Introduction
Malignant pleural mesothelioma (MPM), resulting from
the malignant transformation of pleura mesothelial cells, is an
aggressive, therapy-resistant and mostly fatal cancer strongly
associated with the presence of asbestos fibres (1-3).
Other factors or etiological agents have also been associated with
mesothelioma and among these, the fibrous mineral, erionite,
radiations and Simian Virus 40 are considered the most noteworthy
(1,2,4-8).
The global incidence of MPM has increased over the past decade and
has been predicted to peak sometime before 2030 (1,3).
The long latency period between asbestos exposure and tumour
development implies that multiple, and likely diverse, genetic
alterations are required for the malignant transformation of
mesothelial cells; however, the genetic and epigenetic events
responsible for the development of MPM remain unclear.
Mesothelial cells are relatively undifferentiated
and maintain the ability to differentiate into several cell lines.
Accordingly, some mesotheliomas have an epithelial morphology,
others, termed sarcomatoid, have a spindle cell morphology and
others, termed biphasic mesotheliomas, exhibit a combination of the
two (9). It has long been known
that survival is influenced by the histological subtype: The
epithelial variants are less aggressive than the spindle cell
variants (10,11). Patients with the disease have a
very poor prognosis with a median survival of 4 to 14 months
following diagnosis and a reduced quality of life (11-13). Diagnosis often occurs in the
advanced stage and the disease is often refractory to conventional
therapy. Despite advancements being made in current treatments, the
long-term survival rate of patients has not markedly improved and
this unsatisfactory clinical outcome emphasizes an urgent need for
the development of novel therapeutic approaches to more effectively
manage this lethal disease (1-3).
The research of novel diagnostic or therapeutic
targets for mesothelioma has included the study of microRNAs
(miRNAs or miRs) (14-17). miRNAs are small non-coding RNAs,
which regulate the expression of targeted genes by directly binding
to the 3′-untranslated region (3′-UTR) of mRNAs. This interaction
leads to post-transcriptional repression, resulting in reduced
levels of the corresponding protein or the cleavage of their RNA.
miRNAs can regulate their target genes by imperfect base-pairing to
the 3′-UTR; thus, a single miRNA can target several hundred mRNAs.
Moreover, a single target gene often includes more binding sites
for multiple miRNAs that can bind co-operatively, allowing miRNAs
to form a complex regulatory control network (18).
miRNA expression is widely tissue- and cell-type
specific. miRNAs function as regulators in a number of cellular
processes from basic metabolic maintenance, through
differentiation, cell cycle and proliferation, to death and
consequently, in tumorigenesis, cancer metastasis and drug
resistance (14). A number of
miRNAs involved in oncogenic pathways are downregulated in
mesothelioma, and this is largely due to chromosomal aberrations.
It has thus been hypothesized that the re-expression or the
enhanced expression of these miRNAs in mesothelioma cells may
influence important functions, such as proliferation, migration,
invasion, apoptosis, or chemoresistance (17) and may enhance therapeutic
outcomes (15,16). This approach may also be an
opportunity to personalise such a treatment for each patient's
miRNA tumour profile.
The aberrant expression of miR-486-5p (hereafter
referred to as miR-486) has been observed in different types of
human cancer (19) such as
hepatocellular carcinoma (20,21), lung cancer (22,23), breast cancer (24), oesophageal squamous cell
carcinoma (25) and pancreatic
cancer (26). Its hypoexpression
seems to promote the progression of lung, oesophagus and breast
cancer, while it is usually upregulated in pancreatic tumours,
chronic myeloid leukemia and gliomas (27). It was therefore proposed and
applied as an effective biomarker for the diagnosis, as well as the
prognosis of human cancer (27).
In a previous study, the authors aimed to identify a
pattern of miRNAs as possible diagnostic and prognostic biomarkers
for MPM and asbestosis; all investigated miRNAs, including miR-486,
were downregulated in the plasma of patients with respect to the
healthy subjects (28). Given
the low expression of miR-486 in mesothelioma, it was hypothesized
that miR-486 may act as a tumour suppressor, targeting several
genes. Out of these genes, particular attention was paid to the
Provirus integration site for Moloney murine leukaemia virus 1
(PIM1), for its high expression in human cancers (29-32) and in mesothelioma cell lines
(33), and as PIM1 is a
target of miR-486 (34,35). PIM1 is a serine/threonine
kinase that acts as protooncogene to mediate cell survival and has
been associated with carcinogenesis by promoting tumour cell
proliferation and inhibiting apoptosis.
The present study therefore aimed to restore high
levels of miR-486 in mesothelioma cell lines by miRNA mimic
transfection in order to investigate its regulatory functions and
to evaluate the possible effects of its overexpression on cell
proliferation, cell cycle, apoptosis and on the modulation of
sensitivity to cisplatin (CDDP). CDDP is thus far the only therapy
approved against MPM and miR-486 seems to enhance cell sensitivity
to this drug (36-38).
Materials and methods
Cells, cell culture and treatments
The human mesothelioma cell lines, H28 (CRL 5820
lot. 59191373) and H2052 (CRL 5915 lot. 63445445), were obtained
from the American Type Culture Collection (ATCC) and maintained as
a monolayer culture in the Roswell Park Memorial Institute
(RPMI)-1640 nutrient medium supplemented with 10% foetal bovine
serum (Euroclone S.p.A.) at 37°C in 5% CO2 in a
humidified air atmosphere. Only cells in the logarithmic phase of
growth were used in the experiments and allowed to attach for 24 h
prior to transfection or treatments. For CDDP treatment, cells were
incubated with various concentrations of the drug (0-100 µM)
for 24, 48 or 72 h. An untreated sample was used as a control.
After establishing the IC50 value for the different
lines, this concentration was used in subsequent treatments, where
not otherwise indicated.
miRNA transfection
mirVana™ miR-486-mimic and control-mimic were
designed and synthesized by Life Technologies; Thermo Fisher
Scientific, Inc. Cells were transfected with miR-mimic diluted with
Opti-MEM I reduced serum medium (Gibco-Life Technologies; Thermo
Fisher Scientific, Inc.) using Lipofectamine 2000 (Invitrogen;
Thermo Fisher Scientific, Inc.), according to the manufacturer's
protocol, at the final concentrations of 50 or 10 nM. Cells were
seeded at the number suggested in the manufacturer's protocol for
the different plates. After 24, 48 or 72 h, total RNA was extracted
and the transfection efficiency was verified by reverse
transcription-quantitative PCR (RT-qPCR).
RNA extraction
RNA was isolated from the cultured cells using
TRIzol reagent (Thermo Fisher Scientific, Inc.) according to the
manufacturer's instructions. The extracted RNA was digested using
the DNase I (DNA-free kit; Thermo Fisher Scientific, Inc.) to
remove any genomic DNA contamination and it was quantified using a
NanoDrop spectrophotometer (Thermo Fisher Scientific, Inc.).
miRNA quantification
Total RNA was reverse transcribed using a TaqMan
MicroRNA RT kit (Thermo Fisher Scientific, Inc.), according to the
manufacturer's instruction. The reaction included 3 µl of
stem-loop RT primer 50 nM, 1.5 µl of 10X RT buffer, 0.15
µl of dNTPs 100 mM, 0.19 µl RNase Inhibitor 20
U/µl, 1 µl of MultiScribe reverse transcriptase 50
U/µl and 5 µl of RNA sample in a total volume of 15
µl. The retrotranscription program included a cycle of 30
min at 16°C, a cycle of 30 min at 42°C and a cycle of 5 min at
85°C. Quantitative PCR (qPCR) was performed using the QuantStudio 7
Flex Real-Time PCR System (Applied Biosystems; Thermo Fisher
Scientific, Inc.). A total of 1.33 µl of cDNA solutions were
amplified using TaqMan 2X Universal PCR Master Mix (Life
Technologies; Thermo Fisher Scientific, Inc.) and TaqMan microRNA
assays [(miRNA-486-5p:ID 001268), Thermo Fisher Scientific, Inc.]
in 20 µl of mixture. The reaction consisted of one step at
95°C for 10 min, followed by 40 cycles of 95°C for 15 sec and 60°C
for 1 min. All assays were performed in duplicate, and one
no-template and two interpolate controls were used in each
experiment. The Cq values of the target miRNAs were normalized to
sno-RNU6B [(TaqMan microRNA Assays, ID 001093), Thermo Fisher
Scientific, Inc.] and the fold changes in expression of each miRNA
were calculated using 2−ΔΔCq method (39).
Gene expression
cDNA was synthesized using a commercial kit based on
the use of inverse transcriptase, [High-Capacity RNA-to-cDNA™ kit
(Applied Biosystems; Thermo Fisher Scientific, Inc.)], following
the manufacturer's recommended experimental conditions. RT-qPCR was
performed using the QuantStudio 7 Flex Real-Time PCR System (Thermo
Fisher Scientific, Inc.) employing TaqMan 2X Universal PCR Master
Mix (Life Technologies; Thermo Fisher Scientific, Inc.) and the
following TaqMan gene expression assays (Thermo Fisher Scientific,
Inc.): CCND1 (Hs00765553_m1), CCNE1 (Hs01026536_m1),
CDK4 (Hs00364847_m1) and PIM1 (Hs01065498_m1). The
reactions consisted of one step at 95°C for 10 min, followed by 40
cycles of 95°C for 15 sec and 60°C for 1 min. All assays were
performed in duplicate, and one no-template and two interpolate
controls were used in each experiment. The expression values of
each mRNAs were normalized to the expression of the GAPDH
housekeeping gene [(Hs02758991_g1, Thermo Fisher Scientific, Inc.].
The changes in the expression of each mRNA with respect to the
untreated controls were calculated using the 2−ΔΔCq
method (39).
Cell viability assay
Cells were seeded in 96-well plates (5,000-10,000
cells/well) and incubated at 37°C overnight, and then transfected
with miR-486 mimic or negative control miRNA, respectively,
according to the manufacturer's protocol (Invitrogen; Thermo Fisher
Scientific, Inc.). The proliferation of MPM cells was examined by
3-(4,5-dimethylthiazole)-2, 5-diphenyltetrazoliumbromide (MTT)
assay (Sigma-Aldrich; Merck KGaA). At the fixed time points, 10
µl of MTT reagent (5 mg/ml) was added into each well prior
to incubation at 37°C for 3 h. The reaction was terminated by the
addition of 100 µl of solubilization solution and the
absorbance was recorded using a Multiscan Ascent plate reader
(Thermo Labsystems) at a wavelength of 560 nm. The number of viable
cells was determined using a calibration curve, consisting of a
decreasing number of cells and confirmed by counting viable cells
in a haemocytometer (trypan blue exclusion). All measurements were
performed at least in triplicate.
Cell cycle analysis
Cells were seeded in 6-well plates (200,000
cells/well), transfected and, after 24, 48 or 72 h, flow cytometric
analysis of the cell cycle phase distribution of cells was
performed after staining, overnight at 4°C, the fixed cells with
propidium iodide (PI). At the end of transfection and/or
treatments, the cells were collected by trypsinization, washed
twice with ice-cold PBS, and fixed with 96% ethanol overnight at
4°C. Following fixation, the cells were washed again with PBS,
incubated with RNase and PI at 4°C overnight prior to flow
cytometric analysis. Cell cycle phase distribution was analysed
using a FC500™ flow cytometer (Beckman Coulter, Inc.) and
FlowJo_V10 software (FlowJo, LLC).
Measurement of interleukin (IL-6
release)
Immediately after the transfection period, cell
culture supernatants were collected and centrifuged at 16,000 × g
for 5 min at 4°C to remove cell debris and particles. The IL-6
concentrations were measured using commercially available ELISA
kits (cat. no. KHC0061, Invitrogen; Thermo Fisher Scientific, Inc.)
following the manufacturer's instructions, and were normalized to
the number of cells. The concentrations of IL-6 in the medium from
the treated cells were compared to the basal concentrations
observed in the untreated cells.
Apoptosis
Annexin V staining was performed for the apoptosis
assay. Following transfection and/or treatments, the cells were
harvested by trypsinization, washed with cold PBS three times and
stained with Annexin V-fluorescein isothiocyanate (FITC), PI in the
dark, according to the manufacturer's instructions (Bender
MedSystems GmbH). Cells were immediately sorted in a FC500™ flow
cytometer (Beckman Coulter, Inc.) and data were analysed using
FlowJo_V10 software. The percentages of Annexin V-positive cells
were calculated as the cell apoptotic rate.
To evaluate the activity of caspase-3, a
proluminescent DEVD-aminoluciferin substrate was added to the cell
culture, according to the manufacturer's protocol (Promega
Corporation). The luminescent signal generated by caspase cleavage,
proportional to caspase-3/7 activity, was quantified by means of a
Cary Eclipse fluorescence spectrophotometer (Varian, Inc.).
Western blot analysis
Cells were lysed in RIPA buffer (Thermo Fisher
Scientific, Inc.), and the proteins contents were quantified using
the BCA protein assay kit (Thermo Fisher Scientific, Inc.). 30
µg of proteins were separated by 10% sodium dodecyl
sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), then
transferred onto nitrocellulose membranes (Schleicher &
Schuell). The membranes were incubated in succession with the
blocking buffer (5% BSA in PBS) for 2 h at room temperature, with
the primary mouse monoclonal antibody anti-PIM1 diluted 1:250 (cat.
no. ab54503), rabbit monoclonal antibodies anti-CDK4 diluted
1:1,000 (cat. no. ab108355), anti-cyclin D1 dilute 1:200 (cat. no.
ab16663) or anti-cyclin E1 diluted 1:1,000 (cat. no. ab33911) (all
from Abcam) overnight at 4°C and finally with
horseradish-peroxidase conjugated goat anti-mouse diluted 1:200,000
(cat. no. STAR207P; Bio-Rad Laboratories, Inc.) or anti-rabbit IgG
secondary antibodies diluted 1:200,000 (cat. no. STAR208P, Bio-Rad
Laboratories, Inc.), incubated 2 h at room temperature. Protein
blots were detected using an ECL Chemiluminescent Substrate
(Cyanogen) and the intensity of the bands was quantitatively
analysed using the Fluor-Sä MultiImager (Bio-Rad Laboratories,
Inc.).
JC-1 assay
Mitochondrial membrane potential was evaluated by
JC-1 assay (Biotium, Inc.). At the end of the treatment period, all
cells were recovered and stained at 37°C for 15 min with the
cationic dye according to the manufacturer's instructions. The cell
suspension was finally transferred into the wells of a black
96-well plate and the absorbance was recorded using a Cary Eclipse
fluorescence microplate reader (Varian, Inc.; red fluorescence:
Excitation 550 nm, emissions 600 nm; green fluorescence: Excitation
485 nm, emission 540 nm). Hydrogen peroxide was used as a positive
control. The ratio of the fluorescence of JC-1 aggregates (red) to
monomers (green) was calculated and normalized to the respective
control.
Oxidative stress
The formation of intracellular reactive oxygen
species (ROS) was evaluated by 2,7-dichlorodihydrofluorescein
diacetate (DCFH-DA) (Sigma-Aldrich; Merck KGaA): This non-polar and
non-fluorescent compound can diffuse into the cytoplasm where it is
cleaved by intracellular esterases to yield polar, non-fluorescent
2′,7′-dichlorofluorescein (DCF). DCFH can then react with ROS to
form a highly fluorescent two-electron oxidation product, DCF.
Following miR-486 mimic transfection, the cells were treated for 24
h with CDDP (17.3 µM for the H2052 and 50 µM for the
H28 cells) and then incubated with 10 µM DCFH-DA in PBS at
37°C for 30 min in the dark. Hydrogen peroxide (10 µM) was
used as a positive control for the assay. Cells were then
harvested, washed with PBS and analysed by FC500™ flow cytometer
(Instrumentation Laboratory) and the FlowJo_V10 software.
Statistical analysis
The results are presented as the mean ± standard
deviation of almost three experiments. Statistical analyses were
carried out using ine-way analysis of variance (ANOVA) with
Dunnett's or Turkey's post hoc tests. The differences were
considered statistically significant with values of P<0.05,
P<0.01 and P<0.001.
Results
miR-486 expression following
transfection
In order to investigate the biological roles of
miRNA-486 in the human mesothelioma cell lines, H28 and H2052, its
expression was selectively regulated by miR-486 mimic transfection.
Two concentrations of miR mimic were tested: 10 and 50 nM. The
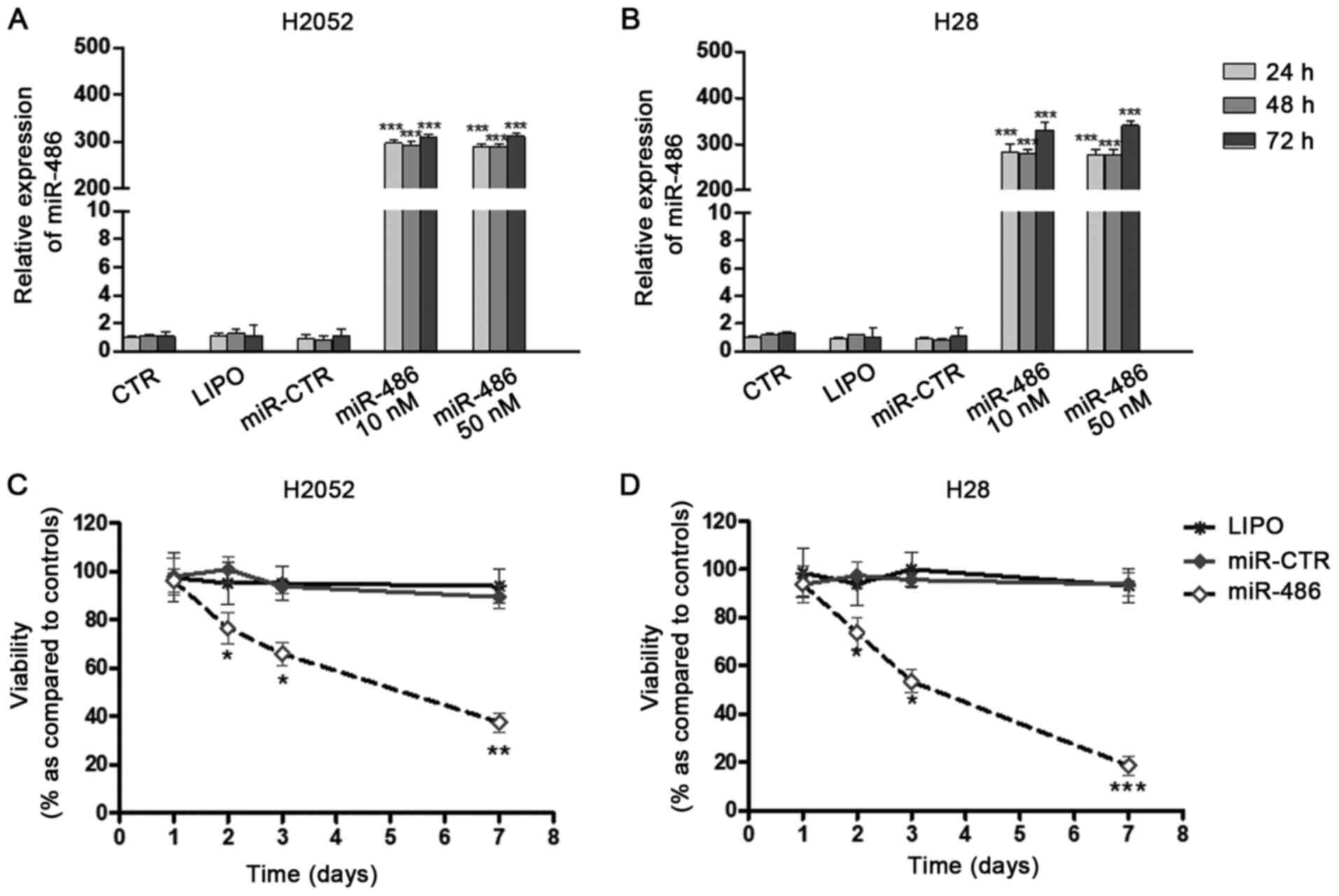
results of RT-qPCR indicated that in both cell lines, the miRNA was
overexpressed in comparison to the controls (untreated,
Lipofectamine-treated or miR-control-transfected cells) beginning
from 24 h following transfection. No significant differences were
observed between the two concentrations (Fig. 1A and B). For this reason, in the
subsequent experiments, the concentration of 10 nM was used.
miR-486 affects cell proliferation, cell
cycle progression and IL-6 release
miR-486 was transiently transfected into H28 and
H2052 cells. It was observed that miR-486 overexpression led to a
reduction in the proliferation of both cell lines (Fig. 1C and D), without significant
lysis or apoptosis. This decrease occurred late (48 h following
transfection) and this was time-dependent. In order to evaluate the
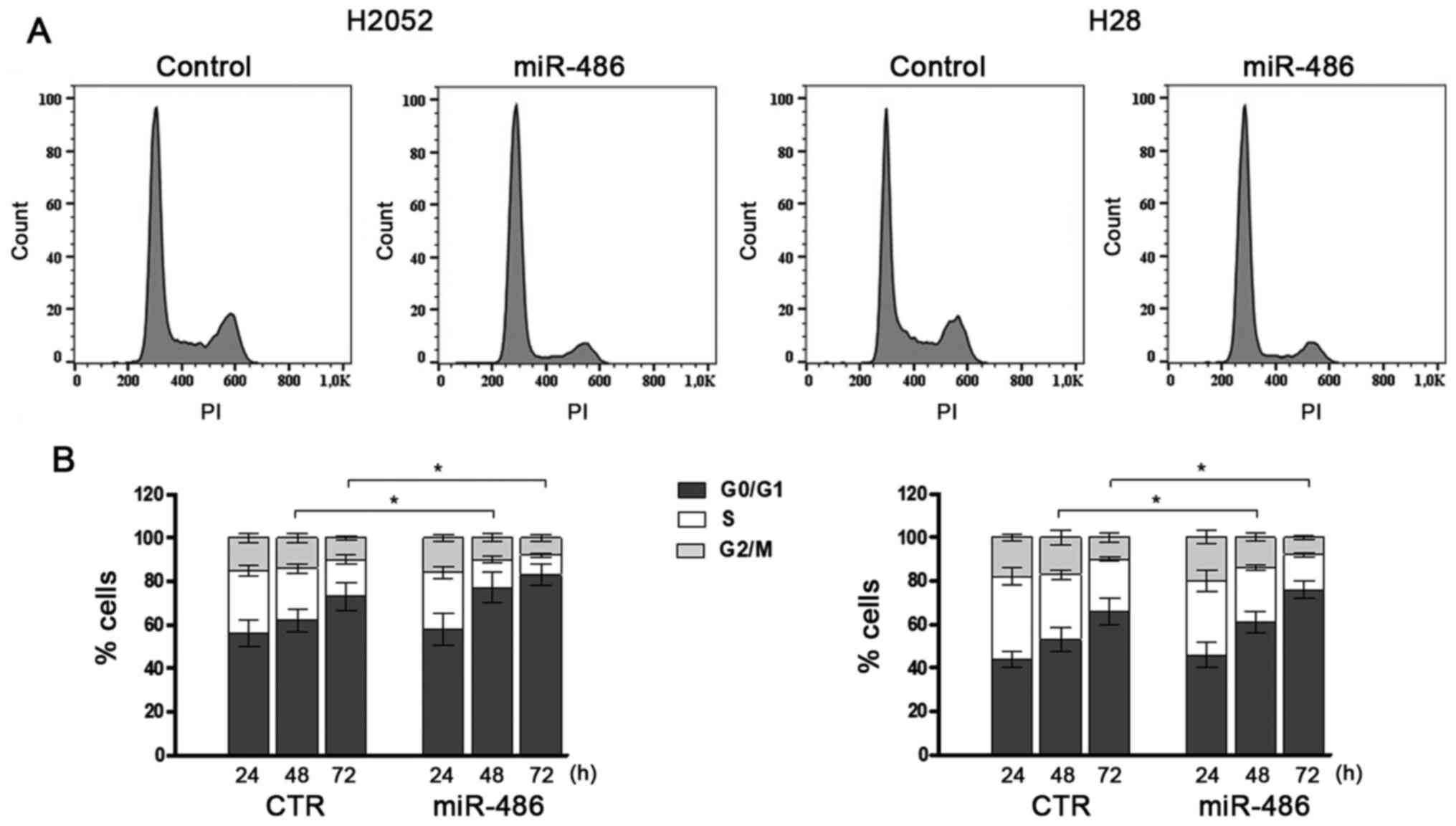
effects of miR-486 on cell proliferation, the percentage of cells
in the different stages of the cell cycle was analysed by flow
cytometry. The results revealed that the overexpression of miR-486
significantly reduced cell proliferation and this was mainly
reflected by the higher percentage of cells arrested in the G0/G1
phase (Fig. 2). The
corresponding proportions of cells in the S phase and in the G2/M
phase decreased significantly. The absence of a subG0/G1 peak
confirmed the absence of a massive apoptotic activation. The
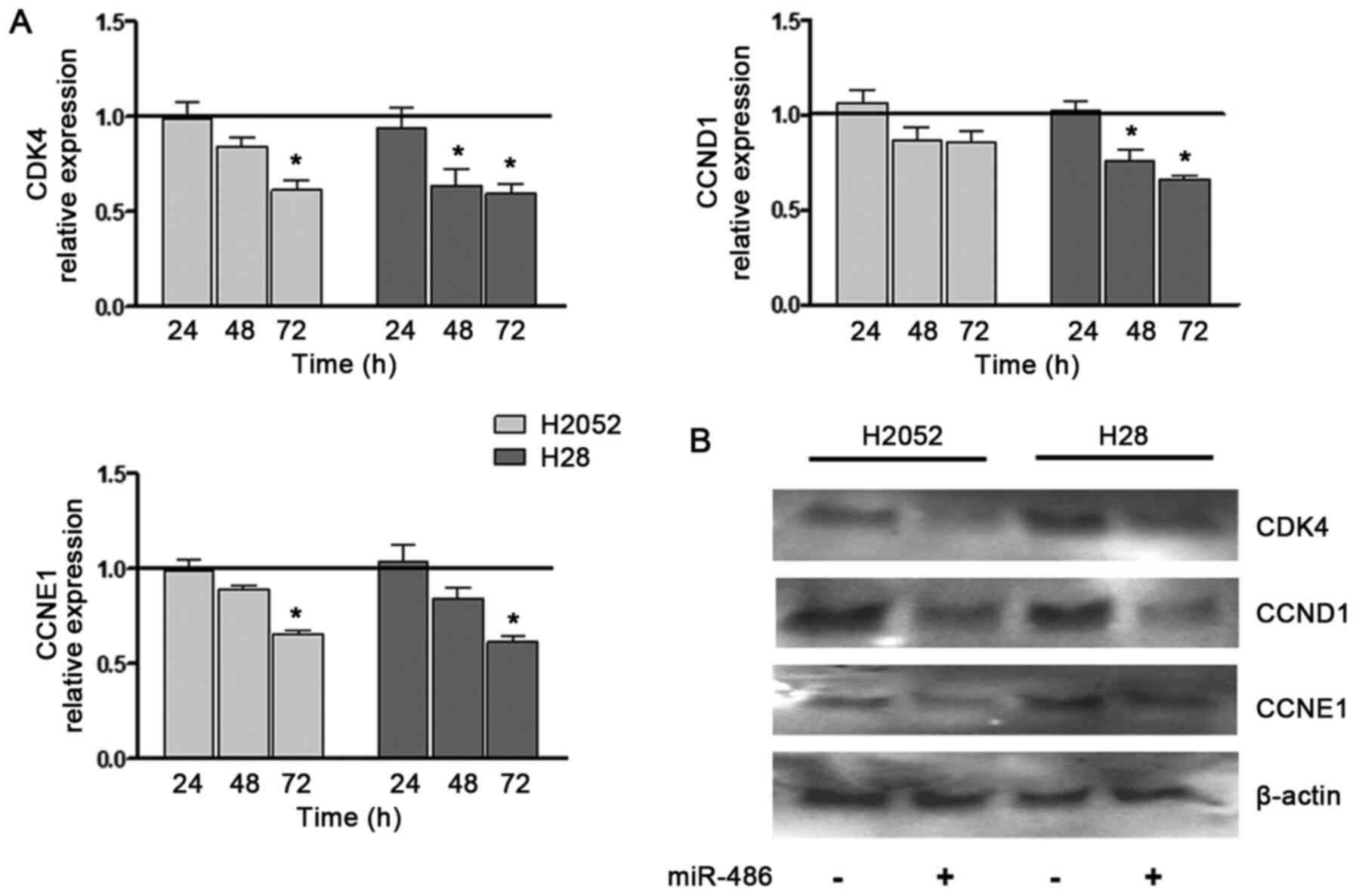
expression of cyclins involved in the G1/S transition (CCNE1
and CCND1 and CDK4) was also examined. These
expression levels significantly decreased in the miR-486
mimic-transfected H28 and H2052 cells, although at different time
points and at varying extents (Fig.
3A). In the H28 cells, the reduction was observed at an earlier
stage, and was more relevant and time-dependent than that observed
in the H2052 cells. The same trend was confirmed by western blot
analysis (Fig. 3B).
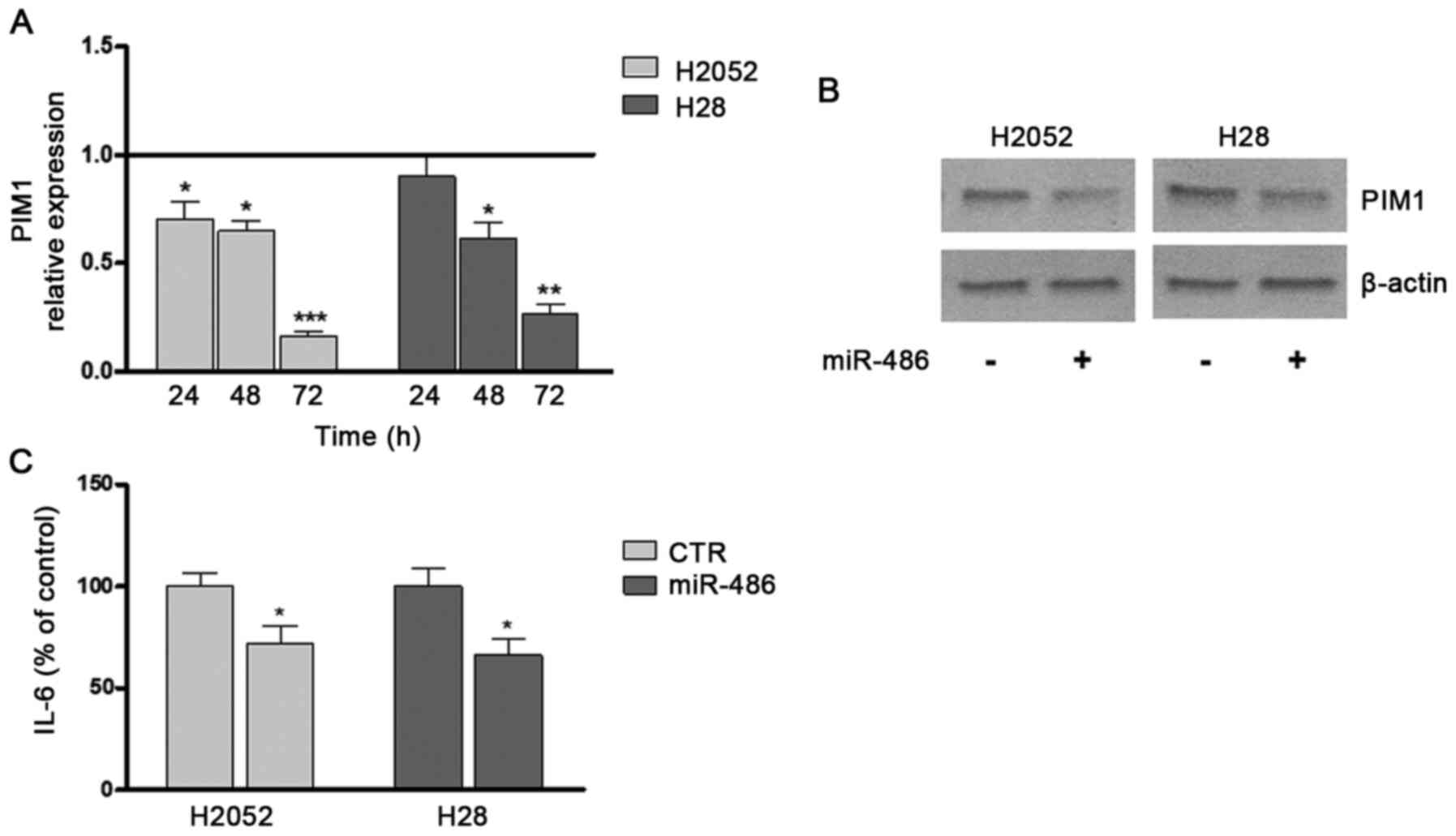
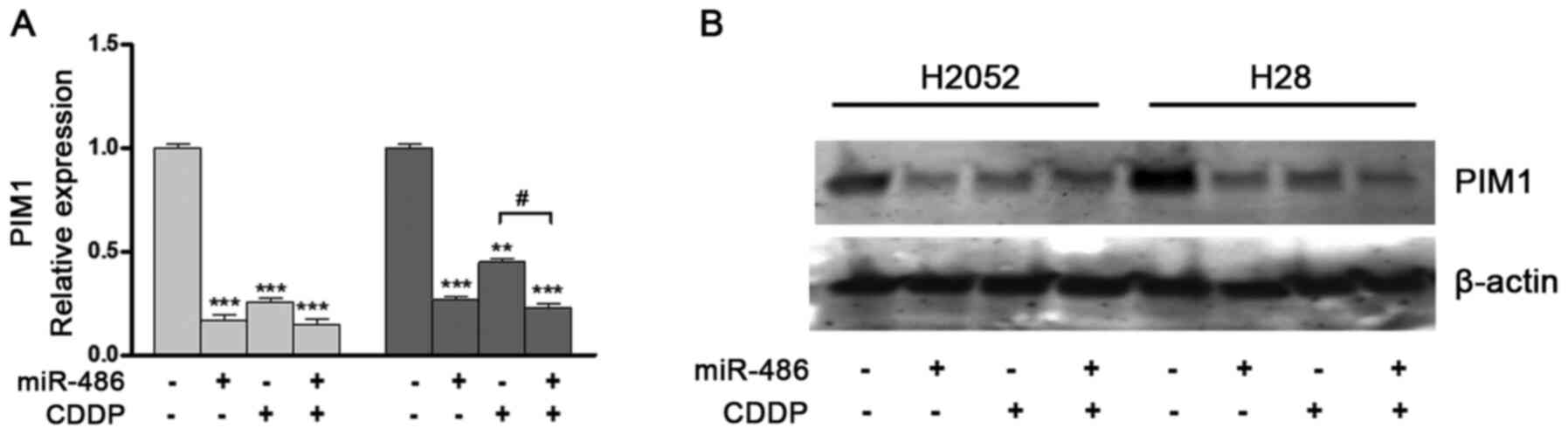
Transfection induced a time-dependent decrease in
PIM1 expression beginning from 24 h and reaching maximum
significance after 72 h compared to the untreated control (Fig. 4A). These results were confirmed
by western blot analysis (Fig.
4B). These effects of miR-486 were also coupled with a
significant decrease, of approximatively 30% compared with the
controls, in the release of the inflammatory molecule, IL-6, into
the culture medium (Fig.
4C).
miR-486 transfection sensitizes MPM cells
to CDDP
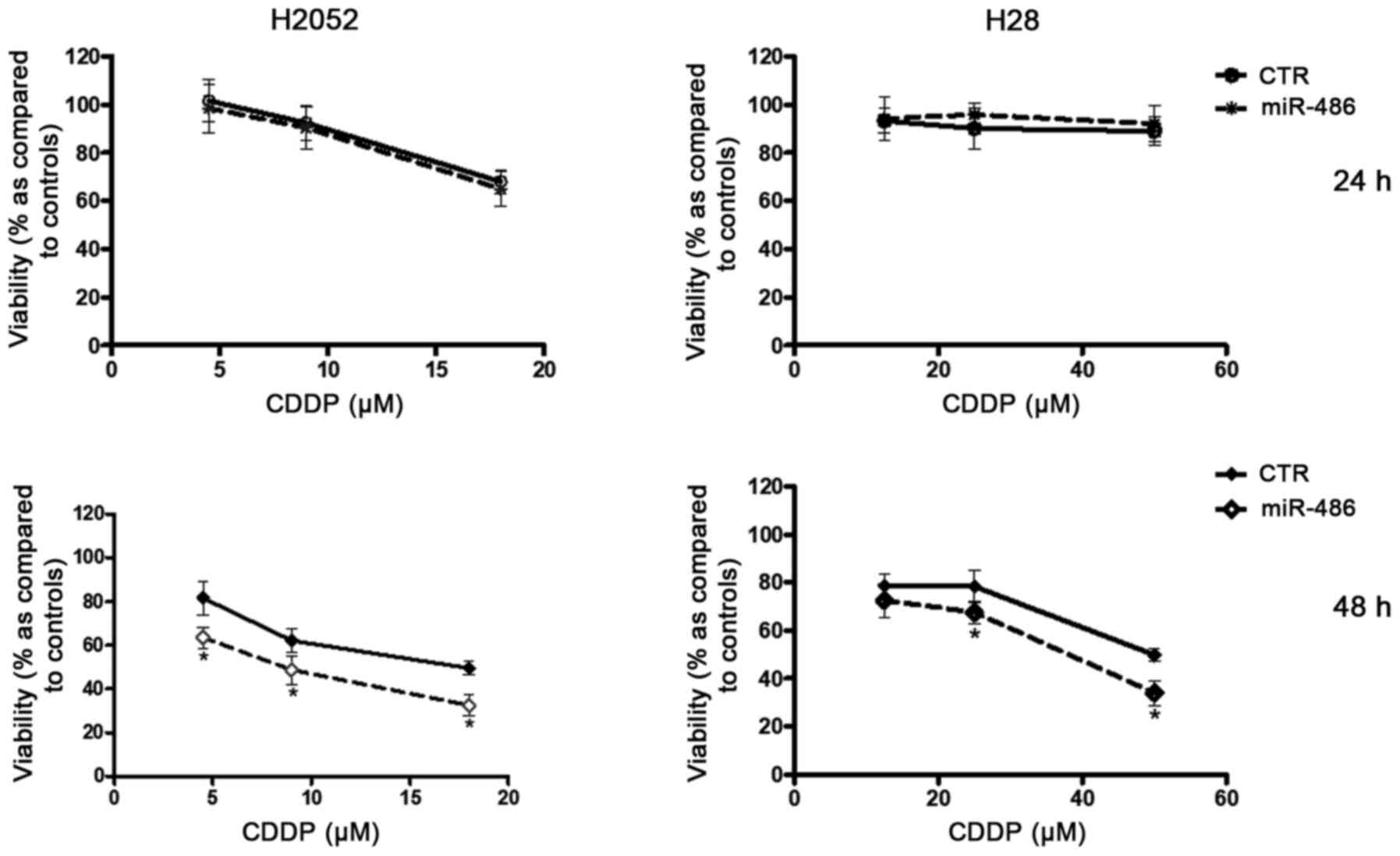
Treatment of the H28 and H2052 cells with various
concentrations of CDDP (range, 1-100 µM) led to a
concentration- and time dependent decrease in the viability of both
cell lines, although the H2052 cells were found to be more
sensitive than the H28 cells: At 48 h, the IC50 value
for the H2052 cells was less than half of that for the H28 cells
(17.83 µM vs. 50 µM). The MPM cells were transfected
with miR-486 and then treated with increasing concentrations of
CDDP. In both lines, miR-486 reduced viability, beginning from 48 h
following CDDP exposure in comparison to the cells treated with the
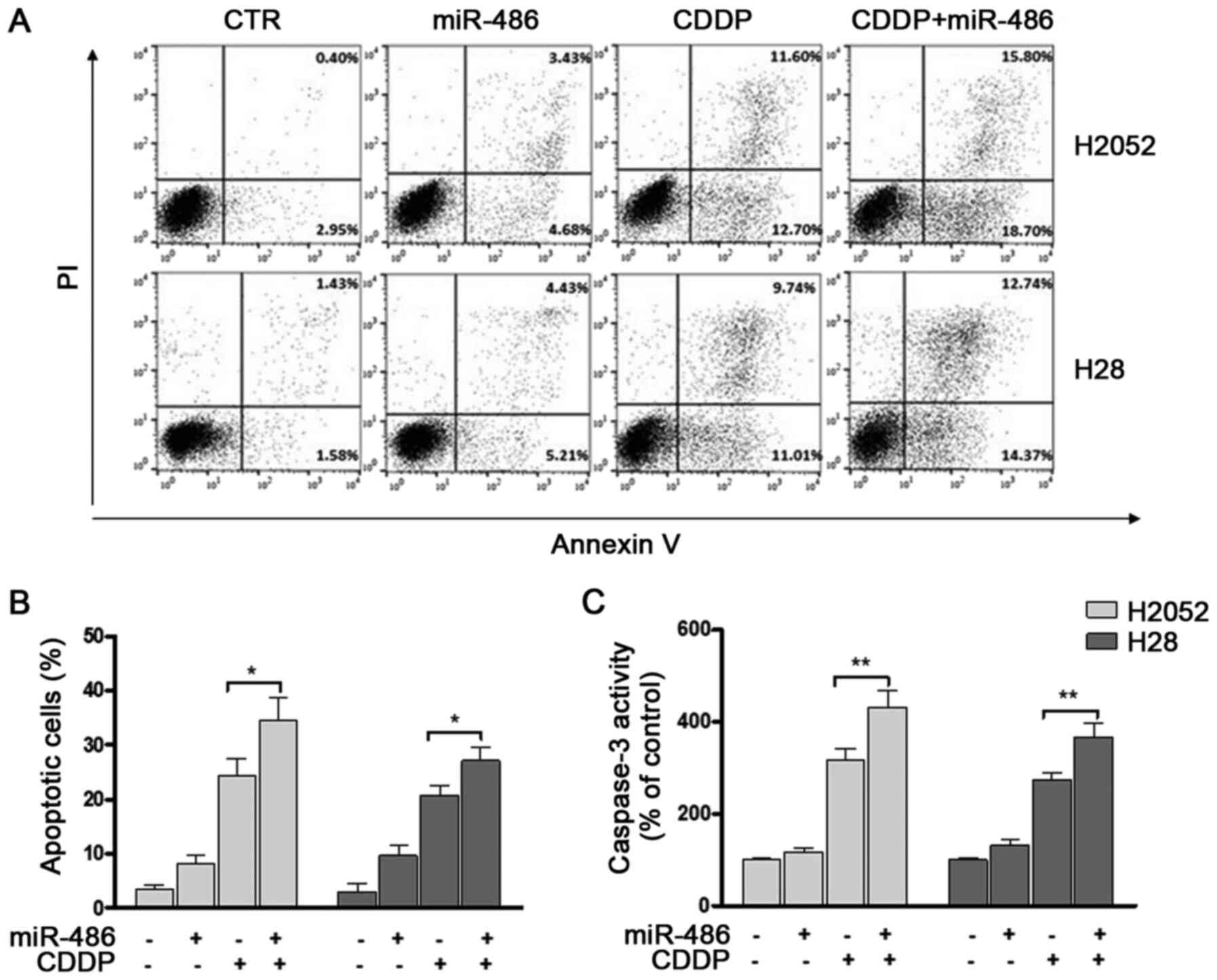
drug alone (Fig. 5).
Correspondingly, significant increases in the numbers of apoptotic
cells were observed (Fig. 6A and
B) and coupled with the stimulation of caspase-3 activity
(Fig. 6C). In cells transfected
with miRNA-486 and treated with CDDP no further decrease in
PIM1 expression was observed compared with the cells
transfected with the mimic only, while it was reduced compared with
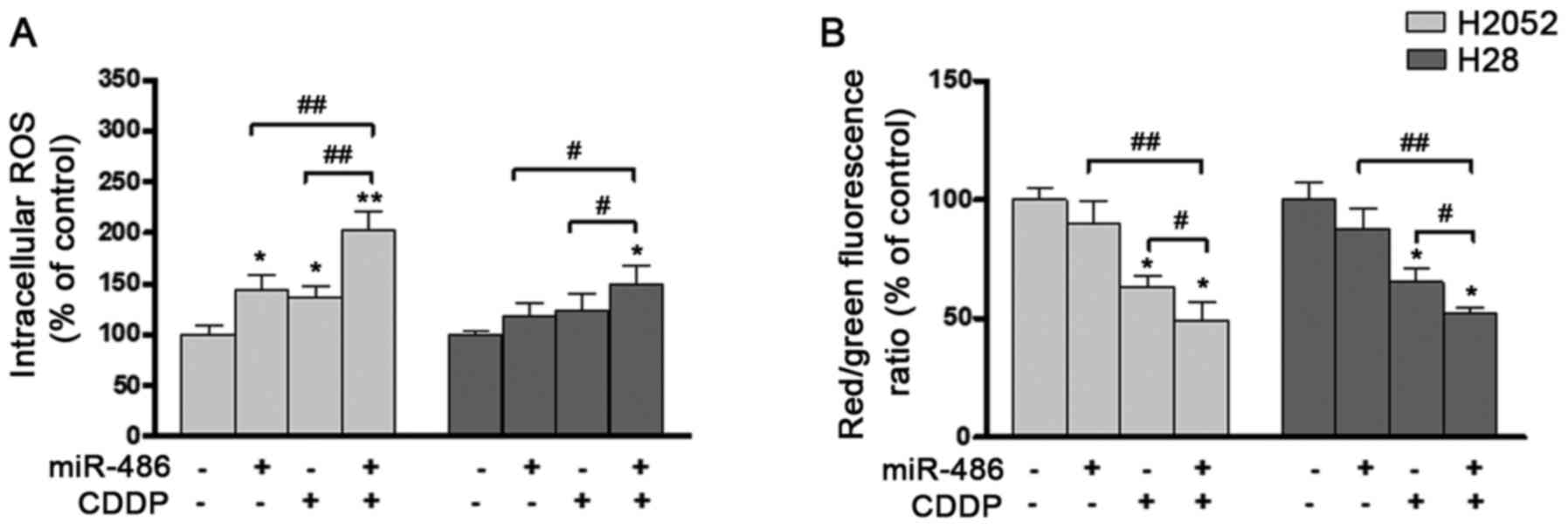
the cells treated with CDDP alone (Fig. 7). At the same time, an increase
in the intracellular ROS levels was observed, and this was
particularly significant in the H2052 cells transfected with
miRNA-486 end treated with CDDP compared with the control (Fig. 8A). In the H28 cells, ROS
accumulation, although present, was lower and not significant
compared with the control. At the 24-h time point, the transfected
H2052 and H28 cells also exhibited a slight mitochondrial
depolarization, as revealed by a decreased red/green fluorescence
ratio of the membrane-permeant JC-1 dye (Fig. 8B).
Discussion
As miRNAs are involved in various biological
processes in cancer, they are expected to play a crucial role in
cancer diagnosis and prognosis surveillance (40). Furthermore, a miRNA-based
therapeutic strategy, aimed to restore tumour suppressor miRNA that
may be lost or expressed at reduced levels in cancer cells, is
expected to constitute a novel potential direction for MPM
treatment, particularly for personalized therapy (41,42). To date, at least 20 miRNAs are
being evaluated in clinical trials; however, the identification of
novel miRNAs is required, not only for therapeutic purposes, but
also to understand the mechanisms of oncogenesis and the evolution
of tumours (43).
miR-486, which is located in the last intron of the
Ankyrin-1 (Ank1) gene on human chromosome 8, was first identified
from a human foetal liver cDNA library (44,45). It has been reported to be
involved in different types of cancer (19-25,27,46,47) and that its expression is differs
markedly between the early and advanced stages of non-small cell
lung cancer (NSCLC) (48).
Subsequently, a meta-analysis indicated that miR-486 may be used as
ideal biomarker for cancer diagnosis, but also concluded that a low
expression of this miRNA did not increase the risk of a poor
outcome (27).
To date, a few potential targets for miR-486 have
been identified (19) and the
mechanistic role for miR-486 as either an oncogene or tumour
suppressor, particularly in MPM, remains largely unknown. In a
previous study (28), the
authors aimed at the identification of diagnostic/prognostic
biomarkers for asbestos-related diseases; the expression of
miRNA-486 was significantly lower in patients with MPM and
asbestosis than that in the controls, and this association was
significant in plasma and tissues. Furthermore, its tissue
expression was positively related to the cumulative survival of
patients with MPM. Based on these conclusions, it was hypothesized
that miR-486 may play an important role in tumorigenesis and tumour
development. Therefore, the present study investigated its
potential function by transfecting miR-486 in two in vitro
models of MPM. Not surprisingly, it was found that miR-486
significantly suppressed cellular growth and cycle progression by
targeting the cyclins and in particular, CDK4.
As CDK4 plays an important role in the G1/S phase
transition, its deregulation is one of the most common alterations
found in human cancers, including MPM. This checkpoint is crucial
in cell survival, ensuring the detection and repair of genetic
damage, as well as the prevention of uncontrolled cell division.
The suppressive effects of miR-486 on CDK4 have already been
observed in oesophageal cancer (49) and NSCLC (48), leading to the conclusion that
miR-486 may act as tumour suppressor. The 3′-UTR of CDK4 is
the direct interaction region between this miRNA and CDK4
(48).
The antiproliferative effects of miR-486 on MPM
cells are pivoted by the reduction in PIM1 expression
observed. In this rapidly growing invasive cancer, the role of
PIM1 downregulation in the suppression of cell proliferation
by cell arrest at the G0/G1 phase and of cell invasion and
migration, has already been demonstrated, as well as the role of
the inhibition of other miRNAs (33,50). The negative correlation between
miR-486 and PIM1 in cancer tissues and the
miR-486/PIM1 axis has already been observed in lung cancer
(35), breast cancer (34) and osteosarcoma (51). The present study confirmed this
association in mesothelioma. Unlike what has been observed in lung
cancer cells, in which the enforced expression of miR-486-5p
resulted in a decrease in PIM1 protein expression not accompanied
by a significant reduction in mRNA expression in cells, suggesting
that miR-486 may inhibit its expression mainly through
post-transcriptional regulation (35), in the present study, a parallel
decrease in PIM1 mRNA and protein expression was observed.
Previous studies have confirmed that PIM1
plays a role in cellular proliferation by regulating cell cycle
progression at multiple points and through various target proteins,
and that its levels change with the stages of the cell cycle,
related to the onset of DNA synthesis (32). This highly conserved protein is
an unusual serine or threonine kinase, as it is constitutively
active and may rightly be considered a cell survival factor. Its
overexpression may contribute to cancer development by protecting
cells from undergoing apoptosis induced by exposure to stress
factors and drugs, by promoting cell proliferation and genomic
instability (52,53). The increased survival was found
to be associated with the maintenance of mitochondrial
transmembrane potential. In the absence or by the reduced levels of
PIM1, not only do cells exhibit a reduced mitochondrial
membrane potential, but they also exhibit an elevated production of
harmful ROS (31,32). Based on current knowledge,
PIM1 overexpression was suggested to be employed as a
reliable diagnostic marker for several types of tumours and, in
some cases, as a prognostic indicator of clinical outcomes
(54-56). On the other hand, this may be a
promising therapeutic target in appropriately selected cancers with
the overexpression of this protein (30,57). This proposal is also supported by
the finding that the elimination of PIM1 activity has no
apparent effect on normal cells and yet is lethal to overexpressing
cancer cells, avoiding the detrimental side-effects of most
conventional treatments (32,57).
Another important result obtained by miR-486
transfection is the reduction in the levels of inflammatory
markers, such as IL-6, and this is very beneficial. Inflammation
and the immune response emerged as key players in driving MPM
progression and represent promising therapeutic targets:
Manipulations of the inflammatory tumour stroma have been suggested
to render MPM susceptible to therapies that have shown relevant
results in other solid and aggressive tumours (2,3).
Previous studies have also provided the rationale for utilising
anti-IL-6 therapeutics alongside standard chemotherapy in an
attempt to prolong survival and alleviate symptoms commonly
witnessed in this disease, such as cachexia, thrombocytosis and
immunosuppression (58-63).
To date, the only systemic treatment approved and
registered by the Food and Drug Administration and the European
Medicines Agency is platinum-based chemotherapy; however, its use
in the treatment of MPM mostly yields frustrating results and the
average life expectancy is usually <1 year with a 5-year
survival of <5% (64-66). Acquired resistance to drugs is
still a major clinical issue. A number of studies have indicated
that several miRNAs, and even miR-486, act as chemo-resistance
regulators, controlling the levels of drug resistance-related genes
(21,26,43). The data of the present study
demonstrated this modulating function of miR-486, enhancing the
inhibitory and apoptotic effects of CDDP, thus suggesting the
potential therapeutic advantage of combined CDDP/miR-486
therapeutic protocols in human MPM.
Of note, the decrease in mitochondrial membrane
potential and the enhancement of ROS production were additively
promoted by the overexpression of miR-486 and CDDP treatment. By
contrast, no additive effect was observed for the decreased
expression of PIM1 due to miR-486 overexpression and CDDP
treatment. When two stressful events, such as CDDP and miR-486
upregulation occur at the same time, a complex crosstalk may be
established and responses to the stressors may be enhanced. This
result seems to be independent of PIM1 hypo-regulation and to
involve other processes controlling redox equilibrium. Due to its
broad target recognition and to its ability to have multiple
effects, miR-486 is more advantageous and preferable than molecules
able only to inhibit the PIM1 activity. Regardless of the
histopathological type or BAP1 mutation, and despite some
differences in the extent of the responses, no discrepant
behaviours have been observed between the two lines differing in
histology and BAP1 mutations: H28 (epithelioid, BAP1
null) and H2052 (sarcomatoid, BAP1 wild-type) (67). The main difference concerns drug
resistance as epithelial H28 cells are more resistant to cisplatin
than sarcomatoid H2052 cells, and this behaviour may be due to
BAP1 mutation, whose loss-of-function is strongly associated
with epithelioid differentiation and may play a role in promoting
survival and in influencing chemosensitivity (68).
The complex biology of MPM requires an appropriate
combination of approaches to overcome the vastly disappointing
responses to chemotherapy. The miR-486 pathway not only has the
potential for a greater understanding of the MPM pathogenesis, but
also for therapeutic intervention, as it was demonstrated to play a
role in regulating, at the same time, multiple mechanisms that are
involved in the development and progression of this disease. A
treatment effectively targeting multiple pathways simultaneously
would be a major advance. In cancer, such an approach should not
only inhibit cell growth more effectively, but should also prevent
the loss of drug efficacy, due to the common emergence of
resistance acting on a single pathway.
Availability of data and materials
The datasets used and/or analysed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
SP, RA and PM conceived the study, and planned and
carried out the experiments. RA wrote the manuscript with support
from SP, PM, GP and DP. DP, DC, MT, MC, MG and GP critically
revised the manuscript for important intellectual content. All
authors (SP, RA, DP, MC, GP, MT, MG, DC and PM) contributed to the
interpretation of the results and revised critically the manuscript
for important intellectual content. SP and PM confirm the
authenticity of all the raw data. All authors have read and
approved the final version of the manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
All authors declare that they have no competing
interests.
Acknowledgments
Not applicable.
Funding
No funding was received.
References
|
1
|
Roe OD and Stella GM: Malignant pleural
mesothelioma: History, controversy and future of a manmade
epidemic. Eur Respir Rev. 24:115–131. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Carbone M and Yang H: Mesothelioma: Recent
highlights. Ann Transl Med. 5:2382017. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Abbott DM, Bortolotto C, Benvenuti S,
Lancia A, Filippi AR and Stella GM: Malignant pleural mesothelioma:
Genetic and microenviromental heterogeneity as an unexpected
reading frame and therapeutic challenge. Cancers (Basel).
12:11862020. View Article : Google Scholar
|
|
4
|
Takagi A, Hirose A, Futakuchi M, Tsuda H
and Kanno J: Dose-dependent mesothelioma induction by
intraperitoneal administration of multi-wall carbon nanotubes in
p53 heterozygous mice. Cancer Sci. 103:1440–1444. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Takagi A, Hirose A, Nishimura T, Fukumori
N, Ogata A, Ohashi N, Kitajima S and Kanno J: Induction of
mesothelioma in p53+/− mouse by intraperitoneal
application of multi-wall carbon nanotube. J Toxicol Sci.
33:105–116. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Goodman JE, Nascarella MA and Valberg PA:
Ionizing radiation: A risk factor for mesothelioma. Cancer Causes
Control. 20:1237–1254. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Pershouse MA, Heivly S and Girtsman T: The
role of SV40 in malignant mesothelioma and other human
malignancies. Inhal Toxicol. 18:995–1000. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Bruno C, Tumino R, Fazzo L, Cascone G,
Cernigliaro A, De Santis M, Giurdanella MC, Nicita C, Rollo PC,
Scondotto S, et al: Incidence of pleural mesothelioma in a
community exposed to fibres with fluoro-edenitic composition in
Biancavilla (Sicily, Italy). Ann Ist Super Sanita. 50:111–118.
2014.PubMed/NCBI
|
|
9
|
Carbone M, Ly BH, Dodson RF, Pagano I,
Morris PT, Dogan UA, Gazdar AF, Pass HI and Yang H: Malignant
mesothelioma: Facts, myths, and hypotheses. J Cell Physiol.
227:44–58. 2012. View Article : Google Scholar
|
|
10
|
Xu LL, Yang Y, Wang Z, Wang XJ, Tong ZH
and Shi HZ: Malignant pleural mesothelioma: Diagnostic value of
medical thoracoscopy and long-term prognostic analysis. BMC Pulm
Med. 18:562018. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zhang W, Wu X, Wu L, Zhang W and Zhao X:
Advances in the diagnosis, treatment and prognosis of malignant
pleural mesothelioma. Ann Transl Med. 3:1822015.PubMed/NCBI
|
|
12
|
Bibby AC, Tsim S, Kanellakis N, Ball H,
Talbot DC, Blyth KG, Maskell NA and Psallidas I: Malignant pleural
mesothelioma: An update on investigation, diagnosis and treatment.
Eur Respir Rev. 25:472–486. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Cao S, Jin S, Cao J, Shen J, Hu J, Che D,
Pan B, Zhang J, He X, Ding D, et al: Advances in malignant
peritoneal mesothelioma. Int J Colorectal Dis. 30:1–10. 2015.
View Article : Google Scholar
|
|
14
|
Slaby O, Laga R and Sedlacek O:
Therapeutic targeting of non-coding RNAs in cancer. Biochem J.
474:4219–4251. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Lo Russo G, Tessari A, Capece M, Galli G,
de Braud F, Garassino MC and Palmieri D: MicroRNAs for the
diagnosis and management of malignant pleural mesothelioma: A
literature review. Front Oncol. 8:6502018. View Article : Google Scholar
|
|
16
|
Tomasetti M, Gaetani S, Monaco F, Neuzil J
and Santarelli L: Epigenetic Regulation of miRNA expression in
malignant mesothelioma: MiRNAs as biomarkers of early diagnosis and
therapy. Front Oncol. 9:12932019. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Birnie KA, Prele CM, Thompson PJ, Badrian
B and Mutsaers SE: Targeting microRNA to improve diagnostic and
therapeutic approaches for malignant mesothelioma. Oncotarget.
8:78193–78207. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Bartel DP: MicroRNAs: Target recognition
and regulatory functions. Cell. 136:215–233. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
ElKhouly AM, Youness RA and Gad MZ:
MicroRNA-486-5p and microRNA-486-3p: Multifaceted pleiotropic
mediators in oncological and non-oncological conditions. Noncoding
RNA Res. 5:11–21. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Huang XP, Hou J, Shen XY, Huang CY, Zhang
XH, Xie YA and Luo XL: MicroRNA-486-5p which is downregulated in
hepatocellular carcinoma, suppresses tumor growth by targeting
PIK3R1. FEBS J. 282:579–594. 2015. View Article : Google Scholar
|
|
21
|
Sun H, Cui C, Xiao F, Wang H, Xu J, Shi X,
Yang Y, Zhang Q, Zheng X, Yang X, et al: MiR-486 regulates
metastasis and chemosensitivity in hepatocellular carcinoma by
targeting CLDN10 and CITRON. Hepatol Res. 45:1312–1322. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zhu J, Zeng Y, Xu C, Qin H, Lei Z, Shen D,
Liu Z and Huang JA: Expression profile analysis of microRNAs and
downregulated miR-486-5p and miR-30a-5p in non-small cell lung
cancer. Oncol Rep. 34:1779–1786. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Wang J, Tian X, Han R, Zhang X, Wang X,
Shen H, Xue L, Liu Y, Yan X, Shen J, et al: Downregulation of
miR-486-5p contributes to tumor progression and metastasis by
targeting protumorigenic ARHGAP5 in lung cancer. Oncogene.
33:1181–1189. 2014. View Article : Google Scholar :
|
|
24
|
Rask L, Balslev E, Sokilde R, Hogdall E,
Flyger H, Eriksen J and Litman T: Differential expression of
miR-139, miR-486 and miR-21 in breast cancer patients
sub-classified according to lymph node status. Cell Oncol (Dordr).
37:215–227. 2014. View Article : Google Scholar
|
|
25
|
Yi Y, Lu X, Chen J, Jiao C, Zhong J, Song
Z, Yu X and Lin B: Downregulated miR-486-5p acts as a tumor
suppressor in esophageal squamous cell carcinoma. Exp Ther Med.
12:3411–3416. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Wang W, Liu B, Sun S, Lan L, Chen Y, Han
S, Li L and Li Z: Downregulation of miR-486-5p enhances the
anti-tumor effect of 5-fluorouracil on pancreatic cancer cells.
Onco Targets Ther. 13:1649–1659. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Jiang M, Li X, Quan X, Yang X, Zhaeng C,
Hao X, Qu R and Zhou B: MiR-486 as an effective biomarker in cancer
diagnosis and prognosis: A systematic review and meta-analysis.
Oncotarget. 9:13948–13958. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Mozzoni P, Ampollini L, Goldoni M, Alinovi
R, Tiseo M, Gnetti L, Carbognani P, Rusca M, Mutti A, Percesepe A
and Corradi M: MicroRNA expression in malignant pleural
mesothelioma and asbestosis: A pilot study. Dis Markers.
2017:96459402017. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Xiang X, Yuan D, Liu Y, Li J, Wen Q, Kong
PY, Gao L, Zhang C, Peng X and Zhang X: PIM1 overexpression in
T-cell lymphomas protects tumor cells from apoptosis and confers
doxorubicin resistance by upregulating c-myc expression. Acta
Biochim Biophys Sin (Shanghai). 50:800–806. 2018. View Article : Google Scholar
|
|
30
|
Cao L, Wang F, Li S, Wang X, Huang D and
Jiang R: PIM1 kinase promotes cell proliferation, metastasis and
tumor growth of lung adenocarcinoma by potentiating the c-MET
signaling pathway. Cancer Lett. 444:116–126. 2019. View Article : Google Scholar
|
|
31
|
Chauhan SS, Toth RK, Jensen CC, Casillas
AL, Kashatus DF and Warfel NA: PIM kinases alter mitochondrial
dynamics and chemosensitivity in lung cancer. Oncogene.
39:2597–2611. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Magnuson NS, Wang Z, Ding G and Reeves R:
Why target PIM1 for cancer diagnosis and treatment? Future Oncol.
6:1461–1478. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Mawas AS, Amatya VJ, Suzuki R, Kushitani
K, Mohi El-Din MM and Takeshima Y: PIM1 knockdown inhibits cell
proliferation and invasion of mesothelioma cells. Int J Oncol.
50:1029–1034. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Zhang G, Liu Z, Cui G, Wang X and Yang Z:
MicroRNA-486-5p targeting PIM-1 suppresses cell proliferation in
breast cancer cells. Tumour Biol. 35:11137–11145. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Pang W, Tian X, Bai F, Han R, Wang J, Shen
H, Zhang X, Liu Y, Yan X, Jiang F and Xing L: Pim-1 kinase is a
target of miR-486-5p and eukaryotic translation initiation factor
4E, and plays a critical role in lung cancer. Mol Cancer.
13:2402014. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Jin X, Pang W, Zhang Q and Huang H:
MicroRNA-486-5p improves nonsmall-cell lung cancer chemotherapy
sensitivity and inhibits epithelial-mesenchymal transition by
targeting twinfilin actin binding protein 1. J Int Med Res.
47:3745–3756. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Blower PE, Chung JH, Verducci JS, Lin S,
Park JK, Dai Z, Liu CG, Schmittgen TD, Reinhold WC, Croce CM, et
al: MicroRNAs modulate the chemosensitivity of tumor cells. Mol
Cancer Ther. 7:1–9. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Salimian J, Baradaran B, Azimzadeh
Jamalkandi S, Moridikia A and Ahmadi A: MiR-486-5p enhances
cisplatin sensitivity of human muscle-invasive bladder cancer cells
by induction of apoptosis and down-regulation of metastatic genes.
Urol Oncol. 38:738.e9–738.e21. 2020. View Article : Google Scholar
|
|
39
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
40
|
Sturchio E, Berardinelli MG, Boccia P,
Zanellato M and Gioiosa S: MicroRNAs diagnostic and prognostic
value as predictive markers for malignant mesothelioma. Arch
Environ Occup Health. 75:471–482. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Reid G, Kao SC, Pavlakis N, Brahmbhatt H,
MacDiarmid J, Clarke S, Boyer M and van Zandwijk N: Clinical
development of TargomiRs, a miRNA mimic-based treatment for
patients with recurrent thoracic cancer. Epigenomics. 8:1079–1085.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Reid G, Johnson TG and van Zandwijk N:
Manipulating microRNAs for the treatment of malignant pleural
mesothelioma: Past, present and future. Front Oncol. 10:1052020.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Seo HA, Moeng S, Sim S, Kuh HJ, Choi SY
and Park JK: MicroRNA-Based Combinatorial cancer therapy: Effects
of MicroRNAs on the efficacy of anti-cancer therapies. Cells.
9:292019. View Article : Google Scholar
|
|
44
|
Fu H, Tie Y, Xu C, Zhang Z, Zhu J, Shi Y,
Jiang H, Sun Z and Zheng X: Identification of human fetal liver
miRNAs by a novel method. FEBS Lett. 579:3849–3854. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Small EM, O'Rourke JR, Moresi V,
Sutherland LB, McAnally J, Gerard RD, Richardson JA and Olson EN:
Regulation of PI3-kinase/Akt signaling by muscle-enriched
microRNA-486. Proc Natl Acad Sci USA. 107:4218–4223. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Mees ST, Mardin WA, Sielker S, Willscher
E, Senninger N, Schleicher C, Colombo-Benkmann M and Haier J:
Involvement of CD40 targeting miR-224 and miR-486 on the
progression of pancreatic ductal adenocarcinomas. Ann Surg Oncol.
16:2339–2350. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
He M, Wang G, Jiang L, Qiu C, Li B, Wang J
and Fu Y: MiR-486 suppresses the development of osteosarcoma by
regulating PKC-δ pathway. Int J Oncol. 50:1590–1600. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Shao Y, Shen YQ, Li YL, Liang C, Zhang BJ,
Lu SD, He YY, Wang P, Sun QL, Jin YX and Ma ZL: Direct repression
of the oncogene CDK4 by the tumor suppressor miR-486-5p in
non-small cell lung cancer. Oncotarget. 7:34011–34021. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Lang B and Zhao S: MiR-486 functions as a
tumor suppressor in esophageal cancer by targeting CDK4/BCAS2.
Oncol Rep. 39:71–80. 2018.
|
|
50
|
Amatya VJ, Mawas AS, Kushitani K, Mohi
El-Din MM and Takeshima Y: Differential microRNA expression
profiling of mesothelioma and expression analysis of miR-1 and
miR-214 in mesothelioma. Int J Oncol. 48:1599–1607. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Liu Y, Zhang J, Xing C, Wei S, Guo N and
Wang Y: MiR-486 inhibited osteosarcoma cells invasion and
epithelial-mesenchymal transition by targeting PIM1. Cancer
Biomark. 23:269–277. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Zhang X, Song M, Kundu JK, Lee MH and Liu
ZZ: PIM Kinase as an executional target in cancer. J Cancer Prev.
23:109–116. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Zhao W, Qiu R, Li P and Yang J: PIM1: A
promising target in patients with triple-negative breast cancer.
Med Oncol. 34:1422017. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Chen J and Tang G: PIM-1 kinase: A
potential biomarker of triple-negative breast cancer. Onco Targets
Ther. 12:6267–6273. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Yang H, He K, Dong W, Fang J, Zhong S,
Tang L and Long L: PIM-1 may function as an oncogene in cervical
cancer via activating the EGFR signaling. Int J Biol Markers.
35:67–73. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Xu J, Xiong G, Cao Z, Huang H, Wang T, You
L, Zhou L, Zheng L, Hu Y, Zhang T and Zhao Y: PIM-1 contributes to
the malignancy of pancreatic cancer and displays diagnostic and
prognostic value. J Exp Clin Cancer Res. 35:1332016. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Asati V, Mahapatra DK and Bharti SK: PIM
kinase inhibitors: Structural and pharmacological perspectives. Eur
J Med Chem. 172:95–108. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Abdul Rahim SN, Ho GY and Coward JI: The
role of interleukin-6 in malignant mesothelioma. Transl Lung Cancer
Res. 4:55–66. 2015.PubMed/NCBI
|
|
59
|
Donnenberg AD, Luketic JD and Donnenberg
VS: Secretome of pleural effusions associated with non-small cell
lung cancer (NSCLC) and malignant mesothelioma: Therapeutic
implications. Oncotarget. 10:6456–6465. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Nakano T, Chahinian AP, Shinjo M, Tonomura
A, Miyake M, Togawa N, Ninomiya K and Higashino K: Interleukin 6
and its relationship to clinical parameters in patients with
malignant pleural mesothelioma. Br J Cancer. 77:907–912. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Acencio MM, Soares B, Marchi E, Silva CS,
Teixeira LR and Broaddus VC: Inflammatory cytokines contribute to
asbestos-induced injury of mesothelial cells. Lung. 193:831–837.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Topley N, Jörres A, Luttmann W, Petersen
MM, Lang MJ, Thierauch KH, Müller C, Coles GA, Davies M and
Williams JD: Human peritoneal mesothelial cells synthesize
interleukin-6: Induction by IL-1 beta and TNF alpha. Kidney Int.
43:226–233. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Adachi Y, Aoki C, Yoshio-Hoshino N,
Takayama K, Curiel DT and Nishimoto N: Interleukin-6 induces both
cell growth and VEGF production in malignant mesotheliomas. Int J
Cancer. 119:1303–1311. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Norbet C, Joseph A, Rossi SS, Bhalla S and
Gutierrez FR: Asbestos-related lung disease: A pictorial review.
Curr Probl Diagn Radiol. 44:371–382. 2015. View Article : Google Scholar
|
|
65
|
Hiddinga BI, Rolfo C and van Meerbeeck JP:
Mesothelioma treatment: Are we on target? A review. J Adv Res.
6:319–330. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Nicolini F, Bocchini M, Bronte G, Delmonte
A, Guidoboni M, CrinO L and Mazz M: Malignant pleural mesothelioma:
State-of-the-Art on current therapies and promises for the future.
Front Oncol. 9:15192020. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Sacco JJ, Kenyani J, Butt Z, Carter R,
Chew HY, Cheeseman LP, Darling S, Denny M, Urbe S, Clague MJ and
Coulson JM: Loss of the deubiquitylase BAP1 alters class I histone
deacetylase expression and sensitivity of mesothelioma cells to
HDAC inhibitors. Oncotarget. 6:13757–13771. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Guazzelli A, Meysami P, Bakker E,
Demonacos C, Giordano A, Krstic-Demonacos M and Mutti L: BAP1
status determines the sensitivity of malignant mesothelioma cells
to gemcitabine treatment. Int J Mol Sci. 20:4292019. View Article : Google Scholar :
|