total numbers of bacteria in culture
positive CL care solutions did not differ significantly between the
subjects with AS and healthy subjects (log10 CFU/ml,
2.82±1.89 vs. 4.95±1.09, P=0.2086, data not shown). Histamine was
detected in 2 subjects with AS (subject 2, 0.032 µg/ml;
subject 7, 0.223 µg/ml).
Microbiome analysis of CL care
solution
The bacterial compositions of CL care solutions were
assessed by 16S rDNA Illumina sequencing (Table SII). Diverse bacteria were
detected in the CL care solutions even in the culture-negative
samples. A BLAST search against the Greengenes database identified
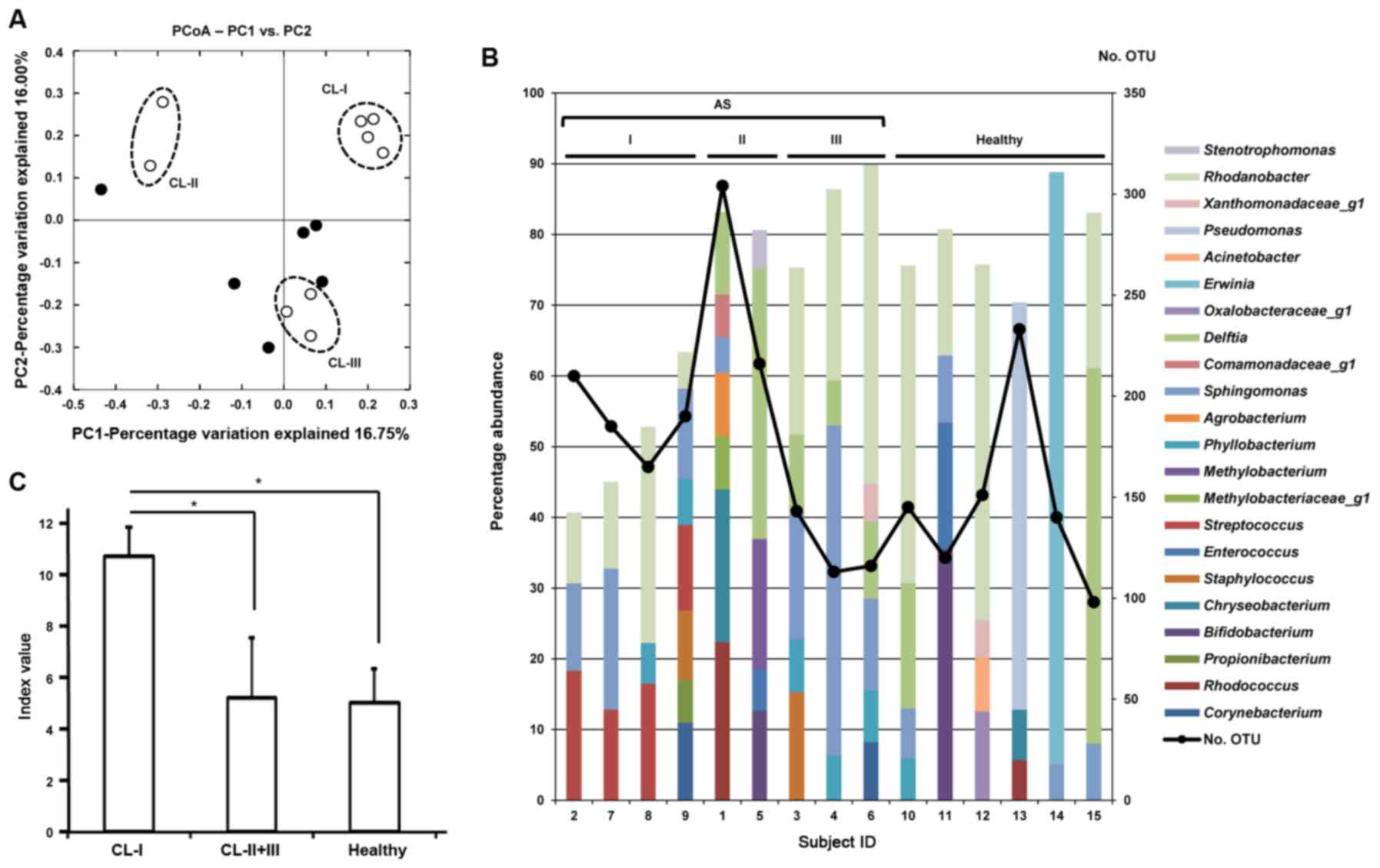
98-304 OTUs/sample. As shown in Fig.
1A, 3 contamination patterns
were observed in the CL care solutions of the subjects with AS by
PCoA. Group CL-I (subjects 2, 7, 8 and 9) exhibited highly similar
contamination patterns, which clearly differed from those of the
healthy subjects. Groups CL-II (subjects 1 and 5) and CL-III
(subjects 3, 4 and 6) were not clearly separated from the healthy
subjects. The abundance of Gram-positive bacteria in the microbiome
of the CL care solutions used by the CL-I group was higher than
that of the healthy subjects (42.24±9.47% vs. 16.85±22.76%
abundance) (Table SII). Fig. 1B shows the representative
bacterial genera with >5% abundance in each CL care solution.
The CL care solutions of the CL-I group uniquely contained
Streptococcus (shown in magenta) as a dominant member of the
contaminants (12.1-18.3% abundance). The total percentage of OTUs
with >5% abundance in the CL-I group was lower (<65%) than
that in the other subjects (>70%), although the observed OTU
number was relatively high (165-210). These findings indicate that
the CL care solutions of the CL-I group contained a diverse minor
population of bacteria. Correspondingly, the Shannon-Weaver index
calculated from the abundance at the genus level was markedly
higher in the CL-I group (10.70±1.14) than in the other AS
(5.19±2.36) or healthy (5.00±1.33) subjects (P<0.01) (Fig. 1C).
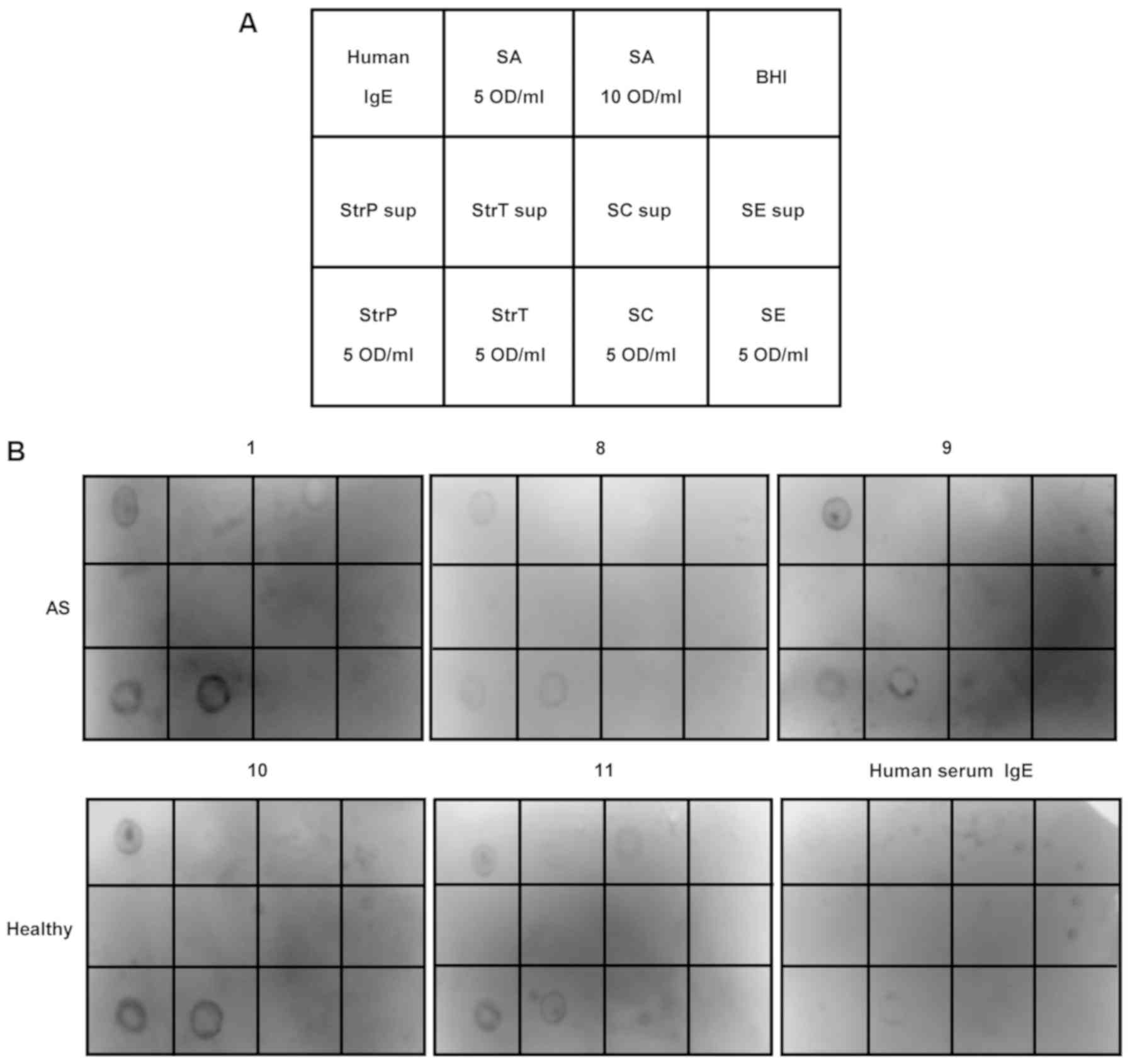 | Figure 3Western dot blot hybridization using
tear fluids. (A) Arrangement of western dot blot hybridization. The
nitrocellulose membrane was divided into 12 sections. Human IgE,
positive control; SA, bacterial cell suspension of
Staphylococcus (S.) aureus; BHI, brain heart infusion;
negative control, StrP sup, culture supernatants of
Streptococcus (Str.) pneumoniae; StrT sup, culture
supernatants of Str. tigurinus; SC sup, culture supernatants
of S. capitis; SE sup, culture supernatants of S.
epidermidis; StrP, bacterial cell suspension of Str.
pneumoniae; StrT, bacterial cell suspension of Str.
tigurinus; SC, bacterial cell suspension of S. capitis;
SE, bacterial cell suspension of S. epidermidis. (B) Images
of nitrocellulose membrane. IgE binding to bacterial cell
suspension of Str. pneumoniae and Str. tigurinus was
detected in tear fluids from subjects with allergic symptoms (AS)
as well as healthy subjects. |
Microbiome analysis of tear fluid
The microbial composition in the tear fluid from the
subjects with AS and healthy subjects was examined (Table SIII). The tear fluid from subject
1 was excluded from the analysis owing to its small volume. A total
of 36-177 OTUs/sample wer detected. Of note, the tear microbiome
analysis revealed similar trends to that of the CL care solutions.
As shown in the PCoA plot, tear fluids from the subjects with AS
clearly separated into groups resembling the microbiome patterns in
CL care solutions (Fig. 2A). The
representative bacterial genera with >5% abundance in each tear
fluid are shown in Fig. 2B. The
total percentage of OTUs with >5% abundance in the CL-I group
was lower (<50%) than that in other subjects (>50%); however,
the observed OTU number was relatively high (91-132). These results
indicated that bacterial diversity in the tear fluids of the CL-I
group was higher than that of the other subjects. Although
Streptococcus was detected in all the tear fluids, apart
from subject 5, its abundance in the CL-I group was significantly
higher than that in the other subjects (19.02±5.50 vs. 3.08±3.35%,
P<0.01). To compare the microbial compositions in the CL care
solutions and tear fluids, the Illumina sequences obtained from
both types of samples were merged and PCoA was performed (Fig. 2C). Although the PCoA plots were
clearly separated depending on the sample type, the plots denoting
tear and CL care solution samples of the CL-I group were close to
each other, as compared with the distance between plots denoting
tear and CL care solution samples in subjects other than the CL-I
group. This was due to the high abundance of Streptococcus
in both types of samples in the CL-I group.
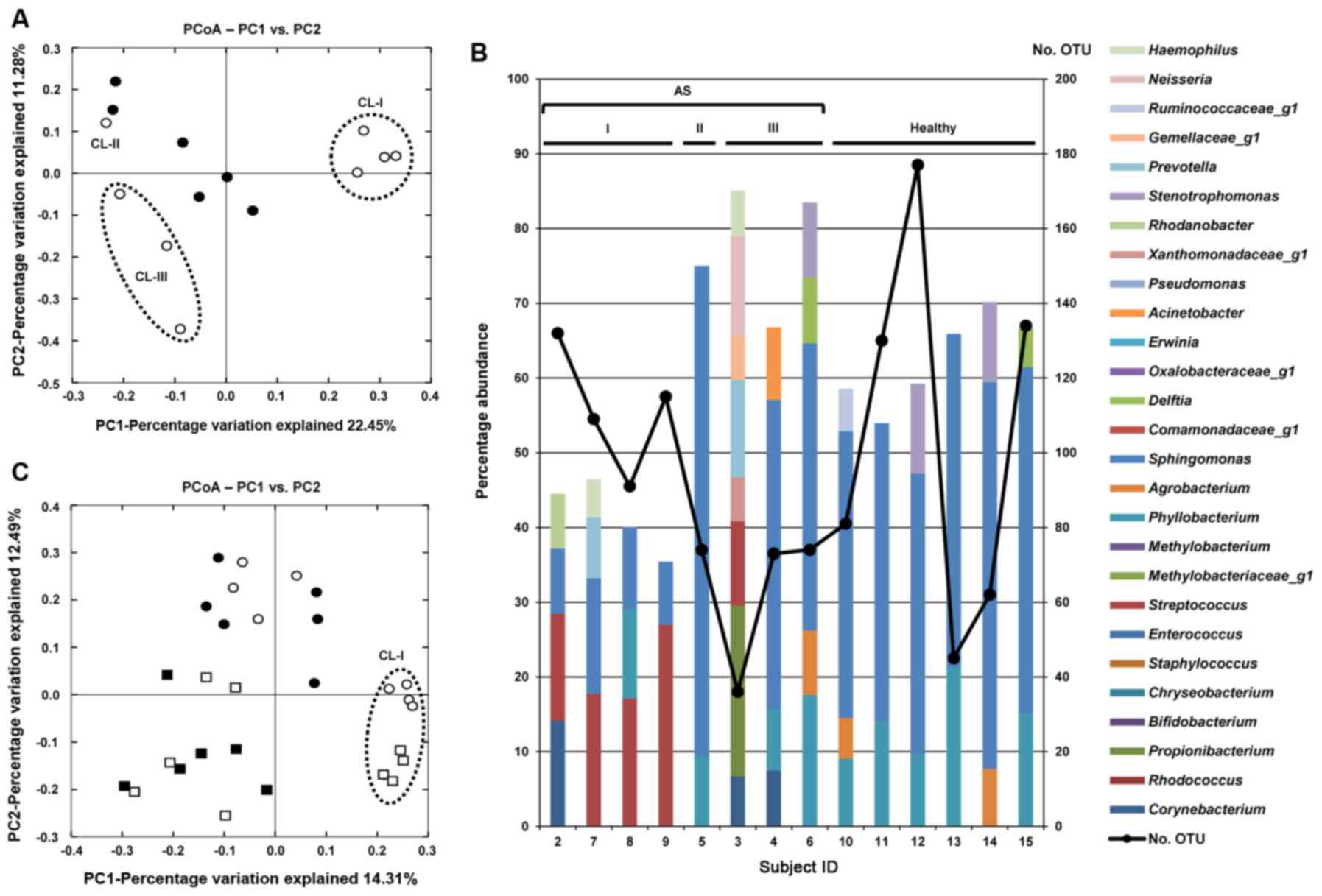 | Figure 2Microbiome analysis of tear fluids.
(A) PCoA of tear fluids. Black dots denote healthy subjects, and
white dots denote allergic symptoms (AS) subjects. Tear fluids from
AS subjects clearly separated according to the grouping based on
the microbiome patterns in contact lens (CL) care solutions. (B)
Representative bacterial genera with >5% abundance in each tear
fluid. Proportion of Streptococcus in the tear fluid of the
CL-I group was significantly higher than in the other subjects. (C)
Comparison of the microbial compositions in CL care solutions and
tear fluids. White circles denote tear fluids of subjects with AS,
black circles denote tear fluids of healthy subjects, white squares
denote CL care solutions of subjects with AS, and black squares
denote CL care solutions of healthy subjects. The microbiomes in CL
care solutions and tear fluids of the CL-I group were clustered
closely by PCoA. PCoA, principal coordinate analysis; CL, contact
lens; AS, allergic symptoms; OTU, operational taxonomic unit; I,
CL-I group; II, CL-II group; III, CL-III group. |
Detection of tear IgE reacting with
Gram-positive cocci
The arrangement of bacterial antigens is presented
in Fig. 3A. As shown in Fig. 3B, IgE binding to the bacterial
cell suspension of Streptococcus pneumoniae and
Streptococcus tigurinus was detected in the tear fluids of 6
subjects with AS (100%) as well as in 2 healthy subjects (100%).
IgE binding to Staphylococcus species was not present except
for subject 11, in which IgE weakly reacted with concentrated
Staphylococcus aureus cells. No signal was detected when the
culture supernatants were used as antigens. Interestingly, IgE from
human serum weakly reacted with Streptococcus tigurinus
cells.
Discussion
A number of studies have reported that CL and the
care accessories are contaminated with microorganisms not only in
patients with infectious keratitis, but also in asymptomatic CL
wearers (22-28). Sample cultivation has identified
the contaminating microorganisms in the CL care accessories, and
the most frequently isolated microorganisms are bacteria (22,26,28). Among CL care accessories, the lens
storage case is the most contaminated (23,24), and there are reports that the
contamination rate exceeds 80% (22,27). It is considered that lens storage
cases are contaminated rapidly, followed by contamination spread to
the CL and CL care solution bottles (24,28). Shin et al analyzed the
ocular surface microbiome of CL wearers using NGS technology and
reported that CL wearing alters the microbiome (5). Considering the possibility that the
source of CL contamination is the lens storage case (24,28), it is reasonable to analyze the
microbiome of the CL care solution in the lens storage case to
identify the bacteria which migrate to the ocular surface by CL
wearing.
In terms of the comparison of the subjects with AS
and the healthy subjects, the present study first investigated both
the CL care solutions in the lens storage cases and the tear fluids
of the CL wearers by culture. It was found that the culture
positive rate of the CL care solutions and the contaminant
bacterial number did not differ significantly between the subjects
with AS and the healthy subjects. As previously reported by
Tagliaferri et al (4),
this result suggests that CL care solutions of the subjects with AS
are not always more contaminated by cultivatable bacteria than
those from the healthy subjects. According to several articles that
investigated lens storage case bacterial contamination,
Staphylococcus species, Serratia species, and
Pseudomonas species are frequently isolated (23,24,26). The present study also mainly
isolated Pseudomonas sp., Microbacterium sp. and
Staphylococcus sp. (Table
I). The contamination sources are considered to be fingers,
eyelids and watery environments near the storage cases (25). However, there are a number of
fastidious bacteria in the environment (6), and the percentage of environmental
bacteria that can be grown in culture is only 1% (29). Attention should also be paid to
the fact that cultivation can only detect viable bacteria. Not only
viable bacteria, but also dead bacteria accumulating in the lens
storage case may be involved in an allergic conjunctivitis onset
(30). Therefore, meta-genomic
analysis is more advantageous for investigating the micro-biome of
lens storage cases than conventional cultivation. With regard to
the tear sample cultures, bacterial isolation failed for all but
one of the subjects. It is reasonable to assume that a few
microliters of the tear fluid contain only a minute number of
bacteria, as the tear fluid plays a role in self-cleaning of the
ocular surface using flow. Since it increases the probability of
detection by amplifying DNA even in a specimen containing a small
number of bacteria, genomic analysis is likely more useful than
culture.
As a result of the present study of the microbiome
in CL care solutions and tear fluids using NGS techniques, it was
found that the bacteria contained in the CL care solutions and the
tear fluids were more diverse than those detected by culture
(Tables SII and SIII). CL wearing may affect the tear
fluid microbiome by circulating the care solutions between the
ocular surface and storage case, which is supported by the fact
that the microbiome of tear fluids in subjects with AS was divided
into 3 clusters, similar to that of CL care solutions (Fig. 2A). It was hypothesized that the
contamination pattern of the CL care solution in each case varies
depending on individual environmental differences. In fact, the
microbiome of CL care solution did not exhibit a specific
contamination pattern in healthy subjects in this study (Fig. 1A). PCoA revealed that the CL-I
group in the subjects with AS was distinguished from the healthy
subjects (Figs. 1A and 2A) and the histamine concentration in
the tear fluids of the 2 subjects clustered in the CL-I group were
elevated (Table I). The CL care
solutions and the tear fluids of the CL-I group were found to be
contaminated with a different population of bacteria as compared
with other subjects (Figs. 1B, C
and 2B). In particular, the
proportion of Streptococcus were significantly higher
(Figs. 1B and 2B), suggesting the possible involvement
of Streptococcus in the etiology of at least certain
subtypes of CL-associated AS. By contrast, in the CL-II and CL-III
groups there was no significant difference in the microbiome of CL
care solutions from the healthy subjects; therefore factors other
than the change in the microbiome of CL care solutions may be
related to AS in the CL-II and III groups. Alternatively, the lack
of an elevation in the histamine concentration in the tear fluids
of subjects in the CL-II and III groups suggests that symptoms in
these subjects were not caused by an allergic response. Patients
with corneal and/or conjunctival disorders may experience itching
even without an allergic conjunctivitis (31,32). The division of subjects with AS
onto 3 clusters according to their microbiome PCoA plotting
patterns may have occurred as the disease that caused itching in
each group differed. However, whether the ocular surface microbiome
would differ based on the disease causing the itching remains to be
elucidated. Although dry eye and atopic dermatitis cases were
excluded, the cause of the symptoms in subjects belonging to the
CL-II and III groups cannot be specified, since all other diseases
that can induce AS were not differentiated. Therefore, the reason
for the 3 different microbiome PCoA plotting patterns cannot be
specified in the present study.
IgE is an important chemical mediator in allergic
diseases. CLPC, the allergic conjunctivitis associated with CL
wear, is considered to be related to type I and type IV allergies
(33). Studies have reported that
IgE of the ocular surface of CLPC patients was significantly
increased, compared with that of healthy CL wearers, owing to the
involvement of type I allergy (34,35). However, Zhao et al examined
only the total IgE eluted from the CL and did not examine
allergen-specific IgE; thus, the antigen has not been clarified
(34). Barishak et al
examined the specific antigen of IgE in tear fluids of CLPC
patients using radioallergosorbent test, and they detected IgE
against house dust mites and cat epithelium only in one of those
(35). In the present study, the
western dot blotting method was used to identify bacteria-specific
IgE in the tear fluids of subjects. In view of the results that a
number of Gram-positive bacteria, particularly Firmicutes, were
detected in the micro-biome (Tables
SII and SIII), the
reactivity of the tear IgE against the Streptococcus and
Staphylococcus was examined. IgE against
Streptococcus was detected in tear fluid not only from
subjects with AS, but also from healthy subjects (Fig. 3B), and the human serum IgE also
responded to Streptococcus, suggesting that
Streptococcus may have a strong influence on the human
immune system. Bisgaard et al reported that the colonization
of Streptococcus pneumoniae in the hypopharynx during the
neonatal period was involved in the development of pediatric asthma
(36). There is also a study
reporting that the colonization rate of Streptococcus
pneumoniae in the oropharynx is higher in asthma patients than
in healthy subjects (37).
Research published in recent years examining the airway microbiome
using 16S rDNA analysis has revealed that early asymptomatic
colonization with Streptococcus is a strong asthma predictor
and the presence of Streptococcus sp. is related to severe
asthma (38-41). Prior infection of the pharynx with
Streptococcus is involved in the onset and exacerbation of
psoriasis, an autoimmune skin disease. The molecular structure of
streptococcal M-protein and keratinocyte protein are similar, so
that streptococci-specific T cells are considered to cause a
cross-reactivation with epidermal self-antigens (42-45). These suggests that
Streptococcus is involved in the development and progression
of several immune diseases, and it possibly plays a similarly
important role in AS.
In the present study, the tear IgE response to
Streptococcus pneumoniae and Streptococcus tigurinus
was tested, both belonging to mitis group of streptococci (Fig. S1). Given that IgE did not react
with the culture supernatant, but with the bacterial cells in the
present study, it appears to react with common antigens of the
mitis group of streptococci that are expressed on the bacterial
surface but not with a substance produced and secreted by
Streptococcus. Since the IgE reacting to streptococcal
antigens is present in tear fluids of a wide range of individuals,
repeated exposure to the mitis group of streptococci may elicit AS
in certain populations.
There are several limitations to the present study.
First, the sample size was small. Patients were recruited who had
not suffered extensively from wearing contact lenses in their daily
life. Therefore, it was difficult to collect a large number of
candidates without providing incentives. Consequently, there was a
marked sex difference, with 12 females and 3 males. Although there
have been several reports of no sex differences regarding the rate
of contamination of CL care accessories (46,47), a biased sex ratio may still be a
confounding factor in the current microbiome analysis. CLPC is more
common in long-term CL wearers. However, in the present study, all
subjects were in their late teens or early 20s, and were using CLs
for <5 years; thus, CLPC was mild in all subjects. If a similar
microbiome study could be conducted by dividing a large number of
subjects based on short- and long-term CL wear experience, then new
findings may be obtained. Although the sample size was tolerable in
the present study, a larger sample size would still be more
reliable. Second, itching can also occur in eye diseases other than
allergies. Therefore, the results of the present study do not
suggest that Streptococcus spp. are associated with allergic
eye diseases, but only that they are associated with their
symptoms. Third, as the antigens have not been comprehensively
examined, the possibility that there may be an antigen related to
AS other than Streptococcus, such as certain aeroallergens
remains. Fourth, from the results of the present study, it was
undetermined whether AS developed due to the high proportion of
Streptococcus in the CL care solution and tear microbiome or
if the microbiome alteration following allergy development resulted
in the high proportion of Streptococcus in these samples.
Finally, in specimens with a low bacterial volume, the results may
be affected by contamination that can occur in all steps from
sample collection to DNA manipulation, although it is impossible to
completely avoid bacterial contamination from all experimental
processes. If the experimental process is the same in all samples,
the contamination level would be expected to be the same. Further
studies with large numbers of allergic conjunctivitis subjects,
diagnosed by serum IgE measurement and conjunctival scraping to
detect eosinophils and lacking bacterial contamination of the
ocular surface microbiome are required to resolve these
limitations. PCoA with an increased sample size may provide new
trends for the species involved in allergic conjunctivitis. If the
presence of specific IgE, in tears and serum, can be confirmed for
that species, the identification of the ones involved in allergic
conjunctivitis may then be possible.
In conclusion, in the present study, the CL care
solutions remaining in lens storage cases and tear fluids of some
CL wearers who suffer from AS exhibited a significantly higher
relative level of Streptococcus, and the IgE reacting to
streptococcal antigens was present in tear fluids. This suggests an
association between Streptococcus contamination of the lens
storage case and AS associated with CL wear. However, further
studies are required to determine one of the true antigens of AS
associated with CL wear.
Supplementary Data
Funding
HE received grant funding from Pfizer Japan Inc. YS
received grant funding from Pfizer Japan Inc. and Daiichi Sankyo Co
Ltd.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request. Sequence data have been deposited in the DDBJ database
(accession nos. DRA010383: https://ddbj.nig.ac.jp/DRASearch/
submission?acc=DRA010383; PRJDB10068: https://ddbj.nig.ac.jp/BPSearch/bioproject?acc=PRJDB10068;SAMD00232908-SAMD00232922:
https://ddbj.nig.ac.jp/DRASearch/experiment?acc=DRX224997,
https://ddbj.nig.ac.jp/DRASearch/experiment?acc=DRX224998,
https://ddbj.nig.ac.jp/DRASearch/experiment?
acc=DRX224999, https://ddbj.nig.ac.jp/DRASearch/
experiment?acc=DRX225000, https://ddbj.nig.ac.jp/DRASearch/ experiment?
acc=DRX225001, https://ddbj.nig.ac.jp/DRASearch/
experiment?acc=DRX225002, https://ddbj.nig.ac.jp/DRASearch/
experiment?acc=DRX225003, https://ddbj.nig.ac.jp/DRASearch/
experiment?acc=DRX225004, https://ddbj.nig.ac.jp/DRASearch/
experiment?acc=DRX225005, https://ddbj.nig.ac.jp/DRASearch/
experiment?acc=DRX225006, https://ddbj.nig.ac.jp/DRASearch/
experiment?acc=DRX225007, https://ddbj.nig.ac.jp/DRASearch/
experiment?acc=DRX225008, https://ddbj.nig.ac.jp/DRASearch/
experiment?acc=DRX225009, https://ddbj.nig.ac.jp/DRASearch/
experiment?acc=DRX225010, https://ddbj.nig.ac.jp/DRASearch/
experiment?acc=DRX225011).
Authors' contributions
HE and TK conceived the study, established the
methodology, and administered the projects. FH, HE, HNI, AT and HY
curated the data. TK and HNI analyzed the data and super-vised the
study. FH, HE, and TK validated the results. FH, HE, and TK wrote
the original draft of the manuscript. HE, TK, YS and SK conceived
and supervised the study, reviewed and edited the original draft.
All authors read and approved the final manuscript.
Ethics approval and consent to
participate
The present study followed the tenets of the
Declaration of Helsinki and was approved by the institutional
review board of Kindai University Faculty of Medicine (Approval no.
28-137). Informed consent was obtained from all subjects after
explanation of the nature and possible consequences of the
study.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgments
The authors would like to thank Mr. Yu Noguchi and
Mr. Takuya Taoka, Tokushima University, School of Medicine, for
providing assistance with collecting the CL storage cases.
Abbreviations:
|
AS
|
allergic symptoms
|
|
CL
|
contact lens
|
|
CLPC
|
contact lens-induced papillary
conjunctivitis
|
|
NGS
|
next-generation sequencing
|
|
BHI
|
brain heart infusion
|
|
TE
|
Tris-ethylenediaminetetraacetic
acid
|
|
HPLC
|
high-performance liquid
chromatography
|
|
PBS
|
phosphate-buffered saline
|
|
OUT
|
operational taxonomic unit
|
|
PCoA
|
principal coordinate analysis
|
|
PFA
|
paraformaldehyde
|
|
PBS-T
|
PBS with Tween-20
|
References
|
1
|
Skotnitsky CC, Naduvilath TJ, Sweeney DF
and Sankaridurg PR: Two presentations of contact lens-induced
papillary conjunctivitis (CLPC) in hydrogel lens wear: Local and
general. Optom Vis Sci. 83:27–36. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Solomon A: Allergic manifestations of
contact lens wearing. Curr Opin Allergy Clin Immunol. 16:492–497.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Tan ME, Demirci G, Pearce D, Jalbert I,
Sankaridurg P and Willcox MD: Contact lens-induced papillary
conjunctivitis is associated with increased albumin deposits on
extended wear hydrogel lenses. Adv Exp Med Biol. 506:951–955. 2002.
View Article : Google Scholar
|
|
4
|
Tagliaferri A, Love TE and Szczotka-Flynn
LB: Risk factors for contact lens-induced papillary conjunctivitis
associated with silicone hydrogel contact lens wear. Eye Contact
Lens. 40:117–122. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Shin H, Price K, Albert L, Dodick J, Park
L and Dominguez-Bello MG: Changes in the eye microbiota associated
with contact lens wearing. MBio. 7:e001982016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Amann RI, Ludwig W and Schleifer KH:
Phylogenetic identification and in situ detection of indivisual
microbial cells without cultivation. Microbiol Rev. 59:143–169.
1995. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Human Microbiome Project Consortium:
Structure, function and diversity of the healthy human microbiome.
Nature. 486:207–214. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ahmed S, Macfarlane GT, Fite A, McBain AJ,
Gilbert P and Macfarlane S: Mucosa-associated bacterial diversity
in relation to human terminal ileum and colonic biopsy samples.
Appl Environ Microbiol. 73:7435–7442. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Grice EA, Kong HH, Renaud G, Young AC;
NISC Comparative Sequencing Program; Bouffard GG, Blakesley RW,
Wolfsberg TG, Turner ML and Segre JA: A diversity profile of the
human skin microbiota. Genome Res. 18:1043–1050. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Dong Q, Brulc JM, Iovieno A, Bates B,
Garoutte A, Miller D, Revanna KV, Gao X, Antonopoulos DA, Slepak VZ
and Shestopalov VI: Diversity of bacteria at healthy human
conjunctiva. Invest Ophthalmol Vis Sci. 52:5408–5413. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Eguchi H, Hotta F, Kuwahara T, Imaohji H,
Miyazaki C, Hirose M, Kusaka S, Fukuda M and Shimomura Y:
Diagnostic approach to ocular infections using various techniques
from conventional culture to next-generation sequencing analysis.
Cornea. 36(Suppl 1): S46–S52. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Huang Y, Yang B and Li W: Defining the
normal core micro-biome of conjunctival microbial communities. Clin
Microbiol Infect. 22:643.e7–643.e12. 2016. View Article : Google Scholar
|
|
13
|
Bakhtiar SM, LeBlanc JG, Salvucci E, Ali
A, Martin R, Langella P, Chatel JM, Miyoshi A, Bermúdez-Humarán LG
and Azevedo V: Implications of the human microbiome in inflammatory
bowel diseases. FEMS Microbiol Lett. 342:10–17. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Turnbaugh PJ, Ley RE, Mahowald MA, Magrini
V, Mardis ER and Gordon JI: An obesity-associated gut microbiome
with increased capacity for energy harvest. Nature. 444:1027–1031.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Chiacchierini E, Restuccia D and Vinci G:
Evaluation of two different extraction methods for chromatographic
determination of bioactive amines in tomato products. Talanta.
69:548–555. 2006. View Article : Google Scholar
|
|
16
|
Morita H, Kuwahara T, Ohshima K, Sasamoto
H, Itoh K, Hattori M, Hayashi T and Takami H: An improved DNA
isolation method for metagenomic analysis of the microbial flora of
the human intestine. Microbes Environ. 22:214–222. 2007. View Article : Google Scholar
|
|
17
|
Klindworth A, Pruesse E, Schweer T,
Peplies J, Quast C, Horn M and Glöckner FO: Evaluation of general
16S ribosomal RNA gene PCR primers for classical and
next-generation sequencing-based diversity studies. Nucleic Acids
Res. 41:e12013. View Article : Google Scholar :
|
|
18
|
Aronesty E: Comparison of sequencing
utility programs. Open Bioinformatics J. 7:1–8. 2013. View Article : Google Scholar
|
|
19
|
Navas-Molina JA, Peralta-Sánchez JM,
González A, McMurdie PJ, Vázquez-Baeza Y, Xu Z, Ursell LK, Lauber
C, Zhou H, Song SJ, et al: Advancing our understanding of the human
microbiome using QIIME. Methods Enzymol. 531:371–444. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Edgar RC: Search and clustering orders of
magnitude faster than BLAST. Bioinformatics. 26:2460–2461. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Kanda Y: Investigation of the freely
available easy-to-use soft-ware 'EZR' for medical statistics. Bone
Marrow Transplant. 48:452–458. 2013. View Article : Google Scholar
|
|
22
|
Dantam J, McCanna DJ, Subbaraman LN,
Papinski D, Lakkis C, Mirza A, Berntsen DA, Morgan P, Nichols JJ
and Jones LW: Microbial contamination of contact lens storage cases
during daily wear use. Optom Vis Sci. 93:925–932. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Boost MV and Cho P: Microbial flora of
tears of orthokeratology patients, and microbial contamination of
contact lenses and contact lens accessories. Optom Vis Sci.
82:451–458. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Yung MS, Boost M, Cho P and Yap M:
Microbial contamination of contact lenses and lens care accessories
of soft contact lens wearers (university students) in Hong Kong.
Ophthalmic Physiol Opt. 27:11–21. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Willcox MD, Power KN, Stapleton F, Leitch
C, Harmis N and Sweeney DF: Potential sources of bacteria that are
isolated from contact lenses during wear. Optom Vis Sci.
74:1030–1038. 1997. View Article : Google Scholar
|
|
26
|
Wu YT, Zhu H, Harmis NY, Iskandar SY,
Willcox M and Stapleton F: Profile and frequency of microbial
contamination of contact lens cases. Optom Vis Sci. 87:E152–E158.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Willcox MD, Carnt N, Diec J, Naduvilath T,
Evans V, Stapleton F, Iskandar S, Harmis N, de la Jara PL and
Holden BA: Contact lens case contamination during daily wear of
silicone hydrogels. Optom Vis Sci. 87:456–464. 2010.PubMed/NCBI
|
|
28
|
McLaughlin-Borlace L, Stapleton F,
Matheson M and Dart JK: Bacterial biofilm on contact lenses and
lens storage cases in wearers with microbial keratitis. J Appl
Microbiol. 84:827–838. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Staley JT and Konopka A: Measurement of in
situ activities of nonphotosynthetic microorganisms in aquatic and
terrestrial habitats. Annu Rev Microbiol. 39:321–346. 1985.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Eguchi H, Hotta F, Kuwahara T,
Nakayama-Imaohji H, Kusaka S and Shimomura Y: Acute
keratoconjunctivitis due to contamination of contact lens care
solution with histamine-producing Raoultella species. A case
report. Medicine (Baltimore). 96:e93102017. View Article : Google Scholar
|
|
31
|
Nichols KK: Patient-reported symptoms in
dry eye disease. Ocul Surf. 4:137–145. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Belmonte C: Pain, dryness, and itch
sensations in eye surface disorders are defined by a balance
between inflammation and sensory nerve injury. Cornea. 38(Suppl 1):
S11–S24. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Stapleton F, Stretton S, Sankaridurg PR,
Chandoha H and Shovlin J: Hypersensitivity responses and contact
lens wear. Cont Lens Anterior Eye. 26:57–69. 2003. View Article : Google Scholar
|
|
34
|
Zhao Z, Fu H, Skotnitsky CC, Sankaridurg
PR and Willcox MD: IgE antibody on worn highly oxygen-permeable
silicone hydrogel contact lenses from patients with contact
lens-induced papillary conjunctivitis (CLPC). Eye Contact Lens.
34:117–121. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Barishak Y, Zavaro A, Samra Z and
Sompolinsky D: An immunological study of papillary conjunctivitis
due to contact lenses. Curr Eye Res. 3:1161–1168. 1984. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Bisgaard H, Hermansen MN, Buchvald F,
Loland L, Halkjaer LB, Bønnelykke K, Brasholt M, Heltberg A,
Vissing NH, Thorsen SV, et al: Childhood asthma after bacterial
colonization of the airway in neonates. N Engl J Med.
357:1487–1495. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Jounio U, Juvonen R, Bloigu A,
Silvennoinen-Kassinen S, Kaijalainen T, Kauma H, Peitso A,
Saukkoriipi A, Vainio O, Harju T and Leinonen M: Pneumococcal
carriage is more common in asthmatic than in non-asthmatic young
men. Clin Respir J. 4:222–229. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Teo SM, Mok D, Pham K, Kusel M, Serralha
M, Troy N, Holt BJ, Hales BJ, Walker ML, Hollams E, et al: The
infant airway micro-biome in health and disease impacts later
asthma development. Cell Host Microbe. 17:704–715. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Zhang Q, Cox M, Liang Z, Brinkmann F,
Cardenas PA, Duff R, Bhavsar P, Cookson W, Moffatt M and Chung KF:
Airway microbiota in severe asthma and relationship to asthma
severity and phenotypes. PLoS One. 11:e01527242016. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Kloepfer KM, Lee WM, Pappas TE, Kang TJ,
Vrtis RF, Evans MD, Gangnon RE, Bochkov YA, Jackson DJ, Lemanske RF
Jr, et al: Detection of pathogenic bacteria during rhinovirus
infection is associated with increased respiratory symptoms and
exacerbations of asthma. J Allergy Clin Immunol. 133:1301–1307.
e1–e3. 2014. View Article : Google Scholar :
|
|
41
|
Green BJ, Wiriyachaiporn S, Grainge C,
Rogers GB, Kehagia V, Lau L, Carroll MP, Bruce KD and Howarth PH:
Potentially pathogenic airway bacteria and neutrophilic
inflammation in treatment resistant severe asthma. PLoS One.
9:e1006452014. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Weisenseel P and Prinz JC: Incidental
detection of S. pyogenes-DNA in psoriatic skin by PCR. Arch
Dermatol Res. 296:573–576. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Valdimarsson H, Thorleifsdottir RH,
Sigurdardottir SL, Gudjonsson JE and Johnston A: Psoriasis-as an
autoimmune disease caused by molecular mimicry. Trends Immunol.
30:494–501. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Prinz JC: Psoriasis vulgaris-a sterile
antibacterial skin reaction mediated by cross-reactive T cells? An
immunological view of the pathophysiology of psoriasis. Clin Exp
Dermatol. 26:326–332. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Gudjonsson JE, Thorarinsson AM,
Sigurgeirsson B, Kristinsson KG and Valdimarsson H: Streptococcal
throat infections and exacerbation of chronic plaque psoriasis: A
prospective study. Br J Dermatol. 149:530–534. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Gray TB, Cursons RT, Sherwan JF and Rose
PR: Acanthamoeba, bacterial, and fungal contamination of contact
lens storage cases. Br J Ophthalmol. 79:601–605. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Devonshire P, Munro FA, Abernethy C and
Clark BJ: Microbial contamination of contact lens cases in the west
of Scotland. Br J Ophthalmol. 77:41–45. 1993. View Article : Google Scholar : PubMed/NCBI
|