Introduction
In orthodontic treatment, force is applied to the
teeth using appliances and transmitted to the periodontal tissues,
which eventually leads to tissue reconstruction of the gingiva,
periodontal ligaments and alveolar bone, thereby moving the
misaligned teeth to their normal positions (1). The adaptive reconstruction of
periodontal tissue is the biological basis of tooth movement
(2). As an important part of the
periodontal tissue, gingiva is often involved in the process of
orthodontic treatment, such as gingiva proliferation, recession and
retraction. Such malformations persist for a number of years after
treatment (3). Gingival
reconstruction not only affects the rate of tooth movement and the
stability after correction (4,5),
but also leads to recurrence after orthodontic treatment (6).
Orthodontic pressure is converted into biological
signals, which are transmitted from the extracellular to the
intracellular environment, inducing a series of associated signal
transduction reactions that affect the process of gingival tissue
remodeling (7). Human gingival
fibroblasts (HGFs) are the main cells in the gingival tissue, where
they receive and transmit signals (8). In previous decades, although a
number of groups have widely studied the reconstruction of alveolar
bone and periodontal membrane under compressive force (9), mechanisms underlying gingival
reconstruction remain largely unexplored. Our previous study showed
that compressive force could promote the expression of transforming
growth factor (TGF)-β1 and the proliferation of HGFs (10). TGF-β is an important growth factor
that regulates cell proliferation and apoptosis (11,12); however, the factors and signaling
pathways involving TGF-β that regulate HGF proliferation and
apoptosis remain unknown.
Smad4, a key downstream factor of TGF-β signaling
pathway, plays a crucial regulatory role (13). In the classical TGF-β pathway,
activated Smad2/3 and Smad4 form the SMAD complex, which
translocates to the nucleus (14). Smad4 is considered to be a tumor
suppressor gene that is closely related to cell apoptosis (15-17). Imbalance of Smad4 expression in
the TGF-β signaling pathway may lead to various diseases, including
cancers, fibrous lesions, metabolic diseases and cardiovascular
diseases (18-21). It has been found that Smad4
affects apoptosis by regulating apoptosis-related proteins; for
example, the low expression of Smad4 in colon cancer can inhibit
cell apoptosis by promoting Bcl-2 and Bcl-w expression, and
decreasing caspase-3 and caspase-9 expression (22). Therefore, it is hypothesized that
Smad4 may play a pivotal role in the proliferation and apoptosis of
HGFs under compressive force.
Biodegradable poly(lactide-co-glycolide) (PLGA) has
become one of the most widely used biological scaffold materials
(23-26). Compared with other scaffold
materials, PLGA possesses good biological properties (27-29). Compared with a two-dimensional
culture, a three-dimensional (3D) culture can more effectively
mimic the natural environment within the human body, and improve
the accuracy and reliability of experimental results. Therefore, a
3D HGF-PLGA culture model was established to explore the roles of
Smad4, caspase-3 and Bcl-2 in regulating proliferation and
apoptosis pathways in HGFs. The present study aimed to further
understand the molecular mechanisms underlying orthodontic gingival
reconstruction.
Materials and methods
3D culture of HGFs
HGFs were cultured in 3D PLGA as previously reported
(10). Normal gingival tissues
were collected from healthy teeth that received orthodontic
extraction or gingival resection. Patients enrolled in the study
had no systemic diseases, and were aged 10-14 years, with 17 males
and 19 females. Normal gingival tissues were cut into small pieces
(1×3 mm) and spread flat in culture flasks. Dulbecco's modified
Eagle's medium (HyClone; Cytiva) with 20% fetal bovine serum (FBS;
Gibco; Thermo Fisher Scientific, Inc.) and 100 U/ml antibiotics
(Gibco; Thermo Fisher Scientific, Inc.) were put into the flasks
for primary culture. After the primary cells had proliferated, they
were digested with 0.25% trypsin (Sigma-Aldrich; Merck KGaA)
containing 100 ng/ml DNase (Roche Diagnostics) for 1 min. The
digestion was stopped by adding Dulbecco's modified Eagle's medium
with 10% FBS and 100 U/ml antibiotics. Cells were cultured at 37°C
with 5% CO2. PLGA scaffolds (molar ratio, 75:25; pore
diameter, 100-200 µm; porosity, 85%; size, 2×2 cm x 300
µm; Shandong Medical Instrument Research Institute) were cut
into 1×1×0.2 cm sizes. All donors and their guardians provided
written informed consent, and samples were collected between March
2018 and March 2019 at the Department of Oral and Maxillofacial
Surgery of the Stomatological Hospital affiliated to Guangxi
Medical University (Nanning, China).
Application of compressive force
After 4-6 generations, HGFs were digested, and then
the cell count was adjusted to 1×105/ml. HGF suspension
(0.5-1 ml) was dripped on each piece of PLGA scaffold to enable
more cells to attach to the scaffold. The scaffold was stored in an
incubator for 2 h at 37°C. After confirming that the cells were
stably attached to the PLGA, the PLGA were cultured in 6-well
plates with 1 ml containing 10% fetal bovine serum medium for a
further 24 h in an incubator with a constant temperature of 37°C.
HGFs were continuously subjected to a compressive stress of 25
g/cm2 for 12, 24, 48 and 72 h by placing glass bottles
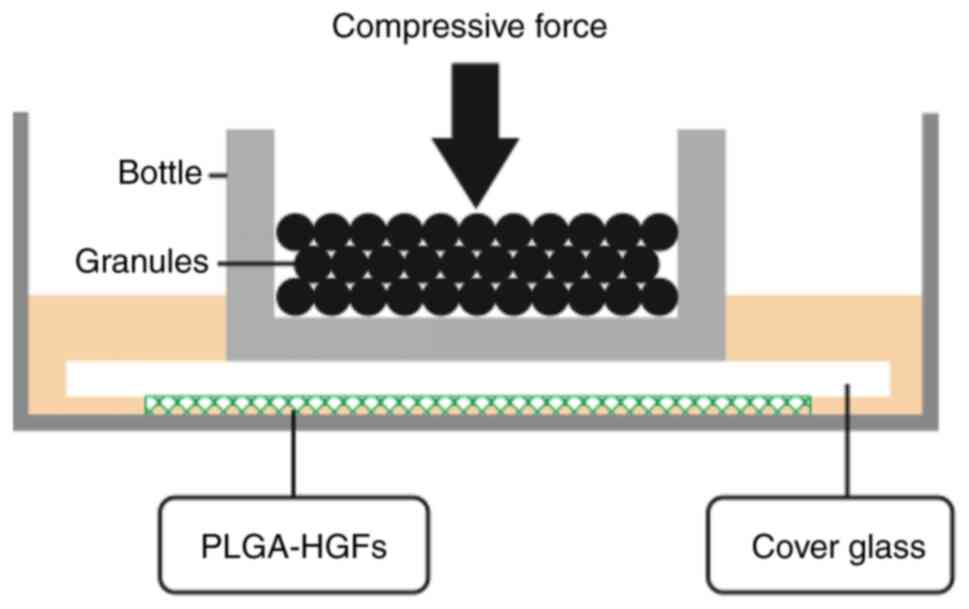
containing lead granules on the top, as showed in Fig. 1.
Cell viability
HGFs were seeded into 96-well plates and the cell
count was adjusted to 5×104 cells/well. Then, Cell
Counting Kit-8 (CCK-8) assays were performed after 0, 12, 24, 48
and 72 h of compressive force. CCK-8 solution (10 µl) was
added to each well (1:10), and the cells were cultured for an
additional 2 h before measuring the absorbance using a microplate
reader (Model 3550; Thermo Fisher Scientific, Inc.) at 450 nm.
Overexpression of Smad4 in HGFs
After 4-6 generations, HGFs were digested, and the
cell count was adjusted. HGFs were seeded in 96-well plates with
5×103 cells/well and cultured for 12 h. Then, lentivirus
(Lv-control or Lv-Smad4; plasmid backbone, GV492; Shanghai GeneChem
Co., Ltd.) and transfection enhancement solution HiTrans A
(Shanghai GeneChem Co., Ltd.) was added (MOI=50). After 24 h,
aspirate the supernatant in the culture plate and replace with a
new serum medium. Additionally, a blank control group constituting
normal HGFs was included in the study. The infection efficiency was
observed via green fluorescence using a fluorescence microscope
(Olympus Corporation) 72 h after infection. Reverse
transcription-quantitative (RT-q)PCR and western blotting (WB) were
performed to analyze Smad4 expression at the mRNA and protein
levels.
RT-qPCR
Total RNA from HGFs was isolated using
TRIzol® reagent (Invitrogen; Thermo Fisher Scientific,
Inc.). Then, RNA quality was tested using an RNA 6000 Nano kit
(Agilent Technologies, Inc.). cDNA was synthesized using an
ExScript RT reagent kit (Takara Bio, Inc.), according to the
manufacturer's protocol (37°C for 15 min and 85°C for 5 sec,
followed by storage at 4°C until further use. The RT-qPCR was
performed on an ABI 7300 Real-Time PCR system (Applied Biosystems;
Thermo Fisher Scientific, Inc.) with SYBR Premix Ex Taq (Takara
Biotechnology Co., Ltd.) at 95°C for 30 sec, followed by 40 cycles
at 95°C for 5 sec and 60°C for 31 sec, after which a melt curve
analysis was performed at 95°C for 15 sec, 60°C for 1 min and 95°C
for 15 sec. The primer sequences were: Smad4 forward, 5′-TGG TAG
GAT TGT GAG GAT TAA ATC AG-3′ and reverse, 5′-ACC ATC CTA ACA CAC
CTA ATT TAG TC-3′; caspase-3 forward, 5′-CCT AGC GGA TGG GTG CTA
TT-3′ and reverse, 5′-CTG AGG TTT GCT GCA TCG AC-3′; Bcl-2 forward,
5′-AAT ATC CAA TCC TGT GCT GCTA-3′ and reverse, 5′-GTC CAC GTT CTT
CAT TGT TAC TTC-3′; and GAPDH forward, 5′-TGT GTC CGT CGT GGA
TCTGA-3′ and reverse, 5′-TTG CTG TTG AAG TCG CAG GAG-3′. All
reactions were performed in triplicate and GAPDH was used as an
internal control. Each experiment was repeated at least three
times. The relative expression level was calculated using the
2−ΔΔCq method (30).
WB
Total protein was extracted from cells using RIPA
lysis buffer with PMSF (1,000:1; Pierce; Thermo Fisher Scientific,
Inc.). The protein concentration was detected using a BCA assay.
Proteins (30 µg/sample) and marker (5 µl) were loaded
onto 10% SDS-PAGE gels. Electrophoresis was conducted under a
constant voltage of 200 V for 45 min. Proteins were transferred to
PVDF membranes at 200 mA for 1.5 h. TBS-0.3% Tween-20 was used to
wash the PVDF membranes 3 times (5 min/wash). Membranes were then
blocked in 5% skimmed milk for 1 h at room temperature before being
incubated with monoclonal antibodies against human Smad4 (1:1,000;
cat. no. ab40759; Abcam), Bcl-2 (1:1,000; cat. no. 4223S; Cell
Signaling Technology, Inc.), caspase-3 (1:5,000; cat. no. ab32351;
Abcam) and GAPDH (1:5,000; cat. no. SAB4300645; Sigma-Aldrich;
Merck KGaA) for overnight incubation at 4°C, followed by incubation
for 1 h at room temperature with secondary anti-mouse/rabbit
horseradish peroxidase-conjugated antibodies (1:5,000; cat. nos.
SA00001-1/SA00001-2; ProteinTech Group, Inc.). After several
washes, protein bands were visualized using an Alpha Chemical
Luminescent Gel Imaging System (Fluor Chem HD2; ProteinSimple) with
ECL luminescence reagent (cat. no. abs920; Absin Bioscience, Inc.),
The relative density of protein expression was analyzed
quantitatively with Image Lab v3.0 software (Bio-Rad Laboratories,
Inc.).
Statistical analysis
SPSS 21 (IBM Corp.) was used for statistical
analysis of experimental data. Data are presented as the mean ± SD
of at least three independent experiments. Data were analyzed using
one-way or two-way ANOVA with post hoc tests (Dunnett's or Tukey)
for multiple comparisons. P<0.05 and α=0.05 were considered to
indicate a statistically significant difference.
Results
Compressive force induces the
proliferation of HGFs
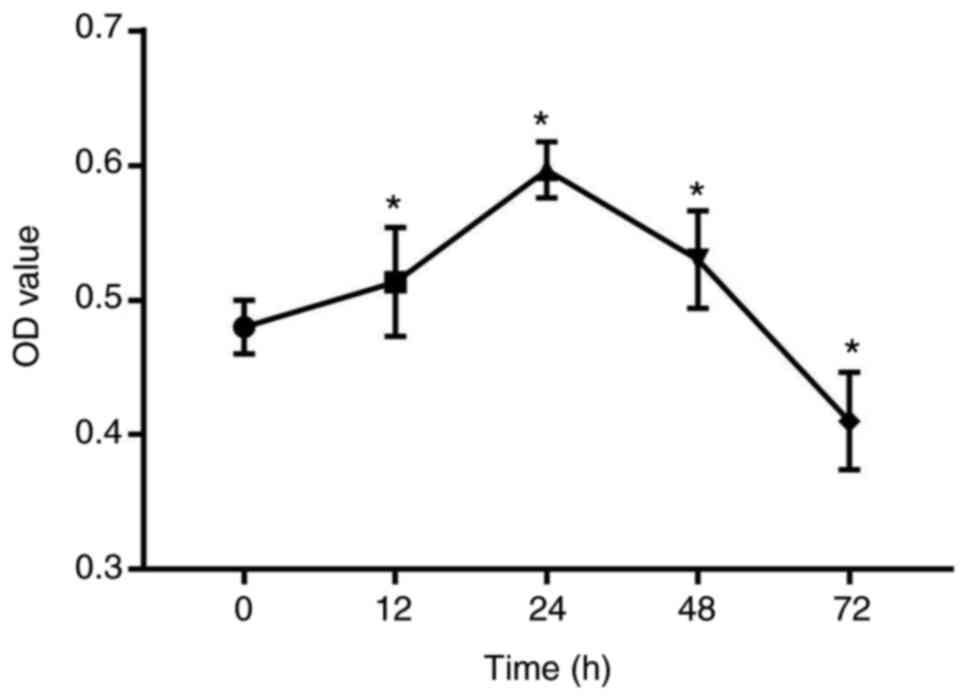
A 3D PLGA-HGFs culture model was previously
established by our group as showed in Fig. 1 (10). Compared with the control group (0
h), the proliferation ability of HGFs significantly increased under
compressive force for 12, 24 and 48 h, peaking at 24 h.
Subsequently, the proliferation of HGFs was inhibited and cell
viability decreased by 72 h (P<0.05; Fig. 2).
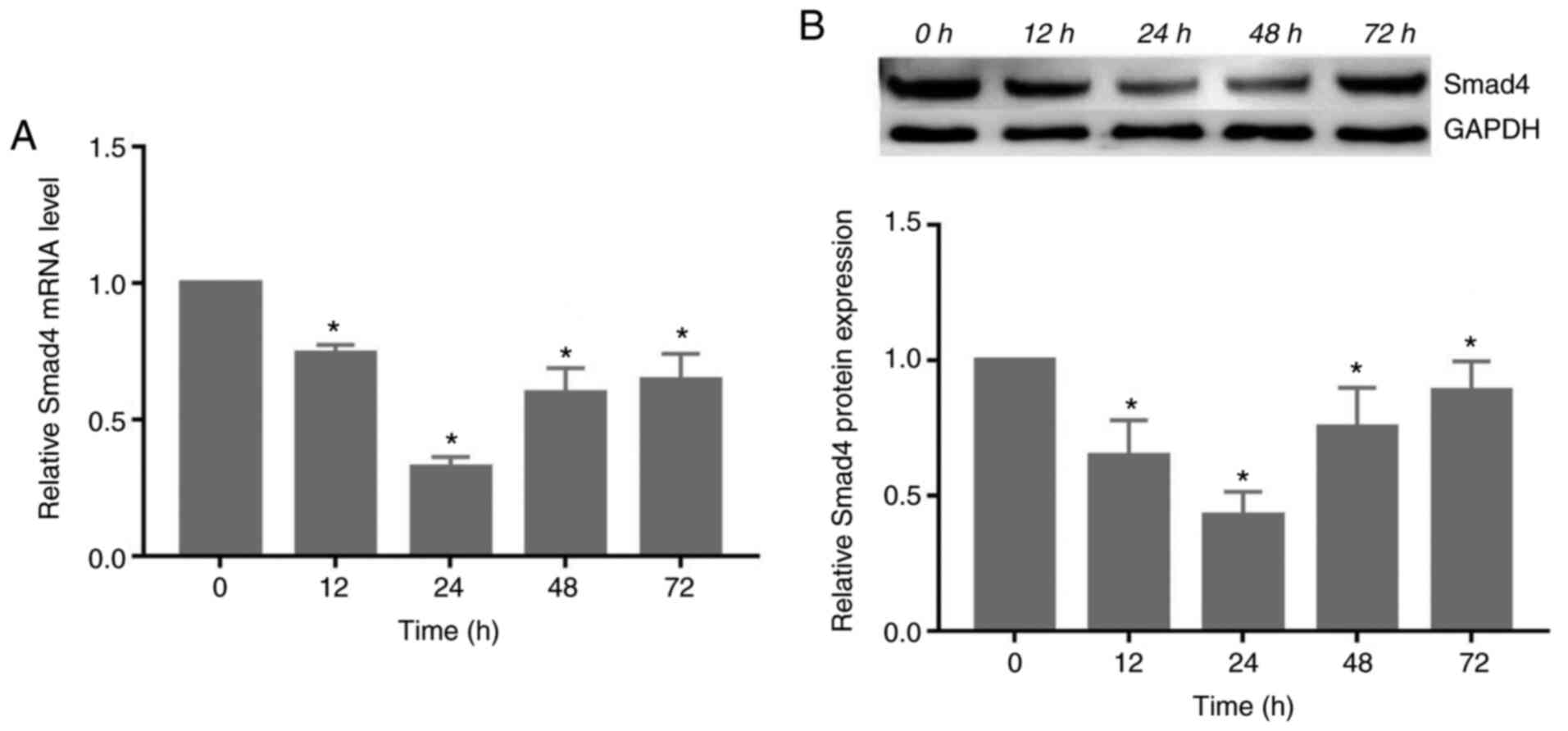
Compressive force decreases the
expression of Smad4 in HGFs
The gene and protein expression of Smad4 decreased
significantly following exposure to compressive force, reaching its
lowest level at 24 h. Subsequently, the expression levels increased
at 48 and 72 h (P<0.05; Fig.
3).
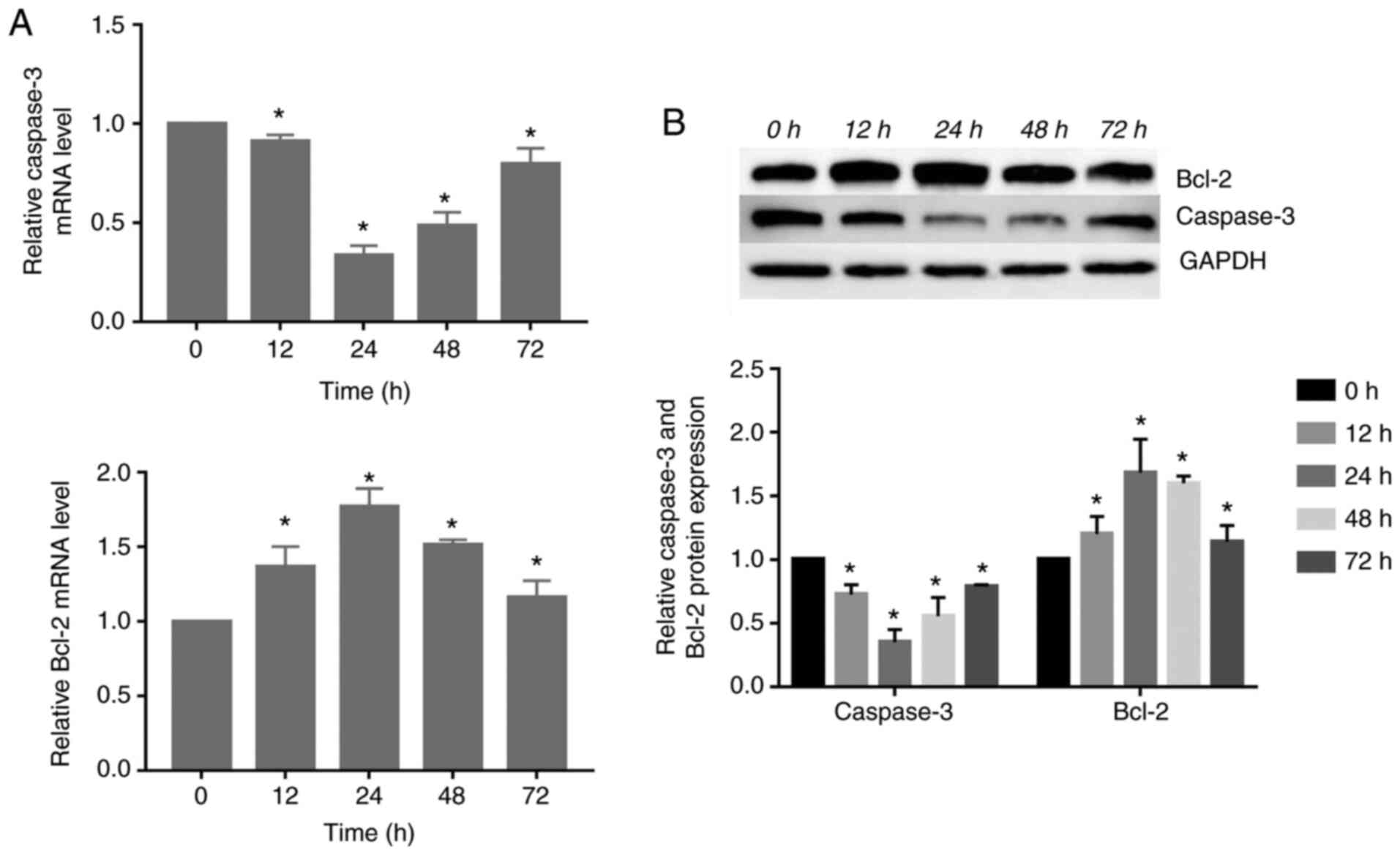
Expression of caspase-3 and Bcl-2 changed
in response to compressive force in HGFs
The gene and protein expression of caspase-3
decreased significantly, reaching its lowest level at 24 h.
Subsequently, its expression was increased at 48 and 72 h. However,
the gene and protein expression of Bcl-2 increased significantly
compared with the control group, peaking at 24 h. Subsequently, the
expression levels gradually decreased (P<0.05; Fig. 4).
Assessment of Smad4 mRNA and protein
levels
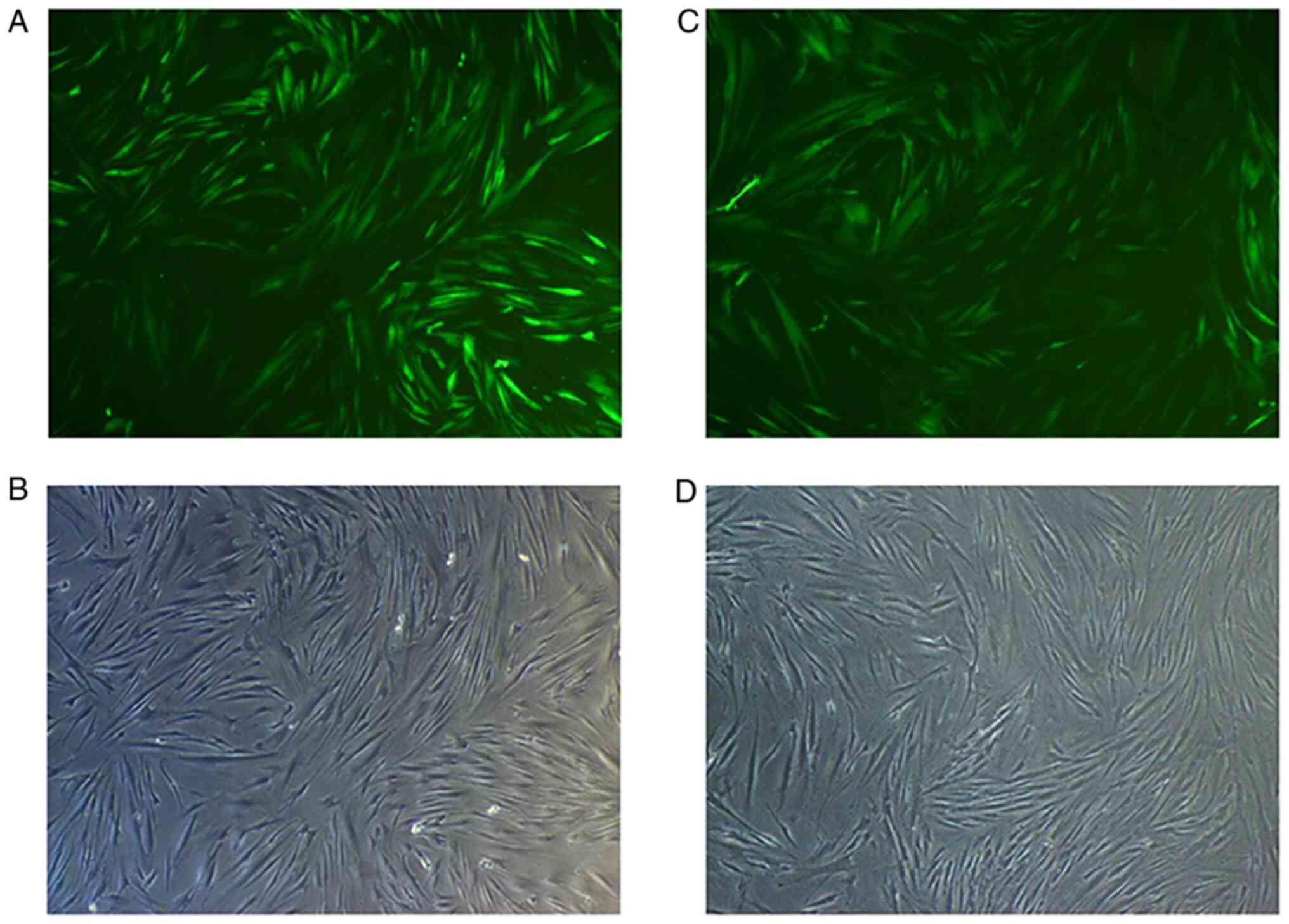
At 72 h after transfection of HGFs with Lv-Smad4,
the expression efficiency of enhanced green fluorescent protein was
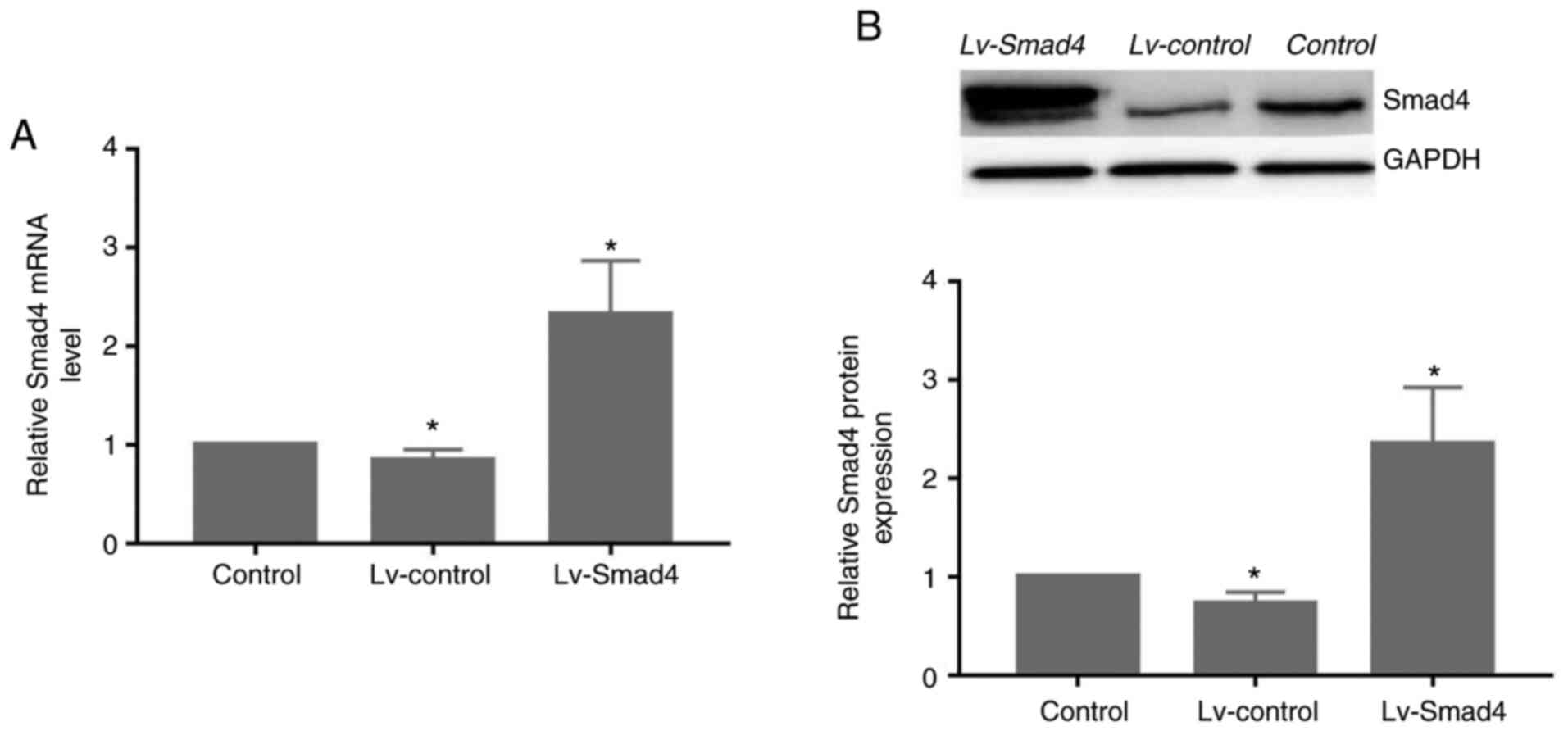
~90% (Fig. 5). Furthermore, gene
and protein expression analysis revealed that the levels of Smad4
(Fig. 6) were significantly
upregulated in the Lv-Smad4 group compared with the Lv-control and
blank control groups (P<0.01). The expression levels in the
Lv-control group significantly decreased compared with the control
cells (P<0.05), which indicated that the lentiviral system
effectively delivered the exogenous Smad4 gene.
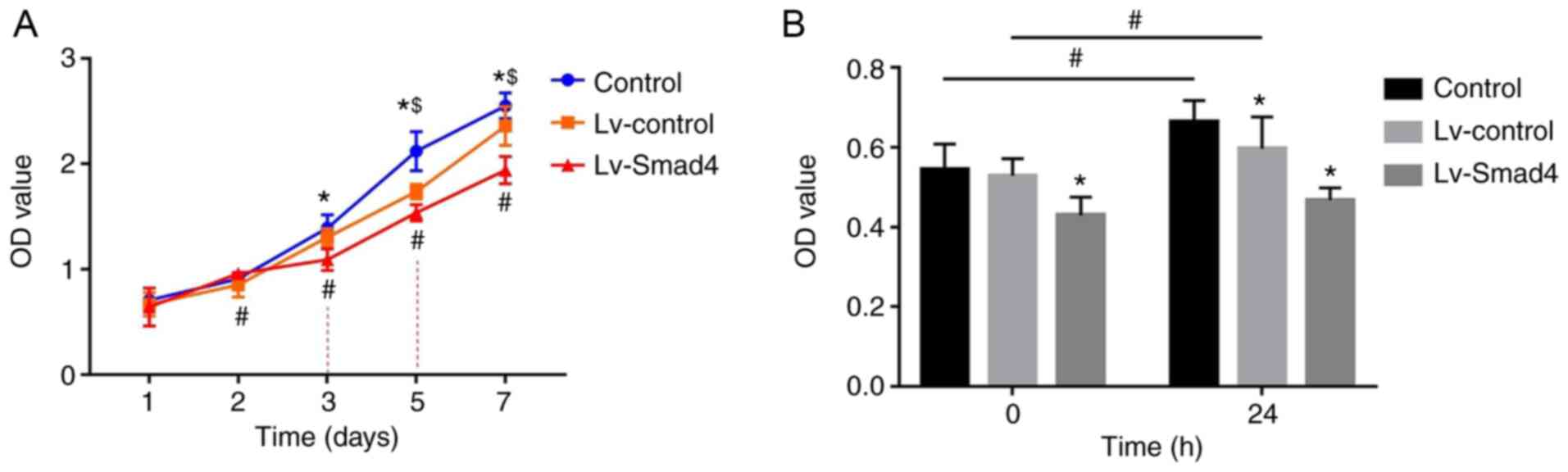
Smad4 overexpression inhibits the effects
of compressive force on HGF proliferation
The CCK-8 assay results showed that the absorbance
values of HGFs increased with the increased culture time
(P<0.05; Fig. 7A). In the
first two days, the difference in optical density (OD) values
between the Lv-Smad4, Lv-control and control groups were not
statistically significant. However, the OD value of the Lv-Smad4
group decreased compared with the Lv-control and control groups
from day 3onwards (P<0.05). Additionally, all three groups were
significantly different to each other from day 5 onwards, with the
highest OD values in the control group and the lowest in the
Lv-Smad4 group (P<0.05). Furthermore, the CCK-8 results revealed
the participation of Smad4 in HGF proliferation induced by
compressive force, as shown in Fig.
7B. There was no significant increase in HGF proliferation in
the Lv-Smad4 group after 24 h of compressive force compared with
before force was applied (P>0.05; Fig. 7B). Conversely, in the Lv-control
and Lv-Smad4 groups, stimulation with continuous compressive force
for 24 h significantly increased HGF proliferation (P<0.05;
Fig. 7B).
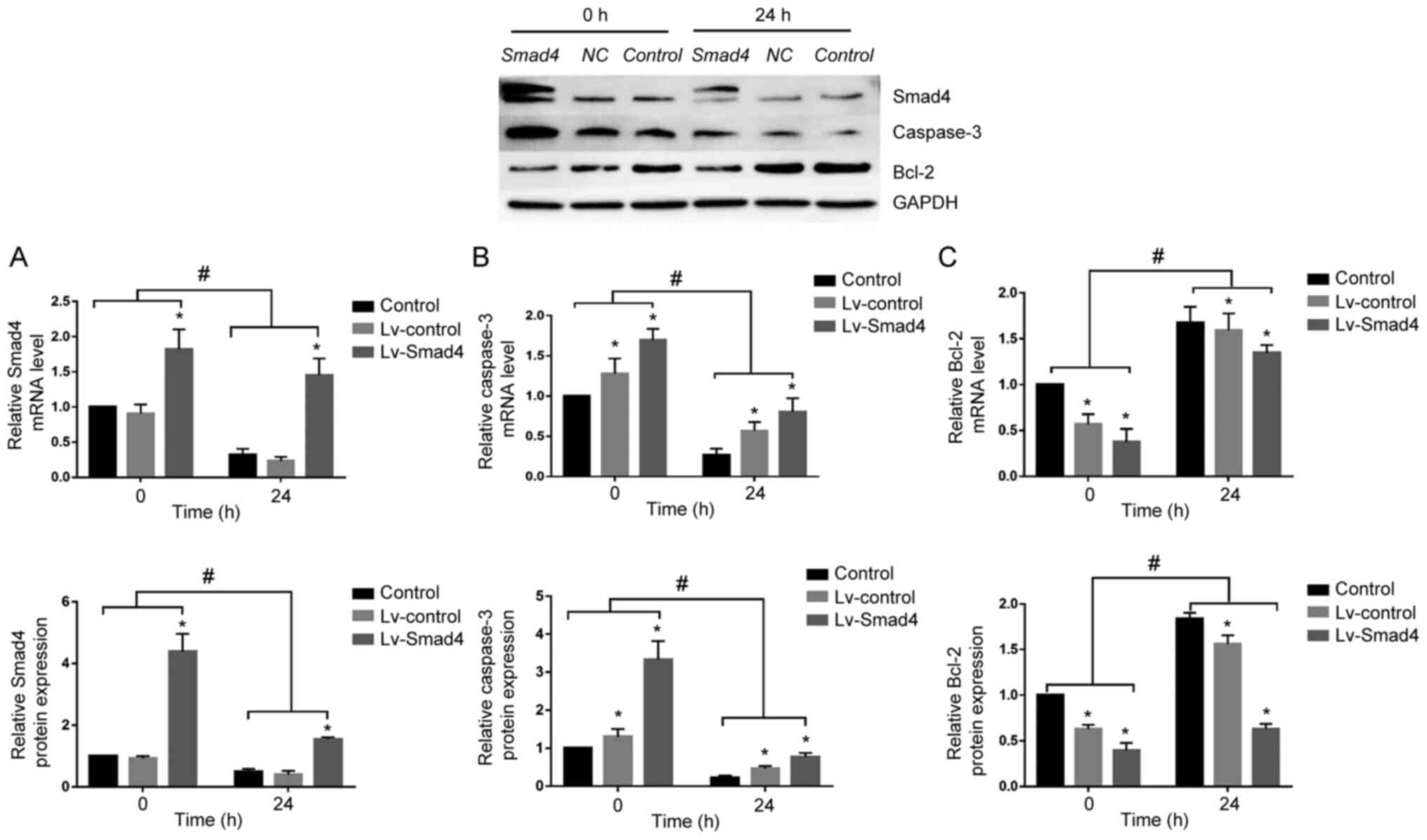
Smad4 overexpression reverses the
expression of caspase-3 and Bcl-2 in HGFs
To determine whether Smad4 mediates caspase-3 and
Bcl-2 expression under compressive force, Smad4 was overexpressed
in HGFs. Although continuous compressive force stimulation for 24 h
significantly decreased the expression of Smad4 in the Lv-Smad4
group, Smad4 expression was upregulated compared with in the
Lv-control and control groups. Furthermore, it was demonstrated
that with or without compressive force stimulation, the expression
levels of Bcl-2 and caspase-3 were downregulated and upregulated,
respectively, in the Lv-Smad4 group compared with the control group
(Fig. 8). These findings
indicated that Smad4 is an important apoptosis-associated factor in
HGFs; compressive force inhibited HGF apoptosis and promoted
proliferation by downregulating the expression of Smad4.
Discussion
Clinically, gingival hyperplasia and accumulation of
the stressed side are commonly found during orthodontic treatment
(1-4). Exploring this phenomenon, Schwarz
(31) reported that when the
orthodontic force did not exceed the end capillary pressure of the
periodontal ligament on the root surface of the tooth, it was most
conducive to the movement of teeth, and exceeding this threshold
results in the necrosis of periodontal tissue. In the human body
and most mammals, this pressure is ~20-26 g/cm2.
Therefore, it has been proposed that under reasonable pressure,
weight loading models can more effectively simulate changes in the
periodontal tissue on the compressed side of the body (32,33). For example, Kook et al
(34) and Li et al
(35) used force-loading models
to demonstrate that the proliferative ability of periodontal
ligament fibroblasts is enhanced under suitable compressive
pressure stimulation. In addition, Baker et al (36) reported that the elastic modulus of
porous PLGA scaffolds is ~4 MPa, which is close to the elastic
modulus of human gingival tissue (37). Therefore, the present study was
conducted based on these findings and early experimental results of
our research group, such as a PLGA-HGFs 3D culture model, an
optimal value of 25 g/cm2 and high expression of TGF-β1
(10).
Studies have found that Smad4 is closely related to
cell proliferation and apoptosis (15-17). It has been found in tumor research
that Smad4 is a tumor suppressor gene, and its decreased or absent
expression in a variety of tumors has been found to lead to the
abnormal proliferation of tissues and cells (19,20); however, it has been found in some
studies of cardiovascular diseases that TGF-β1/Smad4 can inhibit
cell apoptosis. For example, Yin et al (38) silenced Smad4 in human aortic
vascular smooth muscle cells (HASMCs) and found that low expression
of Smad4 decreased the anti-angiotensin stimulation ability of
HASMCs, leading to HASMC migration and apoptosis. Therefore,
studies have suggested that the effect of TGF-β1/Smad4 signaling on
cell apoptosis is dependent on the cell type and environment.
In the present study, HGF proliferation and
expression of Smad4 were time-dependent. The proliferation ability
of HGF enhanced and peaked after 24 h of compressive force, then
began to decrease with longer exposure. Conversely, the expression
of Smad4 decreased and reached a nadir at 24 h, then began to
increase. These results suggested that HGFs underwent a series of
proliferative and apoptotic processes under compressive force that
involved Smad4.
To further explore the relationship between Smad4
and cell proliferation, Smad4 was overexpressed in HGFs. The
results of CCK-8 assays revealed that proliferation in the Lv-Smad4
group was significantly decreased compared with in the Lv-control
and the control groups from day 3, indicating that Smad4 inhibited
the proliferation of HGFs compared with normal cells, which may be
due to increased apoptosis. Additionally, the proliferation of HGFs
under compressive force was significantly inhibited in the Lv-Smad4
group. These results suggested that Smad4 is associated with the
proliferation and apoptosis of HGFs under compressive force, thus
highlighting Smad4 as a pro-apoptotic factor in HGFs.
Apoptosis is a gene-regulated programmed cell death
process; it is a physiological response of cells to adapt to the
environment and maintain the stability of the internal environment
of the body (39). There are two
main pathways of apoptosis: The mitochondrial pathway and the death
receptor pathway. Caspase and Bcl-2 family proteins are involved in
both apoptosis pathways (40,41). Caspase-3 plays a key role in
promoting apoptosis and is considered the executor of apoptosis
(42). The Bcl-2 protein family
is a large complex family, mostly located in the mitochondria,
endoplasmic reticulum and continuous perinuclear membrane, that
mediates the mitochondrial apoptosis pathway (43). Bcl-2 can inhibit apoptosis and is
one of the most important oncogenes in apoptotic research (44). It has been reported that
downregulated expression of Smad4 in breast cancer can inhibit cell
apoptosis by decreasing caspase-3 and caspase-9, and promoting
Bcl-2 expression (45).
Overexpression of Smad4 in human vascular endothelial cells has
shown that Smad4 can inhibit endothelial cell proliferation by
promoting caspase-3 activation and blocking the cell cycle
(46). However, the role of
compressive force and Smad4 in the apoptosis of HGFs, and the
relationship between Smad4, caspase-3 and Bcl-2, remain
undetermined.
In the present study, the expression of Bcl-2 and
caspase-3 were also highly time-dependent. The expression of Bcl-2
was upregulated under compressive force and peaked at 24 h, at
which point it began to decrease. Conversely, the expression of
caspase-3 decreased, reaching its lowest point after 24 h of
compressive force stimulation, at which point it began to increase.
These results suggested that Bcl-2 and caspase-3 were involved in
apoptotic processes in HGFs under compressive force.
It was further shown that, compared with normal
cells, caspase-3 expression was increased in the Lv-Smad4 group,
while Bcl-2 expression was downregulated. This suggested that Smad4
regulated the expression of caspase-3 and Bcl-2 in HGFs without
compressive force. Furthermore, it was found that Lv-Smad4 cells
exhibited downregulated expression of Bcl-2 and upregulated
expression of caspase-3 compared with Lv-control and control group
cells under compressive force, further suggesting that compressive
force regulated the expression of caspase-3 and Bcl-2 via Smad4,
thereby regulating HGF proliferation and apoptosis.
In conclusion, the present findings indicated that
Smad4 promoted HGF proliferation by regulating caspase-3 and Bcl-2
under compressive force. Therefore, Smad4 may be an important
target for preventing gingival hyperplasia after orthodontic
treatment. However, there were certain limitations to the present
study. For example, proliferation and apoptosis were only analyzed
using CCK-8 assays; additional methods should be used, such as flow
cytometry. In addition, the discussion on Smad4 was limited to
enhancing the expression of Smad4; subsequent studies should
further validate these findings by silencing Smad4.
Funding
The present study was supported by grants from the
National Natural Science Foundation of China (grant no. 81070860)
and the Natural Science Foundation of Guangxi Province (grant no.
2013GXNSFAA019183).
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
SZ conceived and coordinated the study, designed,
performed and analyzed the experiments, and wrote the paper. LN,
YW, LW and SM collected and analyzed data, and revised the
manuscript. All authors approved the final version of the
manuscript, LN and SM confirm the authenticity of all the raw
data.
Ethics approval and consent to
participate
The present study was approved by the Hospital
Institutional Review Board (approval no. 20150304-22) of Guangxi
Medical University. All donors and their guardians signed an
informed consent form.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgments
Not applicable.
References
|
1
|
Dutra EH, Ahmida A, Lima A, Schneider S,
Nanda R and Yadav S: The effects of alveolar decortications on
orthodontic tooth movement and bone remodelling in rats. Eur J
Orthod. 40:423–429. 2018. View Article : Google Scholar :
|
|
2
|
Massaeli H, Viswanathan D, Pillai DG and
Mesaeli N: Endoplasmic reticulum stress enhances endocytosis in
calreticulin deficient cells. Biochim Biophys Acta Mol Cell Res.
1866:727–736. 2019. View Article : Google Scholar
|
|
3
|
Redlich M, Shoshan S and Palmon A:
Gingival response to orthodontic force. Am J Orthod Dentofacial
Orthop. 116:152–158. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Erkan M, Pikdoken L and Usumez S: Gingival
response to mandibular incisor intrusion. Am J Orthod Dentofacial
Orthop. 132:143.e9–13. 2007. View Article : Google Scholar
|
|
5
|
Kalra A, Jaggi N, Bansal M, Goel S,
Medsinge SV, Abraham R and Jasoria G: Comparison of rate of canine
retraction into recent extraction site with and without gingival
fiberotomy: A clinical study. J Contemp Dent Pract. 14:419–426.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Krishnan V, Ambili R, Davidovitch Z and
Murphy NC: Gingiva and orthodontic treatment. Semin Orthodont.
13:257–271. 2007. View Article : Google Scholar
|
|
7
|
Bartold PM and McCulloch CA: Information
generation and processing systems that regulate periodontal
structure and function. Periodontol 2000. 63:7–13. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Katsumi A, Naoe T, Matsushita T, Kaibuchi
K and Schwartz MA: Integrin activation and matrix binding mediate
cellular responses to compressive stretch. J Biol Chem.
280:16546–16549. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Jiang N, Guo W, Chen M, Zheng Y, Zhou J,
Kim SG, Embree MC, Songhee Song K, Marao HF and Mao JJ: Periodontal
ligament and alveolar bone in health and adaptation: Tooth
movement. Front Oral Biol. 18:1–8. 2016.
|
|
10
|
Nan L, Zheng Y, Liao N, Li S, Wang Y, Chen
Z, Wei L, Zhao S and Mo S: Mechanical force promotes the
proliferation and extracellular matrix synthesis of human gingival
fibroblasts cultured on 3D PLGA scaffolds via TGF-β expression. Mol
Med Rep. 19:2107–2114. 2019.PubMed/NCBI
|
|
11
|
Aukkarasongsup P, Haruyama N, Matsumoto T,
Shiga M and Moriyama K: Periostin inhibits hypoxia-induced
apoptosis in human periodontal ligament cells via TGF-β signaling.
Biochem Biophys Res Commun. 441:126–132. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Xu H, He Y, Feng JQ, Shu R, Liu Z, Li J,
Wang Y, Xu Y, Zeng H, Xu X, et al: Wnt3α and transforming growth
factor-β induce myofibroblast differentiation from periodontal
ligament cells via different pathways. Exp Cell Res. 353:55–62.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Moustakas A, Souchelnytskyi S and Heldin
CH: Smad regulation in TGF-beta signal transduction. J Cell Sci.
114:4359–4369. 2001.
|
|
14
|
Singh P, Srinivasan R and Wig JD: The Smad
family and its role in pancreatic cancer. Indian J Cancer.
48:351–360. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Mccarthy AJ and Chetty R: Smad4/DPC4. J
Clin Pathol. 71:661–664. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Duda D, Sunamura M, Lefter LP, Furukawa T,
Yokoyama T, Yatsuoka T, Abe T, Inoue H, Motoi F, Egawa S, et al:
Restoration of SMAD4 by gene therapy reverses the invasive
phenotype in pancreatic adenocarcinoma cells. Oncogene.
22:6857–6864. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Hruban RH, Offerhaus GJ, Kern SE, Goggins
M, Wilentz RE and Yeo CJ: Tumor-Suppressor genes in pancreatic
cancer. J Hepatobiliary Pancreat Surg. 5:383–391. 1998. View Article : Google Scholar
|
|
18
|
Michot C, Le Goff C, Mahaut C, Afenjar A,
Brooks AS, Campeau PM, Destree A, Di Rocco M, Donnai D, Hennekam R,
et al: Myhre and LAPS syndromes: Clinical and molecular review of
32 patients. Eur J Hum Genet. 22:1272–1277. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Ahmed S, Bradshaw AD, Gera S, Dewan MZ and
Xu R: The TGF-β/Smad4 signaling pathway in pancreatic
carcinogenesis and its clinical significance. J Clin Med. 6:52017.
View Article : Google Scholar
|
|
20
|
Woodford-Richens KL, Rowan AJ, Gorman P,
Halford S, Bicknell DC, Wasan HS, Roylance RR, Bodmer WF and
Tomlinson IP: SMAD4 mutations in colorectal cancer probably occur
before chromosomal instability, but after divergence of the
microsatellite instability pathway. Proc Natl Acad Sci USA.
98:9719–9723. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Shihab FS, Yamamoto T, Nast CC, Cohen AH,
Noble NA, Gold LI and Border WA: Transforming growth factor-beta
and matrix protein expression in acute and chronic rejection of
human renal allografts. J Am Soc Nephrol. 6:286–294.
1995.PubMed/NCBI
|
|
22
|
Zhang B, Zhang B, Chen X, Bae S, Singh K,
Washington MK and Datta PK: Loss of Smad4 in colorectal cancer
induces resistance to 5-fluorouracil through activating Akt
pathway. Br J Cancer. 110:946–957. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Huang Y, Chen C, Ren J and Ren T: The
preparation of poly(lactide-co-glycolide)(PLGA)/modified
nano-hydroxyapatite(MHA) composite scaffold for tissue engineering.
J Funct Materials. 38:629–632. 2007.In Chinese.
|
|
24
|
Sachar A, Strom TA, San Miguel S, Serrano
MJ, Svoboda KK and Liu X: Cell-matrix and cell-cell interactions of
human gingival fibroblasts on three-dimensional nanofibrous gelatin
scaffolds. J Tissue Eng Regen Med. 8:862–873. 2014. View Article : Google Scholar
|
|
25
|
Wei G, Jin Q, Giannobile WV and Ma PX:
Nano-fibrous scaffold for controlled delivery of recombinant human
PDGF-BB. J Control Release. 112:103–110. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Guo N, Zhang Q, Sun Y and Yang H:
Separation and identification of acylated leuprorelin inside PLGA
microspheres. Int J Pharm. 560:273–281. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Xie H, Gu Z, Li C, Franco C, Wang J, Lia
L, Meredith N, Ye Q and Wang C: A novel bioceramic scaffold
integrating silk fibroin in calcium polyphosphate for bone
tissue-engineering. Ceram Int. 42:2386–2392. 2016. View Article : Google Scholar
|
|
28
|
Alyaa A, Mehjabeen A, Kannan M, Yeb Q and
Blawertc C: Biodegradable polymer for sealing porous PEO layer on
pure magnesium: An in vitro degradation study. Appl Surf Sci.
301:463–467. 2014. View Article : Google Scholar
|
|
29
|
Liu Z, Yin X, Ye Q, He W, Ge M, Zhou X, Hu
J and Zou S: Periodontal regeneration with stem cells-seeded
collagen-hydroxyapatite scaffold. J Biomater Appl. 31:121–131.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
31
|
Schwarz AM: Tissue changes incidental to
orthodontic tooth movement. Int J Orthod Oral Surg Radiog.
18:331–352. 1932. View Article : Google Scholar
|
|
32
|
Otero L, García DA and Wilches-Buitrago L:
Expression and presence of OPG and RANKL mRNA and protein in human
periodontal ligament with orthodontic force. Gene Regul Syst Bio.
10:15–20. 2016.PubMed/NCBI
|
|
33
|
Grant M, Wilson J, Rock P and Chapple I:
Induction of cytokines, MMP9, TIMPs, RANKL and OPG during
orthodontic tooth movement. Eur J Orthod. 35:644–651. 2013.
View Article : Google Scholar
|
|
34
|
Kook SH, Jang YS and Lee JC: Human
periodontal ligament fibroblasts stimulate osteoclastogenesis in
response to compression force through TNF-α-mediated activation of
CD4+ T cells. J Cell Biochem. 112:2891–2901. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Li M, Yi J, Yang Y, Zheng W, Li Y and Zhao
Z: Investigation of optimal orthodontic force at the cellular level
through three-dimensionally cultured periodontal ligament cells.
Eur J Orthod. 38:366–372. 2016. View Article : Google Scholar
|
|
36
|
Baker SC, Rohman G, Southgate J and
Cameron NR: The relationship between the mechanical properties and
cell behaviour on PLGA and PCL scaffolds for bladder tissue
engineering. Biomaterials. 30:1321–1328. 2009. View Article : Google Scholar
|
|
37
|
Subbarayan R, Murugan Girija D, Mukherjee
J, Mamidanna SRR and Ranga Rao S: Comparision of gingival and
umbilical cord stem cells based on its modulus and neuronal
differentiation. J Cell Biochem. 118:2000–2008. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Yin P, Wang Y and Shen Z: SMAD4 gene
silencing promotes human vascular smooth muscle cells migration and
apoptosis. J Jiangsu Univ (Medicine Edition). 28:26–29. 2018.In
Chinese.
|
|
39
|
Grilo AL and Mantalaris A: Apoptosis: A
mammalian cell bioprocessing perspective. Biotechnol Adv.
37:459–475. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Cohen JJ, Duke RC, Fadok VA and Sellins
KS: Apoptosis and programmed cell death in immunity. Annu Rev
Immunol. 10:267–293. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Zhang Y, Alexander PB and Wang XF:
TGF-beta family signaling in the control of cell proliferation and
survival. Cold Spring Harb Perspect Biol. 9:a0221452017. View Article : Google Scholar
|
|
42
|
Mcilwain DR, Berger T and Mak TW: Caspase
functions in cell death and disease. Cold Spring Harb Perspect
Biol. 7:a0267162015. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Ashkenazi A, Fairbrother WJ, Leverson JD
and Souers AJ: From basic apoptosis discoveries to advanced
selective BCL-2 family inhibitors. Nat Rev Drug Discov. 16:273–284.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Taylor RC, Cullen SP and Martin SJ:
Apoptosis: Controlled demolition at the cellular level. Nat Rev Mol
Cell Biol. 9:231–241. 2008. View
Article : Google Scholar
|
|
45
|
Liu N, Li Y, Li R, Sun L and Liu X:
Influence of silencing Smad4 gene in proliferation and apoptosis of
breast carcinoma MCF-7 cells. J Jilin Univ (Medical Edition).
43:887–892. 2017.In Chinese.
|
|
46
|
Demagny H and De Robertis EM: Point
mutations in the tumor suppressor Smad4/DPC4 enhance its
phosphorylation by GSK3 and reversibly inactivate TGF-β signaling.
Mol Cell Oncol. 3:e10251812015. View Article : Google Scholar
|