Introduction
Multiple myeloma (MM) is a B cell malignancy of
transformed plasma cells derived from mature post-follicular B
cells that occurs in older individuals with a median age at
diagnosis of 69 years and a median overall survival (OS) of 6-7
years (1,2). The disease is the second most
common hematological malignancy in high-income countries and its
development usually starts with a monoclonal gammopathy of
undetermined significance (MGUS), followed by smoldering MM and
finally the full MM disease emerges (3). Progression affects a succession of
genetic events and bone marrow (BM) microenvironment modifications,
such as induction of angiogenesis, suppression of cell-mediated
immunity and development of paracrine signaling loops (such as IL6
and VEGF) (4). Myeloma cells
release cytokines that induce osteoclast activity and bone
resorption. The increased bone resorption in turn causes the
release of growth factors that stimulate myeloma cells, thus
establishing a vicious cycle (5,6).
A marked increase in osteoclast activity leading to bone
destruction characterizes MM (5). Patients with symptomatic MM usually
show an increase of calcium, renal insufficiency, anemia or bone
lesions defined as CRAB criteria (7).
Although MM is still an incurable blood cancer,
prognosis today has considerably improved compared with in past
years, and in the last two decades overall response rates (ORR),
progression-free survival (PFS) and OS have improved (1). Between 2011 and 2014, the risk of
death in patients with a new diagnosis of MM was 35% lower than
between 2006 and 2010 (8). Novel
agents and combination therapies, including proteasome inhibitors
(PIs), immunomodulators (IMiDs), anti-human CD38 (daratumumab)
(9,10), autologous stem cell
transplantation (ASCT) (11) and
maintenance therapy (12) have
improved OS rates compared with historical approaches based on
steroids and chemotherapy (13).
Recently, novel agents such as chimeric antigen
receptor T cell and Bi-specific antibodies associated with old and
new drugs have led to a further improvement in the survival of
these patients (14). In this
scenario of combined therapies, the risk of adverse events is
increased, thus making it even more critical to carefully monitor
patients regarding possible side effects to avoid early
complications that may compromise therapeutic outcome (15). Enhancements in OS are not uniform
across patients with MM, especially considering ethnic minorities
(16) and patients diagnosed at
an older age of onset (17).
Moreover, MM continues to represent an incurable disease due to the
frequent development of drug resistance and its intrinsic tendency
to relapse (18). All these
considerations explain why the development of more effective or
synergistic therapeutic approaches undoubtedly represents an unmet
clinical need.
Traditional herbal medicines are a useful reserve of
biologically active natural products with therapeutic effects that
could be used in the treatment of several diseases, including
cancer. Our previous work demonstrated that Inula viscosa
extract has a potent anti-lymphoma activity through inhibition of
cell proliferation and induction of cell apoptosis, suggesting
potential molecular mechanisms that include the downregulation of
genes involved in the control of growth and survival pathways,
known to be deregulated in Burkitt Lymphoma cells (19). Tomentosin and inuviscolide are
sesquiterpene lactones isolated from I. viscosa (L.) Aiton,
which has been applied for a long time in traditional medicine for
its anti-inflammatory (20),
anthelmintic, antipyretic, anti-septic and antiphlogistic
activities (21,22) and in the treatment of diabetes
(23) and lung disorders
(24).
Messaoudi et al (25) performed a chemical composition
analysis of different I. viscosa extracts from three
different regions of Morocco, revealing the presence of
sesquiterpene lactones, as well as tomentosin and inuviscolide, as
major representative compounds, with tomentosin ranging from 22 to
64% and inuviscolide ranging from 0 to 58% (25). Recently, different studies have
reported the potential anti-cancer effects of sesquiterpene
lactones, and shown that tomentosin and inuviscolide exert
antiproliferative effects on human cancer cell lines (26-29). However, so far, the effects of
singular compounds in MM have not been investigated. Based on our
previous results showing antitumoral activity of natural extract
from I. viscosa on Raji cells, the anti-neoplastic effects
of synthetized sesquiterpene lactones, such as inuviscolide and
tomentosin, in human MM cell lines were evaluated, with the aim to
isolate specific bioactive natural products that could be applied
in the treatment of human MM. Moreover, in order to investigate the
mechanisms underlying the activity of tomentosin in this specific
context, an array-based gene expression analysis was performed.
Materials and methods
Cell culture
MM.1S and RPMI-8226 were obtained from Dr Marina
Ferrarini from San Raffaele Telethon Institute for Gene Therapy
(SR-Tiget), San Raffaele Scientific Institute (Milan, Italy). MRC5
cells were obtained from IRCCS University Hospital San Martino-IST
National Institute for Cancer Research (Genova, Italy). MM.1S and
RPMI-8226 cells were cultured in RPMI-1640 (Euroclone SpA)
supplemented with 10% FBS (Euroclone SpA), 2 mM L-Glutamine (Merck
KGaA) and Antibiotic/Antimycotic Solution (Merck KGaA). MRC5 cells
were cultured in DMEM high-glucose (Euroclone SpA) supplemented
with 10% FBS, 2 mM L-Glutamine, non-essential amino acids, 1 mM
sodium pyruvate (Merck KGaA) and Antibiotic/Antimycotic Solution.
Cells were maintained at 37°C in a 95% humidified atmosphere and 5%
CO2 in T25 filtered-flasks.
Cytotoxicity assay
MM.1S, RPMI-8226 and MRC5 cells were seeded in a
384-multiwell microplate at a concentration of 10,000 (MM.1S) or
1,600 (RPMI-8226, MRC5) cells in 25 µl per well. Cells were
chosen based on a preliminary proliferation setting assay. The day
after, cells were treated with inuviscolide (cat. no. P-25946716;
Mcule, Inc.), tomentosin (cat. no. P-25953579; Mcule, Inc.) and
cisplatin (cat. no. C2210000; Merck KGaA) at concentrations of 50,
25, 12.5, 6.25, 3.125, 1.56 and 0.75 µM, or with vehicle
solution (DMSO 0.5%) for 24 h at 37°C. Subsequently, cell viability
was assessed using CellTox™ Green Cytotoxicity (cat. no. G8731;
Promega Corporation) and CellTiter-Glo (cat. no. G7571; Promega
Corporation) assays using sequential multiplex protocol. CellTox
Green Dye was added to samples at the same time of treatment at
manufacturer's indicated concentrations. Plating of cells,
preparation of serial dilutions and addition of compounds to cells
were performed using an automated liquid handling platform (Gilson
Pipetmax; Gilson). After 48 h from treatment, GFI was quantified
with Cytation 5 (BioTek Instruments, Inc.). For each sample, four
images were taken to cover the entire area of the well. Immediately
after, CellTiter-Glo luminescent cell viability assay was performed
according to the manufacturer's instructions. Relative luminescence
units (RLU) were quantified with GloMax Discover (Promega
Corporation). All experiments were performed in triplicate. The
half maximal inhibitory concentration values were calculated using
R software version 3.2.2 (loess/approx functions) (30), given 100 to RLU observed in
samples treated with vehicle solution and 0 to samples without
cells (absolute IC50). Moreover, the IC50
calculation was performed by using drug concentrations adjusted on
compound purity, compound concentrations were multiplied for 0.7×
or 0.93× for the inuviscolide and tomentosin, respectively, and a
corrected analysis was performed.
Apoptotic assay
MM.1S, RPMI-8226 and MRC5 were seeded in a
384-multiwell microplate at the concentration of 20,000 (MM.1S) or
10,000 (RPMI-8226, MRC5) cells in 25 µl per well. The day
after, cells were treated with Tomentosin at concentrations of 50,
25, 12.5 µM, or vehicle solution (DMSO 0.1%). After 24 h of
treatment, Caspase-Glo 3/7 (cat. no. G8090; Promega Corporation),
Caspase-Glo 8 (cat. no. G8200; Promega Corporation), or Caspase-Glo
9 (cat. no. G8210; Promega Corporation) assays were performed
according to the manufacturer's instructions. Luminescence reading
was carried out with GloMax Discover Microplate Reader (Promega
Corporation). All experiments were performed in triplicate.
Cell cycle analysis
Cell cycle analysis was performed by seeding
1×106 RPMI-8226 cells in 5 ml in T25 flask. After 24 h,
cells were treated with tomentosin 25 µM or vehicle solution
(DMSO 0.05%). Samples were collected after 6, 8, 12, 18 and 24 h of
treatment. Samples were washed in PBS, fixed with 70% ice-cold
ethanol and incubated at -20°C overnight. Fixed cells were washed
in PBS and stained with 7-aminoactinomycin (7-AAD; BD Biosciences).
DNA content of at least 10,000 cells for sample was evaluated with
BD FACSCanto II (BD Biosciences), and values are presented as mean
± standard deviation (SD) of the percentage of three independent
experiments. Data were analyzed with R-Bioc Manager software
(flowCore/flowViz package) version 1.56.0 (31).
Gene expression analysis
To perform gene expression analysis 3×106
RPMI-8226 cells were seeded in 15 ml in T75 flask. After 24 h,
cells were treated with 25 µM tomentosin or with vehicle
solution (DMSO 0.05%). Samples were collected after 8 h of
treatment, before the onset of cell cycle perturbation, considering
that the number of cells in the G2/M phase increases at 12 h after
treatment. Then, the gene expression profile of cells treated with
tomentosin before the cell cycle arrest and apoptosis start was
investigated. Three biological replicates were performed.
Expression of 2,559 cancer-related genes was assessed by
high-throughput mRNA quantitation (HTG EdgeSeq Oncology Biomarker
Panel, HTG Molecular Diagnostics; PMID. 30327311). Counts were
normalized and analyzed using the R-Bioc Manager software (DESeq2
package) version 3.0 (32).
Genes with a maximum count value among all samples <84,
corresponding to 10% quantile, were considered as low expressed and
removed from analysis. Genes with fold-change >1.5 or <0.66
in treated samples and false discovery rate (FDR) <0.05 were
considered as differentially expressed genes (DEGs). For Gene Set
Enrichment Analysis (GSEA) (33), normalized counts were uploaded
into the GSEA-Broad Institute website (PMID. 16199517). The maximum
and minimum sizes for gene sets were 500 and 15, respectively. A
total of 2,257 gene sets from the Gene Ontology (GO) (34,35) Biological Processes database
(c5.bp.v7.1.symbols.gmt) were used as a library for analysis (PMID.
21546393). The statistical significance (nominal P-value) of the
normalized enrichment score (NES) was estimated by running 1,000
gene set permutations.
Functional classification and pathway
analysis of DEGs
The Search Tool for the Retrieval of Interacting
Genes (STRING) database (http://www.string-db.org/) was used to investigate the
potential function of the 126 DEGs identified in tomentosin-treated
MM cells, executing GO enrichment analysis and Kyoto Encyclopedia
of Genes and Genomes (KEGG) pathway analysis (36), and to understand how DEGs
interact with each other a genetic interaction network was
constructed. Terms with FDR-corrected enrichment P<0.05 were
considered. KEGG pathway results were displayed using Cytoscape
software version 3.8.2 (http://www.cytoscape.org).
The 126 DEGs were submitted to the Connectivity Map
online web tool (https://www.broadinstitute.org/connectivitymap-cmap),
which supplies a data-driven and systematic approach for
discovering associations among genes, chemicals and diseases.
Statistical data analysis
All data are presented as mean ± SD from experiments
in triplicate, and statistical significance between cell lines
treated and untreated cells was set at P<0.05. Data analysis was
performed by one-way analysis of variance (ANOVA) followed by
Tukey's post hoc test for the statistical analysis of cell
viability and caspase activity, two-way ANOVA and Tukey's post hoc
test were applied for the statistical analysis of cell cycle, using
R software, version 3.2.2 (30).
Results
Cytotoxicity of sesquiterpene lactones on
MM cell lines
The first objective was to characterize the effects
of various concentrations of synthetized sesquiterpene lactones on
the proliferation of two MM and normal fibroblast cell lines (MMS1,
RPMI-8226 and MRC5). All cell lines were treated with increasing
concentrations of inuviscolide, tomentosin and cisplatin (50, 25,
12.5, 6.25, 3.125, 1.56, 0.75 µM) for 48 h and cell
viability was determined. For each cell line the IC50
values of all the three drugs are shown in Table I.
 | Table IIC50 values of
inuviscolide, tomentosin and cisplatin after 48 h of treatment. |
Table I
IC50 values of
inuviscolide, tomentosin and cisplatin after 48 h of treatment.
| Treatment | Cell line | IC50,
mM | 95% confidence
interval, mM |
|---|
| Inuviscolide | RPMI-8226 | 42.53 | 39.99-45.60 |
| MM.1S | >50.00 | ND |
| MRC5 | >50.00 | ND |
| Tomentosin | RPMI-8226 | 26.14 | 22.82-29.90 |
| MM.1S | 26.34 | 24.94-27.73 |
| MRC5 | >50.00 | ND |
| Cispaltin | RPMI-8226 | 17.05 | 15.14-18.92 |
| MM.1S | 8.89 | 8.39-9.42 |
| MRC5 | 14.75 | 13.09-16.47 |
Fig. 1 shows the
impact of dose-dependent cytotoxicity of the drugs on cell
viability at 48 h after treatments (blue lines). Specifically, cell
viability assay showed that inuviscolide and tomentosin treatments
did not induce changes in cell viability at low concentrations (in
the range from 0.75 to 10.0 µM). Whereas, cell viability
notably decreased during treatment, inducing cytotoxic effects
starting from the concentration of 25.0 µM, and increasing
proportionally the damage with the highest concentration.
Specifically, at 50.0 µM MM.1S cells showed higher
sensitivity to tomentosin than other cell lines, and both MM cell
lines at 25.0 µM showed higher sensitivity than MRC5.
Inuviscolide showed toxicity on MRC5 fibroblasts comparable to or
higher than tumor cell lines. These results indicated that
tomentosin exerted preferential antiproliferative activity towards
both MM cell lines compared with a non-tumor cell line.
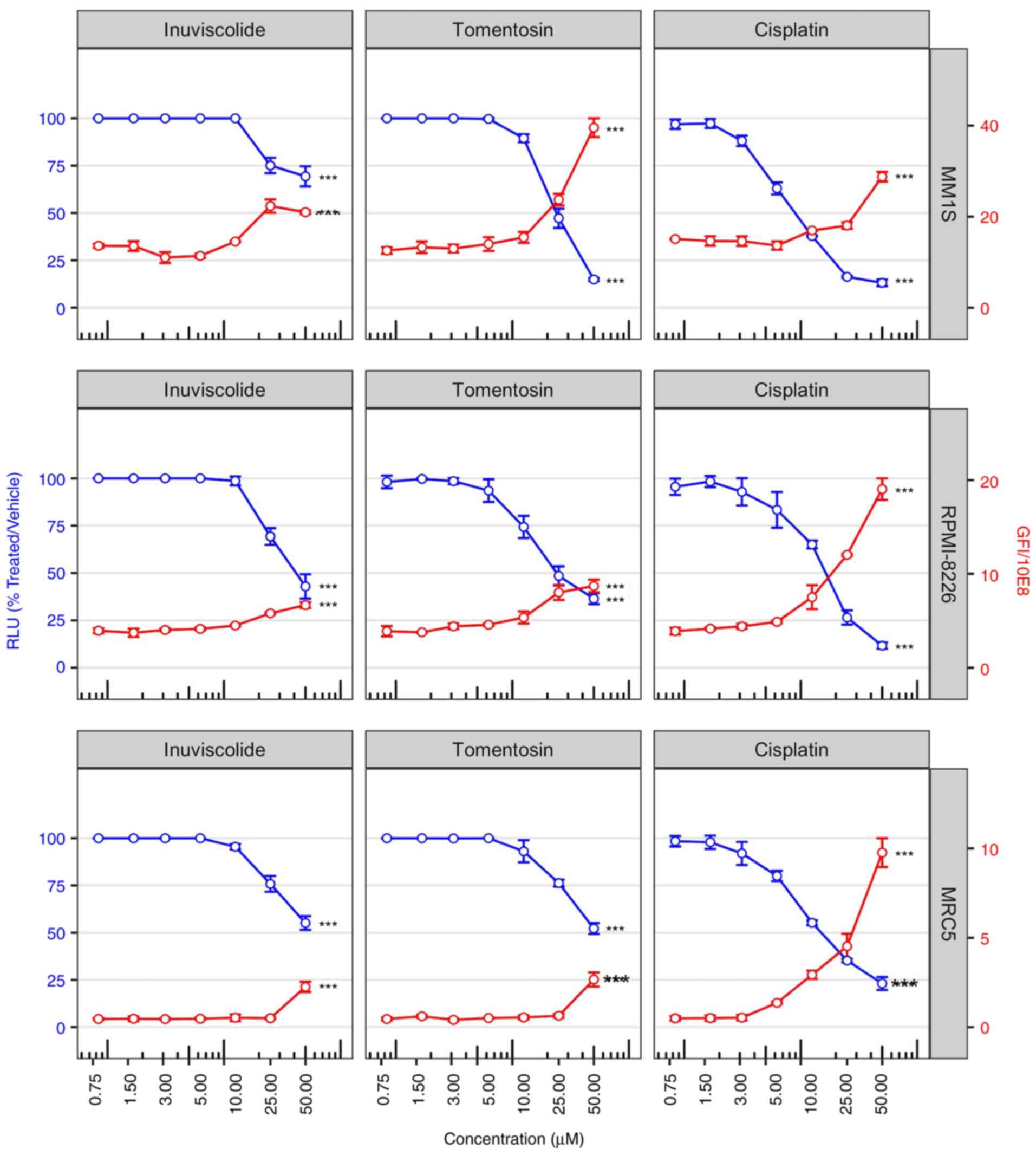 | Figure 1Cell viability is reduced after
inuviscolide and tomentosin treatment. Cell viability scatterplots
were generated after 48 h of treatment with inuviscolide,
tomentosin or cisplatin at 50, 25, 12.5, 6.25, 3.12, 1.56 or 0.8
µM. RLU obtained by the addition of CellTiter-Glo reagent
and representative of the number of living cells were normalized to
samples treated with vehicle solution (left y-axis, blue lines and
dots). Total obtained by addition of CellTox green dye and
representative of the number of dead cells, was divided by 108
(right y-axis, red lines and dots). Mean ± SD values are shown.
One-way ANOVA was applied. ***P<0.001. RLU, relative
luminescence units. |
In order to assess the amount of cell death caused
by drug treatments, selected morphological features were used to
label a cell as dead or alive. A primary feature is the disruption
of the cell membrane, allowing CellTox Green dye to enter into the
cells and bind to DNA. The observed increase in cell death was in
accordance with the proportional decrease in cell viability
following the rise of drug concentrations, as shown by a red line
in Fig. 1.
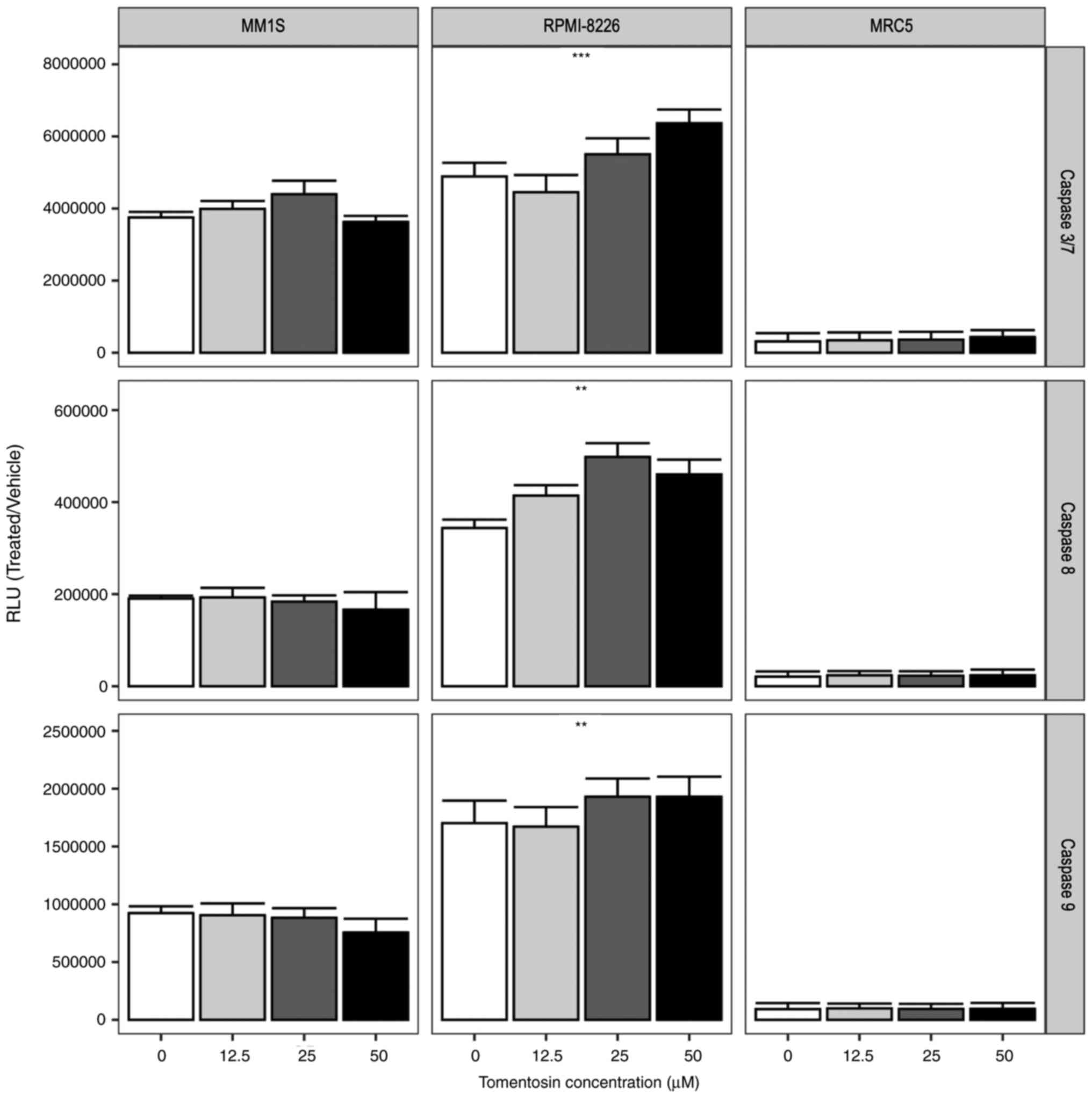
Tomentosin effect on apoptotic cellular
processes in MM cell lines
The induction of the apoptotic cascade is one of the
main mechanisms of therapy-induced tumor-cell death (37). To determine whether the effects
of sesquiterpene lactones depends on its ability to activate the
apoptotic cascade, myeloma cell lines were treated with tomentosin
at concentrations of 50, 25, 12.5 µM for 24 h. Due to the
strong cytotoxicity of inuviscolide on MRC5 fibroblasts, comparable
to or higher than the one observed in tumor cell lines, our
experiments focused on tomentosin treatment.
The activation of enzymatic activities of the main
effector caspases, caspase-3/7 and initiator caspase-8 and -9 was
measured using Caspase-Glo 3/7, 8 and 9 assays. The RLU showed that
tomentosin treatment increases caspase activity (P<0.05), as
shown in Fig. 2, and more
specifically the effector caspase-3/7 activity was significantly
increased in RPMI-8226 cells. Furthermore, the activities of
caspase-8 and -9, which are representative initiator caspases in
the death receptor-mediated and mitochondrial apoptotic pathways,
respectively, were also measured, showing a significant increase in
RPMI-8226 cells (Fig. 2).
Overall, these results suggested that tomentosin promoted
apoptosis-induced death on RPMI-8226 cells, mediated by both the
death receptor and the mitochondrial pathways. Conversely,
non-significant activation of enzymatic activities of caspases were
identified in the MM.1S cell line.
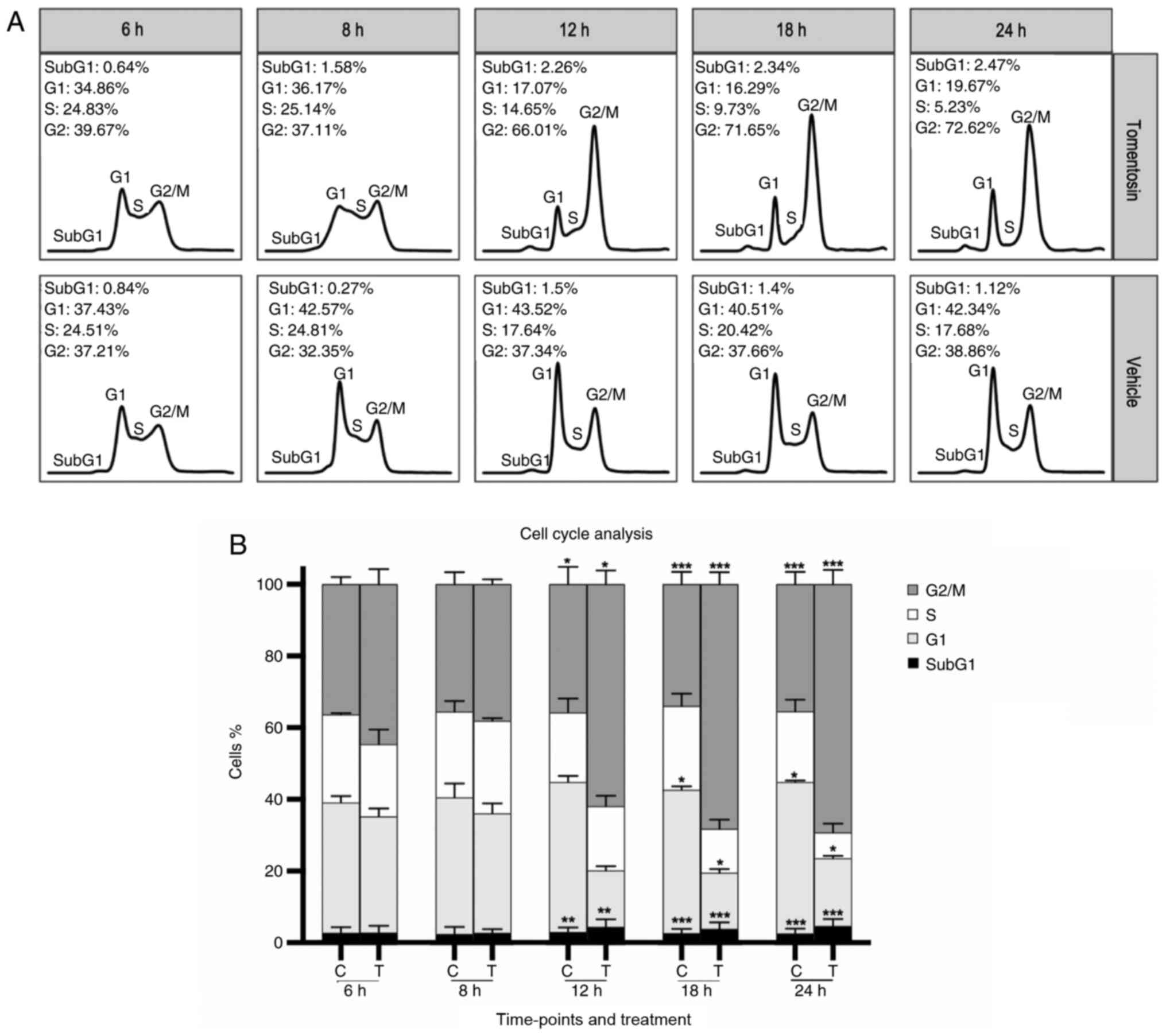
Tomentosin effect on cell cycle in MM
cell lines
To address whether the antiproliferative effect of
tomentosin on MM cells was associated with cell cycle regulation,
DNA cell cycle analysis by flow cytometry was performed in
RPMI-8226 cells treated with tomentosin or vehicle (Fig. 3A). Incubation with tomentosin (25
µM) for 6, 8, 12, 18 and 24 h showed a progressive decrease
of cell number in S-phase at 18 and 24 h (at 24 h, 5.23 vs. 17.68%
in control, P<0.05) and an increase of cell number in G2/M-phase
that started at 12 h (at 24 h, 72.62 vs. 38.86% in control,
P<0.05), accompanied by a corresponding decrease in the
proportion of cells in G0/G1 (at 24 h, 19.67 vs. 42.34% in control,
P<0.05) (Fig. 3B). These
results revealed that MM cells treated with tomentosin were blocked
in the G2/M phase. These data suggested that both cell cycle arrest
and cell apoptosis could explain the antiproliferative effects of
tomentosin and may result in the inhibition of RPMI-8226 cell
viability.
Tomentosin effects on gene expression
profiling in MM cell lines
To assess DEGs contributing to tomentosin effects on
RPMI-8226 cells and identify its possible mechanisms of action, the
HTG EdgeSeq Oncology Biomarker Panel was used to analyze the
expression of 2,559 cancer-related genes by high-throughput mRNA
quantitation analysis. Comparing global gene expression between
RPMI-8226 cells treated with 25 µM tomentosin or untreated
(vehicle), 126 genes were shown to be differentially expressed.
Specifically, 71 genes were upregulated and 53 genes were
downregulated (Table SI).
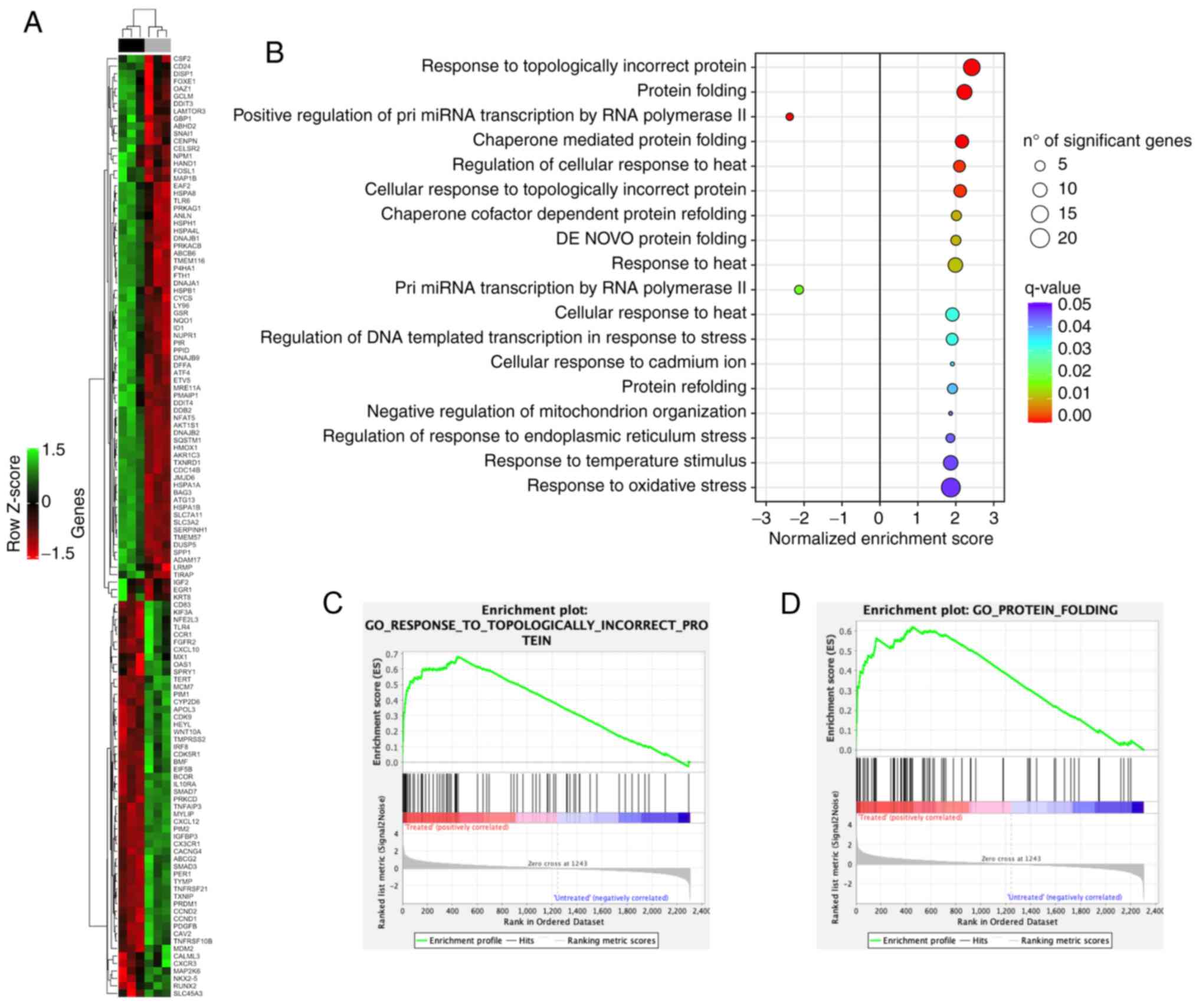
An unsupervised hierarchical clustering analysis of
126 DEGs allowed to clearly separate tomentosin-treated RPMI-8226
cells vs. untreated cells (Fig.
4A). To investigate the biological role of the 126 DEGs
deregulated by tomentosin in RPMI-8226 cells, a functional
enrichment analysis was performed. DEGs identified in
tomentosin-treated RPMI-8226 were significantly involved in
biological processes, such as 'response to topologically incorrect
protein', 'protein folding', 'regulation of response to endoplasmic
reticulum stress', 'response to heat' and 'response to oxidative
stress' (Fig. 4B-D).
Table SII shows
the list of DEGs involved in each biological process. Furthermore,
the significant identified DEGs were enriched in proteins that have
key roles in cellular pathways, such MAPK, p53, PI3K-Akt, Hedgehog,
Hippo, WNT, Apelin and cell cycle, apoptosis, ferroptosis,
immune-system, DNA damage response, protein processing in
endoplasmic reticulum (ER), angiogenesis, osteoclast
differentiation and signaling pathways regulating pluripotency of
stem cells (Table SIII).
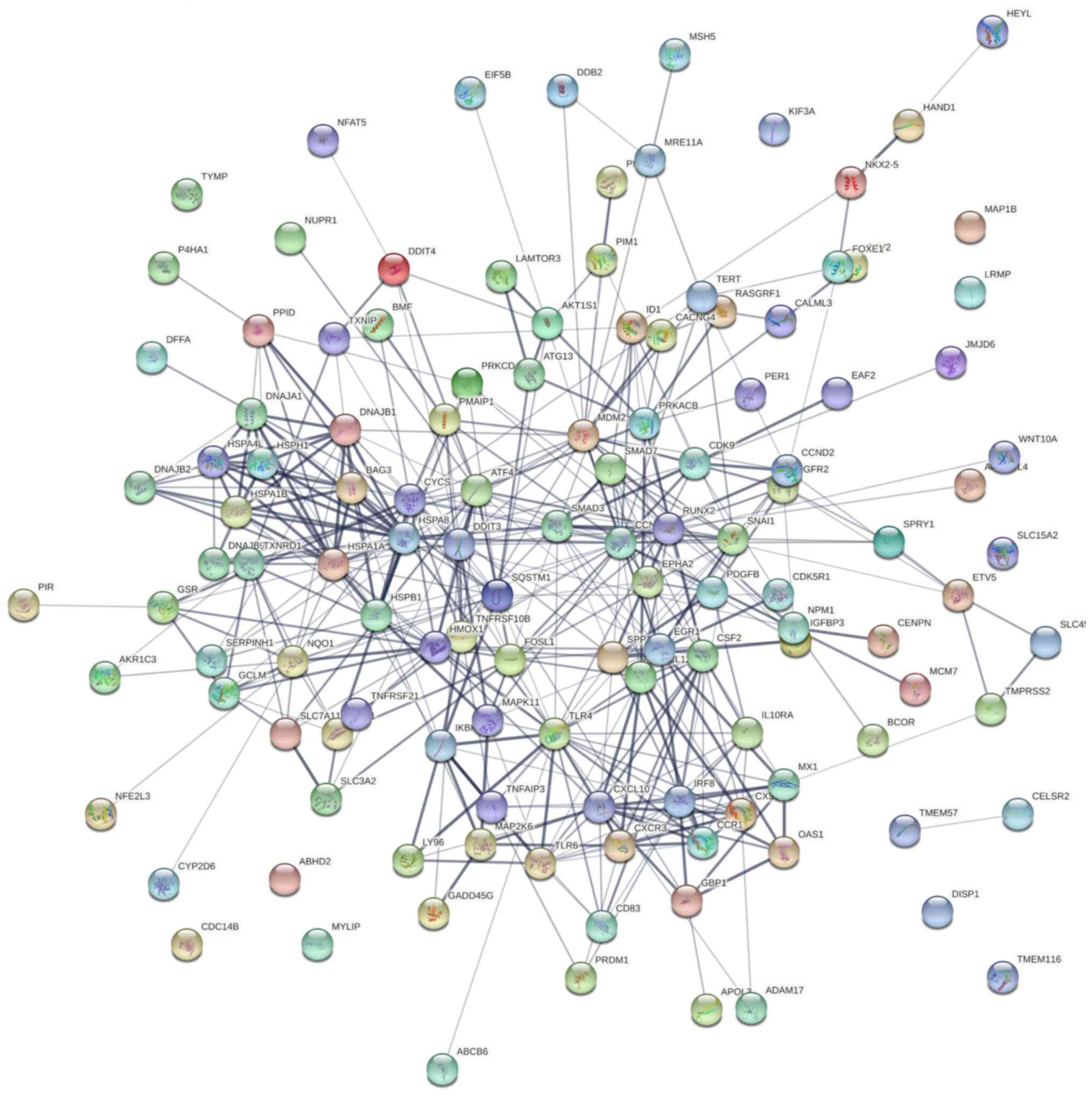
To address the systems biology and identify how
tomentosin deregulates DEGs in MM from a systems perspective, all
DEGs were analyzed using the STRING bioinformatics platform.
Fig. 5 shows the interaction
network of involved tomentosin-deregulated DEGs, represented by 125
nodes and 429 edges, an average node degree of 6.86. Besides,
network analysis also showed that the clustering coefficient and
PPI enrichment P-values were 0.534 and <0.0001, respectively,
thus underlining the reliable robustness of the network.
To discover the functional connections between
drugs, genes and diseases through the transitory features of common
gene-expression changes, the 126 DEGs were submitted to the
Connectivity Map online web tool. The results suggested that the
mechanisms of tomentosin activity on RPMI-8226 cells may be similar
to those exerted by heat shock protein (HSP) inhibitors (Fig. 6), considering that Tomentosin
deregulates the expression of the same genes (ATF4, DDIT3, HSPA4L,
HSPB1, HSPA1A, HSPA8, DNAJB1, DNAJB2, DNAJA1, SERPINH1, HSPH1,
HSPA1B) affected by HSP inhibitors.
Discussion
Improvements in survival for patients with MM with
recent diagnosis have been well-described according to randomized
clinical trials (38-40) and assumed as a result of the
higher use of autologous hematopoietic stem cell transplantation,
as well as of the introduction of novel agents, specifically
proteasome inhibitors and immunomodulatory agents (41-43). A previous population-based study
in MM revealed that OS is increasing also among racial/ethnic
minorities, although OS improvements are consistently limited to
younger patients <70 years of age (38). Substantially, MM continues to
represent an incurable disease due to the frequent development of
drug resistance and to its intrinsic tendency to recur over time.
Accordingly, the identification of further effective and
potentially synergistic therapeutic agents that may overcome the
strong collateral effects of drugs currently utilized in MM and
their substantial inefficacy in elderly patients, still represents
a landmark for patient outcomes.
Tomentosin and inuviscolide are natural potent
bioactive compounds extracted from I. viscosa, identified as
worthwhile therapeutic agents against various types of cancer. Our
previous study demonstrated that I. viscosa extract
exhibited powerful antiproliferative and cytotoxic activities on
Raji cell line, showing a dose- and time-dependent decrease in cell
viability, obtained by cell cycle arrest in the G2/M phase and an
increase in cell apoptosis. The molecular mechanisms underlying
such an antineoplastic activity are based on targeting and on
downregulation of genes involved in cell cycle and apoptosis
(19).
So far, the anti-neoplastic effects of sesquiterpene
lactones on MM have not been studied. Thus, the present study aimed
to investigate if sesquiterpene lactones may be responsible for the
bioactivity of I. viscosa plants. This study showed that
tomentosin exerts antiproliferative and cytotoxic activity against
MM.1S and RPMI-8226 cell lines. In fact, the treatment of these
cell lines with increasing concentrations of tomentosin showed a
dose-dependent decrease in cell viability, paralleled by a
reduction in cell proliferation due to an induction of cell cycle
arrest in the G2/M phase. Moreover, a dose-dependent increase in
cell apoptosis was observed. As demonstrated in the present study,
tomentosin promoted apoptosis-induced death mediated by both the
death receptor and the mitochondrial pathways on RPMI-8226 cells.
These findings suggested that both cell cycle arrest and cell
apoptosis could be the primary mechanisms underlying the
antiproliferative effects of tomentosin, resulting in the
inhibition of RPMI-8226 cell viability. Tomentosin did not promote
apoptosis-induced death mediated by both the death receptor and the
mitochondrial pathways in MM.1S cell lines. The differences
identified between the two MM cell lines should be dependent on
wide heterogeneity that characterized the various MM cell lines
produced. Specifically, RPMI-8226 cells are considered as
well-molecularly characterized MM cells and represent MM at
diagnosis, conversely MM.1S cells are less well-characterized and
represent terminal tumor (44).
Considering these differences, the results should be related to
this heterogeneity, and it should be hypothesized that tomentosin
is more efficient on tumors at the initial stages of
development.
The current results are supported by previous
studies, in which several anticancer properties have been
attributed to tomentosin. Rozenblat et al (26) showed strong pro-apoptotic effects
of both sesquiterpene in human aggressive melanoma cell lines,
suggesting their potential pharmacological value. Moreover,
tomentosin exerts anticancer effects mainly by inducing apoptotic
death through different mechanisms in gastric cancer, cervical
cancer and osteosarcoma (27-29). Recently, Yang et al
(45) demonstrated that
tomentosin induces apoptosis in human leukemia cells via the
caspase-facilitated pro-apoptotic pathway, and inhibition of the
NF-κB-stimulated Bcl-2. Of note, a recent in vivo study
showed the antitumor effects of I. viscosa extract on skin
carcinogenesis induced in mice by the inhibition of proteasome
activity (46). The ingestion of
I. viscosa extract in mice delayed the formation of skin
papilloma and reduced their size and numbers (46).
In order to analyze the molecular mechanisms
involved in the anti-cancer activity of tomentosin, a
high-throughput mRNA quantitation analysis on 2,559 cancer-related
genes was performed in the present study. A total of 126 DEGs
showing different activities in human MM cell lines treated with
tomentosin or untreated were identified.
Deregulation of various biological pathways is a
hallmark in tumor pathogenesis. The current data showed that a
number of DEGs and pivotal pathways involved in MM biology were
deregulated by tomentosin treatment. The PPI network analysis
demonsrtated that tomentosin treatment in human MM cell lines
induced a downregulation of genes involved in pathways known to
lead immune-system processes, such as cytokine-cytokine receptor
interaction, chemokine signaling pathway, NF-κB signaling pathway,
inflammatory mediator regulation of TRP channels, Toll-like
receptor (TLR) signaling pathway and TNF signaling pathway. Of
note, CCR1, CXCR1-3, CXCL10-12 and MAP2K6 were downregulated in
human MM by tomentosin treatment. Recently, Zeissig et al
(47) showed within in
vivo experiments that CCR1 overexpression is a key driver of MM
plasma cells escaping from the BM to circulation during MM
progression. In addition, inhibition of CCR1 via therapeutic
targeting or knockout reduces MM OPM2 or RPMI-8226 dissemination in
intratibial xenograft models (47). Based on the mechanistical role of
CCR1 in human MM, we could hypothesize that CCR1 downregulation
induced by tomentosin treatment could potentially prevent MM
progression and metastasization, thus blocking the development
severe of osteolytic lesions (48). CXCR3 overexpression on tumor
cells and regulatory immune cell populations has been associated
with poor survival in patients affected by several subtypes of
tumors (49-51). During the development of
hematological tumors in BM, CXCR3 activation could induce natural
killer (NK) cell mobilization from BM into peripheral blood,
blocking their approach towards tumor micro-environment. In MM the
upregulation of CXCR3 ligands is associated with the severity of
the disease and with poor survival (51,52). Bonanni et al (53) demonstrated that the inhibition of
CXCR3/CXCL10 axis induces a powerful long-lasting antitumor effect
of NK cell-based adoptive immunotherapy due to an increased
accumulation in BM (53). These
observations support the present results, which showed that
tomentosin treatment inhibited the CXCR3/CXCL10 axis in MM, thus it
may potentially be able to overcome the impairment of NK cell tumor
infiltration and improve their effects within anti-cancer
immunity.
TLRs are sensors for the innate immunity and
initiators of adaptive immunity and are required for normal B
lymphocyte activation (54).
Previous studies have identified the role of TLR systems on
malignant B cell proliferation, differentiation and apoptotic
processes (55). Xu et al
(56) showed that TLR4 and TLR9
overexpression can induce the NF-κB pathway, which produces the
upregulation of IL6, thus promoting MM cell proliferation and
survival. Phosphorylated MAP2K6, in response to inflammatory
cytokines or environmental stress, i.e. after stimulation of the
TLRs by pathogen-associated molecular pattern, activates
p38-MAP-kinase, JNK and ERK pathways controlling cell cycle arrest,
transcription activation and apoptosis (57). The TLR4 and MAP2K6 downregulation
identified after tomentosin treatment in MM could be also
concordant with a reduction of signaling pathways involved in the
inflammatory process. Besides, MAP2K6 is also identified as a poor
prognostic factor in MM (58).
IL10RA is a receptor with potent anti-inflammatory
activity, which also improves survival of progenitor myeloid cells
through the activation of major survival pathways, consisting of
IRS-2 and PI3K/AKT pathways (59,60). The IL10RA downregulation induced
by tomentosin treatment in MM could be considered as likely
responsible for decreasing cell survival.
The PPI network analysis demonstrated the
downregulation of genes involved in pathways known to be implicated
in cellular neoplastic processes, such as growth, proliferation,
migration and invasion. However, the involvement of tomentosin in
MM progression was based exclusively on gene expression profiling
and enrichment analysis, representing a limitation of the present
study. Importantly, these data showed the downregulation of CCND1
and CCND2 genes in MM cells treated with tomentosin, which may
explain the anti-proliferative effects of tomentosin resulting in
the inhibition of RPMI-8226 cell viability. The critical role of
D-type cyclins in governing cell cycle entry and progression and
their almost universal dysregulation in MM propose their central
role in disease pathogenesis and biology (61).
Zhang et al (62) showed that RUNX2 was highly
expressed in non-Hodgkin's lymphoma and MM cell lines promoting the
proliferation and cell adhesion-mediated drug resistance (CAM-DR).
The induction of RUNX2 downregulation reverses CAM-DR. Based on the
current data, tomentosin could be considered as a RUNX2 inhibitor,
thus representing a possible option to overcome drug resistance
(62).
MCM7 is one of the most important subunits of the
miniature chromosome maintenance (MCM) complex that participates in
the cell cycle regulation from G1 to S phase (63) in mRNA transcription and
regulation of DNA damage (64,65). Notably, Tian et al
(66) silencing MCM7 in leukemia
cells in vitro and in vivo experiments led to a
significant decrease in proliferation, inhibition of cell cycle
progression and enhancing apoptosis. Tomentosin could be proposed
as a MCM7 inhibitor, therefore representing a suitable strategy for
growth inhibition and apoptosis induction in MM.
The indirect mechanism to reduce or damage p53
activity is mediated by the overexpression of negative regulators.
The interaction of MDM2 with p53 favors the translocation of p53
from the nucleus to the cytoplasm and its quick degradation by the
proteasome (67). Consequently,
p53 activation can be obtained by inhibiting the association of the
protein with their negative regulators in tumors overexpressing
them. The first molecule identified as a potent inhibitor of the
p53-MDM2 interaction was nutlin (68). Nutlin-3 has a potent antitumor
effect through the induction of apoptosis in different
hematological tumor cells lines, including MM (69-71). The present results suggested that
tomentosin promoted apoptosis-induced death on RPMI-8226 cells
mediated by both the death receptor and the mitochondrial pathways.
Besides, it could be possible that another mechanism of
apoptosis-induced death and growth inhibition induced by Tomentosin
is via deregulation of the MDM2 gene. Of note, the
tomentosin-induced downregulation of SMAD7 and SMAD3 should
strengthen the growth inhibition and apoptosis induction regulated
by the TGF-b family pathway in neoplastic cells (72).
MM cells treated with tomentosin also show
deregulation of CDK9, which belongs to the CDK-transcriptional
regulator sub-group. CDK9 is responsible for the synthesis of Mcl-1
and XIAP antiapoptotic proteins, and maintaining the conditions for
cancer cell survival (73).
Recently, novel 2,4-disubstituted pyrimidine derivatives showing
CDK9 inhibitory and antiproliferative effects on RPMI-8226 were
synthesized (74).
Gene expression profiling analysis of MM cell lines
treated with tomentosin in the present study showed the
upregulation of DnaJ family of proteins and HSP-70, which support
protein folding and misfolded proteins degradation, intracellular
trafficking, modulating signaling pathways and regulating immune
responses (75,76). Considering that cytoplasmic
HSP-70s, inducible Hsp72 and constitutively expressed Hsc70 are
regularly upregulated in myeloma, and Hsp70 inhibition is also
successful in influencing myeloma cell death (77-79), the current data suggested that
tomentosin did not directly affect their expression in our
experimental conditions.
Notably, the current study demonstrated that
tomentosin induces the upregulation of ATF4 and DDIT3 genes in MM
cells, whose proteins are enriched in protein processing in the ER
pathway and protein folding process. Accumulation of misfolded
proteins, typical of MM cells, in the ER lumen causes ER stress and
activation of the unfolded protein response signaling pathway,
which triggers downstream pathways to inhibit protein translation
and increases protein folding activity expanding chaperone
expression levels (80). The
chronic activation of the ER stress response can trigger apoptotic
signals, leading to the expression of downstream signaling, which
damage the target cells (81).
The increased ATF4 and DDIT3 expression induced by
tomentosin in MM may be responsible for the activation of the
unfolded protein response PERK/eIF2a/ATF4/DDIT3 pathway. The
activation of the transcription factor ATF4, which further
activates the proapoptotic CHOP (DDIT3), suggests that in the
presence of tomentosin the protective cell mechanisms activated by
the unfolded protein response signaling is not sufficient to
restore normal ER function and apoptois is induced. This hypothesis
was supported by the results obtained from functional connections
analysis executed using the Connectivity Map tool, which suggested
that the mechanism of tomentosin in RPMI-8226 cells may be similar
to those exerted by HSP inhibitors.
Our future investigations will include in
vivo experiments to analyze the molecular mechanisms by which
tomentosin induces pharmacological effects in human MM, supported
by the gene expression profile and enrichment analyses obtained by
tomentosin treatment on MM cell lines.
To sum up, the present study demonstrated that
tomentosin has a potent antitumor activity on MM cells through
inhibition of cell proliferation and induction of cell apoptosis.
Furthermore, through the execution of a gene expression profiling
analysis and the construction of a genetic interaction network, the
molecular mechanisms responsible for the effects of tomentosin on
MM were explored. Specifically, tomentosin induces the
downregulation of genes enriched in immune-system pathways, as well
as pathways that favor proliferation, growth, migration and
invasion processes. Of note, tomentosin may inhibit the growth of
MM through the induction of apoptosis by upregulation of the
PERK/eIF2a/ATF4/DDIT3 pathway. These results suggested that
tomentosin could be considered a potential natural drug with
limited toxicity and relevant antitumor activity in the therapeutic
armamentarium available for patients with MM.
Supplementary Data
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
PV, GP, CF, LB, MRDM and LP were responsible for
conceptualization of the present study. PV, RM, GG, LS, VB and MRM
performed the experiments. IM, FPF and MRDM conducted the formal
analysis of the data, and wrote the original draft preparation. CF,
LP, GP and LB reviewed and edited the manuscript. GP, CF and LP
were responsible for funding acquisition. IM and FPF confirm the
authenticity of all the raw data. All authors read and approved the
final version of the manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgments
Not applicable.
References
|
1
|
Kumar SK, Dispenzieri A, Lacy MQ, Gertz
MA, Buadi FK, Pandey S, Kapoor P, Dingli D, Hayman SR, Leung N, et
al: Continued improvement in survival in multiple myeloma: Changes
in early mortality and outcomes in older patients. Leukemia.
28:1122–1128. 2014. View Article : Google Scholar :
|
|
2
|
Röllig C, Knop S and Bornhäuser M:
Multiple myeloma. Lancet. 385:2197–2208. 2015. View Article : Google Scholar
|
|
3
|
van de Donk NWCJ, Pawlyn C and Yong KL:
Multiple myeloma. Lancet. 397:410–427. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Kyle RA and Rajkumar SV: Drug therapy:
Multiple myeloma. N Engl J Med. 351:1860–1873. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Roodman GD: Pathogenesis of myeloma bone
disease. Leukemia. 23:435–441. 2009. View Article : Google Scholar
|
|
6
|
Raje NS, Bhatta S and Terpos E: Role of
the RANK/RANKL pathway in multiple myeloma. Clin Cancer Res.
25:12–20. 2019. View Article : Google Scholar
|
|
7
|
Kyle RA, Child JA, Anderson K, Barlogie B,
Bataille R, Bensinger W, Bladé J, Boccadoro M, Dalton W, Dimopoulos
M, et al: Criteria for the classification of monoclonal
gammopathies, multiple myeloma and related disorders: A report of
the international myeloma working group. Br J Haematol.
121:749–757. 2003. View Article : Google Scholar
|
|
8
|
Maiese EM, Evans KA, Chu BC and Irwin DE:
Temporal trends in survival and healthcare costs in patients with
multiple myeloma in the United States. Am Heal Drug Benefits.
11:39–46. 2018.
|
|
9
|
Fonseca R, Abouzaid S, Bonafede M, Cai Q,
Parikh K, Cosler L and Richardson P: Trends in overall survival and
costs of multiple myeloma, 2000-2014. Leukemia. 31:1915–1921. 2017.
View Article : Google Scholar :
|
|
10
|
Durie BGM, Hoering A, Abidi MH, Rajkumar
SV, Epstein J, Kahanic SP, Thakuri M, Reu F, Reynolds CM, Sexton R,
et al: Bortezomib with lenalidomide and dexamethasone versus
lenalidomide and dexamethasone alone in patients with newly
diagnosed myeloma without intent for immediate autologous stem-cell
transplant (SWOG S0777): A randomised, open-label, phase 3 trial.
Lancet. 389:519–527. 2017. View Article : Google Scholar :
|
|
11
|
Child JA, Morgan GJ, Davies FE, Owen RG,
Bell SE, Hawkins K, Brown J, Drayson MT and Selby PJ; Medical
Research Council Adult Leukaemia Working Party: High-dose
chemotherapy with hematopoietic stem-cell rescue for multiple
myeloma. N Engl J Med. 348:1875–1883. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
McCarthy PL, Holstein SA, Petrucci MT,
Richardson PG, Hulin C, Tosi P, Bringhen S, Musto P, Anderson KC,
Caillot D, et al: Lenalidomide maintenance after autologous
stem-cell transplantation in newly diagnosed multiple myeloma: A
meta-analysis. J Clin Oncol. 35:3279–3289. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Bergsagel PL: Where we were, where we are,
where we are going: Progress in multiple myeloma. Am Soc Clin Oncol
Educ Book:. 199:–203. 2014.
|
|
14
|
Teoh PJ and Chng WJ: CAR T-cell therapy in
multiple myeloma: More room for improvement. Blood Cancer J.
11:842021. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Ludwig H, Delforge M, Facon T, Einsele H,
Gay F, Moreau P, Avet-Loiseau H, Boccadoro M, Hajek R, Mohty M, et
al: Prevention and management of adverse events of novel agents in
multiple myeloma: A consensus of the European myeloma network.
Leukemia. 32:1542–1560. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Pulte D, Redaniel MT, Brenner H, Jansen L
and Jeffreys M: Recent improvement in survival of patients with
multiple myeloma: Variation by ethnicity. Leuk Lymphoma.
55:1083–1089. 2014. View Article : Google Scholar
|
|
17
|
Kristinsson SY, Anderson WF and Landgren
O: Improved long-term survival in multiple myeloma up to the age of
80 years. Leukemia. 28:1346–1348. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Giuliani N, Accardi F, Marchica V, Dalla
Palma B, Storti P, Toscani D, Vicario E and Malavasi F: Novel
targets for the treatment of relapsing multiple myeloma. Expert Rev
Hematol. 12:481–496. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Virdis P, Migheli R, Galleri G, Fancello
S, Cadoni MPL, Pintore G, Petretto GL, Marchesi I, Fiorentino FP,
di Francesco A, et al: Antiproliferative and proapoptotic effects
of Inula viscosa extract on Burkitt lymphoma cell line. Tumor Biol.
42:10104283199010612020. View Article : Google Scholar
|
|
20
|
Barbetti P, Chiappini I, Fardella G and
Menghini A: A new eudesmane acid from Dittrichia (Inula) viscosa.
Planta Med. 51:4711985. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Lauro L and Rolih C: Observations and
research on an extract of Inula viscosa Ait. Boll Soc Ital Biol
Sper. 66:829–834. 1990.In Italian. PubMed/NCBI
|
|
22
|
Lev E and Amar Z: Ethnopharmacological
survey of traditional drugs sold in Israel at the end of the 20th
century. J Ethnopharmacol. 72:191–205. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Yaniv Z, Dafni A, Friedman J and Palevitch
D: Plants used for the treatment of diabetes in Israel. J
Ethnopharmacol. 19:145–151. 1987. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Al-Qura'n S: Ethnopharmacological survey
of wild medicinal plants in Showbak, Jordan. J Ethnopharmacol.
123:45–50. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Messaoudi M, Chahmi N, El-Mzibri M, Gmouh
S, Amzazi S, Benbacer L and El-Hassouni M: Cytotoxic Effect and
chemical composition of Inula viscosa from three different regions
of morocco. Eur J Med Plants. 16:1–9. 2016. View Article : Google Scholar
|
|
26
|
Rozenblat S, Grossman S, Bergman M,
Gottlieb H, Cohen Y and Dovrat S: Induction of G2/M arrest and
apoptosis by sesquiterpene lactones in human melanoma cell lines.
Biochem Pharmacol. 75:369–382. 2008. View Article : Google Scholar
|
|
27
|
Yang H, Zhao H, Dong X, Yang Z and Chang
W: Tomentosin induces apoptotic pathway by blocking inflammatory
mediators via modulation of cell proteins in AGS gastric cancer
cell line. J Biochem Mol Toxicol. 34:e225012020. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Merghoub N, El Btaouri H, Benbacer L,
Gmouh S, Trentesaux C, Brassart B, Attaleb M, Madoulet C, Wenner T,
Amzazi S, et al: Tomentosin induces telomere shortening and
caspase-dependant apoptosis in cervical cancer cells. J Cell
Biochem. 118:1689–1698. 2017. View Article : Google Scholar
|
|
29
|
Lee CM, Lee J, Nam MJ, Choi YS and Park
SH: Tomentosin displays anti-carcinogenic effect in human
osteosarcoma MG-63 cells via the induction of intracellular
reactive oxygen species. Int J Mol Sci. 20:15082019. View Article : Google Scholar :
|
|
30
|
Ihaka R and Gentleman R: R: A language for
data analysis and graphics. J Comput Graph Stat. 5:299–314.
1996.
|
|
31
|
Ellis B, Gentleman R, Hahne F, Le Meur N,
Sarkar D and Jiang M: flowViz: Visualization for flow cytometry. R
package version 1.56.0. 2021, https://www.bioconductor.org/packages/release/bioc/html/flowViz.html.
|
|
32
|
Love MI, Huber W and Anders S: Moderated
estimation of fold change and dispersion for RNA-seq data with
DESeq2. Genome Biol. 15:5502014. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Subramanian A, Tamayo P, Mootha VK,
Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub
TR, Lander ES and Mesirov JP: Gene set enrichment analysis: A
knowledge-based approach for interpreting genome-wide expression
profiles. Proc Natl Acad Sci USA. 102:15545–15550. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Ashburner M, Ball CA, Blake JA, Botstein
D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT,
et al: Gene ontology: Tool for the unification of biology. The gene
ontology consortium Nat Genet. 25:25–29. 2000.
|
|
35
|
Carbon S, Douglass E, Good BM, Unni DR,
Harris NL, Mungall CJ, Basu S and Elser J: The gene ontology
resource: Enriching a GOld mine. Nucleic Acids Res. 49D:D325–D334.
2021.
|
|
36
|
Kanehisa M and Goto S: KEGG: Kyoto
encyclopedia of genes and genomes. Nucleic Acids Res. 28:27–30.
2000. View Article : Google Scholar
|
|
37
|
Igney FH and Krammer PH: Death and
anti-death: Tumour resistance to apoptosis. Nat Rev Cancer.
2:277–288. 2002. View
Article : Google Scholar : PubMed/NCBI
|
|
38
|
Costa LJ, Brill IK, Omel J, Godby K, Kumar
SK and Brown EE: Recent trends in multiple myeloma incidence and
survival by age, race, and ethnicity in the United States. Blood
Adv. 1:282–287. 2017. View Article : Google Scholar
|
|
39
|
Cavo M, Tacchetti P, Patriarca F, Petrucci
MT, Pantani L, Galli M, Di Raimondo F, Crippa C, Zamagni E, Palumbo
A, et al: Bortezomib with thalidomide plus dexamethasone compared
with thalidomide plus dexamethasone as induction therapy before,
and consolidation therapy after, double autologous stem-cell
transplantation in newly diagnosed multiple myeloma: A randomised 3
study. Lancet. 376:2075–2085. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Miguel JS, Weisel K, Moreau P, Lacy M,
Song K, Delforge M, Karlin L, Goldschmidt H, Banos A, Oriol A, et
al: Pomalidomide plus low-dose dexamethasone versus high-dose
dexamethasone alone for patients with relapsed and refractory
multiple myeloma (MM-003): A randomised, open-label, phase 3 trial.
Lancet Oncol. 14:1055–1066. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Singhal S, Mehta J, Desikan R, Ayers D,
Roberson P, Eddlemon P, Munshi N, Anaissie E, Wilson C, Dhodapkar
M, et al: Antitumor activity of thalidomide in refractory multiple
myeloma. N Engl J Med. 341:1565–1571. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Palumbo A, Hajek R, Delforge M, Kropff M,
Petrucci MT, Catalano J, Gisslinger H, Wiktor-Jędrzejczak W,
Zodelava M, Weisel K, et al: Continuous lenalidomide treatment for
newly diagnosed multiple myeloma. N Engl J Med. 366:1759–1769.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Dimopoulos M, Spencer A, Attal M, Prince
HM, Harousseau JL, Dmoszynska A, San Miguel J, Hellmann A, Facon T,
Foà R, et al: Lenalidomide plus dexamethasone for relapsed or
refractory multiple myeloma. N Engl J Med. 357:2123–2132. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Drexler HG and MacLeod RAF: Malignant
hematopoietic cell lines: In vitro models for the study of
plasmacytoid dendritic cell leukemia. Leuk Res. 33:1166–1169. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Yang L, Xie J, Almoallim HS, Alharbi SA
and Chen Y: Tomentosin inhibits cell proliferation and induces
apoptosis in MOLT-4 leukemia cancer cells through the inhibition of
mTOR/PI3K/Akt signaling pathway. J Biochem Mol Toxicol.
35:e227192021. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
El Yaagoubi OM, Lahmadi A, Bouyahya A,
Filali H, Samaki H, El Antri S and Aboudkhil S: Antitumor effect of
Inula viscosa extracts on DMBA-induced skin carcinoma are mediated
by proteasome inhibition. Biomed Res Int. 2021:66875892021.
View Article : Google Scholar :
|
|
47
|
Zeissig MN, Hewett DR, Panagopoulos V,
Mrozik KM, To LB, Croucher PI, Zannettino ACW and Vandyke K:
Expression of the chemokine receptor CCR1 promotes the
dissemination of multiple myeloma plasma cells in vivo.
Haematologica. Nov 5–2020.Epub ahead of print. View Article : Google Scholar
|
|
48
|
Dairaghi DJ, Oyajobi BO, Gupta A,
McCluskey B, Miao S, Powers JP, Seitz LC, Wang Y, Zeng Y, Zhang P,
et al: CCR1 blockade reduces tumor burden and osteolysis in vivo in
a mouse model of myeloma bone disease. Blood. 120:1449–1457. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Ganghammer S, Gutjahr J, Hutterer E, Krenn
PW, Pucher S, Zelle-Rieser C, Jöhrer K, Wijtmans M, Leurs R, Smit
MJ, et al: Combined CXCR3/CXCR4 measurements are of high prognostic
value in chronic lymphocytic leukemia due to negative
co-operativity of the receptors. Haematologica. 101:e99–e102. 2016.
View Article : Google Scholar :
|
|
50
|
Mulligan AM, Raitman I, Feeley L,
Pinnaduwage D, Nguyen LT, O'Malley FP, Ohashi PS and Andrulis IL:
Tumoral lymphocytic infiltration and expression of the chemokine
CXCL10 in breast cancers from the ontario familial breast cancer
registry. Clin Cancer Res. 19:336–346. 2013. View Article : Google Scholar :
|
|
51
|
Bolomsky A, Schreder M, Hübl W, Zojer N,
Hilbe W and Ludwig H: Monokine induced by interferon gamma
(MIG/CXCL9) is an independent prognostic factor in newly diagnosed
myeloma. Leuk Lymphoma. 57:2516–2525. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Guillerey C, Huntington ND and Smyth MJ:
Targeting natural killer cells in cancer immunotherapy. Nat
Immunol. 17:1025–1036. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Bonanni V, Antonangeli F, Santoni A and
Bernardini G: Targeting of CXCR3 improves anti-myeloma efficacy of
adoptively transferred activated natural killer cells. J Immunother
Cancer. 7:2902019. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Vijay K: Toll-like receptors in immunity
and inflammatory diseases: Past, present, and future. Int
Immunopharmacol. 59:391–412. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Chiron D, Bekeredjian-Ding I,
Pellat-Deceunynck C, Bataille R and Jego G: Toll-like receptors:
Lessons to learn from normal and malignant human B cells. Blood.
112:2205–2213. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Xu Y, Zhao Y, Huang H, Chen G, Wu X, Wang
Y, Chang W, Zhu Z, Feng Y and Wu D: Expression and function of
toll-like receptors in multiple myeloma patients: Toll-like
receptor ligands promote multiple myeloma cell growth and survival
via activation of nuclear factor-kappaB. Br J Haematol.
150:543–553. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Rossi D: Role of MYD88 in
lymphoplasmacytic lymphoma diagnosis and pathogenesis. Hematology
Am Soc Hematol Educ Program. 2014:113–118. 2014. View Article : Google Scholar
|
|
58
|
de Boussac H, Bruyer A, Jourdan M, Maes A,
Robert N, Gourzones C, Vincent L, Seckinger A, Cartron G, Hose D,
et al: Kinome expression profiling to target new therapeutic
avenues in multiple myeloma. Haematologica. 105:784–795. 2020.
View Article : Google Scholar :
|
|
59
|
Crawley JB, Williams LM, Mander T, Brennan
FM and Foxwell BMJ: Interleukin-10 stimulation of
phosphatidylinositol 3-kinase and p70 S6 kinase is required for the
proliferative but not the antiinflammatory effects of the cytokine.
J Biol Chem. 271:16357–16362. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Zhou JH, Broussard SR, Strle K, Freund GG,
Johnson RW, Dantzer R and Kelley KW: IL-10 inhibits apoptosis of
promyeloid cells by activating insulin receptor substrate-2 and
phosphatidylinositol 3′-kinase. J Immunol. 167:4436–4442. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Zhan F, Huang Y, Colla S, Stewart JP,
Hanamura I, Gupta S, Epstein J, Yaccoby S, Sawyer J, Burington B,
et al: The molecular classification of multiple myeloma. Blood.
108:2020–2028. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Zhang PP, Wang YC, Cheng C, Zhang F, Ding
DZ and Chen DK: Runt-related transcription factor 2 influences cell
adhesion-mediated drug resistance and cell proliferation in B-cell
non-Hodgkin's lymphoma and multiple myeloma. Leuk Res. 92:106340Mar
9–2020. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Luo JH: Oncogenic activity of MCM7
transforming cluster. World J Clin Oncol. 2:120–124. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Cortez D, Glick G and Elledge SJ:
Minichromosome maintenance proteins are direct targets of the ATM
and ATR checkpoint kinases. Proc Natl Acad Sci USA.
101:10078–10083. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Tsao CC, Geisen C and Abraham RT:
Interaction between human MCM7 and Rad17 proteins is required for
replication checkpoint signaling. EMBO J. 23:4660–4669. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Tian L, Liu J, Xia GH and Chen BA:
RNAi-mediated knockdown of MCM7 gene on CML cells and its
therapeutic potential for leukemia. Med Oncol. 34:212017.
View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Moll UM and Petrenko O: The MDM2-p53
interaction. Mol Cancer Res. 1:1001–1008. 2003.
|
|
68
|
Vassilev LT, Vu BT, Graves B, Carvajal D,
Podlaski F, Filipovic Z, Kong N, Kammlott U, Lukacs C, Klein C, et
al: In vivo activation of the p53 pathway by small-molecule
antagonists of MDM2. Science. 303:844–848. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Teoh PJ, Chung TH, Sebastian S, Choo SN,
Yan J, Ng SB, Fonseca R and Chng WJ: P53 haploinsufficiency and
functional abnormalities in multiple myeloma. Leukemia.
28:2066–2074. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Saha MN, Qiu L and Chang H: Targeting p53
by small molecules in hematological malignancies. J Hematol Oncol.
6:232013. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Teoh PJ and Chng WJ: P53 abnormalities and
potential therapeutic targeting in multiple myeloma. Biomed Res
Int. 2014:7179192014. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Halder SK, Beauchamp RD and Datta PK:
Smad7 induces tumorigenicity by blocking TGF-beta-induced growth
inhibition and apoptosis. Exp Cell Res. 307:231–246. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Chen R, Wierda WG, Chubb S, Hawtin RE, Fox
JA, Keating MJ, Gandhi V and Plunkett W: Mechanism of action of
SNS-032, a novel cyclin-dependent kinase inhibitor, in chronic
lymphocytic leukemia. Blood. 113:4637–4645. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Czudor Z, Balogh M, Bánhegyi P, Boros S,
Breza N, Dobos J, Fábián M, Horváth Z, Illyés E, Markó P, et al:
Novel compounds with potent CDK9 inhibitory activity for the
treatment of myeloma. Bioorganic Med Chem Lett. 28:769–773. 2018.
View Article : Google Scholar
|
|
75
|
Hartl FU: Molecular chaperones in cellular
protein folding. Nature. 381:571–580. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Muralidharan S and Mandrekar P: Cellular
stress response and innate immune signaling: Integrating pathways
in host defense and inflammation. J Leukoc Biol. 94:1167–1184.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Chatterjee M, Andrulis M, Stühmer T,
Müller E, Hofmann C, Steinbrunn T, Heimberger T, Schraud H,
Kressmann S, Einsele H and Bargou RC: The PI3K/Akt signaling
pathway regulates the expression of Hsp70, which critically
contributes to Hsp90-chaperone function and tumor cell survival in
multiple myeloma. Haematologica. 98:1132–1141. 2013. View Article : Google Scholar :
|
|
78
|
Zhang L, Fok JJL, Mirabella F, Aronson LI,
Fryer RA, Workman P, Morgan GJ and Davies FE: Hsp70 inhibition
induces myeloma cell death via the intracellular accumulation of
immunoglobulin and the generation of proteotoxic stress. Cancer
Lett. 339:49–59. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Braunstein MJ, Scott SS, Scott CM, Behrman
S, Walter P, Wipf P, Coplan JD, Chrico W, Joseph D, Brodsky JL and
Batuman O: Antimyeloma effects of the heat shock protein 70
molecular chaperone inhibitor MAL3-101. J Oncol. 2011:2320372011.
View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Workman P and Davies FE: A stressful life
(or death): Combinatorial proteotoxic approaches to
cancer-selective therapeutic vulnerability. Oncotarget. 2:277–280.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Tiku V, Tan MW and Dikic I: Mitochondrial
functions in infection and immunity: (Trends in Cell Biology 30,
263-275, 2020). Trends Cell Biol. 30:7482020. View Article : Google Scholar : PubMed/NCBI
|