Introduction
Currently, deaths from non-small cell lung cancer
(NSCLC) account for 85% of lung cancer-related deaths worldwide
(1). The disease is associated
with a high degree of malignancy, and early and extensive
metastasis, followed by a poor clinical prognosis (2). As lung cancers are the most lethal
tumors in North America, their diagnosis and treatment are
attracting increasing attention. Therefore, research on the
molecular mechanisms of NSCLC is crucial for the investigation of
the mechanisms responsible for the development of this tumor and
for the identification of treatment strategies. In recent years,
deoxyribonucleic acid (DNA) methylation, as one of the most common
epigenetic modifications, has been linked to specific sequences of
the CpG islands and plays an important role in the progression of
lung cancer (3-6). Generally, DNA methylation results in
the inactivation of gene expression (7,8),
while demethylation in the promoter region activates gene
transcription levels (9,10).
Reactive oxygen species (ROS), which are normally
produced in and eliminated from all types of cells, exert
physiological and pathological effects (11). In a tumor micro-environment (TME)
of ischemia-hypoxia, ROS levels are commonly higher than in normal
environments, which is crucial for the study of tumorigenesis and
treatments (12,13). Peroxiredoxins (PRDXs) as a class
of antioxidant enzymes that include 6 members, PRDX 1-6, which play
a role in regulating cell proliferation, differentiation and
apoptosis by modulating ROS (14). Recent studies have indicated that
PRDXs participate in tumor progression, and upregulated levels of
PRDXs have been suggested to be responsible for tumor prognosis and
drug resistance (15-17). In the present study, it was found
that PRDX5 was highly expressed in NSCLC tissues, while of
note, there were obvious methyl islands in the promoter region.
Therefore, it was hypothesized that the hypomethylation of the
PRDX5 gene promoter region may be related to the
pathogenesis of NSCLC.
Signal transducers and activators of transcription
(STATs) are a family of transcription factors (TFs) that can be
activated by different cytokines and act as carriers during
interaction between cytokines and receptors, maintaining the
intracellular transmission of signals. Different STATs have their
own specific functions; e.g., STAT4 and STAT1 induce T-helper 1
cell (Th1) differentiation, whole STAT6 mediates
Th2 cell differentiation (18,19). STAT3, as the
first-discovered member of the STAT family, was first defined as an
acute response protein participating in various physiological or
pathological processes; it is widely expressed in the human body
and can be activated by a number of types of cytokines or by
various stressors (20). Recent
studies have demonstrated that the abnormal expression of
STAT3 is also found in a variety of tumors (21-23). However, the mechanisms through
which STAT3 promotes tumor progression remain unclear.
The present study aimed to explore the methylation
status of the PRDX5 gene promoter region in NSCLC. It was
deter-mined that the ROS-mediated hypomethylation of PRDX5
promoted STAT3 binding. Furthermore, the results revealed
that the upregulation of PRDX5 mediated by STAT3
activated the nuclear factor (erythroid-derived 2)-like 2 (Nrf2)
signaling pathway.
Materials and methods
Patients and tissue samples
NSCLC and adjacent non-cancerous tissues were
obtained from 121 patients with NSCLC who underwent surgical
resection at the Affiliated Hospital of Nantong University
(Nantong, China) between January, 2006 and January, 2011. Detailed
clinicopathological parameters of the patients are provided in
Table I. The study protocol was
approved by the Ethics Committee of Affiliated Hospital of Nantong
University. Written informed consent was obtained from patients
prior to collecting samples.
 | Table IClinicopathological characteristics
and PRDX5 methylation in 121 patients with NSCLC. |
Table I
Clinicopathological characteristics
and PRDX5 methylation in 121 patients with NSCLC.
| Clinicopathological
parameters | No. of
patients | PRDX5 methylation
| P-value |
|---|
| Methylated
(n=42) | Unmethylated
(n=79) |
|---|
| Age (years) | | | | 0.879 |
| <60 | 53 | 18 | 35 | |
| ≥60 | 68 | 24 | 44 | |
| Sex | | | | 0.830 |
| Male | 56 | 20 | 36 | |
| Female | 65 | 22 | 43 | |
| Clinical TNM
stage | | | | |
| I-II | 73 | 31 | 42 | 0.027a |
| III-IV | 48 | 11 | 37 | |
| Lymph node
involvement | | | | 0.438 |
| Negative | 49 | 19 | 30 | |
| Positive | 72 | 23 | 49 | |
|
Differentiation | | | | 0.830 |
| Well and
moderate | 65 | 22 | 43 | |
| Poor | 56 | 20 | 36 | |
Cell lines and cell culture
The lung cancer cell lines, A549 (SCSP-503), H1299
(SCSP-589) and H157 (ATCC® CRL5802™), and the normal
bronchial epithelial cell (EC) line, 16HBE (ATCC®
PCS-300-040™), were purchased from the Shanghai Cell Bank of the
Chinese Academy of Sciences or the American Type Culture Collection
(ATCC). Following cell line authentication using short tandem
repeat (STR) profiling, the cells were cultured in Dulbecco's
modified Eagle's medium (DMEM; Gibco; Thermo Fisher Scientific,
Inc.) supplemented with 10% fetal bovine serum (FBS, Gibco; Thermo
Fisher Scientific, Inc.), 50 U/ml penicillin and 50 µg/ml
streptomycin (Invitrogen; Thermo Fisher Scientific, Inc.); they
were then incubated in a humidified atmosphere at 37°C and 5%
CO2.
Establishment of model of oxidative
stress (OS) induced by H2O2
H2O2 (Sigma-Aldrich; Merck
KGaA) was used to the establishment of a model of OS. In brief, the
lung cancer cell lines, A549, H1299 and H157, were pre-treated with
various concentrations of H2O2 (0, 50, 100 or
200 µM) for 30 min of stimulation.
Methylation-specific and bisulfite
sequencing polymerase chain reaction (MSP and BSP)
The CpG island online prediction software
(http://www.urogene.org/index.html)
was used to predict the CpG islands of the PRDX5 gene
promoter. MSP was used to measure the methylation status of CpG
islands in the PRDX5 gene promoter region. The methylated
(M) band indicated that CpG sites were methylated, while the
unmethylated (U) band indicated unmethylated status. The patients
with NSCLC were divided into the PRDX5-methylated and -unmethylated
group (U/M ≥1, unmethylated; U/M <1, methylated). The
demethylation ratio was calculated as U/(M + U). BSP was used to
verify the methylation status of these islands. Extracted DNA
samples isolated from NSCLC tissues and cells were modified with
bisulfite reagents (Zymo Research) as per the manufacturer's
instructions, which specified a change from unmethylated cytosine
to thymine. The MSP primer pairs for PRDX5 were as follows:
PRDX5-MSP-M forward, 5′-GGG GTT GAA TTT TAT AGG GTA GAT AC-3′ and
reverse, 5′-GAC CTA ACG AAA ATT TAT ACG ACG A-3′; PRDX5-MSP-U
forward, 5′-GGG GTT GAA TTT TAT AGG GTA GAT AT-3′ and reverse,
5′-AAC CTA ACA AAA ATT TAT ACA ACA AC-3′. For BSP,
bisulfite-treated DNA was amplified by PCR using the following
primers: PRDX5-BSP forward, 5′-GGG TTG AAT TTT ATA GGG TAG ATA-3′
and reverse, 5′-CTA CTT ACC CAC AAT CTA CTA AAC TC-3′. PCR products
were purified using the Wizard SV Gel and PCR Clean-up System and
then cloned into a pGEM-T Easy Vector System (both from Promega
Corporation). A total of 8 colonies were randomly selected for the
extraction of plasmid DNA using a Promega Spin Mini kit (Promega
Corporation) and the DNA was sequenced on an ABI 3130 Genetic
Analyzer (Applied Biosystems; Thermo Fisher Scientific, Inc.).
Reverse transcription-quantitative PCR
(qRT-PCR)
RT-qPCR was used to determine PRDX5 mRNA
expression. Total RNA was extracted from tissues and cells using
TRIzol reagent (Takara Bio, Inc.) as per manufacturer's
instructions. First-strand complementary deoxyribonucleic acid
(cDNA) was then synthesized using a PrimeScript RT Reagent kit and
SYBR-Green I (Takara Bio, Inc.) was used for the qPCR analysis of
the cDNA as per the manufacturer's instructions. The PCR
thermocycling conditions were as follows: 95°C for 5 min, (95°C for
15 sec, 58°C for 45 sec, and 72°C for 60 sec) 40 cycles, and 72°C
for 1 min. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH)
was used as the internal control. The primer sequences for
PRDX5and GAPDH were as follows: PRDX5 forward,
5′-CCA ATC AAG ACA CAC CTG CC-3′ and reverse, 5′-TCT TGA GAC GTC
GAT TCC CA-3′; GAPDHforward, 5′-GAA GGT GAA GGT CGG AGT C-3′
and reverse, 5′-GAA GAT GGT GAT GGG ATT TC-3′. Quantities were
calculated using the comparative 2−ΔΔCq method (24). All steps were performed in
triplicate.
Western blot analysis
Tissues and cells were lysed using
radioimmunoprecipitation assay (RIPA) buffer containing protease
inhibitors (Promega Corporation) to obtain total protein. The
protein concentrations were then determined using a bicinchoninic
acid (BCA) Protein Assay kit (Bio-Rad Laboratories, Inc.). Protein
samples (containing 30 µg protein) were separated on 10%
sodium dodecyl sulfate polyacrylamide gel electrophoresis
(SDS-PAGE) gels and then transferred onto polyvinylidene difluoride
(PVDF) membranes. The membranes were blocked with 5% skimmed milk
for 1 h at room temperature and then incubated overnight with
primary antibodies diluted to 1:1,000 at 4°C. The following day,
the membranes were further incubated with horseradish peroxidase
(HRP)-conjugated secondary antibodies at room temperature for 1 h
(1:2,000; cat no. 7056, Cell Signaling Technology, Inc.). GAPDH
(1:1,000; cat no. A00227-1, Boster Biological Technology, Ltd.) was
used as an internal reference. The immunoreactive proteins were
detected using an enhanced chemiluminescence kit (Cell Signaling
Technology, Inc.). The blots were then scanned on an Odyssey Fc
Imaging System (Li-COR Biosciences) and the grayscale value was
used for statistical analysis. The antibodies against
PRDX5(1:1,000; cat no. ab180587), STAT3(1:1,000; cat
no. ab76315), E-cadherin (1:1,000; cat no. ab40772), β-actin
(1:1,000; cat no. ab8226), vimentin (1:1,000; cat no. ab92547),
Nrf2(1:1,000; cat no. ab89443) and reduced nicotinamide
adenine dinucleotide phosphate (NADPH) dehydrogenase [quinone] 1
(NQO1) (1:1,000; cat no. ab80588) were all from Abcam.
Construction and transfection of plasmids
and small interfering RNA (siRNA)
The overexpression plasmids (pcDNA3.1-STAT3
and PRDX5) and the control (pcDNA3.1-vector), siRNAs against
STAT3 and PRDX5 [si-STAT3(CGT CAT TAG CAG AAT
CTC ATT) and si-PRDX5(GGA ATC GAC GTC TCA AGA GGT),
respectively] and corresponding negative-control (NC) siRNAs
(si-NC, GCA GAT AGG TAG GCG TTA T) were all designed and
synthesized by Guangzhou RiboBio Co., Ltd. The process of transient
cell transfection was conducted using standard methods as per
manufacturer's instructions. Briefly, cells were maintained in
medium with fetal bovine serum (FBS, 10%) until the confluence
reached 70-80%, and all the oligonucleotides (RNA and DNA) were
then transfected into the cells using Lipofectamine 2000
(Invitrogen; Thermo Fisher Scientific, Inc.). following 48 h of
transfection, the transfection efficiency was detected by western
blot analysis.
Chromatin immunoprecipitation (ChIP)
assay
In order to explore the transcription factors that
may be involved in the regulation of PRDX5 gene expression,
the online software Jaspar (http://jaspar.genereg.net) was used to predict the
transcription factors that may bind to the promoter region. ChIP
assay was then performed using a ChIP kit (magnetic beads; Cell
Signaling Technology, Inc.) as per the manufacturer's instructions.
Briefly, DNA-protein complexes were cross-linked with 1%
formaldehyde, and 1% SDS Lysis Buffer was then added followed by
sonication. The used antibody to immunoprecipitated
anti-STAT3(1:20, ab76315; Abcam) or normal mouse
immunoglobulin G (IgG, 1:200, cat. no. 554002, BD Biosciences) was
added at 4°C for 12 h. DNA was then purified out of the
antibody-protein-DNA complex and used for PCR. Specific ChIP
primers for detailed sequences of the PRDX5 promoter were as
follows: Site 1 forward, TAT TGG ATA GCC AGG AGA ACC and reverse,
GGA ACC TCC TGC TGA GAC G (131 bp); site 2 forward, ATG TCG CCG CAC
AAA CT and reverse, CCC ACA AAC ACG AGA AGT TCC (147 bp); site 3
forward, GAA ACC GCT TTT GGT TTT AAA C and reverse, CCA ACC CTT GAC
CCA ATG AC (94 bp); site 4 forward, CTC AGG GGG TAG GAG AGC A and
reverse, GGT TTA AAA CCA AAA GCG GT (182 bp); and site 5 forward,
CTC TCC TCC CCC TCC TAG GG and reverse, TGG CCT CCA TCC CCT TCC
(188 bp). The following PCR conditions were used: 95°C for 5 min,
95°C for 30 sec, 55-60°C for 30 sec, and 72°C for 30 sec for a
total of 30 cycles, and then 72°C for 10 min. The acquired products
were observed using agarose gel electrophoresis.
Luciferase reporter assay (LRA)
The sequences (760 and 640) from upstream to the
start of the PRDX5gene were cloned into pGL3 luciferase
reporter vector (Promega Corporation), and were transfected into
the cells with Lipofectamine 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.) together with internal reference vectors,
phRL-TK. Luciferase activities were detected using a Dual
Luciferase Assay System (Promega Corporation) at 48 h following
transfection in triplicate, and Renilla luciferase activity
served as the internal control.
DNA methylation in vitro
PGL3-640 and PGL3-760 were treated with DNA
SssI methylase (New England Biolabs) for 4 h at 37°C; and
these plasmids were incubated similarly but without SssI
methylase (unmethylated control). The plasmids were then further
purified with a PCR product clean-up kit (Axygen). The unmethylated
or methylated activities of PGL3-640 and PGL3-760 were measured as
per the above methods.
Cell migration and invasion assay
Cell migration and invasion were detected using
Transwell chambers (Corning, Inc.) coated with or without Matrigel
(no. 356234; BD Biosciences). A total of 2×104 cells in
100 µl were added to the chambers with serum-free DMEM,
while the lower chambers were filled with culture medium containing
10% FBS in a humidified 5% CO2 atmosphere at 37°C. After
48 h, the invading or migrating cells were fixed with 4%
paraformaldehyde and stained with 0.5% crystal violet
(Sigma-Aldrich; Merck KGaA) for 20 min at room temperature. Cells
were counted under a micro-scope (Leica DM2500, Leica Microsystems,
Inc.) at ×200 magnification.
Statistical analysis
SPSS software version 17.0 (IBM Corp.) was used for
statistical analysis. All data are expressed as the means ±
standard deviation (SD) in triplicate. A paired or independent
sample t-test was used to analyze differences between 2 groups. For
the comparison of multiple groups, differences were analyzed by
one-way analysis of variance (ANOVA). When ANOVA detected
significant differences, the data of the variables of each
experimental group were compared with that of the control group
using a Dunnett's post hoc test. An χ2 test was used to
examine the association between the promoter methylation of
PRDX5 and the patient clinicopathological parameters.
Correlation analysis was performed using Spearman's correlation
coefficient (SCC). A value of P<0.05 was considered to indicate
a statistically significant difference.
Results
PRDX5 upregulation in NSCLC tissues is
associated with CpG island demethylation in the promoter
region
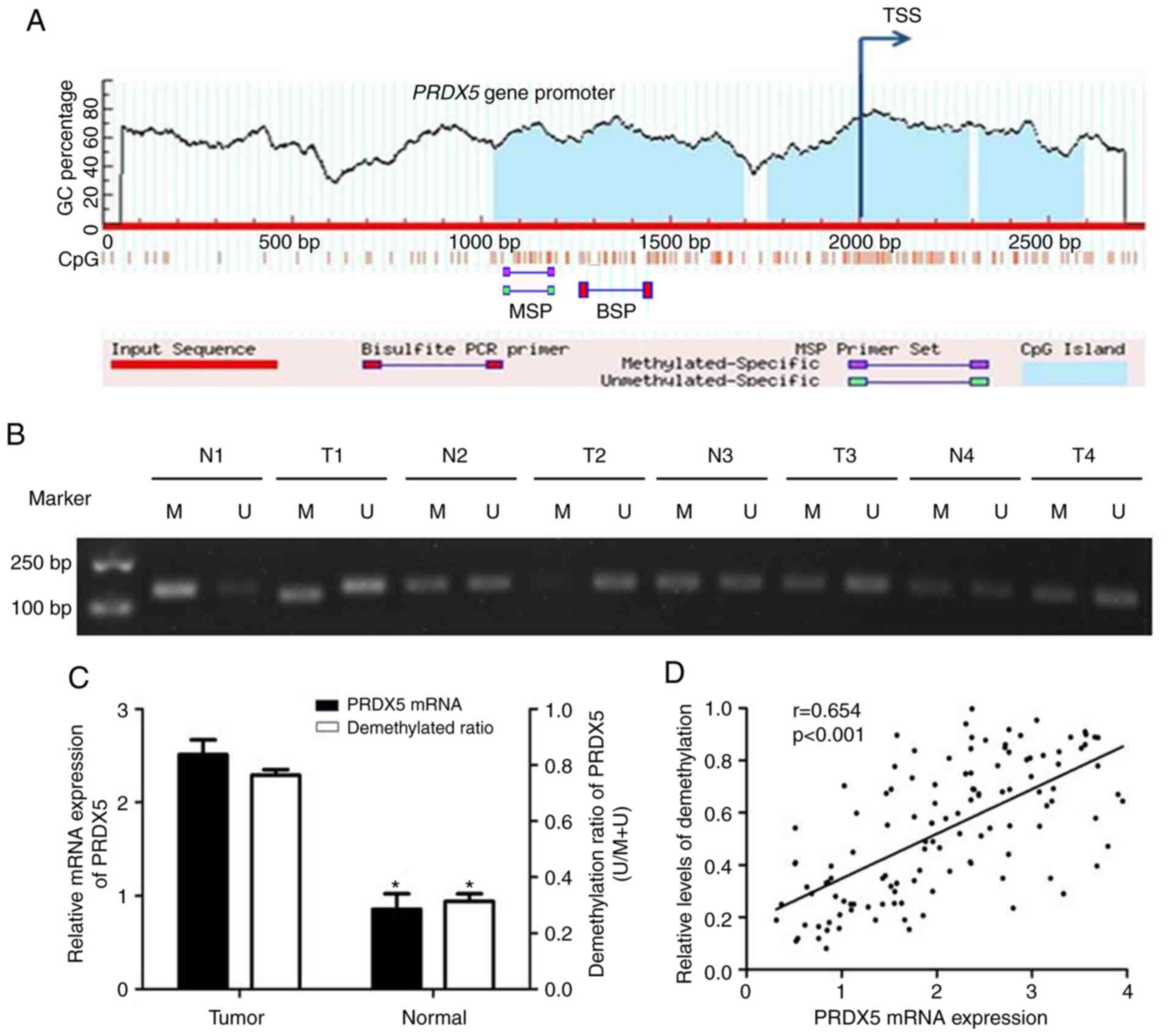
First, to determine whether PRDX5
upregulation in NSCLC tissues was associated with the demethylation
of CpG islands in the promoter region, the CpG island online
prediction software (http://www.urogene.org/index.html) was searched and it
was found that 3 CpG islands existed near the transcription start
site (Fig. 1A). Since the
promoter region upstream of TSS may have more transcription factors
binding to start gene transcription, and the length of the first
CpG island is longer, the 1st one was selected to design primers.
Subsequently, 121 pairs of NSCLC tissues and adjacent non-cancerous
tissues were analyzed to determine the methylation status of CpG
islands in the PRDX5 promoter region by MSP. The results
revealed that 79 of the 121 patients with NSCLC exhibited
PRDX5 promoter demethylation, and that the demethylation was
associated with tumor, node and metastasis (TNM) stage (P=0.027),
but not with age, sex, lymph nodes or differentiation (Table I). A total of 8 representative
cases of MSP results are presented in Fig. 1B. All the 121 tumor tissues
exhibited a significantly higher demethylation ratio in the
PRDX5promoter region compared with that in adjacent
non-cancerous tissues (P<0.05; Fig. 1C). Additionally, the results of
RT-qPCR revealed that PRDX5 mRNA expression was upregulated
in NSCLC tissues compared with adjacent tissues (P<0.05,
Fig. 1C). As shown in Fig. 1D, PRDX5 mRNA expression
positively correlated with the demethylation status of the promoter
region.
Hypomethylation of specific CpG sites
within the PRDX5 promoter region promotes transcriptional
activities under conditions of OS
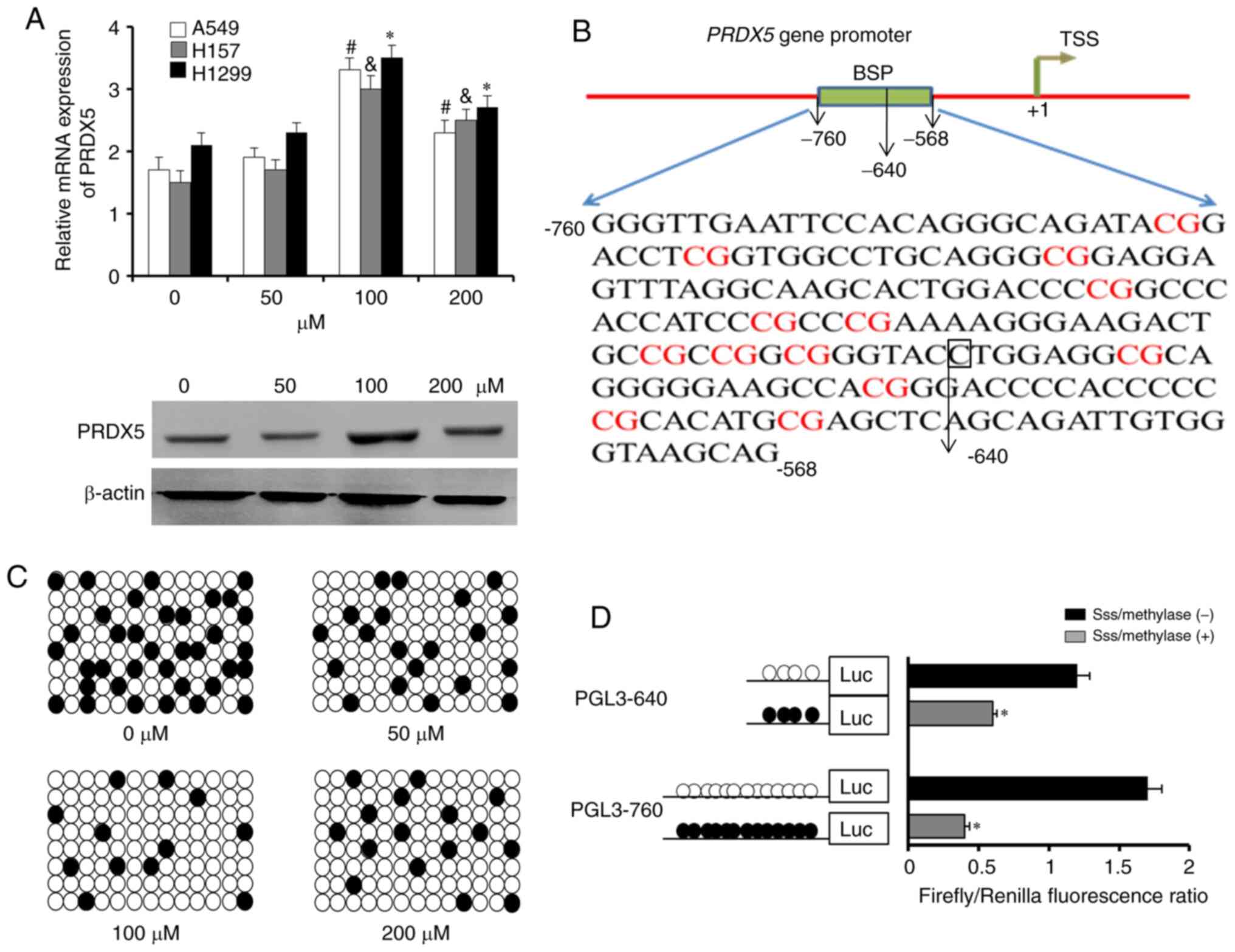
To obtain further details of the demethylation
status of specific CpG sites within the PRDX5 promoter
region under conditions of OS, an in vitro model of ROS was
induced using H2O2 (Sigma-Aldrich; Merck
KGaA). A 193 bp length of PCR products (-760 to -568 bp) was
analyzed following sodium bisulfite treatment as part of the BSP
method. The lung cancer cell lines, A549, H1299 and H157, were
pre-treated with various concentrations of
H2O2 (0, 50, 100 or 200 µM). Following
30 min of stimulation, the results of RT-qPCR for relative
PRDX5 mRNA expression revealed the significant upregulation
of PRDX5 expression in the 100 µM
H2O2 group in the 3 cell lines (Fig. 2A, upper panel); similar results
were obtained for protein expression in the H1299 cells (Fig. 2A, bottom panel). The H1299 cells
that had been treated with 100 µM H2O2
were then selected for BSP. As shown in Fig. 2B, the sequencing region contained
13 CpG sites from -760 to -568 bp. The results of BSP revealed
lower demethylation frequencies in H1299 cells treated with 0
µM H2O2, but maximum demethylation in
those treated with 100 µM H2O2
(Fig. 2C).
Finally, to determine which CpG sites were
responsible for the demethylation-related activation of the
PRDX5 gene under conditions of OS, two PRDX5 gene
promoter regions (PGL3-640 and PGL3-760) were constructed; these
were then treated with SssImethylase in vitro and
transfected into H1299 cells that had been pre-treated with 100
µM H2O2 (Fig. 2D). Compared with the treated
promoter constructs, the untreated constructs exhibited a
significantly greater demethylation and promoter activity. No
marked differences in the promoter activity of PGL3-640 or PGL3-760
were observed between the SssImethylase-treated and
untreated groups. These results indicated that the region of the
CpG sites from -640 to -568 bp may play as an important role in
regulating PRDX5 gene transcription.
Promoted binding of STAT3 to the PRDX5
gene promoter region due to DNA demethylation
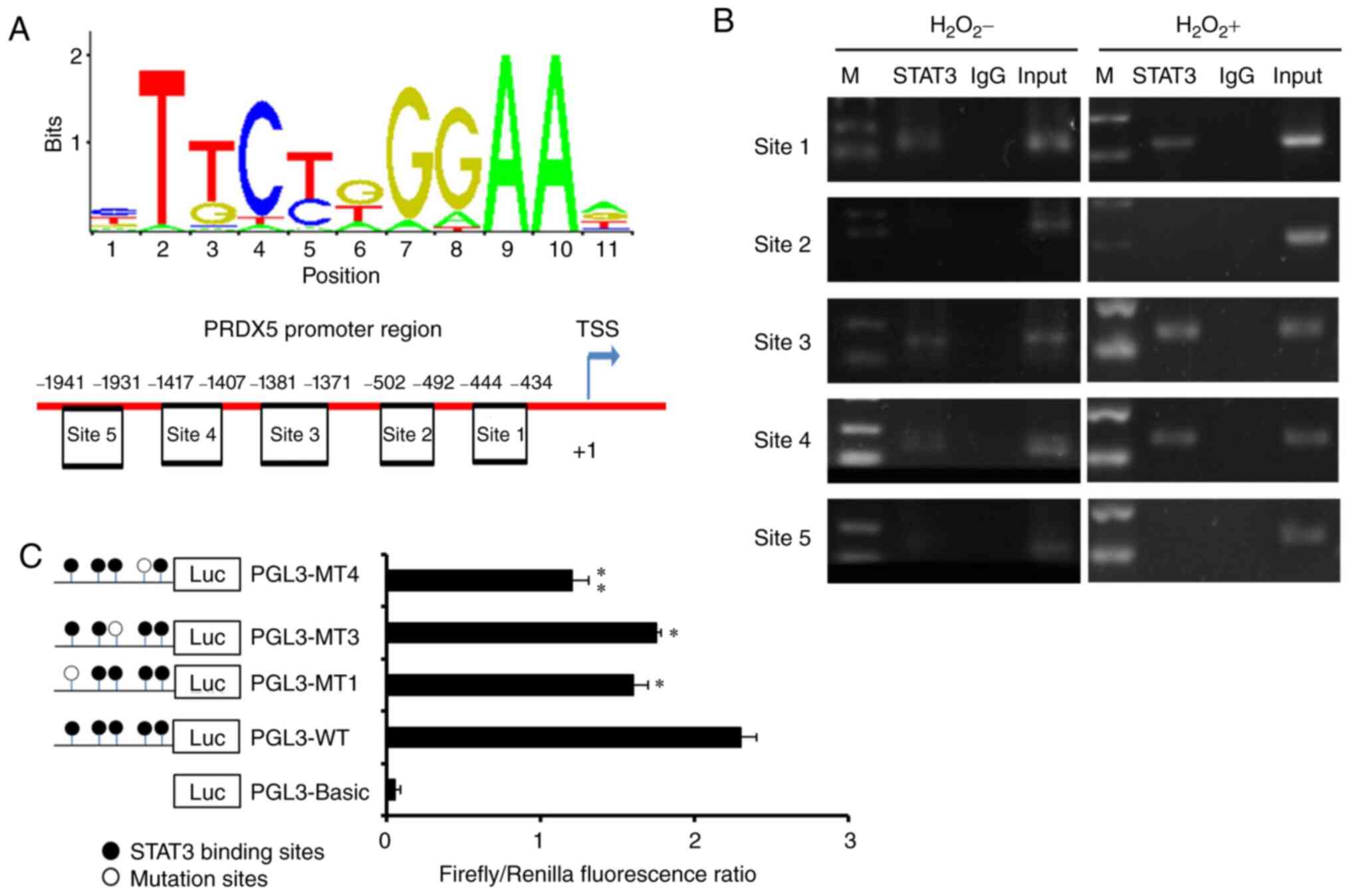
To explore related TFs that may be involved in the
regulation of PRDX5 expression, the TFs that could
potentially bind to the TF binding site near the promoter region
were predicted using the website, http://jaspar.genereg.net/ (Fig. 3A). A total of 5 potential
STAT3 binding sites were screened near the PRDX5
promoter region (site 1, -444 to -434; site 2, -502 to -492; site
3, -1,381 to -1,371; site 4, -1,417 to -1,407; site 5, -1,941 to
-1,931 bp). Subsequently, the actual binding of STAT3 to the
transcriptional binding sites of the DNA promoter region under
conditions of OS was examined by ChIP assay. The results revealed
that STAT3 could obviously bind to sites 1, 3 and 4, but not
to sites 2 or 5 (Fig. 3B and
C).
Finally, to further verify the effective binding
sites indicated by the results of ChIP assay, mutant plasmids that
were directed against each of sites 1, 3 and 4 were constructed.
The results of luciferase detection revealed a significant decrease
in the PGL3-MT1 and PGL3-MT4 regions (Fig. 3C). This indicated that sites 1 and
4 were the effective TF binding sites for PRDX5 gene
transcription.
STAT3-regulated PRDX5 signaling affects
the migration and invasion of lung cancer cells under conditions of
OS
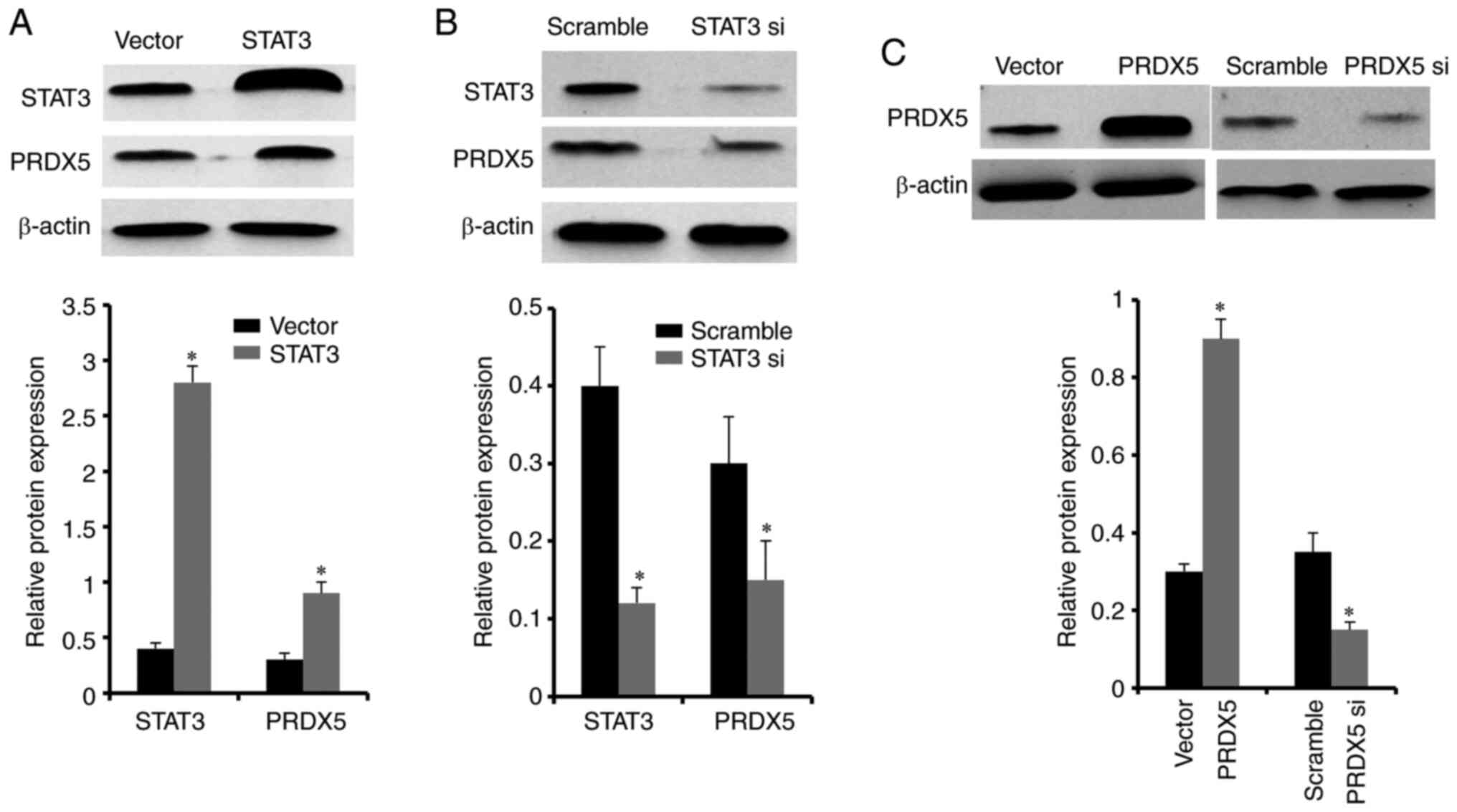
To further demonstrate that STAT3 was
involved in regulating PRDX5 expression, STAT3 siRNA
and the expression plasmid, pcDNA3.1-STAT3 were constructed.
These were then transfected each into H1299 cells pre-treated with
100 µM H2O2. After 48 h, the results
of western blot analysis revealed that STAT3 protein
expression was significantly decreased in the STAT3 siRNA
group compared with the scramble group, while protein expression
levels were significantly increased in the group transfected with
pcDNA3.1-STAT3 compared with the control vector group. In
addition, STAT3 knockdown significantly decreased the
protein expression of PRDX5, while the overexpression of
STAT3 significantly increased the protein level of
PRDX5 (Fig. 4A and B).
To further clarify the effects of PRDX5 on
the migration and invasion of lung cancer cells under conditions of
OS, PRDX5 siRNA and the corresponding overexpression plasmid
pcDNA3.1-PRDX5 were constructed and transfected into H1299
cells pre-treated with 100 µM H2O2.
The results indicated that the PRDX5 protein levels significantly
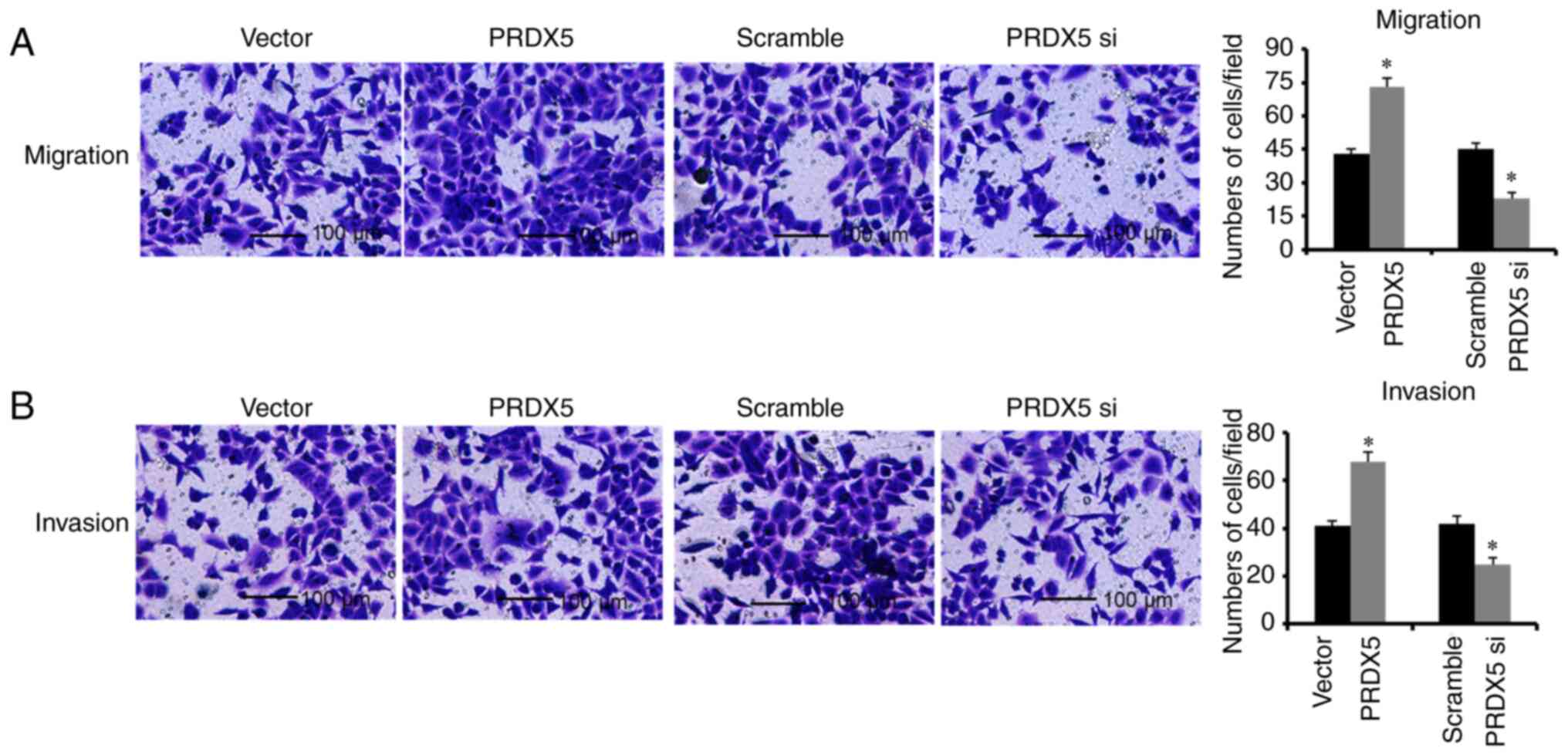
decreased following transfection with PRDX5 siRNA (Fig. 4C). In addition, the cell migratory
and invasive abilities were also significantly suppressed following
the knockdown of PRDX5 (Fig.
5). By contrast, PRDX5 protein expression significantly
increased following transfection with pcDNA3.1-PRDX5
(Fig. 4C), and the cell migratory
and invasive abilities were also increased (Fig. 5). At the same time, when
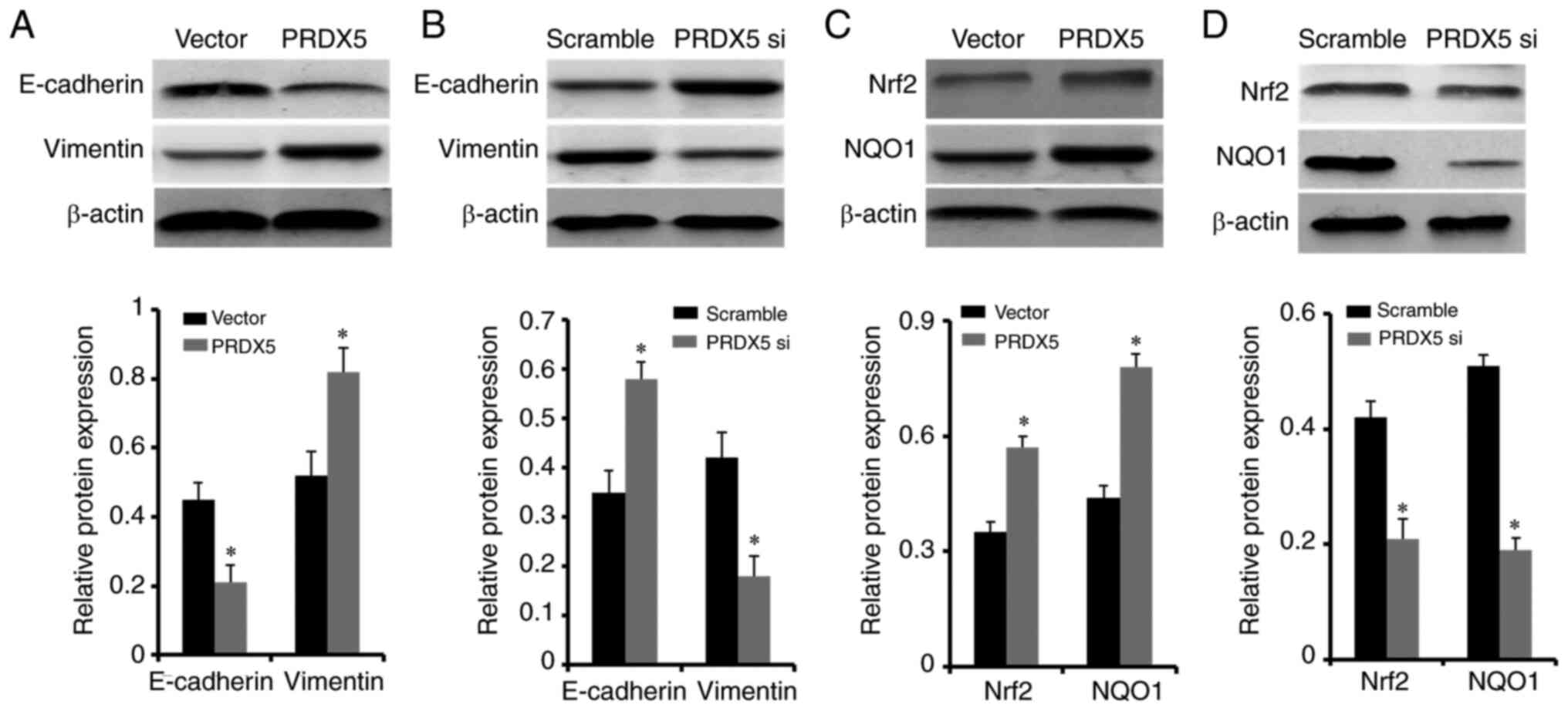
PRDX5 was overexpressed in lung cancer cells under
conditions of OS, the levels of the epithelial-mesenchymal
transition (EMT) biomarkers, E-cadherin and vimentin, were
significantly decreased and increased, respectively (Fig. 6A). Conversely, PRDX5
knockdown resulted in significantly increased E-cadherin and
decreased vimentin protein expression levels (Fig. 6B). The above-mentioned results
indicated that PRDX5 may be regulated by STAT3, and
that it affected the migratory and invasive abilities of lung
cancer cells under conditions of OS, promoting the EMT
phenotype.
PRDX5 activates the Nrf2 signaling
pathway in lung cancer cells under conditions of OS
To verify that PRDX5 can activate Nrf2
signaling under conditions of OS, PRDX5 siRNA and
pcDNA3.1-PRDX5 were transfected into H1299 cells pre-treated
with 100 µM H2O2. The expression of
key molecules of OS, Nrf2 and NQO1, was significantly upregulated
when the cells were transfected with pcDNA3.1-PRDX5
(Fig. 6C). By contrast,
PRDX5 knockdown resulted in significantly decreased protein
expression levels of Nrf2 and NQO1 (Fig. 6D). These results indicated that
PRDX5 may be involved in the activation of the Nrf2
signaling pathway in lung cancer cells under conditions of OS.
Discussion
Lung cancer is currently a malignancy with an
unclear molecular mechanism, particularly as regards NSCLC
(25). As NSCLC progresses, ROS
levels are increased in cancer cells compared with in normal cells,
mainly due to the abnormal metabolic level in tumors; this leads to
the overproduction of ROS in ischemic hypoxic environments. As a
peroxiredoxin family member, PRDX5 plays an important role
in maintaining intracellular ROS or peroxide levels induced by
cytokines (26). In the present
study, H1299 cells were pre-treated with 100 µM
H2O2 to establish the ROS model in
vitro. Although this induced ROS and the upregulated endogenous
expression of PRDX5, this does not have an impact on the
results of subsequent experiments, as it is the result of the
comparison between the two groups. As the results were based on the
comparison between the 2 groups, and are from an epigenetic
perspective, it was clarified that the upregulated expression of
the PRDX5 gene mainly participated in the migration and
invasion of lung cancer cells under conditions of OS and activated
the Nrf2 signaling pathway.
DNA methylation is one of the most common means of
epigenetic regulation. For example, it plays an important role in
the modulation of cancers and inflammation or tissue-damaging pain
(27-29). In the present study, it was found
that PRDX5 was upregulated due to the demethylation of its
promoter region, and at the same time, that different OS levels led
to varying degrees of demethylation and PRDX5 mRNA
expression. This demonstrated that OS promoted PRDX5
expression by demethylating the promoter region. There were 2
possibilities: The one is that the number of samples was still not
sufficient to explain the problem. The other is that it was really
no association between the two.
Using ChIP assay, it was verified that STAT3
functioned as a TF in binding to the PRDX5 gene promoter
region. In addition, to the best of our knowledge, the present
study also demonstrated for the first time that the ability of
STAT3 to bind to this region was markedly enhanced in NSCLC
under conditions of OS. This enhanced affinity promoted
PRDX5 expression, while the mutation of the binding site
between the PRDX5 gene promoter region and STAT3
resulted in a significantly decreased PRDX5expression. From
an epigenetic perspective, these results may have been caused by
the hypomethylation of the PRDX5gene promoter region,
leading to enhanced affinity between binding sites. Choi et
al (30) reported that the
overexpression of PRDX5 suppressed the TGF-β induced
upregulation of STAT3 phosphorylation. Perhaps under certain
specific conditions (tumor or non-tumor conditions), PRDX5
and STAT3 may have a negative feedback regulation, forming a
negative feedback regulation loop to perform specific functions.
The present study did not notice this point; thus, the authors aim
to continue to explore the association between the 2 genes in the
future. In addition, it is hypothesized that the mutation of all 3
binding sites will not disrupt STAT3 binding completely, for
the binding sites are only acquired by prediction and
identification by experiments; it can also not be ruled out that
there may be other binding sites.
EMT is an important biological process in the
migration and invasion of malignant tumor cells derived from ECs
(31). It plays an important role
in embryonic development (32),
chronic inflammation (33),
cancer metastasis (34) and a
variety of fibrotic diseases (35), the main characteristic of which is
a decrease in the expression of cell adhesion molecules (CAMs),
such as E-cadherin. The cytoskeleton of cytokeratin is transformed
into vimentin, which has the morphological characteristics of
mesenchymal cells. Ahn et al reported that PRDX5
promoted EMT in colon cancer (36). In contrast to this aforementioned
study, the present study found that the hypomethylation of
PRDX5 promoted STAT3 binding, and promoted cell
migration, invasion and EMT progression, which manifested as
E-cadherin downregulation and vimentin upregulation. By contrast,
the lower expression of PRDX5 suppressed cell migration,
invasion and EMT. These results indicated that PRDX5 may
affect cell migration and invasion by activating EMT.
The Nrf2 antioxidant-responsive element
(ARE) pathway is one of the most important endogenic anti-OS
pathways to be discovered to date. There is evidence to indicate
that when activated, this pathway can inhibit the degradation of
Nrf2 protein mediated by ubiquitin, stabilize the
concentration of Nrf2 protein in cytoplasm, and enhance the
transcriptional activity of Nrf2 protein (37,38). The results of the present study
revealed that PRDX5 overexpression under conditions of OS
significantly increased the protein levels of Nrf2 and
NQO1, which are key proteins of the Nrf2/ARE
signaling pathway. These results may provide a strategy for the
treatment of NSCLC.
In conclusion, the present study demonstrated that
the ROS-mediated hypomethylation of PRDX5 enhanced
STAT3 binding affinity with the PRDX5 gene promoter
region, and promoted cell migration and invasion, as well as the
activation of the Nrf2 signaling pathway in NSCLC. The
demethylation status of the PRDX5 promoter may be used as an
NSCLC epigenetic biomarker and STAT3/PRDX5 signaling may
prove to be a potential target in the treatment of NSCLC.
Funding
This study was supported by the National Natural
Science Foundation of China (grant no. 8157101409) and the Nantong
Science and Technology Project (grant no. MS12017010-2).
Availability of data and materials
All data generated or analyzed during the current
study are included in this published article.
Authors' contributions
XC and QX designed the study. XC, XMC and WZX
performed the experiments. XMC and BL collected the data and
performed the analysis. BZ and QW collected the data. XC and QX
wrote the article. All authors read and approved the
manuscript.
Ethics approval and consent to
participate
The study protocol was approved by the Ethics
Committee of Affiliated Hospital of Nantong University. Written
informed consent was obtained from all patients prior to sample
collection.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgments
Not applicable.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2017. CA Cancer J Clin. 67:7–30. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Liu SV and Giaccone G: Lung cancer in
2013: Refining standard practice and admitting uncertainty. Nat Rev
Clin Oncol. 11:69–70. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Song X, Zhao C, Jiang L, Lin S, Bi J, Wei
Q, Yu L, Zhao L and Wei M: High PITX1 expression in lung
adenocarcinoma patients is associated with DNA methylation and poor
prognosis. Pathol Res Pract. 214:2046–2053. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Zhang X, Rong X, Chen Y and Su L:
Methylation-mediated loss of SFRP2 enhances invasiveness of
non-small cell lung cancer cells. Hum Exp Toxicol. 37:155–162.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Chen L, Wang Y, Liu F, Xu L, Peng F, Zhao
N, Fu B, Zhu Z, Shi Y, Liu J, et al: A systematic review and
meta-analysis: Association between MGMT hypermethylation and the
clinicopathological characteristics of non-small-cell lung
carcinoma. Sci Rep. 8:14392018. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Raungrut P, Petjaroen P, Geater SL,
Keeratichananont W, Phukaoloun M, Suwiwat S and Thongsuksai P:
Methylation of 14-3-3s gene and prognostic significance of 14-3-3s
expression in non-small cell lung cancer. Oncol Lett. 14:5257–5264.
2017.PubMed/NCBI
|
|
7
|
Zhou Y, Qiu XP, Li ZH, Zhang S, Rong Y,
Yang GH and Fang-Zheng: Clinical significance of aberrant
cyclin-dependent kinase-like 2 methylation in hepatocellular
carcinoma. Gene. 683:35–40. 2019. View Article : Google Scholar
|
|
8
|
Peters I, Dubrowinskaja N, Hennenlotter J,
Antonopoulos WI, Von Klot CAJ, Tezval H, Stenzl A, Kuczyk MA and
Serth J: DNA methylation of neural EGFL like 1 (NELL1) is
associated with advanced disease and the metastatic state of renal
cell cancer patients. Oncol Rep. 40:3861–3868. 2018.PubMed/NCBI
|
|
9
|
Li F, Xue ZY, Yuan Y, Huang SS, Fan YH,
Zhu X and Wei L: Upregulation of CXCR4 through promoter
demethylation contributes to inflammatory hyperalgesia in rats. CNS
Neurosci Ther. 24:947–956. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Jiang C, Guo J, Cheng H and Feng YH:
Induced expression of endogenous CXCR4 in iPSCs by targeted CpG
demethylation enhances cell migration toward the ligand CXCL12.
Inflammation. 42:20–34. 2019. View Article : Google Scholar
|
|
11
|
Kang D and Hamasaki N: Mitochondrial
oxidative stress and mitochondrial DNA. Clin Chem Lab Med.
41:1281–1288. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Nogueira V and Hay N: Molecular pathways:
Reactive oxygen species homeostasis in cancer cells and
implications for cancer therapy. Clin Cancer Res. 19:4309–4314.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Lin S, Li Y, Zamyatnin AA Jr, Werner J and
Bazhin AV: Reactive oxygen species and colorectal cancer. J Cell
Physiol. 233:5119–5132. 2018. View Article : Google Scholar
|
|
14
|
Knoops B, Goemaere J, Van der Eecken V and
Declercq JP: Peroxiredoxin 5: Structure, mechanism, and function of
the mammalian atypical 2-Cys peroxiredoxin. Antioxid Redox Signal.
15:817–829. 2011. View Article : Google Scholar
|
|
15
|
Kim B, Park J, Chang KT and Lee DS:
Peroxiredoxin 5 prevents amyloid-beta oligomer-induced neuronal
cell death by inhibiting ERK-Drp1-mediated mitochondrial
fragmentation. Free Radic Biol Med. 90:184–194. 2016. View Article : Google Scholar
|
|
16
|
Kim B, Kim YS, Ahn HM, Lee HJ, Jung MK,
Jeong HY, Choi DK, Lee JH, Lee SR, Kim JM and Lee DS: Peroxiredoxin
5 overexpression enhances tumorigenicity and correlates with poor
prognosis in gastric cancer. Int J Oncol. 51:298–306. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Byun JM, Kim SS, Kim KT, Kang MS, Jeong
DH, Lee DS, Jung EJ, Kim YN, Han J, Song IS, et al: Overexpression
of peroxiredoxin-3 and -5 is a potential biomarker for prognosis in
endometrial cancer. Oncol Lett. 15:5111–5118. 2018.PubMed/NCBI
|
|
18
|
Baumann C, Bonilla WV, Fröhlich A,
Helmstetter C, Peine M, Hegazy AN, Pinschewer DD and Löhning M:
T-bet- and STAT4-dependent IL-33 receptor expression directly
promotes antiviral Th1 cell responses. Proc Natl Acad Sci USA.
112:4056–4061. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Koch MA, Thomas KR, Perdue NR, Smigiel KS,
Srivastava S and Campbell DJ: T-bet(+) Treg cells undergo abortive
Th1 cell differentiation due to impaired expression of IL-12
receptor β2. Immunity. 37:501–510. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Akira S, Nishio Y, Inoue M, Wang XJ, Wei
S, Matsusaka T, Yoshida K, Sudo T, Naruto M and Kishimoto T:
Molecular cloning of APRF, a novel IFN-stimulated gene factor 3
p91-related transcription factor involved in the gp130-mediated
signaling pathway. Cell. 77:63–71. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Jiang LH, Hao YL and Zhu JW: Expression
and prognostic value of HER-2/neu, STAT3 and SOCS3 in
hepatocellular carcinoma. Clin Res Hepatol Gastroenterol.
43:282–291. 2019. View Article : Google Scholar
|
|
22
|
Fathi N, Rashidi G, Khodadadi A, Shahi S
and Sharifi S: STAT3 and apoptosis challenges in cancer. Int J Biol
Macromol. 117:993–1001. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Chu Y, Wang Y, Peng W, Xu L, Liu M, Li J,
Hu X, Li Y, Zuo J and Ye Y: STAT3 activation by IL-6 from
adipose-derived stem cells promotes endometrial carcinoma
proliferation and metastasis. Biochem Biophys Res Commun.
500:626–631. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
25
|
Raju S, Joseph R and Sehgal S: Review of
checkpoint immunotherapy for the management of non-small cell lung
cancer. Immunotargets Ther. 7:63–75. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Sabharwal SS, Waypa GB, Marks JD and
Schumacker PT: Peroxiredoxin-5 targeted to the mitochondrial
intermembrane space attenuates hypoxia-induced reactive oxygen
species signal-ling. Biochem J. 456:337–346. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Pan Y, Liu G, Zhou F, Su B and Li Y: DNA
methylation profiles in cancer diagnosis and therapeutics. Clin Exp
Med. 18:1–14. 2018. View Article : Google Scholar
|
|
28
|
Zhang HH, Hu J, Zhou YL, Qin X, Song ZY,
Yang PP, Hu S, Jiang X and Xu GY: Promoted interaction of nuclear
factor-kB with demethylated purinergic P2X3 receptor gene
contributes to neuropathic pain in rats with diabetes. Diabetes.
64:4272–4284. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Zhou YL, Jiang GQ, Wei J, Zhang HH, Chen
W, Zhu H, Hu S, Jiang X and Xu GY: Enhanced binding capability of
nuclear factor-kB with demethylated P2X3 receptor gene contributes
to cancer pain in rats. Pain. 156:1892–1905. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Choi HI, Ma SK, Bae EH, Lee JU and Kim SW:
Peroxiredoxin 5 protects TGF-β induced fibrosis by inhibiting stat3
activation in rat kidney interstitial fibroblast cells. PLoS One.
11:e01492662016. View Article : Google Scholar
|
|
31
|
Chen T, You Y, Jiang H and Wang ZZ:
Epithelial-mesenchymal transition (EMT): A biological process in
the development, stem cell differentiation, and tumorigenesis. J
Cell Physiol. 232:3261–3272. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Acloque H, Ocaña OH, Matheu A, Rizzoti K,
Wise C, Lovell-Badge R and Nieto MA: Reciprocal repression between
Sox3 and snail transcription factors defines embryonic territories
at gastrulation. Dev Cell. 21:546–558. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Correa-Costa M, Andrade-Oliveira V, Braga
TT, Castoldi A, Aguiar CF, Origassa CST, Rodas ACD, Hiyane MI,
Malheiros DMAC, Rios FJO, et al: Activation of platelet- activating
factor receptor exacerbates renal inflammation and promotes
fibrosis. Lab Invest. 94:455–466. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Ruscetti M, Quach B, Dadashian EL,
Mulholland DJ and Wu H: Tracking and functional characterization of
epithelial-mesen-chymal transition and mesenchymal tumor cells
during prostate cancer metastasis. Cancer Res. 75:2749–2759. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Grande MT, Sanchez-Laorden B, Lopez-Blau
C, De Frutos CA, Boutet A, Arévalo M, Rowe RG, Weiss SJ,
López-Novoa JM and Nieto MA: Snail1-induced partial
epithelial-to-mesenchymal transition drives renal fibrosis in mice
and can be targeted to reverse established disease. Nat Med.
21:989–997. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Ahn HM, Yoo JW, Lee S, Lee HJ, Lee HS and
Lee DS: Peroxiredoxin 5 promotes the epithelial-mesenchymal
transition in colon cancer. Biochem Biophys Res Commun.
487:580–586. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Stewart D, Killeen E, Naquin R, Alam S and
Alam J: Degradation of transcription factor Nrf2 via the
ubiquitin-proteasome pathway and stabilization by cadmium. J Biol
Chem. 278:2396–2402. 2003. View Article : Google Scholar
|
|
38
|
Nguyen T, Sherratt PJ, Huang HC, Yang CS
and Pickett CB: Increased protein stability as a mechanism that
enhances Nrf2-mediated transcriptional activation of the
antioxidant response element. Degradation of Nrf2 by the 26 S
proteasome. J Biol Chem. 278:4536–4541. 2003. View Article : Google Scholar
|