Introduction
Esophageal cancer is the seventh most common
malignant tumor, with >483,000 new cases and >439,000-related
deaths in 2015 worldwide (1,2).
It has been reported that the global incidence of esophageal cancer
has sharply increased by >6-fold (3,4).
With the development and improvement of esophageal cancer treatment
strategies, the 5-year survival rate of patients with early-stage
esophageal cancer has improved significantly; however, the
prognosis of patients with advanced disease remains dismal
(5). However, due to the lack of
early typical clinical symptoms, the majority of cases are already
in the late stages of the disease at the time of diagnosis.
Moreover, local recurrence or distant metastasis results in the
poor prognosis of patients with esophageal cancer (6). Thus, a novel therapeutic target for
esophageal cancer needs to be identified, and the further
understanding of the pathogenesis of esophageal cancer is urgently
required.
It has been widely accepted that TGFB induced factor
homeobox 1 (TGIF1), a member of the TGIF family, functions as a
transcriptional repressor of TGF-β signaling and retinoid X
receptor (RXR) signaling (7,8).
Recent studies have demonstrated that TGIF is also involved in
Wnt/β-catenin signaling (9,10) and regulates basic energy
metabolism in cells (11).
Consistent with the critical roles of TGIF1 in the regulation of
important signaling pathways, TGIF1 has also been shown to be
involved in tumorigenesis and tumor development, such as in lung
(12), breast (13) and colorectal (14) cancer. Wang et al reported
that the knockdown of TGIF inhibited the proliferation of EC-109
cells (15). However, the roles
of TGIF in the carcinogenesis and metastasis of esophageal cancer
and the underlying mechanisms are not yet fully understood.
Thus, the present study aimed to investigate the
roles of TGIF1 in the growth and metastasis of esophageal cancer.
The data presented herein indicate that TGIF1 is significantly
upregulated in esophageal cancer tissues and cells, and that TGIF1
knockdown reduces the proliferation and tumorigenicity of
esophageal cancer in vivo and in vitro. Moreover, it
is demonstrated that TGIF1 functions as a potential tumor promoter
in esophageal cancer by regulating the Wnt/β-catenin and
Akt/mammaliain target of rapamycin (mTOR) signaling pathways.
Materials and methods
Cells, cell culture and transfection
Human esophageal cancer cell lines (TE-10, KYSE-150,
TE-1, kyse410 and Eca-109) and the human esophageal epithelial cell
line, Het1A, were obtained from Cell Bank of Chinese Academy of
Sciences. Cells were maintained in RPMI-1640 medium supplementing
with 10% fetal bovine serum (FBS) at 37°C. A total of 3 siRNAs
targeting TGIF1 (si-TGIF1-1#/-2#/-3#) were synthesized from
Guangzhou RiboBio Co., Ltd. and transfected (100 nM) into the cells
using Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific,
Inc.) for 48 h, and scramble siRNA sequence was used as the
negative control (si-NC). The siRNA sequences were as follows:
TGIF1-siNC, 5′-UUC UCC GAA CGU GUC ACG UTT-3′; TGIF1-siRNA1, 5′-GAA
AGA UGU CCC UUU CUC UCU-3′; TGIF1-siRNA2, 5′-CCA AAU CAG UUC ACA
AUU UCC-3′; TGIF1-siRNA3, 5′-GUG GAU UUC AGC UUC UAG UGG-3′. The
lentiviral vector overexpressing TGIF1 were established from
Shanghai GeneChem Co., Ltd. (3 µl/6-wells plates), which was
transfected for 48 h. The blank plasmid was used as the negative
control (OE-NC).
Reverse transcription-quantitative PCR
(RT-qPCR)
Total RNA was extracted from the cells using TRIzol
reagent and reverse transcribed into cDNA using the HiFiScript cDNA
Synthesis kit (CW2569M; Beijing CWBio). qPCR was performed using
the Ultra SYBR Mixture kit (CW0956;Beijing CWBio). The PCR
thermocycling conditions were as follows: 95°C for 5 min and 40
cycles of 95°C for 12 sec and 55°C for 40 sec. Primers used in the
present study were as follows: TGIF1 forward, 5′-GAC ATT CCC TTG
GAC CTT TCT-3′ and reverse, 5′-TAC AGC CAA TCC CGA AGA ATC-3′;
β-actin forward, 5′-CCC GAG CCG TGT TTC CT-3′ and reverse, 5′-GTC
CCA GTT GGT GAC GAT GC-3′. Relative gene expression was calculated
by the 2−ΔΔCq method (16).
Analysis of TGIF1 expression in
esophageal cancer
The expression data of TGIF1 was obtained from (Gene
Expression Profiling Interactive Analysis (GEPIA; http://gepia.cancer-pku.cn/index.html)
(17). A total of 182 cases of
esophageal cancer and 286 normal samples were included in the GEPIA
dataset. |log2FC| The cut-off was set to 1. The log2(TPM + 1) was
used for log-scale.
Western blot analysis
Proteins were extracted from the cells with 48 h
following transfection by using RIPA buffer (Beyotime Institute of
Biotechnology) and then quantified using a BCA Protein Assay kit
(Beijing CWBio). Proteins (30 µg) were separated by 10%
SDS-PAGE and then transferred onto PVDF membranes. Following
blocking with skimmed milk for 1 h, the membranes were incubated
with the following primary antibodies (1:1,000) overnight at 4°C:
Anti-TGIF1 (cat. no. ab220965), anti-N-cadherin (cat. no. ab18203),
anti-E-cadherin (cat. no. ab15148) and anti-Snail (cat. no.
ab53519) antibodies were obtained from Abcam. Anti-Bcl-2 (cat. no.
12789-1-AP), anti-Bax (cat. no. 50599-2-Ig), anti-cyclin 1 (cat.
no. 60186-1-Ig) and anti-GAPDH (cat. no. 60004-1-lg) antibodies
were obtained from Proteintech Group, Inc. Anti-cleaved caspase-3
(cat. no. 9661), anti-Wnt3a (cat. no. 2391), anti-β-catenin (cat.
no. 9562), anti-Akt (cat. no. 9272), anti-p-Akt (cat. no. 9271),
anti-mTOR (cat. no. 2972) and anti-p-mTOR (cat. no. 2971)
antibodies were obtained from Cell Signaling Technology, Inc. The
membranes were further incubated with HRP-conjugated secondary
antibodies (1:5,000; cat. no. S A00004-3, Proteintech Group, Inc.)
at room temperature for 1 h. Signals were developed using an ECL
kit (BioVision, Inc.). The data were analyzed using ImageJ software
(version 1.8.0; National Institutes of Health) and GraphPad Prism
software (version 7.04; GraphPad Software, Inc.).
Cell counting kit (CCK-8) assay
Following transfection for 24 h, the cells were
plated in a 96-well plate at a density of 1×103
cells/well. Cell viability was measured using a CCK-8 assay
(Beijing Solarbio Science & Technology Co., Ltd.) at 0, 24, 48
and 72 h, respectively. Prior to detection, the cells were
incubated with 10 µl CCK8 regent at 37°C for 1.5 h, and the
OD450nm value was measured.
Colony formation assay
Cells were seeded in 60-mm plates at 500 cells/plate
and cultured in RPMI-1640 medium supplementing with 10% fetal
bovine serum (FBS) for 10 days at 37°C. Following washing with PBS,
cells were stained with 0.1% crystal violet (cat. no. C0121;
Beyotime Institute of Biotechnology) 30 min at room temperature and
colonies were then counted and imaged under a light microscope
(Leica Microsystems GmbH) at ×100 magnification.
Transwell assay
Transwell chambers with Matrigel gel (BD
Biosciences) or not were used to measure cell invasion and
migration, respectively. Cells were suspended in a serum-free
medium and plated into the upper chamber (1×104
cells/well). The lower chambers were filled with 500 µl
medium containing 10% FBS. Following incubation for 24 h at 37°C,
the migrated or invaded cells were fixed with 4% paraformaldehyde
for 30 min and stained with 0.1% crystal violet for 20 min at room
temperature, which were then counted and imaged under a microscope
(Leica Microsystems GmbH; magnification, ×100).
Flow cytometric assay
Cells were incubated at 37°C in a serum-free medium
for 24 h and suspended in 1X binding buffer. Approximately
1-5×105 cells were incubated with Annexin V/FITC-PI
(Beijing Solarbio Science & Technology Co., Ltd.) for 20 min at
room temperature. The cell samples were then analyzed using a flow
cytometer (BD FACSCanto™ II; BD Biosciences), and the rate of
apoptosis was calculated using BD FACSDiva™ software (BD
Biosciences).
Tumor xenograft assay
A total of 30 female nude mice (18-22 g; 4-6 weeks
old) were purchased from Huafukang Biotechnology Co., Ltd. The
study protocols were approved by the Institutional Animal
Experimentation Committee of Shandong Cancer Hospital (Shandong,
China). All animals were housed at the Animal Care Facility of
Shandong Cancer Hospital at 25°C with a 12/12-h light/dark cycle in
a vivarium with humidified airflow, and were allowed free access to
normal chow and water during the study period. Five different cells
(5×106) in 150 µl of PBS were subcutaneously
injected into the right hind legs of each mouse. To maintain the
transfected characteristics in vivo, cholesterol-modified
TGIF1 siRNA for in vivo RNA delivery was designed and
synthesized by Guangzhou RiboBio Co., Ltd. For the delivery of
cholesterol-conjugated RNA, 10 nmol RNA in 0.1 ml saline buffer
were locally injected into the tumor mass once every 3 days for 3
weeks, as previously described (18). Five different tumor models were
established: i) si-TGIF1-1#; ii) si-TGIF1-2#; iii) si-NC; iv)
OE-NC; v) TGIF1-OE (n=5 per group). Tumor volumes were calculated
every day using a Vernier caliper (volume=length × width × width ×
0.5), as previously described (19). When the volumes of the tumors
reached approximately 100-150 mm3, the tumor growth
delay experiment was performed. As described in a previous study by
the authors (20), the relative
tumor volume (RTV; RTV = Vt/V0, where Vt is the volume of the tumor
at any given time and V0 is the initial volume before treatment)
was calculated and the growth curve was analyzed. The mice were
euthanized by pentobarbital (100 mg/kg) followed by cervical
dislocation at 20 days post-injection or when the tumor volume had
reached 2,000 mm3. Tumors were collected immediately
following the euthanasia of the mice.
Immunohistochemistry of the mouse tumor
tissue
Immunohistochemistry was performed as previously
described (21). The tumor
tissues were sectioned (4-µm-thick) and dewaxed. After
antigen retrieval was performed using 10 mmol/l citrate buffer, the
sections were incubated with 3% H2O2 and
blocked with 5% BSA at 37°C for 30 min. The related detection
protein included the primary antibodies (1:1,000) described above
and anti-Ki67 (1:500, cat. no. ab15580) obtained from Abcam, and
anti-TGIF1 (1:1,000, cat. no. orb47063) obtained from Biorbyt. The
sections were then incubated with primary antibodies at 4°C
overnight. After rewarming at room temperature the following day,
the sections were incubated with secondary antibody (1:200; cat.
no. A-21442; Thermo Fisher Scientific, Inc.) using the two-step
polymer HRP (cat. no. PV-9005; OriGene Technologies, Inc.)
detection system at room temperature for 1 h. The samples were
visualized using 3,3-diaminobenzidine. Images were acquired using a
light microscope (Leica Microsystems GmbH; magnification,
×100).
Statistical analysis
In the present study, data are presented as the
means ± SD from 3 independent experiments and statistically
analyzed using GraphPad Prism7 software. Statistically significant
differences between groups were analyzed using a Student's t-test
or one-way ANOVA followed by Bonferroni's post hoc test. A value of
P<0.05 was considered to indicate a statistically significant
difference.
Results
TGIF1 increases the proliferation of
esophageal cancer cells
The present study first investigated TGIF1 mRNA
expression in esophageal cancer by Gene Expression Profiling
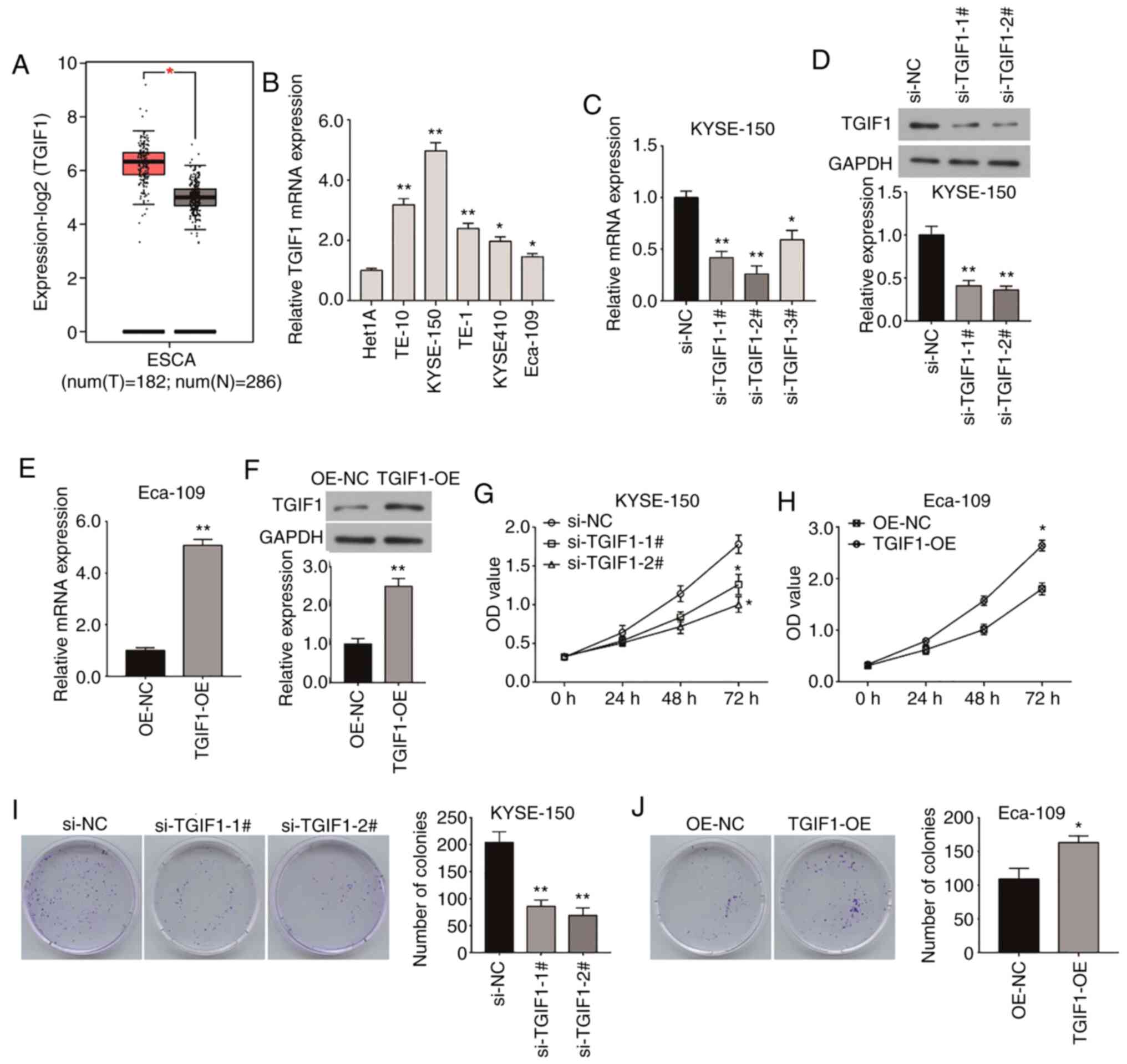
Interactive Analysis. As shown in Fig. 1A, the mRNA expression of TGIF1 in
esophageal cancer tissues (n=182) was higher than that in normal
tissues (n=286). Moreover, RT-qPCR was performed to examine the
mRNA expression of TGIF1 in 5 esophageal cancer cell lines (TE-10,
KYSE-150, TE-1, kyse410 and Eca-109) and the human esophageal
epithelial cell line, Het1A. The results revealed that compared
with the Het1A cells, the mRNA expression of TGIF1 was
significantly upregulated in all esophageal cancer cell lines
(Fig. 1B). Thus, these data
indicated that TGIF1 was upregulated in esophageal cancer tissues
and cells.
To investigate TGIF1 function in the progression of
esophageal cancer, loss-of-function and gain-of-function
experiments were performed. As shown in Fig. 1C, TGIF1 mRNA expression was
significantly knocked down by 3 different siRNA-TGIF1 sequences
(si-TGIF1-1#, -2# and -3#) in the KYSE-150 cells, which exhibited
the highest TGIF1 expression; si-TGIF1-1#, and si-TGIF1-2# were
used in the following experiments due to their greater knockdown
efficiency. Accordingly, the TGIF1 protein level was also
significantly suppressed by si-TGIF1-1# or si-TGIF1-2# (Fig. 1D). As the Eca-109 cells exhibited
the lowest expression of TGIF1, they were transfected with
pcDNA3.1-TGF1 to upregulate its expression at both the mRNA and
protein level (Fig. 1E and F).
Subsequently, CCK-8 and colony formation assays were performed to
examine the proliferative ability of the 2 esophageal cancer cells
following transfection with si-TGIF1 or pcDNA3.1-TGF1 (Fig. 1G-J). As shown in Fig. 1G, the silencing of TGIF1 markedly
decreased the viability of the KYSE-150 cells when compared with
the NC group. Notably, TGIF1 overexpression significantly enhanced
the viability of the Eca-109 cells (Fig. 1H). Moreover, the silencing of
TGIF1 significantly decreased the colony-forming ability of the
KYSE-150 cells, which was increased by TGIF1 overexpression in the
Eca-109 cells (Fig. 1I and
J).
TGIF1 enhances the migratory and invasive
abilities of esophageal cancer cells
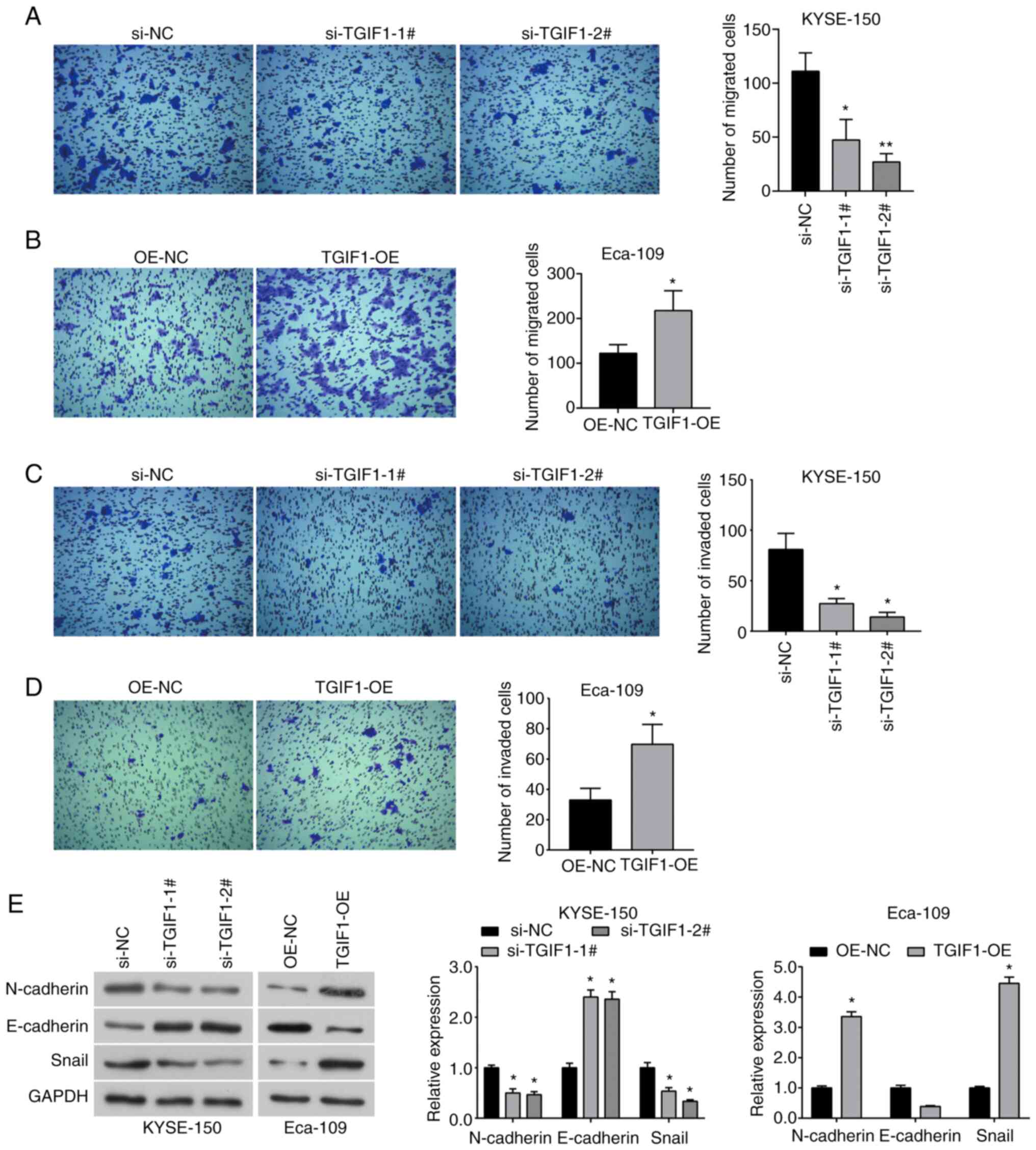
Transwell assay was conducted to detect the
preliminarily effect of TGIF1 on the metastatic capacity of
esophageal cancer cells. The data demonstrated that the silencing
of TGIF1 significantly decreased the migratory ability of the
KYSE-150 cells (Fig. 2A), while
the upregulation of TGIF1 enhanced the migratory ability of the
Eca-109 cells (Fig. 2B).
Correspondingly, the invasive ability of the KYSE-150 cells was
also suppressed by TGIF1 knockdown (Fig. 2C), while in the Eca-109 cells in
which TGIF1 was overexpressed, the opposite results were observed
(Fig. 2D). Furthermore, western
blot analysis was used to examine the expression of
epithelial-mesenchymal transition (EMT)-related proteins in
esophageal cancer cells. The results indicated that the expression
of N-cadherin and Snail was significantly decreased in the KYSE-150
cells transfected with si-TGIF1, while E-cadherin expression was
upregulated (Fig. 2E); however,
the expression of the above-mentioned proteins exhibited an
opposite trend in the TGIF1-overexpressing cells (Fig. 2E). Therefore, it was demonstrated
that TGIF1 can enhance the metastatic potential of esophageal
cancer cells by regulating the EMT process.
TGIF1 impairs the apoptosis of esophageal
cancer cells
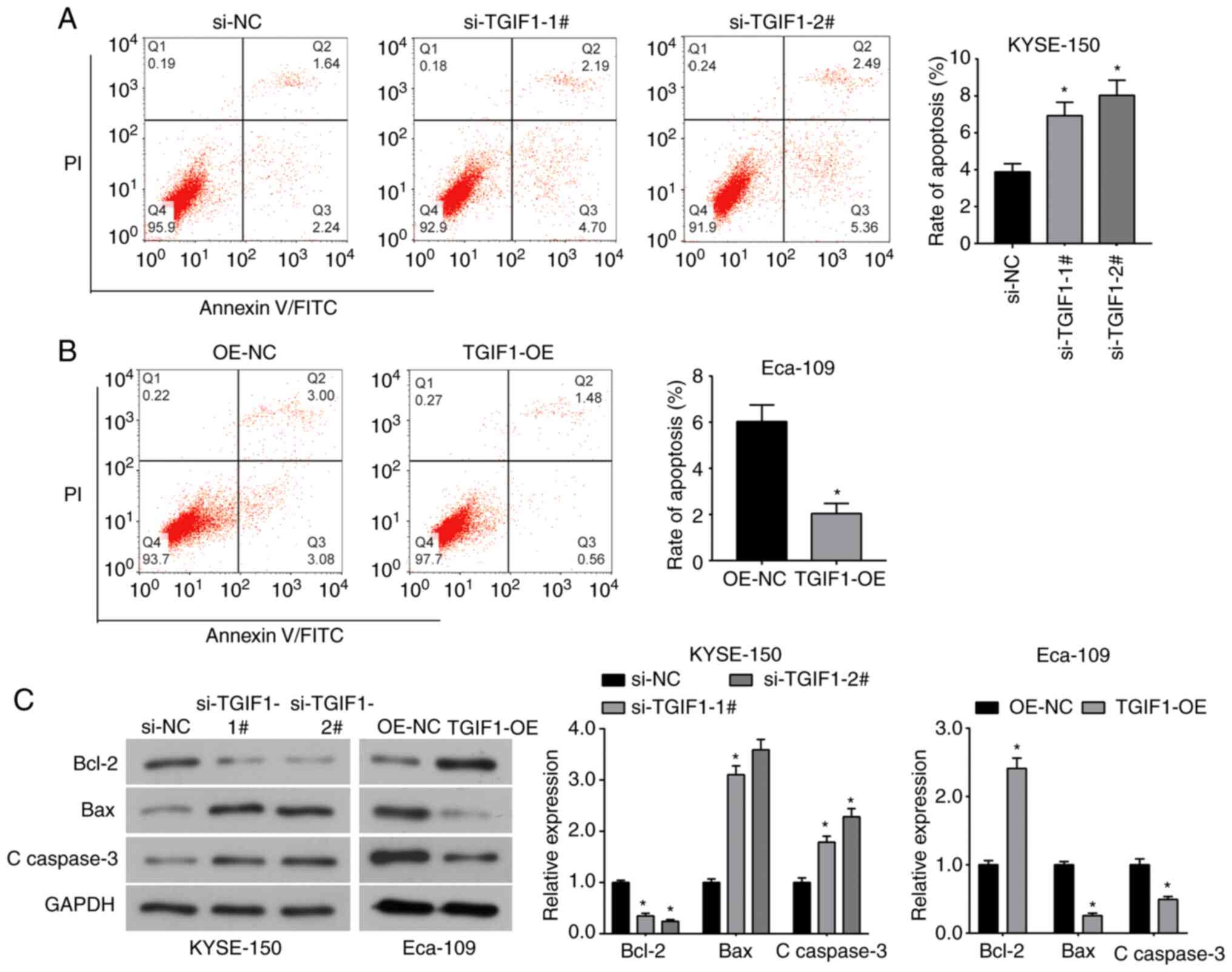
Flow cytometric assay indicated that compared with
the control group, the percentage of apoptotic KYSE-150 cells was
significantly increased by si-TGIF1 (Fig. 3A), while the percentage of
apoptotic Eca-109 cells was significantly decreased by TGIF1
overexpression (Fig. 3B).
Furthermore, western blot analysis revealed that the expression of
the anti-apoptotic protein, Bcl-2, was significantly downregulated
by si-TGIF1, whereas it was upregulated by TGIF1 overexpression.
However, the expression of the pro-apoptotic proteins, Bax and
cleaved caspase-3, was upregulated in the TGIF1-silenced KYSE-150
cells, and decreased in TGIF1-overexpressing Eca-109 cells
(Fig. 3C). Taken together, these
results revealed that the silencing of TGIF1 induced the apoptosis
of esophageal cancer cells by regulating the Bcl-2/Bax axis and
caspase-3 activation.
TGIF1 promotes the activation of
Wnt/β-catenin and Akt/mTOR signaling pathways in esophageal cancer
cells
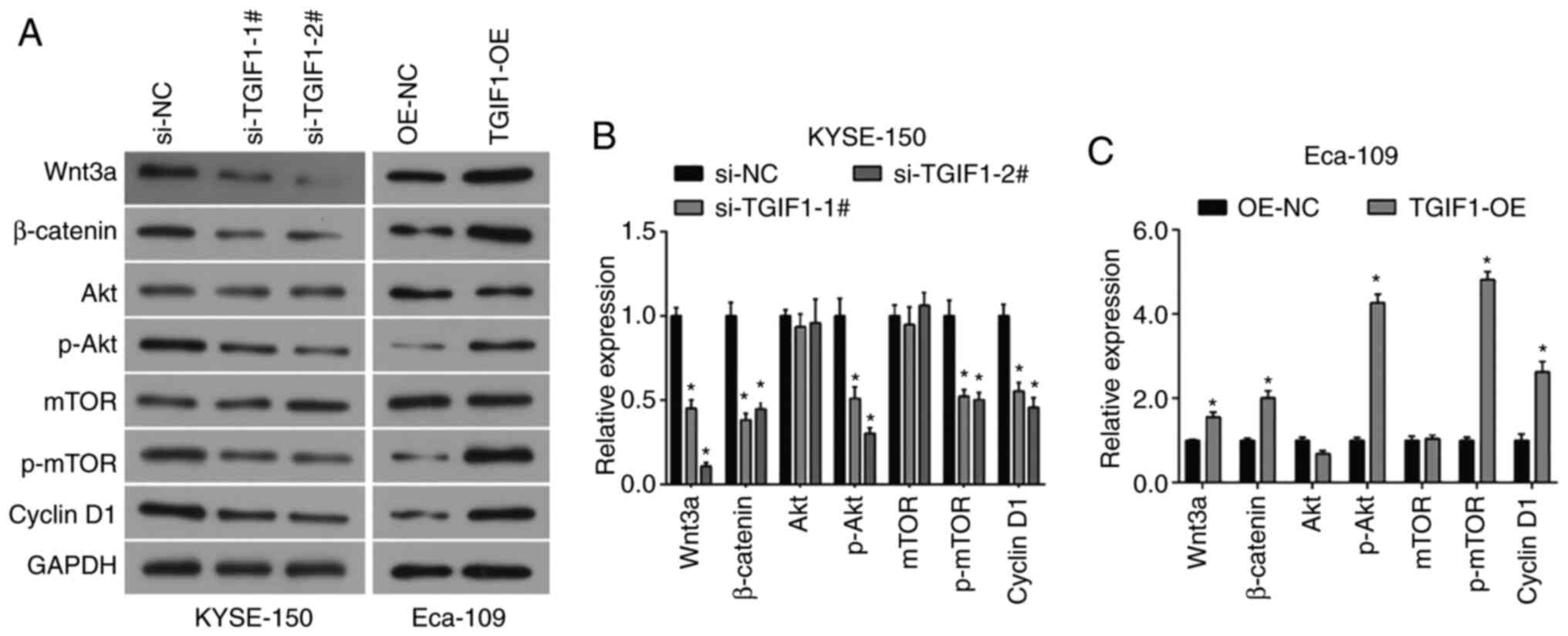
It has been reported that TGIF1 can promote Wnt
signaling in breast cancer cells (10). Herein, as shown in Fig. 4, it was found that the silencing
TGIF1 inhibited the expression of Wnt3a and β-catenin, which was
upregulated in TGIF1-overexpressing cells, suggesting that TGIF1
also promoted Wnt/β-catenin signaling in esophageal cancer cells.
Additionally, it was observed that Akt/mTOR, another classical
signaling pathway involved in cellular functions, was also affected
by TGIF1 in esophageal cancer cells (Fig. 4A-C). TGIF1 knockdown suppressed
the phosphorylation of Akt, and mTOR and cyclin D1 expression in
the KYSE-150 cells, while TGIF1 overexpression increased the
expression of these proteins in the Eca-109 cells (Fig. 4), suggesting that TGIF1 cab
promote the activation of the Akt/mTOR signaling pathway in
esophageal cancer cells and that the knockdown TGIF1 arrested the
cell cycle in the G1 phase. The ratio of phosphorylated to total
protein is presented in Fig.
S1.
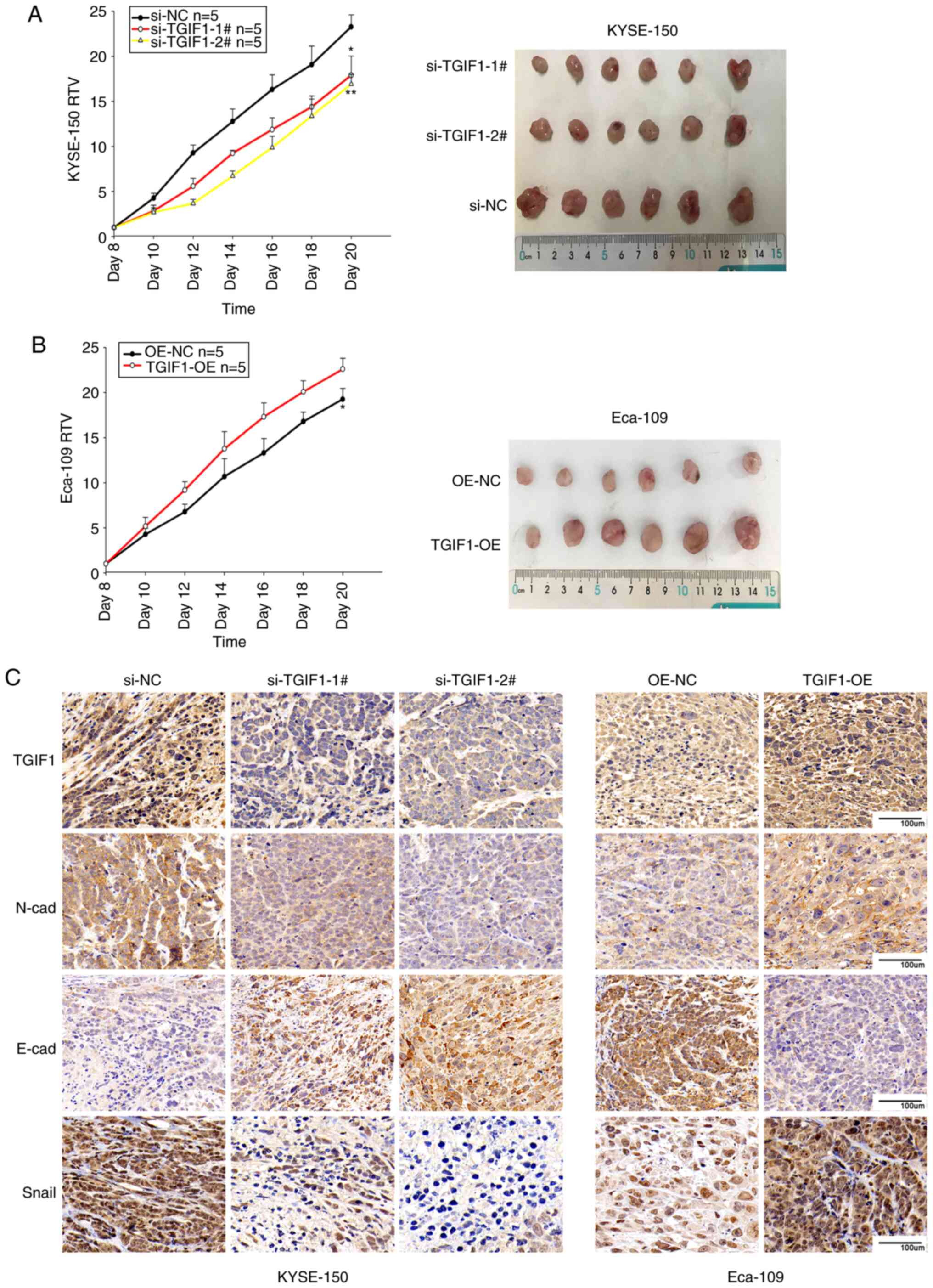
Effects of TGIF1 on the growth of
esophageal cancer in vivo
To verify the effects of TGIF1 on esophageal cancer
in vivo, KYSE-150 (si-NC, si-TGIF1-1#, si-TGIF1-2#) and
Eca-109 (OE-NC,TGIF1-OE) subcutaneous xenograft tumor models were
established using BALB/c nude mice and the relative tumor volume
was measured. Tumor growth was found to be markedly attenuated in
the mice injected with the KYSE-150 cells and TGIF1 siRNA
(si-TGIF1-1#, si-TGIF1-2#) compared with those injected with
KYSE-150 cells and the negative control siRNA (si-NC; si-TGIF1-1#
vs. si-NC group, P<0.05; si-TGIF1-2# vs. si-NC group, P<0.01;
Fig. 5A). Of note, the RTV of
the Eca-109-derived tumors overexpressing TGIF1 (TGIF1-OE) was
larger than that of the Eca-109-derived tumors injected with the
negative control (OE-NC; P<0.05; Fig. 5B). These data suggested that
TGIF1 knockdown suppressed tumor formation and tumor growth in
vivo, while the upregulation of TGIF1 enhanced tumor growth
in vivo.
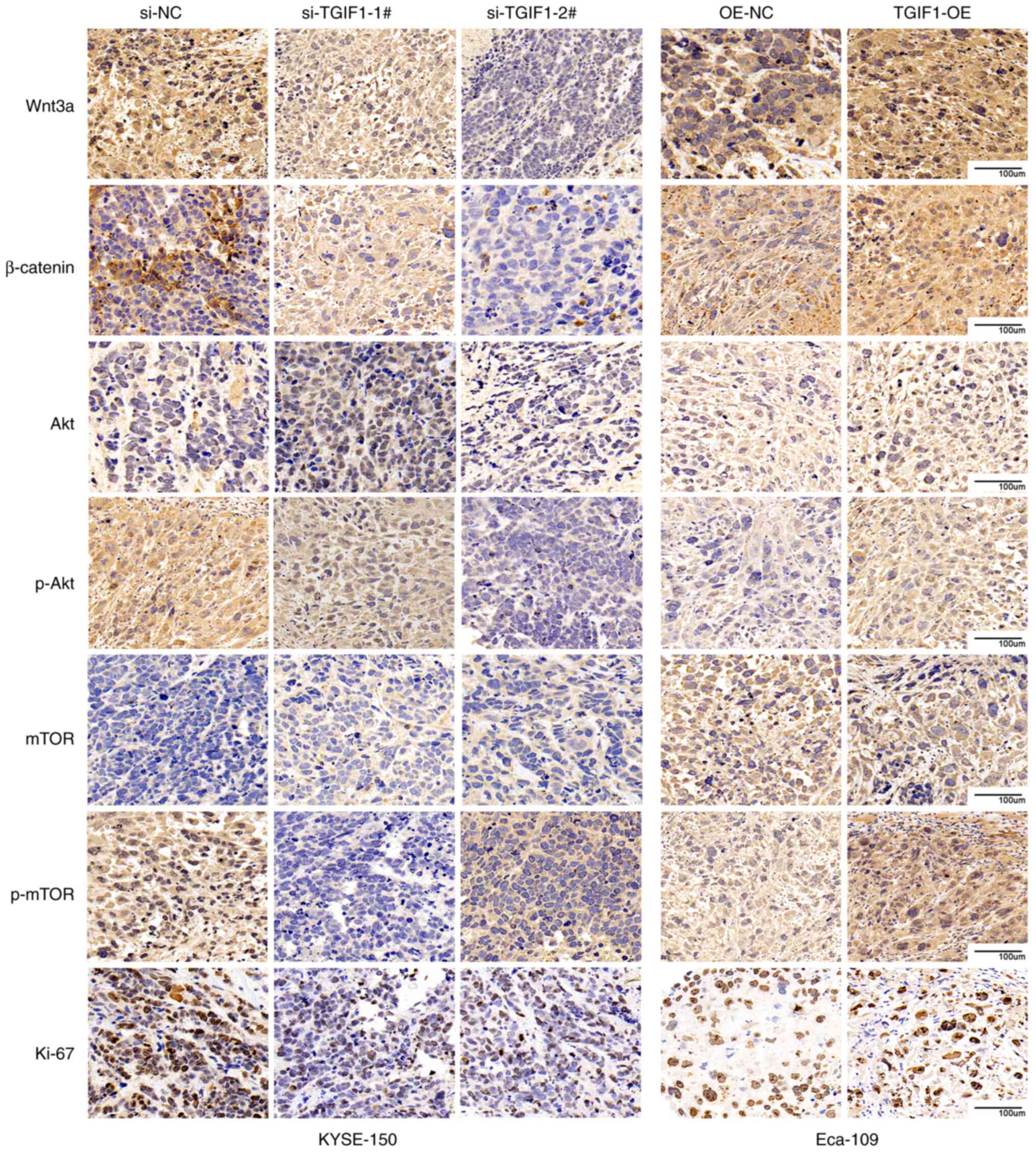
TGIF1 affects proliferation-related genes
and pathways in vivo
The expression levels of TGIF1, E-cadherin,
N-cadherin, Snail, Wnt/β-catenin, Akt/mTOR and Ki-67 in the tumor
xenografts were detected by immunohistochemical staining. As shown
in Figs. 5C and S2, the TGIF1, Snail and N-cadherin
expression levels were downregulated by si-TGIF1 in the
KYSE-150-derived tumor xenografts, whereas these were upregulated
by TGIF1 overexpression in the Eca-109-derived tumor xenografts.
The expression of E-cadherin was upregulated in the TGIF1-silenced
KYSE-150-derived tumors, and was decreased in the
TGIF1-overexpressing Eca-109-derived tumors. Additionally, as shown
in Figs. 6 and S3, the expression levels of Wnt3a and
β-catenin were inhibited by the silencing of TGIF1, whereas they
were upregulated by TGIF1 overexpression. TGIF1 knockdown
suppressed the phosphorylation of Akt/mTOR and Ki-67 expression in
the KYSE-150-deriged tumors, while TGIF1 overexpression increased
the expression of these proteins in the Eca-109-derived tumors. The
downregulation of p-mTOR by the knockdown of TGIF1 may be the key
factor of the classical Akt/mTOR signaling pathway. These results
were consistent with the results obtained in vitro and
confirm that TGIF1 increases the proliferation of esophageal cancer
cells via the Wnt/β-catenin and Akt/mTOR signaling pathways, and
promotes EMT in vivo.
Discussion
Previous studies have reported that TGIF1 plays a
critical role in tumor initiation and progression, and its
overexpression is associated with a poor prognosis of patients with
colorectal (10,11), lung (12), gastric (22) and breast (13,23) cancer. The study by Wang et
al demonstrated that TGIF1 was upregulated in colorectal cancer
and functioned as an oncogene, promoting cancer cell proliferation
and migration (14). Another
study by Wang et al reported that TGIF1 knockdown suppressed
the migration and invasion of breast cancer cells (13). The study by Haider et al
further demonstrated that the lack of TGIF1 also restricted the
progression of breast cancer bone metastases (23). Inconsistent with the oncogenic
role of TGIF1 in tumorigenesis and development reported in previous
studies, Parajuli et al revealed that TGIF1 funcioned as a
tumor suppressor in pancreatic ductal adenocarcinoma (24). Weng et al also found that
the loss of TGIF1 induced the development of pancreatic cancer
(25). The reason for the
inconsistency with other cancer types was that TGIF1 is involved in
different pathways in pancreatic cancer. It has been reported that
the silencing of TGIF inhibits the proliferation and tumorigenicity
of EC109 cells (15). However,
the role of TGIF1 in the metastasis of esophageal cancer and the
underlying mechanisms remain elusive.
Compared with the study by Wang et al
(15), the present study used
different esophageal cancer cell lines and constructed an TGIF1
overexpression model. Furthermore, the present study verified more
functions of TGIF1 as an oncogene and explored the possible
mechanisms. Herein, it was found that TGIF1 was upregulated in
esophageal cancer tissues and cell lines. Importantly, the data
demonstrated that the silencing of TGIF1 significantly inhibited
the proliferation and colony-forming capabilities of the KYSE-150
cells, while TGIF1 overexpression resulted in an opposite phenotype
in the Eca-109 cells, which was consistent with the findings of a
previous study (15). Moreover,
it was found that TGIF1 reduced apoptosis of esophageal cancer
cells by regulating the Bcl-2/Bax axis and caspase-3 activation.
EMT plays a pivotal role in tumor metastasis proven by the
upregulated expression of N-cadherin, while the expression level of
E-cadherin is downregulated (26-30). Furthermore, the present study
demonstrated that TGIF1 increased the migratory and invasive
capabilities of esophageal cancer cells and regulated the
expression of EMT biomarkers; TGIF1 knockdown downregulated the
N-cadherin and Snail expression, and upregulated E-cadherin
expression in KYSE-150 cells and tumors, indicating that TGIF1 may
increase the metastatic potential of esophageal cancer cells by
regulating the EMT process. These findings suggest thatTGIF1
functions as an oncogene in the growth and metastasis of esophageal
cancer.
In addition to its well-known ability to suppress
TGF-β signaling, TGIF1 has also been confirmed to activate Wnt
signaling in breast, colorectal and lung cancer cells (12,14). The Wnt/β-catenin signaling
pathway is involved in various biological processes, such as cell
proliferation, movement, differentiation and cell death (31-34), which can regulate target gene
transcription by β-catenin entering the cell nucleus (35,36). Zhang et al demonstrated
that TGIF promotes β-catenin abundance by diverting Axin1/2 from
the β-catenin destruction complex, and was thus involved in
Wnt1-induced mammary tumor formation (10). Wang et al reported that
TGIF1 promoted the proliferation of colorectal cancer cells by
activating Wnt/β-catenin signaling via its DNA binding ability and
interaction with β-catenin (14). Based on these studies, it was
hypothesized that the Wnt/β-catenin signaling pathway may play a
tumor-promoting role with TGIF1 in esophageal cancer. Herein, it
was found that Wnt3a and β-catenin expression was significantly
downregulated in cells and tumors in which TGIF1 was knocked down,
whereas it was upregulated by TGIF1 overexpression, indicating that
TGIF1 may promote the progression of esophageal cancer partly by
activating the Wnt/β-catenin signaling pathway. Additionally, the
activation of the Akt/mTOR signaling pathway was also upregulated
by TGIF1, by increasing the p-Akt and p-mTOR levels, suggesting
that the Akt/mTOR signaling pathway may be involved in the
tumor-promoting effect of TGIF1. This warrants further exploration
in future studies.
Some limitations of the present study should be
acknowledged. It is known that the incidence of esophageal
adenocarcinoma equals or exceeds the incidence of esophageal
squamous cell carcinoma in the US and other western countries.
Therefore, one limitation of the present study is that esophageal
adenocarcinoma cell lines were not been used. Second, the effects
of the knockdown of TGIF1 on the cell cycle should be further
investigated in future studies. Third, the effects of TGIF
overexpression on the related pathways need to be further explored.
Moreover, a transgenic animal model could be used to assess the
functional role of TGIF in esophageal tumorigenesis.
In conclusion, the present study demonstrated that
the silencing of TGIF1 suppressed esophageal cancer cell
proliferation, migration and invasion, and promoted apoptosis,
suggesting that TGIF1 plays oncogenic role in the progression of
esophageal cancer. Moreover, the Wnt/β-catenin and Akt/mTOR
signaling pathways may be involved in the tumor-promoting effects
of TGIF1. Thus, TGIF1 may be a therapeutic target for the treatment
of esophageal cancer.
Supplementary Data
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
BL designed the study and revised the draft of the
manuscript. YY and LK completed the main experiments and drafted
the manuscript. HG created the figures and performed the
statistical analyses. XL performed the histological examination of
the tumors. WH assisted in the design of the study. FS and LJ
analyzed the data and revised the manuscript. All authors read and
approved the final manuscript.
Ethics approval and consent to
participate
The animal experiments were approved by the Animal
Ethics Committee at the Shandong Cancer Hospital Affiliated to
Shandong University (Jinan, China).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgments
Not applicable.
Funding
The present study was supported by grants from the Academic
promotion program of Shandong First Medical University (no.
2020RC002), the National Natural Science Foundation of China (nos.
81530060, 81874224 and 81671785), the National Key Research and
Development Program of China (no. 2016YFC0105106), the Key Research
and Development Project of Shandong Provincial (no. 2016GSF201123),
and the Foundation of Taishan Scholars (nos. ts20120505,
tsqn201909187 and tsqn201909140).
References
|
1
|
Bollschweiler E, Plum P, Mönig SP and
Hölscher AH: Current and future treatment options for esophageal
cancer in the elderly. Expert Opin Pharmacother. 18:1001–1010.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Simard EP, Ward EM, Siegel R and Jemal A:
Cancers with increasing incidence trends in the United States: 1999
through 2008. CA Cancer J Clin. 62:118–128. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Huang FL and Yu SJ: Esophageal cancer:
Risk factors, genetic association, and treatment. Asian J Surg.
41:210–215. 2018. View Article : Google Scholar
|
|
5
|
He X, Meng F, Qin L, Liu Z, Zhu X, Yu Z
and Zheng Y: KLK11 suppresses cellular proliferation via inhibition
of Wnt/β-catenin signaling pathway in esophageal squamous cell
carcinoma. Am J Cancer Res. 9:2264–2277. 2019.
|
|
6
|
Deng H, Shi H, Chen L, Zhou Y and Jiang J:
Over-expression of Nectin-4 promotes progression of esophageal
cancer and correlates with poor prognosis of the patients. Cancer
Cell Int. 19:1062019. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Hneino M, François A, Buard V, Tarlet G,
Abderrahmani R, Blirando K, Hoodless P, Benderitter M and Milliat
F: The TGF-β/Smad repressor TG-Interacting factor 1 (TGIF1) plays a
role in radiation-induced intestinal injury independently of a Smad
signaling pathway. PLoS One. 7:e356722012. View Article : Google Scholar
|
|
8
|
Guca E, Suñol D, Ruiz L, Konkol A, Cordero
J, Torner C, Aragon E, Martin-Malpartida P, Riera A and Macias MJ:
TGIF1 homeodomain interacts with Smad MH1 domain and represses
TGF-β signaling. Nucleic Acids Res. 46:9220–9235. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Razzaque MS and Atfi A: TGIF function in
oncogenic Wnt signaling. Biochim Biophys Acta. 1865:101–104.
2016.
|
|
10
|
Zhang MZ, Ferrigno O, Wang Z, Ohnishi M,
Prunier C, Levy L, Razzaque M, Horne WC, Romero D, Tzivion G, et
al: TGIF governs a feed-forward network that empowers Wnt signaling
to drive mammary tumorigenesis. Cancer Cell. 27:547–560. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Shah A, Melhuish TA, Fox TE, Frierson HF
Jr and Wotton D: TGIF transcription factors repress acetyl CoA
metabolic gene expression and promote intestinal tumor growth.
Genes Dev. 33:388–402. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Xiang G, Yi Y, Weiwei H and Weiming W:
TGIF1 promoted the growth and migration of cancer cells in nonsmall
cell lung cancer. Tumour Biol. 36:9303–9310. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wang Y, Li L, Wang H, Li J and Yang H:
Silencing TGIF suppresses migration, invasion and metastasis of
MDA-MB-231 human breast cancer cells. Oncol Rep. 39:802–808.
2018.
|
|
14
|
Wang JL, Qi Z, Li YH, Zhao HM, Chen YG and
Fu W: TGFβ induced factor homeobox 1 promotes colorectal cancer
development through activating Wnt/β-catenin signaling. Oncotarget.
8:70214–70225. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wang Y, Pan T, Li L, Wang H, Li J, Zhang D
and Yang H: Knockdown of TGIF attenuates the proliferation and
tumorigenicity of EC109 cells and promotes cisplatin-induced
apoptosis. Oncol Lett. 14:6519–6524. 2017.
|
|
16
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
17
|
Tang Z, Li C, Kang B, Gao G, Li C and
Zhang Z: GEPIA: A web server for cancer and normal gene expression
profiling and interactive analyses. Nucleic Acids Res. 45:W98–W102.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Sang LJ, Ju HQ, Liu GP, Tian T, Ma GL, Lu
YX, Liu ZX, Pan RL, Li RH, Piao HL, et al: LncRNACamK-A Regulates
Ca(2+)-signaling-mediated tumor microenvironment remodeling. Mol
Cell. 72:71–83.e7. 2018. View Article : Google Scholar
|
|
19
|
Yang T, Huang T, Zhang D, Wang M, Wu B,
Shang Y, Sattar S and Ding L: TGF-β receptor inhibitor LY2109761
enhances the radiosensitivity of gastric cancer by inactivating the
TGF-β/SMAD4 signaling pathway. Aging (Albany NY). 11:8892–8910.
2019. View Article : Google Scholar
|
|
20
|
Yu Y, Li X, Liu J, Dong M and Xing L:
Correlation of hypoxia status with radiosensitizing effects of
sodium glycididazole: A preclinical study. Oncol Lett.
15:6481–6488. 2018.PubMed/NCBI
|
|
21
|
Yu Y, Guan H, Jiang L, Li X, Xing L and
Sun X: Nimotuzumab, an EGFR-targeted antibody, promotes
radiosensitivity of recurrent esophageal squamous cell carcinoma.
Int J Onco. l56:945–956. 2020.
|
|
22
|
Jia K, Wen QH, Zhao X, Cheng JM, Cheng L
and Xi M: XTP8 stimulates migration and invasion of gastric
carcinoma through interacting with TGIF1. Eur Rev Med Pharmacol
Sci. 24:2412–2420. 2020.PubMed/NCBI
|
|
23
|
Haider MT, Saito H, Zarrer J,
Uzhunnumpuram K, Nagarajan S, Kari V, Horn-Glander M, Werner S,
Hesse E and Taipaleenmäki H: Breast cancer bone metastases are
attenuated in a Tgif1-deficient bone microenvironment. Breast
Cancer Res. 22:342020. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Parajuli P, Singh P, Wang Z, Li L,
Eragamreddi S, Ozkan S, Ferrigno O, Prunier C, Razzaque MS, Xu K
and Atfi A: TGIF1 functions as a tumor suppressor in pancreatic
ductal adenocarcinoma. EMBO J. 38:e1010672019. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Weng CC, Hsieh MJ, Wu CC, Lin YC, Shan YS,
Hung WC, Chen LT and Cheng KH: Loss of the transcriptional
repressor TGIF1 results in enhanced Kras-driven development of
pancreatic cancer. Mol Cancer. 18:962019. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Sha Y, Haensel D, Gutierrez G, Du H, Dai X
and Nie Q: Intermediate cell states in epithelial-to-mesenchymal
transition. Phys Biol. 16:0210012019. View Article : Google Scholar :
|
|
27
|
Aiello NM and Kang Y: Context-dependent
EMT programs in cancer metastasis. J Exp Med. 216:1016–1026. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Campbell K: Contribution of
epithelial-mesenchymal transitions to organogenesis and cancer
metastasis. Curr Opin Cell Biol. 55:30–35. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Van Staalduinen J, Baker D, Ten Dijke P
and Van Dam H: Epithelial-mesenchymal-transition-inducing
transcription factors: New targets for tackling chemoresistance in
cancer? Oncogene. 37:6195–6211. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Kalluri R: EMT: When epithelial cells
decide to become mesenchymal-like cells. J Clin Invest.
119:1417–1419. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Clevers H: Wnt/beta-catenin signaling in
development and disease. Cell. 127(Pt 3): 469–480. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Hoppler S and Kavanagh CL: Wnt signalling:
Variety at the core. J Cell Sci. 120:385–393. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
MacDonald BT, Tamai K and He X:
Wnt/beta-catenin signaling: Components, mechanisms, and diseases.
Dev Cell. 17:9–26. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Clevers H and Nusse R: Wnt/beta-catenin
signaling and disease. Cell. 149:1192–1205. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Yang Z, Shah K, Busby T, Giles K,
Khodadadi-Jamayran A, Li W and Jiang H: Hijacking a key chromatin
modulator creates epigenetic vulnerability for MYC-driven cancer. J
Clin Invest. 128:3605–3618. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Dejure FR and Eilers M: MYC and tumor
metabolism: Chicken and egg. EMBO J. 36:3409–3420. 2017. View Article : Google Scholar : PubMed/NCBI
|