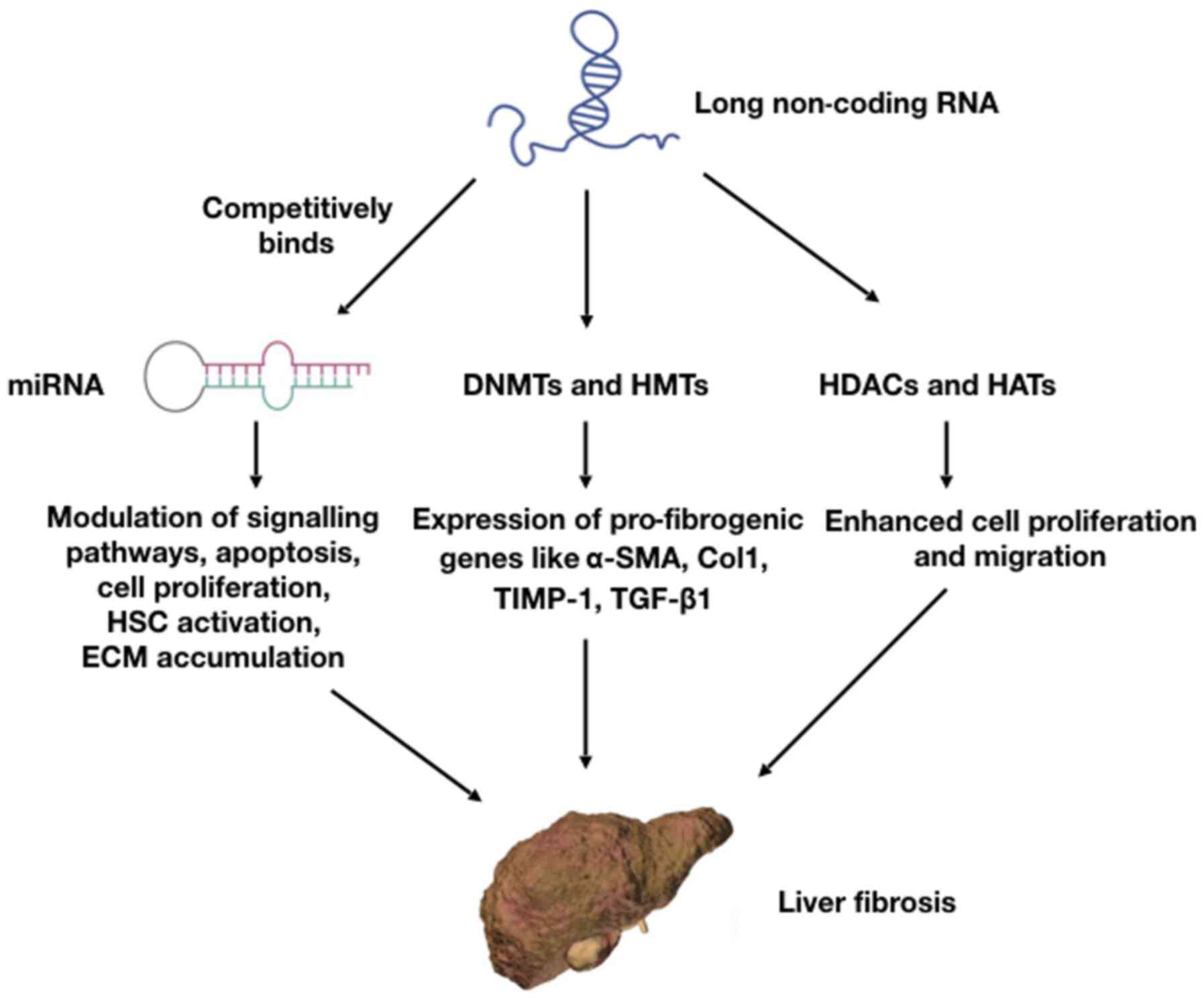
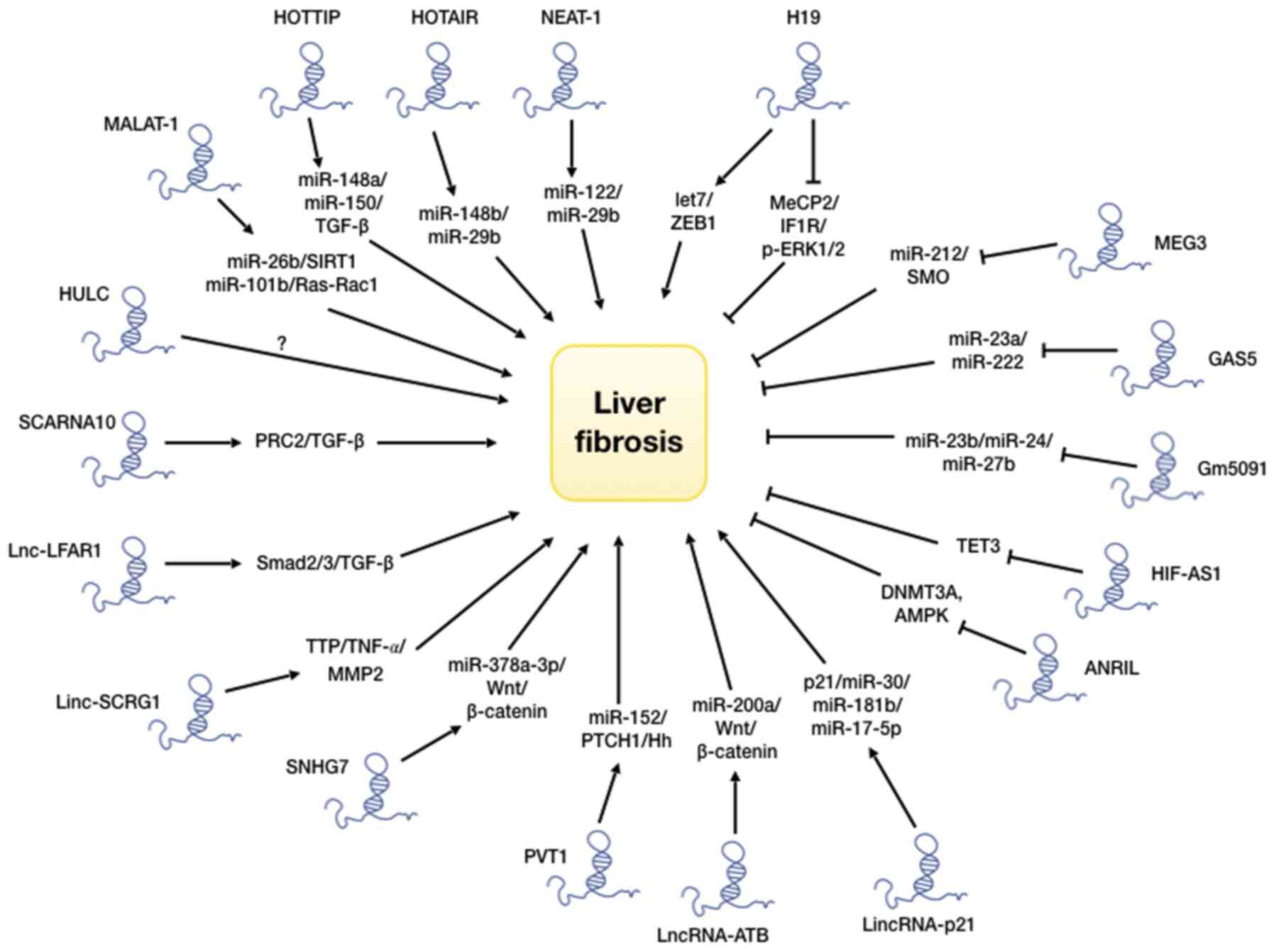
|
1
|
Aydın MM and Akçalı KC: Liver fibrosis.
Turk J Gastroenterol. 29:14–21. 2018. View Article : Google Scholar
|
|
2
|
Lan T, Li C, Yang G, Sun Y, Zhuang L, Ou
Y, Li H, Wang G, Kisseleva T, Brenner D and Guo J: Sphingosine
kinase 1 promotes liver fibrosis by preventing miR-19b-3p-mediated
inhibition of CCR2. Hepatology. 68:1070–1086. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Bataller R and Brenner DA: Liver fibrosis.
J Clin Invest. 115:209–218. 2005. View
Article : Google Scholar : PubMed/NCBI
|
|
4
|
Karsdal MA, Hjuler ST, Luo Y, Rasmussen
DGK, Nielsen MJ, Holm Nielsen S, Leeming DJ, Goodman Z, Arch RH,
Patel K and Schuppan D: Assessment of liver fibrosis progression
and regression by a serological collagen turnover profile. Am J
Physiol Gastrointest Liver Physiol. 316:G25–G31. 2019. View Article : Google Scholar
|
|
5
|
Chen L, Brenner DA and Kisseleva T:
Combatting fibrosis: Exosome-based therapies in the regression of
liver fibrosis. Hepatol Commun. 3:180–192. 2018. View Article : Google Scholar
|
|
6
|
Lledó GM, Carrasco I, Benítez-Gutiérrez
LM, Arias A, Royuela A, Requena S, Cuervas-Mons V and de Mendoza C:
Regression of liver fibrosis after curing chronic hepatitis C with
oral antivirals in patients with and without HIV coinfection. Aids.
32:2347–2352. 2018.PubMed/NCBI
|
|
7
|
Atta HM: Reversibility and heritability of
liver fibrosis: Implications for research and therapy. World J
Gastroenterol. 21:5138–5148. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Dong Z, Li S, Wang X, Si L, Ma R, Bao L
and Bo A: lncRNA GAS5 restrains CCl4-induced hepatic
fibrosis by targeting miR-23a through the PTEN/PI3K/Akt signaling
pathway. Am J Physiol Gastrointest Liver Physiol. 316:G539–G550.
2019. View Article : Google Scholar
|
|
9
|
Dou C, Liu Z, Tu K, Zhang H, Chen C,
Yaqoob U, Wang Y, Wen J, van Deursen J, Sicard D, et al: P300
acetyltransferase mediates stiffness-induced activation of hepatic
stellate cells into tumor-promoting myofibroblasts.
Gastroenterology. 154:2209–2221.e14. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Brandon-Warner E, Benbow JH, Swet JH,
Feilen NA, Culberson CR, McKillop IH, de Lemos AS, Russo MW and
Schrum LW: Adeno-associated virus serotype 2 Vector-mediated
reintroduction of microRNA-19b attenuates hepatic fibrosis. Hum
Gene Ther. 29:674–686. 2018. View Article : Google Scholar :
|
|
11
|
Seki E and Schwabe RF: Hepatic
inflammation and fibrosis: Functional links and key pathways.
Hepatology. 61:1066–1079. 2015. View Article : Google Scholar :
|
|
12
|
Peng H, Wan LY, Liang JJ, Zhang YQ, Ai WB
and Wu JF: The roles of lncRNA in hepatic fibrosis. Cell Biosci.
8:632018. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Campana L and Iredale JP: Regression of
liver fibrosis. Semin Liver Dis. 37:1–10. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Knolle PA and Wohlleber D: Immunological
functions of liver sinusoidal endothelial cells. Cell Mol Immunol.
13:347–353. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Heo MJ, Yun J and Kim SG: Role of
non-coding RNAs in liver disease progression to hepatocellular
carcinoma. Arch Pharm Res. 42:48–62. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wei L, Wang X, Lv L, Liu J, Xing H, Song
Y, Xie M, Lei T, Zhang N and Yang M: The emerging role of microRNAs
and long noncoding RNAs in drug resistance of hepatocellular
carcinoma. Mol Cancer. 18:1472019. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zhou J, Li Y, Liu X, Long Y and Chen J:
LncRNA-regulated autophagy and its potential role in drug-induced
liver injury. Ann Hepatol. 17:355–363. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Yang Z, Jiang S, Shang J, Jiang Y, Dai Y,
Xu B, Yu Y, Liang Z and Yang Y: LncRNA: Shedding light on
mechanisms and opportunities in fibrosis and aging. Ageing Res Rev.
52:17–31. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Klinge CM: Non-coding RNAs in breast
cancer: Intracellular and intercellular communication. Noncoding
RNA. 4:402018.
|
|
20
|
Zoghbi HY and Beaudet AL: Epigenetics and
human disease. Cold Spring Harb Perspect Biol. 8:a0194972016.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Sahu B, Pani S, Swalsingh G and Bal NC:
Non and epigenetic mechanisms in regulation of adaptive
thermogenesis in skeletal muscle. Front Endocrinol (Lausanne).
10:5172019. View Article : Google Scholar
|
|
22
|
Ortuno-Sahagun D, Schleibs R and Pallas M:
Editorial: Epigenetic mechanisms regulating neural plasticity.
Front Cell Neurosci. 13:1182019. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Martinez SR, Gay MS and Zhang L:
Epigenetic mechanisms in heart development and disease. Drug Discov
Today. 20:799–811. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Maldonado L and Hoque MO: Epigenomics and
ovarian carcinoma. Biomark Med. 4:543–570. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Vrtacnik P, Marc J and Ostanek B:
Epigenetic mechanisms in bone. Clin Chem Lab Med. 52:589–608. 2014.
View Article : Google Scholar
|
|
26
|
Wu SC and Zhang Y: Active DNA
demethylation: Many roads lead to Rome. Nat Rev Mol Cell Biol.
11:607–620. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Barcena-Varela M, Colyn L and
Fernandez-Barrena MG: Epigenetic mechanisms in hepatic stellate
cell activation during liver fibrosis and carcinogenesis. Int J Mol
Sci. 20:25072019. View Article : Google Scholar :
|
|
28
|
Lachiondo-Ortega S, Mercado-Gómez M,
Serrano-Maciá M, Lopitz-Otsoa F, Salas-Villalobos TB, Varela-Rey M,
Delgado TC and Martínez-Chantar ML: Ubiquitin-like
post-translational modifications (Ubl-PTMs): Small peptides with
huge impact in liver fibrosis. Cells. 8:15752019. View Article : Google Scholar
|
|
29
|
Dooley S and Ten Dijke P: TGF-β in
progression of liver disease. Cell Tissue Res. 347:245–256. 2012.
View Article : Google Scholar
|
|
30
|
Zhou C, York SR, Chen JY, Pondick JV,
Motola DL, Chung RT and Mullen AC: Long noncoding RNAs expressed in
human hepatic stellate cells form networks with extracellular
matrix proteins. Genome Med. 8:312016. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Li XQ, Ren ZX, Li K, Huang JJ, Huang ZT,
Zhou TR, Cao HY, Zhang FX and Tan B: Key anti-fibrosis associated
long noncoding RNAs identified in human hepatic stellate cell via
transcriptome sequencing analysis. Int J Mol Sci. 19:6752018.
View Article : Google Scholar :
|
|
32
|
Kong Y, Huang T, Zhang H, Zhang Q, Ren J,
Guo X, Fan H and Liu L: The lncRNA NEAT1/miR-29b/Atg9a axis
regulates IGFBPrP1-induced autophagy and activation of mouse
hepatic stellate cells. Life Sci. 237:1169022019. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Li XQ, Zhang QQ, Zhang HY, Guo XH, Fan HQ
and Liu LX: Interaction between insulin-like growth factor binding
protein-related protein 1 and transforming growth factor beta 1 in
primary hepatic stellate cells. Hepatobiliary Pancreat Dis Int.
16:395–404. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Yu F, Jiang Z, Chen B, Dong P and Zheng J:
NEAT1 accelerates the progression of liver fibrosis via regulation
of microRNA-122 and Kruppel-like factor 6. J Mol Med (Berl).
95:1191–1202. 2017. View Article : Google Scholar
|
|
35
|
Fu WM, Zhu X, Wang WM, Lu YF, Hu BG, Wang
H, Liang WC, Wang SS, Ko CH, Waye MM, et al: Hotair mediates
hepatocarcinogenesis through suppressing miRNA-218 expression and
activating P14 and P16 signaling. J Hepatol. 63:886–895. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Yu F, Chen B, Dong P and Zheng J: HOTAIR
epigenetically modulates PTEN expression via MicroRNA-29b: A novel
mechanism in regulation of liver fibrosis. Mol Ther. 25:205–217.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Zheng J, Mao Y, Dong P, Huang Z and Yu F:
Long noncoding RNA HOTTIP mediates SRF expression through sponging
miR-150 in hepatic stellate cells. J Cell Mol Med. 23:1572–1580.
2019. View Article : Google Scholar
|
|
38
|
Li Z, Wang J, Zeng Q, Hu C, Zhang J, Wang
H, Yan J, Li H and Yu Z: Long noncoding RNA HOTTIP promotes mouse
hepatic stellate cell activation via downregulating miR-148a. Cell
Physiol Biochem. 51:2814–2828. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Wu Y, Liu X, Zhou Q, Huang C, Meng X, Xu F
and Li J: Silent information regulator 1 (SIRT1) ameliorates liver
fibrosis via promoting activated stellate cell apoptosis and
reversion. Toxicol Appl Pharmacol. 289:163–176. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Yu F, Lu Z, Cai J, Huang K, Chen B, Li G,
Dong P and Zheng J: MALAT1 functions as a competing endogenous RNA
to mediate Rac1 expression by sequestering miR-101b in liver
fibrosis. Cell Cycle. 14:3885–3896. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Leti F, Legendre C, Still CD, Chu X,
Petrick A, Gerhard GS and DiStefano JK: Altered expression of
MALAT1 lncRNA in nonalcoholic steatohepatitis fibrosis regulates
CXCL5 in hepatic stellate cells. Transl Res. 190:25–39.e21. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Shen X, Guo H, Xu J and Wang J: Inhibition
of lncRNA HULC improves hepatic fibrosis and hepatocyte apoptosis
by inhibiting the MAPK signaling pathway in rats with nonalcoholic
fatty liver disease. J Cell Physiol. 234:18169–18179. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Zhang K, Han Y, Hu Z, Zhang Z, Shao S, Yao
Q, Zheng L, Wang J, Han X, Zhang Y, et al: SCARNA10, a
nuclear-retained long non-coding RNA, promotes liver fibrosis and
serves as a potential biomarker. Theranostics. 9:3622–3638. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Zhang K, Han X, Zhang Z, Zheng L, Hu Z,
Yao Q, Cui H, Shu G, Si M, Li C, et al: The liver-enriched
lnc-LFAR1 promotes liver fibrosis by activating TGFβ and Notch
pathways. Nat Commun. 8:1442017. View Article : Google Scholar
|
|
45
|
Wu JC, Luo SZ, Liu T, Lu LG and Xu MY:
Linc-SCRG1 accelerates liver fibrosis by decreasing RNA-binding
protein tristetraprolin. FASEB J. 33:2105–2115. 2019. View Article : Google Scholar
|
|
46
|
Yu F, Dong P, Mao Y, Zhao B, Huang Z and
Zheng J: Loss of lncRNA-SNHG7 promotes the suppression of hepatic
stellate cell activation via miR-378a-3p and DVL2. Mol Ther Nucleic
Acids. 17:235–244. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Zheng J, Yu F, Dong P, Wu L, Zhang Y, Hu Y
and Zheng L: Long non-coding RNA PVT1 activates hepatic stellate
cells through competitively binding microRNA-152. Oncotarget.
7:62886–62897. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Fu N, Zhao SX, Kong LB, Du JH, Ren WG, Han
F, Zhang QS, Li WC, Cui P, Wang RQ, et al:
LncRNA-ATB/microRNA-200a/β-catenin regulatory axis involved in the
progression of HCV-related hepatic fibrosis. Gene. 618:1–7. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Yu F, Guo Y, Chen B, Shi L, Dong P, Zhou
M, Zheng J, et al: TGF-β-induced hepatocyte lincRNA-p21 contributes
to liver fibrosis in mice. Sci Rep. 7:29572017. View Article : Google Scholar
|
|
50
|
Yu F, Guo Y, Chen B, Shi L, Dong P, Zhou M
and Zheng J: LincRNA-p21 inhibits the Wnt/β-catenin pathway in
activated hepatic stellate cells via sponging MicroRNA-17-5p. Cell
Physiol Biochem. 41:1970–1980. 2017. View Article : Google Scholar
|
|
51
|
Yu F, Lu Z, Chen B, Dong P and Zheng J:
Identification of a novel lincRNA-p21-miR-181b-PTEN signaling
cascade in liver fibrosis. Mediators Inflamm. 2016:98565382016.
View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Zheng J, Dong P, Mao Y, Chen S, Wu X, Li
G, Lu Z and Yu F: Linc RNA-p21 inhibits hepatic stellate cell
activation and liver fibrogenesis via p21. FEBS J. 282:4810–4821.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Yu F, Zhou G, Huang K, Fan X, Li G, Chen
B, Dong P and Zheng J: Serum linc RNA-p21 as a potential biomarker
of liver fibrosis in chronic hepatitis B patients. J Viral Hepat.
24:580–588. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
He Y, Wu YT, Huang C, Meng XM, Ma TT, Wu
BM, Xu FY, Zhang L, Lv XW and Li J: Inhibitory effects of long
noncoding RNA MEG3 on hepatic stellate cells activation and liver
fibrogenesis. Biochim Biophys Acta. 1842:2204–2215. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Chen MJ, Wang XG, Sun ZX and Liu XC:
Diagnostic value of LncRNA-MEG3 as a serum biomarker in patients
with hepatitis B complicated with liver fibrosis. Eur Rev Med
Pharmacol Sci. 23:4360–4367. 2019.PubMed/NCBI
|
|
56
|
Yu F, Geng W, Dong P, Huang Z and Zheng J:
LncRNA-MEG3 inhibits activation of hepatic stellate cells through
SMO protein and miR-212. Cell Death Dis. 9:10142018. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Yu F, Zheng J, Mao Y, Dong P, Lu Z, Li G,
Guo C, Liu Z and Fan X: Long non-coding RNA growth arrest-specific
transcript 5 (GAS5) inhibits liver fibrogenesis through a mechanism
of competing endogenous RNA. J Biol Chem. 290:28286–28298. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Zhou B, Yuan W and Li X: LncRNA Gm5091
alleviates alcoholic hepatic fibrosis by sponging miR-27b/23b/24 in
mice. Cell Biol Int. 42:1330–1339. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Zhang QQ, Xu MY, Qu Y, Hu JJ, Li ZH, Zhang
QD and Lu LG: TET3 mediates the activation of human hepatic
stellate cells via modulating the expression of long non-coding RNA
HIF1A-AS1. Int J Clin Exp Pathol. 7:7744–7751. 2014.
|
|
60
|
Yang JJ and Yang Y, Zhang C, Li J and Yang
Y: Epigenetic silencing of LncRNA ANRIL enhances liver fibrosis and
HSC activation through activating AMPK pathway. J Cell Mol Med.
24:2677–2687. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Giovarelli M, Bucci G, Ramos A, Bordo D,
Wilusz CJ, Chen CY, Puppo M, Briata P and Gherzi R: H19 long
noncoding RNA controls the mRNA decay promoting function of KSRP.
Proc Natl Acad Sci USA. 111:E5023–E5028. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Liang WC, Fu WM, Wang YB, Sun YX, Xu LL,
Wong CW, Chan KM, Li G, Waye MM and Zhang JF: H19 activates Wnt
signaling and promotes osteoblast differentiation by functioning as
a competing endogenous RNA. Sci Rep. 6:201212016. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Li X, Liu R, Huang Z, Gurley EC, Wang X,
Wang J, He H, Yang H, Lai G, Zhang L, et al: Cholangiocyte-derived
exosomal long noncoding RNA H19 promotes cholestatic liver injury
in mouse and humans. Hepatology. 68:599–615. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Yang JJ, She Q, Yang Y, Tao H and Li J:
DNMT1 controls LncRNA H19/ERK signal pathway in hepatic stellate
cell activation and fibrosis. Toxicol Lett. 295:325–334. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Yang JJ, Liu LP, Tao H, Hu W, Shi P, Deng
ZY and Li J: MeCP2 silencing of LncRNA H19 controls hepatic
stellate cell proliferation by targeting IGF1R. Toxicology.
359-360:39–46. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Song Y, Liu C, Liu X, Trottier J, Beaudoin
M, Zhang L, Pope C, Peng G, Barbier O, Zhong X, et al: H19 promotes
cholestatic liver fibrosis by preventing ZEB1-mediated inhibition
of epithelial cell adhesion molecule. Hepatology. 66:1183–1196.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Xiao Y, Liu R, Li X, Gurley EC, Hylemon
PB, Lu Y, Zhou H and Cai W: Long noncoding RNA H19 contributes to
cholangiocyte proliferation and cholestatic liver fibrosis in
biliary atresia. Hepatology. 70:1658–1673. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Liu R, Li X, Zhu W, Wang Y, Zhao D, Wang
X, Gurley EC, Liang G, Chen W, Lai G, et al: Cholangiocyte-derived
exosomal long noncoding RNA H19 promotes hepatic stellate cell
activation and cholestatic liver fibrosis. Hepatology.
70:1317–1335. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Fernandes JCR, Acuña SM, Aoki JI,
Floeter-Winter LM and Muxel SM: Long non-coding RNAs in the
regulation of gene expression: Physiology and disease. Noncoding
RNA. 5:172019.
|
|
70
|
Xu F, Liu C, Zhou D and Zhang L:
TGF-β/SMAD pathway and its regulation in hepatic fibrosis. J
Histochem Cytochem. 64:157–167. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Fabregat I, Moreno-Càceres J, Sánchez A,
Dooley S, Dewidar B, Giannelli G and Ten Dijke P; IT-LIVER
Consortium: TGF-beta signalling and liver disease. FEBS J.
283:2219–2232. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Caja L, Dituri F, Mancarella S,
Caballero-Diaz D, Moustakas A, Giannelli G and Fabregat I: TGF-β
and the tissue microenvironment: Relevance in fibrosis and cancer.
Int J Mol Sci. 19:12942018. View Article : Google Scholar
|
|
73
|
Martens L, Rühle F and Stoll M: LncRNA
secondary structure in the cardiovascular system. Noncoding RNA
Res. 2:137–142. 2017. View Article : Google Scholar
|
|
74
|
Zampetaki A, Albrecht A and Steinhofel K:
Long non-coding RNA structure and function: Is there a link? Front
Physiol. 9:12012018. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
He Z, Yang D, Fan X, Zhang M, Li Y, Gu X
and Yang M: The roles and mechanisms of lncRNAs in liver fibrosis.
Int J Mol Sci. 21:14822020. View Article : Google Scholar :
|
|
76
|
Kuo CC, Hänzelmann S, Sentürk Cetin N,
Frank S, Zajzon B, Derks JP, Akhade VS, Ahuja G, Kanduri C, Grummt
I, et al: Detection of RNA-DNA binding sites in long noncoding
RNAs. Nucleic Acids Res. 47:e322019. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Teng KY and Ghoshal K: Role of noncoding
RNAs as biomarker and therapeutic targets for liver fibrosis. Gene
Expr. 16:155–162. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Roderburg C, Mollnow T, Bongaerts B,
Elfimova N, Vargas Cardenas D, Berger K, Zimmermann H, Koch A,
Vucur M, Luedde M, et al: Micro-RNA profiling in human serum
reveals compartment-specific roles of miR-571 and miR-652 in liver
cirrhosis. PLoS One. 7:e329992012. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Klose RJ and Bird AP: Genomic DNA
methylation: The mark and its mediators. Trends Biochem Sci.
31:89–97. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Shang Z, Yu J, Sun L, Tian J, Zhu S, Zhang
B, Dong Q, Jiang N, Flores-Morales A, Chang C and Niu Y: LncRNA
PCAT1 activates AKT and NF-κB signaling in castration-resistant
prostate cancer by regulating the PHLPP/FKBP51/IKKα complex.
Nucleic Acids Res. 47:4211–4225. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Gujar H, Weisenberger DJ and Liang G: The
roles of human DNA methyltransferases and their isoforms in shaping
the epigenome. Genes (Basel). 10:1722019. View Article : Google Scholar
|
|
82
|
Edwards JR, Yarychkivska O, Boulard M and
Bestor TH: DNA methylation and DNA methyltransferases. Epigenetics
Chromatin. 10:232017. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Arand J, Spieler D, Karius T, Branco MR,
Meilinger D, Meissner A, Jenuwein T, Xu G, Leonhardt H, Wolf V and
Walter J: In vivo control of CpG and non-CpG DNA methylation by DNA
methyltransferases. PLoS Genet. 8:e10027502012. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Okano M, Bell DW, Haber DA and Li E: DNA
methyltransferases Dnmt3a and Dnmt3b are essential for de novo
methylation and mammalian development. Cell. 99:247–257. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Du J, Johnson LM, Jacobsen SE and Patel
DJ: DNA methylation pathways and their crosstalk with histone
methylation. Nat Rev Mol Cell Biol. 16:519–532. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Page A, Mann DA and Mann J: The mechanisms
of HSC activation and epigenetic regulation of HSCs phenotypes.
Curr Pathobiol Rep. 2:163–170. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Wilson CL, Mann DA and Borthwick LA:
Epigenetic reprogramming in liver fibrosis and cancer. Adv Drug
Deliv Rev. 121:124–132. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Dowson C and O'Reilly S: DNA methylation
in fibrosis. Eur J Cell Biol. 95:323–330. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Mann J, Oakley F, Akiboye F, Elsharkawy A,
Thorne AW and Mann DA: Regulation of myofibroblast
transdifferentiation by DNA methylation and MeCP2: Implications for
wound healing and fibrogenesis. Cell Death Differ. 14:275–285.
2007. View Article : Google Scholar
|
|
90
|
Komatsu Y, Waku T, Iwasaki N, Ono W,
Yamaguchi C and Yanagisawa J: Global analysis of DNA methylation in
early-stage liver fibrosis. BMC Med Genomics. 5:52012. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Götze S, Schumacher EC, Kordes C and
Häussinger D: Epigenetic changes during hepatic stellate cell
activation. PLoS One. 10:e01287452015. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
El Taghdouini A, Sørensen AL, Reiner AH,
Coll M, Verhulst S, Mannaerts I, Øie CI, Smedsrød B, Najimi M,
Sokal E, et al: Genome-wide analysis of DNA methylation and gene
expression patterns in purified, uncultured human liver cells and
activated hepatic stellate cells. Oncotarget. 6:26729–26745. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Xiong WJ, Hu LJ, Jian YC, Wang LJ, Jiang
M, Li W and He Y: Wnt5a participates in hepatic stellate cell
activation observed by gene expression profile and functional
assays. World J Gastroenterol. 18:1745–1752. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Miao CG, Yang YY, He X, Huang C, Huang Y,
Zhang L, Lv XW, Jin Y and Li J: Wnt signaling in liver fibrosis:
Progress challenges and potential directions. Biochimie.
95:2326–2335. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Jiang F, Parsons CJ and Stefanovic B: Gene
expression profile of quiescent and activated rat hepatic stellate
cells implicates Wnt signaling pathway in activation. J Hepatol.
45:401–409. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Bian EB, Huang C, Wang H, Chen XX, Zhang
L, Lv XW and Li J: Repression of Smad7 mediated by DNMT1 determines
hepatic stellate cell activation and liver fibrosis in rats.
Toxicol Lett. 224:175–185. 2014. View Article : Google Scholar
|
|
97
|
Bian EB, Huang C, Ma TT, Tao H, Zhang H,
Cheng C, Lv XW and Li J: DNMT1-mediated PTEN hypermethylation
confers hepatic stellate cell activation and liver fibrogenesis in
rats. Toxicol Appl Pharmacol. 264:13–22. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Page A, Paoli PP, Hill SJ, Howarth R, Wu
R, Kweon SM, French J, White S, Tsukamoto H, Mann DA and Mann J:
Alcohol directly stimulates epigenetic modifications in hepatic
stellate cells. J Hepatol. 62:388–397. 2015. View Article : Google Scholar :
|
|
99
|
Tian W, Fan Z, Li J, Hao C, Li M, Xu H, Wu
X, Zhou B, Zhang L, Fang M and Xu Y: Myocardin-related
transcription factor A (MRTF-A) plays an essential role in hepatic
stellate cell activation by epigenetically modulating TGF-β
signaling. Int J Biochem Cell Biol. 71:35–43. 2016. View Article : Google Scholar
|
|
100
|
Gregory GD, Vakoc CR, Rozovskaia T, Zheng
X, Patel S, Nakamura T, Canaani E and Blobel GA: Mammalian ASH1L is
a histone methyltransferase that occupies the transcribed region of
active genes. Mol Cell Biol. 27:8466–8479. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
101
|
Perugorria MJ, Wilson CL, Zeybel M, Walsh
M, Amin S, Robinson S, White SA, Burt AD, Oakley F, Tsukamoto H, et
al: Histone methyltransferase ASH1 orchestrates fibrogenic gene
transcription during myofibroblast transdifferentiation.
Hepatology. 56:1129–1139. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
102
|
Cao R and Zhang Y: The functions of E
(Z)/EZH2-mediated methylation of lysine 27 in histone H3. Curr Opin
Genet Dev. 14:155–164. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
103
|
Mann J, Chu DC, Maxwell A, Oakley F, Zhu
NL, Tsukamoto H and Mann DA: MeCP2 controls an epigenetic pathway
that promotes myofibroblast transdifferentiation and fibrosis.
Gastroenterology. 138:705–714. 714.e1–4. 2010. View Article : Google Scholar :
|
|
104
|
Martin-Mateos R, De Assuncao TM, Arab JP,
Jalan-Sakrikar N, Yaqoob U, Greuter T, Verma VK, Mathison AJ, Cao
S, Lomberk G, et al: Enhancer of zeste homologue 2 inhibition
attenuates TGF-β dependent hepatic stellate cell activation and
liver fibrosis. Cell Mol Gastroenterol Hepatol. 7:197–209. 2019.
View Article : Google Scholar
|
|
105
|
Panebianco C, Oben JA, Vinciguerra M and
Pazienza V: Senescence in hepatic stellate cells as a mechanism of
liver fibrosis reversal: A putative synergy between retinoic acid
and PPAR-gamma signalings. Clin Exp Med. 17:269–280. 2017.
View Article : Google Scholar
|
|
106
|
Schwartz YB and Pirrotta V: Polycomb
silencing mechanisms and the management of genomic programmes. Nat
Rev Genet. 8:9–22. 2007. View Article : Google Scholar
|
|
107
|
Hammond CM, Strømme CB, Huang H, Patel DJ
and Groth A: Histone chaperone networks shaping chromatin function.
Nat Rev Mol Cell Biol. 18:141–158. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
108
|
Mannaerts I, Nuytten NR, Rogiers V,
Vanderkerken K, van Grunsven LA and Geerts A: Chronic
administration of valproic acid inhibits activation of mouse
hepatic stellate cells in vitro and in vivo. Hepatology.
51:603–614. 2010. View Article : Google Scholar
|
|
109
|
Mannaerts I, Eysackers N, Onyema OO, Van
Beneden K, Valente S, Mai A, Odenthal M and van Grunsven LA: Class
II HDAC inhibition hampers hepatic stellate cell activation by
induction of microRNA-29. PLoS One. 8:e557862013. View Article : Google Scholar : PubMed/NCBI
|
|
110
|
Shaker ME, Ghani A, Shiha GE, Ibrahim TM
and Mehal WZ: Nilotinib induces apoptosis and autophagic cell death
of activated hepatic stellate cells via inhibition of histone
deacetylases. Biochim Biophys Acta. 1833:1992–2003. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
111
|
Duarte S, Baber J, Fujii T and Coito AJ:
Matrix metalloproteinases in liver injury, repair and fibrosis.
Matrix Biol. 44-46:147–156. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
112
|
Qin L and Han YP: Epigenetic repression of
matrix metalloproteinases in myofibroblastic hepatic stellate cells
through histone deacetylases 4: Implication in tissue fibrosis. Am
J Pathol. 177:1915–1928. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
113
|
Ding G, Li W, Liu J, Zeng Y, Mao C, Kang Y
and Shang J: LncRNA GHET1 activated by H3K27 acetylation promotes
cell tumorigenesis through regulating ATF1 in hepatocellular
carcinoma. Biomed Pharmacother. 94:326–331. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
114
|
Yang F, Huo XS, Yuan SX, Zhang L, Zhou WP,
Wang F and Sun SH: Repression of the long noncoding RNA-LET by
histone deacetylase 3 contributes to hypoxia-mediated metastasis.
Mol Cell. 49:1083–1096. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
115
|
Pope C, Mishra S, Russell J, Zhou Q and
Zhong XB: Targeting H19, an imprinted long non-coding RNA, in
hepatic functions and liver diseases. Diseases. 5:112017.
View Article : Google Scholar :
|
|
116
|
Zhu XT, Yuan JH, Zhu TT, Li YY and Cheng
XY: Long noncoding RNA glypican 3 (GPC3) antisense transcript 1
promotes hepatocellular carcinoma progression via epigenetically
activating GPC3. FEBS J. 283:3739–3754. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
117
|
Issa JJ, Roboz G, Rizzieri D, Jabbour E,
Stock W, O'Connell C, Yee K, Tibes R, Griffiths EA, Walsh K, et al:
Safety and tolerability of guadecitabine (SGI-110) in patients with
myelodysplastic syndrome and acute myeloid leukaemia: A
multicentre, randomised, dose-escalation phase 1 study. Lancet
Oncol. 16:1099–1110. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
118
|
Jansen YJL, Verset G, Schats K, Van Dam
PJ, Seremet T, Kockx M, Van Laethem JB and Neyns B: Phase I
clinical trial of decitabine (5-aza-2′-deoxycytidine) administered
by hepatic arterial infusion in patients with unresectable
liver-predominant metastases. ESMO Open. 4:e0004642019. View Article : Google Scholar
|
|
119
|
Kuang Y, El-Khoueiry A, Taverna P,
Ljungman M and Neamati N: Guadecitabine (SGI-110) priming
sensitizes hepatocellular carcinoma cells to oxaliplatin. Mol
Oncol. 9:1799–1814. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
120
|
Eckschlager T, Plch J, Stiborova M and
Hrabeta J: Histone deacetylase inhibitors as anticancer drugs. Int
J Mol Sci. 18:14142017. View Article : Google Scholar :
|
|
121
|
Park KC, Park JH, Jeon JY, Kim SY, Kim JM,
Lim CY, Lee TH, Kim HK, Lee HG, Kim SM, et al: A new histone
deacetylase inhibitor improves liver fibrosis in BDL rats through
suppression of hepatic stellate cells. Br J Pharmacol.
171:4820–4830. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
122
|
Liu Y, Wang Z, Wang J, Lam W, Kwong S, Li
F, Friedman SL, Zhou S, Ren Q, Xu Z, et al: A histone deacetylase
inhibitor, largazole, decreases liver fibrosis and angiogenesis by
inhibiting transforming growth factor-β and vascular endothelial
growth factor signalling. Liver Int. 33:504–515. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
123
|
Watanabe T, Tajima H, Hironori H,
Nakagawara H, Ohnishi I, Takamura H, Ninomiya I, Kitagawa H,
Fushida S, Tani T, et al: Sodium valproate blocks the transforming
growth factor (TGF)-β1 autocrine loop and attenuates the
TGF-β1-induced collagen synthesis in a human hepatic stellate cell
line. Int J Mol Med. 28:919–925. 2011.PubMed/NCBI
|
|
124
|
Niki T, Rombouts K, De Bleser P, De Smet
K, Rogiers V, Schuppan D, Yoshida M, Gabbiani G and Geerts A: A
histone deacetylase inhibitor, trichostatin A, suppresses
myofibroblastic differentiation of rat hepatic stellate cells in
primary culture. Hepatology. 29:858–867. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
125
|
Ding D, Chen LL, Zhai YZ, Hou CJ, Tao LL,
Lu SH, Wu J and Liu XP: Trichostatin A inhibits the activation of
Hepatic stellate cells by Increasing C/EBP-α Acetylation in vivo
and in vitro. Sci Rep. 8:43952018. View Article : Google Scholar
|
|
126
|
Smith E and Shilatifard A: The chromatin
signaling pathway: Diverse mechanisms of recruitment of
histone-modifying enzymes and varied biological outcomes. Mol Cell.
40:689–701. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
127
|
Yadav T, Quivy JP and Almouzni G:
Chromatin plasticity: A versatile landscape that underlies cell
fate and identity. Science. 361:1332–1336. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
128
|
Hu B, Gharaee-Kermani M, Wu Z and Phan SH:
Essential role of MeCP2 in the regulation of myofibroblast
differentiation during pulmonary fibrosis. Am J Pathol.
178:1500–1508. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
129
|
Bian EB, Huang C, Wang H, Chen XX, Tao H,
Zhang L, Lv XW and Li J: The role of methyl-CpG binding protein 2
in liver fibrosis. Toxicology. 309:9–14. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
130
|
Mellén M, Ayata P, Dewell S, Kriaucionis S
and Heintz N: MeCP2 binds to 5hmC enriched within active genes and
accessible chromatin in the nervous system. Cell. 151:1417–1430.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
131
|
Bárcena-Varela M, Caruso S, Llerena S,
Aacute;lvarez-Sola G, Uriarte I, Latasa MU, Urtasun R, Rebouissou
S, Alvarez L, Jimenez M, et al: Dual targeting of histone
methyltransferase G9a and DNA-Methyltransferase 1 for the treatment
of experimental hepatocellular carcinoma. Hepatology. 69:587–603.
2019. View Article : Google Scholar
|
|
132
|
Moran-Salvador E, Garcia-Macia M,
Sivaharan A, Sabater L, Zaki MYW, Oakley F, Knox A, Page A, Luli S,
Mann J and Mann DA: Fibrogenic activity of MECP2 is regulated by
phosphorylation in hepatic stellate cells. Gastroenterology.
157:1398–1412.e9. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
133
|
Florean C: Food that shapes you: How diet
can change your epigenome. Science in School. May 13–2014.Epub
ahead of print.
|
|
134
|
Yang JJ, Tao H, Deng ZY, Lu C and Li J:
Non-coding RNA-mediated epigenetic regulation of liver fibrosis.
Metabolism. 64:1386–1394. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
135
|
Jiang X, Tsitsiou E, Herrick SE and
Lindsay MA: MicroRNAs and the regulation of fibrosis. FEBS J.
277:2015–2021. 2010. View Article : Google Scholar : PubMed/NCBI
|