|
1
|
Wakim KG: Physiology of the liver. Am J
Med. 16:256–271. 1954. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Lee S, Mardinoglu A, Zhang C, Lee D and
Nielsen J: Dysregulated signaling hubs of liver lipid metabolism
reveal hepatocellular carcinoma pathogenesis. Nucleic Acids Res.
44:5529–5539. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Han HS, Kang G, Kim JS, Choi BH and Koo
SH: Regulation of glucose metabolism from a liver-centric
perspective. Exp Mol Med. 48:e2182016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Wang Y, Song L, Liu M, Ge R, Zhou Q, Liu
W, Li R, Qie J, Zhen B, Wang Y, et al: A proteomics landscape of
circadian clock in mouse liver. Nat Commun. 9:15532018. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Evarts RP, Hu Z, Fujio K, Marsden ER and
Thorgeirsson SS: Activation of hepatic stem cell compartment in the
rat: Role of transforming growth factor alpha, hepatocyte growth
factor, and acidic fibroblast growth factor in early proliferation.
Cell Growth Differ. 4:555–561. 1993.PubMed/NCBI
|
|
6
|
Kang JH, Toita R and Murata M: Liver
cell-targeted delivery of therapeutic molecules. Crit Rev
Biotechnol. 36:132–143. 2016. View Article : Google Scholar
|
|
7
|
Gao B: Hepatoprotective and
anti-inflammatory cytokines in alcoholic liver disease. J
Gastroenterol Hepatol. 27(Suppl 2): S89–S93. 2012. View Article : Google Scholar
|
|
8
|
Asrani SK, Devarbhavi H, Eaton J and
Kamath PS: Burden of liver diseases in the world. J Hepatol.
70:151–171. 2019. View Article : Google Scholar
|
|
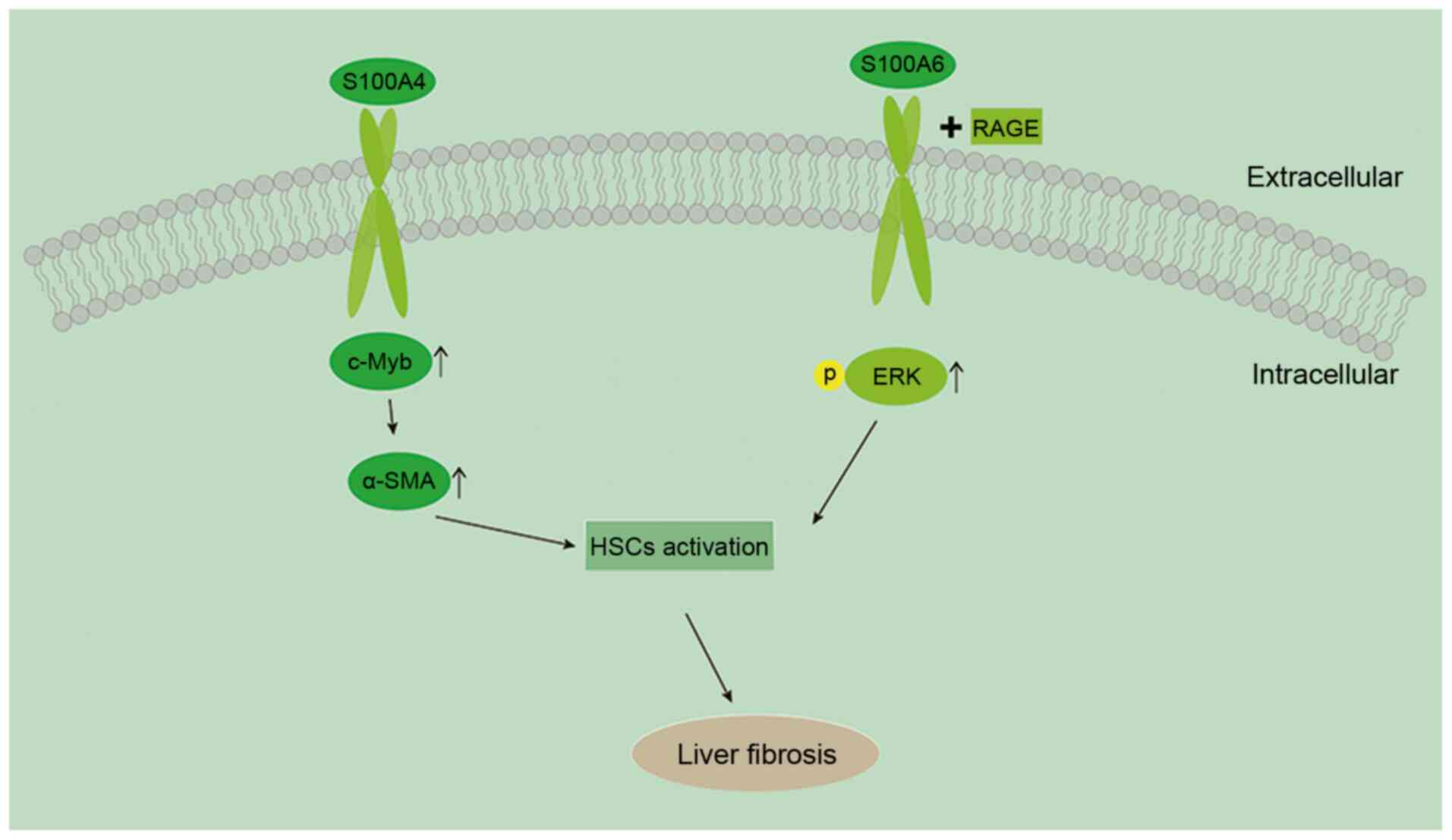
9
|
GBD 2017 Disease and Injury Incidence and
Prevalence Collaborators: Global, regional, and national incidence,
prevalence, and years lived with disability for 354 diseases and
injuries for 195 countries and territories, 1990-2017: A systematic
analysis for the Global Burden of Disease Study 2017. Lancet.
392:1789–1858. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Paik JM, Golabi P, Younossi Y, Mishra A
and Younossi ZM: Changes in the Global Burden of Chronic Liver
Diseases From 2012 to 2017: The Growing Impact of NAFLD.
Hepatology. 72:1605–1616. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wang FS, Fan JG, Zhang Z, Gao B and Wang
HY: The global burden of liver disease: The major impact of China.
Hepatology. 60:2099–2108. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Cooke GS, Andrieux-Meyer I, Applegate TL,
Atun R, Burry JR, Cheinquer H, Dusheiko G, Feld JJ, Gore C,
Griswold MG, et al: Accelerating the elimination of viral
hepatitis: A Lancet Gastroenterology & Hepatology Commission.
Lancet Gastroenterol Hepatol. 4:135–184. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Xiao J, Wang F, Wong NK, He J, Zhang R,
Sun R, Xu Y, Liu Y, Li W, Koike K, et al: Global liver disease
burdens and research trends: Analysis from a Chinese perspective. J
Hepatol. 71:212–221. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Avila MA, Dufour JF, Gerbes AL, Zoulim F,
Bataller R, Burra P, Cortez-Pinto H, Gao B, Gilmore I, Mathurin P,
et al: Recent advances in alcohol-related liver disease (ALD):
Summary of a Gut round table meeting. Gut. 69:764–780. 2020.
View Article : Google Scholar
|
|
15
|
Younossi ZM, Koenig AB, Abdelatif D, Fazel
Y, Henry L and Wymer M: Global epidemiology of nonalcoholic fatty
liver disease-Meta-analytic assessment of prevalence, incidence,
and outcomes. Hepatology. 64:73–84. 2016. View Article : Google Scholar
|
|
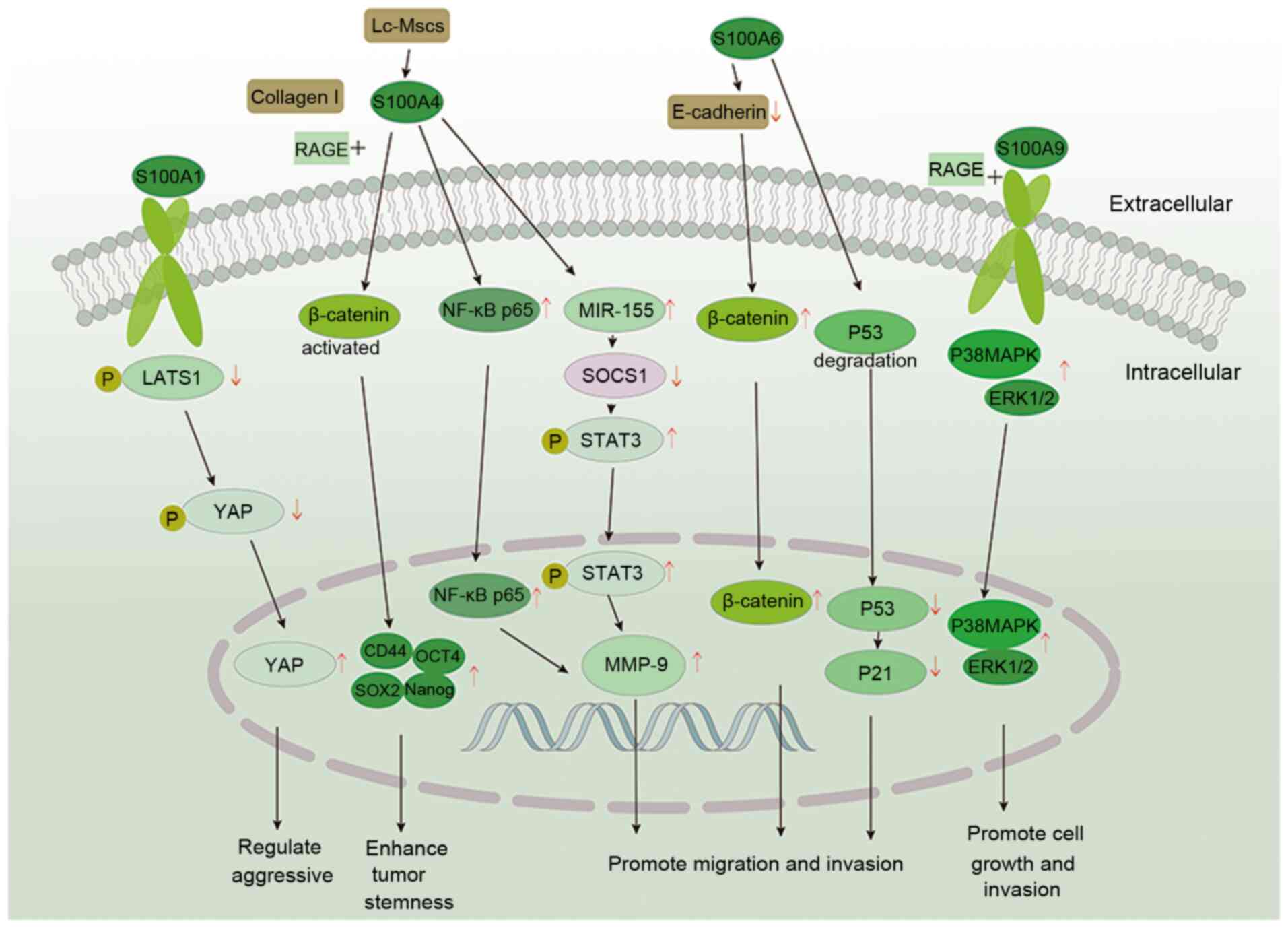
16
|
Donato R, Cannon BR, Sorci G, Riuzzi F,
Hsu K, Weber DJ and Geczy CL: Functions of S100 proteins. Curr Mol
Med. 13:24–57. 2013. View Article : Google Scholar :
|
|
17
|
Chan JK, Roth J, Oppenheim JJ, Tracey KJ,
Vogl T, Feldmann M, Horwood N and Nanchahal J: Alarmins: Awaiting a
clinical response. J Clin Invest. 122:2711–2719. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Kraus C, Rohde D, Weidenhammer C, Qiu G,
Pleger ST, Voelkers M, Boerries M, Remppis A, Katus HA and Most P:
S100A1 in cardiovascular health and disease: Closing the gap
between basic science and clinical therapy. J Mol Cell Cardiol.
47:445–455. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Bresnick AR, Weber DJ and Zimmer DB: S100
proteins in cancer. Nat Rev Cancer. 15:96–109. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Cristóvão JS and Gomes CM: S100 proteins
in Alzheimer's disease. Front Neurosci. 13:4632019. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Austermann J, Spiekermann C and Roth J:
S100 proteins in rheumatic diseases. Nat Rev Rheumatol. 14:528–541.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Foell D, Kucharzik T, Kraft M, Vogl T,
Sorg C, Domschke W and Roth J: Neutrophil derived human S100A12
(EN-RAGE) is strongly expressed during chronic active inflammatory
bowel disease. Gut. 52:847–853. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Yang Z, Tao T, Raftery MJ, Youssef P, Di
Girolamo N and Geczy CL: Proinflammatory properties of the human
S100 protein S100A12. J Leukoc Biol. 69:986–994. 2001.PubMed/NCBI
|
|
24
|
Turnier JL, Fall N, Thornton S, Witte D,
Bennett MR, Appenzeller S, Klein-Gitelman MS, Grom AA and Brunner
HI: Urine S100 proteins as potential biomarkers of lupus nephritis
activity. Arthritis Res Ther. 19:2422017. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Ren ZG, Zhao JD, Gu K, Wang J and Jiang
GL: Hepatic proliferation after partial liver irradiation in
Sprague-Dawley rats. Mol Biol Rep. 39:3829–3836. 2012. View Article : Google Scholar
|
|
26
|
Dixon LJ, Barnes M, Tang H, Pritchard MT
and Nagy LE: Kupffer cells in the liver. Compr Physiol. 3:785–797.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Chen L, Hu X, Wu H, Jia Y, Liu J, Mu X, Wu
H and Zhao Y: Over-expression of S100B protein as a serum marker of
brain metastasis in non-small cell lung cancer and its prognostic
value. Pathol Res Pract. 215:427–432. 2019. View Article : Google Scholar
|
|
28
|
Yan XL, Jia YL, Chen L, Zeng Q, Zhou JN,
Fu CJ, Chen HX, Yuan HF, Li ZW, Shi L, et al: Hepatocellular
carcinoma-associated mesenchymal stem cells promote hepatocarcinoma
progression: Role of the S100A4-miR155-SOCS1-MMP9 axis. Hepatology.
57:2274–2286. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Maletzki C, Bodammer P, Breitrück A and
Kerkhoff C: S100 proteins as diagnostic and prognostic markers in
colorectal and hepatocellular carcinoma. Hepat Mon.
12:e72402012.PubMed/NCBI
|
|
30
|
Liu Z, Liu H, Pan H, Du Q and Liang J:
Clinicopathological significance of S100A4 expression in human
hepatocellular carcinoma. J Int Med Res. 41:457–462. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Moore BW: A soluble protein characteristic
of the nervous system. Biochem Biophys Res Commun. 19:739–744.
1965. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Donato R: S100: A multigenic family of
calcium-modulated proteins of the EF-hand type with intracellular
and extracellular functional roles. Int J Biochem Cell Biol.
33:637–668. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Marenholz I, Heizmann CW and Fritz G: S100
proteins in mouse and man: From evolution to function and pathology
(including an update of the nomenclature). Biochem Biophys Res
Commun. 322:1111–1122. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Chow KH, Park HJ, George J, Yamamoto K,
Gallup AD, Graber JH, Chen Y, Jiang W, Steindler DA, Neilson EG, et
al: S100A4 is a biomarker and regulator of glioma stem cells that
is critical for mesenchymal transition in glioblastoma. Cancer Res.
77:5360–5373. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Dahlmann M, Kobelt D, Walther W, Mudduluru
G and Stein U: S100A4 in cancer metastasis: Wnt signaling-driven
interventions for metastasis restriction. Cancers (Basel).
8:592016. View Article : Google Scholar
|
|
36
|
Donato R: Intracellular and extracellular
roles of S100 proteins. Microsc Res Tech. 60:540–551. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Österreicher CH, Penz-Österreicher M,
Grivennikov SI, Guma M, Koltsova EK, Datz C, Sasik R, Hardiman G,
Karin M and Brenner DA: Fibroblast-specific protein 1 identifies an
inflammatory subpopulation of macrophages in the liver. Proc Natl
Acad Sci USA. 108:308–313. 2011. View Article : Google Scholar :
|
|
38
|
Zhang J, Chen L, Liu X, Kammertoens T,
Blankenstein T and Qin Z: Fibroblast-specific protein
1/S100A4-positive cells prevent carcinoma through collagen
production and encapsulation of carcinogens. Cancer Res.
73:2770–2781. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Zhang J, Chen L, Xiao M, Wang C and Qin Z:
FSP1+ fibroblasts promote skin carcinogenesis by
maintaining MCP-1-mediated macrophage infiltration and chronic
inflammation. Am J Pathol. 178:382–390. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Kuźnicki J, Kordowska J, Puzianowska M and
Woźniewicz BM: Calcyclin as a marker of human epithelial cells and
fibroblasts. Exp Cell Res. 200:425–430. 1992. View Article : Google Scholar
|
|
41
|
Markowitz J and Carson WE III: Review of
S100A9 biology and its role in cancer. Biochim Biophys Acta.
1835:100–109. 2013.
|
|
42
|
Saiki Y and Horii A: Multiple functions of
S100A10, an important cancer promoter. Pathol Int. 69:629–636.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
He H, Li J, Weng S, Li M and Yu Y:
S100A11: Diverse function and pathology corresponding to different
target proteins. Cell Biochem Biophys. 55:117–126. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Guignard F, Mauel J and Markert M:
Identification and characterization of a novel human neutrophil
protein related to the S100 family. Biochem J. 309:395–401. 1995.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Bagheri V: S100A12: Friend or foe in
pulmonary tuberculosis? Cytokine. 92:80–82. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Donato R, Sorci G, Riuzzi F, Arcuri C,
Bianchi R, Brozzi F, Tubaro C and Giambanco I: S100B's double life:
Intracellular regulator and extracellular signal. Biochim Biophys
Acta. 1793:1008–1022. 2009. View Article : Google Scholar
|
|
47
|
Santamaria-Kisiel L, Rintala-Dempsey AC
and Shaw GS: Calcium-dependent and -independent interactions of the
S100 protein family. Biochem J. 396:201–214. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Goyette J and Geczy CL:
Inflammation-associated S100 proteins: New mechanisms that regulate
function. Amino Acids. 41:821–842. 2011. View Article : Google Scholar
|
|
49
|
Pruenster M, Vogl T, Roth J and Sperandio
M: S100A8/A9: From basic science to clinical application. Pharmacol
Ther. 167:120–131. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Lim SY, Raftery MJ and Geczy CL: Oxidative
modifications of DAMPs suppress inflammation: The case for S100A8
and S100A9. Antioxid Redox Signal. 15:2235–2248. 2011. View Article : Google Scholar
|
|
51
|
Averill MM, Kerkhoff C and Bornfeldt KE:
S100A8 and S100A9 in cardiovascular biology and disease.
Arterioscler Thromb Vasc Biol. 32:223–229. 2012. View Article : Google Scholar
|
|
52
|
Gross SR, Sin CG, Barraclough R and
Rudland PS: Joining S100 proteins and migration: For better or for
worse, in sickness and in health. Cell Mol Life Sci. 71:1551–1579.
2014. View Article : Google Scholar
|
|
53
|
Donato R, Sorci G and Giambanco I: S100A6
protein: Functional roles. Cell Mol Life Sci. 74:2749–2760. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Riuzzi F, Sorci G, Arcuri C, Giambanco I,
Bellezza I, Minelli A and Donato R: Cellular and molecular
mechanisms of sarcopenia: The S100B perspective. J Cachexia
Sarcopenia Muscle. 9:1255–1268. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Wang S, Song R, Wang Z, Jing Z, Wang S and
Ma J: S100A8/A9 in inflammation. Front Immunol. 9:12982018.
View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Sun H, Zhao A, Li M, Dong H, Sun Y, Zhang
X, Zhu Q, Bukhari A, Cao C, Su D, et al: Interaction of calcium
binding protein S100A16 with myosin-9 promotes cytoskeleton
reorganization in renal tubulointerstitial fibrosis. Cell Death
Dis. 11:1462020. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Akiyama N, Hozumi H, Isayama T, Okada J,
Sugiura K, Yasui H, Suzuki Y, Kono M, Karayama M, Furuhashi K, et
al: Clinical significance of serum S100 calcium-binding protein A4
in idiopathic pulmonary fibrosis. Respirology. 25:743–749. 2020.
View Article : Google Scholar
|
|
58
|
Zhong A, Xu W, Zhao J, Xie P, Jia S, Sun
J, Galiano RD, Mustoe TA and Hong SJ: S100A8 and S100A9 are induced
by decreased hydration in the epidermis and promote fibroblast
activation and fibrosis in the dermis. Am J Pathol. 186:109–122.
2016. View Article : Google Scholar
|
|
59
|
Chen L, Li J, Zhang J, Dai C, Liu X, Wang
J, Gao Z, Guo H, Wang R, Lu S, et al: S100A4 promotes liver
fibrosis via activation of hepatic stellate cells. J Hepatol.
62:156–164. 2015. View Article : Google Scholar
|
|
60
|
Cancemi P, Buttacavoli M, Di Cara G,
Albanese NN, Bivona S, Pucci-Minafra I and Feo S: A multiomics
analysis of S100 protein family in breast cancer. Oncotarget.
9:29064–29081. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Liu Y, Cui J, Tang YL, Huang L, Zhou CY
and Xu JX: Prognostic roles of mRNA expression of S100 in
non-small-cell lung cancer. Biomed Res Int.
2018:98158062018.PubMed/NCBI
|
|
62
|
Moravkova P, Kohoutova D, Rejchrt S,
Cyrany J and Bures J: Role of S100 proteins in colorectal
carcinogenesis. Gastroenterol Res Pract. 2016:26327032016.
View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Fang D, Zhang C, Xu P, Liu Y, Mo X, Sun Q,
Abdelatty A, Hu C, Xu H, Zhou G, et al: S100A16 promotes metastasis
and progression of pancreatic cancer through FGF19-mediated AKT and
ERK1/2 pathways. Cell Biol Toxicol. Jan 2–2021.Epub ahead of print.
View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Liu Y, Luo G and He D: Clinical importance
of S100A9 in osteosarcoma development and as a diagnostic marker
and therapeutic target. Bioengineered. 10:133–141. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Wang T, Huo X, Chong Z, Khan H, Liu R and
Wang T: A review of S100 protein family in lung cancer. Clin Chim
Acta. 476:54–59. 2018. View Article : Google Scholar
|
|
66
|
Destek S and Gul VO: S100A4 may be a good
prognostic marker and a therapeutic target for colon cancer. J
Oncol. 2018:18287912018. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Sun X, Wang T, Zhang C, Ning K, Guan ZR,
Chen SX, Hong TT and Hua D: S100A16 is a prognostic marker for
colorectal cancer. J Surg Oncol. 117:275–283. 2018. View Article : Google Scholar
|
|
68
|
Yuan W, Goldstein LD, Durinck S, Chen YJ,
Nguyen TT, Kljavin NM, Sokol ES, Stawiski EW, Haley B, Ziai J, et
al: S100a4 upregulation in Pik3caH1047R;Trp53R270H;MMTV-C re-driven
mammary tumors promotes metastasis. Breast Cancer Res. 21:1522019.
View Article : Google Scholar
|
|
69
|
Louka ML and Ramzy MM: Involvement of
fibroblast-specific protein 1 (S100A4) and matrix
metalloproteinase-13 (MMP-13) in CCl4-induced reversible liver
fibrosis. Gene. 579:29–33. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Zadorozhna M, Di Gioia S, Conese M and
Mangieri D: Neovascularization is a key feature of liver fibrosis
progression: Anti-angiogenesis as an innovative way of liver
fibrosis treatment. Mol Biol Rep. 47:2279–2288. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Hernandez-Gea V and Friedman SL:
Pathogenesis of liver fibrosis. Annu Rev Pathol. 6:425–456. 2011.
View Article : Google Scholar
|
|
72
|
Pellicoro A, Ramachandran P, Iredale JP
and Fallowfield JA: Liver fibrosis and repair: Immune regulation of
wound healing in a solid organ. Nat Rev Immunol. 14:181–194. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Song LJ, Yin XR, Mu SS, Li JH, Gao H,
Zhang Y, Dong PP, Mei CJ and Hua ZC: The differential and dynamic
progression of hepatic inflammation and immune responses during
liver fibrosis induced by Schistosoma japonicum or carbon
tetrachloride in mice. Front Immunol. 11:5705242020. View Article : Google Scholar :
|
|
74
|
Parola M and Pinzani M: Liver fibrosis:
Pathophysiology, pathogenetic targets and clinical issues. Mol
Aspects Med. 65:37–55. 2019. View Article : Google Scholar
|
|
75
|
Zhang CY, Yuan WG, He P, Lei JH and Wang
CX: Liver fibrosis and hepatic stellate cells: Etiology,
pathological hallmarks and therapeutic targets. World J
Gastroenterol. 22:10512–10522. 2016. View Article : Google Scholar
|
|
76
|
Blomhoff R, Rasmussen M, Nilsson A, Norum
KR, Berg T, Blaner WS, Kato M, Mertz JR, Goodman DS, Eriksson U, et
al: Hepatic retinol metabolism. Distribution of retinoids, enzymes,
and binding proteins in isolated rat liver cells. J Biol Chem.
260:13560–13565. 1985. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Li D, He L, Guo H, Chen H and Shan H:
Targeting activated hepatic stellate cells (aHSCs) for liver
fibrosis imaging. EJNMMI Res. 5:712015. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Puche JE, Saiman Y and Friedman SL:
Hepatic stellate cells and liver fibrosis. Compr Physiol.
3:1473–1492. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Henderson NC and Iredale JP: Liver
fibrosis: Cellular mechanisms of progression and resolution. Clin
Sci (Lond). 112:265–280. 2007. View Article : Google Scholar
|
|
80
|
Kisseleva T, Cong M, Paik Y, Scholten D,
Jiang C, Benner C, Iwaisako K, Moore-Morris T, Scott B, Tsukamoto
H, et al: Myofibroblasts revert to an inactive phenotype during
regression of liver fibrosis. Proc Natl Acad Sci USA.
109:9448–9453. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Iredale JP, Thompson A and Henderson NC:
Extracellular matrix degradation in liver fibrosis: Biochemistry
and regulation. Biochim Biophys Acta. 1832:876–883. 2013.
View Article : Google Scholar
|
|
82
|
Schaefer B, Rivas-Estilla AM, Meraz-Cruz
N, Reyes-Romero MA, Hernández-Nazara ZH, Domínguez-Rosales JA,
Schuppan D, Greenwel P and Rojkind M: Reciprocal modulation of
matrix metalloproteinase-13 and type I collagen genes in rat
hepatic stellate cells. Am J Pathol. 162:1771–1780. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Uchinami H, Seki E, Brenner DA and
D'Armiento J: Loss of MMP 13 attenuates murine hepatic injury and
fibrosis during cholestasis. Hepatology. 44:420–429. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Miranda KJ, Loeser RF and Yammani RR:
Sumoylation and nuclear translocation of S100A4 regulate
IL-1beta-mediated production of matrix metalloproteinase-13. J Biol
Chem. 285:31517–31524. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Yammani RR, Carlson CS, Bresnick AR and
Loeser RF: Increase in production of matrix metalloproteinase 13 by
human articular chondrocytes due to stimulation with S100A4: Role
of the receptor for advanced glycation end products. Arthritis
Rheum. 54:2901–2911. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Xia P, He H, Kristine MS, Guan W, Gao J,
Wang Z, Hu J, Han L, Li J, Han W and Yu Y: Therapeutic effects of
recombinant human S100A6 and soluble receptor for advanced
glycation end products(sRAGE) on CCl(4)-induced liver fibrosis in
mice. Eur J Pharmacol. 833:86–93. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Baik SJ, Kim TH, Yoo K, Moon IH, Choi JY,
Chung KW and Song DE: Decreased S100B expression in chronic liver
diseases. Korean J Intern Med. 32:269–276. 2017. View Article : Google Scholar :
|
|
88
|
Park JW, Kim MJ, Kim SE, Kim HJ, Jeon YC,
Shin HY, Park SJ, Jang MK, Kim DJ, Park CK and Choi EK: Increased
expression of S100B and RAGE in a mouse model of bile duct
ligation-induced liver fibrosis. J Korean Med Sci. 36:e902021.
View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Lanini S, Ustianowski A, Pisapia R, Zumla
A and Ippolito G: Viral hepatitis: Etiology, epidemiology,
transmission, diagnostics, treatment, and prevention. Infect Dis
Clin North Am. 33:1045–1062. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Thomas DL: Global elimination of chronic
hepatitis. N Engl J Med. 380:2041–2050. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Jie Z, Liang Y, Yi P, Tang H, Soong L,
Cong Y, Zhang K and Sun J: Retinoic acid regulates immune responses
by promoting IL-22 and modulating S100 proteins in viral hepatitis.
J Immunol. 198:3448–3460. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Radaeva S, Wang L, Radaev S, Jeong WI,
Park O and Gao B: Retinoic acid signaling sensitizes hepatic
stellate cells to NK cell killing via upregulation of NK cell
activating ligand RAE1. Am J Physiol Gastrointest Liver Physiol.
293:G809–G816. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Lee YS and Jeong WI: Retinoic acids and
hepatic stellate cells in liver disease. J Gastroenterol Hepatol.
27(Suppl 2): S75–S79. 2012. View Article : Google Scholar
|
|
94
|
Yan LB, Zhang QB, Zhu X, He M and Tang H:
Serum S100 calcium binding protein A4 improves the diagnostic
accuracy of transient elastography for assessing liver fibrosis in
hepatitis B. Clin Res Hepatol Gastroenterol. 42:64–71. 2018.
View Article : Google Scholar
|
|
95
|
Wu R, Zhang Y, Xiang Y, Tang Y, Cui F, Cao
J, Zhou L, You Y and Duan L: Association between serum S100A9
levels and liver necroinflammation in chronic hepatitis B. J Transl
Med. 16:832018. View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Cai J, Han T, Nie C, Jia X, Liu Y, Zhu Z
and Gao Y: Biomarkers of oxidation stress, inflammation, necrosis
and apoptosis are associated with hepatitis B-related
acute-on-chronic liver failure. Clin Res Hepatol Gastroenterol.
40:41–50. 2016. View Article : Google Scholar
|
|
97
|
Li J, Zou B, Yeo YH, Feng Y, Xie X, Lee
DH, Fujii H, Wu Y, Kam LY, Ji F, et al: Prevalence, incidence, and
outcome of non-alcoholic fatty liver disease in Asia, 1999-2019: A
systematic review and meta-analysis. Lancet Gastroenterol Hepatol.
4:389–398. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Sayiner M, Koenig A, Henry L and Younossi
ZM: Epidemiology of nonalcoholic fatty liver disease and
nonalcoholic steatohepatitis in the united states and the rest of
the world. Clin Liver Dis. 20:205–214. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Afonso MB, Rodrigues PM, Simão AL and
Castro RE: Circulating microRNAs as potential biomarkers in
non-alcoholic fatty liver disease and hepatocellular carcinoma. J
Clin Med. 5:302016. View Article : Google Scholar :
|
|
100
|
Zhang YH, Ma Q, Ding P, Li J, Chen LL, Ao
KJ and Tian YY: S100A4 gene is crucial for
methionine-choline-deficient diet-induced non-alcoholic fatty liver
disease in mice. Yonsei Med J. 59:1064–1071. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
101
|
Liu X, Wang Y, Ming Y, Song Y, Zhang J,
Chen X, Zeng M and Mao Y: S100A9: A potential biomarker for the
progression of non-alcoholic fatty liver disease and the diagnosis
of non-alcoholic steatohepatitis. PLoS One. 10:e01273522015.
View Article : Google Scholar : PubMed/NCBI
|
|
102
|
Zhang L, Zhang Z, Li C, Zhu T, Gao J, Zhou
H, Zheng Y, Chang Q, Wang M, Wu J, et al: S100A11 promotes liver
steatosis via FOXO1-mediated autophagy and lipogenesis. Cell Mol
Gastroenterol Hepatol. 11:697–724. 2021. View Article : Google Scholar :
|
|
103
|
Ni HM, Chao X and Ding WX: S100A11
overexpression promotes fatty liver diseases via increased
autophagy? Cell Mol Gastroenterol Hepatol. 11:885–886. 2021.
View Article : Google Scholar :
|
|
104
|
Sobolewski C, Abegg D, Berthou F, Dolicka
D, Calo N, Sempoux C, Fournier M, Maeder C, Ay AS, Clavien PA, et
al: S100A11/ANXA2 belongs to a tumour suppressor/oncogene network
deregulated early with steatosis and involved in inflammation and
hepatocellular carcinoma development. Gut. 69:1841–1854. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
105
|
GBD 2016 Causes of Death Collaborators:
Global, regional, and national age-sex specific mortality for 264
causes of death, 1980-2016: A systematic analysis for the Global
Burden of Disease Study 2016. Lancet. 390:1151–1210. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
106
|
Purohit V, Gao B and Song BJ: Molecular
mechanisms of alcoholic fatty liver. Alcohol Clin Exp Res.
33:191–205. 2009. View Article : Google Scholar :
|
|
107
|
Anstee QM, Daly AK and Day CP: Genetics of
alcoholic and nonalcoholic fatty liver disease. Semin Liver Dis.
31:128–146. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
108
|
Kwon HJ, Won YS, Park O, Chang B, Duryee
MJ, Thiele GE, Matsumoto A, Singh S, Abdelmegeed MA, Song BJ, et
al: Aldehyde dehydrogenase 2 deficiency ameliorates alcoholic fatty
liver but worsens liver inflammation and fibrosis in mice.
Hepatology. 60:146–157. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
109
|
Rehm J, Taylor B, Mohapatra S, Irving H,
Baliunas D, Patra J and Roerecke M: Alcohol as a risk factor for
liver cirrhosis: A systematic review and meta-analysis. Drug
Alcohol Rev. 29:437–445. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
110
|
Roerecke M, Vafaei A, Hasan OSM, Chrystoja
BR, Cruz M, Lee R, Neuman MG and Rehm J: Alcohol consumption and
risk of liver cirrhosis: A systematic review and meta-analysis. Am
J Gastroenterol. 114:1574–1586. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
111
|
Seitz HK, Bataller R, Cortez-Pinto H, Gao
B, Gual A, Lackner C, Mathurin P, Mueller S, Szabo G and Tsukamoto
H: Alcoholic liver disease. Nat Rev Dis Primers. 4:162018.
View Article : Google Scholar : PubMed/NCBI
|
|
112
|
Yuan Q, Hou S, Zhai J, Tian T, Wu Y, Wu Z,
He J, Chen Z and Zhang J: S100A4 promotes inflammation but
suppresses lipid accumulation via the STAT3 pathway in chronic
ethanol-induced fatty liver. J Mol Med (Berl). 97:1399–1412. 2019.
View Article : Google Scholar
|
|
113
|
Llovet JM, Kelley RK, Villanueva A, Singal
AG, Pikarsky E, Roayaie S, Lencioni R, Koike K, Zucman-Rossi J and
Finn RS: Hepatocellular carcinoma. Nat Rev Dis Primers. 7:62021.
View Article : Google Scholar : PubMed/NCBI
|
|
114
|
International Agency for Research on
Cancer: Liver: GLOBOCAN. 2020, https://gco.iarc.fr/today/data/factsheets/cancers/11-Liver-fact-sheet.pdf.
|
|
115
|
Chen Z, Xie H, Hu M, Huang T, Hu Y, Sang N
and Zhao Y: Recent progress in treatment of hepatocellular
carcinoma. Am J Cancer Res. 10:2993–3036. 2020.PubMed/NCBI
|
|
116
|
Allemani C, Matsuda T, Di Carlo V,
Harewood R, Matz M, Nikšić M, Bonaventure A, Valkov M, Johnson CJ,
Estève J, et al: Global surveillance of trends in cancer survival
2000-14 (CONCORD-3): Analysis of individual records for 37 513 025
patients diagnosed with one of 18 cancers from 322 population-based
registries in 71 countries. Lancet. 391:1023–1075. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
117
|
Goyal L, Muzumdar MD and Zhu AX: Targeting
the HGF/c-MET pathway in hepatocellular carcinoma. Clin Cancer Res.
19:2310–2318. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
118
|
Guo Q, Wang J, Cao Z, Tang Y, Feng C and
Huang F: Interaction of S100A1 with LATS1 promotes cell growth
through regulation of the Hippo pathway in hepatocellular
carcinoma. Int J Oncol. 53:592–602. 2018.PubMed/NCBI
|
|
119
|
Tao R, Wang ZF, Qiu W, He YF, Yan WQ, Sun
WY and Li HJ: Role of S100A3 in human hepatocellular carcinoma and
the anticancer effect of sodium cantharidinate. Exp Ther Med.
13:2812–2818. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
120
|
Zhang J, Zhang DL, Jiao XL and Dong Q:
S100A4 regulates migration and invasion in hepatocellular carcinoma
HepG2 cells via NF-κB-dependent MMP-9 signal. Eur Rev Med Pharmacol
Sci. 17:2372–2382. 2013.PubMed/NCBI
|
|
121
|
Zhai X, Zhu H, Wang W, Zhang S, Zhang Y
and Mao G: Abnormal expression of EMT-related proteins, S100A4,
vimentin and E-cadherin, is correlated with clinicopathological
features and prognosis in HCC. Med Oncol. 31:9702014. View Article : Google Scholar : PubMed/NCBI
|
|
122
|
Zhu K, Huang W, Wang W, Liao L, Li S, Yang
S, Xu J, Li L, Meng M, Xie Y, et al: Up-regulation of S100A4
expression by HBx protein promotes proliferation of hepatocellular
carcinoma cells and its correlation with clinical survival. Gene.
749:1446792020. View Article : Google Scholar : PubMed/NCBI
|
|
123
|
Hanahan D and Coussens LM: Accessories to
the crime: Functions of cells recruited to the tumor
microenvironment. Cancer Cell. 21:309–322. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
124
|
Malanchi I, Santamaria-Martínez A, Susanto
E, Peng H, Lehr HA, Delaloye JF and Huelsken J: Interactions
between cancer stem cells and their niche govern metastatic
colonization. Nature. 481:85–89. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
125
|
Tsai KS, Yang SH, Lei YP, Tsai CC, Chen
HW, Hsu CY, Chen LL, Wang HW, Miller SA, Chiou SH, et al:
Mesenchymal stem cells promote formation of colorectal tumors in
mice. Gastroenterology. 141:1046–1056. 2011. View Article : Google Scholar
|
|
126
|
Karnoub AE, Dash AB, Vo AP, Sullivan A,
Brooks MW, Bell GW, Richardson AL, Polyak K, Tubo R and Weinberg
RA: Mesenchymal stem cells within tumour stroma promote breast
cancer metastasis. Nature. 449:557–563. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
127
|
Huang S and He X: The role of microRNAs in
liver cancer progression. Br J Cancer. 104:235–240. 2011.
View Article : Google Scholar :
|
|
128
|
Gallardo M, Kemmerling U, Aguayo F, Bleak
TC, Muñoz JP and Calaf GM: Curcumin rescues breast cells from
epithelial-mesenchymal transition and invasion induced by
anti-miR-34a. Int J Oncol. 56:480–493. 2020.
|
|
129
|
Datta J, Islam M, Dutta S, Roy S, Pan Q
and Teknos TN: Suberoylanilide hydroxamic acid inhibits growth of
head and neck cancer cell lines by reactivation of tumor suppressor
microRNAs. Oral Oncol. 56:32–39. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
130
|
Wang B, Majumder S, Nuovo G, Kutay H,
Volinia S, Patel T, Schmittgen TD, Croce C, Ghoshal K and Jacob ST:
Role of microRNA-155 at early stages of hepatocarcinogenesis
induced by choline-deficient and amino acid-defined diet in C57BL/6
mice. Hepatology. 50:1152–1161. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
131
|
Xie Q, Chen X, Lu F, Zhang T, Hao M, Wang
Y, Zhao J, McCrae MA and Zhuang H: Aberrant expression of microRNA
155 may accelerate cell proliferation by targeting sex-determining
region Y box 6 in hepatocellular carcinoma. Cancer. 118:2431–2442.
2012. View Article : Google Scholar
|
|
132
|
Chen G, Wang D, Zhao X, Cao J, Zhao Y,
Wang F, Bai J, Luo D and Li L: miR-155-5p modulates malignant
behaviors of hepatocellular carcinoma by directly targeting CTHRC1
and indirectly regulating GSK-3β-involved Wnt/β-catenin signaling.
Cancer Cell Int. 17:1182017. View Article : Google Scholar
|
|
133
|
Reya T, Morrison SJ, Clarke MF and
Weissman IL: Stem cells, cancer, and cancer stem cells. Nature.
414:105–111. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
134
|
Li Y, Wang J, Song K, Liu S, Zhang H, Wang
F, Ni C, Zhai W, Liang J, Qin Z and Zhang J: S100A4 promotes
hepatocellular carcinogenesis by intensifying fibrosis-associated
cancer cell stemness. Oncoimmunology. 9:17253552020. View Article : Google Scholar : PubMed/NCBI
|
|
135
|
Kim J, Kim J, Yoon S, Joo J, Lee Y, Lee K,
Chung J and Choe I: S100A6 protein as a marker for differential
diagnosis of cholangiocarcinoma from hepatocellular carcinoma.
Hepatol Res. 23:2742002. View Article : Google Scholar : PubMed/NCBI
|
|
136
|
Hua Z, Chen J, Sun B, Zhao G, Zhang Y,
Fong Y, Jia Z and Yao L: Specific expression of osteopontin and
S100A6 in hepatocellular carcinoma. Surgery. 149:783–791. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
137
|
Tong A, Gou L, Lau QC, Chen B, Zhao X, Li
J, Tang H, Chen L, Tang M, Huang C and Wei YQ: Proteomic profiling
identifies aberrant epigenetic modifications induced by hepatitis B
virus X protein. J Proteome Res. 8:1037–1046. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
138
|
Li Z, Tang M, Ling B, Liu S, Zheng Y, Nie
C, Yuan Z, Zhou L, Guo G, Tong A and Wei Y: Increased expression of
S100A6 promotes cell proliferation and migration in human
hepatocellular carcinoma. J Mol Med (Berl). 92:291–303. 2014.
View Article : Google Scholar
|
|
139
|
Song D, Xu B, Shi D, Li S and Cai Y:
S100A6 promotes proliferation and migration of HepG2 cells via
increased ubiquitin-dependent degradation of p53. Open Med (Wars).
15:317–326. 2020. View Article : Google Scholar
|
|
140
|
Arai K, Yamada T and Nozawa R:
Immunohistochemical investigation of migration inhibitory
factor-related protein (MRP)-14 expression in hepatocellular
carcinoma. Med Oncol. 17:183–188. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
141
|
Németh J, Stein I, Haag D, Riehl A,
Longerich T, Horwitz E, Breuhahn K, Gebhardt C, Schirmacher P, Hahn
M, et al: S100A8 and S100A9 are novel nuclear factor kappa B target
genes during malignant progression of murine and human liver
carcinogenesis. Hepatology. 50:1251–1262. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
142
|
Wu R, Duan L, Ye L, Wang H, Yang X, Zhang
Y, Chen X, Zhang Y, Weng Y, Luo J, et al: S100A9 promotes the
proliferation and invasion of HepG2 hepatocellular carcinoma cells
via the activation of the MAPK signaling pathway. Int J Oncol.
42:1001–1010. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
143
|
Wu R, Duan L, Cui F, Cao J, Xiang Y, Tang
Y and Zhou L: S100A9 promotes human hepatocellular carcinoma cell
growth and invasion through RAGE-mediated ERK1/2 and p38 MAPK
pathways. Exp Cell Res. 334:228–238. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
144
|
Duan L, Wu R, Zhang X, Wang D, You Y,
Zhang Y, Zhou L and Chen W: HBx-induced S100A9 in NF-κB dependent
manner promotes growth and metastasis of hepatocellular carcinoma
cells. Cell Death Dis. 9:6292018. View Article : Google Scholar
|
|
145
|
Wei R, Zhu WW, Yu GY, Wang X, Gao C, Zhou
X, Lin ZF, Shao WQ, Wang SH, Lu M and Qin LX: S100 calcium-binding
protein A9 from tumor-associated macrophage enhances cancer stem
cell-like properties of hepatocellular carcinoma. Int J Cancer.
148:1233–1244. 2021. View Article : Google Scholar
|
|
146
|
Shan X, Miao Y, Fan R, Qian H, Chen P, Liu
H, Yan X, Li J and Zhou F: MiR-590-5P inhibits growth of HepG2
cells via decrease of S100A10 expression and Inhibition of the Wnt
pathway. Int J Mol Sci. 14:8556–8569. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
147
|
Zhao JT, Chi BJ, Sun Y, Chi NN, Zhang XM,
Sun JB, Chen Y and Xia Y: LINC00174 is an oncogenic lncRNA of
hepatocellular carcinoma and regulates miR-320/S100A10 axis. Cell
Biochem Funct. 38:859–869. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
148
|
Takeuchi K and Ito F: EGF receptor in
relation to tumor development: Molecular basis of responsiveness of
cancer cells to EGFR-targeting tyrosine kinase inhibitors. FEBS J.
277:316–326. 2010. View Article : Google Scholar
|
|
149
|
Wheeler SE, Suzuki S, Thomas SM, Sen M,
Leeman-Neill RJ, Chiosea SI, Kuan CT, Bigner DD, Gooding WE, Lai SY
and Grandis JR: Epidermal growth factor receptor variant III
mediates head and neck cancer cell invasion via STAT3 activation.
Oncogene. 29:5135–5145. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
150
|
Luo X, Xie H, Long X, Zhou M, Xu Z, Shi B,
Jiang H and Li Z: EGFRvIII mediates hepatocellular carcinoma cell
invasion by promoting S100 calcium binding protein A11 expression.
PLoS One. 8:e833322013. View Article : Google Scholar :
|
|
151
|
Mitsui Y, Tomonobu N, Watanabe M,
Kinoshita R, Sumardika IW, Youyi C, Murata H, Yamamoto KI, Sadahira
T, Rodrigo AGH, et al: Upregulation of mobility in pancreatic
cancer cells by secreted S100A11 through activation of surrounding
fibroblasts. Oncol Res. 27:945–956. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
152
|
Cai H, Ye BG, Ao JY, Zhu XD, Zhang YY,
Chai ZT, Wang CH and Sun HC: High expression of S100A12 on
intratumoral stroma cells indicates poor prognosis following
surgical resection of hepatocellular carcinoma. Oncol Lett.
16:5398–5404. 2018.PubMed/NCBI
|
|
153
|
Shen H, Wu H, Sun F, Qi J and Zhu Q: A
novel four-gene of iron metabolism-related and methylated for
prognosis prediction of hepatocellular carcinoma. Bioengineered.
12:240–251. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
154
|
Sun XJ, Wang MC, Zhang FH and Kong X: An
integrated analysis of genome-wide DNA methylation and gene
expression data in hepatocellular carcinoma. FEBS Open Bio.
8:1093–1103. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
155
|
Schuppan D and Pinzani M: Anti-fibrotic
therapy: Lost in translation? J Hepatol. 56(Suppl 1): S66–74. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
156
|
Huang CH, Kuo CJ, Liang SS, Chi SW, Hsi E,
Chen CC, Lee KT and Chiou SH: Onco-proteogenomics identifies
urinary S100A9 and GRN as potential combinatorial biomarkers for
early diagnosis of hepatocellular carcinoma. BBA Clin. 3:205–213.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
157
|
Meng J, Gu F, Fang H and Qu B: Elevated
serum S100A9 indicated poor prognosis in hepatocellular carcinoma
after curative resection. J Cancer. 10:408–415. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
158
|
Zhang J, Jiao J, Cermelli S, Muir K, Jung
KH, Zou R, Rashid A, Gagea M, Zabludoff S, Kalluri R and Beretta L:
miR-21 inhibition reduces liver fibrosis and prevents tumor
development by inducing apoptosis of CD24+ progenitor
cells. Cancer Res. 75:1859–1867. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
159
|
Jiao J, González Á, Stevenson HL, Gagea M,
Sugimoto H, Kalluri R and Beretta L: Depletion of
S100A4+ stromal cells does not prevent HCC development
but reduces the stem cell-like phenotype of the tumors. Exp Mol
Med. 50:e4222018. View Article : Google Scholar
|