Introduction
As the most lethal pestilent primary tumor of the
brain, gliomas constitute ~70% of primary malignant cancers in the
human brain. According to the World Health Organization (WHO),
gliomas are classified into an adverse group of tumors, including
pilocytic astrocytomas and glioblastomas (1,2).
Glioblastoma multiforme (GBM), ranks as the maximal grade of benign
glioma. This type of cancer is prominently attributed to the
composition of a highly distinctive type of cells and therefore
displays phenotypic heterogeneity (1,3).
Despite significant improvements in cancer therapeutic strategies
and methods, the overall survival of patients with GBM has not been
considerably improved (4,5).
Therefore, it is required to develop novel biomarkers in order to
improve the diagnostic potential of GBM.
MicroRNAs (miRNAs/miRs) consist of clusters of small
non-coding RNAs of 17-27 nucleotides in length. By binding to
target mRNAs, miRNAs regulate gene expression after transcription
(6). Various studies have
revealed that miRNAs affect the function of a novel cluster of
tumor suppressor genes and oncogenes that contribute to
tumorigenesis, apoptosis, angiogenesis and the invasion of cancer
cells (6-8). miR-124 is the most well-identified
miRNA in the brain and its aberrant expression has been observed in
different malignancies, such as breast cancer (9), gastric cancer (10) and acute lymphoblastic leukemia
(11), which indicates its
potential roles in carcinogenesis (12). Previous studies indicated that
miR-124 expression was downregulated in tissues of patients with
glioma and that it exhibited a negative correlation with the glioma
pathological grade. In addition, in vitro experiments
demonstrated that miR-124 overexpression suppressed the
proliferation and invasion of glioma cells, illustrating its
important role as a tumor suppressor (13,14). The present study explored the
possible effects of miR-124-3p and its potential target, Ras
homology Growth-related (RhoG), in the oncogenic events and
deterioration of GBM.
Materials and methods
Ethical approval
The research protocol was approved by the Medical
Ethics Committee of Kunming Medical University (Kunming, China) and
was performed in accordance with the Declaration of Helsinki
(revised in 2000). GBM tumor tissues were acquired from the
patients following their permission at The Department of
Neurosurgery, First Affiliated Hospital of Kunming Medical
University Province (Kunming, China).
Tissues preparation and cell culture
A total of 73 patients were recruited from the
clinic of the Department of Neurosurgery, First Affiliated Hospital
of Kunming Medical University between February 2018 and December
2019. The patients provided informed consent for their
participation in the study. The patients ranged in age from 41 to
76 years, with a median age of 48 years. There were 36 male and 37
female patients included. Age-specific expression levels of RhoG
and miR-124-3p estimates are described in Tables II and III. Patients who underwent surgery at
our hospital and had not received prior anticancer treatment were
included. Patients excluded were those suffering from cerebral
trauma, recurrent infections, systemic disease and other
immunosuppression conditions as well as those that were
HIV-infected. The resected sections were classified by three
independent senior pathologists according to the histopathological
data. The carcinoma tissues were not obtained for further analysis
until they were verified as the primary GBM. Subsequently, the
primary neoplasm and the adjacent tissue (≥3 cm away from the
neoplasm) (15) were obtained
and divided into two parts for gene or protein expression
detection.
 | Table IIClinicopathologic features of GBM
associated with RhoG levels. |
Table II
Clinicopathologic features of GBM
associated with RhoG levels.
| Features | RhoG mRNA levels
| rs | P-value |
|---|
| Low expression
level n=26 | High expression
level n=47 |
|---|
| Ages (years) | | | 0.113 | 0.21 |
| 40-49 | 9 | 14 | | |
| 50-60 | 10 | 18 | | |
| >60 | 7 | 15 | | |
| Sex | | | 0.128 | 0.19 |
| Male | 14 | 22 | | |
| Female | 12 | 25 | | |
| Tumor size
(cm) | | | 0.612 |
0.0015a |
| <5 cm | 23 | 11 | | |
| ≥5 cm | 3 | 36 | | |
| WHO grade | | | 0.401 |
0.002a |
| I-II | 15 | 8 | | |
| III | 8 | 10 | | |
| IV | 3 | 29 | | |
 | Table IIIAssociations between miR-124-3p, RhoG
levels and clinicopathologic features of GBM patients by
multivariate analysis. |
Table III
Associations between miR-124-3p, RhoG
levels and clinicopathologic features of GBM patients by
multivariate analysis.
| Features | miR-124-3p
expression
| RhoG expression
|
|---|
| Low level (N=44) (≤
median) n (%) | High level (N=29)
(> median) n (%) | Adjusted OR (95%
CI) | Low level (N=26) (≤
median) n (%) | High level (N=47)
(> median) n (%) | Adjusted OR (95%
CI) |
|---|
| WHO grade | | | | | | |
| I-II | 5 (6.8) | 22 (30.1) | 2.24 (0.96,
4.02) | 15 (20.6) | 10 (13.7) | 1.58 (0.98,
4.21) |
| III | 21 (28.8) | 5 (6.8) | 16.87 (12.36,
28.21)a | 8 (10.9) | 29 (39.7) | 14.11 (9.45,
29.2)a |
| IV | 18 (24.7) | 2 (2.8) | 21.87 (12.31,
31.58)a | 3 (4.2) | | 18.42 (0.75,
24.5)a |
| Tumor size
(cm) | | | | | | |
| <5 | 10 (18.9) | 25 (16.3) | 1.82
(1.13-5.01) | 23 (31.5) | 11 (15.1) | 1.23 (0.73,
6.49) |
| ≥5 | 34 (62.1) | 4 (2.7) | 49.03
(29.92-58.12)a | 3 (4.1) | 36 (49.3) | 29.63 (20.4,
42.9)a |
The human GBM cell lines, U87 and U251, and the
human normal skin fibroblast cell line, HF, were provided by The
Cell Bank of Type Culture Collection of The Chinese Academy of
Sciences (Shanghai, China). The U87 and U251 cells were verified by
The Cell Bank of Type Culture Collection of the Chinese Academy of
Sciences. No misidentified and/or contaminated cell lines were used
in the present study. It must be emphasized that the cell line U87
has been reported to be misidentified/contaminated. Therefore, this
cell line is considered a glioblastoma of unknown origin, according
to the Cellosaurus database (https://web.expasy.org/cellosaurus/CVCL_0022).
Therefore, in the present study, the identity of this cell line was
validated using STR profile identification by the Cell Bank of the
Chinese Academy of Sciences. The results revealed that the matching
ratio between the test sample and the ATCC standard data was 94.4%
and that this result confirmed the identity of the cell line used
in this study as U87. The culture of the cells was performed by
using the mixture medium, which contained Dulbecco's Modified
Eagle's Medium (DMEM), 10% fetal bovine serum, 100 U/ml penicillin
and 100 µg/ml streptomycin (all from Thermo Fisher
Scientific, Inc.). The culture conditions were 37°C with a
humidified atmosphere containing 5% CO2.
Cell treatment
The U87 and U251 cell lines were transiently
transfected with either miR-124-3p (1.0 µl; 3 µM) or
miR-124-3p inhibitor (2.5 µl; 3 µM) or the matched
control using Lipofectamine 3000 reagent (Invitrogen; Thermo Fisher
Scientific, Inc.). The miR-124-3p, miR-124-3p inhibitor, miRNA
mimics negative control (miR-NC) and scrambled siRNA negative
control (miR-124 inhibitor NC) were provided by Guangzhou RiboBio
Co., Ltd. The sequences of the miRNAs used were as follows:
miR-124-3p (mimics), CCG UAA GUG GCG CAC GGA AU; miR-NC, AAA GCC
UUA UUC CUU CGU ACG; miR-124-3p inhibitor, CAU UAC GGC CAA UAU GUA
AGG CA; and miR-124 inhibitor NC, AUG GUA CGU GUA GGC CUA CUA UG.
The cells (2×105) were seeded in a plate and transfected
with the aforementioned reagents for 48 h at 37°C as recommended by
the manufacturer's protocol. Subsequently, the treated cells were
harvested for further analyses, including reverse
transcription-quantitative PCR (RT-qPCR), western blotting, cell
proliferation and apoptosis assays.
RT-qPCR
Samples from human GBM cells or from the tissue
sections were used to evaluate the relative expression levels of
miR-124-3p and RhoG by RT-qPCR. The extraction of total RNA was
performed using TRIzol® (Invitrogen; Thermo Fisher
Scientific, Inc.) and an miRNeasy mini kit (Qiagen AB). The
All-in-One™ miRNA quantitative RT-PCR Detection kit was used for
the RT-qPCR, according to the manufacturer's protocol (GeneCopoeia,
Inc.). The miRNA RT-PCR primers of hsa-miR-124-3p and U6 (reference
gene) were provided by Applied Biosystems; Thermo Fisher
Scientific, Inc. The expression levels of the examined samples were
normalized to those of U6 (15).
The sequences of the primers were as follows: U6 forward, 5′-CTC
GCT TCG GCA GCA CA-3′ and reverse, 5′-AAC GCT TCA CGA ATT TGC
GT-3′; hsa-miR-124-3p, forward, 5′-AUU UCC CUC AAU CCU ACU C-3′ and
reverse, 5′-UGA GCT GUC AAC GCC UAU AUC-3′.
To assess the expression of RhoG mRNA, Moloney
Murine Leukemia Virus Reverse Transcriptase (Takara Bio, Inc.) and
2X Mix SYBR Green I (Takara Bio, Inc.) were used. Each PCR reaction
consisted of two parts, including the initial denaturation at 95°C
for 3 min and multiple cycles of amplification (39 cycles at 95°C
for 10 sec, 57°C for 15 sec, and 72°C for 30 sec), followed by a
denaturation step at 95°C for 10 sec, an annealing step at 65°C for
5 sec, and a final extension step at 95°C for 10 sec. GAPDH was
used as the reference control. The primers were provided by
Invitrogen; Thermo Fisher Scientific, Inc. and the sequences were
described as follows: GAPDH forward, 5′-CGA GAT CCC TCC AAA ATC
AA-3′ and reverse, 5′-TTC ACA CCC ATG ACG AAC AT-3′; and RhoG
forward, 5′-CAG AGC ATC AAG TGG TGG TGG TG-3′ and reverse, 5′-GAG
GAT GCA GGA CCG CCC ACG-3′. The relative expressions levels of the
genes examined were evaluated using the 2−ΔΔCq method
(16).
Western blot analysis
The BCA Assay kit (Beyotime Institute of
Biotechnology) was used to determine the protein concentrations of
each sample following total protein extraction by the cell lysis
buffer (Beyotime Institute of Biotechnology). Subsequently, the
expression levels of RhoG, Bcl-2, Bax, caspase-3 and caspase-9
proteins were assessed as previously described (17). Briefly, 40 µg protein per
lane was separated via 10% SDS-PAGE gel and transferred to a
nitrocellulose membrane (Beyotime Institute of Biotechnology). The
membrane was soaked in 5% skimmed milk for 2 h at 37°C and
subsequently incubated with the corresponding primary antibody at
4°C overnight. The details of each antibody are described as
follows: RhoG (1:1,000; cat. no. sc-80015), Bax (1:500; cat. no.
sc-7480) and Bcl-2 (1:800; cat. no. sc-7382; all from Santa Cruz
Biotechnology, Inc.), caspase-9 (1:1,000; product code ab32539;
Abcam), caspase-3 (1:500; product code ab13847; Abcam) and GAPDH
(1:500; cat. no. sc-47724; Santa cruz Biotechnology, Inc.).
Following the primary antibody incubation, the membranes were
washed three times with 0.01 M PBS buffer solution (Yunnan Labreal
Biotechnology Co., Ltd.) and incubated with mouse IgG binding
protein-HRP (1:2,000; cat. no. sc-516102) and mouse anti-rabbit
IgG-HRP (1:1500; cat. no. sc-2357; both from Santa Cruz
Biotechnology, Inc.) secondary antibodies at 37°C for 1 h. ECL
reagent (EMD Millipore) was utilized to observe the protein bands
and GAPDH was used as the reference for protein expression.
Bioinformatics prediction for the target
genes of miR-124-3p
The three computational algorithms, TargetScan
(version 7.2; http://www.targetscan.org) (18), miRanda (http://www.microrna.org) (19) and PicTar (http://pictar.mdc-berlin.de) (20) were employed to investigate the
candidate targets of miR-124-3p involved in GBM progression.
According to the intersection, the target genes were predicted by
the aforementioned databases and the RhoG gene was selected.
Subsequent studies were performed to assess its function as a
potential target of miR-124-3p.
Validation of the miR-124-3p target gene
RhoG
Luciferase reporter assays were performed to
validate the miR-124-3p target gene RhoG in GBM cells. The putative
binding locations at the 3′ untranslated region (3′-UTR) of the
human RhoG mRNA were predicted and wild-type (wt) or the mutant
(mut) of RhoG was established and packaged in the pLUC plasmid
(Guangzhou RiboBio Co., Ltd.). Mutations were produced using the
QuikChange Site-Directed Mutagenesis kit (Stratagene; Agilent
Technologies, Inc.) according to the manufacturer's instructions.
The primers used for the construction of the luciferase reporters
and mutations of binding sites are presented in Table I. A random control luciferase
plasmid (Blank control) was also purchased (Guangzhou RiboBio Co.,
Ltd.). Briefly, 1×106 U87 or U251 cells were seeded into
96-well plates and cultured at 70% confluence 24 h prior to the
transfection. A total of 30 ng of the pLUC-UTR-wt or the
pLUC-UTR-mut corresponding to the RhoG luciferase plasmid (pluc
RhoG 3′-UTR-wt or pluc RhoG 3′-UTR-mut) was mixed with 40 nM
miR-124-3p, miR-124-3p inhibitor or the matched control miR-NC
(miR-NC or miRNA-124-3p inhibitor NC), and transfected with 10 ng
Renilla plasmid into GBM cells using Lipofectamine 3000 at
37°C for 4 h. Following 12 h of cell culture, the relative
luciferase activity was detected using a Dual-Luciferase Reporter
assay system (Promega Corporation) The firefly luciferase activity
was normalized to the Renilla luciferase activity and
collected for analysis using a luminometer (Centro LB 960; Berthold
Technologies GmbH & Co. KG). Three independent examinations
were conducted, which were completed in triplicate.
 | Table IPrimers for RhoG luciferase reporter
constructions. |
Table I
Primers for RhoG luciferase reporter
constructions.
| Gene | Sequence of forward
primer | Sequence of reverse
primer |
|---|
| RhoG |
5′-TTACTAGTCCCTGGCACTTGGCTTGGA-3′ |
5′-TAGAAGCTTGAGTCAGTCAGCAAATGCGT-3′ |
| RhoG mutant | | |
| Position 48-54 |
5′-CTTTTTCTCTGAATACATATTTCTCCTTAAG-3′ |
5′-CTTAAGGAGAAATATGTATTCAGAGAAAAAG-3′ |
| Position 56-62 |
5′-TCCGCCTCAGCTATACATAAAGGACTAATTC-3′ |
5′-GAATTAGTCCTTTATGTATAGCTGAGGCGGA-3′ |
| Position 76-83 |
5′-GCTTATTCTCCGTTCCAAGGCATATCGCGTAA-3′ |
5′-ATATGGCCCTATATTTATAGCGCAGTTTCCAGC-3′ |
| Position
139-145 |
5′-CCCACCAGTTATACATAGGTGCCTTGTCC-3′ |
5′-GGACAAGGCACCTATGTATAACTGGTGGG-3′ |
RhoG knockdown
To validate whether miR-124-3p exerts significant
effects on GBM deterioration by regulating RhoG expression, the
proliferative, migratory and invasive abilities of the tumor cells
were quantified in miR-124-3p gain and loss-function cells. RhoG
small interfering RNA (siRNA/si) was introduced in these cells. The
siRNA sequences aimed at human RhoG (si-RhoG) and the matched
negative control (si-NC) sequences were designed according to
previously validated oligonucleotides (21) and purchased from Guangzhou
RiboBio Co., Ltd. Lentiviral particles (lentivirus-GFP, 5
µg; Shanghai GeneChem Co., Ltd.) were packaged into 293T
cells (Cell Bank of Type Culture Collection of the Chinese Academy
of Sciences) using Lipofectamine 3000 for RhoG gene knockdown. The
2nd generation system was employed. The ratio of the lentiviral
plasmid: Packaging vector was 1:1 according to the manufacturer's
instructions. The multiplicity of infection (MOI) used to infect
cells was 10.45 U/ml penicillin (Thermo Fisher Scientific, Inc.)
and was used to create stable cell lines. GBM cells and normal HF
cells were transfected with recombinant 1×106
lentivirus-transducing units. After 48 h of incubation at 37°C, the
transfection efficiency was conducted by siRNA knockdown and
assessed using RT-qPCR. Subsequently, combined incubation of RhoG
siRNA-transfected U87 and U251 cells with miR-124-3p or its
inhibitor was performed at 37°C. At 24 h post-transfection, the
cells were harvested for further analysis.
Cell proliferation assay
Using an MTT assay kit (Sigma-Aldrich; Merck KGaA),
cell viability was measured. Briefly, the cells were seeded in
96-well plates at a density of 2×104 cells/well. The
cells were cultured for 48 h and 0.1 mg MTT reagent was added per
well and incubated at 37°C. Following incubation for an additional
4 h, the culture medium was replaced with 150 µl of dimethyl
sulfoxide (DMSO) to dissolve the formazan crystals. The absorbance
of each well was detected at 570 nm by using a microplate reader
(Multiskan Mk3; Thermo Fisher Scientific, Inc.).
Apoptosis assay
The Annexin-PE/7AAD kit (BD Pharmingen; BD
Biosciences) was utilized to assess the induction of early and late
apoptosis of GBM cells following various treatments. Labeled cells
were verified and analyzed by a flow cytometer (FACScan; BD
Biosciences). The FACStation software V6.1X (BD Biosciences) was
employed for data analysis. The experiments were performed in
triplicate. In addition, the activities of caspases-3 and/or
caspase-7 were evaluated using the Apo-ONE Homogeneous Caspase-3/7
Assay kit (Promega; Corporation) according to the manufacturer's
instructions.
Migration and invasion detection
The scratch wound assay was performed to assess the
migration of the GBM cells following certain treatments, as
previously described (22).
Briefly, at ~80% confluence, the tumor cells infected with
different treatments were transferred into six-well plates and
cultured at 37°C. Subsequently a scratch wound was created through
the middle of each well plate vertically using a 10-µl tip.
The scratched cells were removed by PBS washing thrice, and fresh
serum-free medium was replaced. At 48 h post-incubation, the cells
were observed by a light microscope at a magnification of ×200
(Olympus Corporation).
In addition, the detection of migration and invasion
detection was accomplished in accordance with a previously
described method (23). Briefly,
for the migration assay, tumor cells were prepared in serum-free
medium at a density of 1×105 cells/ml and seeded in the
upper chamber. A pre-coated chamber with 500 ng/ml Matrigel
solution (BD Biosciences) was employed for the invasion assay.
Following 48 h of incubation, the migratory cells on the surface of
the upper membrane were washed. The remaining cells that had
migrated/invaded to the lower membrane were collected for
statistical analysis following staining with 0.1% crystal violet
for 20 min and fixation with 20% methanol for 30 sec at 20-24°C.
The stained cells were observed, imaged and counted under an IBX3
inverted microscope (Olympus Corporation) at a magnification of
×200.
Statistical analysis
Statistical analyses were performed using the SPSS
version 21.0 software (IBM Corp.). The data are expressed as the
mean ± standard deviation (SD). Multivariate logistic regression
analysis and Student t-test was employed to assess the association
between miR-124-3p levels (low or high), RhoG gene levels (low or
high), and the clinicopathological characteristics of GBM. The
median expression levels of miR-124-3p or RhoG were used as the
cutoff values (24). The odds
ratios (ORs) with 95% confidence intervals were also measured.
Logistic regression analysis was performed to estimate adjusted ORs
for ordinal data. Significant differences between mRNA and proteins
expression levels, cell viability, induction of apoptosis and
migration and invasive activities, among multiple groups were
determined using ANOVA and Student-Newman-Keuls test. P<0.05 was
considered to indicate a statistically significant difference.
Results
Expressional changes of miR-124-3p and
its potential target RhoG in GBM tissues and cells
The expression levels of miR-124-3p and its
predicted target gene RhoG were evaluated in the samples obtained
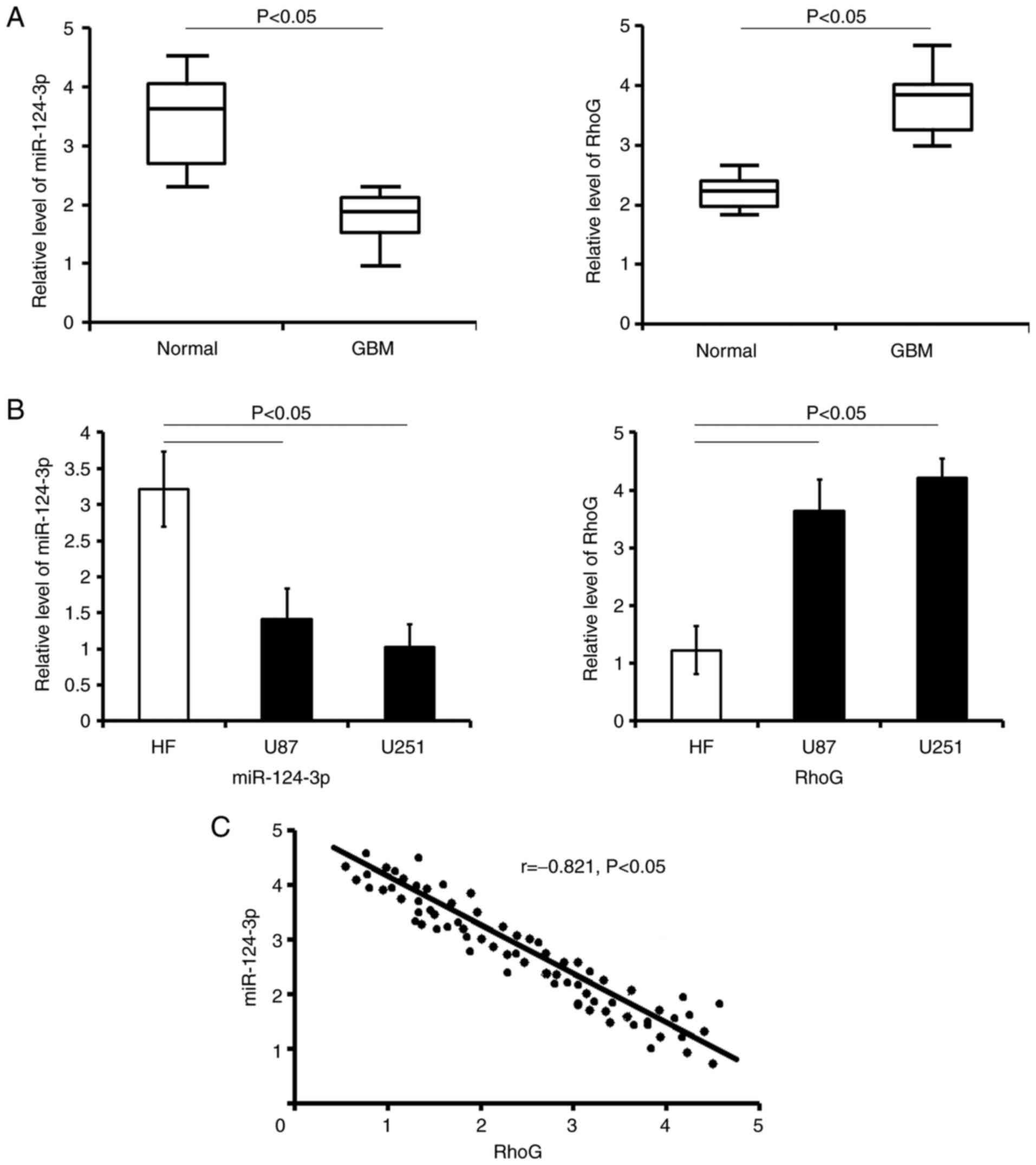
from patients with GBM by RT-qPCR. The results revealed that
miR-124-3p expression in GBM carcinoma tissues was lower compared
with the normal tissues (P<0.05; Fig. 1A). By contrast, the mRNA
expression levels of RhoG in cancer samples were upregulated
compared with those in the normal tissues (P<0.05; Fig. 1A).
In human GBM cells, the data indicated that the
expression pattern of miR-124-3p and RhoG was similar to that
observed in GBM carcinoma cells (vs. HF cells respectively;
P<0.05; Fig. 1B).
Correlation analysis revealed that the downregulated
miR-124-3p expression levels were negatively correlated with RhoG
expression levels in patients with GBM (r=-0.821; P<0.05;
Fig. 1C).
Expression levels of RhoG in tumor
samples are associated with GBM progression
The expression levels of RhoG, a candidate target of
miR-124-3p, were investigated in cancer tissues obtained from
patients with GBM by RT-qPCR. The patients were divided into two
groups according to the miR-124-3p and RhoG expression levels, as
follows: Subjects with miR-124-3p/RhoG expression levels lower than
the mean expression level were classified as the low-expression
level group and subjects with miR-124-3p/RhoG expression levels
higher than the mean expression level were classified as the
high-expression level group (24). The association between the
clinicopathological parameters of patients with GBM and RhoG mRNA
expression levels were analyzed. The results indicated that
increased levels of RhoG were positively associated with tumor size
(r=0.612; P=0.0015) and WHO grade (r=0.401, P=0.002) (Table II). Regression analysis revealed
that the downregulated expression levels of miR-124-3p were
negatively associated with RhoG expression levels during GBM
progression (Table III). The
data indicated that the unusual alterations in miR-124-3p and RhoG
expression levels were detrimental to the prognosis of GBM.
Assessment of the effects of miR-124-3p
or miR-124-3p inhibitor transfection
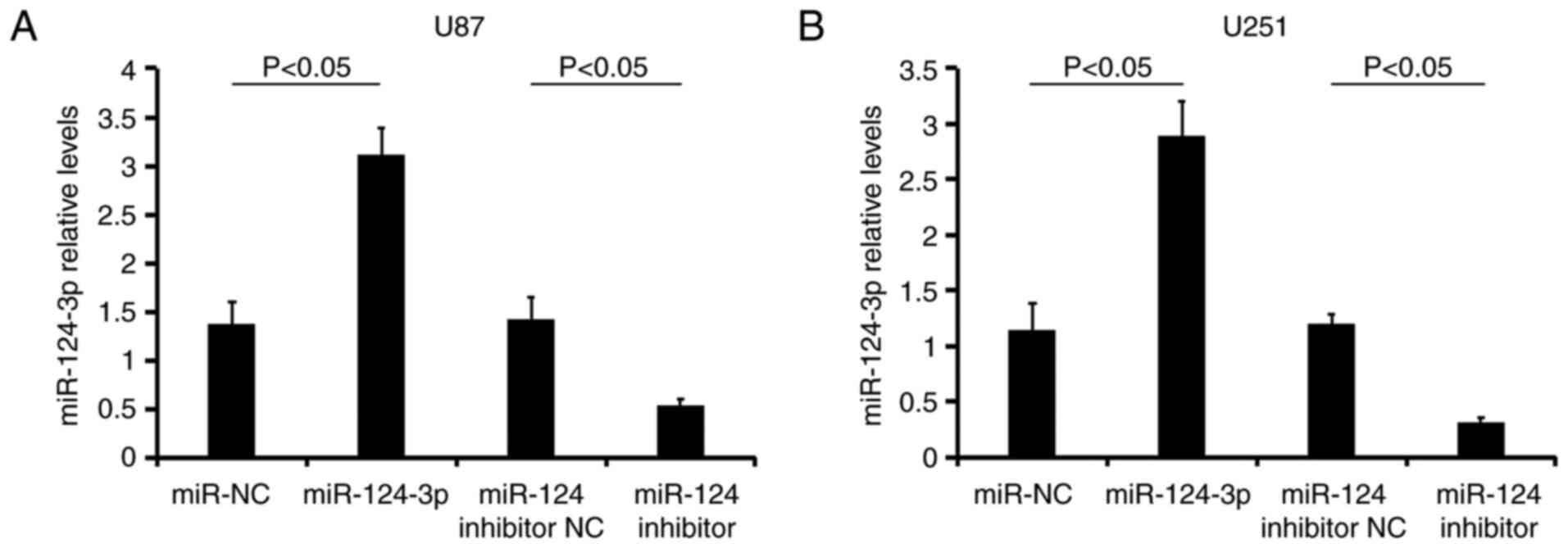
The miR-124-3p expression levels were analyzed in
U87 (Fig. 2A) and U251 (Fig. 2B) cells following various
treatments using RT-qPCR analysis. The transfection of miR-124-3p
into the two types of GBM cells resulted in a significant increase
in the miR-124-3p expression levels compared with
miR-NC-transfected cells (P<0.05). In addition, the treatment of
the cells with the miR-124-3p inhibitor decreased miR-124-3p levels
(vs. the miR-124 inhibitor NC group; P<0.05; Fig. 2). The data revealed that the
transfections were successful.
miR-124-3p targets RhoG at the 3′-UTR
region
miR-124-3p exhibited four putative binding sites
(two sites were conserved between the species rat, mouse, and human
species and others were poorly conserved) for RhoG mRNA on the
3′-UTR (Fig. 3A). The
co-transfection of miR-124-3p packaged with pLUC RhoG 3′-UTR-wt
significantly decreased the relative luciferase activity. This was
not observed in the pLUC RhoG 3′-UTR-mut or in the miR-NC
transfected groups (Fig. 3B and
D). The mutations in the putative miR-124-3p binding sites in
the four 3′-UTRs abrogated the response to miR-124-3p (pluc RhoG
3′-UTR-mut + miR-NC vs. pluc RhoG 3′-UTR-mut + miR-124-3p;
P>0.05; Fig. 3D). Moreover,
the administration of the miR-124-3p inhibitor to the cells
significantly reduced the relative luciferase activity (Fig. 3B). The transfection of miR-124-3p
into the two types of GBM cells resulted in a notable reduction of
RhoG expression levels compared with miR-NC-transfected cells
(P<0.05). By contrast, the treatment of the cells with the
miR-124-3p inhibitor increased RhoG levels (vs. the miR-124
inhibitor NC group; P<0.05; Fig.
3C).
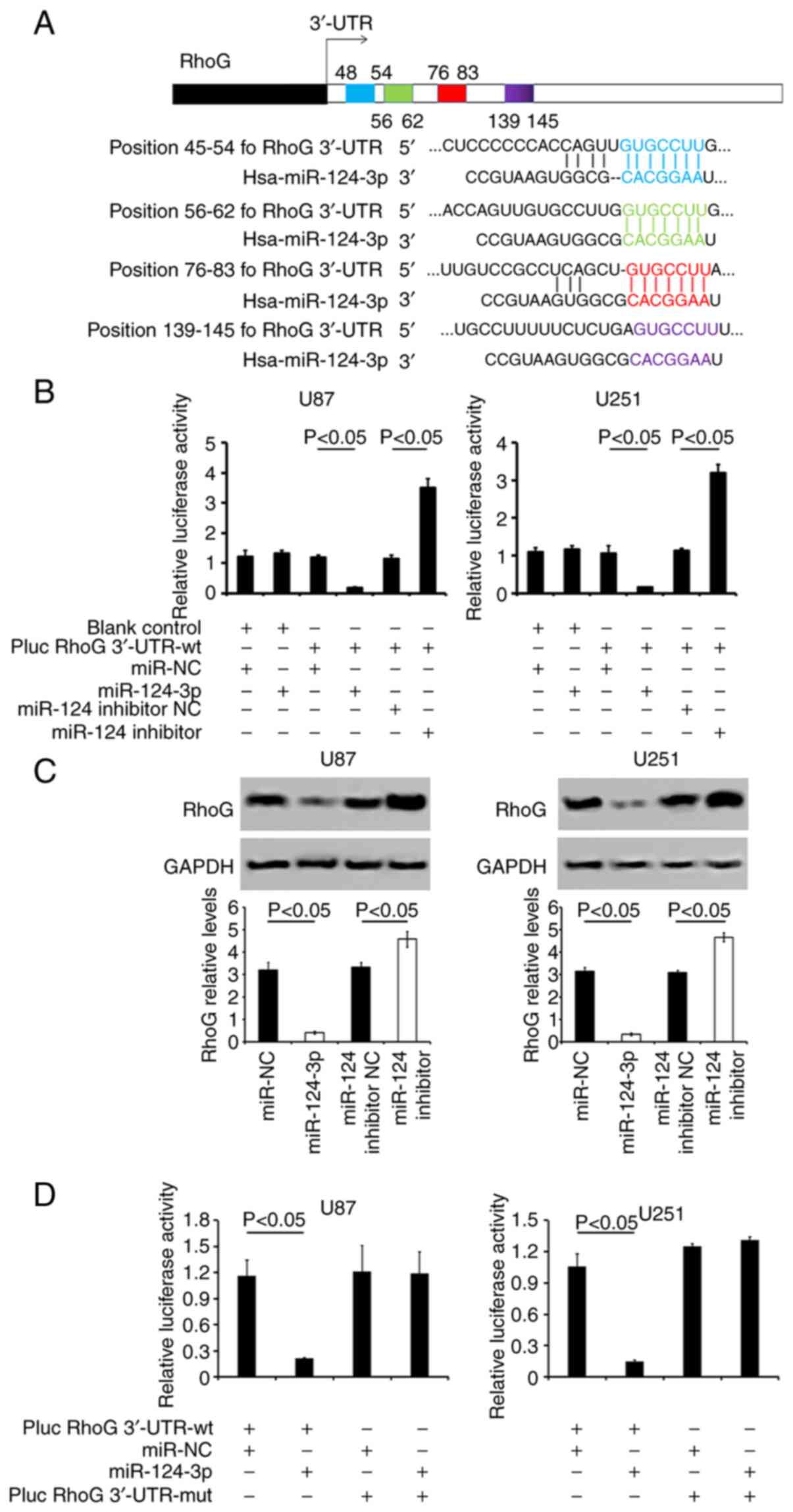 | Figure 3miR-124-3p targets the 3′-UTR seed
site of RhoG. (A) Prediction of the 'seed regions' on RhoG 3′-UTR
regulated by miR-124-3p. The 'seed̓ regions of the two
miR-124-3p-binding sites were located at positions 48-54
(highlighted in blue) and 76-83 (highlighted in red) of the RhoG
3′-UTR. These regions are conserved between the species rat, mouse,
and human. The poorly conserved binding site position 56-62 is
highlighted in green, and another position 139-145 is highlighted
in purple. (B) Examination of the relative luciferase activity in
the cells co-transfected with miR-124-3p or miR-124-3p inhibitor
and pluc RhoG 3′-UTR-wt, using a dual-luciferase reporter assay.
(C) RhoG protein expression levels were analyzed in U87 and U251
cells following various treatments using western blot analysis. The
representative lanes of RhoG protein expression are also presented.
GAPDH served as the control. (D) Detection of the relative
luciferase activity of miR-124-3p co-transfected with pLUC-RhoG
3′-UTR-wt or -mut using a dual-luciferase reporter assay. All the
data are representative of five independent experiments (n=5).
miR-124-3p, microRNA-124-3p; 3′-UTR, 3′-untranslated region; RhoG,
Ras homology Growth-related; wt, wild-type; mut, mutant; SD,
standard deviation; NC, negative control. |
Therefore, these results indicated that miR-124-3p
was directly bound to RhoG 3′ UTR and that it could downregulate
RhoG expression.
Evaluation of the effects caused by RhoG
siRNA transfection
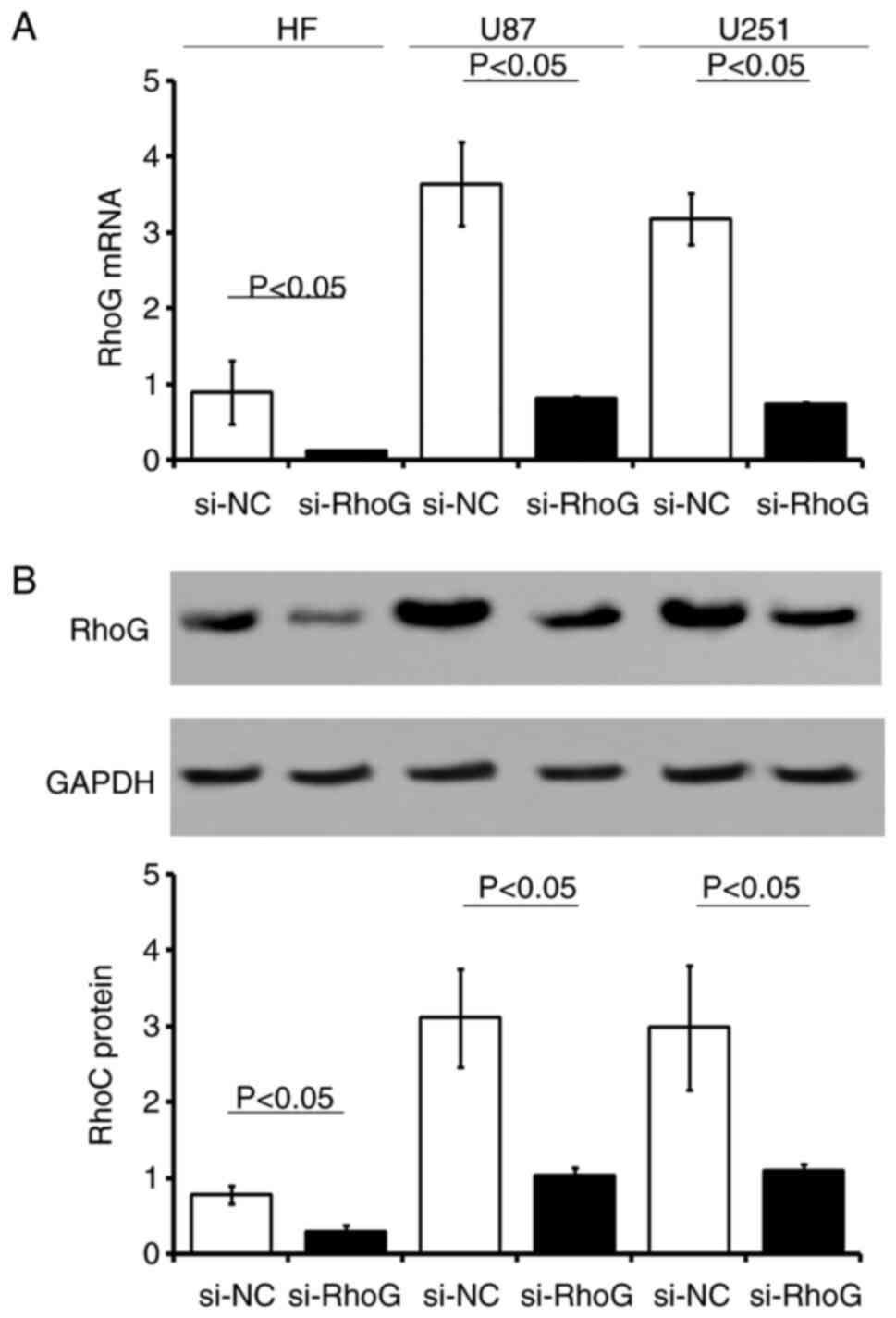
The stable expression of three RhoG siRNA sequences
in HF, U87 and U251 cells resulted in a >75% reduction in RhoG
mRNA and protein expression levels (Fig. 4). The GBM cell lines and normal
HF cells were stably transfected with RhoG siRNA-expressing or
si-NC vector. The expression levels of RhoG mRNA or protein were
significantly downregulated in cells transfected with si-RhoG
compared with those transfected with the si-NC control sequences,
respectively (P<0.05; Fig.
4).
miR-124-3p inhibits the viability and
apoptosis of GBM cells by RhoG regulation
An MTT assay was performed to examine the possible
influence of miR-124-3p or RhoG on the viability of GBM cells at 0,
24, 48, 72 and 96 h post-treatment. Transfection of the cells with
miR-124-3p or RhoG siRNA significantly decreased the cell viability
following 24, 48, 72 and 96 h of transfection in U87 (P<0.05;
Fig. 5A) and U251 cells
(P<0.05; Fig. 5B) compared
with that observed in the NC groups. The functional inhibition of
miR-124-3p by the application of its specific inhibitor resulted in
a marked increase in the cell viability of U87 (P<0.05; Fig. 5A) or U251 cells (P<0.05;
Fig. 5B).
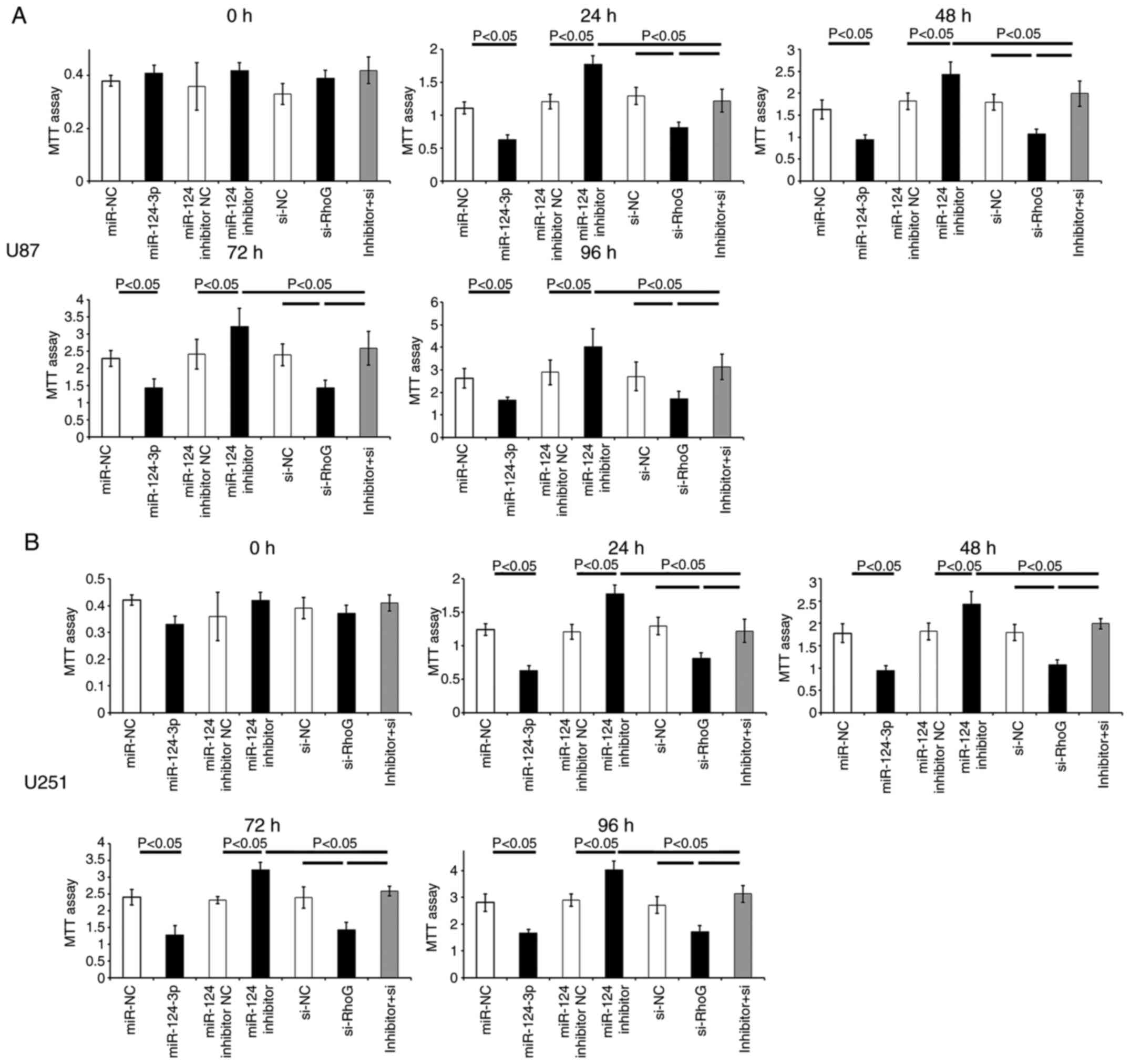 | Figure 5Effects of miR-124-3p/RhoG on the
viability of GBM cells. Cell proliferation of (A) U87 and (B) U251
was detected using an MTT assay at 0, 24, 48, 72 and 96 h
post-treatment. The data are presented as the mean ± SD. All the
data are representative of five independent experiments (n=5).
miR-124-3p, microRNA-124-3p; RhoG, Ras homology Growth-related;
GBM; glioblastoma multiforme; MTT,
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide; SD,
standard deviation; NC, negative control; si-NC, RhoG siRNA
negative control; si-RhoG, RhoG siRNA; inhibitor+si, miR-124-3p
inhibitor + RhoG siRNA. |
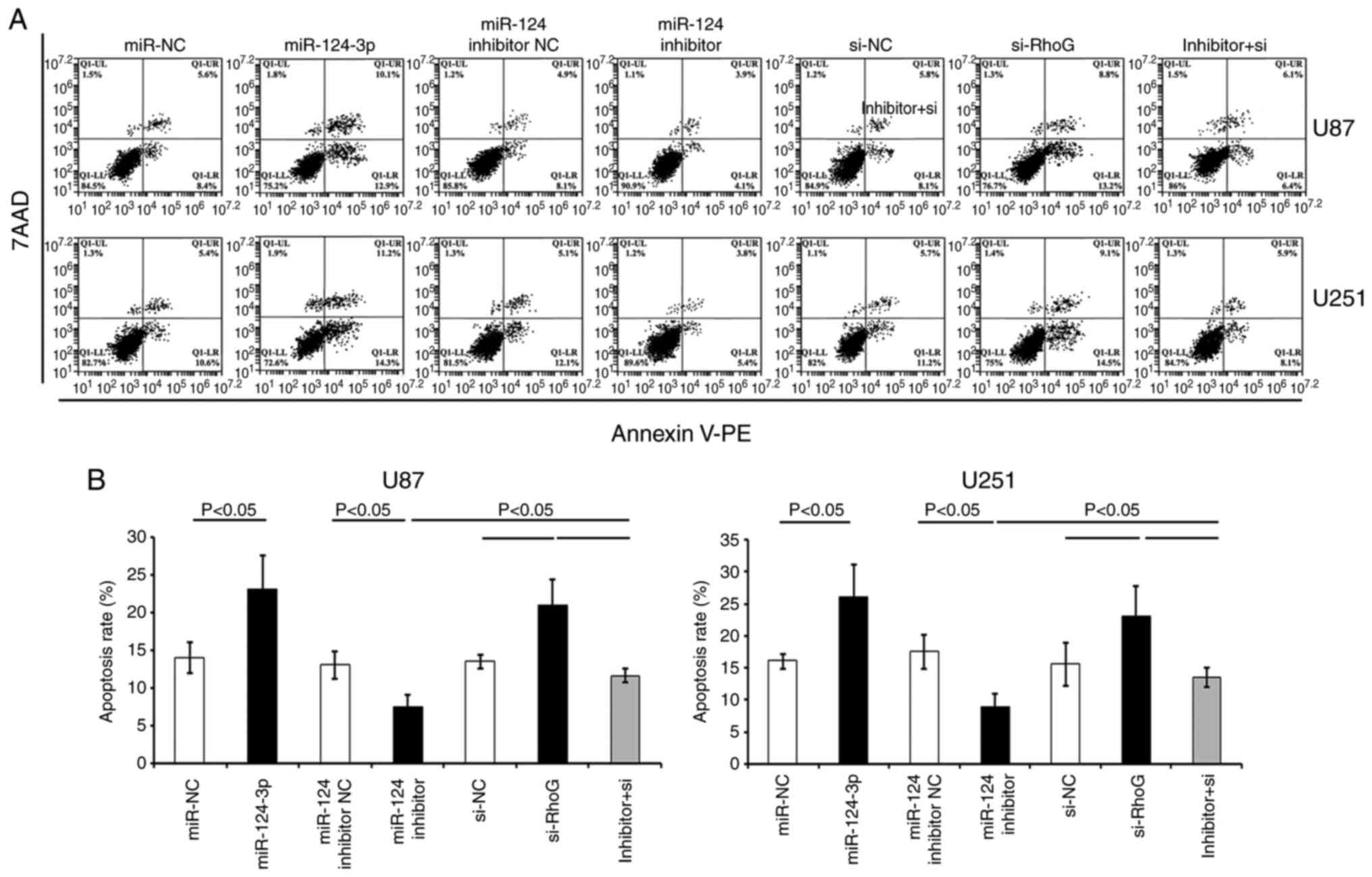
In comparison with the control NC groups (miR-NC or
si-NC), Annexin-PE/7AAD staining indicated that the transfection
with either RhoG siRNA or miR-124-3p significantly increased the
percentage of apoptotic U87 and U251 cells (P<0.05; Fig. 6). GBM cells transfected with
miR-124-3p inhibitor indicated a lower apoptotic rate compared with
the miR-124 inhibitor NC-treated cells (Fig. 6). The transfection of the cells
with RhoG siRNA sequences and the subsequent treatment with the
miR-124-3p inhibitor partially attenuated the effects produced by
the inhibitor (Figs. 5 and
6).
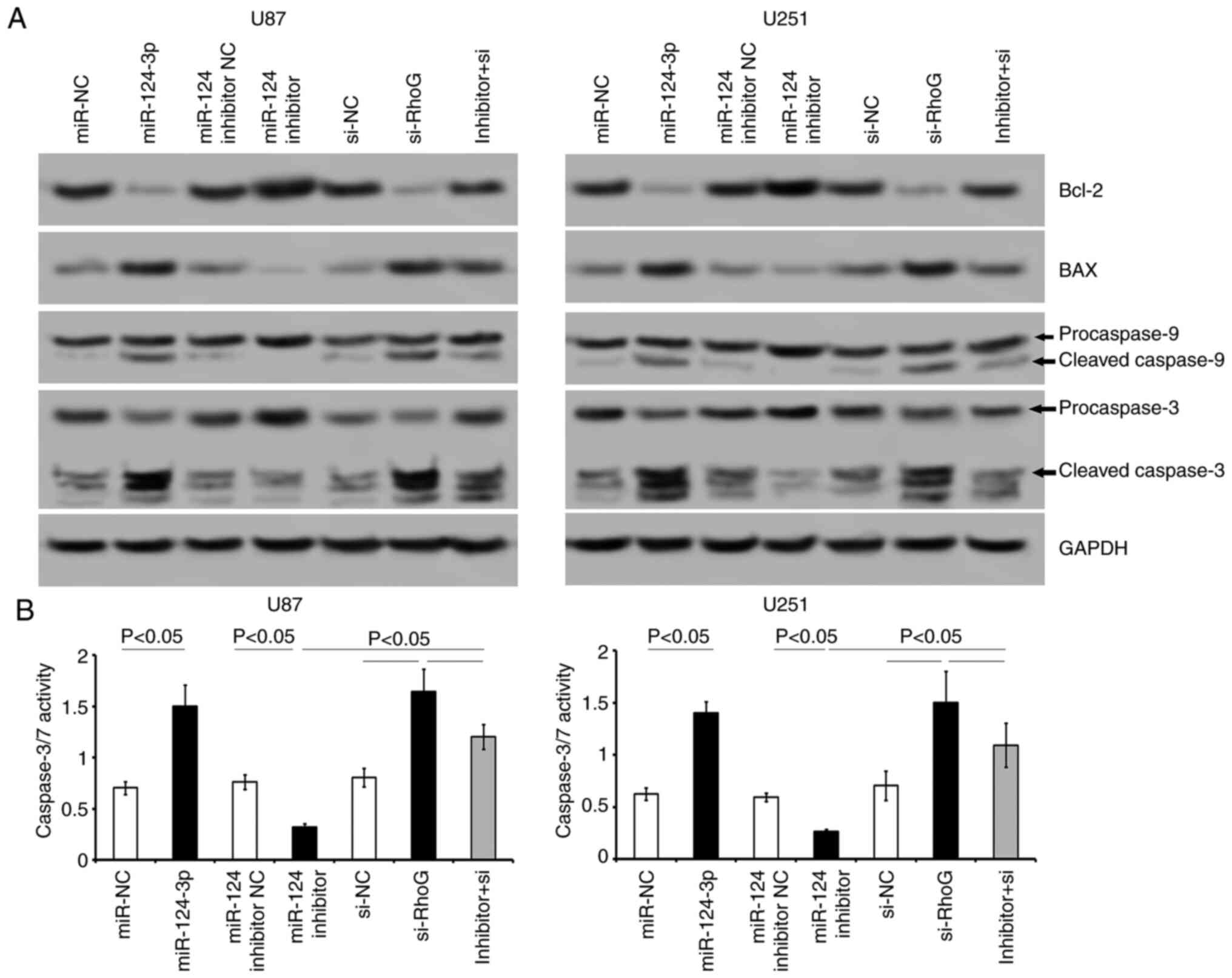
Western blot analysis indicated that the miR-124-3p
or RhoG siRNA-transfected cells had downregulated Bcl-2 expression
levels and higher BAX expression levels compared with that of the
miR-NC or si-NC cell groups (Fig.
7A). Transfection of the cells with either miR-124-3p or RhoG
siRNA sequences promoted the cleavage of procaspase-9 and
procaspase-3 into the corresponding active fragments in both GBM
cell lines (Fig. 7A). Inhibition
of miR-124-3p induced an increase in Bcl-2 levels and a decrease in
BAX, cleaved caspase-9 and caspase-3 levels (Fig. 7A). Caspase-3/7 activity was
significantly increased following the transfection of miR-124-3p or
RhoG siRNA into the cells (P<0.05; Fig. 7B), while the transfection of the
cells with the miR-124-3p inhibitor suppressed the activity of
caspase-3/7 (P<0.05; Fig.
7B). The co-transfection of miR-124-3p inhibitor and RhoG siRNA
to the cells attenuated the apoptotic effects caused by the
miR-124-3p inhibitor transfection alone (Figs. 6 and 7). The data indicated that the
restoration of miR-124-3p levels in GBM cells was conducive for the
prevention of GBM progression, possibly by regulating RhoG.
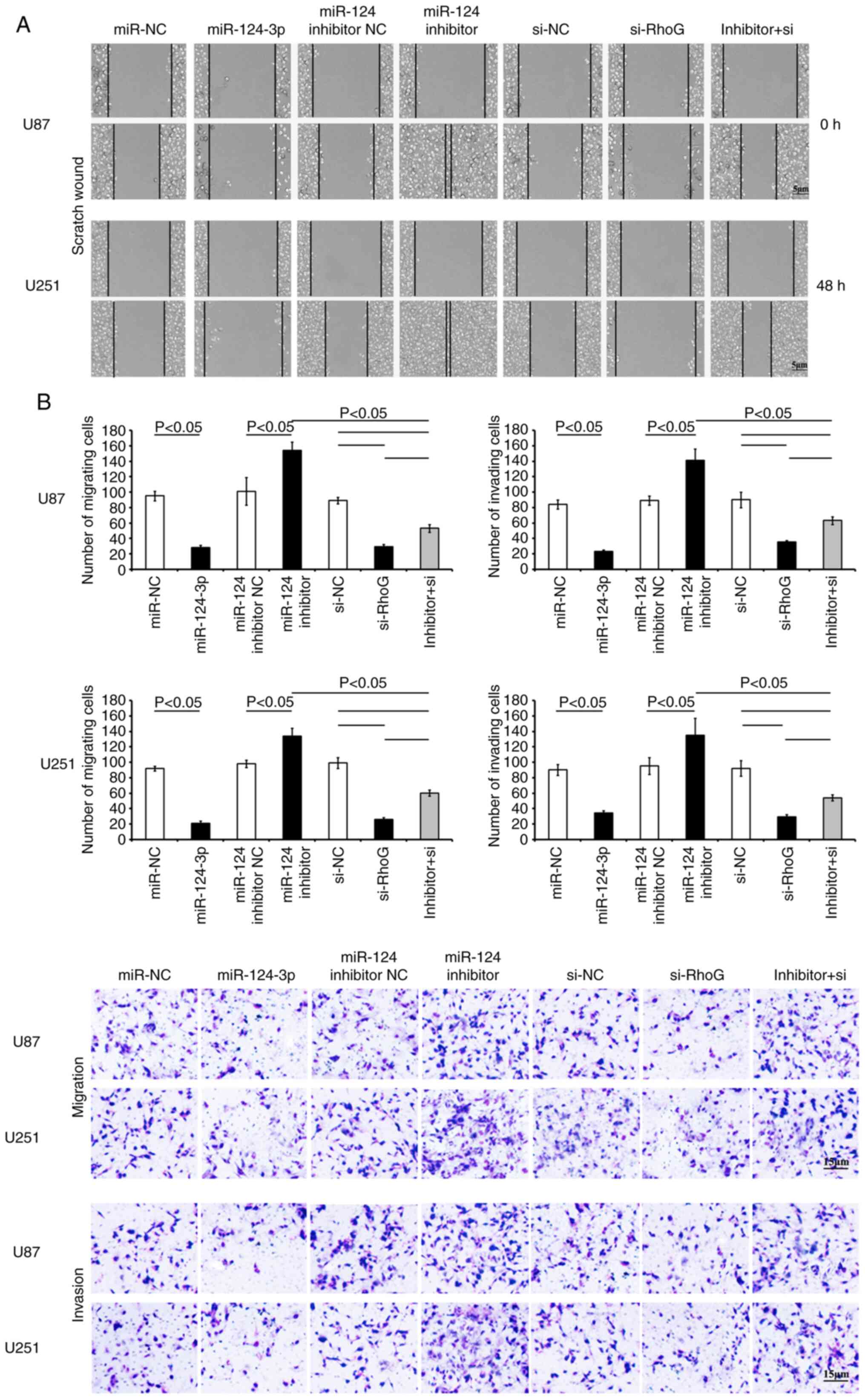
Targeting of RhoG by miR-124-3p inhibits
the migration of GBM cells
Transfection of the cells with miR-124-3p resulted
in a significant suppression of cell migration and invasion
(Fig. 8). The scratch wound
assay revealed that the transfection of the cells with miR-124-3p
markedly reduced the migration of GBM cells compared with that of
the control NC cells (Fig. 8A).
The cells that were stained with crystal violet in the Transwell
migration and invasion assays revealed considerably lower intensity
in the miR-124-3p-transfected group compared with that of the NC
control groups (P<0.05; Fig. 8B
and C). However, the transfection of the cells with miR-124-3p
inhibitor significantly increased the migration and invasion in
both GBM cell lines (vs. NC control; P<0.05, respectively). RhoG
siRNA-treated GBM cells presented a higher reduction in migration
and invasion compared with the si-NC cells (Fig. 8). As anticipated, the
co-transfection of the cells with both the miR-124-3p inhibitor and
RhoG siRNA alleviated the effects on cell migration and invasion
caused by single transfection of the cells with the miR-124-3p
inhibitor (Fig. 8). The data
indicated that miR-124-3p contributed to the reduction in the
migration and invasion of GBM cells, which may be associated with
the regulation of RhoG.
Discussion
The results of the present data revealed that the
downregulated expression levels of miR-124-3p were negatively
correlated with RhoG expression levels during tumor progression in
samples from both patients with GBM and GBM cell lines. In
addition, dual luciferase reporter assays verified that miR-124-3p
bound to the 3′-UTR of RhoG and that this interaction could inhibit
RhoG expression in GBM cells. The overexpression of miR-124-3p also
suppressed cell viability and migration and induced the apoptosis
of GBM cells by regulating RhoG expression. Therefore, these
findings indicated the potential impact of miR-124-3p/RhoG on the
development of GBM.
Glioma is a complicated neuroglial neoplasm, which
consists of various homogeneous subtypes. These cancers are
characterized by distinctive changes in expression levels of
specific miRNAs and their corresponding targets (25). It has been revealed that the
dysregulation of miRNAs contributes to the deterioration of GBM
(26). The present study
demonstrated that the aberrant decrease in miR-124-3p expression
was detected in GBM human tissues and that it was associated with
specific clinicopathological features and disease deterioration.
In vitro assays further revealed that miR-124-3p
upregulation suppressed the viability and migration of GBM cells.
The results indicated the potential role of miR-124-3p in GBM
progression. Similar observations have been reported by other
previous studies, which have revealed the function of miR-124 as a
tumor suppressor. miR-124 has been demonstrated to downregulate the
expression levels of various targets, including Smad2 (13) and p62 (14), and exhibits high potential as a
therapeutic target of glioma.
The Argonaute (AGO) proteins, interact with mature
miRNAs and can form the RNA-induced silencing complex for
silencing, the expression of target genes following transcription
(27,28). It is well known that a single
cohort of miRNAs can target a large number of varied target mRNAs
via interacting with the AGO proteins (29,30).
Various genes have been identified and validated as
targets of miR-124. These genes are involved in different
pathological progresses. RhoG, which is regulated by miR-124, is
involved in neuronal development (31,32). It has also been revealed that
miR-124 is silenced at the post-transcriptional level in a variety
of types of cancer cells and that this process modulates their
proliferation via the regulation of CDK6, CTGF, ITGB1, RhoG and
ROCK1 (9-12).
In the present study, RhoG was identified and
validated as a miR-124-3p target gene implicated in the progression
of GBM. Rho family small GTPases exert essential effects on cell
migration and actin cytoskeleton formation. Moreover, they regulate
various pathways involved in the development of tumors (33-35). Due to its crucial role in the
cytoskeletal reorganization, the small GTPase RhoG has attracted
considerable attention. It has been revealed that RhoG is critical
to phagocytosis and that it has the ability to alter gene
transcription and cell vability (34,35). It is also involved in cell
migration and invasion of pathogens (32). As a vital upstream target of Rac,
RhoG plays a crucial role in the regulation of cell migration by
activating Rac through its effector ELMO (36-38). Previous studies have also
revealed that Src homology 3 domain-containing guanine nucleotide
exchange factor (SGEF)-RhoG is an essential downstream regulator of
migration and invasion in TWEAK/Fn14-induced glioblastoma cells
(39). In the present study,
correlation analysis revealed that the endogenous downregulated
expression levels of miR-124-3p were correlated with RhoG
expression levels in patients with GBM. High levels of RhoG were
associated with the deterioration of GBM. The results of luciferase
reporter assays indicated that miR-124-3p was bound to the 3′-UTR
sequence of the RhoG gene and that it could inhibit RhoG
transcription in GBM cells. The treatment of cells with RhoG siRNA
or miR-124-3p markedly reduced their expansion, migration and
invasion, while it promoted the induction of apoptosis. In contrast
to these findings, the treatment of the cells with the miR-124-3p
inhibitor enhanced the viability and migration of GBM cells.
Furthermore, RhoG siRNA relieved the effects induced by the
miR-124-3p inhibitor, which led to a marked increase in the
viability and migration of GBM cells. The present study
demonstrated that RhoG was involved in the survival, proliferation,
migration and invasion of GBM. This effect was mediated by
miR-124-3p. The data indicated that miR-124-3p acted as a tumor
suppressor and delayed the deterioration of GBM by targeting
RhoG.
Furthermore, miR-124-3p and/or RhoG siRNA increased
in the apoptotic rates of GBM cells and the expression of activated
caspases 3/7, whereas they altered the transcriptions of the
proteins associated with apoptosis. It has been revealed that
phosphatidylinositol 3-kinase (PI3K) plays a vital role in
malignant transformation (40).
The PI3K-induced inhibition of apoptosis is performed by several
proteins regulated by protein kinase B (PKB) and/or the
PKB-enzyme-dependent (such as GSK-3 and ILK) signaling pathway
(40). The direct interaction of
these proteins with PI3K and the activation of Akt and RhoG may
facilitate the induction of apoptosis (41). Therefore, the present study
proposed that the inhibition of tumor cell proliferation induced by
RhoG siRNA may be associated with the anchorage deficit by means of
a PI3K-dependent mechanism. However, further investigation is
required to confirm this hypothesis (41,42).
In addition, the present study demonstrated that the
miR-124-3p inhibitor influenced GBM cell proliferation, viability
and migration, even in the absence of RhoG. These results suggested
that miR-124-3p influenced the carcinogenic transformation of GBM
cells through a mechanism involving RhoG inhibition. Combined with
the research reported by other groups, the present findings
demonstrate that miR-124 may target multiple proteins, including
CTGF, ITGB1, RhoG, ROCK1, CTGF, ROCK2 and/or EZH2, that function
spatiotemporally or in cooperation with different cellular
processes (9,43). These observations provide
evidence that miR-124 acts via the regulation of various proteins,
which are upregulated in several types of human cancer (13,14,43-45). The overexpression of these
proteins is positively correlated with tumor metastasis and/or poor
disease prognosis (13,14,43-45). In addition to the regulation of
RhoG, miR-124-3p was synchronously involved in GBM tumorigenesis
through other regulatory mechanisms. However, further
investigations are necessary to confirm these findings.
Moreover, since the increased expression level of
RhoG was detected in the glioma carcinomas and cell lines, specific
siRNA targeting RhoG was used to suppress the endogenous RhoG
expression and to explore the potential therapeutic effects of RhoG
knockdown in GBM cases. It was also verified whether RhoG was a
target gene for miR-124-3p. Therefore, miR-124-3p inhibitor and
RhoG siRNA were co-transfected to determine whether RhoG siRNA
attenuated the effects induced by miR-124-3p inhibitor. The
miR-124-3p inhibitor induced the recovery in endogenous RhoG
expression, while RhoG siRNA transfection led to RhoG expression
inhibition. These results further verified that miR-124-3p was
involved in the carcinogenic events of GBM by targeting RhoG. In a
future study, the effects of overexpression of RhoG may be
investigated since this is a limitation of the present study.
In conclusion, the findings of the present study
indicated that the downregulated expression levels of miR-124-3p
were conversely correlated with RhoG expression levels during
glioma progression in GBM tissues and cells. In addition, the
results of the dual luciferase reporter assays confirmed that
miR-124-3p interacted with the 3′-UTR of RhoG and inhibited RhoG
transcription in GBM cells. Finally, the overexpression of
miR-124-3p suppressed the expansion and infiltration of GBM cells
and increased their apoptotic rate by RhoG transcriptional
regulation. The present study demonstrated the possible effects of
miR-124-3p on the development of GBM by targeting RhoG.
Availability of data and materials
The datasets used and analyzed in the present study
are available from the corresponding author on reasonable
request.
Authors' contributions
SC, CJS and XPW conceived, designed, wrote,
submitted and replied to review comments of the manuscript. SC, JXL
and YPW collected the tumor tissues and prepared the cell cultures.
CJS, YPW, TY and XPW performed cell treatment and luciferase
reporter assays. SC, CJS, TY, and XPW performed the RT-qPCR,
apoptosis assay and cell viability assay. SC, CJS, JXL and XPW
conducted the western blot detection and scratch wound assay. JXL,
YPW and TY performed the statistical analysis. All authors read and
approved the final manuscript and agree to be accountable for all
aspects of the research in ensuring that the accuracy or integrity
of any part of the work are appropriately investigated and
resolved.
Ethics approval and consent to
participate
The present study complied with the Declaration of
Helsinki (revised in 2000) and approved by the Medical Ethics
Committee of Yunnan province. GBM tumor tissues were acquired from
patients with permission of the Department of Neurosurgery, First
Affiliated Hospital of Kunming Medical University Province,
Kunming, China. The patients provided informed consent for their
participation in the study.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgments
Not applicable.
Funding
The present study was supported in part by grants from the
Health Commission of Yunnan Province Training Plan for Medical
Reserve Talents (grant no. H-2017029), the Yunnan Clinical Medical
Center of Nervous System Diseases (grant no. ZX2019-03-05), and the
Research Innovation Team of Yunnan province (grant no.
2019HC022).
References
|
1
|
Ricard D, Idbaih A, Ducray F, Lahutte M,
Hoang-Xuan K and Delattre JY: Primary brain tumours in adults.
Lancet. 379:1984–1996. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Louis DN, Ohgaki H, Wiestler OD, Cavenee
WK, Burger PC, Jouvet A, Scheithauer BW and Kleihues P: The 2007
WHO classification of tumours of the central nervous system. Acta
Neuropathol. 114:97–109. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
van den Bent MJ: Interobserver variation
of the histopathological diagnosis in clinical trials on glioma: A
clinician's perspective. Acta Neuropathol. 120:297–304. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Joseph JV, Balasubramaniyan V, Walenkamp A
and Kruyt FA: TGF-β as a therapeutic target in high grade
gliomas-promises and challenges. Biochem Pharmacol. 85:478–485.
2013. View Article : Google Scholar
|
|
5
|
Nicolaidis S: Biomarkers of glioblastoma
multiforme. Metabolism. 64(3 Suppl 1): S22–S27. 2015. View Article : Google Scholar
|
|
6
|
Ambros V: The functions of animal
microRNAs. Nature. 431:350–355. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Rolle K: miRNA Multiplayers in glioma.
From bench to bedside. Acta Biochim Pol. 62:353–365. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Bartel DP: MicroRNAs: Target recognition
and regulatory functions. Cell. 136:215–233. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Lv XB, Jiao Y, Qing Y, Hu H, Cui X, Lin T,
Song E and Yu F: miR-124 suppresses multiple steps of breast cancer
metastasis by targeting a cohort of pro-metastatic genes in vitro.
Chin J Cancer. 30:821–830. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ando T, Yoshida T, Enomoto S, Asada K,
Tatematsu M, Ichinose M, Sugiyama T and Ushijima T: DNA methylation
of microRNA genes in gastric mucosae of gastric cancer patients:
Its possible involvement in the formation of epigenetic field
defect. Int J Cancer. 124:2367–2374. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Agirre X, Vilas-Zornoza A, Jiménez-Velasco
A, Martin-Subero JI, Cordeu L, Gárate L, San José-Eneriz E,
Abizanda G, Rodríguez-Otero P, Fortes P, et al: Epigenetic
silencing of the tumor suppressor microRNA Hsa-miR-124a regulates
CDK6 expression and confers a poor prognosis in acute lymphoblastic
leukemia. Cancer Res. 69:4443–4453. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Lujambio A, Ropero S, Ballestar E, Fraga
MF, Cerrato C, Setién F, Casado S, Suarez-Gauthier A,
Sanchez-Cespedes M, Git A, et al: Genetic unmasking of an
epigenetically silenced microRNA in human cancer cells. Cancer Res.
67:1424–1429. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Lv Z and Zhao Y: MiR-124 inhibits cell
proliferation, invasion, and migration in glioma by targeting
Smad2. Int J Clin Exp Pathol. 10:11369–11376. 2017.PubMed/NCBI
|
|
14
|
Deng D, Luo K, Liu H, Nie X, Xue L, Wang
R, Xu Y, Cui J, Shao N and Zhi F: p62 acts as an oncogene and is
targeted by miR-124-3p in glioma. Cancer Cell Int. 19:2802019.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Song S, Fajol A, Tu X, Ren B and Shi S:
miR-204 suppresses the development and progression of human
glioblastoma by targeting ATF2. Oncotarget. 7:70058–70065. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
17
|
Hu T, Li YS, Chen B, Chang YF, Liu GC,
Hong Y, Chen HL and Xiyang YB: Elevated glucose-6-phosphate
dehydrogenase expression in the cervical cancer cases is associated
with the cancerigenic event of high-risk human papillomaviruses.
Exp Biol Med (Maywood). 240:1287–1297. 2015. View Article : Google Scholar
|
|
18
|
Lewis BP, Burge CB and Bartel DP:
Conserved seed pairing, often flanked by adenosines, indicates that
thousands of human genes are microRNA targets. Cell. 120:15–20.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
John B, Enright AJ, Aravin A, Tuschl T,
Sander C and Marks DS: Human MicroRNA targets. PLoS Biol.
2:e3632004. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Krek A, Grün D, Poy MN, Wolf R, Rosenberg
L, Epstein EJ, MacMenamin P, da Piedade I, Gunsalus KC, Stoffel M
and Rajewsky N: Combinatorial microRNA target predictions. Nat
Genet. 37:495–500. 2005. View
Article : Google Scholar : PubMed/NCBI
|
|
21
|
Samson T, Welch C, Monaghan-Benson E, Hahn
KM and Burridge K: Endogenous RhoG is rapidly activated after
epidermal growth factor stimulation through multiple
guanine-nucleotide exchange factors. Mol Biol Cell. 21:1629–1642.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
He Z, Huang C, Lin G and Ye Y:
siRNA-induced TRAF6 knockdown promotes the apoptosis and inhibits
the invasion of human lung cancer SPC-A1 cells. Oncol Rep.
35:1933–1940. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Liang L, Wong CM, Ying Q, Fan DN, Huang S,
Ding J, Yao J, Yan M, Li J, Yao M, et al: MicroRNA-125b
suppressesed human liver cancer cell proliferation and metastasis
by directly targeting oncogene LIN28B2. Hepatology. 52:1731–1740.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
He Z, Xia Y, Liu B, Qi X, Li Z, Wang J,
Chen L and Chen Y: Down-regulation of miR-452 is associated with
poor prognosis in the non-small-cell lung cancer. J Thorac Dis.
8:894–900. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Yang H and Wang Y: Five miRNAs considered
as molecular targets for predicting neuroglioma. Tumour Biol.
37:1051–1059. 2016. View Article : Google Scholar
|
|
26
|
Qiu S, Lin S, Hu D, Feng Y, Tan Y and Peng
Y: Interactions of miR-323/miR-326/miR-329 and
miR-130a/miR-155/miR-210 as prognostic indicators for clinical
outcome of glioblastoma patients. J Transl Med. 11:102013.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Liu X, Fortin K and Mourelatos Z:
MicroRNAs: Biogenesis and molecular functions. Brain Pathol.
18:113–121. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Wang WX, Wilfred BR, Hu Y, Stromberg AJ
and Nelson PT: Anti-Argonaute RIP-Chip shows that miRNA
transfections alter global patterns of mRNA recruitment to
microribonucleoprotein complexes. RNA. 16:394–404. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Baek D, Villén J, Shin C, Camargo FD, Gygi
SP and Bartel DP: The impact of microRNAs on protein output.
Nature. 455:64–71. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Miranda KC, Huynh T, Tay Y, Ang YS, Tam
WL, Thomson AM, Lim B and Rigoutsos I: A pattern-based method for
the identification of MicroRNA binding sites and their
corresponding heteroduplexes. Cell. 126:1203–1217. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Franke K, Otto W, Johannes S, Baumgart J,
Nitsch R and Schumacher S: miR-124-regulated RhoG reduces neuronal
process complexity via ELMO/Dock180/Rac1 and Cdc42 signalling. EMBO
J. 31:2908–2921. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Schumacher S and Franke K:
miR-124-regulated RhoG: A conductor of neuronal process complexity.
Small GTPases. 4:42–46. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Etienne-Manneville S and Hall A: Rho
GTPases in cell biology. Nature. 420:629–635. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Sahai E and Marshall CJ: RHO-GTPases and
cancer. Nat Rev Cancer. 2:133–142. 2002. View Article : Google Scholar
|
|
35
|
Vega FM and Ridley AJ: Rho GTPases in
cancer cell biology. FEBS Lett. 582:2093–2101. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Katoh H and Negishi M: RhoG activates Rac1
by direct interaction with the Dock180-binding protein Elmo.
Nature. 424:461–464. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Hiramoto K, Negishi M and Katoh H: Dock4
is regulated by RhoG and promotes Rac-dependent cell migration. Exp
Cell Res. 312:4205–4216. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Elfenbein, Rhodes JM, Meller J, Schwartz
MA, Matsuda M and Simons M: Suppression of RhoG activity is
mediated by a syndecan 4-synectin-RhoGDI1 complex and is reversed
by PKCalpha in a Rac1 activation pathway. J Cell Biol. 186:75–83.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Fortin Ensign SP, Mathews IT, Eschbacher
JM, Loftus JC, Symons MH and Tran NL: The Src homology 3
domain-containing guanine nucleotide exchange factor is
overexpressed in high-grade gliomas and promotes tumor necrosis
factor-like weak inducer of apoptosis-fibroblast growth
factor-inducible 14-induced cell migration and invasion via tumor
necrosis factor receptor-associated factor 2. J Biol Chem.
288:21887–21897. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Krasilnikov MA: Phosphatidylinositol-3
kinase dependent pathways: The role in control of cell growth,
survival, and malignant transformation. Biochemistry (Mosc).
65:59–67. 2000.
|
|
41
|
Murga C, Zohar M, Teramoto H and Gutkind
JS: Rac1 and RhoG promote cell survival by the activation of PI3K
and Akt, independently of their ability to stimulate JNK and
NF-kappaB. Oncogene. 21:207–216. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Yamaki N, Negishi M and Katoh H: RhoG
regulates anoikis through a phosphatidylinositol 3-kinase-dependent
mechanism. Exp Cell Res. 313:2821–2832. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Zheng F, Liao YJ, Cai MY, Liu YH, Liu TH,
Chen SP, Bian XW, Guan XY, Lin MC, Zeng YX, et al: The putative
tumour suppressor microRNA-124 modulates hepatocellular carcinoma
cell aggressiveness by repressing ROCK2 and EZH2. Gut. 61:278–289.
2012. View Article : Google Scholar
|
|
44
|
Chien W, O'Kelly J, Lu D, Leiter A, Sohn
J, Yin D, Karlan B, Vadgama J, Lyons KM and Koeffler HP: Expression
of connective tissue growth factor (CTGF/CCN2) in breast cancer
cells is associated with increased migration and angiogenesis. Int
J Oncol. 38:1741–1747. 2011.PubMed/NCBI
|
|
45
|
Fowler A, Thomson D, Giles K, Maleki S,
Mreich E, Wheeler H, Leedman P, Biggs M, Cook R, Little N, et al:
miR-124a is frequently down-regulated in glioblastoma and is
involved in migration and invasion. Eur J Cancer. 47:953–963. 2011.
View Article : Google Scholar : PubMed/NCBI
|