Introduction
Osteoarthritis (OA) is a degenerative disease characterized by cartilage destruction and loss, subchondral bone thickening and hardening, and osteophyte formation accompanied by varying degrees of synovitis and joint capsule thickening (1). The occurrence of OA is associated with certain factors, including mechanical instability, age, inflammation, trauma and genetics (2). At present, no effective disease-modifying therapies exist for OA due to the limited understanding of its pathogenesis. Although studies have identified elements that are involved in the progression of OA, the precise mechanism of OA pathogenesis has not yet been elucidated.
Articular cartilage directly participates in maintaining normal joint motor functions and is primarily composed of cartilage extracellular matrix (ECM) and cartilage cells (3). Articular cartilage cells are the only type of cell found in articular cartilage (4), and maintain the metabolic balance of cartilage tissue by producing ECM molecules, including collagens and proteoglycans, as well as matrix-degrading enzymes, such as matrix metalloproteinases (MMPs) (5). The pathological destruction of joint tissues in OA is primarily due to the reduced ability of chondrocytes to generate articular cartilage ECM and the enhanced action of chondrocytes to generate catabolic matrix-degrading enzymes (6). Previous studies have demonstrated that the imbalance in chondrocyte matrix metabolism caused by mechanical instability is a critical factor in the development of OA (7,8). Therefore, exploring the molecular mechanisms by which mechanical instability causes the imbalance in chondrocyte matrix metabolism is important for prevention and treatment of OA.
Circular RNAs (circRNAs/circs) are a novel type of RNA that, unlike linear RNAs, form a covalently closed continuous loop and are highly represented in the eukaryotic transcriptome. A large number of circRNAs are derived from exon sequences and are located in the cytoplasm of eukaryotic cells (9). Numerous studies have demonstrated that some circRNAs are involved in the development of OA, such as circRNA.33186, circSERPINE2 and circCDK14 (10-13). Previous studies have confirmed that the most important regulation method of circRNAs involves competitive endogenous RNAs adsorbing microRNAs (miRNAs/miRs) to regulate the expression of downstream genes (14,15). Thus, exploring the roles of circRNAs and their potential miRNA regulators is critical for identifying the molecular mechanisms of OA.
The present study aimed to investigate whether circRNAs might serve a role in mechanical instability-regulated chondrocyte matrix metabolism in OA. The expression levels of mechanics-related circPhc3 in human and mouse OA cartilage samples were measured using reverse transcriptionquantitative PCR (RT-qPCR) and fluorescence in situ hybridization (FISH). The effects of circPhc3 on chondrocyte ECM metabolism were further investigated by overexpressing and knocking down circPhc3 in OA chondrocytes. The downstream target of circPhc3 was examined by performing a luciferase reporter assay. The results of present study revealed a fundamental role of circPhc3 in the progression of OA and provided a potential drug target for OA therapy.
Materials and methods
Primary OA and traumatic OA specimen selection
Healthy patients (n=10; 6 female patients and 4 male patients; age, 45±18 years) and 10 patients with knee OA (n=10; 6 female patients and 4 male patients; age, 54±17 years) were enrolled in Tongji Hospital affiliated to Tongji University (Shanghai, China) between October 2020 and December 2020. Healthy human knee cartilage samples were obtained from those patients undergoing arthroscopic partial meniscectomy with no evidence for OA. Patients presenting with cartilage chondrosis, bone-marrow lesions/edema and no ligamentous injury > grade I medial-collateral ligament strain were included as donors for healthy human knee cartilage samples. Human OA knee cartilage samples were obtained from patients with a diagnosis of advanced OA who underwent total knee arthroplasty. Knees were assessed by radiographs using the Kellgren-Lawrence (KL) scale (16) for OA. Any knees presenting imaging signs of previous bone fracture, previous knee surgery (including knee replacement procedures, ligamentoplasty and cartilage repair procedures) or poor image quality were excluded from the present study. All individuals provided written informed consent for the use of human specimens for experiments. The present study was approved by the Ethics Committee of Tongji Hospital of Tongji University (approval no. 2020TJYLL287).
Animal experiments
C57BL/6 mice (n=36; age, 8 weeks; weight, 20-30 g) were kept under standard animal room conditions at 25±3°C with 12-h light/dark cycles, 60% relative humidity and ad libitum access to food and water. Mice were divided randomly into the following two groups (n=18 per group): i) Sham operation; and ii) OA. The OA group underwent destabilized medial meniscus (DMM) surgery to trigger OA. Briefly, following induction of anaesthesia using an intraperitoneal injection of 1% sodium pentobarbital (50 mg/kg), the joint capsule was opened. The medial meniscotibial ligament was transected with microscissors, and the medial meniscus was displaced medially. After washing with saline to remove tissue debris, the skin incision was closed. In the Sham operation group, only the joint cavity was opened and then the skin incision was closed with suture. The health and behaviour of OA model mice and Sham mice were monitored once a day. No accidental death of animals was observed during the experiment. After 8 weeks, the mice were euthanized. Rapid weight loss (15-20% drop in one day) and some symptoms of the central nervous system (including head and neck skewing, deformity, epilepsy, turning circle and paralysis) were considered as humane endpoints of the animal experiment. All mice (n=36) were euthanised at the end of the study by cervical dislocation under deep anaesthesia. Subsequently, the joints were collected for mRNA quantification, histological assessments and cell culture. All animal care and experimental procedures were approved by the Institutional Animal Care and Use Committee of Tongji Hospital of Tongji University (approval no. IACUC20200422) and were carried out in accordance with the Guide for Laboratory Animal Use (https://life.tongji.edu.cn/05/e2/c12677a132578/page.htm).
Immunofluorescence staining
Mouse chondrocytes were fixed with 4% paraformaldehyde for 30 min at room temperature and then washed three times with PBS. Subsequently, cells were permeabilized with 0.4% Trion X-100, followed by blocking with 10% donkey serum (Abcam) for 1 h at room temperature. The samples and cells were then incubated with anti-collagen type II, α1 (Col2a1; 1:200; cat. no. sc-52658; Santa Cruz Biotechnology, Inc.), anti-Aggrecan (1:200; cat. no. MA3-16888; Invitrogen; Thermo Fisher Scientific, Inc.) and anti-MMP-3 (1:200; cat. no. ab39012; Abcam) primary antibodies in blocking solution at 4°C overnight. Next, the samples and cells were incubated with the corresponding mouse immunoglobulin G (H+L) (Alexa Fluor 488, 1:1,000; cat. no. A-11001; Invitrogen; Thermo Fisher Scientific, Inc.) and rabbit immunoglobulin G (H+L) secondary antibodies (Alexa Fluor 555; 1:1,000; cat. no. A-21428; Invitrogen; Thermo Fisher Scientific, Inc.) secondary antibodies (both Invitrogen; Thermo Fisher Scientific, Inc.) and DAPI for 1 h at room temperature. A fluorescence microscope (Carl Zeiss AG) was used to observe stained samples.
Histological analysis
Human and mice tissue samples were fixed in 4% paraformaldehyde for 30 min at room temperature and then decalcified with ethylenediaminetetraacetic acid-buffered saline solution (pH 7.4; 0.25 M). After conventional dehydration and paraffin embedding, specimens were sliced into 4-µm sections, and routinely dewaxed and washed with xylene. Hematoxylin and eosin (H&E) staining was performed, in which the slices were stained with hematoxylin for 5 min, followed by staining with 0.3% eosin solution for 3 min (Beyotime Institute of Biotechnology) at room temperature. For safranin-O/fast green staining, the slices were stained with solid green dye solution for 5 min, and then stained with Safranin O stain (Beijing Solarbio Science & Technology Co., Ltd.) for 2 min at room temperature. For toluidine blue staining, the slices were stained with 1% toluidine blue solution for 10 min at room temperature, then washed with distilled water for 5 min. After staining, specimens were dehydrated in an ascending alcohol and sealed with neutral resin. Histological changes from H&E and toluidine blue staining were observed. Proteoglycans/collagen contents were observed with safranin O-solid green staining. Staining was observed using a light microscope.
Chondrocyte culture and tensile strain loading
Cartilage was extracted from OA patients and healthy patients, followed by washing twice with PBS. The tissues were cut into 2 mm2 pieces, and then digested with 1 mg/ml protease K (Sigma-Aldrich; Merck KGaA) for 30 min and 2 mg/ml collagenase overnight at 37°C. Human chondrocytes were collected by centrifugation (10,000 × g for 10 min) at room temperature. Mouse chondrocytes were isolated from articular cartilage in the knee joints of mice as previously described (17). Briefly, articular cartilage tissues were cut into small pieces (<1 mm3), followed by digestion with 0.25% trypsin and 0.2% type II collagenase (Roche Diagnostics GmbH). Following centrifugation (10,000 × g for 10 min) at room temperature, cells were cultured in DMEM/F12 media (Gibco; Thermo Fisher Scientific, Inc.) with 10% fetal bovine serum (FBS; Gibco; Thermo Fisher Scientific, Inc.) and 1% antibiotics at 37°C under 5% CO2. Chondrocytes were identified by Alcian blue staining. Briefly, chondrocytes were fixed in 4% paraformaldehyde for 30 min at room temperature. Subsequently, 1% alcian blue solution was added to the cells for 12 h at room temperature. Cells were washed with distilled water for 5 min and then visualized using a light microscope.
At 70-80% confluence, cells were subjected to cyclic tensile strain with a 0.5 Hz sinusoidal curve at 5-20% elongation for 6-24 h using a Flexcell1 FX-5000 tension system (Flexcell International Corporation). The chondrocytes in the control group were cultured without intervention.
FISH
Mice articular cartilage tissue samples and chondrocytes were fixed in 4% paraformaldehyde for 30 min at room temperature. Mice tissue samples were decalcified with ethylenediaminetetraacetic acid-buffered saline solution (pH 7.4; 0.25 M). After conventional dehydration and paraffin embedding, specimens were sliced into 4-µm sections, and then routinely dewaxed and washed with xylene. An Alexa Fluor 488-labeled circPhc3 probe was designed and synthesized by Invitrogen (Thermo Fisher Scientific, Inc.). The probe signals were tested in articular cartilage tissue and chondrocytes with the Fluorescent in Situ Hybridization kit (Shanghai GenePharma Co., Ltd.) according to the manufacturer's protocol. FISH results were acquired using laser scanning confocal microscopy (LSM800; Carl Zeiss, AG). ImageJ software (National Institutes of Health) was used to analyse the immunofluorescence accumulation optical density of circPhc3 in cartilage samples.
Cell transfection
The overexpression plasmid vector of mouse circPhc3 was constructed by Shanghai GeneChem Co., Ltd. The circPhc3 sequence (1 kb upstream) was inserted into the pcDNA3.1 His c plasmid (Shanghai GeneChem Co., Ltd.). circPhc3 without the downstream reverse sequence was used as the Mock group. A short hairpin RNAs (shRNAs) directed against circPhc3 was constructed into the pGPU6/GFP/Neo vector (Shanghai GeneChem Co., Ltd.) to generate silencing plasmids. A non-specific scrambled shRNA was used as the negative control (NC). miR-93-3p mimic, miR-93-3p mimic-NC, anti-miRNA oligonucleotides (AMO)-93-3p and AMO-NC were purchased from Shanghai GeneChem Co., Ltd. Chondrocytes were seeded (1×105 cells/cm2). At 70% confluence, cells were transfected with 1 µg plasmid DNA or 100 nM miRNA mimic, AMO or corresponding NCs using Lipofectamine® 3000 reagent (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol at 37°C. At 24 h post-transfection, transfection efficiency was determined via RT-qPCR. All functional experiments were performed at 48 h post-transfection. The sequences of the constructs used in the present study were as follows: miR-93-3p mimic, 5′-ACU GCU GAG CAC UUC CCG-3′; AMO-93-3p, 5′-CGG GAA GUG CUC AGC AGU-3′; mimic NC, 5′-UUC UCC GAA CGU GUC ACG UTT-3′; AMO-NC, 5′-CAG UAC UUU UGU GUA GUA CAA-3′; circPhc3 shRNA, 5′-AGG TAA TTC AGC AGG CCT TGC-3′; and shRNA-NC, 5′-TTC TCC GAA CGT GTC ACG-3′.
Western blotting
Chondrocytes were harvested and lysed with RIPA lysis buffer (Beyotime Institute of Biotechnology) as previously described (18). The total protein concentration was quantified using the bicinchoninic acid protein assay kit (Pierce; Thermo Fisher Scientific, Inc.). Proteins (50 µg) were separated via 7.5-10% SDS-PAGE and transferred to nitrocellulose membranes (Cytiva). After blocking with 5% dry milk in TBST (0.1% Tween-20) for 1 h, the membranes were incubated overnight at 4°C with primary antibodies targeted against recombinant MMP-3 (1:1,000; cat. no. ab52915, Abcam), Aggrecan (1:500; cat. no. MA3-16888; Invitrogen; Thermo Fisher Scientific, Inc.), Col2α1 (1:200; cat. no. sc-52658; Santa Cruz Biotechnology, Inc.), FoxO1 (1:1,000; cat. no. ab179450; Abcam), MMP-13 (1:1,000; cat. no. ab39012; Abcam), a disintegrin and metalloprotease with thrombospondin type 1 motif 5 (ADAMTS5; 1:1,000; cat. no. ab41037; Abcam) and β-actin (1:1,000; cat. no. ab8226; Abcam). Subsequently, the membranes were incubated with horseradish peroxidase-conjugated rabbit anti-mouse IgG H&L (1:1,000; cat. no. ab190475; Abcam) and goat anti-rabbit IgG H&L (1:1,000; cat. no. ab205718; Abcam) secondary antibodies for 1 h at room temperature. Target bands were visualized using Pierce™ ECL Western Blotting Substrate (Thermo Fisher Scientific, Inc.). ImageJ software (version 1.8.0; National Institutes of Health) was used to semi-quantify protein expression with β-actin as the loading control.
RT-qPCR
Total RNA was isolated from chondrocytes and purified using the RNeasy Mini Kit (Qiagen GmbH) as previously described (19). Total RNA (1 µg) was reverse transcribed into cDNA using Superscript II reverse-transcriptase (Thermo Fisher Scientific, Inc.). Subsequently, qPCR was performed using the SYBR Premix Ex Tag kit (Takara Bio, Inc.) and an ABI 7500 sequencing-detection system (Applied Biosystems; Thermo Fisher Scientific, Inc.). The following thermocycling conditions were used for qPCR: 95°C for 3 min; 40 cycles of 95°C for 10 sec and 60°C for 30 sec. A melt curve step from 65-95°C was performed in increments of 0.5°C per 5 sec. The sequences of the primers used in the present study are listed in Table I. The relative mRNA and miRNA expression levels were quantified using the 2-ΔΔCq method (20) and normalized to the internal reference genes β-actin and U6, respectively.
 |
Table I
Sequences of primers used for reverse transcription-quantitative PCR.
|
Table I
Sequences of primers used for reverse transcription-quantitative PCR.
| Gene |
Sequence (5′→3′) |
| Mouse Col2α1 |
F: GGGAATGTCCTCTGCGATGAC |
| R: GAAGGGGATCTCGGGGTTG |
| Mouse Aggrecan |
F: CCTGCTACTTCATCGACCCC |
| R: AGATGCTGTTGACTCGAACCT |
| Mouse MMP-3 |
F: ACATGGAGACTTTGTCCCTTTT |
| R: TTGGCTGAGTGGTAGAGTCCC |
| Mouse β-actin |
F: GGCTGTATTCCCCTCCATCG |
| R: CCAGTTGGTAACAATGCCATGT |
| Human Col2α1 |
F: TGGACGCCATGAAGGTTTTCT |
| R: TGGGAGCCAGATTGTCATCTC |
| Human Aggrecan |
F: ACTCTGGGTTTTCGTGACTCT |
| R: ACACTCAGCGAGTTGTCATGG |
| Human MMP-3 |
F: CTGGACTCCGACACTCTGGA |
| R: CAGGAAAGGTTCTGAAGTGACC |
| Human β-actin |
F: CATGTACGTTGCTATCCAGGC |
| R: CTCCTTAATGTCACGCACGAT |
| mmu_circ-0011776 |
F: CATGTACGTTGCTATCCAGGC |
| R: TGTTTGTCATCCCCCAGGGT |
| mmu_circ_0009580 |
F: TTGGCAAGGTCACTGGGAGC |
| R: GGTAATCGGCTGTCCCATCC |
| mmu_circ_0006232 |
F: ATGCCTGTGTGAAGCATCCTG |
| R: AACTTGTCAAAGCGGGCGAA |
| mmu_circ_0000436 |
F: TTCGCCCAGACATGTACCCC |
| R: AACAGCAGCAGCTATCTCAACA |
| mmu_circ_0008444 |
F: AGGCCAAACAACAAGATCAGC |
| R: TCCTTGGGACTGGGCTCTTC |
| mmu_circ_0009975 |
F: TCTGTGTCAGATGACGGAGGG |
| R: AGAGGCCACGAGGTCAAGAA |
| mmu_circ_0000972 |
F: TGCATCTACAAGATGACGGAG |
| R: GCCACGAGGTCAAGAAGCTC |
| mmu_circ_0010621 |
F: CCCAGGTCTGGAATCTCGGA |
| R: GCAAGGCCTGCTGAATTACCT |
| mmu_circ_0008589 |
F: AAGCTGTGATAACGCGTCCG |
| R: TACTAAGCTCGCTGATCCTGC |
| mmu-miR-93-3p |
F: ACACTCCAGCTGGGCAAAGTGCTGTTCGTGC |
| R: CTCAACTGGTGTCGTGGAGTCG GCAATTCAGTTGAGCGGGAAGT |
| mmu-U6 |
F: CTCGCTTCGGCAGCACATATACT |
| R: ACGCTTCACGAATTTGCGTGTC |
Bioinformatics analysis
The bioinformatics database CircNet (http://circnet.mbc.nctu.edu.tw/) was used to predict the potential target miRNAs of circPhc3, which were further predicted by Shanghai Kangcheng Biotech. The miRDB (http://mirdb.org/) and TargetScanHuman (www.targetscan.org) databases were used to predict the potential target genes of miR-93-3p.
Luciferase reporter assays
To reveal the interaction between circPhc3 and miR-93-3p, a circPhc3 sequence containing the complementary sequences of miR-93-3p was chemically synthesized and cloned downstream of the firefly luciferase gene in the pMIR-REPORT™ reporter vector (Thermo Fisher Scientific, Inc.). After incubation in DMEM (Thermo Fisher Scientific, Inc.) supplemented with 0.5% FBS for 48 h, 293T cells (The Cell Bank of Type Culture Collection of The Chinese Academy of Sciences; 1×105 cells/well) were co-transfected with 1 µg pMIR-circPhc3, 50 ng pRL-TK vector (Promega Corporation) and 100 nM miR-93-3p mimic or miR-NC using Lipofectamine 3000 for 48 h at 37°C. At 48 h post-transfection, cells were collected and luciferase activities were measured using a dual luciferase reporter assay kit (Promega Corporation). Firefly luciferase activities were normalized to Renilla luciferase activities.
Statistical analysis
Data are presented as the mean ± SD from three or more independent experiments. A two-sided unpaired Student's t-test was used to analyse the difference between two groups. Comparisons among multiple groups were analysed using one-way ANOVA followed by Tukey's post hoc test. Statistical analyses were performed using the NCSS statistical software (version 21.0.3; NCSS Statistical Software). P<0.05 was considered to indicate a statistically significant difference.
Results
circPhc3 expression is reduced in human and mouse OA cartilage
OA mouse models were established to determine the expression levels of mechanics-related circRNAs in OA cartilage. Alterations in articular cartilage in the DMM-induced mouse model of OA were observed by safranin O (Fig. 1A), H&E (Fig. 1B) and toluidine blue (Fig. 1C) staining. The results showed a greater loss of integrity and more severe destruction in OA mouse cartilage compared with that in Sham mouse cartilage. Subsequently, the expression levels of Col2α1, Aggrecan and MMP-3 in OA mouse cartilage were assessed by RT-qPCR (Fig. 1D). The mRNA expression levels of Col2α1 and Aggrecan were significantly decreased in OA mouse cartilage compared with those in Sham mouse cartilage, whereas MMP-3 mRNA expression was significantly increased in OA mouse cartilage, indicating that the mouse model of OA was successfully established. The expression levels of mechanics-related circRNAs were then measured in OA mouse cartilage, where circ_0011776, circ_0009580, circ_0004001, circ_0008444 and circ_0009975 were significantly upregulated, whereas circ_0006232, circ_0000436, circ_0000972, circ_0010621 (circPhc3) and circ_0008589 were significantly downregulated in OA mouse cartilage compared with that in Sham mouse cartilage (Fig. 1E). circPhc3 displayed the lowest expression levels among the differentially expressed circRNAs. Fluorescence in situ hybridization (FISH) staining showed that the expression of circPhc3 was decreased in OA mouse cartilage, which was evidenced by the significantly reduced green fluorescence of circPhc3 observed in OA mouse cartilage compared with that in Sham mouse cartilage (Fig. 1F and G). Furthermore, the expression of circPhc3 in knee articular cartilage from patients with OA was measured. Safranin-O staining of articular cartilage samples from patients with OA showed that the articular cartilage had suffered serious destruction (Fig. 1H), which was accompanied by significantly increased expression levels of MMP-3 and decreased expression levels of Col2α1 and Aggrecan in the articular cartilage samples of patients with OA compared with those from healthy patients (Fig. 1I). The RT-qPCR results showed that the expression level of circPhc3 in the cartilage of patients with OA was significantly lower compared with that in the cartilage of healthy patients (Fig. 1J).
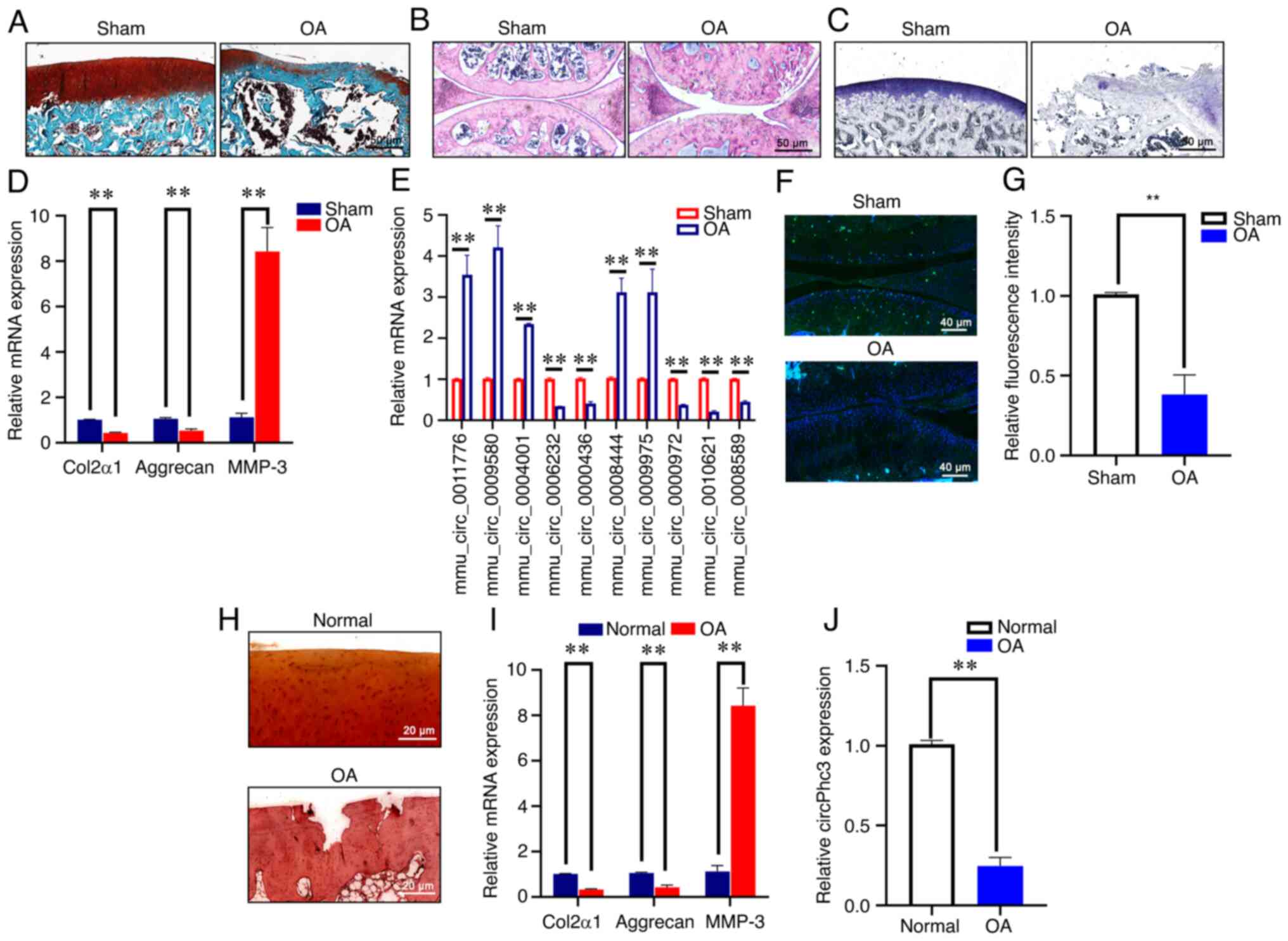 |
Figure 1
circPhc3 expression is reduced in human and mouse OA cartilage. (A-C) Changes in articular cartilage in the destabilized medial meniscus-induced mouse model of OA were observed by (A) safranin O-fast green, (B) hematoxylin and eosin and (C) toluidine blue staining (n=6). (D) mRNA expression levels of Col2α1, Aggrecan and MMP-3 in mouse OA cartilage were analysed by RT-qPCR. **P<0.01 (n=4). (E) Expression levels of mechanics-related circRNAs in mouse OA cartilage were analysed by RT-qPCR. **P<0.01 (n=4). (F) Expression of circPhc3 in mouse OA cartilage was (F) detected by fluorescence in situ hybridization staining and (G) quantified. **P<0.01 (n=4). (H) Changes in articular cartilage in patients with OA were observed by safranin O staining (n=6). (I) mRNA expression levels of Col2α1, Aggrecan and MMP-3 in cartilage tissues of patients with were analysed by RT-qPCR. **P<0.01 (n=4). (J) Expression levels of circPhc3 in cartilage tissues of patients with OA were analysed by RT-qPCR. **P<0.01 (n=4). circ, circular RNA; OA, osteoarthritis; Col2α1, collagen type II, α1; MMP, matrix metalloproteinase; RT-qPCR, reverse transcription-quantitative PCR; mmu, Mus musculus.
|
circPhc3 expression is reduced in chondrocytes in response to mechanical stimuli
Chondrocytes from OA model mice were isolated and identified by chondrocyte-specific Alcian blue staining (Fig. 2A). FISH staining showed that the expression of circPhc3 was markedly decreased in chondrocytes from OA model mice, as evidenced by a reduction in cytoplasmic green fluorescence compared with that in chondrocytes from Sham mice (Fig. 2B). The RT-qPCR results further confirmed that the expression of circPhc3 was significantly lower in chondrocytes from OA model mice compared with that in Sham mice (Fig. 2C). Next, chondrocytes were isolated from patients with OA and subjected to RT-qPCR. The results showed that the expression of circPhc3 was also significantly decreased in the chondrocytes isolated from patients with OA compared with that in chondrocytes isolated from healthy patients (Fig. 2D). In addition, the expression of circPhc3 in chondrocytes exposed to mechanical load of different strengths and for different time courses was assessed. The results demonstrated that mechanical stimuli reduced the expression of circPhc3 in a time- and strength-dependent manner (Fig. 2E and F).
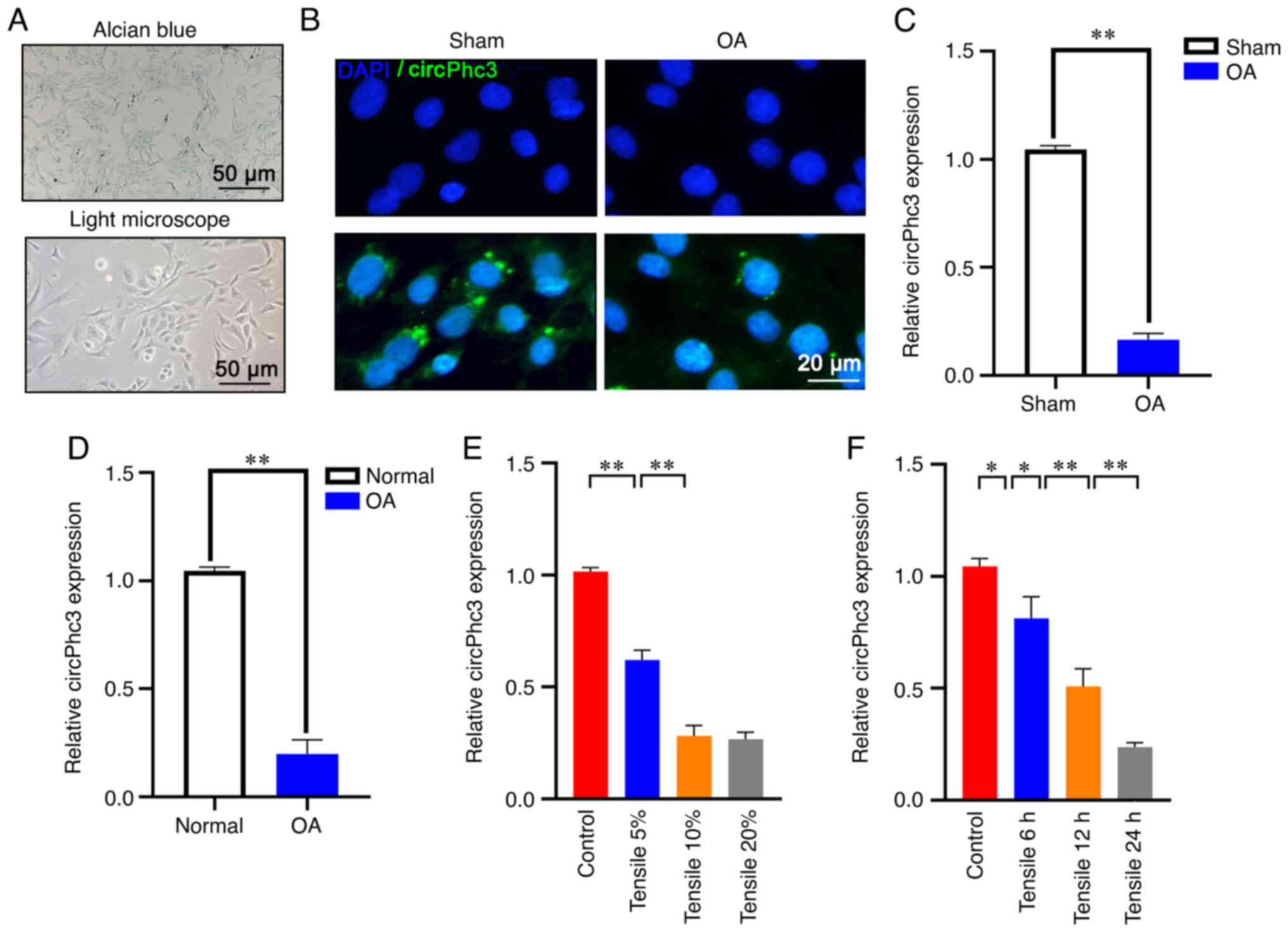 |
Figure 2
circPhc3 expression is reduced in chondrocytes in response to mechanical stimuli. (A) Alcian blue staining was used to identify chondrocytes isolated from OA model mice (n=6). (B) fluorescence in situ hybridization staining demonstrated that circPhc3 expression was decreased in chondrocytes from OA model mice, as evidenced by decreases in cytoplasmic green fluorescence (n=6). (C) circPhc3 expression levels in chondrocytes isolated from OA model mice and Sham mice were analysed by RT-qPCR. **P<0.01 (n=4). (D) circPhc3 expression levels in chondrocytes isolated from patients with OA and healthy patients were analysed by RT-qPCR. **P<0.01 (n=4). (E) circPhc3 expression levels in chondrocytes treated with mechanical load of (E) different strengths or (F) for different time periods. *P<0.05 and **P<0.01 (n=4). circ, circular RNA; OA, osteoarthritis; RT-qPCR, reverse transcription-quantitative PCR.
|
circPhc3 is involved in the secretion of ECM and cartilage degrading enzymes
To investigate the role of circPhc3 in the secretion of ECM and cartilage degrading enzymes, circPhc3 expression was knocked down and overexpressed using shRNA-circPhc3 and circPhc3 overexpression vector, respectively (Fig. 3A and B). The western blotting results showed that silencing circPhc3 significantly increased the expression of MMP-3 and decreased the expression levels of Aggrecan and Col2α1 compared with that in the shRNA-NC group (Fig. 3C-F). By contrast, circPhc3 overexpression displayed the opposite effect on protein expression levels. Similar results observed using RT-qPCR (Fig. 3G-I). Compared with those in the shRNA-NC group, circPhc3-silenced chondrocytes displayed significantly increased mRNA expression levels of MMP-3 and decreased mRNA expression levels of Aggrecan and Col2α1 (Fig. 3G-I). The immunofluorescence staining results revealed that the green fluorescence intensities of Col2α1 and Aggrecan were notably lower in the shRNA-circPhc3 group compared with those in the shRNA-NC group, whereas these fluorescence intensities were obviously stronger in the circPhc3 overexpression group compared with those in the Mock group (Fig. 3J). By contrast, the green fluorescence intensities of MMP-3 were markedly increased in the shRNA-circPhc3 group and lower in the circPhc3-overexpression group compared with those in the shRNA-NC and Mock groups, respectively. Moreover, the effect of circPhc3 on MMP-13 and ADAMTS5 expression levels in chondrocytes was assessed using western blotting. The results showed that no significant changes in MMP-13 and ADAMTS5 expression levels were observed in circPhc3-overexpression or -silenced chondrocytes (Fig. S1). Taken together, these results indicated that circPhc3 promoted the secretion of ECM and inhibited the production of cartilage degrading enzymes.
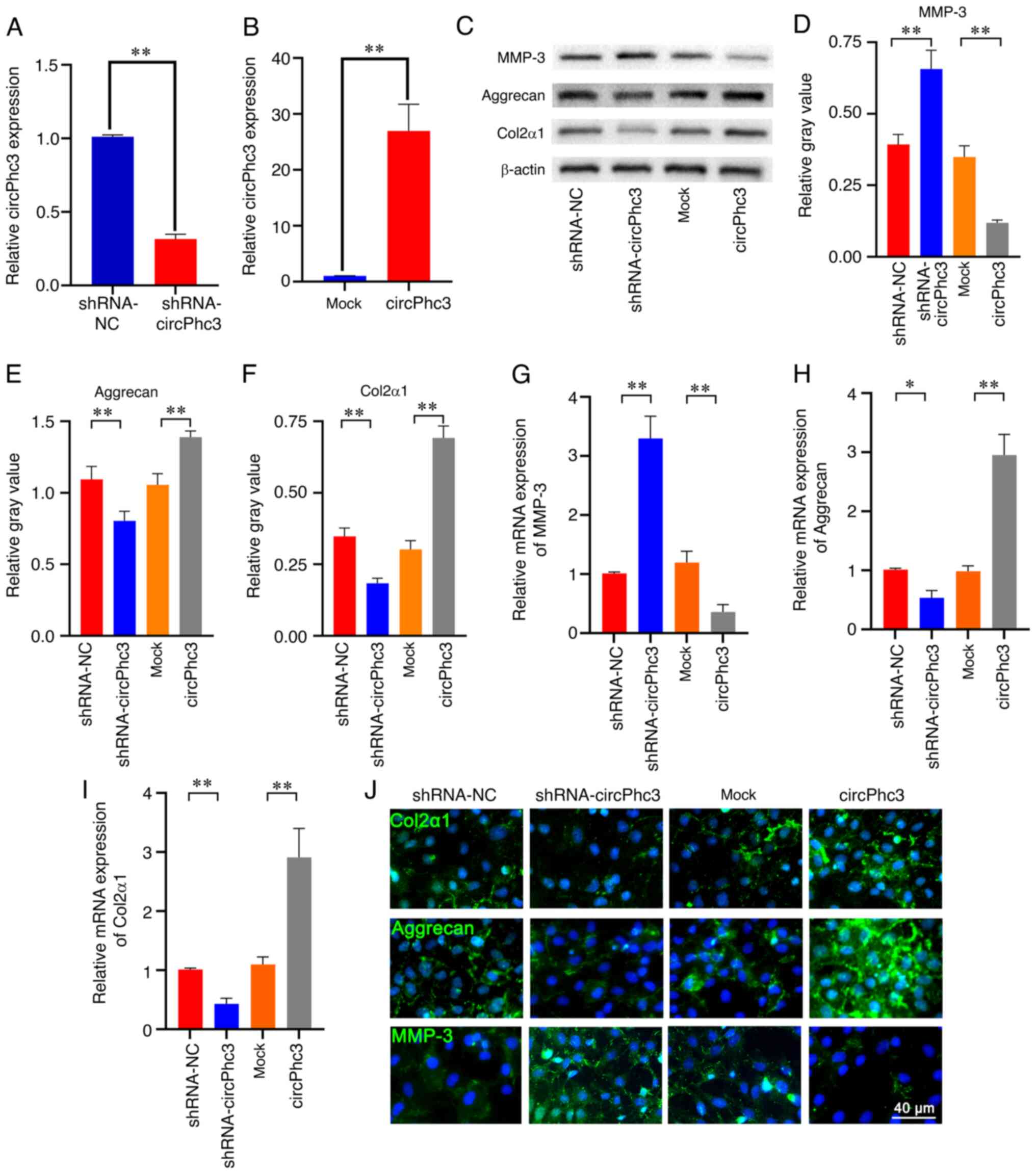 |
Figure 3
circPhc3 is involved in the secretion of extracellular matrix and cartilage-degrading enzymes. (A) circPhc3 expression levels in chondrocytes transfected with shRNA-NC or shRNA-circPhc3 were analysed by RT-qPCR. The results showed that circPhc3 silencing was effective. **P<0.01 (n=4). (B) circPhc3 expression levels in chondrocytes transfected with Mock and circPhc3 overexpression vector were analysed by RT-qPCR. The results showed that circPhc3 overexpression was effective. **P<0.01 (n=4). Protein expression levels were (C) determined by western blotting and semi-quantified for (D) MMP-3, (E) Aggrecan and (F) Col2α1 in chondrocytes transfected with shRNA-NC, shRNA-circPhc3, Mock or circPhc3 overexpression vector. **P<0.01 (n=4). mRNA expression levels of (G) MMP-3, (H) Aggrecan and (I) Col2α1 in chondrocytes transfected with shRNA-NC, shRNA-circPhc3, Mock or circPhc3 overexpression vector were detected by RT-qPCR. *P<0.05 and **P<0.01 (n=4). (J) Col2α1, Aggrecan and MMP-3 expression in chondrocytes transfected with shRNA-NC, shRNA-circPhc3, Mock and circPhc3 overexpression vector were detected by immunofluorescence staining (n=4). circ, circular RNA; shRNA, short hairpin RNA; NC, negative control; RT-qPCR, reverse transcription-quantitative PCR; MMP, matrix metalloproteinase; Col2α1, collagen type II, α1.
|
circPhc3 is involved in mechanical loading-regulated production of ECM and cartilage degrading enzymes
The western blotting results showed that the expression of MMP-3 was significantly increased and the expression levels of Aggrecan and Col2α1 were significantly decreased in chondrocytes treated with 10% tensile for 24 h compared with those in the Control group (Fig. 4A-D). These tensile-induced effects were significantly attenuated by circPhc3 overexpression in chondrocytes. Similar results were observed following immunofluorescence staining (Fig. 4E). The RT-qPCR results further confirmed that mechanical loading significantly promoted the expression of MMP-3 and inhibited the expression levels of both Aggrecan and Col2α1 in chondrocytes compared with those in the Control group (Fig. 4F-H). However, circPhc3 overexpression alleviated the effects of mechanical loading on MMP-3, Aggrecan and Col2α1 expression levels in chondrocytes. These results suggested that circPhc3 was involved in mechanical loading-regulated production of ECM and cartilage degrading enzymes.
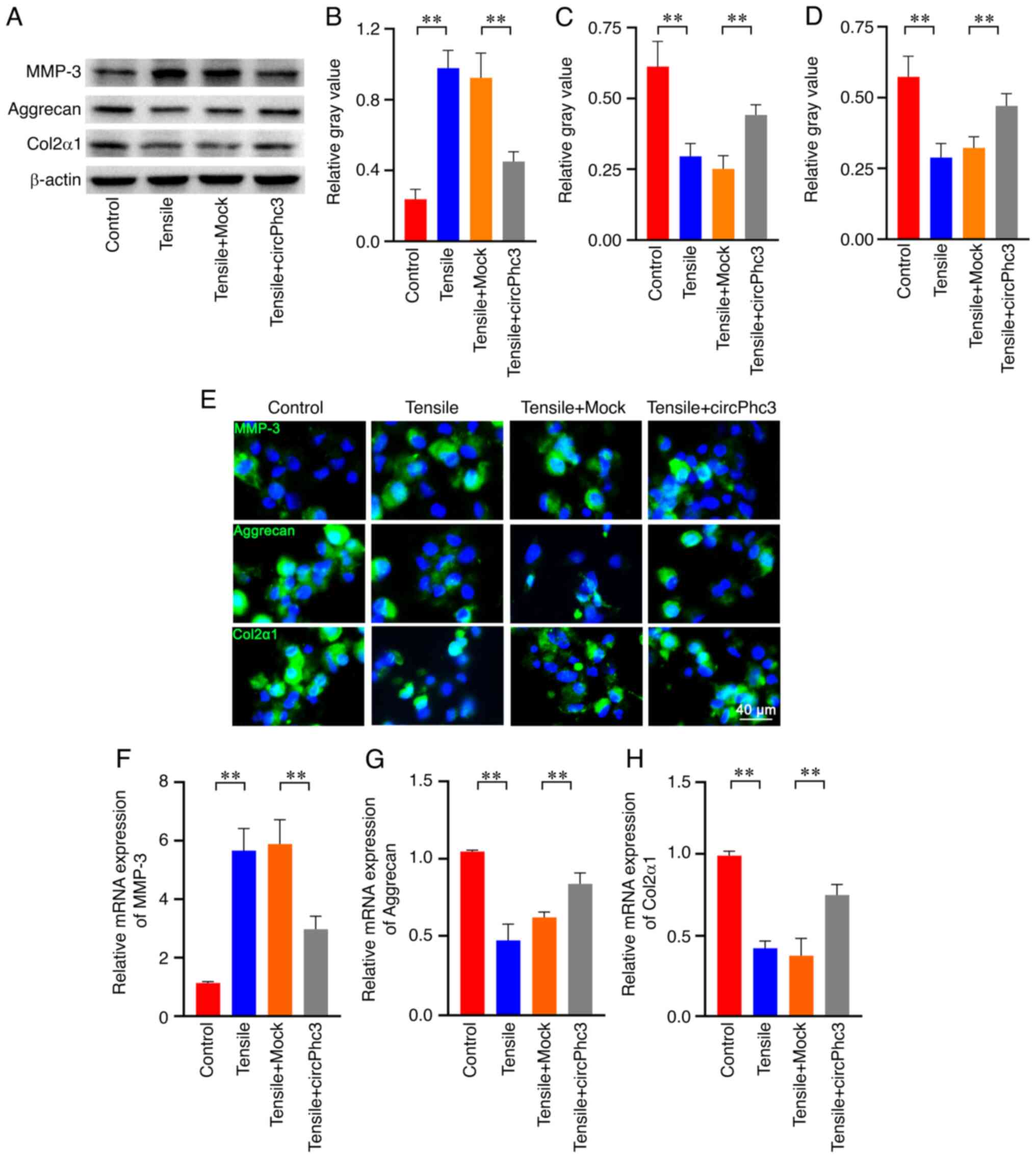 |
Figure 4
circPhc3 is involved in mechanical loading-regulated production of extracellular matrix and cartilage-degrading enzymes. Protein expression levels were (A) determined by western blotting and semi-quantified for (B) MMP-3, (C) Aggrecan and (D) Col2α1 in chondrocytes transfected with Mock or circPhc3 overexpression vector, followed by treatment with 10% tensile for 48 h. **P<0.01 (n=4). (E) MMP-3, Aggrecan and Col2α1 expression and location in chondrocytes transfected with Mock or circPhc3 overexpression vector under 10% tensile were detected by immunofluorescence staining (n=4). mRNA expression levels of (F) MMP-3, (G) Aggrecan and (H) Col2α1 in chondrocytes transfected with Mock or circPhc3 overexpression vector under 10% tensile. **P<0.01 (n=4). circ, circular RNA; MMP, matrix metalloproteinase; Col2α1, collagen type II, α1.
|
miR-93-3p targeted by circPhc3 is involved in the secretion of ECM and cartilage degrading enzymes
To explore the molecular mechanisms by which circPhc3 participates in the secretion of ECM and cartilage degrading enzymes in chondrocytes, algorithms were used to predict the putative targeting miRNAs of circPhc3. According to bioinformatics analysis data provided by Shanghai Kangcheng Biotech, circPhc3 contained multiple binding sites for miR-93-3p (Fig. 5A). The dual luciferase reporter assay results showed that firefly luciferase activity was significantly decreased following co-transfection with pMIR-circPhc3 and miR-93-3p mimics (Fig. 5B). Subsequently, the expression of miR-93-3p in chondrocytes overexpressing or silencing circPhc3 was assessed using RT-qPCR. The results showed that miR-93-3p expression was significantly increased in circPhc3-silenced chondrocytes compared with that in shRNA-NC-transfected chondrocytes, whereas miR-93-3p expression was significantly decreased in circPhc3-overexpression chondrocytes compared with those in the Mock group (Fig. 5C). These results indicated that miR-93-3p may be a high affinity target of circPhc3 in chondrocytes.
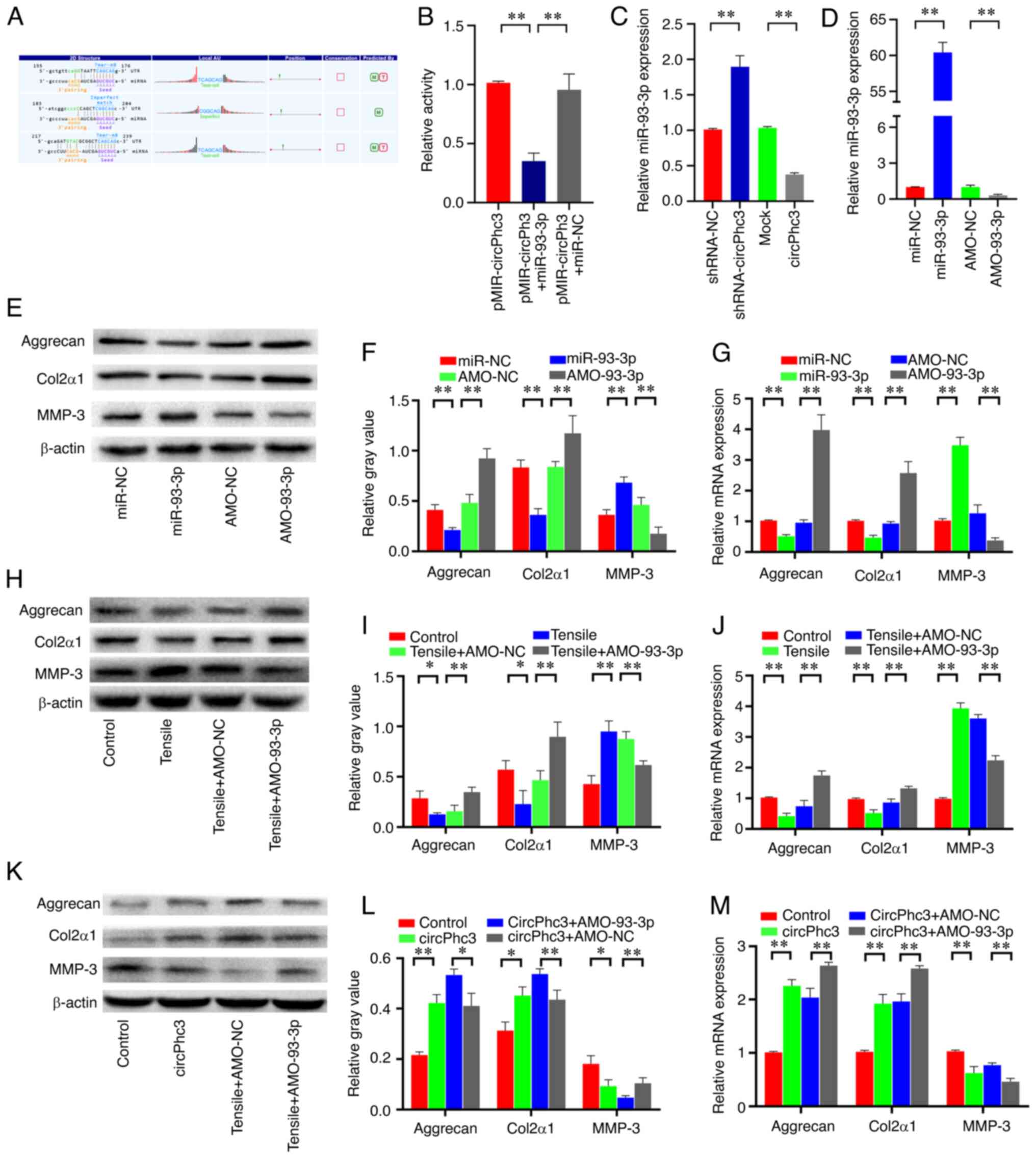 |
Figure 5
miR-93-3p targeted by circPhc3 is involved in the secretion of extracellular matrix and cartilage-degrading enzymes. (A) Bioinformatics analysis demonstrated the circPhc3 had multiple binding sites for miR-93-3p. (B) Dual luciferase reporter assay results showed that the firefly luciferase activity was significantly decreased when the cells were transfected with pMIR-circPhc3 and miR-93-3p mimics compared with those transfected with pMIR-circPh3 and miR-NC. **P<0.01 (n=4). (C) miR-93-3p expression levels in chondrocytes transfected with shRNA-NC, shRNA-circPhc3, Mock or circPhc3 overexpression vector were detected by RT-qPCR. **P<0.01 (n=4). (D) miR-93-3p expression levels in chondrocytes transfected with miR-NC, miR-93-3p mimic, AMO-NC or AMO-93-3p were detected by RT-qPCR. **P<0.01 (n=4). Protein expression levels were (E) determined by western blotting and (F) semi-quantified for Aggrecan, Col2α1 and MMP-3 in chondrocytes transfected with miR-NC, miR-93-3p mimics, AMO-NC or AMO-93-3p. **P<0.01 (n=4). (G) mRNA expression levels of Aggrecan, Col2α1 and MMP-3 in chondrocytes transfected with miR-NC, miR-93-3p mimics, AMO-NC or AMO-93-3p were detected by RT-qPCR. **P<0.01 (n=4). Protein expression levels were (H) detected by western blotting and (I) semi-quantified for Aggrecan, Col2α1 and MMP-3 expression in chondrocytes transfected with AMO-NC or AMO-93-3p under 10% tensile for 48 h. *P<0.05 and **P<0.01 (n=4). (J) mRNA expression levels of Aggrecan, Col2α1 and MMP-3 in chondrocytes transfected with AMO-NC or AMO-93-3p under 10% tensile for 48 h were detected by RT-qPCR. **P<0.01 (n=4). Protein expression levels were (K) determined by western blotting and (L) semi-quantified for Aggrecan, Col2α1 and MMP-3 in chondrocytes transfected with circPhc3 overexpression vector and AMO-NC or AMO-93-3p. *P<0.05 and **P<0.01 (n=4). (M) mRNA expression levels of Aggrecan, Col2α1 and MMP-3 in chondrocytes transfected with circPhc3 overexpression vector and AMO-NC or AMO-93-3p were detected by RT-qPCR. **P<0.01 (n=4). miR, microRNA; circ, circular RNA; NC, negative control; shRNA, short hairpin RNA; NC, negative control; RT-qPCR, reverse transcription-quantitative PCR; AMO, anti-miR oligonucleotides; Col2α1, collagen type II, α1; MMP, matrix metalloproteinase.
|
To investigate whether miR-93-3p was involved in the secretion of ECM and cartilage degrading enzymes, miR-93-3p was overexpressed or silenced in chondrocytes by transfection with miR-93-3p mimic or AMO-93-3p, respectively (Fig. 5D). The western blotting results showed that miR-93-3p mimic significantly decreased the expression levels of Aggrecan and Col2α1 and increased the expression levels of MMP-3 in chondrocytes compared with those in the miR-NC group (Fig. 5E and F). By contrast, AMO-93-3p displayed the opposite effect. Similar results were observed via RT-qPCR (Fig. 5G). To further examine whether miR-93-3p was involved in the regulation of matrix synthesis and catabolism by mechanical loading in chondrocytes, the expression levels of Aggrecan, Col2α1 and MMP-3 were measured in chondrocytes transfected with AMO-93-3p, followed by 10% tensile treatment for 48 h. The western blotting and RT-qPCR results showed that miR-93-3p knock-down significantly alleviated mechanical loading-mediated upregulation of MMP-3 expression in chondrocytes (Fig. 5H-J). Mechanical loading-mediated downregulation of Aggrecan and Col2α1 expression levels were also attenuated by AMO-93-3p in chondrocytes. To further confirm whether circPhc3 could bind to miR-93-3p to regulate chondrocyte matrix metabolism, the expression level of miR-93-3p was altered in chondrocytes using miR-93-3p mimic and AMO-93-3p. The results showed that circPhc3 overexpression significantly increased the expression levels of Aggrecan and Col2α1 and decreased the expression levels of MMP-3 in chondrocytes compared with those in the Control group (Fig. 5K-M). These changes were further enhanced by AMO-93-3p in chondrocyte (Fig. 5K-M). These results indicated that circPhc3 targeting of miR-93-3p was involved in the regulation of chondrocyte matrix metabolism by mechanical loading.
miR-93-3p regulates chondrocyte matrix metabolism by targeting FoxO1
Based on the aforementioned results, miR-93-3p was confirmed to be involved in chondrocyte matrix metabolism. Thus, miR-93-3p might target several regulatory factors related to chondrocyte matrix metabolism. To address this issue, computation and bioinformatics-based approaches were used to predict the putative targets of miR-93-3p using TargetScan (www.targetscan.org). These analyses identified FoxO1 as a candidate target of miR-93-3p (Fig. 6A). The western blotting results demonstrated that the expression of FoxO1 was significantly decreased in the articular cartilage from OA model mice compared with that from Sham mice (Fig. 6B and C). Furthermore, the expression of FoxO1 was also significantly downregulated in chondrocytes treated with 10% tensile for 48 h compared with that in Control chondrocytes (Fig. 6D and E). To observe the effect of miR-93-3p on FoxO1 expression in chondrocytes, chondrocytes were transfected with miR-93-3p or AMO-93-3p to overexpress or knock down the expression of miR-93-3p, respectively. The western blotting results showed that miR-93-3p overexpression significantly decreased the expression of FoxO1 in chondrocytes compared with that in the miR-NC group (Fig. 6F and G). However, FoxO1 expression was significantly increased in chondrocytes transfected with AMO-93-3p compared with that in AMO-NC chondrocytes. Further analysis revealed that miR-93-3p overexpression enhanced the inhibitory effect of mechanical loading on FoxO1 expression, whereas AMO-93-3p alleviated mechanical loading-mediated FoxO1downregulation (Fig. 6H and I). Subsequently, the effects of circPhc3 on FoxO1 expression in chondrocytes were assessed. Compared with the Mock and shRNA-NC groups, circPhc3 overexpression significantly increased the expression of FoxO1, whereas circPhc3 knockdown significantly downregulated FoxO1 expression in chondrocytes, respectively (Fig. 6J and K). A luciferase reporter assay was performed to investigate whether miR-93-3p bound to the 3′-UTR of FoxO1. The results showed that luciferase activity was significantly decreased in the reporter containing the 3′-UTR of FoxO1 treated with miR-93-3p compared with those treated with miR-NC, suggesting that FoxO1 was the target gene of miR-93-3p (Fig. 6L).
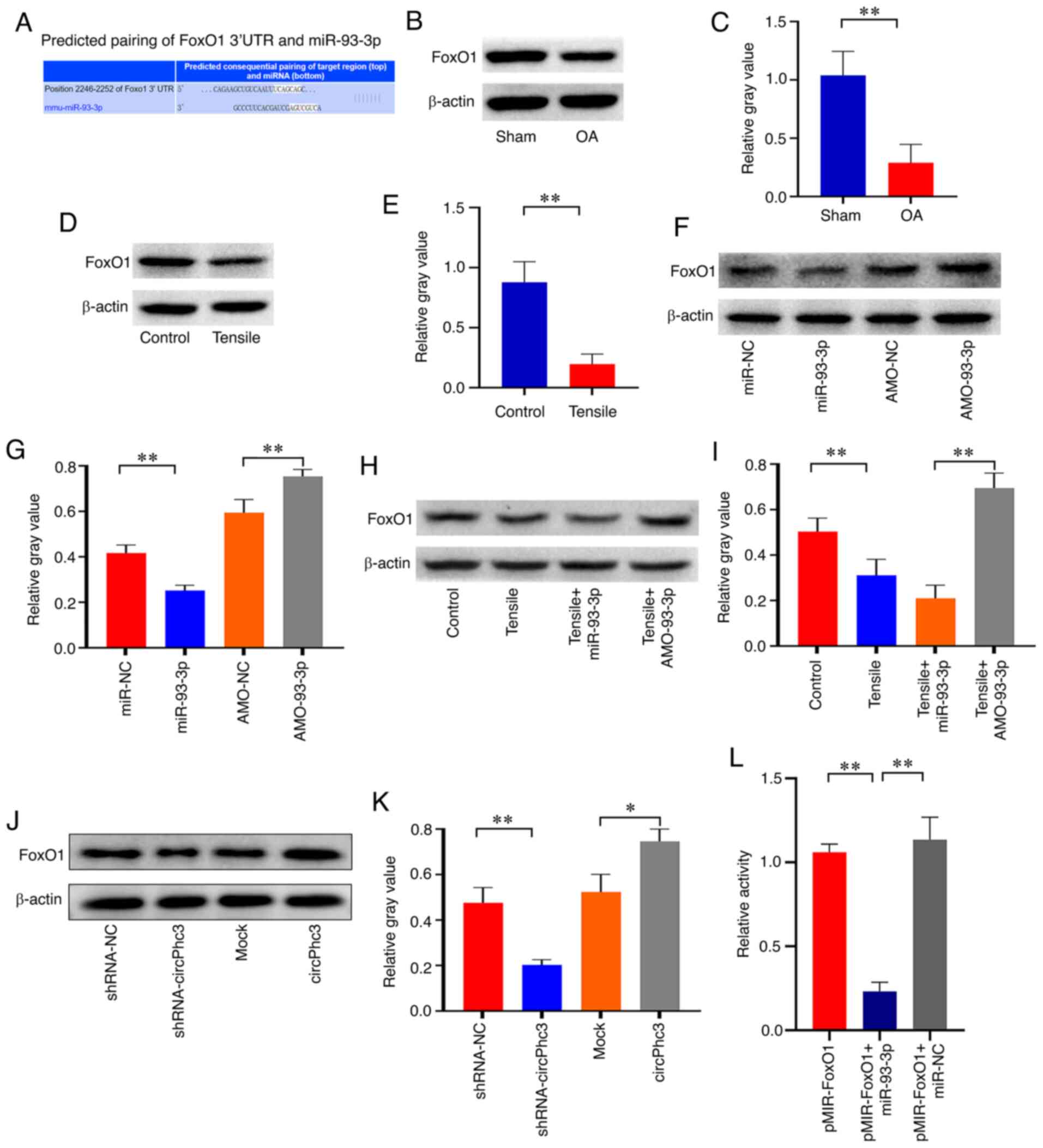 |
Figure 6
miR-93-3p regulates chondrocyte matrix metabolism by targeting FoxO1. (A) Complimentary sequences between miR-93-3p and FoxO1. FoxO1 protein expression in the articular cartilage of OA model mice and Sham mice was (B) determined by western blotting and (C) semi-quantified. **P<0.01 (n=4). FoxO1 protein expression in chondrocytes treated with 10% tensile for 48 h was (D) determined by western blotting and (E) semi-quantified. **P<0.01 (n=4). FoxO1 protein expression in chondrocytes transfected with miR-NC, miR-93-3p mimics, AMO-NC or AMO-93-3p was (F) determined by western blotting and (G) semi-quantified. **P<0.01 (n=4). FoxO1 protein expression in chondrocytes transfected with miR-93-3p mimics or AMO-93-3p under 10% tensile for 48 h was (H) determined by western blotting and (I) semi-quantified. **P<0.01 (n=4). FoxO1 protein expression in chondrocytes transfected with shRNA-NC, shRNA-circPhc3, Mock or circPhc3 overexpression vector was (J) determined by western blotting and (K) semi-quantified. *P<0.05 and **P<0.01 (n=4). (L) Luciferase activity of the reporter containing the 3′-UTR of FoxO1 treated with miR-93-3p mimic was significantly decreased compared with that in the pMIR-FoxO1 group. **P<0.01 (n=6). miR, microRNA; OA, osteoarthritis; NC, negative control; AMO, anti-miR oligonucleotides; shRNA, short hairpin RNA; mmu, Mus musculus; UTR, untranslated region.
|
Discussion
Mechanical instability is a critical factor for the initiation and progression of OA (21). Previous studies have reported that mechanical instability could cause disorders of cartilage cell synthesis and matrix metabolism (7,8,22). The present study demonstrated that mechanical instability downregulated the expression of circPhc3 in articular cartilage tissue, and circPhc3 participated in the production of ECM and cartilage-degrading enzymes primarily via sponging miR-93-3p to regulate FoxO1 expression. Therefore, the present study provided a novel theoretical basis for understanding the pathogenesis of OA induced by mechanical instability.
Recent studies have confirmed that circRNAs serve an important role in the pathogenesis of OA (23-26). Zhou et al (10) found that interleukin-1β increased the expression of circRNA_33186 in chondrocytes in a time- and dose-dependent manner. Silencing circRNA_33186 in chondrocytes could effectively alleviate interleukin-1β-induced apoptosis of chondrocytes, suggesting that circRNA_33186 is involved in the process of inflammatory factor-induced chondrocyte apoptosis in OA. Liu et al (27) reported that an increased expression of circCER was accompanied by increasing expression levels of interleukin-1 and tumour necrosis factor in chondrocytes. Silencing circCER with small interfering RNA suppressed MMP-13 expression and increased ECM formation. Furthermore, Li et al (28) confirmed that hsa_circ_0045714 could regulate ECM synthesis, as well as the proliferation and apoptosis of chondrocytes by promoting the expression of miR-193b. To the best of our knowledge, the present study demonstrated for the first time that circPhc3 was involved in mechanical instability-regulated chondrocyte matrix metabolism in OA.
The chondrocytes in damaged cartilage are particularly susceptible to mechanical stress, and the tensile properties of damaged cartilage are lost due to the disrupted collagen network (22). Previous studies have found that the regulation of circRNAs by mechanical stress is involved in the pathogenesis of numerous diseases, such as intervertebral disk degeneration, orthodontic tooth movement and endplate cartilage degeneration (29-31). Xiang et al (30) reported that circCOG8 expression was obviously downregulated by mechanical stress in disk nucleus pulposus cells. The circCOG8/miR-182-5p/FOXO3 pathway may also be considered an important underlying mechanism in the involvement of compression during the progression of intervertebral disc degeneration (30). Xiao et al (32) detected tension-sensitive circRNA_0058097 molecules in human endplate chondrocytes, which might lead to novel preventative and therapeutic strategies for endplate cartilage degeneration. The present study demonstrated that circPhc3 was susceptible to mechanical stress, which is a critical factor for the maintenance of the balance of ECM in OA. However, the exact mechanisms by which mechanical stress regulates circPhc3 expression in chondrocytes requires further investigation.
circRNAs serve a crucial role in regulating gene expression by acting as miRNA sponges (33). In the present study, miR-93-3p was confirmed to be a high-affinitive target of circPhc3 in chondrocytes, which was involved in the regulation of ECM metabolism by circPhc3 in OA. Numerous studies have shown that miR-93-3p participates in the pathogenesis of inflammatory-related diseases. miR-93-3p could alleviate lipopolysaccharide-induced inflammation and apoptosis in H9c2 cardiomyocytes by inhibiting toll-like receptor 4 (34). Butler et al (35) found that miR-93-3p was correlated with the level of C-reactive protein, a marker of inflammation. Furthermore, miR-93-3p was also confirmed to be involved in the regulation of bone metabolism. Peng et al (36) suggested that long non-coding RNA-NTF3-5/miR-93-3p/RUNX family transcription factor 2 could regulate the osteogenic differentiation of mesenchymal stem cells and might serve as a therapeutic target for bone regeneration in the posterior maxilla. Moreover, Mäkitie et al(37) confirmed that circulating levels of miR-93-3p were related to patients with osteoporosis susceptible to defective PLS3 function. However, to the best of our knowledge, no previous studies have demonstrated that miR-93-3p is an important regulator of OA; therefore, the present study was the first to investigate the role of miR-93-3p in OA.
The results of the present study showed that miR-93-3p regulated chondrocyte matrix metabolism by targeting FoxO1. Previous studies have indicated that FoxO1 is involved in maintaining the balance of anabolism and catabolism of cartilage matrix. Wang et al (38) found that FoxO1 was a crucial mediator of TGF-β/TAK1 signalling and protected against OA by maintaining articular cartilage homeostasis. Lee et al (39) reported that FoxO1 supported homeostasis and protected against meniscus damage in response to mechanical overuse and during aging and OA. FoxO1 is also a transcription factor as a regulator of chondrocyte oxidative stress resistance and tissue homeostasis (40). In the present study, the results confirmed that FoxO1 was a high-affinitive target of miR-93-3p in OA chondrocytes. Although the results demonstrated that FoxO1 was a target gene of miR-93-3p in OA chondrocytes, a number of previous studies have reported that miR-93-3p has multiple target genes (34,41,42). Therefore, whether other miR-93-3p-targeting genes are also involved in maintaining the balance of chondrocyte matrix metabolism should be explored in future studies.
In conclusion, the results of the present study suggested that the circPhc3/miR-93-3p/FoxO1 pathway might be involved in the regulation of chondrocyte matrix metabolism by mechanical loading in OA. The present study aimed to establish a molecular mechanism of mechanical instability-induced OA to provide novel insights into the development of pharmacological and physical therapies that can modify the course of OA.
Supplementary Data
Availability of data and materials
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
All authors contributed to the design of the study. ZW, XW, CJ, YY and ZR collected the data and performed the analysis. YY led the draft of the manuscript. ZW and YY confirmed the authenticity of all the raw data. All authors read and approved the final manuscript.
Ethics approval and consent to participate
All individuals provided written informed consent for the use of human specimens for experiments. The present study was approved by the Ethics Committees of Tongji Hospital of Tongji University (approval no. 2020TJYLL287). All animal care and experimental procedures were approved by the Tongji Hospital Animal Study Committee (approval no. IACUC20200422) and were carried out in accordance with the Guide for Laboratory Animal Use.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Acknowledgments
Not applicable.
Funding
No funding was received.
Abbreviations:
|
circRNA
|
circular RNA
|
|
OA
|
osteoarthritis
|
|
ECM
|
extracellular matrix
|
|
MMP
|
matrix metalloproteinase
|
|
DMM
|
destabilized medial meniscus
|
|
H&E
|
hematoxylin and eosin
|
|
NC
|
negative control
|
|
AMO
|
anti-microRNA oligonucleotides
|
|
RT-qPCR
|
reverse transcription-quantitative PCR
|
References
|
1
|
Jeon OH, Kim C, Laberge RM, Demaria M, Rathod S, Vasserot AP, Chung JW, Kim DH, Poon Y, David N, et al: Local clearance of senescent cells attenuates the development of post-traumatic osteoarthritis and creates a pro-regenerative environment. Nat Med. 23:775–781. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Palazzo C, Nguyen C, Lefevre-Colau MM, Rannou F and Poiraudeau S: Risk factors and burden of osteoarthritis. Ann Phys Rehabil Med. 59:134–138. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Sakata R, Iwakura T and Reddi AH: Regeneration of articular cartilage surface: Morphogens, cells, and extracellular matrix scaffolds. Tissue Eng Part B Rev. 21:461–473. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Murata D, Fujimoto R and Nakayama K: Osteochondral regeneration using adipose tissue-derived mesenchymal stem cells. Int J Mol Sci. 21:35892020. View Article : Google Scholar :
|
|
5
|
Vela-Anero A, Hermida-Gomez T, Gato-Calvo L, Vaamonde-García C, Díaz-Prado S, Meijide-Faílde R, Blanco FJ and Burguera EF: Long-term effects of hydrogen sulfide on the anabolic-catabolic balance of articular cartilage in vitro. Nitric Oxide. 70:42–50. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Anderson DE, Markway BD, Weekes KJ, McCarthy HE and Johnstone B: Physioxia promotes the articular chondrocyte-like phenotype in human chondroprogenitor-derived self-organized tissue. Tissue Eng Part A. 24:264–274. 2018. View Article : Google Scholar :
|
|
7
|
Guilak F: Biomechanical factors in osteoarthritis. Best Pract Res Clin Rheumatol. 25:815–823. 2011. View Article : Google Scholar
|
|
8
|
Onitsuka K, Murata K, Kokubun T, Fujiwara S, Nakajima A, Morishita Y and Kanemura N: Effects of controlling abnormal joint movement on expression of MMP13 and TIMP-1 in osteoarthritis. Cartilage. 11:98–107. 2020. View Article : Google Scholar :
|
|
9
|
Kristensen LS, Andersen MS, Stagsted LVW, Ebbesen KK, Hansen TB and Kjems J: The biogenesis, biology and characterization of circular RNAs. Nat Rev Genet. 20:675–691. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zhou ZB, Huang GX, Fu Q, Han B, Lu JJ, Chen AM and Zhu L: circRNA.33186 contributes to the pathogenesis of osteoarthritis by sponging miR-127-5p. Mol Ther. 27:531–541. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Shen S, Wu Y, Chen J, Xie Z, Huang K, Wang G, Yang Y, Ni W, Chen Z, Shi P, et al: CircSERPINE2 protects against osteoarthritis by targeting miR-1271 and ETS-related gene. Ann Rheum Dis. 78:826–836. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Shen P, Yang Y, Liu G, Chen W, Chen J, Wang Q, Gao H, Fan S, Shen S, Zhao X, et al: CircCDK14 protects against osteoarthritis by sponging miR-125a-5p and promoting the expression of Smad2. Theranostics. 10:9113–9131. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Yu CX and Sun S: An emerging role for circular RNAs in osteoarthritis. Yonsei Med J. 59:349–355. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Thomson DW and Dinger ME: Endogenous microRNA sponges: Evidence and controversy. Nat Rev Genet. 17:272–283. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer M, et al: Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 495:333–338. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Kellgren JH and Lawrence JS: Radiological assessment of osteo-arthrosis. Ann Rheum Dis. 16:494–502. 1957. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Jeon OH, Wilson DR, Clement CC, Rathod S, Cherry C, Powell B, Lee Z, Khalil AM, Green JJ, Campisi J, et al: Senescence cell-associated extracellular vesicles serve as osteoarthritis disease and therapeutic markers. JCI Insight. 4:e1250192019. View Article : Google Scholar
|
|
18
|
Yang F, Huang R, Ma H, Zhao X and Wang G: miRNA-411 regulates chondrocyte autophagy in osteoarthritis by targeting hypoxia-inducible factor 1 alpha (HIF-1α). Med Sci Monit. 26:e9211552020.
|
|
19
|
Weiss S, Hennig T, Bock R, Steck E and Richter W: Impact of growth factors and PTHrP on early and late chondrogenic differentiation of human mesenchymal stem cells. J Cell Physiol. 223:84–93. 2010.PubMed/NCBI
|
|
20
|
Livak KJ and Schmittgen TD: Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001. View Article : Google Scholar
|
|
21
|
Blalock D, Miller A, Tilley M and Wang J: Joint instability and osteoarthritis. Clin Med Insights Arthritis Musculoskelet Disord. 8:15–23. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Guilak F, Fermor B, Keefe FJ, Kraus VB, Olson SA, Pisetsky DS, Setton LA and Weinberg JB: The role of biomechanics and inflammation in cartilage injury and repair. Clin Orthop Relat Res. 423:17–26. 2004. View Article : Google Scholar
|
|
23
|
Wang T, Hao Z, Liu C, Yuan L, Li L, Yin M, Li Q, Qi Z and Wang Z: LEF1 mediates osteoarthritis progression through circRNF121/miR-665/MYD88 axis via NF κB signaling pathway. Cell Death Dis. 11:5982020. View Article : Google Scholar
|
|
24
|
Chen C: Serum hsa_circ_101178 as a potential biomarker for early prediction of osteoarthritis. Clin Lab. View Article : Google Scholar
|
|
25
|
Shen S, Yang Y, Shen P, Ma J, Fang B, Wang Q, Wang K, Shi P, Fan S and Fang X: circPDE4B prevents articular cartilage degeneration and promotes repair by acting as a scaffold for RIC8A and MID1. Ann Rheum Dis. 80:1209–1219. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Ehrlich GD: Circular RNAs as diagnostic biomarkers for osteoarthritis. Genet Test Mol Biomarkers. 23:701–702. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Liu Q, Zhang X, Hu X, Dai L, Fu X, Zhang J and Ao Y: Circular RNA related to the chondrocyte ECM regulates MMP13 Expression by functioning as a miR-136 'Sponge' in human cartilage degradation. Sci Rep. 6:225722016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Li BF, Zhang Y, Xiao J, Wang F, Li M, Guo XZ, Xie HB, Xia H and Chen B: Hsa_circ_0045714 regulates chondrocyte proliferation, apoptosis and extracellular matrix synthesis by promoting the expression of miR-193b target gene IGF1R. Hum Cell. 30:311–318. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Liu Q, Zhang X, Hu X, Yuan L, Cheng J, Jiang Y and Ao Y: Emerging roles of circRNA related to the mechanical stress in human cartilage degradation of osteoarthritis. Mol Ther Nucleic Acids. 7:223–230. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Xiang Q, Kang L, Zhao K, Wang J, Hua W, Song Y, Feng X, Li G, Lu S, Wang K, et al: CircCOG8 downregulation contributes to the compression-induced intervertebral disk degeneration by targeting miR-182-5p and FOXO3. Front Cell Dev Biol. 8:5819412020. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Wang H, Feng C, Jin Y, Tan W and Wei F: Identification and characterization of circular RNAs involved in mechanical force-induced periodontal ligament stem cells. J Cell Physiol. 234:10166–10177. 2019. View Article : Google Scholar
|
|
32
|
Xiao L, Ding B, Xu S, Gao J, Yang B, Wang J and Xu H: circRNA_0058097 promotes tension-induced degeneration of endplate chondrocytes by regulating HDAC4 expression through sponge adsorption of miR-365a-5p. J Cell Biochem. 121:418–429. 2020. View Article : Google Scholar
|
|
33
|
Zhu X, Du J and Gu Z: Circ-PVT1/miR-106a-5p/HK2 axis regulates cell growth, metastasis and glycolytic metabolism of oral squamous cell carcinoma. Mol Cell Biochem. 474:147–158. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Tang B, Xuan L, Tang M, Wang H, Zhou J, Liu J, Wu S, Li M, Wang X and Zhang H: miR-93-3p alleviates lipopolysaccharide-induced inflammation and apoptosis in H9c2 cardiomyocytes by inhibiting toll-like receptor 4. Pathol Res Pract. 214:1686–1693. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Butler AE, Ramachandran V, Hayat S, Dargham SR, Cunningham TK, Benurwar M, Sathyapalan T, Najafi-Shoushtari SH and Atkin SL: Expression of microRNA in follicular fluid in women with and without PCOS. Sci Rep. 9:163062019. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Peng W, Zhu SX, Wang J, Chen LL, Weng JQ and Chen SL: Lnc-NTF3-5 promotes osteogenic differentiation of maxillary sinus membrane stem cells via sponging miR-93-3p. Clin Implant Dent Relat Res. 20:110–121. 2018. View Article : Google Scholar
|
|
37
|
Mäkitie RE, Hackl M, Weigl M, Frischer A, Kämpe A, Costantini A, Grillari J and Mäkitie O: Unique, gender-dependent serum microRNA profile in PLS3 gene-related osteoporosis. J Bone Miner Res. 35:1962–1973. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Wang C, Shen J, Ying J, Xiao D and O'Keefe RJ: FoxO1 is a crucial mediator of TGF-β/TAK1 signaling and protects against osteoarthritis by maintaining articular cartilage homeostasis. Proc Natl Acad Sci USA. 117:30488–30497. 2020. View Article : Google Scholar
|
|
39
|
Lee KI, Choi S, Matsuzaki T, Alvarez-Garcia O, Olmer M, Grogan SP, D'Lima DD and Lotz MK: FOXO1 and FOXO3 transcription factors have unique functions in meniscus development and homeostasis during aging and osteoarthritis. Proc Natl Acad Sci USA. 117:3135–3143. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Akasaki Y, Alvarez-Garcia O, Saito M, Carames B, Iwamoto Y and Lotz MK: FoxO transcription factors support oxidative stress resistance in human chondrocytes. Arthritis Rheumatol. 66:3349–3358. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Miao X, Xi Z, Zhang Y, Li Z, Huang L, Xin T, Shen R and Wang T: Circ-SMARCA5 suppresses colorectal cancer progression via downregulating miR-39-3p and upregulating ARID4B. Dig Liver Dis. 52:1494–1502. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Zhu Y, Wang Y, Zhao J, Shen J, Wang Z, Bai M, Fan Y, Yin R, Mao Y and Bai W: CircRNA-1967 participates in the differentiation of goat SHF-SCs into hair follicle lineage by sponging miR-93-3p to enhance LEF1 expression. Anim Biotechnol. 22:1–13. 2021. View Article : Google Scholar
|




















