Introduction
Breast cancer is one of the most common malignant
types of tumor among women. In advanced stages, cancer cells
metastasize to the liver, brain, lung and bone tissue, along the
lymphatic and blood vessels. Metastasis is a complex process
involving the dysregulation of multiple genes (1). Abnormally expressed microRNAs
(miRNAs or miRs) are closely related to the development of breast
cancer (2). Breast
cancer-associated miRNAs (let-7, miR-155, miR-21, miR-510, miR-192,
miR-200, etc.) have been shown to be involved in the regulation of
cell proliferation, differentiation, apoptosis and the maintenance
of breast cancer stem cells (3,4).
Although a number of oncogenes and tumor suppressor genes have been
implicated in the development and progression of breast cancer, the
underlying molecular mechanisms remain poorly understood.
miRNAs are endogenous short-chain RNAs, of
approximately 22 nucleotides in length (5). miRNAs are important
post-transcriptional regulators that induce mRNA degradation and
the translational repression of genes by binding to the 3′
untranslated regions (UTRs) of their target mRNAs (6). The dysregulation of miRNA expression
contributes to the development of multiple types of cancers.
miRNA-28-5p is an intronic miRNA of the lipoma-preferred partner
gene (LPP), which is located on chromosome 3q27-28.
miR-28-5p directly binds to the LPP mRNA and suppresses its
expression, which subsequently inhibits cell migration and adhesion
(7). miR-28-5p has been found to
function as a tumor suppressor, as it has been shown to be
downregulated in various types of human malignancies, such as
hepatocellular carcinoma (8),
renal cancer (9), colorectal
cancer (10) and in
nasopharyngeal cancer cells (11). miR-28-5p is involved in the
regulation of cell proliferation, apoptosis and metastasis. For
example, it has been shown to inhibit the proliferation and
migration of renal cancer cell lines by suppressing the expression
of Ras-related protein Rap-1b (RAP1B) (9); the expression of miR-28-5p is
associated with the metastasis, recurrence and the poor prognosis
of liver cancer (8). The
overexpression of miR-28-5p in HCT116, RKO and SW480 cells has been
shown to reduce cell migration and invasion (10). In addition, miR-28-5p
overexpression has been shown to suppress nasopharyngeal cancer
cell proliferation and induce cell cycle arrest and apoptosis
(11). In the present study,
bioinformatics analysis using the Human MicroRNA Expression
Database (HMED) also revealed that miR-28-5p was expressed at a low
level in breast cancer. However, the function of miR-28-5p in
breast cancer cell metastasis, and the target genes of miR-28-5p in
breast cancer are poorly understood.
The present study thus aimed to investigate the role
of miR-28-5p in breast cancer migration, identify the target genes
of miR-28-5p, and elucidate the molecular mechanisms through which
miR-28-5p regulates its target genes. The results of the present
study may enhance our under-standing of the role of miR-218-5p in
the development of breast cancer.
Materials and methods
Plasmid construction
The human WD repeat and SOCS box containing 2 (WSB2)
coding sequence (CDS) (NM_018639.5) was amplified from MCF-7 cell
cDNA using PCR with the following primers: Forward,
5′-TTCAAGCTTATGGAGGCCGGAGAGGAA-3′ and
reverse, 5′-TTAGGATCCTTAAAAAGTCCTGTATGTG-3′.
The PCR fragments were recovered, digested with HindIII III
(AAGCTT; indicated by underlined text above) and BamHI
(GGATCC; indicated by underlined text above), and then cloned into
the pEGFP-C3 vector (BD Biosciences).
The 3′-UTR of WSB2 was amplified from MCF-7
cell cDNA by PCR with the following primers: Forward,
5′-TCGCTCGAGATGACTATTCAGATGGCTAC-3′
and reverse, 5′-AGAGCGGCCGCCTCCATAAAGCACCGATT-3′.
The PCR fragments were recovered, digested with XhoI
(CTCGAG; indicated by underlined text above) and NotI
(GCGGCCGCC; indicated by underlined text above), and then cloned
into the psiCHECK™-2 vector (Promega Corporation).
WSB2-3′-UTR target site-directed mutagenesis
was performed using a Quik Change Site-Directed Mutagenesis kit
(Stratagene; Agilent Technologies, Inc.) with the forward primer,
5′-TTCAACTCTACTGCGAAACAAAAATAACCCATTAAAAGTACTGTTCTCCTTCAGTG-3′
and the reverse primer, 5′-CACTGAAGGAGAACAGTACTTTTAATGGGTTATTTTTGTTTCGCAGTAGAGTTGAA-3′.
Underlined base pairs indicate mutation sites.
Cells and cell culture
Human mammary epithelial cells (HMECs), and the
T-47D, ZR-75-30, MDA-MB-231 and MCF-7 cells (ATCC) were maintained
in Dulbecco's modified Eagle's medium (DMEM)-high glucose medium
(Gibco; Thermo Fisher Scientific, Inc.) supplemented with 10% fetal
bovine serum (HyClone; GE Healthcare Life Sciences), 100 U/ml
penicillin and streptomycin, and incubated at 37°C with 5%
CO2.
Cell transfection
The MCF-7 cells were transfected with the plasmids
using Lipofectamine™ 2000 (Invitrogen, Thermo Fisher Scientific,
Inc.), according to the manufacturer's instructions.
The MCF-7 cells were seeded at a density of
1×104 cells/well in 96-well plates, and transfected with
40 nM of miR-28-5p mimics, the miR-28-5p inhibitor, or controls
(Shanghai GenePharma Co., Ltd.) for the migration assay. The cells
were co-transfected with 40 nM of miR-28-5p mimics, or control,
with 100 ng/well pEGFP-C3-WSB2 or pEGFP-C3 for the migration assay.
The cells were co-transfected with 100 ng/well of WSB2 3′UTR-wt or
WSB2 3′UTR-mut and 40 nM of miR-28-5p mimics, the miR-28-5p
inhibitor, and controls for a dual-luciferase reporter (DLR)
assay.
The MCF-7 cells were seeded at a density of
7×104 cells/well in 24-well plates. The cells were
transfected with 200 ng/well pEGFP-C3-WSB2, or pEGFP-C3 for
subcellular localization and western blot analysis. The cells
transfected with 40 nM of miR-28-5p mimics, the miR-28-5p
inhibitor, or controls for reverse transcription-quantitative PCR
(RT-qPCR) and western blotting. The cells were transfected with 20,
40 or 60 nM of miR-28-5p mimics and control for RT-qPCR. The cells
were transfected with 40 nM of miR-28-5p mimics and control for
gene chip assay.
RT-qPCR. Total RNA was extracted, purified
and used for first-strand cDNA synthesis; RT-qPCR reagents and
procedures were as previously described (12). Quantification was performed using
the 2−ΔΔCq method (13).
The specific product of human miR-28-5p was
amplified by RT-qPCR using the following primers: miR-28-5p
forward, 5′-GCG CAT TGC ACT TGT CTC G-3′ and reverse, 5′-AGT GCA
GGG TCC GAG GTA TT-3′; and U6 forward, 5′-CTC GCT TCG GCA GCA
CATA-3′ and reverse, 5′-CGA ATT TGC GTG TCA TCCT-3′. U6 was used as
an internal control.
MCF-7 cells were transfected as aforementioned and
at 12, 24 and 48 h following transfection, total RNA was extracted.
The specific products of human RAP1B, WSB2 and
VEGFA were amplified by RT-qPCR using the following primers:
RAP1B forward, 5′-AAG AAA GTC CAA AG-3′ and reverse, 5′-TTT
CCT TCA ACA-3′; WSB2 forward, 5′-GTT AAT TCG GAA GCT AGA
GG-3′ and reverse, 5′-CAA AGC CCA TTG GTC ATA-3′; VEGFA
forward, 5′-ATT GGA GCC TTG CCT TGC-3′ and reverse, 5′-TCC ACC AGG
GTC TCG ATT G-3′; and glyceraldehyde-3-phosphate dehydrogenase
(GAPDH) forward, 5′-TGA CTT CAA CAG CGA CAC CCA-3′ and reverse,
5′-CAC CCT GTT GCT GTA GCC AAA-3′. GAPDH was used as an
internal control.
Cell migration assay
The MCF-7 cells were transfected as described above.
After 24 h, a cell migration assay was performed as previously
described using Transwell chambers (14). After 16 h, the cells passing the
membrane were stained using 0.25% crystal violet (Sigma-Aldrich) at
37°C for 10 min, and the crystal violet-stained cells were washed
off using 33% acetic acid and measured on a spectrophotometer
(Infinite M200; Tecan Group, Ltd.) at 570 nm.
DLR assay
MCF-7 cells were transfected as described above.
After 24 h, DLR assay was performed using the DLR Assay system
(Promega Corp.; Turner BioSystems, Inc.) according to the
manufacturer's protocol. Luciferase activity was normalized to
Renilla luciferase activity. Experiments were performed in
triplicate.
Subcellular localization
The MCF-7 cells were transfected as described above.
At 24 h following transfection, the cell nuclei were stained using
4′,6-diamidino-2-phenylindole (DAPI) for 30 min at 37°C and
visualized under an inverted fluorescence microscope (TE 2000-U;
Nikon Corporation), as previously described (15).
Western blot analysis
The MCF-7 cells were transfected as described above.
At 24 h post-transfection, western blot analysis was performed as
previously described (16).
Anti-green fluorescent protein (GFP) antibody (SC8334; Santa Cruz
Biotechnology, Inc.; 1:1,000 dilution), anti-WSB2 antibody
(ab127176; Abcam; 1:1,000 dilution), anti-β-actin antibody (ab8227;
Abcam; 1:10,000 dilution), and a secondary antibody (goat
anti-rabbit IgG H&L) conjugated to horseradish peroxidase (HRP)
(ab6721, Abcam, 1:10,000 dilution) were used. All anti-bodies were
incubated with the membranes for 90 min at 37°C.
Gene chip assay
The MCF-7 cells were transfected as described above.
At 24 h post-transfection, RNA was extracted using TRIzol
(15596-018; Thermo Fisher Scientific, Inc.), purified using an
RNeasy mini kit (74106; Qiagen GmbH), amplified and labeled using
the low input quick amp labeling kit (5190-2305; Agilent
Technologies, Inc.); each slide was then hybridized, scanned; and
data were extracted, and normalized as previously described
(12,17). Two samples were used for gene chip
assay. Differentially expressed mRNAs were identified as |log2(fold
change)|≥1.5. The Kyoto Encyclopedia of Genes and Genomes (KEGG)
(http://www.genome.jp/kegg, release
number 91.0; July, 2019) pathway analysis was used to characterize
the differentially expressed genes (DEGs).
Bioinformatics analysis
miR-28-5p expression in breast tumor and normal
breast tissue was analyzed using HMED (http://bioinfo.life.hust.edu.cn/web/GEDS/) (18). The target gene of miR-28-5p was
predicted using TargetScan7.1 (http://www.targetscan.org/), PicTar (http://pictar.mdc-berlin.de/) and miRDB (http://www.mirdb.org/) soft-ware. The association
between miR-28-5p and WSB2 was analyzed using bc-GenExMiner v4.4
(http://bcgenex.centregauducheau.fr/BC-GEM/GEM-requete.php).
The expression of WSB2 in breast invasive carcinoma (BRCA)
based on The Cancer Genome Atlas (TCGA) samples, was examined
through UALCAN (http://ualcan.path.uab.edu/cgi-bin/TCGAExResultNew2.pl)
(19). KMPLOT analysis of the
association of WSB2 with BRCA patient survival was also examined
through UALCAN (19). KMPLOT
analysis of the association of miR-28-5p with BRCA patient survival
was based on the Molecular Taxonomy Of Breast Cancer International
Consortium (METABRIC) (http://kmplot.com/analysis/index.php?p=service&cancer=breast_mirna)
(20).
Immunohistochemical analysis
A microarray containing samples from 40 cases of
breast cancer and matching adjacent normal tissue was obtained
(BR804a, Avilabio). Anti-WSB2 antibody (ab187987; Abcam, 1:50
dilution) was applied, and immunohistochemical analysis was
performed as previously described (14). The results of staining were
analyzed using ImageJ software version 1.46.
Statistical analysis
SPSS 9.0 software (SPSS, Inc.) was used for
statistical analysis. Data were analyzed using the Student's
t-test, and one-way analysis of variance followed by a Dunnett's
test or Tukey's post hoc test. Pearson's correlation analysis was
used to investigate the correlation between miR-28-5p and WSB2
expression. A P-value <0.05 was considered to indicate a
statistically significant difference.
Results
miR-28-5p inhibits the migration of MCF-7
cells
miR-28-5p expression in breast tumor and normal
breast tissue was analyzed using HMED (18). miR-28-5p exhibited a lower
expression in breast cancer tissue than in normal tissue (Fig. 1A). Kaplan-Meier analysis revealed
that a low expression level of miR-28-5p was associated with a poor
survival in breast cancer (Fig.
1B). Subsequently, RT-qPCR was performed to examine the
expression levels of miR-28-5p in 4 human breast cancer cell lines,
and HMECs. As shown in Fig. 1C,
the expression of miR-28-5p was significantly lower in the 4 human
breast cancer cell lines (ZR-75-30, MDA-MB-231, T-47D and MCF-7)
compared with that in HMECs, suggesting that the decreased
expression of miR-28-5p is associated with the development of
breast cancer. The expression level of miR-28-5p in MCF-7 cells was
higher than that in the other 3 breast cancer cell lines (Fig. 1C). Furthermore, the effect of
miR-28-5p on the migration of MCF-7 cells was examined by a
Transwell chamber assay. MCF-7 cells were transfected with
miR-28-5p mimics, miR-28-5p inhibitor, or the respective controls.
The expression level of miR-28-5p was confirmed by RT-qPCR
(Fig. 1D). It was found that the
MCF-7 cells transfected with miR-28-5p mimics exhibited a decrease
in cell migration compared with that in the control, whereas
transfection with miR-28-5p inhibitor resulted in an increased
mobility of the MCF-7 cells compared with that in the control
(Fig. 1E). The cells passing
through the membrane were further quantified using crystal violet
staining (Fig. 1F).
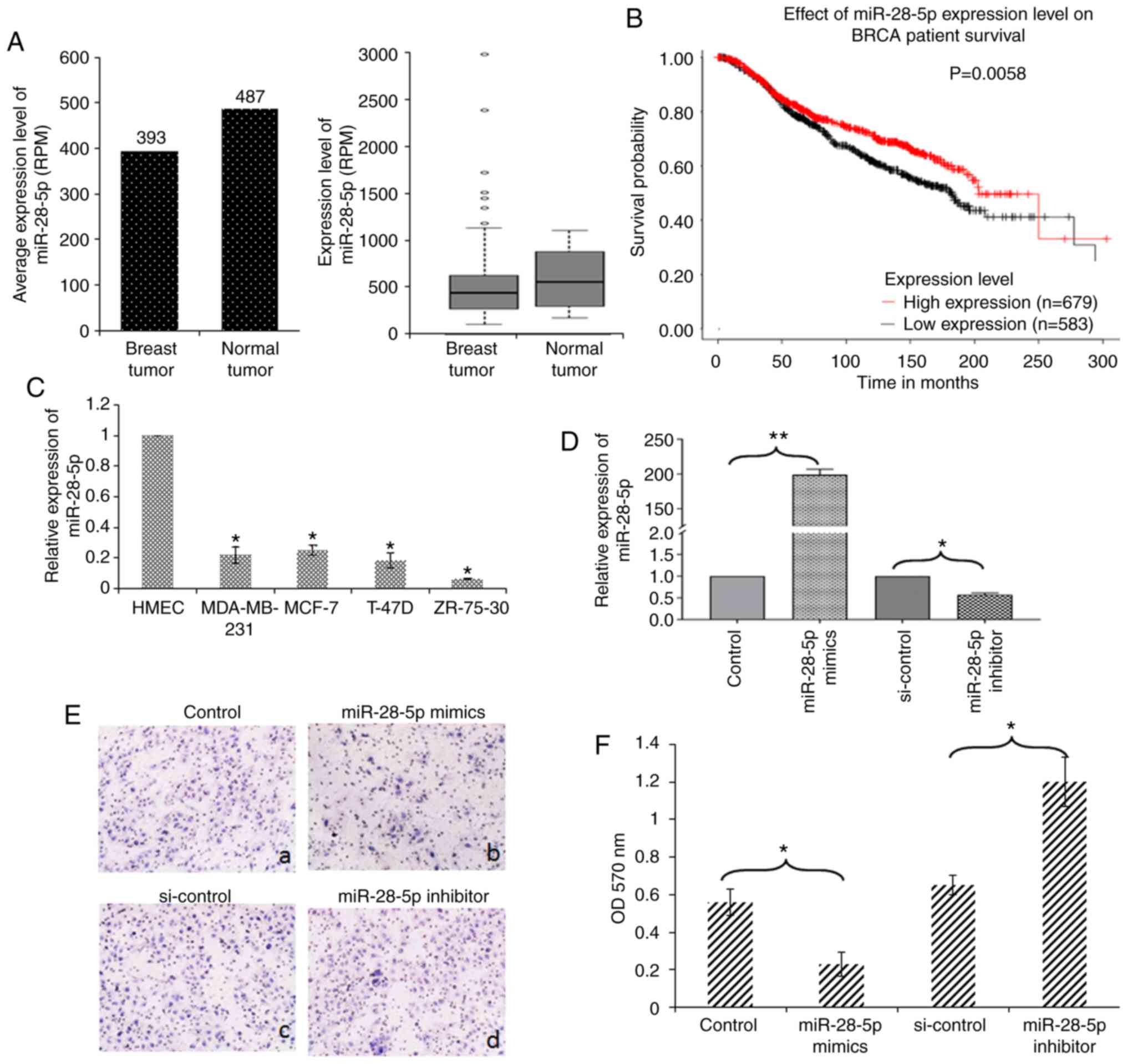 | Figure 1miR-28-5p inhibits the migration of
MCF-7 cells. (A) miR-28-5p expression in breast tumor and normal
breast tissue was analyzed using HMED. (B) Kaplan-Meier analysis of
the effect of the miR-28-5p expression level on BRCA patient
survival was analyzed using KMPLOT. (C) The expression levels of
miR-28-5p in ZR-75-30, MDA-MB-231, T-47D, MCF-7 cells and HMECs
were determined by RT-qPCR. *P<0.05 vs. HMECs. (D)
MCF-7 cells were transfected with control, miR-28-5p mimics,
si-control, or the miR-28-5p inhibitor, and the expression levels
of miR-28-5p were determined using RT-qPCR. *P<0.05,
**P<0.01 vs. the respective control. (E and F) MCF-7
cells were transfected with (a) control, (b) miR-28-5p mimics, (c)
si-control, or (d) miR-28-5p inhibitor and subjected to Transwell
assays. After 16 h, the cells passing the membrane were dyed using
0.25% crystal violet and assessed spectrophoto-metrically.
*P<0.05 vs. the respective control. |
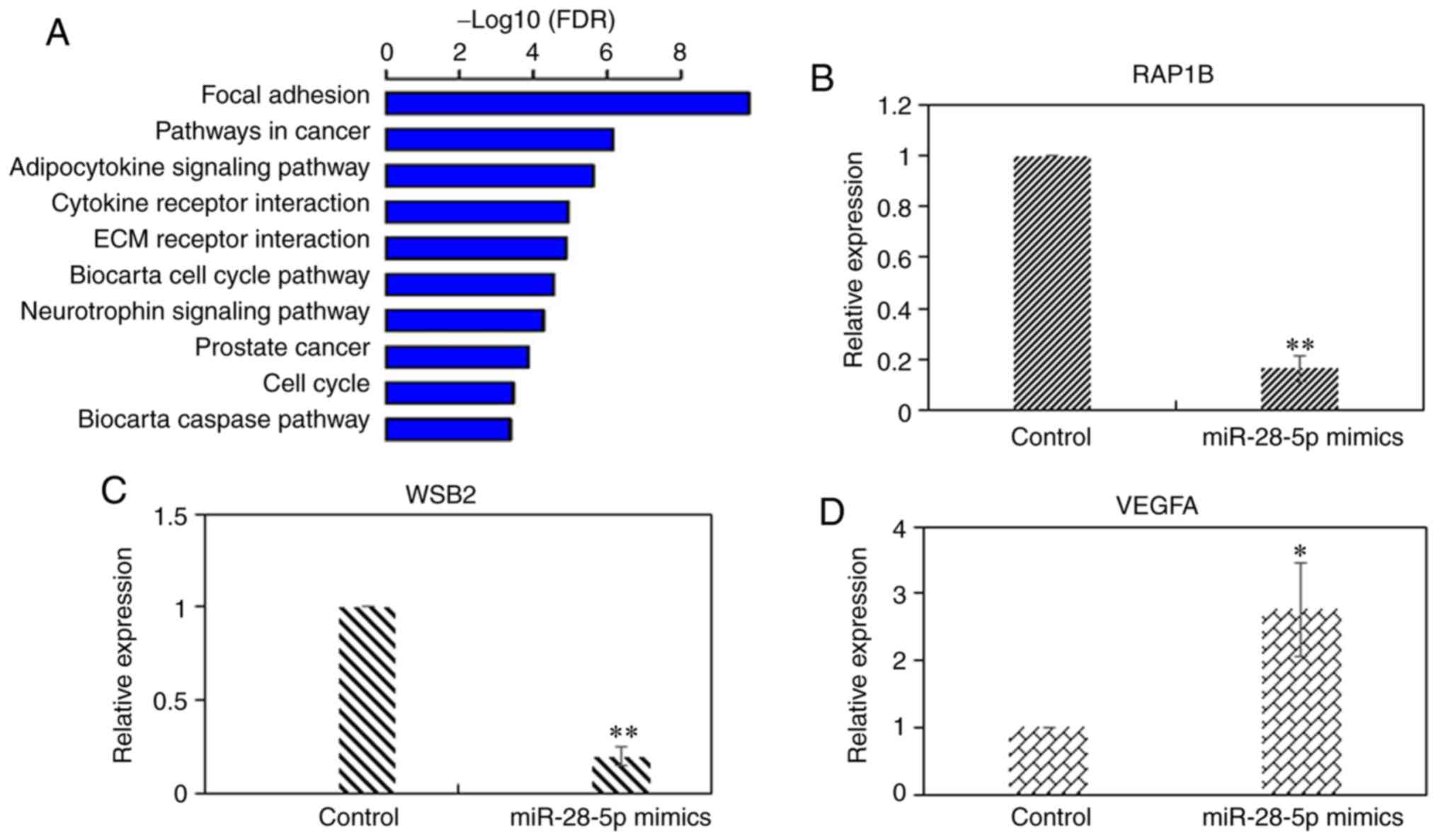
Target genes regulated by miR-28-5p
miRNAs regulate gene expression by binding to the
3′UTRs of their target mRNAs (6).
In the present study, to determine the target genes of miR-28-5p,
gene chip analysis we used to identify the DEGs between MCF-7 cells
overexpressing miR-28-5p and the negative control miRNA. The
overexpression of miR-28-5p was confirmed by RT-qPCR (Fig. 1D). The results of gene chip assay
revealed that 648 genes were differentially expressed, including
283 upregulated genes and 365 downregulated genes in MCF-7 cells
overexpressing miR-28-5p (Table
SI). The DEGs were analyzed using KEGG pathway analysis. The
top 10 significantly enriched pathways are shown in Fig. 2A. RAP1B and VEGFA
are involved in the 'focal adhesion' and 'pathway in cancer'
pathways, respectively. WSB2 is an unannotated gene in KEGG,
but was predicted as a target gene of miR-28-5p by TargetScan7.1,
PicTar, and miRDB software. The alteration in the expression levels
of RAP1B, VEGFA and WSB2 was observed in
breast cancer (Table I). RT-qPCR
was performed to experimentally validate these findings (Fig. 2B-D). The results indicated that
RAP1B, VEGFA and WSB2 were regulated by
miR-28-5p.
 | Table IGenes regulated by miR-28-5p. |
Table I
Genes regulated by miR-28-5p.
| Entrez | Symbol | Description | Fold change miR-28
vs. control |
|---|
| 7422 | VEGFA | Vascular
endothelial growth factor A | 2.0758 |
| 5908 | RAP1B | RAP1B, member of
RAS oncogene family | −1.7277 |
| 55884 | WSB2 | WD repeat and SOCS
box containing 2 | −1.5569 |
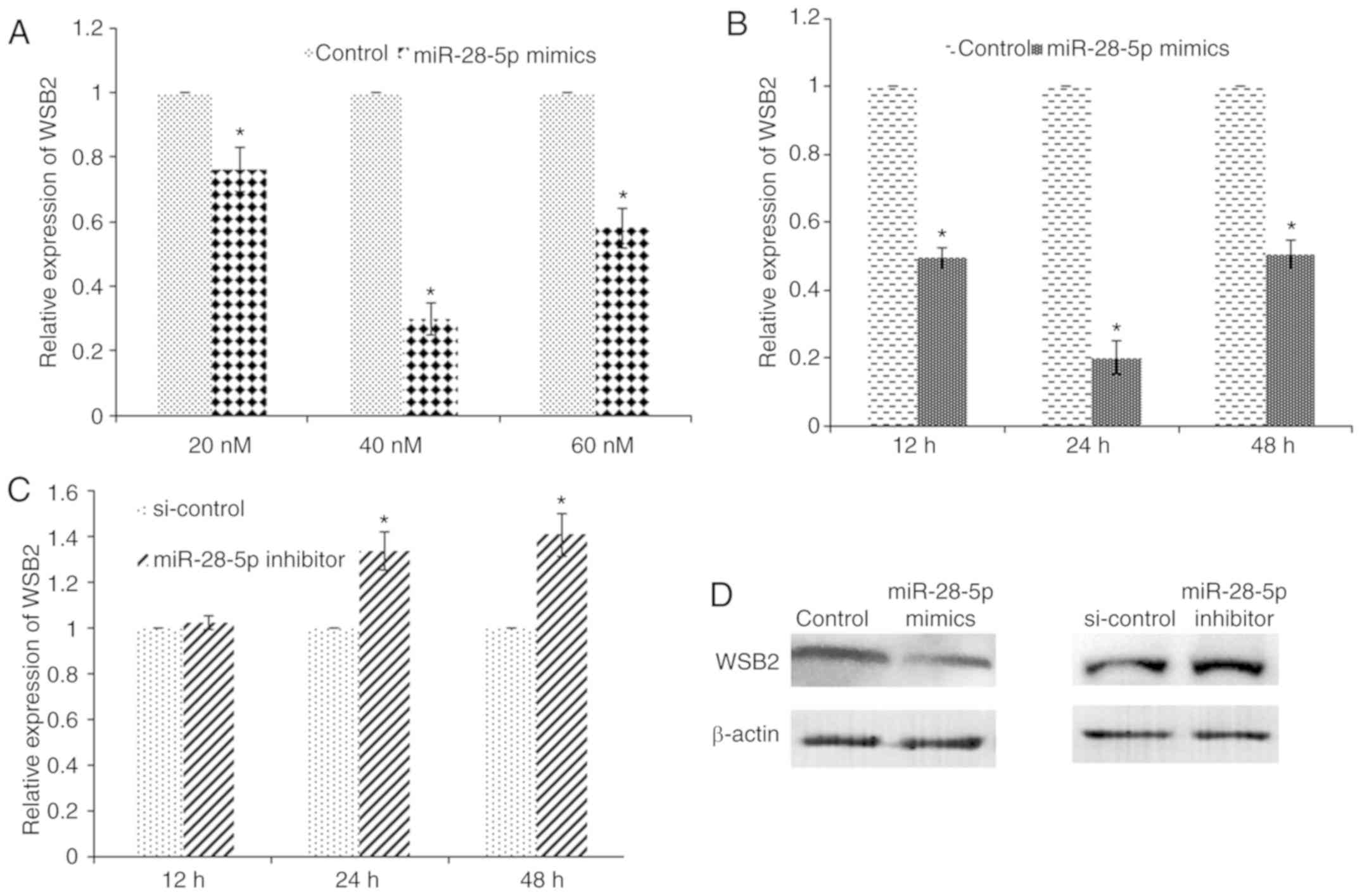
miR-28-5p targets the 3′UTR of WSB2
mRNA
To examine the effects of miR-28-5p on the
expression of its target genes, MCF-7 cells were transfected with
20, 40 and 60 nM miR-28-5p mimics for 24 h. RT-qPCR analysis
revealed that transfection with miR-28-5p mimics led to the
downregulation of WSB2 expression in a dose-dependent manner
(Fig. 3A) Furthermore, MCF-7
cells were transfected with 40 nM miR-28-5p mimics, and WSB2
mRNA expression was measured at 12, 24 and 48 h post-transfection.
RT-qPCR analysis revealed that transfection with miR-28-5p mimics
led to the downregulation of WSB2 expression in a
time-dependent manner (Fig. 3B).
miR-28-5p mimics led to a 2-fold decrease in WSB2 mRNA
expression at 12 h, and a 5-fold decrease at 24 h, compared with
the control, respectively (Fig.
3B). By contrast, transfection with miR-28-5p inhibitor
increased the expression of WSB2 (Fig. 3C). At the protein level, the level
of WSB2 decreased following transfection with miR-28-5p mimics, and
increased following transfection with miR-28-5p inhibitor (Fig. 3D). The analysis of the correlation
between miR-28-5p and WSB2 revealed that there was a weak negative
correlation between miR-28-5p and WSB2 (R=-0.35) in triple-negative
breast cancer (TNBC) (Fig.
S1).
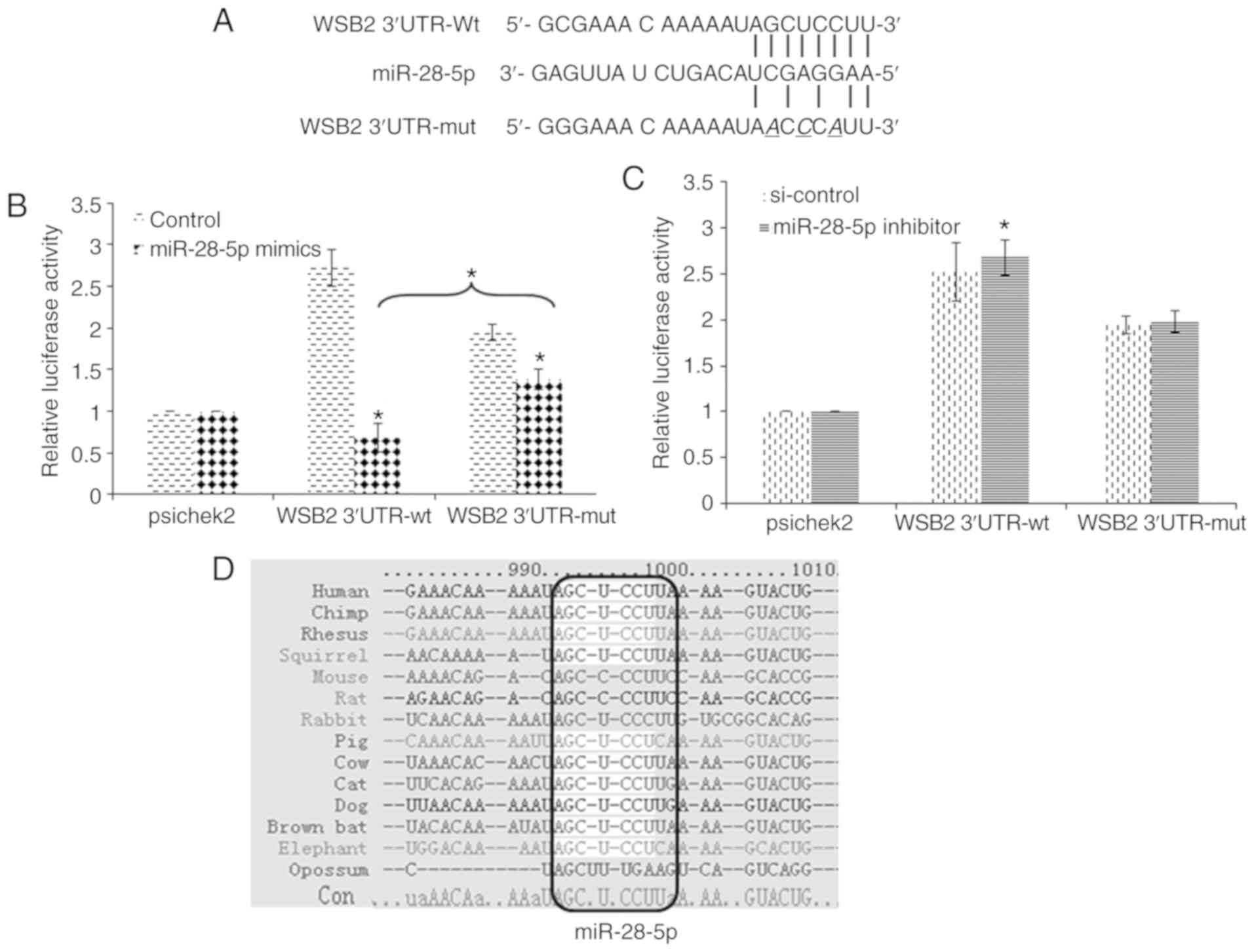
The binding sites for miR-28-5p were predicted in
the 3′UTR of WSB2. We cloned 3′UTR of the WSB2 gene
into vector psiCheck-2 for a luciferase reporter assay (Fig. 4A). The luciferase reporter
activity of the wild-type 3′UTR of WSB2 decreased upon
transfection with miR-28-5p mimics (Fig. 4B). By contrast, transfection with
miR-28-5p inhibitor led to an increase in reporter activity
(Fig. 4C). Furthermore,
site-directed mutagenesis of the 3′UTR of WSB2 was carried
out, and the luciferase reporter activity of the mutant 3′UTR
(WSB2-3′UTR-mut) was assessed (Fig.
4A). The regulatory effect of miR-28-5p mimics was
significantly weakened in the mutant reporter, although the
luciferase reporter activity of WSB2-3′UTR-mut decreased upon
transfection with miR-28-5p mimics (Fig. 4B). The regulatory effect of
miR-28-5p inhibitor was abolished in the mutant reporter (Fig. 4C). The 3′UTR of WSB2 of
human, chimp, rhesus, squirrel, mouse, rat, rabbit, pig, cat, dog,
brown bat, elephant and opossum was analyzed using TargetScan7.1 to
predict miR-28-5p binding sites. The binding site is conserved in
multiple species, and the core consensus sequence is 5′-AGCUCCUU-3′
(Fig. 4D). These results
indicated that WSB2 is the direct target gene of miR-28-5p,
and this mechanism may be conserved in other species.
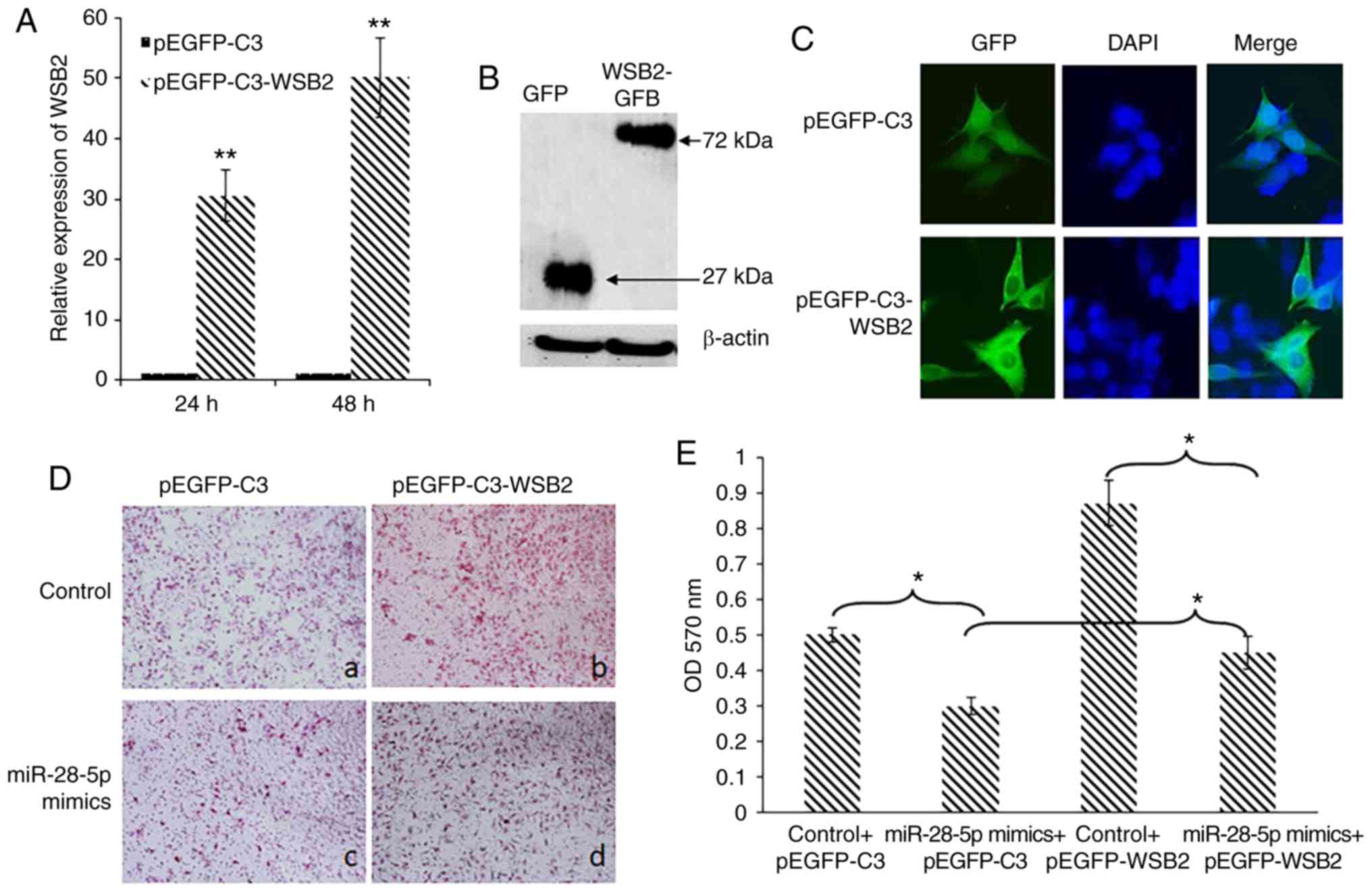
Overexpression of WSB2 attenuates the
inhibitory effects on MCF-7 cell migration induced by
miR-28-5p
The effect of WSB2 on the migration of MCF-7 cells
was assessed using a migration assay. MCF-7 cells were transfected
with empty pEGFP-C3 vector or pEGFP-C3-WSB2 that overexpresses
WSB2. The overexpression of WSB2 mRNA was confirmed
by RT-qPCR (Fig. 5A). Western
blot analysis revealed that pEGFP-C3-WSB2 expressed a 72-kDa
WSB2-GFP fusion protein (Fig.
5B), which was primarily localized in the cytoplasm (Fig. 5C). Migration assays revealed that
MCF-7 cells transfected with miR-28-5p mimics exhibited a decrease
in migration compared with the control (Fig. 5D, panels a and c), which was
attenuated by the overexpression of WSB2 (Fig. 5D, panels b and d). Crystal violet
staining further confirmed that WSB2 overexpression
attenuated the inhibitory effects of miR-28-5p on MCF-7 cell
migration (Fig. 5E). These
findings support the view that WSB2 is a target of
miR-28-5p, contributing to a better understanding of the mechanism
through which miR-28-5p inhibits the migration of MCF-7 cells.
Expression level of WSB2 in breast cancer
tissues
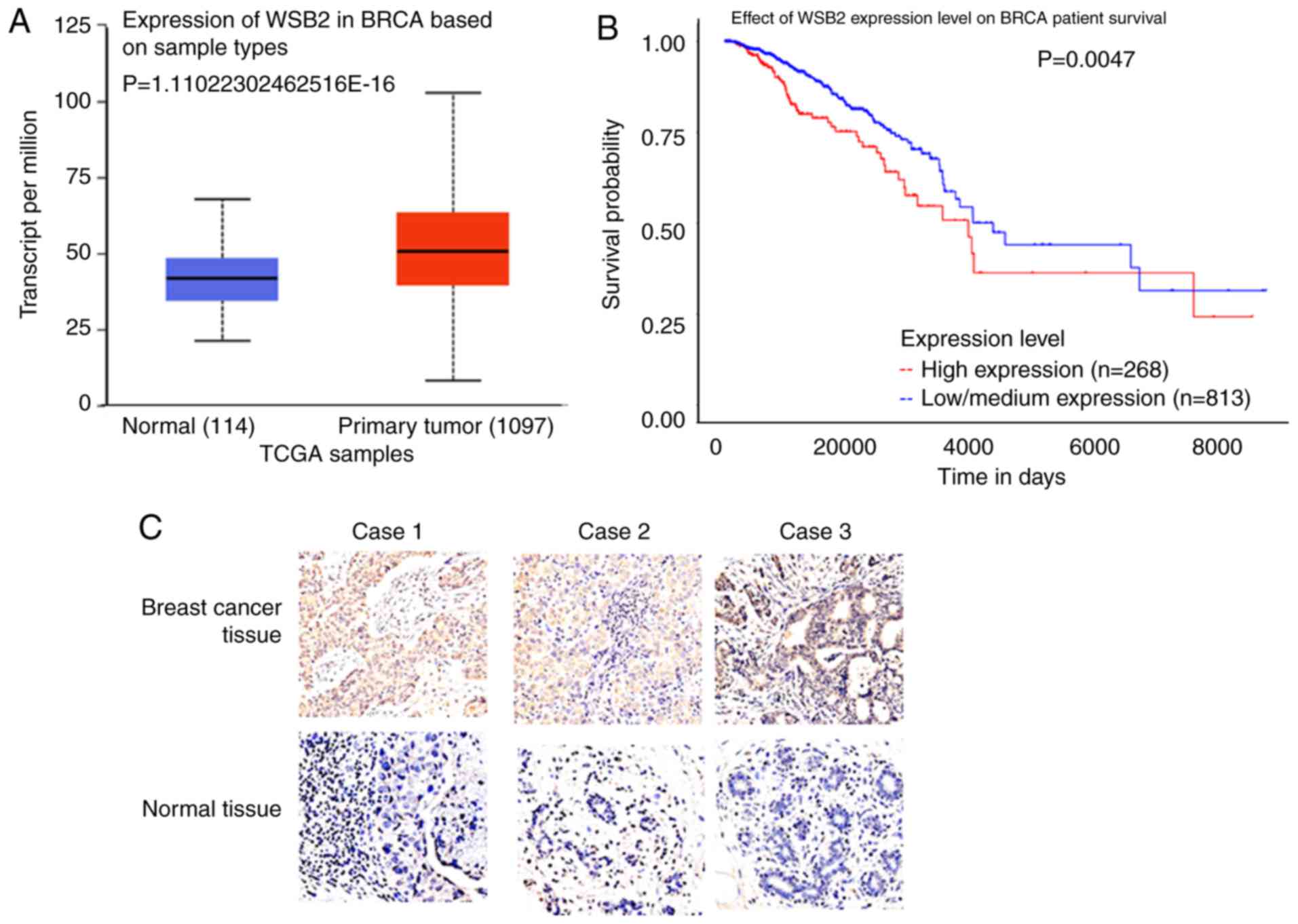
UALCAN analysis revealed that WSB2 was
significantly upregulated in primary breast tumor tissues, compared
with that in normal tissues (Fig.
6A). Kaplan-Meier analysis revealed that a high expression
level of WSB2 was associated with a poor survival in breast cancer
(Fig. 6B). Furthermore,
immunohistochemistry revealed that the integrated optical density
of WSB2 was significantly higher in the malignant breast tumor
tissue compared with that in the adjacent normal breast tissue
(Fig. 6C and Table II). These results indicate that
WSB2 promotes the malignancy of breast cancer cells.
 | Table IIExpression of WSB2 in breast cancer
and normal breast tissues specimens. |
Table II
Expression of WSB2 in breast cancer
and normal breast tissues specimens.
| Specimens | Number | Optical
density | Integrated optical
density |
|---|
| Breast cancer | 40 | 0.018±0.001a | 87±8.51a |
| Normal | 40 | 0.01±0.001 | 49±5.02 |
Discussion
miRNAs regulate tumor development by modulating gene
expression. The downregulation of miR-28-5p is involved in the
development and progression of several types of human cancer
(8-11,21). The present study demonstrated that
miR-28-5p, which has a lower expression in breast cancer tissue and
breast cancer cell lines, inhibited the migration of breast cancer
cells. These data support the view that miR-28-5p functions as a
tumor suppressor miRNA.
miRNAs regulate gene expression by binding to the
3′UTRs of their target mRNAs, and a single miRNA can regulate the
expression of multiple genes. Studies have demonstrated that
LPP (7), RAP1B
(9), NRF2 (22) and ZEB1 (23) are direct target genes of
miR-28-5p. The present study revealed that RAP1B was
downregulated by miR-28-5p, which was consistent with the findings
of a previous study (9). The
present study demonstrated that WSB2 was a previously
unknown target gene of miR-28-5p, which targets the 3′UTR of
WSB2. However, the mechanisms through which miR-28-5p
inhibits WSB2 require further investigation. Uncovering the
miR-28-5p regulatory network is a challenging task that requires
large-scale methods to identify miRNA targets.
WSB2 contains WD repeats and a SOCS box, which
mediate intracellular signal transduction (24). Its homolog, WSB1, has been
reported as an important regulator of aggressive metastasis in
hormone receptor-negative breast cancer (25). It has been demonstrated that the
expression of WSB2 is higher in human melanoma tissues;
consistently, cell cycle progression and migration of A375 and G361
melanoma cells were shown to be significantly inhibited by
WSB2 knockdown (26). The
present study demonstrated that WSB2 was upregulated in the breast
cancer tissue, which was associated with a poor survival in breast
cancer. Moreover, WSB2 promotes the migration of MCF-7 cells, which
was consistent with previous findings; for example, WSB2 exerts a
negative regulatory effect on IL-21R expression and signal
transduction (27); the knockdown
of WSB2 decreased the levels of c-Myc, β-catenin, p-Rb, CDK4
and Cyclin D3 in G361 melanoma cells (26); WSB1 knockdown has been
shown to be associated with decreased matrix metalloproteinase
(MMP) activity (25). However,
the mechanisms through which WSB2 regulates breast cancer
progression warrant further investigation.
In conclusion, the present study demonstrated that
miR-28-5p exhibits a low expression in breast cancer tissues and
breast cancer cell lines, and it inhibits the migration of breast
cancer cells by regulating WSB2 expression. WSB2 mRNA
is the direct target of miR-28-5p, containing an evolutionarily
conserved binding site for miR-28-5p in its 3′UTR. WSB2 is highly
expressed in breast cancer tissues, and its overexpression
attenuates the inhibitory effects of miR-28-5p on MCF-7 cell
migration, supporting the view that WSB2 functions down-stream of
miR-28-5p. In the future, this miR-28-5p/WSB2 axis may become a
novel therapeutic target in breast cancer.
Supplementary Data
Acknowledgments
Not applicable.
Funding
The present was supported by grants from the Natural
Science Foundation of Hebei Province (no. H2018209140) and the
National Natural Science Foundation of China (no. 81772834).
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
LM performed the RT-qPCR and western blot analyses.
YZ and FH analyzed the gene chip data, and performed the
immunohistochemical analysis. FH was a major contributor to the
writing of the manuscript. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
Patient data used in the present study were obtained
from a microarray, as well as TCGA. Patient survival was analyzed
with KMPLOT through UALCAN.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Hunter KW, Crawford NP and Alsarraj J:
Mechanisms of metastasis. Breast Cancer Res. 10:S22008. View Article : Google Scholar :
|
|
2
|
Shi M and Guo N: MicroRNA expression and
its implications for the diagnosis and therapeutic strategies of
breast cancer. Cancer Treat Rev. 35:328–334. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Teoh SL and Das S: The role of MicroRNAs
in diagnosis, prognosis, metastasis and resistant cases in breast
cancer. Curr Pharm Des. 23:1845–1859. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Bertoli G, Cava C and Castiglioni I:
MicroRNAs: New biomarkers for diagnosis, prognosis, therapy
prediction and therapeutic tools for breast cancer. Theranostics.
5:1122–1143. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Pei Y, Wang X and Zhang X: Predicting the
fate of microRNA target genes based on sequence features. J Theor
Biol. 261:17–22. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Schneider C, Setty M, Holmes AB, Maute RL,
Leslie CS, Mussolin L, Rosolen A, Dalla-Favera R and Basso K:
MicroRNA 28 controls cell proliferation and is down-regulated in
B-cell lymphomas. Proc Natl Acad Sci USA. 111:8185–8190. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zhou SL, Hu ZQ, Zhou ZJ, Dai Z, Wang Z,
Cao Y, Fan J, Huang XW and Zhou J: Mir-28-5p-IL-34-macrophage
feed-back loop modulates hepatocellular carcinoma metastasis.
Hepatology. 63:1560–1575. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wang C, Wu C, Yang Q, Ding M, Zhong J,
Zhang CY, Ge J, Wang J and Zhang C: Mir-28-5p acts as a tumor
suppressor in renal cell carcinoma for multiple antitumor effects
by targeting RAP1B. Oncotarget. 7:73888–73902. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Almeida MI, Nicoloso MS, Zeng L, Ivan C,
Spizzo R, Gafà R, Xiao L, Zhang X, Vannini I, Fanini F, et al:
Strand-Specific miR-28-5p and miR-28-3p have distinct effects in
colorectal cancer cells. Gastroenterology. 142:886–896. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Lv Y, Yang H, Ma X and Wu G:
Strand-Specific miR-28-3p and miR-28-5p have differential effects
on nasopharyngeal cancer cells proliferation, apoptosis, migration
and invasion. Cancer Cell Int. 19:1872019. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Hu F, Zhang Y, Li M, Bai Y and Zhang X:
Expression and role of HEPIS in breast cancer. Oncol Lett.
18:6648–6656. 2019.PubMed/NCBI
|
|
13
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
14
|
Hu F, Zhang Y, Li M, Zhao L, Chen J, Yang
S and Zhang X: BMP-6 inhibits the metastasis of MDA-MB-231 breast
cancer cells by regulating MMP-1 expression. Oncol Rep.
35:1823–1830. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Hu F, Gou L, Liu Q, Zhang W, Luo M and
Zhang X: G-Patch domain containing 2, a gene highly expressed in
testes, inhibits nuclear factor-kappaB and cell proliferation. Mol
Med Rep. 11:1252–1257. 2015. View Article : Google Scholar
|
|
16
|
Liu Q, Song YJ, Meng LJ, Hu F, Gou LX, Jia
CH, Tang HM, Wang WJ, Li M, Zhang XJ and Jia MC: Role of LM23 in
cell proliferation and apoptosis and its expression during the
testis development. Asian J Androl. 15:539–544. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zhao F, Pan S, Gu Y, Guo S, Dai Q, Yu Y
and Zhang W: Small activating RNA restores the activity of the
tumor suppressor HIC-1 on breast cancer. PLoS One. 9:e864862014.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Gong J, Wu Y, Zhang X, Liao Y, Sibanda VL,
Liu W and Guo AY: Comprehensive analysis of human small RNA
sequencing data provides insights into expression profiles and
miRNA editing. RNA Biol. 11:1375–1385. 2014. View Article : Google Scholar
|
|
19
|
Chandrashekar DS, Bashel B, Balasubramanya
SA, Creighton CJ, Ponce-Rodriguez I, Chakravarthi BV and Varambally
S: UALCAN: A portal for facilitating tumor subgroup gene expression
and survival analyses. Neoplasia. 19:649–658. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Lanczky A, Nagy A, Bottai G, Munkácsy G,
Szabó A, Santarpia L and Győrffy B: MiRpower: A web-tool to
validate survival-associated miRNAs utilizing expression data from
2178 breast cancer patients. Breast Cancer Res Treat. 160:439–446.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Xiao F, Cheng Z, Wang P, Gong B, Huang H,
Xing Y and Liu F: MicroRNA-28-5p inhibits the migration and
invasion of gastric cancer cells by suppressing AKT
phosphorylation. Oncol Lett. 15:9777–9785. 2018.PubMed/NCBI
|
|
22
|
Yang M, Yao Y, Eades G, Zhang Y and Zhou
Q: MiR-28 regulates Nrf2 expression through a Keap1-independent
mechanism. Breast Cancer Res Treat. 129:983–991. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Bao Y, Wang S, Xie Y, Jin K, Bai Y and
Shan S: MiR-28-5p relieves neuropathic pain by targeting Zeb1 in
CCI rat models. J Cell Biochem. 119:8555–8563. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Neer EJ, Schmidt CJ, Nambudripad R and
Smith TF: The ancient regulatory-protein family of WD-repeat
proteins. Nature. 371:297–300. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Poujade FA, Mannion A, Brittain N,
Theodosi A, Beeby E, Leszczynska KB, Hammond EM, Greenman J,
Cawthorne C and Pires IM: WSB-1 regulates the metastatic potential
of hormone receptor negative breast cancer. Br J Cancer.
118:1229–1237. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zhang Y, Li Z, Zhao W, Hu H, Zhao L, Zhu
Y, Yang X, Gao B, Yang H, Huang Y and Song X: WD repeat and SOCS
box containing protein 2 in the proliferation, cycle progression,
and migration of melanoma cells. Biomed Pharmacother.
116:1089742019. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Nara H, Onoda T, Rahman M, Araki A,
Juliana FM, Tanaka N and Asao H: Regulation of interleukin-21
receptor expression and its signal transduction by WSB-2. Biochem
Biophys Res Commun. 392:171–177. 2010. View Article : Google Scholar : PubMed/NCBI
|