The level of histone acetylation is of importance
for nuclear stability, chromatin structure, gene expression and
physiological functions in hepatocytes (1,2).
Histone acetyltransferases (HATs) and histone deacetylases (HDACs)
are antagonistic proteases that serve regulatory roles in the
balance of histone acetylation and deacetylation in nucleosomes
(3). HATs, which favor of
histone acetylation, transfer acetyl groups from acetyl-CoA to the
ε-NH2 group of lysine residue side chains and neutralize
the positive charge of histone tails, thus making chromatin
structure more loose and conducive to active transcription
(4–6). HDACs, which facilitate histone
deacetylation, remove the acetyl group from the ε-amino group of
lysine residue side chains, reconstitute positive charge on the
surface of lysine and increase binding affinity with the negatively
charged surface of DNA (5,6).
Furthermore, interactions between histone and DNA result in the
formation of compacted and inactive chromatin that restrains gene
transcription (7–10).
The dysfunction of histone deacetylation is
associated with the occurrence and development of liver disease
(11). HDACs are emerging as
next-generation drug targets, thus gaining increasing attention and
recognition (12–15). HDAC2 is responsible for the
deacetylation of the N-terminal of histone H3 and H4, leading to a
more compacted chromatin structure and transcriptional gene
silencing (16,17). HDAC2 participates in the genesis
and development of renal, cardiovascular, neurological and lung
disease (18–23). Furthermore, small molecular
compounds, peptides and other biological agents that inhibit HDAC2
show potential in the treatment of cancer (24–26), as well as degenerative and
inflammatory immune disease (27–29). In particular, evidence has also
highlighted the key role of HDAC2 in the pathological process of
liver disease (30–32).
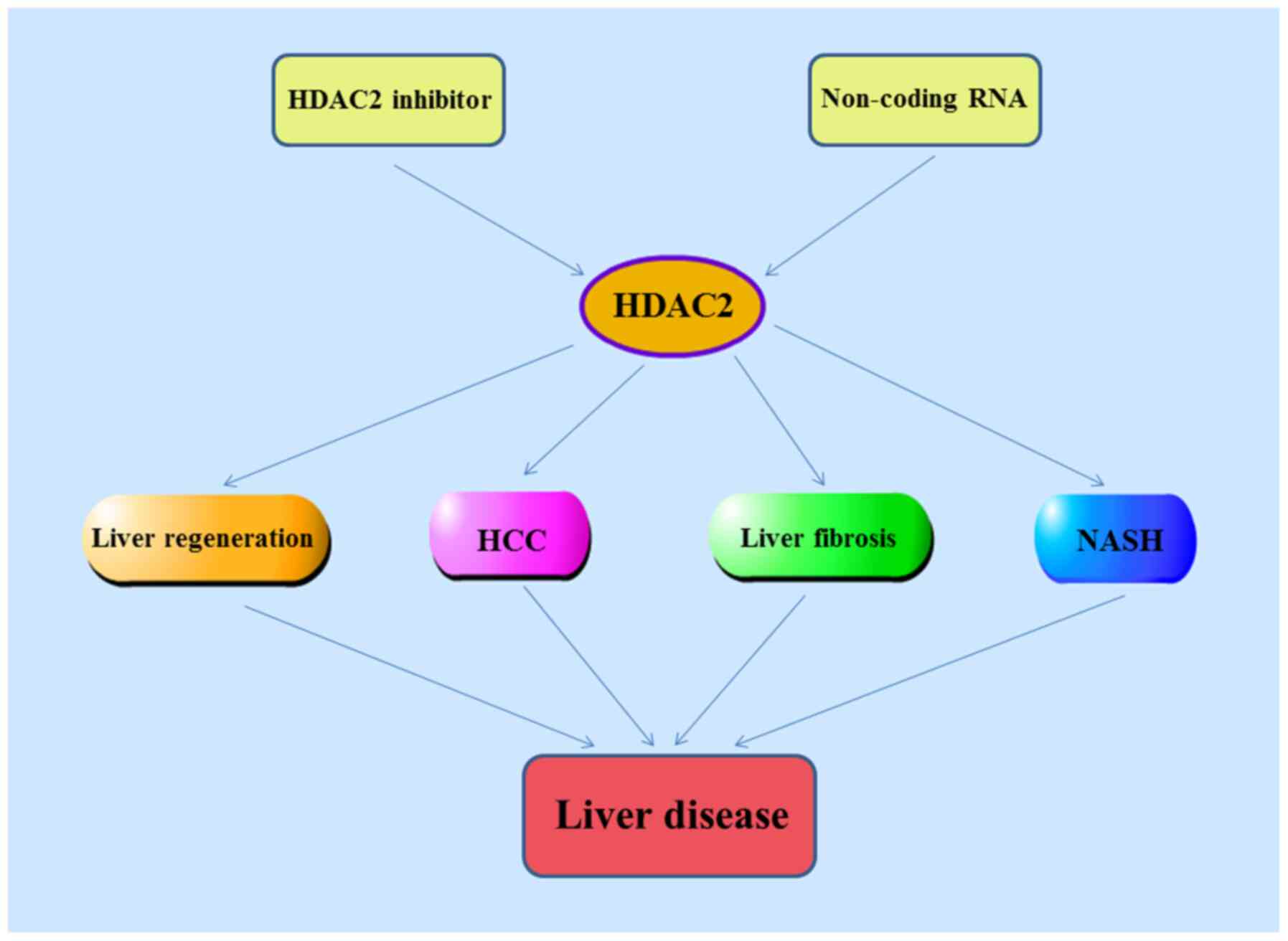
The present review introduces gene localization,
structural information and biological functions of HDAC2, as well
as the pharmacological role of HDAC2, its expression level in a
variety of liver diseases and effects on hepatocyte apoptosis and
proliferation, liver regeneration, hepatocellular carcinoma (HCC),
liver fibrosis and non-alcoholic steatohepatitis (NASH). Finally, a
number of selective HDAC2 inhibitors and non-coding (nc)RNAs
relevant for HDAC2 expression in liver disease are reviewed
(Fig. 1).
Histone proteins serve structural and functional
roles almost in all nuclear processes (33,34). Histones, DNA and a number of
different protein complexes form chromatin to facilitate dynamic
changes that occur during DNA replication, cell-cycle progression,
transcription process and post-transcription events (35). Changes in chromatin that do not
involve a change in DNA sequence are defined as epigenetic
modification. One of the earliest known types of chromatin
epigenetic modifications is histone acetylation, although its
potential role in cell fate determination has not been fully
elucidated (36). Acetylation
has been widely studied and its potential roles and regulatory
mechanisms have been revealed (37,38).
HDAC2 belongs to class I HDAC family, which is
primarily located in the nucleus. HDAC2 is a specific enzyme with
high activity and enantioselectivity to histone substrates
(60). HDAC2 shares high
structural homology and a common catalytic mechanism with other
class I HDACs, particularly HDAC1. Similar to other class I HDACs,
HDAC2 comprises a conserved deacetylase domain with short amino-
and carboxy-terminal extensions, which are key for localization and
maintaining their stability and function (48). HDAC1 and HDAC2 have notable amino
acid homology. In large-scale gene expression analysis of brain and
heart tissue, they affect different target gene sets by forming the
same compressor complex (61).
The crystal structure of human HDAC2 protein in the presence of
hydroxamates has been revealed. The HDAC2 catalytic site is made up
of a 'foot pocket', a lipophilic 'tube' and a catalytic
Zn2+ 8 Å deep. More specifically, the 'foot pocket',
tightly adjacent to the zinc binding site, is primarily formed by
Tyr29, Met35, Phe114 and Leu144. The lipophilic tube, leading from
the surface to the zinc binding site, is surrounded by Gly154,
Phe155, His183, Phe210 and Leu276 (62,63). Furthermore, the zinc ion is
accompanied by Asp181, His183, and Asp269 (62–64). HDAC2 inhibitors typically have a
pharmacophore comprising three sectors: A zinc-binding group, a
linker portion and a hydrophobic cap group (65–67). Based on molecular docking and
virtual screening techniques, a series of compounds with novel
skeletal structures have been identified as HDAC inhibitors, and
their inhibitory activities and clinical therapeutic effects have
been investigated (68,69). HDAC2 is the most thoroughly
studied member of the HDAC family, which can be modulated by
post-translational modifications, such as phosphorylation (70,71), acetylation (72), ubiquitination (73) and sumoylation (74). In particular, post-translational
phosphorylation of HDAC2 negatively regulates its deacetylase
activity and serves an active role in chronic inflammation
(75). Furthermore, HDAC2
possesses several phosphorylation sites at the C-terminal, which
are concentrated on serine residue (76).
Due to its anatomical location and complex
intersection, the liver is vulnerable to a variety of toxic,
metabolic, inflammatory and necrotic stimuli (77,78). Hepatocytes are highly sensitive
to death receptor-mediated apoptosis due to ubiquitous expression
of death receptors in the liver (79). It has been demonstrated that the
abnormal death of hepatocytes is a key trigger factor for the
occurrence and development of both acute and chronic liver disease
(77). It is therefore necessary
to investigate the underlying pathogenesis and therapeutic targets
of liver disease and the expression levels and pharmacological
activity of HDAC2. There is increasing evidence that HDAC2 is
involved in the regulation of hepatocyte death, thereby implying
that HDAC2 serves a key role in the pathogenesis of liver disease
(80,81). Here, experimental evidence for
the effects of HDAC2 on hepatocyte apoptosis will be discussed.
TGF-β, a pleiotropic growth factor, has been shown
to induce apoptosis in primary rat and AML-12 murine hepatocytes,
consequently leading to liver injury and regeneration termination
(80,81). Through transfection of HDAC2 RNA
interference (RNAi) plasmid, Lei et al (80) found that HDAC2 serves as a
significant anti-apoptotic factor in TGF-β1-induced apoptosis of
AML-12. Lei et al further found that downregulation of HDAC2
significantly induces spontaneous cell apoptosis and increases the
apoptotic response following TGF-β1 treatment. Moreover, ERK1/2 has
been shown to be significantly inhibited in cells transfected with
HDAC2i. In HDAC2i-transfected cells, low baseline levels of
phosphorylated ERK1/2 were concomitant with decreased
TGF-β1-induced apoptosis, suggesting that negative regulation of
ERK1/2 is associated with the role of HDAC2 in apoptosis. In sum,
these findings highlighted that silencing HDAC2 may induce
spontaneous apoptosis of AML12 hepatocytes by promoting ERK1/2
expression and pharmacological activity. Taken together, this
supports the important physiological role of HDAC2 in hepatocyte
apoptosis. The anti-apoptotic effect of HDAC2 overexpression may be
a promising therapeutic strategy for treatment of liver
disease.
Impaired proliferation of hepatocytes is associated
with the occurrence and progression of liver disease (82). However, little is known about the
underlying mechanisms that lead to defective hepatocyte
proliferation. HDAC2 serves critical roles in cell proliferation
and tissues regeneration (83).
HDAC2 knockout inhibits proliferation and induces senescence of
MCF7 cells by enhancing the binding activity and interaction of
p53-DNA (84). HDAC2 deficiency
usually results in different cellular phenotypes, suggesting that
HDAC2 has a cell-type-specific role that may be relevant to the
cell proliferative status (30).
Turgeon et al (85)
demonstrated that defects in tissue structure and perturbation of
microenvironment homeostasis are accompanied by inhibition of cell
proliferation when HDAC1/2 is knocked out. There is increasing
evidence that HDAC2 promote the proliferation of liver cancer cell
lines, although there is no indication of HDAC2 involvement in the
proliferation of normal liver cells (86–88). Ler et al (86) found that the combined knock out
of HDAC2 and HDAC1 decreases cell proliferation and improves
survival of patients with HCC. HDAC2 overexpression is routinely
detected in cancer cells, and HDAC2 deficiency and inhibition lead
to HCC cell apoptosis (87). The
role of HDAC2 in cell proliferation was previously observed in the
development of cardiac and B cells; HDAC2 and HDAC1 jointly inhibit
cell cycle protein-dependent kinase p21 (WAF1/CIP1) and p57KIP2
transcription and promote progression from G1 to S phase
(88). By contrast, HDAC2
suppresses transcription of p21WAF1/CIP1 via binding to Sp1-binding
site enriched proximal region of the p21WAF1/CIP1 promoter
(89).
Liver regeneration is of clinical significance in
various types of liver disease (90,91). In the event of massive hepatocyte
loss or damage, the intrinsic regenerative capacity of hepatocytes
is activated by endogenous molecule-mediated signaling pathways
(92). Rapid synchronous
compensatory regeneration occurs following 2/3 partial hepatectomy
(PH), and regenerated hepatocytes immediately enter the cell cycle
and proliferate rapidly, restoring their original quality and
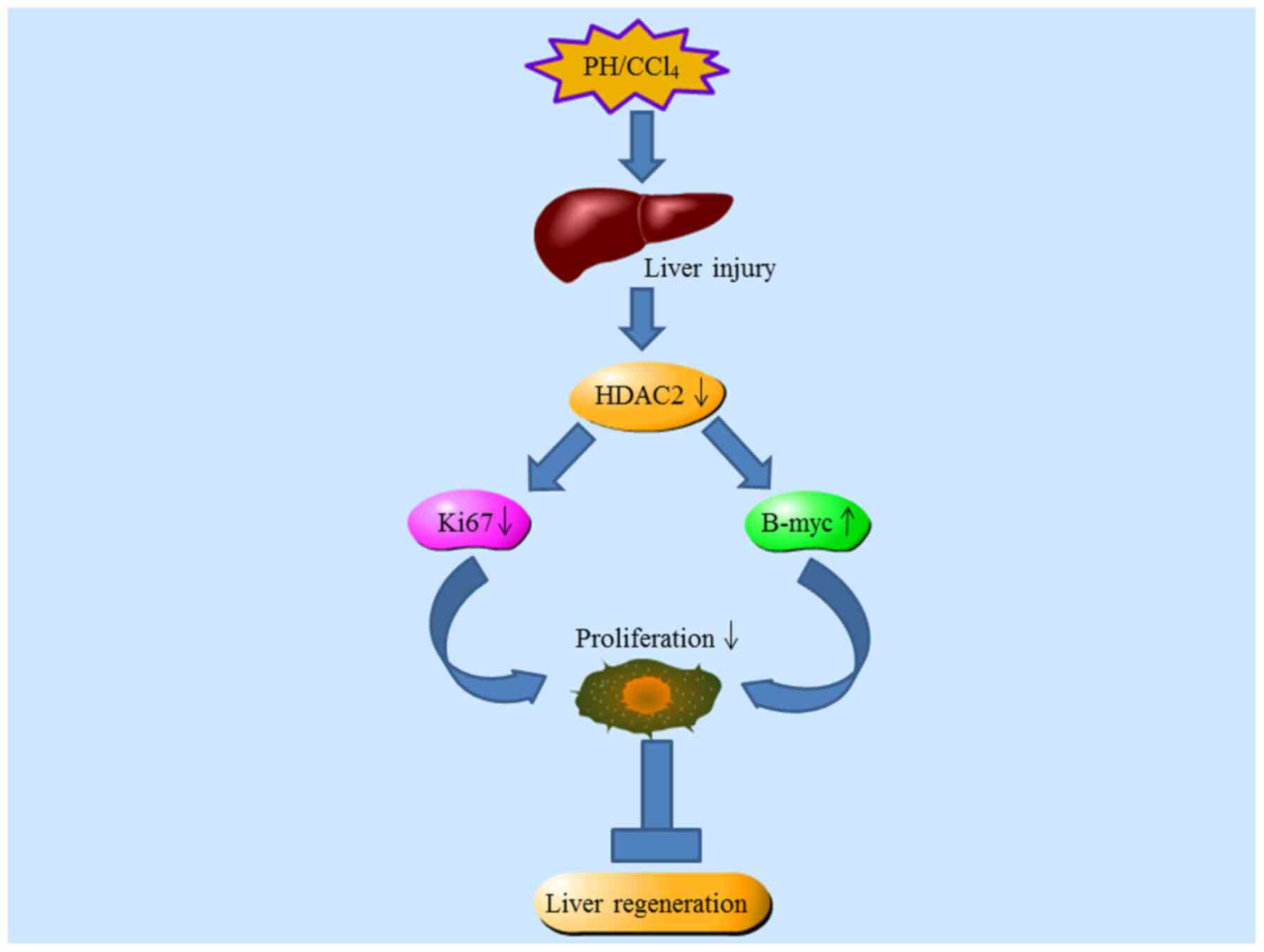
function (93). Studies on the
deficiency of HDAC2 in mouse liver regeneration have confirmed the
key role of HDAC2 in liver regeneration (94,95). Following PH, the liver/body
weight ratio is significantly lower in hepatocyte-selective
HDAC2-/- mice compared with wild-type mice;
HDAC2-/- mice also show more severe liver damage.
Additionally, the expression of HDAC2 gradually increases within
0.5 to 3.0 days in mouse post-hepatectomy livers at an early stage
of regeneration. Ki67, a mitotic marker, is decreased by ~30–70% in
HDAC2 knockout mice, subsequently leading to defective mitosis.
Decreased cyclinD1 and CDK2 in HDAC2-deficient hepatocytes suggests
that HDAC2 liver-specific knockout triggers downregulation of cell
cycle proteins and blocks cell cycle progression. Studies have
shown that HDAC2 is expressed differently in male and female mice,
and HDAC2 can directly bind to the promoter of B-myc (93,95). The expression of B-myc in the
female liver is higher than in the male liver, which may be
potential mechanism for the significantly slower rate of
replication and quality reconstruction of individual female
hepatocytes following PH. In conclusion, the altered metabolic
pattern in HDAC2 knockout mice is consistent with the well-known
regenerative characteristic of hepatocytes. This evidence also
confirms a key role for HDAC2 in the metabolic response following
PH (Fig. 2).
Liver fibrosis, primarily characterized by excessive
accumulation of extracellular matrix proteins, is a worldwide
medical problem with increasing annual morbidity (96,97). The majority cases of liver
fibrosis arise in the context of various etiology of liver damage,
such as chronic viral infections (98), excessive alcohol consumption
(99), metabolic disorder
(100) or autoimmune disease
(101). The regression and
improvement of liver fibrosis are primarily attributed to
inactivation and apoptosis of activated hepatic stellate cells
(HSCs) (102). Notably,
emerging evidence has revealed the potential features and roles of
HDACs in the progression of liver fibrosis (103,104). It was also reported that
several HDACs are involved in the activation of HSCs and the
progression of hepatic fibrosis (105,106). In addition, accumulating
evidence has highlighted the key role of HDAC2 in the development
of renal fibrosis and pulmonary fibrosis (18,107,108). In this regard, it is worth
verifying the functional role of HDAC2 in the occurrence and
reversal of liver fibrosis.
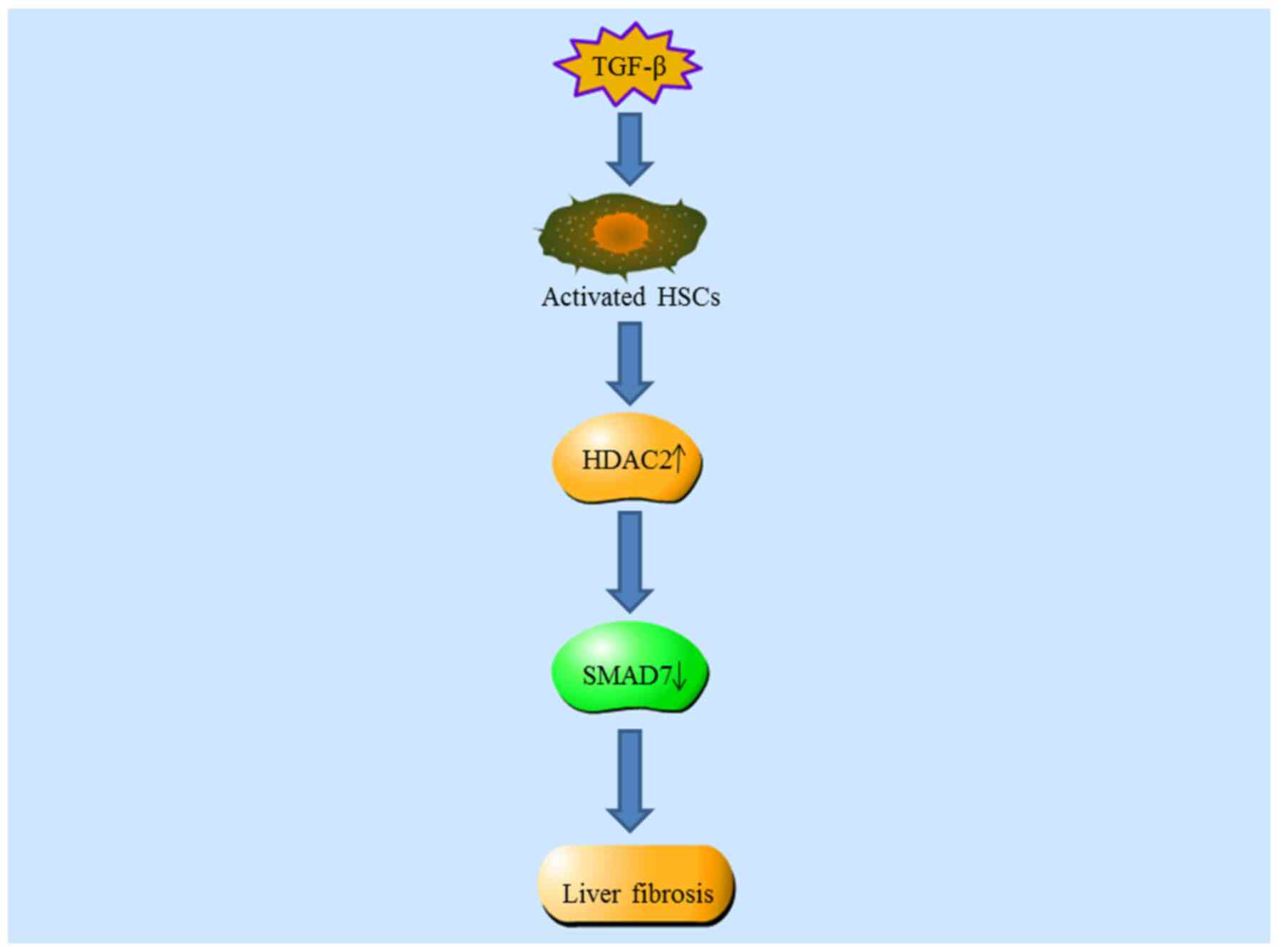
In order to reveal the possible association between
HDAC2 and liver fibrosis, a CCl4-induced mouse liver
fibrosis and its spontaneous reversal model have been successively
established (119); this
supported the view that aberrant HDAC expression and activity
participate in the occurrence and development of liver fibrosis.
Expression of HDAC2 is increased during CCl4-induced
liver fibrosis and significantly decreased during its reversal.
Similarly, the expression of HDAC2 is also significantly increased
in human hepatic fibrosis. Exposure of HSC-T6 cells to TGF-β1
results in increased HDAC2 expression in a dose- and time-dependent
manner. Loss-of-function analyses have confirmed that loss of HDAC2
induces cell cycle arrest and inhibits the expression of collagen
1α1 and α-smooth muscle actin protein in HSC-T6 cells activated by
TGF-β1 (110–112). Mechanistically, it has been
widely reported that HDAC2-small interfering (si)RNA leads to
increased expression of SMAD7 compared with scramble
siRNA-transfected groups (111,112). Collectively, these results
suggest that HDAC2 activates HSCs and promotes the occurrence of
liver fibrosis by suppressing SMAD7 expression. In conclusion,
these findings may demonstrate the role of HDAC2 in the progression
and reversal of liver fibrosis, and therefore have significant
implications for the development of novel treatment strategies for
liver fibrosis (Fig. 3).
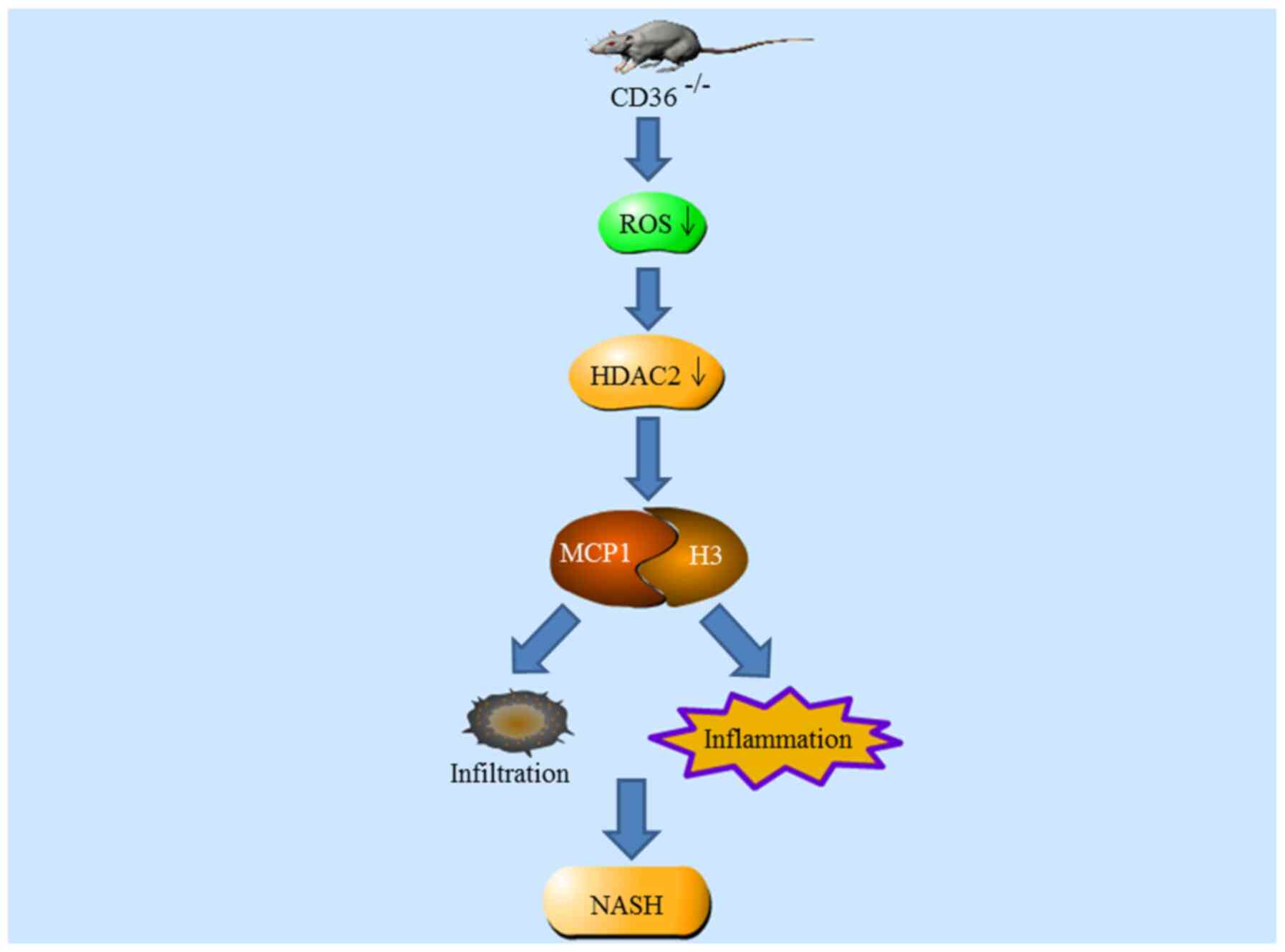
NASH, a more aggressive form of non-alcoholic fatty
liver disease, is pathologically characterized by cell damage,
inflammatory cell infiltration and hepatocyte ballooning (113,115). Sustained accumulation of
reactive oxygen species (ROS) and resultant oxidative stress,
mitochondrial dysfunction and accumulation of triglyceride and
lipotoxic metabolites have been identified as contributing factors
to NASH (115). To date, there
are no current Food and Drug Administration (USA)-approved
effective therapies to manage NASH (116). The inhibitory modulation of
HDAC2 may contribute to the prevention of NASH (117).
HCC is one of the most common types of solid
malignancy and is driven by different molecular mechanisms
(118,119). Researchers have linked gene
expression signatures with the occurrence and prognosis of HCC and
investigated gene expression patterns and potential therapeutic
targets (120). Evidence
suggests that HDAC2 is overexpressed in tumors, and HDAC2
downregulation leads to high expression levels of cell cycle
circuit elements, including p21WAF1/Cip1, which is a
well-characterized regulatory factor that serves a key role in cell
senescence (121,122). With regard to liver cancer,
HDAC2 promotes proliferation, and its aberrant expression may be a
prognostic indicator of HCC (86).
A study found that HDAC2 overexpression is
associated with poor survival of patients with low-grade and
early-stage tumors, suggesting that HDAC2 is an independent and
reliable predictor of survival of patients with HCC (123). Noh et al (89) assessed the tumorigenic potential
of HDAC2, evaluated abnormal HDAC2 expression and investigated its
regulatory mechanism in HCC; abnormal regulation of HDAC2 served a
key role in HCC progression by regulating cell cycle regulatory
components at the transcriptional level. Their data also showed
that HDAC2 overexpression was not associated with Wnt and c-myc
signaling pathways, which play an important role in malignant cell
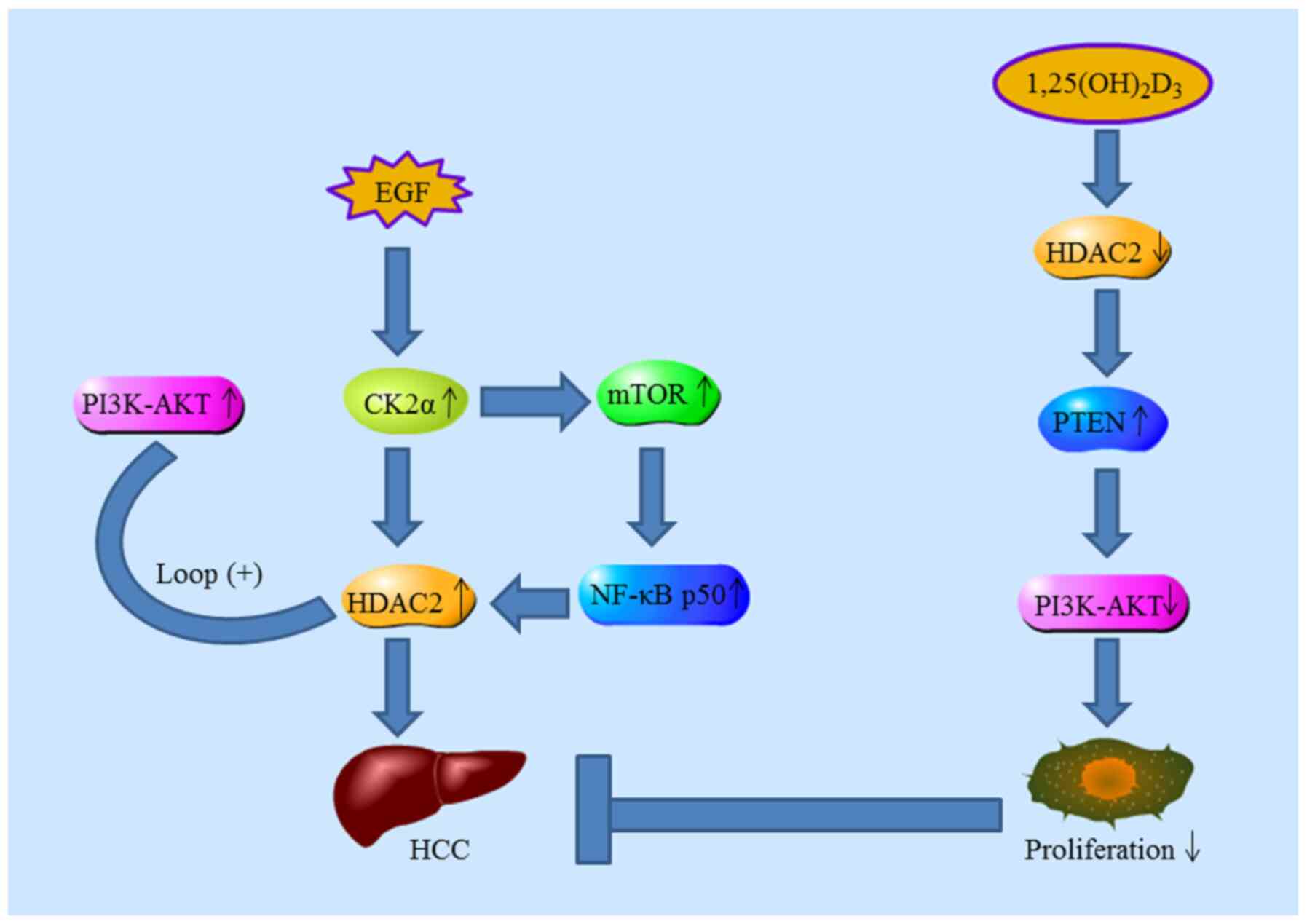
proliferation. Kim et al (124) investigated the underlying
mechanism of HDAC2 in tumorigenesis; increased expression of casein
kinase II (CK2α) was positively correlated with HDAC2. They
proposed a regulatory mechanism whereby increased HDAC2 expression
in HCC is primarily caused by the activation of CK2α/AKT pathways
mediated by EGF. Lee et al (108) indicated that systemic delivery
of HDAC2 siRNA encapsulated in lipid nanoparticles is sufficient to
inhibit HCC progression. In addition, mTORC1 activation and NF-κB
p50 nuclear translocation are essential for the transcriptional
activation of oncogenic HDAC2 in HCC (125). Furthermore,
1,25(OH)2D3 inhibits the progression of HCC
by downregulating HDAC2 (126,127). Consistently, Wang et al
(128) also found that high
levels of HDAC2 expression are negatively correlated with PTEN
expression in HCC patients with poor prognosis (Fig. 5).
Merck60 selectively inhibits HDAC1 and HDAC2,
thereby increasing histone acetylation and disrupting core gene
regulatory architecture in rhabdomyosarcoma (129). Methot et al (130) investigated novel selective
HDAC1/HDAC2 inhibitors (SHI-1:2), which incorporate a biaryl
zinc-binding motif into a nicotinyl scaffold; the optimized SHI-1:2
structure exhibited notable inhibitory activity against HDAC1 and
HDAC2, and its specific selectivity for HDAC1/HDAC2 was 100 times
higher than that for other HDACs (131). N-(2-amino-5-substituted phenyl)
benzamide significantly induced HCT116 cell death by specifically
targeting HDAC2 (62). In
addition, the effects of C15 urushiol and its triazole derivatives
on the apoptosis of liver cancer cells have been qualitatively and
quantitatively verified (132).
Venturelli et al (133)
reported that 6- and 8-prenylnaringenin enter into the 'foot
pocket' of HDAC2 and combine with zinc ion of their catalytic
center, subsequently inhibiting excessive proliferation of melanoma
cells. N-[4-(Hydrazinecarbonyl)phenyl]-3,5,6-trimethylpyr-
azine-2-carboxamide exhibits notable anticancer activity in
vivo (IC50=1.60 μM) (134) Among squaramide-based
derivatives, the lead compound 42 exhibits good druggability by
specifically inhibiting HDAC2 (67). Isopropyl derivative 5 and
tert-butyl derivative 6, is which derived from the lead compound
NSC746457, exhibit a significantly inhibitory effect on HDAC2
(63). Novel indazole and
pyrazolo(3,4-b) pyridine derivatives have been designed and
synthesized via fragment-based virtual screening; biological
evaluation showed that compounds 15k and 15m possess distinctly
inhibitory effect towards HDAC2 (66). Rosmarinic acid has been
demonstrated to downregulate HDAC2 expression, subsequently leading
to cell cycle arrest and apoptosis (135).
N-(2-aminophenyl)–4-[(4-fluorophenoxy)methyl] benzamide exhibits
antitumor activity by inhibiting HDAC2 at an IC50 of
3.84 μM (65). In
addition, a series of 2-aminobenzamide-based compounds exhibit
highly inhibitory effects on solid cancer cell lines and low
cytotoxicity against normal cells (Table II) (68).
In summary, these results demonstrate that HDAC2
possesses carcinogenic properties. These studies also suggest that
the development of novel compounds or ncRNAs may be a promising
therapeutic modality for liver cancer by selectively targeting
HDAC2.
Epigenetic modifications serve prime regulatory
roles in genetic events, such as transcriptional activation and
silencing (141). The effects
of epigenetic modifications have been recognized, although their
specific roles may still be controversial. Histone acetylation
contributes to gene expression, while histone deacetylation leads
to suppression of gene transcription (142). Studies have shown that relative
levels of histone acetylation and deacetylation are of significance
for the regulation of pathophysiological processes, including
proliferation, cell-cycle progression, differentiation, immune
evasion, inflammatory lesion, apoptosis and death (143–145). Pharmacological inhibition of
HDAC activity or expression alters chromatin acetylation levels,
subsequently confusing boundaries between transcriptionally active
and quiescent chromatin (146–148).
The increasing incidence of liver disease requires
novel effective therapeutic interventions. HDAC2 exhibits
attractive pharmacological effects in hepatocyte loss or injury,
HCC and NASH by modulating hepatocyte death and regulating cell
cycle components. In the past decades, researchers have
characterized HDAC classification, structure and subcellular
localization (47–50,52,54). The malignant or beneficial role
of HDAC2 in liver fibrosis, non-alcoholic fatty liver disease and
liver cancer has been revealed. Nonetheless, the mechanism of HDAC2
in the development of liver disease has not been elucidated and
more investigations are needed in future. In summary, these
properties of HDAC2 make it an appealing therapeutic target by
regulating expression of HDAC2 and HDAC2-dependent signaling
pathways.
HDAC2 presents favorable characteristics as a
potential drug target. Consequently, pharmacological agents that
inhibit HDAC2 may be a prospective treatment for liver ailments.
However, the few known HDAC2 inhibitors are broad spectrum
inhibitors that simultaneously inhibit several members of HDACs
family and thus may have more potential adverse side effects.
Therefore, it is necessary to develop novel HDAC2 inhibitors with
higher targeting selectivity. High homology and cellular
co-localization of multiple HDACs makes development and use of
HDAC2 inhibitors difficult. The expression and pharmacological
activity of HDAC2 is important for the prediction, diagnosis and
prognosis of liver disease. NF-κB, c-Myc, Sp1 and Sp3 can bind to
the HDAC2 promoter, thereby augmenting HDAC2 transcription
(149–151). These findings suggest that the
HDAC2 promoter may also be a potential target for pharmacological
intervention.
The present study reviewed the specific roles of
HDAC2 and the potential application of HDAC2 inhibitors in liver
disease. Increasing evidence has highlighted the key role of HDAC2
in the occurrence and development of liver disease and demonstrated
that HDAC2 inhibitor therapy may be a therapeutic approach. Better
understanding of the potential roles and regulatory mechanisms of
HDAC2 in liver disease may improve the ability to predict the pace
of liver disease progression and exploit specific targeted
therapeutic strategies.
Not applicable.
YRL, JL and JQW conceived and designed the study and
wrote the manuscript. ZGH, RNC and XC prepared the figures. DCZ and
HXY prepared the tables. QX, XRW and HYZ revised the manuscript.
All authors read and approved the final version of the manuscript.
Data authentication is not applicable.
Not applicable.
Not applicable.
The authors declare that they have no competing
interests.
Not applicable.
The present study was supported by the National Natural Science
Foundation of China (grant no. 81770609).
|
1
|
Cai C, Yu H, Huang G, Du X, Yu X, Zhou Y
and Shen W: Histone modifications in fatty acid synthase modulated
by carbohydrate responsive element binding protein are associated
with non-alcoholic fatty liver disease. Int J Mol Med.
42:1215–1228. 2018.PubMed/NCBI
|
|
2
|
Ferriero R, Nusco E, De Cegli R, Carissimo
A, Manco G and Brunetti-Pierri N: Pyruvate dehydrogenase complex
and lactate dehydrogenase are targets for therapy of acute liver
failure. J Hepatol. 69:325–335. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Kanyal A, Rawat M, Gurung P, Choubey D,
Anamika K and Karmodiya K: Genome-wide survey and phylogenetic
analysis of histone acetyltransferases and histone deacetylases of
Plasmodium falciparum. FEBS J. 285:1767–1782. 2018. View Article : Google Scholar
|
|
4
|
Berger SL: The complex language of
chromatin regulation during transcription. Nature. 447:407–412.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Khangura RK, Bali A, Jaggi AS and Singh N:
Histone acetylation and histone deacetylation in neuropathic pain:
An unresolved puzzle? Eur J Pharmacol. 795:36–42. 2017. View Article : Google Scholar
|
|
6
|
Kouzarides T: Chromatin modifications and
their function. Cell. 128:693–705. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Leipe DD and Landsman D: Histone
deacetylases, acetoin utilization proteins and acetylpolyamine
amidohydrolases are members of an ancient protein superfamily.
Nucleic Acids Res. 25:3693–3697. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
West AC and Johnstone RW: New and emerging
HDAC inhibitors for cancer treatment. J Clin Invest. 124:30–39.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ibi D and Gonzalez-Maeso J: Epigenetic
signaling in schizophrenia. Cell Signal. 27:2131–2136. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Levenson JM, O'Riordan KJ, Brown KD, Trinh
MA, Molfese DL and Sweatt JD: Regulation of histone acetylation
during memory formation in the hippocampus. J Biol Chem.
279:40545–40559. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Cao YN, Xue Y, Xue L, Jiang X, Wang X,
Zhang Z, Yang J, Lu J, Zhang C, Wang W and Ning G: Hepatic menin
recruits SIRT1 to control liver steatosis through histone
deacetylation. J Hepatol. 59:1299–1306. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Simões-Pires C, Zwick V, Nurisso A,
Schenker E, Carrupt PA and Cuendet M: HDAC6 as a target for
neurodegenerative diseases: What makes it different from the other
HDACs? Mol Neurodegener. 8:72013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Kim HJ and Bae SC: Histone deacetylase
inhibitors: Molecular mechanisms of action and clinical trials as
anti-cancer drugs. Am J Transl Res. 3:166–179. 2011.PubMed/NCBI
|
|
14
|
Glauben R, Batra A, Stroh T, Erben U,
Fedke I, Lehr HA, Leoni F, Mascagni P, Dinarello CA, Zeitz M and
Siegmund B: Histone deacetylases: Novel targets for prevention of
colitis-associated cancer in mice. Gut. 57:613–622. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Dokmanovic M, Clarke C and Marks PA:
Histone deacetylase inhibitors: Overview and perspectives. Mol
Cancer Res. 5:981–989. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Marchion DC, Bicaku E, Turner JG, Schmitt
ML, Morelli DR and Munster PN: HDAC2 regulates chromatin plasticity
and enhances DNA vulnerability. Mol Cancer Ther. 8:794–801. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Jahan S, Sun JM, He S and Davie JR:
Transcription-dependent association of HDAC2 with active chromatin.
J Cell Physiol. 233:1650–1657. 2018. View Article : Google Scholar
|
|
18
|
Noh H, Oh EY, Seo JY, Yu MR, Kim YO, Ha H
and Lee HB: Histone deacetylase-2 is a key regulator of diabetes-
and transforming growth factor-beta1-induced renal injury. Am J
Physiol Renal Physiol. 297:F729–F739. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Yang FQ, Liu M, Yang FP, Che J, Li W, Zhai
W, Wang GC, Zheng JH and Li X: VPA inhibits renal cancer cell
migration by targeting HDAC2 and down-regulating HIF–1α. Mol Biol
Rep. 41:1511–1518. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Fritzsche FR, Weichert W, Roske A, Gekeler
V, Beckers T, Stephan C, Jung K, Scholman K, Denkert C, Dietel M
and Kristiansen G: Class I histone deacetylases 1, 2 and 3 are
highly expressed in renal cell cancer. BMC Cancer. 8:3812008.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Shang L, Pin L, Zhu S, Zhong X, Zhang Y,
Shun M, Liu Y and Hou M: Plantamajoside attenuates
isoproterenol-induced cardiac hypertrophy associated with the HDAC2
and AKT/GSK–3β signaling pathway. Chem Biol Interact. 307:21–28.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Datta M, Staszewski O, Raschi E, Frosch M,
Hagemeyer N, Tay TL, Blank T, Kreutzfeldt M, Merkler D,
Ziegler-Waldkirch S, et al: Histone deacetylases 1 and 2 regulate
microglia function during development, homeostasis, and
neurodegeneration in a context-dependent manner. Immunity.
48:514–529.e6. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Bin YF, Wu LJ, Sun XJ, Liang Y, Bai J,
Zhang JQ, Li MH, Zhong XN, Liang YJ and He ZY: Expression of GR-α
and HDAC2 in steroid-Sensitive and steroid-Insensitive interstitial
lung disease. Biomed Pharmacother. 118:1093802019. View Article : Google Scholar
|
|
24
|
Mahady L, Nadeem M, Malek-Ahmadi M, Chen
K, Perez SE and Mufson EJ: HDAC2 dysregulation in the nucleus
basalis of Meynert during the progression of Alzheimer's disease.
Neuropathol Appl Neurobiol. 45:380–397. 2019. View Article : Google Scholar
|
|
25
|
Lin CL, Tsai ML, Lin CY, Hsu KW, Hsieh WS,
Chi WM, Huang LC and Lee CH: HDAC1 and HDAC2 double knockout
triggers cell apoptosis in advanced thyroid cancer. Int J Mol Sci.
20:4542019. View Article : Google Scholar :
|
|
26
|
Stojanovic N, Hassan Z, Wirth M, Wenzel P,
Beyer M, Schäfer C, Brand P, Kroemer A, Stauber RH, Schmid RM, et
al: HDAC1 and HDAC2 integrate the expression of p53 mutants in
pancreatic cancer. Oncogene. 36:1804–1815. 2017. View Article : Google Scholar
|
|
27
|
Tang W, Zhou W, Xiang L, Wu X, Zhang P,
Wang J, Liu G, Zhang W, Peng Y, Huang X, et al: The
p300/YY1/miR-500a-5p/HDAC2 signalling axis regulates cell
proliferation in human colorectal cancer. Nat Commun. 10:6632019.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Lai T, Wu M, Zhang C, Che L, Xu F, Wang Y,
Wu Y, Xuan N, Cao C, Du X, et al: HDAC2 attenuates airway
inflammation by suppressing IL-17A production in HDM-challenged
mice. Am J Physiol Lung Cell Mol Physiol. 316:L269–L279. 2019.
View Article : Google Scholar
|
|
29
|
Barnes PJ: Corticosteroid resistance in
patients with asthma and chronic obstructive pulmonary disease. J
Allergy Clin Immunol. 131:636–645. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Wilting RH, Yanover E, Heideman MR, Jacobs
H, Horner J, van der Torre J, DePinho RA and Dannenberg JH:
Overlapping functions of Hdac1 and Hdac2 in cell cycle regulation
and haematopoiesis. EMBO J. 29:2586–2597. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Wang Y, Yang F, Jiao FZ, Chen Q, Zhang WB,
Wang LW and Gong ZJ: Modulations of histone deacetylase 2 offer a
protective effect through the mitochondrial apoptosis pathway in
acute liver failure. Oxid Med Cell Longev.
2019:81730162019.PubMed/NCBI
|
|
32
|
Wu J, Zhu P, Lu T, Du Y, Wang Y, He L, Ye
B, Liu B, Yang L, Wang J, et al: The long non-coding RNA LncHDAC2
drives the self-renewal of liver cancer stem cells via activation
of Hedgehog signaling. J Hepatol. 70:918–929. 2019. View Article : Google Scholar
|
|
33
|
Verdone L, Agricola E, Caserta M and Di
Mauro E: Histone acetylation in gene regulation. Brief Funct
Genomic Proteomic. 5:209–221. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Millard CJ, Fairall L, Ragan TJ, Savva CG
and Schwabe JWR: The topology of chromatin-binding domains in the
NuRD deacetylase complex. Nucleic Acids Res. 48:12972–12982. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Verdone L, Caserta M and Di Mauro E: Role
of histone acetylation in the control of gene expression. Biochem
Cell Biol. 83:344–353. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Brownell JE, Zhou J, Ranalli T, Kobayashi
R, Edmondson DG, Roth SY and Allis CD: Tetrahymena histone
acetyltransferase A: A homolog to yeast Gcn5p linking histone
acetylation to gene activation. Cell. 84:843–851. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Kurdistani SK and Grunstein M: Histone
acetylation and deacetylation in yeast. Nat Rev Mol Cell Biol.
4:276–284. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Wang D, Kon N, Lasso G, Jiang L, Leng W,
Zhu WG, Qin J, Honig B and Gu W: Acetylation-regulated interaction
between p53 and SET reveals a widespread regulatory mode. Nature.
538:118–122. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Wang Z, Zang C, Cui K, Schones DE, Barski
A, Peng W and Zhao K: Genome-wide mapping of HATs and HDACs reveals
distinct functions in active and inactive genes. Cell.
138:1019–1031. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Abel T and Zukin RS: Epigenetic targets of
HDAC inhibition in neurodegenerative and psychiatric disorders.
Curr Opin Pharmacol. 8:57–64. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
de Ruijter AJ, van Gennip AH, Caron HN,
Kemp S and van Kuilenburg AB: Histone deacetylases (HDACs):
Characterization of the classical HDAC family. Biochem J. 370(Pt
3): 737–749. 2003. View Article : Google Scholar
|
|
42
|
Gregoretti IV, Lee YM and Goodson HV:
Molecular evolution of the histone deacetylase family: Functional
implications of phylogenetic analysis. J Mol Biol. 338:17–31. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Kiweler N, Brill B, Wirth M, Breuksch I,
Laguna T, Dietrich C, Strand S, Schneider G, Groner B, Butter F, et
al: The histone deacetylases HDAC1 and HDAC2 are required for the
growth and survival of renal carcinoma cells. Arch Toxicol.
92:2227–2243. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Bush EW and McKinsey TA: Protein
acetylation in the cardiorenal axis: The promise of histone
deacetylase inhibitors. Circ Res. 106:272–284. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Yang XJ and Seto E: The Rpd3/Hda1 family
of lysine deacetylases: From bacteria and yeast to mice and men.
Nat Rev Mol Cell Biol. 9:206–218. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Yang WM, Tsai SC, Wen YD, Fejer G and Seto
E: Functional domains of histone deacetylase-3. J Biol Chem.
277:9447–9454. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Martin M, Kettmann R and Dequiedt F: Class
IIa histone deacetylases: Regulating the regulators. Oncogene.
26:5450–5467. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Haberland M, Montgomery RL and Olson EN:
The many roles of histone deacetylases in development and
physiology: Implications for disease and therapy. Nat Rev Genet.
10:32–42. 2009. View Article : Google Scholar
|
|
49
|
Guardiola AR and Yao TP: Molecular cloning
and characterization of a novel histone deacetylase HDAC10. J Biol
Chem. 277:3350–3356. 2002. View Article : Google Scholar
|
|
50
|
Grozinger CM, Hassig CA and Schreiber SL:
Three proteins define a class of human histone deacetylases related
to yeast Hda1p. Proc Natl Acad Sci USA. 96:4868–4873. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Marks PA and Breslow R: Dimethyl sulfoxide
to vorinostat: Development of this histone deacetylase inhibitor as
an anti-cancer drug. Nat Biotechnol. 25:84–90. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Johnstone RW: Histone-deacetylase
inhibitors: Novel drugs for the treatment of cancer. Nat Rev Drug
Discov. 1:287–299. 2002. View
Article : Google Scholar : PubMed/NCBI
|
|
53
|
Michan S and Sinclair D: Sirtuins in
mammals: Insights into their biological function. Biochem J.
404:1–13. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Abbas A and Gupta S: The role of histone
deacetylases in prostate cancer. Epigenetics. 3:300–309. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Yamamoto H, Schoonjans K and Auwerx J:
Sirtuin functions in health and disease. Mol Endocrinol.
21:1745–1755. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Xu WS, Parmigiani RB and Marks PA: Histone
deacetylase inhibitors: Molecular mechanisms of action. Oncogene.
26:5541–5552. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Gao L, Cueto MA, Asselbergs F and Atadja
P: Cloning and functional characterization of HDAC11, a novel
member of the human histone deacetylase family. J Biol Chem.
277:25748–25755. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Verdin E and Ott M: 50 years of protein
acetylation: From gene regulation to epigenetics, metabolism and
beyond. Nat Rev Mol Cell Biol. 16:258–264. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Gong F and Miller KM: Mammalian DNA
repair: HATs and HDACs make their mark through histone acetylation.
Mutat Res. 750:23–30. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Brunmeir R, Lagger S and Seiser C: Histone
deacetylase HDAC1/HDAC2-controlled embryonic development and cell
differentiation. Int J Dev Biol. 53:275–289. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Montgomery RL, Hsieh J, Barbosa AC,
Richardson JA and Olson EN: Histone deacetylases 1 and 2 control
the progression of neural precursors to neurons during brain
development. Proc Natl Acad Sci USA. 106:7876–7881. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Bressi JC, Jennings AJ, Skene R, Wu Y,
Melkus R, De Jong R, O'Connell S, Grimshaw CE, Navre M and Gangloff
AR: Exploration of the HDAC2 foot pocket: Synthesis and SAR of
substituted N-(2-aminophenyl)benzamides. Bioorg Med Chem Lett.
20:3142–3145. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Hou J, Feng C, Li Z, Fang Q, Wang H, Gu G,
Shi Y, Liu P, Xu F, Yin Z, et al: Structure-based optimization of
click-based histone deacetylase inhibitors. Eur J Med Chem.
46:3190–3200. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Zhou H, Wang C, Ye J, Chen H and Tao R:
Design, virtual screening, molecular docking and molecular dynamics
studies of novel urushiol derivatives as potential HDAC2 selective
inhibitors. Gene. 637:63–71. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Xie R, Yao Y, Tang P, Chen G, Liu X, Yun
F, Cheng C, Wu X and Yuan Q: Design, synthesis and biological
evaluation of novel hydroxamates and 2-aminobenzamides as potent
histone deacetylase inhibitors and antitumor agents. Eur J Med
Chem. 134:1–12. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Liu J, Zhou J, He F, Gao L, Wen Y, Gao L,
Wang P, Kang D and Hu L: Design, synthesis and biological
evaluation of novel indazole-based derivatives as potent HDAC
inhibitors via fragment-based virtual screening. Eur J Med Chem.
192:112–189. 2020. View Article : Google Scholar
|
|
67
|
Fournier JF, Bhurruth-Alcor Y, Musicki B,
Aubert J, Aurelly M, Bouix-Peter C, Bouquet K, Chantalat L, Delorme
M, Drean B, et al: Squaramides as novel class I and IIB histone
deacetylase inhibitors for topical treatment of cutaneous t-cell
lymphoma. Bioorg Med Chem Lett. 28:2985–2992. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Yun F, Cheng C, Ullah S, He J, Zahi MR and
Yuan Q: Thioether-based 2-aminobenzamide derivatives: Novel HDAC
inhibitors with potent in vitro and in vivo antitumor activity. Eur
J Med Chem. 176:195–207. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Alsawalha M, Rao Bolla S, Kandakatla N,
Srinivasadesikan V, Veeraraghavan VP and Surapaneni KM: Molecular
docking and ADMET analysis of hydroxamic acids as HDAC2 inhibitors.
Bioinformation. 15:380–387. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Ford J, Ahmed S, Allison S, Jiang M and
Milner J: JNK2-dependent regulation of SIRT1 protein stability.
Cell cycle. 7:3091–3097. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Sun JM, Chen HY and Davie JR: Differential
distribution of unmodified and phosphorylated histone deacetylase 2
in chromatin. J Biol Chem. 282:33227–33236. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Ashktorab H, Belgrave K, Hosseinkhah F,
Brim H, Nouraie M, Takkikto M, Hewitt S, Lee EL, Dashwood RH and
Smoot D: Global Histone H4 Acetylation and HDAC2 expression in
colon adenoma and carcinoma. Dig Dis Sci. 54:2109–2117. 2009.
View Article : Google Scholar :
|
|
73
|
Krämer OH, Zhu P, Ostendorff HP,
Golebiewski M, Tiefenbach J, Peters MA, Brill B, Groner B, Bach I,
Heinzel T and Göttlicher M: The histone deacetylase inhibitor
valproic acid selectively induces proteasomal degradation of HDAC2.
EMBO J. 22:3411–3420. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Brandl A, Wagner T, Uhlig KM, Knauer SK,
Stauber RH, Melchior F, Schneider G, Heinzel T and Krämer OH:
Dynamically regulated sumoylation of HDAC2 controls p53
deacetylation and restricts apoptosis following genotoxic stress. J
Mol Cell Biol. 4:284–293. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Adenuga D and Rahman I: Protein kinase
CK2-mediated phosphorylation of HDAC2 regulates co-repressor
formation, deacetylase activity and acetylation of HDAC2 by
cigarette smoke and aldehydes. Arch Biochem Biophys. 498:62–73.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Tsai SC and Seto E: Regulation of histone
deacetylase 2 by protein kinase CK2. J Biol Chem. 277:31826–31833.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Chen PJ, Cai SP, Huang C, Meng XM and Li
J: Protein tyrosine phosphatase 1B (PTP1B): A key regulator and
therapeutic target in liver diseases. Toxicology. 337:10–20. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Kan C, Ungelenk L, Lupp A, Dirsch O and
Dahmen U: Ischemia-Reperfusion injury in aged Livers-The energy
metabolism, inflammatory response, and autophagy. Transplantation.
102:368–377. 2018. View Article : Google Scholar
|
|
79
|
Guicciardi ME, Malhi H, Mott JL and Gores
GJ: Apoptosis and necrosis in the liver. Compr Physiol. 3:977–1010.
2013.PubMed/NCBI
|
|
80
|
Lei WW, Zhang KH, Pan XC, Wang DM, Hu Y,
Yang YN and Song JG: Histone deacetylase 1 and 2 differentially
regulate apoptosis by opposing effects on extracellular
signal-regulated kinase 1/2. Cell Death Dis. 1:e442010. View Article : Google Scholar
|
|
81
|
Romero-Gallo J, Sozmen EG, Chytil A,
Russell WE, Whitehead R, Parks WT, Holdren MS, Her MF, Gautam S,
Magnuson M, et al: Inactivation of TGF-beta signaling in
hepatocytes results in an increased proliferative response after
partial hepatectomy. Oncogene. 24:3028–3041. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Raven A, Lu WY, Man TY, Ferreira-Gonzalez
S, O'Duibhir E, Dwyer BJ, Thomson JP, Meehan RR, Bogorad R,
Koteliansky V, et al: Cholangiocytes act as facultative liver stem
cells during impaired hepatocyte regeneration. Nature. 547:350–354.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Willis-Martinez D, Richards HW, Timchenko
NA and Medrano EE: Role of HDAC1 in senescence, aging, and cancer.
Exp Gerontol. 45:279–285. 2010. View Article : Google Scholar :
|
|
84
|
Harms KL and Chen X: Histone deacetylase 2
modulates p53 transcriptional activities through regulation of
p53-DNA binding activity. Cancer Res. 67:3145–3152. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Turgeon N, Blais M, Gagne JM, Tardif V,
Boudreau F, Perreault N and Asselin C: HDAC1 and HDAC2 restrain the
intestinal inflammatory response by regulating intestinal
epithelial cell differentiation. PLoS One. 8:e737852013. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Ler SY, Leung CH, Khin LW, Lu GD,
Salto-Tellez M, Hartman M, Iau PT, Yap CT and Hooi SC: HDAC1 and
HDAC2 independently predict mortality in hepatocellular carcinoma
by a competing risk regression model in a Southeast Asian
population. Oncol Rep. 34:2238–2250. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Noh JH, Chang YG, Kim MG, Jung KH, Kim JK,
Bae HJ, Eun JW, Shen Q, Kim SJ, Kwon SH, et al: MiR–145 functions
as a tumor suppressor by directly targeting histone deacetylase 2
in liver cancer. Cancer Lett. 335:455–462. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Makar AB, Mcmartin KE, Palese M and Tephly
TR: Formate assay in body fluids: Application in methanol poisoning
App. Biochem Med. 13:117–126. 1975. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Noh JH, Jung KH, Kim JK, Eun JW, Bae HJ,
Xie HJ, Chang YG, Kim MG, Park WS, Lee JY and Nam SW: Aberrant
Regulation of HDAC2 Mediates proliferation of hepatocellular
carcinoma cells by deregulating expression of G1/S cell cycle
proteins. PLoS One. 6:e281032011. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Yuan X, Yan S, Zhao J, Shi D, Yuan B, Dai
W, Jiao B, Zhang W and Miao M: Lipid metabolism and peroxisome
proliferator-activated receptor signaling pathways participate in
late-phase liver regeneration. J Proteome Res. 10:1179–1190. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Michalopoulos GK and Bhushan B: Liver
regeneration: Biological and pathological mechanisms and
implications. Nat Rev Gastroenterol Hepatol. 18:40–55. 2021.
View Article : Google Scholar
|
|
92
|
Michalopoulos GK: Principles of liver
regeneration and growth homeostasis. Compr Physiol. 3:485–513.
2013.PubMed/NCBI
|
|
93
|
Li L, Guo J, Chen Y, Chang C and Xu C:
Comprehensive CircRNA expression profile and selection of key
CircRNAs during priming phase of rat liver regeneration. BMC
Genomics. 18:802017. View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Xia J, Zhou Y, Ji H, Wang Y, Wu Q, Bao J,
Ye F, Shi Y and Bu H: Loss of Histone Deacetylases 1 and 2 in
hepatocytes impairs murine liver regeneration through Ki67
depletion. Hepatology. 58:2089–2098. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Wang Y, Ye F, Ke Q, Wu Q, Yang R and Bu H:
Gender-dependent histone deacetylases injury may contribute to
differences in liver recovery rates of male and female mice.
Transplant Proc. 45:463–473. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Bansal R, Nagorniewicz B and Prakash J:
Clinical advancements in the targeted therapies against liver
fibrosis. Mediators Inflamm. 2016:76297242016. View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Aydın MM and Akçalı KC: Liver fibrosis.
Turk J Gastroenterol. 29:14–21. 2018. View Article : Google Scholar
|
|
98
|
Gounder PP, Haering C, Bruden DJ,
Townshend-Bulson L, Simons BC, Spradling PR and McMahon BJ: Does
incorporating change in APRI or FIB–4 indices over time improve the
accuracy of a single index for identifying liver fibrosis in
persons with chronic hepatitis C virus infection? J Clin
Gastroenterol. 52:60–66. 2018. View Article : Google Scholar
|
|
99
|
Bilal U, Lau B, Lazo M, McCaul ME, Hutton
HE, Sulkowski MS, Moore RD and Chander G: Interaction between
alcohol consumption patterns, antiretroviral therapy type, and
liver fibrosis in persons living with HIV. AIDS Patient Care STDS.
30:200–207. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Lainé F, Bendavid C, Moirand R, Tessier S,
Perrin M, Guillygomarc'h A, Guyader D, Calon E, Renault A, Brissot
P, et al: Prediction of liver fibrosis in patients with features of
the metabolic syndrome regardless of alcohol consumption.
Hepatology. 39:1639–1646. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
101
|
Sunami Y, Leithäuser F, Gul S, Fiedler K,
Güldiken N, Espenlaub S, Holzmann KH, Hipp N, Sindrilaru A, Luedde
T, et al: Hepatic activation of IKK/NFκB signaling induces liver
fibrosis via macrophage-mediated chronic inflammation. Hepatology.
56:1117–1128. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
102
|
Zhang H, Wu P, Chen F, Hao Y, Lao Y, Ren
L, Sun L, Sun W, Wei H, Chan DW, et al: SILAC-based quantitative
proteomic analysis of secretome between activated and reverted
hepatic stellate cells. Proteomics. 14:1977–1986. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
103
|
Mannaerts I, Eysackers N, Onyema OO, Van
Beneden K, Valente S, Mai A, Odenthal M and van Grunsven LA: Class
II HDAC inhibition hampers hepatic stellate cell activation by
induction of microRNA–29. PLoS One. 8:e557862013. View Article : Google Scholar
|
|
104
|
Pannem RR, Dorn C, Hellerbrand C and
Massoumi R: Cylindromatosis gene CYLD regulates hepatocyte growth
factor expression in hepatic stellate cells through interaction
with histone deacetylase 7. Hepatology. 60:1066–1081. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
105
|
Mannaerts I, Nuytten NR, Rogiers V,
Vanderkerken K, van Grunsven LA and Geerts A: Chronic
administration of valproic acid inhibits activation of mouse
hepatic stellate cells in vitro and in vivo. Hepatology.
51:603–614. 2010. View Article : Google Scholar
|
|
106
|
Qin L and Han YP: Epigenetic repression of
matrix metalloproteinases in myofibroblastic hepatic stellate cells
through histone deacetylases 4: Implication in tissue fibrosis. Am
J Pathol. 177:1915–1928. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
107
|
Huang SK, Scruggs AM, Donaghy J, Horowitz
JC, Zaslona Z, Przybranowski S, White ES and Peters-Golden M:
Histone modifications are responsible for decreased Fas expression
and apoptosis resistance in fibrotic lung fibroblasts. Cell Death
Dis. 4:e6212013. View Article : Google Scholar : PubMed/NCBI
|
|
108
|
Lee YH, Seo D, Choi KJ, Andersen JB, Won
MA, Kitade M, Gómez-Quiroz LE, Judge AD, Marquardt JU, Raggi C, et
al: Antitumor effects in hepatocarcinoma of isoform-selective
inhibition of HDAC2. Cancer Res. 74:4752–4761. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
109
|
Li X, Wu XQ, Xu T, Li XF, Yang Y, Li WX,
Huang C, Meng XM and Li J: Role of histone deacetylases(HDACs) in
progression and reversal of liver fibrosis. Toxicol Appl Pharmacol.
306:58–68. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
110
|
Dooley S, Hamzavi J, Breitkopf K,
Wiercinska E, Said HM, Lorenzen J, Ten Dijke P and Gressner AM:
Smad7 prevents activation of hepatic stellate cells and liver
fibrosis in rats. Gastroenterology. 125:178–191. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
111
|
Dooley S, Hamzavi J, Ciuclan L, Godoy P,
Ilkavets I, Ehnert S, Ueberham E, Gebhardt R, Kanzler S, Geier A,
et al: Hepatocyte-specific Smad7 expression attenuates
TGF-beta-mediated fibrogenesis and protects against liver damage.
Gastroenterology. 135:642–659. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
112
|
Hamzavi J, Ehnert S, Godoy P, Ciuclan L,
Weng H, Mertens PR, Heuchel R and Dooley S: Disruption of the Smad7
gene enhances CCI4-dependent liver damage and fibrogenesis in mice.
J Cell Mol Med. 12(5B): 2130–2144. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
113
|
Oseini AM and Sanyal AJ: Therapies in
non-alcoholic steatohepatitis (NASH). Liver Int. 37(Suppl 1):
S97–S103. 2017. View Article : Google Scholar
|
|
114
|
Utsunomiya H, Yamamoto Y, Takeshita E,
Tokumoto Y, Tada F, Miyake T, Hirooka M, Abe M, Kumagi T, Matsuura
B, et al: Upregulated absorption of dietary palmitic acids with
changes in intestinal transporters in non-alcoholic steatohepatitis
(NASH). J Gastroenterol. 52:940–954. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
115
|
Fukushima J, Kamada Y, Matsumoto H,
Yoshida Y, Ezaki H, Takemura T, Saji Y, Igura T, Tsutsui S, Kihara
S, et al: Adiponectin prevents progression of steatohepatitis in
mice by regulating oxidative stress and Kupffer cell phenotype
polarization. Hepatol Res. 39:724–738. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
116
|
Afrin R, Arumugam S, Rahman A, Wahed MI,
Karuppagounder V, Harima M, Suzuki H, Miyashita S, Suzuki K,
Yoneyama H, et al: Curcumin ameliorates liver damage and
progression of NASH in NASH-HCC mouse model possibly by modulating
HMGB1-NF-κB translocation. Int Immunopharmacol. 44:174–182. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
117
|
Zhong S, Zhao L, Wang Y, Zhang C, Liu J,
Wang P, Zhou W, Yang P, Varghese Z, Moorhead JF, et al: CD36
deficiency aggravates macrophage infiltration and hepatic
inflammation by up-regulating MCP–1 expression of hepatocytes
through HDAC2-dependant pathway. Antioxid Redox Signal. Aug
1–2017.Epub ahead of print. View Article : Google Scholar
|
|
118
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
119
|
Chen Z, Xie H, Hu M, Huang T, Hu Y, Sang N
and Zhao Y: Recent progress in treatment of hepatocellular
carcinoma. Am J Cancer Res. 10:2993–3036. 2020.PubMed/NCBI
|
|
120
|
Lee JS, Chu IS, Heo J, Calvisi DF, Sun Z,
Roskams T, Durnez A, Demetris AJ and Thorgeirsson SS:
Classification and prediction of survival in hepatocellular
carcinoma by gene expression profiling. Hepatology. 40:667–676.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
121
|
Ropero S and Esteller M: The role of
histone deacetylases (HDACs) in human cancer. Mol Oncol. 1:19–25.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
122
|
Bayat S, Mansoori Derakhshan S, Mansoori
Derakhshan N, Shekari Khaniani M and Alivand MR: Downregulation of
HDAC2 and HDAC3 via oleuropein as a potent prevention and
therapeutic agent in MCF–7 breast cancer cells. J Cell Biochem.
120:9172–9180. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
123
|
Quint K, Agaimy A, Di Fazio P, Montalbano
R, Steindorf C, Jung R, Hellerbrand C, Hartmann A, Sitter H,
Neureiter D and Ocker M: Clinical significance of histone
deacetylases 1, 2, 3, and 7: HDAC2 is an independent predictor of
survival in HCC. Virchows Arch. 459:129–139. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
124
|
Kim HS, Chang YG, Bae HJ, Eun JW, Shen Q,
Park SJ, Shin WC, Lee EK, Park S, Ahn YM, et al: Oncogenic
potential of CK2α and its regulatory role in EGF-induced HDAC2
expression in human liver cancer. FEBS J. 281:851–861. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
125
|
Noh JH, Bae HJ, Eun JW, Shen Q, Park SJ,
Kim HS, Nam B, Shin WC, Lee EK, Lee K, et al: HDAC2 provides a
critical support to malignant progression of hepatocellular
carcinoma through feedback control of mTORC1 and AKT. Cancer Res.
74:1728–1738. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
126
|
Huang J, Yang G, Huang Y, Kong W and Zhang
S: 1,25(OH)2D3 inhibits the progression of hepatocellular carcinoma
via down-regulating HDAC2 and upregulating P21(WAFI/CIP1). Mol Med
Re. 13:1373–1380. 2016. View Article : Google Scholar
|
|
127
|
Huang J, Yang G, Huang Y and Zhang S:
Inhibitory effects of 1,25(OH)2D3 on the proliferation of
hepatocellular carcinoma cells through the downregulation of HDAC2.
Oncol Rep. 38:1845–1850. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
128
|
Wang H, Kohashi K, Yoshizumi T, Okumura Y,
Tanaka Y, Shimokawa M, Iwasaki T, Aishima S, Maehara Y and Oda Y:
Coexpression of SALL4 with HDAC1 and/or HDAC2 is associated with
underexpression of PTEN and poor prognosis in patients with
hepatocellular carcinoma. Hum Pathol. 64:69–75. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
129
|
Gryder BE, Pomella S, Sayers C, Wu XS,
Song Y, Chiarella AM, Bagchi S, Chou HC, Sinniah RS, Walton A, et
al: Histone hyperacetylation disrupts core gene regulatory
architecture in rhabdomyosarcoma. Nat Genet. 51:1714–1722. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
130
|
Methot JL, Hamblett CL, Mampreian DM, Jung
J, Harsch A, Szewczak AA, Dahlberg WK, Middleton RE, Hughes B,
Fleming JC, et al: SAR profiles of spirocyclic nicotinamide derived
selective HDAC1/HDAC2 inhibitors (SHI–1:2). Bioorg Med Chem Lett.
18:6104–6109. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
131
|
Methot JL, Chakravarty PK, Chenard M,
Close J, Cruz JC, Dahlberg WK, Fleming J, Hamblett CL, Hamill JE,
Harrington P, et al: Exploration of the internal cavity of histone
deacetylase (HDAC) with selective HDAC1/HDAC2 inhibitors (SHI-1:2).
Bioorg Med Chem Lett. 18:973–978. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
132
|
Qi Z, Wang C, Jiang J and Wu C: Novel C15
Triene Triazole, D-A derivatives anti-HepG2, and as HDAC2
inhibitors: A synergy study. Int J Mol Sci. 19:31842018. View Article : Google Scholar :
|
|
133
|
Venturelli S, Niessner H, Sinnberg T,
Berger A, Burkard M, Urmann C, Donaubauer K, Böcker A, Leischner C,
Riepl H, et al: 6– and 8-Prenylnaringenin, novel natural histone
deacetylase inhibitors found in hops, exert antitumor activity on
melanoma cells. Cell Physiol Biochem. 51:543–556. 2018. View Article : Google Scholar
|
|
134
|
Al-Sanea MM, Gotina L, Mohamed MF, Grace
Thomas Parambi D, Gomaa HA, Mathew B, Youssif BG, Alharbi KS,
Elsayed ZM, Abdelgawad MA and Eldehna WM: Design, synthesis and
biological evaluation of new HDAC1 and HDAC2 inhibitors endowed
with ligustrazine as a novel cap moiety. Drug Des Devel Ther.
14:497–508. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
135
|
Jang YG, Hwang KA and Choi KC: Rosmarinic
acid, a component of rosemary tea, induced the cell cycle arrest
and apoptosis through modulation of HDAC2 expression in prostate
cancer cell lines. Nutrients. 10:17842018. View Article : Google Scholar :
|
|
136
|
Deng L, Tang J, Yang H, Cheng C, Lu S,
Jiang R and Sun B: MTA1 modulated by miR–30e contributes to
epithelial-to-mesenchymal transition in hepatocellular carcinoma
through an ErbB2-dependent pathway. Oncogene. 36:3976–3985. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
137
|
Buurman R, Gürlevik E, Schäffer V, Eilers
M, Sandbothe M, Kreipe H, Wilkens L, Schlegelberger B, Kühnel F and
Skawran B: Histone deacetylases activate hepatocyte growth factor
signaling by repressing MicroRNA–449 in hepatocellular carcinoma
cells. Gastroenterology. 143:811–820.e15. 2012. View Article : Google Scholar
|
|
138
|
He QL, Qin SY, Tao L, Ning HJ and Jiang
HX: Prognostic value and prospective molecular mechanism of
miR–100-5p in hepatocellular carcinoma: A comprehensive study based
on 1,258 samples. Oncol Lett. 18:6126–6142. 2019.PubMed/NCBI
|
|
139
|
Kim HS, Lee KS, Bae HJ, Eun JW, Shen Q,
Park SJ, Shin WC, Yang HD, Park M, Park WS, et al: MicroRNA–31
functions as a tumor suppressor by regulating cell cycle and
epithelial-mesenchymal transition regulatory proteins in liver
cancer. Oncotarget. 6:8089–8102. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
140
|
Dai W, Dai JL, Tang MH, Ye MS and Fang S:
lncRNA-SNHG15 accelerates the development of hepatocellular
carcinoma by targeting miR–490-3p/histone deacetylase 2 axis. World
J Gastroenterol. 25:5789–5799. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
141
|
Turner BM: Cellular memory and the histone
code. Cell. 111:285–291. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
142
|
Khan SN and Khan AU: Role of histone
acetylation in cell physiology and diseases: An update. Clin Chim
Acta. 411:1401–1411. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
143
|
Budillon A, Di Gennaro E, Bruzzese F,
Rocco M, Manzo G and Caraglia M: Histone deacetylase inhibitors: A
new wave of molecular targeted anticancer agents. Recent Pat
Anticancer Drug Discov. 2:119–134. 2007. View Article : Google Scholar
|
|
144
|
Wade PA: Transcriptional control at
regulatory checkpoints by histone deacetylases: Molecular
connections between cancer and chromatin. Hum Mol Genet.
10:693–698. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
145
|
Forsberg EC and Bresnick EH: Histone
acetylation beyond promoters: Long-range acetylation patterns in
the chromatin world. Bioessays. 23:820–830. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
146
|
Zhang L, Qiu Z, Hu Y, Yang F, Yan S, Zhao
L, Li B, He S, Huang M, Li J and Li L: ABA treatment of germinating
maize seeds induces VP1 gene expression and selective
promoter-associated histone acetylation. Physiol Plant.
143:287–296. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
147
|
Tian XL, Lu X, Feng JB, Cai TJ, Li S, Tian
M and Liu QJ: Alterations in histone acetylation following exposure
to 60Co ү-rays and their relationship with chromosome
damage in human lymphoblastoid cells. Radiat Environ Biophys.
57:215–222. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
148
|
Heinz KS, Rapp A, Casas-Delucchi CS,
Lehmkuhl A, Romero-Fernández I, Sánchez A, Krämer OH, Marchal JA
and Cardoso MC: DNA replication dynamics of vole genome and its
epigenetic regulation. Epigenetics Chromatin. 12:182019. View Article : Google Scholar : PubMed/NCBI
|
|
149
|
Ibi D, de la Fuente Revenga M, Kezunovic
N, Muguruza C, Saunders JM, Gaitonde SA, Moreno JL, Ijaz MK,
Santosh V, Kozlenkov A, et al: Antipsychotic-induced Hdac2
transcription via NF-kB leads to synaptic and cognitive side
effects. Nat Neurosci. 20:1247–1259. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
150
|
Bhandari DR, Seo KW, Jung JW, Kim HS, Yang
SR and Kang KS: The regulatory role of c-MYC on HDAC2 and PcG
expression in human multipotent stem cells. J Cell Mol Med.
15:1603–1614. 2011. View Article : Google Scholar
|
|
151
|
Yang H, Salz T, Zajac-Kaye M, Liao D,
Huang S and Qiu Y: Overexpression of histone deacetylases in cancer
cells is controlled by interplay of transcription factors and
epigenetic modulators. FASEB J. 28:4265–4279. 2014. View Article : Google Scholar : PubMed/NCBI
|