Introduction
Pulmonary arterial hypertension (PAH) is a rare
vascular disorder with an estimated prevalence of ~15 cases per
million individuals (1,2). The pathogenesis of PAH is extremely
complex, involving multiple pathological changes, including
pulmonary vascular endothelial dysfunction, pulmonary arteriolar
contraction and pulmonary vascular remodelling (3). Hypoxic pulmonary hypertension (HPH)
is characterized by increasing pulmonary artery blood pressure and
remodelling of structural peripheral pulmonary arteries (4). Pulmonary artery smooth cell (PASMC)
hyperproliferation plays a key role in pulmonary vascular
remodelling in HPH.
At present, the treatment of PAH includes endothelin
receptor antagonists, phosphodiesterase type-5 inhibitors, soluble
guanylate cyclase stimulators, prostacyclins and prostacyclin
receptor agonists (5). There is
widespread acceptance that new drugs should focus on genetically
determined targets, epigenetic modifications, DNA damage, growth
factors, oestrogen signalling and hypoxic stress (6). In the SERAPHIN and GRIPHON studies,
whilst recent accessible treatments for PAH have advanced
substantially, the disease presents high mortality rates (7,8).
As the proliferation of PASMCs plays a key role in the pathogenesis
of PAH, further elucidation of the molecular mechanisms via which
PASMCs participate in PAH is critical for effective treatment.
Unlike other RNAs, circular RNAs (circRNAs/circs)
are non-coding RNAs, which are covalently enclosed contiguous loops
and are expressed at high levels in the eukaryotic transcriptome
(9). Typically, circRNAs are
widely expressed in eukaryotic cells and could potentially be
associated with the diagnosis and treatment of diseases (10,11). Studies have shown that circRNAs
play important roles in the pathogenesis of atherosclerosis,
myocardial infarction, heart failure, cardiomyopathy, pulmonary
hypertension and cell aging (12). circRNAs are also involved in
regulating several biological processes, including the control of
angiogenesis and smooth muscle cell functions (13). Recently, studies have revealed
the function of hsa_circ_0022342 and hsa_circ_0002062 in the
development of chronic thromboembolic pulmonary hypertension
(14,15). mmu_circ_0000790 is associated
with the structural remodelling of pulmonary vessels in mice with
HPH via miR-374c/FOXC1/Notch axis (16). However, only a few reports have
focused on the function of circRNAs in HPH. Therefore, the present
study aimed to identify novel functional circRNAs and understand
their roles in a hypoxia-induced proliferation cellular model.
Studies have shown that circRNAs play crucial roles
in regulating gene expression at the transcriptional level by
interacting with RNA-binding proteins (17-19). Furthermore, it has been revealed
that Grm1 influenced the biological functions of PASMCs via the ERK
pathway (20-23). In the present study, the
RNA-binding protein (RBP) FUS interacted with circ-Grm1. circ-Grm1
was selected as the representative to study the role of circRNAs in
vascular remodelling of hypoxic PAH, and the molecular mechanism of
its targeted regulation of Grm1 combined with FUS was evaluated,
thereby providing a new molecular target in the study of HPH.
Materials and methods
Isolation of PASMCs
Male, 10-12-week-old C57BL/6 mice (weight, 21-25 g)
were purchased from Vital River Bioscience Co., Ltd. (Beijing,
China). The mice were housed in a quiet room with a 12-h light and
dark cycle, at 22±2°C and 60% relative humidity, with free access
to food and water. This study was approved by the Ethical Animal
Care and Use of Laboratory Animals Committee of Qilu Hospital,
Cheeloo College of Medicine, Shandong University (approval no.
KYLL-2020-002).
After 14 days, the mice were sacrificed via cervical
dislocation, and the aortas were collected. The criteria used to
confirm death included the cessation of heartbeat for >5 min and
no pupillary reflex to strong light. The isolation of PASMCs from
mice was conducted under a stereomicroscope, following previously
described methods (24,25). Under aseptic conditions, the
thoracic cavity of mice was opened, and the pulmonary artery trunk
and left and right pulmonary arteries were removed. The pulmonary
artery vessels were isolated and washed with PBS to remove excess
blood. Then, the artery vessels were scraped on both the internal
and external sides of the vessel wall to remove endothelial cells.
The tissue was then rinsed several times with DMEM/F12 (cat. no.
D6421; Sigma-Aldrich; Merck KGaA) supplemented with 1% penicillin
and streptomycin. The collected tissues were cut into
1-mm3 blocks, washed with D-Hank's solution and
transferred into a culture platform. PASMCs were digested by adding
1 mg/ml collagenase (2 h at 37°C; Sigma-Aldrich; Merck KGaA) and
collected via filtration using nylon nets with a 70-mm diameter
(Sigma-Aldrich; Merck KGaA). Then, the cells were centrifuged at
1,200 × g for 10 min at 4°C, washed with PBS and resuspended in
DMEM containing 10% FBS (cat. no. 10099141; Gibco; Thermo Fisher
Scientific, Inc.). After passage at a 1:3 ratio, the cells at
passage 3-5 were selected for subsequent experimentation.
PASMCs were cultured at 37°C under 5% CO2
in DMEM containing 10% FBS (cat. no. 11965092; Gibco; Thermo Fisher
Scientific, Inc.) and 100 U/ml penicillin/streptomycin (cat. no.
P1400; Beijing Solarbio Science & Technology Co., Ltd.). 293
cells were purchased from the American Type Culture Collection and
incubated in DMEM containing 10% FBS (cat. no. 10100147; Gibco;
Thermo Fisher Scientific, Inc.) and 100 U/ml
penicillin/streptomycin.
Normoxia or hypoxia exposure
Before any intervention, PASMCs were cultured in a
serum-free medium for 24 h to synchronize the cell cycles. PASMCs
were also cultured under hypoxia conditions (3% O2, 5%
CO2, 92% N2), and the cells were used to
conduct the subsequent experiments. For normoxia exposure, cells
were maintained in a humidified incubator with 5% CO2
and 21% O2 at 37°C.
High-throughput transcriptome
sequencing
Total RNA from PASMCs in the normoxia (n=2) and
hypoxia groups (n=3) was isolated using TRIzol® reagent
(Invitrogen; Thermo Fisher Scientific, Inc.). circRNA and mRNA
expression levels were profiled using the SEQuoia RiboDepletion kit
(Bio-Rad Laboratories, Inc.) at Shanghai Fulgent Genetics Co., Ltd.
Then, raw data were summarized and normalized at the transcript
level using R (version 3.3.2.; https://cran.r-project.org/bin/windows/base/old/3.2.2/),
and the limma package of R was used for pre-processing and
statistical analysis. Transcripts showing absolute log2-fold of
>1 were considered differentially expressed, and P<0.05
indicated a statistically significant difference between
experimental and control groups.
Plasmid construction and
transfection
The circ-Grm1 overexpression vector (oe-circ-Grm1)
was designed, and the mouse full-length circ-Grm1 was inserted into
a pCD5-ciR vector by Guangzhou Geneseed Biotech Co., Ltd. The mock
vector [oe-negative control (NC)] without circ-Grm1 sequence was
served as a control. Small interfering (si)RNA targeting circ-Grm1
(si-circ-1 and si-circ-2) and their correspond negative controls
(si-NCs) were purchased from Shanghai GenePharma Co., Ltd. si-FUS,
pcDNA3.1 (p)-FUS (p-FUS), p-Grm1 and NCs (si-NC and p-NC) were
purchased from Sangon Biotech Co., Ltd. The sequences were as
follows: si-circ-1, 5′-GGA GGT CTG GTT CGA TGA GAA-3′; si-NC1,
5′-GCG GGA AGT AGT ACG TGG TAT-3′; si-circ-2, 5′-GCT GAA TAT CGA
TGA TTA CAA-3′; si-NC2, 5′-GCT TGA AGA CGT ACA ATT ATA-3′; si-FUS,
5′-GAC CCA TCC TAA CCT ACT CAT-3′; si-NC3, 5′-GCC ACT TCA CAC TCT
CCT AAA-3′.
si-circ-1 (50 nM), si-circ-2 (50 nM), si-FUS (20
µl/ml), p-FUS (4 µg), oe-circ-Grm1 (50 nM), p-Grm1 (4
µg) and NCs (si-NC, oe-NC and p-NC) were transfected in
PASMCs using Lipofectamine® 3000 transfection reagent
(cat. no. L3000015; Invitrogen; Thermo Fisher Scientific, Inc.) at
37°C for 48 h, according to the manufacturer's procedures, when
cells were at a density of 1×106. After 48 h of hypoxia,
cells were collected for subsequent analyses.
Cell proliferation
The proliferative abilities of the PASMCs were
detected using Cell Counting Kit-8 (CCK-8; cat. no. CK04; Dojindo
Laboratories, Inc.), according to the manufacturer's instructions.
The cells were seeded in 96-well plates (2×103
cells/well) in 10 ml DMEM containing 5% FBS. After 48 h of
transfection, 10 µl CCK-8 solution was added to each well at
0, 24, 48, 72 and 96 h. Then, the cells were cultured with CCK-8
solution for 4 h at 37°C. The CCK-8 reagents and stopping solution
were added to each well, and then a microplate reader (Multiskan;
Thermo Fisher Scientific, Inc.). was used to determine the optical
density (OD) of each well at 450 nm.
The EdU assay was carried out with the EdU
labelling/detection kit (Guangzhou RiboBio Co., Ltd.) according to
the manufacturer's instructions. Cells in the logarithmic growth
phase were seeded into 96-well plates with 4×103 cells
per well in 10 ml DMEM containing 10% FBS. Each well was incubated
with 50 µM EdU solution for 2 h and washed with PBS twice
for 5 min. Then, PBS containing 4% paraformaldehyde was used to fix
cells at room temperature for 30 min, and the cells in each well
were incubated in 1X Apollo staining solution for 30 min at room
temperature. Finally, 1X Hoechst 33342 solution was added to each
well and incubated at room temperature in the dark for 30 min to
stain DNA for observation under a fluorescence microscope (Olympus
Corporation, magnification, ×100).
Cell migration
The migration of PASMCs was measured using a wound
healing assay, wherein 2.5×104 cells/cm2
PASMCs were seeded into six-well plates and cultured overnight in
the starvation medium (DMEM containing 5% FBS) for 24 h. Once the
cells reached ~80% confluence (or more), the monolayers were
scratched using a sterile 200-µl pipette tip. Then the
prepared and filled plates were washed with 3-5 ml PBS solution.
Next, images at 0 and 24 h were obtained under a stereomicroscope
(magnification, ×100; Thermo Fisher Scientific, Inc.). ImageJ
software v2.1.4.7 (National Institutes of Health) was used to
detect the width of the scratch, and the migratory abilities of
PASMCs were determined by the number of cells that crossed into the
wound area from their reference point at 0 h.
RNA extraction and reverse
transcription-quantitative (RT-q)PCR
Total RNA was extracted from cultured cells using
TRIzol reagent. RNA (1 µg) was reverse transcribed to cDNA
for a final volume of 20 µl using the PrimeScript RT Reagent
kit (Takara Biotechnology Co., Ltd.) at 16°C for 30 min, 42°C for
30 min and 85°C for 5 min. qPCR analyses were performed with the
SYBR® Premix Ex Taq™ kit (cat. no. RR420A; Takara
Biotechnology Co., Ltd.) using the ABI PRISM® 7500
Sequence Detection system (Applied Biosystems; Thermo Fisher
Scientific, Inc.). The following thermocycling conditions were used
for qPCR: Initial denaturation at 95°C for 10 min, followed by 40
cycles at 95°C for 10 sec, 60°C for 20 sec and 72°C for 30 sec.
Changes in the target genes were determined based on the relative
values of 2−ΔΔCq (26). The primer sequences used are as
follows: circ-Grm1 forward, 5′-GTC TGG AGA GGA GGT CTG GT-3′ and
reverse, 5′-GGC ATA CGT AGC CGA GGA AAA-3′; Grm1 forward, 5′-GAG
CTG AGG TGT CTG CGA AC-3′ and reverse, 5′-GCC ATA AGC TGG ACG CTG
AG-3′; FUS forward, 5′-CAA TAA ATT TGG TGG TCC TCG G-3′ and
reverse, 5′-GGC CTT GCA CGA AGA TGG TA-3′; GAPDH forward, 5′-GAG
GGA TGC TGC CCT TAC CC-3′ and reverse, 5′-TTG TCT ACG GGA CGA GGA
AAC-3′.
Western blot analysis
Protein samples were extracted and quantified using
RIPA buffer (BioVision, Inc.) and a protein concentration detection
kit (Guangzhou Yingdante Science & Technology Co., Ltd.). These
proteins (50 µg) were separated via 12% SDS-PAGE (cat. no.
P0690; Beyotime Institute of Biotechnology) and transferred to a
PVDF membrane (cat. no. SF1J090I08; MilliporeSigma). After being
blocked with 5% BSA (Thermo Fisher Scientific, Inc.) for 2 h at
25°C, the membrane was incubated with the following specific
primary antibodies overnight at 4°C: Anti-cyclin A (1:1,000; cat.
no. 4656), anti-proliferating cell nuclear antigen (PCNA; 1:1,000;
cat. no. 2586), anti-GAPDH (1:1,000; cat. no. 97166), anti-ERK1
(1:1,000; cat. no. 4372), anti-Rap1 (1:1,000; cat. no. 2326; all
from Cell Signaling Technology, Inc.), anti-phosphorylated
(ph)-ERK1 (1:1,000; cat. no. BS-1522R; Thermo Fisher Scientific,
Inc.), anti-Grm1 (1:1,000; cat. no. ab183712), anti-CDK1 (1:1,000;
cat. no. ab265590) and anti-Ki67 (1:1,000; cat. no. ab16667; all
from Abcam). This was followed by incubation with the HRP-labeled
secondary antibody at room temperature for 2 h (1:1,000; mouse
secondary antibody, cat. no. ab150113; 1:1,000; rabbit secondary
antibody, cat. no. ab6721; both from Abcam). Finally, the proteins
were visualized using a ECL reagent (cat. no. P0020; Beyotime
Institute of Biotechnology) to obtain images. The relative
expression of target proteins was calculated using ImageJ software
v2.1.4.7 (National Institutes of Health).
RNA immunoprecipitation (RIP) assay
The EZ-Magna RIP kit (MilliporeSigma) was used for
the RIP assay, following the manufacturer's protocol. PASMCs were
cross-linked by treating with formaldehyde for 10 min at 37°C.
After washing with cold PBS, cells were incubated in 4 ml cell
lysis buffer for 15 min in ice. After nuclear extraction using a
Dounce homogenizer (Wheaton; DWK Life Sciences), the chromatin was
sheared by sonication (25% power, 4.5 sec impact, 9 sec clearance,
14 times) at 37°C. Then, PASMCs were incubated with RIP buffer
containing magnetic beads conjugated with anti-FUS or control
antibodies (IgG), at 4°C for 6 h by following DNase treatment for
30 min at room temperature. The beads were washed using the washing
buffer, and the proteins were removed from the compounds via
incubation with 0.1% SDS/0.5 mg/ml Proteinase K (30 min, 55°C). The
RNA concentrations were detected using NanoDrop spectrophotometry
(Thermo Fisher Scientific, Inc.), and the RNA qualities were
analysed using a bioanalyzer (Agilent Technologies, Inc.).
Pull-down assay
The RNA pull-down assay was used to identify the
circ-Grm1 interaction with FUS using a Magnetic RNA-protein
pull-down kit (Thermo Fisher Scientific, Inc.). Briefly, before
harvesting, PASMCs were transfected with 50 nM biotinylated DNA
probe complementary to circ-Grm1 or negative control (Genepharm,
Inc.) probe for 48 h. Then, the PASMCs were washed with PBS and
incubated for 10 min in the cell lysis buffer (Thermo Fisher
Scientific, Inc.) on ice. Part of the lysates were aliquoted for
input. The remaining lysates were incubated with streptavidin
magnetic beads (Thermo Fisher Scientific, Inc.) precoated with 5%
RNase-free BSA (Thermo Fisher Scientific, Inc.) and yeast transfer
RNA (MilliporeSigma) at 4°C for 30 min. After washing and elution
of RNA-binding protein complexes, the proteins in the pull-down
materials were analysed via western blotting using an antibody
recognizing FUS (1:1,000; cat. no. ab124923; Abcam).
Actinomycin D inhibition assay
The RNA stability of Grm1 mRNA was determined using
an Actinomycin D inhibition assay as described previously (27,28). Firstly, the circ-Grm1 and FUS
siRNAs were transfected into PASMCs, which were then treated with
actinomycin D (5 µg/ml; Sigma-Aldrich; Merck KGaA) at 37°C
for 0, 3, 6 and 9 h. The total RNA was isolated, and Grm1 mRNA
expression was quantified via RT-qPCR, as aforementioned. RNA
half-life (t1/2) was calculated using linear regression
analysis as described previously (27,28).
Bioinformatics analyses
The relationships of circ-Grm1 and Grm1 with FUS
were predicted by the bioinformatical analysis tool starBase v 2.0
(http://starbase.sysu.edu.cn/index.php) though
'RBP-target' plate. Bioinformatical (RPISeq Version 1.0; http://pridb.gdcb.iastate.edu/RPISeq/)
tools were used to predict the interaction probabilities of
circ-Grm1 and Grm1 with FUS. For Kyoto Encyclopedia of Genes and
Genomes (KEGG, https://www.genome.jp/kegg/pathway.html) analyses,
Grm1 expression was treated as a numeric variable. The Pearson
correlation coefficient of other genes and Grm1 expression was
calculated, and then the genes were sequenced according to the
correlation coefficient. KEGG analyses based on the Grm1-related
gene were conducted using the clusterProfiler R package (https://bioconductor.org/packages/release/bioc/html/clusterProfiler.html,
version no. 4.0.5) (29)
(organism='mmu', P-value Cutoff=0.05) to evaluate the potential
biological mechanisms mediating Grm1-related HPH.
Statistical analysis
All experimental values are shown as the mean ± SD
obtained using SPSS 26.0 statistical analysis (IBM Corp.). All
experiments were performed independently in triplicate. The
two-tailed Student's t-test was used to compare the differences
among two groups, whereas comparison among multiple groups was
analysed using a one-way ANOVA with a Tukey's post hoc test.
P<0.05 was considered to indicate a statistically significant
difference.
Results
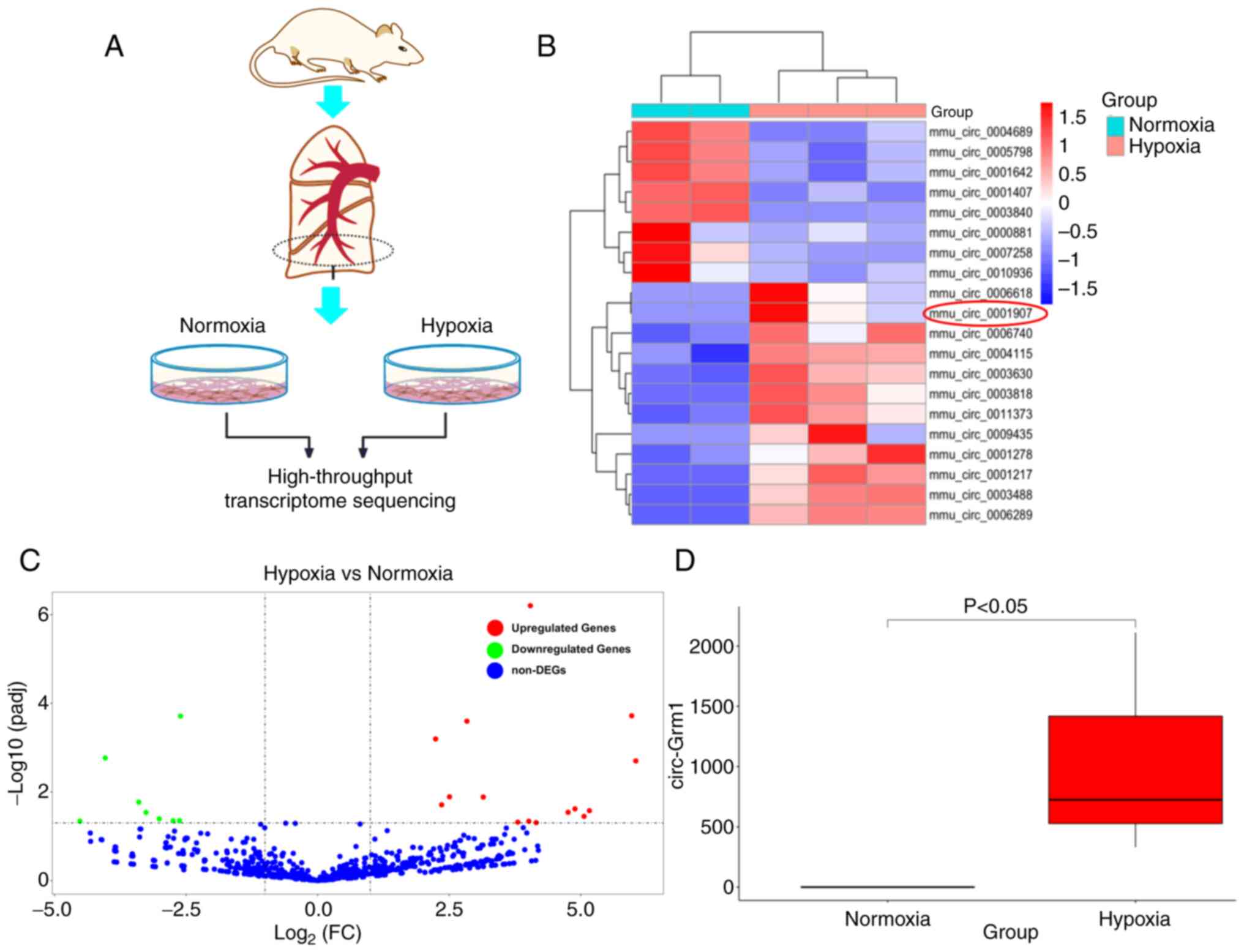
circ-Grm1 is associated with the
pathogenesis of PAH in hypoxic PASMCs
To evaluate the altered phenotype of PAH-PASMCs,
cell libraries of primary cultured PASMCs from mice lung tissues
were established (Fig. 1A). The
results of high-throughput transcriptome sequencing demonstrated
that circ-Grm1 expression increased in the hypoxic groups and
showed higher expression levels compared with the control group
(Fig. 1B and C). Additionally,
the high-throughput transcriptome sequencing analyses showed that
the expression of circ-Grm1 was elevated in the hypoxic PASMCs
compared with the normxia group (Fig. 1D). These results suggested that
hypoxia could promote the expression level of circ-Grm1 in
PASMCs.
Silencing of circ-Grm1 suppresses the
proliferation and migration of hypoxic PASMCs
To assess the significance of circ-Grm1 in hypoxic
PASMCs, the expression of circ-Grm1 was knocked down using the RNA
silencing technique. The results of RT-qPCR analysis demonstrated
the efficiency of RNA silencing, which revealed successful
downregulation of circ-Grm1 at the RNA level (Fig. 2A). Then, the effects of circ-Grm1
silencing on the proliferative ability of hypoxic PASMCs were
detected using CCK-8 and EdU assays (Fig. 2B-D). Additionally, the expression
levels of proliferation-associated proteins (cyclin A, PCNA, CDK1
and Ki67) were estimated via western blot analysis. As shown in
Fig. 2D, the expression levels
of these proteins were significantly downregulated after the
knockdown of circ-Grm1 compared with the si-NC group. These
findings suggested that the knockdown of circ-Grm1 could inhibit
the proliferation of PASMCs.
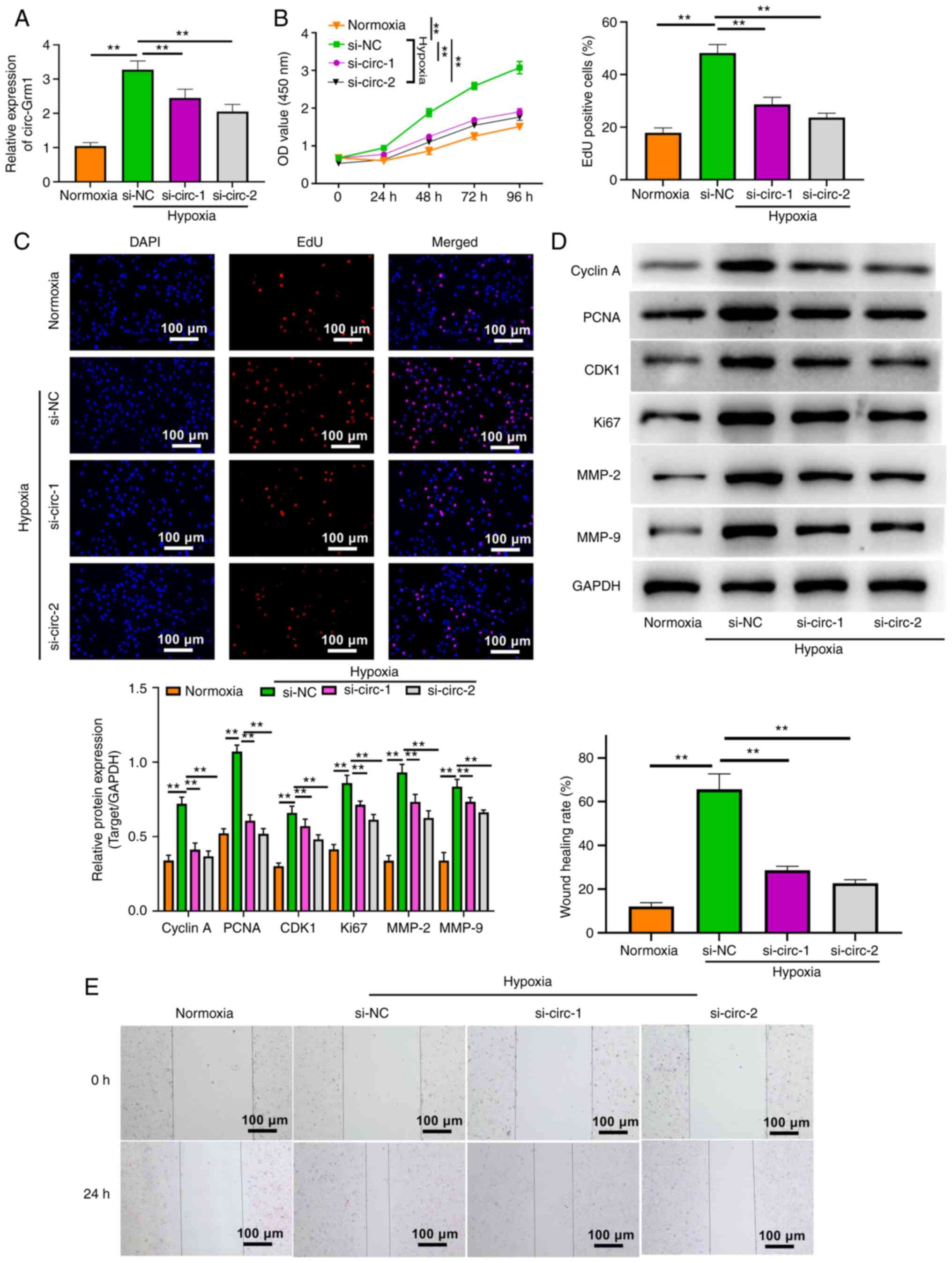 | Figure 2Silencing of circ-Grm1 suppresses the
proliferation and migration of hypoxic PASMCs. (A) circ-Grm1
expression was detected via reverse transcription-quantitative PCR
analysis in cells of normoxia, si-NC and si-circ-Grm1groups. (B)
Cell Counting Kit-8 assay and (C) EdU assays were performed to
determine the proliferative ability of PASMCs in the normoxia,
si-NC and si-circ-Grm1 groups. (D) Western blot analysis was
performed to measure the expression levels of cyclin A, PCNA, CDK1,
Ki67, MMP-2 and MMP-9 in cells from the normoxia, si-NC and
si-circ-Grm1 groups. (E) Wound healing assay was performed to
determine the migratory abilities of PASMCs. Scale bar, 100
µm. The experiments were independently conducted three
times, and values are shown as the mean ± SD.
**P<0.01. EdU, 5-ethynyl-2-deoxyuridine; si, small
interfering RNA; NC, negative control; PASMCs, pulmonary artery
smooth muscle cells; circ, circular RNA; circ-Grm1, circular RNA
glutamate metabotropic receptor 1; PCNA, proliferating cell nuclear
antigen; OD, optical density. |
As shown in Fig.
2E, the results of the wound healing assay indicated that
knockdown of circ-Grm1 significantly decreased the migration of
PASMCs. Furthermore, the effects of circ-Grm1 knockdown on
migration were also confirmed by detecting the expression levels of
migration-associated proteins, MMP-2 and MMP-9. The results
demonstrated that the knockdown of circ-Grm1 significantly
decreased MMP-2 and MMP-9 expression in PASMCs (Fig. 2D). Collectively, it was suggested
that silencing of circ-Grm1 inhibited the proliferation and
migration of hypoxic PASMCs.
Grm1 is targeted by circ-Grm1 and
circ-Grm1 can bind with FUS
Fig. 3A and B
show the volcano map and heat map analyses measured via
high-throughput transcriptome sequencing, respectively. The results
of high-throughput transcriptome sequencing suggested that the
expression level of Grm1 was significantly lower in the hypoxic
group compared with that of the normxia group (Fig. 3C) and varied inversely with the
expression of circ-Grm1 (Fig.
3D) in hypoxic and normxia group. The bioinformatical analysis
tool starBase v 2.0 (http://starbase.sysu.edu.cn/index.php) was used to
predict whether GRM1 could bind to FUS. FUS is a multifunctional
RBP that plays a vital role in various cellular processes,
including transcription, cell cycle progression, angiogenesis and
apoptosis (30-34). The prediction of the binding
abundance between FUS and Grm1 was also determined using
bioinformatics (http://pridb.gdcb.iastate.edu/RPISeq/). The binding
scores of FUS and GRM1 are shown in Fig. 3E. The RIP assay revealed that
Grm1 was highly bound to FUS in PASMCs (Fig. 3F).
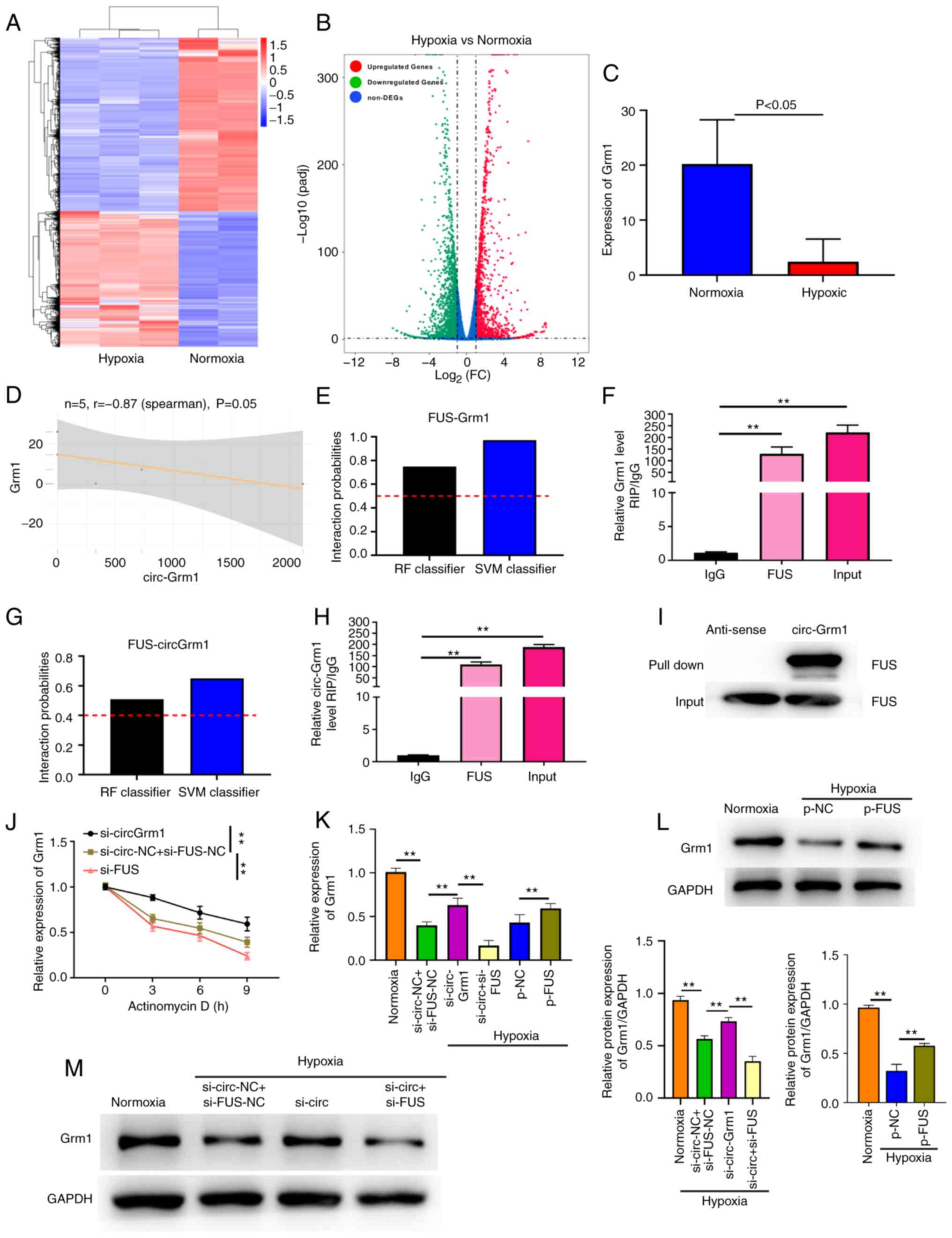 | Figure 3circ-Grm1 binds to FUS and suppresses
Grm1 translation. (A) Volcano map and (B) heat map analysis
measured via high-throughput transcriptome sequencing in normxia
and hypoxic PASMCs. (C) Analysis of Grm1 expression in PASMCs via
high-throughput transcriptome sequencing analysis. (D) Scatter
plots were used to depict the correlations between circ-Grm1 and
Grm1. (E) Interaction probabilities of Grm1 with FUS were predicted
using bioinformatics analysis (predictions with P>0.5 were
considered 'positive'). (F) RIP assay showed that FUS precipitated
with Grm1 in PASMCs. (G) Interaction probabilities of circ-Grm1
with FUS were predicted using bioinformatics analysis, which
indicated that circ-Grm1 and FUS were likely to interact
(predictions with P>0.5 were considered 'positive'). (H) RIP
assays were conducted to detect the combination of circ-Grm1 and
FUS, and then RT-qPCR was performed to detect the co-precipitated
RNA. (I) Pull-down assay indicated that biotin-labelled circ-Grm1
interacted with FUS. (J) Actinomycin D was used to disturb RNA
synthesis in PASMCs, and RT-qPCR was performed to detect the
degradation rates of the RNA every 3 h. (K) After transfection with
si-circ-Grm1, si-FUS and p-FUS, RT-qPCR was used to detect Grm1
expression in PASMCs. After transfection with (L) p-FUS, (M)
si-circ-Grm1 and si-FUS, western blotting was used to detect Grm1
expression in PASMCs. Values were presented as the mean ± SD from
three independent experiments. **P<0.01. RIP, RNA
immunoprecipitation; RT-qPCR, reverse transcription-quantitative
PCR; si, small interfering RNA; NC, negative control; PASMCs,
pulmonary artery smooth muscle cells; circ, circular RNA;
circ-Grm1, circular RNA glutamate metabotropic receptor 1; p-,
pcDNA3.1; FC, fold change; RF, Random Forest; SVM, Support Vector
Machine. |
The significance of the circRNA-protein interaction
has recently been revealed, including via a RBP (35,36). The binding scores of FUS and
circ-Grm1 are shown in Fig. 3G,
and this this result was verified using a RIP assay. The RIP and
pull-down assays demonstrated that circ-Grm1 showed a significant
binding ability to FUS in PASMCs (Fig. 3H and I).
circ-Grm1 recruits FUS to inhibit Grm1
mRNA stability and expression
To further examine whether the circ-Grm1/FUS complex
could modulate the downstream genes of Grm1, the stability of the
Grm1 mRNA was investigated in the presence of actinomycin D, a
transcription inhibitor. The results of RNA stability assay
indicated that the degradation rate of Grm1 mRNA was increased and
decreased in PASMCs upon circ-Grm1 and FUS knockdown, respectively,
compared with that of the control group at 0, 3, 6 and 9 h
(Fig. 3J). These findings
confirmed that circ-Grm1 could sponge FUS and suppress Grm1 mRNA
stability. As shown in Fig. 3K,
RT-qPCR was used to verify the expression level of Grm1 after FUS
overexpression or knockdown and circ-Grm1 knockdown. The
transfection efficiency of circ-Grm1 knockdown and FUS
overexpression or knockdown was shown in Figs. 2A and S1A and B, respectively. It was found
that the knockdown effect of si-circ-2 group was higher. Therefore,
si-circ-2 was selected for use in subsequent experiments
experiment.
The results demonstrated that overexpression of FUS
significantly increased the expression level of Grm1 in PASMCs
(Fig. 3L), thereby suggesting
that FUS expression was positively associated with Grm1 expression
in PASMCs. Additionally, it was determined that the knockdown of
circ-Grm1 significantly elevated Grm1 expression in PASMCs
(Fig. 3M). Compared with the
si-circ-Grm1 group, the co-transfection of si-circ-Grm1 and si-FUS
significantly decreased the expression level of Grm1 (Fig. 3M). These findings indicated that
the circ-Grm1/FUS complex could lead to the prohibition of mRNA
stability and expression of Grm1.
Grm1 reverses the promoting role of
circ-Grm1 in the hypoxia PASMCs
To investigate the effects of GRM1 in response to
circ-Grm1, Grm1 was overexpressed in hypoxic PASMCs. The results of
western blotting assays demonstrated the overexpression efficiency
in the cells, which showed upregulation of GRM1 at the protein
level (Fig. 4A). Moreover, the
transfection efficiency of circ-Grm1 overexpression and Grm1
overexpression was examined via RT-qPCR (Fig. S1C and D). The CCK-8 and EdU
assays results demonstrated that the cell proliferative ability was
inhibited after the co-transfection of oe-circ-Grm1 and
pcDNA3.1-Grm1 compared with the co-transfection of oe-circ-Grm1 and
pcDNA3.1-NC at 0, 24, 48, 72 and 96 h (Fig. 4B and C). Furthermore, the
expression levels of proliferation-associated proteins (cyclin A,
PCNA, CDK1 and Ki67) were detected via western blot analysis. As
shown in Fig. 4D, the expression
levels of these proteins were significantly upregulated after
overexpression of circ-Grm1, whereas they were downregulated after
the co-transfection of oe-circ-Grm1 and p-Grm1. Additionally, the
wound healing assay results suggested that overexpression of
circ-Grm1 significantly increased the migration of PASMCs. However,
after the co-transfection of oe-circ-Grm1 and p-Grm1, the migratory
ability of PASMCs was impaired (Fig.
4E). MMPs modulate extracellular matrix composition and
integrity, and MMP-2 and 9 play key roles in cleaving the
extracellular matrix components that contribute to cell migration
and vascular remodelling (37,38). Thus, the migration results were
also verified by detecting the expression levels of
migration-associated proteins, MMP-2 and MMP-9. These findings
suggested that Grm1 could inhibit the function of circ-Grm1 with
the promotion of the proliferative and migratory abilities in
hypoxic PASMCs.
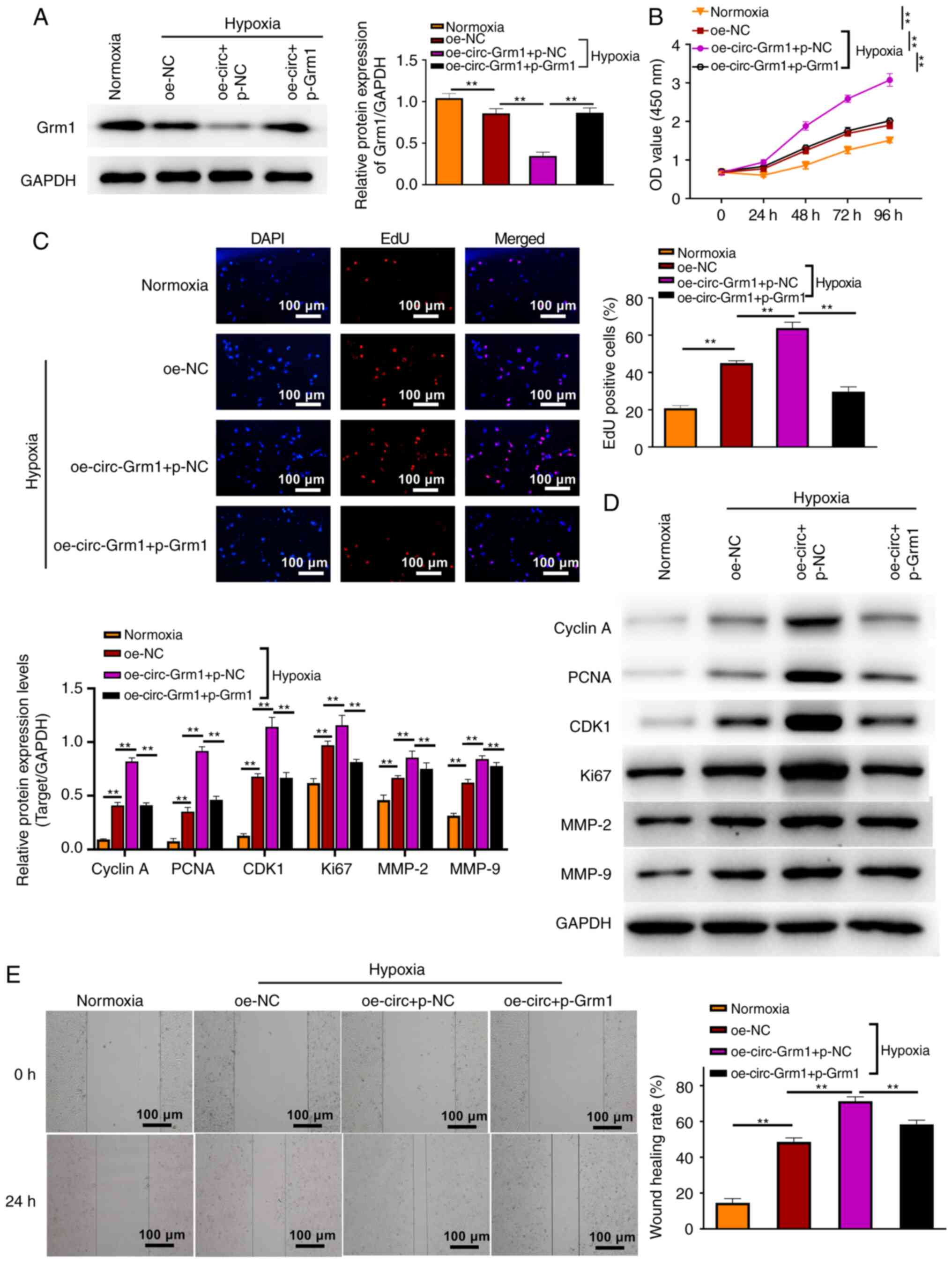 | Figure 4Grm1 reverses the promoting role of
circ-Grm1 in the hypoxia PASMCs. (A) GRM1 expression in PASMCs from
all groups was detected via western blotting. (B) Cell Counting
Kit-8 and (C) EdU assays exhibited the proliferative ability of
PASMCs in different groups. Scale bar, 100 µm. (D) Western
blot analysis was performed to measure the expression levels of
cyclin A, PCNA, CDK1, Ki67, MMP-2 and MMP-9 in normoxia and hypoxia
PASMCs. (E) Wound healing assay was performed to detect the
migratory ability of PASMCs. Values are shown as the mean ± SD of
three independent tests. **P<0.01. NC, negative
control; PASMCs, pulmonary artery smooth muscle cells; circ,
circular RNA; circ-Grm1, circular RNA glutamate metabotropic
receptor 1; p-, pcDNA3.1; EdU, 5-ethynyl-2-deoxyuridine; OD,
optical density; oe, overexpression; PCNA, proliferating cell
nuclear antigen. |
circ-Grm1 and Grm1 are involved in
function of PASMCs by regulating the Rap1 signalling pathway
The effects of circ-Grm1 and Grm1 on the signalling
pathways were determined using KEGG analysis and western blotting.
The 'Rap1 signalling pathway' was significantly associated with the
circ-Grm1 and Grm1 in HPH (Fig.
5A). Additionally, the western blotting results indicated that
the protein expression levels of Grm1 and Rap1b in the hypoxia
groups were lower compared with those of the normoxia group,
whereas these were increased in cells overexpressing Grm1 compared
with oe-circ-Grm1 group (Fig.
5B). Conversely, the expression level of ph-ERK1 in the hypoxia
groups was higher compared with the control group, whereas it was
lower in cells with overexpressing Grm1 compared with oe-circ-Grm1
group (Fig. 5B). In addition,
the co-transfection of oe-circ-Grm1 and p-Grm1 reversed the effects
of Grm1 overexpression on the expression levels of Grm1, Rap1b and
ph-ERK1. Therefore, it was suggested that circ-Grm1 could inhibit
the biological functionalities of the Rap1 signalling pathway in
PASMCs in HPH and promote the phosphorylation of ERK1.
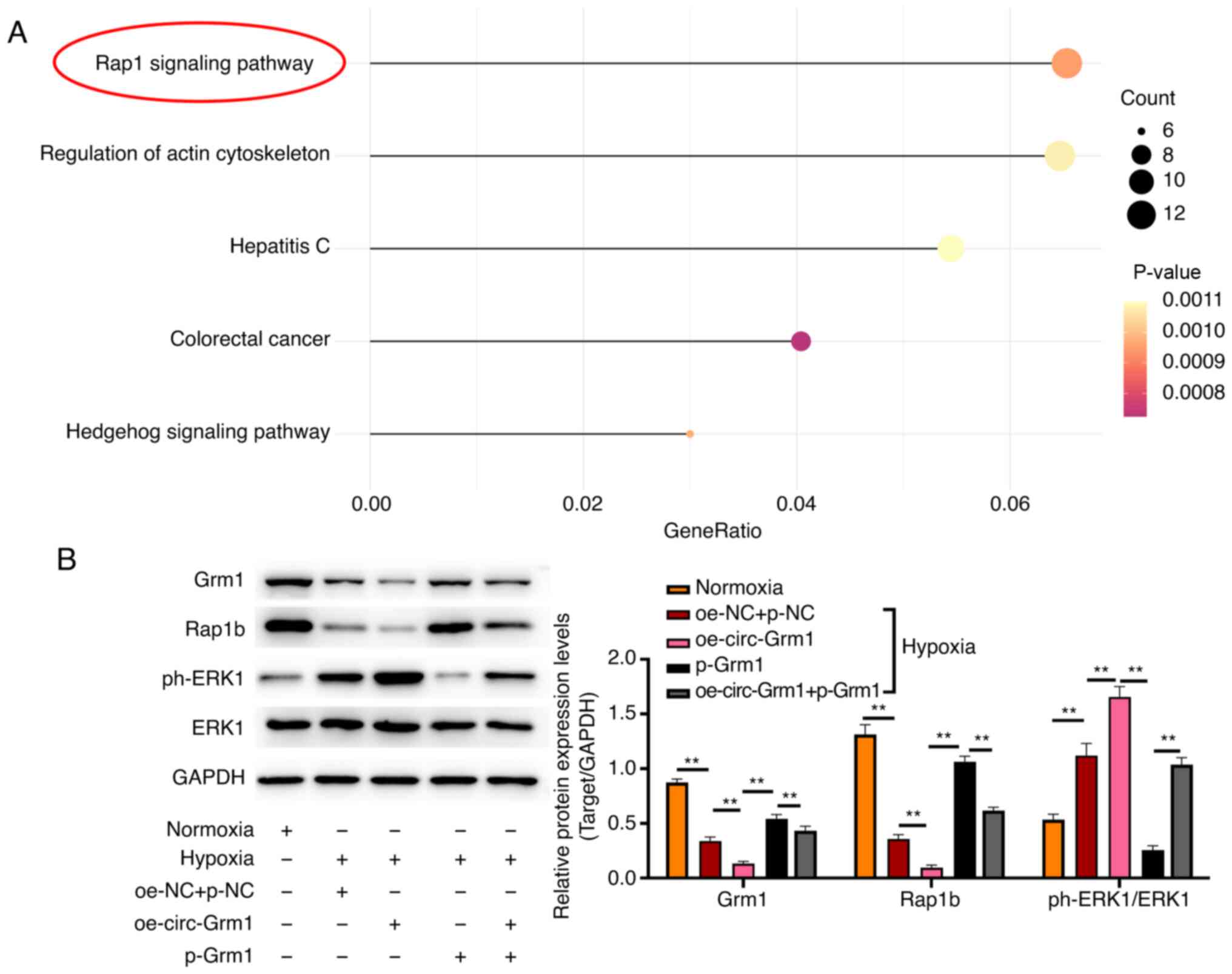 | Figure 5circ-Grm1 and Grm1 are associated
with the function of PASMCs by regulating the Rap1 signalling
pathway. (A) Kyoto Encyclopedia of Genes and Genomes (https://www.genome.jp/kegg/pathway.html) analysis was
performed using the clusterProfiler R package. (B) Expression
levels of Grm1, Rap1b, p-ERK1 and ERK1 protein obtained in PASMCs
from normoxia, hypoxia, oe-NC + p-NC, oe-circ-Grm1, p-Grm1 and
oe-circ-Grm1 + p-Grm1 groups were detected via western blot
analysis. Data are shown as the mean ± SD based on three
independent experiments. **P<0.01. NC, negative
control; PASMCs, pulmonary artery smooth muscle cells; circ,
circular RNA; circ-Grm1, circular RNA glutamate metabotropic
receptor 1; p-, pcDNA3.1; ph-, phosphorylated. |
Discussion
A previous study revealed that hypoxia-induced
pulmonary hypertension was mainly caused by hypoxic pulmonary
vasoconstriction, pulmonary vascular remodelling and polycythaemia
(39). Pulmonary vascular
remodelling is a key feature of hypoxia-induced pulmonary
hypertension associated with the dysfunction of endothelial cells,
smooth muscle cells and fibroblasts (40). Previous studies have shown that
circRNAs are involved in PASMC proliferation, migration and
apoptosis, leading to pulmonary vascular remodelling (4,41). In the present study, to the best
of our knowledge, it was demonstrated for the first time that
circ-Grm1 was involved in HPH and the mechanisms via which circRNA
regulated PASMC proliferation and migration were shown in Fig. 6.
As a non-coding RNA, circRNA was recently
discovered. Previous studies have reported that circRNAs are
associated with vascular dysfunction and multiple processes, such
as vascular development, growth and remodelling (42-48). The present study performed a
high-throughput transcriptome sequencing analysis of the PASMCs to
investigate the differently expressed circRNAs and finally
identified the upregulated circ-Grm1. circ-Grm1 was first found to
be upregulated in hypoxic PASMC models, as determined by
high-throughput transcriptome sequencing. The functional cellular
experiments revealed that the knockdown of circ-Grm1 could
effectively inhibit the proliferation and migration of PASMCs in
vitro. Therefore, these results indicated that circ-Grm1
participates in the regulation of the biological function of
PASMCs.
circRNA plays a vital role in regulating downstream
molecules, such as Grm1. As a G-protein-coupled receptor, Grm1 is
normally localized to the central and peripheral nervous systems.
Grm1 is correlated with cell proliferation, migration and invasion
in prostate cancer (49). It has
been shown that Grm1 activation is coupled with ERK1/2 (50). Moreover, activation of ERK1/2 via
phosphorylation on tyrosine and threonine residues plays a pivotal
role in intracellular signalling and can mediate cellular
processes, such as cell proliferation, invasion, metastasis,
survival angiogenesis and apoptosis (51,52).
As aforementioned, the high-throughput transcriptome
sequencing analysis results identified that circ-Grm1 was
upregulated in hypoxic PASMC models, but the expression of its
cognate mRNA (Grm1) was significantly decreased. To clarify this
phenomenon, we hypothesized that FUS could mediate the expression
of circ-Grm1 and Grm1. Therefore, bioinformatics analysis was used
to predict whether circ-Grm1 and Grm1 may bind to FUS. The RIP and
pull-down assays revealed that FUS could interact with both
circ-Grm1 and Grm1. Based on the observed FUS binding to circ-Grm1
and Grm1 mRNA, Grm1 expression following circ-Grm1 knockdown or FUS
overexpression was increased, but decreased following FUS
knockdown. Cui and Placzek (53)
and Zhao et al (54)
reported that mRNA translation and mRNA stability were major
factors contributing to protein expression levels. Thus, to test
this, si-NC, circ-Grm1 and si-FUS transfected cells were treated
with actinomycin D to block transcription, and incremental RNA
samples were harvested to assess Grm1 mRNA half-life. The RNA
stability assay demonstrated that linear regression decay curves of
Grm1 mRNA showed an increase and decrease in half-life for
si-circ-Grm1 and si-FUS transfected PASMCs under hypoxia,
respectively. These results suggested that circ-Grm1 could sponge
FUS and suppresses Grm1 mRNA stability and Grm1 protein
expression.
The present study found that Grm1 reversed the
boosting function of circ-Grm1 in the progression of HPH. Previous
reports have shown that circ-0000790 monitors FOXC1 by binding to
miR-374c, and affects the biological functionalities of PASMCs in
mice in the HPH model, whereas miR-374c reversed the enhanced
functionalities of mmu_circ_0000790 of silencing-RNA groups with
the progression of HPH (4).
Therefore, these results indicated that circ-Grm1 participates in
the regulation of PASMCs' biological function.
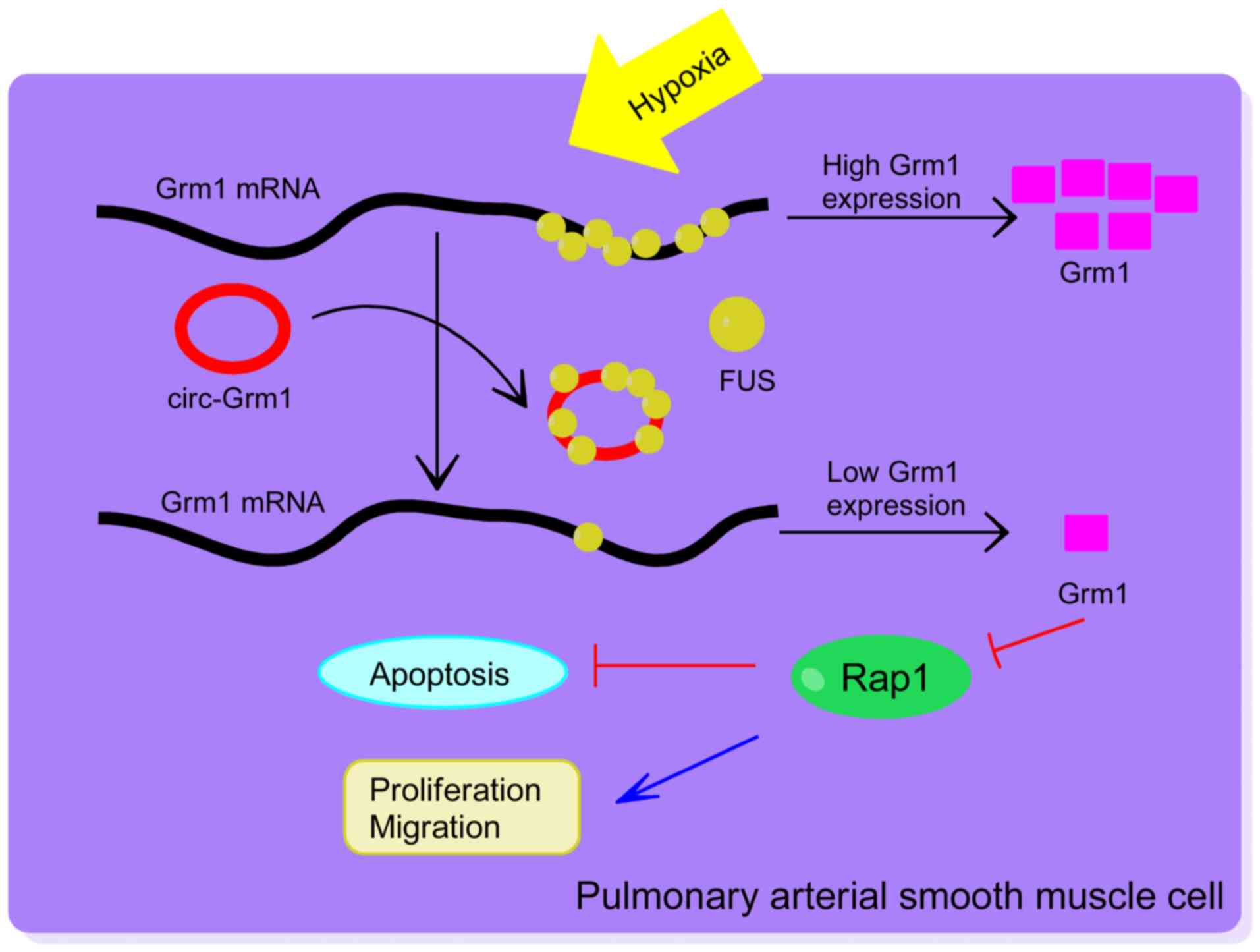
circRNAs act as molecular connectors for proteins,
which were combined with corresponding pathways. In the present
study, circ-Grm1 was upregulated in hypoxic PASMCs and competed
with binding of FUS (an RBP), which could inhibit the expression of
downstream transcripts of Grm1. Then, the Rap1 signalling pathway
was inhibited and accompanied by activation of the ERK signalling
pathway. Rap1 is associated with the mass of cellular signal
transduction pathways, which have been associated with
proliferative and migratory cell phenotypes (55), and the signalling pathway of ERK
acts as a significant modulator in proliferation, differentiation,
migration, senescence and apoptosis in cells (56,57). Nikam et al (58) reported that reduced p-ERK
expression could have been responsible for the slower growth rate
of HMVECs following the suppression of Rap1a or -1b expression.
Studies have also shown that Rap1 activated GAP-GTP by immediately
restraining the reactivities of the phosphorylation of ERK to
promote proliferation and migration in VSMCs (59), and this was also demonstrated in
the present study. The current results demonstrated that the
circ-Grm1/Grm1/Rap1 signalling axis regulated PASMC proliferation
and migration in hypoxia-induced PAH, thereby providing a new
potential target for the treatment and diagnosis of this
disease.
In conclusion, the present study identified that
circ-Grm1 was involved in vascular remodelling in PAH by promoting
the proliferation and migration of PASMCs via suppression of GRM1
expression by FUS. Furthermore, it was found that the Rap1
signalling pathway was also involved in this process. Taken
together, the current results suggest that circ-Grm1 may be used as
a novel biomarker in the diagnosis and treatment of PAH.
Supplementary Data
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request. The RNA-seq datasets in this manuscript have been
deposited in the National Center for Biotechnology Information
Sequence Read Archive (http://www.ncbi.nlm.nih.gov/sra) under BioProject no.
PRJNA753221 (accession nos. SRR15440746 and SRR15403718).
Authors' contributions
Conception and design: SS. Performed research: QK,
ZC and MW. Data analysis and interpretation: SS, HZ and CZ.
Manuscript writing: SS. The authenticity of the raw data has been
assessed by SS and CZ. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
This experiment was conducted with approval of the
Animal Ethics Committee of Qilu Hospital of Shandong University
(approval no. KYLL-2020-002). All animal experiments in this study
were in strict accordance with the protocols stated in the Guide
for the Care and Use of Laboratory Animals published by the US
National Institutes of Health. Appropriate measures were taken to
minimize the number and suffering of animals.
Patient consent for publication
Not applicable.
Competing interests
The authors declared no potential conflicts of
interest with respect to the research, authorship, and/or
publication of this article.
Acknowledgments
Not applicable.
Funding
This research was supported by Shandong Key Research and
Development Plan (grant nos. 2019GSF108186 and 2014GSF118066),
Shandong, China.
References
|
1
|
Lau EMT, Giannoulatou E, Celermajer DS and
Humbert M: Epidemiology and treatment of pulmonary arterial
hypertension. Nat Rev Cardiol. 14:603–614. 2017. View Article : Google Scholar
|
|
2
|
Thenappan T, Ryan JJ and Archer SL:
Evolving epidemiology of pulmonary arterial hypertension. Am J
Respir Crit Care Med. 186:707–709. 2012. View Article : Google Scholar
|
|
3
|
Zolty R: Pulmonary arterial hypertension
specific therapy: The old and the new. Pharmacol Ther.
214:1075762020. View Article : Google Scholar
|
|
4
|
Xu L, Ma Y, Zhang H, Lu QJ, Yang L, Jiang
GN and Liao WL: HMGA2 regulates circular RNA ASPH to promote tumor
growth in lung adenocarcinoma. Cell Death Dis. 11:5932020.
View Article : Google Scholar
|
|
5
|
Mulvaney EP, Reid HM, Bialesova L,
Mendes-Ferreira P, Adão R, Brás-Silva C and Kinsella BT: Efficacy
of the thromboxane receptor antagonist NTP42 alone, or in
combination with sildenafil, in the sugen/hypoxia-induced model of
pulmonary arterial hypertension. Eur J Pharmacol. 889:1736582020.
View Article : Google Scholar
|
|
6
|
Sitbon O, Gomberg-Maitland M, Granton J,
Lewis MI, Mathai SC, Rainisio M, Stockbridge NL, Wilkins MR,
Zamanian RT and Rubin LJ: Clinical trial design and new therapies
for pulmonary arterial hypertension. Eur Respir J. 53:18019082019.
View Article : Google Scholar
|
|
7
|
McLaughlin VV, Hoeper MM, Channick RN,
Chin KM, Delcroix M, Gaine S, Ghofrani HA, Jansa P, Lang IM, Mehta
S, et al: Pulmonary arterial hypertension-related morbidity is
prognostic for mortality. J Am Coll Cardiol. 71:752–763. 2018.
View Article : Google Scholar
|
|
8
|
Galiè N, Humbert M, Vachiery JL, Gibbs S,
Lang I, Torbicki A, Simonneau G, Peacock A, Noordegraaf AV,
Beghetti M, et al: 2015 ESC/ERS guidelines for the diagnosis and
treatment of pulmonary hypertension. Rev Esp Cardiol (Engl Ed).
69:1772016. View Article : Google Scholar
|
|
9
|
Chaabane M, Andreeva K, Hwang JY, Kook TL,
Park JW and Cooper NGF: seekCRIT: Detecting and characterizing
differentially expressed circular RNAs using high-throughput
sequencing data. PLoS Comput Biol. 16:e10083382020. View Article : Google Scholar
|
|
10
|
Du H, He Z, Feng F, Chen D, Zhang L, Bai
J, Wu H, Han E and Zhang J: Hsa_circ_0038646 promotes cell
proliferation and migration in colorectal cancer via
miR-331-3p/GRIK3. Oncol Lett. 20:266–274. 2020.
|
|
11
|
Song R, Li Y, Hao W, Yang L, Chen B, Zhao
Y, Sun B and Xu F: Circular RNA MTO1 inhibits gastric cancer
progression by elevating PAWR via sponging miR-199a-3p. Cell Cycle.
19:3127–3139. 2020. View Article : Google Scholar
|
|
12
|
Altesha MA, Ni T, Khan A, Liu K and Zheng
X: Circular RNA in cardiovascular disease. J Cell Physiol.
234:5588–5600. 2019. View Article : Google Scholar
|
|
13
|
Boon RA, Jaé N, Holdt L and Dimmeler S:
Long noncoding RNAs: From clinical genetics to therapeutic targets?
J Am Coll Cardiol. 67:1214–1226. 2016. View Article : Google Scholar
|
|
14
|
Miao R, Wang Y, Wan J, Leng D, Gong J, Li
J, Liang Y, Zhai Z and Yang Y: Microarray expression profile of
circular RNAs in chronic thromboembolic pulmonary hypertension.
Medicine (Baltimore). 96:e73542017. View Article : Google Scholar
|
|
15
|
Wang Y, Tan X, Wu Y, Cao S, Lou Y, Zhang L
and Hu F: Hsa_ circ_0002062 promotes the proliferation of pulmonary
artery smooth muscle cells by regulating the Hsa-miR-942-5p/CDK6
signaling pathway. Front Genet. 12:–673229. 2021.
|
|
16
|
Yang L, Liang H, Meng X, Shen L, Guan Z,
Hei B, Yu H, Qi S and Wen X: mmu_circ_0000790 is involved in
pulmonary vascular remodeling in mice with HPH via
microRNA-374c-mediated FOXC1. Mol Ther Nucleic Acids. 20:292–307.
2020. View Article : Google Scholar
|
|
17
|
Abdelmohsen K, Panda AC, Munk R,
Grammatikakis I, Dudekula DB, De S, Kim J, Noh JH, Kim KM,
Martindale JL and Gorospe M: Identification of HuR target circular
RNAs uncovers suppression of PABPN1 translation by CircPABPN1. RNA
Biol. 14:361–369. 2017. View Article : Google Scholar
|
|
18
|
Du WW, Fang L, Yang W, Wu N, Awan FM, Yang
Z and Yang BB: Induction of tumor apoptosis through a circular RNA
enhancing Foxo3 activity. Cell Death Differ. 24:357–370. 2017.
View Article : Google Scholar
|
|
19
|
Wang J, Song YX, Ma B, Wang JJ, Sun JX,
Chen XW, Zhao JH, Yang YC and Wang ZN: Regulatory roles of
non-coding RNAs in colorectal cancer. Int J Mol Sci.
16:19886–19919. 2015. View Article : Google Scholar
|
|
20
|
Fan YN, Li C, Huang L, Chen L, Tang Z, Han
G and Liu Y: Characterization of group I metabotropic glutamate
receptors in rat and human adrenal glands. Front Physiol.
11:4012020. View Article : Google Scholar
|
|
21
|
Khan AJ, LaCava S, Mehta M, Schiff D,
Thandoni A, Jhawar S, Danish S, Haffty BG and Chen S: The glutamate
release inhibitor riluzole increases DNA damage and enhances
cytotoxicity in human glioma cells, in vitro and in vivo.
Oncotarget. 10:2824–2834. 2019. View Article : Google Scholar
|
|
22
|
Namkoong J, Martino JJ and Chen S: From
existing therapies to novel targets: A current view on melanoma.
Front Biosci. 11:2081–2092. 2006. View
Article : Google Scholar
|
|
23
|
Yip D, Le MN, Chan JL, Lee JH, Mehnert JA,
Yudd A, Kempf J, Shih WJ, Chen S and Goydos JS: A phase 0 trial of
riluzole in patients with resectable stage III and IV melanoma.
Clin Cancer Res. 15:3896–3902. 2009. View Article : Google Scholar
|
|
24
|
Wang R, Xu YJ, Liu XS, Zeng DX and Xiang
M: Knockdown of connective tissue growth factor by plasmid-based
short hairpin RNA prevented pulmonary vascular remodeling in
cigarette smoke-exposed rats. Arch Biochem Biophys. 508:93–100.
2011. View Article : Google Scholar
|
|
25
|
Wang R, Xu YJ, Liu XS, Zeng DX and Xiang
M: CCN2 promotes cigarette smoke-induced proliferation of rat
pulmonary artery smooth muscle cells through upregulating cyclin D1
expression. J Cell Biochem. 113:349–359. 2012. View Article : Google Scholar
|
|
26
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
27
|
Kong W, Wei J, Abidi P, Lin M, Inaba S, Li
C, Wang Y, Wang Z, Si S, Pan H, et al: Berberine is a novel
cholesterol-lowering drug working through a unique mechanism
distinct from statins. Nat Med. 10:1344–1351. 2004. View Article : Google Scholar
|
|
28
|
Muller C, Goubin F, Ferrandis E,
Cornil-Scharwtz I, Bailly JD, Bordier C, Bénard J, Sikic BI and
Laurent G: Evidence for transcriptional control of human mdr1 gene
expression by verapamil in multidrug-resistant leukemic cells. Mol
Pharmacol. 47:51–56. 1995.
|
|
29
|
Yu G, Wang LG, Han Y and He QY:
clusterProfiler: An R package for comparing biological themes among
gene clusters. OMICS. 16:284–287. 2012. View Article : Google Scholar
|
|
30
|
Deng WG, Kawashima H, Wu G, Jayachandran
G, Xu K, Minna JD, Roth JA and Ji L: Synergistic tumor suppression
by coexpression of FUS1 and p53 is associated with down-regulation
of murine double minute-2 and activation of the apoptotic
protease-activating factor 1-dependent apoptotic pathway in human
non-small cell lung cancer cells. Cancer Res. 67:709–717. 2007.
View Article : Google Scholar
|
|
31
|
Hesson LB, Cooper WN and Latif F:
Evaluation of the 3p213 tumour-suppressor gene cluster. Oncogene.
26:7283–7301. 2007. View Article : Google Scholar
|
|
32
|
Ji L and Roth JA: Tumor suppressor FUS1
signaling pathway. J Thorac Oncol. 3:327–330. 2008. View Article : Google Scholar
|
|
33
|
Lin J, Sun T, Ji L, Deng W, Roth J, Minna
J and Arlinghaus R: Oncogenic activation of c-Abl in non-small cell
lung cancer cells lacking FUS1 expression: Inhibition of c-Abl by
the tumor suppressor gene product Fus1. Oncogene. 26:6989–6996.
2007. View Article : Google Scholar
|
|
34
|
Zou Z, Ma T, He X, Zhou J, Ma H, Xie M,
Liu Y, Lu D, Di S and Zhang Z: Long intergenic non-coding RNA 00324
promotes gastric cancer cell proliferation via binding with HuR and
stabilizing FAM83B expression article. Cell Death Dis. 9:7172018.
View Article : Google Scholar
|
|
35
|
Du WW, Zhang C, Yang W, Yong T, Awan FM
and Yang BB: Identifying and characterizing circRNA-protein
interaction. Theranostics. 7:4183–4191. 2017. View Article : Google Scholar
|
|
36
|
Hentze MW and Preiss T: Circular RNAs:
Splicing's enigma variations. EMBO J. 32:923–925. 2013. View Article : Google Scholar
|
|
37
|
Li YX, Run L, Shi T and Zhang YJ: CTRP9
regulates hypoxia-mediated human pulmonary artery smooth muscle
cell proliferation, apoptosis and migration via TGF-β1/ERK1/2
signaling pathway. Biochem Biophys Res Commun. 490:1319–1325. 2017.
View Article : Google Scholar
|
|
38
|
You B, Liu Y, Chen J, Huang X, Peng H, Liu
Z, Tang Y, Zhang K, Xu Q, Li X, et al: Vascular peroxidase 1
mediates hypoxia-induced pulmonary artery smooth muscle cell
proliferation, apoptosis resistance and migration. Cardiovasc Res.
114:188–199. 2018. View Article : Google Scholar
|
|
39
|
Naeije R and Dedobbeleer C: Pulmonary
hypertension and the right ventricle in hypoxia. Exp Physiol.
98:1247–1256. 2013. View Article : Google Scholar
|
|
40
|
Ghofrani HA, Voswinckel R, Reichenberger
F, Weissmann N, Schermuly RT, Seeger W and Grimminger F: Hypoxia-
and non-hypoxia-related pulmonary hypertension-established and new
therapies. Cardiovasc Res. 72:30–40. 2006. View Article : Google Scholar
|
|
41
|
Zhou S, Jiang H, Li M, Wu P, Sun L, Liu Y,
Zhu K, Zhang B, Sun G, Cao C and Wang R: Circular RNA
hsa_circ_0016070 is associated with pulmonary arterial hypertension
by promoting PASMC proliferation. Mol Ther Nucleic Acids.
18:275–284. 2019. View Article : Google Scholar
|
|
42
|
Beltrán-García J, Osca-Verdegal R,
Nácher-Sendra E, Cardona-Monzonís A, Sanchis-Gomar F, Carbonell N,
Pallardó FV, Lavie CJ and García-Giménez JL: Role of non-coding
RNAs as biomarkers of deleterious cardiovascular effects in sepsis.
Prog Cardiovasc Dis. Jul 13–2021.Epub ahead of print. View Article : Google Scholar
|
|
43
|
Li R, Jiang Q and Zheng Y: Circ_0002984
induces proliferation, migration and inflammation response of VSMCs
induced by ox-LDL through miR-326-3p/VAMP3 axis in atherosclerosis.
J Cell Mol Med. 25:8028–8038. 2021. View Article : Google Scholar
|
|
44
|
Wang F and Zhang M: Circ_001209 aggravates
diabetic retinal vascular dysfunction through regulating
miR-15b-5p/COL12A1. J Transl Med. 19:2942021. View Article : Google Scholar
|
|
45
|
Zhu QQ, Pu XB, Chen TC, Qiu CY, Wu ZH,
Tian L, He YY, Wang XH, Shang T, Wang X, et al: Hsa_circ_0008360
sponges miR-186-5p to target CCND2 to modulate high glucose-induced
vascular endothelial dysfunction. Cell Cycle. 20:1389–1401. 2021.
View Article : Google Scholar
|
|
46
|
Guo HM and Liu ZP: Up-regulation of
circRNA_0068481 promotes right ventricular hypertrophy in PAH
patients via regulating miR-646/miR-570/miR-885. J Cell Mol Med.
25:3735–3743. 2021. View Article : Google Scholar
|
|
47
|
Hong L, Ma X, Liu J, Luo Y, Lin J, Shen Y
and Zhang L: Circular RNA-HIPK3 regulates human pulmonary artery
endothelial cells function and vessel growth by regulating
microRNA-328-3p/STAT3 axis. Pulm Circ. 11:204589402110002342021.
View Article : Google Scholar
|
|
48
|
Yang T, Long T, Du T, Chen Y, Dong Y and
Huang ZP: Circle the cardiac remodeling with circRNAs. Front
Cardiovasc Med. 8:7025862021. View Article : Google Scholar
|
|
49
|
Ali S, Shourideh M and Koochekpour S:
Identification of novel GRM1 mutations and single nucleotide
polymorphisms in prostate cancer cell lines and tissues. PLoS One.
9:e1032042014. View Article : Google Scholar
|
|
50
|
Thandi S, Blank JL and Challiss RAJ:
Group-I metabotropic glutamate receptors, mGlu1a and mGlu5a, couple
to extracellular signal-regulated kinase (ERK) activation via
distinct, but overlapping, signalling pathways. J Neurochem.
83:1139–1153. 2002. View Article : Google Scholar
|
|
51
|
Inamdar GS, Madhunapantula SRV and
Robertson GP: Targeting the MAPK pathway in melanoma: Why some
approaches succeed and other fail. Biochem Pharmacol. 80:624–637.
2010. View Article : Google Scholar
|
|
52
|
Pearson G, Robinson F, Beers Gibson T, Xu
BE, Karandikar M, Berman K and Cobb MH: Mitogen-activated protein
(MAP) kinase pathways: Regulation and physiological functions.
Endocr Rev. 22:153–183. 2001.
|
|
53
|
Cui J and Placzek WJ: PTBP1 modulation of
MCL1 expression regulates cellular apoptosis induced by antitubulin
chemotherapeutics. Cell Death Differ. 23:1681–1690. 2016.
View Article : Google Scholar
|
|
54
|
Zhao Y, Liu Y, Lin L, Huang Q, He W, Zhang
S, Dong S, Wen Z, Rao J, Liao W and Shi M: The lncRNA MACC1-AS1
promotes gastric cancer cell metabolic plasticity via AMPK/Lin28
mediated mRNA stability of MACC1. Mol Cancer. 17:692018. View Article : Google Scholar
|
|
55
|
Zhu B, Cao A, Li J, Young J, Wong J,
Ashraf S, Bierzynska A, Menon MC, Hou S, Sawyers C, et al:
Disruption of MAGI2-RapGEF2-Rap1 signaling contributes to podocyte
dysfunction in congenital nephrotic syndrome caused by mutations in
MAGI2. Kidney Int. 96:642–655. 2019. View Article : Google Scholar
|
|
56
|
Liang D, Xiang L, Yang M, Zhang X, Guo B,
Chen Y, Yang L and Cao J: ZnT7 can protect MC3T3-E1 cells from
oxidative stress-induced apoptosis via PI3K/Akt and MAPK/ERK
signaling pathways. Cell Signal. 25:1126–1135. 2013. View Article : Google Scholar
|
|
57
|
Ma H, Han F, Yan X, Qi G, Li Y, Li R, Yan
S, Yuan C, Song K and Kong B: PBK promotes aggressive phenotypes of
cervical cancer through ERK/c-Myc signaling pathway. J Cell
Physiol. 236:2767–2781. 2020. View Article : Google Scholar
|
|
58
|
Nikam VS, Wecker G, Schermuly R, Rapp U,
Szelepusa K, Seeger W and Voswinckel R: Treprostinil inhibits the
adhesion and differentiation of fibrocytes via the cyclic adenosine
monophosphate-dependent and Ras-proximate protein-dependent
inactivation of extracellular regulated kinase. Am J Respir Cell
Mol Biol. 45:692–703. 2011. View Article : Google Scholar
|
|
59
|
Li Q, Teng Y, Wang J, Yu M, Li Y and Zheng
H: Rap1 promotes proliferation and migration of vascular smooth
muscle cell via the ERK pathway. Pathol Res Pract. 214:1045–1050.
2018. View Article : Google Scholar
|