Introduction
Acute kidney injury (AKI) is a frequent clinical
emergency associated with various aetiologies and
pathophysiological processes leading to decreased glomerular
filtration function, and is considered an independent predictor of
mortality in hospitalized patients, especially for high-risk
patients. Approximately 1.7 million individuals succumb to AKI
worldwide every year (1,2). The most common causes of AKI
include ischemia, hypoxia, or nephrotoxicity (3). An underlying feature is a rapid
decline in glomerular filtration rate usually associated with
decreases in renal blood flow. The precise cause underlying the
pathology of the disease remains unknown. Identification of gene
signatures associated with disease onset and progression would help
to improve understanding of the molecular mechanisms involved in
the disease pathogenesis.
Pathogenically, AKI is generally described as the
injury of renal tubular epithelial cells and vasculature,
accompanied by the activation of a robust inflammatory response
(4). Inflammation response has
been recognized to play an important role in ischemic and toxic
models of AKI (5,6). In experimental models, tubular cell
injury and death have been revealed to be directly prevented by
suppressing inflammation (7).
Alternative splicing (AS) allows mRNA to produce
different protein subtypes, which represents an important
post-transcriptional regulatory mechanism for gene expression and
greatly expands gene coding capabilities, protein diversity and
biology function in eukaryotic organisms (8). AS is a highly-regulated process
performed by an intricate molecular machine-spliceosome; any fault
in this regulation can result in cellular dysfunction and disease
(9). More than 2,000 splicing
mutation disease entities are known, resulting in 370 diseases and
involving 303 genes (10). An
increasing number of splice isoforms have been previously reported
to be associated with various kidney diseases (11). Alternative splicing events (ASEs)
are largely controlled by RNA-binding proteins (RBPs) that
recognize specific regulatory sequences embedded in the pre-mRNA
transcripts and coordinate with the function of context-dependent
genetic mechanisms (12).
Inflammation, AS and RBPs in cisplatin and
ischemia-reperfusion (IR)-induced AKI was identified. In
vitro experiments verified that the RBP gene RNA binding fox-1
homolog 1 (RBFOX1) was involved in the pathogenesis of AKI. This
comprehensive analysis may reveal the expression of inflammation,
AS and RBPs in AKI and the role of RBFOX1 in this disease. In
conclusion, the present study provided novel insights into the
pathogenesis of AKI.
Materials and methods
Establishment of the AKI mouse model
A total of 30 male C57BL/6J mice (six to eight weeks
old; 20-25 g) were purchased from Beijing Vital River Laboratory
Animal Technology Co., Ltd. All mice were adaptively reared for one
week in a standardized environment with a 12-h light/dark cycle and
a controlled 20-26°C temperature and 40-70% humidity. Access to
food and water was ad libitum. Mice were randomly divided
into three groups (n=10/group): Control, cisplatin and IR. After
the C57BL/6J mice were adapted to the environment for one week, the
AKI model was established by a single injection of cisplatin and
IR. In accordance with a previous study on the dosage and duration
of cisplatin (13), a
cisplatin-induced AKI model was established by intraperitoneal
injection (at approximately 0.5 cm on both sides of the midline of
the hypogastrium) of cisplatin (cat. no. HY-17394; MedChemExpress)
dissolved in physiological saline at a single dose of 30 mg/kg body
weight for 24 h. Under a 3-4% induction dose and 1-2% maintenance
dose of isoflurane anesthesia, the bilateral renal pedicles were
clamped through a midline abdominal incision for 30 min and blood
flow was restored for 24 h to establish an AKI model induced by
severe IR injury (14). The
control mice were not given special intervention under the same
feeding conditions. The vital signs of the treated mice were
observed every 6 h. Within 24 h, 3 mice in the cisplatin group and
1 mouse in the IR group succumbed. Death was verified by applying
pressure on the mouse nail bed (toe-pinch reflex). After 24 h, all
mice were sacrificed under isoflurane anesthesia, 0.5-0.6 ml blood
was collected from the heart and both kidneys were dissected after
cardiac perfusion with physiological saline solution. All animal
procedures in the present study were approved (approval no.
WDRM-20200904) by the Ethics Review Committee of Animal Welfare of
Renmin Hospital of Wuhan University (Wuhan, China) and in
accordance with the National Institutes of Health Guide for the
Care and Use of Laboratory Animals.
Renal function assessment
The supernatant was collected after the blood
samples were centrifuged at 4°C and 1,000 × g for 15 min. According
to the protocol of the kits provided by the manufacturer (cat. nos.
K066 and K072a; Changchun Huili Biotech Co., Ltd.), serum
creatinine (Cr) and blood urea nitrogen (BUN) levels, respectively,
were detected using an automatic biochemical analyzer (Rayto Life
and Analytical Sciences Co., Ltd.).
Histopathological studies and
immunohistochemical staining
The kidney tissue was fixed in 4% paraformaldehyde
for 24 h at room temperature and paraffin embedding and then cut
into 4-µm-thick sections and placed on a glass slide. For
periodic acid-Schiff (PAS) staining, the slices were placed in PAS
dye solution staining for 45 min at 37°C. In accordance with the
terminal deoxynucleotidyl transferase dUTP nick-end labeling
(TUNEL) kit protocol provided by the manufacturer (cat. no. G1507;
Wuhan Servicebio Technology Co., Ltd.), the tissue sections were
incubated in the reaction solution at 37°C for 1 h. At 37°C, the
nucleus was stained in 3,3N-diaminobenzidine tertrahydrochloride
and developed for 30 min followed by counterstaining in hematoxylin
staining solution for 1 min. The sections were then briefly dried
and mounted with resin mounting medium. The tissue and cell
sections were blocked with 5% goat serum at room temperature for 30
min and then incubated with rabbit anti-RBFOX1 (1:1,000; cat. no.
ab254413; Abcam) overnight at 4°C and goat anti-rabbit secondary
antibody (1:100; cat. no. GB21303; Wuhan Servicebio Technology Co.,
Ltd.) labeled with sulfo-Cyanine3 dye at room temperature for 30
min; finally, the nuclei were counterstained using 2 µg/ml
4′,6-diamidino-2-phenylindole at room temperature for 10 min. A
total of 10 fields of view were randomly selected from each slice
under an optical fluorescence microscope for evaluation and
analysis (Olympus Corporation; magnification, ×200). The percentage
of damaged renal tubules was calculated in accordance with the
scoring criteria as follows: 0, no damage; 1, <25%; 2, 25-49%;
3, 50-75%; 4, >75% (15).
Semiquantitative analysis of the positive staining area of
apoptotic cells was conducted using ImageJ software (v1.8.0;
National Institutes of Health).
RNA extraction and sequencing
The kidney tissues were ground into fine powder
prior to RNA extraction. Total RNA was extracted with
TRIzol® (cat. no. 15596-026; Invitrogen; Thermo Fisher
Scientific, Inc.). The RNA was further purified with two
phenol-chloroform treatments and then treated with RQ1 DNase (cat.
no. M6101; Promega Corporation) to remove DNA. The quality and
quantity of the purified RNA were redetermined by measuring the
absorbance at 260 nm/280 nm (A260/A280) using Smartspec Plus
(Bio-Rad Laboratories, Inc.). The integrity of RNA was further
verified by 1.5%-agarose gel electrophoresis.
For each sample, 1 µg of the total RNA was
used for RNA sequencing (RNA-seq) library preparation by VAHTS
Stranded mRNA-seq Library Prep kit (cat. no. NR602; Vazyme Biotech,
Co., Ltd.). Polyadenylated mRNAs were purified and fragmented and
then converted into double strand cDNA. After the step of end
repair and a-tailing, the DNAs were ligated to VAHTS RNA adapters
(cat. no. NR803; Vazyme Biotech Co., Ltd.). Purified ligation
products corresponding to 200-500 bps were digested with
heat-labile uracil-DNA glycosylase (UDG) and the single-strand cDNA
was amplified, purified, quantified and stored at -80°C prior to
sequencing.
For high-throughput sequencing, the libraries were
prepared following the manufacturer's protocol and were applied to
Illumina HiSeq X Ten system for 150 nucleotides (nt) paired-end
sequencing by HiSeq X Ten Reagent kit v2.5 (cat. no. FC-501-2501;
Illumina, Inc.) (GEO accession code: GSE142138 (16); URL: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE142138).
RNA-Seq raw data clean and alignment
Raw reads containing more than 2-N bases were
initially discarded. Then, adaptors and low-quality bases were
trimmed from raw sequencing reads using FASTX-Toolkit (v0.0.13)
(http://hannonlab.cshl.edu/fastx_toolkit/). The short
reads less than 16 nt were also dropped. Subsequently, clean reads
were aligned to the GRch38 genome by TopHat2, allowing four
mismatches (17). Uniquely
mapped reads were used for gene read number counting and fragments
per kilobase of exon model per million mapped fragment (FPKM)
calculation (18).
Differentially expressed gene
analysis
The R Bioconductor package edgeR was utilized to
screen out the differentially expressed genes (DEGs) and common
DEGs (co-DEGs) (19). A false
discovery rate (FDR) <0.05 and fold change >2 or <0.5 were
set as the cut-off criteria for identifying DEGs. The intersections
of upregulated and downregulated DEGs in AKI tissues induced by
cisplatin and IR were obtained for further functional enrichment
analysis.
AS analysis
The ASEs and regulated alternative splicing events
(RASEs) between the samples were defined and quantified using the
ABLas pipeline as previously described (20). In summary, ABLas detection of 10
types of ASEs was based on the splice junction reads, including
exon skipping (ES), alternative 5′ splice site (A5SS), alternative
3′ splice site (A3SS), intron retention, mutually exclusive exons,
mutually exclusive 5′UTRs, mutually exclusive 3′UTRs, cassette
exon, A3SS&ES and A5SS&ES.
To assess the dysregulation of ASEs, an unpaired
Student's t-test was performed to evaluate the significance of the
ratio alteration of AS events. Events, which were significant at
P-value cutoff corresponding to a false discovery rate cutoff of
5%, were considered dysregulated ASEs. The overlapping regulated
alternative splicing genes (RASGs) induced by cisplatin and IR were
obtained using Venn diagram tool.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR) validation of DEGs and
ASEs
RT-qPCR was performed for certain DEGs. The primer
sequences are presented in Table
I. Total RNA remaining from RNA-seq library preparation was
used for RT-qPCR. RNA was reverse transcribed into cDNA using an
M-MLV reverse transcriptase assay kit according to the
manufacturer's protocol (cat. no. R021-01; Vazyme Biotech Co.,
Ltd.). qPCR was performed with the StepOne real-time PCR system
using the SYBR Green PCR Reagent kit (cat. no. 11143ES50; Shanghai
Yeasen Biotechnology Co., Ltd.). The thermocycling conditions were
as follows: Initial denaturation at 95°C for 10 min, 40 cycles of
denaturation at 95°C for 15 sec and annealing and extension at 60°C
for 1 min. PCR amplifications were performed in triplicate for each
sample. The RNA expression levels of all the genes were normalized
against those of GAPDH using the 2−∆∆Cq method (21).
 | Table IPrimers used for reverse
transcription-quantitative PCR. |
Table I
Primers used for reverse
transcription-quantitative PCR.
| Gene | Primer sequence
(5′-3′) |
|---|
| KIM-1 | F:
CCAGGCGCTGTGGATTCTTA |
| R:
TGTACCGACTGCTCTTCTGATAGG |
| NGAL | F:
CAGAGCTACAATGTGCAAGTGGC |
| R:
CAGCTCCTTGGTTCTTCCATACA |
| RBFOX1 | F:
TCATTTAGAGCAAGTGGGTG |
| R:
TCTGGGTTTGCAGATTGAGT |
| CSF-1 | F:
GCACACAGGGGCGGCT |
| R:
GCATTCTCTACCCCTCCACCT |
| IL-1β | F:
ATGTGCTGGTGCTTCATTCA |
| R:
AGCCCGCACTGAGGTCTTTC |
| IL-6 | F:
CCCCAATTTCCAATGCTCTCC |
| R:
CGCACTAGGTTTGCCGAGTA |
| TLR2 | F:
AGTCAGGAACTGGGTGGAGA |
| R:
ACCAAGACCTACCTGGAGTG |
| CXCL1 | F:
CTGTGCTAGTAGAAGGGTGTTGT |
| R:
ACGAGACCAGGAGAAACAGG |
| IL-34 | F:
GATACGGCATTTGGTTGGTC |
| R:
GGCTGGTTGCTATCCCTTACA |
| CXCL10 | F:
ACCCATTGATACATACTTGA |
| R:
GTAAGGACGAACTTAACCAC |
| LGALS3 | F:
TAACCACGCCATGATCTAAG |
| R:
GCAAAGTTTCCCACTCCTAA |
| IKBKB | M-F:
TTCACCTGTCGCCTGATTGTGC |
| AS-F:
CCTCTCAAGAACCTGATTGTGC |
| M/AS-R:
GCCTGGGAAATGAAAGAACG |
| CRK | M/AS-F:
TCAAGGCAGGGTAGTGGAGT |
| AS-R:
AACCATCAGGAGCTCCAATCAG |
| M-R:
CAGCTCACCGACCTCCAATCAG |
| ADK | M/AS-F:
GGCAGAGCCAAACTACGGTG |
| AS-R:
GACCCAGATAATCTCAGCGCTT |
| M-R:
AGCACATTTTCACTCAGCGCTT |
| PAK2 | M-F:
CTTTTTACTTCCTTTTTCTGTGC |
| AS-F:
CAGTCCAGGCCTTTTTCTGTGC |
| M/AS-R:
TCCAGTTCGGATGAGCAGTA |
| CSNK1A1 | M/AS-F:
AGCCTCTTCCAGTGGGCAGGGTCA |
| AS-R:
TCTTTAATACCTGTGGGGGT |
| M-R:
GCTTAGAAACCTGTGGGGGT |
| BCL2L1 | M/AS-F:
GCTCACTTACTGGGTCTGCT |
| AS-R:
AGCAATTCTGAACCTTATCT |
| M-R:
GCAGTCAGCCAGAACCTTATCT |
| SHF | M/AS-F:
TTTTGCTGGTCTCACTGTTG |
| AS-R:
CTGGAAAACCAGCTGGTACCAC |
| M-R:
GGAAAACCAGGTCTGGTACCAC |
| FGF1 | M-F:
CTGTCCCTTGTCCCATCCAC |
| AS-R:
ATATCTGACCTGTGCTGAGCCT |
| M/AS-R:
CCAGTTCTTCAGTGCTGAGCCT |
| ZDHHC16 | M/AS-F:
TAACCATCGCTACTTCTTCTC |
| AS-R:
GTTTCATTTTCTCAATGGCA |
| M-R:
CTGGTGGTACGTCTCAATGGCA |
| GAPDH | F:
GGAGATGCTCAGTGTTGG |
| R:
TGACAATGAATACGGCTACA |
RT-qPCR assay was also performed for ASE validation.
A boundary-spanning primer was used for the sequence encompassing
the junction of the constitutive exon and alternative exon as well
as an opposing primer in a constitutive exon to detect alternative
isoforms. The boundary-spanning primer of alternative exon was
designed according to 'model exon' to detect model splicing or
'altered exon' to detect altered splicing (22).
Functional enrichment analysis
Gene Ontology (GO) terms (23) and Kyoto Encyclopedia of Genes and
Genomes (KEGG) pathways (24)
were identified using KOBAS 2.0 server (25) to sort out functional categories
of DEGs. Hypergeometric test and Benjamini-Hochberg false discovery
rate (FDR)-controlling procedure were used to define the enrichment
of each term.
Protein-protein interaction (PPI) network
construction and module analysis
The Search Tool for the Retrieval of Interacting
Genes (http://www.string-db.org) was used to
analyze the association between DEGs that are co-, up- or
downregulated by cisplatin and IR. The PPI network was visualized
and constructed using Cytoscape v3.8.2 (26).
Cell culture and hypoxia/reoxygenation
(H/R) model preparation
Human renal proximal tubular epithelial cells (HK-2
cells) were obtained from Procell Life Science and Technology Co.,
Ltd. The identification of the HK-2 cell lines was conducted at the
China Centre for Type Culture Collection. The HK-2 cells were
cultured in DMEM/F12 medium (Gibco; Thermo Fisher Scientific, Inc.)
containing 10% fetal bovine serum in a humidified atmosphere of 95%
O2 and 5% CO2 at 37°C. The HK-2 cells were
placed on pre-hypoxia-treated glucose-free and serum-free medium
and then cultured in a three-gas incubator (1% O2, 4%
CO2 and 95% N2) at 37°C for 24 h (27). Finally, the cells were cultured
in a normal medium and then reoxygenated in a normal incubator for
1, 3 and 6 h (28).
Cell transfection experiment
The RBFOX1 plasmid [pEnC
MV-RBFOX1(human)-3xFLAG-SV40-Neo] and the corresponding vector
control (pEnCMV-MCS-3xFLAG-SV40-Neo) were obtained from Wuhan
MiaoLing Biotechnology, Co., Ltd. (www.miaolingbio.com). HK-2 cells (1×106)
were seeded in 6-well culture plates. After the cells reached
60-70% confluence, RBFOX1 plasmid or vector control (5 µg)
was mixed with 5 µl Lipofectamine® 2000
transfection reagent (Invitrogen; Thermo Fisher Scientific, Inc.)
in serum-free medium to prepare the transfection complex. The
mixing system was incubated at room temperature for 15 min and then
uniformly added to each group of cells at 37°C for 24 h. The
transfected cells were treated with normal fresh medium for 4-6 h
and then subjected to further experiments. The transfection
efficiency was determined using western blotting.
Flow cytometric analysis
Following digestion with trypsin, the cells reached
70-80% confluence in 6-well culture plates were centrifuged at 300
× g for 5 min at room temperature. After washing with PBS, 100
µl of 1X binding buffer was added to resuspend the cells.
Then, 5 µl Annexin V-FITC and 10 µl PI (BD
Biosciences) staining solution were added. The cells were incubated
at room temperature for 20 min in the dark. Finally, 400 µl
of 1X binding buffer was added to resuspend the cells in the
solution. Flow cytometric detection was completed within 1 h using
CytoFLEX flow cytometer (Beckman Coulter, Inc.). FlowJo software
(version 10.8.0; BD Biosciences) was used for flow cytometric data
analysis.
ELISA
The levels of inflammatory factors in the cell
supernatant were measured by TNF-α, IL-6 and IL-1β ELISA kits (cat.
nos. BMS223-4, BMS213-2 and BMS224-2, respectively; eBiosience;
Thermo Fisher Scientific, Inc.) in accordance with the
manufacturer's protocol.
Measurement of reactive oxygen species
(ROS) production
The cells were collected and treated with diluted 10
µmol/l DCFH-DA probe (Beyotime Institute of Biotechnology)
and then incubated at 37°C for 20 min. The difference in ROS
content in each group was determined by flow cytometry.
Measurement of the levels of superoxide
dismutase (SOD) and malondialdehyde (MDA) activity
Activities of SOD and MDA were detected following
the protocols of the manufacturers of the corresponding kits (cat.
nos. A001-3-2 and A003-1-2; Nanjing Jiancheng Bioengineering
Institute). Absorbance values at wavelengths of 550 and 532 nm were
measured using a microplate reader to detect the activities of SOD
and MDA in the cells.
Western blot analysis
Total protein was extracted from the kidney tissue
or HK-2 cells using RIPA lysis buffer (BioSharp Life Sciences,
Inc.) and the protein concentration was determined using BCA
Protein assay reagent (Thermo Fisher Scientific, Inc.). The same
amount of protein (15-30 µg) in each well was subjected to
10-12% SDS-PAGE. The transfer polyvinylidene difluoride (PVDF)
(cat. no. IPVH00010; Millipore, Inc.) membrane was blocked with 5%
non-fat milk in Tris-buffered saline containing 0.1% Tween-20 at
room temperature for 1 h and then incubated with primary antibodies
at 4°C overnight: anti-RBFOX1 (1:1,000; cat. no. ab254413),
anti-β-actin (1:1,000; cat. no. ab8226; both from Abcam),
anti-NF-κB (p65; 1:1,000; cat. no. AF5006), anti-phospho-NF-κB
(p-p65; 1:1,000; cat. no. AF2006; both from Affinity Biosciences),
anti-NRF2 (1:1,000; cat. no. 16396-1-AP) and anti-HO-1 (1:1,000;
cat. no. 10701-1-AP; all from ProteinTech Group, Inc.). After
washing thrice with Tris Buffered Saline with Tween for 5 min each
time, the membranes were incubated with the horseradish
peroxidase-conjugated goat anti-mouse IgG (1:1,000; cat. no.
ab205719) and goat anti-rabbit IgG (1:1,000; cat. no. ab205718;
both from Abcam) secondary antibodies at room temperature for 1 h.
Target bands were visualized using enhanced chemiluminescent (ECL)
kit (cat. no. BL523B; Biosharp Life Sciences, Inc.). The ChemiDoc
Imaging System (Bio-Rad Laboratories, Inc.) was used to detect
chemiluminescence blots. ImageJ software (version 1.8.0; National
Institutes of Health) was used to semi-quantify protein expression
with β-actin as the loading control.
Statistical analysis
Data are presented as the mean ± SD from three or
more independent experiments. GraphPad Prism 7 (GraphPad Software,
Inc.) was used for statistical analysis. Continuous variables were
analyzed using one-way ANOVA and unpaired Student's t-test.
Enumerated variables were analyzed using chi-square and Fisher's
exact tests. Bonferroni's test was used for post hoc comparisons.
All experiments were independently repeated in triplicate. A
two-sided P<0.05 was considered to indicate a statistically
significant difference.
Results
Construction and genome-wide profiling of
AKI model induced by cisplatin and IR
Mouse AKI models induced by cisplatin and IR were
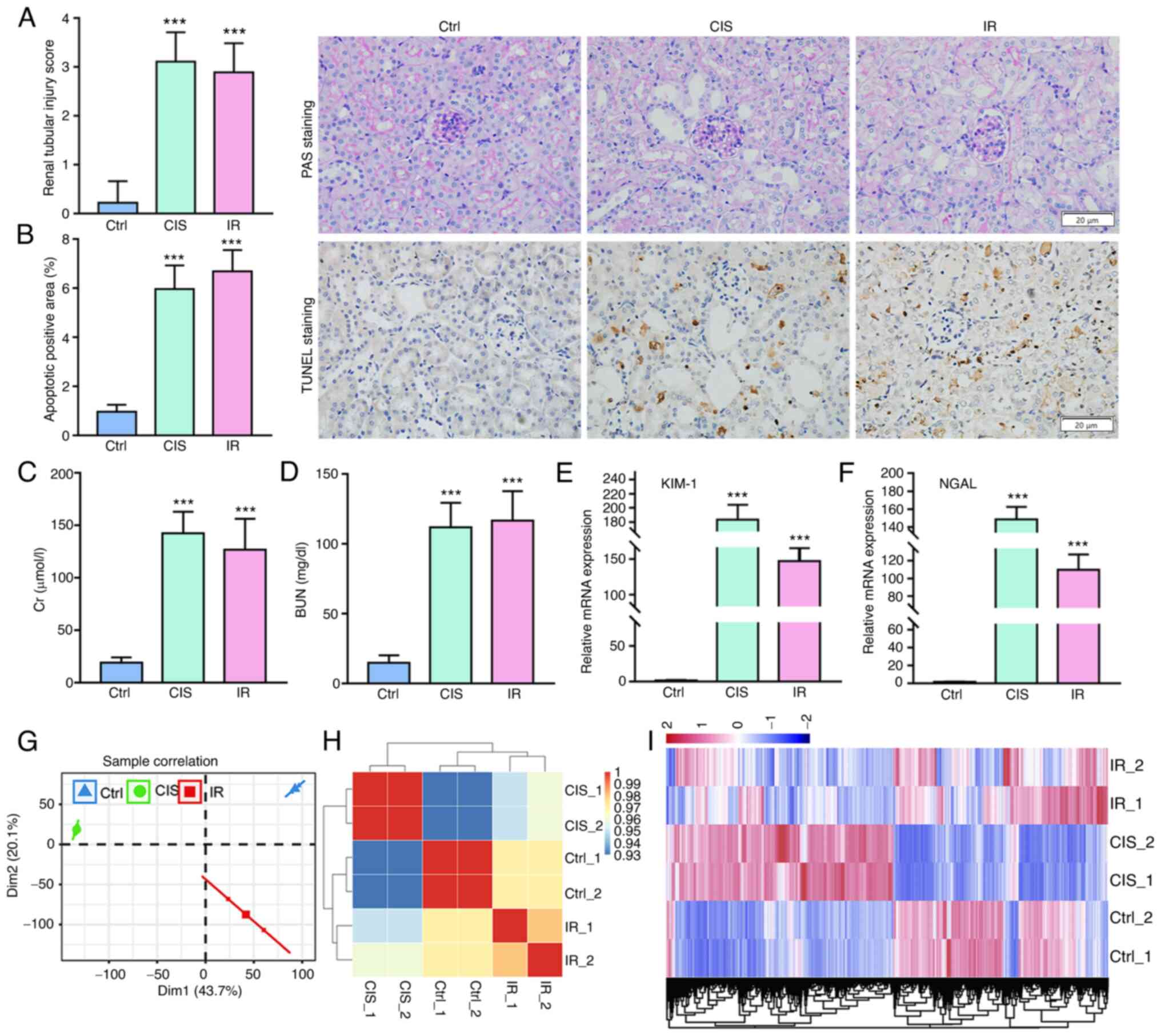
successfully constructed. It was revealed that cisplatin and IR
caused the kidney tissue to become severely damaged and increase in
cell apoptosis (Fig. 1A and B).
The levels of Cr and BUN and the mRNA expression of KIM-1 and NGAL
in kidney tissue were significantly increased, indicating that
cisplatin and IR caused serious damage to the kidney function of
mice compared with those in the controls (Fig. 1C-F).
The principal component analysis and sample
correlation analysis revealed that samples treated with cisplatin
and IR can be separated from control samples (Fig. 1G and H). The hierarchical
clustering heat map revealed that the gene expression profiles of
the cisplatin group and the IR group were evidently different from
those of the control group (Fig.
1I).
Co-DEGs in AKI induced by cisplatin and
IR are involved in the inflammatory response pathway
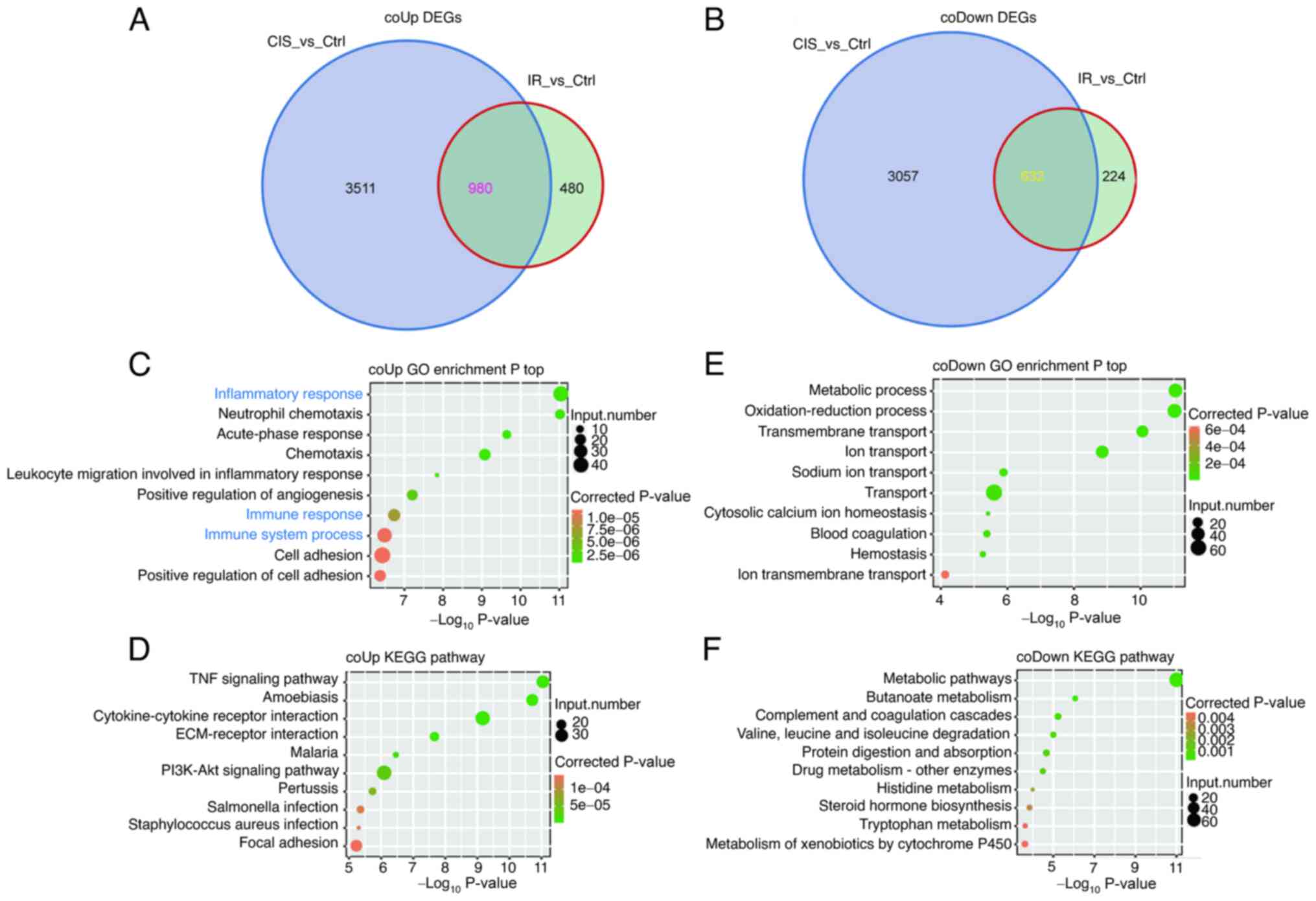
A total of 980 co-upregulated DEGs and 632
co-downregulated DEGs were identified in AKI induced by cisplatin
and IR (Fig. 2A and B). The
co-upregulated DEGs were mainly enriched in acute immune and
inflammatory response (Fig. 2C and
D). The co-downregulated DEGs were mainly related to cell
metabolism and functional damage (Fig. 2E and F).
The PPI network of co-upregulated DEGs revealed that
IL-6, TNF, MMP9, TLR2, IL-1β, TIMP1, CCL2, CXCL1, CXCL10, CSF-1 and
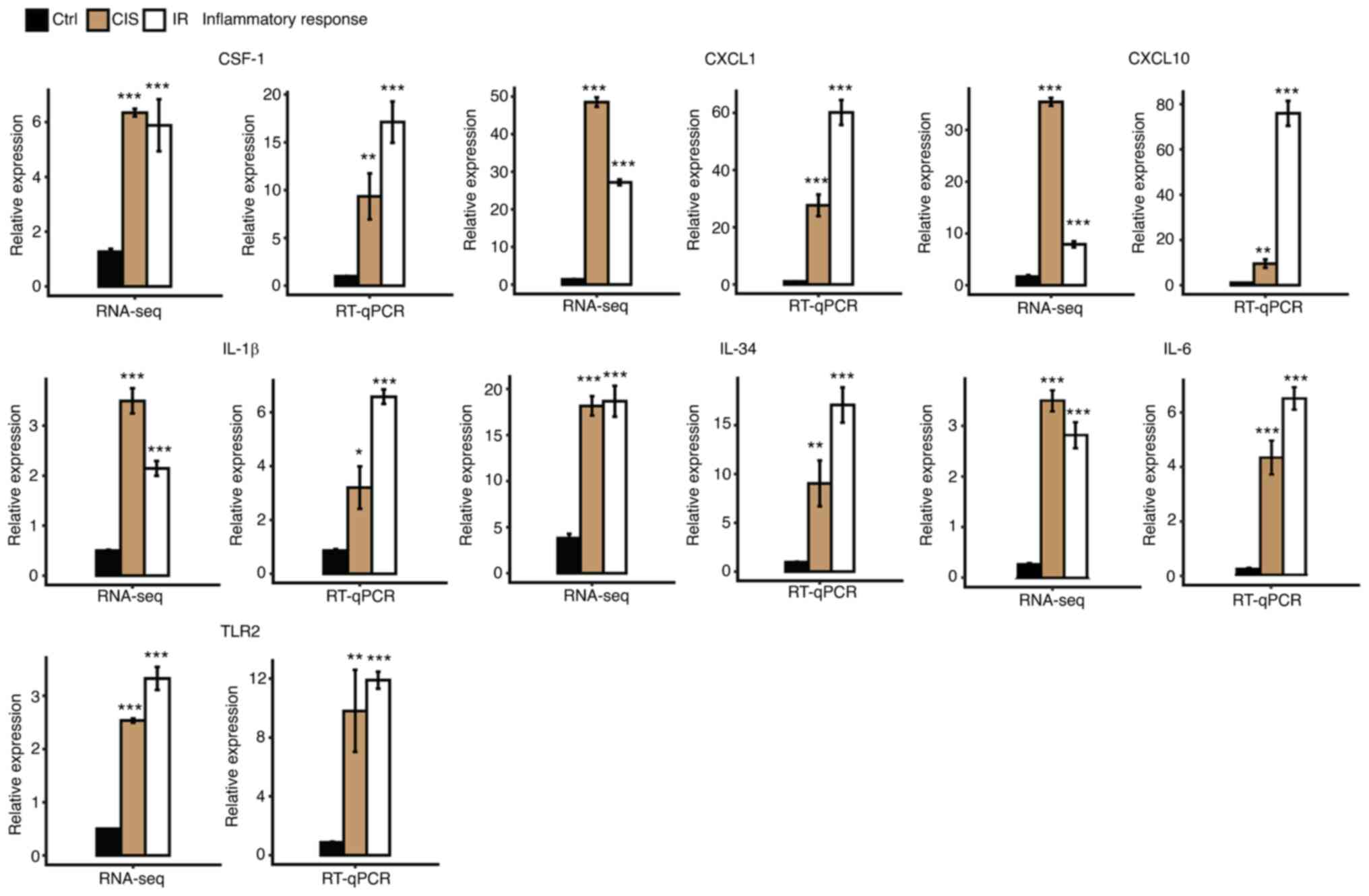
LGALS3 were among the top 40 hub genes (Fig. S1). In addition, further RT-qPCR
results revealed that the inflammatory genes CSF-1, CXCL1, CXCL10,
IL-1β, IL-34, IL-6 and TLR2 were upregulated in AKI induced by
cisplatin and IR (Fig. 3). These
findings suggested that the inflammation response pathway plays an
important role in the pathogenesis of AKI. In addition, the PPI
network of co-downregulated DEGs suggested multiple key genes
including RBFOX1 (Fig. S2).
Transcriptome analysis of AS in AKI
induced by cisplatin and IR
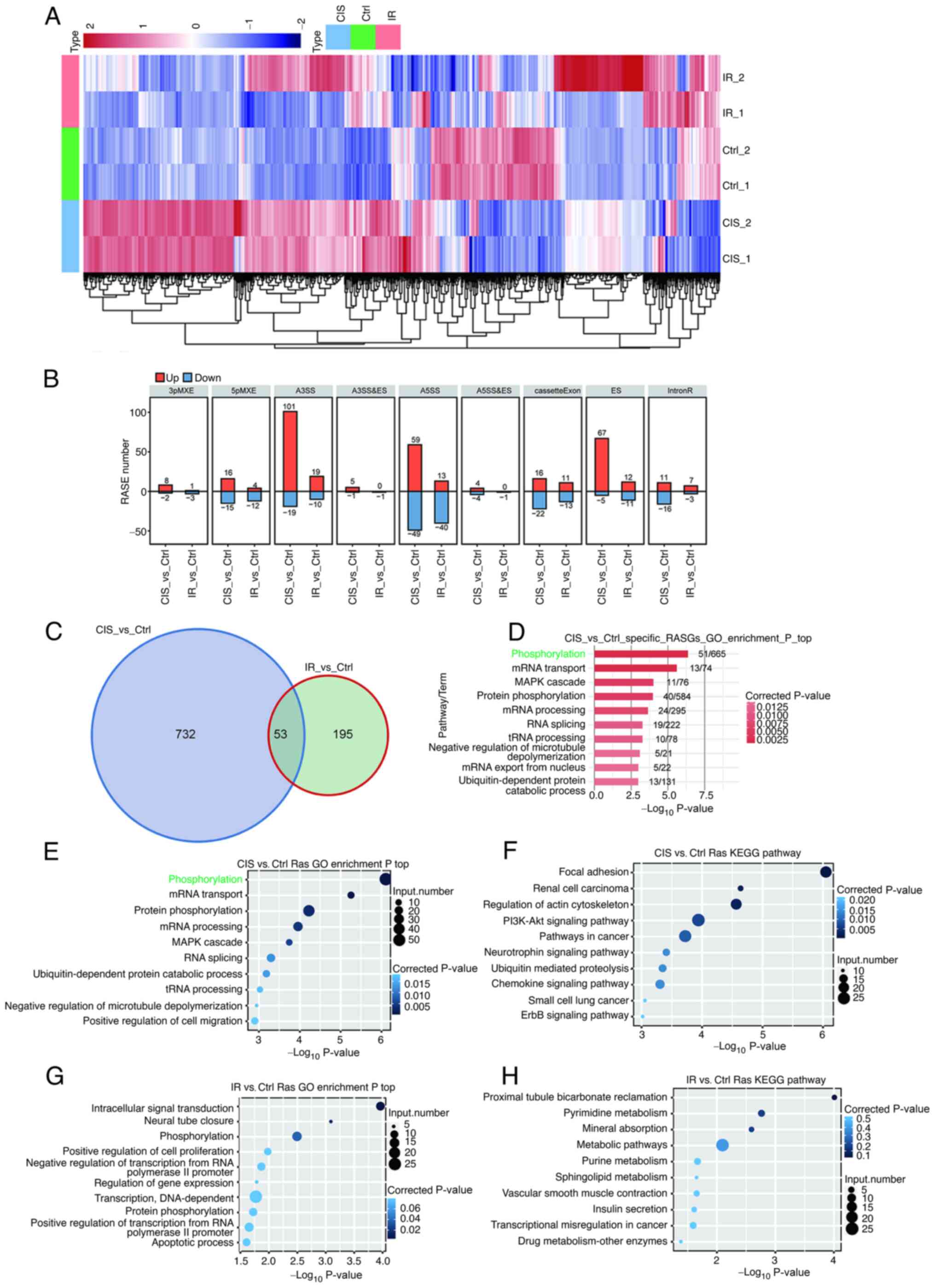
The heat map of the transcriptome analysis revealed
that the differentially expressed profile of RASGs induced by
cisplatin was evidently different from that of IR (Fig. 4A). The types of RASEs were mainly
concentrated in A3SS, A5SS, ES and cassette exon events (Fig. 4B). The number of differences in
RASGs between the cisplatin and IR groups was markedly higher than
the number of intersections (Fig.
4C). Cisplatin-induced RASGs were highly enriched in
phosphorylation and related cell signaling pathways (Fig. 4D-F) and IR-induced RASGs were
related to cell metabolism, apoptosis and phosphorylation (Fig. 4G and H). These findings indicated
that DEGs regulated by AS may play an important role in AKI through
the phosphorylation and apoptosis pathways.
Examples and verification of ASEs in AKI
induced by cisplatin and IR
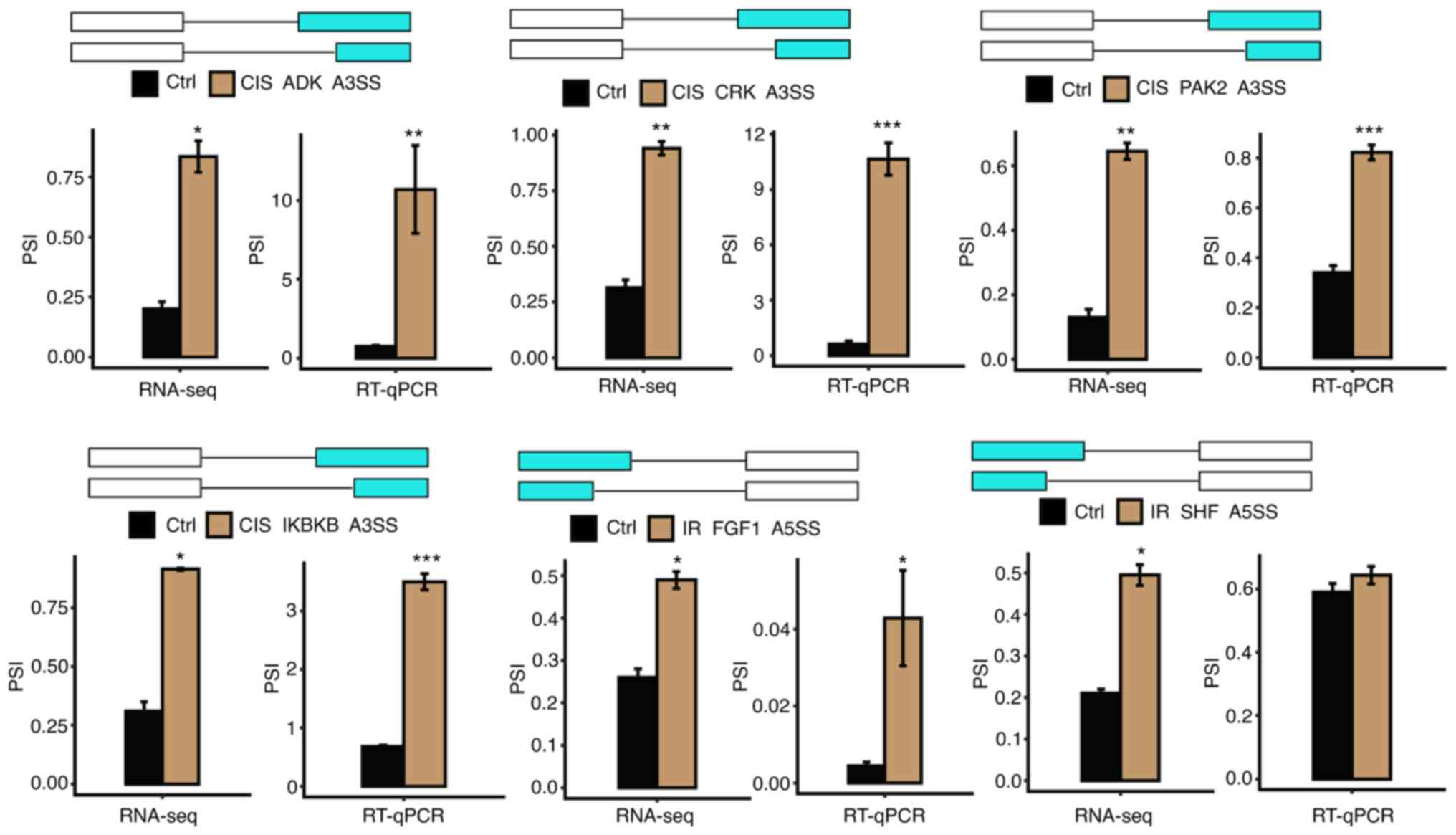
The specific structures and processes of the A3SS,
cassette exon and A5SS events were demonstrated and the expression
of nine mRNA splicing isoforms in cisplatin and IR-induced AKI was
verified by RNA-seq quantification and RT-qPCR (Figs. 5 and 6). Specifically, cisplatin induced
upregulation of CSNK1A1, ADK, CRK, PAK2 and IKBKB genes, which
underwent A3SS events and IR caused upregulation of ZDHHC16
(cassette exon), BCL2L1 (A5SS) and FGF1 (A5SS) genes. RNA-seq
showed that IR induced the upregulation of SHF (A5SS) gene, but
this finding was not confirmed in subsequent RT-qPCR.
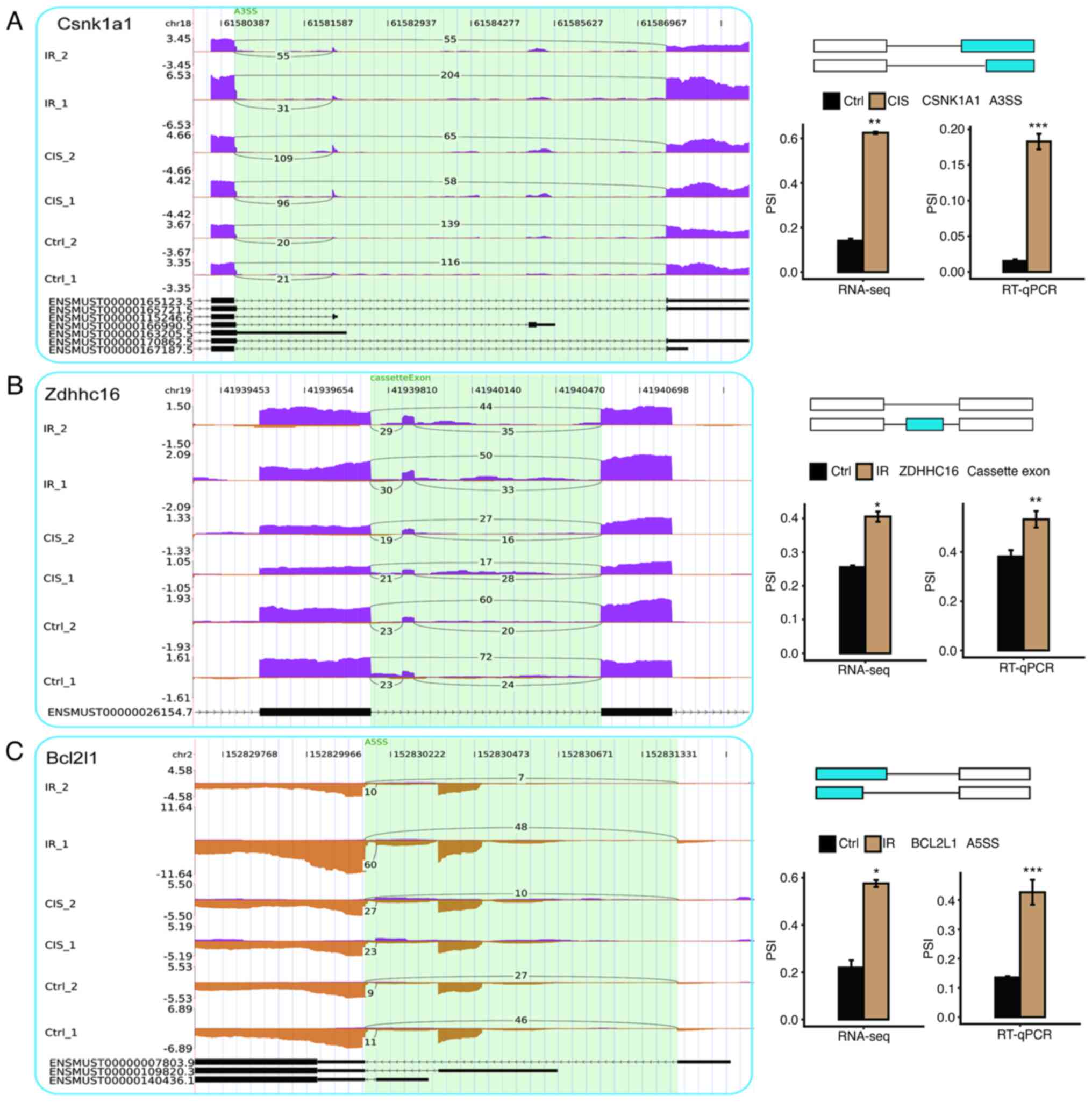 | Figure 5Examples and validation of CIS or
IR-affected ASEs. (A) Alternative 3′ splicing sites (A3SS), (B)
cassette exon, and (C) alternative 5′ splicing site (A5SS) events
were shown in the IGV-sashimi plot of CSNK1A1, ZDHHC16 and BCL2L1
genes. Read distribution of each ASE was plotted in the left panel
with the transcripts of each gene shown below. The schematic
demonstrated the structures of ASEs. The constitutive exons are
denoted by white boxes, intron sequences by a horizontal line
(right panel, top), and alternative exons by blue boxes (right
panel, bottom). RNA-seq quantification and reverse
transcription-quantitative PCR were used to verify the ASEs.
*P<0.05, **P<0.01 and
***P<0.001 vs. Ctrl. ASEs, alternative splicing
events; CIS, cisplatin; IR, ischemia-reperfusion; PSI, percent
spliced in. |
Expression profile of RBPs and in vivo
validation of RBFOX1 in cisplatin and IR-induced AKI
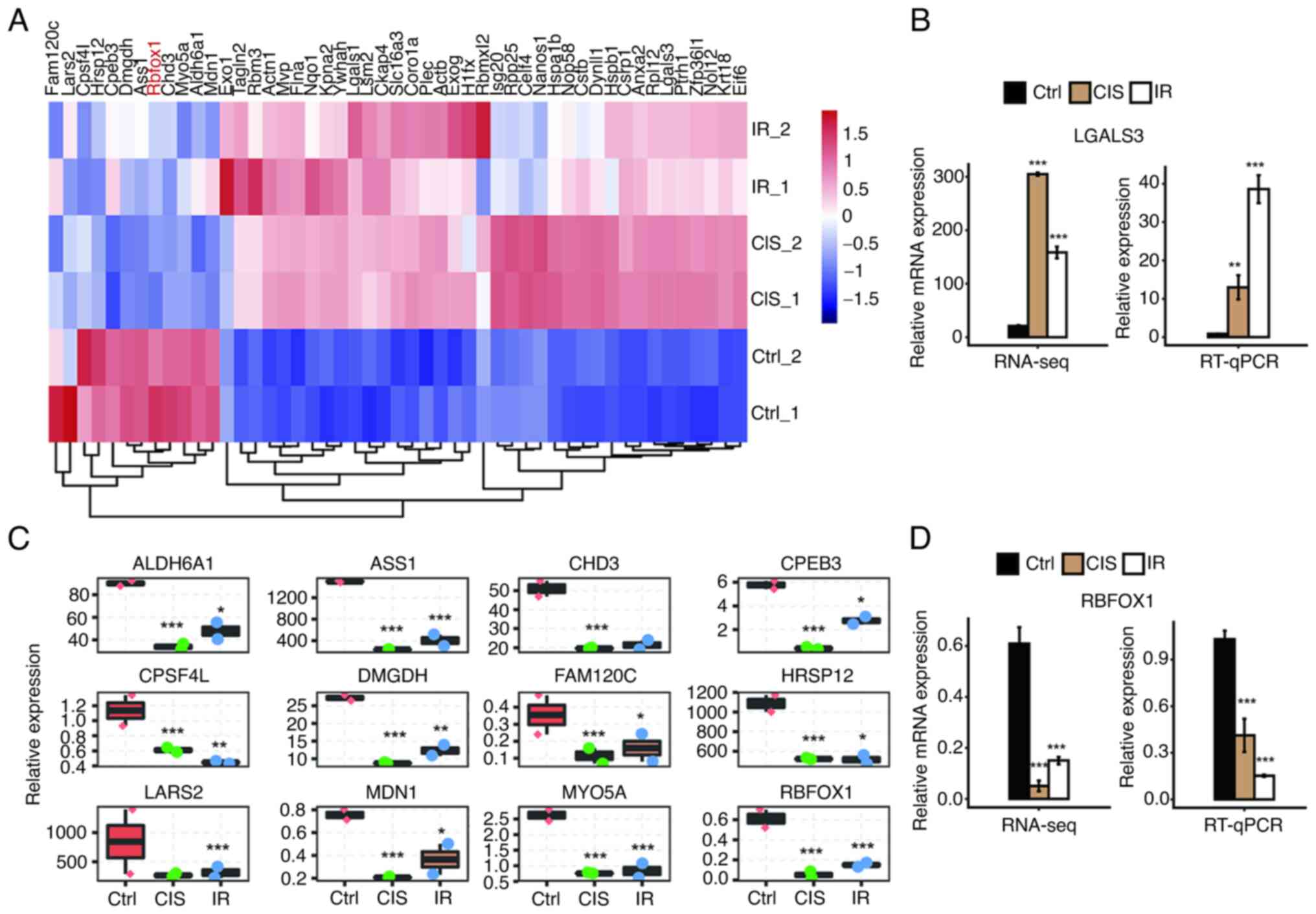
As demonstrated in the heat map, 49 RBP-related
genes were found in co-DEGs induced by cisplatin and IR (Fig. 7A). Among the upregulated genes,
the fold change of LGALS3 was in the forefront, and the expression
of LGALS3 was further validated by RT-qPCR (Fig. 7B). Compared with the control
group, the fold change of RBFOX1 was the highest among the
downregulated RBP genes, whether in the cisplatin group or the IR
group (Fig. 7C). Subsequently,
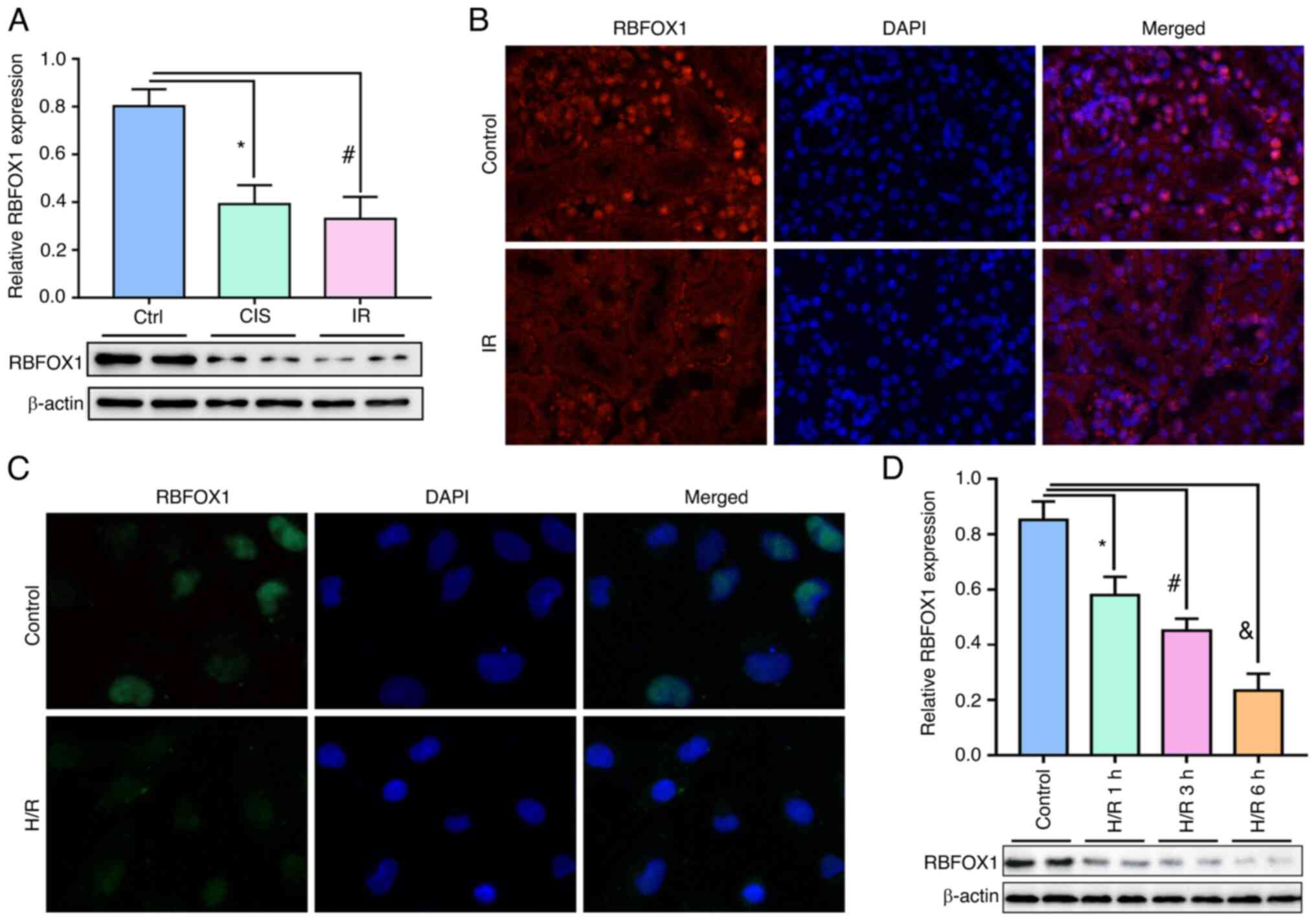
this RBFOX1 trend was verified in vivo. The relative levels
of RBFOX1 in mRNA and protein were significantly downregulated by
cisplatin and IR (Figs. 7D and
8A). The cisplatin and
IR-induced downregulation of RBFOX1 occurred in the nuclei of mouse
renal tubular epithelial cells (Fig.
8B).
RBFOX1 inhibits the expression of
inflammatory cytokines and the level of oxidative stress in HK-2
cells caused by H/R
Consistent with the results of animal experiments,
localization analysis of the HK-2 cells revealed that H/R inhibited
the expression of RBFOX1 in the nucleus (Fig. 8C). H/R caused the downregulation
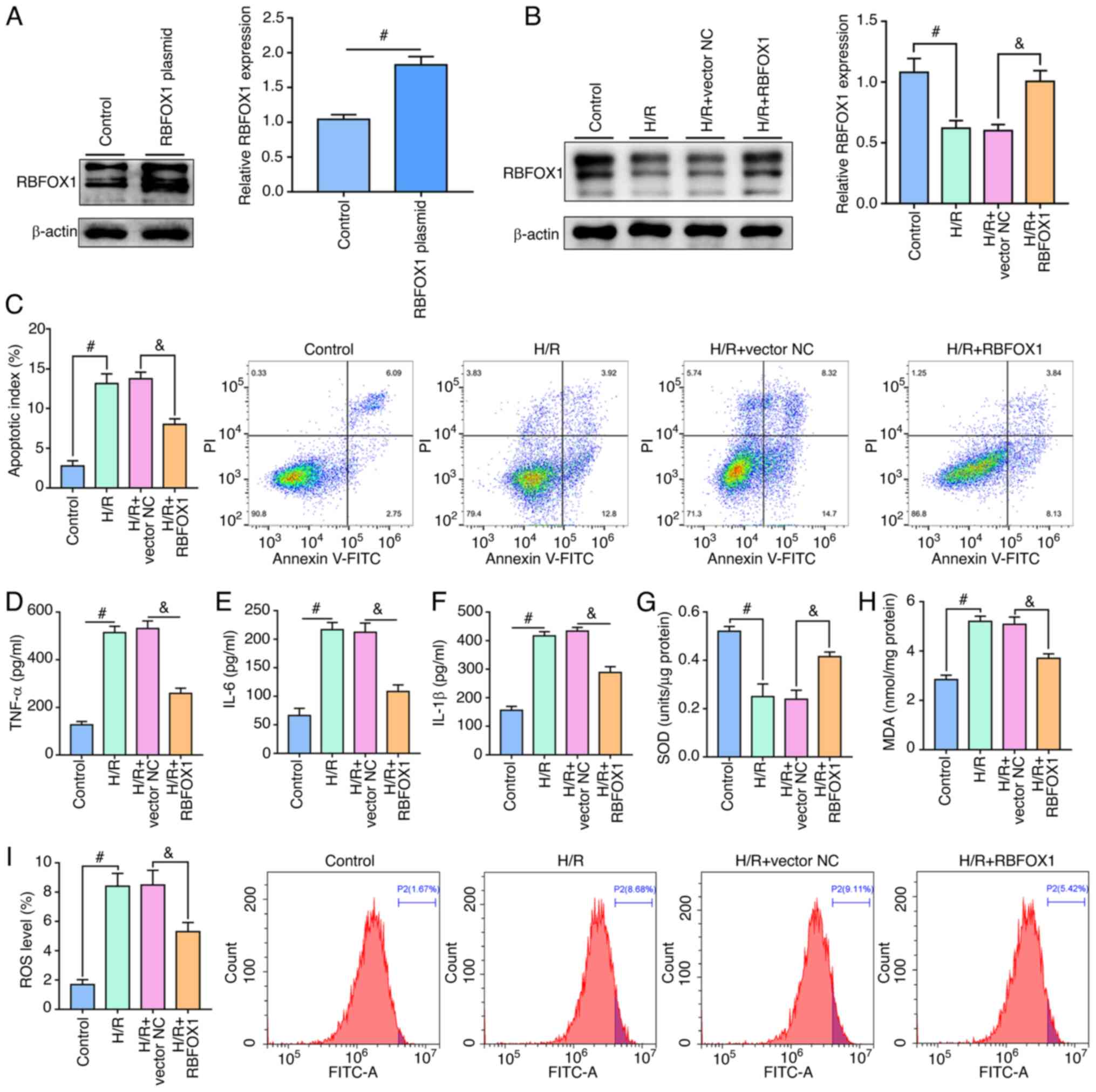
of RBFOX1 in the HK-2 cells in a time-dependent manner (Fig. 8D). The transfection efficiency of
RBFOX1 plasmid in untreated and H/R-treated HK-2 cells was verified
(Fig. 9A and B). Flow cytometric
analysis revealed that upregulation of RBFOX1 reduced H/R-induced
apoptosis in the HK-2 cells (Fig.
9C). In the present study, the effects of RBFOX1 on the
inflammatory response and oxidative stress caused by H/R in the
HK-2 cells were analyzed. Exogenous RBFOX1 inhibited the expression
of the proinflammatory cytokines TNF-α, IL-6 and IL-1β induced by
H/R (Fig. 9D-F). In addition,
transfection of the RBFOX1 plasmid increased the resistance of the
HK-2 cells to H/R-induced oxidative stress as manifested by the
rebound in SOD activity and the decrease in MDA levels and ROS
production (Fig. 9G-I).
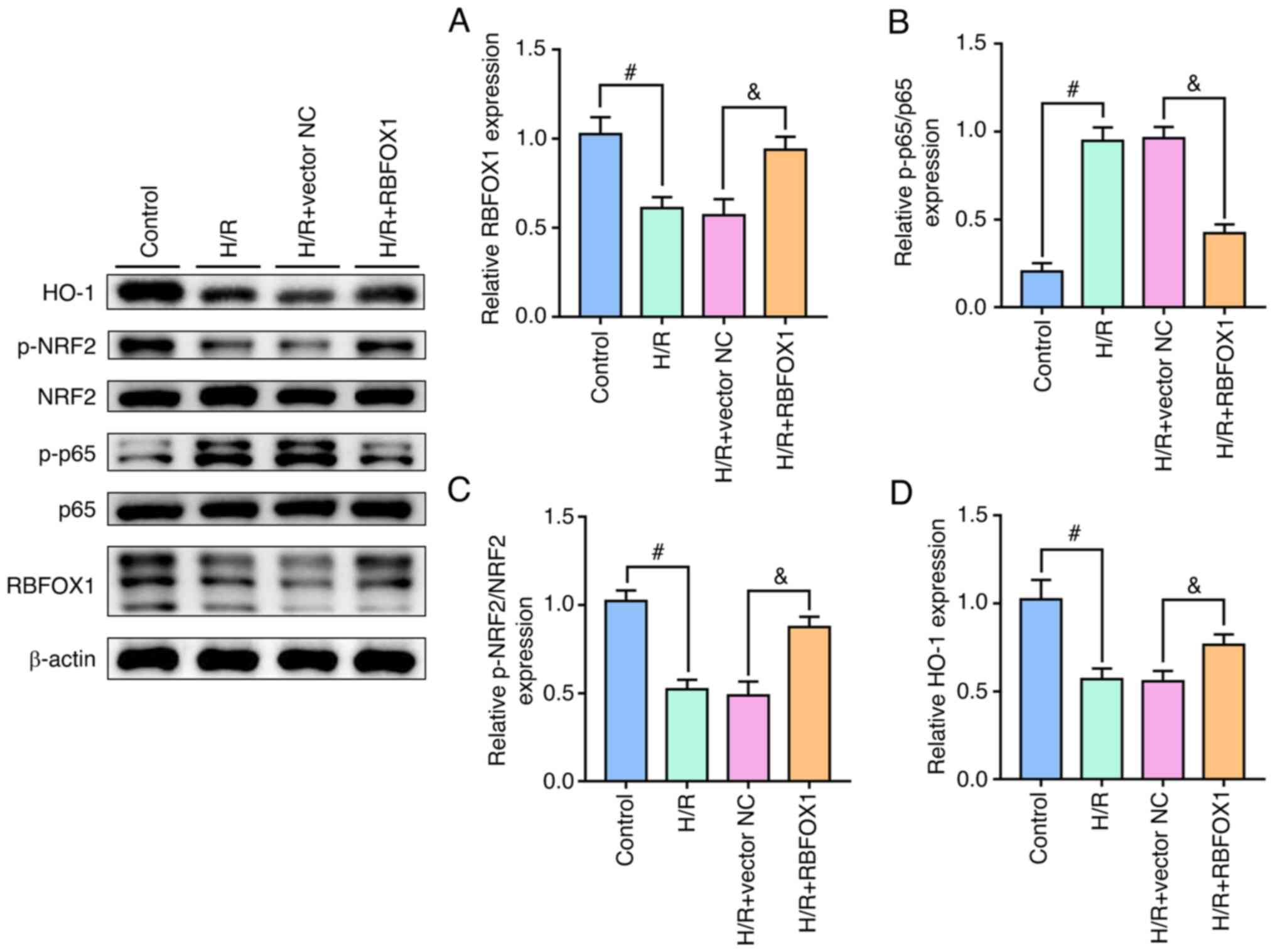
RBFOX1 is capable of inhibiting NF-κB and
activating the NRF2/HO-1 signaling pathway in H/R-induced HK-2
cells
In the present study, the effects of RBFOX1 on NF-κB
and the NRF2 signaling pathway in the HK-2 cells induced by H/R
were further analyzed. The results revealed that H/R activated the
expression of NF-κB and inhibited the NRF2/HO-1 signaling pathway
in the HK-2 cells. Furthermore, exogenous RBFOX1 inhibited NF-κB
but activated the NRF2/HO-1 signaling pathway (Fig. 10). These results suggested that
RBFOX1 may play a protective role against inflammation and
oxidative stress in H/R-induced HK-2 cells by inhibiting NF-κB and
activating the NRF2/HO-1 pathway.
Discussion
AKI is a high-incidence renal disease with systemic
effects and its pathogenesis is extremely complex and still not
fully understood. The genome-wide bioinformatics analysis provides
data basis for systematically revealing the underlying molecular
mechanism of AKI and its internal associations (29). In the present study, the
upregulated co-DEGs were mainly enriched in immune and inflammatory
pathways, including hub genes CSF-1, CXCL1, CXCL10, IL-1β, IL-34,
IL-6 and TLR2. Importantly, the transcriptome analysis of AS and
RBPs in AKI was reported, to the best of our knowledge, for the
first time. Cisplatin-induced AS was enriched in the
phosphorylation pathway, including DEGs CSNK1A1, PAK2, CRK, ADK and
IKBKB regulated by A3SS event. IR-induced AS was enriched in the
apoptosis and proliferation pathways, including RASGs FGF1 (A5SS),
BCL2L1 (A5SS) and ZDHHC16 (cassette exon). Differentially expressed
RBP genes may be involved in the pathogenesis of AKI. LGALS3 was
the top upregulated RBP gene, whereas RBFOX1 was the top
downregulated RBP gene. RBFOX1 was downregulated in the mouse
kidney tissues induced by cisplatin and IR and in the HK-2 cells
induced by H/R. Further experiments suggested that exogenous RBFOX1
may play a protective role against the inflammation and oxidative
stress induced by H/R in the HK-2 cells. This phenomenon may be
related to the inhibition of NF-κB and the activation of the
NRF2/HO-1 signaling pathway.
AKI is described as an inflammatory disease and
inflammatory factors are involved in its damage and repair process
(30). TLR2 was originally
considered to be a proinflammatory factor, but a recent study
revealed that it can play a protective role in AKI by activating
autophagy (31). IL-34 and CSF-1
share the same membrane receptor but have very different functions.
IL-34 mediates renal tubular damage by recruiting macrophages in
the acute phase of AKI and CSF-1 promotes the repair process by
inducing the polarization of M2 macro-phages (32,33). IL-1β activated by inflammasomes
promotes extensive inflammation in various acute and chronic kidney
diseases, but the therapeutic effect of inhibiting IL-1β in AKI
remains to be determined (34).
IL-6 and CXCL1 can promote inflammatory response by inducing
neutrophil infiltration in renal tissue and animal studies have
shown that inhibition of IL-6 or CXCL1 has a protective effect in
AKI (35,36). Inflammatory factors are often
used as biomarkers of AKI. Various clinical data indicated that
IL-6, CXCL1 and CXCL10 in serum and urine of patients have
predictive value in the early diagnosis and prognostic evaluation
of AKI (37,38).
The splice isoforms of CSNK1A1, ADK, CRK, PAK2 and
IKBKB participate in the process of phosphorylation modification
after protein translation. CSNK1A1, PAK2, and IKBKB have
serine/threonine protein kinase activity and play an important role
in intracellular signal transduction. Studies have demonstrated
that CSNK1A1 and PAK2 phosphorylate β-catenin at Ser45 and Ser675,
respectively, thereby inhibiting the WNT signaling pathway
(39,40). IKBKB regulates the immune
response by inducing IkB-a phosphorylation at Ser32 and Ser36
through the classical activation pathway to promote NF-κB signal
transduction (41). Targeted
silencing of IKBKB can reduce IR-induced kidney inflammation
(42). ADK regulates
intracellular adenosine levels through dephosphorylation to
maintain energy homeostasis. Inhibition of ADK appears to improve
cisplatin-induced AKI (43). The
CRK adaptor protein is the main confluence point of the
phosphotyrosine kinase signaling pathway and plays a key role in
maintaining the morphology and function of glomerular podocytes
(44). The splice isoforms of
ZDHHC16, BCL2L1 and FGF1 are closely related to cell proliferation
and apoptosis. ZDHHC16 participates in post-translational
modification through S-acylation and has anti-apoptotic properties
that promote cell proliferation and regulate DNA damage (45). BCL-X(L) and BCL-X(S), as the
splicing isoforms of BCL2L1 (BCL-X), play anti-apoptotic and
pro-apoptotic effects, respectively (46). The upregulation of BCL-X(L) in
the early stage of AKI indicates the existence of adaptive
resistance to apoptosis (47).
FGF1 mediates a precise signal cascade through
autocrine/paracrine-dependent manners, participates in the
regulation of cell proliferation, energy homeostasis and tissue
repair and plays an anti-inflammatory effect in chronic kidney
disease (48).
The strict regulation of AS requires the recruitment
of RBPs. RBP can bind to pre-mRNA through a specific binding domain
and plays a key role in almost all aspects of posttranscriptional
regulation (49). The key RBP
genes LGALS3 and RBFOX1 may play an important role in AKI. LGALS3
is a biomarker of AKI, which can mediate inflammatory damage by
regulating the migration, proliferation and activation of renal
inflammatory cells (50). RBFOX1
is a highly versatile RBP composed of different splicing isomers,
which can regulate transcription and affect the stability of mRNA
by regulating the AS of target exons closely related to the binding
motif UGCAUG (51).
Inflammation and oxidative stress play an important
role in cisplatin and IR-induced AKI and studying their regulated
mechanisms is of great significance to the prevention and treatment
of AKI. Inhibiting NF-κB and activating the NRF2/HO-1 signaling
pathway are important approaches by which protective genes exert
anti-inflammatory and antioxidative stress effects to reduce the
severity of AKI (52). As an
important signal transduction factor in cells, NF-κB can initiate
transcriptional regulation through nuclear translocation after
being stimulated by external sources to promote the expression of
key downstream inflammatory factors (53). The role of NF-κB is regulated by
various genes. For example, SIRT1 directly inhibits NF-κB signaling
by deacetylating the p65 subunit of the NF-κB complex (54). NRF2 is a key transcription factor
of antioxidant response that induces the expression of diverse
genes driven by antioxidant response elements, such as SOD and
HO-1, through translocating to the nucleus after deubiquitination
(55). In addition to the
classic KEAP1 activation, the function of NRF2 is affected by
different genes. p62 can chelate KEAP1 into autophagosomes to
prevent NRF2 degradation (56).
It was identified that RBFOX1 is mainly expressed in the nucleus.
Interestingly, both NF-κB and NRF2 must regulate the transcription
and expression of related genes through nuclear translocation. It
was hypothesized that their role in the nucleus may be regulated by
RBFOX1.
The present study has certain limitations. Although
both cisplatin and IR were used to establish an AKI model, further
increasing the amount of sequencing samples may help in revealing
more possible mechanisms involved in the pathogenesis of AKI.
Transcriptome sequencing and bioinformatics tools revealed the
differential expression patterns of AS and RBP genes in AKI.
Further molecular mechanism studies will help to link the changes
in the expression of AS and RBP genes with the dysfunction caused
by AKI. In the present study, the RBP gene RBFOX1, as an AS
regulator, was observed to affect the expression of NF-κB and NRF2,
but the specific mechanism of this effect remains unknown. Our
future research involves the use of in vivo delivery
experiments to further determine the regulatory mechanism of
RBFOX1.
In the present study, the inflammatory response, AS,
and RBPs in cisplatin and IR-induced AKI models were analyzed via
whole transcriptome sequencing. Several hub genes, such as CSF-1,
CXCL1, CXCL10, IL-1β, IL-34, IL-6 and TLR2, were found to be mainly
enriched in the immune inflammatory response and related signaling
pathways. The role of AS events of the CSNK1A1, PAK2, CRK, ADK,
IKBKB, ZDHHC16, BCL2L1 and FGF1 genes in the pathogenesis of AKI
induced by cisplatin and IR warrants further investigation. As
splicing regulators, the RBP genes, such as LGALS3 and RBFOX1, were
found to be differentially expressed in the cisplatin and
IR-induced AKI. RBFOX1 was downregulated in AKI and it was observed
to inhibit the damage caused by inflammation and oxidative stress
by affecting NF-κB and the NRF2/HO-1 signaling pathway. These
findings provided a new perspective on the mechanisms of AKI.
Future studies should further elucidate the mechanistic aspects of
dysregulated AS in various diseases, such as intervention in the
RBP genes, as this information will provide the theoretical basis
for the development of novel classes of drug splicing-modifying
therapeutics.
Supplementary Data
Availability of data and materials
All data generated or analyzed during this study are
included in this published article. The sequencing data discussed
in this publication are available under GEO Series accession number
GSE142138.
Authors' contributions
FL designed the study. FL, LX and RY performed the
experiments and drafted the manuscript. SH, JX, KJ and BL
participated in data analysis. WY, TR, XZ and FC were involved in
the discussion and interpretation of the results. LX, RY, XZ and FC
were responsible for confirmation of the authenticity of the data.
All authors have read and approved the final manuscript.
Ethics approval and consent to
participate
All animal protocols were approved (approval no.
WDRM-20200904) by the Animal Care and Use Committee of Renmin
Hospital of Wuhan University (Wuhan, China) and all animal
experiments were conducted in accordance with the National
Institutes of Health Guide for the Care and Use of Laboratory
Animals.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgments
Not applicable.
Funding
The present study was supported by grants from the National
Natural Science Foundation of China (grant nos. 81870471 and
81800617) and the Science and Technology Major Project of Hubei
Province (grant nos. 2019AEA170 and 2020BCB017).
References
|
1
|
Li Q, Zhao M and Wang X: The impact of
transient and persistent acute kidney injury on short-term outcomes
in very elderly patients. Clin Interv Aging. 12:1013–1020. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Zhu H, Ren A, Zhou K, Chen Q, Zhang M and
Liu J: Impact of dexmedetomidine infusion on postoperative acute
kidney injury in elderly patients undergoing major joint
replacement: A retrospective cohort study. Drug Des Devel Ther.
14:4695–4701. 2020. View Article : Google Scholar :
|
|
3
|
Brandenburger T, Salgado Somoza A, Devaux
Y and Lorenzen JM: Noncoding RNAs in acute kidney injury. Kidney
Int. 94:870–881. 2018. View Article : Google Scholar
|
|
4
|
Hoste EAJ, Kellum JA, Selby NM, Zarbock A,
Palevsky PM, Bagshaw SM, Goldstein SL, Cerdá J and Chawla LS:
Global epidemiology and outcomes of acute kidney injury. Nat Rev
Nephrol. 14:607–625. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Rabb H, Griffin MD, McKay DB, Swaminathan
S, Pickkers P, Rosner MH and Kellum JA: Inflammation in AKI:
Current understanding, key questions, and knowledge gaps. J Am Soc
Nephrol. 27:371–379. 2016. View Article : Google Scholar :
|
|
6
|
Andrade-Oliveira V, Foresto-Neto O,
Watanabe IKM, Zatz R and Camara NOS: Inflammation in renal
diseases: New and old players. Front Pharmacol. 10:11922019.
View Article : Google Scholar :
|
|
7
|
Guo Y, Ni J, Chen S, Bai M, Lin J, Ding G,
Zhang Y, Sun P, Jia Z, Huang S, et al: MicroRNA-709 mediates acute
tubular injury through effects on mitochondrial function. J Am Soc
Nephrol. 29:449–461. 2018. View Article : Google Scholar
|
|
8
|
Baralle FE and Giudice J: Alternative
splicing as a regulator of development and tissue identity. Nat Rev
Mol Cell Biol. 18:437–451. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Black AJ, Gamarra JR and Giudice J: More
than a messenger: Alternative splicing as a therapeutic target.
Biochim Biophys Acta Gene Regul Mech. 1862:1943952019. View Article : Google Scholar :
|
|
10
|
Ule J and Blencowe BJ: Alternative
splicing regulatory networks: Functions, mechanisms, and evolution.
Mol Cell. 76:329–345. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Stevens M and Oltean S: Alternative
splicing in CKD. J Am Soc Nephrol. 27:1596–1603. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Yee BA, Pratt GA, Graveley BR, Van
Nostrand EL and Yeo GW: RBP-Maps enables robust generation of
splicing regulatory maps. RNA. 25:193–204. 2019. View Article : Google Scholar
|
|
13
|
Xu Y, Ma H, Shao J, Wu J, Zhou L, Zhang Z,
Wang Y, Huang Z, Ren J, Liu S, et al: A role for tubular
necroptosis in cisplatin-induced AKI. J Am Soc Nephrol.
26:2647–2658. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Xiao L, Zhou D, Tan RJ, Fu H, Zhou L, Hou
FF and Liu Y: Sustained activation of Wnt/beta-catenin signaling
drives AKI to CKD progression. J Am Soc Nephrol. 27:1727–1740.
2016. View Article : Google Scholar
|
|
15
|
Yu X, Meng X, Xu M, Zhang X, Zhang Y, Ding
G, Huang S, Zhang A and Jia Z: Celastrol ameliorates cisplatin
nephrotoxicity by inhibiting NF-κB and improving mitochondrial
function. EBioMedicine. 36:266–280. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zhou X, Jiang K, Luo H, Wu C, Yu W and
Cheng F: Novel lncRNA XLOC_032768 alleviates cisplatin-induced
apoptosis and inflammatory response of renal tubular epithelial
cells through TNF-α. Int Immunopharmacol. 83:1064722020. View Article : Google Scholar
|
|
17
|
Kim D, Pertea G, Trapnell C, Pimentel H,
Kelley R and Salzberg SL: TopHat2: Accurate alignment of
transcriptomes in the presence of insertions, deletions and gene
fusions. Genome Biol. 14:R362013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Trapnell C, Williams BA, Pertea G,
Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ and Pachter
L: Transcript assembly and quantification by RNA-Seq reveals
unannotated transcripts and isoform switching during cell
differentiation. Nat Biotechnol. 28:511–515. 2010. View Article : Google Scholar
|
|
19
|
Robinson MD, McCarthy DJ and Smyth GK:
edgeR: A Bioconductor package for differential expression analysis
of digital gene expression data. Bioinformatics. 26:139–140. 2010.
View Article : Google Scholar
|
|
20
|
Song Q, Yi F and Zhang Y, Li DK, Wei Y, Yu
H and Zhang Y: CRKL regulates alternative splicing of
cancer-related genes in cervical cancer samples and HeLa cell. BMC
Cancer. 19:4992019. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
22
|
Tu Y, Wu X, Yu F, Dang J, Wang J, Wei Y,
Cai Z, Zhou Z, Liao W, Li L and Zhang Y: Tristetraprolin
specifically regulates the expression and alternative splicing of
immune response genes in HeLa cells. BMC Immunol. 20:132019.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Ashburner M, Ball CA, Blake JA, Botstein
D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT,
et al: Gene ontology: Tool for the unification of biology. The gene
ontology consortium. Nat Genet. 25:25–29. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Kanehisa M, Furumichi M, Sato Y,
Ishiguro-Watanabe M and Tanabe M: KEGG: Integrating viruses and
cellular organisms. Nucleic Acids Res. 49:D545–D551. 2021.
View Article : Google Scholar :
|
|
25
|
Xie C, Mao X, Huang J, Ding Y, Wu J, Dong
S, Kong L, Gao G, Li CY and Wei L: KOBAS 2.0: A web server for
annotation and identification of enriched pathways and diseases.
Nucleic Acids Res. 39:W316–W322. 2011. View Article : Google Scholar :
|
|
26
|
Shannon P, Markiel A, Ozier O, Baliga NS,
Wang JT, Ramage D, Amin N, Schwikowski B and Ideker T: Cytoscape: A
software environment for integrated models of biomolecular
interaction networks. Genome Res. 13:2498–2504. 2003. View Article : Google Scholar
|
|
27
|
Liu C, Chen K, Wang H, Zhang Y, Duan X,
Xue Y, He H, Huang Y, Chen Z, Ren H, et al: Gastrin attenuates
renal ischemia/reperfusion injury by a PI3K/Akt/bad-mediated
anti-apoptosis signaling. Front Pharmacol. 11:5404792020.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Zhang Y, Zhang JJ, Liu XH and Wang L: CBX7
suppression prevents ischemia-reperfusion injury-induced
endoplasmic reticulum stress through the Nrf-2/HO-1 pathway. Am J
Physiol Renal Physiol. 318:F1531–F1538. 2020. View Article : Google Scholar
|
|
29
|
Marx D, Metzger J, Pejchinovski M, Gil RB,
Frantzi M, Latosinska A, Belczacka I, Heinzmann SS, Husi H,
Zoidakis J, et al: Proteomics and metabolomics for AKI diagnosis.
Semin Nephrol. 38:63–87. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Sabapathy V, Venkatadri R, Dogan M and
Sharma R: The Yin and Yang of alarmins in regulation of acute
kidney injury. Front Med (Lausanne). 7:4412020. View Article : Google Scholar
|
|
31
|
Andrade-Silva M, Cenedeze MA, Perandini
LA, Felizardo RJF, Watanabe IKM, Agudelo JSH, Castoldi A, Gonçalves
GM, Origassa CST, Semedo P, et al: TLR2 and TLR4 play opposite role
in autophagy associated with cisplatin-induced acute kidney injury.
Clin Sci (Lond). 132:1725–1739. 2018. View Article : Google Scholar
|
|
32
|
Baek JH, Zeng R, Weinmann-Menke J,
Valerius MT, Wada Y, Ajay AK, Colonna M and Kelley VR: IL-34
mediates acute kidney injury and worsens subsequent chronic kidney
disease. J Clin Invest. 125:3198–3214. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Zhang MZ, Yao B, Yang S, Jiang L, Wang S,
Fan X, Yin H, Wong K, Miyazawa T, Chen J, et al: CSF-1 signaling
mediates recovery from acute kidney injury. J Clin Invest.
122:4519–4532. 2012. View Article : Google Scholar
|
|
34
|
Anders HJ: Of inflammasomes and alarmins:
IL-1β and IL-1α in kidney disease. J Am Soc Nephrol. 27:2564–2575.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Nechemia-Arbely Y, Barkan D, Pizov G,
Shriki A, Rose-John S, Galun E and Axelrod JH: IL-6/IL-6R axis
plays a critical role in acute kidney injury. J Am Soc Nephrol.
19:1106–1115. 2008. View Article : Google Scholar
|
|
36
|
Liu P, Li X, Lv W and Xu Z: Inhibition of
CXCL1-CXCR2 axis ameliorates cisplatin-induced acute kidney injury
by mediating inflammatory response. Biomed Pharmacother.
122:1096932020. View Article : Google Scholar
|
|
37
|
Zhang WR, Garg AX, Coca SG, Devereaux PJ,
Eikelboom J, Kavsak P, McArthur E, Thiessen-Philbrook H, Shortt C,
Shlipak M, et al: Plasma IL-6 and IL-10 concentrations predict AKI
and long-term mortality in adults after cardiac surgery. J Am Soc
Nephrol. 26:3123–3132. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Erez DL, Denburg MR, Afolayan S, Jodele S,
Wallace G, Davies SM, Seif AE, Bunin N, Laskin BL and Sullivan KE:
Acute kidney injury in children after hematopoietic cell
transplantation is associated with elevated urine CXCL10 and CXCL9.
Biol Blood Marrow Transplant. 26:1266–1272. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Jiang S, Zhang M, Sun J and Yang X: Casein
kinase 1α: Biological mechanisms and theranostic potential. Cell
Commun Signal. 16:232018. View Article : Google Scholar
|
|
40
|
Peng X, Lai KS, She P, Kang J, Wang T, Li
G, Zhou Y, Sun J, Jin D, Xu X, et al: Induction of Wnt signaling
antagonists and p21-activated kinase enhances cardiomyocyte
proliferation during zebrafish heart regeneration. J Mol Cell Biol.
13:41–58. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Hinz M and Scheidereit C: The IκB kinase
complex in NF-κB regulation and beyond. EMBO Rep. 15:46–61. 2014.
View Article : Google Scholar
|
|
42
|
Wan X, Fan L, Hu B, Yang J, Li X, Chen X
and Cao C: Small interfering RNA targeting IKKβ prevents renal
ischemia-reperfusion injury in rats. Am J Physiol Renal Physiol.
300:F857–F863. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Cao W, Yuan Y, Liu X, An X, Huang Z, Wu L,
Zhang B, Zhang A and Xing C: Adenosine kinase inhibition protects
against cisplatin-induced nephrotoxicity. Am J Physiol Renal
Physiol. 317:F107–F115. 2019. View Article : Google Scholar
|
|
44
|
Du J, Meng L, Pang L, Jin B, Duan N, Huang
C, Huang H and Li H: Crk1/2 and CrkL play critical roles in
maintaining podocyte morphology and function. Exp Cell Res.
394:1121352020. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Cao N, Li JK, Rao YQ, Liu H, Wu J, Li B,
Zhao P, Zeng L and Li J: A potential role for protein
palmitoylation and zDHHC16 in DNA damage response. BMC Mol Biol.
17:122016. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Stevens M and Oltean S: Modulation of the
apoptosis gene Bcl-x function through alternative splicing. Front
Genet. 10:8042019. View Article : Google Scholar :
|
|
47
|
Valdes F, Pasaro E, Diaz I, Centeno A,
López E, García-Doval S, González-Roces S, Alba A and Laffon B:
Segmental heterogeneity in Bcl-2, Bcl-xL and Bax expression in rat
tubular epithelium after ischemia-reperfusion. Nephrology
(Carlton). 13:294–301. 2008. View Article : Google Scholar
|
|
48
|
Liang G, Song L, Chen Z, Qian Y, Xie J,
Zhao L, Lin Q, Zhu G, Tan Y, Li X, et al: Fibroblast growth factor
1 ameliorates diabetic nephropathy by an anti-inflammatory
mechanism. Kidney Int. 93:95–109. 2018. View Article : Google Scholar :
|
|
49
|
Corley M, Burns MC and Yeo GW: How
RNA-binding proteins interact with RNA: Molecules and mechanisms.
Mol Cell. 78:9–29. 2020. View Article : Google Scholar :
|
|
50
|
Sun H, Jiang H, Eliaz A, Kellum JA, Peng Z
and Eliaz I: Galectin-3 in septic acute kidney injury: A
translational study. Crit Care. 25:1092021. View Article : Google Scholar :
|
|
51
|
Conboy JG: Developmental regulation of RNA
processing by Rbfox proteins. Wiley Interdiscip Rev RNA. 8:
View Article : Google Scholar : 2017.
|
|
52
|
Xiang H, Xue W, Li Y, Zheng J, Ding C, Dou
M and Wu X: Knockdown of ANGPTL2 protects renal tubular epithelial
cells against hypoxia/reoxygenation-induced injury via suppressing
TLR4/NF-κB signaling pathway and activating Nrf2/HO-1 signaling
pathway. Cell Transplant. 29:9636897209466632020. View Article : Google Scholar
|
|
53
|
Song N, Thaiss F and Guo L: NFκB and
kidney injury. Front Immunol. 10:8152019. View Article : Google Scholar
|
|
54
|
Yeung F, Hoberg JE, Ramsey CS, Keller MD,
Jones DR, Frye RA and Mayo MW: Modulation of NF-kappaB-dependent
transcription and cell survival by the SIRT1 deacetylase. EMBO J.
23:2369–2380. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Wei W, Ma N, Fan X, Yu Q and Ci X: The
role of Nrf2 in acute kidney injury: Novel molecular mechanisms and
therapeutic approaches. Free Radic Biol Med. 158:1–12. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Komatsu M, Kurokawa H, Waguri S, Taguchi
K, Kobayashi A, Ichimura Y, Sou YS, Ueno I, Sakamoto A, Tong KI, et
al: The selective autophagy substrate p62 activates the stress
responsive transcription factor Nrf2 through inactivation of Keap1.
Nat Cell Biol. 12:213–223. 2010. View Article : Google Scholar : PubMed/NCBI
|