Introduction
Nasopharyngeal carcinoma (NPC) is one of the most
common types of head and neck tumor (1,2).
The incidence of NPC varies significantly worldwide, but is
primarily prevalent in East Africa, North Africa, East Asia and
Southeast Asia (3). At present,
platinum chemotherapy is a widely used treatment strategy for NPC,
and cisplatin (DDP) is a common platinum compound that has been
reported to be effective in the treatment of cancer (4). However, drug resistance often leads
to the failure of NPC chemotherapy (5). Therefore, understanding the
mechanism underlying DDP resistance in NPC might aid with improving
the prognosis of patients with NPC.
Long non-coding RNAs (lncRNAs), a class of
transcripts >200 nucleotides in length, lack protein-coding
capacity (6) and serve as
regulatory factors in multiple biological processes, including
apoptosis, metabolism and cell proliferation (7,8).
Increasing evidence has demonstrated that lncRNAs confer
chemoresistance in various types of cancer, such as renal and
hepatocellular cancer (9,10).
Several lncRNAs serve a vital role in drug resistance of NPC. For
instance, lncRNA MAGI2 antisense RNA 3 conferred DDP resistance in
NPC cells via regulating the microRNA
(miRNA/miR)-218-5p/glycerophosphodiester phosphodiesterase domain
containing 5 axis (11). lncRNA
nuclear paraspeckle assembly transcript 1 promoted DDP resistance
in NPC cells via the let-7a-5p/remodeling and spacing factor 1 axis
(12). Testis associated
oncogenic lncRNA knockdown inhibited tumorigenicity and attenuated
DDP resistance in NPC cells (13). However, the function of KCNQ1OT1
in mediating chemoresistance in NPC is not completely
understood.
miRNAs, small non-coding RNAs that are 20-22
nucleotides in length, serve important roles in multiple cellular
processes, including cell proliferation and survival, of human
tumors (14). Previous studies
have revealed that dysregulated miRNAs could mediate the
sensitivity of NPC cells to DDP by targeting mRNAs. For example,
miR-19b served an important role in inhibiting cancer progression
and promoting NPC sensitivity to DDP via inhibiting KRAS (15). Furthermore, miR-205-5p
facilitated cell proliferation and DDP resistance in NPC by
repressing PTEN (16). miR-139
overexpression suppressed DDP-induced progression and enhanced
apoptosis in NPC cells (17).
Nevertheless, the mechanism underlying miR-139 in tumorigenesis and
chemosensitivity in NPC is not completely understood.
The present study investigated the regulatory role
of KCNQ1OT1 in DDP resistance of NPC, and the results of the
present study might provide a novel therapeutic strategy for
DDP-resistant NPC.
Materials and methods
Patients
A total of 50 patients (31 male patients and 19
female patients; age range, 26-65 years; mean age, 46 years) with
NPC (29 DDP-resistant and 21 DDP-sensitive) at Zhuji Central
Hospital (Zhuji, China) were recruited between August 2017 and
December 2019. The inclusion criteria were as follows: i) Diagnosed
with NPC; and ii) had not received preoperative radiotherapy,
chemotherapy or other adjuvant treatments. The exclusion criteria
were as follows: i) Diagnosed with other diseases; and ii) failed
to cooperate with researchers. All specimens were immediately
frozen in liquid nitrogen and stored at -80°C. The present study
was approved by the Ethical Committee of Zhuji Central Hospital.
Written consent was obtained from all patients prior to starting
the study.
Cell lines and culture
NPC cell lines (5-8F and SUNE-1) were purchased from
The Cell Bank of Type Culture Collection of The Chinese Academy of
Sciences. 5-8F and SUNE-1 cells were cultured in RPMI-1640 (Thermo
Fisher Scientific, Inc.) supplemented with 10% FBS (Gibco; Thermo
Fisher Scientific). To establish DDP-resistant NPC cells (5-8F/DDP
and SUNE-1/DDP), cells were treated with 0.5 µg/ml DDP (Qilu
Pharmaceutical Co., Ltd.) at 37°C for 3 weeks and then exposed to
gradually increasing concentrations of DDP (2, 4, 6 or 8
µg/ml) every 3 weeks up to a final concentration of 10
µg/ml for a total 15 weeks. 5-8F/DDP and SUNE-1/DDP cells
were cultured in RPMI-1640 supplemented with 5 µg/ml DDP and
10% FBS. All cells were cultured at 37°C with 5%
CO2.
Cell transfection
Short hairpin (sh)RNA targeting KCNQ1OT1
(shKCNQ1OT1; 0.8 µg; 5′-GCA GAA CCA UCG AUG GUG CGU-3′),
shRNA targeting USP47 (shUSP47; 0.8 µg; 5′-GCC UUU GCA GAC
UCU CAU UUA-3′), shRNA-negative control (NC; shNC; 0.8 µg;
scrambled; 5′-AGU GCU GCG CAC GUG UCU CAU-3′), miR-454 mimics (100
nM; 5′-UAG UGC AAU AUU GCU UAU AGG GU-3′), NC mimics (100 nM;
5′-UUG UAC UAC ACA AAA GUA CUG-3′), miR-454 inhibitor (100 nM;
5′-ACC CUA UAA GCA AUA UUG CAC UA-3′) and NC inhibitor (100 nM;
5′-CAG UAC UUU UGU GUA GUACAA-3′) were purchased from Shanghai
GenePharma Co., Ltd. To overexpress KCNQ1OT1 or USP47, the
full-length KCNQ1OT1 sequence was inserted into the pcDNA3.1 vector
(Shanghai GenePharma Co., Ltd.). 5-8F/DDP and SUNE-1/DDP cells
(1×105) were transfected with shRNA, miR-mimics,
miR-inhibitors, overexpression vectors and the corresponding NCs
using Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.) at 37°C. At 48 h post-transfection, subsequent
experiments were performed.
Reverse transcription-quantitative PCR
(RT-qPCR)
Total RNA was extracted from tissues and cells using
TRIzol® reagent (Invitrogen; Thermo Fisher Scientific,
Inc.). Total RNA was reverse transcribed into cDNA using the
PrimeScript RT reagent kit (Takara Bio, Inc.) according to the
manufacturer's protocol. Subsequently, qPCR was performed using
SYBR® Premix Ex Taq™ II (Takara Bio, Inc.) on the ABI
7500 real-time PCR system (Applied Biosystems; Thermo Fisher
Scientific, Inc.). The following thermocycling conditions were used
for qPCR: Initial denaturation at 95°C for 15 sec; 40 cycles of
denaturation at 94°C for 30 sec, annealing at 60°C for 20 sec and
extension at 72°C for 40 sec. The following primers were used for
qPCR: KCNQ1OT1 forward, 5′-TTG GTA GGA TTT TGT TGA GG-3′ and
reverse, 5′-CAA CCT TCC CCT ACT ACC-3′; miR-454 forward, 5′-TAG TGC
AAU ATT GCT TAU AGG GT-3′ and reverse, 5′-CCU AUA AGC AAU ATT GCA
CTA TT-3′; USP47 forward, 5′-GGC AGG ACG CTC ATT AGG T-3′ and
reverse, 5′-GCA CAA CAT GAT TCC AAG TCA A-3′; GAPDH forward, 5′-TCA
AGG CTG AGA ACG GGA AG-3′ and reverse, 5′-TGG ACT CCA CGA CGT ACT
CA-3′; and U6 forward, 5′-CTC GCT TCG GCA GCA CAT ATA CT-3′ and
reverse, 5′-CGC TTC ACG AAT TTG CGT GT-3′. miRNA and mRNA
expression levels were quantified using the 2−ΔΔCq
method (18) and normalized to
the internal reference genes U6 and GAPDH, respectively.
Colony forming assay
Transfected 5-8F/DDP and SUNE-1/DDP cells were
trypsinized and seeded (1×103 cells/well) into 6-well
plates. Subsequently, cells were cultured in RPMI-1640 supplemented
with 10% FBS for 2-3 weeks. Visible colonies were fixed in 4%
paraformaldehyde for 10 min at room temperature and stained with
0.1% crystal violet for 10 min at room temperature (Sigma-Aldrich;
Merck KGaA). Stained colonies (>30 cells) were counted using a
light microscope (magnification, ×40) and the colony formation rate
was calculated.
Transwell assay
Transwell chambers (pore size, 8 µm; Corning,
Inc.) were used to perform the Transwell assay. To assess cell
migration, 5-8F/DDP and SUNE-1/DDP cells (1×105
cells/well) in serum-free RPMI-1640 were plated into the upper
chamber. Medium supplemented with 10% FBS was plated into the lower
chamber. Following incubation for 24 h at 37°C, migratory cells
were fixed with 4% paraformaldehyde at room temperature for 30 min
and stained with 0.1% crystal violet at 37°C for 2 h. Stained cells
were counted in five randomly selected fields of view using a light
microscope (magnification, ×200). To assess cell invasion, the
upper chambers were precoated with Matrigel for 30 min at 37°C and
then the aforementioned protocol was performed.
Cell Counting Kit-8 (CCK-8) assay
Cells were seeded (5×103 cells/well) into
96-well plates. Cells were treated with different concentrations of
DDP (0, 2, 4, 6, 8 or 10 µg/ml) for 24 h at 37°C.
Subsequently, 10 µl CCK-8 reagent (Beyotime Institute of
Biotechnology) was added to each well and incubated for 2 h at 37°C
with 5% CO2. Absorbance was measured at a wavelength of
450 nm using a microplate reader. IC50 values were
calculated as the concentration of DDP resulting in 50% inhibition
of cell viability, with higher IC50 values suggesting
higher drug resistance potential.
Western blotting
Total protein was extracted from cells and tissues
using RIPA buffer (Beyotime Institute of Biotechnology). Protein
concentrations were determined using a BCA protein assay kit
(Beyotime Institute of Biotechnology). Proteins (10 µg) were
separated via 10% SDS-PAGE and transferred to PVDF membranes (EMD
Millipore). After blocking with 5% non-fat dry milk at room
temperature for 2 h, the membranes were incubated overnight at 4°C
with primary antibodies targeted against: USP47 (1:1,000; cat. no.
ab72143; Abcam) and GAPDH (1:1,000; cat. no. ab9485; Abcam).
Subsequently, the membranes were incubated with a HRP-conjugated
secondary antibody (1:1,000; cat. no. ab6721; Abcam) for 1.5 h at
37°C. Protein bands were visualized using an
enhanced-chemiluminescence reagent (Beyotime Institute of
Biotechnology). Protein expression levels were semi-quantified
using Image Lab software (version 4.1; Bio-Rad Laboratories, Inc.)
with GAPDH as the loading control.
Luciferase reporter assay
StarBase (starbase.sysu.edu.cn/) and TargetScan (www.targetscan.org/vert_72/) were used to predict
the binding sites between miR-454 and KCNQ1OT1 or USP47.
pmirGLO-KCNQ1OT1-wild-type (WT)/mutant (Mut) and
pmirGLO-USP47-WT/Mut reporter plasmids were provided by Shanghai
GenePharma Co., Ltd. 5-8F/DDP and SUNE-1/DDP cells
(1×105) were co-transfected with 0.6 µg
pmirGLO-KCNQ1OT1-Wt/Mut or pmirGLO-USP47-Wt/Mut plasmid and 100 nM
NC mimics or miR-454 mimics using Lipofectamine® 2000 reagent
(Invitrogen; Thermo Fisher Scientific, Inc.) at 37°C. At 48 h
post-transfection, luciferase activities were assessed using the
dual-luciferase reporter assay system (Promega Corporation).
Firefly luciferase activities were normalized to Renilla
luciferase activities.
RNA immunoprecipitation (RIP) assay
RIP assays were performed to investigate whether
KCNQ1OT1 and miR-454 were in the same RNA-induced silencing
complex. The RIP assay was performed using the Magna RIP
RNA-binding protein immunoprecipitation kit (EMD Millipore).
5-8F/DDP and SUNE-1/DDP cells were lysed using RIP lysis buffer
(Beyotime Institute of Biotechnology) and incubated with 50
µl A/G magnetic beads conjugated with argonaute RISC
catalytic component 2 (Ago2) (EMD Millipore) and IgG (EMD
Millipore). Subsequently, immunoprecipitated RNA was extracted.
KCNQ1OT1 and miR-454 expression levels were measured via
RT-qPCR.
Statistical analysis
Statistical analyses were performed using SPSS
software (version 16.0; SPSS, Inc.). Data are presented as the mean
± SD. Each experiment was performed in triplicate. Comparisons
among multiple groups were analyzed using one-way ANOVA followed by
Tukey's post hoc test. Comparisons between two groups were analyzed
using an unpaired Student's t-test. Pearson's correlation analysis
was used to evaluate the correlation between the expression levels
of miR-454 and KCNQ1OT1 or USP47. P<0.05 was considered to
indicate a statistically significant difference.
Results
High expression of KCNQ1OT1 in
DDP-resistant NPC tissues and cells
The RT-qPCR results demonstrated that KCNQ1OT1
expression was significantly upregulated in the 29 DDP-resistant
NPC tissues compared with the 21 DDP-sensitive NPC tissues
(Fig. 1A). Subsequently,
DDP-resistant NPC cells (5-8F/DDP and SUNE-1/DDP) were established.
Cell viability and the IC50 values of DDP in 5-8F/DDP
and SUNE-1/DDP cells were significantly higher compared with 5-8F
and SUNE-1 cells, respectively (Fig.
1B and C). Furthermore, KCNQ1OT1 expression levels were
significantly upregulated in 5-8F/DDP and SUNE-1/DDP cells compared
with parental NPC cells (Fig.
1D). The results indicated that KCNQ1OT1 might be involved in
DDP sensitivity and resistance in NPC cells.
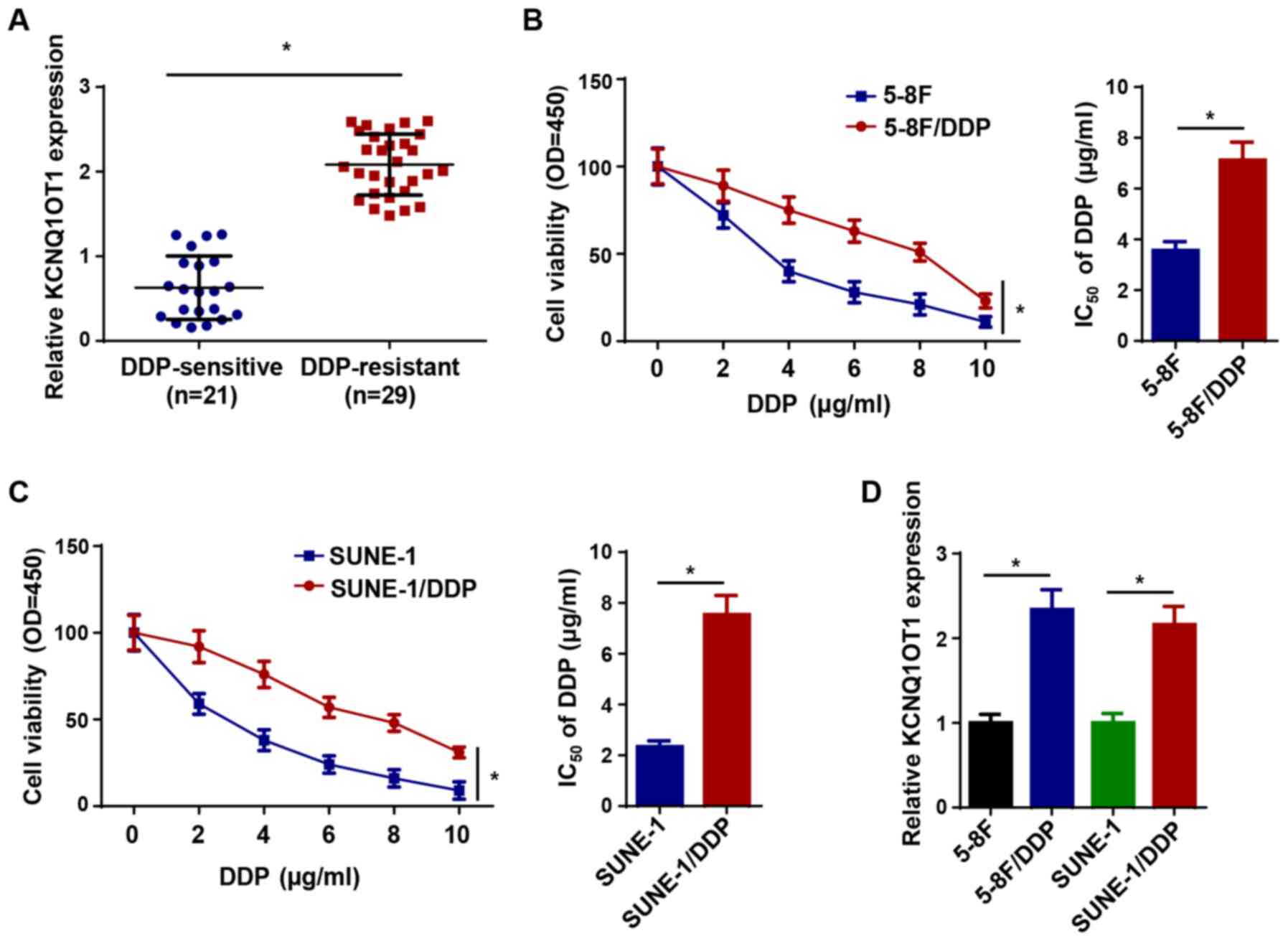 | Figure 1High expression of KCNQ1OT1 in
DDP-resistant NPC tissues and cells. (A) KCNQ1OT1 expression levels
in DDP-sensitive and DDP-resistant NPC tissues. Cell Counting Kit-8
assays were performed to detect cell viability and calculate the
IC50 values of DDP in (B) 5-8F/DDP, 5-8F, (C) SUNE-1/DDP
and SUNE-1 cells. (D) KCNQ1OT1 expression levels in 5-8F/DDP, 5-8F,
SUNE-1/DDP and SUNE-1 cells. Data are presented as the mean ± SD.
*P<0.05. KCNQ1OT1, KCNQ1 opposite strand/antisense
transcript 1; DDP, cisplatin; NPC, nasopharyngeal carcinoma;
RT-qPCR, reverse transcription-quantitative PCR; Cis, cisplatin;
OD, optical density. |
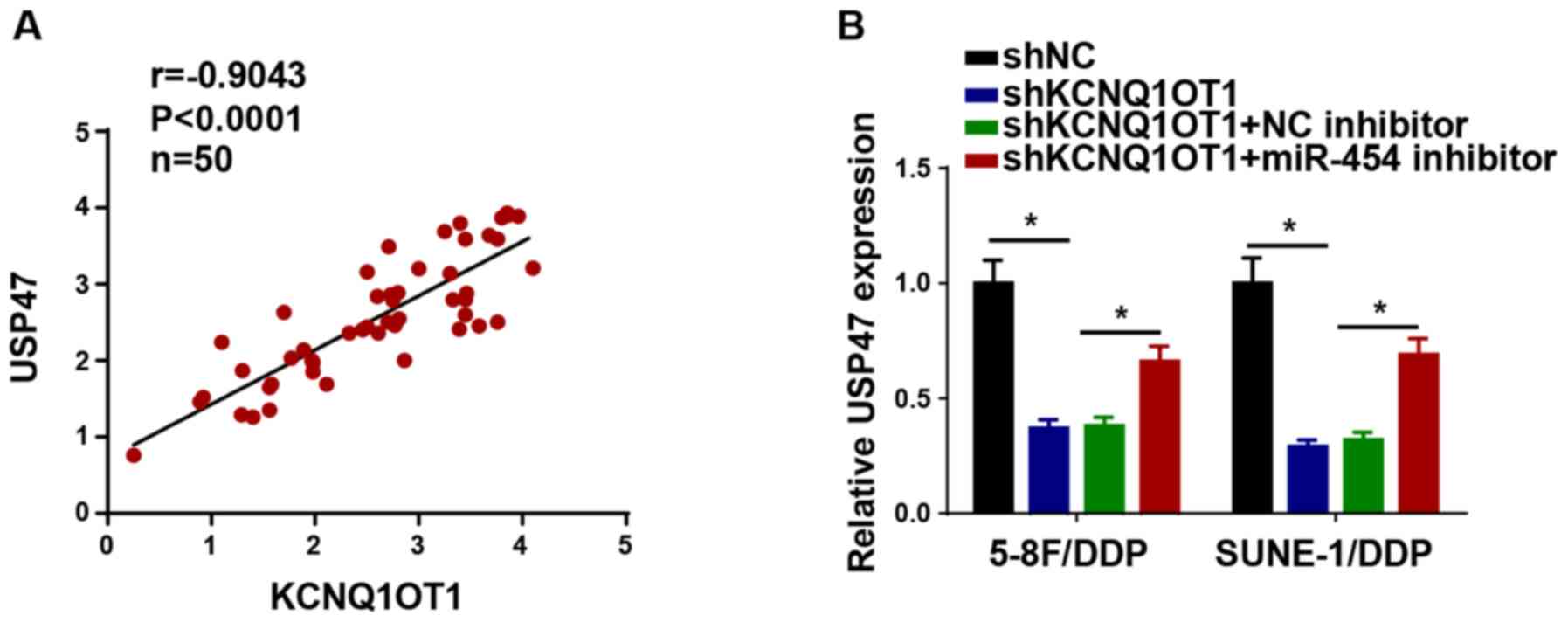
KCNQ1OT1 knockdown increases DDP
sensitivity in NPC cells
To explore the effect of KCNQ1OT1 knockdown on DDP
resistance in NPC cells, shKCNQ1OT1 was transfected into 5-8F/DDP
and SUNE-1/DDP cells to knock down KCNQ1OT1 expression (Fig. 2A). Compared with the shNC group,
KCNQ1OT1 knockdown significantly decreased the IC50
value of DDP in 5-8F/DDP and SUNE-1/DDP cells (Fig. 2B). The CCK-8 assay results
indicated that KCNQ1OT1 knockdown also significantly attenuated
5-8F/DDP and SUNE-1/DDP cell viability compared with the shNC group
(Fig. 2C). In addition, the
colony forming and Transwell assay results indicated that 5-8F/DDP
and SUNE-1/DDP cell proliferation, migration and invasion were
significantly inhibited by KCNQ1OT1 knockdown compared with the
shNC group (Fig. 2D-F).
Therefore, the results indicated that KCNQ1OT1 knockdown decreased
DDP resistance in NPC cells.
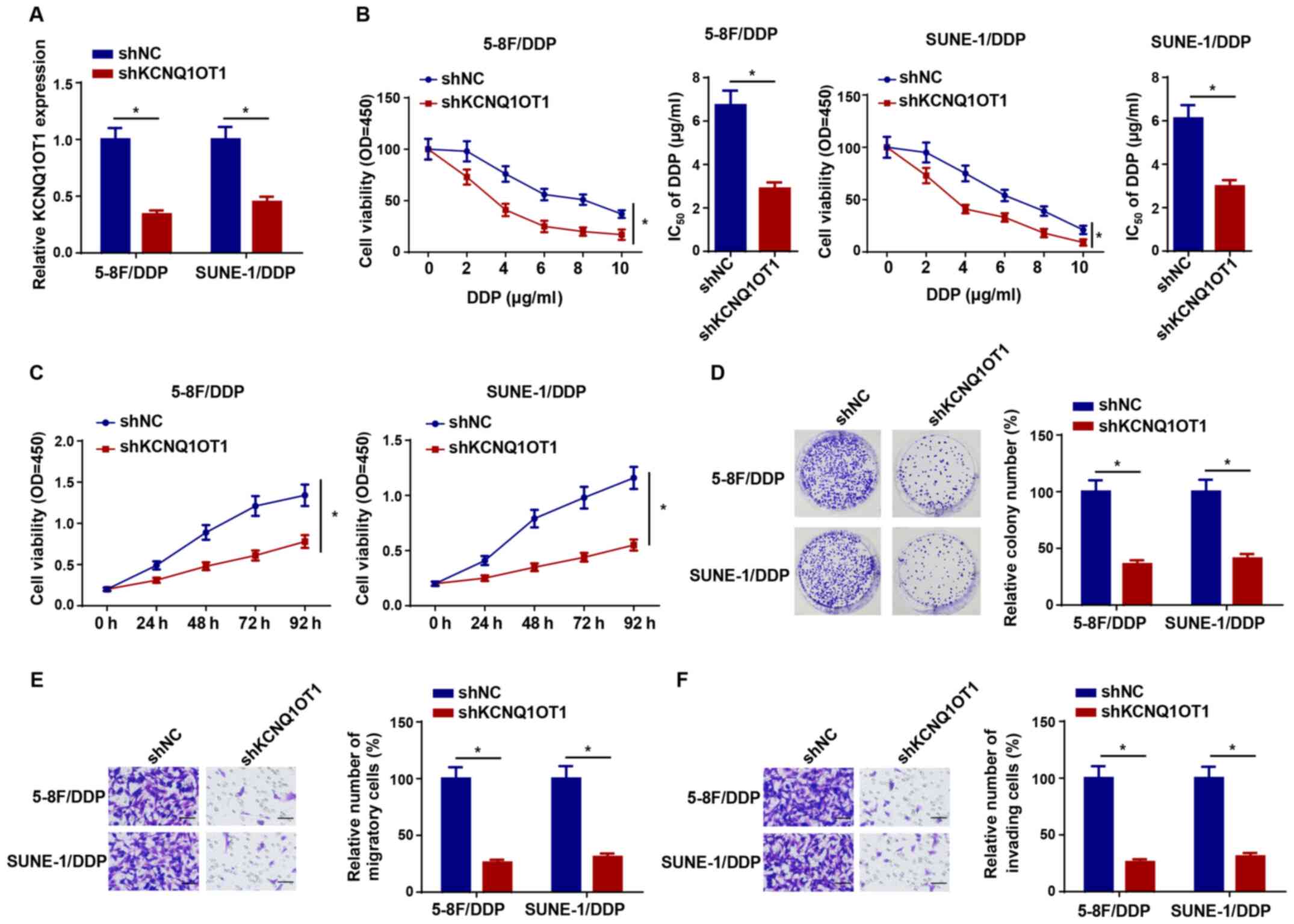 | Figure 2KCNQ1OT1 knockdown increases
nasopharyngeal carcinoma cell DDP sensitivity. (A) Transfection
efficiency of shKCNQ1OT1 in 5-8F/DDP and SUNE-1/DDP cells. (B)
CCK-8 assays were performed to detect cell viability and calculate
the IC50 values of DDP in 5-8F/DDP and SUNE-1/DDP cells
transfected with shNC or shKCNQ1OT1. (C) CCK-8 assays were
performed to detect the cell viability of 5-8F/DDP and SUNE-1/DDP
cells transfected with shKCNQ1OT1. (D) Colony forming
(magnification, ×40), (E) Transwell migration and (F) Transwell
invasion assays were performed to assess cell proliferation,
migration and invasion in 5-8F/DDP and SUNE-1/DDP cells transfected
with shNC or shKCNQ1OT1 (scale bar, 100 µm). Data are
presented as the mean ± SD. *P<0.05. KCNQ1OT1, KCNQ1
opposite strand/antisense transcript 1; DDP, cisplatin; sh, short
hairpin RNA; CCK-8, Cell Counting Kit-8; NC, negative control; OD,
optical density. |
KCNQ1OT1 directly interacts with
miR-454
Increasing evidence has suggested that lncRNAs
mediate their effects via competitively binding to miRNAs (19-22). Therefore, star-Base was used to
predict the miRNA targets of KCNQ1OT1. The results suggested that
KCNQ1OT1 contained binding sites complementary to miR-454 (Fig. 3A). To verify the interaction,
luciferase reporter and RIP assays were conducted. Compared with
the corresponding NC groups, miR-454 mimics and miR-454 inhibitor
significantly reduced and increased the luciferase activity of the
KCNQ1OT1-WT reporter in 5-8F/DDP and SUNE-1/DDP cells,
respectively, whereas the luciferase activity of the KCNQ1OT1-Mut
reporter was not significantly altered (Fig. 3B). Moreover, the RIP assay
results indicated that KCNQ1OT1 and miR-454 expression levels were
significantly enriched in Ago2 compared with IgG (Fig. 3C). miR-454 expression was
significantly higher in DDP-sensitive NPC tissues compared with
DDP-resistant NPC tissues (Fig.
3D). Similarly, miR-454 expression was significantly
downregulated in 5-8F/DDP and SUNE-1/DDP cells compared with
parental NPC cells (Fig. 3E).
The expression levels of KCNQ1OT1 were negatively correlated with
miR-454 expression levels in NPC tissues (Fig. 3F). To explore whether KCNQ1OT1
regulated miR-454 expression, pcDNA3.1-KCNQ1OT1 and shKC-NQ1OT1
were transfected into 5-8F/DDP and SUNE-1/DDP cells to overexpress
or knock down KCNQ1OT1 expression, respectively (Fig. 3G), and then miR-454 expression
levels were measured. The results demonstrated that KCNQ1OT1
overexpression significantly downregulated miR-454 expression
compared with the pcDNA3.1 group, whereas KCNQ1OT1 knockdown
significantly increased miR-454 expression levels compared with the
shNC group (Fig. 3H).
Collectively, the results indicated that KCNQ1OT1 served as a
molecular sponge for miR-454.
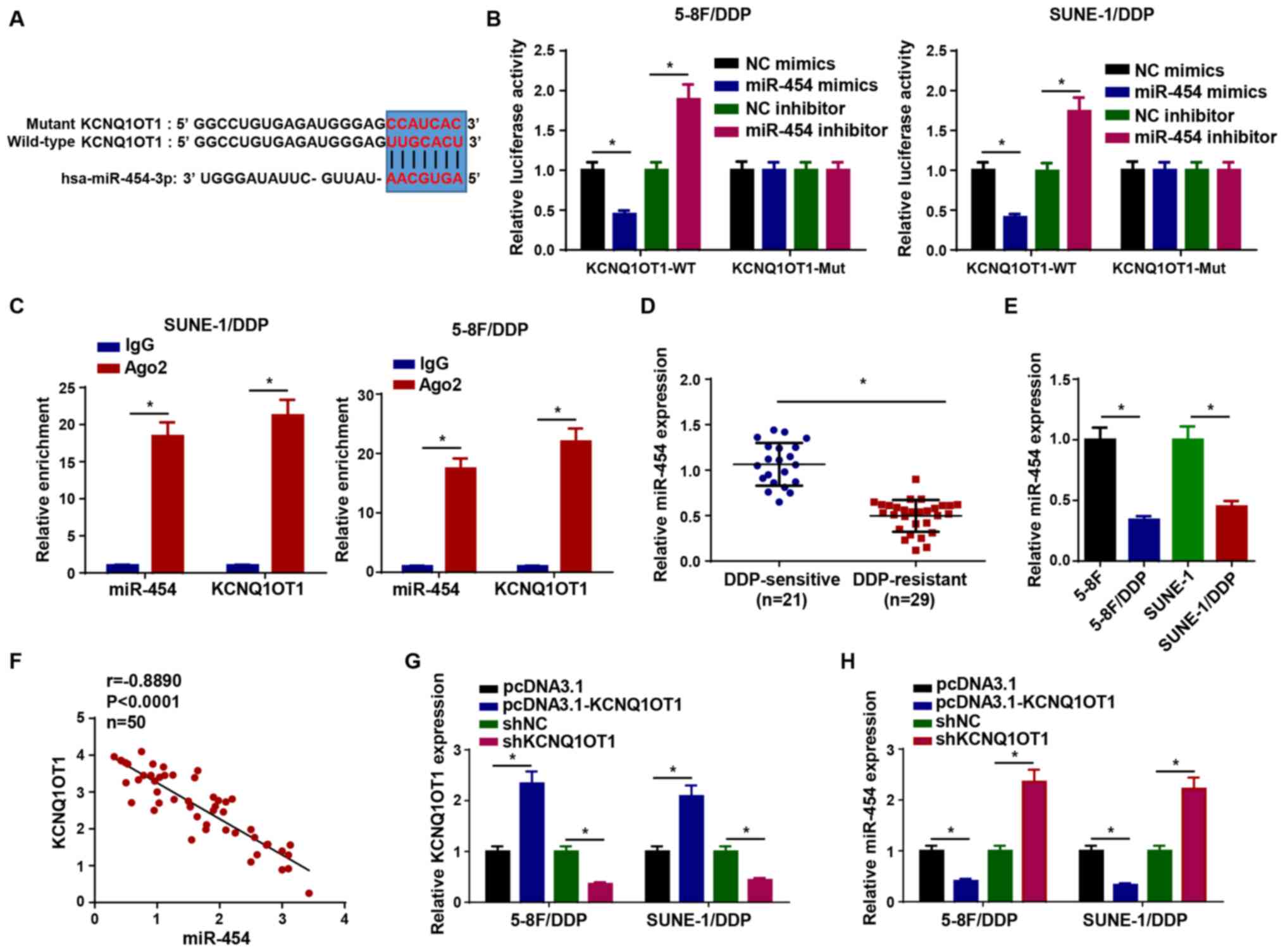 | Figure 3KCNQ1OT1 directly interacts with
miR-454. (A) Binding sites between miR-454 and KCNQ1OT1. (B)
Luciferase activity of KCNQ1OT1-WT and KCNQ1OT1-Mut in 5-8F/DDP and
SUNE-1/DDP cells following transfection with NC mimics, miR-454
mimics, NC inhibitor or miR-454 inhibitor. (C) RNA
immunoprecipitation assays were performed to assess the interaction
between KCNQ1OT1 and miR-454. (D) miR-454 expression levels in
DDP-sensitive and DDP-resistant NPC tissues. (E) miR-454 expression
levels in 5-8F/DDP, 5-8F, SUNE-1/DDP and SUNE-1 cells. (F)
Correlation between KCNQ1OT1 expression and miR-454 expression in
NPC tissues. (G) Transfection efficiency of pcDNA3.1-KCNQ1OT1 and
shKCNQ1OT1. (H) miR-454 expression levels in 5-8F/DDP and
SUNE-1/DDP cells transfected with pcDNA3.1, pcDNA3.1-KCNQ1OT1, shNC
or shKCNQ1OT1. Data are presented as the mean ± SD. *P<0.05.
KCNQ1OT1, KCNQ1 opposite strand/antisense transcript 1; miR,
microRNA; WT, wild-type; Mut, mutant; NC, negative control; DDP,
cisplatin; NPC, nasopharyngeal carcinoma; sh, short hairpin RNA;
Ago2, argonaute RISC catalytic component 2; Cis, cisplatin. |
KCNQ1OT1 modulates the sensitivity of
DDP-resistant NPC cells by downregulating miR-454
To further investigate whether KCNQ1OT1 exerted its
function in the DDP resistance of NPC cells by regulating miR-454
expression, miR-454 was overexpressed in 5-8F/DDP and SUNE-1/DDP
cells by transfection with miR-454 mimics (Fig. 4A). Subsequently, 5-8F/DDP and
SUNE-1/DDP cells were transfected with NC mimics, miR-454 mimics,
miR-454 mimics + pcDNA3.1 or miR-454 mimics + pcDNA3.1-KCNQ1OT1.
The CCK-8 assay results indicated that compared with the NC mimics
group, miR-454 overexpression significantly suppressed cell
viability, which was significantly reversed by co-transfection with
pcDNA3.1-KCNQ1OT1 (Fig. 4B).
Furthermore, the colony forming and Transwell assay results
demonstrated that pcDNA3.1-KCNQ1OT1 significantly reversed miR-454
overexpression-induced inhibition of 5-8F/DDP and SUNE-1/DDP cell
proliferation, migration and invasion (Fig. 4C-E). Collectively, the results
indicated that KCNQ1OT1 enhanced DDP resistance in NPC cells by
downregulating miR-454 expression.
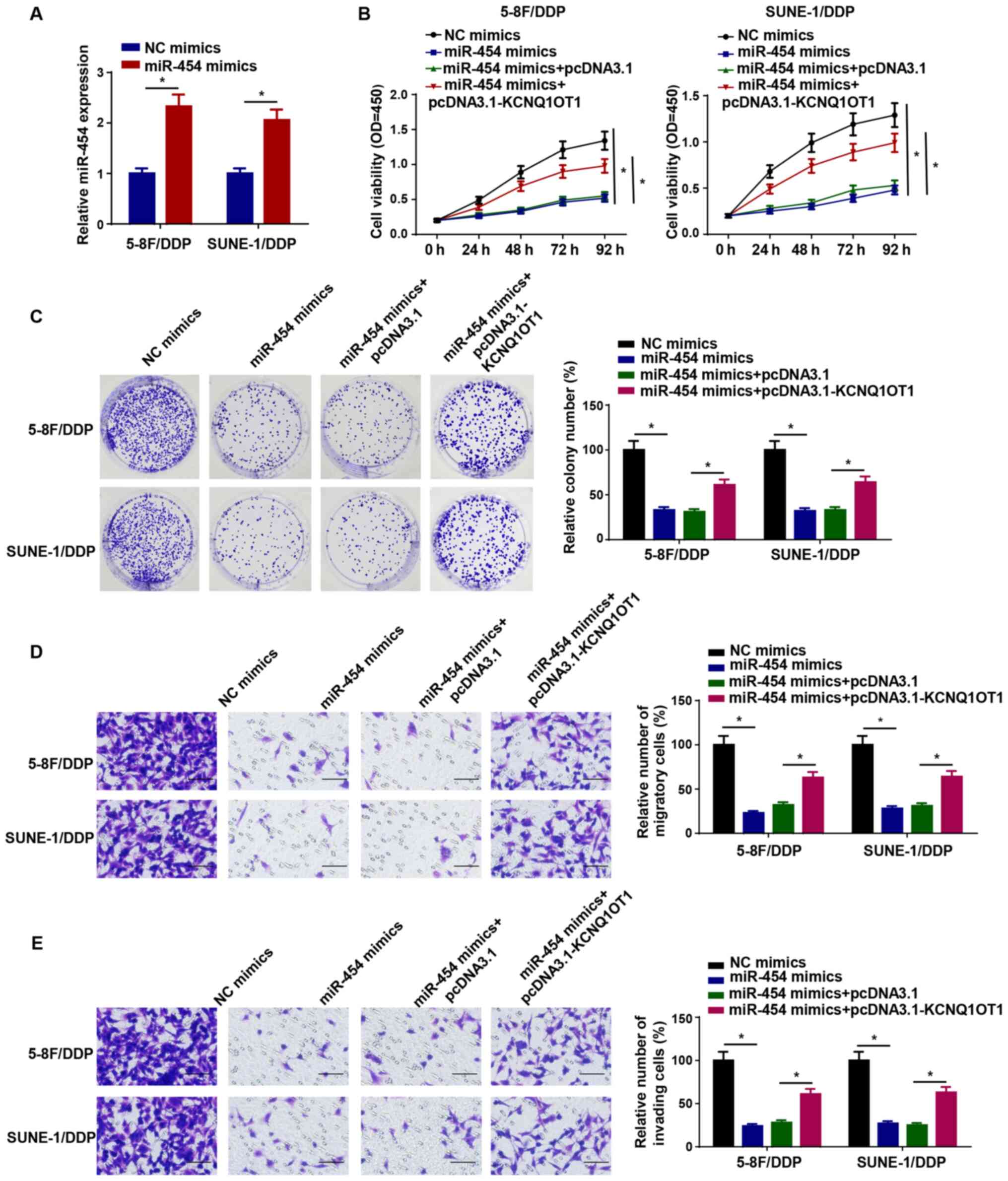 | Figure 4KCNQ1OT1 modulates the sensitivity of
DDP-resistant nasopharyngeal carcinoma cells by downregulating
miR-454. (A) Transfection efficiency of miR-454 mimics. (B) Cell
Counting Kit-8 assays were performed to detected the cell viability
of 5-8F/DDP and SUNE-1/DDP cells transfected with NC mimics,
miR-454 mimics, miR-454 mimics + pcDNA3.1 or miR-454+
pcDNA3.1-KCNQ1OT1. (C) Colony forming (magnification, ×40), (D)
Transwell migration and (E) Transwell invasion assays were
performed to assess cell proliferation, migration and invasion in
5-8F/DDP and SUNE-1/DDP cells transfected with NC mimics, miR-454
mimics, miR-454 mimics + pcDNA3.1 or miR-454+ pcDNA3.1-KCNQ1OT1
(scale bar, 100 µm). Data are presented as the mean ± SD.
*P<0.05. KCNQ1OT1, KCNQ1 opposite strand/antisense
transcript 1; DDP, cisplatin; miR, microRNA; NC, negative control;
OD, optical density. |
USP47 is directly targeted by
miR-454
The binding sites of miR-454 on the 3′-UTR of USP47
were predicted using TargetScan software (Fig. 5A). Compared with the
corresponding NC groups, miR-454 mimics significantly inhibited the
luciferase activity of USP47-WT, whereas miR-454 inhibitor
significantly increased the luciferase activity of USP47-WT in
5-8F/DDP and SUNE-1/DDP cells (Fig.
5B). The RIP assay results demonstrated that miR-454 and USP47
expression levels were significantly enriched in Ago2 compared with
IgG (Fig. 5C). In addition,
USP47 expression was significantly upregulated in DDP-resistant NPC
tissues compared with DDP-sensitive NPC tissues (Fig. 5D). The RT-qPCR and western
blotting results indicated that USP47 mRNA and protein expression
levels were increased in DDP-resistant NPC cells compared with
parental NPC cells (Fig. 5E and
F). Pearson's correlation analysis suggested that USP47
expression was negatively correlated with miR-454 expression in NPC
tissues (Fig. 5G). The
aforementioned results indicated that USP47 was a target of
miR-454.
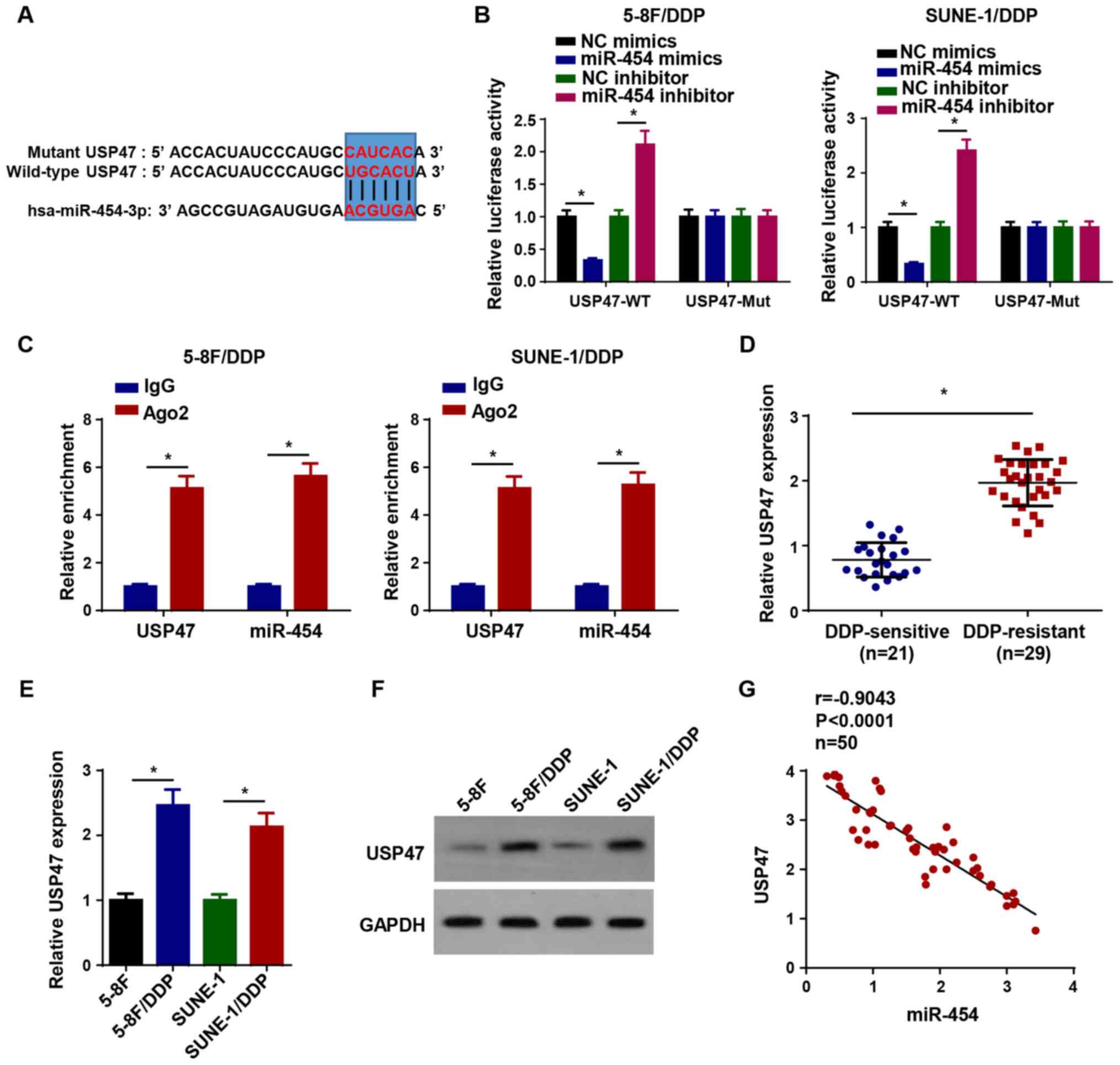 | Figure 5USP47 is directly targeted by
miR-454. (A) Binding sites between miR-454 and the 3′-untranslated
region of USP47. (B) Luciferase activity of USP47-WT and USP47-Mut
in 5-8F/DDP and SUNE-1/DDP cells following transfection with NC
mimics, miR-454 mimics, NC inhibitor or miR-454 inhibitor. (C) RNA
immunoprecipitation assays were performed to assess the interaction
between USP47 and miR-454. (D) USP47 expression levels in
DDP-resistant and DDP-sensitive NPC tissues. USP47 (E) mRNA and (F)
protein expression levels in DDP-resistant NPC cells. (G)
Correlation between USP47 expression and miR-454 expression in NPC
tissues. Data are presented as the mean ± SD.
*P<0.05. USP47, ubiquitin specific peptidase 47; miR,
microRNA; WT, wild-type; Mut, mutant; DDP, cisplatin; NC, negative
control; NPC, nasopharyngeal carcinoma; Ago2, argonaute RISC
catalytic component 2. |
KCNQ1OT1 confers DDP resistance in NPC
cells via upregulating USP47 expression by sponging miR-454
Subsequently, the RT-qPCR results demonstrated that
miR-454 and USP47 expression levels were significantly decreased in
5-8F/DDP and SUNE-1/DDP cells following transfection with miR-454
inhibitor or shUSP47 compared with the NC inhibitor and shNC
groups, respectively, which suggested that miR-454 inhibitor and
shUSP47 downregulated miR-454 and USP47 expression levels in
DDP-resistant NPC cells, respectively (Fig. 6A and B). In addition, 5-8F/DDP
cells were transfected with shNC, shUSP47, shUSP47 + NC inhibitor
or shUSP47 + miR-454 inhibitor. The CCK-8 assay results
demonstrated that USP47 knockdown significantly reduced cell
viability compared with the shNC group, which was significantly
reversed by co-transfection with miR-454 knockdown in 5-8F/DDP and
SUNE-1/DDP cells (Fig. 6C). The
colony forming and Transwell assay results demonstrated that
miR-454 knockdown significantly reversed shUSP47-induced inhibitory
effects on DDP-resistant NPC cell proliferation, migration and
invasion (Fig. 6D-F).
Collectively, the results demonstrated that miR-454 participated in
DDP resistance in NPC via USP47.
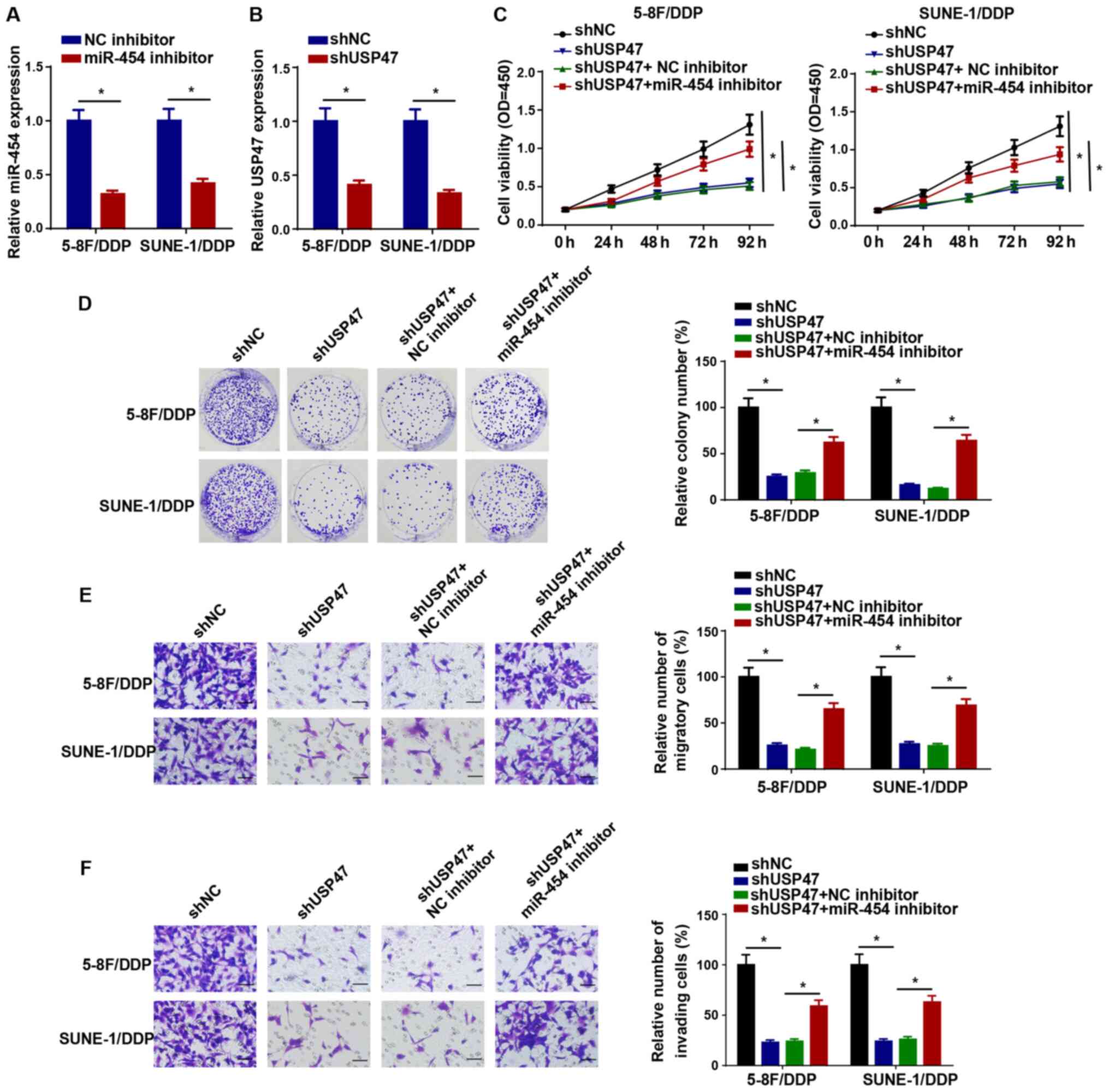 | Figure 6KCNQ1OT1 confers NPC cell DDP
resistance via upregulating USP47 expression in NPC cells by
sponging miR-454. (A) Transfection efficiency of miR-454 inhibitor.
(B) USP47 expression levels in 5-8F/DDP and SUNE-1/DDP cells
transfected with shNC or shUSP47. (C) Cell Counting Kit-8 assays
were performed to detect the cell viability of 5-8F/DDP and
SUNE-1/DDP cells transfected with shNC, shUSP47, shUSP47 + NC
inhibitor or shUSP47 + miR-454 inhibitor. (D) Colony forming
(magnification, ×40), (E) Transwell migration and (F) Transwell
invasion assays were performed to assess cell proliferation,
migration and invasion in 5-8F/DDP and SUNE-1/DDP cells transfected
with shNC, shUSP47, shUSP47 + NC inhibitor or shUSP47 + miR-454
inhibitor (scale bar, 100 µm). Data are presented as the
mean ± SD. *P<0.05. KCNQ1OT1, KCNQ1 opposite
strand/antisense transcript 1; NPC, nasopharyngeal carcinoma; DDP,
cisplatin; USP47, ubiquitin specific peptidase 47; miR, microRNA:
sh, short hairpin RNA; NC, negative control. |
USP47 expression is regulated by KCNQ1OT1
and miR-454
Pearson's correlation analysis suggested that USP47
expression was positively correlated with KCNQ1OT1 expression in
NPC tissues (Fig. 7A). To
further determine whether KCNQ1OT1 mediated its effects via
regulating the miR-454/USP47 axis, 5-8F/DDP and SUNE-1/DDP cells
were transfected with shNC, shKCNQ1OT1, shKCNQ1OT1+ NC inhibitor or
shKC-NQ1OT1 + miR-454 inhibitor. The RT-qPCR results suggested that
USP47 expression was significantly downregulated in
KCNQ1OT1-knockdown DDP-resistant NPC cells compared with the shNC
group, and miR-454 knockdown significantly reversed KCNQ1OT1
knockdown-mediated downregulation of USP47 expression levels
(Fig. 7B). The results indicated
that KCNQ1OT1 contributed to DDP resistance in NPC cells via
regulating the miR-454/USP47 axis.
Discussion
Increasing evidence has indicated that abnormal
lncRNAs might lead to drug resistance in various types of cancer
(23-25), including NPC (26,27). The present study demonstrated
that compared with the shNC group, KCNQ1OT1 knockdown significantly
suppressed cell viability and promoted DDP sensitivity in
DDP-resistant NPC cells by sponging miR-454 via downregulating
USP47. Moreover, the results suggested that the
KCNQ1OT1/miR-454/USP47 axis participated in the regulation of DDP
resistance in NPC.
KCNQ1OT1 is an imprinted antisense lncRNA located at
11p15.5 (28,29). Recent studies have suggested that
KCNQ1OT1 is closely associated with the drug resistance of various
tumors. For example, DDP-induced KCNQ1OT1 regulated the
chemoresistance of tongue cancer via regulating the
miR-124-3p/tripartite motif containing 14 axis (30). The lncRNA KCNQ1OT1/miR-34a axis
increased the chemoresistance of colon cancer cells by targeting
autophagy related 4B cysteine peptidase (31). Moreover, KCNQ1OT1 knockdown
sensitized osteosarcoma cells to DDP and inhibited cell invasion
via the potassium voltage-gated channel subfamily Q member 1/DNA
methyltransferase 1 axis (32).
In the present study, the functions and mechanisms underying
KCNQ1OT1-induced DDP resistance in NPC were investigated. The
results indicated that KCNQ1OT1 expression was significantly
increased in DDP-resistant NPC compared with DDP-sensitive NPC, and
KCNQ1OT1 knockdown significantly suppressed NPC cell viability and
DDP resistance compared with the shNC group.
Previous studies have revealed that lncRNAs might
serve as competitive endogenous RNAs for miRNAs to alter the
binding of miRNAs and miRNA target gene expression levels (33-36). The present study demonstrated
that KCNQ1OT1 directly interacted with miR-454. miR-454 has been
reported to serve vital roles in tumor proliferation, apoptosis and
metastasis in various types of cancer. For example, miR-454
remarkably inhibited bladder cancer cell invasion and migration via
targeting ZEB2 antisense RNA 1 (37). miR-454 overexpression accelerated
apoptosis and suppressed proliferation of glioblastoma cells by
downregulating nuclear factor of activated T cells 2 (38). Moreover, miR-454 inhibition
decreased the repressive effects of HOXA11 antisense RNA knockdown
on DDP-resistant non-small cell lung cancer cells (39). In the present study, compared
with the NC mimics group, miR-454 overexpression significantly
inhibited cell viability and promoted sensitivty to DDP in
DDP-resistant NPC cells, but KCNQ1OT1 overexpression reversed the
effects of miR-454 overexpression on cell viability and DDP
sensitivity.
USP47 is a member of the deubiquitinating enzyme
family (40). Previous studies
have revealed that USP47 was abnormally expressed in multiple types
of cancer (41-43). For example, USP47 overexpression
accelerated ovarian cancer cell development (44), whereas USP47 knockdown inhibited
gastric cancer cell proliferation (45). USP47 facilitated osteosarcoma
cell invasion and migration, and suppressed apoptosis (46). In the present study, USP47 was
identified as a target of miR-454, and USP47 knockdown
significantly inhibited cell viability and DDP resistance in NPC
cells compared with the shNC group. Furthermore, miR-454 knockdown
significantly reversed shUSP47-mediated effects in DDP-resistant
NPC cells. Compared with the shNC group, KCNQ1OT1 knockdown
significantly downregulated USP47 expression, and miR-454 knockdown
significantly reversed shKCNQ1OT1-mediated effects on USP47
expression.
However, the present study had a number of
limitations. Firstly, other mRNAs should be explored to detemine
the downstream regulatory mechanism underlying the KCNQ1OT1/miR-454
axis. Secondly, the present study lacked in vivo
experiments, which should be performed in future studies to further
the understanding of the mechanism under-lying NPC.
In conclusion, the results of the present study
suggested that KCNQ1OT1 facilitated cell viability and DDP
resistance in NPC cells via regulating the miR-454/USP47 axis.
Therefore, KCNQ1OT1 might serve as a potential target for
overcoming DDP resistance in the chemotherapy of NPC.
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
FY, ZZ and XY designed the study. FY and ZL
performed the experiments. FY and ZL analysed the data and prepared
the figures. FY and ZZ drafted the initial manuscript. FY and XY
reviewed and revised the manuscript. All authors read and approved
the final manuscript. ZZ and XY confirm the authenticity of all the
raw data.
Ethics approval and consent to
participate
The present study was approved by the Ethical
Committee of Zhuji Central Hospital. Written consent were obtained
from all patients prior to starting the study.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgments
Not applicable.
References
|
1
|
Wang M, Ji YQ, Song ZB, Ma XX, Zou YY and
Li XS: Knockdown of lncRNA ZFAS1 inhibits progression of
nasopharyngeal carcinoma by sponging miR-135a. Neoplasma.
66:939–945. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Kong YG, Cui M, Chen SM, Xu Y, Xu Y and
Tao ZZ: LncRNA-LINC00460 facilitates nasopharyngeal carcinoma
tumorigenesis through sponging miR-149-5p to up-regulate IL6. Gene.
639:77–84. 2018. View Article : Google Scholar
|
|
3
|
Lee VH, Lam KO, Chang AT, Lam TC, Chiang
CL, So TH, Choi CW and Lee AW: Management of nasopharyngeal
carcinoma: Is adjuvant therapy needed? J Oncol Pract. 14:594–602.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Bergamini A, Pisano C, Di Napoli M,
Arenare L, Della Pepa C, Tambaro R, Facchini G, Gargiulo P,
Rossetti S, Mangili G, et al: Cisplatin can be safely administered
to ovarian cancer patients with hypersensitivity to carboplatin.
Gynecol Oncol. 144:72–76. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Peng X, Li W and Tan G: Reversal of taxol
resistance by cisplatin in nasopharyngeal carcinoma by upregulating
thromspondin-1 expression. Anticancer Drugs. 21:381–388. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Fang Z, Zhao J, Xie W, Sun Q, Wang H and
Qiao B: LncRNA UCA1 promotes proliferation and cisplatin resistance
of oral squamous cell carcinoma by sunppressing miR-184 expression.
Cancer Med. 6:2897–2908. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Wang Y, Chen F, Zhao M, Yang Z, Li J,
Zhang S, Zhang W, Ye L and Zhang X: The long noncoding RNA HULC
promotes liver cancer by increasing the expression of the HMGA2
oncogene via sequestration of the microRNA-186. J Biol Chem.
292:15395–15407. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wu X, Zhang P, Zhu H, Li S, Chen X and Shi
L: Long noncoding RNA FEZF1-AS1 indicates a poor prognosis of
gastric cancer and promotes tumorigenesis via activation of Wnt
signaling pathway. Biomed Pharmacother. 96:1103–1108. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Qu L, Ding J, Chen C, Wu ZJ, Liu B, Gao Y,
Chen W, Liu F, Sun W, Li XF, et al: Exosome-transmitted lncARSR
promotes sunitinib resistance in renal cancer by acting as a
competing endogenous RNA. Cancer Cell. 29:653–668. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zhang Y, Song X, Wang X, Hu J and Jiang L:
Silencing of LncRNA HULC enhances chemotherapy induced apoptosis in
human gastric cancer. J Med Biochem. 35:137–143. 2016. View Article : Google Scholar
|
|
11
|
Cao C, Zhou S and Hu J: Long noncoding RNA
MAGI2-AS3/miR-218 5p/GDPD5/SEC61A1 axis drives cellular
proliferation and migration and confers cisplatin resistance in
nasopharyngeal carcinoma. Int Forum Allergy Rhinol. 10:1012–1023.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Liu F, Tai Y and Ma J: LncRNA
NEAT1/let-7a-5p axis regulates the cisplatin resistance in
nasopharyngeal carcinoma by targeting Rsf-1 and modulating the
Ras-MAPK pathway. Cancer Biol Ther. 19:534–542. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Gao L, Cheng XL and Cao H: LncRNA THOR
attenuates cisplatin sensitivity of nasopharyngeal carcinoma cells
via enhancing cells stemness. Biochimie. 152:63–72. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Song H, Xu Y, Shi L, Xu T, Fan R, Cao M,
Xu W and Song J: LncRNA THOR increases the stemness of gastric
cancer cells via enhancing SOX9 mRNA stability. Biomed
Pharmacother. 108:338–346. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhang Y, Zhao Y, Liu L, Su H, Dong D, Wang
J, Zhang Y, Chen Q and Li C: MicroRNA-19b promotes nasopharyngeal
carcinoma more sensitive to cisplatin by suppressing KRAS. Technol
Cancer Res Treat. 17:15330338187936522018. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zhang P, Lu X, Shi Z, Li X, Zhang Y, Zhao
S and Liu H: miR-205-5p regulates epithelial-mesenchymal transition
by targeting PTEN via PI3K/AKT signaling pathway in
cisplatin-resistant nasopharyngeal carcinoma cells. Gene.
710:103–113. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Shao Q, Zhang P, Ma Y, Lu Z, Meng J, Li H,
Wang X, Chen D, Zhang M, Han Y, et al: MicroRNA-139-5p affects
cisplatin sensitivity in human nasopharyngeal carcinoma cells by
regulating the epithelial-to-mesenchymal transition. Gene.
652:48–58. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
19
|
Zheng S, Jiang F, Ge D, Tang J, Chen H,
Yang J, Yao Y, Yan J, Qiu J, Yin Z, et al: LncRNA
SNHG3/miRNA-151a-3p/RAB22A axis regulates invasion and migration of
osteosarcoma. Biomed Pharmacother. 112:1086952019. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Li FP, Lin DQ and Gao LY: LncRNA TUG1
promotes proliferation of vascular smooth muscle cell and
atherosclerosis through regulating miRNA-21/PTEN axis. Eur Rev Med
Pharmacol Sci. 22:7439–7447. 2018.PubMed/NCBI
|
|
21
|
Liu H, Deng H, Zhao Y, Li C and Liang Y:
LncRNA XIST/miR-34a axis modulates the cell proliferation and tumor
growth of thyroid cancer through MET-PI3K-AKT signaling. J Exp Clin
Cancer Res. 37:2792018. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Ma J, Yan H, Zhang J, Tan Y and Gu W:
Long-chain non-coding RNA (lncRNA) MT1JP suppresses biological
activities of lung cancer by regulating miRNA-423-3p/Bim Axis. Med
Sci Monit. 25:5114–5126. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Hu M, Zhang Q, Tian XH, Wang JL, Niu YX
and Li G: lncRNA CCAT1 is a biomarker for the proliferation and
drug resistance of esophageal cancer via the miR-143/PLK1/BUBR1
axis. Mol Carcinog. 58:2207–2217. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Fu D, Lu C, Qu X, Li P, Chen K, Shan L and
Zhu X: LncRNA TTN-AS1 regulates osteosarcoma cell apoptosis and
drug resistance via the miR-134-5p/MBTD1 axis. Aging (Albany NY).
11:8374–8385. 2019. View Article : Google Scholar
|
|
25
|
Zhuang J, Shen L, Yang L, Huang X, Lu Q,
Cui Y, Zheng X, Zhao X, Zhang D, Huang R, et al: TGFβ1 promotes
gemcitabine resistance through regulating the
LncRNA-LET/NF90/miR-145 signaling axis in bladder cancer.
Theranostics. 7:3053–3067. 2017. View Article : Google Scholar :
|
|
26
|
Li H, Huang J, Yu S and Lou Z: Long
non-coding RNA DLEU1 Up-regulates BIRC6 expression by competitively
sponging miR-381-3p to promote cisplatin resistance in
nasopharyngeal carcinoma. Onco Targets Ther. 13:2037–2045. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Lin FJ, Lin XD, Xu LY and Zhu SQ: Long
Noncoding RNA HOXA11-AS modulates the resistance of nasopharyngeal
carcinoma cells to cisplatin via miR-454-3p/c-Met. Mol Cells.
43:856–869. 2020.PubMed/NCBI
|
|
28
|
Kang Y, Jia Y, Wang Q, Zhao Q, Song M, Ni
R and Wang J: Long Noncoding RNA KCNQ1OT1 promotes the progression
of non-small cell lung cancer via regulating miR-204-5p/ATG3 Axis.
Onco Targets Ther. 12:10787–10797. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Lu X, Wang F, Fu M, Li Y and Wang L:
[ARTICLE WITHDRAWN] Long Noncoding RNA KCNQ1OT1 Accelerates the
Progression of Ovarian Cancer via MicroRNA-212-3/LCN2 Axis. Oncol
Res. 28:135–146. 2020. View Article : Google Scholar
|
|
30
|
Qiao CY, Qiao TY, Jin H, Liu LL, Zheng MD
and Wang ZL: LncRNA KCNQ1OT1 contributes to the cisplatin
resistance of tongue cancer through the KCNQ1OT1/miR-124-3p/TRIM14
axis. Eur Rev Med Pharmacol Sci. 24:200–212. 2020.PubMed/NCBI
|
|
31
|
Li Y, Li C, Li D, Yang L, Jin J and Zhang
B: lncRNA KCNQ1OT1 enhances the chemoresistance of oxaliplatin in
colon cancer by targeting the miR-34a/ATG4B pathway. Onco Targets
Ther. 12:2649–2660. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Qi X, Yu XJ, Wang XM, Song TN, Zhang J,
Guo XZ, Li GJ and Shao M: Knockdown of KCNQ1OT1 suppresses cell
invasion and sensitizes osteosarcoma cells to CDDP by upregulating
DNMT1-Mediated Kcnq1 expression. Mol Ther Nucleic Acids.
17:804–818. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Wang P, Chen D, Ma H and Li Y: LncRNA MEG3
enhances cisplatin sensitivity in non-small cell lung cancer by
regulating miR-21-5p/SOX7 axis. Onco Targets Ther. 10:5137–5149.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Chen J, Yu Y, Li H, Hu Q, Chen X, He Y,
Xue C, Ren F, Ren Z, Li J, et al: Long non-coding RNA PVT1 promotes
tumor progression by regulating the miR-143/HK2 axis in gallbladder
cancer. Mol Cancer. 18:332019. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Liu ZZ, Tian YF, Wu H, Ouyang SY and Kuang
WL: LncRNA H19 promotes glioma angiogenesis through
miR-138/HIF-1α/VEGF axis. Neoplasma. 67:111–118. 2020. View Article : Google Scholar
|
|
36
|
Fu C, Li D, Zhang X, Liu N, Chi G and Jin
X: LncRNA PVT1 facilitates tumorigenesis and progression of glioma
via regulation of MiR-128-3p/GREM1 Axis and BMP signaling pathway.
Neurotherapeutics. 15:1139–1157. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Wang S, Zhang G, Zheng W, Xue Q, Wei D,
Zheng Y and Yuan J: MiR-454-3p and miR-374b-5p suppress migration
and invasion of bladder cancer cells through targetting ZEB2.
Biosci Rep. 38:BSR201814362018. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Zuo J, Yu H, Xie P, Liu W, Wang K and Ni
H: miR-454-3p exerts tumor-suppressive functions by down-regulation
of NFATc2 in glioblastoma. Gene. 710:233–239. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Zhao X, Li X, Zhou L, Ni J, Yan W, Ma R,
Wu J, Feng J and Chen P: LncRNA HOXA11-AS drives cisplatin
resistance of human LUAD cells via modulating miR-454-3p/Stat3.
Cancer Sci. 109:3068–3079. 6;2018. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Yu L, Dong L, Wang Y, Liu L, Long H, Li H,
Li J, Yang X, Liu Z, Duan G, et al: Reversible regulation of SATB1
ubiquitination by USP47 and SMURF2 mediates colon cancer cell
proliferation and tumor progression. Cancer Lett. 448:40–51. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Yan S, Yue Y, Wang J, Li W, Sun M, Gu C
and Zeng L: LINC00668 promotes tumorigenesis and progression
through sponging miR-188-5p and regulating USP47 in colorectal
cancer. Eur J Pharmacol. 858:1724642019. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Naghavi L, Schwalbe M, Ghanem A and
Naumann M: Deubiquitinylase USP47 promotes RelA phosphorylation and
survival in gastric cancer cells. Biomedicines. 6:622018.
View Article : Google Scholar :
|
|
43
|
Cho J, Park J, Shin SC, Jang M, Kim JH,
Kim EE and Song EJ: USP47 promotes tumorigenesis by negative
regulation of p53 through deubiquitinating ribosomal protein S2.
Cancers (Basel). 12. pp. 11372020, View Article : Google Scholar
|
|
44
|
Hu L, Kolibaba H, Zhang S, Cao M, Niu H,
Mei H, Hao Y, Xu Y and Yin Q: MicroRNA-204-5p inhibits ovarian
cancer cell proliferation by down-regulating USP47. Cell
Transplant. 28(1 suppl): S51–S58. 2019. View Article : Google Scholar
|
|
45
|
Zhang B, Yin Y, Hu Y, Zhang J, Bian Z,
Song M, Hua D and Huang Z: MicroRNA-204-5p inhibits gastric cancer
cell proliferation by downregulating USP47 and RAB22A. Med Oncol.
32:3312015. View Article : Google Scholar
|
|
46
|
Zhang S, Ding L, Gao F and Fan H: Long
non-coding RNA DSCAM-AS1 upregulates USP47 expression through
sponging miR-101-3p to accelerate osteosarcoma progression. Biochem
Cell Biol. 98:600–611. 2020. View Article : Google Scholar : PubMed/NCBI
|