Introduction
Autophagy is a cellular metabolic process in which
cells are stimulated by relevant signals to produce an autophagic
membrane. Subsequently, the membrane encapsulates cellular
materials, including misfolded/aggregated proteins or damaged
organelles, inducing their degradation inside autophagic bodies.
Subsequently, autophagic bodies fuse with lysosomes to form
autolysosomes, which are sites of degradation (1,2).
Several studies have reported that autophagy, which is initiated
and regulated by complex gene transcription control networks and
post-translational modifications, is implicated in the occurrence
and progression of a number of diseases (3-6).
Long-chain non-coding RNAs (lncRNAs) have gained increasing
attention in autophagy-associated disease research (7-10). However, the elucidation of the
role of lncRNAs in autophagy remains at an initial early stage. The
macrophage, a type of immune cell that functions as the body's
'scavenger' through the phagocytosis and digestion of cellular
contents, is involved in various cellular and molecular immune
processes (11). It is well
known that autophagy plays a critical role in macrophage-mediated
digestion. Therefore, it is of great value to explore the
regulatory mechanisms employed by lncRNAs in macrophage
autophagy.
The lncRNA growth arrest-specific transcript 5
(GAS5) is 2,554 bp in length (GeneBank no. NR_002840.2, of
mouse origin) and located on chromosome 1 (12). Although the homology between
human and mouse lncRNA GAS5 is relatively low, its function
as a tumor suppressor is highly conserved (13). It has been recently reported that
lncRNA GAS5 also plays an important role in autoimmune
disease, inflammation and autophagy (14-17). Although several studies have
demonstrated that lncRNA GAS5 is able to shift macrophage
polarization towards an M1 phenotype (18,19), the role of lncRNA GAS5 in
macrophage autophagy remains largely unknown. In a previously
published study, it was revealed that the expression of lncRNA
GAS5 in the RAW264.7 macrophage cell line increased
significantly during starvation-induced autophagy (20). However, the specific role of
lncRNA GAS5 in autophagy regulation remains unclear. It has
been demonstrated that lncRNAs can participate in the regulation of
cell biological processes through a number of mechanisms. A
generally recognized theory is that lncRNAs can adsorb microRNAs
(miRNAs/miRs) by functioning as molecular sponges (21). Of note, in the present study,
bioinformatics analysis revealed that miR-181c-5p used the same
binding sequence to interact with lncRNA GAS5 and
autophagy-related gene (ATG)5. Similarly, the binding
site sequence used by miR-1192 to interact with autophagy related
gene 12 (ATG12) was the same as the one used to bind lncRNA
GAS5. According to this theory, lncRNAs can regulate gene
expression through the competitive sponging of its downstream
target miRNAs (22,23). Thus, it was hypothesized that
miR-181c-5p and miR-1192 may competitively bind to lncRNA
GAS5, and directly target the 3′-UTRs of ATG5 and
ATG12, respectively, having the same binding sequence.
In the present study, it was demonstrated for the
first time, to the best of our knowledge, that lncRNA GAS5
expression was significantly enhanced during starvation-induced
autophagy, and that lncRNA GAS5 overexpression markedly
increased the formation of autophagic bodies and the expression of
autophagic markers in the RAW264.7 macrophage cell line. It was
also confirmed that lncRNA GAS5 may promote autophagy by
sponging miR-181c-5p and miR-1192, as well as through the
upregulation of ATG5 and ATG12 expression. Thus, a
novel role of lncRNA GAS5 in autophagy regulation was
defined and a theoretical basis was provided concerning the need to
further study the effects of lncRNA GAS5 on
autophagy-associated disease through macrophages.
Materials and methods
Cells and cell culture
The mouse peritoneal macrophage cell line, RAW264.7
(cat. no. TCM13), was purchased from the Cell Bank of Chinese
Academy Science and cultured in Dulbecco's modified Eagle's medium
(DMEM; Biological Industries), supplemented with 10% fetal bovine
serum (FBS; Biological Industries). The RAW264.7 cells were
incubated at 37°C with 5% CO2. For the determination of
macrophage autophagy in vitro, RAW264.7 cells were cultured
in Earle's balanced salt solution (EBSS; Gibco; Thermo Fisher
Scientific, Inc.) for 12 h, in order to induce macrophage autophagy
(starvation induction). In addition, RAW264.7 cells cultured in
either DMEM containing 10 nM 3-methyladenine (3-MA; Sigma-Aldrich;
Merck KGaA) for 12 h or in DMEM supplemented with 50 nM rapamycin
(RAPA; Sigma-Aldrich; Merck KGaA) for 2 h, were used as cell models
of autophagy inhibition.
RNA isolation and reverse
transcription-quantitative PCR (RT-qPCR)
Total cellular RNA was extracted using
TRIzol® reagent (Invitrogen; Thermo Fisher Scientific,
Inc.) and reverse transcribed into cDNA using a reverse
transcription kit (cat. no. K1622; Thermo Fisher Scientific, Inc.).
The RT-qPCR detection was performed using the SYBR Select Master
Mix (Applied Biosystems; Thermo Fisher Scientific, Inc.) using 500
ng cDNA and 10 pM of each primer and quantified with the Step-One
Plus PCR system (Applied Biosystems; Thermo Fisher Scientific,
Inc.). The following amplifying conditions were used: 95°C for 4
min; 40 cycles of 95°C for 25 sec, 60°C for 25 sec, and 70°C for 25
sec. lncRNA GAS5, miR-181c-5p/miR-1192 and
ATG5/ATG12 relative expression was calculated using
the 2−ΔΔCq method (24). The mouse GAPDH gene was
used for lncRNA GAS5 and ATG5/ATG12
normalization, and the small nuclear RNA U6 was utilized for
miR-181c-5p/miR-1192 normalization. All primers were synthesized by
Sangon Biotech Co., Ltd. The lncRNA GAS5-1 primers were used
for non-transfected cell detection, whereas lncRNA GAS5-2
primers were used for the detection of cells transfected with
exogenous lncRNA GAS5 or its short hairpin RNA (shRNA) form.
The primer sequences used in the present study are listed in
Table I.
 | Table ISequences of the primers used in the
present study. |
Table I
Sequences of the primers used in the
present study.
| Name | Sequence |
|---|
| U6-F |
5′-CTCGCTTCGGCAGCACA-3′ |
| U6-R |
5′-AACGCTTCACGAATTTGCGT-3′ |
| GAPDH-F |
5′-GTCAACGGATTTGGTCTGTATT-3′ |
| GAPDH-R |
5′-AGTCTTCTGGGTGGCAGTGAT-3′ |
| lncRNA
GAS5-1 F |
5′-ATTGGGTTTTGGTCTGGACA-3′ |
| lncRNA
GAS5-1 R |
5′-GCTCTGCCATCAGAATCGTT-3′ |
| lncRNA
GAS5-2 F |
5′-CAATGGCAAATGAGCACTAA-3′ |
| lncRNA
GAS5-2 R |
5′-TCCTCAGATACGCAGAAACA-3′ |
| miR-181c-5p
Stem-loop primer |
5′-CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGACTCACCG-3′ |
| miR-181c-5p F |
5′-ACACTCCAGCTGGGAACATTCAACCTGTCG-3′ |
| miR-181c-5p R |
5′-CTCAACTGGTGTCGTGGA-3′ |
| miR-1192 Stem-loop
primer |
5′-CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGAATTTGGT-3′ |
| miR-1192 F | 5′-ACACTCCAGCTGGG
AAACAAACAAACAGAC-3′ |
| miR-1192 R |
5′-CTCAACTGGTGTCGTGGA-3′ |
| ATG5 F |
5′-TTGGAGACTCCTGCTGTTGA-3′ |
| ATG5 R |
5′-TCATCTTTTAGCATACTCAGATGGG-3′ |
| ATG12 F |
5′-CTCCCAACCCTCACTTCTCG-3′ |
| ATG12 R |
5′-GGAGAGATGCAGCTCAGCAA-3′ |
| lncRNA GAS5
wt/mut F |
5′-CAAGCTTGTTCTGTGGCAAAGGAGGAT-3′ |
| lncRNA GAS5
wt R |
5′-GACGCGTTTCCCACCCACTCCTCTATC-3′ |
| lncRNA GAS5
mut (miR-181c-5p) R |
5′-GACGCGTTTACATGTTGTGTGGGTTGAGGGATCTT-3′ |
| lncRNA GAS5
mut (miR-1192) R |
5′-GACGCGTTACTATACCTAGTTAAAGCTGCCCGGTTA-3′ |
| ATG5 wt/mut
F |
5′-CAAGCTTGTATGCCAAGTATCTGTCTATG-3′ |
| ATG5 wt
R |
5′-GACGCGTTAGCATACTCAGATGGGTTG-3′ |
| ATG5 mut
R |
5′-GACGCGTAATGTACATGTGGACAGCAAGCTAGCTC-3′ |
| ATG12 wt/mut
F |
5′-CAAGCTTCATTGTGATCCATACCTGCT-3′ |
| ATG12 wt
R |
5′-GACGCGTCTACATAGTGAGACCCAGCTT-3′ |
| ATG12 mut
R |
5′-GACGCGTTATTGAACAAAAAAAGCATAACAAAAC-3′ |
Fluorescence in situ hybridization
assay
The distribution of lncRNA GAS5 in RAW264.7
cells was detected using the fluorescence in situ
hybridization (FISH) assay. The lncRNA GAS5 detection probe
and detection kit were purchased from Guangzhou RiboBio Co., Ltd.
FISH assay was performed according to the manufacturer's
instructions. Briefly, RAW264.7 cells were seeded in 24-well
plates. Following a 24-h incubation, at 37°C, cells were treated
with EBSS medium or 10 nM 3-MA for a further 12 h to induce or
inhibit autophagy. Subsequently, all cells were fixed,
pre-hybridized and immersed in the hybridization solution
containing the lncRNA GAS5 probe marked with cyanine 3 (Cy3)
and were incubated at 37°C overnight in the dark. After washing,
the cells were stained with DAPI (Beyotime Institute of
Biotechnology) and viewed using the LSM 710 laser scanning confocal
microscope (Carl Zeiss AG).
Bioinformatics analysis
The UCSC database (http://genome.ucsc.edu/) was analyzed lncRNA GAS5
transcripts. Multiple Experiment Viewer version 4.9.0 was used to
perform clustering and functional annotation for autophagy-related
lncRNAs and miRNAs. Significantly differentially expressed lncRNA
GAS5 and miRNAs were screened out. microRNA. org (http://www.microrna.org/microrna/home.do) was used to
predict the autophagy-related miRNAs that may be targeted by GAS5.
The potential target genes of miRNAs were predicted using miRDB
(http://www.mirdb.org/miRDB/), miRWalk2.0
(http://zmf.umm.uni-heidelberg.de/apps/zmf/mirwalk2/)
and TargetScan (http://www.targetscan.org/). The GAS5/miRNAs/autophagy
protein interaction network was drawn using cytoscape 3.4
(https://cytoscape.org/download.html);
miR-181C-5p and miR-1192 that overlapped with sequencing and were
significantly differentially expressed were screened out.
Cell transfection
For lncRNA GAS5 overexpression, the
GAS5 sequence containing miR-181c-5p and miR-1192 MRE of
GAS5 complementary DNA with the NotI and BamHI
restriction sites was subcloned into the LV5 lentivirus vector 12
µg (GenePharma Co., Ltd.) and co-transfected with pGag/Pol
10 µg (GenePharma Co., Ltd.), pRev 4 µg (GenePharma
Co., Ltd.) and pVSV-G 6 µg (GenePharma Co., Ltd.) into 293T
cells (cat. no. GNHu17, The Cell Bank of Type Culture Collection of
the Chinese Academy of Sciences) to generate LV5-GAS5
lentivirus, with the use of Lipofectamine 3000® (Thermo
Fisher Scientific, Inc.). Following a 72-h incubation period at
37°C, the cell supernatant was collected at 4°C, 3,000 × g for 4
min, and filtered with a 0.45 µm filter. The filtrate was
collected at 4°C, 48,000 × g for 2 h. The infection concentration
was 3×108 TU/ml (MOI, 100/1) for LV5-GAS5 and
5×108 TU/ml (MOI, 100/1) for sh-GAS5.
LV5-GAS5-NC and sh-GAS5-NC were used as controls, at
37°C for 72 h. To determine the potential function of miR-181c-5p
and miR-1192, the miR-181c-5p mimic (50 nM) or miR-181c-5p
inhibitor (50 nM) (GenePharma Co., Ltd.), miR-1192 mimic (50 nM) or
miR-1192 inhibitor (50 nM) (GenePharma Co., Ltd.), or their
negative controls (GenePharma Co., Ltd.) were transfected into
RAW264.7 cells using Lipofectamine 3000® (Invitrogen;
Thermo Fisher Scientific, Inc.). Following a 48-h incubation period
at 37°C, the infection efficiency was detected. Target gene
sequences are shown in Table
II.
 | Table IISynthetic sequences of target
genes. |
Table II
Synthetic sequences of target
genes.
| Name | Sequence |
|---|
| sh-GAS5 |
5′-GGUGUUUCUUUCGCGAUCATT-3′ |
|
sh-GAS5-NC |
5′-UUCUCCGAACGUGUCACGUTT-3′ |
| miR-181c-5p
mimics |
5′-AACAUUCAACCUGUCGGUGAGU-3′ |
| miR-1192
mimics |
5′-AAACAAACAAACAGACCAAAUU-3′ |
| Mimics-NC |
5′-UUCUCCGAACGUGUCACGUTT-3′ |
| miR-181c-5p
inhibitor |
5′-ACUCACCGACAGGUUGAAUGUU-3′ |
| miR-1192
inhibitor |
5′-AAUUUGGUCUGUUUGUUUGUUU-3′ |
| Inhibitor-NC |
5′-CAGUACUUUUGUGUAGUACAA-3′ |
Confocal microscopy
To evaluate the role of lncRNA GAS5 in
autophagy regulation, laser confocal microscopy was performed to
measure the dot-like aggregation of the microtubule-associated
proteins 1A/1B light chain 3B (LC3) protein in the cytoplasm, which
is known to be relatively increased during autophagy (25). The detection procedure was
performed as follows: Cells mounted on a slide were fixed with 4%
paraformaldehyde for 25 min and permeabilized using 0.2% Triton
X-100. The slides were then immersed in blocking buffer containing
5% BSA at room temperature for 30 min and were incubated with an
anti-LC3 rabbit polyclonal antibody (1:500 dilution; cat. no.
43566; Cell Signaling Technology, Inc.) overnight at 4°C. After
washing, the slides were incubated with Alexa Fluor 488-conjugated
goat anti-rabbit IgG (1:1,000; cat. no. SA00006-1; ProteinTech
Group, Inc.) for 2 h at room temperature. Finally, the nuclei were
stained with DAPI (Beyotime Institute of Biotechnology); 150
µl DAPI were added to each slide, stain for 1 h, and soaked
in PBS 4 times for 4 min each at room temperature. Cell morphology
was observed and photographed under a disc scanning unit (DSU)
spinning disk confocal microscope (Olympus Corporation).
Transmission electron microscopy
(TEM)
TEM was carried out to determine morphology and
autophagosome quantity in RAW264.7 cells. Cell collection, specimen
preparation and image acquisition were performed as previously
described (26). For each
experimental group, at least 20 cellular cross-sections were
examined using ITEM digital imaging software (Hitachi, Ltd.).
Dual-luciferase reporter assay
To ascertain the target specificity of miR-181c-5p
or miR-1192 to lncRNA GAS5 and ATG5 or ATG12,
a dual-luciferase reporter assay was performed. Briefly, the target
sequences of lncRNA GAS5 and ATG5 or ATG12 to
miR-181c-5p or miR-1192, and their mutant sequences (prsented in
Table I), were amplified and
sub-cloned into the pMIR-Luc reporter plasmid (Promega Corporation)
to generate the corresponding recombinant plasmids. All the
recombinant plasmids were verified by sequencing. The recombinant
plasmids, including pMIR-wt-lncRNA GAS5(100 ng),
pMIR-mut-lncRNA GAS5(100 ng), pMIR-wt-ATG5(100 ng),
or pMIR-mut-ATG5(100 ng), were then co-transfected with
miR-181c-5p mimics (50 nM), miR-181c-5p (50 nM) inhibitors or their
negative controls into RAW264.7 cells, with the use of
Lipofectamine 3000® (Thermo Fisher Scientific, Inc.).
Similarly, pMIR-wt-lncRNA GAS5(100 ng), pMIR-mut-lncRNA
GAS5(100 ng), pMIR-wt-ATG12(100 ng), or
pMIR-mut-ATG12(100 ng), were co-transfected with miR-1192
mimics (50 nM), miR-1192 inhibitors (50 nM) or their negative
controls into RAW264.7 cells, with the use of Lipofectamine
3000®. Following a 48-h incubation period at 37°C, the
luciferase activities in the cells in each group were determined
using a dual-luciferase reporter assay kit (TransGen Biotech Co.,
Ltd.) according to the manufacturer's instructions. Firefly
luciferase activity was normalized to Renilla luciferase
gene activity with a dual-luciferase reporter assay.
Western blot analysis
Protein was extracted using RIPA lysis buffer
(Nanjing KeyGen Biotech Co., Ltd.) and the protein concentration
was measured with a protein BCA assay kit (Nanjing KeyGen Biotech
Co., Ltd.). Subsequently, 30 µg protein was separated on a
12% SDS-PAGE gel by electrophoresis and transferred to 0.45
µm PVDF membranes (Merck KGaA). The membranes were immersed
in a blocking solution containing 5% non-fat dry milk at room
temperature for 2 h and then incubated overnight at 4°C with an
antiserum containing antibodies against LC3 (1:1,000; cat. no.
2775; Cell Signaling Technology, Inc.), p62 (1:1,000 dilution; cat.
no. 5114; Cell Signaling Technology, Inc.) ATG5 (1:500 dilution;
cat. no. 10181-2-AP; ProteinTech Group, Inc.) and ATG12 (1:500
dilution; cat. no. 10088-2-AP; ProteinTech Group, Inc.).
Subsequently, the membranes were washed and incubated with a
peroxidase-conjugated secondary antibody (1:3,000 dilution; cat.
no. TA130023; OriGene Technologies, Inc.) at room temperature for 2
h. Finally, the protein bands were visualized with an ECL detection
reagent (Thermo Fisher Scientific, Inc.) and the results were
quantified with the use of ImageJ software J2x (Rawak Software
Inc.). An anti-GAPDH antibody (1:1,000 dilution; cat. no. 2118;
Cell Signaling Technology, Inc.) was used as a control and the
results were presented as the ratio of density of target protein to
the GAPDH value.
Statistical analysis
All data in the present study were analyzed and
plotted using SPSS 19.0 (IBM Corp.) and GraphPad Prism 7.0
(GraphPad Software, Inc.). The results are presented as the mean ±
SD of independent experiments. The differences between the groups
were tested using one-way analysis of variance (ANOVA), followed
Tukey's post hoc test.
Results
lncRNA GAS5 expression is associated with
macrophage autophagy
In a previous study using high-throughput sequencing
and cluster analysis, it was observed that lncRNA GAS5 was
selectively upregulated when the RAW264.7 cells were subjected to
starvation treatment (20). Of
note, up to 25 transcripts of lncRNA GAS5 could be
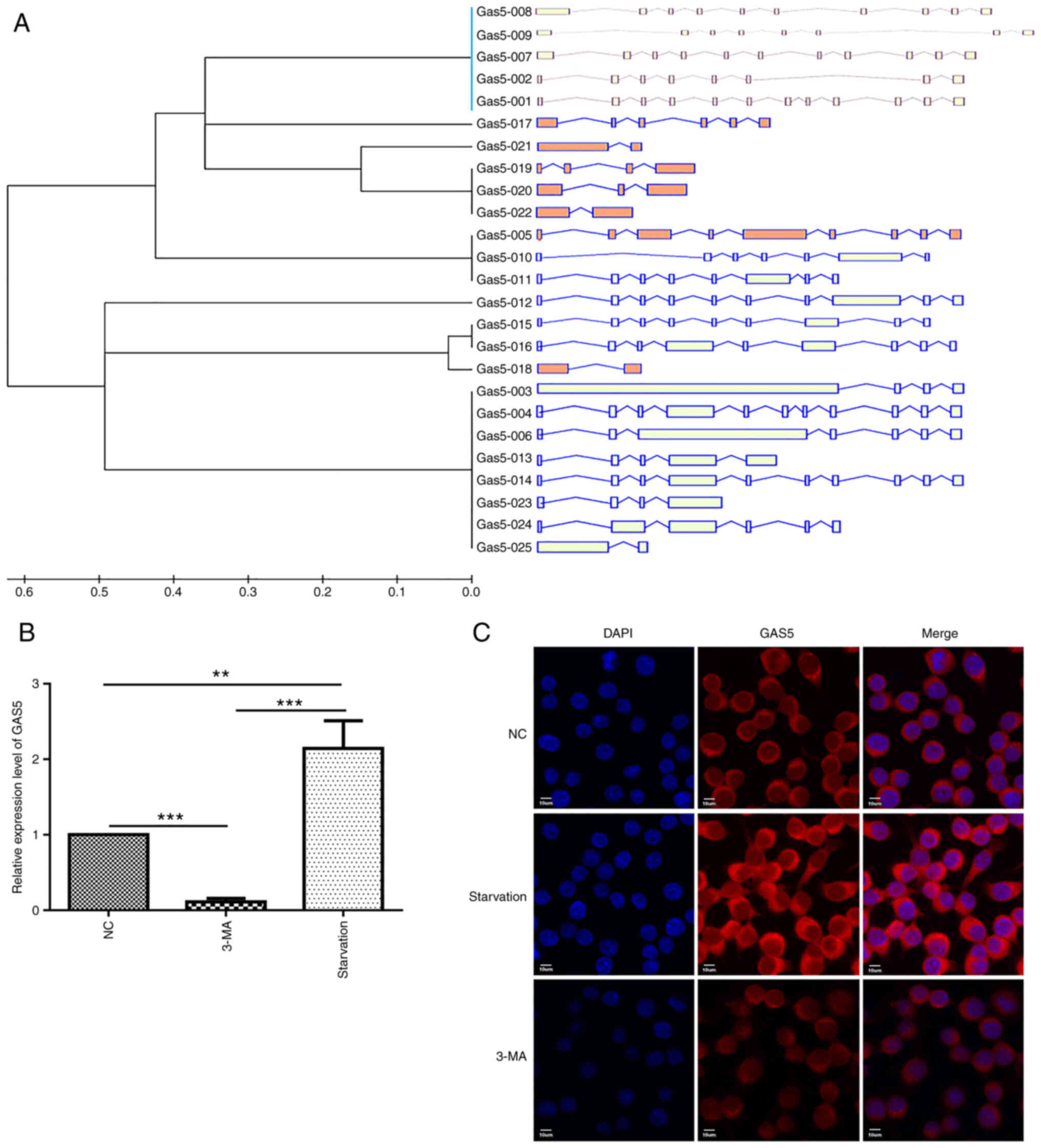
identified by means of UCSC database analysis (http://genome.ucsc.edu/), with lncRNA GAS5-003
being the longest one (2,554 bp) and containing almost all of the
key transcript regions (Fig.
1A). Taking this into account, lncRNA GAS5-003 was used
to explore the role of lncRNA GAS5 in macrophage autophagy.
Firstly, it was validated through RT-qPCR that lncRNA GAS5
expression was significantly upregulated following cell starvation.
As was initially hypothesized, lncRNA GAS5 expression was
downregulated when autophagy was inhibited by 3-MA (Fig. 1B). Additionally, FISH assay was
utilized to localize lncRNA GAS5 expression in the RAW264.7
cells; it was observed that lncRNA GAS5 was confined to the
cytoplasm, and when autophagy occurred, its levels were markedly
increased, and this effect was significantly reversed with the use
of 3-MA (Fig. 1C).
lncRNA GAS5 promotes macrophage
autophagy
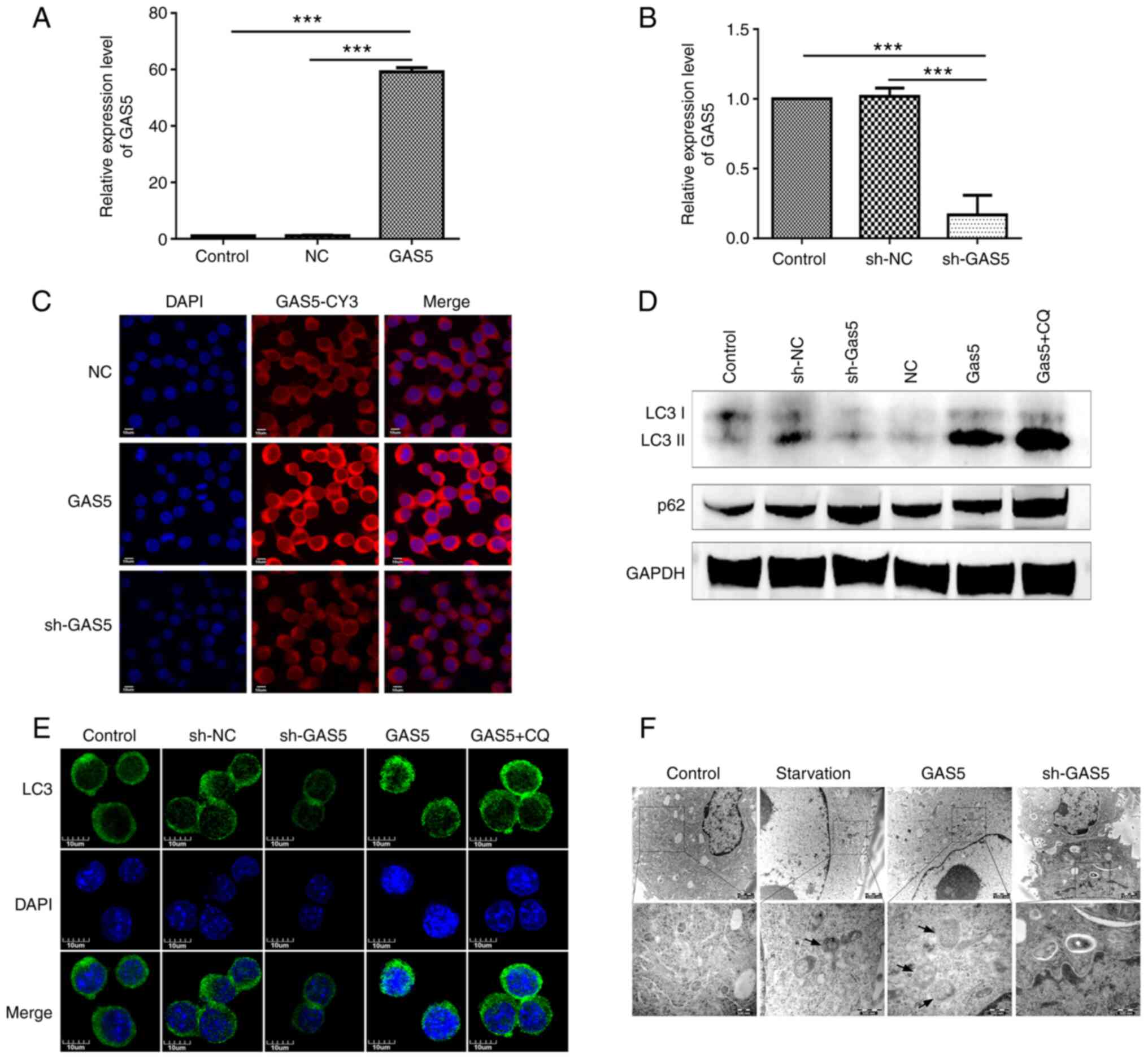
To explore the role of lncRNA GAS5 in
macrophage autophagy, cell models were established in vitro
by transfecting LV5-lncRNA GAS5 (LV5-GAS5) or
LV3-shRNA-lnc GAS5 (sh-GAS5) into RAW264.7 cells in
order to overexpress or knockdown lncRNA GAS5 expression
(Fig. 2A and B). Furthermore,
FISH assay confirmed that the overexpression or knockdown of lncRNA
GAS5 expression through LV5-GAS5 or sh-GAS5
maintained the same intracellular localization pattern with cells
containing wild-type (wt) lncRNA GAS5 (Fig. 2C). Subsequently, the effects of
lncRNA GAS5 on cell autophagy were examined. lncRNA
GAS5 silencing reduced the LC3II to LC3 ratio, a marker of
autophagy, while lncRNA GAS5 overexpression exerted the
opposite effect (Fig. 2D). The
effect of lncRNA GAS5 on autophagy was also verified through
laser confocal microscopy, in order to confirm that the level of
lncRNA GAS5 was associated with LC3 aggregation, a typical
autophagy activation marker (Fig.
2E). In addition, the role of lncRNA GAS5 in the
formation of autophagic bodies was further explored, using TEM. The
overexpression of lncRNA GAS5 exerted the same effect as
that observed with starvation, increasing the numbers of autophagic
bodies in the cytoplasm, whereas lncRNA GAS5 silencing
exerted the opposite effect (Fig.
2F).
lncRNA GAS5 targets miR-181c-5p and
miR-1192
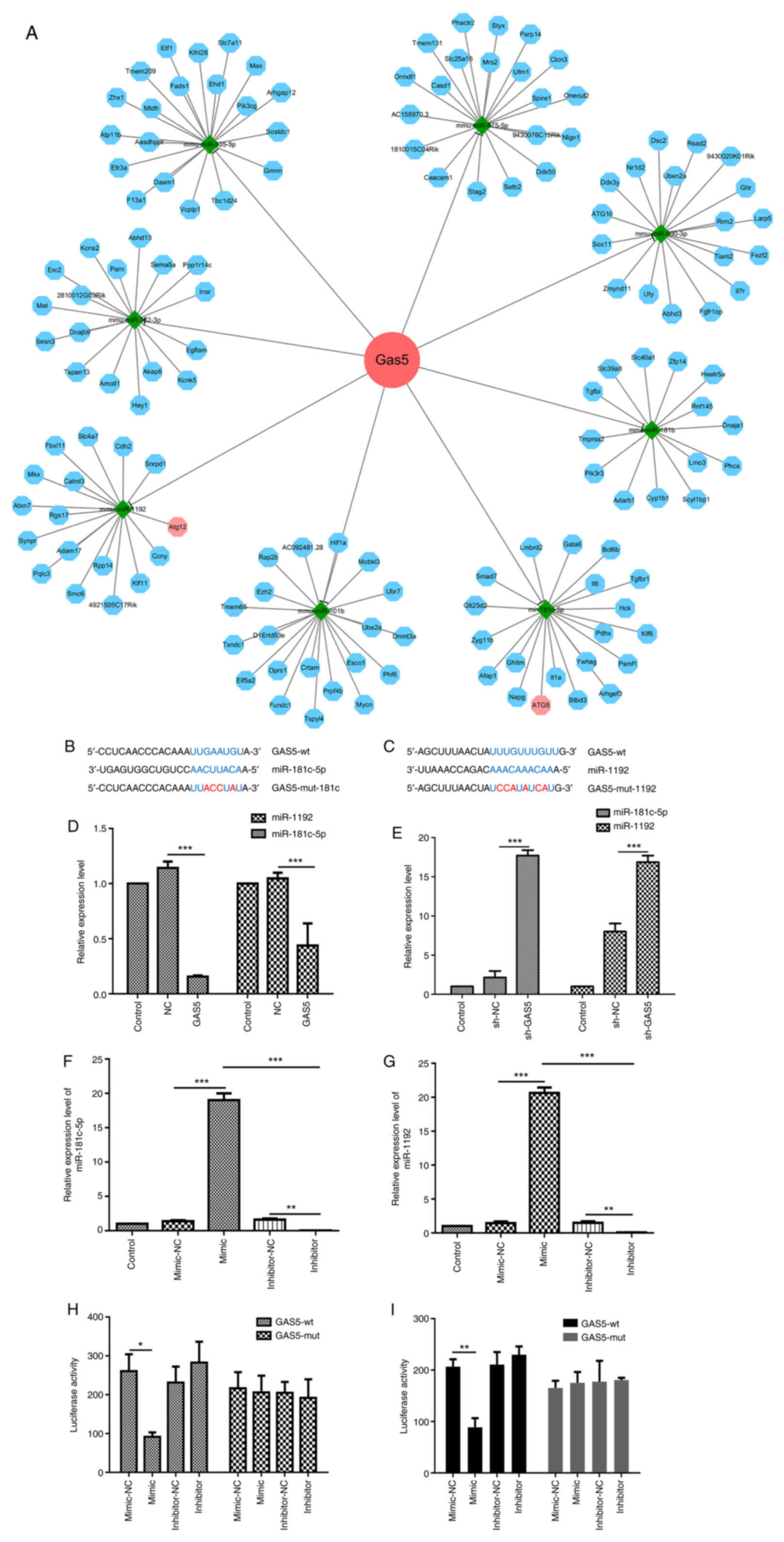
To further explore the molecular mechanisms through
which lncRNA GAS5 promotes autophagy, its potential miRNA
binding partners were analyzed using bioinformatics software,
including StarBase V3.0 (http://starbase.sysu.edu.cn/), and TargetScan
(http://www.targetscan.org/vert_71/).
The analysis identified eight miRNAs selectively targeted by lncRNA
GAS5. Of note, two of these miRNAs, miR-181c-5p and
miR-1192, can also target key proteins of the autophagy pathway
(Fig. 3A-C). RT-qPCR was then
used in order to confirm the association between lncRNA GAS5
and miR-181c-5p or miR-1192. lncRNA GAS5 overexpression
significantly downregulated miR-181c-5p and miR-1192 expression in
RAW264.7 cells (Fig. 3D),
whereas the silencing of lncRNA GAS5 markedly increased the
expression of both miRNAs (Fig.
3E). Since the regulation of lncRNA/miRNAs/autophagy
interaction is a regulatory network, miRNA expression involves a
time-course of events, and the same lncRNA can simultaneously
regulate multiple miRNAs. At the same time, a miRNA can regulate
multiple genes. Therefore, there are certain differences in the
expression of miR-1192, leading to a slight increase in expression
with the sh-NC control. Overall, the experimental results revealed
that silencing GAS5 promoted the expression of miR-1192, and the
increased expression of sh-NC control did not affect the conclusion
of the study. In view of the results of both bioinformatics
analysis and RT-qPCR, it was preliminarily surmised that lncRNA
GAS5 negatively regulates miR-181c-5p and miR-1192
expression.
Subsequently, the effects of miRNA mimic and
inhibitor on RAW 264.7 cells were validated. miR-181c-5p mimic was
able to increase miR-181c-5p expression, while miR-181c-5p
inhibitor markedly decreased the expression level of miR-181c-5p in
RAW 264.7 cells (Fig. 3F).
Similarly, the miR-1192 expression level was significantly
increased by the miR-1192 mimic and decreased by the miR-1192
inhibitor (Fig. 3G). Using these
tools and a luciferase reporter with either a wt or mutated
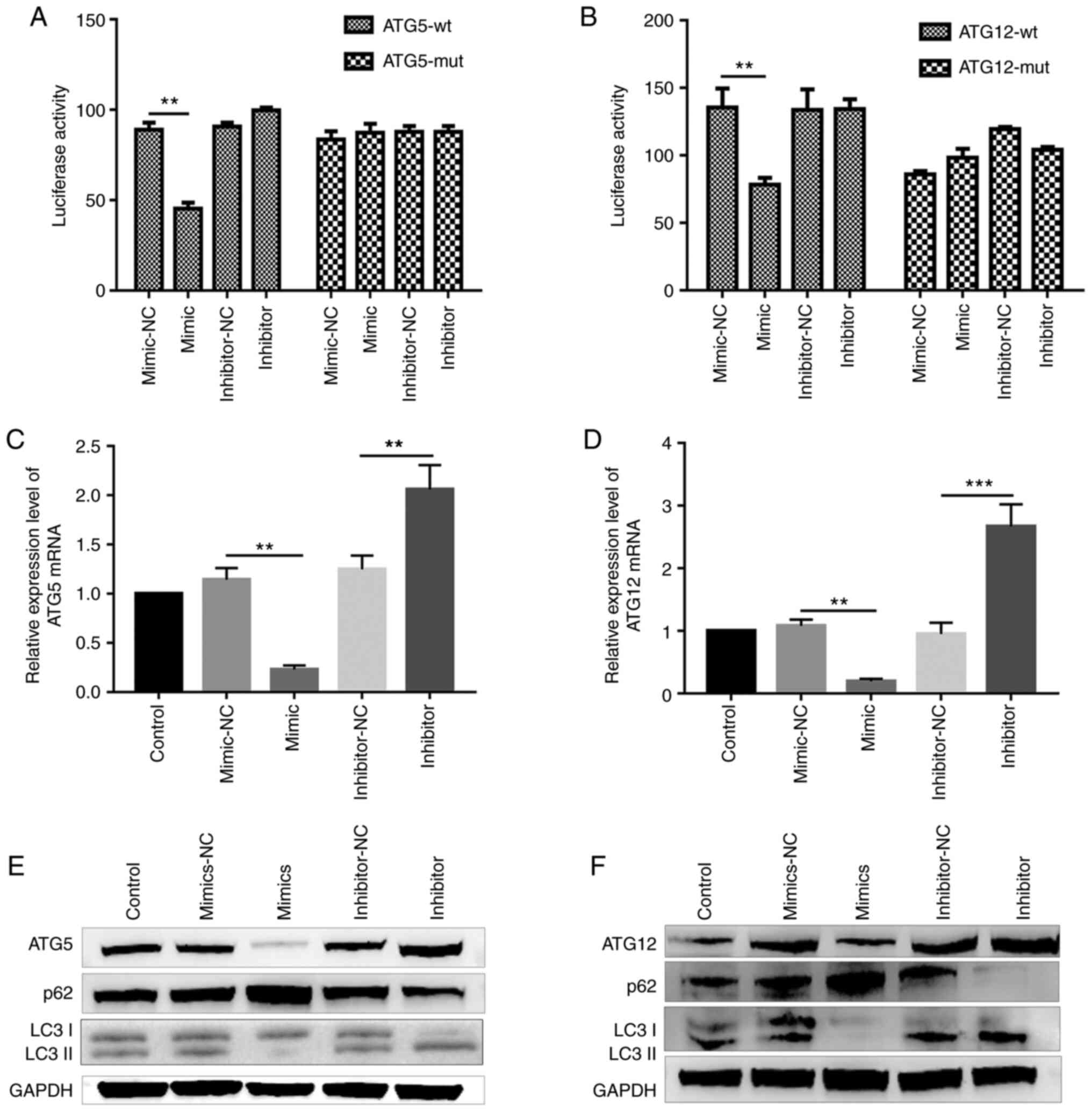
miR-181c-5p or miR-1192 lncRNA-GAS5 binding site (Fig. 3B and C), the direct interaction
between both miRNAs and lncRNA GAS5 was examined.
Dual-luciferase reporter assay revealed a significant decrease in
luciferase activity in the cells co-transfected with the
miR-181c-5p mimic and report-GAS5-wt. By contrast, the
luciferase activity was almost unaltered when the cells were
co-transfected with report-GAS5-mut (miR-181c-5p) and
miR-181c-5p mimic (Fig. 3H).
Co-transfection with miR-1192 mimic and report-GAS5-wt
significantly decreased luciferase activity, while the luciferase
activity was unaltered following co-transfection with miR-1192
mimic and report-GAS5-mut (miR-1192) (Fig. 3I).
miR-181c-5p and miR-1192 inhibit
macrophage autophagy by targeting ATG5 and ATG12, respectively
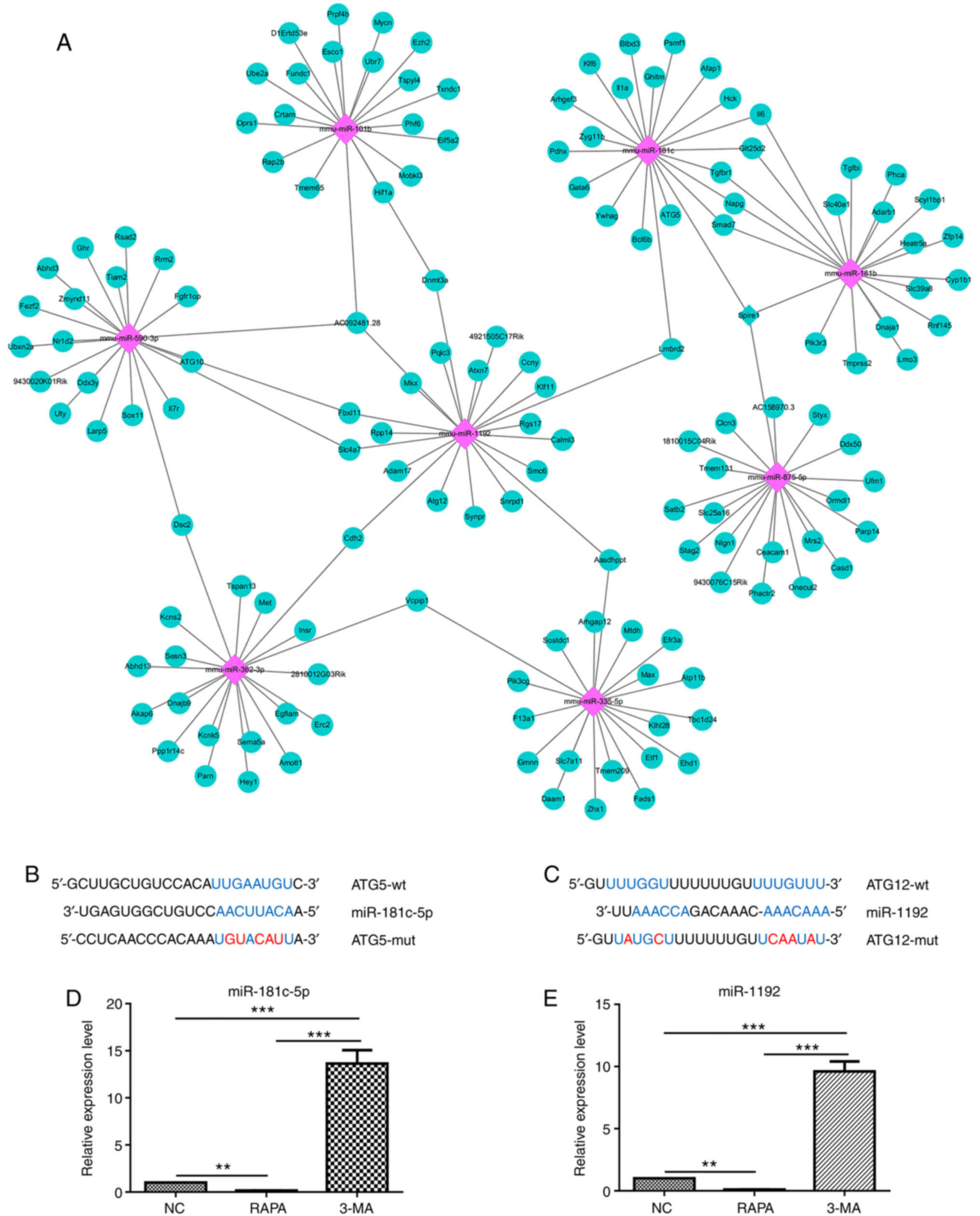
Initially, bioinformatics analysis revealed that
ATG5 was a potential target gene of miR-181c-5p (Fig. 4A and B), while ATG12was a
potential target gene of miR-1192 (Fig. 4A and C). Subsequently, it was
demonstrated that miR-181c-5p and miR-1192 expression was
significantly decreased following treatment with RAPA (a potent
autophagy stimulus), whereas it was significantly increased as a
result of 3-MA autophagy inhibitor treatment (Fig. 4D and E). These results thus
suggested that miR-181c-5p and miR-1192 were involved in macrophage
autophagy.
To confirm the roles of these miRNAs in the
regulation of autophagy, dual-luciferase reporter assay was
performed. It was revealed that the luciferase activity decreased
significantly in the cells co-transfected with the miR-181c-5p
mimic and report-ATG5-wt; however, luciferase activity
remained unaltered in the mutation control group (Fig. 5A). As was initially anticipated,
miR-1192 mimic transfection resulted in a decrease in luciferase
activity when wt ATG12 vector was used. However, this
inhibitory effect was abolished following transfection with the
mutant miR-1192-targeting sequences in their 3′-UTR (Fig. 5B). Thus, miR-181c-5p and miR-1192
could directly target the key autophagy factors ATG5 and
ATG12, respectively. In addition, ATG5 expression was
significantly decreased in the cells transfected with the
miR-181c-5p mimic, both at the mRNA and protein level. By contrast,
the mRNA and protein expression of ATG5was increased in the
miR-181c-5p inhibitor-transfected group (Fig. 5C and E). Furthermore, the
LC3II/LC3I ratio decreased and p62 expression increased in the
miR-181c-5p mimic-transfected group. The opposite effect was
observed in the miR-181c-5p inhibitor-treated group, indicating
that miR-181c-5p plays an inhibitory role in macrophage autophagy
by targeting ATG5, a key autophagic protein. Simultaneously,
RT-qPCR analysis revealed that ATG12 was negatively
regulated by miR-1192 (Fig. 5D),
and western blot analysis confirmed that miR-1192 inhibited
ATG12 expression (Fig.
5F).
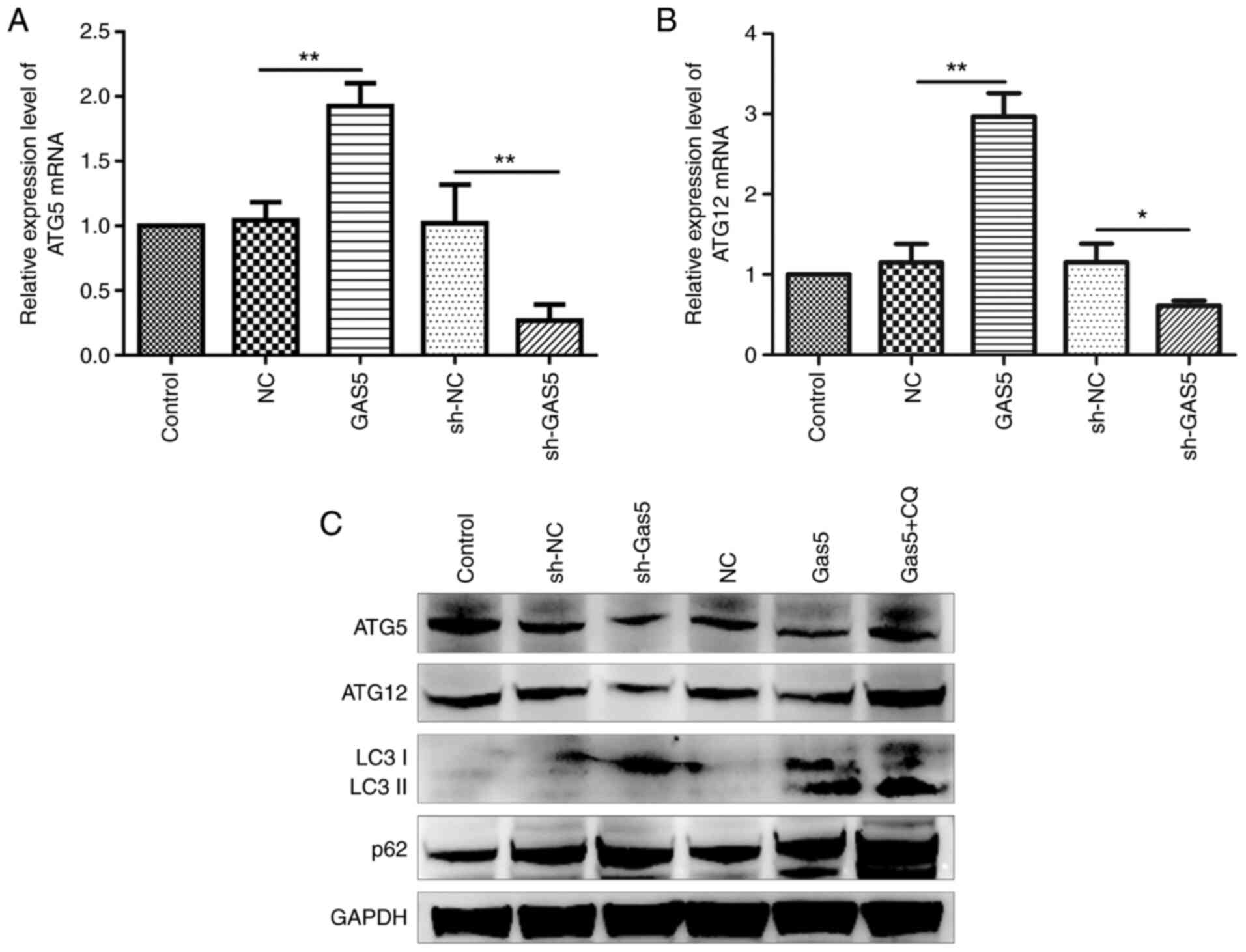
lncRNA GAS5 preserves ATG5 and ATG12
expression
It was confirmed lncRNA GAS5 negatively
regulated both miR-181c-5p and miR-1192 and therefore positively
affected autophagy through the preservation of ATG5 and
ATG12 levels, which are two autophagy factors suppressed by
the aforementioned miRNAs. To this end, the precise mechanisms of
the regulation of autophagy by lncRNA GAS5 were
investigated. It was revealed that ATG5 and ATG12
mRNA and protein expression was significantly enhanced as a result
of lncRNA GAS5 overexpression, whereas this was markedly
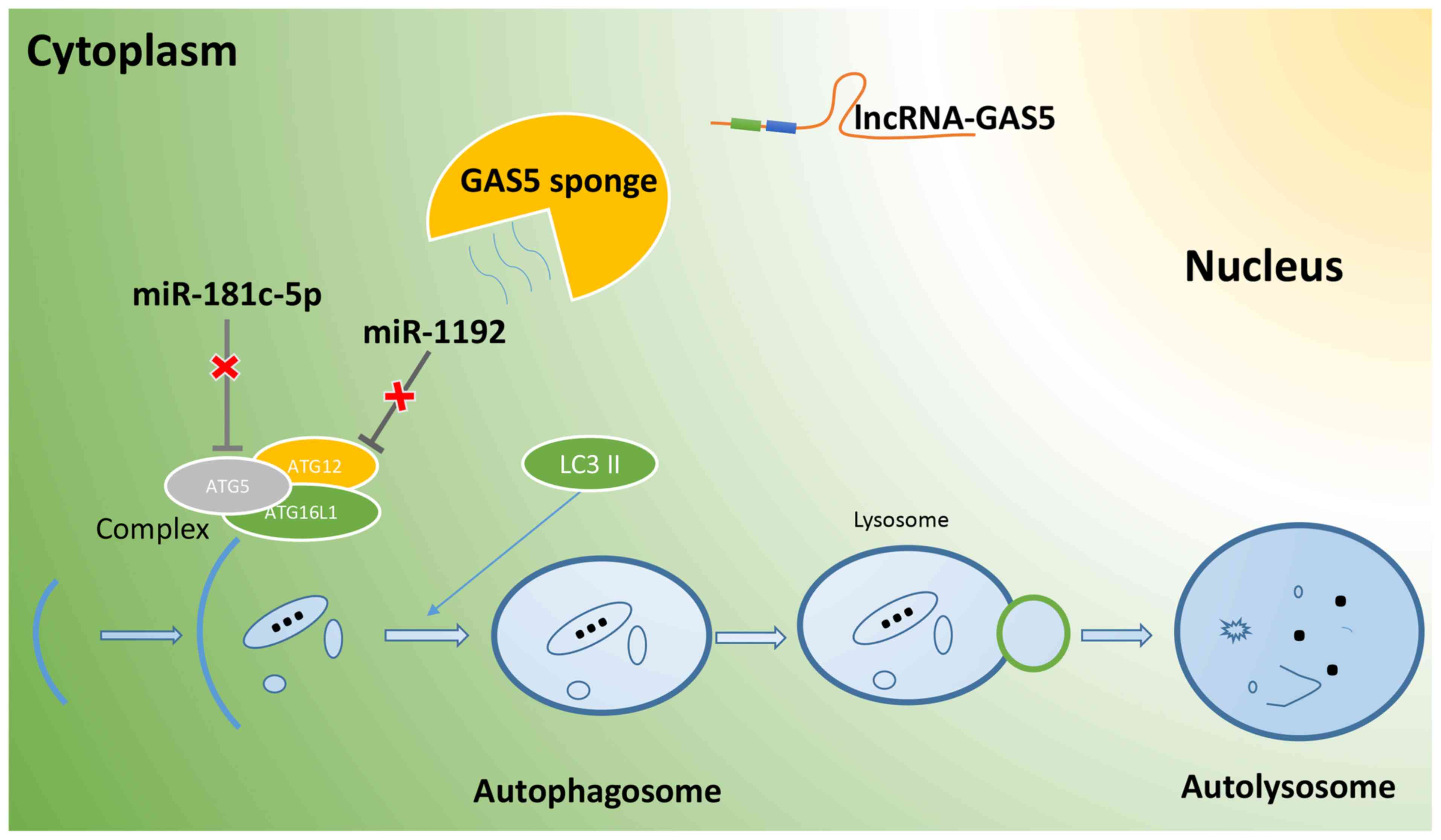
decreased following the silencing of lncRNA GAS5 (Fig. 6). On the whole, lncRNA GAS5
functions as a ceRNA in regulating Atg5 and Atg12 expression by
binding to miR-181c-5p and miR-1192, thus promoting the autophagy
of RAW264.7 cells (Fig. 7).
Discussion
lncRNA GAS5, located on chromosome 1, has
been reported to participate in biological processes, such as cell
proliferation, apoptosis, migration and invasion (27-29). Long regarded as a tumor
suppressor, lncRNA GAS5 has also been found to be involved
in autoimmune disease, inflammation and autophagy. It has been
revealed that there is a 5′-TOP sequence in exon 1 of the lncRNA
GAS5 gene, whose transcription is regulated by the mTOR
signaling pathway. When cell growth is suppressed by starvation,
the activity of mTOR signaling decreases and the translation of
lncRNA GAS5 is blocked. Subsequently, the degradation of
lncRNA GAS5 -regulated by the NMD pathwayis reduced, which
finally culminates in lncRNA GAS5 aggregation and increased
expression (30).
In the present study, cell models of autophagy and
autophagy inhibition were established using the EBSS medium (for
starvation induction) and complete medium supplementing with 3-MA,
respectively. It was demonstrated that lncRNA GAS5
expression was significantly higher than that of other lncRNAs in
the autophagy group. In addition, the lncRNA GAS5-003
contained almost all the key regions of the lncRNA GAS5 gene
within its 25 transcripts. Therefore, lncRNA GAS5-003 was
selected for use in subsequent experiments. The increased
expression of lncRNA GAS5 in the starvation-induced
autophagy group was confirmed through RT-qPCR and FISH assay.
lncRNA GAS5 overexpression markedly increased the
autophagy-associated protein LC3 levels and promoted autophagic
body formation, while the opposite results were observed in the
group which was subjected to lncRNA GAS5 silencing. Thus, it
was hypothesized that lncRNA GAS5 may be implicated in
macrophage-mediated autophagy regulation.
To further explore the autophagy-promoting function
of lncRNA GAS5, its potential target miRNAs were evaluated
using bioinformatics. miR-181c-5p and miR-1192 were revealed as
putative targets of lncRNA GAS5. Although in a previous
study, it was demonstrated that lncRNA GAS5 antagonized the
chemoresistance of pancreatic cancer cells through the regulation
of miR-181c-5p expression (31),
the role of lncRNA GAS5 and miR-181c-5p in macrophage
autophagy remains unknown. In the present study, it was found that
the overexpression of lncRNA GAS5 in RAW264.7 cells
significantly downregulated the expression of miR-181c-5p and
miR-1192, via their direct interaction. Therefore, it was confirmed
that lncRNA GAS5 acts as a sponge for miR-181c-5p and
miR-1192. Additionally, it has been noted that miR-181c-5p is
involved in the regulation of inflammation (32), cell proliferation (33) and apoptosis (34). Moreover, miR-1192 has been proven
to be implicated in myocardial infarction (35), inflammation (36) and muscle injury (37).
In the present study, it was demonstrated that
miR-181c-5p and miR-1192 expression could be regulated by RAPA and
3-MA. Of note, miR-181c-5p and miR-1192 reduced the LC3II/LC3I
ratio and promoted p62 expression. It was hypothesized that these
may also be implicated in RAW264.7 cell autophagy. Furthermore, it
was revealed that ATG5 and ATG12 were potential
targets of miR-181c-5p and miR-1192, respectively, as confirmed by
luciferase assay, RT-qPCR and western blot analysis. To date,
>40 ATGs have been found in yeast and mammals, which are highly
conserved among species and participate in the autophagy regulation
via complex networks (38). When
autophagy is initiated, ATG5, a key protein in the initial
stage of autophagy, is recruited by ATG16L1 to form the
ATG5-ATG12-ATG16L1 complex, which provides
interaction sites for other proteins involved in the formation of
the autophagy membrane, accelerates the extension of the autophagy
membrane, and promotes the transformation of LC3I into LC3II
(39). Therefore, it was
suggested that lncRNA GAS5 may regulate ATG5 and
ATG12expression, by interacting with miR-181c-5p and
miR-1192, respectively. However, in the present study, only a small
part of the autophagic pathway was analyzed. In order to better
define the mechanistic role of lncRNA GAS5 in macrophage
autophagy, a more selective approach may be necessary, including
the design of 'Target Protectors' (40) for the definition of the selective
role of a particular miRNA-targeted mRNA in a specific phenotypic
readout. This may be a future research approach based on currently
presented results.
Based on the experimental results presented herein,
it can be concluded that lncRNA GAS5 promotes macrophage
autophagy by targeting the miR181c-5p/ATG5 and
miR-1192/ATG12 axes. In a follow-up study, the authors aim
to construct in vivo animal models of lncRNA GAS5
overexpression and interference through lentiviral transduction,
and to examine the mechanisms and application of lncRNA GAS5
in diseases. These findings may provide a theoretical basis for the
further exploration of the mechanisms of autophagy-associated
diseases, such as intracellular bacterial and virus infections and
tumorigenesis.
Availability of data and materials
The datasets used and/or analyzed during the
current study are available from the corresponding author on
reasonable request.
Authors' contributions
TX and GX participated in the design and
coordination of the study. TX, XX and YC performed the experiments.
XX and DJ performed the statistical analyses. TX and XX wrote the
manuscript. TX, XX and GX reviewed and edited the manuscript. XX
and GX confirm the authenticity of all the raw data. All authors
have read and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgments
Not applicable.
References
|
1
|
Gozuacik D, Akkoc Y, Ozturk DG and Kocak
M: Autophagy-regulating microRNAs and cancer. Front Oncol.
7:652017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Hurley JH and Young LN: Mechanisms of
autophagy initiation. Annu Rev Biochem. 86:225–244. 2017.
View Article : Google Scholar :
|
|
3
|
Ravanan P, Srikumar IF and Talwar P: The
spotlight for cellular stress responses. Life Sci. 188:53–67. 2017.
View Article : Google Scholar
|
|
4
|
Mainz L and Rosenfeldt MT: Autophagy and
cancer-insights from mouse models. FEBS J. 285:792–808. 2018.
View Article : Google Scholar
|
|
5
|
Cui B, Lin H, Yu J, Yu J and Hu Z:
Autophagy and the immune response. Adv Exp Med Biol. 1206:595–634.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zhang C, Liu A, Su G and Chen Y: Effect of
rapamycin on the level of autophagy in rats with early heart
failure. J Cell Biochem. 120:4065–4070. 2019. View Article : Google Scholar
|
|
7
|
Guo L, Zhao Y, Yang S, Zhang H and Chen F:
An integrated analysis of miRNA, lncRNA, and mRNA expression
profiles. Biomed Res Int. 2014:3456052014.PubMed/NCBI
|
|
8
|
Su Y, Lu J, Chen X, Liang C, Luo P, Qin C
and Zhang J: Long non-coding RNA HOTTIP affects renal cell
carcinoma progression by regulating autophagy via the
PI3K/Akt/Atg13 signaling pathway. J Cancer Res Clin Oncol.
145:573–588. 2019. View Article : Google Scholar
|
|
9
|
Lu D, Yang C, Zhang Z, Cong Y and Xiao M:
Knockdown of Linc00515 inhibits multiple myeloma autophagy and
chemoresistance by upregulating miR-140-5p and downregulating
ATG14. Cell Physiol Biochem. 48:2517–2527. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Guo J, Ma Y, Peng X, Jin H and Liu J:
lncRNA CCAT1 promotes autophagy via regulating ATG7 by sponging
miR-181 in hepatocellular carcinoma. J Cell Biochem.
120:17975–17983. 2019. View Article : Google Scholar
|
|
11
|
Wu MY and Lu JH: Inflammatory response and
phagocytosis. Cells. 9:702019. View Article : Google Scholar
|
|
12
|
Hu J, Zhang L, Liechty C, Zgheib C, Hodges
MM, Liechty KW and Xu J: Long non-coding RNA GAS5 regulates
macrophage polarization and diabetic wound healing. J Invest
Dermatol. 140:1629–1638. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ji J, Dai X, Yeung SJ and He X: The role
of long non-coding RNA GAS5 in cancers. Cancer Manag Res.
11:2729–2737. 2019. View Article : Google Scholar :
|
|
14
|
Mayama T, Marr AK and Kino T: Differential
expression of glucocorticoid receptor noncoding RNA repressor GAS5
in autoimmune and inflammatory diseases. Horm Metab Res.
48:550–557. 2016. View Article : Google Scholar
|
|
15
|
Li F, Sun J, Huang S, Su G and Pi G:
lncRNA GAS5 overexpression reverses LPS-induced inflammatory injury
and apoptosis through up-regulating KLF2 expression in ATDC5
chondrocytes. Cell Physiol Biochem. 45:1241–1251. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Liu L, Wang HJ, Meng T, Lei C, Yang XH,
Wang QS, Jin B and Zhu JF: lncRNA GAS5 inhibits cell migration and
invasion and promotes autophagy by targeting miR-222-3p via the
GAS5/PTEN-signaling pathway in CRC. Mol Ther Nucleic Acids.
17:644–656. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Gu J, Wang Y, Wang X, Zhou D, Wang X, Zhou
M and He Z: Effect of the lncRNA GAS5-miR-23a-ATG3 axis in
regulating autophagy in patients with breast cancer. Cell Physiol
Biochem. 48:194–207. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Chi X, Ding B, Zhang L, Zhang J, Wang J
and Zhang W: lncRNA GAS5 promotes M1 macrophage polarization via
miR-455-5p/SOCS3 pathway in childhood pneumonia. J Cell Physiol.
234:13242–13251. 2019. View Article : Google Scholar
|
|
19
|
Ito I, Asai A, Suzuki S, Kobayashi M and
Suzuki F: M2b macrophage polarization accompanied with reduction of
long noncoding RNA GAS5. Biochem Biophys Res Commun. 493:170–175.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Ma Z, Zhang J, Xu X, Qu Y, Dong H, Dang J,
Huo Z and Xu G: lncRNA expression profile during autophagy and
Malat1 function in macrophages. PLoS One. 14:e02211042019.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Tay Y, Rinn J and Pandolfi PP: The
multilayered complexity of ceRNA crosstalk and competition. Nature.
505:344–352. 2014. View Article : Google Scholar :
|
|
22
|
Schmitz SU, Grote P and Herrmann BG:
Mechanisms of long noncoding RNA function in development and
disease. Cell Mol Life Sci. 73:2491–2509. 2016. View Article : Google Scholar :
|
|
23
|
Yoon JH, Abdelmohsen K and Gorospe M:
Functional interactions among microRNAs and long noncoding RNAs.
Semin Cell Dev Biol. 34:9–14. 2014. View Article : Google Scholar
|
|
24
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
25
|
Guo L, Zhao J, Qu Y, Yin R, Gao Q, Ding S,
Zhang Y, Wei J and Xu G: microRNA-20a inhibits autophagic process
by targeting ATG7 and ATG16L1 and favors mycobacterial survival in
macrophage cells. Front Cell Infect Microbiol. 6:1342016.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Guo L, Zhou L, Gao Q, Zhang A, Wei J, Hong
D, Chu Y, Duan X, Zhang Y and Xu G: MicroRNA-144-3p inhibits
autophagy activation and enhances Bacillus Calmette-Guérin
infection by targeting ATG4a in RAW264.7 macrophage cells. PLoS
One. 12:e01797722017. View Article : Google Scholar
|
|
27
|
Zhang XY, Tang XY, Li N, Zhao LM, Guo YL,
Li XS, Tian CJ, Cheng DJ, Chen ZC and Zhang LX: GAS5 promotes
airway smooth muscle cell proliferation in asthma via controlling
miR-10a/BDNF signaling pathway. Life Sci. 212:93–101. 2018.
View Article : Google Scholar
|
|
28
|
Liu SD, Meng WX, Xu L, Chi C, Sun X and
Liu HY: GAS5 promotes myocardial apoptosis in myocardial
ischemia-reperfusion injury via upregulating LAS1 expression. Eur
Rev Med Pharmacol Sci. 22:8447–8453. 2018.PubMed/NCBI
|
|
29
|
Zeng B, Li Y, Jiang F, Wei C, Chen G,
Zhang W, Zhao W and Yu D: lncRNA GAS5 suppresses proliferation,
migration, invasion, and epithelial-mesenchymal transition in oral
squamous cell carcinoma by regulating the miR-21/PTEN axis. Exp
Cell Res. 374:365–373. 2019. View Article : Google Scholar
|
|
30
|
Pickard MR and Williams GT: Molecular and
cellular mechanisms of action of tumour suppressor GAS5 lncRNA.
Genes (Basel). 6:484–499. 2015. View Article : Google Scholar
|
|
31
|
Gao ZQ, Wang JF, Chen DH, Ma XS, Yang W,
Zhe T and Dang XW: Long non-coding RNA GAS5 antagonizes the
chemoresistance of pancreatic cancer cells through down-regulation
of miR-181c-5p. Biomed Pharmacother. 97:809–817. 2018. View Article : Google Scholar
|
|
32
|
Cao DW, Liu MM, Duan R, Tao YF, Zhou JS,
Fang WR, Zhu JR, Niu L and Sun JG: The lncRNA Malat1 functions as a
ceRNA to contribute to berberine-mediated inhibition of HMGB1 by
sponging miR-181c-5p in post-stroke inflammation. Acta Pharmacol
Sin. 41:22–33. 2020. View Article : Google Scholar
|
|
33
|
Sun ZY, Li Y, Wang H, Cai M, Gao SS, Liu
J, Tong LC, Hu ZB, Wang YX, Wang K, et al: miR-181c-5p mediates
simulated microgravity-induced impaired osteoblast proliferation by
promoting cell cycle arrested in the G2 phase. J Cell Mol Med.
23:3302–3316. 2019. View Article : Google Scholar :
|
|
34
|
Ge L, Cai Y, Ying F, Liu H, Zhang DW, He
YJ, Pang L, Yan D, Xu AM, Ma H and Xia Z: miR-181c-5p exacerbates
hypoxia/reoxygenation-induced cardiomyocyte apoptosis via targeting
PTPN4. Oxid Med Cell Longev. 2019:19579202019. View Article : Google Scholar :
|
|
35
|
Wang Y, Tian MM, Mi CJ, Chen KL, Ji YC,
Wang L, Zhang J and Cheng K: Exercise protects the heart against
myocardial infarction through upregulation of miR-1192. Biochem
Biophys Res Commun. 521:1061–1069. 2020. View Article : Google Scholar
|
|
36
|
Liu J, Wang HZ and Sun Y: Inhibition of
CXCR4 by microRNA-1192 reduces the activation of Th17 cells and
expression of inflammation factors in a mouse model of vulvovaginal
candidiasis. Cell Physiol Biochem. 50:893–910. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Dormoy-Raclet V, Cammas A, Celona B, Lian
XJ, van der Giessen K, Zivojnovic M, Brunelli S, Riuzzi F, Sorci G,
Wilhelm BT, et al: HuR and miR-1192 regulate myogenesis by
modulating the translation of HMGB1 mRNA. Nat Commun. 4:23882013.
View Article : Google Scholar
|
|
38
|
Shibutani ST and Yoshimori T: A current
perspective of autophagosome biogenesis. Cell Res. 24:58–68. 2014.
View Article : Google Scholar :
|
|
39
|
Bento CF, Renna M, Ghislat G, Puri C,
Ashkenazi A, Vicinanza M, Menzies FM and Rubinsztein DC: Mammalian
autophagy: How does it work? Annu Rev Biochem. 85:685–713. 2016.
View Article : Google Scholar
|
|
40
|
Staton AA and Giraldez AJ: Use of target
protector morpholinos to analyze the physiological roles of
specific miRNA-mRNA pairs in vivo. Nat Protoc. 6:2035–49. 2011.
View Article : Google Scholar : PubMed/NCBI
|