Introduction
A functionally and structurally intact vascular
endothelium is essential for maintaining the normal function and
activity of the cardiovascular system. Continuous stimulation
(e.g., stress, inflammation and hypoxia) can cause vascular
endothelial activation and injury, eventually leading to
atherosclerosis (1,2). Angiogenesis, mainly manifested as
endothelial cell (EC) proliferation, migration and tube formation,
is an essential pathological process of atherosclerosis (3). Understanding the regulatory
mechanisms of angiogenesis during endothelial activation and injury
is crucial for controlling the progression of atherosclerosis.
Exosomes, with a size of 50 to 150 nm, are produced
by most cell types and contain a wide range of functional proteins,
lipids, messenger RNAs (mRNAs) and microRNAs (miRNAs or miRs)
(4). Increasing evidence has
indicated that the physiological functions of EC can be regulated
by exosomes shed from various types of cells (5-9).
In particular, ECs secrete functional exosomes, which can in turn
affect the physiological behavior of recipient ECs by delivering
miRNAs or proteins (10,11). In spite of extensive studies, the
effects and mechanisms underlying the communication between
vascular ECs during exosome-mediated angiogenesis remain to be
fully elucidated. As one of the key pathogenetic factors of
angiogenesis during atherosclerosis, oxidative stress can induce
angiogenesis via vascular endothelial growth
factor-dependent/independent signaling pathways (12). However, the role and mechanisms of
exosomes secreted by oxidative stress-stimulated ECs in
angiogenesis remain unclear.
miRNAs are a class of small non-coding RNAs that
post-transcriptionally suppress gene expression by binding to the
3′ untranslated region (3′UTR) of target mRNAs. Several miRNAs have
been shown to regulate vascular endothelial function and
angiogenesis (13), and miR-92a
is most closely related to these processes (14-19). The overexpression of miR-92a in
ECs has been shown to block angiogenesis by targeting several
pro-angiogenic proteins (14). In
the present study, through high-throughput screening, it was found
that miR-92a-3p was markedly downregulated and was the most
abundant miRNA differentially expressed in ECs treated with
oxidative stress-stimulated EC-derived exosomes. It was also
demonstrated that exosomes shed from oxidative stress-stimulated
ECs enhanced EC proliferation, migration and angiogenesis by
decreasing miR-92a-3p expression in target ECs. Moreover, it was
confirmed that tissue factor (TF) was a novel target gene of
miR-92a-3p, which may mediate the regulatory role of miR-92a-3p in
angiogenesis.
Materials and methods
Cells and cell culture
Human umbilical vein ECs (HUVECs) and 293 cells were
purchased from the Shanghai Institutes for Biological Sciences
(CAS). HUVECs were cultured in endothelial cell medium (ECM)
supplemented with 5% fetal bovine serum (FBS), 1% endothelial cell
growth supplement (ECGS), penicillin (100 U/ml) and streptomycin
(100 mg/ml) (Sciencell, Inc.). When HUVEC confluency reached
approximately 70-80%, FBS in ECM was replaced by 5% exosome-free
FBS (System Biosciences) and HUVECs were stimulated with or without
100 µM H2O2 for 24 h to produce
exosomes. HUVECs incubated with exosomes were all cultured in basal
medium (without FBS). 293 cells were cultured in Dulbecco's
modified Eagle's medium (DMEM) containing 10% FBS, penicillin (100
U/ml) and streptomycin (100 µg/ml). All the cells were
cultured in a 5% CO2 incubator at 37°C (Thermo Fisher
Scientific, Inc.).
miRNA transfection
HUVECs at 70-80% confluency were transfected with
miR-92a-3p inhibitor (66.7 nM), miR-92a-3p mimic (40.0 nM), a
negative control (NC) inhibitor or NC mimic (Suzhou Genepharma Co.,
Ltd.), respectively, using Lipofectamine 2000 (2.7 µg/ml for
inhibitor, 1.3 µg/ml for mimic) (Invitrogen; Thermo Fisher
Scientific, Inc.). The sequences of miR-92a-3p/NC mimic or
inhibitor were as follows: miR-92a-3p inhibitor, 5′-ACA GGC CGG GAC
AAG UGC AAU A-3′; miR-92a-3p mimic, 5′-UAU UGC ACU UGU CCC GGC CUG
U-3′ and 5′-AGG CCG GGA CAA GUG CAA UAU U-3′; NC inhibitor, 5′-CAG
UAC UUU UGU GUA GUA CAA-3′; NC mimic, 5′-UUC UCC GAA CGU GUC ACG
UTT-3′ and 5′-ACG UGA CAC GUU CGG AGA ATT-3′. Following 24 or 48 h
of transfection, the cells were harvested or further treated with
exosomes according to the different experimental purposes.
Exosome isolation
Endothelial exosomes in the conditioned medium were
isolated by differential centrifugation, as previously described
(20). The medium was centrifuged
at 300 × g for 10 min, 2,000 × g for 10 min and 10,000 × g for 30
min at 4°C. The supernatant was then filtered through a
0.22-µm filter (EMD Millipore) to remove cellular debris,
followed by ultracentrifugation at 100,000 × g and 4°C for 70 min
(WX+ Ultra series; Thermo Fisher Scientific, Inc.). The exosome
pellets were washed with phosphate-buffered saline (PBS), followed
by a second ultracentrifugation at 100,000 × g and 4°C for 70 min,
after which the exosomes were resuspended in PBS and stored at
−80°C.
Transmission electron microscopy
Exosome morphology was observed using a transmission
electron microscope (TEM). A total of 20 µl of samples were
dropped onto a carbon-coated grid, which was then baked at 60°C for
5 min using a polymerizer (ZB-J0010; Beijing Zhongxing Bairui
Technology Co., Ltd.). The excess liquid was absorbed using filter
paper. The grid was subsequently stained with 2% tungstophosphoric
acid for 5 min. Following 2 washes with distilled water, the grid
was baked again at 60°C and visualized using a TEM at 80 kV
(JEM-1400; Jeol, Ltd.).
Nanoparticle tracking analysis
The particle size of the exosomes was analyzed by
nanoparticle tracking analysis (NTA) using ZetaView PMX110
(Particle Metrix GmbH) in the size mode. Exosome samples were
diluted to the working range
(106-109particles/ml) of the detecting system
with PBS. The video of the Brownian motion of particles was
captured at 11 positions and the particle size was measured using
ZetaView 8.02.28 software.
Exosome labeling
Exosomes were labeled with the red fluorescent dye,
PKH26, according to manufacturer′s instructions with minor
modifications (Sigma-Aldrich; Merck KGaA) (20). A total of 2 µg of exosomes
were rapidly mixed with 1 ml of PKH26 solution (PKH26 dye: Diluent
C, 1:50 dilution). Following 5 min of incubation at 37°C, 5 ml of
whole ECM medium containing 5% exosome-free FBS were added to
terminate the labeling reaction. The labeled exosomes were washed
with PBS and centrifuged at 100,000 × g and 4°C for 1 h. Exosome
pellets were resuspended in 200 µl of PBS. Subsequently, the
labeled exosomes were added to HUVECs and incubated for 12 h in a
5% CO2 incubator at 37°C (Thermo Fisher Scientific,
Inc.). Following incubation, the HUVECs were washed twice and fixed
with 4% paraformaldehyde at room temperature for 15 min. The cells
were washed 3 times and treated with 0.2% Triton X-100 at 37°C for
15 min. Following 2 more washes, the cells were stained with 10
µg/ml of 4,6 diamidino-2-phenylindole (DAPI) for 5 min and
then imaged under a confocal laser scanning microscope (Leica TCS
SP5, Leica Microsystems GmbH).
Small RNA sequencing analysis
Following 24 h of incubation with exosomes, HUVECs
were collected and total RNA were extracted using the miRNeasy Mini
kit (Qiagen, Inc.). NEBNext®Multiplex Small RNA Library
Prep Set for Illumina® (New England Biolabs, Inc.) was
used to generate the sequencing library according to the
manufacturer′s instructions. Briefly, NEB 3′ SR adaptors were
ligated to the 3′ end of small RNA and SR RT Primer hybridized to
the excessive 3′ SR adaptor. The 5′ adapters were then ligated to
the 5′ ends of small RNA. M-MuLV Reverse Transcriptase (RNase
H-) was used to synthesize the first-strand cDNA.
LongAmp Taq 2X Master Mix, SR Primer for illumina and index (X)
primer were used for PCR amplification. The PCR products
corresponding to 140-160 bp were enriched to generate the cDNA
library. Library quality was assessed on an Agilent 2200 system
with DNA High Sensitivity Chips. The clustering of index-coded
samples was carried out on a cBot Cluster Generation System using
the TruSeq SR Cluster kit v3-cBot-HS (Illumina, Inc.) following the
manufacturer′s instructions. The sequencing (for 50-base read
length) of cDNA library was performed on an Illumina Hiseq 2500.
The clean data were obtained by trimming adaptor sequences and
removing low-quality reads. The small RNA tags were aligned using
Bowtie and mapped to the human genome reference (version: GRCh38
NCBI). miRBase20.0 was used as reference to obtain known miRNAs.
Software mirdeep2 and miREvo were integrated to identify novel
miRNAs. Differentially expressed miRNAs were analyzed using the
DESeq R package (1.8.3). miRNAs with a P-value <0.05 were
considered to exhibit a significant differential expression. miRNAs
with similar expression pattern were clustered and displayed as a
heatmap.
Messenger RNA (mRNA) sequencing
analysis
HUVECs were transfected with miR-92a-3p inhibitor
for 48 h and then total RNA were isolated using the miRNeasy Mini
kit (Qiagen, Inc.). The NEBNext® Ultra™ Directional RNA
Library Prep kit for Illumina® (New England Biolabs,
Inc.) was used to generate the sequencing library according to the
manufacturer′s instructions. Briefly, mRNA was fragmented into
150-200 bp using divalent cations at 94°C for 8 min. The cleaved
mRNA fragments were reverse-transcribed into first-strand cDNA, and
the fragments were end repaired and ligated with indexed adapters.
Target bands were harvested through AMPure XP Beads (Beckman
Coulter, Inc.). The products were purified and enriched by PCR to
generate the final cDNA libraries and quantified by Agilent 2200.
The tagged cDNA libraries were pooled in equal ratio and used for
150 bp paired-end sequencing in a single lane of the Illumina
HiSeqXTen. To obtain clean data, raw reads were processed by
removing the adaptor sequences, reads with >5% ambiguous bases
and low-quality reads containing >20% of bases with qualities of
<20. The sequencing data were aligned to human genome (version:
GRCh38 NCBI) using the hisat2 algorithm. HTseq was used to count
gene and the reads per kilobase per million mapped reads (RPKM)
method was used to determine gene expression. Differentially
expressed genes were analyzed using the DESeq2 algorithm with a
fold change of >1.5 or <0.5, a P-value <0.05 and a false
discovery rate of <0.05.
Reverse transcription-quantitative PCR
(RT-qPCR)
Total RNA was isolated from the HUVECs or exosomes
using the miRNeasy Mini kit (Qiagen, Inc.). miRNA-specific
stem-loop primers and the TaqMan MicroRNA Reverse Transcription kit
(Applied Biosystems, Inc.) were used for the amplification of
mature miRNAs. The expression level of mature miR-92a-3p was
normalized to the expression levels of RNU6B in HUVECs and
synthetic C. elegans miR-39 (cel-miR-39) (10 fmol/sample)
(Qiagen, Inc.) in exosomes, respectively. The ImProm-II™
Transcription System and GoTaq 2-Step RT-qPCR System (Promega
Corporation) were used for TF amplification. The TF expression
level was normalized to the expression of GAPDH. The sequences of
the primers were as follows: TF forward, 5′-GCC AGG AGA AAG GGG
AAT-3′ and reverse, 5′-CAG TGC AAT ATA GCA TTT GCA GTA GC-3′; GAPDH
forward, 5′-GAG TCA ACG GAT TTG GTC GT-3′ and reverse, 5′-GAC AAG
CTT CCC GTT CTC AG-3′. The amplifications were performed as
previously described (21). For
the quantification of primary miR-92a, the levels of pri-miR-92a-1
and pri-miR-92a-2 were measured using the High Capacity cDNA
Reverse Transcription kit (Applied Biosystems, Inc.), TaqMan
Pri-miRNA assays (pri-miR-92a-1 assay ID: Hs03302603_pri,
pri-miR-92a-2 assay ID: Hs03295977_pri) and TaqMan Gene Expression
Master Mix (Applied Biosystems, Inc.) according to the
manufacturer′s instructions. The amplification conditions were
pre-incubation at 95°C for 10 min, followed by 40 cycles of 95°C
for 15 sec and 60°C for 1 min. The results were normalized to the
expression of GAPDH (TaqMan assay ID: Hs02758991_g1). qPCR was
performed on an Applied Biosystems system (ViiA7) and all data are
expressed as 2−ΔΔCq(22).
Western blot analysis
HUVECs or exosome samples were lysed using a RIPA
buffer (Solarbio, Inc.) and then centri-fuged at 12,000 × g and 4°C
for 15 min. The supernatant was collected and the protein
concentration was measured using the bicinchoninic acid (BCA)
protein assay kit (Thermo Fisher Scientific, Inc.) according to the
manufacturer′s instructions. A total of 10-20 µg of proteins
were separated on a 10% SDS-polyacrylamide gel and
electrophoretically transferred to PVDF membranes (EMD Millipore).
The membranes were incubated with one of the following primary
antibodies: TF (1:1,000; cat. no. 55147S, Cell Signaling
Technology, Inc.), GAPDH (1:2,000; cat. no. sc-32233; Santa Cruz
Biotechnology, Inc.), Flotillin-1 (1:200; cat. no. sc25506; Santa
Cruz Biotechnology, Inc.), GM130 (1:250; cat. no. 610822, BD
Biosciences), Lamin A/C (1:500; cat. no. sc-7292; Santa Cruz
Biotechnology, Inc.) and Tom20 (1:500; cat. no. sc-17764, Santa
Cruz Biotechnology, Inc.) at 4°C overnight and were then incubated
with HRP-conjugated goat-anti-rabbit (1:2,000; cat. no. sc-2004,
Santa Cruz Biotechnology, Inc.) or mouse (1:4,000 for GAPDH or
1:2,000 for the rest, cat. no. sc-2005; Santa Cruz Biotechnology,
Inc.) IgG secondary antibodies at room temperature for 2 h. The
membranes were visualized using an enhanced-chemiluminescence
system (Thermo Fisher Scientific, Inc.). The bands were quantified
using Image J 1.52a software. The expression level of TF was
normalized to that of GAPDH.
EdU incorporation assay
Proliferating HUVECs were identified using the
Click-iT Plus EdU Imaging kit (Life tech-nologies; Thermo Fisher
Scientific, Inc.). Following co-culture with exosomes or
transfection with miRNA inhibitor, the HUVECs were incubated with
10 µM EdU for 16 h, fixed with 4% paraformaldehyde for 15
min at room temperature and washed with PBS containing 3% bovine
serum albumin (BSA). The cells were then treated according to the
following steps: 20 min in 0.5% Triton X-100, 30 min in the
Click-iT Plus reaction cocktail, and 30 min in 5 µg/ml of
Hoechst 33342 at room temperature. Cell images were captured using
an Olympus IX70 microscope (Olympus Corporation). The ratio of
EdU-positive cells to total cells was analyzed by counting
approximately 1,000 cells in several randomly selected fields.
Scratch wound migration assay
The assessment of HUVEC migration was performed as
previously described (23). A
confluent layer of HUVECs in 24-well plates was scratched with a
sterile 10-µl tip and the detached cells were removed by
washing with PBS. The adherent cells were then incubated with 5
µg/ml exosomes for 20 h before cell migration was captured
using a Leica DM IL LED microscope (Leica Microsystems GmbH) and
quantified by measuring the size of recovered area using ImageJ
1.52a software.
Tube formation assay
Matrigel Matrix Growth Factor Reduced (BD
Biosciences) (300 µl/well) was coated on 24-well plates and
incubated at 37°C for 30 min. Approximately 8×104
HUVECs/well were then seeded onto the gel and cultured in FBS-free
ECM for 20 h to allow tube formation. Tube formation was observed
using a Leica DM IL LED microscope (Leica Microsystems GmbH). The
averages of the total number of branching points and total number
of loops in 4 representative fields were analyzed using ImageJ
1.52a software.
In vivo Matrigel plug assay
A total of 8 female, 8-week-old, BALB/c nude mice
(Charles River Laboratories, Inc.) were used in this study. All the
mice were housed in specific pathogen-free conditions under a
controlled temperature (23±3°C), humidity (60±15%) and 12 h
dark/light cycle, with sterile rodent chow and water provided ad
libitum. The mice were kept in specific pathogen-free grade
filter-top cages and infectious diseases were not detected.
Following transfection with miR-92a-3p inhibitor or NC inhibitor
for 48 h, the HUVECs were mixed with Matrigel Matrix High
Concentration (BD Biosciences) at ratio of 1:1 (V:V). Subsequently,
1 ml of mixture containing 1×107 HUVECs was
subcutaneously injected into the dorsal surface of each nude mouse
(n=4/group) using a 25-gauge needle. After 2 weeks, the mice were
euthanized by an intraperitoneal injection of an overdose of
pentobarbital (120 mg/kg). The Matrigel plugs were harvested,
transferred to ice-cold PBS and embedded in OCT compound. Serial
5-µm-thick sections were cut and stained with anti-CD31
antibody (1:100; cat. no. ab28364, Abcam) at 4°C overnight. The
sections were observed under a Jenoptik fluorescence microscope. A
representative surface of blood vessels was analyzed by analyzing
the CD31-positive region (24).
The animal experimental protocol was approved by Peking University
People′s Hospital Ethics Committee (approval no. 2016PHC072).
Luciferase reporter assay
Luciferase reporter assay was performed as
previously described (21). The
sequence (1,284 bp) of TF 3′UTR containing the miR-92a-3p binding
site was synthesized and cloned into a Firefly luciferase reporter
plasmid pMIR-REPORT™ (Ambion; Thermo Fisher Scientific, Inc.). To
construct the mutant plasmid, the predicted target nucleotides of
miR-92a-3p in TF 3′UTR were changed to opposite bases. 293 cells
were co-transfected with 0.06 pmol/µl of miRNA mimic, 0.3
ng/µl of Firefly luciferase reporter plasmid, as well as
0.01 ng/µl of Renilla luciferase as the control
(pRLTK; Promega Corporation). Following 24 h of transfection, the
luciferase activity was measured using the Dual Luciferase Assay
System (Promega Corporation). Each measured value of Firefly
luciferase activity was normalized to that of Renilla
luciferase activity.
Statistical analysis
The data are presented as the means ± stan-dard
error of the mean (SEM). The Student′s t-test was used to compare
differences between 2 groups, and one-way ANOVA followed by Tukey′s
post hoc test was used to compare differ-ences among multiple
groups. A two-sided value of P<0.05 was considered to indicate a
statistically significant difference. Prism 5.0 was used for all
statistical analyses.
Results
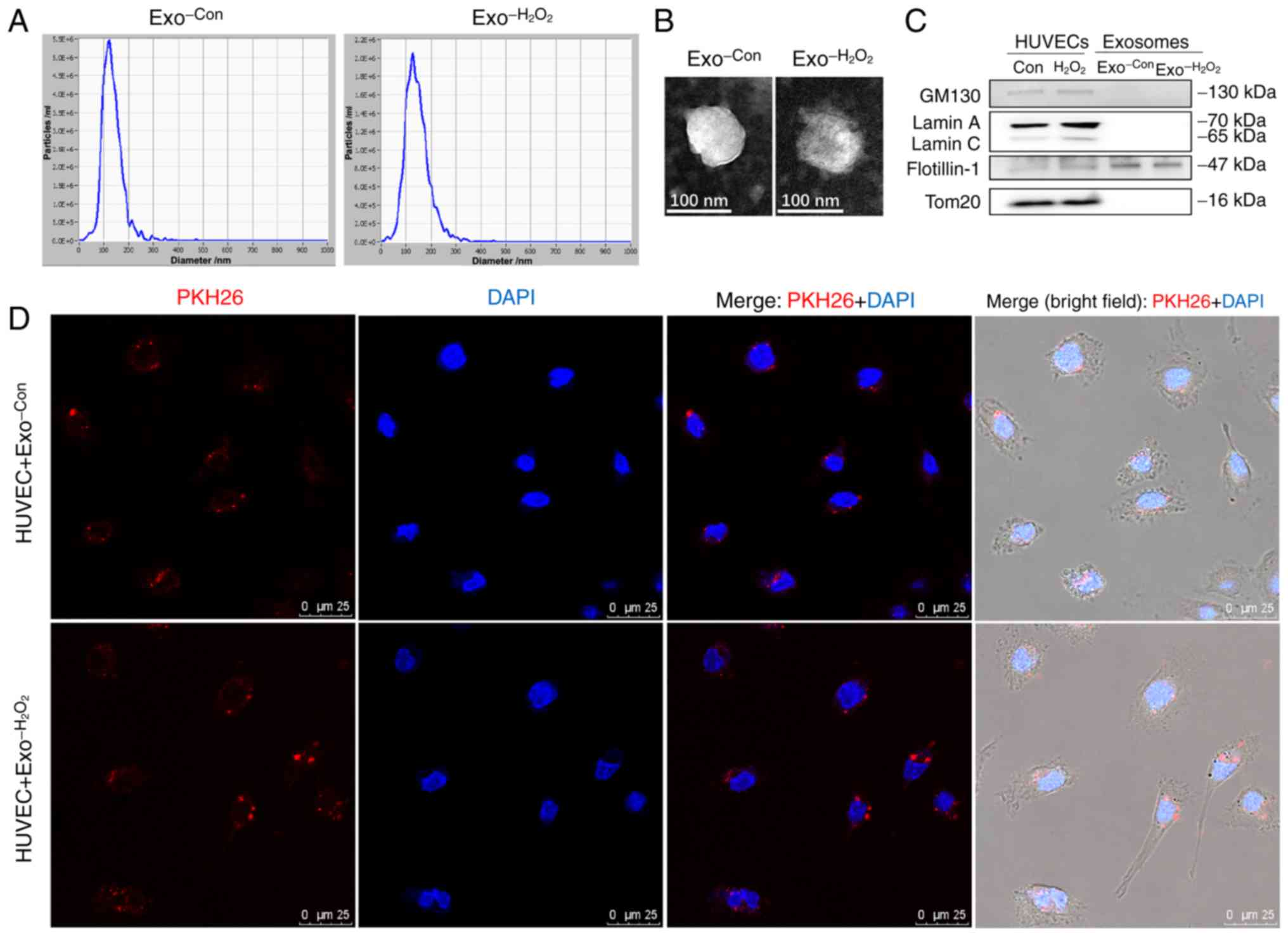
Characterization and internalization of
exosomes derived from ECs
To obtain exosomes from oxidative stress-stimu-lated
ECs, HUVECs were cultured in ECM containing 5% of exosome-free FBS
and treated with 100 µM of H2O2 (these
exosomes were termed Exo−H2O2).
The control exosomes were obtained from HUVECs without
H2O2 stimulation (these exosomes were termed
Exo-Con). Following 24 h of incubation, the conditioned
medium was collected and the exosomes were isolated. The results of
NTA revealed that the particle size distribution of the exosomes
was 50-140 nm in both groups (Fig.
1A); TEM analysis revealed the cup-shaped appearance of the
exosomes (Fig. 1B). The results
of western blot analysis demonstrated that the exosomes (marker,
flotillin 1) (10,25) were not contaminated with Golgi
(marker, GM130), nuclear (marker, Lamin A/C) and mitochondrial
(marker, Tom20) substances (Fig.
1C).
To examine the uptake of exosomes by recipient ECs,
exosomes were labeled with PKH26 and incubated with HUVECs.
Confocal laser scanning microscopic analysis revealed that the
PKH26-labled exosomes were efficiently internalized by the HUVECs
(Fig. 1D).
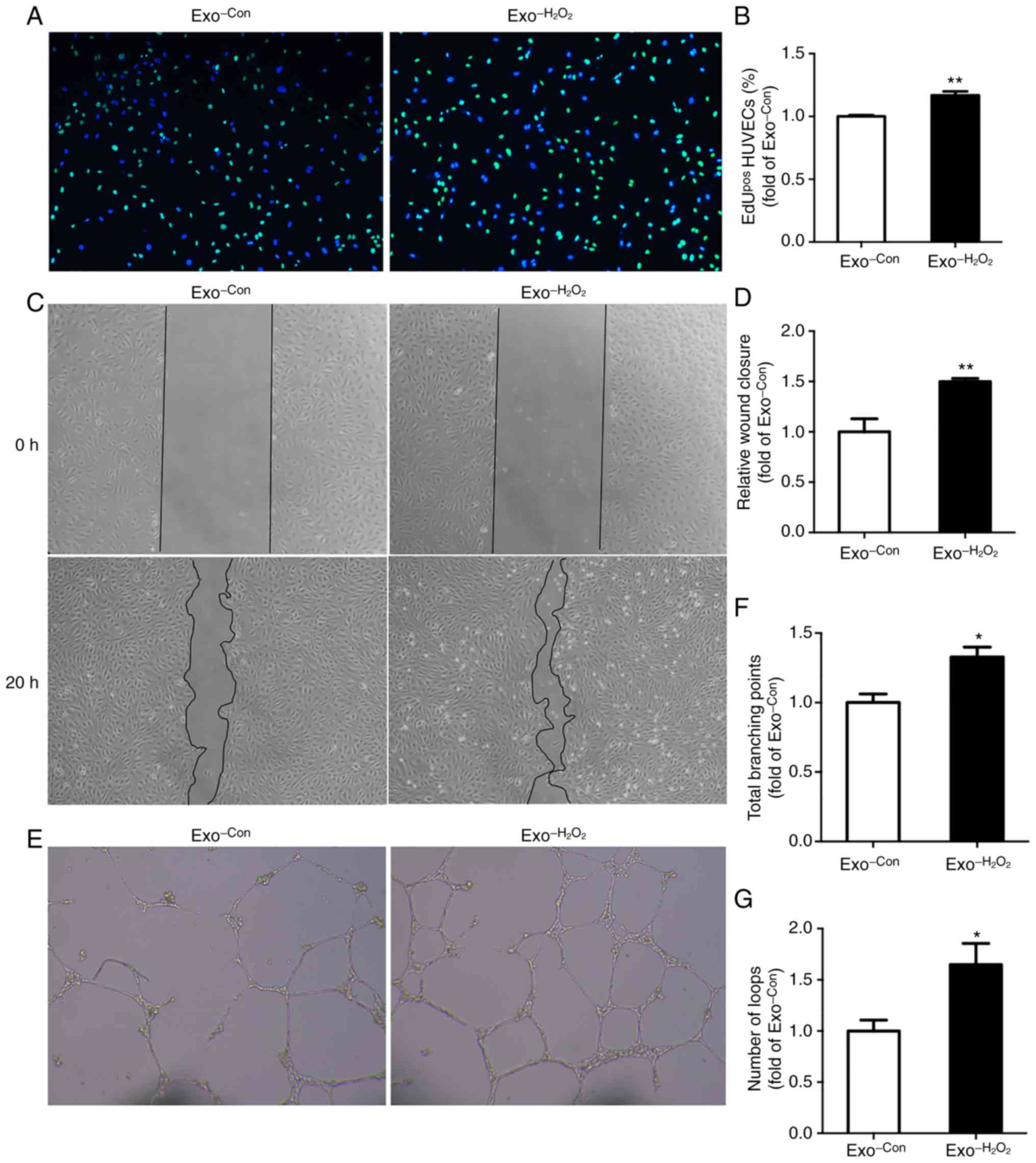
Oxidative stress-induced ECs promote EC
proliferation, migration and angiogenesis
EdU incorporation assay, scratch wound migration
assay and tube formation assay were performed to examine the
effects of exosomes released by oxidative stress-stimulated ECs on
the proliferation, migration and angiogenesis of recipient ECs,
respectively. For the EdU incorporation assay, the HUVECs were
incubated with 5 µg/ml exosomes ( Exo−H2O2 or Exo-Con). Following
48 h of incubation, HUVECs were treated with 10 µM EdU for
16 h. The proliferating cells were incorporated with EdU (green)
and nuclei were stained with Hoechst 33342 (blue). HUVEC
proliferation was increased by 1.2-fold by Exo−H2O2 compared with that by
Exo-Con(Fig. 2A and
B). For scratch wound migration assay, the HUVECs were
scratched and incubated with exosomes for 20 h. HUVEC migration was
enhanced by 1.5-fold by Exo−H2O2 compared with that by
Exo-Con(Fig. 2C and
D). For tube formation assay, the HUVECs were treated with
exosomes for 24 h and then seeded in the Matrigel, and endothelial
networks could be observed after 20 h. The numbers of total
branching points and loops increased by 1.3- and 1.6-fold,
respectively, as compared to the control (Fig. 2E-G).
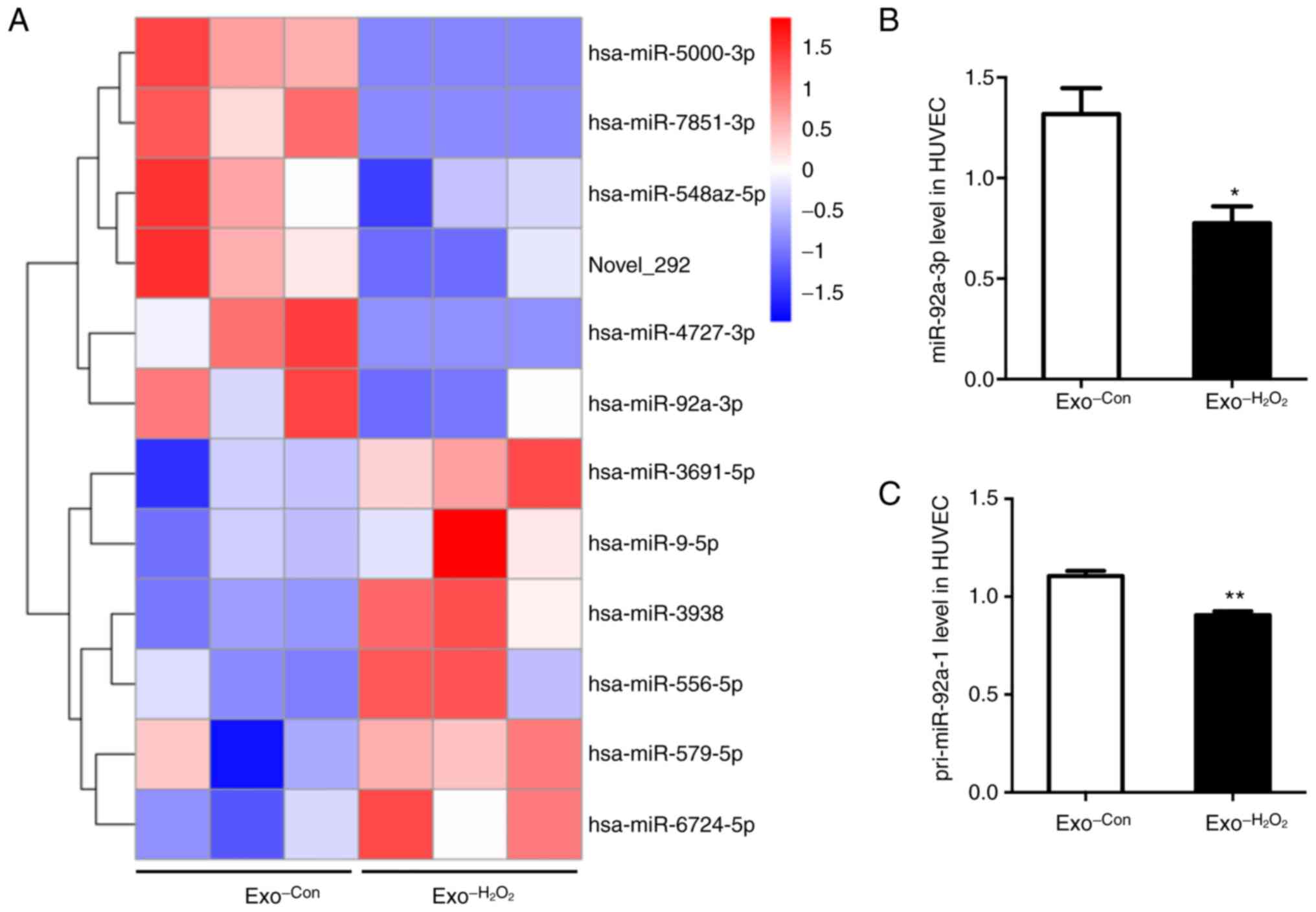
miR-92a-3p expression is inhibited by
Exo−H2O2 in recipient ECs
To explore whether miRNAs mediate the effects of
Exo−H2O2 on recipient ECs,
miRNAs differentially expressed in Exo−H2O2 - and Exo-Con-treated
HUVECs were identified by small RNA sequencing. Following 24 h of
incubation with exosomes, total RNA was extracted from the HUVECs.
The results indicated the presence of 12 miRNAs differentially
expressed between the 2 groups (Fig.
3A and Table SI). Among the
12 miRNAs, miR-92a-3p was the most abundant and has been reported
to play a key role in regulating angio-genesis (13). Therefore, miR-92a-3p was selected
for further validation by RT-qPCR. The results revealed that
miR-92a-3p expression was decreased by 40% in the Exo−H2O2 -treated HUVECs compared with that
in the Exo-Con-treated group (Fig. 3B). Furthermore, to reveal the
cause for the decrease in miR-92a-3p expression in recipient
HUVECs, the expression of primary miR-92a (pri-miR-92a-1 and
pri-miR-92a-2) in HUVECs co-cultured with exosomes was detected by
RT-PCR. The results revealed that pri-miR-92a-1 expression was
decreased by 20% (Fig. 3C) in the
Exo−H2O2 -treated HUVECs
compared with that in the control, and pri-miR-92a-2 was almost
undetectable (data not shown) in both groups. Taken together, these
results suggested that the Exo−H2O2-mediated stimulation of EC
proliferation, migration and angiogenesis may be achieved via the
inhibition of miR-92a-3p in recipient cells.
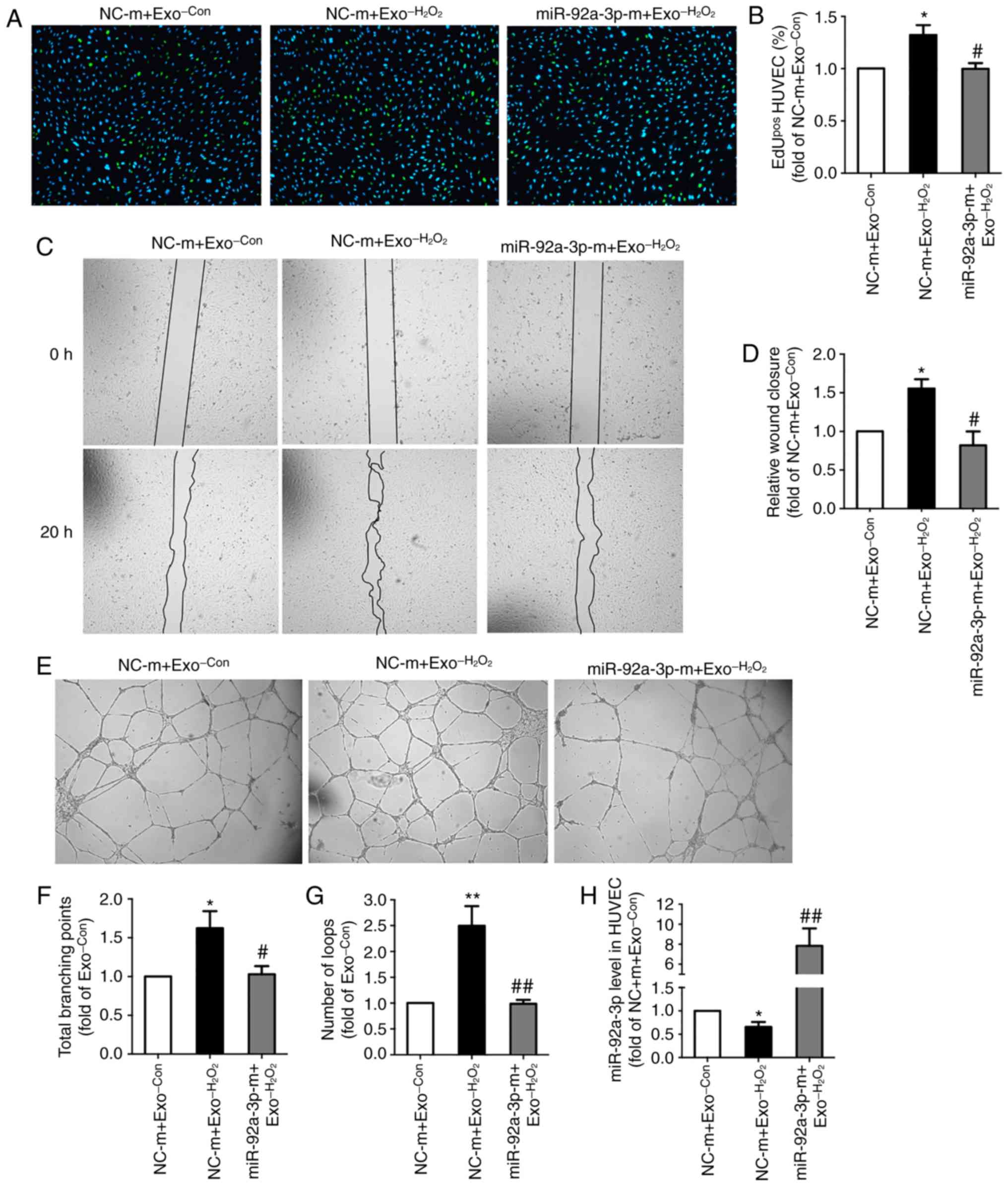
miR-92a-3p overexpression blocks the
effects of Exo−H2O2 on
recipient ECs
To investigate whether miR-92a-3p is involved in the
effects of Exo−H2O2 on target
ECs, miR-92a-3p expression was increased by transfecting the cells
with miR-92a-3p mimic (miR-92a-3p-m) for 24 h prior to the addition
of exosomes. The results of RT-qPCR revealed that miR-92a-3p
expression was significantly upregulated (Fig. 4H). EdU incorporation assay,
scratch wound migration assay and tube formation assay were
performed after incubating the HUVECs with exosomes for 24 h.
Compared with Exo-Con(group, NC-m + Exo-Con)
(set as 1), Exo−H2O2 (group,
NC-m + Exo−H2O2) promoted HUVEC
proliferation (1.3-fold) (Fig. 4A and
B), migration (1.6-fold) (Fig. 4C
and D) and angiogenic capacity (total branching points,
1.6-fold; number of loops, 2.5-fold) (Fig. 4E-G). The above-mentioned effects
induced by Exo−H2O2 were all
abrogated by the overexpression of miR-92a-3p (group, miR-92a-3p-m
+ Exo−H2O2) (Fig. 4A-G). These results indicated that
downregulated expression of miR-92a-3p mediated the role of
Exo−H2O2 in promoting
endothelial proliferation, migration and angiogenesis.
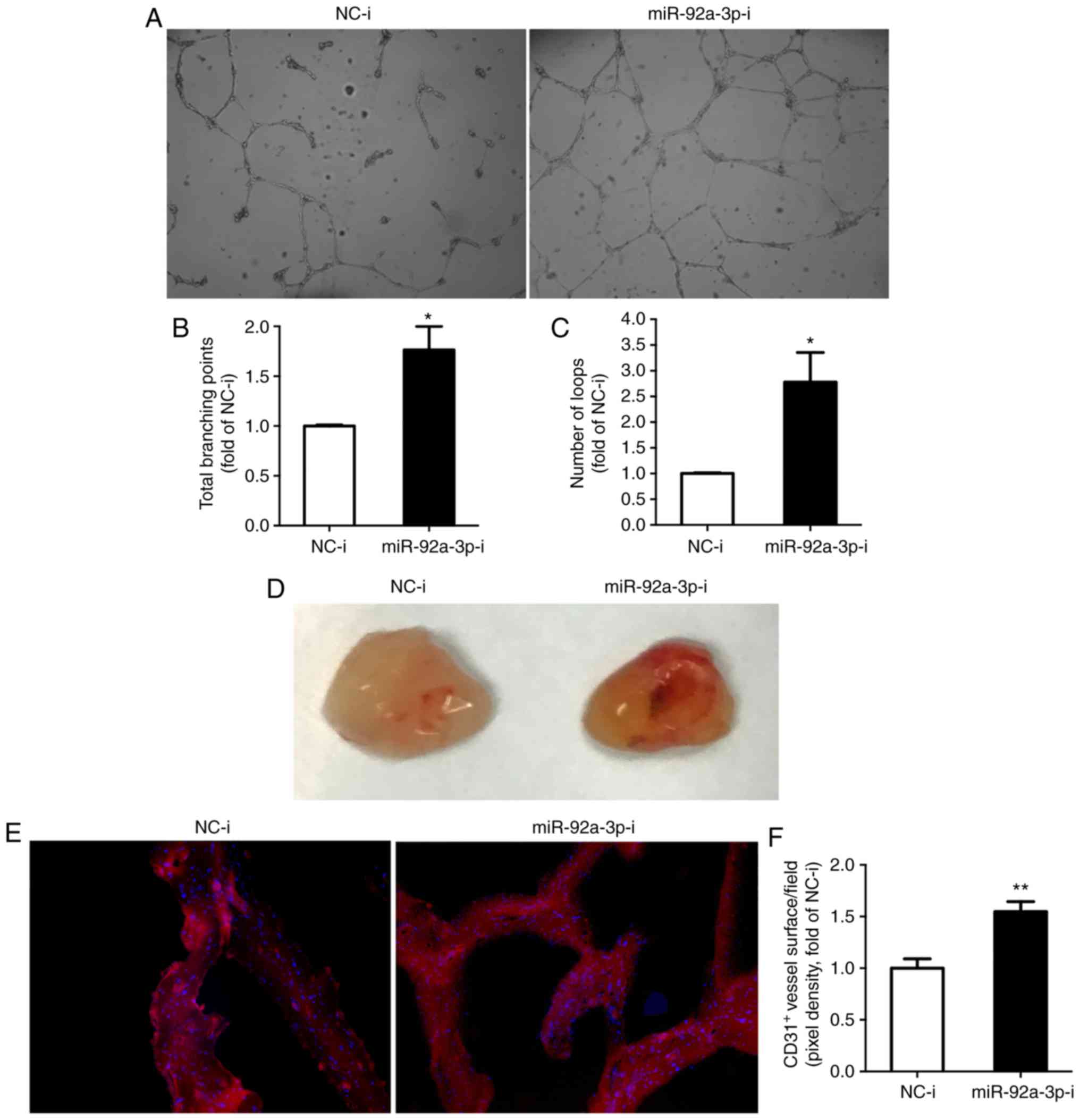
Inhibition of miR-92a-3p induces
angiogenesis
To further determine the role of miR-92a-3p in
angiogenesis, miR-92a-3p expression was directly inhibited by
transfecting the HUVECs with miR-92a-3p inhibitor for 48 h. The
decreased expression of miR-92a-3p induced EC proliferation
(1.2-fold) (Fig. S1A and B),
migration (2.0-fold) (Fig. S1C and
D) and angiogenesis in vitro (total branching points,
1.7-fold; number of loops, 2.8-fold) (Fig. 5A-C). Moreover, the in vivo
Matrigel plug assay revealed that in vivo blood vessel
formation was enhanced by 1.5-fold in the miR-92a-3p inhibitor
group (Fig. 5D-F). Taken
together, these results further indicated that Exo−H2O2 promoted angiogenesis by
decreasing miR-92a-3p expression in recipient HUVECs.
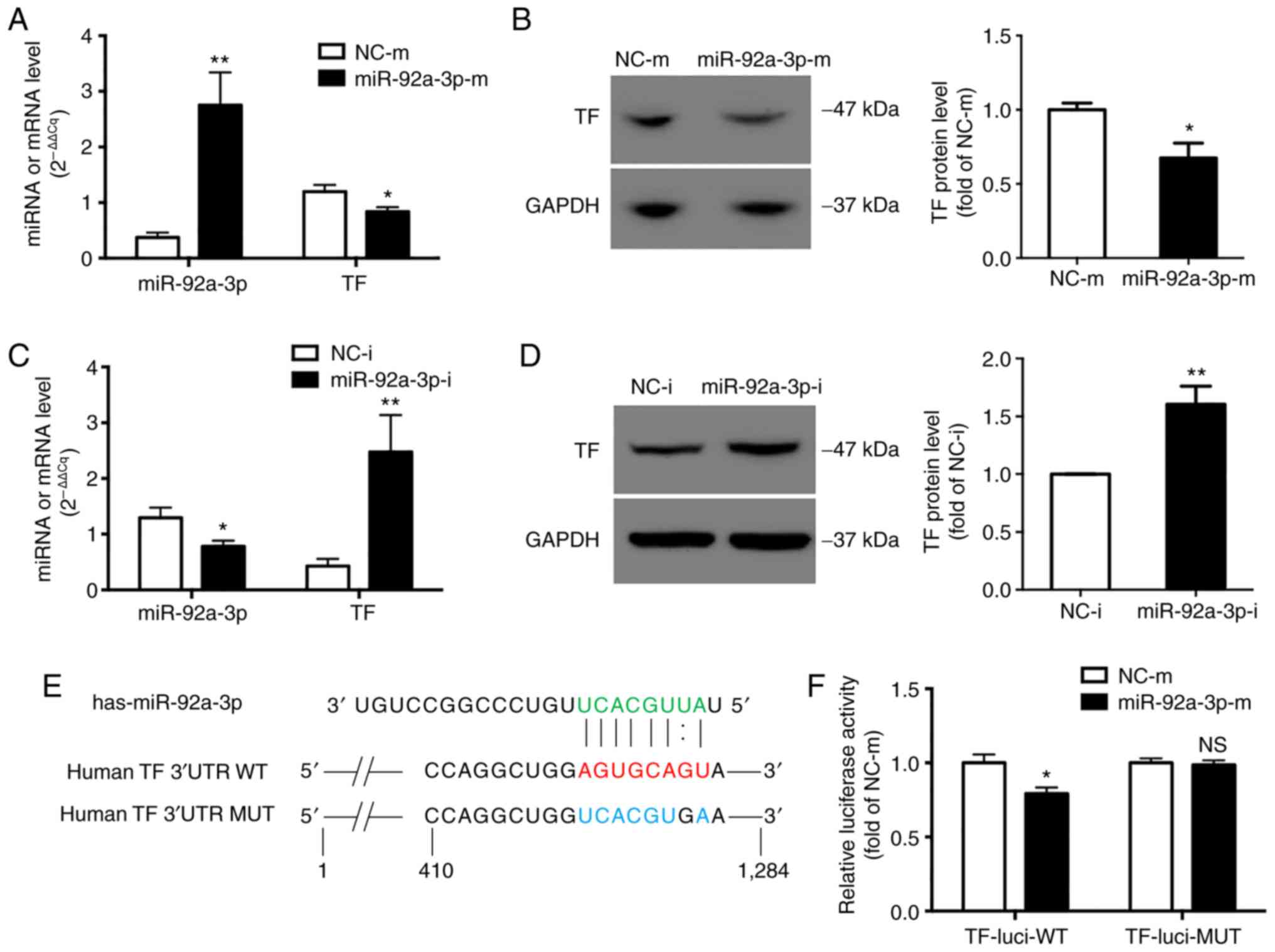
miR-92a-3p inhibits TF expression in
ECs
To identify the targets of miR-92a-3p involved in
angiogenesis, differentially expressed genes were identified in
HUVECs transfected for 48 h with miR-92a-3p inhibitor or
NC-inhibitor. The results identified 197 differentially expressed
mRNAs, among which 91 mRNAs were upregulated (Fig. S2). In addition, 4,167 targets of
miR-92a-3p were predicted using the RNAhybrid program (26), including 21 upregulated genes that
were also identified by mRNA sequencing (Fig. S2 and Table I). Among the genes closely related
to angiogenesis, TF (F3) was most significantly upregulated.
 | Table IPotential targets of miR-92a-3p in
HUVECs. |
Table I
Potential targets of miR-92a-3p in
HUVECs.
| Gene | Fold change
(miR-92a-3p-i vs. NC-i) | FDR |
|---|
| TNFRSF9 | 3.5 | 4.1E-05 |
| NCOA7 | 1.9 | 5.5E-11 |
| TNFAIP2 | 3.7 | 0.0E+00 |
| IL1R1 | 1.6 | 1.2E-02 |
| NUAK2 | 2.3 | 8.2E-13 |
| INSR | 1.6 | 2.2E-04 |
| GUCY1A3 | 2.7 | 4.9E-11 |
| F3 (TF) | 7.0 | 0.0E+00 |
| AQP1 | 1.8 | 1.5E-04 |
| C2CD4A | 13.0 | 0.0E+00 |
| SOD2 | 2.0 | 1.3E-12 |
| SLC16A7 | 2.0 | 4.0E-03 |
| SPAST | 1.6 | 1.3E-03 |
| SGPP2 | 11.6 | 2.8E-05 |
| CX3CL1 | 6.7 | 0.0E+00 |
| TRAF1 | 1.6 | 5.5E-03 |
| TNIP3 | 4.9 | 3.4E-09 |
| C8orf4 | 2.2 | 2.7E-12 |
| ZC2HC1A | 2.3 | 1.5E-02 |
| ICAM1 | 2.5 | 1.2E-15 |
| CD83 | 2.1 | 2.8E-04 |
To determine whether TF is a novel target of
miR-92a-3p, gain- and loss-of-function experiments were performed
by transfecting HUVECs for 48 h with miR-92a-3p mimic or inhibitor,
respectively. The results of RT-qPCR revealed that miR-92a-3p was
successfully overexpressed (Fig.
6A) or inhibited (Fig. 6C).
Accordingly, the upregulation of miR-92a-3p inhibited TF expression
by 30% at the mRNA level (Fig.
6A) and by 33% at the protein level (Fig. 6B), whereas the down-regulation of
miR-92a-3p increased TF expression by 6.4-fold at the mRNA level
(Fig. 6C) and by 1.6-fold at the
protein level (Fig. 6D).
TF is a direct target of miR-92a-3p
To examine whether miR-92a-3p acts directly on TF
3′UTR, 2 luciferase reporter plasmids containing wild-type
(TF-luci-WT) or mutant TF 3′UTR (TF-luci-MUT) (Fig. 6E) were constructed. The results of
luciferase activity assay revealed that miR-92a-3p inhibited the
luciferase activity of TF-luci-WT constructs by approxi-mately 20%
(Fig. 6F), but failed to decrease
the luciferase activity of TF-luci-MUT (Fig. 6F).
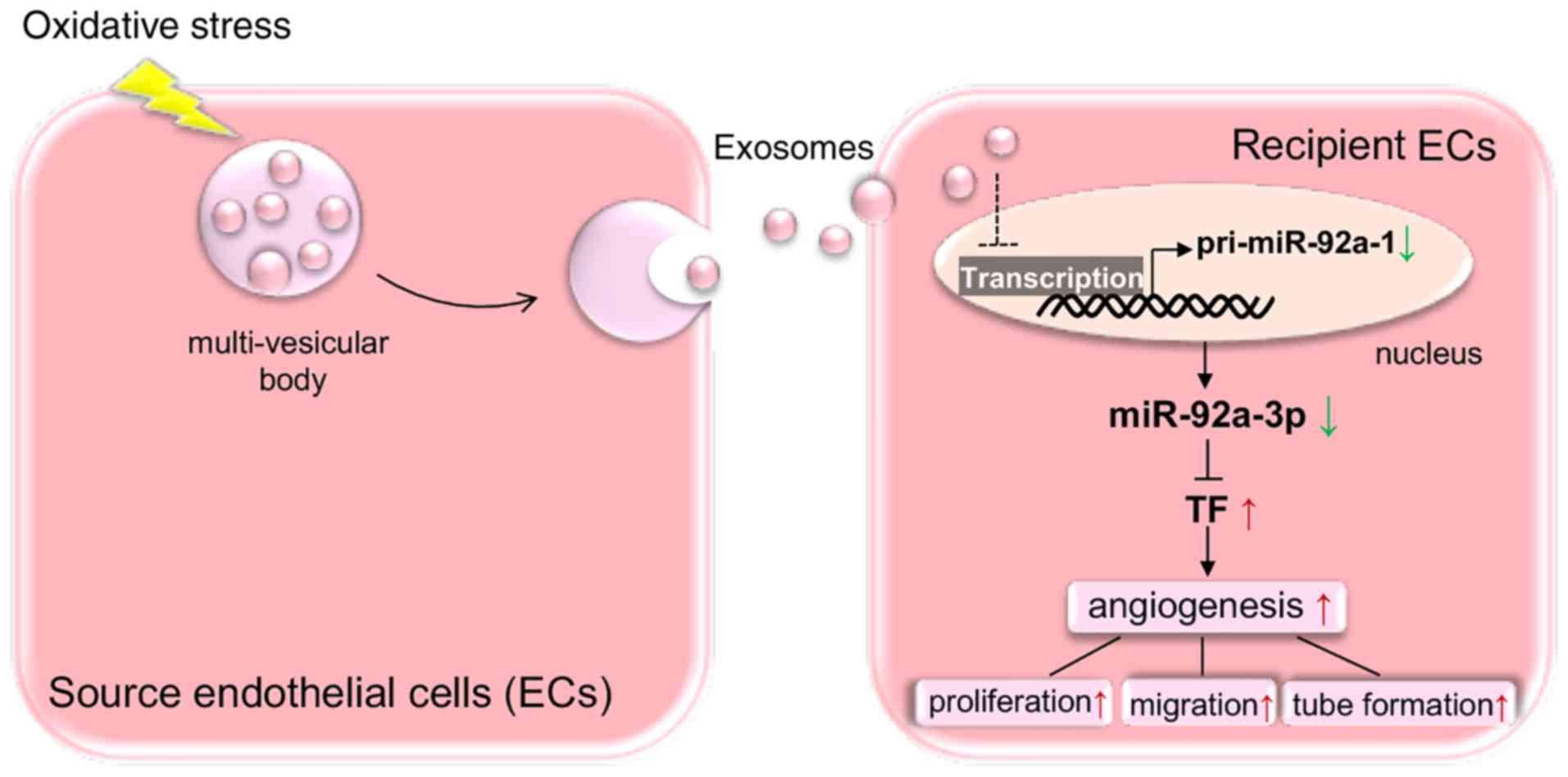
Taken together, the present study revealed a novel
proangiogenic mechanism between ECs mediated by oxidative
stress-stimulated endothelial exosomes by inhibiting the expression
of miR-92a-3p in target ECs (Fig.
7).
Discussion
Various types of pathological stimulation can lead
to vascular endothelial activation and injury. Activated or injured
ECs release characteristic extracellular vesicles (EVs), including
exosomes, microvesicles and apoptotic bodies. The content of
exosomes may partly depend on the stimuli and the state of donor
cells, which can exert distinct biological effects on target cells
(4). The roles of non-EC-derived
EVs in angiogenesis have previously been investigated (3), whereas the regulation of endothelial
exosomes in angiogenesis remains to be fully determined.
The present study aimed to investigate the role of
exosomes shed by oxidative stress-activated ECs in angio-genesis.
It was found that the exosomes derived from oxidative
stress-activated ECs promoted EC proliferation, migration and
angiogenesis, which were mediated by the downregulation of
miR-92a-3p in recipient ECs. Moreover, TF, a procoagulant and
proangiogenic gene (27), was
identified as a novel target of miR-92a-3p, and may be involved in
the role of activated endothelial exosomes in promoting
angiogenesis.
Several studies have revealed the signaling
transfer and proangiogenic effects of exosomes between non-ECs and
ECs (5-9). As regards EC-to-EC communication, it
has been reported that exosomes isolated from ECs cultured in
exosome-free medium or stimulated by interleukin-3 can promote
angiogenesis (10,11). Similarly, it was observed in the
present study that H2O2-activated endothelial
exosomes were effectively taken up by target ECs and led to an
enhanced cell proliferation, migration and angiogenesis. Although
the exosomes isolated from ECs exposed to different stimuli and in
a different state exerted similar effects, the mechanisms reported
in these studies differ. The exosomes isolated from ECs cultured in
an exosome-free medium promoted angiogenesis by suppressing the
expression of ataxia telangiectasia mutated (ATM) via transferring
miR-214 into recipient ECs. Endothelial exosomes generated in
response to interleukin-3 stimulation enhanced angiogenesis by
delivering miR-126-3p and pSTAT5 into recipient ECs, leading to the
activation of the pro-angiogenic pathway and the suppression of
antiangiogenic signal molecules (10,11). Herein, it was confirmed that the
exosomes isolated from H2O2-activated ECs
stimulated angiogenesis by decreasing miR-92a-3p expression in
recipient cells. The effect was similar to that observed with the
direct inhibition of miR-92a-3p in ECs observed in the present
study and other studies (14-18). Conversely, a recent study reported
that endothelial microvesicles (100-1,000 nm in size), which were
derived from ECs exposed to oxidized low-density lipoprotein
(oxLDL) stimulation, promoted angiogenesis by transferring
upregulated miR-92a-3p to recipient cells (19). The controversial role of
miR-92a-3p in regulating angio-genesis may change depending on the
experimental model and condition.
In previous findings on cell-to-cell
communications, the dysregulation of miRNAs in recipient cells has
been found to be usually caused by the delivery of miRNAs from
donor cells. In addition, intercellular protein transfer can
regulate the activity of specific signal pathways in recipient
cells (28). In the present
study, it was found that the decreased expression of miR-92a-3p in
Exo−H2O2-treated HUVEC may be
attributed to the transcriptional inhibition of miR-92a-3p, which
may be caused by the cargo in exosomes. The potential active
molecules or signaling path-ways involved in this process warrant
further investigation. miR-9-5p has also been reported to be
closely associated with angiogenesis (29-32). In the small RNA sequencing data of
the present study, miR-9-5p was significantly upreg-ulated in
Exo−H2O2-treated HUVECs. It was
also found that miR-9-5p expression was increased in both
H2O2-stimulated HUVEC and their exosomes
(data not shown). These results suggested that miR-9-5p, delivered
by Exo−H2O2-induced ECs, may
also be involved in the regulation of the biological effects on
target HUVECs, and the molecular mechanisms of miR-9-5p
upregulation differ from those of miR-92a-3p downregulation in
HUVECs caused by Exo−H2O2
exposure. miR-9-5p and miR-92a-3p may synergistically regulate EC
angiogenesis induced by Exo−H2O2, and this hypothesis is still
under investigation.
There are multiple known mRNA targets of miR-92a in
regulating cell proliferation, migration and angiogenesis, such as
Kruppel-like factor 2 (KLF2) (33), KLF4 (15), integrin subunit alpha5 (ITGA5)
(14,17) and sirtuin 1 (17). The mRNA expression profiles
obtained in the present study suggested that 91 of 197 differently
expressed genes were markedly upregulated by the inhibition of
miR-92a-3p in target ECs. A to9tal of 21 of the 91 genes were
candidate targets of miR-92a-3p predicted by the RNAhybrid program.
Among these 21 genes, TF is not only an initiator of blood
coagulation, but also an activator of angiogenesis, and may mediate
the effects of miR-92a-3p observed in the present study. There are
2 natural isoforms of TF, membrane-bound full-length (fl)TF and
soluble alternatively spliced (as)TF. Both isoforms have been found
to affect cell proliferation, migration and angiogenesis (27). It has been reported that the
inhibition of miR-19a and miR-126 promoted flTF and asTF synthesis
in EC (34). flTF has been shown
to indirectly promote angiogenesis via the induction of
proangiogenic factor expression by the FVIIa/protease-activated
receptors (PAR)-2 dependent signaling (35), whereas asTF has been reported to
directly increase the proangiogenic activity of EC independently of
the PAR-2 signaling via integrin ligation (36). In the present study, TF was found
as a new target of miR-92a-3p by gain and loss of function assays
and luciferase reporter assays.
There are some limitations to the present study.
The present study did not i) validate the pro-angiogenic effect of
oxidative stress-activated endothelial exosomes through in
vivo experiments; ii) determine the molecules or signaling
pathway, inducing the downregulation of miR-92a-3p in target EC
incubated with oxidative stress-activated endothelial exosomes;
iii) clarify the synergistically proangiogenic role of miR-9-5p
from oxidative stress-activated endothelial exosomes and miR-92a-3p
in target ECs. These questions remain to be investigated in future
studies.
In conclusion, the findings of the present study
provide new mechanistic insight into the regulation of
angiogenesis. In response to oxidative stress, exosomes are
released from ECs and convey potentially proangiogenic signals to
target ECs, eventually leading to a decreased miR-92a-3p expression
and subsequent angiogenesis. Moreover, TF may mediate the
biological effects of miR-92a-3p on ECs. Finally, this study
revealed a pro-angiogenic mechanism in ECs mediated by the
suppression of miR-92a-3p expression via oxidative
stress-stimulated endothelial exosomes (Fig. 7).
Supplementary Data
Funding
The present study was funded by grants from the
National Natural Science Foundation of China (nos. 81600340,
81770356 and 81970301), Capital Health Research and Development of
Special Funds (no. 2020-2-4084) and Peking University People′s
Hospital Research and Development Funds (no. RDY2018-26).
Availability of data and materials
The datasets used and/or analyzed in the current
study are available from the corresponding author upon reasonable
request.
Authors′ contributions
HC and SL designed the study. SL performed
experiments, analyzed data and wrote the manuscript. LY performed
experiments and analyzed data. LS, ZL, CL, FZ, YC and MW performed
experiments. HC and CL critically revised the manuscript. All
authors read and approved the final manuscript.
Ethics approval and consent to
participate
The animal experimental protocol was approved by
Peking University People′s Hospital Ethics Committee (approval no.
2016PHC072).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgments
Not applicable.
References
|
1
|
Potente M and Mäkinen T: Vascular
heterogeneity and specialization in development and disease. Nat
Rev Mol Cell Biol. 18:477–494. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Münzel T, Camici GG, Maack C, Bonetti NR,
Fuster V and Kovacic JC: Impact of oxidative stress on the heart
and vasculature: Part 2 of a 3-part series. J Am Coll Cardiol.
70:212–229. 2017. View Article : Google Scholar
|
|
3
|
Todorova D, Simoncini S, Lacroix R,
Sabatier F and Dignat-George F: Extracellular vesicles in
angiogenesis. Circ Res. 120:1658–1673. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Mathieu M, Martin-Jaular L, Lavieu G and
Théry C: Specificities of secretion and uptake of exosomes and
other extracellular vesicles for cell-to-cell communication. Nat
Cell Biol. 21:9–17. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Li J, Zhang Y, Liu Y, Dai X, Li W, Cai X,
Yin Y, Wang Q, Xue Y, Wang C, et al: Microvesicle-mediated transfer
of microRNA-150 from monocytes to endothelial cells promotes
angiogenesis. J Biol Chem. 288:23586–23596. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zheng B, Yin WN, Suzuki T, Zhang XH, Zhang
Y, Song LL, Jin LS, Zhan H, Zhang H, Li JS and Wen JK:
Exosome-mediated miR-155 transfer from smooth muscle cells to
endothelial cells induces endothelial injury and promotes
atherosclerosis. Mol Ther. 25:1279–1294. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Deregibus MC, Cantaluppi V, Calogero R, Lo
Iacono M, Tetta C, Biancone L, Bruno S, Bussolati B and Camussi G:
Endothelial progenitor cell derived microvesicles activate an
angiogenic program in endothelial cells by a horizontal transfer of
mRNA. Blood. 110:2440–2448. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Gong M, Yu B, Wang J, Wang Y, Liu M, Paul
C, Millard RW, Xiao DS, Ashraf M and Xu M: Mesenchymal stem cells
release exosomes that transfer miRNAs to endothelial cells and
promote angiogenesis. Oncotarget. 8:45200–45212. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Umezu T, Tadokoro H, Azuma K, Yoshizawa S,
Ohyashiki K and Ohyashiki JH: Exosomal miR-135b shed from hypoxic
multiple myeloma cells enhances angiogenesis by targeting
factor-inhib-iting HIF-1. Blood. 124:3748–3757. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
van Balkom BW, de Jong OG, Smits M,
Brummelman J, den Ouden K, de Bree PM, van Eijndhoven MA, Pegtel
DM, Stoorvogel W, Würdinger T and Verhaar MC: Endothelial cells
require miR-214 to secrete exosomes that suppress senescence and
induce angiogenesis in human and mouse endothelial cells. Blood.
121:3997–4006. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Lombardo G, Dentelli P, Togliatto G, Rosso
A, Gili M, Gallo S, Deregibus MC, Camussi G and Brizzi MF:
Activated Stat5 trafficking via endothelial cell-derived
extracellular vesicles controls IL-3 pro-angiogenic paracrine
action. Sci Rep. 6:256892016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kim YW and Byzova TV: Oxidative stress in
angiogenesis and vascular disease. Blood. 123:625–631. 2014.
View Article : Google Scholar :
|
|
13
|
Sun LL, Li WD, Lei FR and Li XQ: The
regulatory role of microRNAs in angiogenesis-related diseases. J
Cell Mol Med. 22:4568–4587. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Bonauer A, Carmona G, Iwasaki M, Mione M,
Koyanagi M, Fischer A, Burchfield J, Fox H, Doebele C, Ohtani K, et
al: MicroRNA-92a controls angiogenesis and functional recovery of
ischemic tissues in mice. Science. 324:1710–1713. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Iaconetti C, Polimeni A, Sorrentino S,
Sabatino J, Pironti G, Esposito G, Curcio A and Indolfi C:
Inhibition of miR-92a increases endothelial proliferation and
migration in vitro as well as reduces neointimal proliferation in
vivo after vascular injury. Basic Res Cardiol. 107:2962012.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Hinkel R, Penzkofer D, Zühlke S, Fischer
A, Husada W, Xu QF, Baloch E, van Rooij E, Zeiher AM, Kupatt C and
Dimmeler S: Inhibition of microRNA-92a protects against
ischemia/reperfusion injury in a large-animal model. Circulation.
128:1066–1075. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Daniel JM, Penzkofer D, Teske R, Dutzmann
J, Koch A, Bielenberg W, Bonauer A, Boon RA, Fischer A, Bauersachs
J, et al: Inhibition of miR-92a improves re-endothelialization and
prevents neointima formation following vascular injury. Cardiovasc
Res. 103:564–572. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Bellera N, Barba I, Rodriguez-Sinovas A,
Ferret E, Asín MA, Gonzalez-Alujas MT, Pérez-Rodon J, Esteves M,
Fonseca C, Toran N, et al: Single intracoronary injection of
encapsulated antagomir-92a promotes angiogenesis and prevents
adverse infarct remodeling. J Am Heart Assoc. 3:e0009462014.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Liu Y, Li Q, Hosen MR, Zietzer A, Flender
A, Levermann P, Schmitz T, Frühwald D, Goody P, Nickenig G, et al:
Atherosclerotic conditions promote the packaging of functional
MicroRNA-92a-3p into endothelial microvesicles. Circ Res.
124:575–587. 2019. View Article : Google Scholar
|
|
20
|
Bang C, Batkai S, Dangwal S, Gupta SK,
Foinquinos A, Holzmann A, Just A, Remke J, Zimmer K, Zeug A, et al:
Cardiac fibroblast-derived microRNA passenger strand-enriched
exosomes mediate cardiomyocyte hypertrophy. J Clin Invest.
124:2136–2146. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Li S, Chen H, Ren J, Geng Q, Song J, Lee
C, Cao C, Zhang J and Xu N: MicroRNA-223 inhibits tissue factor
expression in vascular endothelial cells. Atherosclerosis.
237:514–520. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
23
|
Li S, Geng Q, Chen H, Zhang J, Cao C,
Zhang F, Song J, Liu C and Liang W: The potential inhibitory
effects of miR-19b on vulnerable plaque formation via the
suppression of STAT3 transcriptional activity. Int J Mol Med.
41:859–867. 2018.
|
|
24
|
Rabiolo A, Bignami F, Rama P and Ferrari
G: VesselJ: A new tool for semiautomatic measurement of corneal
neovascularization. Invest Ophthalmol Vis Sci. 56:8199–8206. 2015.
View Article : Google Scholar
|
|
25
|
van Balkom BW, Eisele AS, Pegtel DM,
Bervoets S and Verhaar MC: Quantitative and qualitative analysis of
small RNAs in human endothelial cells and exosomes provides
insights into localized RNA processing, degradation and sorting. J
Extracell Vesicles. 4:267602015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Rehmsmeier M, Steffen P, Hochsmann M and
Giegerich R: Fast and effective prediction of microRNA/target
duplexes. RNA. 10:1507–1517. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Eisenreich A and Rauch U: Regulation and
differential role of the tissue factor isoforms in cardiovascular
biology. Trends Cardiovasc Med. 20:199–203. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Raposo G and Stahl PD: Extracellular
vesicles: A new communication paradigm? Nat Rev Mol Cell Biol.
20:509–510. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Chen X, Yang F, Zhang T, Wang W, Xi W, Li
Y, Zhang D, Huo Y, Zhang J, Yang A and Wang T: MiR-9 promotes
tumorigenesis and angiogenesis and is activated by MYC and OCT4 in
human glioma. J Exp Clin Cancer Res. 38:992019. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zhuang G, Wu X, Jiang Z, Kasman I, Yao J,
Guan Y, Oeh J, Modrusan Z, Bais C, Sampath D and Ferrara N:
Tumour-secreted miR-9 promotes endothelial cell migration and
angiogenesis by activating the JAK-STAT pathway. EMBO J.
31:3513–3523. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Madelaine R, Sloan SA, Huber N, Notwell
JH, Leung LC, Skariah G, Halluin C, Paşca SP, Bejerano G, Krasnow
MA, et al: MicroRNA-9 couples brain neurogenesis and angiogenesis.
. Cell Rep. 20:1533–1542. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Zhang H, Qi M, Li S, Qi T, Mei H, Huang K,
Zheng L and Tong Q: MicroRNA-9 targets matrix metalloproteinase 14
to inhibit invasion, metastasis, and angiogenesis of neuroblastoma
cells. Mol Cancer Ther. 11:1454–1466. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Shyu KG, Wang BW, Pan CM, Fang WJ and Lin
CM: Hyperbaric oxygen boosts long noncoding RNA MALAT1 exosome
secretion to suppress microRNA-92a expression in therapeutic
angiogenesis. Int J Cardiol. 274:271–278. 2019. View Article : Google Scholar
|
|
34
|
Eisenreich A, Bolbrinker J and Leppert U:
Tissue factor: A conventional or alternative target in cancer
therapy. Clin Chem. 62:563–570. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Versteeg HH, Schaffner F, Kerver M,
Petersen HH, Ahamed J, Felding-Habermann B, Takada Y, Mueller BM
and Ruf W: Inhibition of tissue factor signaling suppresses tumor
growth. Blood. 111:190–199. 2008. View Article : Google Scholar
|
|
36
|
van den Berg YW, van den Hengel LG, Myers
HR, Ayachi O, Jordanova E, Ruf W, Spek CA, Reitsma PH, Bogdanov VY
and Versteeg HH: Alternatively spliced tissue factor induces
angiogenesis through integrin ligation. Proc Natl Acad Sci USA.
106:19497–19502. 2009. View Article : Google Scholar : PubMed/NCBI
|