Introduction
Trophoblasts are a specific type of placental cells,
that play important roles in embryo implantation and in the
formation of the maternal-fetal interface (1). After the blastocyst implants into
the endometrium, trophoblasts differentiate into extravillous and
chorionic trophoblasts. Extravillous trophoblasts play a key role
in invading the decidual stroma and blood vessels of the uterus
(2). There are two types of
chorionic trophoblasts: Syncytiotrophoblasts in the outer layer and
cytotrophoblasts in the inner layer (3). Syncytiotrophoblasts are located on
the surface of the chorion and directly participate in the material
exchange between the mother and fetus (4). Cytotrophoblasts, which are located
in the inner layer, have a greater proliferative ability and under
certain conditions, can differentiate into extravillous
trophoblasts and syncytiotrophoblasts. The dynamic balance between
trophoblast proliferation and apoptosis is critical for maintaining
pregnancy (5), and the disruption
of this balance can lead to complications, such as preeclampsia or
abortion. Studies in China and abroad have demonstrated that the
trophoblast apoptotic index is significantly higher in cases of
spontaneous abortion (SA) than in normal pregnancies (6-9).
Fibronectin 1 (FN1) is a high molecular weight
glycoprotein that has numerous biological functions (10). FN1 is present in the extracellular
matrix and plays key roles in cell adhesion, growth, migration and
differentiation, and is involved in maintaining cell morphology
(11). It is also a major blood
protein, and functions as a crucial non-specific modulin in tissue
repair, and promotes the survival of neurons in ischemic brains.
FN1 binds to cells in the salivary gland and regulates their
molecular structure by binding to collagen, fibrinogen and fibrin
(12-14). In addition, FN1 exhibits
chemo-tactic activity and attracts monocytes, and is involves in
both blood coagulation and wound healing (11). FN1-knockout mice have an embryonic
lethal phenotype that is character-ized by severe cardiac and
vascular disorders (15),
indicating that FN1 is essential for embryonic development. As FN1
is highly expressed in tumor vessels and mediates angiogenesis
during tumorigenesis, it also plays important roles in cancer
progression. It has been reported that FN1 is highly expressed in
breast cancer (16),
nasopharyngeal carcinoma (17),
oral squamous cell carcinoma (11) and thyroid carcinoma (18), and plays a regulatory role in the
major inflammatory cells present in the tumor microenvironment. In
addition, FN1 expression is significantly upregulated in uterine
leiomyoma and is associated with trophoblast invasion, uterine
proliferation and adhesion function (19). However, the effects of FN1 on
trophoblasts and the underlying molecular mechanisms remain
unknown.
Materials and methods
Data collection
The gene expression profiles of the GSE127170
dataset were obtained from the GEO database (https://www.ncbi.nlm.nih.gov/). The GSE127170 dataset
includes data from 12 samples from the Agilent GPL10739 platform
(Affymetrix Human Genome U133 Plus 2.0 Array). It includes
replicates (n=3) for the following 3 groups: Untreated JEG-3 cells
vs. forkolin-treated JEG-3 cells (gene set-1), untreated BeWo cells
vs. forkolin-treated BeWo cells (gene set-2), and untreated JEG-3
and BeWo cells vs. forkolin-treated JEG-3 and BeWo cells (gene
set-3).
Screening results of differentially
expressed genes (DEGs)
The series matrix file dataset of GSE127170 was
downloaded, and the gene probes of the platform were subsequently
transformed to gene names by referencing the GPL10739 platform.
Both the normalization of the data and the screening of the DEGs
were performed using the 'limma' package in R language (version
3.5.1). To identify DEGs, the screening criteria were set to filter
genes with a fold change (FC) value of (|log2FC|) >1 and a
P-value of <0.05.
Construction of the PPI network and
identification of hub genes
STRING is a public database that contains
interactions between known and predicted proteins (https://string-db.org/). PPI is essential for
examining protein function since it is helpful for elucidating the
regulatory functions among proteins. The top 300 DEGs of the
GSE127170 dataset (gene set1-3) were uploaded onto the STRING
website to obtain the interrelations between proteins and to set
the minimum required interaction score to 0.15 to visualize the
interaction networks with Cytoscape (version 3.7.2). cytoHubba, the
plug-in of Cytoscape, was used to screen the hub genes based on the
'Degree' and 'MCC' method.
GO and KEGG pathway analyses for the
identification of overlapping DEGs
The overlapping DEGs from gene set1, gene set2 and
gene set3 were obtained using the Draw Venn Diagram (http://bioinformatics.psb.ugent.be/webtools/Venn/)
online tool. The Metascape website (http://metascape.org/gp/index.html#/main/step1) was
used to perform GO enrichment analysis. KEGG pathway database
(https://www.kegg.jp/kegg/pathway.html) was used to
perform KEGG pathway analysis. P-values of <0.05 were considered
to indicate statistically significant differences.
Chorionic villus specimens
Chorionic villus tissue specimens from 65 patients
diagnosed with SA (mean age, 26.48±3.91 years) and 65 patients with
induced abortion (IA; mean age, 27.41±2.86 years) (termed normal
intrauterine early pregnancy) were obtained between January, 2015
and December, 2018 at The First Affiliated Hospital of Soochow
University. The clinical information pertaining to the patients
diagnosed with SA is presented in Table I. Each patient was examined by two
pathologists and chorionic villus tissue was confirmed based on the
histopathological assessment. Following retrieval, each tissue
specimen was snap-frozen in liquid nitrogen immediately and
preserved at -80°C until further experimentation. The study
protocol was approved by the Ethics Committee of the First
Affiliated Hospital of Soochow University. A written informed
consent was obtained for each sample, which was then analyzed
anonymously. The present study was performed in accordance with the
Declaration of Helsinki.
 | Table IAssociation between FN1 expression
and clinical characteristics of patients diagnosed with SA. |
Table I
Association between FN1 expression
and clinical characteristics of patients diagnosed with SA.
| FN1 expression
| P-value |
|---|
| Cases (n=65) | High (n=29) | Low (n=36) |
|---|
| Age (years) | | | | 0.314 |
| ≥22 and
<30 | 16 | 9 | 7 | |
| ≥30 and
<35 | 20 | 10 | 10 | |
| ≥35 | 29 | 10 | 19 | |
| Pregnancy cycle
(weeks) | | | | 0.718 |
| ≤6 | 33 | 14 | 19 | |
| >6 and ≤10 | 32 | 15 | 17 | |
Inclusion and exclusion criteria
The inclusion criteria in the present study were as
follows: i) A positive result for urine human chorionic
gonadotropin (HCG); ii) B-ultrasound exhibited an embryonic
heartbeat; iii) patients did not take hormonal drugs prior to
surgery. The exclusion criteria were as follows: i) Abnormal
pregnancy; ii) intrauterine device; iii) genitalia infection and
uterine malformation.
Animal models of SP
For the purposes of the experiment, 8- to
10-week-old C57BL/6 male mice, C57BL/6 female mice and FN1 gene
knockout (FN1−/−) C57B/6L female mice were purchased
from Shanghai Slac Laboratory Animal Co., Ltd. The mice were housed
in an environment controlled for temperature (22-24°C) and
conditions of light (12 h light and 12 h darkness), with free
access to standard mouse food and water. A total of 2 female mice
were caged with 1 male mouse every other day, and vaginal plugs
were examined the following morning as a sign of mating behavior.
The day of plug detection was defined as embryonic day (E) 0.5. The
pregnant mice were randomly divided into the normal pregnancy mouse
group (NP), the SA mouse group (SA), the normal pregnancy
FN1−/− mouse group (FN1−/−) and the SA
FN1−/− mouse group (FN1−/−_SA), with 6 mice
in each group. Lipopolysaccharide (LPS; Sigma-Aldrich; Merck KGaA)
at a dose of 2.5 µg dissolved in saline was
intraperitoneally injected into the pregnant C57BL/6 female mice or
FN1−/− C57BL/6 female mice at E 7.5 to induce abortion,
as previously described (20).
Mice in the control group received the same volume of saline as the
vehicle. At E10.5, the mice were sacrificed, and the embryo
resorption rate were calculated by dividing the number of resorbed
embryos by the number of implantations, and the decidua were
collected for use in further experiments. All the experiments
involving animals in present study were carried out in accordance
with the protocols of the Guidelines for the Care and Use of
Laboratory Animals published by the United States National
Institutes of Health (NIH Publication, revised 2011) and the
Guidelines for the Care and Use of Laboratory Animals of the
Chinese Animal Welfare Committee. The procedures were approved by
the Animal Use Committees of Soochow University.
Immunohistochemistry (IHC)
For human, IHC staining was performed for chorionic
villus tissue specimen from IA group and SA group. For mice, IHC
staining was performed for chorionic villus tissue specimen from
the NP group, SA group, FN1−/− group and
FN1−/−_SA group. Each section of chorionic villus tissue
specimen from the patients was subjected to overnight incubation
with one of the following primary antibodies, including anti-FN1
(1:500 dilution; ab2413, Abcam), and each section of chorionic
villus tissue specimen form mice was subjected to overnight
incubation with one of the following primary antibodies, including
anti-cleaved caspase-3 (1:500 dilution; ab2302, Abcam), at
4°C. Each section was then incubated in PBS and washed with
PBS thrice at 37°C. Subsequently, each section was incubated
with goat anti-rabbit immunoglobulin G secondary antibody (1:1,000;
cat. no. BS10043; Bioworld Technology, Inc.) at 37°C for 60
min. The Olympus BX51 light microscope (Olympus Corporation;
magnification, x100) was used to examine the sections following IHC
staining. All specimens were then assigned with scores based on the
cytoplasmic staining intensity (with 0 indicating no staining; 1
indicating weak staining, 2 suggesting moderate staining, and 3
representing strong staining), and the extent of stained cells (0
indicating 0%, 1 suggesting 1-24%, 2 representing 25-49%, 3 was
indicative of 50-74%, and 4 being indicative of 75-100%). Moreover,
the intensity score was multiplied with the score of the stained
cell extent to determine the eventual immunoreactive score, which
ranged from 0 (minimal) to 12 (maximal). In addition, 0, 1-6, and
≥8 points were defined as negative, weak positive and strong
positive expression, separately. Each experiment was carried out in
triplicate.
Histopathological evaluation
The chorionic villus tissues form patients or mice
were immersed in normal 10% neutral buffered formalin. The sections
(5-µm-thick) were cut after paraffin embedding and stained
with hematoxylin and eosin (H&E) at 37°C for 2 min
according to a previous description (21). You can analysis the pathological
changes under a light microscope.
Cells and cell culture
The human choriocarcinoma cell lines, JEG-3 (cat.
no. HTB-36, ATCC) and BeWo (cat. no. CCL-98, ATCC) were cultured in
Dulbecco's modified Eagle's medium (DMEM), 10% (v/v) fetal bovine
serum (FBS), 100 µg/ml penicillin, 100 µg/ml
streptomycin (Invitrogen; Thermo Fisher Scientific, Inc.) at
37°C in a humidified atmosphere with 5% CO2. The
cells were harvested and seeded in 6-well plates at
2×105 cells/well.
Cell transfection
The siRNA sequences targeting FN1 were synthesized
from Shanghai Gene-Pharma Co. Nonsense siRNA was used as the
negative control. The siRNA-FN1 sequence was as follows: 5′-TAC GAA
TCC CCA GGC CCC GGG CCC G-3′. siRNA- FN1 (40 nm) was transfected
into the JEG-3 and BeWo cells using Lipofectamine 2000 (Invitrogen;
Thermo Fisher Scientific, Inc.) to reduce the mRNA and protein
levels of FN1. The FN1 cDNA sequence was cloned into the pcDNA3.1
vector (Shanghai enzyme research Co., Ltd.) to increase the mRNA
and protein levels of FN1. Cells were cultured to 80% confluence in
6-well plates, then either pcDNA3.1 vector (3.0 µg per
1×104 cells/well) or pcDNA3.1-FN1 were transfected into
the cells using Lipofectamine 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.) according to the manufacturer's instructions.
After transfection for 48 h, the mRNA and protein levels of FN1
were detected by RT-qPCR and western blot analysis.
Grouping
JEG-3 or BeWo cells transfected with nonsense siRNA
were identified as the siNC group; siRNA-FN1-transfected JEG-3 or
BeWo cells were identified as the siFN1 group; pcDNA3.1
vector-transfected JEG-3 or BeWo cells were identified as the NC
group; pcDNA3.1-FN1-transfected JEG-3 or BeWo cells were identified
as the FN1 group. LY294002 (PI3K/Akt signaling pathway inhibitor)
was purchased from MedChem Express and insulin-like growth factor-1
(IGF-1; PI3K/Akt signaling pathway activator) was purchased from
PeproTech, Inc. After the JEG-3 cells were pre-treated with
LY294002 (5 µM) for 4 h, pcDNA3.1-FN1 were then transfected
into the JEG-3 cells (FN1 + LY294002). After the BeWo cells were
pre-treated with IGF-1 (5 µM) for 4 h, siRNA-FN1 was then
transfected into the BeWo cells (siFN1 + IGF-1).
Cell Counting kit 8 (CCK-8) assay
The JEG-3 or BeWo cells in the logarithmic phase
(8,000 cells/well) were trypsinized and added into 96-well plates
for a 24-h pre-incubation at 37°C. Following incubation for
4 h at 37°C, CCK8 solution (10 µl) was injected to
each well. The cells at 0 and 48 h were harvested using the CCK-8
kit (cat. no. C0037, Beyotime Institute of Biotechnology) according
to the manufacturer's protocol. The absorption at different stages
was recorded at 450 nm using a microplate reader (Thermo Fisher
Scientific, Inc.).
Cell apoptosis assay
Cells in the different groups were cultured in a
6-well plate at a density of 2×104 cells/well for 24 h.
The cells were then centrifuged, collected and washed with PBS
twice. The supernatant was discarded, and the cells were
resuspended in 400 µl of 1X Binding Buffer and incubated
with 5 µl of Annexin V-FITC for 15 min in the dark. Cells
were mixed thoroughly with 10 µl of PI staining solution
(CA1020, Beijing Solarbio Science & Technology Co., Ltd.) and
incubated for 5 min in the dark. The proportion of cell apoptosis
was assessed using a flow cytometer (CytoFLEX, Beckman Coulter,
Inc.).
RNA extraction and reverse
transcription-quantitative PCR (RT-qPCR) assays
Total RNA was isolated from the
pcDNA3.1-FN1-transfected JEG-3 and BeWo cells and
siRNA-FN1-transfected JEG-3 and BeWo cells using TRIzol reagent
(Invitrogen; Thermo Fisher Scientific, Inc.) following the
manufacturer's instructions. Total RNA was quantified using a
NanoDrop ND-100 Spectrophotometer (NanoDrop Technologies; Thermo
Fisher Scientific, Inc.) at 260 nm. Total RNA (2 µg) was
reverse transcribed into cDNA using SuperScript III (Invitrogen;
Thermo Fisher Scientific, Inc.) according to the manufacturer's
instructions. qPCR was performed using Fast SYBR Green Master Mix
(Applied Biosystems) with an ABI PRISM7900 Sequence Detection
System. The primers used for amplification were as follows: FN1
sense, 5′-TGG TAT TCA GCT TCC TGG CA-3′ and antisense, 5′-CGG GTA
TGG TCT TGG CCT AT-3′; and GAPDH sense, 5′-ACC CAG AAG ACT GTG GAT
GG-3′ and antisense, 5′-TCA GCT CAG GGA TGA CCT TG-3′. qPCR
reaction included an initial denaturation step at 95°C for
10 min, followed by 40 cycles at 95°C for 15 sec and
60°C for 60 sec, and at 75°C to 95°C; the
temperature rises at 1°C per 20 sec. The relative mRNA
expression was normalized to GAPDH, which was calculated based on
the Cq value according to the equation: 2−ΔΔCq (22). Each sample was analyzed in
triplicate.
Western blot analysis
Total proteins were extracted from the cells in the
different groups using RIPA lysis buffer (cat. no. P0013B; Beyotime
Institute of Biotechnology), according to the manufacturer's
instructions. The protein concentration was measured using a BCA
kit (Beyotime). A total of 10 µg each sample was separated
on a 10% SDS-PAGE, transferred to PVDF membranes (EMD Millipore)
and then immunoblotted with antibodies against FN1 (1:800 dilution;
ab2413, Abcam), cleaved caspase-3 (1:1,000 dilution; ab2302,
Abcam), Bax (1:500 dilution; ab32503, Abcam), Bcl-2 (1:800
dilution; ab32124, Abcam), proliferating cell nuclear antigen
(PCNA; 1:1,000 dilution; ab92552, Abcam), Ki67 (1:1,000 dilution;
ab92742, Abcam), phosphorylated (p)-PI3K (1:500, abs130868, Absin
Bioscience Inc.), PI3K (1:1,000, abs119725, Absin Bioscience,
Inc.), p-Akt (1:800 dilution; ab38449, Abcam), Akt (1:800 dilution;
ab8805, Abcam) and GAPDH (1:2,000 dilution; ab181602, Abcam),
followed by incubation with HRP goat anti-rabbit IgG (AS014;
ABclonal). As for FN1, cleaved caspase-3, Bax, Bcl-2, PCNA and Ki67
the dilution of the HRP goat anti-rabbit IgG antibody was 1:2,000;
as for GAPDH, the dilution was 1:10,000. The primary antibodies
FN1, cleaved caspase-3, Bax, Bcl-2, PCNA, Ki67 and GAPDH were
incubated with the membranes at 4°C overnight. The secondary
antibody HRP goat anti-rabbit IgG was incubated with the membranes
at 37°C for 2 h. The results were visualized using the Image
Lab Imaging System software v4.0 (Bio-Rad Laboratories, Inc.).
Statistical analysis
The mean value of FN1 expression in the SA group of
>2.43 was defined as a high expression. The associations of FN1
expression with the clinicopathological features of the patients
were examined using the χ2 test. Each experiment was
repeated at least 3 times. The statistical analyses of the
experimental data were performed using a two-tailed Student's
paired t-test and one-way ANOVA followed by Tukey's post hoc test.
Statistical significance was assessed at least three independent
experiments and a P-value <0.05 was considered to indicate a
statistically significant difference.
Results
DEGs and hub genes identified in gene
set1 of the GSE127170 dataset
The GSE127170 dataset was downloaded from the GEO
database. The analysis of the dataset revealed 903 differentially
expressed genes (DEGs) (410 P<0.05 and |logFC|>1), including
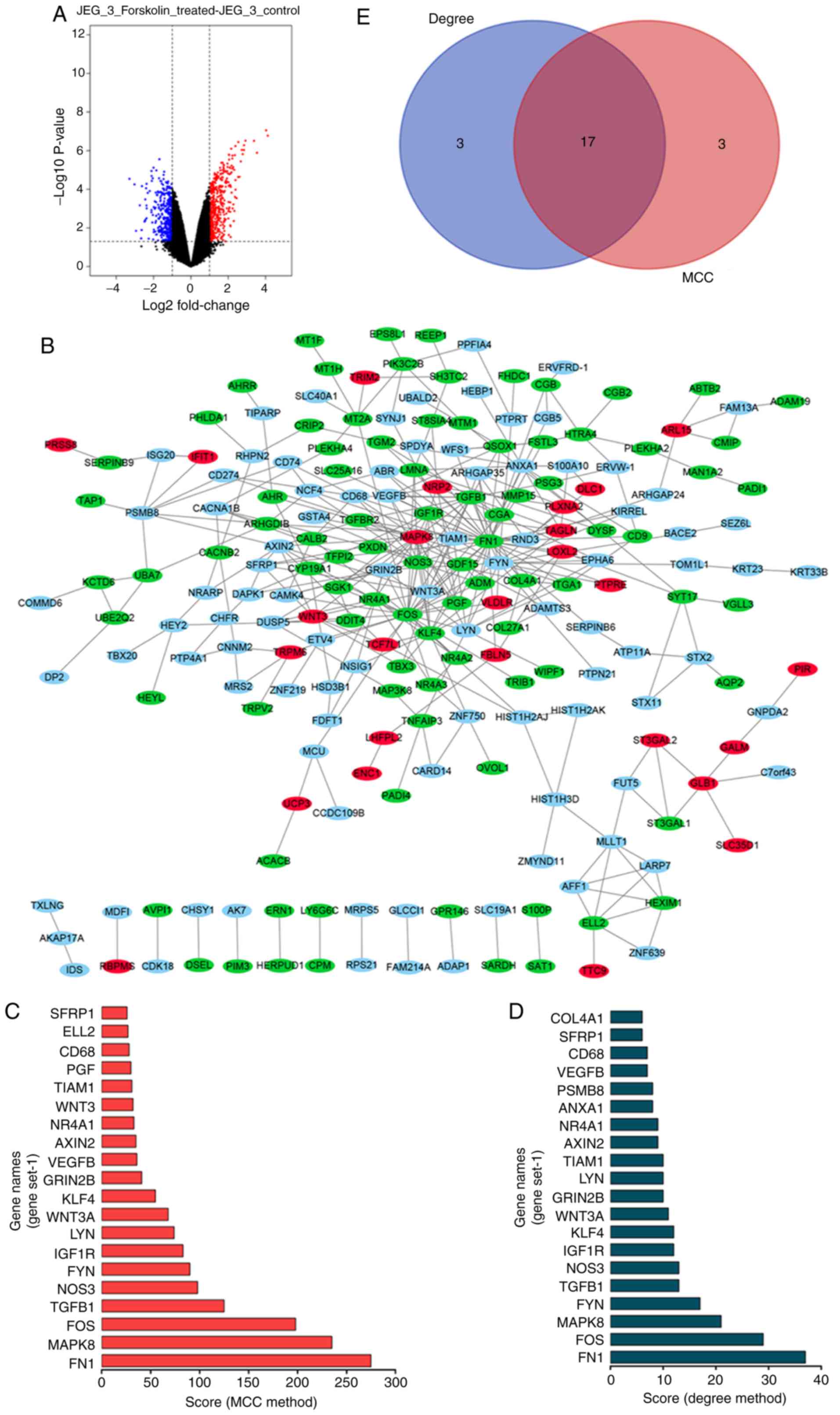
493 upregulated and 410 downregulated genes (Fig. 1A). The top 300 DEGs were used to
construct a PPI network using the STRING website, which was
visualized using Cytoscape (Fig.
1B). The top 20 hub genes were identified by the MCC and Degree
method using the cytoHubba plug-in of Cytoscape (Fig. 1C and D). As shown in Fig. 1E, a total of 17 overlapping hub
genes were identified: LYN proto-oncogene, scr family tyrosine
kinase (LYN), Wnt family member 3A (WNT3A), Fos proto-oncogene,
AP-1 transcription factor subunit (FOS), transforming growth factor
beta 1 (TGFB1), mitogen-activated protein kinase 8 (MAPK8), CD68
molecule (CD68), insulin like growth factor 1 receptor (IGF1R),
glutamate ionotropic receptor nmDA type subunit 2B (GRIN2B),
nuclear receptor subfamily 4 group A member 1 (NR4A1), vascular
endothelial growth factor B (VEGFB), Kruppel like factor 4 (KLF4),
TIAM Rac1 associated GEF 1 (TIAM1), nitric oxide synthase 3 (NOS3),
FYN proto-oncogene, Src family tyrosine kinase (FYN), secreted
frizzled related protein 1 (SFRP1), AXIN2 and FN1.
DEGs and hub genes identified in gene
set2 of the GSE127170 dataset
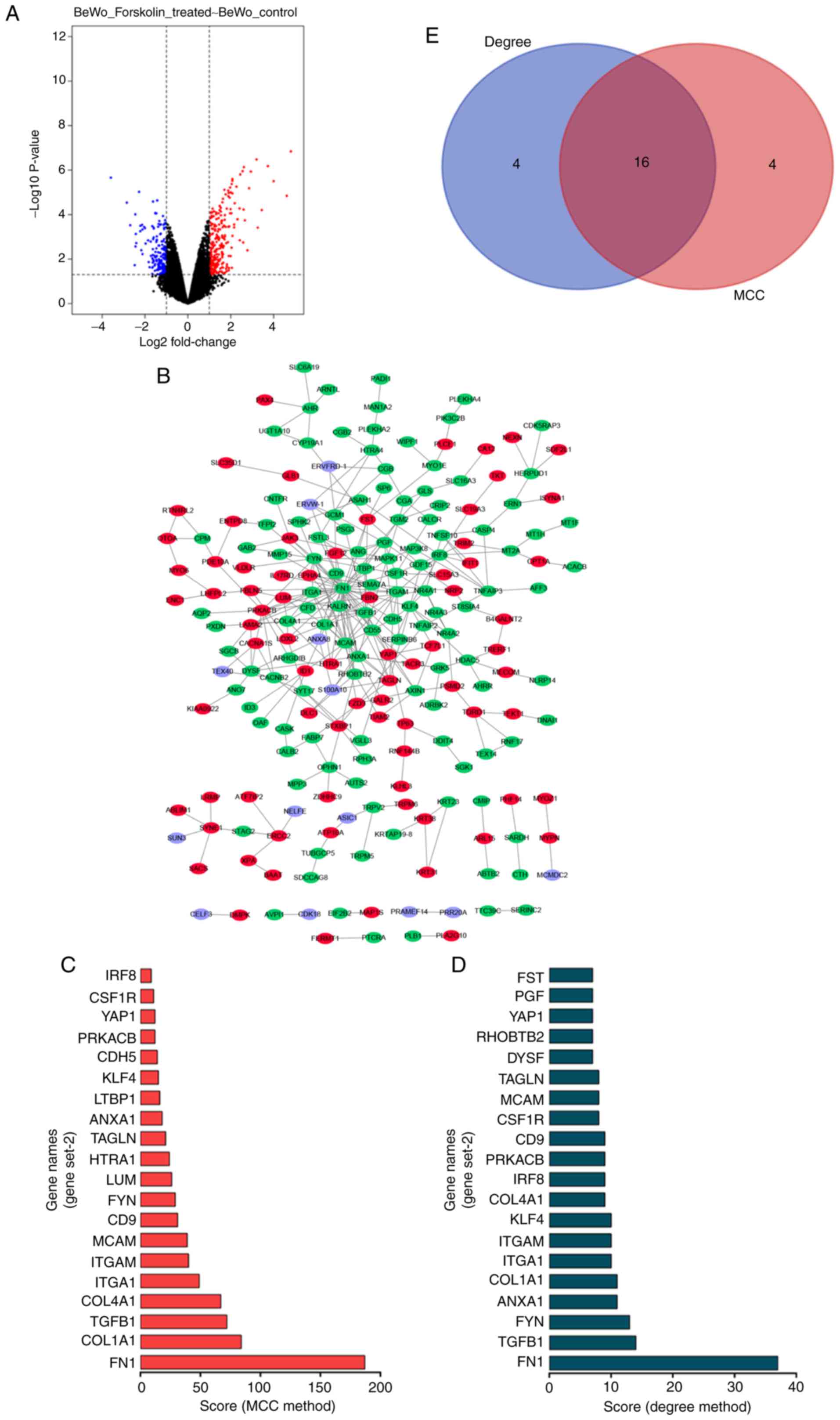
The GSE127170 dataset was downloaded from the GEO
database. The analysis of the dataset revealed 440 differential
genes (DEGs), including 260 upregulated and 180 downregulated genes
(Fig. 2A). The top 300 DEGs were
used to construct a PPI network using the STRING website, which was
visualized using Cytoscape (Fig.
2B). The top 20 hub genes were identified by the MCC and Degree
methods using the cytoHubba plug-in of Cytoscape (Fig. 2C and D). As shown in Fig. 2E, a total of 16 overlapping hub
genes were identified: Yes associated protein 1 (YAP1), integrin
subunit alpha 1 (ITGA1), colony stimulating factor 1 receptor
(CSF1R), CD9 molecule (CD9), melanoma cell adhesion molecule
(MCAM), integrin subunit alpha M (ITGAM), collagen type I alpha 1
chain (COL1A1), TGFB1, protein kinase cAMP-activated catalytic
subunit beta (PRKACB), interferon regulatory factor 8 (IRF8),
annexin A1 (ANXA1), transgelin (TAGLN), KLF4, FYN, collagen type IV
alpha 1 chain (COL4A1) and FN1.
DEGs and hub genes identified in gene
set3 of the GSE127170 dataset
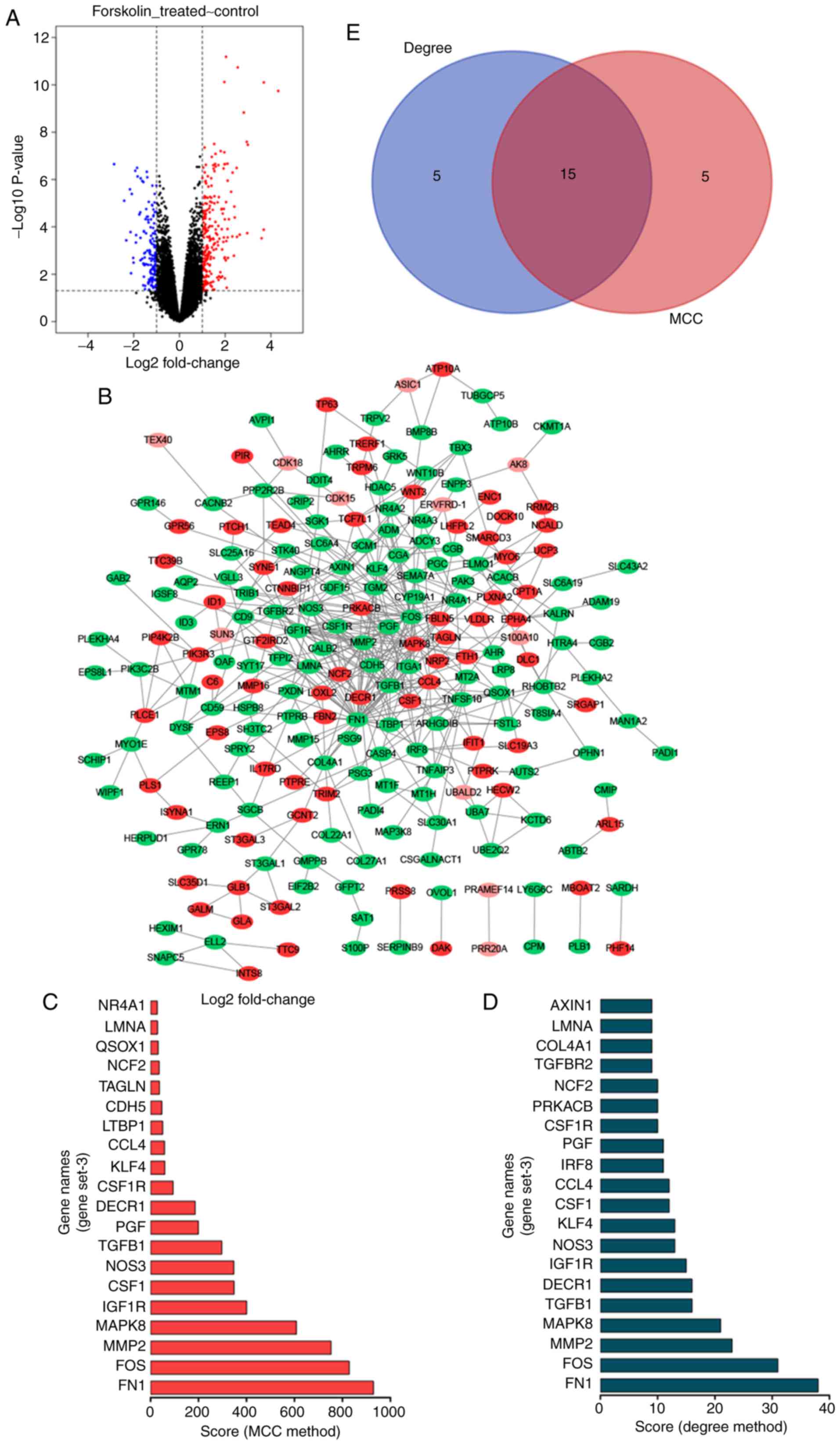
The GSE127170 dataset was downloaded from the GEO
database. The analysis of the dataset revealed 388 differentially
expressed genes (DEGs), including 238 upregulated and 150
downregulated genes (Fig. 3A).
The top 300 DEGs were used to construct a PPI network using the
STRING website, which was visualized in Cytoscape (Fig. 3B). The top 20 hub genes were
identified by the MCC and Degree methods using the cytoHubba
plug-in of Cytoscape (Fig. 3C and
D). As shown in Fig. 3E, a
total of 15 overlapping hub genes were identified: Placental growth
factor (PGF), colony stimulating factor 1 receptor (CSF1R), matrix
metallopeptidase 2 (MMP2), amin A/C (LMNA), FOS, TGFB1, C-C motif
chemokine ligand 4 (CCL4), MAPK8, IGF1R, colony stimulating factor
1 (CSF1), KLF4, nitric oxide synthase 3 (NOS3), neutrophil
cytosolic factor 2 (NCF2), 2,4-dienoyl-CoA reductase 1 (DECR1) and
FN1.
GO and KEGG enrichment analysis
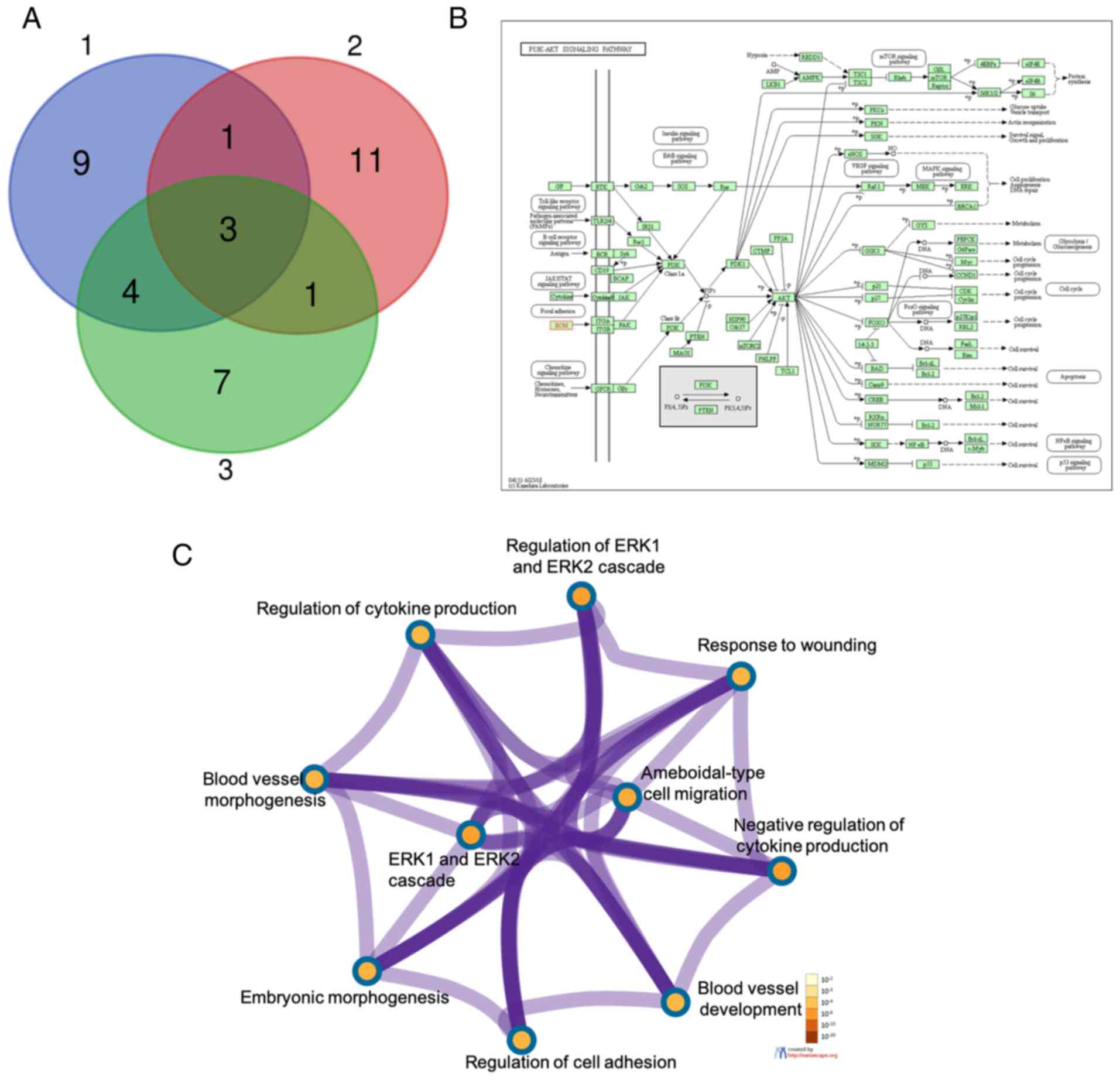
TGFB1, FN1 and KLF4 were overlapping hub genes from
gene set1, gene set2 and gene set3 (Fig. 4A). FN1 was mainly enriched in the
PI3K/Akt signaling pathway (Fig.
4B). TGFB1, FN1 and KLF4 were mainly enriched in the biological
process (BP) term, including the regulation of the ERK1 and ERK2
cascade, the response to wounding, ameboidal-type cell migration,
the negative regulation of cytokine production, blood vessel
development, the regulation of cell adhesion, embryonic
morphogenesis, the ERK1 and ERE2 cascade, blood vessel
morphogenesis and the regulation of cytokine production using the
Metascape website (Fig. 4C).
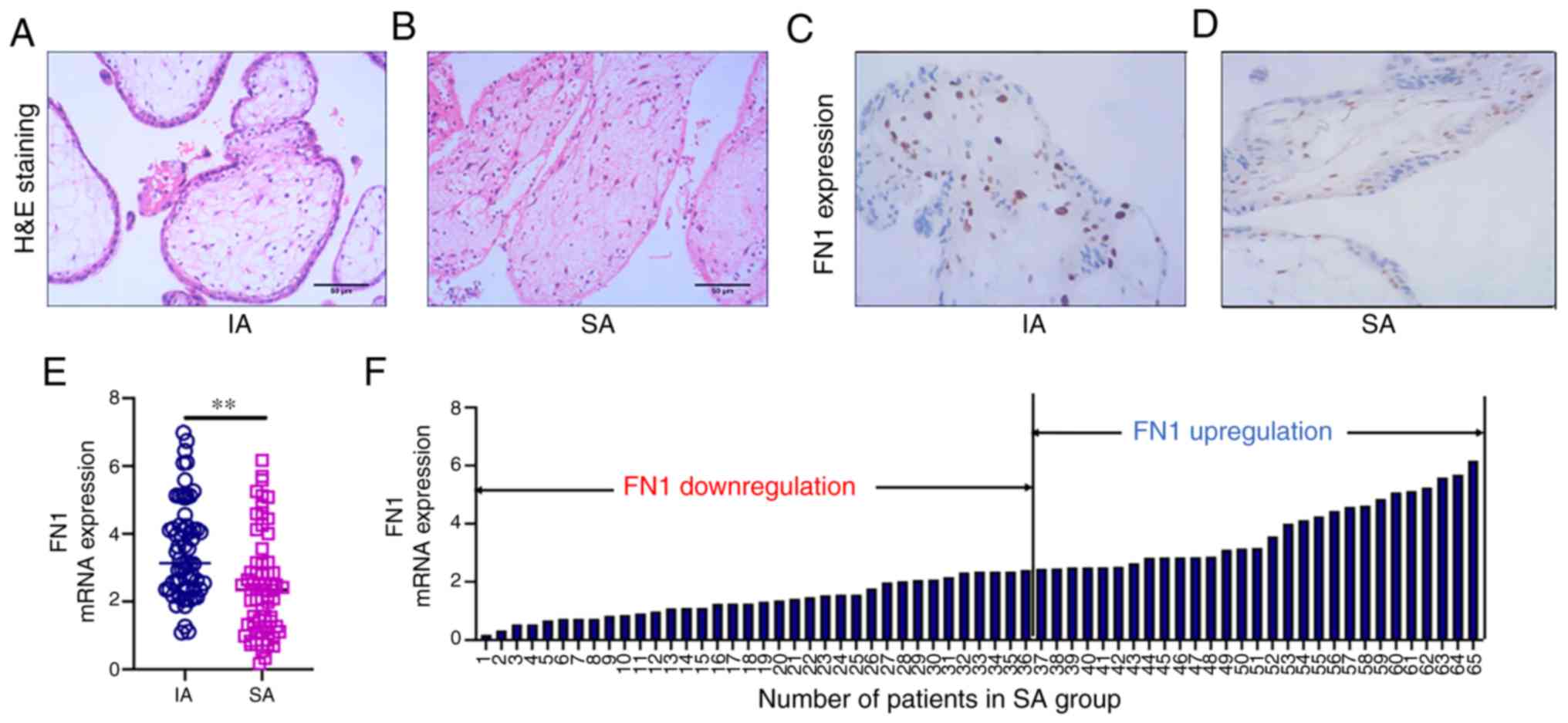
FN1 expression is downregulated in
chorionic villus tissues from the placentas of patients diagnosed
with SA
No change was observed in the morphology of the
chorionic villus tissues of patients in the IA group. The
cytotrophoblasts in the inner layer were cubic, polygonal, or oval,
with clear boundaries and a transparent cytoplasm. The outer
syncytiotrophoblasts contained multiple nuclei. Stellate
mesenchymal cells, myxoid matrix and round or oval cells were
observed in the chorionic villus tissue and there were no blood
vessels in the chorionic villus tissue (Fig. 5A). By contrast, the morphology of
chorionic villus tissues in the SA group were significantly
altered. Microscopic examination revealed different degrees of
degenerative changes, proliferation and degeneration in the
chorionic villus tissues. The number of cytotrophoblasts and
syncytiotrophoblasts were significantly decreased or were absent.
In addition, mesenchymal celluloid degeneration, structural
destruction, homogeneous shaped cells, matrix fiber-like
degeneration, and myxoid degeneration were observed. The basement
membrane of the villous trophoblast cells and blood capillaries was
slightly thickened (Fig. 5B). IHC
revealed FN1 staining in the chorionic villi and surrounding
tissues in the IA group (Fig.
5C). By contrast, the chorionic villi and surrounding tissues
in the SA group exhibited only weak FN1 staining (Fig. 5D). The FN1 mRNA levels in the sera
of patients in the SA group were significantly lower than that
those in the IA group (Fig. 5E).
FN1 expression was <2.43 in 36 patients in the SA group
(Fig. 5F). The analysis of the
association between FN1 expression and the patient characteristics
revealed that FN1 was not associated with age (P=0.314) or
pregnancy cycle (P=0.718; Table
I).
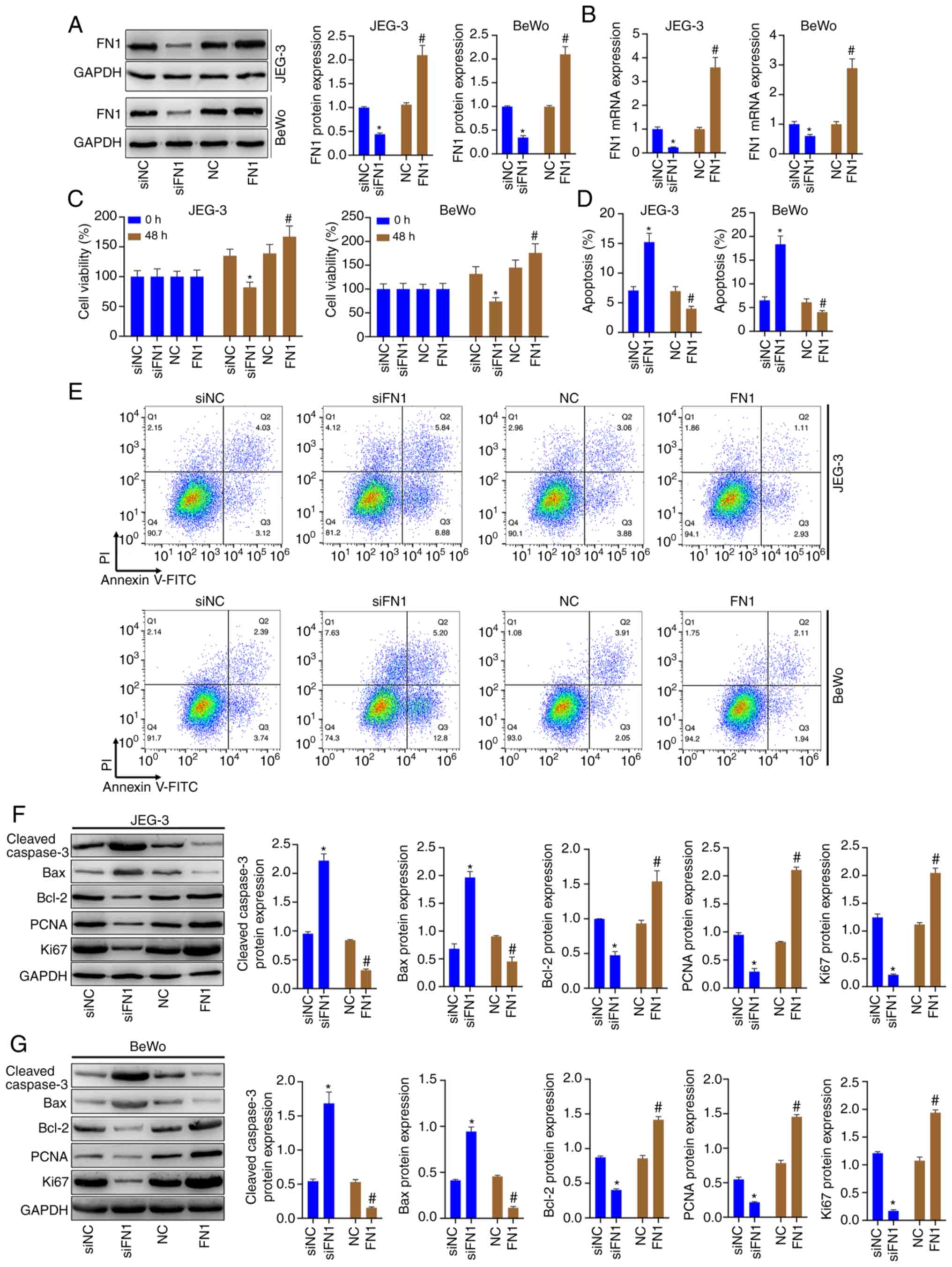
FN1 regulates the viability and apoptosis
of JEG-3 and BeWo cells by modulating the expression of
proliferation- and apoptosis-related proteins
JEG-3 and BeWo cells were transfected with siRNA-FN1
or pcDNA-FN1, and western blot analysis and RT-qPCR were then
conducted to assess the FN1 levels. The results revealed that FN1
protein and mRNA levels were significantly decreased in the
siFN1-transfected cells and were significantly increased in
pcDNA-FN1-transfected cells (Fig. 6A
and B). The results of CCK-8 assay revealed that the knockdown
of FN1 prominently decreased the viability of the JEG-3 and BeWo
cells, whereas the overexpression of FN1 notably increased cell
viability (Fig. 6C). The results
also revealed that FN1 knockdown promoted cell apoptosis, whereas
FN1 overexpression inhibited the apoptosis of JEG-3 and BeWo cells
(Fig. D and E). FN1 silencing
resulted in increased protein levels of cleaved caspase-3 and Bax,
and decreased protein levels of Bcl-2. It also resulted in
significantly decreased protein levels of PCNA and Ki67 in the
JEG-3 and BeWo cells. However, FN1 overexpression markedly
decreased cleaved caspase-3 and Bax protein levels, and increased
Bcl-2, PCNA and Ki67 protein levels in the JEG-3 and BeWo cells, as
shown by western blot analysis (Fig.
6F and G).
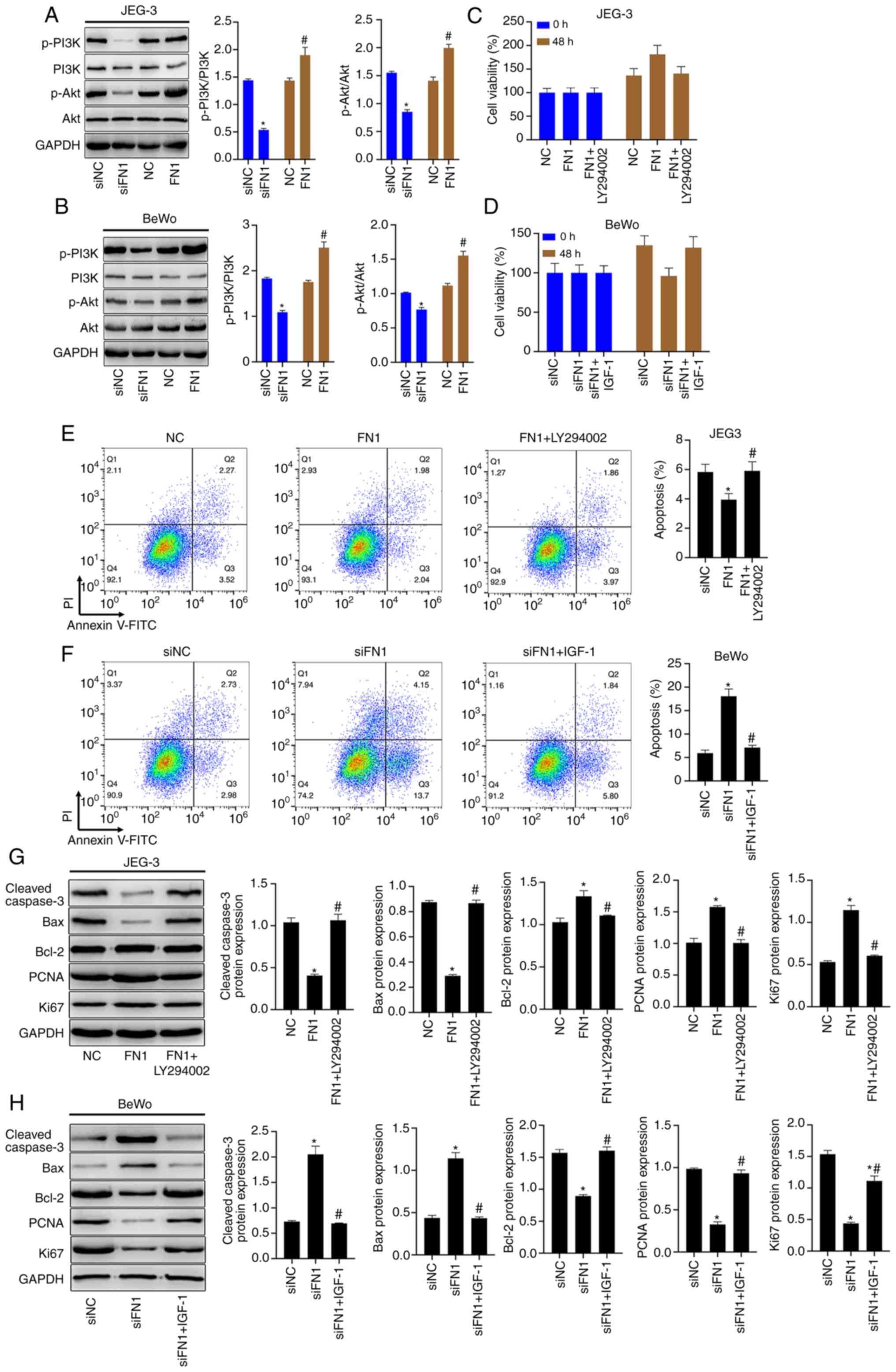
FN1 regulates the viability and the
apoptosis of JEG-3 and BeWo cells by affecting the activation of
the PI3K/Akt signaling pathway
The PI3K and Akt phosphorylation levels were
significantly decreased by FN1 silencing and were notably increased
by FN1 overexpression in the JEG-3 and BeWo cells (Fig. 7A and B). The results of CCK-8
assay revealed that the promoting effects of FN1 overexpression on
the viability of the JEG-3 cells were blocked by the PI3K/Akt
signaling pathway inhibitor, LY294002 (Fig. 7C), and the inhibitory effects of
FN1 silencing on BeWo cell viability were reversed by the PI3K/Akt
signaling pathway activator, IGF-1 (Fig. 7D). LY294002 significantly promoted
the apoptosis of FN1-overexpressing JEG-3 cells (Fig. 7E), and IGF-1 significantly blocked
the induction of apoptosis of the FN1-silenced BeWo cells (Fig. 7F). In addition, the FN1
overexpression-mediated decrease in the levels of cleaved caspase-3
and Bax, and the increase in the protein levels of Bcl-2, PCNA and
Ki67 in the JEG-3 cells were reversed by LY294002 (Fig. 7G). The positive effects of FN1
inhibition on cleaved caspase-3 and Bax protein levels, and the
negative effects on Bcl-2, PCNA and Ki67 protein levels in the BeWo
cells were reversed by IGF-1 (Fig.
7H).
FN1 knockout increases the apoptosis of
trophoblasts in chorionic villus tissues from mice subjected to
SA
The histo-pathological changes in the chorionic
villus tissues of mice in the NP, FN1−/−, SA, and
FN1−/−_SA groups were observed by H&E staining. The
organization of the chorionic villus tissues of mice in the NP
group were normal, and the cell arrangement was tight. However,
there were more histopathological changes in the chorionic villus
tissues of mice in the FN1−/−group than in those of the
NP group, but less than in those of the SA and FN1−/−-SA
groups (Fig. 8A). In addition, a
morphological examination revealed that the pathological changes in
the chorionic villus tissues of mice in the FN1−/−-SA
group were greater than the changes observed in the SA group. A
comparison of the FN1 expression levels in the chorionic villus
tissues from the mice in these groups revealed a gradual decrease
across the NP, SA, FN1−/−, and FN1−/−_SA
groups (Fig. 8B). SA or FN1
knockout alone increased the levels of cleaved caspase-3 in
chorionic villus tissues, and FN1 knockout further increased the
cleaved caspase-3 levels in the chorionic villus tissues of mice
subjected to SA (Fig. 8C).
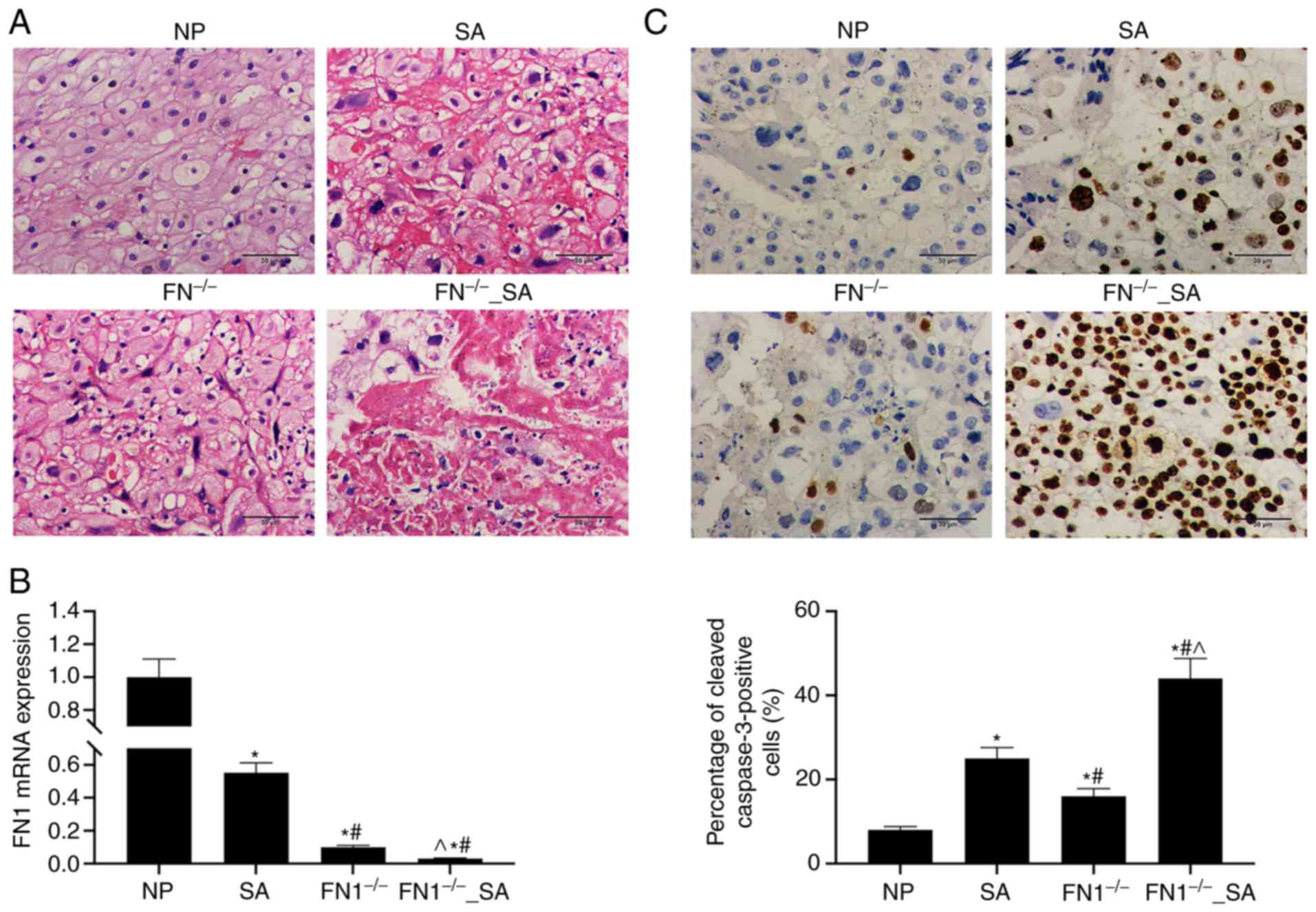 | Figure 8FN1 knockdown aggravates the
apoptosis of trophoblasts in the chorionic villus tissues obtained
from mice subjected to SA. (A) H&E staining was used to detect
histopathological changes in the chorionic villus tissues of mice
in the NP, SA, FN1−/−, and FN1−/−_SA groups.
(B) RT-qPCR was used to determine the mRNA level of FN1 in
chorionic villus tissues of mice in the NP, SA, FN1−/−
and FN1−/−_SA groups. (C) IHC assay was used to detect
FN1 expression in chorionic villus tissues of mice in the NP, SA,
FN1−/− and FN1−/−_SA groups. GAPDH was used
as a loading control. Data are presented as the means ± standard
deviation. *P<0.05 vs. NP group;
#P<0.05 vs. SA group; and ^P<0.05 vs.
FN1−/− group. FN1, fibronectin 1; SA, spontaneous
abortion; NP, normal pregnancy. |
Discussion
Pregnancy is a complex biological process. The
successful implantation of the embryo is closely related to the
development of the blastocyst, the growth of trophoblasts, and the
connection between the maternal-fetal interface and immune
regulation (23). Trophoblasts
are a specific type of placental cells that play important roles in
embryo implantation and the formation of the maternal-fetal
interface (1). Apoptosis is a
necessary physiological process for uterine decidual
reconstruction, trophoblast invasion of the luteus and maintaining
the immune tolerance balance at the maternal-fetal interface
(5). Animal studies have
indicated that compared with normal pregnancy CBA/JxBALB/c model
mice, the SA CBA/JxDBA/2 model mice exhibit greater apoptosis in
the villi and decidual tissues, and the apoptotic index is
significantly higher than that of normal pregnancy model mice
(24,25). The excessive apoptosis of
trophoblasts leads to intracellular dysfunction and abortion. The
present bioinformatics analysis of the GSE127170 dataset revealed
that TGFB1, FN1 and KLF4 are hub genes.
It has been reported that the disruption of the
binding between TGFB1 and its receptor, TGFBR, can affect TGFB1
signal transduction, thus blocking nitric oxide synthase activity,
leading to a decrease in nitric oxide synthesis, an increase in
blood pressure and thrombosis, maternal hypoxia, an insufficient
nutrient supply, and neovascularization (26-28). It has also been reported that FN1
is highly expressed in chorionic leukocytes and is related to the
invasion, proliferation and adhesion of trophoblasts in the uterus
(29,30). In addition, FN1 was the top hub
gene in 3 PPI networks generated using the MCC and Degree methods.
KLF4 is considered to be a tumor suppressor gene, and its
overexpression can promote the apoptosis of endometrial cancer
cells (31). GO and KEGG analysis
revealed that FN1 mainly affects the PI3K/Akt signaling pathway and
is involved in cell proliferation and apoptosis (Fig. 4). Therefore, the effects of FN1 on
trophoblasts has become the primary focus of our research
group.
The results of the present study revealed that the
morphology of the chorionic villi in the SA group differed
significantly from that in the IA group. FN1 mRNA and protein
levels were significantly higher in the chorionic villus and
surrounding tissues in the IA group than in the SA group. In
addition, the FN1 mRNA level was not significantly associated with
the age of the pregnancy cycle of patients with SA. This may be due
to the insufficient number of cases and/or the similarity of
patient statuses (Fig. 5 and
Table I). The present study
examined the effect of FN1 on the proliferation and apoptosis of
trophoblasts by CCK-8 and flow cytometric assays. The results
revealed that the silencing of FN1 significantly decreased cell
viability and promoted cell apoptosis. By contrast, FN1
overexpression significantly increased cell viability and inhibited
apoptosis (Fig. 5). Caspase-3,
Bax and Bcl-2 are critical apoptosis-related genes (32), and studies have demonstrated that
Bcl-2 and Bax not only act as upstream regulatory factors of
caspase-3, but also function as direct substrates of caspase-3. The
present experimental results revealed that FN1 silencing
significantly upregulated the protein levels of cleaved caspase-3,
Bax and Bcl-2, which was consistent with the observed effect of FN1
silencing on cell apoptosis. PCNA and Ki67 reflects the
proliferative activity of cells. FN1 silencing significantly
reduced the protein levels of PCNA and Ki67, which was consistent
with the observed inhibition of cell viability induced by FN1
silencing and the opposite effect of FN1 overexpression (Fig. 6). These results suggest that FN1
regulates the proliferation and apoptosis of trophoblasts.
The PI3K/Akt pathway is an important pathway in the
regulation of cell proliferation and apoptosis (33). Akt is a serine/threonine kinase
and an important target of PI3K. Activated Akt activates or
inhibits its downstream target proteins, such as Bcl-2 and
caspase-3, to regulate cell proliferation and apoptosis (34). The results of the present study
confirmed that in FN1-silenced cells, the apoptotic levels
increased, and the PI3K and Akt phosphorylation levels
significantly decreased. By contrast, in FN1-overexpressing cells,
the apoptotic levels decreased, and PI3K and Akt phosphorylation
levels significantly increased. This suggests that an activated
PI3K/Akt signaling pathway exerts a protective effect on
trophoblasts. Further analyses revealed that the effects of FN1
silencing on cell apoptosis were reversed by a PI3K/Akt signaling
pathway agonist, while the effect of FN1 overexpression on cell
apoptosis was reversed by a PI3K/Akt signal pathway inhibitor
(Fig. 7). These results suggest
that FN1 inhibits trophoblast apoptosis by activating the PI3K/Akt
signaling pathway. The results of the animal experiment revealed
that FN1 knockdown damaged the organization of the chorionic villus
tissues in NP mice and induced cleaved caspase-3 (Fig. 8). More importantly, FN1 knockdown
increased the SA-induced apoptosis of trophoblasts and exacerbated
chorionic villus tissue injury. Thus, FN1 downregulation was
detected in patients with SA and promoted the SA-induced apoptosis
of trophoblasts. In addition, FN1 knockdown induced trophoblast
cell apoptosis in NP mice. These results revealed that FN1 plays an
important role in maintaining the dynamic balance of proliferation
and apoptosis of trophoblasts during pregnancy. However, it is
worth noting that FN1 has been reported to be upregulated in
cervical cancer tissues and to promote cell proliferation (35). Therefore, the results of the
present study only suggest that the overexpression of FN1 may be
helpful for inhibiting the SA-induced apoptosis of trophoblasts,
but do not indicate that the overexpression of FN1 would be helpful
in other diseases.
In conclusion, the present study demonstrated that
FN1 expression was significantly downregulated in the chorionic
villus tissues of patients diagnosed with and mice subjected to SA.
The silencing of FN1 promoted trophoblast apoptosis and inhibited
cell viability; the overexpression of FN1 inhibited trophoblast
apoptosis and promoted cell viability. The activation of the
PI3K/Akt signaling pathway protected the trophoblasts. The
overexpression of FN1 inhibited apoptosis and promoted cell
viability by activating the PI3K/Akt signaling pathway.
Funding
The present study was supported by the scientific
research project youth project of Nantong's Health and family
planning commission (no. QA2019005).
Availability of data and materials
The GSE127170 dataset was used to perform the
present study (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE127170).
All data generated and/or analyzed during the present study are
included in this article.
Authors' contributions
JJ and LC wrote the main manuscript and analyzed the
data. JJ, LC, YZ and YH performed all the experiments and collected
the data. WT and FX conceived and designed the study. All authors
read and approved the final manuscript and agree to be accountable
for all aspects of the research in ensuring that the accuracy or
integrity of any part of the work are appropriately investigated
and resolved.
Ethics approval and consent to
participate
The study protocol was approved by the ethics
committee of the First Affiliated Hospital of Soochow University. A
written informed consent was obtained for each sample, which was
then analyzed anonymously. This study was performed in accordance
with the Declaration of Helsinki. All of the experiments involving
animals in this study were carried out in accordance with the
protocols of the Guidelines for the Care and Use of Laboratory
Animals published by the United States National Institutes of
Health (NIH Publication, revised 2011) and the Guidelines for the
Care and Use of Laboratory Animals of the Chinese Animal Welfare
Committee. The procedures were approved by the Animal Use
Committees of Soochow University.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgements
Not applicable.
References
|
1
|
Moser G, Windsperger K, Pollheimer J, de
Sousa Lopes SC and Huppertz B: Human trophoblast invasion: New and
unexpected routes and functions. Histochem Cell Biol. 150:361–370.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Takahashi H, Ogoyama M, Nagayama S, Suzuki
H, Ohkuchi A, Matsubara S and Takizawa T: Extravillous trophoblast
invasion accelerated by WNT3A, 5A, and 10B via CD44. J Matern Fetal
Neonatal Med. 1–9. 2019.PubMed/NCBI
|
|
3
|
Walentin K, Hinze C and Schmidt-Ott KM:
The basal chorionic trophoblast cell layer: An emerging coordinator
of placenta development. Bioessays. 38:254–265. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Ohgaki R, Ohmori T, Hara S, Nakagomi S,
Kanai-Azuma M, Kaneda-Nakashima K, Okuda S, Nagamori S and Kanai Y:
Essential roles of L-Type amino acid transporter 1 in
syncytiotrophoblast development by presenting fusogenic 4F2hc. Mol
Cell Biol. 37:e00427–16. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Gu Y, Zhao S, Wan J, Meng J, Zuo C, Wang
S, Zhou Y, Li H and Wang X: Hsa-miRNA-125b may induce apoptosis of
HTR8/SVneo cells by targeting MCL1. Reprod Biol. 19:368–373. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ding J, Yin T, Yan N, Cheng Y and Yang J:
FasL on decidual macrophages mediates trophoblast apoptosis: A
potential cause of recurrent miscarriage. Int J Mol Med.
43:2376–2386. 2019.PubMed/NCBI
|
|
7
|
Liu HN, Tang XM, Wang XQ, Gao J, Li N,
Wang YY and Xia HF: MiR-93 inhibits trophoblast cell proliferation
and promotes cell apoptosis by targeting BCL2L2 in recurrent
spontaneous abortion. Reprod Sci. 27:152–162. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Tian S, Yu J, Zhang Y, Bian Y, Ma J and
Yan J: Overexpression of PTEN regulated by miR-19b and miR-494 in
the villous of recurrent spontaneous abortion patients. J Reprod
Immunol. 140:1031332020. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Yang D, Ding J, Wang Y, Yuan M, Xian S,
Zhang L, Liu S, Dai D, Wang F, Zheng Y, et al: YY1-PVT1 affects
trophoblast invasion and adhesion by regulating mTOR
pathway-mediated autophagy. J Cell Physiol. 235:6637–6646. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Xu TP, Huang MD, Xia R, Liu XX, Sun M, Yin
L, Chen WM, Han L, Zhang EB, Kong R, et al: Decreased expression of
the long non-coding RNA FENDRR is associated with poor prognosis in
gastric cancer and FENDRR regulates gastric cancer cell metastasis
by affecting fibronectin1 expression. J Hematol Oncol. 7:632014.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Chen Z, Tao Q, Qiao B and Zhang L:
Silencing of LINC01116 suppresses the development of oral squamous
cell carcinoma by up-regulating microRNA-136 to inhibit FN1. Cancer
Manag Res. 11:6043–6059. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Dong Z, Lin C, Liu Y, Jin H, Wu H, Li Z,
Sun L, Zhang L, Hu X, Wei Y, et al: Upregulation of sestrins
protect atriums against oxidative damage and fibrosis in human and
experimental atrial fibrillation. Sci Rep. 7:463072017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Satoh A, Niwano S, Niwano H, Kishihara J,
Aoyama Y, Oikawa J, Fukaya H, Tamaki H and Ako J: Aliskiren
suppresses atrial electrical and structural remodeling in a canine
model of atrial fibrillation. Heart Vessels. 32:90–100. 2017.
View Article : Google Scholar
|
|
14
|
Tian L, Lu ZP, Cai BB, Zhao LT, Qian D, Xu
QC, Wu PF, Zhu Y, Zhang JJ, Du Q, et al: Activation of pancreatic
stellate cells involves an EMT-like process. Int J Oncol.
48:783–792. 2016. View Article : Google Scholar
|
|
15
|
Wang Y, Fu Y, Yan Z, Zhang XB and Pei M:
Impact of fibronectin knockout on proliferation and differentiation
of human infrapatellar fat pad-derived stem cells. Front Bioeng
Biotechnol. 7:3212019. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Hou J, Li L, Zhu H, Chen H, Wei N, Dai M,
Ni Q and Guo X: Association between breast cancer cell migration
and radiosensitivity in vitro. Oncol Lett. 18:6877–6884.
2019.PubMed/NCBI
|
|
17
|
Gao R, Feng Q and Tan G: microRNA-613
exerts anti-angiogenic effect on nasopharyngeal carcinoma cells
through inactivating the AKT signaling pathway by down-regulating
FN1. Biosci Rep. 39:BSR201821962019. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zhang S, Wang Q, Han Q, Han H and Lu P:
Identification and analysis of genes associated with papillary
thyroid carcinoma by bioinformatics methods. Biosci Rep.
39:BSR201900832019. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Litovkin KV, Ivanova OV, Bauer A, Hoheisel
JD, Bubnov VV and Zaporozhan VN: Microarray study of gene
expression in uterine leiomyoma. Exp Oncol. 30:106–111.
2008.PubMed/NCBI
|
|
20
|
Mu Y, Zhou DN, Yan NN, Ding JL and Yang J:
Upregulation of ADAMTS7 and downregulation of COMP are associated
with spontaneous abortion. Mol Med Rep. 19:2620–2626.
2019.PubMed/NCBI
|
|
21
|
Lu X, Cui J, Cui L, Luo Q, Cao Q, Yuan W
and Zhang H: The effects of human umbilical cord-derived
mesenchymal stem cell transplantation on endometrial receptivity
are associated with Th1/Th2 balance change and uNK cell expression
of uterine in autoimmune premature ovarian failure mice. Stem Cell
Res Ther. 10:2142019. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
23
|
Zipori Y, Haas J, Berger H and Barzilay E:
Multifetal pregnancy reduction of triplets to twins compared with
non-reduced triplets: A meta-analysis. Reprod Biomed Online.
35:296–304. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Lorek D, Kedzierska AE, Slawek A and
Chelmonska-Soyta A: Expression of Toll-like receptors and
costimulatory molecules in splenic B cells in a normal and
abortion-prone murine pregnancy model. Am J Reprod Immunol.
82:e131482019. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Xu WM, Xiao ZN, Wang XB and Huang Y: IL-17
induces fetal loss in a CBA/JxBALB/c mouse model, and an anti-IL-17
antibody prevents fetal loss in a CBA/JxDBA/2 mouse model. Am J
Reprod Immunol. 75:51–58. 2016. View Article : Google Scholar
|
|
26
|
Ciaramella V, Sasso FC, Di Liello R, Corte
CM, Barra G, Viscardi G, Esposito G, Sparano F, Troiani T,
Martinelli E, et al: Activity and molecular targets of pioglitazone
via blockade of proliferation, invasiveness and bioenergetics in
human NSCLC. J Exp Clin Cancer Res. 38:1782019. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Moon JR, Oh SJ, Lee CK, Chi SG and Kim HJ:
TGF-β1 protects colon tumor cells from apoptosis through XAF1
suppression. Int J Oncol. 54:2117–2126. 2019.PubMed/NCBI
|
|
28
|
Tang RZ, Gu SS, Chen XT, He LJ, Wang KP
and Liu XQ: Immobilized transforming growth factor-Beta 1 in a
stiffness-tunable artificial extracellular matrix enhances
mechanotransduction in the epithelial mesenchymal transition of
hepatocellular carcinoma. ACS Appl Mater Interfaces.
11:14660–14671. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Cox J, Malik M, Britten J, Lewis T and
Catherino WH: Ulipristal acetate and extracellular matrix
production in human leiomyomas in vivo: A laboratory analysis of a
randomized placebo controlled trial. Reprod Sci. 25:198–206. 2018.
View Article : Google Scholar :
|
|
30
|
Graubner FR, Boos A, Aslan S, Kucukaslan I
and Kowalewski MP: Uterine and placental distribution of selected
extracellular matrix (ECM) components in the dog. Reproduction.
155:403–421. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Szostek-Mioduchowska AZ, Lukasik K,
Skarzynski DJ and Okuda K: Effect of transforming growth factor -β1
on alpha-smooth muscle actin and collagen expression in equine
endometrial fibroblasts. Theriogenology. 124:9–17. 2019. View Article : Google Scholar
|
|
32
|
Hao G, Zhai J, Jiang H, Zhang Y, Wu M, Qiu
Y, Fan C, Yu L, Bai S, Sun L and Yang Z: Acetylshikonin induces
apoptosis of human leukemia cell line K562 by inducing S phase cell
cycle arrest, modulating ROS accumulation, depleting Bcr-Abl and
blocking NF-κB signaling. Biomed Pharmacother. 122:1096772019.
View Article : Google Scholar
|
|
33
|
Zhang C, Liang R, Gan X, Yang X, Chen L
and Jian J: MicroRNA-384-5p/Beclin-1 as potential indicators for
epigallocatechin gallate against cardiomyocytes ischemia
reperfusion injury by inhibiting autophagy via PI3K/Akt pathway.
Drug Des Devel Ther. 13:3607–3623. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Lv CL, Zhang T, Yan TZ, Yi GK and Gao K:
MicroRNA-448 inhibits the regeneration of spinal cord injury
through PI3K/AKT/Bcl-2 axis. Eur Rev Med Pharmacol Sci.
23:2719–2726. 2019.PubMed/NCBI
|
|
35
|
Wang S, Gao B, Yang H, Liu X, Wu X and
Wang W: MicroRNA-432 is downregulated in cervical cancer and
directly targets FN1 to inhibit cell proliferation and invasion.
Oncol Lett. 18:1475–1482. 2019.PubMed/NCBI
|