Introduction
Ovarian cancer (OVA) is a common malignancy of the
female reproductive system, and is associated with the highest
mortality rate of all gynecological malignant diseases (1). Although advancements have been made
in surgical treatments and chemotherapy, the 5-year survival of
patients with OVA is only approximately 30% due to frequent
recurrence (2,3). Thus, a main objective for the
treatment of OVA is the identification of novel prognostic
biomarkers, which can be used to distinguish patients at a high
risk of relapse and to detect biomarkers that are candidate
therapeutic targets. Previous studies have suggested that circular
RNAs (circRNAs) play an important role in the progression and
development of a number of types of cancer, and have promising
therapeutic and prognostic value (4,5).
However, the molecular mechanisms of action of circRNAs in OVAs
remain largely unclear.
circRNAs belong to a family of regulatory non-coding
RNA (ncRNA) molecules that consist of covalently-closed and
continuous loop structures without a 5′ cap and 3′ polyA tail
(6). The majority of circRNAs are
produced from a precursor mRNA backsplicing (7). The involvement of circRNAs in cancer
pathology has been extensively investigated. It has been reported
that circRNAs negatively modulate microRNA (miRNA or miR)
expression by interacting with binding sites, which subsequently
affects the downstream levels of mRNAs (6). A major circRNA function is to
function as a miRNA sponge (8-10).
The dysregulation of the circRNA/miRNA/mRNA axis in signaling
pathways plays a role in various types of cancer, including OVA
(11-13). For example, circPLEKHM3 functions
as a tumor suppressor via the regulation of the
miR-9/DNAJB6/BRCA1/KLF4/AKT1 axis in OVA (14). circRNA1656 has been shown to be
downregulated in high-grade serous OVA tissues and cell lines, and
has been shown to be a novel biomarker (15). circRNA UBAP2 has been shown to
promote OVA progression by sponging miR-144 (16).
The present study investigated the functional role
of hsa_circ_0026123 and found that hsa_circ_0026123 expression was
upregulated in OVA tissues and cell lines; however,
hsa_circ_0026123 silencing suppressed cell proliferation and
migration, and decreased the expression of markers associated with
cancer stem cell (CSC) differentiation via the regulation of the
miR-124-3p/enhancer of zeste homolog 2 (EZH2) signaling pathway.
These results may provide a viable reference for the clinical
diagnosis and treatment of OVA.
Materials and methods
Animal ethics statement
The present study used 12 BALB/c female nude mice (4
weeks old; weighing 15-20 g; SLARC). The mice were housed in a
temperature-controlled (25°C) room under a 12-h light/dark cycle to
mimic the normal physiological the day/night cycle. Standard chow
and water were freely available to the animals following
sterilization. The Ethics Committee of Shanghai Tongji Hospital,
Tongji University, Shanghai, China approved all animal
experiments.
Tissue specimens
OVA tissues (n=20) and healthy fallopian tube tissue
(n=20) were obtained (from patients aged 48-74 years) undergoing
surgery (healthy fallopian tube tissues were from the unilateral
removal of fallopian tubes) at the Shanghai Tongji Hospital from
January, 2019 to July, 2019 were sequentially analyzed. The Ethics
Committee of Shanghai Tongji Hospital approved the present study.
Informed consent was obtained from each patient prior to the
analyses of the tissues. All cases were confirmed by post-operative
pathological diagnosis. Patients who had received neoadjuvant
chemotherapy or radiation therapy prior to surgery were excluded
from the present study. A total of 20 high-grade ovarian cancer
tissue samples (labeled A1-3) and 20 healthy fallopian tube tissue
samples (labeled B1-3) were collected.
Cells, cell culture and transfection
Human OVA cell lines (A2780, TOV112D, SKOV3, and
OVCAR-3) and the normal ovarian cell line (ISOE80) (from the
American Type Culture Collection) were cultured in Dulbecco's
modified Eagle's medium (DMEM; Gibco; Thermo Fisher Scientific,
Inc.) consisting of 10% fetal bovine serum (FBS; Gibco; Thermo
Fisher Scientific, Inc.) and penicillin in a humidified incubator
with 5% CO2 at 37°C. Small interfering RNAs (siRNAs) for
hsa_circ_0026123 (si-circ0026123; 5′-AAG GAG AGG AAT ACT AAT TAT
CC-3′) (Shanghai GenePharma Co., Ltd.), miR-124-3p mimics (5′-UAA
GGC ACG CGG UGA AUG CC-3′) (Shanghai GenePharma Co., Ltd.),
miR-124-3p inhibitors (5′-CGU GUU CAC AGC GGA CCU UGA U-3′), EZH2
overexpression vector (the sequence was inserted into the pcDNA3.1
vector) (Shanghai GenePharma Co., Ltd.), and their negative
controls (Shanghai GenePharma Co., Ltd.) were transfected into
cultured SKOV3 cells using Lipofectamine 2000 (Invitrogen; Thermo
Fisher Scientific, Inc.) with 50 nM according to standard
procedures. Following transfection for 48 h, SKOV3 cells were
collected for use in the experiments. To further validate the
effects of hsa_circ_0026123 in the in vivo experiments,
lentiviral stabilized SKOV3 cells (lenti-viral vector:
pCDH-CMV-MCS-EF1-copGFP; Novagen) in which hsa_circ_0026123 was
silenced were constructed. GFP detection was performed at 72 h
following infection, and when the green fluorescence was >95%,
the transfection was considered successful.
Bioinformatics analysis
The circRNA/miRNA target genes were predicted using
Interactome (https://circinteractome.nia.nih.gov/). The interactive
association between miR-124-3 and EZH2 was predicted using the
Targetscan (http://starbase.sysu.edu.cn/).
Reverse transcription-quantitative PCR
(RT-qPCR)
Total RNA was extracted from tissues and cells using
TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.). The
RNA concentration was measured using an ultraviolet
spectrophotometer (Hitachi Corporation). Subsequently, the
extracted total RNA was reverse transcribed into cDNA using a
Reverse Transcription lit (Takara Biotechnology, Inc.). The
thermo-cycling conditions were as follows: 30 sec at 95°C, 5 sec
for 40 cycles at 95°C, and 35 sec at 60°C. SYBR-Green (Thermo
Fisher Scientific, Inc.) was used for qPCR detection. Relative
expression was calculated using the 2−ΔΔCq method
(17). GAPDH and U6 were utilized
as the internal references. The experiments were repeated 3 times.
The primer sequences were as follows: hsa_circ_0026123 forward,
5′-CAT CAT ATC TCA AAG TAA AGT C-3′ and reverse, 5′-CCA AGA AGC CCT
GAA GAC CG-3′; miR-124-3p forward, 5′-ACA CTC CAG CTG GGT AAG GCA
CGC GGT GAA-3′ and reverse, 5′-CTC AAC TGG TGT CGT GGA GTC GGC AAT
TCA GTT GAG GGC ATT CAC-3′; GAPDH forward, 5′-TGT TCG TCA TGG GTG
TGA AC-3′ and reverse, 5′-ATG GCA TGG ACT GTG GTC AT-3′; U6
forward, 5′-CTC GCT TCG GCA GCA CA-3′ and reverse, 5′-AAC GCT TCA
CGA ATT TGC GT-3′.
Western blot analysis
Cells or tissues were resuspended in
radioimmunoprecipitation assay (RIPA) lysis buffer (Beyotime
Institute of Biotechnology, Inc.) on ice for 30 min. The samples
were centrifuged at 15,000 × g for 20 min at 4°C, and the
supernatants were collected and boiled for 5 min. Protein
concentrations were quantified using the Bradford protein assay
(Thermo Fisher Scientific, Inc.). Denatured proteins (15 µg)
were separated by 12% SDS-PAGE and then electro-transferred onto
polyvinylidene difluoride membranes (EMD Millipore). Following
blocking with 5% non-fat milk blocking buffer for 1 h at room
temperature, the membranes were incubated with primary antibodies
(all from Abcam) against OCT4 (cat. no. ab200834; 1:500), Nanog
(cat. no. ab109250; 1:500) and glyceraldehyde 3-phosphate
dehydrogenase (GAPDH) (cat. no. ab8245; 1:1,000) overnight at 4°C.
The membranes were subsequently washed with PBS 3 times and
incubated with secondary antibodies (cat. no. ab6728; 1:5,000;
Abcam) at room temperature for 1 h. The labeled bands were
visualized with an enhanced chemiluminescence reagent using the
Bio-Rad Gel Doc 2000 system (Bio-Rad Laboratories, Inc.), and
quantified with the QuantityOne software (v4.6; Bio-Rad
Laboratories, Inc.). Protein expression levels were normalized to
GAPDH.
Fluorescence in situ hybridization
(FISH)
Specific probes to hsa_circ_0026123 (Dig-5′-CAT TAA
CAT CAA GCT GAC CAG TGC ACC GG-3′-Dig) were prepared by Geneseed
Biotech, Inc. Signals were detected by FITC-conjugated anti-biotin
antibodies (Jackson ImmunoResearch, Inc.). Nuclei were
counterstained with 4,6-diamidino-2-phenylindole (DAPI; Shanghai
Yisheng Biotechnology Co., Ltd.) at room temperature for 15 min.
Finally, images were obtained on a Zeiss LSM 700 confocal
microscope (Carl Zeiss GmbH).
Cloning formation and cell proliferation
assays
A Cell Counting Kit-8 (CCK-8) assay was used to
determine cell proliferation. Transfected cells were seeded into
96-well plates at a density of 2,000 cells/well in triplicate
wells. Cell viability was measured using the CCK-8 system
(Invitrogen; Thermo Fisher Scientific, Inc.) at 0, 24, 48, 72, and
96 h after seeding, following standard procedures.
For the colony formation assay, transfected cells
were seeded into 6-well plates at a density of 2,000 cells/well and
maintained them in DMEM containing 10% FBS for 10 days. The
colonies were imaged with Nikon camera D610 (Nikon Corporation) and
counted after fixing with 4% paraformaldehyde (Shanghai Yisheng
Biotechnology Co., Ltd.) and staining with 0.1% crystal violet
(Shanghai Yisheng Biotechnology Co., Ltd.) at room temperature for
15 min.
Cell migration assay
Cell migration was measured using 24-well
Transwell® chambers (8 µm pore membrane; BD
Biosciences). Cells (1×105) were plated into the upper
chamber with 200 µl of serum-free medium and the bottom
chamber was filled with 500 µl DMEM (Gibco; Thermo Fisher
Scientific, Inc.) with 20% FBS (Gibco; Thermo Fisher Scientific,
Inc.). Following culture under 37°C for 1 day, the cells in the
bottom chamber were fixed with 4% paraformaldehyde for 30 min, and
stained them with 0.1% crystal violet (Shanghai Yisheng
Biotechnology Co., Ltd.) for 10 min at room temperature. The cells
were observed and photographed using Axio Observer D1 microscope
(magnification, ×200; Zeiss AG).
Tumor xenograft formation and metastasis
assays
Viable wild-type (WT) or si-circ0026123 SKOV3 cells
(2×107) (mentioned above) were injected into the right
flanks of the nude mice. Tumor sizes were measured every 5 days for
30 consecutive days using a Vernier caliper, and the tumor volume
was calculated using the following formula: Volume=length ×
width2 ×0.5.
Dual luciferase reporter assay
Hsa_circ_0026123-wild type (WT), EZH2 3′-UTR-wild
type (WT), hsa_ circ_0026123-mutant (Mut), EZH2 3′-UTR-mutant (Mut)
were constructed using pGL3-Basic luciferase vectors (Promega
Corporation) and transfected (50 nM) into 239T cells (American Type
Culture Collection) with or without NC (50 nM) or miR-124-3p mimics
(50 nM), respectively. After 48 h, a dual-luciferase reporter assay
kit (Promega Corporation) was used to determine luciferase
activity. Renillaluciferase was used for normalization. The
luciferase activities were measured using a luciferase assay kit
(Promega Corporation). Three independent experiments were performed
in triplicate.
Statistical analysis
Differences between groups were assessed using
one-way ANOVA method with Tukey's post hoc test. Results were
presented as the means ± SEM. P-values <0.05 were considered to
indicate statistically significant differences. Statistical
analyses were performed using GraphPad Prism 5.02 software
(GraphPad, Inc.).
Results
High hsa_circ_0026123 expression in OVA
cells and tissues and its role in migration, proliferation and CSC
differentiation
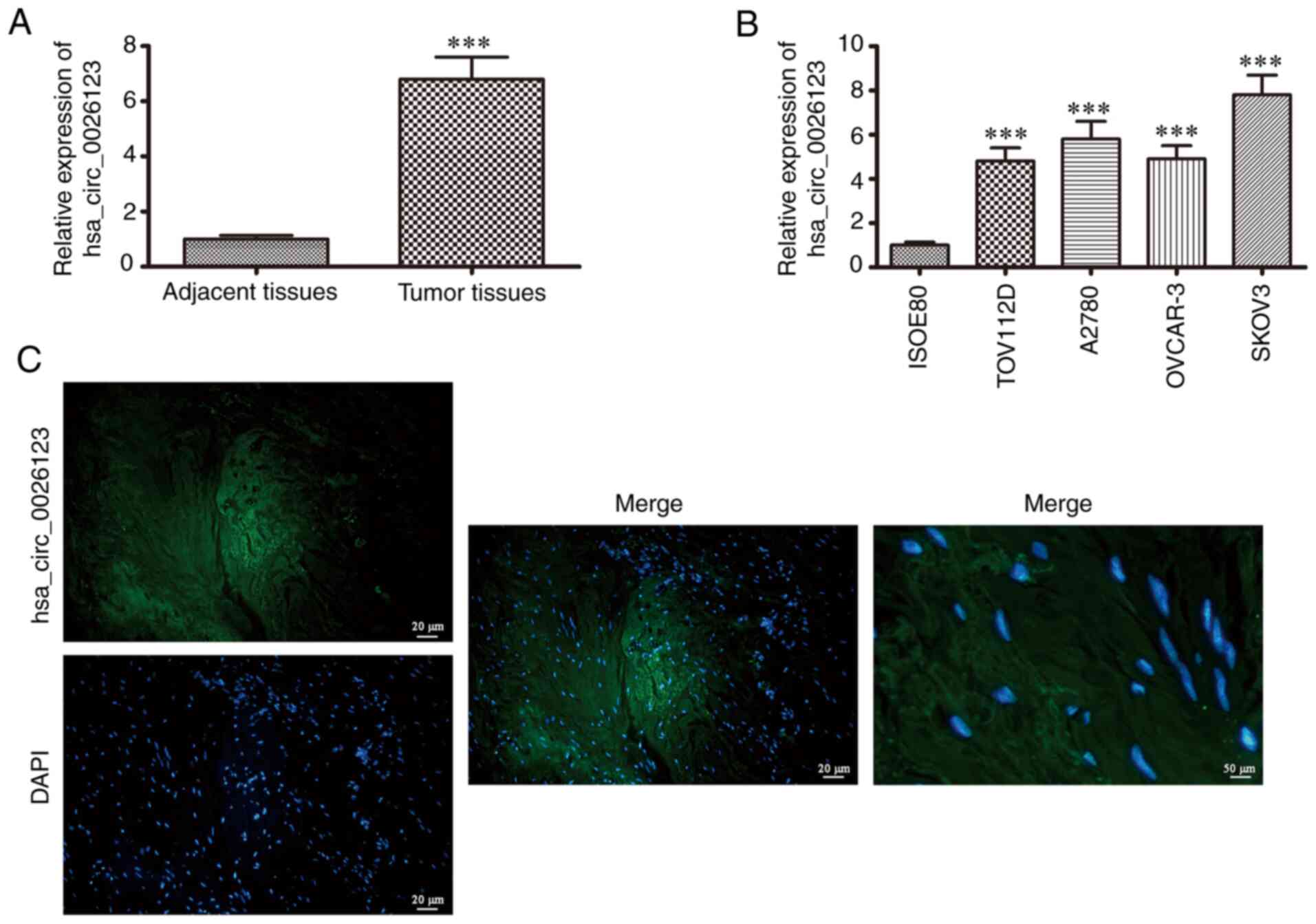
hsa_circ_0026123 expression was detected in 20 OVA
tissue samples and corresponding adjacent normal tissues by
RT-qPCR. The data revealed that hsa_circ_0026123 expression was
significantly increased in OVA tissues when compared with adjacent
normal tissues (Fig. 1A),
suggesting that hsa_circ_0026123 plays a role in OVA progression.
RT-qPCR also revealed that hsa_circ_0026123 expression in human OVA
cell lines (A2780, TOV112D, OVCAR-3 and SKOV3) was increased when
compared with that in the normal ovarian cell line (ISOE80;
Fig. 1B). The SKOV3 cells
exhibited the highest hsa_circ_0026123 expression; thus the SKOV3
cells were selected for use in the following experiments. FISH was
performed to detect hsa_circ_0026123 subcellular localization. The
results revealed that hsa_circ_0026123 was localized in the
cytoplasm (Fig. 1C).
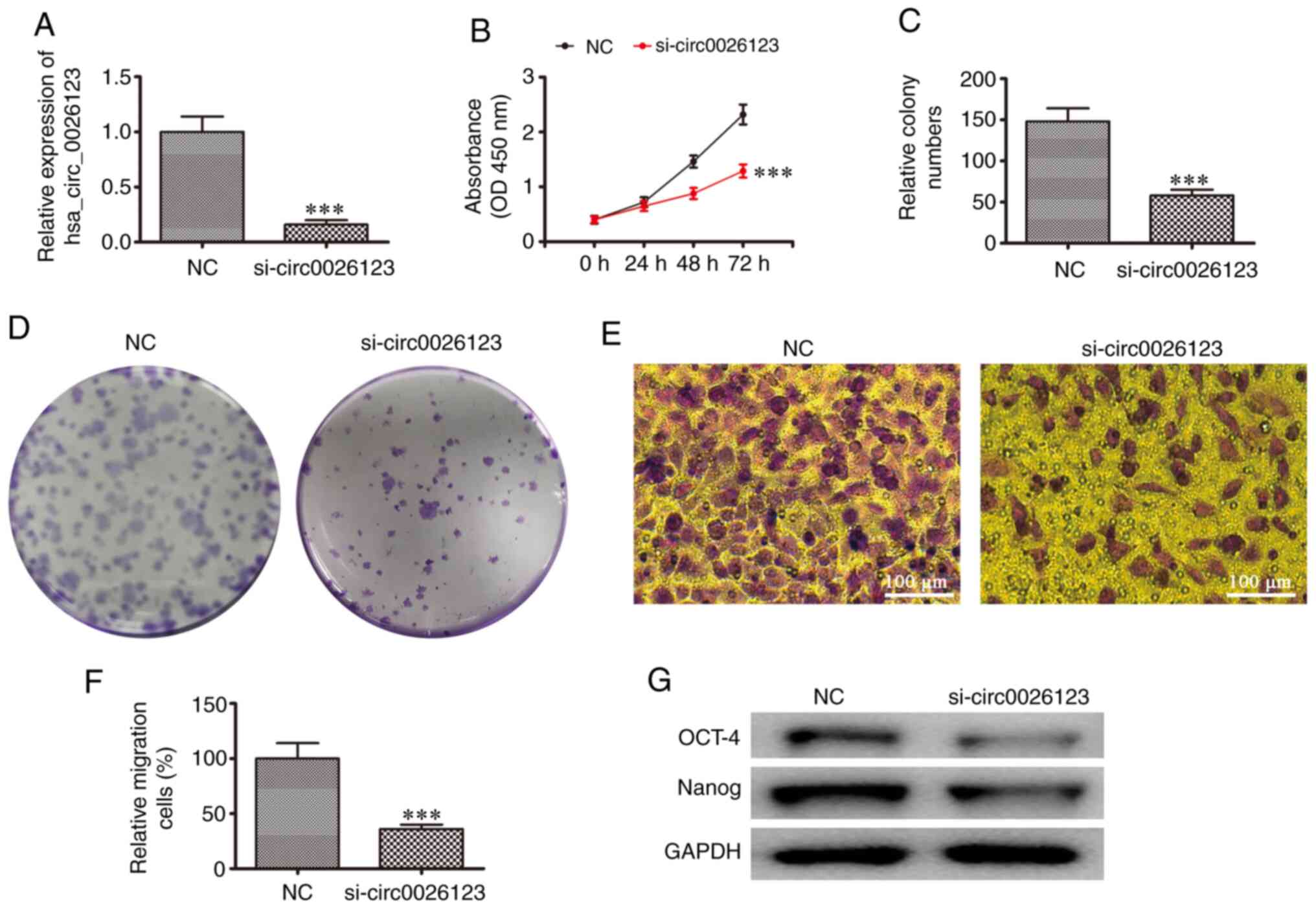
To confirm that hsa_circ_0026123 plays a role in the
progression of OVA, siRNAs against hsa_circ_0026123 were
constructed. The results of RT-qPCR revealed that hsa_circ_0026123
expression in SKOV3 significantly decreased following transfection
with siRNA against hsa_circ_0026123 (si-circ0026123) when compared
with the negative control (NC; Fig.
2A). CCK-8 (Fig. 2B) and
colony formation (Fig. 2C and D)
assays revealed that SKOV3 cell proliferation was decreased
following the downregulation of hsa_circ_0026123. The results of
Transwell assay also demonstrated that the silencing of
hsa_circ_0026123 suppressed SKOV3 cell migration (Fig. 2E and F). In addition, western blot
analyses revealed that the silencing of hsa_circ_0026123 decreased
the protein expression levels of markers (OCT-4 and Nanog) related
to CSC differentiation. These results suggested that the
downregulation of hsa_circ_0026123 suppressed OVA cell
proliferation, migration and CSC differentiation.
miR-124-3p and EZH2 are the downstream
targets of hsa_circ_0026123
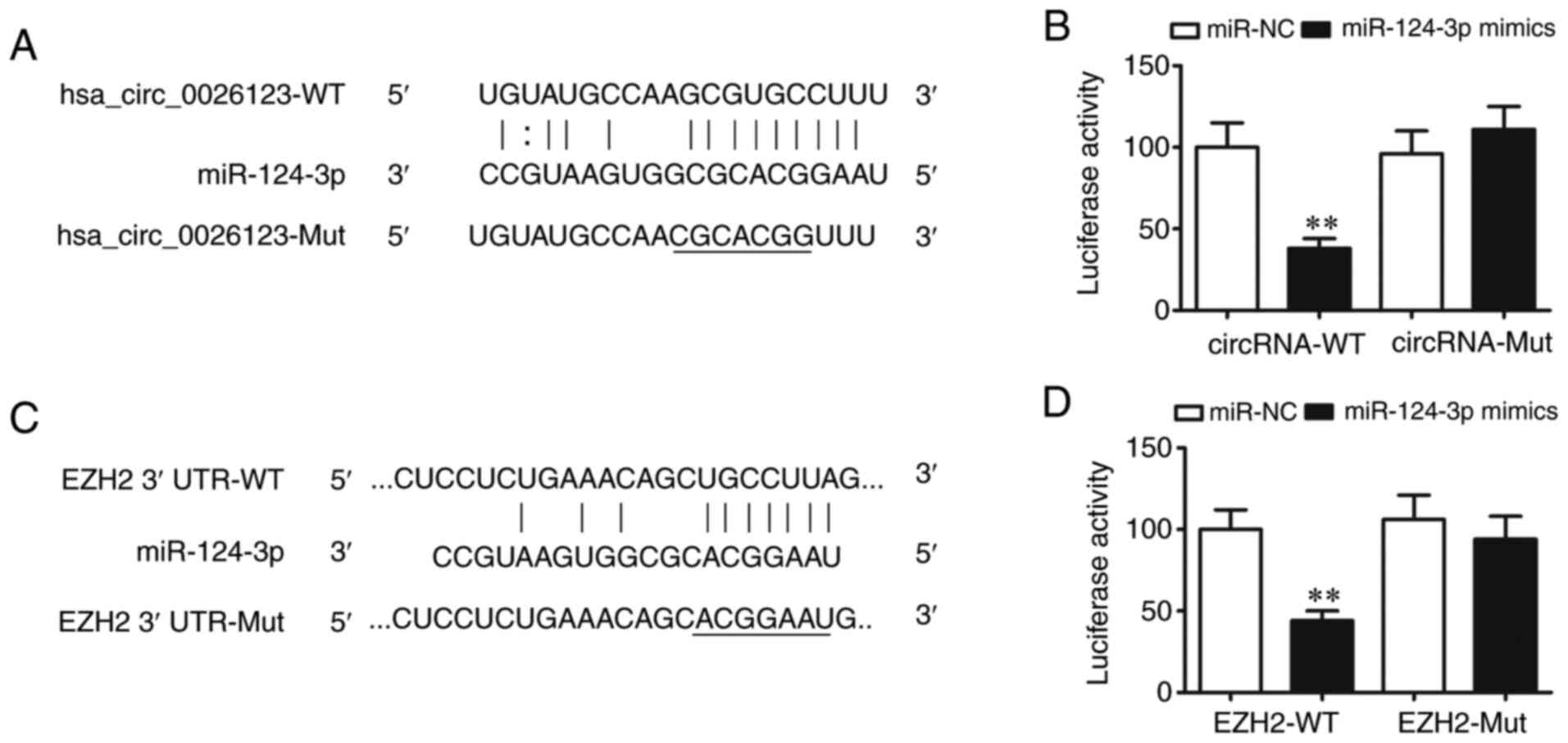
Bioinformatics analysis confirmed that
hsa_circ_0026123 targeted miR-124-3p. To further verify this
observation, a luciferase reporter vector was constructed. The
luciferase reporter assay results revealed that miR-124-3p
inhibited luciferase activity following transfection with the WT
luciferase reporter vector, while it had no effects on luciferase
activity following transfection with a mutated (MUT) luciferase
reporter vector, which indicated that miR-124-3p was a target of
hsa_circ_0026123 (Fig. 3A and
B).
Bioinformatics analysis (http://starbase.sysu.edu.cn/) revealed that a number
of genes were the downstream targets of miR-124-3p, including
MYLIP, WEE1, BTG2 and EZH2. However, only EZH2 has two binding
sites. This indicated that miR-124-3p directly interacted with the
EZH2 3′-UTR and suppressed the post-translational EZH2 expression
(Fig. 3C). The luciferase
reporter assay revealed that miR-124-3p inhibited luciferase
activity following transfection with a WT luciferase reporter
vector, whereas luciferase activity was not altered after
transfection with a MUT luciferase reporter vector, which indicated
that EZH2 was the miR-124-3p target (Fig. 3D). Taken together, the results
demonstrated that hsa_circ_0026123 knockdown inhibited OVA
metastasis and growth by targeting the miR-124-3p/EZH2 axis.
hsa_circ_0026123 knockdown inhibits OVA
cell proliferation, migration and CSC differentiation via the
regulation of the miR-124-3p/EZH2 axis in vitro
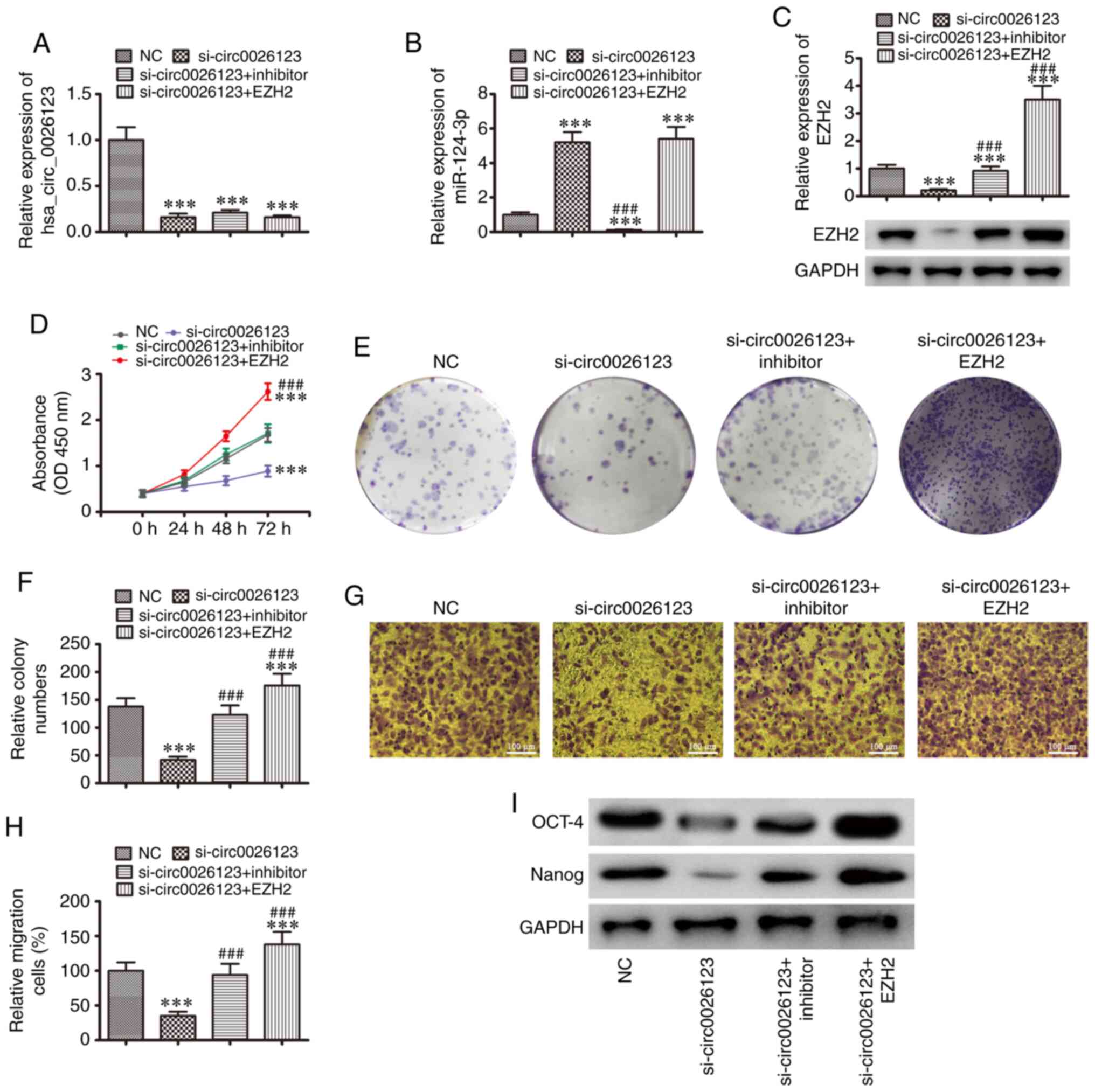
To further explore the regulatory mechanisms, SKOV3
cells were transfected with a hsa_circ_0026123 silencing vector
(si-circ0026123), combined with an EZH2 overexpression vector, or
were transfected with an miR-124-3p inhibitor. The results
confirmed that hsa_circ_0026123 expression was downregulated
following transfection with si-circ0026123, as well as by the
overexpression of EZH2 or the downregulation of miR-124-3p, which
did not 'rescue' hsa_circ_0026123 expression (Fig. 4A). The results RT-qPCR revealed
that the downregulation of hsa_ circ_0026123 promoted miR-124-3p
expression. Transfection with the miR-124-3p specific inhibitor
suppressed miR-124-3p expression. However, the overexpression of
EZH2 did not affect miR-124-3p expression following the silencing
of hsa_circ_0026123 in the SKOV3 cell line (Fig. 4B). Western blot analysis confirmed
that the downregulation of hsa_ circ_0026123 suppressed EZH2
expression. The inhibition of miR-124-3p 'rescued' EZH2 expression,
and EZH2 expression was increased following transfection with the
EZH2 overexpression vector (Fig.
4C). Overall, these results demonstrated that EZH2 was a
miR-124-3p downstream target.
The results of CCK-8 (Fig. 4D) and cloning formation (Fig. 4E and F) assays revealed that the
overexpression of EZH2 or the downregulation of miR-124-3p
'rescued' SKOV3 cell proliferation following the silencing of
hsa_circ_0026123. The results of Transwell migration assay also
revealed that the overexpression of EZH2 or the downregulation of
miR-124-3p restored SKOV3 cell migration following the silencing of
hsa_circ_0026123 (Fig. 4G and H).
Western blot analysis also demonstrated that the overexpression of
EZH2 or the downregulation of miR-124-3p restored the expression
levels of CSC differentiation-related markers (OCT-4 and Nanog)
after the silencing of hsa_circ_0026123, suggesting that
hsa_circ_0026123 knockdown inhibited OVA cell migration and
proliferation, and decreased the levels of CSC
differentiation-related markers via the regulation of the
miR-124-3p/EZH2 axis in vitro.
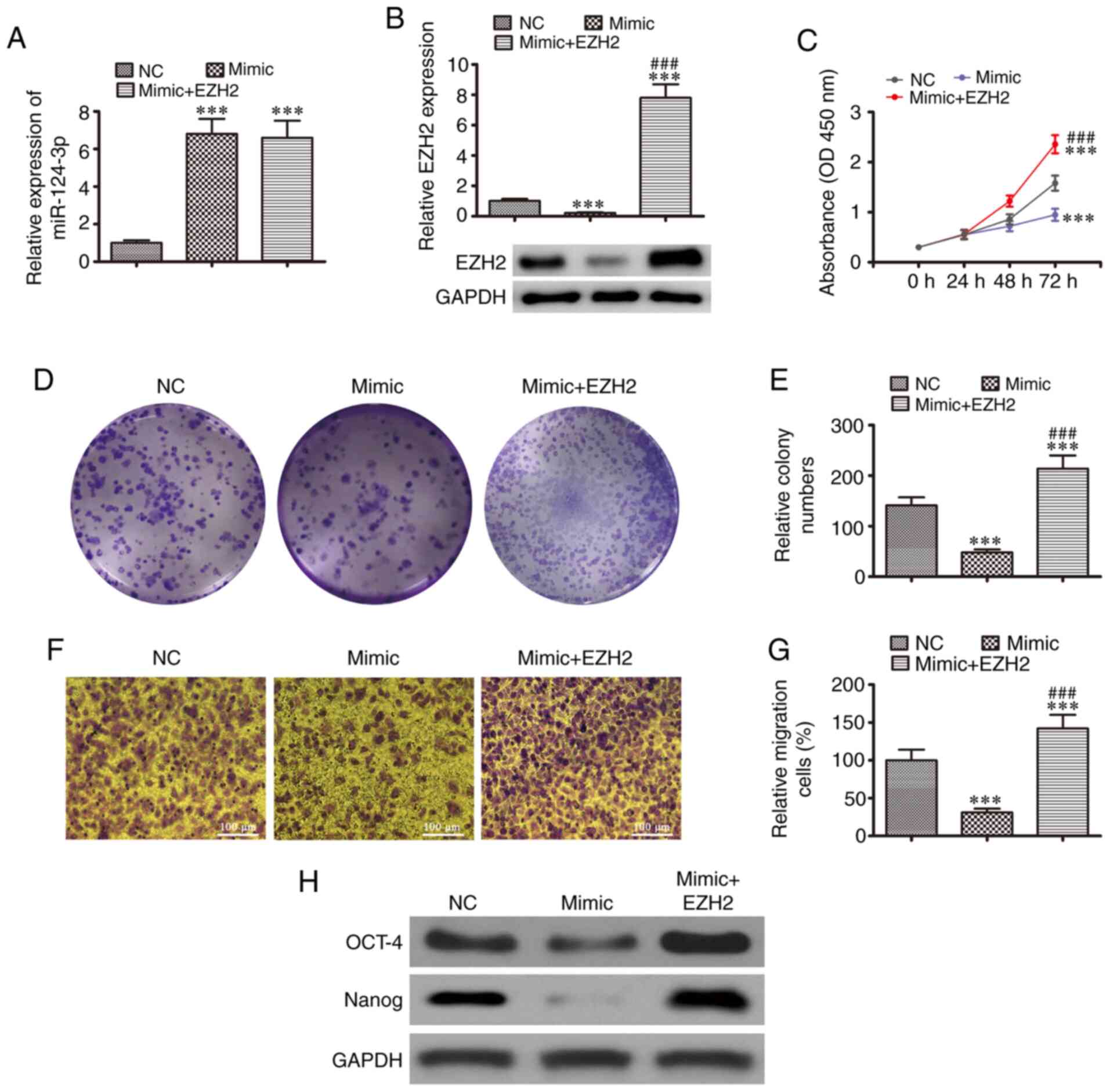
Overexpression of EZH2 restores cell
proliferation and migration, and CSC differentiation following the
upregulation of miR-124-3p in vitro
To determine the association between miR-124-3p and
EZH2, SKOV3 cells were transfected with miR-124-3p mimics combined
with or without the EZH2 overexpression vector. The results of
RT-qPCR revealed that miR-124-3p expression was upregulated in
SKOV3 cell lines following transfection with miR-124-3p mimic, and
EZH2 overexpression had no effect on miR-124-3p expression
(Fig. 5A). Western blot analysis
revealed that EZH2 expression was downregulated following the
overexpression of miR-124-3p, which was restored following
transfection with the EZH2 overexpression vector (Fig. 5B). The results of CCK-8 (Fig. 5C) and cloning formation (Fig. 5D and E) assay revealed that cell
proliferation was suppressed by miR-124-3p overexpression; however,
EZH2 overexpression restored and enhanced the SKOV3 cell
proliferation. The results of Transwell migration assays revealed
that miR-124-3p expression inhibited cell migration, while EZH2
upregulation increased the migration of SKOV3 cells (Fig. 5F and G). Western blot analysis
also demonstrated that miR-124-3p expression suppressed the levels
of markers related to CSC differentiation (OCT-4 and Nanog);
however, EZH2 overexpression promoted CSC differentiation, as
evidenced by the increased levels of OCT-4 and Nanog, even after
miR-124-3p overexpression. These results indicated that EZH2
overexpression recovered the growth, migration and the levels of
CSC differentiation-related markers following the upregulation of
miR-124-3p using in vitro.
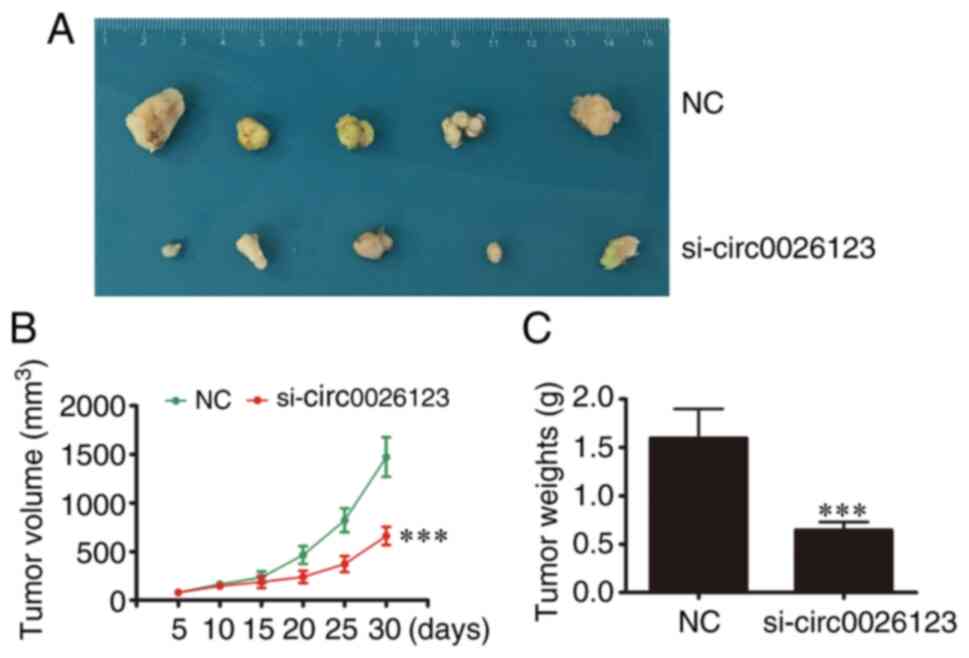
hsa_circ_ 0026123 downregulation
suppresses tumor growth in nude mouse xenografts
To confirm whether hsa_circ_0026123 plays a role in
OVA progression, lentiviral stable SKOV3 cell strains were
constructed in which hsa_circ_0026123 was knocked down. The data
indicated that hsa_circ_0026123 knockdown suppressed the volume and
weight of tumors, when compared with the NC group (Fig. 6).
Discussion
The present study confirmed that hsa_circ_0026123
expression was increased in both OVA tissues and OVA cell lines.
hsa_circ_0026123 is 1,155 bp in length and is constructed with part
of the TUBA1B gene exon. The downregulation of
hsa_circ_0026123 suppressed cell migration and proliferation, and
CSC differentiation, suggesting that hsa_circ_0026123 plays a role
in OVA progression. Previous studies have reported that circRNA
sponges are characterized by increased expression levels and miRNA
binding sites, which are likely to be effective sponges, when
compared with linear ones (6,18).
The sponging activity is the main function of some circRNAs; thus,
in tumor development, circRNA/miRNA/mRNA inter-action networks may
play important roles (19,20).
In the present study, to further identify the
downstream miRNA, bioinformatics analysis was used to demonstrated
that miR-124-3p was a target of hsa_circ_0026123. Luciferase
reporter assays confirmed that hsa_circ_0026123 interacted with
miR-124-3p. The downregulation of hsa_circ_0026123 promoted
miR-124-3p expression, and the silencing of miR-124-3p restored the
proliferation and migration, and the levels of CSC
differentiation-related markers following the silencing of
hsa_circ_0026123, suggesting that miR-124-3p exerts an antitumor
effect. Previous studies have demonstrated that miR-124-3p
upregulation inhibits cancer cell activity, including that of
nasopharyngeal carcinoma, bladder cancer, hepatocellular carcinoma,
esophageal squamous cell carcinoma and endometrial cancer (21-25). Therefore, hsa_ circ_0026123
expression promoted the progression of OVA by sponging
miR-124-3p.
The present study demonstrated that miR-124-3p
inter-acted with the EZH2 3′-UTR. The results of luciferase
reporter assays suggested that miR-124-3p interacted with the
3′-UTR of EZH2. The overexpression of EZH2 restored the
proliferation and migration, and the levels of markers related to
CSC differentiation following the overexpression of miR-124-3p. It
has been reported that the inhibition of EZH2 results in a
suppressed epithelial-mesenchymal transition in cancer cells
(26). The gene product is a
matricellular protein, which stimulates endothelial cell migration
and proliferation, as well as the angiogenic activity (27,28). The downregulation of EZH2
suppresses CSC proliferation and differentiation (29-31). Numerous chemical entities have
been regarded as EZH2 inhibitors in recent decades, many of which
underwent the cancer clinical trials (32,33). The results of the present study
demonstrated that hsa_circ_0026123 promoted OVA cell migration and
proliferation by sponging miR-124-3p and enhancing EZH2
expression.
In conclusion, the present study provides evidence
that hsa_circ_0026123 promotes the proliferation of OVA cells
potentially via activating miR-124-3p/EZH2 signaling. The present
study suggests that hsa_circ_0026123 is a candidate biomarker for
the prognosis and diagnosis of OVA, which extends the drug
applications targeting hsa_circ_0026123, suggesting a promising
role of hsa_circ_0026123 in the treatment of OVA.
Abbreviations:
|
OVA
|
ovarian cancer
|
|
circRNAs
|
circular RNAs
|
|
miRNA/miR
|
microRNA
|
|
FBS
|
fetal bovine serum
|
|
siRNA
|
small interfering RNA
|
|
NC
|
negative control
|
|
CSC
|
cancer stem cell
|
Funding
The present study was funded by a grant from the
National Natural Science Foundation of China (no. 81702745).
Availability of data and materials
The datasets used and analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
XY and XT designed the research and revised the
manuscript. JW and YS performed the experiments and drafted the
manuscript. HL performed data analysis. XY and YS revised the
manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to
participate
The Ethics Committee of Shanghai Tongji Hospital
approved this study. We obtained informed consent from each patient
prior to the analyses of tissues. The Ethics Committee of Shanghai
Tongji Hospital, Tongji University, Shanghai, China approved all
animal experiments.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgments
Not applicable.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2018. CA Cancer J Clin. 68:7–30. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Rustin G, van der Burg M, Griffin C, Qian
W and Swart AM: Early versus delayed treatment of relapsed ovarian
cancer. Lancet. 377:380–381. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Doubeni CA, Doubeni AR and Myers AE:
Diagnosis and management of ovarian cancer. Am Fam Physician.
93:937–944. 2016.PubMed/NCBI
|
|
4
|
Li Y, Zheng Q, Bao C, Li S, Guo W, Zhao J,
Chen D, Gu J, He X and Huang S: Circular RNA is enriched and stable
in exosomes: A promising biomarker for cancer diagnosis. Cell Res.
25:981–984. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Zhang SJ, Chen X, Li CP, Li XM, Liu C, Liu
BH, Shan K, Jiang Q, Zhao C and Yan B: Identification and
characterization of circular RNAs as a new class of putative
biomarkers in diabetes retinopathy. Invest Ophthalmol Vis Sci.
58:6500–6509. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wilusz JE and Sharp PA: Molecular biology.
A circuitous route to noncoding RNA. Science. 340:440–441. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Kumar L, Shamsuzzama, Haque R, Baghel T
and Nazir A: Circular RNAs: The emerging class of non-coding RNAs
and their potential role in human neurodegenerative diseases. Mol
Neurobiol. 54:7224–7234. 2017. View Article : Google Scholar
|
|
8
|
Piwecka M, Glazar P, Hernandez-Miranda LR,
Memczak S, Wolf SA, Rybak-Wolf A, Filipchyk A, Klironomos F, Jara
CAC, Fenske P, et al: Loss of a mammalian circular RNA locus causes
miRNA deregulation and affects brain function. Science.
357:eaam85262017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Shan K, Liu C, Liu BH, Chen X, Dong R, Liu
X, Zhang YY, Liu B, Zhang SJ, Wang JJ, et al: Circular noncoding
RNA HIPK3 mediates retinal vascular dysfunction in diabetes
mellitus. Circulation. 136:1629–1642. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Hsiao KY, Lin YC, Gupta SK, Chang N, Yen
L, Sun HS and Tsai SJ: Noncoding effects of circular RNA CCDC66
promote colon cancer growth and metastasis. Cancer Res.
77:2339–2350. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Guo Q, He Y, Sun L, Kong C, Cheng Y and
Zhang G: In silico detection of potential prognostic circRNAs
through a re-annotation strategy in ovarian cancer. Oncol Lett.
17:3677–3686. 2019.PubMed/NCBI
|
|
12
|
Xiong DD, Dang YW, Lin P, Wen DY, He RQ,
Luo DZ, Feng ZB and Chen G: A circRNA-miRNA-mRNA network
identification for exploring underlying pathogenesis and therapy
strategy of hepatocellular carcinoma. J Transl Med. 16:2202018.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Guan YJ, Ma JY and Song W: Identification
of circRNA-miRNA-mRNA regulatory network in gastric cancer by
analysis of microarray data. Cancer Cell Int. 19:1832019.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhang L, Zhou Q, Qiu Q, Hou L, Wu M, Li J,
Li X, Lu B, Cheng X, Liu P, et al: CircPLEKHM3 acts as a tumor
suppressor through regulation of the miR-9/BRCA1/DNAJB6/KLF4/AKT1
axis in ovarian cancer. Mol Cancer. 18:1442019. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Gao Y, Zhang C, Liu Y and Wang M: Circular
RNA profiling reveals circRNA1656 as a novel biomarker in high
grade serous ovarian cancer. Biosci Trends. 13:204–211. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Sheng M, Wei N, Yang HY, Yan M, Zhao QX
and Jing LJ: CircRNA UBAP2 promotes the progression of ovarian
cancer by sponging microRNA-144. Eur Rev Med Pharmacol Sci.
23:7283–7294. 2019.PubMed/NCBI
|
|
17
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
18
|
Li J, Yang J, Zhou P, Le Y, Zhou C, Wang
S, Xu D, Lin HK and Gong Z: Circular RNAs in cancer: Novel insights
into origins, properties, functions and implications. Am J Cancer
Res. 5:472–480. 2015.PubMed/NCBI
|
|
19
|
Liu J, Song S, Lin S, Zhang M, Du Y, Zhang
D, Xu W and Wang H: Circ-SERPINE2 promotes the development of
gastric carcinoma by sponging miR-375 and modulating YWHAZ. Cell
Prolif. 52:e126482019. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Liu G, Shi H, Deng L, Zheng H, Kong W, Wen
X and Bi H: Circular RNA circ-FOXM1 facilitates cell progression as
ceRNA to target PPDPF and MACC1 by sponging miR-1304-5p in
non-small cell lung cancer. Biochem Biophys Res Commun.
513:207–212. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Liu C, Zhang H and Liu H: Long noncoding
RNA UCA1 accelerates nasopharyngeal carcinoma cell progression by
modulating miR-124-3p/ITGB1 axis. Onco Targets Ther. 12:8455–8466.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Fu W, Wu X, Yang Z and Mi H: The effect of
miR-124-3p on cell proliferation and apoptosis in bladder cancer by
targeting EDNRB. Arch Med Sci. 15:1154–1162. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Cui RJ, Fan JL, Lin YC, Pan YJ, Liu C, Wan
JH, Wang W, Jiang ZY, Zheng XL, Tang JB and Yu XG: miR-124-3p
avail-ability is antagonized by LncRNA-MALAT1 for Slug-induced
tumor metastasis in hepatocellular carcinoma. Cancer Med.
8:6358–6369. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Lv Y, Chen S, Wu J, Lin R, Zhou L, Chen G,
Chen H and Ke Y: Upregulation of long non-coding RNA OGFRP1
facilitates endometrial cancer by regulating miR-124-3p/SIRT1 axis
and by activating PI3K/AKT/GSK-3β pathway. Artif Cells Nanomed
Biotechnol. 47:2083–2090. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zeng B, Zhang X, Zhao J, Wei Z, Zhu H, Fu
M, Zou D, Feng Y, Luo H and Lei Y: The role of
DNMT1/hsa-miR-124-3p/BCAT1 pathway in regulating growth and
invasion of esophageal squamous cell carcinoma. BMC Cancer.
19:6092019. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zhao M, Hu X, Xu Y, Wu C, Chen J, Ren Y,
Kong L, Sun S, Zhang L, Jin R and Zhou X: Targeting of EZH2
inhibits epithelialmesenchymal transition in head and neck squamous
cell carcinoma via regulating the STAT3/VEGFR2 axis. Int J Oncol.
55:1165–1175. 2019.PubMed/NCBI
|
|
27
|
Nishimoto S, Hamajima Y, Toda Y, Toyoda H,
Kitamura K and Komurasaki T: Identification of a novel smooth
muscle associated protein, smap2, upregulated during neointima
formation in a rat carotid endarterectomy model. Biochim Biophys
Acta. 1576:225–230. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Maier S, Paulsson M and Hartmann U: The
widely expressed extracellular matrix protein SMOC-2 promotes
keratinocyte attachment and migration. Exp Cell Res. 314:2477–2487.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Liu H, Sun Q, Sun Y, Zhang J, Yuan H, Pang
S, Qi X, Wang H, Zhang M, Zhang H, et al: MELK and EZH2 cooperate
to regulate medulloblastoma cancer stem-like cell proliferation and
differentiation. Mol Cancer Res. 15:1275–1286. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Chen JF, Luo X, Xiang LS, Li HT, Zha L, Li
N, He JM, Xie GF, Xie X and Liang HJ: EZH2 promotes colorectal
cancer stem-like cell expansion by activating p21cip1-Wnt/β-catenin
signaling. Oncotarget. 7:41540–41558. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Karami Madani G, Rad A, Molavi M, Ardalan
Khales S, Abbaszadegan MR and Forghanifard MM: Predicting the
correlation of EZH2 and cancer stem cell markers in esophageal
squamous cell carcinoma. J Gastrointest Cancer. 49:437–441. 2018.
View Article : Google Scholar
|
|
32
|
Zhang H, Qi J, Reyes JM, Li L, Rao PK, Li
F, Lin CY, Perry JA, Lawlor MA, Federation A, et al: Oncogenic
deregulation of EZH2 as an opportunity for targeted therapy in lung
cancer. Cancer Discov. 6:1006–1021. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Stazi G, Zwergel C, Mai A and Valente S:
EZH2 inhibitors: A patent review (2014-2016). Expert Opin Ther Pat.
27:797–813. 2017. View Article : Google Scholar : PubMed/NCBI
|