Introduction
As the most commonly occurring form of primary renal
tumor, renal cell carcinoma (RCC) is a malignancy accompanied by a
high mortality rate, with approximately 202,000 newly diagnosed
cases and 102,000-related deaths worldwide annually (1–3).
Surgical resection remains the only known curative treatment for
localized RCC (2). Additionally,
radiotherapy and chemotherapy are considered in some cases
(4). However, cancer relapse in a
number of patients occurs frequently due to resistance, resulting
in a 12% 5-year survival rate for patients with metastatic disease
(5,6). Therefore, recurrence remains the
main reason for the poor prognosis. However, the underlying
mechanisms responsible for RCC recurrence have not yet been fully
elucidated.
Steroid receptor RNA activator (SRA) functions as
coactivator in various types of cancer, such as breast cancer
(7,8) ovarian and prostate cancers (7,8).
Compared to adjacent normal tissues, the levels of SRA have been
found to be elevated in breast cancer tissues (9). Patients with breast cancer with a
high SRA level have a significantly worse survival rate than
patients with a low SRA level. It has been indicated that SRA may
serve as a novel prognostic marker for patients with estrogen
receptor (ER)-positive breast cancer (10). Moreover, SRA is a potent inducer
and regulator of gene expression, particularly of genes associated
with epithelial-mesenchymal transition (EMT) and the Notch
signaling pathway in cervical cancer (11). Although there is limited evidence
linking SRA to RCC, the immunohistochemistry (IHC) of primary RCC
samples has revealed that a high androgen receptor expression is
significantly associated with a lower tumor stage and a better
prognosis (12,13). Additionally, the expression levels
of glucocorticoid and progestin receptor are increased in RCC
tissues (14).
In the present study, the role and underlying
mechanisms of SRA in RCC were investigated. Mechanistic analysis
demonstrated that SRA suppressed the stem-like features of RCC
cells. However, the ERK signaling pathway was found potentially not
be involved in the SRA-induced migration and invasion of RCC cells.
Taken together, SRA may be a novel and promising therapeutic target
in RCC therapy.
Materials and methods
Human tissue samples
To compare the expression levels of SRA among RCC
development, RCC tissues (14 cases) and adjacent non-tumorous
tissues (14 cases) were obtained. The tissues were obtained from
the Department of Cancer Resource Base at Sun Yat-sen University
(SYSUCC) Cancer Center 7 years ago. All the fresh specimens were
stored in liquid nitrogen until use. These tissues were confirmed
by doctors from the Department of Pathology, SYSUCC. All the
recruited patients provided their oral informed consent prior to
their inclusion. Ethical consent was granted for the study by the
Committees for Ethical Review of Hunan University of Chinese
Medicine. The ethics committee agreed with informed consent being
obtained orally in the present study.
Cells and cell culture
RCC cell lines (786-O, SN12C, SKRC39, A498 and
Caki-1), a papillary renal cell carcinoma cell line (Caki-2) and
the HK-2 cell line were obtained from SYSUCC. The RCC and Caki-2
cells were cultured in RPMI-1640 (Gibco; Thermo Fisher Scientific,
Inc.) and the HK-2 cells were cultured in Dulbecco's modified
Eagle's medium (DMEM; Gibco; Thermo Fisher Scientific, Inc.). All
cells were cultured with 10% fetal bovine serum (FBS; Gibco; Thermo
Fisher Scientific, Inc.) and 1% penicillin/streptomycin
(Invitrogen; Thermo Fisher Scientific, Inc.) in a 5% CO2
humidified atmosphere at 37°C. The short 6 tandem repeat (STR)
testing was used to authenticate all cell lines and all cells were
maintained mycoplasma-free. Cells were treated with 5 μM
PD98059 (MedChemExpress LLC) for 24 h.
Stable knockdown of SRA in SKRC39
cells
The SKRC39 cells were divided into the following
groups: The short hairpin RNA (shSRA) group and the scramble group
(with the same amount of nonsense RNA). Plasmids (0.6 g) in the
shRNA and scramble groups were diluted in 250 μl Opti-MEM
(Thermo Fisher Scientific, Inc.). Similarly, Lipofectamine 2000
(Invitrogen; Thermo Fisher Scientific, Inc.) was first diluted in
250 μl Opti-MEM before being added in a dropwise matter to
the plasmid solution. The mixture was incubated at 25°C for 20 min
prior to transfection into the SKRC39 cells. The SKRC39 cells were
transfected for 48 h. The following are the target sequences of the
shRNA plasmid (Guang Zhou Fu Neng) applied: Non-target shRNA
sequence, 5′-GCTTCGCGCCGTAGTCTTA-3′; sequence 1 (for SKRC39-shSRA1
cells), 5′-GGAGGAAAGTTGTCAATAC-3′; sequence 2 (for SKRC39-shSRA3
cells), 5′-TCA GTG GAT GGT AGG AGT T-3′.
Stable overexpression of SRA in A498
cells
Lentiviral plasmid was packaged and used for cell
transfection according to the manufacturer's instructions
(Invitrogen; Thermo Fisher Scientific, Inc.). The clone-A498 cells
were transfected with 1 ng/μl pLenti6/V5-SRA or 1
ng/μl pLenti6/V5 (Invitrogen; Thermo Fisher Scientific,
Inc.) using Lipofectamine 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.). Puromycin (Sigma-Aldrich; Merck KGaA) was added
to select stably transfected clones for 21 days.
MTS assay
Cell proliferation was assessed every day of the
week upon the use of 3-(4,5-dimethylthiazol-2-yl)-5-(3-carb
oxymethoxyphenyl)-2-(4-sulfophenyl)-2H tetrazolium inner salt (MTS)
(Sigma-Aldrich; Merck KGaA). The optical absorbance at 490 nm was
measured using ElX 800 microplate reader (BioTek Instruments
Inc.).
Colony formation assays
Cells were seeded in 6-well plates at 50% confluence
and then transfected with SRA overexpression plasmid or shSRA. AT
24 h post-transfection, the cells were seeded in 6-well plates and
cultured for 15 days. Colonies on each plate were counted using a
TH4-200 microscope (Olympus Corporation) following fixation with 4%
paraformaldehyde and staining with 0.1% crystal violet (Beijing
Solarbio Science & Technology Co., Ltd.) at 25°C for 15
min.
Transwell cell migration assay
Transwell chambers (Costar, Corning Inc.) coated
without Matrigel (BD Biosciences) on the pore size of an 8
μm membrane were used to investigate cell migration. The
cells were starved for 24 h. Following cells digested with 0.25%
trypsin-EDTA (Thermo Fisher Scientific Inc.) and centrifugated at
300 × g for 5 min at 25°C, the cells were resuspended in serum-free
medium, and the cells were counted under CKX41 light microscope
(Olympus Corporation). The cell concentration was adjusted to
2x104 cells/200 μl, and the cell suspension was
placed in the bottom of a 24-well plate. This was followed by the
addition of 800 μl of 10% FBS medium to each well. Following
incubation at 37°C for 24 h, the cells in the internal compartment
were removed using a cotton swab. The migrated cells were fixed in
methanol and stained with a 1% crystal violet solution at 25°C for
15 min. Under CKX41 light microscope (Olympus Corporation), 3
randomly selected fields of view at ×200 magnification were
counted, and images were captured using Olympus Stream system
(Olympus Corporation).
Transwell cell invasion assay
Transwell invasive chambers coated with Matrigel on
the upper surface of an 8 μm (pore size) membrane were used
to determine cell invasion. Cells were plated in new 24-well
plates, 500 μl of serum-free medium were added to the upper
and lower chambers, and a serum-free cell suspension was prepared
following culture at 37°C for 24 h. The cells were counted under a
microscope and were then adjusted to 4x104 cells/200
μl. The cells were all transferred to a new well plate, the
upper chamber medium was removed, 200 μl cell suspension was
added, and the lower chamber was filled with 800 μl of 10%
FBS medium. Following culture at 37°C for 24 h, the cells in the
inner chamber were not wiped off using a cotton swab. The invasive
cells were fixed and stained with 1% crystal violet solution at
25°C for 15 min. Under CKX41 light microscope (Olympus
Corporation), 3 fields (x200 magnification) were selected randomly
for counting and photographing to compare the differences in cell
invasiveness between the 2 groups.
Western blot analysis and antibodies
Cell lysates were extracted with
radio-immunoprecipitation assay (RIPA, Beyotime Institute of
Biotechnology, Inc.) buffer with protease inhibitor cocktail
(CWBiotech). Protein concentrations were determined using the BCA™
Protein Assay kit (CWBiotech) according to the protocol provided by
the manufacturer. Proteins (20 μl) loaded onto each lane
were subjected to 10% sodium dodecyl sulfate-polyacrylamide gel
electrophoresis (SDS-PAGE), and then transferred to a
polyvinylidene difluoride (PVDF, EMD Millipore) membrane. After
blocking with 5% skimmed milk for 4 h, PVDF membranes were
incubated overnight at 4°C with the following primary antibodies:
Cyclin D1 (1:1,000; cat. no. ab16663, Abcam), cyclin E (1:1,000;
cat. no. 4129, Cell Signaling Technology, Inc.), cyclin-dependent
kinase (CDK)4 (1:1,000; cat. no. 11026-1-AP), CDK6 (1:1,000; cat.
no. 14052-1-AP) (both from Proteintech), poly(ADP-ribose)
polymerase (PARP; 1:1,000; cat. no. 9532), cleaved-PARP (1:1,000;
cat. no. 5625), p53 (1:1,000; cat. no. 2527), p21 (1:1,000; cat.
no. 2947), β-actin (1:1,000; cat. no. 4970) (all from Cell
Signaling Technology, Inc.), desmoplakin (1:1,000; cat. no.
ab109445, Abcam), E-cadherin (1:1,000, cat. no. 3195), fibronectin
(1:1,000; cat. no. 26836), N-cadherin (1:1,000; cat. no. 13116),
Vimentin (1:1,000; cat. no. 5741), Snail (1:1,000; cat. no. 3879),
p-ERK1/2 (1:1,000; cat. no. 4370), ERK1/2 (1:1,000; cat. no. 4695),
p-JNK (1:1,000; cat. no. 4668), JNK (1:1,000; cat. no. 9252),
p-STAT3 (1:2,000, cat. no. 9145), STAT3 (1:1,000; cat. no. 30835)
and GAPDH (1:1,000; cat. no. 5174) (all from Cell Signaling
Technology, Inc.). After rinsing with TBST 3 times for 10 min each
time, the PVDF membranes were incubated with anti-rabbit antibody
(1:2,000; cat. no. 7074, Cell Signaling Technology, Inc.) or
anti-mouse (1:2,000; cat. no. 14709, Cell Signaling Technology,
Inc.) at room temperature for 1.5 h. Subsequently, the membranes
were rinsed with TBST 3 times (10 min each time) and visualized
using Pierce™ ECL Western Blotting Substrate (Pierce; Thermo Fisher
Scientific, Inc.) (15). The
blots developed with chromogenic substrates were stripped of
antibodies by using stripping buffer solutions (Beyotime Institute
of Biotechnology Inc.). The blots were then re-probed to detect a
second protein with the same or very similar molecular weights
through chemiluminescence.
Reverse transcription-quantitative PCR
(RT-qPCR)
Total cellular RNA and tissue RNA were extracted
using TRIzol reagent (Life Technologies; Thermo Fisher Scientific,
Inc.). Total RNA was quantified using a Nanodrop 1000
spectrophotometer (Thermo Fisher Scientific, Inc.) and equal
amounts (1 μg) were transcribed into cDNA using the iScript™
cDNA Synthesis kit (cat. no. 1708891, Bio-Rad Laboratories, Inc.)
according to the manufacturer's protocol. The appropriate size of
the qPCR products was confirmed by agarose gel electrophoresis.
Subsequently, the samples were amplified and analyzed using a SYBR
Premix Ex Taq™ kit (cat. no. RR820A; Takara Bio, Inc.) and an
Applied Biosystems 7500 system (Thermo Fisher Scientific, Inc.).
The thermocycling conditions were as follows: 95°C for 10 min,
followed by 40 cycles of 95°C for 30 sec, 55°C for 30 sec and 72°C
for 1 min. All PCR assays were replicated at least 3 times. The
fold change in gene expression was calculated using the
2−ΔΔCq method (16). A
melting curve analysis was performed for each amplicon to verify
the specificity of each amplification step. β-actin was used as the
normalization controls for RCC cell lines. The gene primers used
for amplification are presented in Table I.
 | Table ISequences of primers used for
RT-qPCR. |
Table I
Sequences of primers used for
RT-qPCR.
| Gene name | Forward | Reverse |
|---|
| SRA |
5′-GGCTCTACTGGTGCAAGAGC-3′ | 5′-GGAAGCCTGGT
ATGGTATGG-3′ |
| β-actin |
5′-CATCCGCAAAGACCTGTACG-3′ |
5′-CCTGCTTGCTGATCCACA TC-3′ |
| Snail |
5′-CCCAATCGGAAGCCTAACTA-3′ |
5′-GCTGGAAGGTAAACTCTGGA-3′ |
| E-cadherin |
5′-CCTGGGACTCCACCTACAGA-3′ |
5′-CAACATACCTGATGGGGCGG-3′ |
| Vimentin |
5′-GCTGAATGACCGCTTCGCCAAC-3′ |
5′-GCTCCCGCATCTCCTCCTCGTA-3′ |
Flow cytometry
The cell cycle was analyzed with a commercial kit
(Beyotime Institute of Biotechnology, Inc.) according to the
manufacturer's instructions. Briefly, the cells were collected and
fixed with pre-cooling 70% alcohol for 2 h at 4°C. After discarding
the supernatant, the precipitate was washed with PBS and
centrifuged at 300 × g for 5 min at 25°C. A total of 500 μl
of staining buffer, including 25 μl propidium iodide (PI)
staining solution (Beijing Solarbio Science & Technology Co.,
Ltd.), 100 μl RNase A (Thermo Fisher Scientific, Inc.) and
0.2% Triton X-100 (Beijing Solarbio Science & Technology Co.,
Ltd.) were added to the wells, followed by incubation for 30 min at
37°C. The samples were then detected immediately using a flow
cytometer (Beckman Coulter, Inc.) (17).
Apoptosis assays were performed with the Annexin
VFITC/propidium iodide (PI) apoptosis detection kit from KeyGen
Biotech according to manufacturer's instructions. RCC cells were
collected and fixed in 75% ethanol overnight. The cells were
subsequently incubated with 500 μl of binding buffer, 5
μl of Annexin V-FITC and 5 μl propidium Iodide for 15
min at 25°C in the dark. A flow cytometer (Beckman Coulter, Inc.)
and Kaluza software 2.0 (Beckman Coulter, Inc.) were used to detect
and analyze the percentage of apoptotic and necrotic cells.
CD44+/CD24− cell population
analysis was performed by flow cytometry. Allophycocyanin
(APC)-conjugated mouse anti-human CD44 monoclonal antibody (cat.
no. BD 559942) and phycoerythrin/cyanine 7 (PE/Cy7)-conjugated
mouse anti-human CD24 monoclonal antibody (cat. no. BD 561646) were
purchased from BD Pharmingen (BD Biosciences). CD24 and CD44
expression was analyzed in cells derived from monolayer cultures
following dissociation in trypsin-EDTA at 37°C. Cells
(1x105) were centrifuged at 300 × g for 5 min at 25°C.
The cells were washed in PBS, resuspended with anti-CD24-PE/Cy7
(1:20 dilution) and anti-CD44-APC (1:20 dilution). Samples were
incubated for 30 min at 4°C in the dark. The labeled cells were
washed using PBS and analyzed using a BD FACSAria II flow cytometer
and software DiVa (BD Biosciences). The negative fraction was
determined using appropriate isotype controls.
Statistical analysis
The results are expressed as the means ± SD. A
two-tailed Student's t-test was used for statistical comparisons. A
paired t-test was used for patient tissue comparisons, and an
unpaired t-test was used for cell line analyses. One-way analysis
of variance (ANOVA) was used to analyze the significance among
groups. The least significant difference (LSD) was used as a post
hoc test was used following a significant one-way ANOVA result when
analyzing datasets containing 3 groups. For datasets containing
>3 groups, the results were analyzed by one-way ANOVA, followed
by Tukey's post-hoc test. P-value <0.05 was considered to
indicate a statistically significant difference.
Results
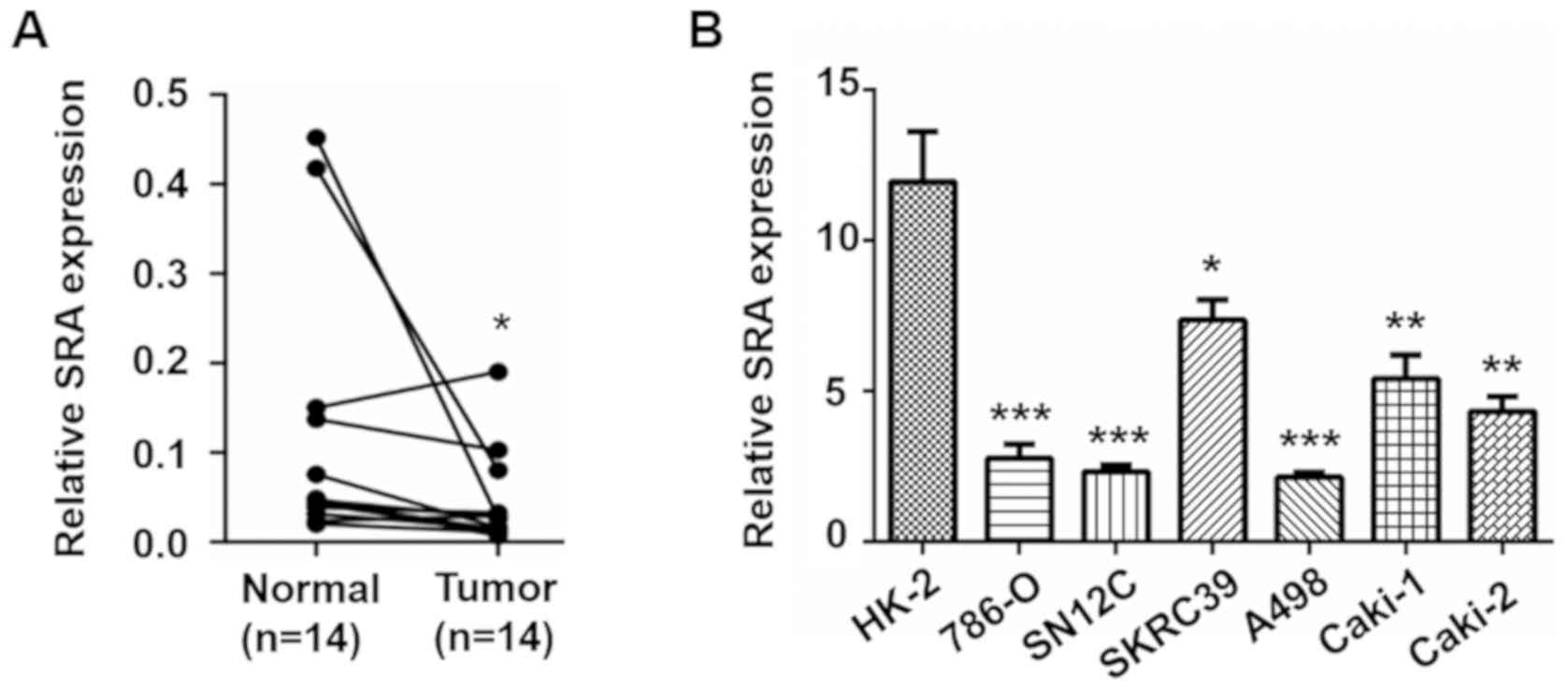
SRA is downregulated in both RCC clinical
samples and cell lines
To explore the clinical significance of SRA in human
RCC, RT-qPCR was applied to detect the expression of SRA in RCC
tissues and adjacent tissues from 14 patients (Fig. 1A). Compared to the matched normal
renal tissues, the expression levels of SRA were significantly
lower in RCC tissues. To confirm this result, the expression levels
of SRA were evaluated in a panel of RCC cell lines (786-O, SN12C,
SKRC39, A498, Caki-1 and Caki-2 cells) and renal tubular epithelial
cells (HK-2 cells). Consistent with the results obtained with the
clinical samples, the expression levels of SRA were significantly
lower in the RCC cell lines than in the HK-2 cells (Fig. 1B). Furthermore, among the RCC cell
lines, the SKRC39 cells, the metastatic cell line, Caki-1, and the
papillary renal cell carcinoma cell line, Caki-2, exhibited higher
SRA levels than non-the metastatic cell lines, 786-O, SN12C and
A498 (Fig. 1B). These data
suggested a potential role of SRA in the progression of RCC.
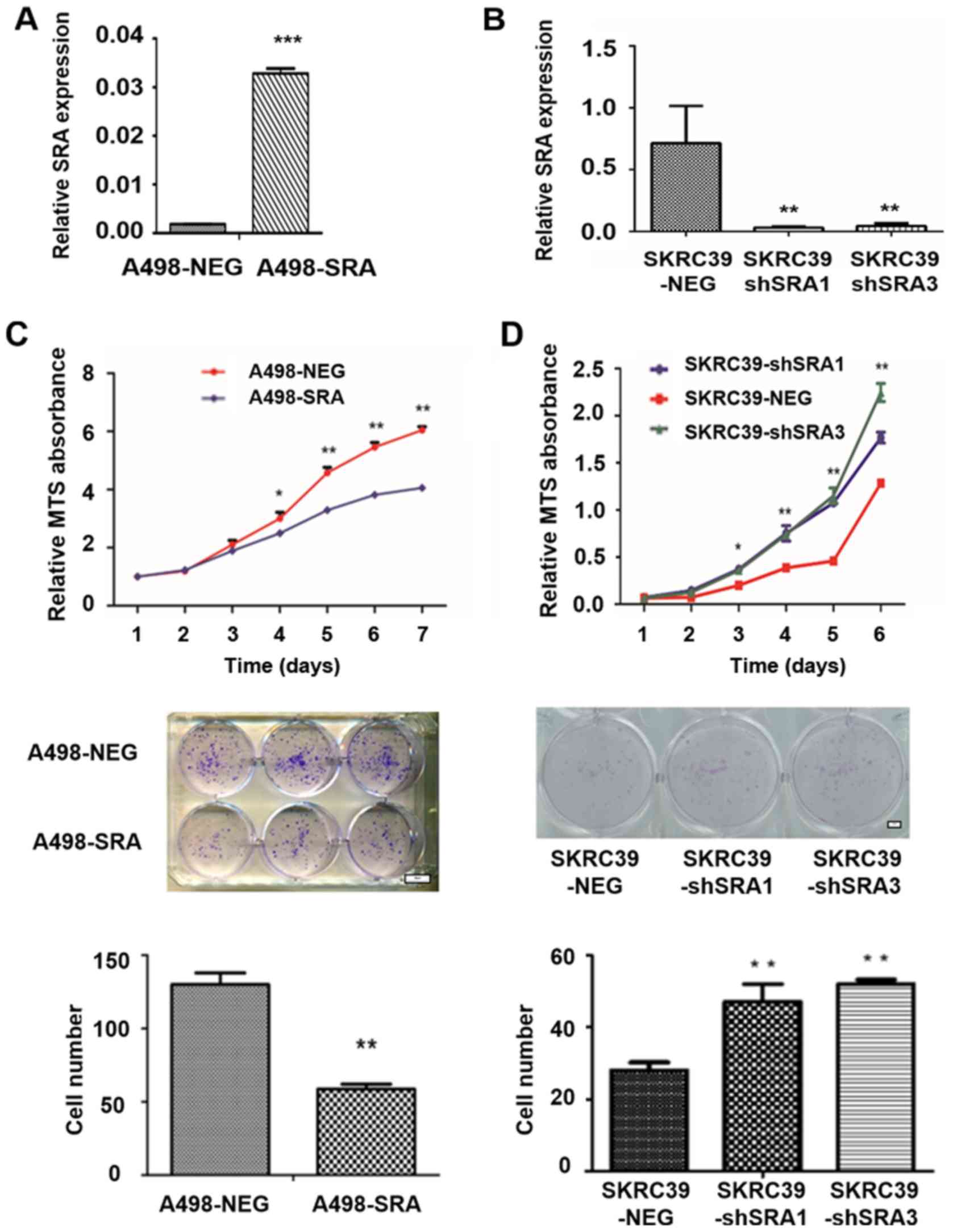
SRA inhibits the viability and
proliferation of RCC cell lines
Based on the above-mentioned results, the A498 cells
with a low SRA level and SKRC39 cells with a high SRA level, were
used to verify the hypothesis of the role of SRA in RCC. Several
cell clones with genetically manipulated SRA were obtained as
follows: i) A498 cells stably overexpressing SRA (A498-SRA) and a
negative control A498 (A498-NEG) (Fig. 2A); and ii) SKRC39 cells subjected
to the shRNA-mediated knockdown of SRA (SKRC39-shSRA1 and
SKRC39-shSRA3), and expressing non-targeting shRNA (SKRC39-NEG)
(Fig. 2B). As was expected, the
overexpression of SRA significantly inhibited the proliferation of
the A498-SRA cells, compared with that of the A498-NEG cells
(Fig. 2C, upper panel). However,
the knockdown of SRA in the SKRC39 cells exerted opposite effects
(Fig. 2D, upper panel).
Additionally, SRA overexpression reduced the colony formation
ability of the A498 cells (Fig.
2C, lower panels), while the downregulation of SRA promoted the
colony formation of SKRC39 cells (Fig. 2D, lower panels).
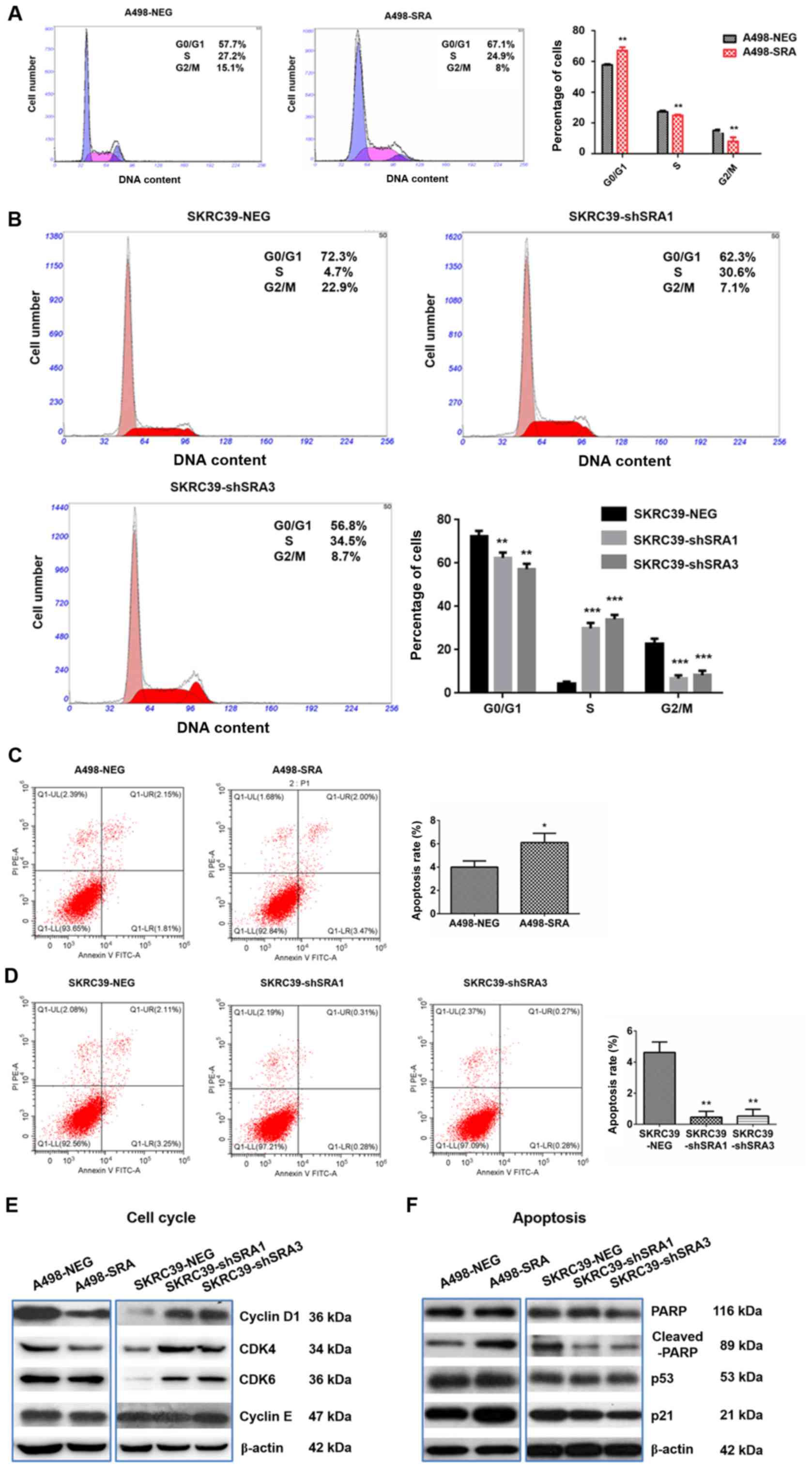
SRA inhibits the cell cycle and promotes
the apoptosis of RCC cell lines
It is known that cell proliferation is a cell
cycle-dependent process during cancer development (18). In the present study, to further
examine the effects of SRA on RCC proliferation, a cell cycle
analysis was performed using flow cytometry. A significant
accumulation of A498-SRA cells in the G0/G1 phase accompanied by a
reduction in the number of cells in the S phase was observed
(Fig. 3A). However, SRA knockdown
in the SKRC39 cells promoted cell entry into the S phase (Fig. 3B). These data indicated that SRA
inhibited RCC cell proliferation possibly by inducing cell cycle
arrest at the G0/G1 phase. Subsequently, the expression levels of
the cell cycle-related proteins were detected. Cyclin D1, CDK4,
CDK6 and cyclin E are key proteins regulating cell cycle
progression from the G1 to the S phase (19). The present study found that SRA
overexpression markedly downregulated cyclin D1 and CDK4
expression, whereas it exerted no obvious effects on CDK6 and
cyclin E expression (Fig. 3E).
Conversely, the knockdown of SRA upregulated the levels of cyclin
D1, CDK4 and CDK6 (Fig. 3E).
The anti-apoptotic ability is one of the important
characteristics of tumor cells. The present study thus examined the
effects of SRA on apoptosis using flow cytometry. The results
revealed that the apoptotic rate of the A498 cells transfected with
SRA overexpression plasmid was significantly increased compared
with that of the control cells (Fig.
3C). The knockdown of SRA markedly reduced the percentage of
apoptotic cells (Fig. 3D). PARP1
is involved in the initial response to DNA damage, which is also
inactivated by caspase-3 cleavage in p53-dependent manner (20). In the present study, the
over-expression of SRA markedly increased the cleavage of PARP in
the A498 cells, while it also increased the levels of p53 and p21
(Fig. 3F). The knockdown of SRA
exerted opposite effects on the SKRC39 cells (Fig. 3F). Thus, SRA inhibited RCC cell
proliferation by interfering with cell cycle progression and
promoting cell apoptosis.
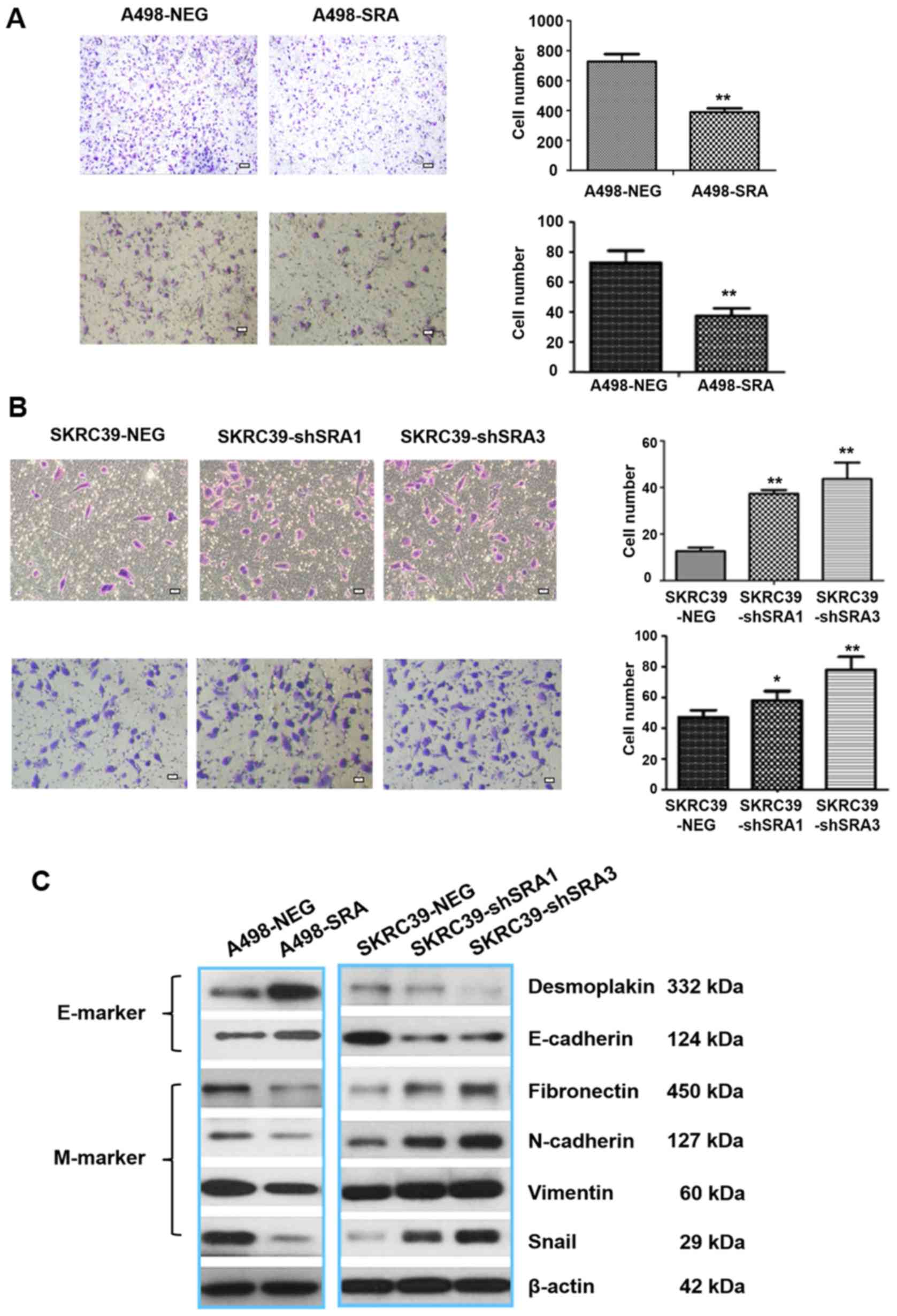
SRA inhibits the migration, invasion and
EMT of RCC cells
To examine the effects of SRA on cell motility,
Transwell assay was performed. It was found that the stable
overexpression of SRA significantly inhibited the migration and
invasion of A498 cells (Fig. 4A).
By contrast, the knockdown of SRA enhanced the migratory and
invasive capabilities of the SKRC39 cells (Fig. 4B). During cancer migration and
invasion, EMT is triggered by multiple signal pathways. Moreover,
the activation of EMT further facilitates stem cell-like properties
in cancers (21,22). Snail, a zinc-finger transcription
factor, is directly involved in the repression of epithelial
marker, E-cadherin, transcription and promotes the acquisition of a
mesenchymal phenotype during EMT process (23,24). The findings of the present study
demonstrated that the overexpression of SRA in the A498 cells
resulted in an increment in the levels of epithelial markers
(desmoplakin and E-cadherin), and led to a decrease in the levels
of mesenchymal markers, such as fibronectin, N-cadherin, vimentin
and Snail. Conversely, the SKRC39-shSRA cells exhibited opposite
effects (Fig. 4C).
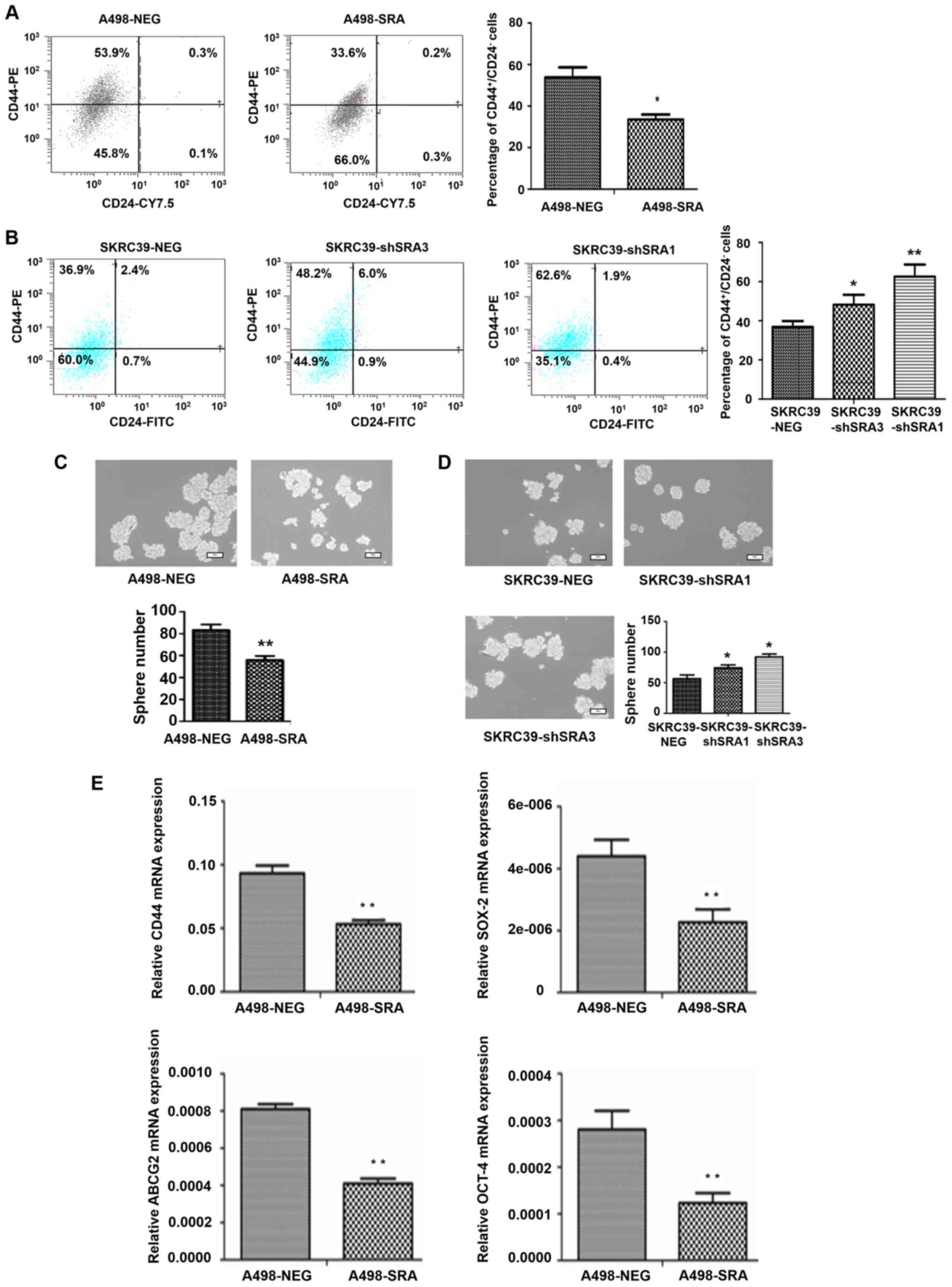
SRA inhibits the stem-like
characteristics of RCC cells
It has been reported that cancer cell phenotype and
biology of CD44+/CD24− cells are often
applied to identify stem-like cells in tumorigenesis of RCC
(25). Herein, flow cytometric
analysis was performed to detect the percentages of
CD44+/CD24− cells in the A498-SRA cells and
SKRC39-shSRA cells. As shown in Fig.
5A, the overexpression of SRA significantly decreased the
percentage of CD44+/CD24− in the A498 cells
(Fig. 5A), while the knockdown of
SRA in the SKRC39 cells exerted an opposite effect (Fig. 5B). As previously reported,
sphere-forming assays can reflect the ability of cellular
self-renewal and proliferation, resulting in the acquisition of
stem cell properties (26). The
present study found that the overexpression of SRA reduced the
sphere-forming effi-ciency of A498 cells (Fig. 5C), while the knockdown of SRA
induced SKRC39 cell to form spheres (Fig. 5D). These findings were consistent
with those of previous studies (27–30) and the present data, demonstrating
that SRA decreased the expression levels of the stemness markers,
CD44, sex determining region Y-box 2 (SOX-2), ATP-binding cassette
transporter G2 (ABCG2) and octamer-binding transcription factor
4(OCT-4) (Fig. 5E). The
above-mentioned results suggested that SRA regulated the stem-like
characteristics of RCC cells.
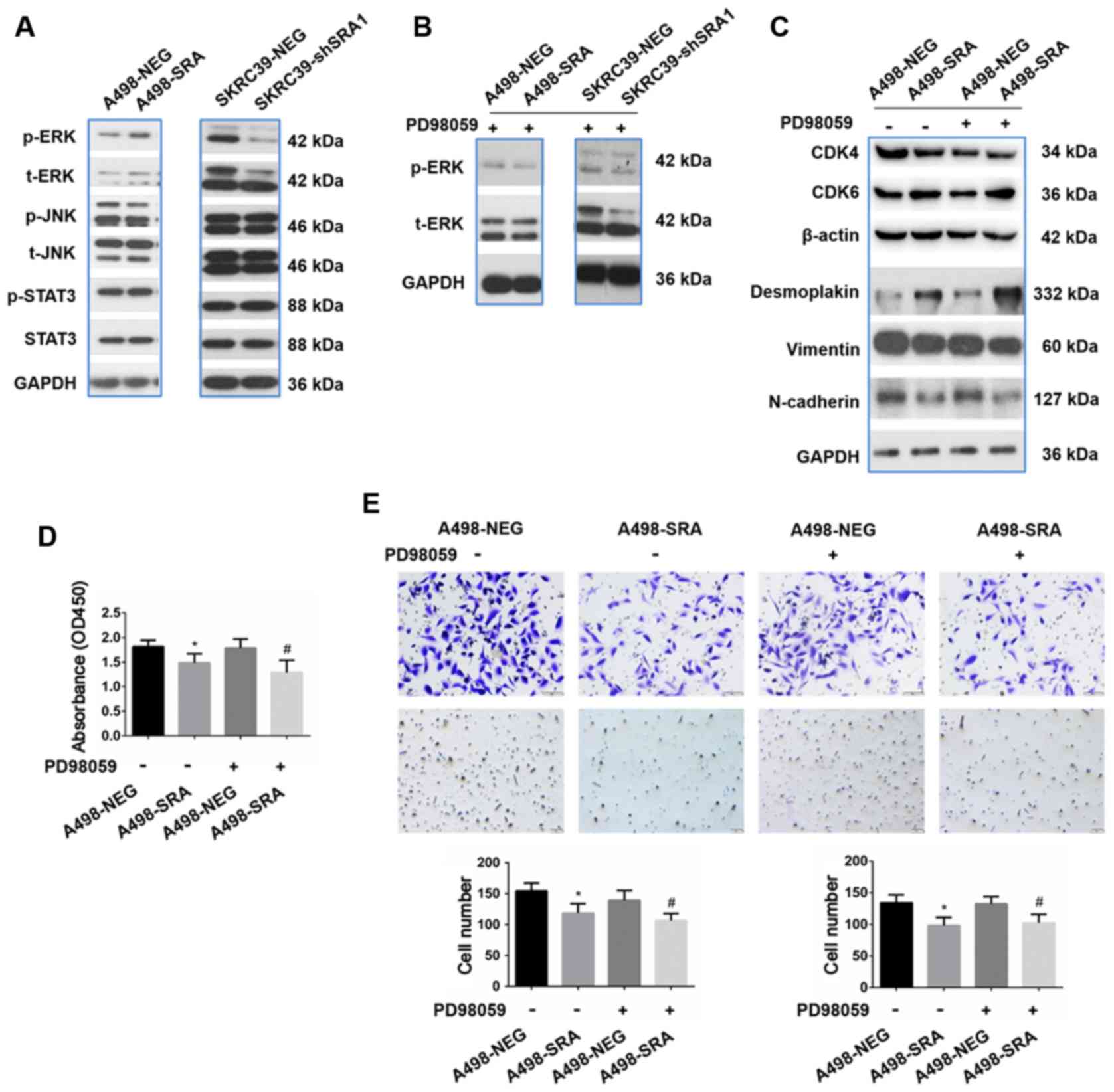
The ERK signaling pathway is not required
for the inhibition of the proliferation, migration and invasion of
RCC cells by SRA
To further investigate the preliminary mechanisms of
EMT regulated by SRA in RCC cells, the effects of SRA on several
classical signaling pathways, including the ERK, JNK and STAT3
pathways were detected. Among these signaling pathways, it was
found that the overexpression of SRA signifi-cantly upregulated the
levels of phospho-ERK1/2 in the A498 cells, while the knockdown of
SRA markedly inhibited ERK1/2 phosphorylation in the SKRC39 cells
(Fig. 6A). Furthermore, the ERK
inhibitor, PD98059, abolished ERK1/2 phosphorylation in both the
A498-SRA and SKRC39-shRNA cells (Fig.
6B). The potential role of ERK in proliferation and EMT markers
was also investigated in the A498-SRA cells and SKRC39-shRNA cells.
As shown in Fig. 6C, the
abrogation of the ERK1/2 pathway did not affect the expression
levels of proliferation- and EMT-related markers mediated by SRA.
Although SRA attenuated the abilities of proliferation, migration
and invasion, PD98059 exerted no obvious effects on A498-NEG cells
or A498-SRA cells compared with the untreated A498-NEG cells or
A498-SRA cells (Fig. 6D and
E).
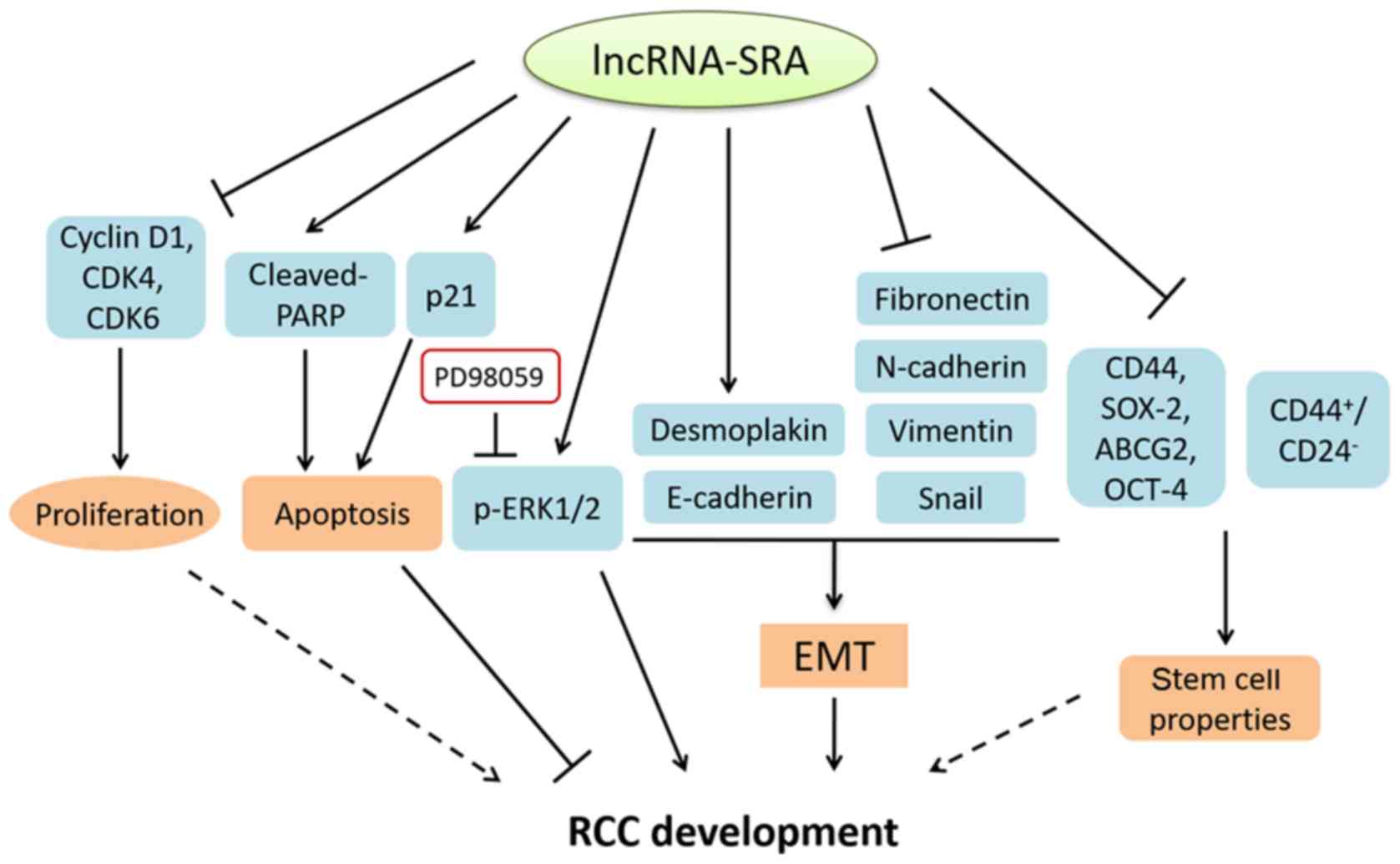
The above-mentioned results indicated that SRA
inhibited the cell proliferation, promoted apoptosis, suppressed
EMT and stemness characteristics, while ERK signaling pathway was
dispensable for the roles of SRA in RCC progression (Fig. 7).
Discussion
RCC is the most common type of urological
malignancy. The incidence of RCC has been increasing over the past
two decades (3). The lack of
effective biomarkers and therapies has led to a high human
morbidity (31). It is worth
noting that among all hallmarks of cancer, sustaining proliferative
signaling, activating migration and invasion, and gaining stem cell
characteristics are most important factors responsible for systemic
spreading and treatment resistance (32).
SRA was initially identified in 1999 and functions
to increase the activity of steroid receptors (33). Although a number of studies on the
structure and molecular functions of SRA are emerging, the
physiological roles of SRA have not yet been fully elucidated. The
present study found SRA was downregulated in both RCC clinical
samples and cell lines. SRA inhibited the proliferation, migration,
invasion and stemness of RCC cells via suppressing the cell cycle,
intervening the EMT process and altering stemness markers, which
indicated a potential tumor suppressive role of SRA in RCC.
However, in contrast to the findings of the present study, several
studies have defined SRA as an oncogene, since it is overexpressed
in ovarian and breast cancers (34,35). It was therefore considered that
SRA may exhibit a tissue-specific expression pattern and function
as an oncogene or tumor suppressor in different types of cancers.
Moreover, intracellular signaling pathways, the tumor
microenvironment, as well as energy metabolism in different cancer
types are also closely related to the functions of SRA. For
example, Lanz et al demonstrated that the overexpression of
SRA exhibited potent promoting activities on cellular proliferation
and differentiation via the aberrant stimulation of ER activity in
breast cancer (9). SRA plays an
anticancer role in osteosarcoma by sponging miR-208a, as it reduces
cell migration, invasion and proliferation, while it promotes cell
apoptosis (36). In the present
study, the expression levels of SRA were found to be significantly
down-regulated in RCC tissues in comparison with the matched normal
tissues. The overexpression of SRA inhibited the proliferation,
colony formation, migration, invasion and stemness of RCC cells.
Conversely, the knockdown of SRA markedly promoted cell growth and
the aggressive behaviors in vitro. These findings revealed
that SRA played a beneficial role in suppressing RCC proliferation
and cell motility.
RCC cells are shown to have an intermediate EMT
phenotype with low levels of desmoplakin and E-cadherin, and
mesenchymal features, such as fibronectin, N-cadherin, vimentin and
Snail (37,38). As was expected, the findings of
the present study demonstrated that SRA significantly increased the
expression levels of epithelial-specific markers and decreased the
expression levels of mesenchymal-associated regulators. The loss of
E-cadherin is considered to be a crucial event in EMT, whereas
N-cadherin promotes trans-endothelial migration, which subsequently
leads to a diminished in inter-cellular adhesion, and ultimately
triggers cancer invasion and migration. The EMT status renders make
cells more prone to exhibit cancer stem cell-like properties
(39). As regards the existence
of cancer stem cells, accumulating evidence has been indicated that
stem-like cells are contributed to cancer initiation and
progression (40). Similarly, the
present study demonstrated that SRA reduced the percentage of
CD44+/CD24− cells, which was associated with
increased stem-like characteristics and a poor prognosis for
patients with RCC. Consistently, SRA inhibited the self-renewal
capability and reduced the expression levels of SOX-2, CD44, ABCG2
and OCT-4, which play important roles in maintaining cancer cell
stemness (41,42). Herein, SRA promoted RCC
progression by regulating EMT markers, stemness properties and stem
cell-like protein expressions.
Considering the crucial roles of several signaling
pathways, such as ERK, JNK and STAT pathway in cancer progression
(43–45), the present study investigated the
alterations of the above-mentioned pathways in RCC cells. The
results revealed that SRA markedly activated the ERK signaling
pathway. However, the inhibition of the ERK pathway with PD98059
had minimal effects on proliferation, migration and invasion
inhibited by SRA. Therefore, the specific role of the ERK pathway
on proliferation and invasion inhibited by SRA warrant further
investigation. During RCC progression mediated by SRA, ERK
phosphorylation may lead to the proteasome-dependent degradation of
multiple phosphoproteins required for cell proliferation and
invasion. lncRNA sequences convey functions by binding to DNA or
protein. In a previous study, it was found that SRA induced the
upregulation of phospho-cAMP response element binding protein
(p-CREB) expression via the MEK-ERK signaling pathway in
vascular smooth muscle cell proliferation (46). Thus, CREB may be a direct
interacting molecule of SRA in the proliferation and stemness of
RCC cells. As a ubiquitous transcription factor, CREB carries out
its role by binding to the promoter containing a CRE motif. The
molecular mechanisms of SRA in regulating the promoter containing
CRE motif warrant further investigation.
In conclusion, the findings of the present study
demonstrated the inhibitory effects of SRA on RCC proliferation,
migration and invasion through the regulation of the cell cycle,
EMT and stemness characteristics, which are potentially independent
of the ERK signaling pathway (Fig.
7). Therefore, determining the protective functions of SRA may
provide a promising therapeutic target for the diagnosis and
therapy of RCC.
Funding
The present study was supported by grants from the
National Natural Sciences Foundation of China to LQ (nos. 81973668,
81774130, 81270359 and 81000946), the National Natural Sciences
Foundation of China to DL (no. 81670268), the National Science Fund
of Hunan Province for Distinguished Young Scholars to LQ (no.
2018JJ1018), the Open Funds of State Key Laboratory of Oncology in
South China to LQ (no. HN2019-07) and the First-Class Discipline of
Pharmaceutical Science of Hunan.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
LQ conceived and designed the study. CJZ, NZ and CL
analyzed the data and wrote the manuscript. YTY performed the
experiments. HTW provided the clinical samples. YS and DL
interpreted the data for the study. All authors read and approved
the final manuscript.
Ethics approval and consent to
participate
All procedures performed in studies involving human
participants were in accordance with the ethical standards of the
institutional and national research committee and with the 1964
Helsinki Declaration and its later amendments or comparable ethical
standards. The study was approved by the Committee for Ethical
Review of Hunan University of Chinese Medicine. Informed consent
was obtained prior to patient enrollment.
Patient consent for publication
Not applicable.
Competing interests
All authors declare that they have no competing
interests.
Acknowledgements
The authors would like to express their sincere
gratitude to Professor Chao-Nan Qian of the SYSUCC Cancer Center
for his support. The authors would also like to thank the SYSUCC
Cancer Center for providing kind assistance. In addition, the
authors are grateful to Dr Yajing Liu of the University of Michigan
for her kind revision of the article.
References
|
1
|
Koul H, Huh JS, Rove KO, Crompton L, Koul
S, Meacham RB and Kim FJ: Molecular aspects of renal cell
carcinoma: A review. Am J Cancer Res. 1:240–254. 2011.PubMed/NCBI
|
|
2
|
Ferlay J, Steliarova-Foucher E,
Lortet-Tieulent J, Rosso S, Coebergh JW, Comber H, Forman D and
Bray F: Cancer incidence and mortality patterns in Europe Estimates
for 40 countries in 2012. Eur J Cancer. 49:1374–1403. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2017. CA Cancer J Clin. 67:7–30. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Unverzagt S, Moldenhauer I, Nothacker M,
Roßmeißl D, Hadjinicolaou AV, Peinemann F, Greco F and Seliger B:
Immunotherapy for metastatic renal cell carcinoma. Cochrane
Database Syst Rev. 5:Cd0116732017.PubMed/NCBI
|
|
5
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2015. CA Cancer J Clin. 65:5–29. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ridge CA, Pua BB and Madoff DC:
Epidemiology and staging of renal cell carcinoma. Semin Intervent
Radiol. 31:3–8. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Murphy LC, Simon SL, Parkes A, Leygue E,
Dotzlaw H, Snell L, Troup S, Adeyinka A and Watson PH: Altered
expression of estrogen receptor coregulators during human breast
tumorigenesis. Cancer Res. 60:6266–6271. 2000.PubMed/NCBI
|
|
8
|
Liu C, Wu HT, Zhu N, Shi YN, Liu Z, Ao BX,
Liao DF, Zheng XL and Qin L: Steroid receptor RNA activator:
Biologic function and role in disease. Clin Chim Acta. 459:137–146.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Lanz RB, Chua SS, Barron N, Soder BM,
DeMayo F and O'Malley BW: Steroid receptor RNA activator stimulates
proliferation as well as apoptosis in vivo. Mol Cell Biol.
23:7163–7176. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Yan Y, Skliris GP, Penner C,
Chooniedass-Kothari S, Cooper C, Nugent Z, Blanchard A, Watson PH,
Myal Y, Murphy LC and Leygue E: Steroid Receptor RNA Activator
Protein (SRAP): A potential new prognostic marker for estrogen
receptor-positive/node-negative/younger breast cancer patients.
Breast Cancer Res. 11:R672009. View
Article : Google Scholar : PubMed/NCBI
|
|
11
|
Eoh KJ, Paek J, Kim SW, Kim HJ, Lee HY,
Lee SK and Kim YT: Long non-coding RNA, steroid receptor RNA
activator (SRA), induces tumor proliferation and invasion through
the NOTCH pathway in cervical cancer cell lines. Oncol Rep.
38:3481–3488. 2017.PubMed/NCBI
|
|
12
|
Bennett NC, Rajandram R, Ng KL and Gobe
GC: Evaluation of steroid hormones and their receptors in
development and progression of renal cell carcinoma. J Kidney
Cancer VHL. 1:17–25. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Langner C, Ratschek M, Rehak P, Schips L
and Zigeuner R: Steroid hormone receptor expression in renal cell
carcinoma: An immunohistochemical analysis of 182 tumors. J Urol.
171:611–614. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Czarnecka AM, Niedzwiedzka M, Porta C and
Szczylik C: Hormone signaling pathways as treatment targets in
renal cell cancer (Review). Int J Oncol. 48:2221–2235. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Qin L, Zhang X, Zhang L, Feng Y, Weng GX,
Li MZ, Kong QL, Qian CN, Zeng YX, Zeng MS, et al: Downregulation of
BMI-1 enhances 5-fluorouracil-induced apoptosis in nasopharyngeal
carcinoma cells. Biochem Biophys Res Commun. 371:531–535. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
17
|
Qin L, Yang YB, Yang YX, Gong YZ, Li XL,
Li GY, Luo HD, Xie XJ, Zheng XL and Liao DF: Inhibition of smooth
muscle cell proliferation by ezetimibe via the cyclin D1-MAPK
pathway. J Pharmacol Sci. 125:283–291. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Deng M, Zeng C, Lu X, He X, Zhang R, Qiu
Q, Zheng G, Jia X, Liu H and He Z: miR-218 suppresses gastric
cancer cell cycle progression through the CDK6/Cyclin D1/E2F1 axis
in a feedback loop. Cancer Lett. 403:175–185. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Darzynkiewicz Z, Zhao H, Zhang S, Lee MY,
Lee EY and Zhang Z: Initiation and termination of DNA replication
during S phase in relation to cyclins D1, E and A, p21WAF1, Cdt1
and the p12 subunit of DNA polymerase delta revealed in individual
cells by cytometry. Oncotarget. 6:11735–11750. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Shamanna RA, Hoque M, Pe'ery T and Mathews
MB: Induction of p53, p21 and apoptosis by silencing the NF90/NF45
complex in human papilloma virus-transformed cervical carcinoma
cells. Oncogene. 32:5176–5185. 2013. View Article : Google Scholar
|
|
21
|
Micalizzi DS, Haber DA and Maheswaran S:
Cancer metastasis through the prism of epithelial-to-mesenchymal
transition in circulating tumor cells. Mol Oncol. 11:770–780. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zhou P, Li B, Liu F, Zhang M, Wang Q, Liu
Y, Yao Y and Li D: The epithelial to mesenchymal transition (EMT)
and cancer stem cells: Implication for treatment resistance in
pancreatic cancer. Mol Cancer. 16:522017. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Thaper D, Vahid S, Nip KM, Moskalev I,
Shan X, Frees S, Roberts ME, Ketola K, Harder KW, Gregory-Evans C,
et al: Targeting Lyn regulates Snail family shuttling and inhibits
metastasis. . Oncogene. 36:3964–3975. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Yu CC, Lo WL, Chen YW, Huang PI, Hsu HS,
Tseng LM, Hung SC, Kao SY, Chang CJ and Chiou SH: Bmi-1 regulates
Snail expression and promotes metastasis ability in head and neck
squamous cancer-derived ALDH1 positive cells. J Oncol.
2011:6092592011. View Article : Google Scholar
|
|
25
|
Yuan ZX, Mo J, Zhao G, Shu G, Fu HL and
Zhao W: Targeting strategies for renal cell carcinoma: From renal
cancer cells to renal cancer stem cells. Front Pharmacol.
7:4232016. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Xiao W, Gao Z, Duan Y, Yuan W and Ke Y:
Notch signaling plays a crucial role in cancer stem-like cells
maintaining stemness and mediating chemotaxis in renal cell
carcinoma. J Exp Clin Cancer Res. 36:412017. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Kim Y, Yeon M and Jeoung D: DDX53
regulates cancer stem cell-like properties by binding to SOX-2. Mol
Cells. 40:322–330. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Hu J, Li J, Yue X, Wang J, Liu J, Sun L
and Kong D: Expression of the cancer stem cell markers ABCG2 and
OCT-4 in right-sided colon cancer predicts recurrence and poor
outcomes. Oncotarget. 8:28463–28470. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Wang MC, Jiao M, Wu T, Jing L, Cui J, Guo
H, Tian T, Ruan ZP, Wei YC, Jiang LL, et al: Polycomb complex
protein BMI-1 promotes invasion and metastasis of pancreatic cancer
stem cells by activating PI3K/AKT signaling, an ex vivo, in vitro,
and in vivo study. Oncotarget. 7:9586–9599. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Senbanjo LT and Chellaiah MA: CD44: A
multifunctional cell surface adhesion receptor is a regulator of
progression and metastasis of cancer cells. Front Cell Dev Biol.
5:182017. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Su D, Stamatakis L, Singer EA and
Srinivasan R: Renal cell carcinoma: Molecular biology and targeted
therapy. Curr Opin Oncol. 26:321–327. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Hanahan D and Weinberg RA: Hallmarks of
cancer: The next generation. Cell. 144:646–674. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Lanz RB, McKenna NJ, Onate SA, Albrecht U,
Wong J, Tsai SY, Tsai MJ and O'Malley BW: A steroid receptor
coactivator, SRA, functions as an RNA and is present in an SRC-1
complex. Cell. 97:17–27. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Cooper C, Guo J, Yan Y,
Chooniedass-Kothari S, Hube F, Hamedani MK, Murphy LC, Myal Y and
Leygue E: Increasing the relative expression of endogenous
non-coding Steroid Receptor RNA Activator (SRA) in human breast
cancer cells using modified oligonucleotides. Nucleic Acids Res.
37:4518–4531. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Lin K, Zhan H, Ma J, Xu K, Wu R, Zhou C
and Lin J: Silencing of SRA1 regulates ER expression and attenuates
the growth of stromal cells in ovarian endometriosis. Reprod Sci.
24:836–843. 2017. View Article : Google Scholar
|
|
36
|
Guo W, Jiang H, Li H, Li F, Yu Q, Liu Y,
Jiang W and Zhang M: LncRNA-SRA1 suppresses osteosarcoma cell
proliferation while promoting cell apoptosis. Technol Cancer Res
Treat. 18:15330338198414382019. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Son H and Moon A: Epithelial-mesenchymal
transition and cell invasion. Toxicol Res. 26:245–252. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Lee JY and Kong G: Roles and epigenetic
regulation of epithelial-mesenchymal transition and its
transcription factors in cancer initiation and progression. Cell
Mol Life Sci. 73:4643–4660. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Nieto MA, Huang RY, Jackson RA and Thiery
JP: EMT: 2016. Cell. 166:21–45. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Cheng B, Yang G, Jiang R, Cheng Y, Yang H,
Pei L and Qiu X: Cancer stem cell markers predict a poor prognosis
in renal cell carcinoma: A meta-analysis. Oncotarget.
7:65862–65875. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Sabnis NG, Miller A, Titus MA and Huss WJ:
The Efflux transporter ABCG2 maintains prostate stem cells. Mol
Cancer Res. 15:128–140. 2017. View Article : Google Scholar :
|
|
42
|
Qin L, Yin YT, Zheng FJ, Peng LX, Yang CF,
Bao YN, Liang YY, Li XJ, Xiang YQ, Sun R, et al: WNT5A promotes
stemness characteristics in nasopharyngeal carcinoma cells leading
to metastasis and tumorigenesis. Oncotarget. 6:10239–10252. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Chiu LY, Hsin IL, Yang TY, Sung WW, Chi
JY, Chang JT, Ko JL and Sheu GT: The ERK-ZEB1 pathway mediates
epithelial-mesenchymal transition in pemetrexed resistant lung
cancer cells with suppression by vinca alkaloids. Oncogene.
36:242–253. 2017. View Article : Google Scholar
|
|
44
|
Wang SY, Gao K, Deng DL, Cai JJ, Xiao ZY,
He LQ, Jiao HL, Ye YP, Yang RW, Li TT, et al: TLE4 promotes
colorectal cancer progression through activation of JNK/c-Jun
signaling pathway. Oncotarget. 7:2878–2888. 2016. View Article : Google Scholar :
|
|
45
|
Chuang CH, Greenside PG, Rogers ZN, Brady
JJ, Yang D, Ma RK, Caswell DR, Chiou SH, Winters AF, Grüner BM, et
al: Molecular definition of a metastatic lung cancer state reveals
a targetable CD109-Janus kinase-Stat axis. Nat Med. 23:291–300.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Zhang CJ, Liu C, Wang YX, Zhu N, Hu ZY,
Liao DF and Qin L: Long non-coding RNA-SRA promotes neointimal
hyperplasia and vascular smooth muscle cells proliferation via
MEK-ERK-CREB pathway. Vascul Pharmacol. 116:16–23. 2019. View Article : Google Scholar : PubMed/NCBI
|