Introduction
For years, the pathological evaluation of tumor tissue has been essential for cancer diagnosis and care. The field of pathology has been becoming more and more complex, requiring the pathologists to integrate new techniques and develop new skills. With the advent of targeted therapies, this has included the use of molecular biology techniques. The application of molecular biology to cancer diagnostics started 15-20 years ago with the fluorescence in situ hybridization analysis of the ERBB2 gene in breast cancer (1), followed by sequence analysis of EGFR and KRAS in lung and colon cancers (2). At present, an increasing number of genes are analyzed, irreversibly transforming 'classic' pathology into modern molecular pathology. However, one of the major challenges of the molecular diagnosis of patients with cancer is to be able to fit the suitable target analysis into the right (from a clinical standpoint) timing. The latter should allow the diagnosticians to obtain objective (uninfluenced by the first-line treatment) molecular results. Concurrently, the results must be provided early enough to guide treatment decisions integrating precision medicine solutions in time to obtain favorable outcomes in metastatic patients.
Recent years have witnessed tremendous technological development which has revolutionized cancer diagnostics. In particular, the development of next-generation sequencing (NGS) has significantly increased analytical capacities, which has enabled laboratories to analyze increasing numbers of targets and indicators. Molecular analyses have become standard for diagnosis of several solid cancers in Europe. These analyses cover common and different target genes that could be mutated in tumors: ALK (3), BRAF (4), EGFR (5), ERBB2 (6), KRAS (7), and MET (8) in lung cancers, BRAF (9), KRAS (10), and NRAS (11) in colorectal tumors, BRAF (12), KIT (13), and NRAS (14) in melanoma, as well as KIT and PDGFRA in gastrointestinal stromal tumors (GISTs) (15). These genes are known to be mutated at hotspots, but the analytical performance of the NGS technology allows us to analyze larger gene regions, or even whole coding parts of these genes, which may result in important clinical implications.
In the present study, an NGS custom panel which was designed and validated for molecular diagnosis of lung cancer, colorectal cancer, melanoma and GISTs based on the analysis of solid and liquid biopsies was presented. This NGS custom panel has been used successfully in our clinical practice for three years.
Materials and methods
Patients and samples
The present study was conducted on solid tumor samples from 2,289 patients diagnosed at the Georges Francois-Leclerc Cancer Center (CGFL; Dijon, France) between January 2017 and December 2019. All tumor samples were sent to our lab for analyses as part of the routine clinical diagnosis procedure. They included 1,299 lung cancer samples, 790 colorectal cancer samples, 158 melanomas and 42 GISTs. Formalin-fixed paraffin-embedded tumor blocks were obtained from different pathology laboratories. Additionally, blood samples (liquid biopsies) were collected into special 10-ml cell free DNA collection tubes whenever possible for plasma DNA extraction and analysis. The present study on patient samples was conducted in accordance with the Declaration of Helsinki and was approved (approval no. 00010311) by the Ethics Committee of the Georges-François Leclerc Cancer Center (Dijon, France) and by the Consultative Committee of Burgundy (Dijon, France) for the Protection of Persons Participating in Biomedical Research (Comité Consultatif de Protection des Personnes en Recherche Biomédicale de Bourgogne). Written informed consent was provided by all patients.
DNA extraction
Tumor slides prepared from formalin-fixed paraffin-embedded tumor specimens were analyzed by a pathologist to determine tumor cell content. Only specimens containing more than 5% of tumor cells were used for DNA extraction. DNA was extracted from five 8-µm-thick tumor slides per patient using the Maxwell-16 formalin-fixed paraffin-embedded Plus LEV DNA purification kit (Promega Corporation) according to the manufacturer's protocol and quantified using a fluorimetric assay on a Qubit device 4 (Life Technologies; Thermo Fisher Scientific, Inc.).
In order to separate blood plasma, liquid biopsies were centrifuged at 1,150 × g for 6 min at room temperature. Then, the supernatant was transferred into a new tube and centrifuged at 12,000 × g for 15 min at 4°C. Next, DNA was extracted from ~2 to 4 ml of plasma using the QIAamp circulating nucleic acid kit (Qiagen GmbH) according to the manufacturer's protocol.
Panel design and library preparation
A panel focusing on coding regions (-5/+5 nucleotides in flanking introns except for MET exon 14 flanking introns for which -80/+80 nucleotides were analyzed) of ten genes related to approved treatments for lung and colorectal cancers, melanomas and GISTs was developed: ALK, BRAF, EGFR, ERBB2, KIT, KRAS, MAP2K1, MET, NRAS, and PDGFRA (Table I). Coding regions of the TP53 gene (detectable in liquid biopsies) as well as five regions containing single nucleotide polymorphisms (SNPs) were also included to control for sample cross-contamination (Table II) (16). The panel design was patented under the number WO2019/197541.
 |
Table I
Genes included in our next-generation sequencing panel and clinical impact of their activating variants.
|
Table I
Genes included in our next-generation sequencing panel and clinical impact of their activating variants.
| Gene |
Tumor type |
Associated treatment or prognostic effect |
Impact |
| ALK |
Lung cancer |
ALK TKI |
Sensitivity of resistance to ALK inhibitor |
| BRAF |
Colon cancer |
No therapeutics |
MSI Testing |
| Lung cancer |
BRAF inhibitor |
Sensitivity only for the p.(Val600Glu) variant |
| Melanoma |
BRAF inhibitor |
Sensitivity only for p.(Val600X) variants |
| EGFR |
Lung cancer |
EGFR TKI |
Sensitivity or resistance (secondary mutations, insertions exon 20) |
| ERBB2 |
Lung cancer |
Mobocertinib |
Sensitivity |
| KIT |
GIST |
Imatinib |
Sensitivity or resistance (secondary mutations) |
| Melanoma |
Imatinib |
Sensitivity |
| KRAS |
Colon cancer |
EGFR inhibitor |
Resistance |
| Lung cancer |
Sotorasib |
Sensitivity only for the p.(Gly12Cys) variant |
| MAP2K1 |
Melanoma |
BRAF inhibitor |
Resistance |
| MEK inhibitor |
Sensitivity or resistance |
| MET |
Lung cancer |
MET TKI |
Sensitivity |
| NRAS |
Colon cancer |
EGFR inhibitor |
Resistance |
| Melanoma |
Prognosis |
/ |
| PDGFRA |
GIST |
Imatinib |
Sensitivity or resistance (secondary mutations) |
 |
Table II
List of five regions containing SNPs integrated in our panel for detecting putative cross-contamination between samples.
|
Table II
List of five regions containing SNPs integrated in our panel for detecting putative cross-contamination between samples.
| Gene |
Amino acid variation |
Nucleotide variation |
European population frequency (%)a |
African population frequency (%)a |
| EGFR |
Q787Q |
c.2361G/A |
40.59/59.41 |
59.18/40.82 |
| EIF1AY |
Gene present only on the Y chromosome |
|
|
|
| KDR |
Q472H |
c.1416A/T |
76.03/23.97 |
88.19/11.81 |
| SLC28A1 |
V189I |
c.565G/A |
64.85/35.15 |
78.62/21.38 |
| TP53 |
R72P |
c.215G/C |
26.34/73.66 |
68.54/31.46 |
The panel performances were evaluated on 15 criteria required to obtain the ISO15189 accreditation (ISO15189:2012). The latter is necessary to obtain the accreditation by the French Accreditation Committee (COFRAC), obligatory for routine molecular diagnosis laboratories in France.
For library preparation, 400 ng of DNA from solid tumors were fragmented with a Covaris LE220-plus device (Covaris, Inc.) to obtain fragments ~300 bp-long. For liquid biopsies, 30 µl of DNA were directly engaged in the process, without shearing. Then, libraries were prepared with the SureSelectXT (cat. no. G9642B) technology (Agilent Technologies, Inc.) following the manufacturer's instructions. Paired-end (2×111 bases) sequencing was performed on a MiSeq device (Illumina, Inc.) twice a week. For solid samples, all hotspot mutations were confirmed by allelic discrimination, fragment analysis or Sanger sequencing for identity checking.
Bioinformatic analysis
Reads in FASTQ format were aligned to the reference human genome GRCh37 using the Burrows-Wheeler aligner (BWA v.0.7.15) (17). Local realignment was performed using the Genome Analysis Toolkit (GATK v.3.6) (18-20). Duplicate reads were removed using Picard v.2.5 (21). Outlyzer (v1.0) (22) was used to identify variants. These variants were annotated using Annovar (Annovar2016Feb01) (23) and SnpEff (v4.3i) (24). Quality controls were performed using fastQC (v0.11.8) (25), Samtools (v1.9) (26), and Qualimap (v2.2.1) (27) information through multiQC (v1.7) software (28).
Variants with a frequency superior to 1% in the general population were filtered and did not appear in results tables analyzed by the biologists.
Statistical analysis
Analysis was performed with GraphPad Prism version 8.3.0 (GraphPad Software, Inc.). No statistical analyses were performed for Figs. 1A and 4 and S1B. Concerning Fig. 4C and D, non-parametric Kruskal-Wallis test was used followed by Dunn's test. For Fig. 4C, n=287 in the lung subset, n=78 in the colon subset and n=18 in the melanoma subset. For Fig. 4D, n=81 for EGFR, n=50 for RAS/BRAF/MAP2K1 group, and n=81 for TP53 subset. For Fig. 4E, non-parametric Mann-Whitney U-test was used; the EGFR-sensitive mutation subset had 85 cases and resistance mutation sets had 27 cases. P<0.05 was considered to indicate a statistically significant difference.
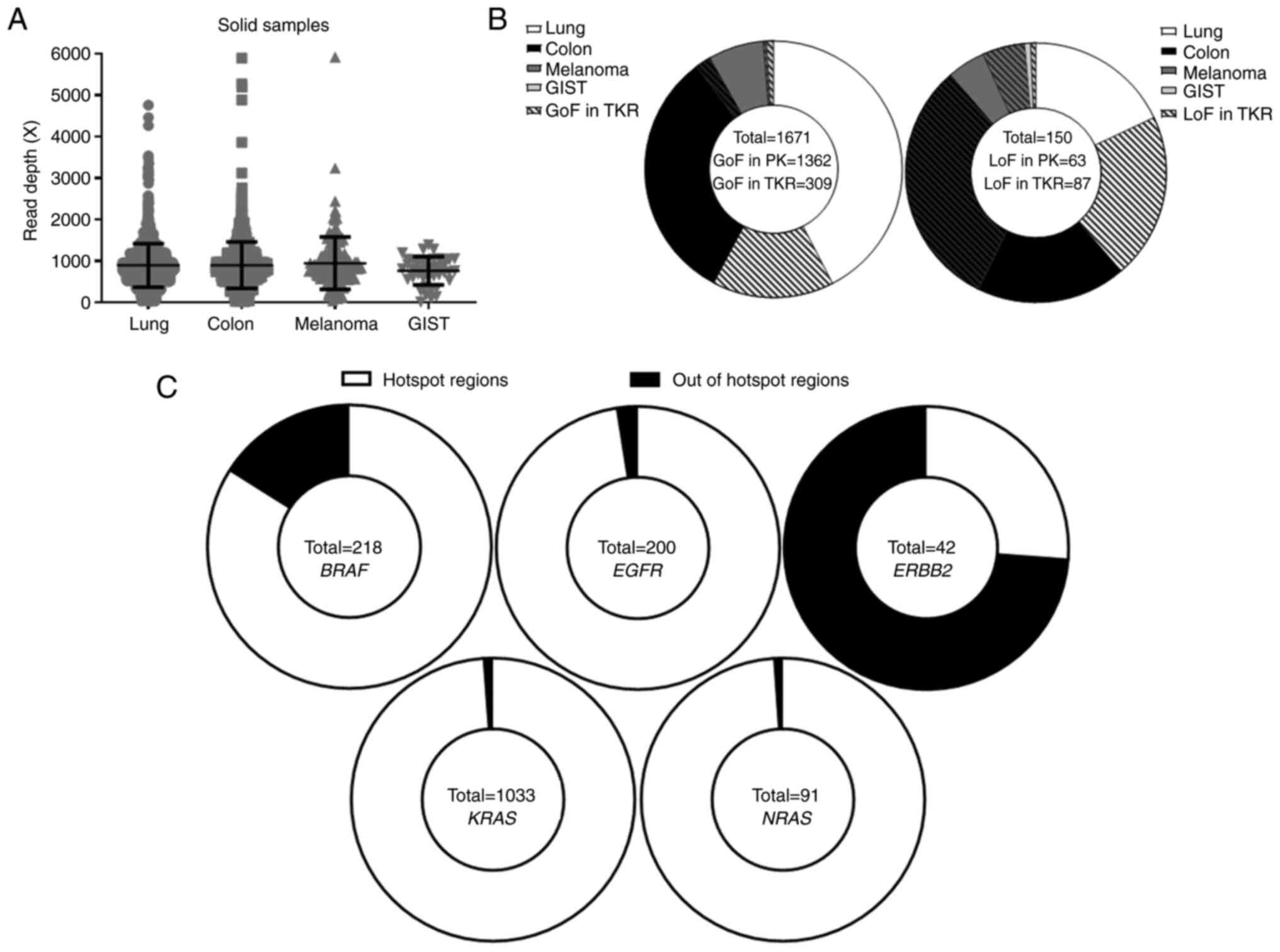 |
Figure 1
Landscape of variants detected in tumor samples from four tumor types (lung carcinoma, colon carcinoma, melanoma and GIST by our custom next-generation sequencing panel. (A) Read depth coverage obtained for solid biopsies by tumor type. (B) Distribution of GoF variants (left) and LoF variants (right) between genes encoding PK and those encoding TKR, by tumor type. (C) Distribution of GoF mutations in BRAF, EGFR, ERBB2, KRAS and NRAS between hotspot and non-hotspot regions. GIST, gastrointestinal stromal tumor; GoF, gain-of-function; LoF, loss-of-function; PK, protein kinase; TKR, tyrosine kinase receptor.
|
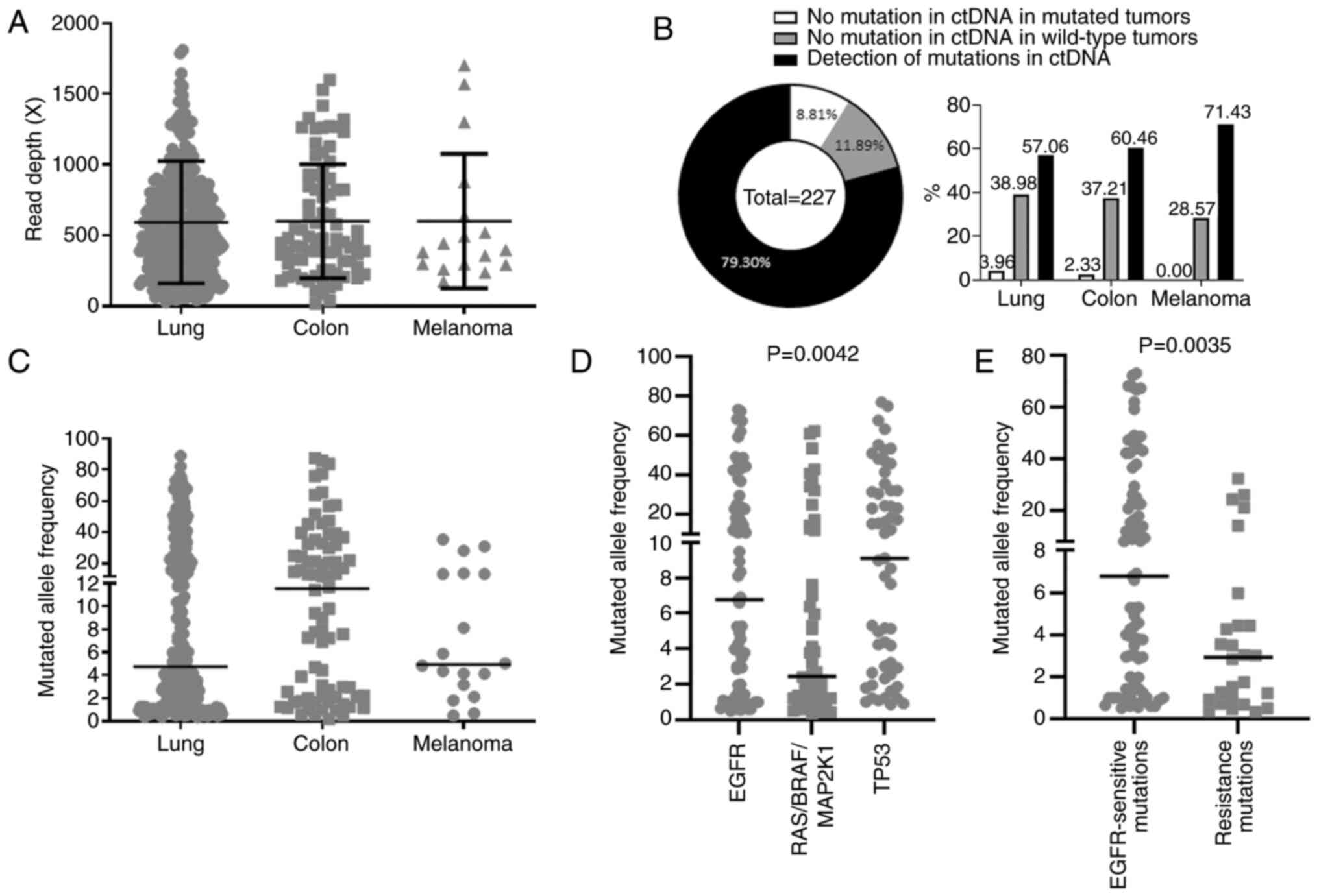 |
Figure 4
Performance of our custom next-generation sequencing panel in liquid biopsies. (A) Read depth coverage obtained for liquid biopsies of lung and colorectal cancers, and of melanomas. (B) Left, detection of variants in 227 liquid biopsies compared with results obtained for matched tumor samples; right, representation of these 227 liquid biopsies depending on their origin. (C) Mutant allele frequencies obtained for liquid biopsies from patients with lung cancer, colorectal cancer and melanoma. (D) Mutant allele frequencies for EGFR variants, for KRAS, NRAS, BRAF and MAP2K1 (RAS/BRAF/MAP2K1) variants, as well as TP53 variants in liquid biopsies from patients with lung cancer. (E) Mutant allele frequencies in liquid biopsies from EGFR-mutant lung cancers for variants sensitizing to EGFR tyrosine kinase inhibitor and those inducing resistance to this treatment. ctDNA, circulating tumor DNA.
|
Results
Panel description and performances
An NGS-based panel for routine molecular diagnosis of four tumor types was developed: Lung carcinoma, colon carcinoma, melanoma and GIST. Our panel includes the coding regions of ten genes related to approved treatments for these four tumor types (Table I). It also includes the coding regions of the TP53 gene to detect tumor material in liquid biopsies as well as five regions containing SNPs to control for sample cross-contamination (29).
In order to comply with the French regulations on routine molecular diagnosis laboratories, our panel was assessed for 15 items according to the ISO15189 standards. Our method met all the requirements for the ISO15189 accreditation (Table III). Repeatability was calculated using five replicates of the Horizon HD728 internal control (AmpliTech Sarl) with a specific focus on two-point variants: G719S in the EGFR gene and G12A in KRAS. The EGFR G719S mutation had a mean detection frequency of 17.6±1.8% (expected frequency, 16.7%) with a variation coefficient of 10.2%, while KRAS G12A was detected with a mean frequency of 3.9±0.8% (expected frequency, 5%) with a variation coefficient of 20.2%. Intermediate fidelity was assessed on the same variants by analyzing 20 replicates. EGFR G719S was detected with a mean frequency of 17.2±0.9% (expected frequency, 16.7%) with a variation coefficient of 5.1% and KRAS G12A had a mean detection frequency of 4.1±0.6% (expected frequency, 5%) with a variation coefficient of 13.5%. Using the same 20 replicates, the rightness of our panel was determined to obtain a bias of 3% for the EGFR G719S mutation and -18% for the KRAS G12A mutation. Subsequently, to calculate the accuracy of our technique, nine samples received from the 2017 European Molecular Genetics Quality Network (EMQN) campaign were used. The expected genotype was detected in all samples, thus showing a 100% accuracy of the technique for genotype detection and an 89% accuracy for the alternative allelic frequency (Fig. S1A). The alternative allelic frequency of eight variants from the Horizon HD728 control on 231 consecutive runs was also examined (Fig. S1B). Except for the BRAF V600E mutation for which the alternative allelic frequency was slightly overestimated, all alternative allelic frequencies were equal to expected values. Based on the analysis of the same 2017 EMQN campaign samples, a sensitivity and specificity of 100% was obtained. These values were confirmed by benchmarking using 231 runs with the Horizon HD728 control. Additionally, during the three-year period analyzed, the presence of all variants of interest (n=1,505) found in ALK (n=1), BRAF (n=51), EGFR (n=166), ERBB2 (n=11), KIT (n=30), KRAS (n=1018), MET (n=34), NRAS (n=90), and PDGFRA (n=4) was confirmed by other targeted technical approaches. Finally, our very low background (<1%) enabled us to set the detection limit at 1% (particularly for hotspot regions). For liquid biopsies, the systematic visualization of raw data (bam files) decreased the detection limit below 0.5%.
 |
Table III
Fifteen items assessed and the results obtained for our next-generation sequencing method validation in accordance with the ISO15189 standard.
|
Table III
Fifteen items assessed and the results obtained for our next-generation sequencing method validation in accordance with the ISO15189 standard.
| Item tested |
Result |
| Repeatability |
Coefficient of variation < or =20.2% |
| Intermediate fidelity |
Coefficient of variation < or =13.5% |
| Rightness |
−18% < Bias <3% |
| Accuracy |
100% for the genotype; 89% for allelic frequency |
| Sensitivity |
100% |
| Specificity |
100% |
| Benchmarking |
100% concordance |
| Background |
<1% |
| Coverage |
100% of regions of interest |
| Limit of detection |
1% of the mutated allele |
| Intra-run contamination |
No contamination detected |
| Inter-run contamination |
No contamination detected |
| Robustness |
Continuously managed with a positive control |
| Stability of reagents |
All reagents and libraries are stable under our laboratory conditions of use |
Mutational landscape in tumor samples
From January 2017 to December 2019, 2,289 solid tumor samples from lung (n=1,299), colon/rectum (n=790), melanoma (n=158), and GIST (n=42) were sequenced with our custom NGS panel. The median read depth obtained was 828.52X [lung cancer, 832.94X; colorectal cancer, 818.16X; melanoma, 842.36X; and GIST, 800.10X (Fig. 1A)]. A total of 1,180 different variations that were classified as benign (n=67), unknown (n=797), gain-of-function (GoF; n=196) or loss-of-function (LoF; n=110) were observed for each gene (Tables IV and SI). In total, 1,671 of the detected variants were GoF variants. They were distributed between protein kinase-coding genes (81.5% of variants for all tumor types together, including 42.43% in lung cancers, 32.14% in colon cancers, 6.88% in melanomas and 0.05% in GISTs) and those encoding tyrosine kinase receptors (18.5% for all the four tumor types: 15.26% in lung cancers, 1.98% in colon cancers, 0.36% in melanomas and 0.9% in GISTs; Figs. 1B left and S2). LoF variants in protein kinase-coding genes (42% for all tumors types: 18% in lung cancers, 18.67% in colon cancers, 4.66% in melanomas, and 0.67% in GISTs) were also observed, mostly in the BRAF gene which harbored 89.23% of all LoF variants in protein kinase-coding genes. Surprisingly, 58% of LoF variants occurred in genes encoding the tyrosine kinase receptor (20.67, 31.33, 5.33 and 0.67% for lung, colon, melanoma and GIST, respectively; Figs. 1B right and S2). The location, number, and tissue distribution of GoF and LoF variants for each gene are presented in Fig. S3. Notably, for BRAF, EGFR, ERBB2, KRAS and NRAS genes, certain of the detected GoF variants were different from known hotspot variants (Fig. 1C). BRAF and ERBB2 genes were particularly affected. BRAF hotspot is located at codon 600 where activating variants for 183 (83.94%) patients were detected. A total of 35 patients (16.06%) were carriers of activating variants in the BRAF gene, located at codons 257, 464, 469, 483, 485, 486, 499, 597, 599, 601, and 731 (Table SI). Concerning ERBB2, the analyzed hotpot region was exon 20, known in particular for frequent insertions in lung adenocarcinoma. A total of 11 (26.19%) insertions were detected. As expected, all of them were found in lung adenocarcinomas. Surprisingly, 31 (73.81%) point variants out of exon 20 (except for one-point mutation) were detected, concerning mainly colon adenocarcinomas (n=24; 77.42%; Fig. S2). For EGFR, KRAS, and NRAS genes, GoF variants were mostly in hotspot regions (Fig. 1C).
 |
Table IV
Distribution of the four classes of genetic variants in the ten genes included in our next-generation sequencing panel for each tumor type.
|
Table IV
Distribution of the four classes of genetic variants in the ten genes included in our next-generation sequencing panel for each tumor type.
| Variant significance |
Lung cancer (n=1299) (%) |
Colon cancer (n=790) (%) |
Melanoma (n=158) (%) |
Gastrointestinal stromal tumor (n=42) (%) |
| ALK variants |
|
|
|
|
| Benign |
9 (0.69) |
10 (1.27) |
1 (0.63) |
0 (0) |
| Unknown |
96 (7.39) |
68 (8.61) |
23 (15.46) |
1 (2.38) |
| Gain-of-function |
1 (0.08) |
0 (0) |
0 (0) |
0 (0) |
| Loss-of-function |
12 (0.92) |
11 (1.39) |
1 (0.63) |
0 (0) |
| BRAF variants |
|
|
|
|
| Benign |
2 (0.15) |
2 (0.25) |
2 (1.27) |
0 (0) |
| Unknown |
39 (3) |
16 (2.03) |
13 (8.23) |
2 (4.76) |
| Gain-of-function |
54 (4.16) |
103 (13.04) |
61 (38.61) |
0 (0) |
| Loss-of-function |
25 (1.92) |
24 (3.04) |
9 (5.70) |
0 (0) |
| EGFR variants |
|
|
|
|
| Benign |
10 (0.77) |
3 (0.38) |
1 (0.63) |
0 (0) |
| Unknown |
59 (4.54) |
36 (4.56) |
20 (12.66) |
2 (4.76) |
| Gain-of-function |
196 (15.09) |
6 (0.76) |
0 (0) |
0 (0) |
| Loss-of-function |
4 (0.31) |
3 (0.38) |
1 (0.63) |
0 (0) |
| ERBB2 variants |
|
|
|
|
| Benign |
54 (4.16) |
33 (4.18) |
3 (1.9) |
2 (4.76) |
| Unknown |
53 (4.08) |
26 (3.29) |
12 (7.59) |
2 (4.76) |
| Gain-of-function |
18 (1.39) |
24 (3.04) |
0 (0) |
0 (0) |
| Loss-of-function |
1 (0.08) |
2 (0.25) |
0 (0) |
0 (0) |
| KIT variants |
|
|
|
|
| Benign |
6 (0.46) |
7 (0.89) |
0 (0) |
0 (0) |
| Unknown |
58 (4.46) |
38 (4.81) |
9 (5.7) |
8 (19.05) |
| Gain-of-function |
2 (0.15) |
1 (0.13) |
5 (3.16) |
31 (73.81) |
| Loss-of-function |
3 (0.23) |
6 (0.76) |
2 (1.27) |
1 (2.38) |
| KRAS variants |
|
|
|
|
| Benign |
2 (0.15) |
0 (0) |
1 (0.63) |
0 (0) |
| Unknown |
5 (0.38) |
3 (0.38) |
4 (2.53) |
0 (0) |
| Gain-of-function |
627 (48.27) |
396 (50.13) |
9 (5.7) |
1 (2.38) |
| Loss-of-function |
0 (0) |
2 (0.25) |
0 (0) |
0 (0) |
| MAP2K1 variants |
|
|
|
|
| Benign |
1 (0.08) |
0 (0) |
0 (0) |
0 (0) |
| Unknown |
17 (1.31) |
5 (0.63) |
1 (0.63) |
0 (0) |
| Gain-of-function |
9 (0.69) |
8 (1.01) |
3 (1.9) |
0 (0) |
| Loss-of-function |
2 (0.15) |
0 (0) |
0 (0) |
0 (0) |
| MET variants |
|
|
|
|
| Benign |
43 (3.31) |
16 (2.03) |
2 (1.27) |
1 (2.38) |
| Unknown |
72 (5.54) |
42 (5.32) |
25 (15.82) |
0 (0) |
| Gain-of-function |
38 (2.93) |
1 (0.13) |
1 (0.63) |
0 (0) |
| Loss-of-function |
6 (0.46) |
15 (1.90) |
3 (1.9) |
0 (0) |
| NRAS variants |
|
|
|
|
| Benign |
1 (0.08) |
0 (0) |
0 (0) |
0 (0) |
| Unknown |
11 (0.85) |
2 (0.25) |
3 (1.9) |
0 (0) |
| Gain-of-function |
19 (1.46) |
30 (3.80) |
42 (26.58) |
0 (0) |
| Loss-of-function |
0 (0) |
2 (0.25) |
1 (0.63) |
0 (0) |
| PDGFRA variants |
|
|
|
|
| Benign |
58 (4.46) |
46 (5.82) |
6 (3.8) |
1 (2.38) |
| Unknown |
65 (5) |
48 (6.08) |
15 (9.49) |
0 (0) |
| Gain-of-function |
0 (0) |
1 (0.13) |
0 (0) |
4 (9.52) |
| Loss-of-function |
4 (0.31) |
10 (1.27) |
0 (0) |
0 (0) |
Distribution of activating variants by tumor type
It was identified that 51.61% of the 1,299 lung adenocarcinomas harbored activating variants in the RAS/RAF/MEK pathway, 19.41% had a GoF mutation in a tyrosine-kinase domain receptor, and 25.94% were wild-type. The most frequently altered gene was KRAS (45.3%), followed by EGFR (15.09%), BRAF (4.16%), MET (2.93%), NRAS (1.46%), ERBB2 (1.39%), MAP2K1 (0.69%), and ALK (only one case; Fig. 2A). Out of the 790 analyzed colorectal tumors, 67.97% harbored activating variants in the RAS/RAF/MEK pathway, 3.04% had a GoF mutation in a tyrosine-kinase domain receptor and 28.99% were wild-type. The most frequently altered gene was KRAS (50.13%) followed by BRAF (13.03%), NRAS (3.80%), ERBB2 (3.04%) and MAP2K1 (1.01%; Fig. 2B). As regards melanoma, 72.79% harbored activating mutations in the RAS/RAF/MEK pathway, 3.16% had a GoF mutation in a tyrosine-kinase domain receptor, and 24.05% were wild-type. The most frequently altered gene was BRAF (38.61%), followed by NRAS (26.58%), KRAS (5.7%), KIT (3.16%), and MAP2K1 (1.9%; Fig. 2C). Finally, none of the GISTs were mutated in the RAS/RAF/MEK pathway, while GoF variants in a tyrosine-kinase domain receptor were detected in 83.33% of these tumors and 16.67% of tumors were wild-type. Only two genes were mutated in GISTs, the majority of tumors carrying mutations in KIT (73.81% mutant tumors), followed by PDGFRA (only 9.52%; Fig. 2D).
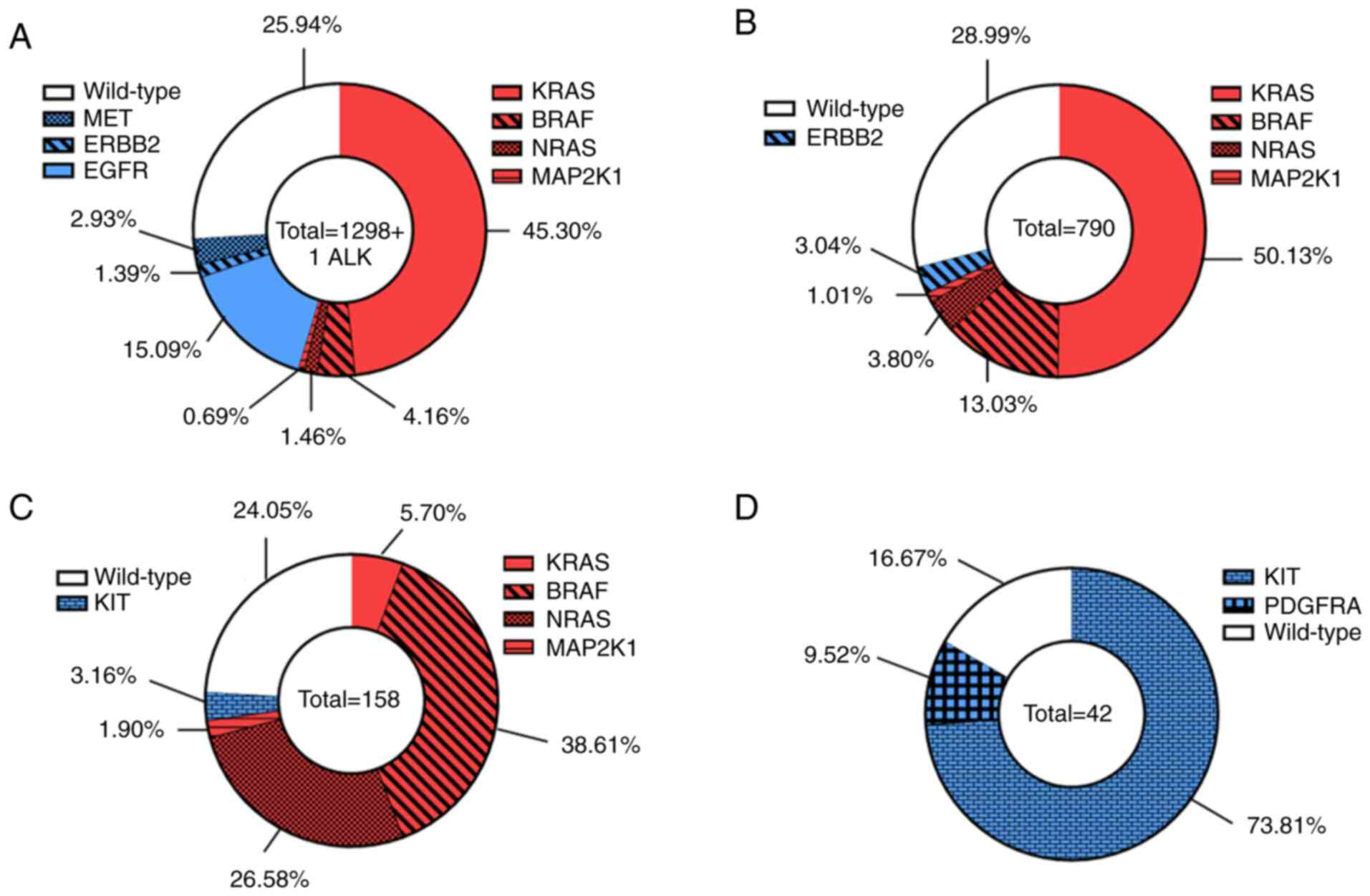 |
Figure 2
Distribution of activating variants between different genes of our custom next-generation sequencing panel. (A) Lung cancers, (B) colorectal cancers, (C) melanomas, and (D) gastrointestinal stromal tumors. Wild-type samples appear in white, protein kinase-coding genes in red and those encoding tyrosine kinase receptors in blue.
|
Exon distribution of activating variants in the different genes
Since GoF variants are often located in hotspot regions, the exon distribution for all the genes on the panel was examined, except for ALK which had only one activating variant (Table SII). The BRAF gene harbored 87.15% (n=190) of activating variants in exon 15, followed by 11.47% (n=25) in exon 11, and 1.38% in exons 6, 12 and 18 (one variant in each exon; Fig. 3A). For the EGFR gene, 1.49% of activating variants were in exons 6 and 7 (n=1 for each), 4.46% (n=9) in exon 18, 41.58% (n=84) in exon 19, 17.82% (n=36) in exon 20 and 34.65% (n=70) in exon 21 (Fig. 3B). Activating variants were distributed among seven different exons in the ERBB2 gene, with 14.29% (n=6) in exon 8, 21.43% (n=9) in exon 12, 21.43% (n=9) in exons 17 (n=5), 18 (n=1), and 19 (n=3), 28.56% (n=12) in exon 20 and 14.29% (n=6) in exon 21 (Fig. 3C). The 39 GoF variants in the KIT gene were located in five exons, with 5.13% (n=2) in exons 8 and 9, 87.18% (n=34) in exon 11, and 7.69% (n=3) in exons 14 (n=1) and 17 (n=2) (Fig. 3D). The most often mutated gene, KRAS, had 89.84% (n=928) of GoF variants in exon 2, 6% in exon 3 (n=62) and 4.16% (n=43) in exon 4 (Fig. 3E). The 20 GoF variants of the MAP2K1 gene were mostly in exon 2 (65%, n=13), followed by 25% (n=5) in exon 3, and 10% (n=2) in exon 6 (Fig. 3F). Only three exons harbored activating variants in the MET gene, with 92.50% (n=37) located in exon 14 under the form of splicing variants, and 7.50% in exons 16 (n=2) and 17 (n=1; Fig. 3G). NRAS harbored 28.57% (n=26) variants in exon 2 and 71.43% (n=65) in exon 3 (Fig. 3H). The least mutated gene, PDGFRA, had 60% of variants in exons 11 (n=1) and 12 (n=3), and 40% (n=2) in exon 18 (Fig. 3I).
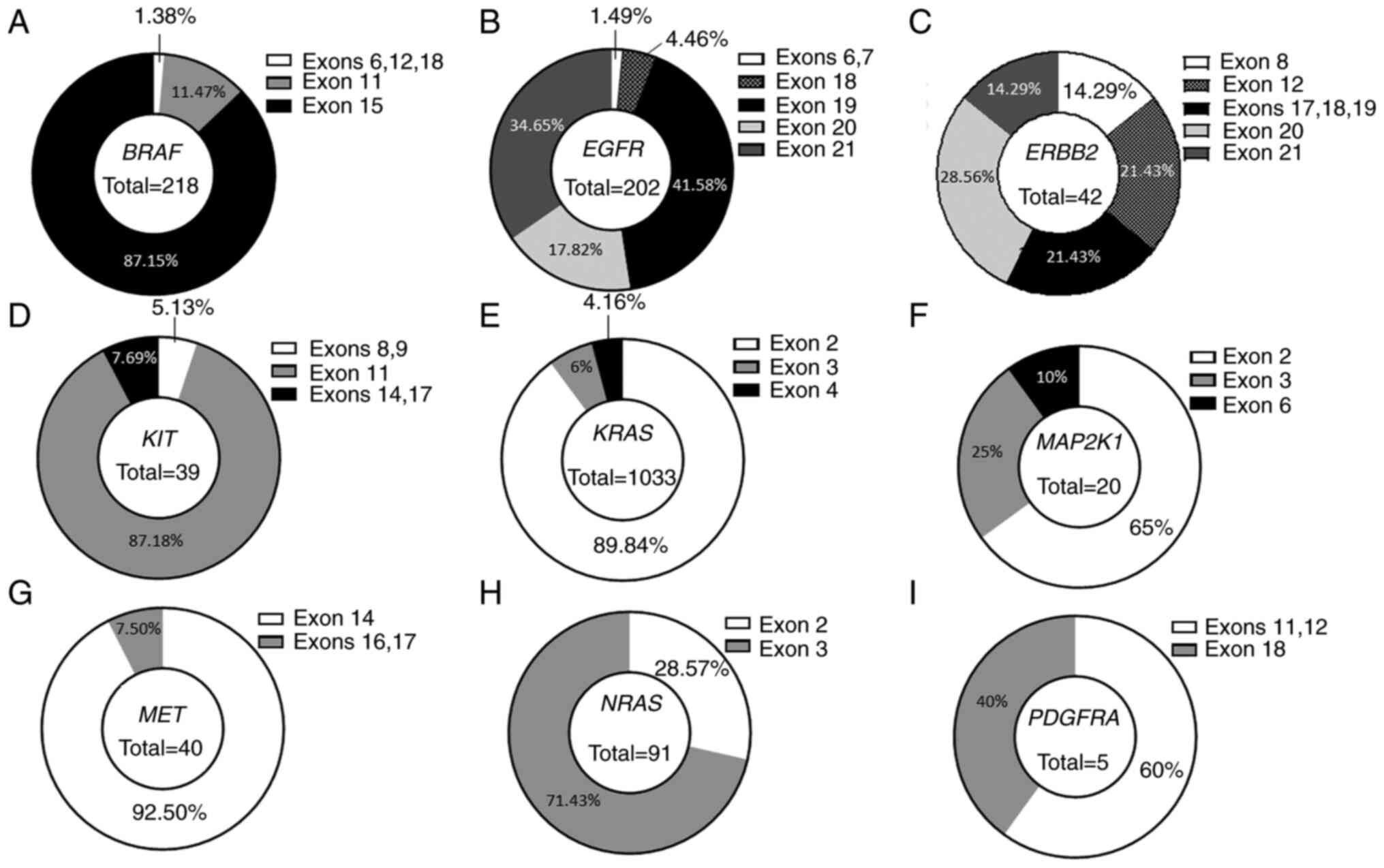 |
Figure 3
Distribution of activating variants between exons of genes analyzed with our custom next-generation sequencing panel. Percentages of activating variants detected in (A) BRAF, (B) EGFR, (C) ERBB2, (D) KIT, (E) KRAS, (F) MAP2K1, (G) MET, (H) NRAS and (I) PDGFRA are indicated for each exon.
|
Use of the panel for analyzing liquid biopsies
In recent years, liquid biopsies have been routinely used in clinical practice, particularly for monitoring patients with EGFR-mutant lung cancer treated with an EGFR tyrosine kinase inhibitor. They are also used for primary diagnosis of lung cancer and monitoring of disease progression in patients with other cancer types. In order to keep fluidity in result release, three years ago the analyses of liquid biopsy samples were integrated in the same wet lab flow as those of solid samples. During these three years, 454 liquid samples from 340 patients were analyzed, obtaining a mean coverage of 592.8X for lung cancer samples (n=356), 600.8X for colorectal cancer samples (n=81) and 601.1X for melanoma samples (n=17; Fig. 4A). Hotspot or LoF TP53 variants were detected in 180 out of 340 patients. Among the 160 patients in whom no variants were found, 27 had tumors with no alterations in the analyzed genes, 20 had mutant tumors and 113 had tumors of unknown genotype. Paired solid and liquid biopsy material was available for 227 patients with lung cancer, colon cancer, or melanoma (no liquid biopsies available for GIST patients). Concordant results were found for the biopsy pairs for as numerous as 91.19% of these patients, with 79.30% of them harboring gene variants which were detected in both tumor DNA and circulating tumor DNA (ctDNA) and 11.89% having wild-type tumors and no variants detected in liquid biopsies. Only 8.81% of patients had no variants detectable in ctDNA, whereas they had mutant tumors (Fig. 4B, left panel). When focus was addressed on each cancer type, it was observed that liquid biopsies from melanoma were the most positive, followed by colon and lung cancer. Mutation in ctDNA was not detected in 2.33% of patients with colon cancer and in 3.96% of patients with lung cancer (Fig. 4B, right panel). Finally, between liquid biopsies and solid tumors, a concordance was obtained in 100% of cases with melanomas, 97.67% with colon cancers and 96.04% with lung cancer.
For further analysis of liquid biopsy samples with ctDNA variants, mutant allele frequency of these variants was compared between the different tumor types (lung, colon, and melanoma). Similar median mutant allele frequency values were found for lung cancer samples and melanomas (4.74 and 4.935%, respectively), whereas they were higher (11.53%) for colon tumors (Fig. 4C). Next, focus was addressed on lung cancer samples which were the most numerous. It appeared that the mutant allele frequencies were significantly different between the three subsets (P=0.0042). For the EGFR and TP53 genes, mutant allele frequencies were not significantly different (6.778 and 9.113%, respectively; P=0.6255). Mutant allele frequencies in EGFR and RAS/BRAF/MAPK1 genes were almost significantly different (6.778 and 2.445%, P=0.0634). This difference was significant between RAS/BRAF/MAPK1 and TP53 genes (2.445 and 9.113%; P=0.0033) (Fig. 4D). Finally, comparing mutant allele frequencies of primary EGFR variants (driver variants) with those of new variants appearing after progression under treatment (resistance variants) it was identified that mutated allele frequencies of driver variants were significantly higher than those of resistance variants (P=0.0035; Fig. 4E).
Discussion
The increase in the incidence of solid cancers combined with the development of targeted therapies require molecular diagnosis laboratories to genotype an increasing number of genes in increasing numbers of samples, with more or less redundant targets. A possibility to satisfy this need in a reduced amount of time emerged with the application of NGS which enables molecular biologists to analyze numerous targets for numerous patients simultaneously and within timing compatible with patient care. However, providing reliable results quickly, at an affordable cost, and in accordance with quality standards remains one of the major challenges of molecular diagnostics. In order to tackle this challenge, certain manufacturers developed commercial kits for analyzing dozens of genes (from 20 to 50, or even more) known to be involved in the development of solid and liquid cancers. These gene panels are usually based on capture technology and focus on hotspot-containing regions of selected genes. These selections do not appear to be compatible with clinical practice (30). In fact, routine molecular diagnosis should first and foremost help guide decisions on prescribing approved treatments. Therefore, the laboratories do not have to screen for other targets which would be necessary for inclusion in clinical trials or for off-label prescription. Both of these activities require specific knowledge and expertise in a regulated environment (clinical trials), which is not necessary in the context of daily routine diagnostic services. For all these reasons, a ten-gene custom NGS panel was developed for routine diagnosis of four cancer types including lung cancers, colorectal cancers, melanomas and GISTs. In the present study, the design and performance of this panel was described. Our three-year experience of using it routinely in our diagnostic practice was also reported.
Our panel was validated on 15 items according to the ISO15189 standards, obtaining the accreditation required for its use in routine diagnostic laboratories in France. It exhibited very favorable analytical performances, with the 100% accuracy for genotyping hotspot regions with a relatively low coverage. It was revealed that a minimal coverage of 80X on each nucleotide was sufficient to obtain a sensitivity and specificity of 100% in our experimental conditions (tumor cell content >5%). In real life, and particularly for liquid biopsies and hotspot regions, the minimal coverage obtained is widely over 80X. Real life sensitivity obtained for the analysis of liquid biopsies was 91.19%, which remains higher than that reported in recent studies (31-33). It is important to note that hotspot visualization on raw data for liquid biopsies is essential to obtain such sensitivity.
The analysis of the whole coding sequences of ten genes with a theranostic potential revealed that 73.22% of the observed variants (frequency <1% in the general population) were either benign (5.68%), or of unknown significance (67.54%). Consequently, a small proportion of the observed variants could be classified as GoF (16.61%) or LoS variants (9.32%). Given that only GoF variants have a therapeutic impact, only a small number of variants are clinically important, and those are localized in specific exons. A total of 156 exons were analyzed and it was identified that only 37 exons harbored clinically pertinent variants. Nevertheless, despite the low impact of non-hotspot regions, it is important to cover them to accommodate for future applications of gene amplification analysis. Indeed, it appears that copy number amplifications could be useful as markers predicting response to treatments (34,35) or as theranostic biomarkers (8,36,37). Therefore, the analysis of the entire gene, or at least of its whole coding sequence, warrants a more reliable analysis.
It was decided to not include intron regions specific to fusion of ALK, ROS1 or RET in the panel. In fact, research of fusions from DNA requires the knowledge of the specific partners and their specific breakpoints, rendering it impossible to detect 100% of fusions. It is considered that targeted RNAseq is more appropriate for fusion detection. Nevertheless, it is totally possible to add these intron sequences and/or other coding regions of new genes to the panel. New target genes could be identified at any time depending on the results of clinical trials. Based on this fact, our research group selected capture technology that allows the addition of new probes without disturbing the specificity of the validated panel backbone.
Through the analysis of lung tumor samples, hotspot variants were detected in ~74% of samples, with more than a half of them harboring a variant in the KRAS (45%) or BRAF gene (4%), markedly higher than described to date (38,39). The analysis of MAP2K1 and NRAS revealed that 2% of the tumors harbored a GoF mutation that could be targetable by MEK inhibitors (40). Variants in tyrosine kinase domain receptors were found in 19% of patients, with the highest prevalence of EGFR mutations (15%), the latter being slightly higher than expected for Caucasian patients with lung cancer based on literature data (41,42).
In colorectal cancer, KRAS has been reported as the most often mutated gene (40%), followed by BRAF (10%), and NRAS (4%) (43). With our panel, it was found that 50% of colorectal cancers were KRAS-mutant and 13% had variants in BRAF, indicating that our panel may be more sensitive in detecting variants in these genes. The NRAS mutation prevalence identified with our panel (3.8%) was very close to that reported in the literature (4%) (43,44). Notably, ~2% of the analyzed colorectal cancers had GoF variants in the MAP2K1 gene, which may give rise to resistance to anti-EGFR therapies (44). Finally, due to the inclusion of the ERBB2 gene in the panel it was revealed that 3% of colorectal cancers had a GoF variant, a finding which could help identify new therapeutic strategies for these patients (45).
In melanoma, a prevalence of BRAF mutations was obtained in accordance with that reported in a recent study (38.61% of mutant tumors), whereas the prevalence of NRAS variants (26.58%) was higher than reported (16.4%), and that of KIT variants was lower (3.16% vs. 10%) (46). The low prevalence of KIT variants could be explained by the lower number of mucosal melanomas in our series, whereas the higher prevalence of NRAS variants may be explained by the fact that our patients were mostly Caucasians. In fact, NRAS variants were more often reported in populations from Italy, Sweden, Spain, and the USA (46), predominantly Caucasian, similar to the population analyzed in the present study.
Finally, even if the number of GISTs included in our analysis was quite low, it is noteworthy that 83% of them harbored a KIT or a PDGFRA GoF variant, and that PDGFRA variants occurred in ~30% of KIT-wild-type locally-advanced GISTs, as described in previous studies (47,48). In contrast to other analyzed cancers, GISTs did not harbor any GoF variants in other genes of the panel. LoF variants were also found in certain oncogenes. BRAF LoF variants have already been described as inducing constitutive activation of CRAF and downstream protein kinases (49). However, in our series, LoF variants were also present in tyrosine kinase receptors. The absence of loss of heterozygosity detection did not enable us to conclude on the complete loss of the protein. To date, no study had investigated the impact of such variants on tumor cells. These may be merely passenger variants but they may also have an activating impact on tumor cells. The latter hypothesis is supported by the fact that certain variants are clearly associated with particular cancer types. For example, EGFR mutations generating a stop codon in the tyrosine kinase domain were only detected in lung cancers, whereas MET and PDGFRA LoF variants were mainly present in colon cancers. Further studies are required to clarify this issue.
Our clinical practice demonstrated that processing liquid biopsies at the same time as solid samples produced favorable analytical performances (owing to the reading of hotspots from raw data and the use of LoF variants of the TP53 gene). In addition, in liquid biopsies median mutant allele frequencies differed between tumor types. They were significantly higher for colorectal cancers than for lung cancers or melanomas, suggesting that progressing colorectal cancer may release more cell-free DNA. When focusing on lung cancer samples, it was also found that mutated allele frequencies were different between altered genes. TP53 and EGFR variants had significantly higher mutated allele frequencies than variants in the RAS/BRAF/MAP2K1 genes. Finally, it was also found that in lung cancers progressing under TKI, variants inducing resistance to treatment had significantly lower allele frequencies than the original sensitizing variants.
In conclusion, our three-year practical experience using a custom ten-gene NGS panel for the molecular diagnosis of four solid cancers produced very positive results. It was demonstrated that this panel has favorable analytical performances and can be used for analyzing not only solid but also liquid biopsies. With our work process, it was possible to release clinical reports in less than five working days from the receipt of samples. The use of our panel can provide for a more relevant biomarker analysis and could be an effective way to counter the problem of low molecular diagnosis. The latter should not be underestimated. It was reported that less than 50% of patients had no molecular results before receiving the first line of treatment (50). In our opinion, the use of relatively small dedicated NGS panels is the optimal choice to rapidly obtain relevant results before first-line treatment. Such a method should also be developed for routine diagnosis of other cancer types, whereas the use of large panels should be reserved for patients in therapeutic failure.
Supplementary Data
Availability of data and materials
Genomic data could be shared upon reasonable request to the corresponding author in accordance to French law for genomic data.
Authors' contributions
SC designed the panel and performed the experiments. AB, MC, ACh, AG and MJ performed the experiments. HM and CT performed bioinformatics analyses. FB, CCB and LA qualified solid samples. JA, ACo, VD and VG interpreted the routine results. RB designed the panel, interpreted the routine results, analyzed the data, designed the study and wrote the manuscript. RB and VG confirmed the authenticity of all the raw data. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The present study on patient samples was conducted in accordance with the Declaration of Helsinki and was approved (approval no. 00010311) by the Ethics Committee of the Georges-François Leclerc Cancer Center (Dijon, France) and by the Consultative Committee of Burgundy (Dijon, France) for the Protection of Persons Participating in Biomedical Research (Comité Consultatif de Protection des Personnes en Recherche Biomédicale de Bourgogne). Written informed consent was provided by all patients.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Acknowledgments
The authors would like to thank Satt Sayens (Dijon, France) for the support in patenting the panel design and Dr Katarzyna Szymanska for editing the manuscript.
Funding
No funding was received.
References
|
1
|
Kallioniemi OP, Kallioniemi A, Kurisu W, Thor A, Chen LC, Smith HS, Waldman FM, Pinkel D and Gray JW: ERBB2 amplification in breast cancer analyzed by fluorescence in situ hybridization. Proc Natl Acad Sci USA. 89:5321–5325. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Marks JL, Broderick S, Zhou Q, Chitale D, Li AR, Zakowski MF, Kris MG, Rusch VW, Azzoli CG, Seshan VE, et al: Prognostic and therapeutic implications of EGFR and KRAS mutations in resected lung adenocarcinoma. J Thorac Oncol. 3:111–116. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Choi YL, Soda M, Yamashita Y, Ueno T, Takashima J, Nakajima T, Yatabe Y, Takeuchi K, Hamada T, Haruta H, et al: EML4-ALK mutations in lung cancer that confer resistance to ALK inhibitors. N Engl J Med. 363:1734–1739. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Nguyen-Ngoc T, Bouchaab H, Adjei AA and Peters S: BRAF alterations as therapeutic targets in non-small-cell lung cancer. J Thorac Oncol. 10:1396–1403. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Lynch TJ, Bell DW, Sordella R, Gurubhagavatula S, Okimoto RA, Brannigan BW, Harris PL, Haserlat SM, Supko JG, Haluska FG, et al: Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 350:2129–2139. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Chuang JC, Stehr H, Liang Y, Das M, Huang J, Diehn M, Wakelee HA and Neal JW: ERBB2-Mutated metastatic non-small cell lung cancer: Response and resistance to targeted therapies. J Thorac Oncol. 12:833–842. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Skoulidis F, Li BT, Dy GK, Price TJ, Falchook GS, Wolf J, Italiano A, Schuler M, Borghaei H, Barlesi F, et al: Sotorasib for lung cancers with KRAS G12C mutation. N Engl J Med. 384:2371–2381. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wolf J, Seto T, Han JY, Reguart N, Garon EB, Groen HJM, Tan DSW, Hida T, de Jonge M, Orlov SV, et al: Capmatinib in MET Exon 14-Mutated or MET-Amplified non-small-cell lung cancer. N Engl J Med. 383:944–957. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Sanz-Garcia E, Argiles G, Elez E and Tabernero J: BRAF mutant colorectal cancer: Prognosis, treatment, and new perspectives. Ann Oncol. 28:2648–2657. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Karapetis CS, Khambata-Ford S, Jonker DJ, O'Callaghan CJ, Tu D, Tebbutt NC, Simes RJ, Chalchal H, Shapiro JD, Robitaille S, et al: K-ras mutations and benefit from cetuximab in advanced colorectal cancer. N Engl J Med. 359:1757–1765. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Douillard JY, Oliner KS, Siena S, Tabernero J, Burkes R, Barugel M, Humblet Y, Bodoky G, Cunningham D, Jassem J, et al: Panitumumab-FOLFOX4 treatment and RAS mutations in colorectal cancer. N Engl J Med. 369:1023–1034. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Flaherty KT, Puzanov I, Kim KB, Ribas A, McArthur GA, Sosman JA, O'Dwyer PJ, Lee RJ, Grippo JF, Nolop K and Chapman PB: Inhibition of mutated, activated BRAF in metastatic melanoma. N Engl J Med. 363:809–819. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Carvajal RD, Antonescu CR, Wolchok JD, Chapman PB, Roman RA, Teitcher J, Panageas KS, Busam KJ, Chmielowski B, Lutzky J, et al: KIT as a therapeutic target in metastatic melanoma. JAMA. 305:2327–2334. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Johnson DB and Puzanov I: Treatment of NRAS-mutant melanoma. Curr Treat Options Oncol. 16:152015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Lasota J and Miettinen M: KIT and PDGFRA mutations in gastrointestinal stromal tumors (GISTs). Semin Diagn Pathol. 23:91–102. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Sherry ST, Ward MH, Kholodov M, Baker J, Phan L, Smigielski EM and Sirotkin K: dbSNP: The NCBI database of genetic variation. Nucleic Acids Res. 29:308–311. 2001. View Article : Google Scholar :
|
|
17
|
Li H: Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv 1303.3997v2. 2013.
|
|
18
|
McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M and DePristo MA: The genome analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20:1297–1303. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, Philippakis AA, del Angel G, Rivas MA, Hanna M, et al: A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 43:491–498. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Van der Auwera GA, Carneiro MO, Hartl C, Poplin R, Del Angel G, Levy-Moonshine A, Jordan T, Shakir K, Roazen D, Thibault J, et al: From FastQ data to high confidence variant calls: The genome analysis toolkit best practices pipeline. Curr Protoc Bioinformatics. 43:11 10 11–11 10 33. 2013.
|
|
21
|
Toolkit Picard: Broad Institute, GitHub Repository. 2019, https://broadinstitute.github.io/picard/.
|
|
22
|
Muller E, Goardon N, Brault B, Rousselin A, Paimparay G, Legros A, Fouillet R, Bruet O, Tranchant A, Domin F, et al: OutLyzer: Software for extracting low-allele-frequency tumor mutations from sequencing background noise in clinical practice. Oncotarget. 7:79485–79493. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Wang K, Li M and Hakonarson H: ANNOVAR: Functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 38:e1642010. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Cingolani P, Platts A, Wang le L, Coon M, Nguyen T, Wang L, Land SJ, Lu X and Ruden DM: A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin). 6:80–92. 2012. View Article : Google Scholar
|
|
25
|
Andrews S: FastQC: A quality control tool for high thoughput sequence data. 2010, http://www.bioinformatics.babraham.ac.uk/projects/fastqc.
|
|
26
|
Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G and Durbin R: 1000 Genome Project Data Processing Subgroup: The sequence alignment/map format and SAMtools. Bioinformatics. 25:2078–2079. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Okonechnikov K, Conesa A and Garcia-Alcalde F: Qualimap 2: Advanced multi-sample quality control for high-throughput sequencing data. Bioinformatics. 32:292–294. 2016.
|
|
28
|
Ewels P, Magnusson M, Lundin S and Kaller M: MultiQC: Summarize analysis results for multiple tools and samples in a single report. Bioinformatics. 32:3047–3048. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Boidot R, Végran F and Ghiringhelli F: Detection method of somatic genetic anomalies, combination of capture probes and kit of detection. EP3553182. Filed April 11, 2018 issued October 16, 2019.
|
|
30
|
Jennings LJ, Arcila ME, Corless C, Kamel-Reid S, Lubin IM, Pfeifer J, Temple-Smolkin RL, Voelkerding KV and Nikiforova MN: Guidelines for validation of next-generation sequencing-based oncology panels: A joint consensus recommendation of the association for molecular pathology and college of american pathologists. J Mol Diagn. 19:341–365. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Papadopoulou E, Tsoulos N, Tsantikidi K, Metaxa-Mariatou V, Stamou PE, Kladi-Skandali A, Kapeni E, Tsaousis G, Pentheroudakis G, Petrakis D, et al: Clinical feasibility of NGS liquid biopsy analysis in NSCLC patients. PLoS One. 14:e02268532019. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Esagian SM, Grigoriadou GI, Nikas IP, Boikou V, Sadow PM, Won JK and Economopoulos KP: Comparison of liquid-based to tissue-based biopsy analysis by targeted next generation sequencing in advanced non-small cell lung cancer: A comprehensive systematic review. J Cancer Res Clin Oncol. 146:2051–2066. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Garcia J, Kamps-Hughes N, Geiguer F, Couraud S, Sarver B, Payen L and Ionescu-Zanetti C: Sensitivity, specificity, and accuracy of a liquid biopsy approach utilizing molecular amplification pools. Sci Rep. 11:107612021. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Shen WD, Chen HL and Liu PF: EGFR gene copy number as a predictive biomarker for resistance to anti-EGFR mono-clonal antibodies in metastatic colorectal cancer treatment: A meta-analysis. Chin J Cancer Res. 26:59–71. 2014.PubMed/NCBI
|
|
35
|
Bahcall M, Awad MM, Sholl LM, Wilson FH, Xu M, Wang S, Palakurthi S, Choi J, Ivanova EV, Leonardi GC, et al: Amplification of Wild-type KRAS imparts resistance to crizotinib in MET exon 14 mutant non-small cell lung cancer. Clin Cancer Res. 24:5963–5976. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Randon G, Yaeger R, Hechtman JF, Manca P, Fucà G, Walch H, Lee J, Élez E, Seligmann J, Mussolin B, et al: EGFR amplification in metastatic colorectal cancer. J Natl Cancer Inst. 113:1561–1569. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Suryavanshi M, Shah A, Kumar D, Panigrahi MK, Metha A and Batra U: MET amplification and response to MET inhibitors in stage IV lung adenocarcinoma. Oncol Res Treat. 40:198–202. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Chen D, Zhang LQ, Huang JF, Liu K, Chuai ZR, Yang Z, Wang YX, Shi DC, Liu Q, Huang Q and Fu WL: BRAF mutations in patients with non-small cell lung cancer: A systematic review and meta-analysis. PLoS One. 9:e1013542014. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Lohinai Z, Klikovits T, Moldvay J, Ostoros G, Raso E, Timar J, Fabian K, Kovalszky I, Kenessey I, Aigner C, et al: KRAS-mutation incidence and prognostic value are metastatic site-specific in lung adenocarcinoma: Poor prognosis in patients with KRAS mutation and bone metastasis. Sci Rep. 7:397212017. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Arcila ME, Drilon A, Sylvester BE, Lovly CM, Borsu L, Reva B, Kris MG, Solit DB and Ladanyi M: MAP2K1 (MEK1) mutations define a distinct subset of lung adenocarcinoma associated with smoking. Clin Cancer Res. 21:1935–1943. 2015. View Article : Google Scholar :
|
|
41
|
Barlesi F, Mazieres J, Merlio JP, Debieuvre D, Mosser J, Lena H, Ouafik L, Besse B, Rouquette I, Westeel V, et al: Routine molecular profiling of patients with advanced non-small-cell lung cancer: Results of a 1-year nationwide programme of the French cooperative thoracic intergroup (IFCT). Lancet. 387:1415–1426. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Zhang YL, Yuan JQ, Wang KF, Fu XH, Han XR, Threapleton D, Yang ZY, Mao C and Tang JL: The prevalence of EGFR mutation in patients with non-small cell lung cancer: A systematic review and meta-analysis. Oncotarget. 7:78985–78993. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Bellio H, Fumet JD and Ghiringhelli F: Targeting BRAF and RAS in colorectal cancer. Cancers (Basel). 13:22012021. View Article : Google Scholar
|
|
44
|
Chuang J, Wang C, Guo Y, Valenzuela V, Wu J and Fakih M: MAP2K1 mutations in advanced colorectal cancer predict poor response to anti-EGFR therapy and to vertical targeting of MAPK pathway. Clin Colorectal Cancer. 20:72–78. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Ross JS, Fakih M, Ali SM, Elvin JA, Schrock AB, Suh J, Vergilio JA, Ramkissoon S, Severson E, Daniel S, et al: Targeting HER2 in colorectal cancer: The landscape of amplification and short variant mutations in ERBB2 and ERBB3. Cancer. 124:1358–1373. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Gutierrez-Castaneda LD, Nova JA and Tovar-Parra JD: Frequency of mutations in BRAF, NRAS, and KIT in different populations and histological subtypes of melanoma: A systemic review. Melanoma Res. 30:62–70. 2020. View Article : Google Scholar :
|
|
47
|
Mei L, Smith SC, Faber AC, Trent J, Grossman SR, Stratakis CA and Boikos SA: Gastrointestinal stromal tumors: The GIST of precision medicine. Trends Cancer. 4:74–91. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Heinrich MC, Corless CL, Duensing A, McGreevey L, Chen CJ, Joseph N, Singer S, Griffith DJ, Haley A, Town A, et al: PDGFRA activating mutations in gastrointestinal stromal tumors. Science. 299:708–710. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Sen B, Peng S, Tang X, Erickson HS, Galindo H, Mazumdar T, Stewart DJ, Wistuba I and Johnson FM: Kinase-impaired BRAF mutations in lung cancer confer sensitivity to dasatinib. Sci Transl Med. 4:136ra702012. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Robert NJ, Nwokeji ED, Espirito JL, Chen L, Karhade M, Evangelist MC, Spira AI, Neubauer MA, Bullock SA, Coleman RL, et al: Biomarker tissue journey among patients (pts) with untreated metastatic non-small cell lung cancer (mNSCLC) in the U.S. Oncology Network community practices. J Clin Oncol. 39(Suppl 15)9004:2021. View Article : Google Scholar
|


















