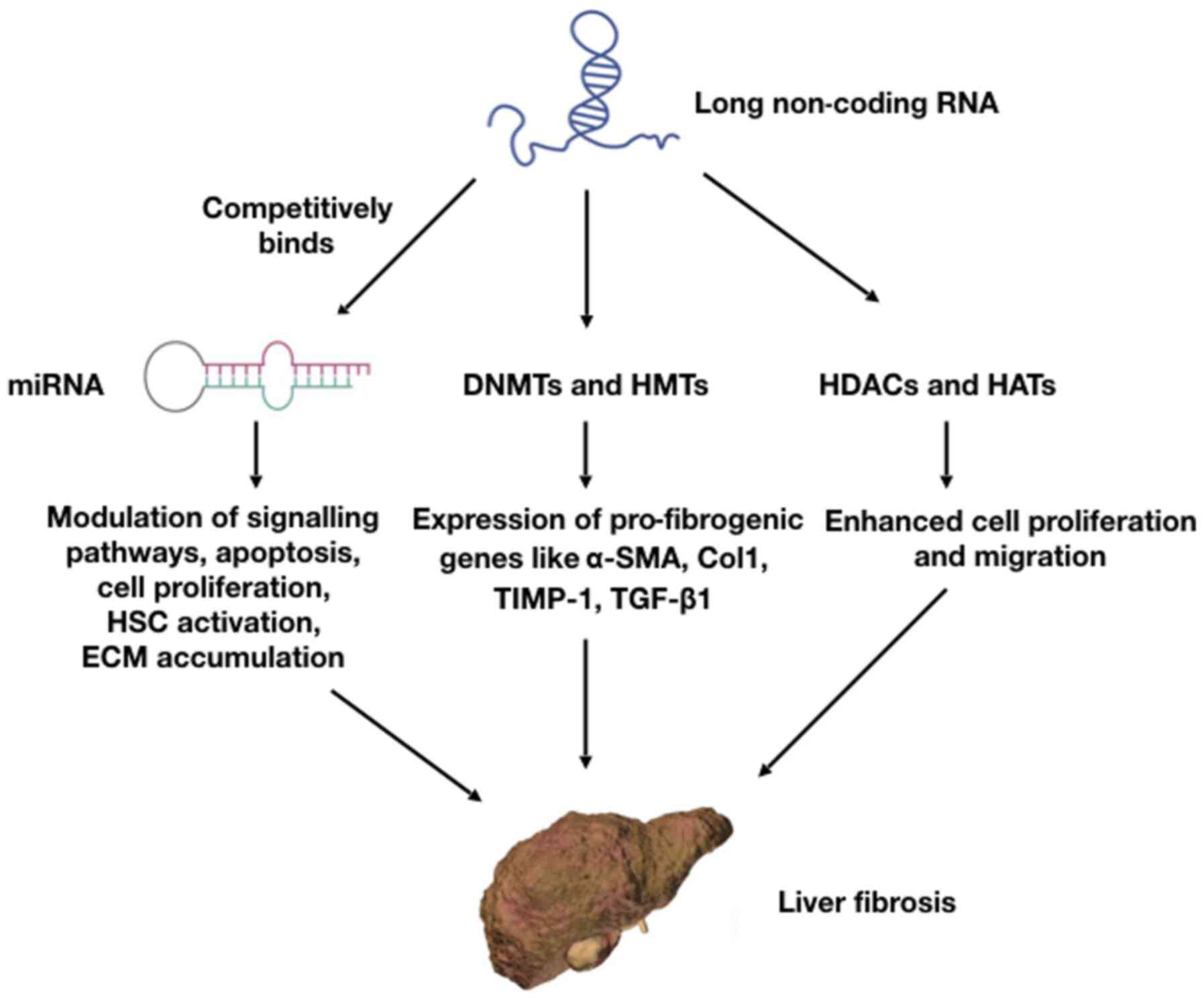
Hepatic fibrosis results from an aberrant wound
healing process in the liver. In general, fibrosis results from an
accumulation of extracellular matrix (ECM) proteins.
Pathogenetically, apart from the abnormal deposition of ECM
proteins, the improper degradation of ECM proteins also occurs
(1,2). Liver fibrosis results from
persistent wound repair and regenerative processes due to continued
hepatic injury caused by chronic liver diseases, such as
non-alcoholic steatohepatitis (NASH), the excessive consumption of
alcohol, hepatitis B and C infection, obstruction in the bile
ducts, etc. (3,4), ultimately leading to cirrhosis and
predisposing affected patients towards the development of
malignancies, such as hepatocellular carcinoma (HCC) (4-7).
Although extensive research has been made into liver fibrosis (as
discussed below), the mechanisms governing the process are not yet
fully understood.
Hepatic stellate cells (HSCs), which are primarily
involved in growth, regeneration and differentiation comprise 5-8%
of total liver cells (8). The
activation of HSCs is a key step in liver fibrosis, as it leads to
the accumulation of the ECM proteins, α-smooth muscle actin (α-SMA)
and type I collagen (9,10). Non-activated or quiescent HSCs
lose their vitamin A stores and transdifferentiate into fibrogenic
myofibroblast-like cells following liver injury (11-14). Therefore, the suppression of HSC
activation is crucial for the therapy of liver fibrosis.
Transcriptome analysis, gene expression profiling by
microarray and RNA sequencing have increasingly pointed to the role
of non-coding RNAs (ncRNAs) as crucial regulators of gene
expression and signal transduction (15). In liver carcinogenesis and
fibrosis, ncRNAs have emerged as key regulators of gene expression
(16-18). Therefore, ncRNAs may also be
viewed as putative biomarkers for diagnosis and therapeutic targets
for the treatment of liver fibrosis. Among the ncRNAs, major
players include microRNAs (miRNAs or miRs) and long non-coding RNAs
(lncRNAs). lncRNAs with sizes of ≥200 nucleotides, are categorized
into 4 subgroups as follows: Exonic, intronic, intergenic and
bidirectional, depending on their placement and orientation with
respect to protein coding genes. In liver fibrosis, lncRNAs may act
as competitive endogenous RNAs (ceRNA), sponging miRNAs. This
action releases the inhibition imposed by miRNAs on mRNA
translation, thereby indirectly regulating gene expression. Other
members of the ncRNA group include circular RNAs, PIWI-interacting
RNAs, etc. (19).
Epigenetics encompass the changes in gene
expression, that do not involve mutations in the DNA sequence.
Epigenetic alterations are long-lasting and are heritable.
Epigenetic alterations bring about changes in DNA properties along
with transcriptional regulation. The epigenetic regulation of genes
is significant in terms of gene expression and physiological
function. Furthermore, epigenetic alterations are key pathogenetic
mechanisms involved in various, if not all chronic diseases
(20) involving all organs, such
as skeletal muscle (21), the
brain (22), heart (23), ovaries (24) and bones (25).
Epigenetic regulators can turn 'on or off' the
expression of genes, depending on the prevailing microenvironment.
Along with ncRNAs, other epigenetic processes include DNA and
histone methylations and other biochemical mechanisms, causing
alterations of DNA and histones. Such processes control several
biological processes, such as genome imprinting, transposon
mediated gene silencing, X chromosome inactivation, etc. (26). DNA methylation is catalyzed by DNA
methyltransferases (DNMT). There are 3 DNMTs, namely DNMT1, DNMT3a
and DNMT3b. Histone methylation is carried out as a balancing act
between histone methyltransferase and histone dimethyl transferase
(26). Additional epigenetic
histone modification involves the addition/removal of acetyl groups
on histone proteins. Histone acetylation is carried out by a group
of enzymes known as histone acetyl transferases (HATs) and
deacetylation is carried by histone deacetylases (HDACs). These 2
enzymes play important regulatory roles in HSC activation and the
development of liver fibrosis (27). Other less well-studied mechanisms
include ubiquitination, NEDDylation and SUMOylation. The targeting
of ubiquitin enzymes has been shown to improve the physiology in
liver fibrosis (28).
Transforming growth factor-β (TGF-β) is a cytokine secreted by the
immune system. It has been demonstrated that the TGF-β signaling
pathway plays a crucial role in the development and progression of
liver fibrosis (29). The dynamic
interactions of lncRNAs with various pathways, such as miRNAs, DNA,
histone methylation and histone acetylation are illustrated in
Fig. 1.
The present review focuses on the role of lncRNAs in
the promotion and inhibition of liver fibrosis. Furthermore, the
interaction of lncRNAs with signaling pathways and the regulation
of liver fibrosis through DNA methylation and acetylation is
explored. The present review also discusses possible therapeutic
implications of such processes in the treatment of liver
fibrosis.
The transcriptome profiling of quiescent and
activated HSCs lead to a better understanding of the mechanistic
roles of lncRNAs in the promotion or inhibition of liver fibrosis
(30,31). A large volume of data have been
obtained from the genome-wide screening of HSCs and tissues from
human and animal models with respect to the expression patterns of
lncRNAs. Understanding these expression patterns and their
resulting regulation of cellular pathways are important for the
development of targeted therapies for liver fibrosis. As will be
evident from the discussions below, lncRNAs play diverse roles in
mediating liver fibrosis. Some function as activators of liver
fibrosis, whereas others may exert inhibitory effects. In addition,
some of them may also play a dual role.
lncRNAs which activate liver fibrosis are usually
upregulated or overexpressed in liver tissues affected by fibrosis.
These lncRNAs activate HSCs and promote ECM protein overexpression.
Several of these lncRNAs also activate the TGF-β signaling pathway.
As they are all upregulated, a potential therapeutic approach under
consideration is lncRNA blockade using a small molecule or siRNA.
The discussions below focus on specific lncRNAs and their roles in
such processes.
The lncRNA, nuclear paraspeckle assembly
transcript-1 (NEAT-1), may act as a miRNA sequestrating agent. In a
recent study, it was found that NEAT-1 regulated the localization
of miR-29b to the cytoplasm (32). Insulin like growth factor binding
protein related protein 1 (IGFBPrP1) interacts with the TGF-β
signaling pathway to promote liver fibrosis (33). IGFBPrP1 induces the activation and
autophagy of HSCs, which is regulated by Atg9a and NEAT-1, while
miR-29b inhibits such a process (31), indicating that the
NEAT-1/Atg9a/miR-29b pathway regulates liver fibrosis. NEAT-1
overexpression and its downstream effects on liver fibrosis are
further prevented by Kruppel like factor 6 (KLF6) knockdown and
miR122 overexpression (34).
Therefore, NEAT-1/miR-122/KLF6 may function as another regulatory
mechanism for liver fibrosis, modulating multiple signaling
pathways.
Hox transcript antisense RNA (HOTAIR) has been found
to be overexpressed in carbon tetrachloride (CCL4)-induced liver
fibrosis in human and mouse models (30). HOTAIR binds to miR-148b and
regulates the DNMT1/MEG3/p53 pathway in HSCs (35). It has also been reported that the
HOTAIR-mediated downregulation of miR-29b attenuates its epigenetic
control, inducing the hypermethylation of the phosphatase and
tensin homolog (PTEN) gene, resulting in the progression of liver
fibrosis (36). It is of further
interest to note that one miRNA; i.e, miR29b, may regulate
multiple, seemingly unrelated lncRNAs, such as NEAT-1 and HOTAIR,
leading to the development of hepatic fibrosis.
Hox A distal transcript (HOTTIP) is a lncRNA which
promotes HSC activation and fibrosis by functiong as a competitive
endogenous RNA (ceRNA) for miR-148a and miR-150, and inducing the
expression of serum response factor (SRF) (37). High HOTTIP levels reduce the
expression of miR-148a, removing its inhibitory effects and thus
increasing levels of TGF-β receptor type 1 (TGFBR1) and receptor 2
(TGFBR2), thereby promoting liver fibrosis (38).
Metastasis-associated lung adenocarcinoma transcript
1 (MALAT1) expression has been found to be upregulated in liver
fibrosis; MALAT1 regulates the expression of sirtuin 1 (SIRT1),
which deacetylates Smad3, a mediator of the TGF-β signaling pathway
and inhibits its binding to its target genes like collagen type 1
promoter (39). Through such an
activation of SIRT1, the downregulation of the TGF-β signaling
pathway and the inhibition of liver fibrosis may occur. MALAT1 also
induces mouse HSC activation via the regulation of RAS-Rac1 by
functioning as a ceRNA for miR-101b (40). In patients with NASH, MALAT1 has
been found to be highly upregulated compared to normal tissues
(41). MALAT1 can aggravate liver
fibrosis through the generation of inflammatory chemokines, such as
CXCL5 (41). Therefore, MALAT1,
through its effects on the inflammatory processes, may modulate
liver fibrosis.
Highly upregulated in liver cancer (HULC) plays an
important role in liver fibrosis. HULC promotes liver fibrosis and
is upregulated in tissues affected by non-alcoholic fatty liver
disease (NAFLD) in rat models (42). The inhibition of HULC by siRNA
results in the improvement of liver fibrosis and reduces the
apoptosis of hepatocytes through the inhibition of the MAP kinase
(MAPK) signaling pathway (42).
Small cajal body specific RNA 10 (SCARNA10) is a
lncRNA which is elevated in serum and tissues of patients with
liver fibrosis. It has also been shown to be upregulated in liver
tissues in a mouse model of liver fibrosis (43). SCARNA10 is a positive regulator of
the TGF-β signaling pathway and inhibits polycomb repressive
complex 2 (PRC2), which blocks TGF-β signaling (43). TGF-β, TGF-βR1, Smad2, Smad3 and
KLF6 levels decline upon SCARNA10 silencing (43).
Liver enriched fibrosis associated lncRNA 1 (LFAR1)
is known to promote liver fibrosis. It has been demonstrated that
TGF-β-induced hepatocyte apoptosis, CCL4-induced fibrosis and HSC
activation are reduced upon lnc-LFAR1 silencing (44). LFAR1 directly binds to the Smad2/3
complex in the cytoplasm, and activates the TGF-β and NOTCH
pathways (44). Another lncRNA,
SCRG1, is also highly upregulated in fibrotic human liver tissues
(45). SCRG1 binds to RNA binding
protein tristetraprolin (TTP) and accelerates liver fibrosis
(45). The overexpression of TTP
leads to the reduction of lncRNA SCRG1 and increases the
degradation of matrix metallopeptidase 2 (MMP2) and TNF-α mRNAs,
and vice versa (45).
In humans, small nuclear RNA host gene 7 (SNHG7) is
significantly upregulated in fibrotic liver tissues (45). The knockdown of SNHG7 prevents HSC
activation and collagen overproduction (46). SNHG7 binds to miR-378a-3p.
Blocking miR-378a-3p production releases its inhibitory effects on
liver fibrosis induced by SNHG silencing. SNHG7 further activates
the Wnt/β-catenin pathway to promote liver fibrosis. miR-378a-3p
targets disheveled segment polarity 2 (DVL2) protein, which is a
component of the SNHG7-mediated Wnt/β-catenin pathway of liver
fibrosis (46).
Plasmocytoma variant translocation 1 (PVT1) is a
lncRNA which is upregulated in fibrotic liver tissues and activates
HSCs (47). PVT1 knockdown
reduces type I collagen and α-SMA levels and inhibits HSC
activation (47). PVT1
competitively binds to miR-152 and inhibits patched 1 (PTCH1)
expression, which in turn activates the Hedgehog pathway, and
promotes cell migration and liver fibrosis (47). Activated by transforming growth
factor beta (ATB) is a lncRNA which is upregulated in hepatitis C
virus (HCV)-mediated liver fibrosis. ATB silencing induces the
aberrant expression of miR-200a and reduces β-catenin expression,
thereby suppressing HSC activation (48).
There are several lncRNAs which act as suppressors
of liver fibrosis; these are usually downregulated in liver
fibrosis, and inhibit HSC activation and migration. Conceptually,
they can be used as biomarkers. Furthermore, therapeutic strategies
involving the overexpression of these RNAs may potentially be a
therapeutic approach for liver fibrosis.
Maternally expressed gene 3 (MEG3) is a lncRNA which
is significantly downregulated in human liver fibrosis and
CCL4-induced fibrosis in experimental models (54). MEG3 prevents fibrosis following
TGF-β1-induced LX-2 cell activation. The overexpression of MEG3 in
TGF-β1-stimulated LX-2 cells induces p53 activation, cytochrome
c release and apoptosis (54). The overexpression of MEG3 also
reduces α-SMA and type I collagen levels. Furthermore, as
circulating serum MEG3 levels are negatively associated with liver
fibrosis in patients with hepatitis, it may further be a useful
diagnostic marker (55).
Smoothened (SMO) is involved in Hedgehog pathway for EMT; MEG3
binds to SMO and reduces EMT and thereby inhibits HSC activation
(56). miR-212 targets MEG3 and
acts as a negative regulator of MEG3. Hence, MEG3 possibly
regulates EMT and HSC activation through SMO and miR-212. Growth
arrested specific transcript 5 (GAS5) is a lncRNA, the expression
of which is reduced in fibrotic liver tissues of mice, rats and
humans (57). The overexpression
of GAS5 inhibits collagen production and HSC activation. GAS5 acts
as a ceRNA for miR-222 in the cytoplasm and increases the levels of
p27 tumor suppressor protein, leading to suppressed HSC activation
and proliferation (57). GAS5
also acts as a ceRNA for miR-23 in CCL4-induced liver fibrosis and
increases miR-23 expression, leading to PTEN inhibition, resulting
in the activation of the phosphatidyl-3 kinase/protein kinase
B/mammalian target of rapamycin/Snail (PI3K/Akt/mTOR/Snail) pathway
(8). Overall, it appears that
GAS5/miR-222/p27 and GAS5/miR-23/PI3K/Akt/mTOR/Snail pathways play
regulatory roles in this process.
Gm5091 is an intergenic lncRNA which is
downregulated in mouse hepatic stellate cells during
alcohol-induced hepatic fibrosis (58). Gm5091 binds to miR-27b/23b/24 and
negatively regulates cell migration, ROS production, IL-1β
secretion, type I collagen expression and HSC activation markers,
such α-SMA and Desmin (58).
lncRNA hypoxia inducible factor 1A-antisense RNA 1 (HIF1A-AS1)
levels also significantly increase when ten-eleven translocation
(TET) family protein TET3 is silenced in HSCs (59). The silencing of HIF1A-AS1 further
promotes the proliferation of HSCs and suppresses apoptosis
(59). Antisense noncoding RNA in
the INK4 Locus (ANRIL) is an inhibitor of liver fibrosis and is
negatively regulated by DNMT3A (60). In activated HSCs and liver
fibrotic tissues, ANRIL is downregulated. The silencing of ANRIL by
DNMT3A results in HSC activation and liver fibrosis (60). Conversely, the overexpression of
ANRIL suppresses HSC activation and reduces the expression levels
of DNMT3a, Col1a1 and α-SMA, and inhibits adenosine
mono-phosphate-activated protein kinase (AMPK) signaling (60).
lncRNA H19 is involved in the regulation of cell
proliferation and differentiation. It is genetically imprinted,
maternally expressed and conserved in mice and humans (61). H19 is upregulated in human liver
diseases and animal models of critical limb ischemia (62,63). H19 plays a dual role liver
fibrosis (both an inhibitory and promoting role) through distinct
cellular targets. The inhibitory roles of H19 in liver fibrosis
involve its effects on DNA methylation. In activated HSCs and rat
liver fibrotic tissue, the reduced expression of H19, coupled with
the elevated expression of DNMT1 and the increased methylation of
H19 promoter are present (64).
The silencing of H19 in HSCs increases the expression of
phospho-extracellular signal regulated kinases 1/2 (p-ERK1/2)
(65). Methyl CpG binding protein
2 (MeCp2) is upregulated liver fibrosis and in activated HSCs. In a
rat model of liver fibrosis, the overexpression of MeCp2 has shown
to reduce H19 levels and the knockdown of MeCp2 leads to increased
levels of H19 (65). The
silencing of MeCp2 blocks HSC proliferation. Furthermore, the
overexpression of H19 downregulates insulin like growth factor 1
receptor (IGF1R) and vice versa (65). Therefore, MeCp2 possibly targets
IGF1R to promote liver fibrosis by negatively regulating H19 and
the MeCp2/H19/IGF1R pathway (64,65). H19 is further involved in the
regulation of liver fibrosis in the context of cholestasis by
modulating cell migration. Epithelial cell adhesion molecule
(EpCAM) is negatively regulated by E box binding homeobox 1 (ZEB1)
protein. In a mouse model of cholestatic liver fibrosis, H19 was
shown to bind to ZEB1 to inhibit EpCAM expression and promote cell
migration (66). In patients with
biliary atresia (BA)-related liver fibrosis, H19 promotes
cholangiocyte proliferation and cholestatic liver fibrosis via the
regulation of sphingosine 1 phosphate receptor 2
(SIPR2)/sphingosine kinase 2 (SphK2) and the let-7/high mobility
group AT-hook 2 (HMGA2) pathway (67). Cholangiocyte-derived H19-enriched
exosomes induce the activation and differentiation of cultured HSCs
and HSC-derived fibroblasts (68). Hence, the mechanisms of liver
fibrosis involving H19 warrant further thorough investigations in
order to develop therapeutic interventions for liver fibrosis
modulating such processes. Various lncRNAs, their cellular targets
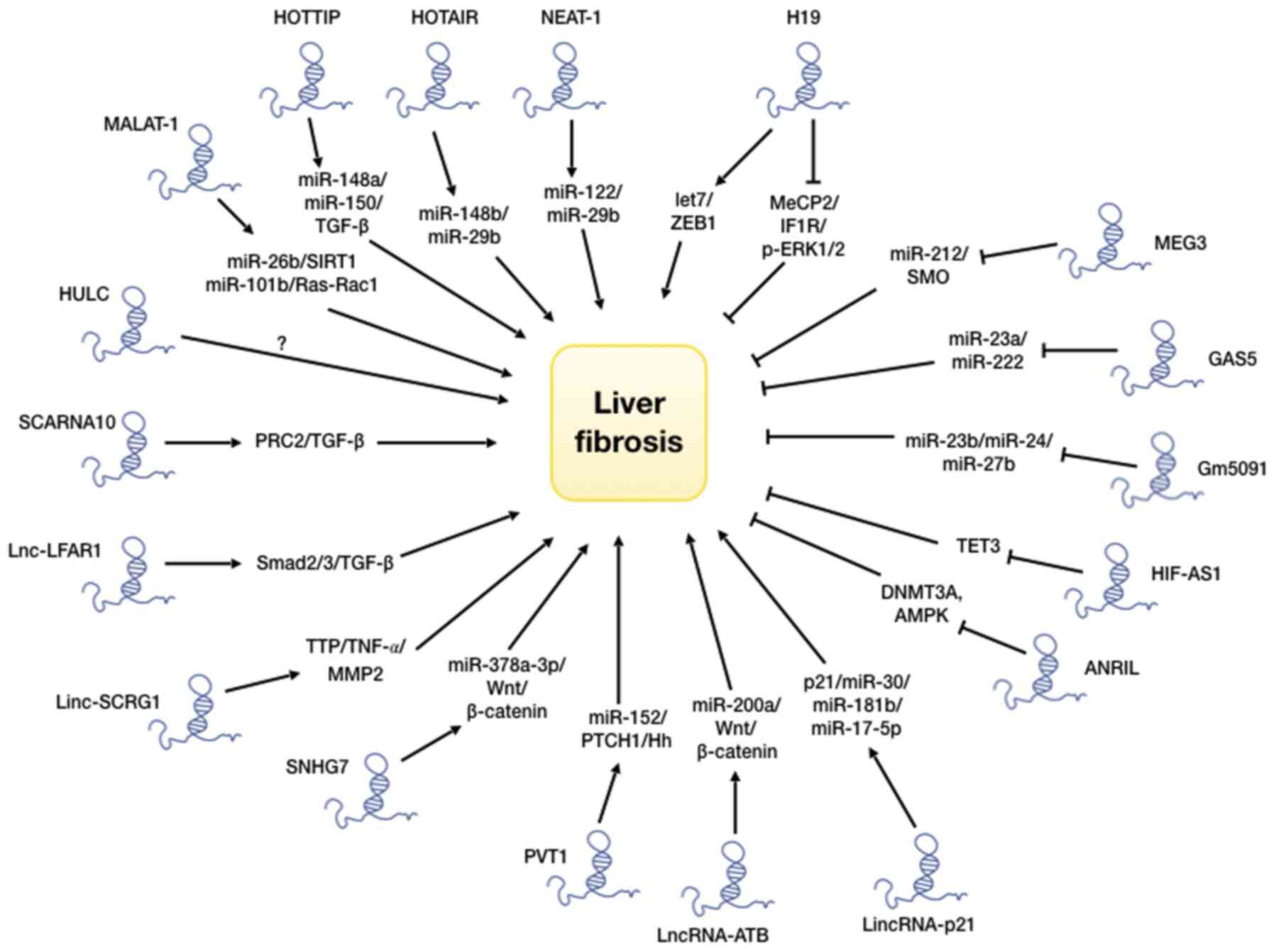
and their roles in liver fibrosis are presented in Table I.
As discussed above, lncRNAs are known to interact
with and regulate multiple signaling pathways, such as TGF-β, p53,
Hedgehog, PI3 kinase/Akt/mToR, Wnt/β-catenin and MAP kinase, which
dictate the initiation and progression of liver fibrosis (12,69). The above-mentioned discussions
outline some such pathways. The most well-studied pathway for liver
fibrosis is the TGF-β signaling pathway, which has been shown to
interact with a number of lncRNAs. TGF-β and its downstream
effectors, such as Smad proteins control the progression of liver
fibrosis, beginning from initial liver injury and inflammation
(70-72). The regulation of liver
fibrosis-related pathways by lncRNAs is illustrated in Fig. 2. Some of these pathways promote
liver fibrosis, while others suppress it.
The mechanisms of action by lncRNAs are usually
directed through its complex secondary structures, and the ability
to bind to other RNA and protein molecules. This endows them with
both unique and diverse traits with which to regulate other
molecules (73-76). There are broadly 2 types of
mechanisms through which lncRNAs regulate the activity of the
target molecule; firstly, acting as a ceRNA to bind to a miRNA
through complementary base pairing and inhibiting the normal
function of the miRNA; secondly, by directly binding to a signaling
protein through its complex secondary structure and
blocking/activating the protein function. An additional mechanism
of lncRNA functions includes the binding to DNA to form a triple
helical structure. This helical structure can function as a docking
site for binding of proteins which can regulate gene expression
(77). Recent advances in ncRNA
studies have enabled their use as a diagnostic tool (78,79). Circulating ncRNAs from patients
with liver fibrosis may be quantified and monitored over the course
of time as surrogate markers for disease progression (80). Studies on lncRNA-based therapy
have also been successfully implemented in preclinical models
(81). However, clinical trials
to verify the efficacy and toxicity of these therapies remain to be
performed, at least to the best of our knowledge. Nonetheless,
lncRNAs may represent a novel molecular therapeutic approach for
liver fibrosis.
The epigenetic control of gene expression is a
well-studied area. The present review focuses on DNA methylation,
as well as histone methylation and acetylation. DNA methylation is
one of the better understood epigenetic mechanisms. The methylation
of DNA occurs at the cytosine residue located 5′ to guanosine
residue in a stretch of CpG dinucleotides termed CpG islands. The
methylation of CpG stretches in the promoter region of a gene and
is known to inhibit or repress gene expression. The methylation of
cytosine residues prevents the binding of DNA binding proteins
required for transcription (82).
DNA methylation is carried out by a group of enzymes known as DNA
methyltransferases (DNMTs), which are divided into 2 subcategories:
Those involved in the de novo methylation of DNA (DNMT3a and
DNMT3b) and those involved in maintenance of methylation patterns
post-DNA replication (DNMT1) (83-87).
During the development of liver fibrosis, changes in
patterns of DNA methylation, altered activity and the expression of
associated enzymes have been observed. These changes are crucial
for the establishment and progression of liver fibrosis (88-90). As previously demonstrated,
activated cultured rat HSCs, when treated with the DNA methylation
inhibitor, 5-aza, 2-deoxycytidine, have failed to differentiate
into myofibroblasts (91). During
HSC activation or trans-differentiation, a global DNA
hypomethylation is observed (92,93). Although there is an overall
demethylation during HSC activation, genome-wide methylation
studies have revealed that hypo- or hypermethylation occuring in
the promoter region is specific to each gene (92). During HSC activation, the Wnt
signaling pathway has been found to be epigenetically regulated;
Apc2 and Wnt5a are upregulated upon promoter
hypomethylation (93-95). Similarly, pro-fibrogenic genes,
such as Actg2, Loxl1, Loxl2 and Col4a1
are activated upon the hypomethylation of their respective
promoters (87). Conversely, DNA
hypermethylation causes the transcriptional repression in genes,
such as Adamts9 and Mmp15 (95), which produce matrix
metalloproteinases involved in ECM breakdown and cell migration.
Similarly, Smad7, TGF-βR1 antagonist (96) and pro-apoptotic PTEN
(97) are also negatively
regulated by DNA hypermethylation.
DNMTs are not the only methylating agents in cells.
The methylation of histones is another form of epigenetic
regulatory mechanism. There are 2 types of histone regulatory
enzymes; histone methyl transferases (HMTs) and histone
demthyltransferases (HDMTs). Recent studies have indicated the
roles of these enzymes in liver fibrosis. The exposure of HSCs to
ethanol increases the expression of histone-3 lysine-4 (H3K4)
methyl transferase, MLL1, which is recruited the promoter of the
pro-fibrogenic elastin gene (98). The TGF-β stimulation of mouse
embryonic fibroblasts (MEFs) increases dimethylated (H3K4me2) and
trimethylated (H3K4me3) histone levels in the promoters of the
pro-fibrogenic genes, Col1a1 and Col1a2. H3K4me2 and H3K4me3
histone signatures are imprinted by a complex protein associated
with Set1 (COMPASS). This protein complex COMPASS contains HMTs,
such as ASH2 and SET1, which binds to the promoters of
pro-fibrogenic genes upon the TGF-β stimulation of HSCs and MEFs
(98). ASH1 is also a HMT, which
targets H3K4 methylation and is upregulated during HSC
transdifferentiation (99). ASH1
binds to the promoters of α-SMA, Col1a1, TIMP-1 and TGF-β1, and
causes the transcriptional activation of these genes (100,101).
Enhancer of zeste homologue 2 (EZH2) further acts as
a HMT, which is part of polycomb repressor complex 2 (PRC2) that
cause H3K27 methylation and promotes the progression of liver
fibrosis (102). In
CCL4-mediated liver injury, EZH2 expression is increased in
activated HSCs (103). The TGF-β
stimulation of HSCs can also induce EZH2 (104). The increased expression of EZH2
in activated HSCs causes H3K27 methylation in exons A1 and A2 of
peroxisome proliferator activator receptor-γ (PPAR-γ) causing
transcriptional repression of PPAR-γ (105,106). The methylation of exons leads to
the recruitment of polycomb repressor complex 1 (PRC1) and causes
chromatin condensation, thus preventing the transcriptional
elongation of downstream exons (107). The downregulation of PPAR-γ is
necessary to enable HSC transdifferentiation into myofibroblasts.
The epigenetic regulation of genes involved in liver fibrosis
through methylation is presented in Table II. Histone
acetylation/deacetylation is an important epigenetic mechanism
which regulates gene expression and action. Studies have suggested
a greater role of HDACs compared to HATs in HSC activation and
fibrogenesis. Based on their structure and mechanisms of action,
HDACs are grouped into 4 classes: Class I-HDAC1, 2, 3, 8; Class
II-HDAC4, 5, 6, 7, 9, 10; Class III-Sirt 1-7; Class IV-HDAC11.
During HSC activation, HDAC1 and 2 protein levels have been found
to be downregulated (108). In a
previous study, in the culture of primary HSCs, HDAC5 and 6 levels
peaked at day 4 before decreasing back to baseline levels (109). It was further observed that the
pharmacological or siRNA-mediated inhibition of class II HDACs
resulted in the upregulation of anti-fibrotic miR-29 (110). The inhibition of HDACs reduced
the levels of HSC activation markers e.g., α-SMA, collagen and
lysyl oxidase, and also inhibited cell proliferation (109). The inhibition of HDAC1, 2 and 4
by nilotinib has been shown to result in the increased apoptosis
and autophagy of activated rat HSCs and human LX-2 cells (110). Cell death occurs in activated
HSCs only, not quiescent ones. MMPs are metallopeptidases, which
play an important role in liver injury and ECM accumulation
(111). MMPs are produced by
quiescent HSCs, which release growth factors required for wound
healing and ECM production (112). However, their levels decrease
during HSC transdifferentiation and in liver fibrosis. An
interesting link has been found between MMPs and HDACs, which
indicates that the ectopic expression of HDAC4 in quiescent HSCs
suppresses Mmp9 and Mmp13 gene expression (112). As lncRNAs are regulated in a
similar manner as protein-coding genes, a regulatory asssociation
of lncRNAs through acetylation is present (113-116). These studies have demonstrated
that HDACs play a regulatory role in liver fibrosis and HSC
activation, and may be considered as drug targets.
Pharmacological inhibitors of DNMTs, such as
5-azacytidine, decitabine, guadecitabine, etc. are being tested for
the treatment of hematological cancers and for solid tumors, such
as hepatocellular carcinoma (HCC) (117-119). These drugs have been shown to be
reasonably effective in safety trials and may have potential for
use in the treatment of liver fibrosis. Inhibitors of HDACs are
also being used to treat malignancies. These inhibitors block cell
proliferation and induce apoptosis, and also suppress HSC
activation (120,121). Largazole, an HDAC inhibitor, has
been shown to suppress liver fibrogenesis and angiogenesis by
inhibiting TGF-β and VEGF signaling (18,122). It has also been shown that
sodium valproate, a class I and II HDAC inhibitor, when added to
cultured HSCs, blocked TGF-β signaling and downregulated TGF-β
induced Col1α1 expression (123). Trichostatin A (TSA), another
HDAC inhibitor, has been shown to suppress HSC differentiation into
myfibroblasts, and to downregulate α-SMA, and type I and III
collagen (124). In CCL4-induced
liver fibrosis, TSA has been found to suppress HSC activation
through the acetylation of CAAT/enhancer binding protein α
(C/EBP-α) (125). These
epigenetic inhibitors provide promising therapeutic targets for
liver fibrosis.
The histone code is defined as the epigenetic
signature (methylation and/or acetylation) imprinted on histones.
The expression of genes is regulated by chromatin remodeling, which
in turn is regulated by epigenetic modifications. The interaction
between DNA methylation and histone code represents a functional
regulatory mechanism which influences chromatin structure (126,127). The epigenetic modifications on
DNA provides docking sites for many transcriptional regulatory
proteins which determine gene expression.
Methyl CpG binding protein-2 (MeCP2) binds to
methylated cytosine residues on stretches of CpG dinucleotides.
MeCP2 regulates the expression of the α-SMA gene. MeCP2
knockout mice exhibit a reduced α-SMA expression and
collagen deposition in alveolar cells (128,129). Similar effects have been
observed in liver fibrosis (103), establishing the role of MeCP2 in
myofibroblast differentiation and liver fibrosis. One of the key
steps in HSC transdifferentiation is the epigenetic repression of
the PPAR-γ gene, which inhibits HSC apoptosis and promotes
fibrogenesis. MeCP2 binds to the PPAR-γ promoter and
methylates histone H3K9, thereby causing the transcriptional
silencing of PPAR-γ (103). MeCP2 also induces the expression
of EZH2 histone methyl transferase, which methylates histone H3K27
as a part of PRC2 causing the silencing of PPAR-γ (102). MeCP2 also induces the expression
of histone methyl transferase ASH1, which activates the
transcription of profibrogenic genes, such as α-SMA, Col1a1, TIMP-1
and TGF-β1 during myofibroblast differentiation (103). Hence, a coordinated crosstalk
between DNA methylation and the histone code, and the epigenetic
enzymes can cause transcriptional activation or repression,
depending on the specific molecular interactions near the promoters
of target genes. MeCP2 causes the transcriptional repression of
genes when it binds to 5-methyl cytosine whereas it activates gene
expression when it binds to 5-hydroxymethyl cytosine (130,131). It has been demonstrated that
both DNMT1 and H3K9 methyl transferase inhibits PPAR-γ by
binding to its promoter during HSC activation (131), whereas the inhibition of DNMT1
and G9a results in PPAR-γ upregulation and the suppression
of HSC activation (131).
It is of interest to note that the production and/or
action of lncRNAs are also regulated by DNA and histone
methylation. For example, DNMT1 regulates the lncRNA H19/ERK
signaling pathway, which promotes HSC activation and liver
fibrogenesis (64). ANRIL is
regulated by DNMT3a during HSC activation. DNMT3a suppresses ANRIL
expression in activated HSCs with a concomitant increase in the
expression of α-SMA, Col1a1 and AMPK and p-AMPK in rat and human
model of liver fibrosis (60).
MeCP2 as discussed above is an epigenetic regulator of pro and
anti-fibrogenic genes. However, MeCP2 has also been reported to
silence the expression of lncRNA H19 through the IGF1R pathway and
to promote HSC proliferation (65). The pro-fibrogenic activity of
MeCP2 is regulated by phosphorylation at Serine 80 and its deletion
in HSCs results in the altered expression of 284 mRNAs and 244
lncRNAs (132). The
understanding of the crosstalk between epigenetic regulators and
lncRNAs may help to broaden our understanding of the pathogenesis
and subsequent potential therapeutic approaches for liver fibrosis
(12,132). EZH2, which is a HMT, methylates
H3K27 histone within the exons of PPARγ (102). The methylation of histones leads
to the recruitment of the PRC1 complex, which causes the
reorganization of the chromatin structure. The binding of PRC1
causes chromatin condensation, which prevents the binding of RNA
polymerase II to the DNA template, causing premature termination of
transcription (106), indicating
that epigenetic mechanisms indirectly regulate gene expression by
changing chromatin dynamics.
One of the main external stimuli that may affect
epigenetic changes is the food that is consumed by an individual.
The type of food, as well as the method through which food is
prepared, may influence DNA and or histone damage, and subsequent
epigenetic alterations, leading to alterations in gene expression.
In addition, exposure to environmental toxins, such as smoking and
other pollutants may also affect cells through epigenetic
mechanisms (133). However, the
detailed analyses of such an etiology and how the aforesaid changes
are influenced, are beyond the scope of the present review.
Similarly, as the present review focused on lncRNAs, miRNAs and
their effects on liver fibrosis either directly or distally through
an exosome-mediated mechanism were not addressed. A previously
published review article has addressed this topic (134,135).
The present review aimed to highlight the importance
of lncRNAs in the initiation and progression of liver fibrosis. The
present review also outlined how such processes are regulated by
histone and DNA methylation and histone acetylation. The recent
impetus into the study of lncRNAs has provided a clearer picture
into their roles in HSC activation and liver fibrosis. However, the
complex regulatory network of lncRNAs and their associations with a
plethora of molecules, renders the elucidation of the underlying
mechanisms challenging. A diagrammatic outline is illustrated in
Fig. 2. As there may also be
disease-specific variation, a therapeutic strategy may involve a
combination of disease-specific targets. Furthermore, the majority
of the studies have focused on HSCs and the potential contribution
by other cells may also have to be investigated in order to obtain
a better understanding of the mechanisms involved.
NG received A fellowship from ICMR no. INDOiFRC/452
I (Y-U) t 201 g2UIHD, Government of India. SC received funding from
CIHR grant no. 425360, Government of Canada.
Not applicable.
Both authors (NG and SC) contributed equally to the
conception, design, writing, and critical reviewing and reading of
the manuscript. Both authors read and approved the final
manuscript.
Not applicable.
Not applicable.
The authors declare that they have no competing
interests.
Not applicable.
|
1
|
Aydın MM and Akçalı KC: Liver fibrosis.
Turk J Gastroenterol. 29:14–21. 2018. View Article : Google Scholar
|
|
2
|
Lan T, Li C, Yang G, Sun Y, Zhuang L, Ou
Y, Li H, Wang G, Kisseleva T, Brenner D and Guo J: Sphingosine
kinase 1 promotes liver fibrosis by preventing miR-19b-3p-mediated
inhibition of CCR2. Hepatology. 68:1070–1086. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Bataller R and Brenner DA: Liver fibrosis.
J Clin Invest. 115:209–218. 2005. View
Article : Google Scholar : PubMed/NCBI
|
|
4
|
Karsdal MA, Hjuler ST, Luo Y, Rasmussen
DGK, Nielsen MJ, Holm Nielsen S, Leeming DJ, Goodman Z, Arch RH,
Patel K and Schuppan D: Assessment of liver fibrosis progression
and regression by a serological collagen turnover profile. Am J
Physiol Gastrointest Liver Physiol. 316:G25–G31. 2019. View Article : Google Scholar
|
|
5
|
Chen L, Brenner DA and Kisseleva T:
Combatting fibrosis: Exosome-based therapies in the regression of
liver fibrosis. Hepatol Commun. 3:180–192. 2018. View Article : Google Scholar
|
|
6
|
Lledó GM, Carrasco I, Benítez-Gutiérrez
LM, Arias A, Royuela A, Requena S, Cuervas-Mons V and de Mendoza C:
Regression of liver fibrosis after curing chronic hepatitis C with
oral antivirals in patients with and without HIV coinfection. Aids.
32:2347–2352. 2018.PubMed/NCBI
|
|
7
|
Atta HM: Reversibility and heritability of
liver fibrosis: Implications for research and therapy. World J
Gastroenterol. 21:5138–5148. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Dong Z, Li S, Wang X, Si L, Ma R, Bao L
and Bo A: lncRNA GAS5 restrains CCl4-induced hepatic
fibrosis by targeting miR-23a through the PTEN/PI3K/Akt signaling
pathway. Am J Physiol Gastrointest Liver Physiol. 316:G539–G550.
2019. View Article : Google Scholar
|
|
9
|
Dou C, Liu Z, Tu K, Zhang H, Chen C,
Yaqoob U, Wang Y, Wen J, van Deursen J, Sicard D, et al: P300
acetyltransferase mediates stiffness-induced activation of hepatic
stellate cells into tumor-promoting myofibroblasts.
Gastroenterology. 154:2209–2221.e14. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Brandon-Warner E, Benbow JH, Swet JH,
Feilen NA, Culberson CR, McKillop IH, de Lemos AS, Russo MW and
Schrum LW: Adeno-associated virus serotype 2 Vector-mediated
reintroduction of microRNA-19b attenuates hepatic fibrosis. Hum
Gene Ther. 29:674–686. 2018. View Article : Google Scholar :
|
|
11
|
Seki E and Schwabe RF: Hepatic
inflammation and fibrosis: Functional links and key pathways.
Hepatology. 61:1066–1079. 2015. View Article : Google Scholar :
|
|
12
|
Peng H, Wan LY, Liang JJ, Zhang YQ, Ai WB
and Wu JF: The roles of lncRNA in hepatic fibrosis. Cell Biosci.
8:632018. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Campana L and Iredale JP: Regression of
liver fibrosis. Semin Liver Dis. 37:1–10. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Knolle PA and Wohlleber D: Immunological
functions of liver sinusoidal endothelial cells. Cell Mol Immunol.
13:347–353. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Heo MJ, Yun J and Kim SG: Role of
non-coding RNAs in liver disease progression to hepatocellular
carcinoma. Arch Pharm Res. 42:48–62. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wei L, Wang X, Lv L, Liu J, Xing H, Song
Y, Xie M, Lei T, Zhang N and Yang M: The emerging role of microRNAs
and long noncoding RNAs in drug resistance of hepatocellular
carcinoma. Mol Cancer. 18:1472019. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zhou J, Li Y, Liu X, Long Y and Chen J:
LncRNA-regulated autophagy and its potential role in drug-induced
liver injury. Ann Hepatol. 17:355–363. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Yang Z, Jiang S, Shang J, Jiang Y, Dai Y,
Xu B, Yu Y, Liang Z and Yang Y: LncRNA: Shedding light on
mechanisms and opportunities in fibrosis and aging. Ageing Res Rev.
52:17–31. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Klinge CM: Non-coding RNAs in breast
cancer: Intracellular and intercellular communication. Noncoding
RNA. 4:402018.
|
|
20
|
Zoghbi HY and Beaudet AL: Epigenetics and
human disease. Cold Spring Harb Perspect Biol. 8:a0194972016.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Sahu B, Pani S, Swalsingh G and Bal NC:
Non and epigenetic mechanisms in regulation of adaptive
thermogenesis in skeletal muscle. Front Endocrinol (Lausanne).
10:5172019. View Article : Google Scholar
|
|
22
|
Ortuno-Sahagun D, Schleibs R and Pallas M:
Editorial: Epigenetic mechanisms regulating neural plasticity.
Front Cell Neurosci. 13:1182019. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Martinez SR, Gay MS and Zhang L:
Epigenetic mechanisms in heart development and disease. Drug Discov
Today. 20:799–811. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Maldonado L and Hoque MO: Epigenomics and
ovarian carcinoma. Biomark Med. 4:543–570. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Vrtacnik P, Marc J and Ostanek B:
Epigenetic mechanisms in bone. Clin Chem Lab Med. 52:589–608. 2014.
View Article : Google Scholar
|
|
26
|
Wu SC and Zhang Y: Active DNA
demethylation: Many roads lead to Rome. Nat Rev Mol Cell Biol.
11:607–620. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Barcena-Varela M, Colyn L and
Fernandez-Barrena MG: Epigenetic mechanisms in hepatic stellate
cell activation during liver fibrosis and carcinogenesis. Int J Mol
Sci. 20:25072019. View Article : Google Scholar :
|
|
28
|
Lachiondo-Ortega S, Mercado-Gómez M,
Serrano-Maciá M, Lopitz-Otsoa F, Salas-Villalobos TB, Varela-Rey M,
Delgado TC and Martínez-Chantar ML: Ubiquitin-like
post-translational modifications (Ubl-PTMs): Small peptides with
huge impact in liver fibrosis. Cells. 8:15752019. View Article : Google Scholar
|
|
29
|
Dooley S and Ten Dijke P: TGF-β in
progression of liver disease. Cell Tissue Res. 347:245–256. 2012.
View Article : Google Scholar
|
|
30
|
Zhou C, York SR, Chen JY, Pondick JV,
Motola DL, Chung RT and Mullen AC: Long noncoding RNAs expressed in
human hepatic stellate cells form networks with extracellular
matrix proteins. Genome Med. 8:312016. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Li XQ, Ren ZX, Li K, Huang JJ, Huang ZT,
Zhou TR, Cao HY, Zhang FX and Tan B: Key anti-fibrosis associated
long noncoding RNAs identified in human hepatic stellate cell via
transcriptome sequencing analysis. Int J Mol Sci. 19:6752018.
View Article : Google Scholar :
|
|
32
|
Kong Y, Huang T, Zhang H, Zhang Q, Ren J,
Guo X, Fan H and Liu L: The lncRNA NEAT1/miR-29b/Atg9a axis
regulates IGFBPrP1-induced autophagy and activation of mouse
hepatic stellate cells. Life Sci. 237:1169022019. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Li XQ, Zhang QQ, Zhang HY, Guo XH, Fan HQ
and Liu LX: Interaction between insulin-like growth factor binding
protein-related protein 1 and transforming growth factor beta 1 in
primary hepatic stellate cells. Hepatobiliary Pancreat Dis Int.
16:395–404. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Yu F, Jiang Z, Chen B, Dong P and Zheng J:
NEAT1 accelerates the progression of liver fibrosis via regulation
of microRNA-122 and Kruppel-like factor 6. J Mol Med (Berl).
95:1191–1202. 2017. View Article : Google Scholar
|
|
35
|
Fu WM, Zhu X, Wang WM, Lu YF, Hu BG, Wang
H, Liang WC, Wang SS, Ko CH, Waye MM, et al: Hotair mediates
hepatocarcinogenesis through suppressing miRNA-218 expression and
activating P14 and P16 signaling. J Hepatol. 63:886–895. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Yu F, Chen B, Dong P and Zheng J: HOTAIR
epigenetically modulates PTEN expression via MicroRNA-29b: A novel
mechanism in regulation of liver fibrosis. Mol Ther. 25:205–217.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Zheng J, Mao Y, Dong P, Huang Z and Yu F:
Long noncoding RNA HOTTIP mediates SRF expression through sponging
miR-150 in hepatic stellate cells. J Cell Mol Med. 23:1572–1580.
2019. View Article : Google Scholar
|
|
38
|
Li Z, Wang J, Zeng Q, Hu C, Zhang J, Wang
H, Yan J, Li H and Yu Z: Long noncoding RNA HOTTIP promotes mouse
hepatic stellate cell activation via downregulating miR-148a. Cell
Physiol Biochem. 51:2814–2828. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Wu Y, Liu X, Zhou Q, Huang C, Meng X, Xu F
and Li J: Silent information regulator 1 (SIRT1) ameliorates liver
fibrosis via promoting activated stellate cell apoptosis and
reversion. Toxicol Appl Pharmacol. 289:163–176. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Yu F, Lu Z, Cai J, Huang K, Chen B, Li G,
Dong P and Zheng J: MALAT1 functions as a competing endogenous RNA
to mediate Rac1 expression by sequestering miR-101b in liver
fibrosis. Cell Cycle. 14:3885–3896. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Leti F, Legendre C, Still CD, Chu X,
Petrick A, Gerhard GS and DiStefano JK: Altered expression of
MALAT1 lncRNA in nonalcoholic steatohepatitis fibrosis regulates
CXCL5 in hepatic stellate cells. Transl Res. 190:25–39.e21. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Shen X, Guo H, Xu J and Wang J: Inhibition
of lncRNA HULC improves hepatic fibrosis and hepatocyte apoptosis
by inhibiting the MAPK signaling pathway in rats with nonalcoholic
fatty liver disease. J Cell Physiol. 234:18169–18179. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Zhang K, Han Y, Hu Z, Zhang Z, Shao S, Yao
Q, Zheng L, Wang J, Han X, Zhang Y, et al: SCARNA10, a
nuclear-retained long non-coding RNA, promotes liver fibrosis and
serves as a potential biomarker. Theranostics. 9:3622–3638. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Zhang K, Han X, Zhang Z, Zheng L, Hu Z,
Yao Q, Cui H, Shu G, Si M, Li C, et al: The liver-enriched
lnc-LFAR1 promotes liver fibrosis by activating TGFβ and Notch
pathways. Nat Commun. 8:1442017. View Article : Google Scholar
|
|
45
|
Wu JC, Luo SZ, Liu T, Lu LG and Xu MY:
Linc-SCRG1 accelerates liver fibrosis by decreasing RNA-binding
protein tristetraprolin. FASEB J. 33:2105–2115. 2019. View Article : Google Scholar
|
|
46
|
Yu F, Dong P, Mao Y, Zhao B, Huang Z and
Zheng J: Loss of lncRNA-SNHG7 promotes the suppression of hepatic
stellate cell activation via miR-378a-3p and DVL2. Mol Ther Nucleic
Acids. 17:235–244. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Zheng J, Yu F, Dong P, Wu L, Zhang Y, Hu Y
and Zheng L: Long non-coding RNA PVT1 activates hepatic stellate
cells through competitively binding microRNA-152. Oncotarget.
7:62886–62897. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Fu N, Zhao SX, Kong LB, Du JH, Ren WG, Han
F, Zhang QS, Li WC, Cui P, Wang RQ, et al:
LncRNA-ATB/microRNA-200a/β-catenin regulatory axis involved in the
progression of HCV-related hepatic fibrosis. Gene. 618:1–7. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Yu F, Guo Y, Chen B, Shi L, Dong P, Zhou
M, Zheng J, et al: TGF-β-induced hepatocyte lincRNA-p21 contributes
to liver fibrosis in mice. Sci Rep. 7:29572017. View Article : Google Scholar
|
|
50
|
Yu F, Guo Y, Chen B, Shi L, Dong P, Zhou M
and Zheng J: LincRNA-p21 inhibits the Wnt/β-catenin pathway in
activated hepatic stellate cells via sponging MicroRNA-17-5p. Cell
Physiol Biochem. 41:1970–1980. 2017. View Article : Google Scholar
|
|
51
|
Yu F, Lu Z, Chen B, Dong P and Zheng J:
Identification of a novel lincRNA-p21-miR-181b-PTEN signaling
cascade in liver fibrosis. Mediators Inflamm. 2016:98565382016.
View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Zheng J, Dong P, Mao Y, Chen S, Wu X, Li
G, Lu Z and Yu F: Linc RNA-p21 inhibits hepatic stellate cell
activation and liver fibrogenesis via p21. FEBS J. 282:4810–4821.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Yu F, Zhou G, Huang K, Fan X, Li G, Chen
B, Dong P and Zheng J: Serum linc RNA-p21 as a potential biomarker
of liver fibrosis in chronic hepatitis B patients. J Viral Hepat.
24:580–588. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
He Y, Wu YT, Huang C, Meng XM, Ma TT, Wu
BM, Xu FY, Zhang L, Lv XW and Li J: Inhibitory effects of long
noncoding RNA MEG3 on hepatic stellate cells activation and liver
fibrogenesis. Biochim Biophys Acta. 1842:2204–2215. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Chen MJ, Wang XG, Sun ZX and Liu XC:
Diagnostic value of LncRNA-MEG3 as a serum biomarker in patients
with hepatitis B complicated with liver fibrosis. Eur Rev Med
Pharmacol Sci. 23:4360–4367. 2019.PubMed/NCBI
|
|
56
|
Yu F, Geng W, Dong P, Huang Z and Zheng J:
LncRNA-MEG3 inhibits activation of hepatic stellate cells through
SMO protein and miR-212. Cell Death Dis. 9:10142018. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Yu F, Zheng J, Mao Y, Dong P, Lu Z, Li G,
Guo C, Liu Z and Fan X: Long non-coding RNA growth arrest-specific
transcript 5 (GAS5) inhibits liver fibrogenesis through a mechanism
of competing endogenous RNA. J Biol Chem. 290:28286–28298. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Zhou B, Yuan W and Li X: LncRNA Gm5091
alleviates alcoholic hepatic fibrosis by sponging miR-27b/23b/24 in
mice. Cell Biol Int. 42:1330–1339. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Zhang QQ, Xu MY, Qu Y, Hu JJ, Li ZH, Zhang
QD and Lu LG: TET3 mediates the activation of human hepatic
stellate cells via modulating the expression of long non-coding RNA
HIF1A-AS1. Int J Clin Exp Pathol. 7:7744–7751. 2014.
|
|
60
|
Yang JJ and Yang Y, Zhang C, Li J and Yang
Y: Epigenetic silencing of LncRNA ANRIL enhances liver fibrosis and
HSC activation through activating AMPK pathway. J Cell Mol Med.
24:2677–2687. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Giovarelli M, Bucci G, Ramos A, Bordo D,
Wilusz CJ, Chen CY, Puppo M, Briata P and Gherzi R: H19 long
noncoding RNA controls the mRNA decay promoting function of KSRP.
Proc Natl Acad Sci USA. 111:E5023–E5028. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Liang WC, Fu WM, Wang YB, Sun YX, Xu LL,
Wong CW, Chan KM, Li G, Waye MM and Zhang JF: H19 activates Wnt
signaling and promotes osteoblast differentiation by functioning as
a competing endogenous RNA. Sci Rep. 6:201212016. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Li X, Liu R, Huang Z, Gurley EC, Wang X,
Wang J, He H, Yang H, Lai G, Zhang L, et al: Cholangiocyte-derived
exosomal long noncoding RNA H19 promotes cholestatic liver injury
in mouse and humans. Hepatology. 68:599–615. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Yang JJ, She Q, Yang Y, Tao H and Li J:
DNMT1 controls LncRNA H19/ERK signal pathway in hepatic stellate
cell activation and fibrosis. Toxicol Lett. 295:325–334. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Yang JJ, Liu LP, Tao H, Hu W, Shi P, Deng
ZY and Li J: MeCP2 silencing of LncRNA H19 controls hepatic
stellate cell proliferation by targeting IGF1R. Toxicology.
359-360:39–46. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Song Y, Liu C, Liu X, Trottier J, Beaudoin
M, Zhang L, Pope C, Peng G, Barbier O, Zhong X, et al: H19 promotes
cholestatic liver fibrosis by preventing ZEB1-mediated inhibition
of epithelial cell adhesion molecule. Hepatology. 66:1183–1196.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Xiao Y, Liu R, Li X, Gurley EC, Hylemon
PB, Lu Y, Zhou H and Cai W: Long noncoding RNA H19 contributes to
cholangiocyte proliferation and cholestatic liver fibrosis in
biliary atresia. Hepatology. 70:1658–1673. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Liu R, Li X, Zhu W, Wang Y, Zhao D, Wang
X, Gurley EC, Liang G, Chen W, Lai G, et al: Cholangiocyte-derived
exosomal long noncoding RNA H19 promotes hepatic stellate cell
activation and cholestatic liver fibrosis. Hepatology.
70:1317–1335. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Fernandes JCR, Acuña SM, Aoki JI,
Floeter-Winter LM and Muxel SM: Long non-coding RNAs in the
regulation of gene expression: Physiology and disease. Noncoding
RNA. 5:172019.
|
|
70
|
Xu F, Liu C, Zhou D and Zhang L:
TGF-β/SMAD pathway and its regulation in hepatic fibrosis. J
Histochem Cytochem. 64:157–167. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Fabregat I, Moreno-Càceres J, Sánchez A,
Dooley S, Dewidar B, Giannelli G and Ten Dijke P; IT-LIVER
Consortium: TGF-beta signalling and liver disease. FEBS J.
283:2219–2232. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Caja L, Dituri F, Mancarella S,
Caballero-Diaz D, Moustakas A, Giannelli G and Fabregat I: TGF-β
and the tissue microenvironment: Relevance in fibrosis and cancer.
Int J Mol Sci. 19:12942018. View Article : Google Scholar
|
|
73
|
Martens L, Rühle F and Stoll M: LncRNA
secondary structure in the cardiovascular system. Noncoding RNA
Res. 2:137–142. 2017. View Article : Google Scholar
|
|
74
|
Zampetaki A, Albrecht A and Steinhofel K:
Long non-coding RNA structure and function: Is there a link? Front
Physiol. 9:12012018. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
He Z, Yang D, Fan X, Zhang M, Li Y, Gu X
and Yang M: The roles and mechanisms of lncRNAs in liver fibrosis.
Int J Mol Sci. 21:14822020. View Article : Google Scholar :
|
|
76
|
Kuo CC, Hänzelmann S, Sentürk Cetin N,
Frank S, Zajzon B, Derks JP, Akhade VS, Ahuja G, Kanduri C, Grummt
I, et al: Detection of RNA-DNA binding sites in long noncoding
RNAs. Nucleic Acids Res. 47:e322019. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Teng KY and Ghoshal K: Role of noncoding
RNAs as biomarker and therapeutic targets for liver fibrosis. Gene
Expr. 16:155–162. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Roderburg C, Mollnow T, Bongaerts B,
Elfimova N, Vargas Cardenas D, Berger K, Zimmermann H, Koch A,
Vucur M, Luedde M, et al: Micro-RNA profiling in human serum
reveals compartment-specific roles of miR-571 and miR-652 in liver
cirrhosis. PLoS One. 7:e329992012. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Klose RJ and Bird AP: Genomic DNA
methylation: The mark and its mediators. Trends Biochem Sci.
31:89–97. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Shang Z, Yu J, Sun L, Tian J, Zhu S, Zhang
B, Dong Q, Jiang N, Flores-Morales A, Chang C and Niu Y: LncRNA
PCAT1 activates AKT and NF-κB signaling in castration-resistant
prostate cancer by regulating the PHLPP/FKBP51/IKKα complex.
Nucleic Acids Res. 47:4211–4225. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Gujar H, Weisenberger DJ and Liang G: The
roles of human DNA methyltransferases and their isoforms in shaping
the epigenome. Genes (Basel). 10:1722019. View Article : Google Scholar
|
|
82
|
Edwards JR, Yarychkivska O, Boulard M and
Bestor TH: DNA methylation and DNA methyltransferases. Epigenetics
Chromatin. 10:232017. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Arand J, Spieler D, Karius T, Branco MR,
Meilinger D, Meissner A, Jenuwein T, Xu G, Leonhardt H, Wolf V and
Walter J: In vivo control of CpG and non-CpG DNA methylation by DNA
methyltransferases. PLoS Genet. 8:e10027502012. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Okano M, Bell DW, Haber DA and Li E: DNA
methyltransferases Dnmt3a and Dnmt3b are essential for de novo
methylation and mammalian development. Cell. 99:247–257. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Du J, Johnson LM, Jacobsen SE and Patel
DJ: DNA methylation pathways and their crosstalk with histone
methylation. Nat Rev Mol Cell Biol. 16:519–532. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Page A, Mann DA and Mann J: The mechanisms
of HSC activation and epigenetic regulation of HSCs phenotypes.
Curr Pathobiol Rep. 2:163–170. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Wilson CL, Mann DA and Borthwick LA:
Epigenetic reprogramming in liver fibrosis and cancer. Adv Drug
Deliv Rev. 121:124–132. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Dowson C and O'Reilly S: DNA methylation
in fibrosis. Eur J Cell Biol. 95:323–330. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Mann J, Oakley F, Akiboye F, Elsharkawy A,
Thorne AW and Mann DA: Regulation of myofibroblast
transdifferentiation by DNA methylation and MeCP2: Implications for
wound healing and fibrogenesis. Cell Death Differ. 14:275–285.
2007. View Article : Google Scholar
|
|
90
|
Komatsu Y, Waku T, Iwasaki N, Ono W,
Yamaguchi C and Yanagisawa J: Global analysis of DNA methylation in
early-stage liver fibrosis. BMC Med Genomics. 5:52012. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Götze S, Schumacher EC, Kordes C and
Häussinger D: Epigenetic changes during hepatic stellate cell
activation. PLoS One. 10:e01287452015. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
El Taghdouini A, Sørensen AL, Reiner AH,
Coll M, Verhulst S, Mannaerts I, Øie CI, Smedsrød B, Najimi M,
Sokal E, et al: Genome-wide analysis of DNA methylation and gene
expression patterns in purified, uncultured human liver cells and
activated hepatic stellate cells. Oncotarget. 6:26729–26745. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Xiong WJ, Hu LJ, Jian YC, Wang LJ, Jiang
M, Li W and He Y: Wnt5a participates in hepatic stellate cell
activation observed by gene expression profile and functional
assays. World J Gastroenterol. 18:1745–1752. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Miao CG, Yang YY, He X, Huang C, Huang Y,
Zhang L, Lv XW, Jin Y and Li J: Wnt signaling in liver fibrosis:
Progress challenges and potential directions. Biochimie.
95:2326–2335. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Jiang F, Parsons CJ and Stefanovic B: Gene
expression profile of quiescent and activated rat hepatic stellate
cells implicates Wnt signaling pathway in activation. J Hepatol.
45:401–409. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Bian EB, Huang C, Wang H, Chen XX, Zhang
L, Lv XW and Li J: Repression of Smad7 mediated by DNMT1 determines
hepatic stellate cell activation and liver fibrosis in rats.
Toxicol Lett. 224:175–185. 2014. View Article : Google Scholar
|
|
97
|
Bian EB, Huang C, Ma TT, Tao H, Zhang H,
Cheng C, Lv XW and Li J: DNMT1-mediated PTEN hypermethylation
confers hepatic stellate cell activation and liver fibrogenesis in
rats. Toxicol Appl Pharmacol. 264:13–22. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Page A, Paoli PP, Hill SJ, Howarth R, Wu
R, Kweon SM, French J, White S, Tsukamoto H, Mann DA and Mann J:
Alcohol directly stimulates epigenetic modifications in hepatic
stellate cells. J Hepatol. 62:388–397. 2015. View Article : Google Scholar :
|
|
99
|
Tian W, Fan Z, Li J, Hao C, Li M, Xu H, Wu
X, Zhou B, Zhang L, Fang M and Xu Y: Myocardin-related
transcription factor A (MRTF-A) plays an essential role in hepatic
stellate cell activation by epigenetically modulating TGF-β
signaling. Int J Biochem Cell Biol. 71:35–43. 2016. View Article : Google Scholar
|
|
100
|
Gregory GD, Vakoc CR, Rozovskaia T, Zheng
X, Patel S, Nakamura T, Canaani E and Blobel GA: Mammalian ASH1L is
a histone methyltransferase that occupies the transcribed region of
active genes. Mol Cell Biol. 27:8466–8479. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
101
|
Perugorria MJ, Wilson CL, Zeybel M, Walsh
M, Amin S, Robinson S, White SA, Burt AD, Oakley F, Tsukamoto H, et
al: Histone methyltransferase ASH1 orchestrates fibrogenic gene
transcription during myofibroblast transdifferentiation.
Hepatology. 56:1129–1139. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
102
|
Cao R and Zhang Y: The functions of E
(Z)/EZH2-mediated methylation of lysine 27 in histone H3. Curr Opin
Genet Dev. 14:155–164. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
103
|
Mann J, Chu DC, Maxwell A, Oakley F, Zhu
NL, Tsukamoto H and Mann DA: MeCP2 controls an epigenetic pathway
that promotes myofibroblast transdifferentiation and fibrosis.
Gastroenterology. 138:705–714. 714.e1–4. 2010. View Article : Google Scholar :
|
|
104
|
Martin-Mateos R, De Assuncao TM, Arab JP,
Jalan-Sakrikar N, Yaqoob U, Greuter T, Verma VK, Mathison AJ, Cao
S, Lomberk G, et al: Enhancer of zeste homologue 2 inhibition
attenuates TGF-β dependent hepatic stellate cell activation and
liver fibrosis. Cell Mol Gastroenterol Hepatol. 7:197–209. 2019.
View Article : Google Scholar
|
|
105
|
Panebianco C, Oben JA, Vinciguerra M and
Pazienza V: Senescence in hepatic stellate cells as a mechanism of
liver fibrosis reversal: A putative synergy between retinoic acid
and PPAR-gamma signalings. Clin Exp Med. 17:269–280. 2017.
View Article : Google Scholar
|
|
106
|
Schwartz YB and Pirrotta V: Polycomb
silencing mechanisms and the management of genomic programmes. Nat
Rev Genet. 8:9–22. 2007. View Article : Google Scholar
|
|
107
|
Hammond CM, Strømme CB, Huang H, Patel DJ
and Groth A: Histone chaperone networks shaping chromatin function.
Nat Rev Mol Cell Biol. 18:141–158. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
108
|
Mannaerts I, Nuytten NR, Rogiers V,
Vanderkerken K, van Grunsven LA and Geerts A: Chronic
administration of valproic acid inhibits activation of mouse
hepatic stellate cells in vitro and in vivo. Hepatology.
51:603–614. 2010. View Article : Google Scholar
|
|
109
|
Mannaerts I, Eysackers N, Onyema OO, Van
Beneden K, Valente S, Mai A, Odenthal M and van Grunsven LA: Class
II HDAC inhibition hampers hepatic stellate cell activation by
induction of microRNA-29. PLoS One. 8:e557862013. View Article : Google Scholar : PubMed/NCBI
|
|
110
|
Shaker ME, Ghani A, Shiha GE, Ibrahim TM
and Mehal WZ: Nilotinib induces apoptosis and autophagic cell death
of activated hepatic stellate cells via inhibition of histone
deacetylases. Biochim Biophys Acta. 1833:1992–2003. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
111
|
Duarte S, Baber J, Fujii T and Coito AJ:
Matrix metalloproteinases in liver injury, repair and fibrosis.
Matrix Biol. 44-46:147–156. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
112
|
Qin L and Han YP: Epigenetic repression of
matrix metalloproteinases in myofibroblastic hepatic stellate cells
through histone deacetylases 4: Implication in tissue fibrosis. Am
J Pathol. 177:1915–1928. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
113
|
Ding G, Li W, Liu J, Zeng Y, Mao C, Kang Y
and Shang J: LncRNA GHET1 activated by H3K27 acetylation promotes
cell tumorigenesis through regulating ATF1 in hepatocellular
carcinoma. Biomed Pharmacother. 94:326–331. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
114
|
Yang F, Huo XS, Yuan SX, Zhang L, Zhou WP,
Wang F and Sun SH: Repression of the long noncoding RNA-LET by
histone deacetylase 3 contributes to hypoxia-mediated metastasis.
Mol Cell. 49:1083–1096. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
115
|
Pope C, Mishra S, Russell J, Zhou Q and
Zhong XB: Targeting H19, an imprinted long non-coding RNA, in
hepatic functions and liver diseases. Diseases. 5:112017.
View Article : Google Scholar :
|
|
116
|
Zhu XT, Yuan JH, Zhu TT, Li YY and Cheng
XY: Long noncoding RNA glypican 3 (GPC3) antisense transcript 1
promotes hepatocellular carcinoma progression via epigenetically
activating GPC3. FEBS J. 283:3739–3754. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
117
|
Issa JJ, Roboz G, Rizzieri D, Jabbour E,
Stock W, O'Connell C, Yee K, Tibes R, Griffiths EA, Walsh K, et al:
Safety and tolerability of guadecitabine (SGI-110) in patients with
myelodysplastic syndrome and acute myeloid leukaemia: A
multicentre, randomised, dose-escalation phase 1 study. Lancet
Oncol. 16:1099–1110. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
118
|
Jansen YJL, Verset G, Schats K, Van Dam
PJ, Seremet T, Kockx M, Van Laethem JB and Neyns B: Phase I
clinical trial of decitabine (5-aza-2′-deoxycytidine) administered
by hepatic arterial infusion in patients with unresectable
liver-predominant metastases. ESMO Open. 4:e0004642019. View Article : Google Scholar
|
|
119
|
Kuang Y, El-Khoueiry A, Taverna P,
Ljungman M and Neamati N: Guadecitabine (SGI-110) priming
sensitizes hepatocellular carcinoma cells to oxaliplatin. Mol
Oncol. 9:1799–1814. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
120
|
Eckschlager T, Plch J, Stiborova M and
Hrabeta J: Histone deacetylase inhibitors as anticancer drugs. Int
J Mol Sci. 18:14142017. View Article : Google Scholar :
|
|
121
|
Park KC, Park JH, Jeon JY, Kim SY, Kim JM,
Lim CY, Lee TH, Kim HK, Lee HG, Kim SM, et al: A new histone
deacetylase inhibitor improves liver fibrosis in BDL rats through
suppression of hepatic stellate cells. Br J Pharmacol.
171:4820–4830. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
122
|
Liu Y, Wang Z, Wang J, Lam W, Kwong S, Li
F, Friedman SL, Zhou S, Ren Q, Xu Z, et al: A histone deacetylase
inhibitor, largazole, decreases liver fibrosis and angiogenesis by
inhibiting transforming growth factor-β and vascular endothelial
growth factor signalling. Liver Int. 33:504–515. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
123
|
Watanabe T, Tajima H, Hironori H,
Nakagawara H, Ohnishi I, Takamura H, Ninomiya I, Kitagawa H,
Fushida S, Tani T, et al: Sodium valproate blocks the transforming
growth factor (TGF)-β1 autocrine loop and attenuates the
TGF-β1-induced collagen synthesis in a human hepatic stellate cell
line. Int J Mol Med. 28:919–925. 2011.PubMed/NCBI
|
|
124
|
Niki T, Rombouts K, De Bleser P, De Smet
K, Rogiers V, Schuppan D, Yoshida M, Gabbiani G and Geerts A: A
histone deacetylase inhibitor, trichostatin A, suppresses
myofibroblastic differentiation of rat hepatic stellate cells in
primary culture. Hepatology. 29:858–867. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
125
|
Ding D, Chen LL, Zhai YZ, Hou CJ, Tao LL,
Lu SH, Wu J and Liu XP: Trichostatin A inhibits the activation of
Hepatic stellate cells by Increasing C/EBP-α Acetylation in vivo
and in vitro. Sci Rep. 8:43952018. View Article : Google Scholar
|
|
126
|
Smith E and Shilatifard A: The chromatin
signaling pathway: Diverse mechanisms of recruitment of
histone-modifying enzymes and varied biological outcomes. Mol Cell.
40:689–701. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
127
|
Yadav T, Quivy JP and Almouzni G:
Chromatin plasticity: A versatile landscape that underlies cell
fate and identity. Science. 361:1332–1336. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
128
|
Hu B, Gharaee-Kermani M, Wu Z and Phan SH:
Essential role of MeCP2 in the regulation of myofibroblast
differentiation during pulmonary fibrosis. Am J Pathol.
178:1500–1508. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
129
|
Bian EB, Huang C, Wang H, Chen XX, Tao H,
Zhang L, Lv XW and Li J: The role of methyl-CpG binding protein 2
in liver fibrosis. Toxicology. 309:9–14. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
130
|
Mellén M, Ayata P, Dewell S, Kriaucionis S
and Heintz N: MeCP2 binds to 5hmC enriched within active genes and
accessible chromatin in the nervous system. Cell. 151:1417–1430.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
131
|
Bárcena-Varela M, Caruso S, Llerena S,
Aacute;lvarez-Sola G, Uriarte I, Latasa MU, Urtasun R, Rebouissou
S, Alvarez L, Jimenez M, et al: Dual targeting of histone
methyltransferase G9a and DNA-Methyltransferase 1 for the treatment
of experimental hepatocellular carcinoma. Hepatology. 69:587–603.
2019. View Article : Google Scholar
|
|
132
|
Moran-Salvador E, Garcia-Macia M,
Sivaharan A, Sabater L, Zaki MYW, Oakley F, Knox A, Page A, Luli S,
Mann J and Mann DA: Fibrogenic activity of MECP2 is regulated by
phosphorylation in hepatic stellate cells. Gastroenterology.
157:1398–1412.e9. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
133
|
Florean C: Food that shapes you: How diet
can change your epigenome. Science in School. May 13–2014.Epub
ahead of print.
|
|
134
|
Yang JJ, Tao H, Deng ZY, Lu C and Li J:
Non-coding RNA-mediated epigenetic regulation of liver fibrosis.
Metabolism. 64:1386–1394. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
135
|
Jiang X, Tsitsiou E, Herrick SE and
Lindsay MA: MicroRNAs and the regulation of fibrosis. FEBS J.
277:2015–2021. 2010. View Article : Google Scholar : PubMed/NCBI
|