Introduction
Lung cancer is the most common type of cancer and is
the leading cause of cancer-related mortality around the world
(1). Annually, ~2.1 million
patients are diagnosed with lung cancer worldwide and 1.8 million
patients die of this malignancy (2). Non-small cell lung cancer (NSCLC)
is the predominant pathological type of lung cancer, comprising
over 80% of lung cancer cases (3) and NSCLC contributes to the vast
majority of cases for both morbidity and mortality (4). At present, the primary therapeutic
techniques for NSCLC are surgical resection, chemoradiotherapy,
targeted therapy and immunotherapy (5). Due to the lack of typical symptoms
and hidden symptoms of NSCLC, patients have advanced NSCLC at the
time of clinical diagnosis and thus miss the optimal treatment
period (6). Commendable
advancements in diagnostic and treatment methods have been made;
however, the efficacy of treatment for NSCLC remains unfavorable,
highlighting the importance of developing more efficacious
therapies (7). According to the
data from 2019, the 5-year survival rate of NSCLC globally was low,
<18%, which is largely attributed to a high relapse rate and
metastasis (8). Therefore, a
comprehensive understanding of NSCLC pathogenesis may aid in the
identification of potential diagnostic biomarkers and treatment
strategies.
Long non-coding (lnc)RNAs are a family of gene
transcripts, >200 nucleotides in length, which lack long open
reading frames (9). lncRNAs were
once considered useless; however, recent studies have confirmed
that lncRNAs participate in a number of crucial and fundamental
cellular biological processes (10,11). lncRNAs control gene expression at
different transcriptional levels via distinctly different
mechanisms (12). Aberrant
expression of lncRNAs has been proved in nearly all types of human
cancer, such as bladder (13),
cervical (14), gastric
(15) and pancreatic cancers
(16). Several lncRNAs have been
found to be increased or decreased in NSCLC and have been
identified as important indicators of cancer progression, such as
WTA-AS (17), B4GALT1-AS1
(18) and CCAT1 (19). These lncRNAs have tumor-promoting
or tumor-inhibiting effects in NSCLC and have been associated with
tumor biological behavior (20).
MicroRNAs (miRNAs/miR), which are single-stranded
RNA molecules, 17-21 nucleotides in length, are another type of nc
RNA (21). miRNAs can decrease
gene expression by base pairing with the 3′-untranslated region
(UTR) of their downstream genes, ultimately degrading mRNAs or
suppressing translation (22).
Accumulating evidence has confirmed the importance of miRNAs in
carcinogenesis and the progression of NSCLC (23-25). lncRNAs with miRNA response
elements can function together as competing endogenous (ce) RNAs by
sponging miRNAs, thereby sequestering miRNAs from their target
mRNAs (26). Consequently,
investigation of tumor-associated lncRNAs and miRNAs in NSCLC may
increase the understanding into the mechanisms that contribute to
NSCLC progression and aid in the development of diagnostic and
therapeutic targets.
Multiple lncRNAs have been associated with
tumorigenesis of NSCLC, such as GAS5 (27), LINC02678 (28) and CCDC144NL-AS1 (29); however, whether LINC01748 plays a
crucial regulatory role requires further investigation, as to the
best of our knowledge the expression level and function of
LINC01748 in other types of cancer has not been analyzed.
Therefore, the aim of the present study was to investigate the
expression profile, clinical status and biological functions of
LINC01748 in NSCLC. Furthermore, different experiments were
utilized to understand the mechanisms behind the carcinogenic
effects of LINC01748 in NSCLC cell lines.
Materials and methods
Patient samples
Paired NSCLC and adjacent normal tissues were
collected from 57 patients (33 males, 24 females; age range, 32-75
years; mean age, 56.7±7.4 years) from the Weifang Yidu Central
Hospital (Weifang, China) between February 2014 and December 2015.
The clinicopathological parameters of these patients, including
tumor size, TNM stage and lymph node metastasis, were also
collected. Adjacent normal tissues were collected 2 cm away from
the tumor tissues. All patients with NSCLC were diagnosed using
histopathology. The tissues were immediately washed with PBS and
the tissues were stored in liquid nitrogen until RNA extraction.
The inclusion criteria were as follows: i) Diagnosed with NSCLC;
ii) was not treated with anticancer therapies prior to surgery; and
iii) agreed to take part in the research. The exclusion criteria
were as follows: i) Patients with a history of radiotherapy,
chemotherapy, or targeted therapy; and ii) patients who suffered
from other types of human cancer, such as colorectal, gastric or
prostate cancers. The Ethics Committee of Weifang Yidu Central
Hospital approved the present research. Written informed consent
was obtained from all participants prior to surgery.
Cell culture
The BEAS-2B (CRL-9609™) human normal bronchial
epithelial cell line was cultured in bronchial epithelial cell
growth medium (Lonza Group, Ltd.) containing 10% FBS (Gibco; Thermo
Fisher Scientific, Inc.). The H522 (CRL-5810™) and NCI-H460 (H460;
CVCL_01459) (both from American Type Culture Collection) NSCLC cell
lines were cultured in RPMI-1640 medium supplemented with 10% FBS
(both from Gibco; Thermo Fisher Scientific, Inc.).
Another two NSCLC cell lines, SK-MES-1 and A549,
were purchased from the National Collection of Authenticated Cell
Cultures and maintained in MEM and F-12K medium, respectively and
both supplemented with 10% FBS (all from Gibco; Thermo Fisher
Scientific, Inc.). Penicillin-streptomycin (1%) was also used in
the culture medium for all the cells. The H460 cell line is a large
cell lung cancer cell line, while the H522 and A549 cell lines are
lung adenocarcinoma (LUAD) cell lines. The SK-MES-1 cell line is a
lung squamous cell carcinoma (LUSC) cell line. All the cells were
cultured at 37°C in a humidified incubator with 5%
CO2.
Transfection assay
Small interfering (si) RNAs targeting LINC01748
(si-LINC01748#1 and si-LINC01748#2) were purchased from Shanghai
GenePharma Co., Ltd., with scrambled negative control (NC) siRNA
(si-NC) as the control. The si-LINC017481#1 sequence was 5′-GTC ATT
TTA AGC TAT TTA ATT TT-3′; the si-LINC017481#2 sequence was 5′-ATG
TAT TTA CAT CGT TTT ATA CA-3′; and the si-NC sequence was 5′-CAC
GAT AAG ACA ATG TAT TT-3′. Guangzhou RiboBio Co., Ltd., designed
and synthesized the following miRNA oligonucleotides: MiR-520a-5p
mimic, NC mimic, miR-520a-5p inhibitor and NC inhibitor. The
miR-520a-5p mimic sequence was 5′-UCU UUC AUG AAG GGA GAC CUC-3′
and the NC mimic sequence was 5′-UUG UAC UAC ACA AAA GUA CUG-3′.
The miR-520a-5p inhibitor sequence was 5′-AGA AAG UAC UUC CCU CUG
GAG-3′ and the NC inhibitor sequence was 5′-ACU ACU GAG UGA CAG UAG
A-3′. High mobility group AT-hook 1 (HMGA1)-overexpression vector
pcDNA3.1-HMGA1 and empty pcDNA3.1 control was obtained from
GenScript. The cells were transfected with siRNAs (100 pmol), miRNA
mimic (100 pmol), miRNA inhibitor (100 pmol) or vector (4
µg) and Lipofectamine® 2000 (Invitrogen; Thermo
Fisher Scientific, Inc.) at 37°C with 5% CO2 for 6 h. A
Cell Counting Kit-8 (CCK-8) assay was performed after 24 h, while
at 48 h post-transfection, reverse transcription-quantitative PCR
(RT-qPCR), flow cytometry, migration and invasion assays, and
western blot analysis were performed.
RT-qPCR
Total RNA was extracted from tissues or cells using
TRIzol® (Invitrogen; Thermo Fisher Scientific, Inc.) and
reverse transcribed into first-strand cDNA with a PrimeScript
Reagent kit with gDNA Eraser (Takara Biotechnology Co., Ltd.). RT
was performed at 37°C for 15 min, then 85°C for 5 sec. The mRNA
expression levels of LINC01748 and HMGA1 were detected using a TB
Green® Premix Ex Taq™ (Takara Biotechnology Co., Ltd.)
and normalized to the expression level of GAPDH. The following
thermocycling conditions were used: Initial denaturation at 95°C
for 30 sec, followed by 40 cycles at 95°C for 3 sec, 60°C for 30
sec and 72°C for 30 sec.
RNAiso for small RNA (Takara Biotechnology Co.,
Ltd.) was used to extract small RNA from the tissues and cells. The
total RNA was reverse transcribed using the miR-X miRNA
First-Strand Synthesis kit (Takara Biotechnology Co., Ltd.). Next,
the expression level of miR-520a-5p was detected using a miR-X
miRNA RT-qPCR TB Green® kit (Takara Biotechnology Co.,
Ltd.). RT was performed at 37°C for 60 min, then 85°C for 5 sec.
The following thermocycling conditions were used: Initial
denaturation at 95°C for 10 sec; 95°C for 5 sec and 60°C for 20 sec
for 40 cycles; then 95°C for 60 sec, 55°C for 30 sec and 95°C for
30 sec. The small nuclear RNA, U6 was used as the reference control
for miR-520a-5p. The 2−ΔΔCq method (30) was used for gene expression
calculation.
The following primers were used: LINC01748 forward,
5′-TGC TAC ATG GGT AAG GGA GGG-3′ and reverse, 5′-TTC TGG TAG ACG
CAC TCG TCC T-3′; HMGA2 forward, 5′-CGC ACT TCA GCC CAG GGA CAA-3′
and reverse, 5′-CTT CAG CCC AGG GAC AAC GGT CTC TTA GGA GAG GGC TCA
C-3′; GAPDH forward, 5′-CGG AGT CAA CGG ATT TGG TCG TAT-3′ and
reverse, 5′-AGC CTT CTC CAT GGT GGT GAA GAC-3′; miR-520a-5p
forward, 5′-TCG GCA GGC UCC AGA GGG AAG U-3′ and reverse, 5′-CAC
TCA ACT GGT GTC GTG GA-3′; and U6 forward, 5′-CTC GCT TCG GCA GCA
CA-3′ and reverse, 5′-AAC GCT TCA CGA ATT TGC GT-3′.
CCK-8 assay
Cellular proliferative ability was determined using
a CCK-8 assay. In short, the transfected cells were seeded, at a
concentration of 2×103 cells/well into 96-well plates.
After culturing for different periods of time (0, 24, 48 and 72 h),
the cells were cultured at 37°C for a further 2 h with 10 µl
CCK-8 solution (Dojindo Molecular Technologies, Inc.). A microplate
reader was used to measure the absorbance at 450 nm. Finally, the
collective absorbance values were used to plot a cell growth
curve.
Flow cytometry analysis
To determine the apoptotic rate, the NSCLC cells,
transfected with different factors, were treated with EDTA-free
trypsin, washed twice with ice cooled PBS and centrifuged at room
temperature at 1,000 × g for 5 min. The collected cells were then
resuspended in 195 µl Annexin V-FITC binding buffer from an
Annexin V-FITC Apoptosis Detection kit (Beyotime Institute of
Biotechnology). Next, the cell suspension was supplemented with 5
µl Annexin V-FITC and 10 µl PI, followed by a 20-min
incubation at 20°C without light. Finally, the apoptotic cells were
evaluated using a FACSCalibur flow cytometer (BD Biosciences).
CellQuest software v.2.9 (BD Biosciences) was used for data
analysis.
Migration and invasion assays
Transwell chambers (8.0 µm; Corning, Inc.)
were used for both assays. For the migration assay, the cells were
detached, counted and prepared as a single-cell suspension in
FBS-free basal medium (Gibco; Thermo Fisher Scientific, Inc.).
Next, 200 µl cell suspension, containing 5×104
cells, was seeded into the upper chambers. The basolateral chambers
were loaded with 600 µl 20% FBS-supplemented culture medium.
The Transwell chambers were incubated for 24 h at 37°C, then
fixated with 4% paraformaldehyde at room temperature for 30 mins.
The migrated cells that had passed through the pores and attached
to the underside of the membrane were stained with 0.5% crystal
violet at room temperature for 30 mins. The cells which had not
migrated or invaded were gently cleaned with a cotton bud. Finally,
images of the migrated cells were captured using an inverted light
microscope (×200 magnification; Olympus Corp.). The number of
migrated cells in five random fields of view was counted using an
inverted light microscope. For the invasion assay, Matrigel
(Corning, Inc.) was used to coat the upper chambers and polymerized
by incubating at 37°C for 2 h, and experimental procedures were
repeated in the same fashion as the migration test.
Tumor xenograft model
Lentiviruses were produced using a second-generation
lentiviral system. LINC01748 short hairpin RNA (shRNA;
sh-LINC01748) and non-targeting scramble shRNA (sh-NC) were
purchased from Shanghai GenePharma Co., Ltd., and inserted into the
pLKO.1 vector (Addgene, Inc.). The sh-LINC01748 sequence was 5′-CCG
GGT CAT TTT AAG CTA TTT AAT TTT CTC GAG AAA ATT AAA TAG CTT AAA ATG
ACT TTT TG-3′ and the sh-NC sequence was 5′-CCG GCACGA TAA GAC AAT
GTA TTT CTC GAG AAA TAC ATT GTC TTA TCG TGT TTT TG-3′. The
resulting vectors, alongside psPAX2 and pMD2.G (both from Addgene
Inc.), were transfected into 293T cells (The National Collection of
Authenticated Cell Cultures) using Lipofectamine® 2000
(Invitrogen; Thermo Fisher Scientific, Inc.). In total, 30
µg plasmids were used for transfection, and the ratio of
each plasmid (psPAX2: pMD2.G: pLKO.1) was 1:1:2. After incubation
for 5 h at 37°C with 5% CO2, the culture medium was
replaced with fresh DMEM medium containing 10% FBS, 1% glutamax, 1%
non-essential amino acids and 1% sodium pyruvate solution (all from
Gibco; Thermo Fisher Scientific, Inc.). Subsequent to incubation
for 48 h at 37°C with 5% CO2, the lentiviruses harboring
either sh-LINC01748 or sh-NC were collected by ultracentrifugation
at 4°C for 2 h (1,000 × g), mixed with polybrene (5 µg/ml;
Sigma-Aldrich; Merck KGaA) and F-12K medium, were transfected into
the A549 cells with a multiplicity of infection 5. The A549 cells
were incubated with 2 µg/ml puromycin to select stably
transfected cells.
The animal experiment was approved by the Ethics
Committee of Animal Experiments at Weifang Yidu Central Hospital
(Shandong, China). Male BALB/c nude mice (mean weight ± SD,
20.4±1.1 g), aged 4-6 weeks, were purchased from Beijing HFK
Bioscience (Beijing, China). A total of 6 mice were used, and
housed under specific pathogen-free conditions at 25°C and 50%
humidity, with a 10:14 light/dark cycle and ad libitum
access to food and water. The flanks of the mice were
subcutaneously inoculated with 2×106 A549 cells
expressing sh-LINC01748 or sh-NC, which were resuspended in 100
µl PBS. The size of the tumors was detected every 5 days
with a calliper. On day 30 after cell inoculation, the mice were
euthanized by cervical dislocation, and the tumor xenografts were
resected, weighed and imaged. The tumor volume was calculated using
the following formula: Volume=0.5 × length × width2.
Cellular fractionation assay
A Cytoplasmic and Nuclear RNA Purification kit
(Norgen Biotek Corp.) was used to separate the cytoplasmic and
nuclear fractions of the NSCLC cells, which was followed by
determination of the relative LINC01748 mRNA expression level in
both fractions using RT-qPCR.
Bioinformatics analysis
TCGA database (https://portal.gdc.cancer.gov/) was used to examine
mRNA expression level of LINC01748, miR-374a-5p, miR-374b-5p,
miR-520a-5p, miR-525-5p and HMGA1 in LUAD and LUSC. The prediction
of LINC01748 target miRNAs was performed using The Encyclopedia of
RNA Interactomes (ENCORI) and StarBase online tool (starBase 3.0;
http://starbase.sysu.edu.cn/). ENCORI
and TargetScan (http://www.targetscan.org/), bioinformatic tools were
used to analyze miRNAs and identify the downstream targets of
miR-520a-5p.
Luciferase reporter assay
The HMGA1 3′-UTR and LINC01748 fragments with the
wild-type (WT) miR-520a-5p-binding site were prepared by Shanghai
GenePharma Co., Ltd., and inserted into the psiCHECK2 luciferase
reporter vector (Promega Corporation), yielding the HMGA1-WT and
LINC01748-WT reporter vectors. Similarly, mutant (Mut) reporter
plasmids containing HMGA1-Mut and LINC01748-Mut were constructed by
Shanghai GenePharma Co., Ltd. For the reporter assay,
Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.) was used to co-transfect the generated WT or Mut
reporter vectors and miR-520a-5p/NC mimic into the NSCLC cells.
Luciferase activity was detected using a Dual-Luciferase Reporter
Assay system (Promega Corporation) 2 days later. Renilla
luciferase activity was normalized to the firefly luciferase
activity.
RNA immunoprecipitation (RIP) assay
RIP experiments were conducted utilizing an EZ-Magna
RIP RNA binding protein immunoprecipitation kit (EMD Millipore).
The NSCLC cells were treated with RIP cell lysis buffer and the
cell lysates were collected. The supernatant was obtained after
centrifugation at 4°C at 1,000 × g for 15 min. The supernatant
(10%) was used as the 'input', and the rest was incubated in RIP
buffer containing magnetic beads conjugated with Argonaut 2 (Ago2)
or normal IgG control antibodies (EMD Millipore). The magnetic
beads were processed with protease K to detach the proteins, then
incubated overnight at 4°C. After the extraction of the
immunoprecipitated RNA, the enrichment of LINC01748, miR-520a-5p
and HMGA2 were analyzed using RT-qPCR.
Western blot analysis
Total protein was isolated from cells or tumor
xenografts utilizing ice-cold RIPA lysis buffer (Beyotime Institute
of Biotechnology) and quantified using a bicinchoninic acid assay
kit (Beyotime Institute of Biotechnology). Following 10% SDS-PAGE,
the separated proteins (30 µg/lane) were transferred onto
PVDF membranes, which were then blocked for 2 h at room temperature
with 5% skimmed milk diluted with TBS containing 0.1% Tween-20.
Subsequently, the membranes were incubated overnight at 4°C with
primary antibodies targeting HMGA1 (cat. no. ab129153; 1:1,000
dilution) or GAPDH (cat. no. ab128915; 1:1,000 dilution) (both from
Abcam). After incubation for 12 h, the membranes were further
incubated at room temperature with a HRP-labelled secondary
antibody (cat. no. ab205718; 1:5,000 dilution; Abcam) for 1 h.
Finally, the immunoblots were visualized using an ECL Western
Blotting Substrate kit (Abcam).
Statistical analysis
All experiments were repeated three times and all
the results are presented as the mean ± SD. Significant differences
between two groups were analyzed using a paired or unpaired
Student's t-test. Comparisons between multiple groups were analyzed
using one-way analysis of variance followed by a Tukey's post hoc
test. Pearson's correlation analysis was used to detect the
correlation between gene expression levels. According to the median
expression value of LINC01748, all patients with NSCLC were divided
into low or high LINC01748 expression groups. The association
between LINC01748/miR-520a-5p/HMGA1 expression level and the
clinicopathological parameters in patients with NSCLC was assessed
using a χ2 test. Kaplan-Meier analysis and the log-rank
test were utilized for analysis of overall survival time. P<0.05
was considered to indicate a statistically significant
difference.
Results
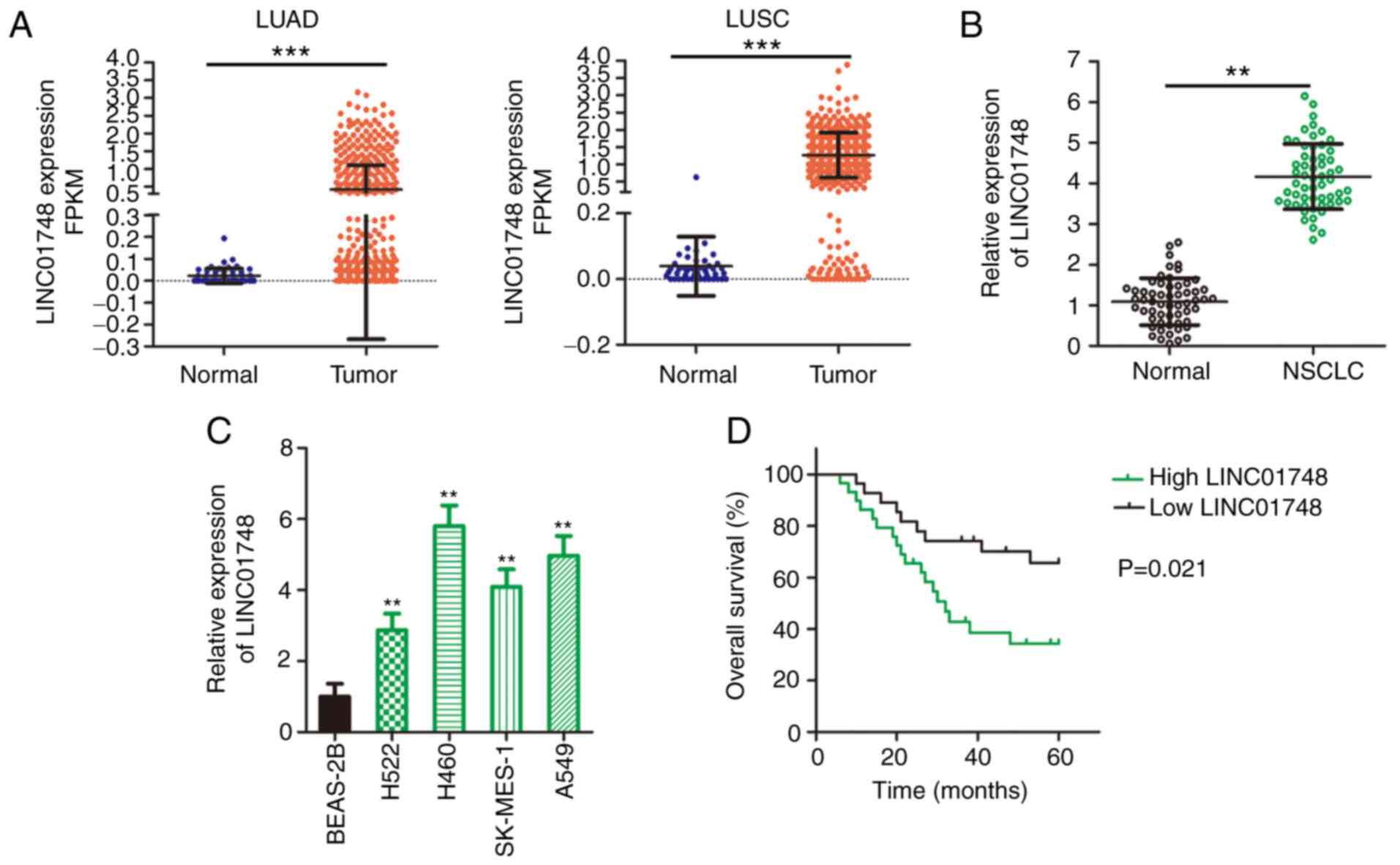
LINC01748 is upregulates in NSCLC and is
associated with poor prognosis
LINC01748 mRNA expression status in patients with
LUAD and LUSC was first analyzed using data from TCGA database. As
compared with that in normal tissues from healthy individuals,
LINC01748 was increased in LUAD and LUSC tissues (Fig. 1A). NSCLC and adjacent normal
tissues were collected from patients with NSCLC recruited into the
present study and LINC01748 mRNA expression level was detected
using RT-qPCR. The LINC01748 mRNA expression level was increased in
the NSCLC tissues compared with that in the adjacent normal tissues
(Fig. 1B). An increase in
LINC01748 mRNA expression level was associated with a larger tumor
size, TNM stage and lymph node metastasis (Table I). In addition, LINC01748 mRNA
expression level in the NSCLC cell lines was increased compared
with that in the normal BEAS-2B cell line (Fig. 1C). Furthermore, patients with
NSCLC and high LINC01748 mRNA expression levels showed shorter
overall survival time compared with that in patients with low
LINC01748 mRNA expression level (Fig. 1D). These findings suggested that
LINC01748 may contribute to cancer progression in NSCLC.
 | Table IAssociation between LINC01748 mRNA
expression level and the clinicopathological factors in patients
with non-small cell lung cancer. |
Table I
Association between LINC01748 mRNA
expression level and the clinicopathological factors in patients
with non-small cell lung cancer.
| Clinicopathological
factors | LINC01748 mRNA
expression level
| P-valuea |
|---|
| High (n=29) | Low (n=28) |
|---|
| Sex | | | 0.596 |
| Male | 18 | 15 | |
| Female | 11 | 13 | |
| Age, years | | | 0.792 |
| <60 | 14 | 12 | |
| ≥60 | 15 | 16 | |
| Tumor size, cm | | | 0.033 |
| <3 | 12 | 20 | |
| ≥3 | 17 | 8 | |
| TNM stage | | | 0.031 |
| I+II | 13 | 21 | |
| III+IV | 16 | 7 | |
| Lymph node
metastasis | | | 0.035 |
| Negative | 10 | 18 | |
| Positive | 19 | 10 | |
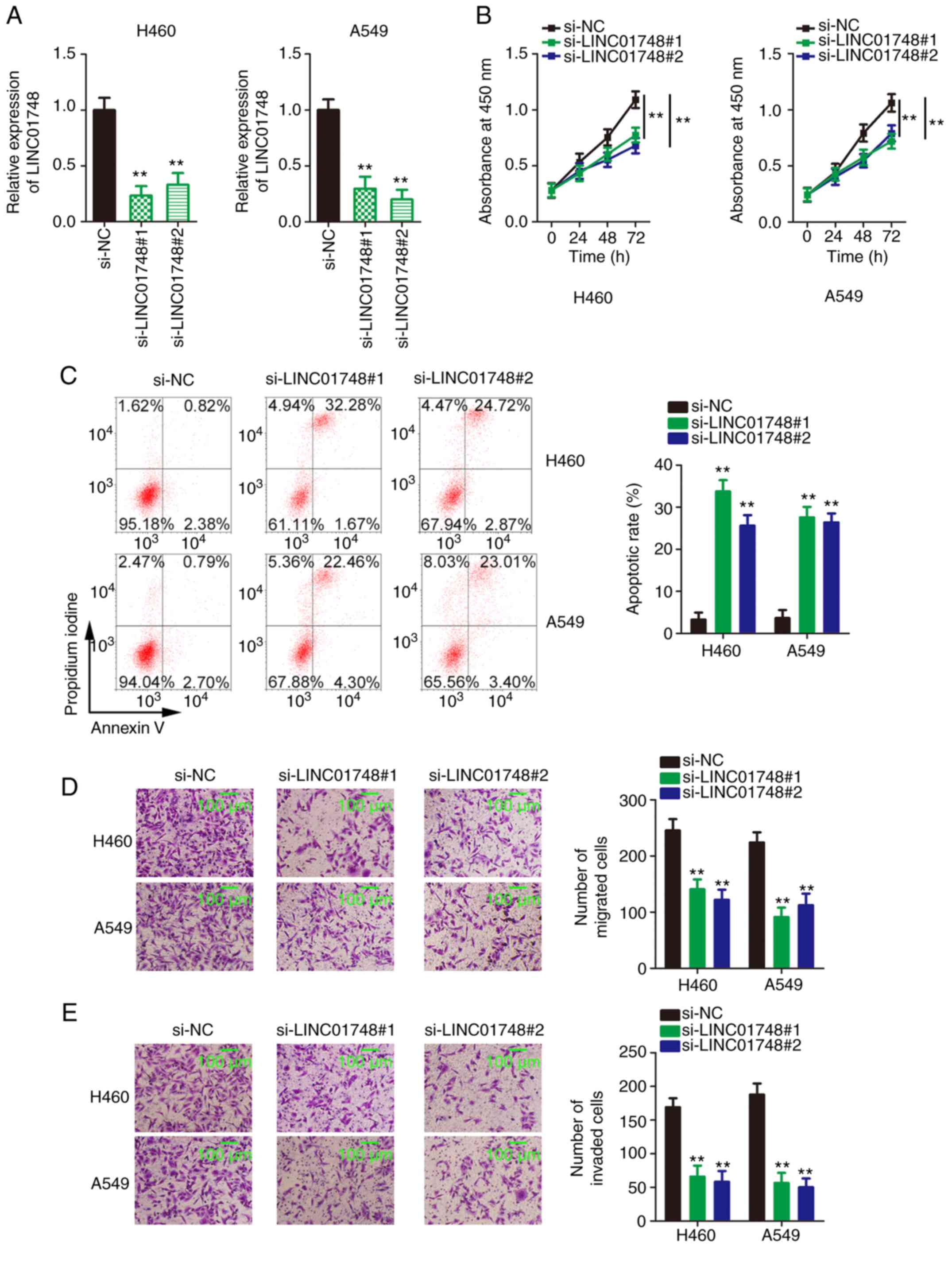
LINC01748 knockdown decreases NSCLC cell
proliferation, migration and invasion but augmented NSCLC cell
apoptosis
The RT-qPCR data, shown in Fig. 1C, revealed that among the 4 NSCLC
cell lines, LINC01748 mRNA expression level was higher in the H460
and A549 cell lines compared with that in the other 2 NSCLC cell
lines. Therefore, the H460 and A549 cell lines were used as models
for subsequent functional assays. A total of 2 siRNAs targeting
LINC01748 were used to avert off-target effects and transfection of
the two different siRNAs decreased LINC01748 expression in NSCLC
cell lines (Fig. 2A). The
proliferative ability of the NSCLC cells significantly decreased
following LINC01748 knockdown (Fig.
2B). In addition, the apoptosis of the NSCLC cells was
increased following LINC01748 knockdown (Fig. 2C). Furthermore, the migration
(Fig. 2D) and invasion (Fig. 2E) of the NSCLC cell lines was
hindered following transfection with si-LINC01748. Taken together,
LINC01748 could have carcinogenic effects and be integral to the
malignancy of the NSCLC cells.
LINC01748 serves as a miR-520a-5p
sponge
To understand the mechanisms involved for the
function of LINC01748, the subcellular location of LINC01748 in the
NSCLC cells was determined. Using a cellular fractionation assay,
most of the LINC01748 was found in the cytoplasm of the NSCLC cells
(Fig. 3A), suggesting that
LINC01748 could act as a ceRNA or miRNA sponge. Bioinformatics
prediction was performed using ENCORI, which identified 11 miRNAs
complementary to LINC01748 (Fig.
3B). Using data from TCGA database, miR-374a-5p, miR-374b-5p,
miR-520a-5p and miR-525-5p (Fig.
3C-F) were found to be downregulated in LUAD and LUSC;
therefore, they were selected for subsequent investigation. After
LINC01748 knockdown, miR-520a-5p was clearly increased, but the
levels of the other 3 miRNAs were unchanged (Fig. 3G). Thus, miR-520a-5p could be the
downstream target of LINC01748.
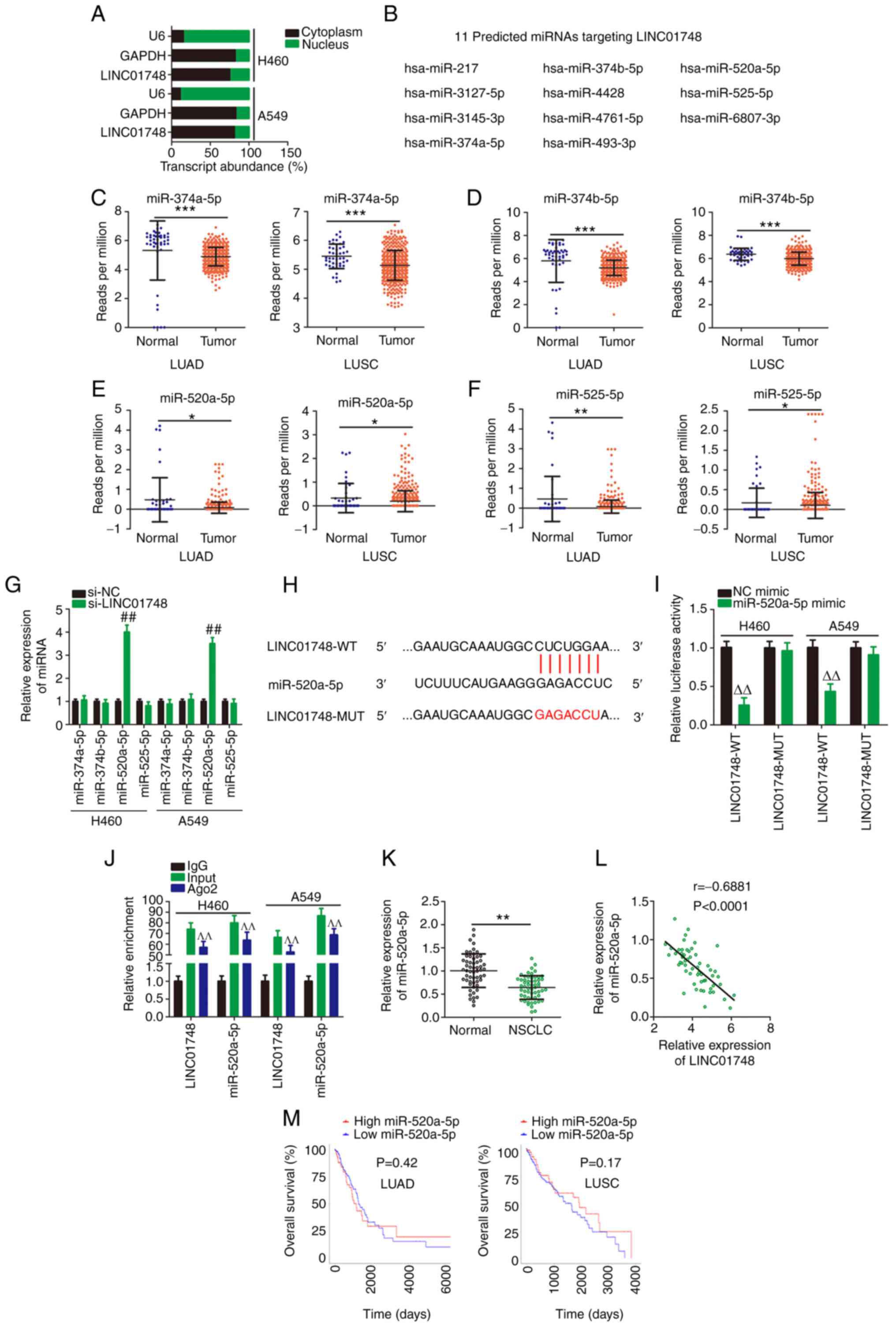 | Figure 3LINC01748 sponges miR-520a-5p. (A)
Cellular location of LINC01748 in NSCLC cells analyzed using
cellular fractionation. (B) In total, 11 predicted target miRNAs of
LINC01748 were identified using The Encyclopedia of RNA
Interactomes. Using TCGA database, the mRNA expression levels of
(C) miR-374a-5p, (D) miR-374b-5p, (E) miR-520a-5p and (F)
miR-525-5p in LUAD and LUSC were analyzed. (G) Following knockdown
of LINC01748, the mRNA expression level of miR-374a-5p,
miR-374b-5p, miR-520a-5p and miR-525-5p was determined using
reverse transcription-quantitative PCR. ##P<0.01
compared with si-NC. (H) The binding site in LINC01748 was
complementary to miR-520a-5p. (I) The measurement of luciferase
activity was analyzed in the NSCLC cells following cotransfection
with LINC01748-WT or LINC01748-Mut and miR-520a-5p mimics or NC
mimic. ∆∆P<0.01 compared with NC mimic. (J) RIP
assays demonstrated that LINC01748 and miR-520a-5p could be
precipitated by the anti-Ago2 antibody. ^^P<0.01
compared with IgG. (K) Relative miR-520a-5p mRNA expression level
in NSCLC tissues. (L) The correlation between miR-520a-5p and
LINC01748 expression in NSCLC tissues. (M) The association between
miR-520a-5p expression and overall survival time in NSCLC was
analyzed using TCGA data. *P<0.05,
**P<0.01 and ***P<0.001. LUSC, lung
squamous cell carcinoma; LUAD, lung adenocarcinoma; NC, negative
control; si, short inhibiting; miR, microRNA; WT, wild-type; Mut,
mutant. |
Luciferase reporter and RIP assays were then
performed to confirm the direct binding between miR-520a-5p and
LINC01748 (Fig. 3H). The
luciferase activity of LINC01748-WT was reduced following
miR-520a-5p overexpression, while LINC01748-Mut activity was
unaltered following miR-520a-5p mimic co-transfection (Fig. 3I). In the RIP assay, both
miR-520a-5p and LINC01748 were enriched in the Ago2-containing
beads but not in the IgG-containing beads (Fig. 3J). Subsequently, miR-520a-5p mRNA
expression levels were lower in NSCLC tissues compared with that in
adjacent normal tissues (Fig.
3K). The decreased miR-520a-5p mRNA expression level was
associated with TNM stage and lymph node metastasis (Table II). In addition, the mRNA
expression levels of miR-520a-5p and LINC01748 showed an inverse
correlation in the NSCLC tissues from 57 patients (Fig. 3L). Data from TCGA was also used
to analyze the association between miR-520a-5p mRNA expression
level and patient survival rate in patients with NSCLC. However,
miR-520a-5p mRNA expression level was not associated with overall
survival rate in patients with LUAD and LUSC (Fig. 3M). Overall, LINC01748 could act
as a miR-520a-5p sponge in NSCLC.
 | Table IIAssociation between miR-520a-5p mRNA
expression level and clinicopathological factors in patients with
non-small cell lung cancer. |
Table II
Association between miR-520a-5p mRNA
expression level and clinicopathological factors in patients with
non-small cell lung cancer.
| Clinicopathological
factors | miR-520a-5p mRNA
expression level
| P-valuea |
|---|
| High (n=29) | Low (n=28) |
|---|
| Sex | | | 0.790 |
| Male | 16 | 17 | |
| Female | 13 | 11 | |
| Age, years | | | 0.113 |
| <60 | 11 | 15 | |
| ≥60 | 18 | 13 | |
| Tumor size, cm | | | 0.596 |
| <3 | 15 | 17 | |
| ≥3 | 14 | 11 | |
| TNM stage | | | 0.016 |
| I+II | 22 | 12 | |
| III+IV | 7 | 16 | |
| Lymph node
metastasis | | | 0.017 |
| Negative | 19 | 9 | |
| Positive | 10 | 19 | |
miR-520a-5p targets HMGA1 in NSCLC
cells
To understand the roles of miR-520a-5p in NSCLC,
miR-520a-5p mimic was transfected into the H460 and A549 cell lines
to upregulate miR-520a-5p (Fig.
4A), followed by a series of functional experiments. Compared
with that in the NC mimic-transfected NSCLC cells, cell
proliferation (Fig. 4B),
migration and invasion (Fig. 4C)
was decreased, while cell apoptosis (Fig. 4D) was increased in cells
transfected with miR-520a-5p mimics.
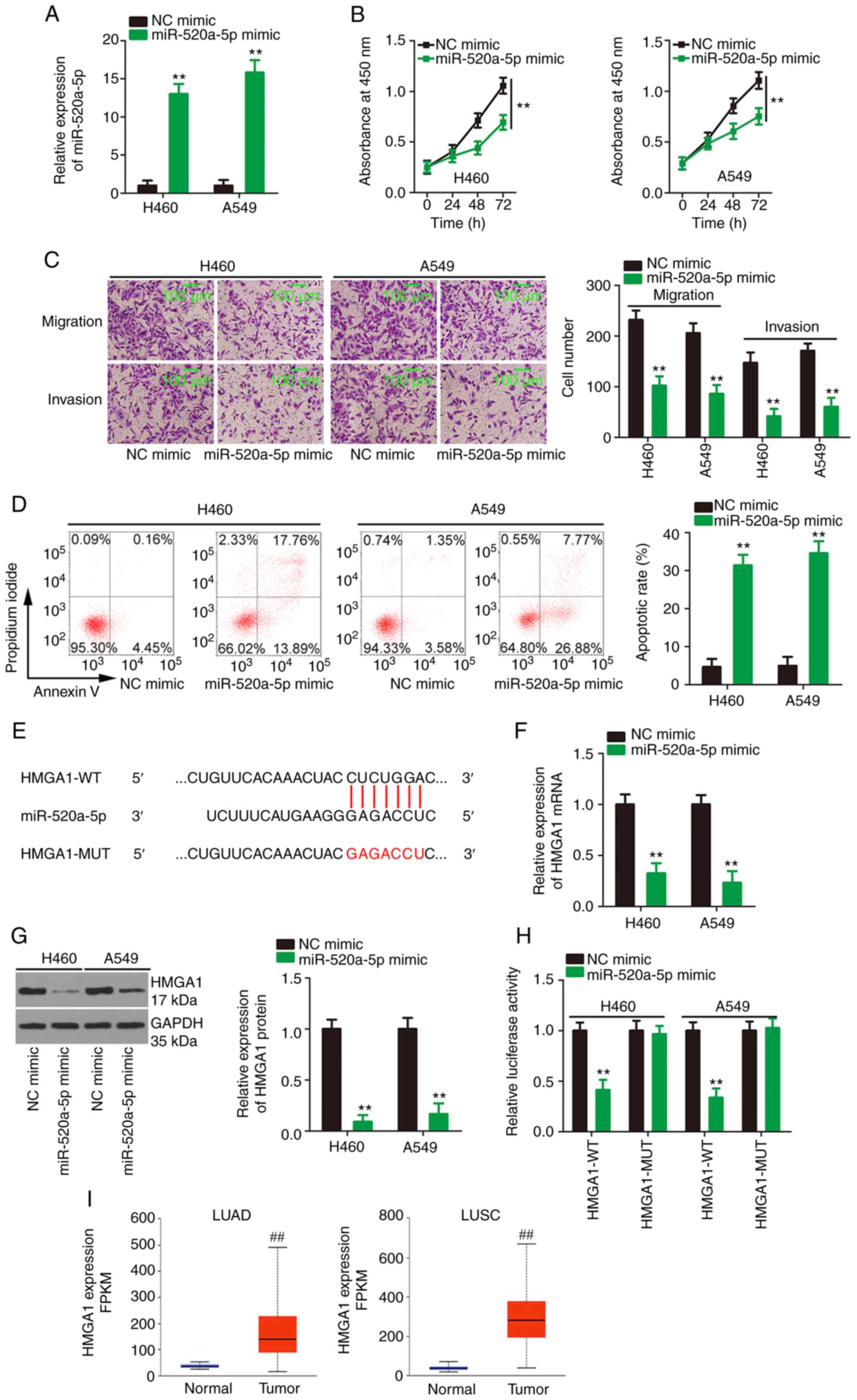 | Figure 4miR-520a-5p directly targets HMGA1.
(A) The transfection efficiency of miR-520a-5p mimic. After
miR-520a-5p was overexpressed, (B) cell proliferation (C) migration
and invasion and (D) apoptosis was analyzed in the NSCLC cell
lines. (E) The binding site for miR-520a-5p within the HMGA1 3′-UTR
was predicted. HMGA1 (F) mRNA and (G) protein expression level in
miR-520a-5p-overexpressing NSCLC cells. (H) Luciferase activity was
measured in the NSCLC cells following cotransfection with HMGA1-WT
or HMGA1-Mut and miR-520a-5p mimic or NC mimic. (I) The mRNA
expression level of HMGA1 in LUAD and LUSC was analyzed in The
Cancer Genome Atlas datasets. The cut-off value is the average
value of HMGA1 level in LUAD and LUSC. **P<0.01 and
##P<0.01. NSCLC, non-small cell lung cancer; LUSC,
lung squamous cell carcinoma; LUAD, lung adenocarcinoma; WT,
wild-type; Mut, mutant; miR, microRNA; NC, negative control; FPKM,
fragments per kilobase of transcript per million mapped reads;
HMGA1, high mobility group AT-hook 1. |
Using bioinformatics tools, HMGA1 (Fig. 4E) was identified as a potential
candidate target gene of miR-520a-5p and was selected for
experimental confirmation as a result of its crucial regulatory
effects on NSCLC progression (31-34). To investigate whether miR-520a-5p
could affect HMGA1 mRNA and protein expression level, NSCLC cells
were transfected with miR-520a-5p mimic or NC mimic and HMGA1 mRNA
and protein expression level was analyzed in the cells.
Transfection with the miR-520a-5p mimic resulted in a significant
decrease in HMGA1 mRNA and protein expression level (Fig. 4F and G, respectively) in the
NSCLC cells. Furthermore, luciferase activity in the NSCLC cells
expressing HMGA1-WT with miR-520a-5p mimics was decreased, but this
effect was counteracted when the binding sequences were mutated
(Fig. 4H). Utilizing TCGA data,
it was found that HMGA1 mRNA expression level was significantly
upregulated in LUAD and LUSC tissues (Fig. 4I). Altogether, miR-520a-5p could
directly target HMGA1 in NSCLC and could have antioncogenic
activity.
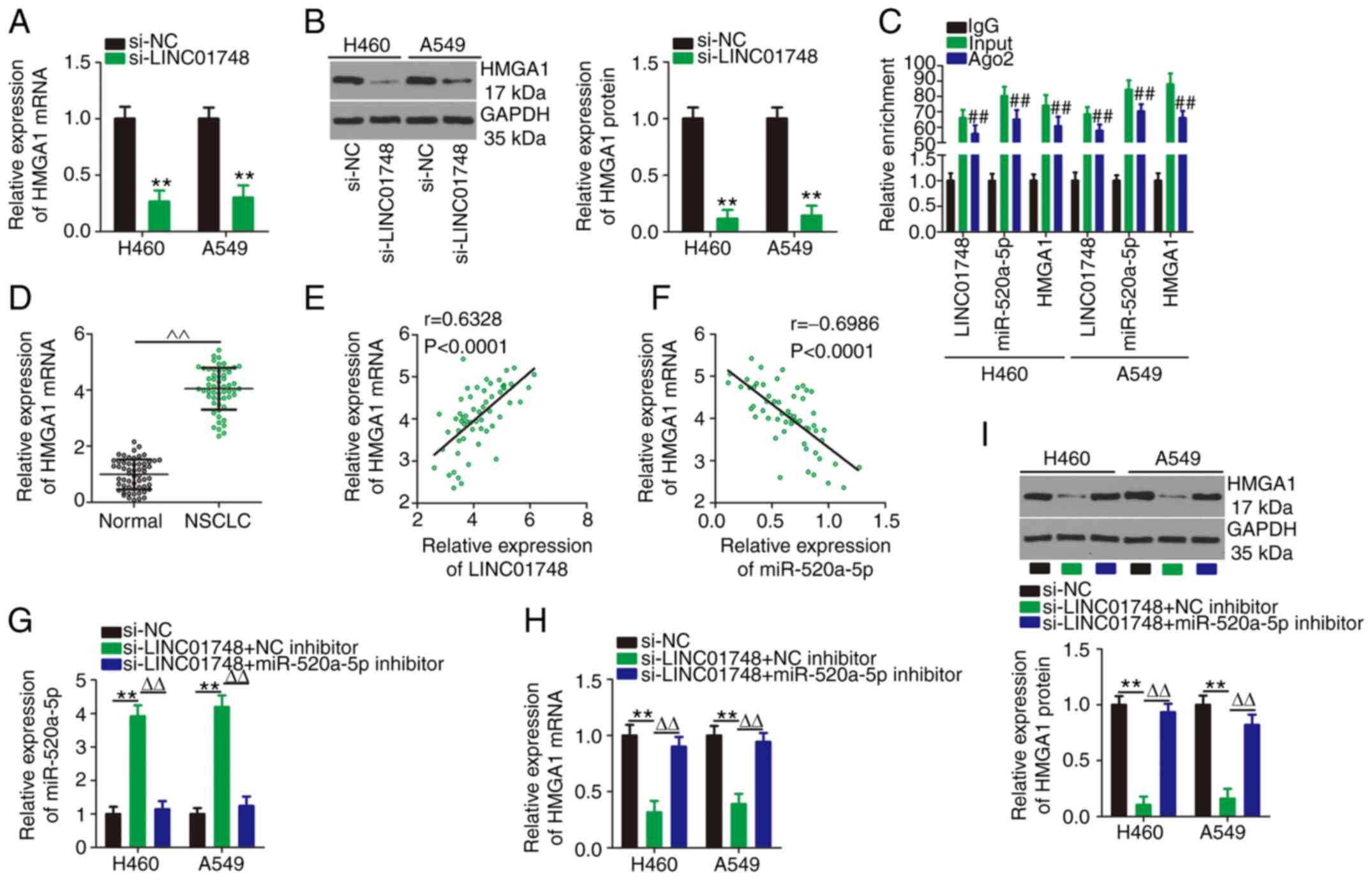
LINC01748 regulates the miR-520a-5p/HMGA1
axis in NSCLC
After identifying that miR-520a-5p could target
HMGA1 in NSCLC, the associations between LINC01748, miR-520a-5p and
HMGA1 were further investigated in NSCLC cell lines. HMGA1 mRNA and
protein expression levels (Fig. 5A
and B, respectively) was significantly reduced in the NSCLC
cells following LINC01748 knockdown. RIP assays confirmed that
LINC01748, miR-520a-5p and HMGA1 were enriched in the
Ago2-containing beads compared with that in the IgG control
(Fig. 5C). In addition, HMGA1
mRNA expression level was increased in NSCLC tissues (Fig. 5D) and was associated with tumor
size, TNM stage or lymph node metastasis (Table III). An increase in HMGA1 mRNA
expression level was positively correlated with LINC01748 mRNA
expression level (Fig. 5E). In
addition, there was a negative correlation between HMGA1 and
miR-520a-5p mRNA expression levels in NSCLC tissues (Fig. 5F). Notably, rescue experiments
illustrated that the increased miR-520a-5p mRNA (Fig. 5G) and decreased HMGA1 mRNA and
protein (Fig. 5H and I)
expression levels in LINC01748 knockdown NSCLC cells were restored
following transfection with the miR-520a-5p inhibitor. Overall,
these data indicated that LINC01748 could positively control HMGA1
expression in the NSCLC cells by acting as a decoy for
miR-520a-5p.
 | Table IIIAssociation between HMGA1 mRNA
expression level and clinicopathological factors in patients with
non-small cell lung cancer. |
Table III
Association between HMGA1 mRNA
expression level and clinicopathological factors in patients with
non-small cell lung cancer.
| Clinicopathological
factors | HMGA1 mRNA
expression level
| P-valuea |
|---|
| High (n=29) | Low (n=28) |
|---|
| Sex | | | 0.182 |
| Male | 14 | 19 | |
| Female | 15 | 9 | |
| Age, years | | | >0.9999 |
| <60 | 10 | 16 | |
| ≥60 | 19 | 12 | |
| Tumor size, cm | | | 0.001 |
| <3 | 10 | 22 | |
| ≥3 | 19 | 6 | |
| TNM stage | | | 0.003 |
| I+II | 11 | 23 | |
| III+IV | 18 | 5 | |
| Lymph node
metastasis | | | 0.008 |
| Negative | 9 | 19 | |
| Positive | 20 | 9 | |
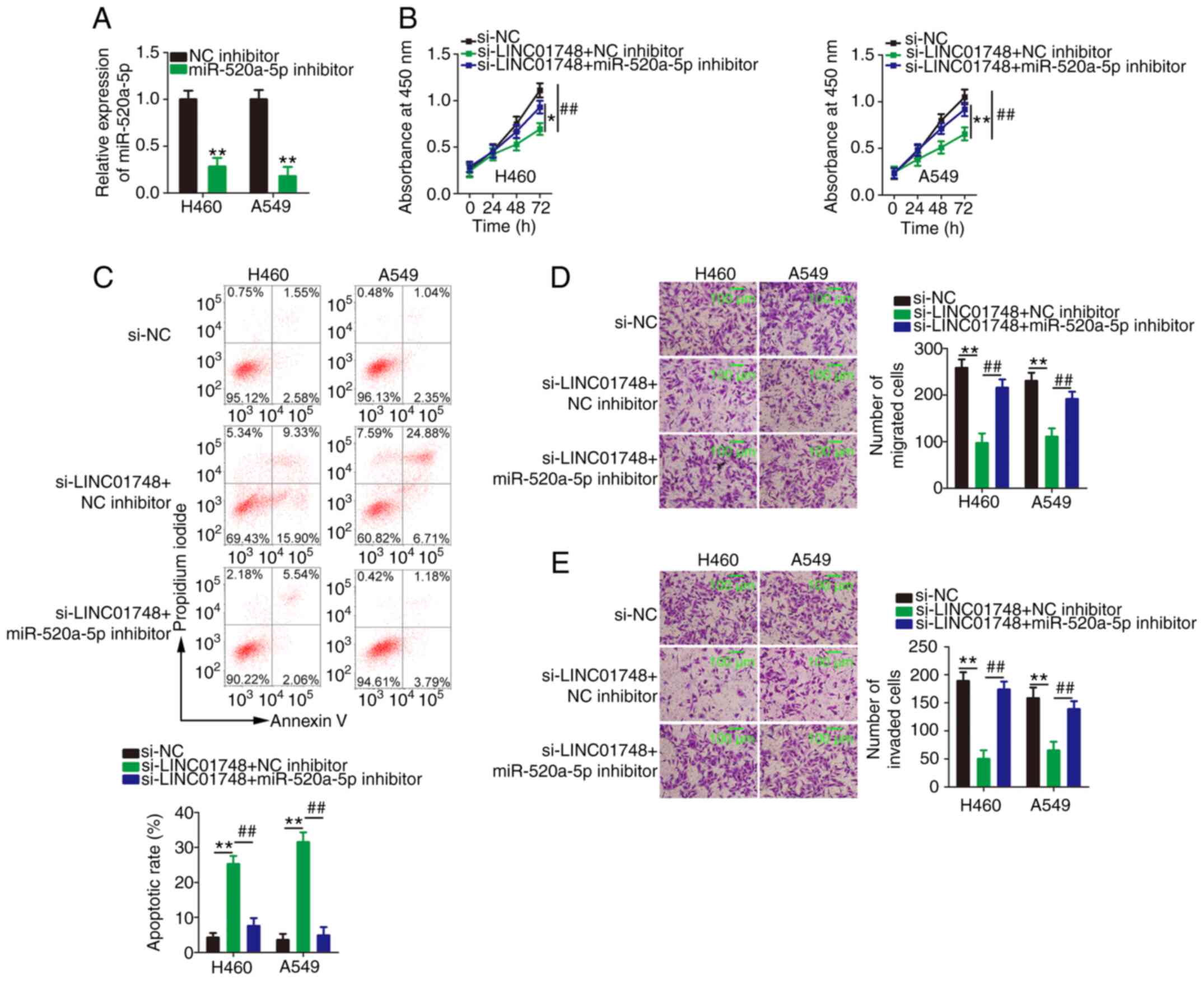
LINC01748 knockdown represses NSCLC
progression via the miR-520a-5p/HMGA1 axis
Lastly, rescue experiments were conducted to
ascertain whether LINC01748 regulated the malignancy of NSCLC cells
via the miR-520a-5p/HMGA1 axis. Initially, the efficiency of the
miR-520a-5p inhibitor in the NSCLC cells was confirmed using
RT-qPCR (Fig. 6A). NSCLC cells
with LINC01748 knocked down were further transfected with
miR-520a-5p inhibitor or NC inhibitor. The proliferation of the
NSCLC cells was significantly suppressed following LINC01748
knockdown and this effect was partly rescued by miR-520a-5p
downregulation (Fig. 6B). In
addition, the knockdown of LINC01748 promoted NSCLC cell apoptosis
and this effect was neutralized by the miR-520a-5p inhibitor
(Fig. 6C). Furthermore,
migration and invasion assays revealed a decrease in the migration
and invasion (Fig. 6D and E) of
the NSCLC cells, but transfection with miR-520a-5p inhibitor offset
these repressive effects.
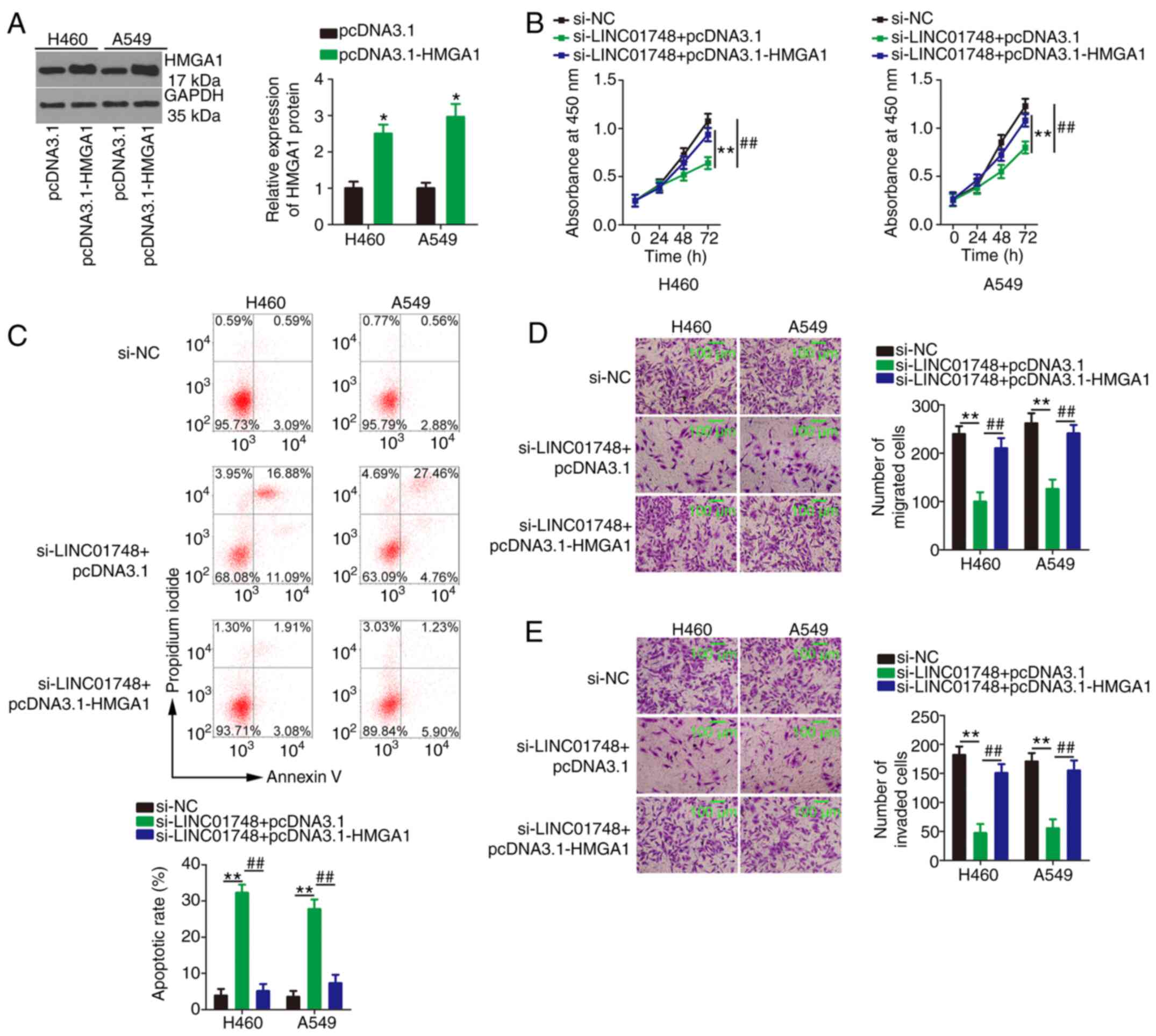
Meanwhile, pcDNA3.1-HMGA1 (Fig. 7A) or empty pcDNA3.1 plasmid was
transfected alongside si-LINC01748 into the NSCLC cells.
Proliferation (Fig. 7B) was
significantly reduced, whereas cell apoptosis (Fig. 7C) was increased in the LINC01748
knockdown NSCLC cells, and these effects were abolished following
transfection with pcDNA3.1-HMGA1. In addition, overexpression of
HMGA1 abrogated the suppressive effect of si-LINC01748 on NSCLC
cell migration (Fig. 7D) and
invasion (Fig. 7E). In short,
the aforementioned results suggested that the miR-520a-5p/HMGA1
axis mediated LINC01748-induced promotion of NSCLC malignancy.
Targeting LINC01748 suppresses NSCLC
tumor growth in vivo
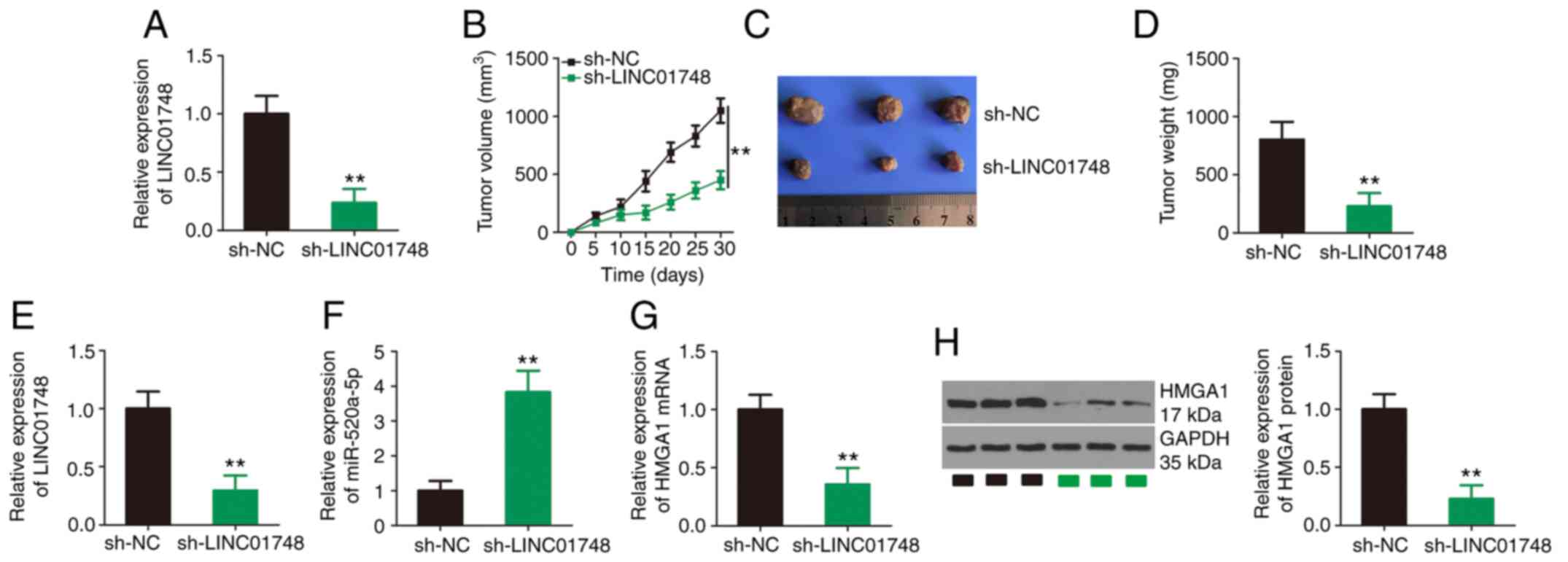
To investigate the effect of LINC01748 on tumor
growth in vivo, a tumor xenograft model was established
using A549 cells expressing sh-LINC01748 or sh-NC. Initially, the
efficiency of sh-LINC01748 transfection was assessed using RT-qPCR
and the results are shown in Fig.
8A. Compared with that in the sh-NC group, the sh-LINC01748
group showed substantially reduced tumor growth (Fig. 8B) with a notably lower tumor size
(Fig. 8C) and weight (Fig. 8D). Expression analysis of the
tumor xenografts revealed that LINC01748 was notably downregulated
in xenografts from the sh-LINC01748 group (Fig. 8E), suggesting the successful and
sustained silencing of LINC01748. Furthermore, the miR-520a-5p mRNA
expression level was increased (Fig.
8F), while the HMGA1 mRNA and protein expression levels
(Fig. 8G and H, respectively)
were decreased in the tumor xenografts originating from
sh-LINC01748-transfected A549 cells. Thus, the knockdown of
LINC01748 inhibited NSCLC tumor growth in vivo.
Discussion
The regulatory activities of lncRNAs in NSCLC
pathogenesis have received increasing attention and recognition
(35-37). Thus, investigating the specific
functions of lncRNAs would be instrumental in developing effective
targets for NSCLC therapy. The important roles of lncRNAs in the
malignancy of NSCLC have been increasingly highlighted (38-40); however, the majority of lncRNA
expression patterns and their exact roles are not completely
understood. The present study increased the understanding of the
lncRNA-miRNA-mRNA network by revealing that LINC01748 could act as
a ceRNA of miR-520a-5p, whereas miR-520a-5p directly targeted HMGA1
to exacerbate the oncogenesis of NSCLC.
Numerous lncRNAs are dysregulated in NSCLC and have
been associated with the regulation of malignant tumor
characteristics (17,41,42). For example, MALAT1 (43), DGCR5 (44) and NCK1-AS1 (45) were found to be increased in NSCLC
tissues and were associated with the oncogenicity of NSCLC. By
contrast, LINC01089 (46),
LINC01089 (47) and DHRS4-AS1
(48) are found to be decreased
in NSCLC and could play anti-carcinogenic roles. However, the
detailed roles of LINC01748 in NSCLC remain unclear; thus, there is
an urgent requirement for further investigation. In the present
study, there was an increase in the mRNA expression level of
LINC01748 in NSCLC tissues in data from TCGA database and in
patients with NSCLC recruited into the current study. Compared with
that in patients with low LINC01748mRNA expression level, patients
with high LINC01748 mRNA expression level displayed shorter overall
survival time. Then, knockdown of LINC01748 revealed that LINC01748
could maintain the malignancy of the NSCLC cells. The knockdown of
LINC01748 attenuated NSCLC cell proliferation, migration and
invasion, but promoted apoptosis in vitro. In addition,
LINC01748 knockdown decreased tumor growth in vivo.
The molecular mechanisms behind the tumor-promoting
activities of LINC01748 in NSCLC require further investigation. The
roles played by lncRNAs are complex and have not yet been entirely
clarified; however, the subcellular location of lncRNAs determines
their mechanism of action (49).
The ceRNA theory is recognized as a novel mechanism of cytoplasmic
lncRNAs. In this theory, lncRNAs exert 'sponge-like' activity for
miRNAs, consequently lowering the regulatory effects of miRNAs on
downstream genes (50).
Next, the subcellular location of LINC01748 in the
NSCLC cells was investigated and whether LINC01748 functioned as a
ceRNA or natural miRNA sponge. The results revealed that LINC01748
was a cytoplasmic lncRNA, which could directly interact with
miR-520a-5p. Subsequent mechanistic experiments, such as luciferase
reporter assays, RIP assays, molecular analysis and rescue
experiments showed that LINC01748 could act as a ceRNA to sponge
miR-520a-5p, thereby increasing the mRNA expression level of the
miR-520a-5p target, HMGA1, in NSCLC cells. Specifically, to the
best of our knowledge, a new ceRNA pathway, the
LINC01748/miR-520a-5p/HMGA1 pathway, in NSCLC was identified for
the first time.
miR-520a-5p has been investigated in numerous
studies and was found to be expressed at low levels in chronic
myelogenous leukemia (51) and
breast cancer (52), and have
anticarcinogenic effects (51,52). A previous study has reported low
miR-520a-5p mRNA expression level in NSCLC (53). The results in the present study
further corroborate the tumor-inhibiting effect of miR-520a-5p in
NSCLC cells. CDK4 (53), GOT-2
(54), ORMDL3 (52) and STAT3 are proven targets of
miR-520a-5p. Therefore, the putative target of miR-520a-5p was
investigated and HMGA1 was selected for experimental confirmation
as a result of its crucial regulatory effects on NSCLC progression
(31-34). Increased HMGA1 mRNA expression
levels in NSCLC were found to be significantly associated with
clinical stage, differentiation, TNM stage and poor prognosis
(33). Furthermore, HMGA1 was
verified as an independent predictive factor for the prognosis of
patients with NSCLC (33). HMGA1
exerts pro-oncogenic activities in NSCLC and was associated with
aggressive phenotypes (31-34).
Previously, 2 lncRNAs, VPS9D1-AS1 (55) and TRPM2-AS (56), were associated with the
regulation of HMGA1 in NSCLC cell lines. In the present study, to
the best of our knowledge, it was confirmed for the first time that
miR-520a-5p was sponged by LINC01748 and was negatively regulated
by this lncRNA in NSCLC cell lines. In addition, HMGA1 was found to
be under control of the LINC01748/miR-520a-5p axis in 2 NSCLC cell
lines at the posttranscriptional level. Furthermore, the effects of
LINC01748 knockdown in 2 NSCLC cells lines were negated by
decreasing the expression levels of miR-520a-5p/HMGA1. Therefore,
the carcinogenic role of LINC01748 in the NSCLC cell lines was
accomplished by sequestering miR-520a-5p and weakening the
inhibitory effect of the latter on HMGA1.
In the current study, it was found that the
proliferation, apoptosis, migration and invasion of the NSCLC cell
lines were affected by LINC01748 however, the expression levels of
the proliferation-, apoptosis-, migration- and invasion-related
proteins were not examined in the NSCLC cell lines following
LINC01748 knockdown. Furthermore, the effect of the
LINC01748/miR-520a-5p/HMGA1 axis on cell colony formation and wound
healing were not determined. In addition, the regulatory effects of
HMGA1 on the proliferation, apoptosis, migration and invasion
abilities of the NSCLC cells were not investigated. These
limitations will be investigated in further studies.
In the present study, the oncogenic activities of
the LINC01748/miR-520a-5p/HMGA1 axis was investigated in 2 NSCLC
cell lines. LINC01748 was found to be overexpressed in NSCLC
tissues and was associated with poor patient prognosis. This lncRNA
sponged miR-520a-5p, leading to HMGA1 overexpression; thus,
exacerbating the aggressiveness of the NSCLC cell lines.
Accordingly, targeting the LINC01748/miR-520a-5p/HMGA1 pathway may
be beneficial as a NSCLC therapy. The role of LINC01748 in other
types of cancer have never been examined, which highlights the
novelty of the present study.
Availability of data and materials
The datasets used and/or analyzed in the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
This study was designed by YT and JC, and confirm
the authenticity of the data. JC analyzed the data. All the
experiments were performed by YT, FX, LX and JC. YT and JC wrote
the manuscript. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
The Ethics Committee of Weifang Yidu Central
Hospital (Shandong, China) approved the present research. The
animal experiment was approved by the Ethics Committee of Animal
Experiments at Weifang Yidu Central Hospital (Shandong, China).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgments
Not applicable.
Funding
This research was supported by Weifang Yidu Central
Hospital.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2020. CA Cancer J Clin. 70:7–30. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Travis WD: Lung cancer pathology: Current
concepts. Clin Chest Med. 41:67–85. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Lemjabbar-Alaoui H, Hassan OU, Yang YW and
Buchanan P: Lung cancer: Biology and treatment options. Biochim
Biophys Acta. 1856:189–210. 2015.PubMed/NCBI
|
|
5
|
Herbst RS, Morgensztern D and Boshoff C:
The biology and management of non-small cell lung cancer. Nature.
553:446–454. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
de Mello RA, Neves NM, Tadokoro H, Amaral
GA, Castelo-Branco P and Zia VAA: New target therapies in advanced
non-small cell lung cancer: A review of the literature and future
perspectives. J Clin Med. 9:35432020. View Article : Google Scholar :
|
|
7
|
Alexander M, Kim SY and Cheng H: Update
2020: Management of non-small cell lung cancer. Lung. 198:897–907.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Duma N, Santana-Davila R and Molina JR:
Non-small cell lung cancer: Epidemiology, screening, diagnosis, and
treatment. Mayo Clin Proc. 94:1623–1640. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Batista PJ and Chang HY: Long noncoding
RNAs: Cellular address codes in development and disease. Cell.
152:1298–1307. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Bhan A, Soleimani M and Mandal SS: Long
noncoding RNA and cancer: A new paradigm. Cancer Res. 77:3965–3981.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Qin T, Li J and Zhang KQ: Structure,
regulation, and function of linear and circular long non-coding
RNAs. Front Genet. 11:1502020. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
He RZ, Luo DX and Mo YY: Emerging roles of
lncRNAs in the post-transcriptional regulation in cancer. Genes
Dis. 6:6–15. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Gao T and Ji Y: Long noncoding RNA
LINC00707 accelerates tumorigenesis and progression of bladder
cancer via targeting miR-145/CDCA3 regulatory loop. Urol Int. Jun
30–2021.Epub ahead of print. View Article : Google Scholar
|
|
14
|
Cao S, Li H and Li L: LncRNA SNHG17
contributes to the progression of cervical cancer by targeting
microRNA-375-3p. Cancer Manag Res. 13:4969–4978. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wang S, Han H, Meng J, Yang W, Lv Y and
Wen X: Long non-coding RNA SNHG1 suppresses cell migration and
invasion and upregulates SOCS2 in human gastric carcinoma. Biochem
Biophys Rep. 27:1010522021.PubMed/NCBI
|
|
16
|
Xu J, Wang J, He Z, Chen P, Jiang X, Chen
Y, Liu X and Jiang J: LncRNA CERS6-AS1 promotes proliferation and
metastasis through the upregulation of YWHAG and activation of ERK
signaling in pancreatic cancer. Cell Death Dis. 12:6482021.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wan Y, Yao D, Fang F, Wang Y, Wu G and
Qian Y: LncRNA WT1-AS downregulates lncRNA UCA1 to suppress
non-small cell lung cancer and predicts poor survival. BMC Cancer.
21:1042021. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Lin JH, Chen FN, Wu CX, Hu SQ and Ma J:
Long non-coding RNA B4GALT1-Antisense RNA 1/microRNA-30e/SRY-box
transcription factor 9 signaling axis contributes to non-small cell
lung cancer cell growth. Oncol Lett. 20:2842020. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Pang L, Zhang Q, Wu Y, Yang Q, Zhang J,
Liu Y and Li R: Long non-coding RNA CCAT1 promotes non-small cell
lung cancer progression by regulating the miR-216a-5p/RAP2B axis.
Exp Biol Med (Maywood). 246:142–152. 2021. View Article : Google Scholar
|
|
20
|
Ye R, Tang R, Gan S, Li R, Cheng Y, Guo L,
Zeng C and Sun Y: New insights into long non-coding RNAs in
non-small cell lung cancer. Biomed Pharmacother. 131:1107752020.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Calin GA and Croce CM: MicroRNA signatures
in human cancers. Nat Rev Cancer. 6:857–866. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Guo H, Ingolia NT, Weissman JS and Bartel
DP: Mammalian microRNAs predominantly act to decrease target mRNA
levels. Nature. 466:835–840. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Castro D, Moreira M, Gouveia AM, Pozza DH
and De Mello RA: MicroRNAs in lung cancer. Oncotarget.
8:81679–81685. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zang H, Wang W and Fan S: The role of
microRNAs in resistance to targeted treatments of non-small cell
lung cancer. Cancer Chemother Pharmacol. 79:227–231. 2017.
View Article : Google Scholar
|
|
25
|
Florczuk M, Szpechcinski A and
Chorostowska-Wynimko J: MiRNAs as biomarkers and therapeutic
targets in non-small cell lung cancer: Current perspectives. Target
Oncol. 12:179–200. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Salmena L, Poliseno L, Tay Y, Kats L and
Pandolfi PP: A ceRNA hypothesis: The Rosetta stone of a hidden RNA
language? Cell. 146:353–358. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Fu Y, Liu L, Zhan J, Zhan H and Qiu C:
LncRNA GAS5 expression in non-small cell lung cancer tissues and
its correlation with Ki67 and EGFR. Am J Transl Res. 13:4900–4907.
2021.PubMed/NCBI
|
|
28
|
Jia D, Xing Y, Zhan Y, Cao M, Tian F, Fan
W, Huang J, Cui Y, Gu R, Cui Y, et al: LINC02678 as a novel
prognostic marker promotes aggressive non-small-cell lung cancer.
Front Cell Dev Biol. 9:6869752021. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Zhang L, Chi B, Chai J, Qin L, Zhang G,
Hua P and Jin C: LncRNA CCDC144NL-AS1 serves as a prognosis
biomarker for non-small cell lung cancer and promotes cellular
function by targeting miR-490-3p. Mol Biotechnol. Jun 11–2021.Epub
ahead of print. View Article : Google Scholar
|
|
30
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
31
|
Sarhadi VK, Wikman H, Salmenkivi K, Kuosma
E, Sioris T, Salo J, Karjalainen A, Knuutila S and Anttila S:
Increased expression of high mobility group A proteins in lung
cancer. J Pathol. 209:206–212. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Cao YD, Huang PL, Sun XC, Ma J, Jin ZL,
Cheng HY, Xu RZ, Li F, Qin SK, Deng YX and Ge XL: Silencing of high
mobility group A1 enhances gemcitabine chemosensitivity of lung
adenocarcinoma cells. Chin Med J (Engl). 124:1061–1068. 2011.
|
|
33
|
Zhang Z, Wang Q, Chen F and Liu J:
Elevated expression of HMGA1 correlates with the malignant status
and prognosis of non-small cell lung cancer. Tumour Biol.
36:1213–1219. 2015. View Article : Google Scholar
|
|
34
|
Ma Y, Li X, Chen S, Du B and Li Y:
MicroRNA-4458 suppresses migration and epithelial-mesenchymal
transition via targeting HMGA1 in non-small-cell lung cancer cells.
Cancer Manag Res. 11:637–649. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Peng W, Wang J, Shan B, Peng Z, Dong Y,
Shi W, He D, Cheng Y, Zhao W, Zhang C, et al: Diagnostic and
prognostic potential of circulating long non-coding RNAs in non
small cell lung cancer. Cell Physiol Biochem. 49:816–827. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Lu T, Wang Y, Chen D, Liu J and Jiao W:
Potential clinical application of lncRNAs in non-small cell lung
cancer. Onco Targets Ther. 11:8045–8052. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Fang C, Wang L, Gong C, Wu W, Yao C and
Zhu S: Long non-coding RNAs: How to regulate the metastasis of
non-small-cell lung cancer. J Cell Mol Med. 24:3282–3291. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Chen J, Wang R, Zhang K and Chen LB: Long
non-coding RNAs in non-small cell lung cancer as biomarkers and
therapeutic targets. J Cell Mol Med. 18:2425–2436. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Zhan Y, Zang H, Feng J, Lu J, Chen L and
Fan S: Long non-coding RNAs associated with non-small cell lung
cancer. Oncotarget. 8:69174–69184. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Wang L, Ma L, Xu F, Zhai W, Dong S, Yin L,
Liu J and Yu Z: Role of long non-coding RNA in drug resistance in
non-small cell lung cancer. Thorac Cancer. 9:761–768. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Zhao MM, Ge LY, Yang LF, Zheng HX, Chen G,
Wu LZ, Shi SM, Wang N and Hang YP: LncRNA NEAT1/miR-204/NUAK1 Axis
is a potential therapeutic target for non-small cell lung cancer.
Cancer Manag Res. 12:13357–13368. 2020. View Article : Google Scholar
|
|
42
|
Guo K, Qi D and Huang B: LncRNA MEG8
promotes NSCLC progression by modulating the
miR-15a-5p-miR-15b-5p/PSAT1 axis. Cancer Cell Int. 21:842021.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Wang S, Wang T, Liu D and Kong H: LncRNA
MALAT1 aggravates the progression of non-small cell lung cancer by
stimulating the expression of COMMD8 via targeting miR-613. Cancer
Manag Res. 12:10735–10747. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Wang J, Shu HZ, Xu CY and Guo SG: LncRNA
DGCR5 promotes non-small cell lung cancer progression via sponging
miR-218-5p. Eur Rev Med Pharmacol Sci. 24:103032020.PubMed/NCBI
|
|
45
|
Li J, Wu X, Cao W and Zhao J: Long
non-coding RNA NCK1-AS1 promotes the proliferation, migration and
invasion of non-small cell lung cancer cells by acting as a ceRNA
of miR-137. Am J Transl Res. 12:6908–6920. 2020.PubMed/NCBI
|
|
46
|
Zhang D, Cai X, Cai S, Chen W and Hu C:
Long intergenic non-protein coding RNA 01089 weakens tumor
proliferation, migration, and invasion by sponging miR-3187-3p in
non-small cell lung cancer. Cancer Manag Res. 12:12151–12162. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Li X, Lv F, Li F, Du M, Liang Y, Ju S, Liu
Z, Zhou B, Wang B and Gao Y: LINC01089 inhibits tumorigenesis and
epithelial-mesenchymal transition of non-small cell lung cancer via
the miR-27a/SFRP1/Wnt/β-catenin axis. Front Oncol. 10:5325812020.
View Article : Google Scholar
|
|
48
|
Yan F, Zhao W, Xu X, Li C, Li X, Liu S,
Shi L and Wu Y: LncRNA DHRS4-AS1 inhibits the stemness of NSCLC
cells by sponging miR-224-3p and upregulating TP53 and TET1. Front
Cell Dev Biol. 8:5852512020. View Article : Google Scholar
|
|
49
|
Qi X, Zhang DH, Wu N, Xiao JH, Wang X and
Ma W: ceRNA in cancer: Possible functions and clinical
implications. J Med Genet. 52:710–718. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Wang L, Cho KB, Li Y, Tao G, Xie Z and Guo
B: Long noncoding RNA (lncRNA)-mediated competing endogenous RNA
networks provide novel potential biomarkers and therapeutic targets
for colorectal cancer. Int J Mol Sci. 20:57582019. View Article : Google Scholar :
|
|
51
|
Kaymaz BT, Cetintas VB, Aktan C and Kosova
B: MicroRNA-520a-5p displays a therapeutic effect upon chronic
myelogenous leukemia cells by targeting STAT3 and enhances the
anticarcinogenic role of capsaicin. Tumour Biol. 35:8733–8742.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Liu X, Song J, Kang Y, Wang Y and Chen A:
Long noncoding RNA SOX21-AS1 regulates the progression of
triple-negative breast cancer through regulation of
miR-520a-5p/ORMDL3 axis. J Cell Biochem. 121:4601–4611. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Xu X, Tao R, Sun L and Ji X:
Exosome-transferred hsa_circ_0014235 promotes DDP chemoresistance
and deteriorates the development of non-small cell lung cancer by
mediating the miR-520a-5p/CDK4 pathway. Cancer Cell Int.
20:5522020. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Jin M, Shi C, Hua Q, Li T, Yang C, Wu Y,
Zhao L, Yang H, Zhang J, Hu C and Huang G: High circ-SEC31A
expression predicts unfavorable prognoses in non-small cell lung
cancer by regulating the miR-520a-5p/GOT-2 axis. Aging (Albany NY).
12:10381–10397. 2020. View Article : Google Scholar
|
|
55
|
Liu H, Zhang X, Jin X, Yang Y, Liang G, Ma
Y and Wang B: Long Noncoding RNA VPS9D1-AS1 Sequesters
microRNA-525-5p to promote the oncogenicity of colorectal cancer
cells by upregulating HMGA1. Cancer Manag Res. 12:9915–9928. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Huang B, Chang C, Wang BL and Li H:
ELK1-induced upregulation of lncRNA TRPM2-AS promotes tumor
progression in gastric cancer by regulating miR-195/HMGA1 axis. J
Cell Biochem. 120:16921–16933. 2019. View Article : Google Scholar : PubMed/NCBI
|