Introduction
Pancreatic adenocarcinoma (PAAD) is one of the most
lethal types of malignancy and has an extremely poor prognosis,
with an overall 5-year survival rate of <5% (1,2).
The main reason for the poor prognosis is that it is difficult to
diagnose PAAD at an early stage, because the cancer-specific
symptoms usually only occur at an advanced stage (3). In addition, there are currently no
effective treatments available for PAAD. Therefore, novel
strategies to diagnose and prevent PAAD are required urgently.
Human far upstream element-binding protein 1 (FUBP1)
was discovered to be an important regulator of transcription, mRNA
splicing and translation by binding to far upstream element (FUSE),
and is located in the reverse strand of chromosome 1p31.1 (4). Upon binding to FUSE, FUBP1 was
reported to upregulate the expression levels of the oncogene Myc,
which subsequently promoted cell growth and metastasis, and
functioned as an oncogene by modulating the FUBP1/FUSE/Myc feedback
loop (4-6). Notably, FUBP1 has also been
reported to exert oncogenic roles in a variety of tumor types,
including liver cancer (7,8),
glioma (9), neuroblastoma
(10), renal cell carcinoma
(11), lung cancer (12), tongue squamous cell carcinoma
(13), esophageal squamous cell
carcinoma (14), gastric cancer
(15,16) and colorectal cancer (17). A previous study demonstrated that
FUBP1 served as an oncogene and was associated with a poor
prognosis in patients with PAAD (18). In addition, FUBP1 regulated the
immune response by upregulating the expression levels of programmed
death-ligand 1 in PAAD cells (18). However, the biological functions
and molecular mechanisms of FUBP1 in PAAD remain unknown and
require further investigations.
The present study analyzed the expression levels of
FUBP1 in PAAD and adjacent normal tissues and determined the
association between FUBP1 and clinical prognosis of PAAD. Moreover,
the role of FUBP1 on the biological functions of PAAD cells, in
addition to the potential mechanisms, were investigated in
vitro.
Materials and methods
Patient studies
From September 2017 to December 2019, a total of 7
patients including 5 males and 2 females, with a mean age of 49.5
years (range, 40-66 years), diagnosed with PAAD at the Shanghai
Fengxian District Central Hospital (Shanghai, China) were enrolled
in the present study. All patients provided written informed
consent prior to participation. The 7 pairs of PAAD and adjacent
normal tissues (at least 3 cm away from the margin of PAAD tissues)
were resected and collected following surgery. All the patients had
not received radiotherapy or chemotherapy before the operation and
were diagnosed with PAAD according to the pathological results. The
patients with other malignant tumors were excluded. The study
protocol was approved by the Ethics Committee of the Shanghai
Fengxian District Central Hospital (approval no. 2017-Ethical
Review-KY-05) and was performed in accordance with the principles
of the Declaration of Helsinki.
Expression dataset
Gene expression RNAseq data of FUBP1 were downloaded
from the UCSC Xena database (http://xena.ucsc.edu/) (19), which included PAAD samples [The
Cancer Genome Atlas (TCGA), n=183] and normal samples [Genotype
Tissue Expression (GTEx), n=165]. The correlation between gene
expression and overall survival was downloaded from the starBase
database (http://starbase.sysu.edu.cn/index.php) (20), which contained 176 PAAD
patients.
Cell lines and culture
Human PAAD cell lines (BxPC-3, PaTu8988, PANC-1 and
SW1990) were conserved at the Central Laboratory of Shanghai
Fengxian District Central Hospital, and were cultured in DMEM
supplemented with 10% FBS (both from Gibco; Thermo Fisher
Scientific, Inc.), and maintained in a humidified incubator with 5%
CO2 at 37°C.
Reverse transcription-quantitative PCR
(RT-qPCR)
Total RNA was extracted from tissues and cells using
TRIzol® reagent (Invitrogen; Thermo Fisher Scientific,
Inc.). Total RNA was reverse transcribed into cDNA using a
PrimeScript™ RT Reagent kit (Takara Bio, Inc.) with the conditions
of 15 min at 37°C and 5 sec at 85°C. qPCR was subsequently
performed on an ABI 7300 Real-Time PCR system (Applied Biosystems;
Thermo Fisher Scientific, Inc.) using a SYBR® Premix
Dimmer Eraser kit (Takara Bio, Inc.). Thermocycling conditions for
the qPCR were as follows: Initial denaturation at 95°C for 30 sec;
40 cycles of denaturation at 95°C for 5 sec; annealing at 58°C for
30 sec, and extension at 72°C for 30 sec. The following primers
pairs were used for the qPCR: FUBP1 forward,
5′-GGACAACACCCGAAAGGATA-3′ and reverse, 5′-ATGTTCCAGTTGCCTTGACC-3′;
and GAPDH forward, 5′-TTGGTATCGTGGAAGGACTCA-3′ and reverse,
5′-TGTCATCATATTTGGCAGGTT-3′ (all purchased from Sangon Biotech Co.,
Ltd.). The relative mRNA expression levels were quantified using
the 2−ΔΔCq method (21). GAPDH was used as the internal
control.
Cell transfection
Cells were transfected with small interfering RNA
(siRNA/si) targeting FUBP1 (si-FUBP1), sinegative control (NC),
FUBP1 overexpression (OE) vector (FUBP1-OE) or control vector using
Lipofectamine® 2000 reagent (Invitrogen; Thermo Fisher
Scientific, Inc.) and Opti-MEM (Thermo Fisher Scientific, Inc.),
according to the manufacturer's protocol. Briefly, PaTu8988 cells
with a confluence of 50-60% in a 6-well plate were transfected with
100 pmol si-FUBP1 or an equal volume of si-NC. For the transfection
of plasmids, the cell density of SW1990 was 70-80% before
transfection, with 5 µg FUBP1-OE or VECTOR plasmids
transfected in each well of a 6-well plate. Subsequently, the
mixture of transfection reagents was replaced with 10% FBS-DMEM
after 5 h at 37°C. Then, the following assays were performed after
post-transfected cells were incubated at 37°C for 48 h. The
sequences of the siRNAs (all from GenePharma, Co., Ltd.) were as
follows: si-FUBP1 forward, 5′-GGUGCUGACAAACCUCUUATT-3′ and reverse,
5′-UAAGAGGUUUGUCAGCACCTT-3′; si-NC forward,
5′-UUCUCCGAACGUGUCACGUTT-3′ and reverse,
5′-ACGUGACACGUUCGGAGAATT-3′. The OE-FUBP1 plasmid contained
full-length amplified sequences of FUBP1 cloned into a pCD513B
plasmid (purchased from Biogot Biotechnology Co., Ltd).
Cell Counting Kit-8 (CCK-8) assay
Cell proliferation was analyzed using a CCK-8 assay
(Dojindo Molecular Technologies, Inc.), according to the
manufacturer's protocol. Briefly, following transfection, cells
were seeded into 96-well plates at a density of 5×103
cells/well and cultured for 10, 24, 48 or 72 h. After the cells
were washed twice with cold PBS, the mixture of 10 µl CCK-8
and 100 µl serum-free DMEM was added into each well and the
plates were incubated at 37°C for 45 min. Subsequently, the
absorbance was measured at a wavelength of 450 nm using a Wellscan
MK3 microplate reader (Thermo Labsystems, Inc.).
Cell migration and invasion assays
Cell migration and invasion assays were performed in
8.0-µm pore insert Transwell chambers (EMD Millipore) and
Transwell invasion Matrigel chambers (Corning, Inc.), respectively.
For the invasion assay, the Transwell membrane of the Matrigel
chambers was precoated for 1 h at 37°C using serum-free DMEM.
Briefly, DMEM supplemented with 10% FBS was plated into the lower
chambers and 5×104 cells in serum-free DMEM were plated
into the upper chambers. Following incubation at 37°C for 24 h, the
non-migratory and non-invasive cells remaining in the upper
chambers were gently removed with a cotton bud. The invasive and
migratory cells in the lower chambers of the Transwell plates were
stained with 0.1% crystal violet for 30 min at room temperature.
The migratory and invasive cells were counted under a bright-field
fluorescence microscope (Olympus Corporation; magnification, ×200)
in five randomly selected fields of view.
Wound healing assay
Upon transfected cells reaching 95% confluence, an
artificial wound was made in the cell monolayer by creating a
scratch with a 200-µl pipette tip. Since it was determined
the pancreatic cells grow quickly, and, with the condition of 5%
FBS medium it was revealed that the scratch wound could not be
measured at 48 h. Thus, this assay was conducted by using 2% FBS
medium to culture the cells (22). The cells were subsequently
cultured in DMEM supplemented with 2% FBS at 37°C after washing
with PBS three times. Images of the wound were captured at 0, 24
and 48 h using an OLYMPUS inverted microscope at a magnification of
×100, and measured by the ImageJ software (version 1.52) (National
Institutes of Health), respectively, to determine the cell
migratory ability.
Western blotting
Protein was extracted from transfected cells using
RIPA buffer (Beyotime Institute of Biotechnology), and protein
concentration was determined using a BCA kit (Beyotime Institute of
Biotechnology). Briefly, 40-60 µg equal amounts of protein
were loaded per lane and separated by 10% SDS-PAGE, and transferred
to PVDF membranes (EMD Millipore). The PVDF membranes were blocked
using 5% non-fat milk in TBST for 1 h at 37°C and then incubated
with primary antibodies at 4°C overnight. Following several washes
using TBST, the PVDF membranes were incubated with specific
secondary antibodies [(HRP-labeled goat anti-mouse IgG; cat. no.
S0002; 1:5,000) or (HRP-labeled goat anti-rabbit IgG, cat. no.
S0001; 1:5,000; both from from Affinity Biosciences)] for 2 h at
room temperature. Finally, after washing the membranes three times
with TBST, the protein bands were visualized using an ECL kit (EMD
Millipore) and the bands were quantified using ImageJ software
(version 1.52) (National Institutes of Health). The following
antibodies were used: Anti-FUBP1 (68 kDa; product code ab192867,
1:1,000; Abcam), anti-E-cadherin (135 kDa; product no. 3195;
1:1,000), anti-N-cadherin (140 kDa; product no. 13116; 1:1,000),
anti-vimentin (57 kDa; product no. 5741; 1:1,000), anti-β-catenin
(92 kDa; product no. 8480; 1:1,000), anti-Smad2/3 (52/60 kDa;
product no. 3102; 1:1,000), anti-phosphorylated (p)-Smad2/3 (52/60
kDa; product no. 8828; 1:1,000), anti-TGFβ1 (12 kDa; product no.
3709; 1:1,000), and anti-β-tubulin (55 kDa; product no. 2128;
1:1,000; all from Cell Signaling Technology, Inc.) and anti-β-actin
(42 kDa; cat. no. T0022, 1:5,000; Affinity Biosciences). β-actin
and β-tubulin primary antibodies were used as the loading
controls.
Immunofluorescence analysis
Following transfection with si-FUBP1 for 48 h, cells
were cultured on glass coverslips for 48 h. The cells were
subsequently fixed with 4% paraformaldehyde for 15 min at room
temperature and blocked with 3% BSA (Beijing Solarbio Science &
Technology Co., Ltd.) for 1 h at room temperature. The cells were
then incubated with the primary antibody overnight at 4°C prior to
incubation with a Cy3-conjugated Goat Anti-Rabbit IgG secondary
antibody (1:500; cat. no. A0516; Beyotime Institute of
Biotechnology) in a dark room for 1 h at room temperature. The cell
nuclei were counterstained with DAPI (Beyotime Institute of
Biotechnology) in a dark room for 15 min at room temperature.
Stained cells were visualized with a fluorescence microscope
(Olympus Corporation; magnification, ×200). The following primary
antibodies were used: Anti-E-cadherin (cat. no. 3195; 1:200) and
anti-vimentin (cat. no. 5741; 1:100; both from Cell Signaling
Technology, Inc.).
Statistical analysis
Statistical analysis was performed using GraphPad
Prism 5.0 software (GraphPad Software, Inc.). All data are
presented as the mean ± SD of ≥3 independent experiments.
Statistical differences were analyzed using a paired Student's
t-test and one-way ANOVA followed by a Bonferroni's post hoc test.
P<0.05 was considered to indicate a statistically significant
difference.
Results
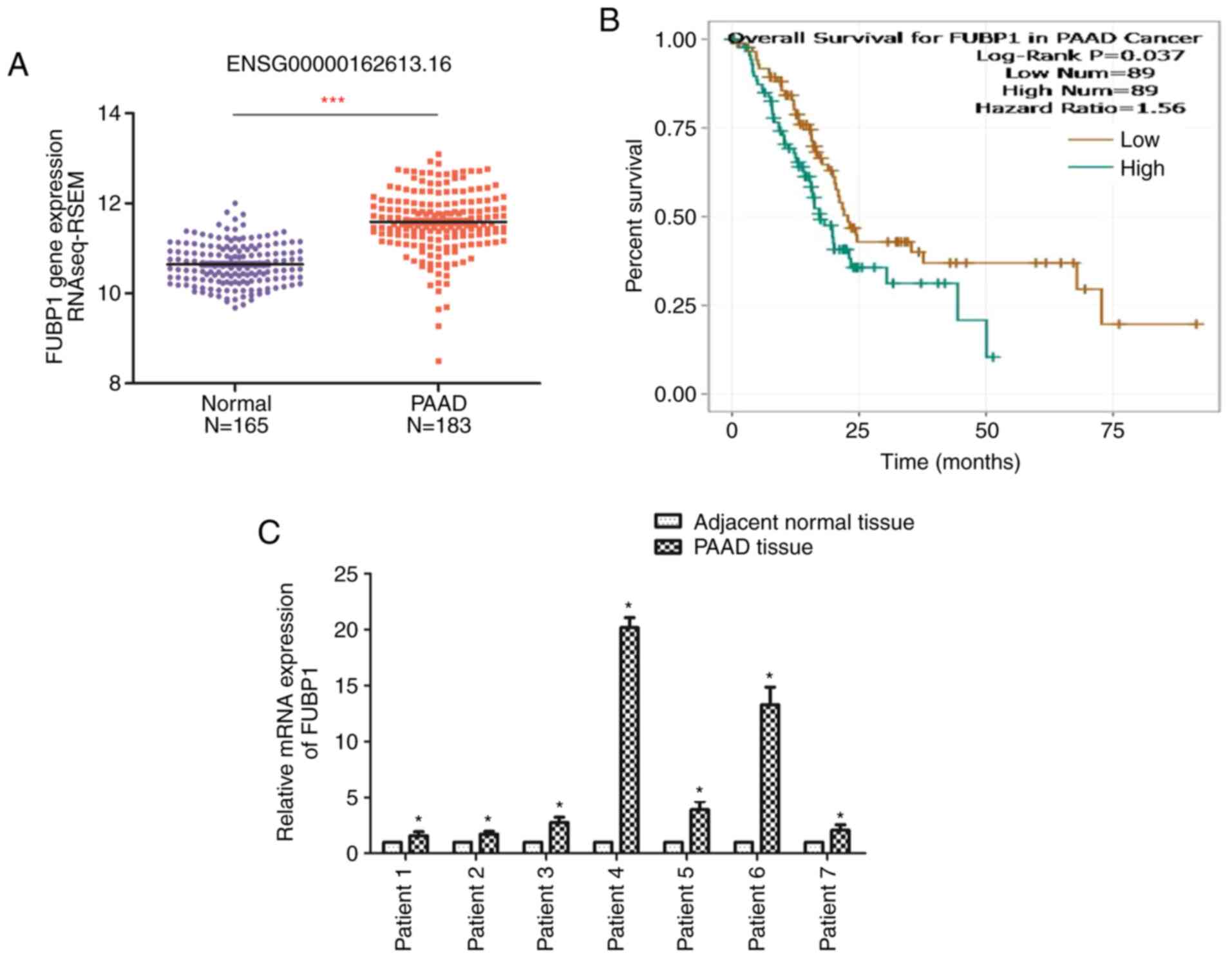
FUBP1 expression levels are upregulated
in PAAD and positively associated with clinical prognosis
The expression levels of FUBP1 in PAAD and normal
tissues were analyzed using data from TCGA database (19). The results revealed that the
expression levels of FUBP1 in PAAD tissues were significantly
upregulated compared with normal pancreatic tissues (Fig. 1A). Thus, the clinical
significance of FUBP1 in PAAD was determined using the starBase
database (20). The long-term
overall survival was determined to be associated with the
expression levels of FUBP1 (Fig.
1B). Thus, the patients with PAAD with upregulated FUBP1
expression levels were at a higher risk of prognosis compared with
patients with lower expression levels of FUBP1. Similarly, RT-qPCR
analysis of seven pairs of PAAD and adjacent normal tissues
revealed that FUBP1 mRNA expression levels were significantly
upregulated in PAAD tissues compared with the corresponding
adjacent normal tissues (Fig.
1C). These findings indicated that FUBP1 may serve as an
oncogene and may be associated with a poor prognosis in patients
with PAAD.
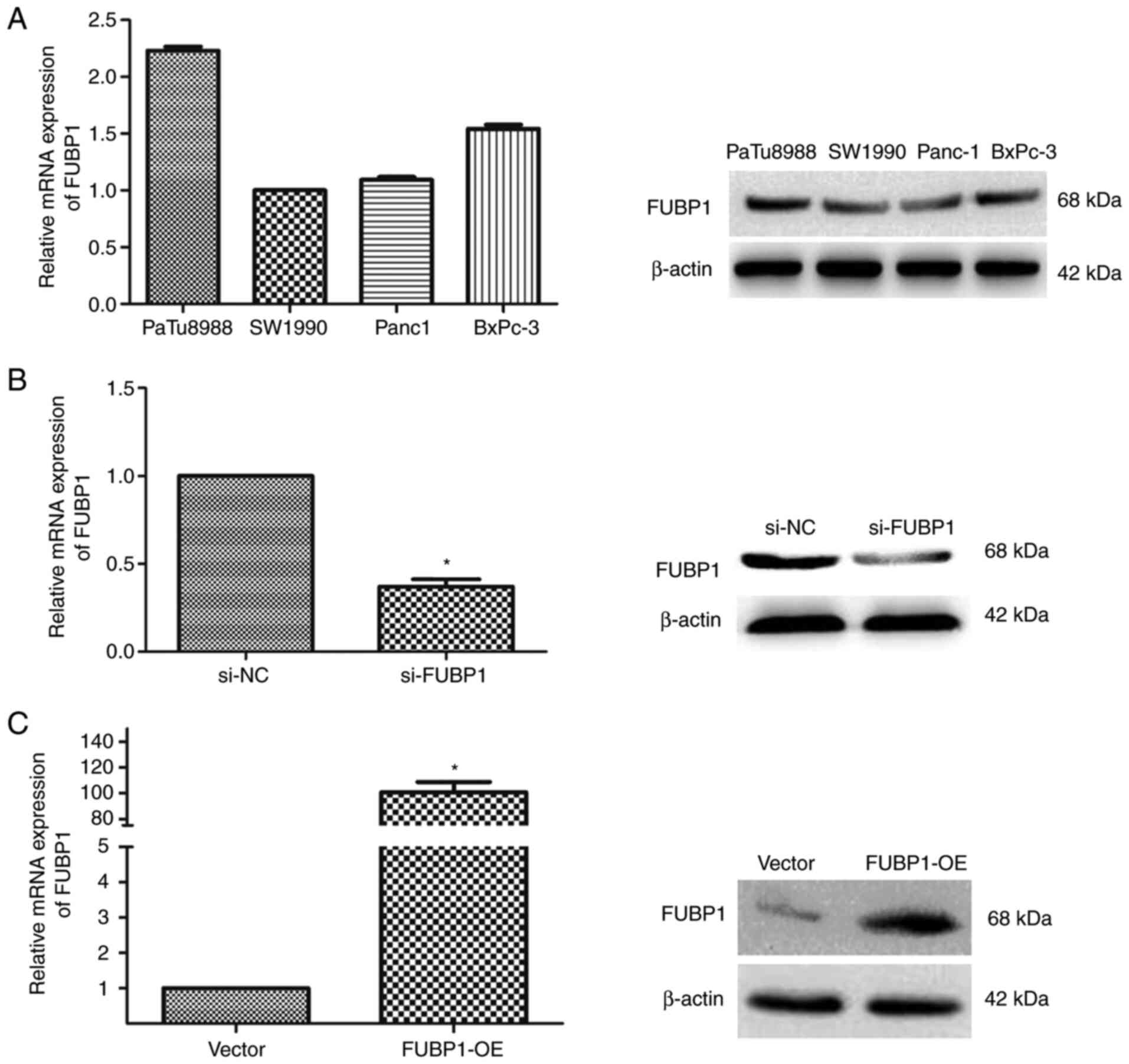
FUBP1 promotes PAAD cell proliferation,
migration and invasion
In human PAAD cell lines, PaTu8988, SW1990, BxPC-3
and Panc-1, the mRNA and protein expression levels of FUBP1 were
analyzed using RT-qPCR and western blotting. Among the four human
PAAD cell lines, the highest expression levels of FUBP1 were
observed in PaTu8988 cells, whereas the lowest expression levels
were recorded in SW1990 cells (Fig.
2A). Thus, PaTu8988 cells were selected to further investigate
the effects of the knockdown of FUBP1 expression in PAAD, while
SW1990 cells were selected to determine the effects of the
overexpression of FUBP1. The transfection efficiencies of cells
transfected with si-FUBP1 or FUBP1-OE were determined using RT-qPCR
and western blotting (Fig. 2B and
C).
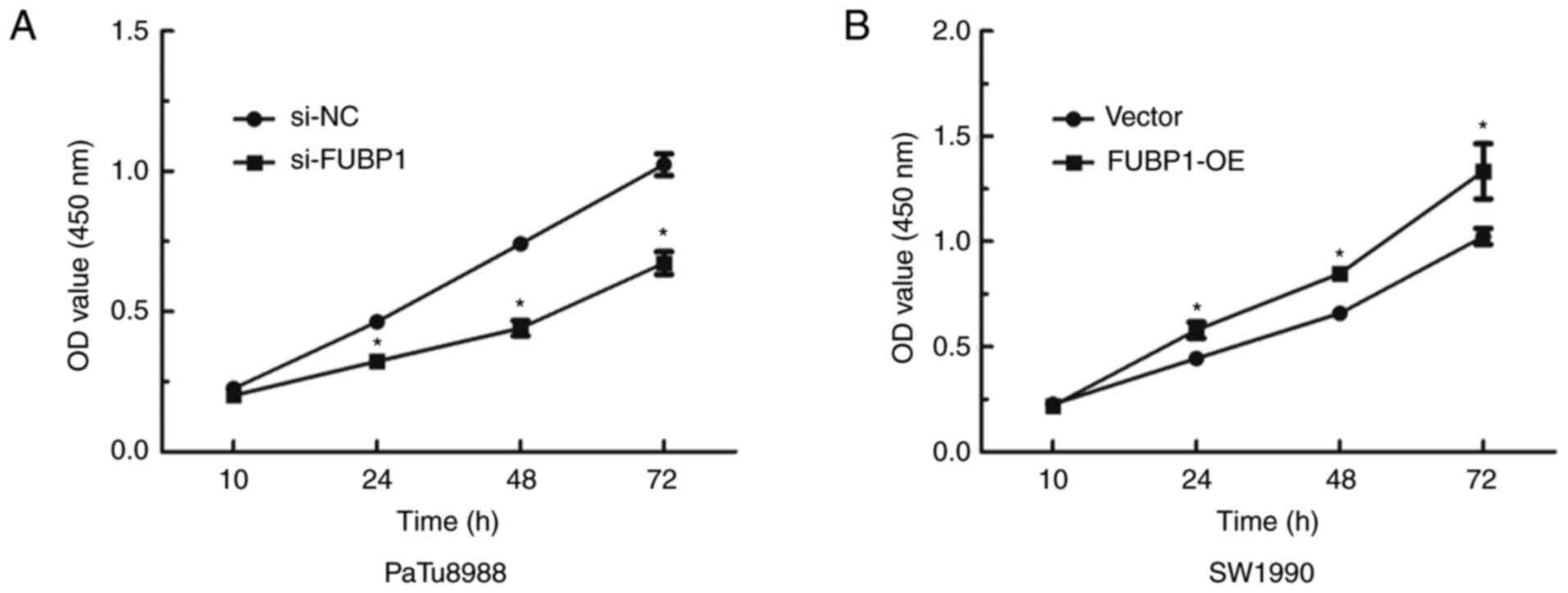
To investigate whether FUBP1 was associated with
biological functions in PAAD, a CCK-8 assay was used to determine
the effect of FUBP1 on cell proliferation, while wound healing and
Transwell assays were conducted to determine the role of FUBP1 on
cell migration and invasion, respectively. The data demonstrated
that the knockdown FUBP1 expression significantly inhibited the
proliferative ability of PaTu8988 cells (Fig. 3A), whereas the overexpression of
FUBP1 promoted proliferation in SW1990 cells (Fig. 3B). Following the knockdown of
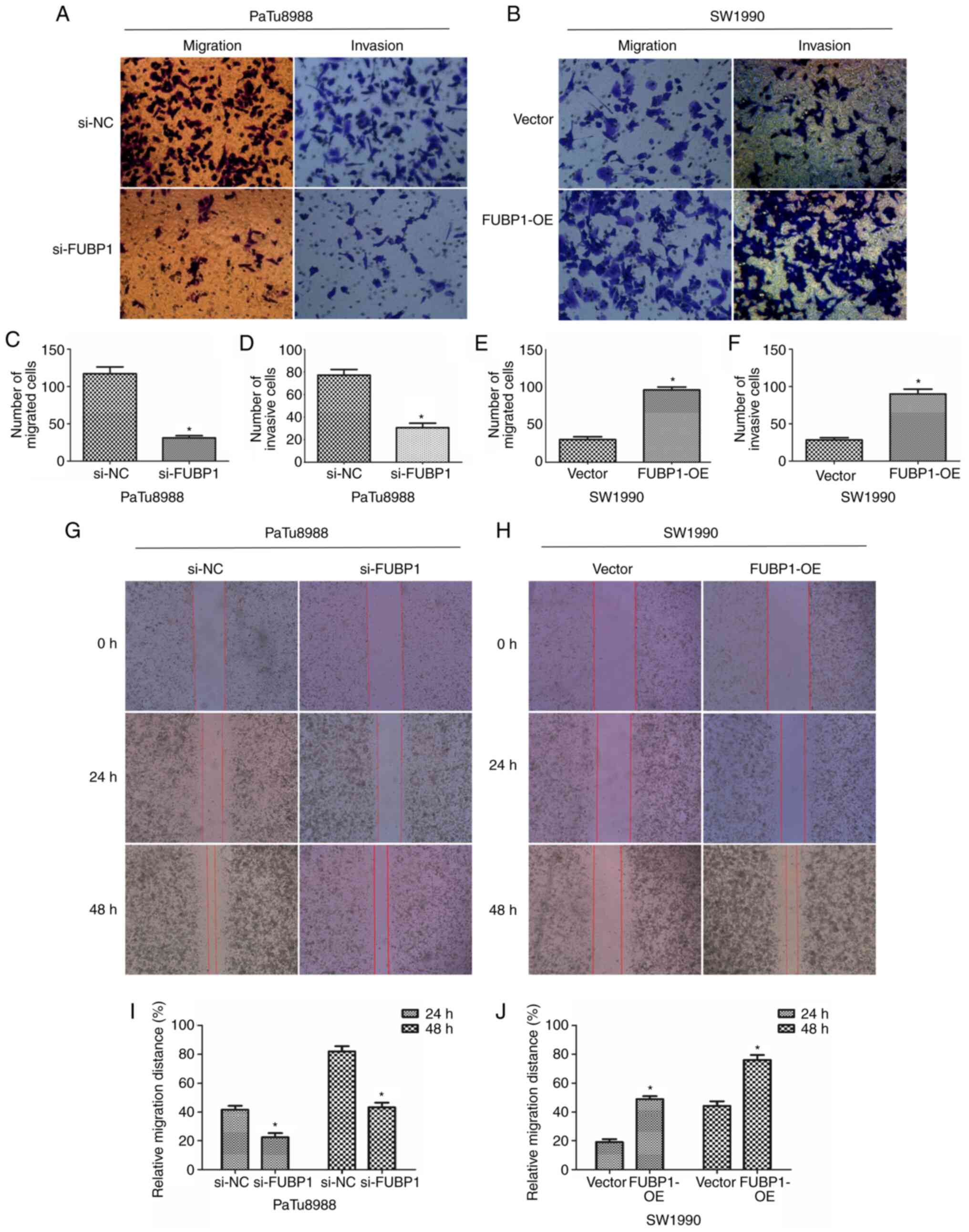
FUBP1 expression levels in PaTu8988 cells, the results of the
Transwell assays revealed that the number of migratory and invasive
cells was significantly decreased (Fig. 4A, C and D). Notably, following
the upregulation of FUBP1, the number of migratory and invasive
cells was significantly increased in SW1990 cells (Fig. 4B, E and F). Similarly, the
results of the wound healing assay also illustrated that the
knockdown of FUBP1 expression inhibited the migratory ability of
PaTu8988 cells (Fig. 4G and I).
By contrast, the overexpression of FUBP1 increased the migratory
ability of SW1990 cells (Fig. 4H and
J). These data suggested that FUBP1 may promote the
proliferation, migration and invasion of PAAD cells, which implied
that FUBP1 may facilitate the adhesion of tumor cells to the
extracellular matrix.
FUBP1 activates epithelial-mesenchymal
transition (EMT) via the TGFβ/Smad signaling pathway in PAAD
cells
During tumor development, tumor cells constantly
communicate with the surrounding microenvironment, which guides
tumor cells undergoing EMT (23,24). EMT is regarded as a pivotal step
for promoting tumor invasion and metastasis and has been discovered
to serve a role in the progression of PAAD (22). Since FUBP1 was revealed to
promote PAAD cell migration and invasion, it was hypothesized that
FUBP1 may be involved in EMT and influence cancer metastasis. To
further determine the effect of FUBP1 on the progression of PAAD,
the expression levels of EMT-related genes and transcription
factors were investigated using western blotting and
immunofluorescence assays. Following the transfection of PaTu8988
cells with si-FUBP1, western blotting revealed that the expression
levels of E-cadherin were significantly upregulated, while the
expression levels of N-cadherin, vimentin and β-catenin were
downregulated (Fig. 5A). In
contrast, the overexpression of FUBP1 in SW1990 cells demonstrated
the opposite trend (Fig. 5B).
Similarly, the results of the immunofluorescence assay revealed
that the knockdown of FUBP1 expression upregulated E-cadherin
expression and downregulated vimentin expression (Fig. 5C). These observations suggested
that FUBP1 may promote the transition of PAAD cells from an
epithelial to mesenchymal phenotype.
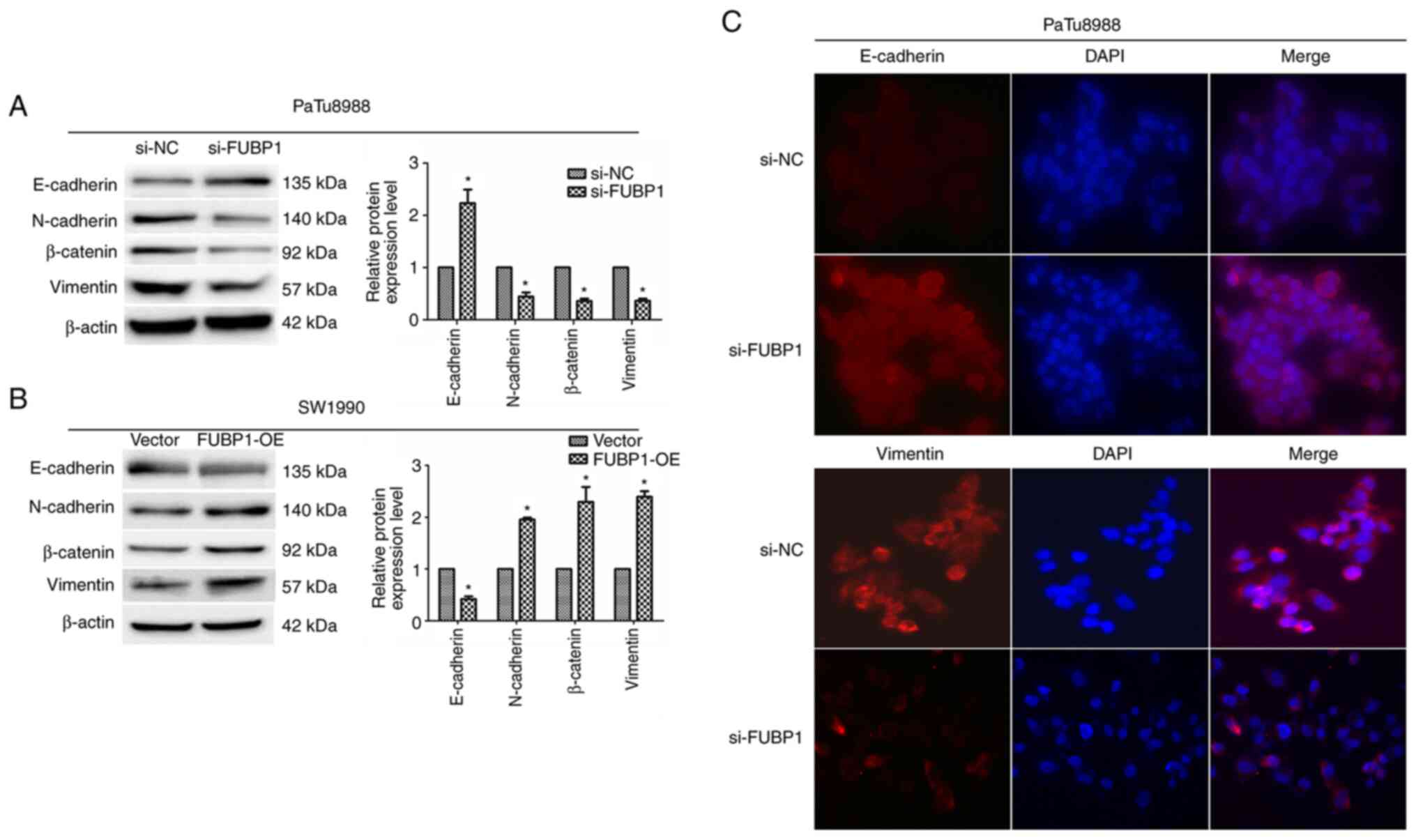 | Figure 5FUBP1 promotes EMT in pancreatic
adenocarcinoma cells. Western blotting was used to analyze the
expression levels of the EMT-related proteins, E-cadherin,
N-cadherin, β-catenin and vimentin, in (A) PaTu8988 cells
transfected with si-NC or si-FUBP1 and (B) SW1990 cells transfected
with empty or FUBP1-OE vectors. β-actin was used as the internal
loading control. (C) Immunofluorescence assay was used to determine
E-cadherin and vimentin expression levels in PaTu8988 cells
transfected with si-NC or si-FUBP1 (magnification, ×200). Data are
presented as the mean ± SD from three independent experiments.
*P<0.05. FUBP1, far upstream element-binding protein
1; EMT, epithelial-mesenchymal transition; si, small interfering
RNA; NC, negative control; OE, overexpression. |
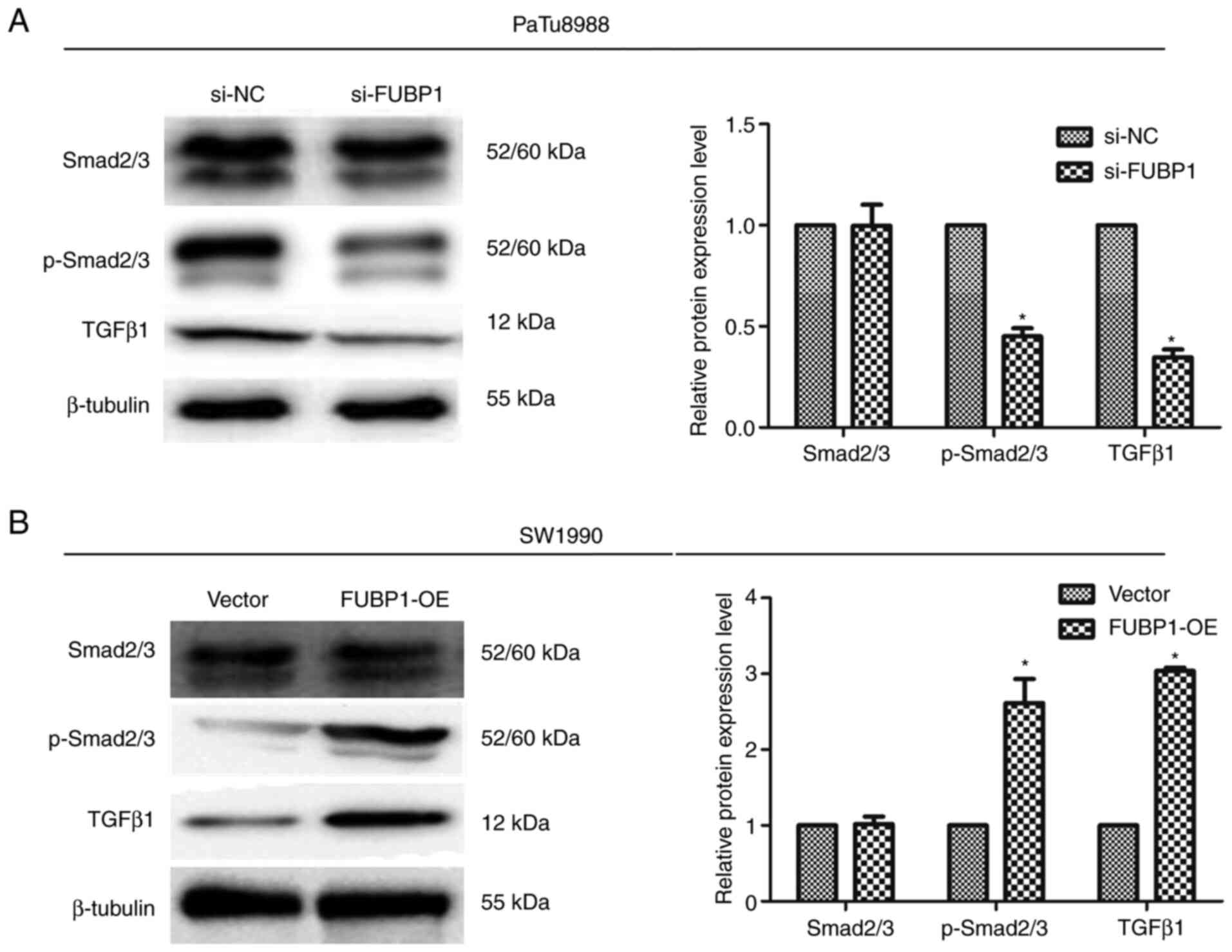
Multiple previous studies have demonstrated that the
TGFβ/Smad signaling pathway was a central regulator of cancer cell
proliferation, metastasis and the EMT process (22,25,26). Thus, the present study
subsequently aimed to determine the molecular mechanisms through
which FUBP1 exerted its functions. In a previous study, Kyoto
Encyclopedia of Genes and Genomes signaling pathway enrichment
analysis identified that FUBP1 was associated with the TGFβ
signaling pathway, and FUBP1 promoted the EMT of hepatocellular
carcinoma cells by activating the TGFβ/Smad signaling pathway
(8). Thus, whether FUBP1 could
regulate EMT through the TGFβ/Smad signaling pathway in human PAAD
cells was investigated. The expression levels of p-Smad2/3 and
TGFβ1, which are pivotal signaling molecules in the TGFβ/Smad
signaling pathway (27), were
analyzed using western blotting. Notably, FUBP1 expression levels
were found to be positively correlated with the expression levels
of TGFβ/Smad signaling pathway target genes. The transfection of
PaTu8988 cells with si-FUBP1 significantly downregulated p-Smad2/3
and TGFβ1 expression levels (Fig.
6A), while the overexpression of FUBP1 in SW1990 cells
significantly upregulated the expression levels of p-Smad2/3 and
TGFβ1 (Fig. 6B). Collectively,
these data indicated that FUBP1 may activate EMT through TGFβ/Smad
signaling in human PAAD cells.
Discussion
The results of the present study revealed that the
expression levels of FUBP1 were upregulated in human PAAD tissues
compared with adjacent normal tissues in both clinical tissues and
data from TCGA database. Survival analysis using the starBase
database also demonstrated that patients with PAAD with upregulated
FUBP1 expression levels were at a higher risk of PAAD-related
mortality compared with patients with lower expression levels of
FUBP1. These data suggested that FUBP1 expression levels may be
upregulated in patients with PAAD and may be associated with a poor
prognosis, which is consistent with the findings of Fan et
al (18). The results of the
CCK-8 assay revealed that FUBP1 promoted cell proliferation.
Moreover, wound healing and Transwell migration and invasion assays
revealed that FUBP1 promoted PAAD cell migration and invasion in
vitro. During EMT, epithelial cells transition from an
epithelial to mesenchymal phenotype, which causes cells to lose
their adhesive ability and acquire migratory and invasive
properties. Characteristic changes during EMT include the
downregulation of the expression levels of epithelial markers, such
as E-cadherin, and the upregulation of mesenchymal markers,
including N-cadherin, vimentin and EMT-related transcription
factors, such as Snail, twist family bHLH transcription factor 1
and zinc finger E-box binding homeobox 1 (24). The results of the present study
revealed that the knockdown of FUBP1 upregulated the expression
levels of E-cadherin, downregulated vimentin, N-cadherin and
β-catenin expression levels, and reversed the progression of EMT.
The TGFβ/Smad signaling pathway is well established as a central
regulator of cancer cell proliferation, metastasis and the EMT
process (22-25). For example, a previous study
demonstrated that FUBP1 promoted EMT by activating the TGFβ/Smad
signaling pathway in hepatocellular carcinoma cells (8). However, to the best of our
knowledge, the role of FUBP1 in TGFβ/Smad signaling
pathway-mediated EMT has not been reported. Therefore, the present
study hypothesized that FUBP1 may regulate the EMT process through
the TGFβ/Smad signaling pathway. To address this hypothesis, the
expression levels of proteins involved downstream of the TGFβ/Smad
signaling pathway cascade were investigated in vitro. The
results revealed that the knockdown of FUBP1 expression decreased
p-Smad2/3 and TGFβ1 expression levels, while the overexpression of
FUBP1 increased p-Smad2/3 and TGFβ1 expression levels. These
findings suggested that FUBP1 may activate EMT via the TGFβ/Smad
signaling pathway in human PAAD cells in vitro.
In conclusion, the results of the present study
revealed that FUBP1 expression levels were upregulated in patients
with PAAD and the upregulated expression levels predicted a poor
clinical prognosis. Moreover, the findings indicated that FUBP1 may
promote the proliferation, invasion and metastasis of PAAD cells.
To the best of our knowledge, the present study was the first to
identify that FUBP1 served as a pivotal regulator of EMT in PAAD
cells by regulating the TGFβ/Smad signaling pathway. Thus, FUBP1
may serve as an oncogene that promotes PAAD cell proliferation and
progression, and may serve as a clinically relevant prognostic
biomarker or represent a novel therapeutic target for PAAD. Further
preclinical studies and clinical trials will be required to
determine whether FUBP1 can predict the benefit of prognosis in
PAAD. In addition, further studies on tissues of PAAD patients are
required to thoroughly understand the clinical features of FUBP1.
In the future, the detailed mechanism of FUBP1, as well as the
underlying effects of other genes regulated by FUBP1 in PAAD
progression will be investigated.
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
XZ, HC and JC conceived and designed the experiments
and wrote the paper. YZ, XZ participated in all the experiments. HC
and JC contributed to the design of the study and interpretation of
experimental results. NZ and YL performed and analyzed the
experiments of western blotting. JL performed and analyzed the
experiments of immunofluorescence. XX analyzed the data and revised
this manuscript. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
All patients provided written informed consent, and
the study was approved by the Ethics Committee of Shanghai Fengxian
District Central Hospital (Shanghai, China).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgments
Not applicable.
Funding
The present study was supported by Shanghai Municipal Health
Commission Foundation (grant no. 20204Y0181) and Shanghai
University of Medicine and Health Sciences Seed Foundation (grant
no. SFP-18-22-15-002).
Abbreviations:
|
FUBP1
|
far upstream element-binding protein
1;
|
|
PAAD
|
pancreatic adenocarcinoma;
|
|
RT-qPCR
|
reverse-transcription- quantitative
PCR;
|
|
EMT
|
epithelial-mesenchymal transition;
|
|
DAB
|
diaminobenzidine;
|
|
FBS
|
fetal bovine serum;
|
|
DMEM
|
Dulbecco's modified Eagle's
medium;
|
|
CCK-8
|
Cell Counting Kit-8
|
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2020. CA Cancer J Clin. 70:7–30. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Miller KD, Nogueira L, Mariotto AB,
Rowland JH, Yabroff KR, Alfano CM, Jemal A, Kramer JL and Siegel
RL: Cancer treatment and survivorship statistics, 2019. CA Cancer J
Clin. 69:363–385. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Stornello C, Archibugi L, Stigliano S,
Vanella G, Graglia B, Capalbo C, Nigri G and Capurso G: Diagnostic
delay does not influence survival of pancreatic cancer patients.
United European Gastroenterol J. 8:81–90. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Zhang J and Chen QM: Far upstream element
binding protein 1: A commander of transcription, translation and
beyond. Oncogene. 32:2907–2916. 2013. View Article : Google Scholar
|
|
5
|
Debaize L and Troadec MB: The master
regulator FUBP1: Its emerging role in normal cell function and
malignant development. Cell Mol Life Sci. 76:259–281. 2019.
View Article : Google Scholar
|
|
6
|
Frost JR, Mendez M, Soriano AM, Crisostomo
L, Olanubi O, Radko S and Pelka P: Adenovirus 5 E1A-Mediated
Suppression of p53 via FUBP1. J Virol. 92:e00439–18. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Rabenhorst U, Thalheimer FB, Gerlach K,
Kijonka M, Böhm S, Krause DS, Vauti F, Arnold HH, Schroeder T,
Schnütgen F, et al: Single-stranded DNA-binding transcriptional
regulator FUBP1 is essential for fetal and adult hematopoietic stem
cell self-renewal. Cell Rep. 11:1847–1855. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Fu PY, Hu B, Ma XL, Tang WG, Yang ZF, Sun
HX, Yu MC, Huang A, Hu JW, Zhou CH, et al: Far upstream
element-binding protein 1 facilitates hepatocellular carcinoma
invasion and metastasis. Carcinogenesis. 41:950–960. 2020.
View Article : Google Scholar
|
|
9
|
Baumgarten P, Harter PN, Tonjes M, Capper
D, Blank AE, Sahm F, von Deimling A, Kolluru V, Schwamb B,
Rabenhorst U, et al: Loss of FUBP1 expression in gliomas predicts
FUBP1 mutation and is associated with oligodendroglial
differentiation, IDH1 mutation and 1p/19q loss of heterozygosity.
Neuropathol Appl Neurobiol. 40:205–216. 2014. View Article : Google Scholar
|
|
10
|
Jiang P, Huang M, Qi W, Wang F, Yang T,
Gao T, Luo C, Deng J, Yang Z, Zhou T, et al: FUBP1 promotes
neuroblastoma proliferation via enhancing glycolysis-a new possible
marker of malignancy for neuroblastoma. J Exp Clin Cancer Res.
38:4002019. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Duan J, Bao X, Ma X, Zhang Y, Ni D, Wang
H, Zhang F, Du Q, Fan Y, Chen J, et al: Upregulation of far
upstream element-binding protein 1 (FUBP1) promotes tumor
proliferation and tumorigenesis of clear cell renal cell carcinoma.
PLoS One. 12:e01698522017. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Muller B, Bovet M, Yin Y, Stichel D, Malz
M, González-Vallinas M, Middleton A, Ehemann V, Schmitt J, Muley T,
et al: Concomitant expression of far upstream element (FUSE)
binding protein (FBP) interacting repressor (FIR) and its splice
variants induce migration and invasion of non-small cell lung
cancer (NSCLC) cells. J Pathol. 237:390–401. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Chen Y, Liu J, Geng N and Feng C:
Upregulation of far upstream element-binding protein 1 (FUBP1)
promotes tumor proliferation and unfavorable prognosis in tongue
squamous cell carcinoma. Int J Biol Markers. 35:56–65. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Yang L, Zhu JY, Zhang JG, Bao BJ, Guan CQ,
Yang XJ, Liu YH, Huang YJ, Ni RZ and Ji LL: Far upstream
element-binding protein 1 (FUBP1) is a potential c-Myc regulator in
esophageal squamous cell carcinoma (ESCC) and its expression
promotes ESCC progression. Tumour Biol. 37:4115–4126. 2016.
View Article : Google Scholar
|
|
15
|
Venturutti L, Cordo Russo RI, Rivas MA,
Mercogliano MF, Izzo F, Oakley RH, Pereyra MG, De Martino M,
Proietti CJ, Yankilevich P, et al: MiR-16 mediates trastuzumab and
lapatinib response in ErbB-2-positive breast and gastric cancer via
its novel targets CCNJ and FUBP1. Oncogene. 35:6189–6202. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zhang F, Tian Q and Wang Y: Far upstream
element-binding protein 1 (FUBP1) is overexpressed in human gastric
cancer tissue compared to non-cancerous tissue. Onkologie.
36:650–655. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Jia MY and Wang YJ: Far upstream
element-binding protein 1 (FUBP1) expression differs between human
colorectal cancer and non-cancerous tissue. Neoplasma. 61:533–540.
2014. View Article : Google Scholar
|
|
18
|
Fan P, Ma J and Jin X: Far upstream
element-binding protein 1 is up-regulated in pancreatic cancer and
modulates immune response by increasing programmed death ligand 1.
Biochem Biophys Res Commun. 505:830–836. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
20
|
Goldman MJ, Craft B, Hastie M, Repečka K,
McDade F, Kamath A, Banerjee A, Luo Y, Rogers D, Brooks AN, et al:
Visualizing and interpreting cancer genomics data via the Xena
platform. Nat Biotechnol. 38:675–678. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Li JH, Liu S, Zhou H, Qu LH and Yang JH:
StarBase v2.0: Decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA
interaction networks from large-scale CLIP-Seq data. Nucleic Acids
Res. 42:D92–97. 2014. View Article : Google Scholar
|
|
22
|
van Staalduinen J, Baker D, Ten Dijke P
and van Dam H: Epithelial-mesenchymal-transition-inducing
transcription factors: New targets for tackling chemoresistance in
cancer? Oncogene. 37:6195–6211. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Lamouille S, Xu J and Derynck R: Molecular
mechanisms of epithelial-mesenchymal transition. Nat Rev Mol Cell
Biol. 15:178–196. 2014. View
Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zhang X, Feng W, Zhang J, Ge L, Zhang Y,
Jiang X, Peng W, Wang D, Gong A and Xu M: Long noncoding RNA PVT1
promotes epithelialmesenchymal transition via the TGFβ/Smad pathway
in pancreatic cancer cells. Oncol Rep. 40:1093–1102.
2018.PubMed/NCBI
|
|
25
|
Zhao L, Liu S, Che X, Hou K, Ma Y, Li C,
Wen T, Fan Y, Hu X, Liu Y and Qu X: Bufalin inhibits TGF-β-induced
epithelial-to-mesenchymal transition and migration in human lung
cancer A549 cells by downregulating TGF-β receptors. Int J Mol Med.
36:645–652. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Park JH, Yoon J, Lee KY and Park B:
Effects of geniposide on hepatocytes undergoing
epithelial-mesenchymal transition in hepatic fibrosis by targeting
TGFβ/Smad and ERK-MAPK signaling pathways. Biochimie. 113:26–34.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Valcourt U, Kowanetz M, Niimi H, Heldin CH
and Moustakas A: TGF-beta and the Smad signaling pathway support
transcriptomic reprogramming during epithelial-mesenchymal cell
transition. Mol Biol Cell. 16:1987–2002. 2005. View Article : Google Scholar : PubMed/NCBI
|