Introduction
Osteosarcoma (OS) is a highly aggressive malignant
bone cancer that primarily affects children and adolescents
(1). Osteosarcoma accounts for
about 3-4% of all pediatric tumors and 30% of malignant bone tumors
(2). It often appears in the
long bones of limbs and in the growth plate of a metaphysis
(3). Several risk factors,
including alkylating agents, ionizing radiation and hereditary
retinoblastoma, are reported to contribute to the occurrence of OS
(4). Currently, surgery,
chemotherapy, immunotherapy and gene therapy are the main
treatments for patients with OS; however, despite the relative
effectiveness of these methods, the mortality and distant
metastasis rates remain extremely high (5-7).
Overall, 20-30% of patients present with metastatic osteosarcoma,
most commonly to the lungs, lymph nodes or other bones (8). The 5-year survival rate of patients
with OS without any local or distant metastasis is 60-70%, whereas
the 5-year survival rate of patients with OS with metastasis is
markedly low (<30%) (9).
Therefore, it is of great significance to fully explore the
molecular mechanisms for the development of OS to develop promising
treatment strategies and improve clinical outcomes.
Long non-coding RNAs (lncRNAs) are a novel class of
protein non-coding transcripts that are >200 nucleotides in
length (10). Increasing
evidence has revealed that lncRNAs serve important roles in the
regulation of various types of cancer (11,12). For example, lncRNA SNHG5
accelerates OS progression by sponging microRNA (miRNA/miR)-212-3p
and regulating serum and glucocorticoid regulated kinase 3
(13). Through the activation of
the phosphatidylinositol 3-kinase/AKT signaling pathway, lncRNA
SNHG12 accelerates gastric cancer progression (14). lncRNA EPIC1 modulates the cell
cycle and promotes the proliferation of pancreatic cancer cells by
interacting with Yes-associated protein-1 (15). Emerging evidence has revealed
that lncRNA LINC00467 functions as a tumor-promoting gene involved
in various types of cancer. For example, LINC00467 accelerates lung
adenocarcinoma cell proliferation via the miR-20b-5p/cyclin D1
(CCND1) axis (16) and
facilitates cervical cancer tumorigenesis by sponging miR-107
(17). However, the biological
function and molecular regulatory mechanism of LINC00467 in OS
remain unclear. Therefore, the purpose of the present study was to
investigate the role of LINC00467 in OS progression.
Materials and methods
Cell lines
The human normal osteoblast Hfob1.19 cell line and
human OS cell lines (Saos2, MG63, U2OS and HOS) were obtained from
The Cell Bank of Type Culture Collection of the Chinese Academy of
Sciences. The cells were cultivated in Dulbecco's modified Eagle's
medium (DMEM) supplemented with fetal bovine serum (FBS) (both
Gibco; Thermo Fisher Scientific, Inc.), 100 U/ml penicillin and 100
µg/ml streptomycin (both Sigma-Aldrich; Merck KGaA) and
maintained in a humidified incubator at 37°C with 5%
CO2.
Tissue samples
A total of 36 paired OS and adjacent normal tissues
(>3 cm from the tumor tissue) were collected from patients with
OS (15 males and 21 females; age range, 26-57 years; mean age ± SD,
47.23±4.58 years) at The Affiliated Huaian Hospital of Xuzhou
Medical University (Huai'an, China) between May 2012 and July 2014.
The collected tissues were immediately frozen in liquid nitrogen
and stored at −80°C for subsequent experiments. None of the
patients had received any preoperative anticancer treatment. The
study protocol was approved by the Ethics Committee of The
Affiliated Huai'an Hospital of Xuzhou Medical University. Written
informed consent was obtained from all patients.
Transfection
To downregulate LINC00467 expression in cells, short
hairpin RNAs (shRNAs) specifically targeting LINC00467
(sh-LINC00467#1, 5′-GAC TCA TGA AAC CAA TCT TCA-3′; sh-LINC00467#2,
5′-GAT GCT CTG TAA ACC ACA TAA-3′) and a scrambled control shRNA
(sh-NC, 5′-CAA CAA GAT GAA GAG CAC CAA-3′) were designed and
synthesized by Shanghai GenePharma Co., Ltd. The sequences of
karyopherin subunit α4 (KPNA4) (Data S1) were synthesized and
subcloned into pcDNA3.1 (Invitrogen; Thermo Fisher Scientific,
Inc.) plasmids to generate pcDNA3.1/KPNA4. The pcDNA3.1 vector
(Invitrogen; Thermo Fisher Scientific, Inc.) carrying the LINC00467
sequence (hereafter LINC00467) was used to overexpress LINC00467.
An empty vector was used as a negative control. The miR-217 mimic,
miR-217 inhibitor and corresponding negative non-targeting controls
(NC mimics, 5′-UUC UCC GAA CGU GUC ACG UTT-3; miR-217 mimics,
5′-UAC UGC AUC AGG AAC UGA CUG G-3′; NC inhibitor, 5′-UAG AACU UGCA
UUG CAA CCG G-3′; miR-217 inhibitor, 5′-CCA GUC AGU UCC UGA UGC
AGUA-3′) were purchased from Shanghai GenePharma Co., Ltd. All
plasmids were transfected into U2OS and HOS cells using
Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.) according to the manufacturer's protocol.
Briefly, 100 pmol plasmids (final concentration, 50 nM) were
diluted in 250 µl serum-free Opti-MEM (cat. no. 51985042;
Gibco; Thermo Fisher Scientific, Inc.) and cultured at room
temperature for 5 min. Meanwhile, another 5 µl Lipofectamine
2000 was diluted in 250 µl serum-free Opti-MEM and incubated
for 5 min at room temperature. These two solutions were mixed and
incubated at room temperature for 20 min before they were added to
the cell culture wells. The transfection effects were assessed by
reverse transcription-quantitative PCR (RT-qPCR) after transfection
for 24 h. The cells were then subjected to further functional
assays.
Cell Counting Kit-8 (CCK-8) assay
The CCK-8 assay (Dojindo Molecular Technologies,
Inc.) was utilized to test cell viability at 0, 24, 48 and 72 h
after transfection. Briefly, the cells were seeded in a 96-well
culture plate (2×104 cells/ml; 100 µl/well) and
cultured for 0, 24, 48 and 72 h. Subsequently, 10 µl CCK-8
reagent was added to each well, and the cells were cultured for 2 h
at 37°C. The absorbance values were measured at 450 nm on a
microplate reader (EL340; BioTek Instruments, Inc.; Agilent
Technologies, Inc.).
Colony formation assay
The transfected U2OS and HOS cells were plated in a
6-well plate and maintained in culture medium, which was replaced
every 2 days for ~2 weeks. Proliferating colonies were fixed with
10% methanol for 30 min at room temperature and stained with 1%
crystal violet (Beyotime Institute of Biotechnology) for 15 min at
room temperature. The viable cell colonies (>50 cells) were
counted and photographed under a light microscope (Olympus
Corporation).
Western blot analysis
Total protein content from OS cells was extracted
using RIPA lysis buffer (Beyotime Institute of Biotechnology)
supplemented with phenylmethylsulfonyl fluoride. The concentration
of total protein was measured using a BCA Protein assay kit
(Nanjing KeyGen Biotech Co., Ltd.). The proteins (30
µg/lane) were separated via 12% SDS-PAGE and transferred to
PVDF membranes. The PVDF membranes were blocked in 5% skimmed milk
for 1 h at room temperature and incubated with primary antibodies
(Abcam) at 4°C overnight. After washing the membranes with 0.1%
PBS-Tween 20, the membranes were further incubated with
HRP-conjugated secondary antibodies (1:2,000; cat. no. ab205718;
Abcam) for 2 h at room temperature. The protein bands were detected
using an Enhanced Chemiluminescence detection system (Thermo Fisher
Scientific, Inc.). GAPDH was used as the internal control. The
primary antibodies were as follows: E-cadherin (1:1,000; cat. no.
ab1416), N-cadherin (1:1,000; cat. no. ab76011), vimentin (1:1,000;
cat. no. ab92547), Slug (1:1,000; cat. no. ab51772), Twist
(1:1,000; cat. no. ab50581) and GAPDH (1:2,000; cat. no.
ab8245).
Transwell assays
For the migration assays, 2×104
transfected cells/well were plated in the upper chamber of
Transwell inserts with 200 µl serum-free DMEM. For the
invasion assay, Transwell chambers (Corning, Inc.) with 8-µm
pores were used. The upper chamber was pre-coated with Matrigel (50
µg/ml; BD Biosciences) at 37°C for 30 min before the cells
were plated. Next, 500 µl DMEM with 10% FBS was added to the
lower chamber. After incubation for 24 h at 37°C, the cells that
migrated to the lower surface of the filter were stained with 1%
crystal violet for 30 min at room temperature. The cells were
imaged using an inverted light microscope (magnification, ×100;
Leica Microsystems GmbH).
Subcellular fractionation location
A PARIS kit (Thermo Fisher Scientific, Inc.) was
used to isolate the nuclear and cytosolic portions of OS cells
according to the manufacturer's protocol. RT-qPCR assays were
performed to detect the expression levels of LINC00467, GAPDH and
U6 in the cytoplasm and nuclear components. GAPDH functioned as a
cytoplasmic control, while U6 functioned as a nuclear control. The
relative proportions of LINC00467, GAPDH and U6 in the cytoplasm or
nucleus are shown as percentages of the total RNA.
RNA pull-down assay
The biotinylated RNAs (LINC00467 probe-biotin and
LINC00467 probe no-biotin) were synthesized by Shanghai GenePharma
Co., Ltd. U2OS and HOS cells were collected and lysed in specific
lysis buffer containing RNase inhibitor (Beyotime Institute of
Biotechnology). Subsequently, the lysate was subjected to
centrifugation at 12,000 × g for 12 min at 4°C. LINC00467
probe-biotin (200 pmol) or LINC00467 probe no-biotin (200 pmol) was
added to the supernatant and incubated for 2 h at 4°C, followed by
incubation with M-280 streptavidin beads (100 µl;
Sigma-Aldrich; Merck KGaA) pre-coated with RNase-free BSA and yeast
tRNA (cat. no. TRNABAK-RO; Sigma-Aldrich; Merck KGaA) for 1 h at
4°C. The beads were subsequently washed twice with cold lysis
buffer, three times with low-salt buffer and once with high-salt
buffer. After purification using TRIzol®(Invitrogen;
Thermo Fisher Scientific, Inc.), the enrichment of miRNAs was
determined by RT-qPCR.
RT-qPCR
TRIzol® reagent was used to extract the
total RNA content from OS tissues and cells. Total RNA was reverse
transcribed into cDNA using a PrimeScript RT reagent kit (cat. no.
RRO36A; Takara Biotechnology Co., Ltd.) according to the
manufacturer's protocol. A SYBR® Premix Ex Taq™ II
reagent kit (cat. no. RR820A: Takara Biotechnology Co., Ltd.) was
used for qPCR with an ABI7500 real-time qPCR system (Applied
Biosystems; Thermo Fisher Scientific, Inc.) under the following
conditions: 95°C for 30 sec, followed by 45 cycles at 95°C for 5
sec and 60°C for 30 sec. U6 was regarded as the internal reference
for miR-217. GAPDH was used as an internal reference for LINC00467
and KPNA4. Relative quantification was analyzed using the
2−ΔΔCq method (18).
The primers used were as follows: LINC00467 forward, 5′-CCT TCT TCC
TCA TCA TCG TC-3′ and reverse, 5′-CCC AGT TTC AGT CCC TCT TG-3′;
miR-217 forward, 5′-CGC TCT ACT GCA TCA GGA ACT GA-3′ and reverse,
5′-GTG CAG GGT CCG AGG T-3′; KPNA4 forward, 5′-TGT GAG CAA GCA GTG
TGG GCA-3′ and reverse, 5′-TGG TGG TGG GTC TTT GTG GCG-3′; GAPDH
forward, 5′-CCC ACT CCT CCA CCT TTGAC-3′ and reverse, 5′-GGA TCT
CGC TCC TGG AA GAT G-3′; U6 forward, 5′-GCU UCG GCA GCA CAU AUA CUA
AAA U-3′ and reverse, 5′-CGC UUC ACG AAU UUG CGU GUC AU-3′.
Luciferase reporter assay
The miRNAs with potential binding sites for
LINC00467 were predicted using the starBase website (http://starbase.sysu.edu.cn/). Fragments of the
wild-type (Wt) and mutant (Mut) reporters of LINC00467
(LINC00467-Wt and LINC00467-Mut) and KPNA4 (KPNA4-Wt and KPNA4-Mut)
sharing putative binding sites with miR-217 were synthesized by
Shanghai GenePharma Co., Ltd., and cloned into pcDNA3.1 vectors
(Invitrogen; Thermo Fisher Scientific, Inc.). For the luciferase
assay, Wt or Mut reporters were co-transfected with the miR-217
mimic or NC mimic into U2OS and HOS cells, as aforementioned. After
48 h of transfection, luciferase activity, as the ratio of firefly
luciferase activity and Renilla luciferase activity, was
determined using a Dual Luciferase Reporter Assay kit (Promega
Corporation). The sequence information of LINC00467-Wt/Mut and
KPNA4-Wt/Mut is shown in Data S1.
Statistical analysis
SPSS 20.0 software (IBM Corp.) was used for data
analysis. The differences among groups were compared via paired or
unpaired Student's t-test, and one-way ANOVA followed by Tukey's
post-hoc test. Spearman's correlation analysis was used to analyze
the correlation among target genes. The survival rate was evaluated
using Kaplan-Meier analysis and compared using the log-rank test.
All variables are shown as the mean ± SD. All experiments were
repeated ≥3 times. P<0.05 was considered to indicate a
statistically significant difference.
Results
LINC00467 expression is elevated in OS
tissues and cells, and high LINC00467 expression is associated with
a poor prognosis
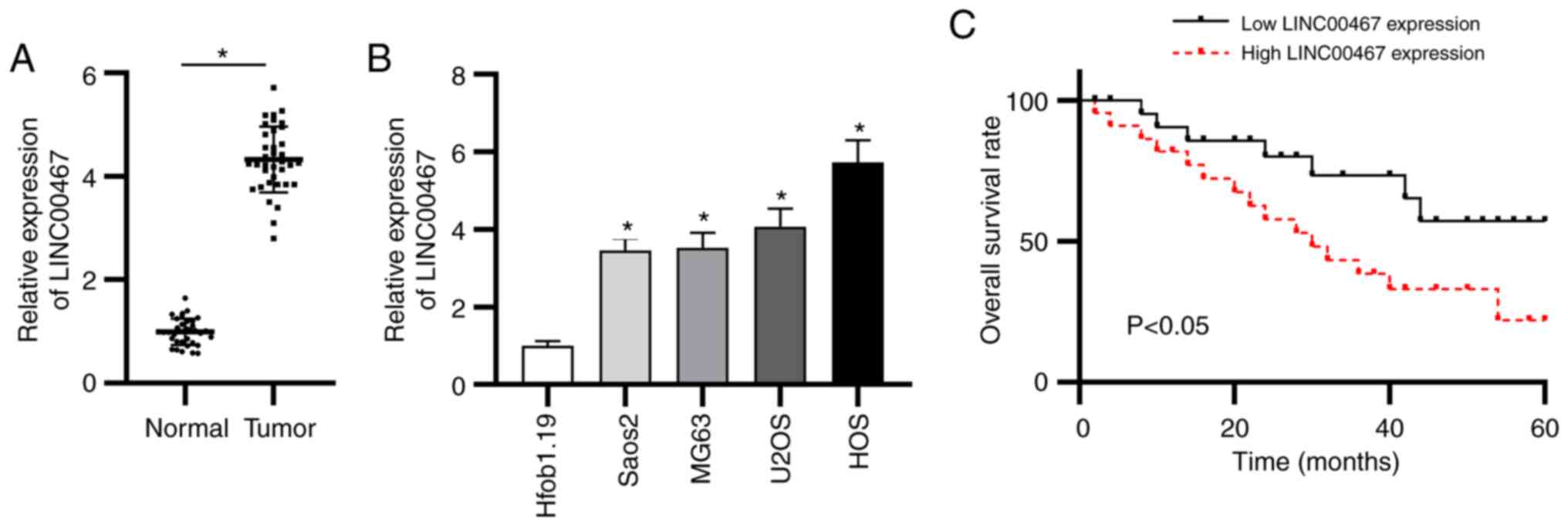
To confirm the function of LINC00467 in OS, the
expression levels of LINC00467 were examined in OS and adjacent
normal tissue samples (n=36). LINC00467 expression was upregulated
in OS tissues compared with in adjacent normal tissues as shown by
RT-qPCR (Fig. 1A). In addition,
LINC00467 expression was quantified in the four OS cell lines
(Saos2, MG63, U2OS and HOS) and in normal osteoblast cells
(Hfob1.19). The results revealed that LINC00467 expression was
significantly upregulated in the four OS cell lines compared with
in Hfob1.19 cells (Fig. 1B).
Additionally, according to the median value of LIC00467 expression
in OS, patients with OS were equally assigned to the low LINC00467
expression group or the high LINC00467 expression group to evaluate
the clinical value of LINC00467. Notably, the overall survival time
of patients with OS with high LINC00467 expression was shorter than
that of patients with low LINC00467 expression (Fig. 1C). These results indicated that
LINC00467 expression was elevated in OS tissues and cells, and that
LINC00467 was closely associated with an adverse prognosis.
LINC00467 inhibition represses OS cell
viability, migration and invasion
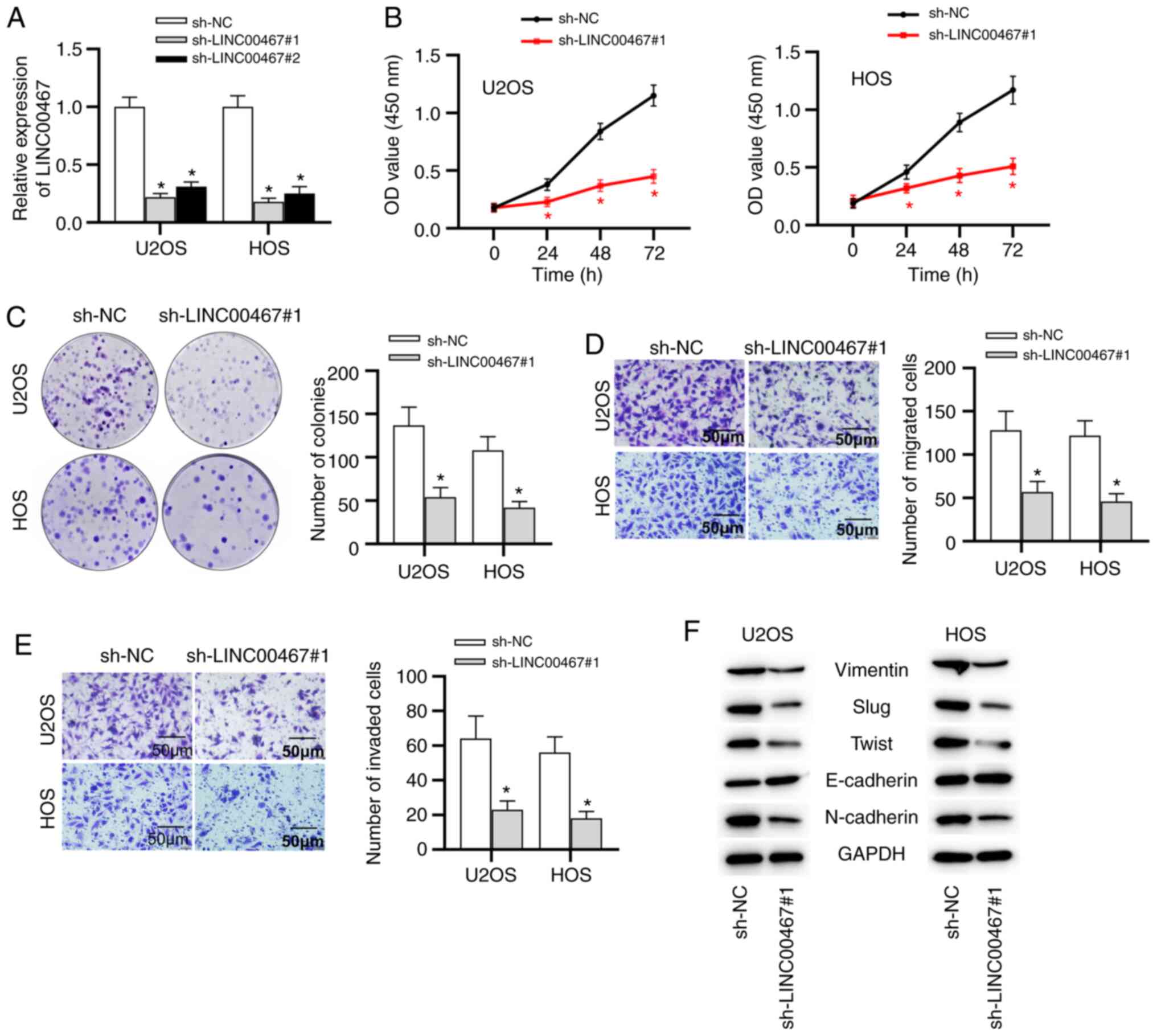
Having examined LINC00467 upregulation in OS, the
function of LINC00467 in OS progression was explored. Considering
that the U2OS and HOS cell lines presented higher LINC00467
expression than that in the other two OS cell lines, U2OS and HOS
cells were selected for follow-up experiments and were transfected
with shRNAs against LINC00467 (sh-LINC00467#1 or sh-LINC00467#2) or
a negative control shRNA (sh-NC). LINC00467 expression was
significantly knocked down in U2OS and HOS cells after transfection
with sh-LINC00467#1/2, especially with sh-LINC00467#1 (Fig. 2A). Subsequently, a CCK-8 assay
was performed to assess the influence of LINC00467-knockdown on OS
cell viability. Compared with the sh-NC group, sh-LINC00467#1
transfection significantly decreased the viability of U2OS and HOS
cells (Fig. 2B). Colony
formation assays revealed that LINC00467-knockdown triggered a
significant decrease in the colony number in U2OS and HOS cells
(Fig. 2C). In addition,
Transwell assays revealed that LINC00467 deficiency significantly
inhibited the migration and invasion of U2OS and HOS cells
(Fig. 2D and E). Moreover,
further experiments suggested that the protein expression levels of
epithelial-mesenchymal transition (EMT)-associated transcription
factors (Slug and Twist), as well as mesenchymal markers
(N-cadherin and Vimentin), were markedly decreased by silencing
LINC00467 (Fig. 2F). However,
the expression levels of the epithelial marker E-cadherin were
markedly upregulated in sh-LINC00467#1-transfected cells compared
with in sh-NC-transfected cells (Fig. 2F). Overall, these findings
suggested that LINC00467 inhibition restrained OS progression by
inhibiting cell viability, migration and invasion.
LINC00467 serves as a molecular sponge
for miR-217 in OS
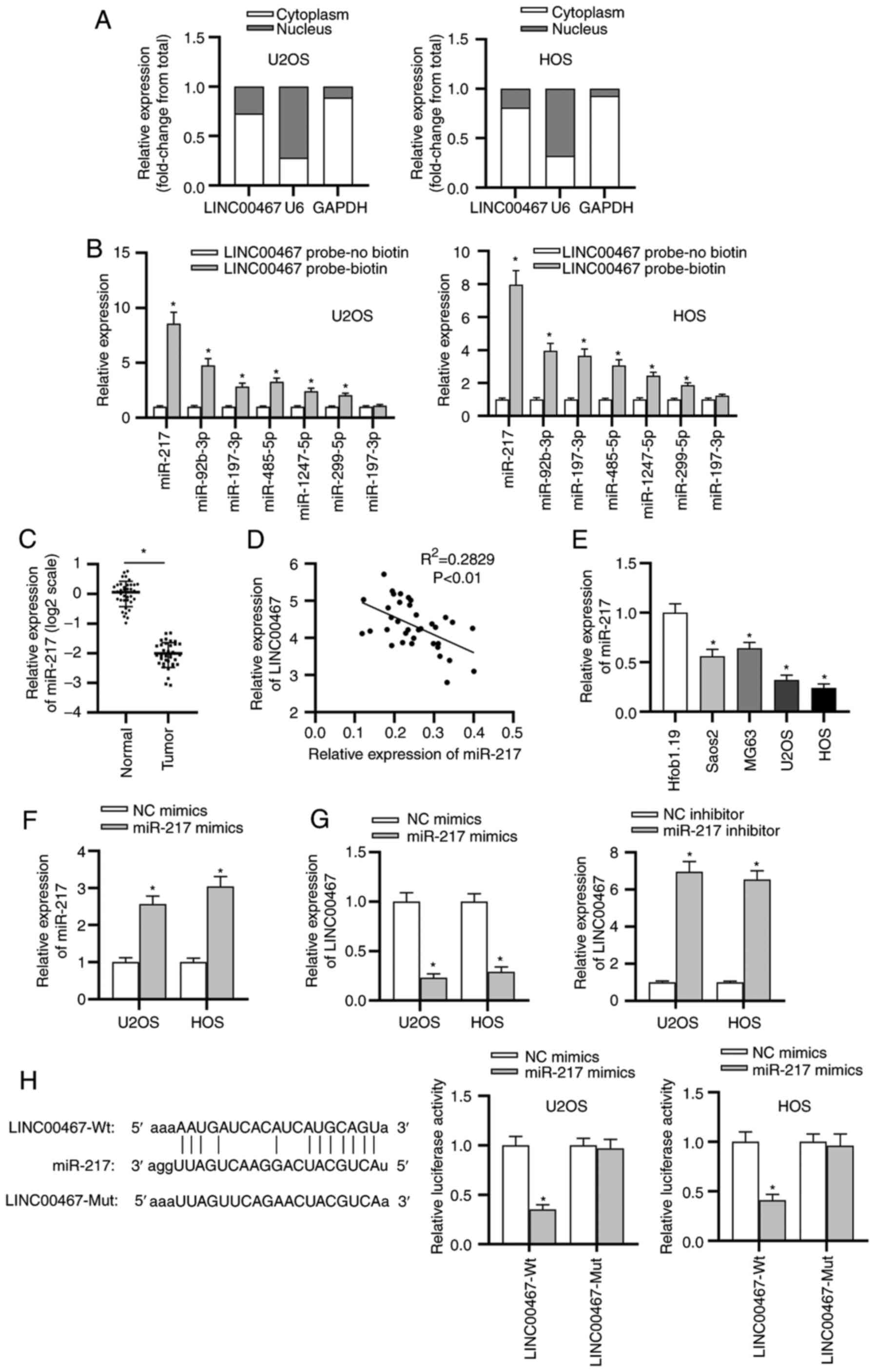
To study the LINC00467-mediated regulatory mechanism
involved in OS progression, a subcellular fractionation location
assay was implemented to confirm the location of LINC00467 in OS
cells. The results delineated that LINC00467 was mainly distributed
in the cytoplasm of OS cells (Fig.
3A). Subsequently, the candidate miRNAs (miR-217, miR-92b-3p,
miR-197-3p, miR-485-5p, miR-1247-5p, miR-299-5p and miR-197-3p)
that may be associated with LINC00467 were predicted using the
starBase website. In the LINC00467 probe-biotin group, miR-217
exhibited the most enrichment compared with other miRNAs (Fig. 3B). RT-qPCR was performed to
examine miR-217 expression, and the results revealed that miR-217
expression was significantly downregulated in OS tissues compared
with that in adjacent normal bone tissues (Fig. 3C). Subsequently, Spearman's
correlation analysis confirmed the negative correlation between
LINC00467 and miR-217 expression in OS tissues (Fig. 3D). Consistently, miR-217
expression was significantly decreased in OS cell lines (Saos2,
MG63, U2OS and HOS) compared with that in normal osteoblast
Hfob1.19 cells (Fig. 3E).
Transfection with miR-217 mimics was used to overexpress miR-217 in
U2OS and HOS cells (Fig. 3F).
Additionally, the effect of miR-217 overexpression was investigated
on LINC00467 expression, revealing through RT-qPCR that the miR-217
mimic significantly decreased LINC00467 expression, while LINC00467
expression was significantly increased by the miR-217 inhibitor
(Fig. 3G). The knockdown
efficiency of the miR-217 inhibitor in cells was validated by
RT-qPCR (Fig. S1A).
Furthermore, the binding capacity between LINC00467 and miR-217 was
analyzed. A luciferase reporter assay demonstrated that the
luciferase activity of LINC00467-Wt reporters was significantly
decreased in the miR-217 mimic-transfected cells; however, no
significant difference was observed in luciferase activity among
LINC00467-Mut groups (Fig. 3H).
Overall, these findings implied that LINC00467 served as a
molecular sponge for miR-217 in OS.
KPNA4 is a downstream target gene of
miR-217
miRNAs have been reported to elicit their influence
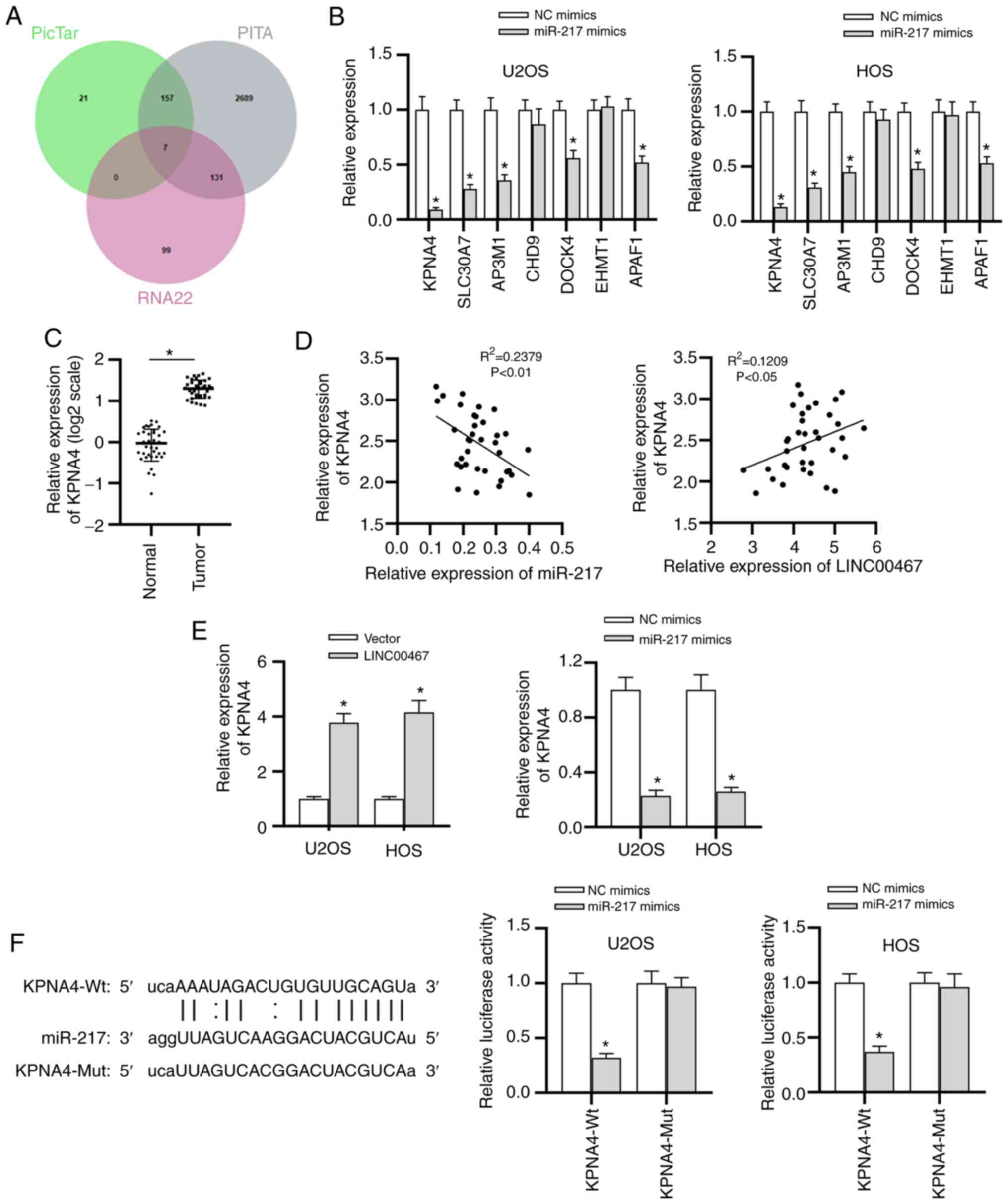
on cancer progression by targeting specific genes (19,20). To determine the potential targets
of miR-217, the underlying genes were analyzed using the starBase
website (search parameters, PicTar, PITA and RNA22), identifying 7
potential mRNAs (KPNA4, SLC30A7, AP3M1, CHD9, DOCK4, EHMT1 and
APAF1) using a Venn diagram (Fig.
4A). Compared with the expression levels of other candidate
mRNAs, KPNA4 in U2OS and HOS cells exhibited the lowest expression
after overexpression of miR-217 (Fig. 4B). Therefore, KPNA4 was selected
from candidate mRNAs for subsequent investigations. KPNA4
expression was significantly elevated in the OS tissue samples
compared with that in adjacent normal tissues (Fig. 4C). In addition, a negative
correlation between KPNA4 and miR-27 expression, as well as a
positive correlation between KPNA4 and LINC00467 expression, was
observed by Spearman's correlation analysis (Fig. 4D). Moreover, the overexpression
of LINC00467 significantly increased KPNA4 expression, while the
miR-217 mimic significantly decreased KPNA4 expression (Fig. 4E). The overexpression efficiency
of LINC00467 in cells was validated by RT-qPCR (Fig. S1B). As illustrated in Fig. 4F, only the luciferase activity of
the KPNA4-Wt reporters was significantly decreased by the miR-217
mimic, confirming that KPNA4 was the downstream target gene of
miR-217 and that they could bind with each other. In summary, the
current findings validated that KPNA4 mRNA was a direct target of
miR-217 in OS.
LINC00467 promotes OS progression by
upregulating KPNA4
Finally, the present study investigated whether
LINC00467 could accelerate OS progression via the miR-217/KPNA4
axis. The results revealed that KPNA4 expression was upregulated in
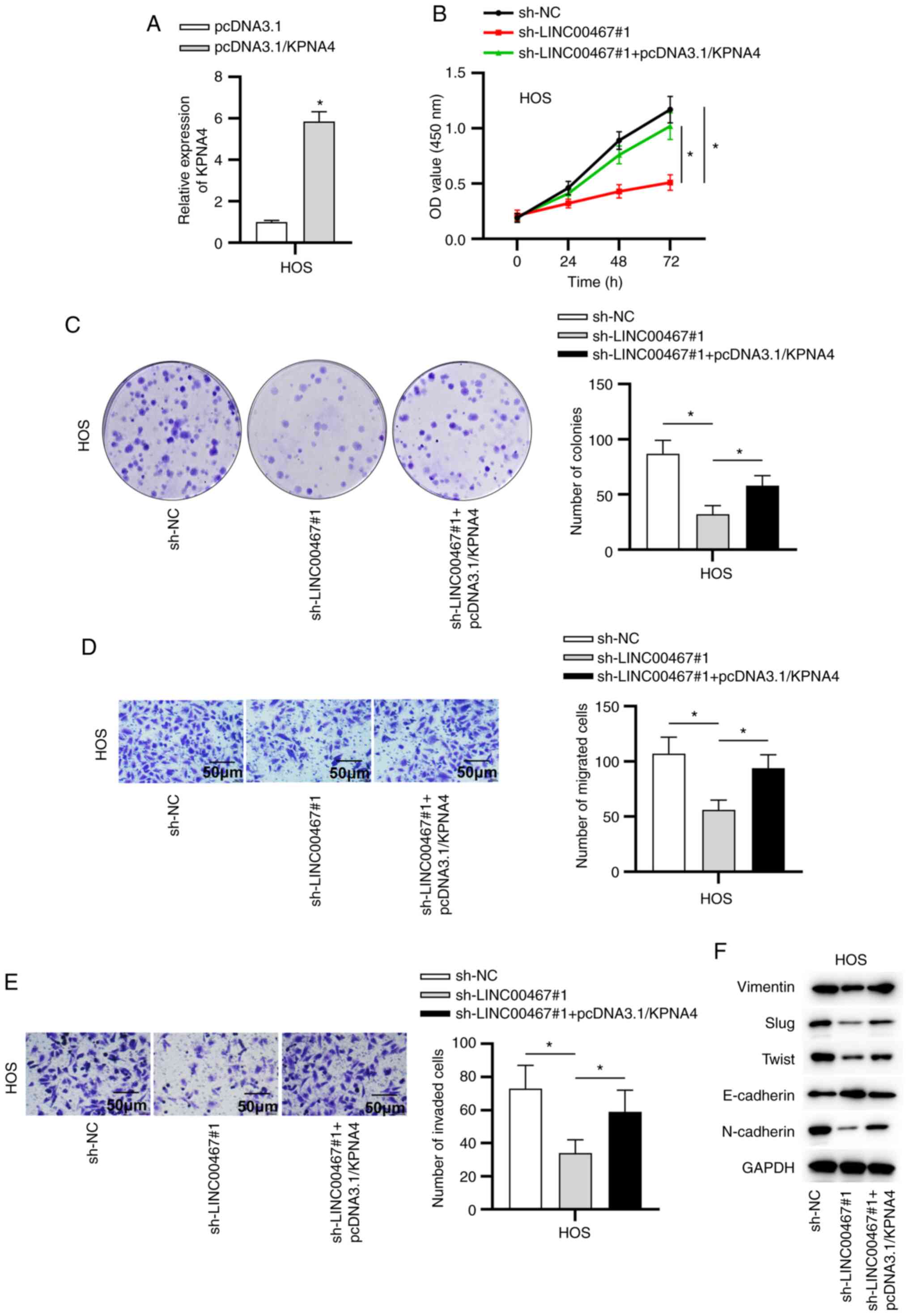
pcDNA3.1/KPNA4-transfected cells (Fig. 5A). Additionally, transfection of
pcDNA3.1/KPNA4 reversed the inhibitory role of sh-LINC00467#1 on
the viability, migration and invasion of OS cells (Fig. 5B-E). Moreover, compared with
sh-LINC00467#1-transfected cells, the protein expression levels of
EMT-associated transcription factors (Slug and Twist), as well as
mesenchymal markers (N-cadherin and Vimentin), were increased in
the sh-LINC00467#1 and pcDNA3.1/KPNA4 co-transfected cells
(Fig. 5F). Furthermore, data
from western blot analysis suggested that KPNA4 overexpression
countervailed the LINC00467-knockdown-mediated inhibitory effect on
the expression levels of the epithelial marker E-cadherin (Fig. 5F). Overall, the current data
revealed that LINC00467 facilitated OS progression by targeting the
miR-217/KPNA4 axis.
Discussion
To date, numerous studies have revealed abnormal
expression levels of lncRNAs in OS (21-23). The dysregulation of lncRNAs has
been verified to strongly affect the progression of OS by acting as
either tumor-suppressive or oncogenic genes (23-25). Hence, identifying the specific
functions of lncRNAs in OS is urgently required to validate
promising diagnostic biomarkers and therapeutic targets for OS. It
has been reported that LINC00467 serves as a cancer-promoting gene
in some types of cancer, such as lung adenocarcinoma, colorectal
cancer and hepatocellular carcinoma (16,26,27). In the present study, it was
revealed that LINC00467 expression was significantly elevated in OS
tissues and cell lines. High LINC00467 expression was closely
associated with a poor prognosis in patients with OS. Knockdown of
LINC00467 repressed the viability, migration, invasion and EMT of
OS cells.
miRNAs are short non-coding RNA molecules with 20-24
nucleotides that serve important roles in the posttranscriptional
regulation of gene expression (28,29). Recently, lncRNAs have been
reported to serve as specific miRNA sponges to regulate various
types of cancer (30,31). For instance, the lncRNA LUCAT1
facilitates ovarian cancer malignancy by regulating the
miR-612/human class I homeobox A13 signaling pathway (32). Through sponging miR-541-3p and
thus regulating CCND1 expression, the lncRNA LOXL1-AS1 modulates
prostate cancer cell proliferation and the cell cycle (33). It has been reported that
LINC00467 acts as an oncogenic gene to sponge miR-20b-5p in lung
adenocarcinoma (16).
Nevertheless, the interaction between LINC00467 and miRNAs in OS
remains unclear. In the present study, miR-217 was chosen from the
potential miRNAs for LINC00467. miR-217 has been reported to have
antitumor effects in multiple types of cancer (34,35). For example, miR-217 inhibits EMT
transition by targeting PTPN14 in gastric carcinoma (36) and suppresses HeLa cell migration
and invasion by regulating MAPK1 (37). The current study validated the
binding between LINC00467 and miR-217, as well as their negative
correlation in OS.
It has been reported that KPNA4 exerts carcinogenic
functions in human cancer (38).
KPNA4 promotes angiogenesis and progression of lung cancer by
knocking down MCM3AP antisense RNA 1 expression (39) and facilitates cutaneous squamous
cell carcinoma tumorigenesis by inhibiting miR-3619-5p expression
(40). The current study
demonstrated that miR-217 could directly bind to KPNA4 and that
there was a negative correlation between KPNA4 and miR-217
expression. Finally, rescue assays indicated that KPNA4
overexpression partially reversed the inhibitory effect of
LINC00467 deficiency on OS progression.
In summary, the present study proved that LINC00467
silencing restrained the progression of OS by targeting the
miR-217/KPNA4 axis, indicating that the LINC00467/miR-217/KPNA4
axis may become a new biological diagnostic and therapeutic target
for OS.
Supplementary Data
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
JY and QZ conceived and designed the study. JY, TF
and MZ performed the experiments. JY and QZ analyzed the data. JY
wrote the manuscript. QZ edited the manuscript. All authors read
and approved the final manuscript.
Ethics approval and consent to
participate
The study protocol was approved by the Ethics
Committee of The Affiliated Huaian Hospital of Xuzhou Medical
University (Huaian, China). Written informed consent was obtained
from all patients.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgments
Not applicable.
References
|
1
|
Mirabello L, Troisi RJ and Savage SA:
Osteosarcoma incidence and survival rates from 1973 to 2004: Data
from the surveillance, epidemiology, and end results program.
Cancer. 115:1531–1543. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Wycislo KL and Fan TM: The immunotherapy
of canine osteosarcoma: A historical and systematic review. J Vet
Intern Med. 29:759–769. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Morris CD, Teot LA, Bernstein ML, Marina
N, Krailo MD, Villaluna D, Janeway KA, DuBois SG, Gorlick RG and
Randall RL: Assessment of extent of surgical resection of primary
high-grade osteosarcoma by treating institutions: A report from the
children's oncology group. J Surg Oncol. 113:351–354. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Xie X, Li YS, Xiao WF, Deng ZH, He HB, Liu
Q and Luo W: MicroRNA-379 inhibits the proliferation, migration and
invasion of human osteosarcoma cells by targetting EIF4G2. Biosci
Rep. 37:BSR201605422017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Zhang Y, He Z, Li Y, Yang Y, Shi J, Liu X,
Yuan T, Xia J, Li D, Zhang J and Yang Z: Selection of surgical
methods in the treatment of upper tibia osteosarcoma and prognostic
analysis. Oncol Res Treat. 40:528–532. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Lee JA, Paik EK, Seo J, Kim DH, Lim JS,
Yoo JY and Kim MS: Radiotherapy and gemcitabine-docetaxel
chemotherapy in children and adolescents with unresectable
recurrent or refractory osteosarcoma. Jpn J Clin Oncol. 46:138–143.
2016.
|
|
7
|
Ferrari S and Serra M: An update on
chemotherapy for osteosarcoma. Expert Opin Pharmacother.
16:2727–2736. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Geller DS and Gorlick R: Osteosarcoma: A
review of diagnosis, management, and treatment strategies. Clin Adv
Hematol Oncol. 8:705–718. 2010.
|
|
9
|
Adamopoulos C, Gargalionis AN, Basdra EK
and Papavassiliou AG: Deciphering signaling networks in
osteosarcoma pathobiology. Exp Biol Med (Maywood). 241:1296–1305.
2016. View Article : Google Scholar
|
|
10
|
Nagano T and Fraser P: No-nonsense
functions for long noncoding RNAs. Cell. 145:178–181. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Tehrani SS, Karimian A, Parsian H,
Majidinia M and Yousefi B: Multiple functions of long non-coding
RNAs in oxidative stress, DNA damage response and cancer
progression. J Cell Biochem. 119:223–236. 2018. View Article : Google Scholar
|
|
12
|
Wilusz JE, Sunwoo H and Spector DL: Long
noncoding RNAs: Functional surprises from the RNA world. Genes Dev.
23:1494–1504. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ju C, Zhou R, Sun J, Zhang F, Tang X, Chen
KK, Zhao J, Lan X, Lin S, Zhang Z and Lv XB: LncRNA SNHG5 promotes
the progression of osteosarcoma by sponging the miR-212-3p/SGK3
axis. Cancer Cell Int. 18:1412018. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhang R, Liu Y, Liu H, Chen W, Fan HN,
Zhang J and Zhu JS: The long non-coding RNA SNHG12 promotes gastric
cancer by activating the phosphatidylinositol 3-kinase/AKT pathway.
Aging (Albany NY). 11:10902–10922. 2019. View Article : Google Scholar
|
|
15
|
Xia P, Liu P, Fu Q, Liu C, Luo Q, Zhang X,
Cheng L, Qin T and Zhang H: Long noncoding RNA EPIC1 interacts with
YAP1 to regulate the cell cycle and promote the growth of
pancreatic cancer cells. Biochem Biophys Res Commun. 522:978–985.
2020. View Article : Google Scholar
|
|
16
|
Ding H, Luo Y, Hu K, Liu P and Xiong M:
Linc00467 promotes lung adenocarcinoma proliferation via sponging
miR-20b-5p to activate CCND1 expression. Onco Targets Ther.
12:6733–6743. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Li GC, Xin L, Wang YS and Chen Y: Long
intervening noncoding 00467 RNA contributes to tumorigenesis by
acting as a competing endogenous RNA against miR-107 in cervical
cancer cells. Am J Pathol. 189:2293–2310. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
19
|
Majid S, Dar AA, Saini S, Yamamura S,
Hirata H, Tanaka Y, Deng G and Dahiya R: MicroRNA-205-directed
transcriptional activation of tumor suppressor genes in prostate
cancer. Cancer. 116:5637–5649. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Yang Z, Chen J, Xie H, Liu T, Chen Y, Ma
Z, Pei X, Yang W and Li L: Androgen receptor suppresses prostate
cancer metastasis but promotes bladder cancer metastasis via
differentially altering miRNA525-5p/SLPI-mediated vasculogenic
mimicry formation. Cancer Lett. 473:118–129. 2020. View Article : Google Scholar
|
|
21
|
Chen J, Wu Z and Zhang Y: LncRNA SNHG3
promotes cell growth by sponging miR-196a-5p and indicates the poor
survival in osteosarcoma. Int J Immunopathol Pharmacol.
33:20587384188207432019. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Li G and Zhu Y: Effect of lncRNA ANRIL
knockdown on proliferation and cisplatin chemoresistance of
osteosarcoma cells in vitro. Pathol Res Pract. 215:931–938. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Su P, Mu S and Wang Z: Long noncoding RNA
SNHG16 promotes osteosarcoma cells migration and invasion via
sponging miRNA-340. DNA Cell Biol. 38:170–175. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Liu B, Zhao H, Zhang L and Shi X:
Silencing of long-non-coding RNA ANCR suppresses the migration and
invasion of osteosarcoma cells by activating the p38MAPK signalling
pathway. BMC Cancer. 19:11122019. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Song K, Yuan X, Li G, Ma M and Sun J: Long
noncoding RNA CASC11 promotes osteosarcoma metastasis by
suppressing degradation of snail mRNA. Am J Cancer Res. 9:300–311.
2019.PubMed/NCBI
|
|
26
|
Li Z, Liu J, Chen H, Zhang Y, Shi H, Huang
L, Tao J, Shen R and Wang T: Ferritin light chain (FTL) competes
with long noncoding RNA Linc00467 for miR-133b binding site to
regulate chemoresistance and metastasis of colorectal cancer.
Carcinogenesis. 41:467–477. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Jiang W, Cheng X, Wang T, Song X, Zheng Y
and Wang L: LINC00467 promotes cell proliferation and metastasis by
binding with IGF2BP3 to enhance the mRNA stability of TRAF5 in
hepatocellular carcinoma. J Gene Med. 22:e31342020. View Article : Google Scholar
|
|
28
|
D'Angelo B, Benedetti E, Cimini A and
Giordano A: MicroRNAs: A puzzling tool in cancer diagnostics and
therapy. Anticancer Res. 36:5571–5575. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Srivastava SK, Bhardwaj A, Leavesley SJ,
Grizzle WE, Singh S and Singh AP: MicroRNAs as potential clinical
biomarkers: Emerging approaches for their detection. Biotech
Histochem. 88:373–387. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Cui Y, Yi L, Zhao JZ and Jiang YG: Long
noncoding RNA HOXA11-AS functions as miRNA sponge to promote the
glioma tumorigenesis through targeting miR-140-5p. DNA Cell Biol.
36:822–828. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Li T, Meng XL and Yang WQ: Long noncoding
RNA PVT1 Acts as a 'sponge' to inhibit microRNA-152 in gastric
cancer cells. Dig Dis Sci. 62:3021–3028. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Yu H, Xu Y, Zhang D and Liu G: Long
noncoding RNA LUCAT1 promotes malignancy of ovarian cancer through
regulation of miR-612/HOXA13 pathway. Biochem Biophys Res Commun.
503:2095–2100. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Long B, Li N, Xu XX, Li XX, Xu XJ, Liu JY
and Wu ZH: Long noncoding RNA LOXL1-AS1 regulates prostate cancer
cell proliferation and cell cycle progression through miR-541-3p
and CCND1. Biochem Biophys Res Commun. 505:561–568. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Jiang B, Zhu SJ, Xiao SS and Xue M:
MiR-217 inhibits M2-like macrophage polarization by suppressing
secretion of inter-leukin-6 in ovarian cancer. Inflammation.
42:1517–1529. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Yin Z and Ren W: MicroRNA-217 acts as a
tumor suppressor and correlates with the chemoresistance of
cervical carcinoma to cisplatin. Onco Targets Ther. 12:759–771.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Chen G, Yang Z, Feng M and Wang Z:
microRNA-217 suppressed epithelial-to-mesenchymal transition
through targeting PTPN14 in gastric cancer. Biosci Rep.
40:BSR201931762020. View Article : Google Scholar :
|
|
37
|
Zhu L, Yang S and Wang J: miR-217 inhibits
the migration and invasion of HeLa cells through modulating MAPK1.
Int J Mol Med. 44:1824–1832. 2019.PubMed/NCBI
|
|
38
|
Yang J, Lu C, Wei J, Guo Y, Liu W, Luo L,
Fisch G and Li X: Inhibition of KPNA4 attenuates prostate cancer
metastasis. Oncogene. 36:2868–2878. 2017. View Article : Google Scholar :
|
|
39
|
Li X, Yu M and Yang C: YY1-mediated
overexpression of long noncoding RNA MCM3AP-AS1 accelerates
angiogenesis and progression in lung cancer by targeting
miR-340-5p/KPNA4 axis. J Cell Biochem. 121:2258–2267. 2020.
View Article : Google Scholar
|
|
40
|
Zhang M, Luo H and Hui L: MiR-3619-5p
hampers proliferation and cisplatin resistance in cutaneous
squamous-cell carcinoma via KPNA4. Biochem Biophys Res Commun.
513:419–425. 2019. View Article : Google Scholar : PubMed/NCBI
|