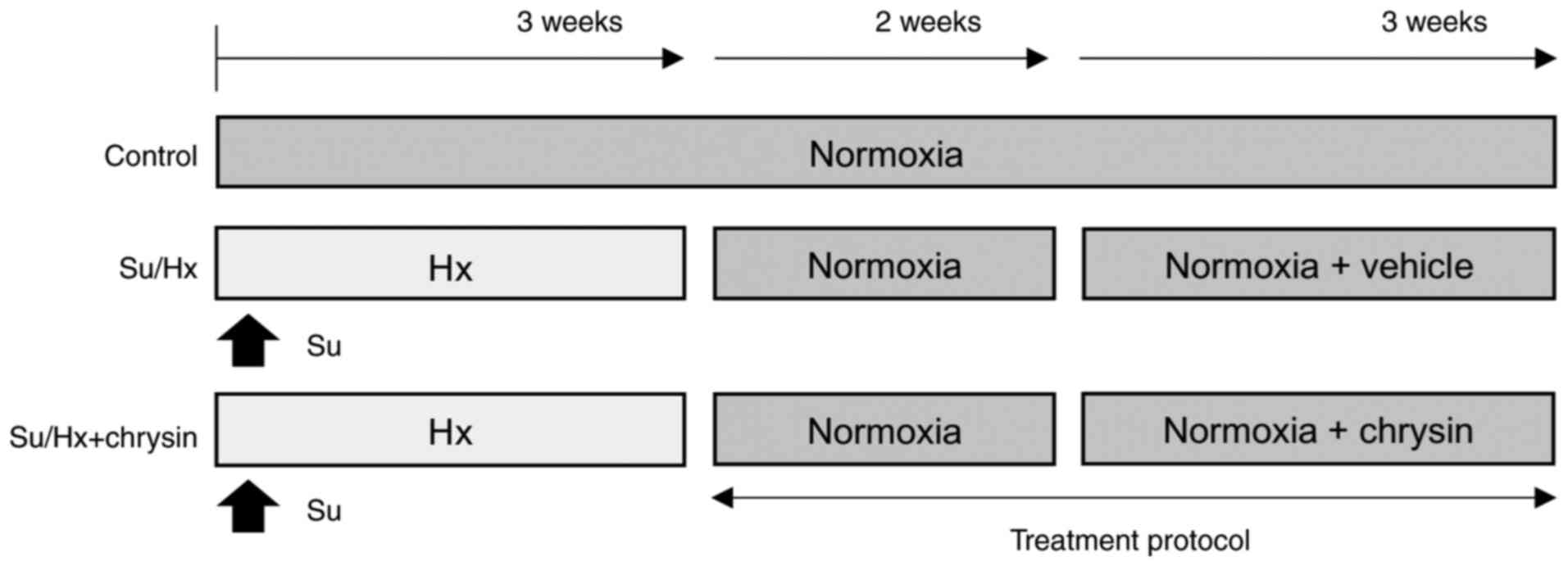
Introduction
Pulmonary arterial hypertension (PAH) is a
progressive disease characterized by pulmonary vascular remodeling,
leading to increased pulmonary vascular resistance and right
ventricle (RV) hypertrophy, resulting in RV failure (1). Although PAH is considered to be a
disease of the lungs, RV adaptation to high PA pressure is the most
important determinant of prognosis and right heart failure remains
a predominant cause of death in PAH (2). RV dysfunction is a complex process
that leads to cardiomyocyte hypertrophy, fibrosis, inflammation and
metabolic change, such as increased glycolysis (3-6).
However, the underlying molecular mechanism of adverse RV
remodeling and dysfunction is poorly understood and, to the best of
our knowledge, there are currently no approved therapies that
improve RV function.
Flavonoids are natural products found in plants and
exhibit diverse biological activity and are involved in regulation
of cell cycle, DNA repair and metabolic gene expression (7). Moreover, flavonoids exert
pharmacological effects, such as anti-cancer, anti-oxidation and
anti-inflammatory effects (8).
Chrysin (5,7-dihydroxyflavone; CH), a phytochemical, is one of the
most active flavonoids and has been shown to exert cardioprotective
effects via its antioxidant and anti-inflammatory activity in
multiple experimental models, including myocardial
ischemia-reperfusion injury and hypoxic rats (9,10). Recent study suggested that CH
attenuates bleomycin-induced pulmonary fibrosis via
anti-inflammatory and anti-oxidative effects and the activity of CH
as a peroxisome proliferator-activated receptor γ (PPAR-γ) agonist
has direct protective effects against inflammation and oxidative
stress (11). Therefore, it was
hypothesized that CH may exert a therapeutic effect in PAH.
The SU5416/hypoxia (Su/Hx) protocol is based on
blockade of vascular endothelial growth factor receptor via
tyrosine kinase inhibitor Su and is commonly used to induce animal
models of PAH (12,13). In the Su/Hx model, animals
receive a single subcutaneous injection of Su in combination with
chronic exposure to Hx followed by normoxia, resulting in severe
PAH. Therefore, Su/Hx models have been considered as a key
preclinical model of PAH due to formation of occlusive pulmonary
vascular lesions in the lung and RV dysfunction with marked cardiac
hypertrophy, which resembles human idiopathic PAH (14,15).
To determine the molecular mechanisms underlying the
effects of PH and CH on RV, gene expression changes in RV of Su/Hx
model rats in the presence and absence of CH were analyzed via RNA
sequencing (RNA-seq). Metabolomic profiling of RVs was performed to
determine metabolic regulation by CH. Pathophysiological changes in
Su/Hx + CH PAH rats were observed and pressure-volume curve was
constructed using a Millar catheter.
Materials and methods
Experimental design and animals
A total of 30 5-week-old male Sprague-Dawley rats
weighing 130-150 g were purchased from Japan SLC (Shizuoka, Japan).
All rats were housed in the animal experimental facility of Chiba
University and had free access to drinking water and food. The rats
were kept at 24°C under a 12 h light/dark cycle in the animal
experimental facility of Chiba University as described previously
(16). All animal procedures
were approved by the Review Board for Animal Experiments of Chiba
University (approval no. 30-126) and were performed in accordance
with the guidelines of the Animal Research Committee of Laboratory
Animal Center, Graduate School of Medicine, Chiba University
(17). The rats were divided
into three groups (n= 10 rats per group) as follows: i) Untreated
control (CTRL); ii) Su/Hx + vehicle (PBS) and iii) Su/Hx + CH
administration (Fig. 1).
Su/Hx rat model and CH
administration
The Su/Hx rat model was established as previously
described (18,19). Briefly, Su (20 mg/kg; R&D
Systems, Inc.), a vascular endothelial growth factor receptor
inhibitor, was dissolved in 1 ml carboxymethyl cellulose and the
suspension was subcutaneously injected into rats. The rats were
maintained under normobaric Hx (10% O2) for 3 weeks then
returned to normoxic conditions (21% O2) for 2
weeks.
CH was obtained from Santa Cruz Biotechnology, Inc.,
dissolved in PBS and administered to Su/Hx rats (100 mg/kg)
once/day for 3 weeks, beginning at the end of the 2 week normoxic
period (Fig. 1).
RNA-seq
A total of ~500 ng total RNA from the heart and lung
was ribosomal RNA-depleted using the NEBNext rRNA Depletion kit and
converted to an Illumina sequencing library using the NEBNext Ultra
Directional RNA Library Prep kit for Illumina (both New England
BioLabs, Inc.). The libraries were validated using a Bioanalyzer
(Agilent Technologies, Inc.) and 1.5 pM (final loading
concentration) of libraries sequenced using the NextSeq 500
(Illumina, Inc.) with the paired-end 36-base read option. Reads
were mapped to Rattus norvegicus Rnor_6.0 (rn6) reference genome
and quantified using the CLC Genomics Workbench (version 12.0,
Qiagen GmbH). RNA-seq datasets were deposited in National Center
for Biotechnology Information Gene Expression Omnibus database
(ncbi.nlm.nih.gov/geo; accession no.
GSE186989).
Identification of differentially
expressed genes (DEGs)
Read counts were normalized by calculating the
number of reads per kilobase per million reads (RPKM) for each
transcript in individual samples using CLC Genomics Workbench
software (version 11.0.1, Qiagen GmbH). DEGs were identified using
fold change ≥2 or ≤−2 filtering analysis with a false discovery
rate (FDR) of P<0.05. Gene expression was visualized using
principal components analysis (PCA) plot and clustering heat map
analyses. Volcano plots were used to visualize changes in gene
expression between −log10 P-value and log2
fold change using CLC Genomics Workbench software (version 12.0,
Qiagen GmbH).
Functional enrichment analysis
An unsupervised method for clustering genes into
groups based on their expression pattern across all samples,
K-means clustering and functional enrichment analysis of DEGs were
visualized using integrated Differential Expression and Pathway
analysis (iDEP) (bioinformatics.sdstate.edu/idep/) (20). Gene Ontology (GO) terms of
biological process (http://www.geneontology.org/) and Kyoto Encyclopedia
of Genes and Genomes (KEGG) pathway (http://www.genome.jp/kegg/) enrichment of DEGs between
sample groups were analyzed and assigned using the web-based
Enrichr suite software (maayanlab.cloud/Enrichr/) (21).
Metabolome analysis
Based on the results of transcriptomic analysis,
mitochondrial and metabolic reprogramming was investigated.
Metabolite extraction from the right heart of Su/Hx and Su/Hx + CH
rats and metabolome analysis were performed according to Human
Metabolome Technologies, Inc. Instructions, as previously described
(Appendix S1) (22).
Reverse transcription-quantitative
(RT-q)PCR
Total RNA from RV tissue was extracted using the
Rneasy Fibrous Tissue kit and isolated RNA (0.2-0.5 µg) was
reverse-transcribed using an RT2 First Strand kit (both Qiagen
GmbH) according to the manufacturer's instructions. qPCR was
performed using SYBR® Green ROX qPCR Master Mix (Qiagen
GmbH) in an ABI 7300 system (Applied Biosystems; Thermo Fisher
Scientific, Inc.). RT2 First Strand kit (cat. no. 330404; Qiagen
GmbH) were used to analyze expression of Pparγ, carnitine palmitoyl
transferase 1 B (Cpt1b), cluster of differentiation 36 (Cd36),
mitofusin1 (Mfn1), Mfn2, optic atrophy 1 (Opa1), mitochondrial
transcription factor A (Tfam) and dynamin-related protein 1 (Drp1).
The primers are listed in Appendix S2. PCR was performed as
follows: 10 min at 95°C, followed by 40 cycles of 15 sec at 95°C
and 1 min at 60°C. Data analysis was performed using the
comparative Cq method and ABI software (SDS version 1.4). β-actin
was used for normalization of mRNA expression levels (23).
Invasive right heart catheterization
Pulmonary hemodynamics were measured in rats lightly
anesthetized with isoflurane (4% for induction, 1-2% for
maintenance) after blood pressure was measured. Anesthetized rats
underwent tracheostomy and were placed on a ventilator (Harvard
Apparatus). A 2.0-F microtip pressure transducer (Millar
Instruments) was inserted into the RV. RV systolic pressure (SP),
stroke volume, heart rate and differential of pressure with time
(dP/dt) index were continuously monitored for >10 min and data
were analyzed using a Power Lab system (AD Instruments). At the end
of monitoring, arterial elastance (Ea) and end-systolic elastance
(Ees) were measured with vena cava compression. Following
hemodynamic measurement, rats were euthanized with pentobarbital
sodium (150 mg/kg). Hearts were removed and the RV free wall was
dissected from the left ventricle and septum (LV + S). The RV:LV +
S weight was calculated as an index of RV hypertrophy. Sections of
the right heart and lung were stored in RNAprotect Tissue Reagent
(Qiagen GmbH) and formalin for RT-qPCR and pathological analysis,
respectively.
Pathological analysis
Paraffin-embedded RV and lung specimens were
sectioned (3 µm each). Myocyte cross-sectional area (CSA)
was visualized using hematoxylin and eosin (H&E) staining.
H&E staining was performed with Mayer's Hematoxylin solution
for 5 min and eosin solution (Muto Pure Chemical Co., Ltd.) for 2
min at room temperature. Images of H&E staining were captured
and CSA was calculated using ImageJ software (version 1.53k;
National Institutes of Health). A total of five sections were
randomly selected from five rats/group. A total of ≥30 myocytes
were observed/group. RV specimens were stained with Masson's
trichrome stain. Masson's trichrome stain was performed with the
mordant solution for 25 min, 0.75% orange G solution (Muto Pure
Chemical Co., Ltd.) for 1 min, Masson B stain solution for 25 min,
2.5% phosphotungstic acid solution for 20 min, and aniline blue
(Muto Pure Chemical Co., Ltd.) for 8 min at room temperature. The
stained RV myocardial slides were observed in five randomly
selected fields of view at 400× magnification. All slides were
examined using a light microscope Eclipse 55i (Nikon
Corporation).
The fibrotic area dyed blue was quantified using
ImageJ. Lung specimens were stained with Elastica van Gieson (EVG)
stain. EVG staining was performed with Maeda modified
resorcin-fuchsin solution for 50 min, Weigert's Iron Hematoxylin
Staining (both Muto Pure Chemical Co., Ltd.) for 10 min and Van
Gieson Solution (FUJIFILM Wako Pure Chemical Co., Ltd.) for 2 min,
at room temperature. The extent of pulmonary vessel luminal
obstruction was evaluated as previously reported: Grade 0, no
evidence of neointimal formation; grade 1, partial (≤50%) luminal
occlusion; and grade 2, severe (>50%) luminal occlusion. All
pulmonary arteries (outer diameter, <100 µm) in one
section/animal were counted (14).
Statistical analysis
To assess differences between groups, unpaired
two-tailed t-test and one-way ANOVA were used due to potential
assumption violations (equal variance) when using more standard
tests. When significant differences were detected, individual mean
values were compared using post-hoc tests that allowed for multiple
comparisons with adequate type I error control (Bonferroni).
Statistical analysis was performed using GraphPad Prism version 9.1
(GraphPad Software, Inc.) Data are presented as the mean ± SEM of
≥3 independent repeats. P<0.05 was considered to indicate a
statistically significant difference.
Results
Transcriptomic changes in RV of Su/Hx +
CH rats
To determine CH-mediated alteration of gene
expression in RV of Su/Hx rats, RNA-seq was used to analyze the
transcriptome of heart tissue from CTRL, Su/Hx and Su/Hx + CH rats.
PCA was performed to assess the reproducibility of biological
replicates between CTRL, Su/Hx and Su/Hx + CH groups. Su/Hx and
Su/Hx + CH samples were notably separated from CTRL samples
(Fig. 2A). PCA plot showed that
the Su/Hx + CH group was partially separated from the Su/Hx
group.
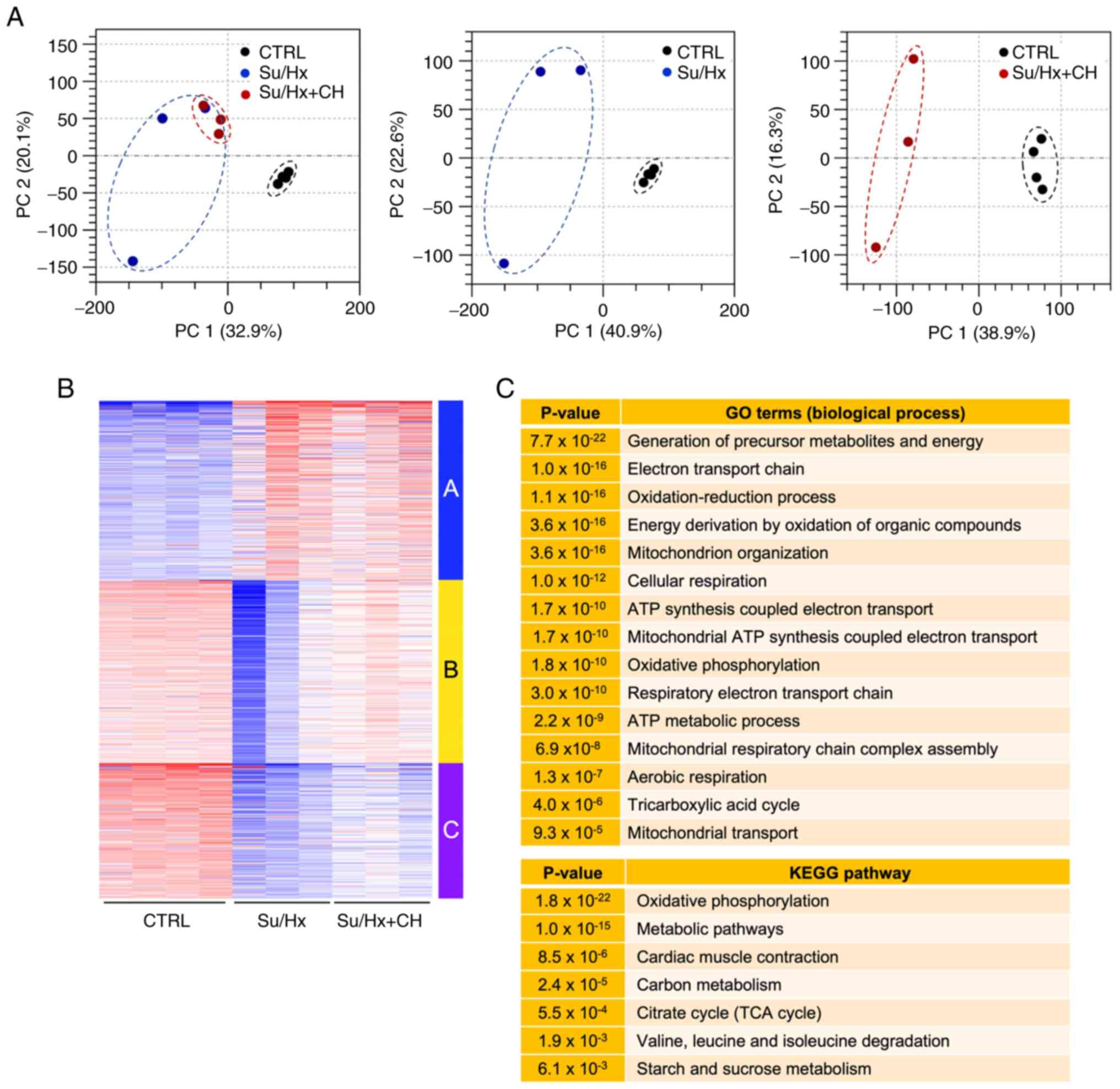 | Figure 2Analysis of right heart tissue
transcriptome. (A) PCA of RNA-sequencing data. PC1 and 2 represent
32.9 and 20.1% (left), 40.9 and 22.6% (middle), and 38.9 and 16.3%
(right) of total variation, respectively. Each dot denotes a single
biological replicate; dashed circles represent three
replicates/individual sample. (B) K-means clustering of 2,000
variable genes. Genes were classified based on similar expression
patterns (red, high; white, intermediate, blue, low). Individual
samples are displayed on the vertical axis, and genes on the
horizontal axis. (C) Functional enrichment analysis of genes in
Cluster B. Enriched GO terms associated with biological process and
KEGG pathways. PCA, principal components analysis; GO, Gene
Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes; CTRL,
control; Su, SU5416; Hx, hypoxia; CH, chrysin. |
K-means clustering of 2,000 variable genes was
performed based on similar expression patterns (Fig. 2B). Cluster B, expression of which
was downregulated in Su/Hx and restored in the Su/Hx + CH group,
was used for functional enrichment analysis (Fig. 2C). Compared with CTRL,
downregulated genes in the Su/Hx group were associated with
mitochondrial energy metabolism, such as 'generation of precursor
metabolites and energy', 'electron transport chain', 'mitochondrion
organization', 'ATP synthesis coupled electron transport',
'mitochondrial ATP synthesis coupled electron transport',
'oxidative phosphorylation' and 'tricarboxylic acid cycle', and
these expression patterns were restored in Su/Hx + CH group. These
results indicated that CH was involved in regulation of
mitochondrial and metabolic gene expression in the RV of Su/Hx
rats.
DEG profiles
Difference in RPKM was calculated and DEGs were
identified using FDR <0.05 and fold change ≥2 or ≤-2. Compared
with CTRL, hierarchical clustering analysis and volcano plot showed
that 2,694 (1,376 up- and 1,318 downregulated) and 1,640 (906 up-
and 734 downregulated) unique genes were significantly changed in
Su/Hx and Su/Hx + CH groups, respectively (Fig. 3A-D). DEGs in the Su/Hx and Su/Hx
+ CH groups were compared to identify genes upon which CH exerted
cardioprotective effects. A total of 619 DEGs upregulated in Su/Hx
but not Su/Hx + CH compared with CTRL were identified (Fig. 3E). In addition, among
transcripts, 1,318 DEGs in the Su/Hx group were downregulated
relative to CTRL; of these, 782 transcripts were not downregulated
in the Su/Hx + CH group (Fig.
3F).
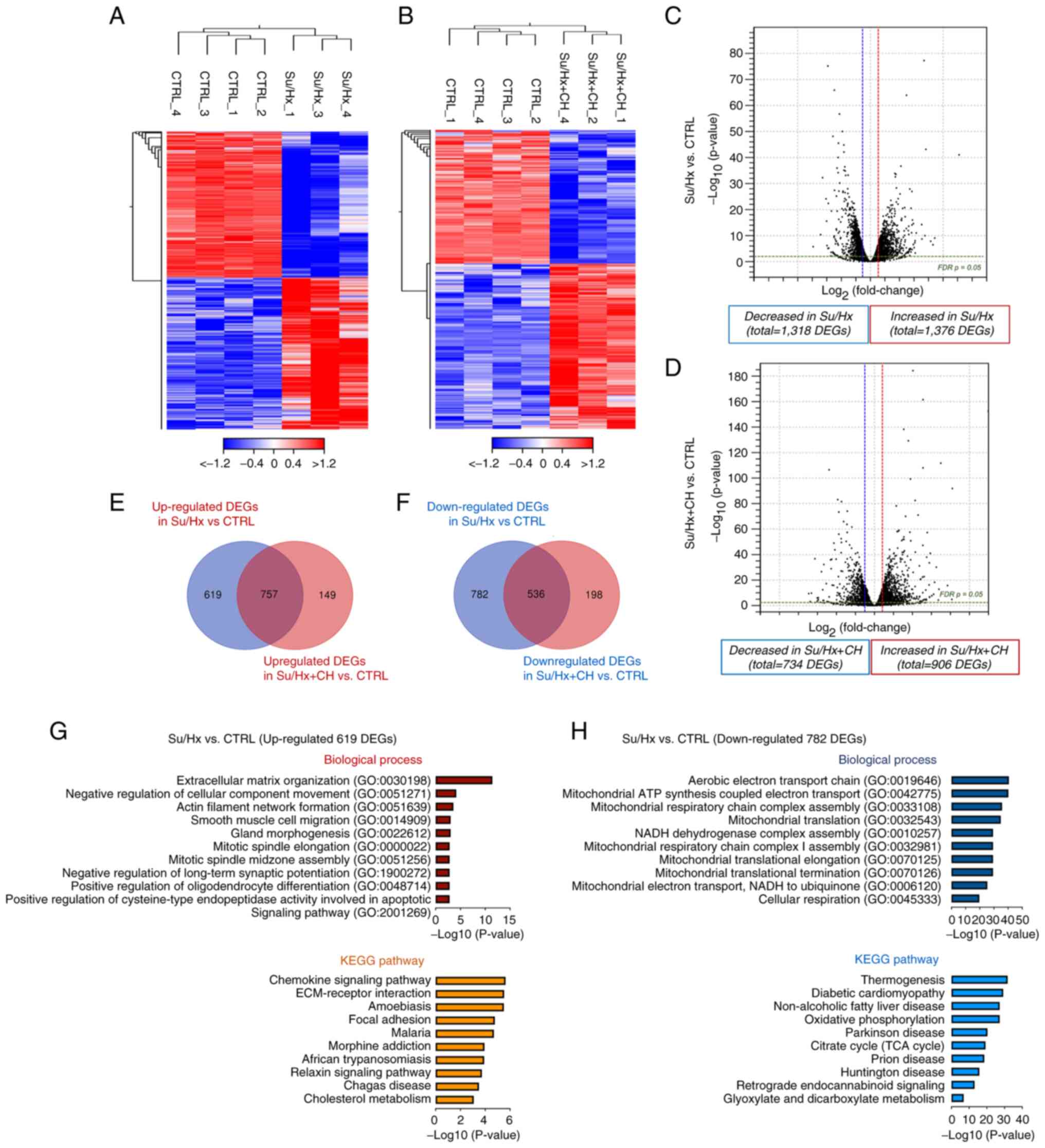 | Figure 3Expression profiling of right heart
tissue. Hierarchical clustering of differential expression profiles
between CTRL and (A) Su/Hx and (B) and Su/Hx + CH. Individual
samples are shown in columns, and genes in rows. Heatmaps represent
relative expression (red, high; white, intermediate; blue, low).
Volcano plots representing DEGs between CTRL and (C) SU/Hx and (D)
Su/Hx + CH. Vertical lines, log2 fold change ≥2 or ≤-2; horizontal
line, significance cut-off (false discovery rate, P=0.05). Venn
diagram identifying differences in (E) up- and (F) downregulated
DEGs. (G) Functional enrichment analysis of 619 upregulated DEGs in
Su/Hx vs. CTRL. Upregulated GO terms are hypothesized to be the
disease etiology in the Su/Hx model and suppressed by CH. (H)
Functional enrichment analysis of 782 downregulated DEGs in Su/Hx
vs. CTRL. Downregulated GO terms are hypothesized to be the disease
etiology in the Su/Hx model and restored by CH. CTRL, control; Su,
SU5416; Hx, hypoxia; CH, chrysin; DEG, differentially expressed
gene; GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and
Genomes. |
To characterize the functional features of 619 up-
and 782 downregulated DEGs in the Su/Hx group, enrichment analysis
for GO annotation and KEGG pathway enrichment was performed. In
biological process, the top ten terms were associated with cardiac
fibrosis, such as 'extracellular matrix organization' (GO:0030198),
'negative regulation of cellular component movement' (GO:0051271)
and 'actin smooth muscle migration' (GO:0014909; Fig. 3G). KEGG pathway analysis showed
the terms 'chemokine signaling pathway', 'extracellular
matrix-receptor interaction' and 'focal adhesion'. These results
suggested that increased expression of fibrosis- and
inflammation-associated genes positively regulated RV dysfunction
in Su/Hx rats and CH may protect against upregulation of these
genes. Biological process mapping of 782 downregulated DEGs
revealed significant association with mitochondrial function,
including 'aerobic electron transport chain' (GO:0019646),
'mitochondrial ATP synthesis coupled electron transport'
(GO:0042775), 'mitochondrial respiratory chain complex assembly'
(GO:0033108) and 'mitochondrial translation' (GO:0032543) in the
Su/Hx group (Fig. 3H). These
data indicate that CH may improve mitochondrial biogenesis in the
RV of Su/Hx PAH rats.
Metabolic changes in right heart of Su/Hx
+ CH rats
To evaluate whether CH affected cardiac metabolism
in PAH, metabolic changes in the heart of Su/Hx rats in the
presence or absence of CH were determined via mass
spectrometry-based metabolic analysis. After isotopic and fragment
peaks were removed, capillary electrophoresis time-of-flight mass
spectrometry detected peaks in all samples. These peaks were
identified with metabolite standards by matching the m/z values and
quantified using the normalized migration time. PC1 and PC2 of
metabolites in ten samples accounted for 36.0 and 21.2% of total
metabolites, respectively. However, PCA plots revealed that Su/Hx
samples exhibited greater within-group variability than CH and
overlapped with Su/Hx samples (Fig.
S1A). By contrast, hierarchical heatmap clustering analysis
showed that the biological replicates were separated between Su/Hx
and Su/Hx + CH groups (Fig.
S1B). These results indicated differences in the amounts of
certain metabolites between the Su/Hx and Su/Hx + CH groups.
Whole glycolysis, lipid metabolism, and TCA cycle
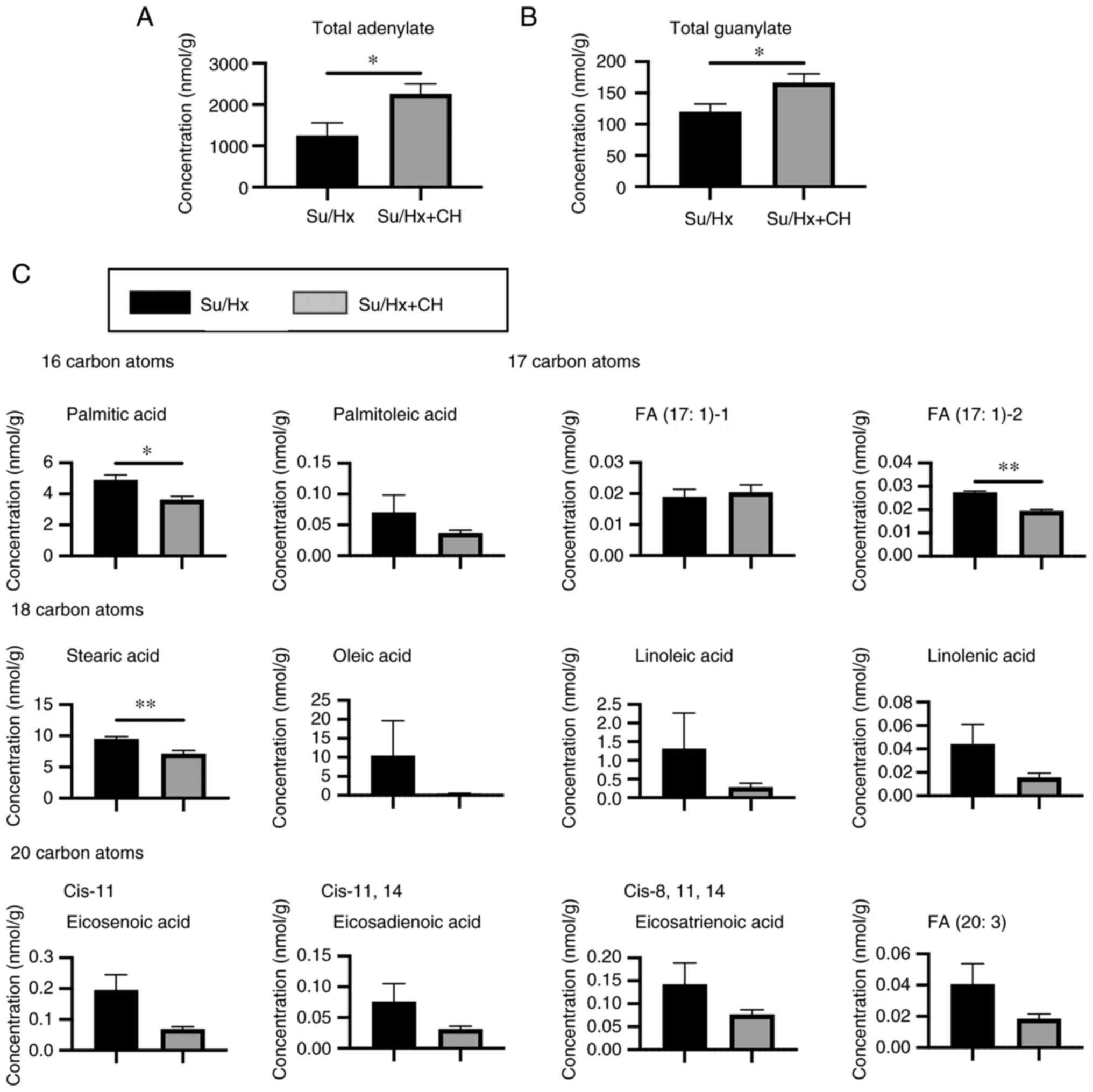
data are presented in Fig. S2.
No significant differences were observed in glycolysis, TCA cycle,
glutathione, NADH and NADPH, which indicated that the glycolytic
metabolic pathway was not significantly affected by CH. By
contrast, the total concentration of adenylate was significantly
higher in Su/Hx + CH than Su/Hx, indicating that CH improved energy
production (Fig. 4A). Total
concentration of guanylate was also significantly greater in Su/Hx
+ CH than Su/Hx (Fig. 4B).
LC-TOFMS showed that concentration of long-chain fatty acids
(LCFAs), such as palmitic acid/stearic acid, was higher in Su/Hx
and lower in Su/Hx + CH (Fig.
4C).
RT-qPCR analysis of right heart
tissue
Genes associated with mitochondrial energy
production were analyzed using RT-qPCR in right heart tissue
(n=5/group). RT-qPCR revealed that RNA expression of Pparg, Cd36
and Cpt1b in the Su/Hx + CH group was higher than in the Su/Hx
group (fold regulation, 1.93, 4.61 and 7.73, respectively; Fig. 5A-C). RNA expression levels of
genes associated with mitochondrial biogenesis, such as Mfn1, Mfn2,
Opa1 and Tfam, were upregulated in Su/Hx + CH compared with Su/Hx
(fold regulation, 3.47, 3.41, 2.78 and 2.94, respectively; Fig. 5D-G). No significant difference
was observed between groups regarding RNA expression of genes
associated with mitochondrial fission, such as Drp1 (fold
regulation, 2.61; Fig. 5H).
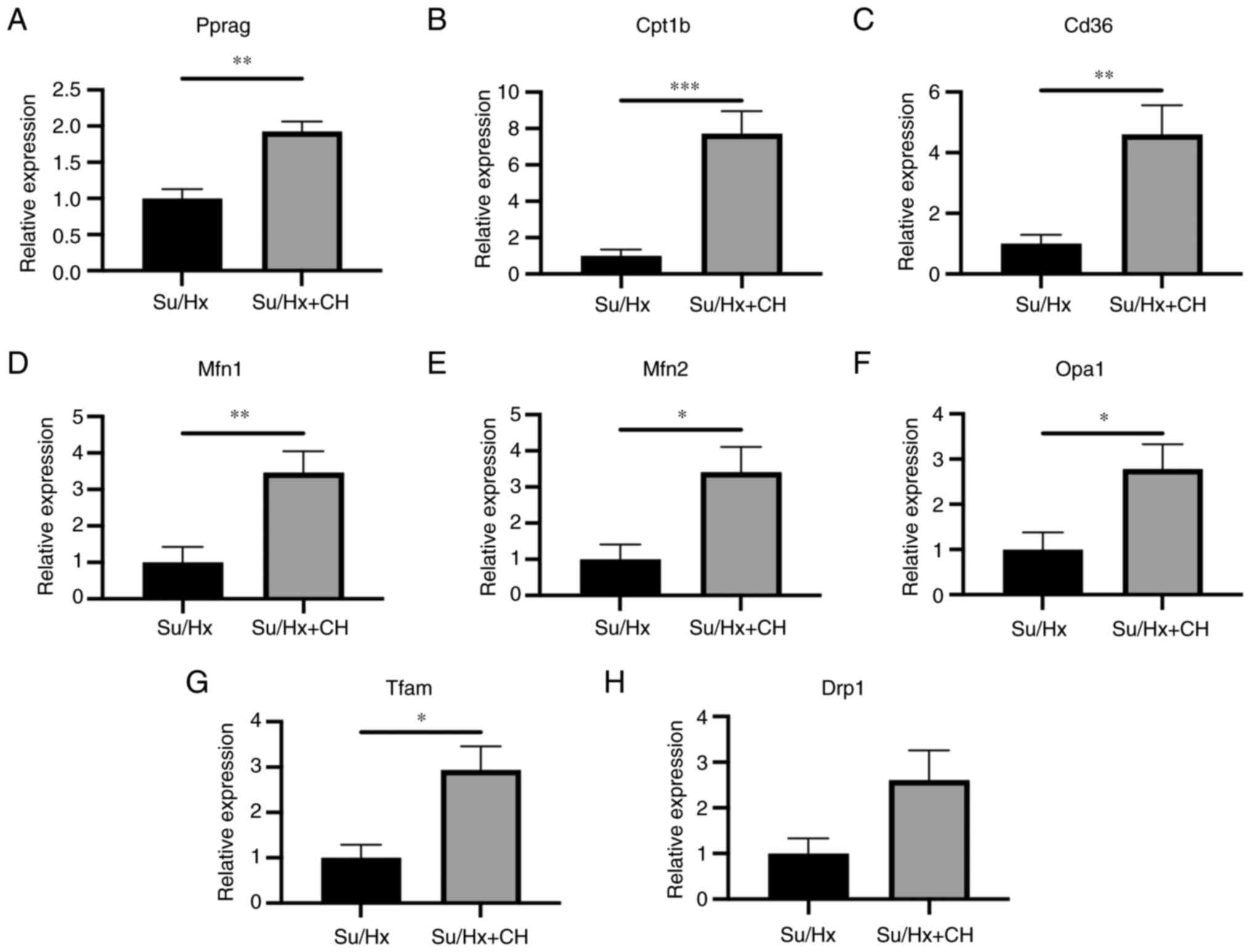 | Figure 5RT-qPCR analysis of right heart
tissue. Genes associated with PPARγ, lipid metabolism and
mitochondrial dynamics were analyzed using RT-qPCR in the right
heart tissue. RNA expression levels of (A) Pparg, (B) Cd36 and (C)
Cpt1B were higher in Su/Hx + CH than Su/Hx (fold regulation, 1.93,
4.61 and 7.73, respectively). Among genes associated with fusion
and biogenesis dynamics, (D) Mfn1, (E) Mfn2, (F) Opa1 and (G) Tfam
expression levels were higher in Su/Hx + CH than Su/Hx (fold
regulation, 3.47, 3.41, 2.78 and 2.94, respectively). (H) No
significant difference was observed between Su/Hx and Su/Hx + CH
groups regarding Drpl expression. *P<0.05,
**P<0.01, ***P<0.001 vs. Su/Hx. Cd36,
cluster of differentiation 36; Cpt1b, carnitine palmitoyl
transferase 1 B; Drp1, dynamin-related protein 1; Mfn, mitofusin;
Opa1, optic atrophy 1; Pparg, peroxisome proliferator-activated
receptor γ; Tfam, mitochondrial transcription factor A; RT-q,
reverse transcription-quantitative; Su, SU5416; Hx, hypoxia; CH,
chrysin. |
Hemodynamic analysis using Millar
Mikro-Tip® catheter
Hemodynamic parameters were measured using Millar
Mikro-Tip catheter system (n=5-8). Representative pressure-volume
curve is presented in Fig. 6A.
RV:LV + S was significantly lower in Su/Hx + CH than in Su/Hx
(Fig. 6B). RVSP was 82.0±5.5 in
Su/Hx and 49.4±4.0 mmHg in Su/Hx + CH (Fig. 6C). Maximum and minimum dP/dt,
indices of RV systolic and diastolic performance, were lower in
Su/Hx and higher in Su/Hx + CH (Fig.
6D-E). No significant differences were observed in stroke
volume and cardiac index (Fig. 6F
and G). Ees/Ea, a clinical index of ventricle-artery functional
coupling, was higher in Su/Hx + CH than Su/Hx group (Fig. 6H and I).
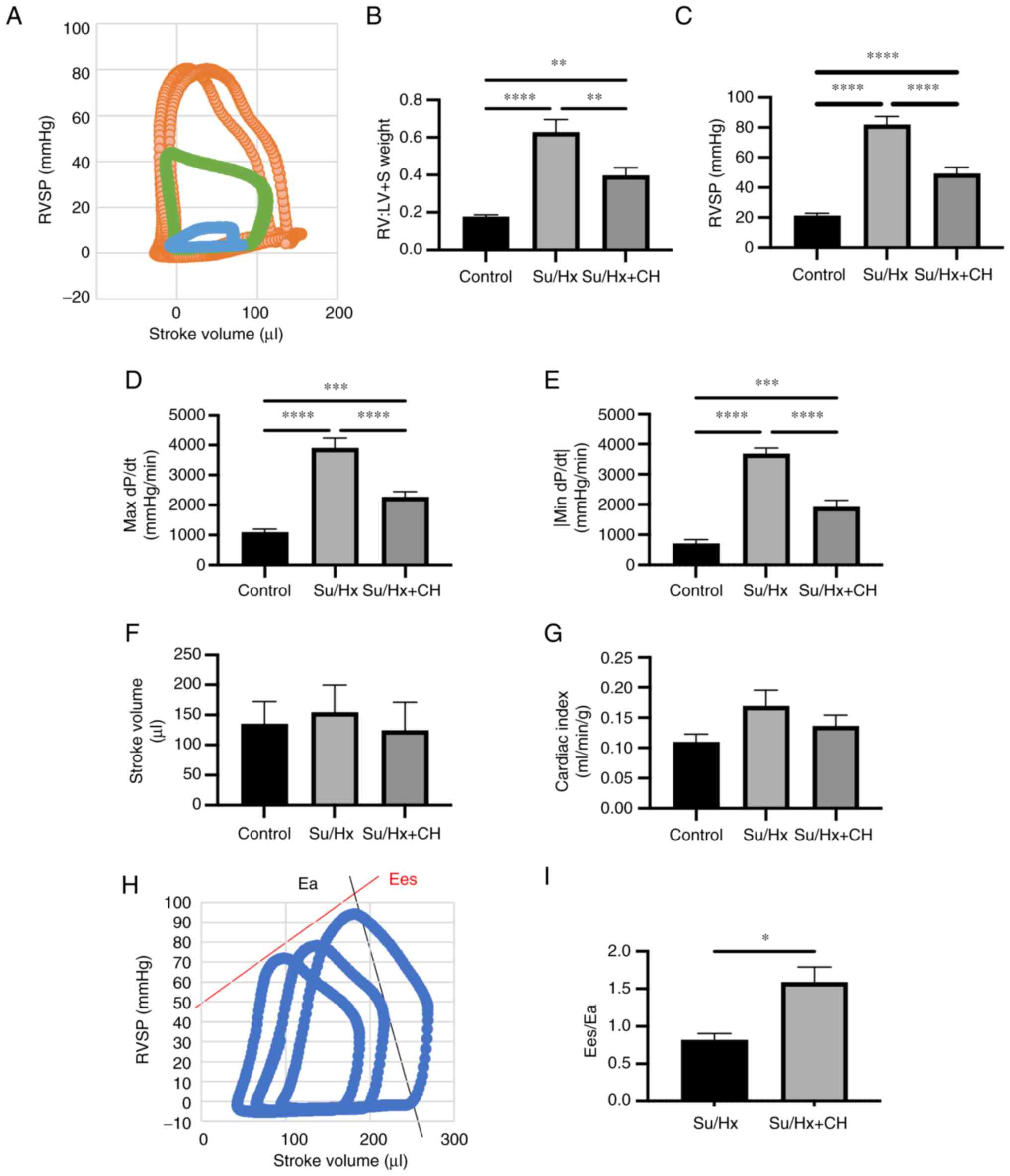 | Figure 6Hemodynamic analysis using Millar
Mikro-Tip® catheter. (A) Representative pressure-volume
curve. Orange, Su/Hx; green, Su/Hx + CH; blue, CTRL. (B) RV:LV + S
weight. (C) RVSP, (D) max dP/dt, and (E) absolute value of min
dP/dt were lower in Su/Hx + CH group than Su/Hx. (F) Stroke volume.
(G) Cardiac index. (H) Calculation of Ea/Ees using pressure-volume
curve. (I) Ea/Ees in Su/Hx and Su/Hx + CH. *P<0.05,
**P<0.01, ***P<0.001,
****P<0.0001. Ea, arterial elastance; Ees,
end-systolic elastance; LV + S, left ventricule + septum; RV, right
ventricle; SP, systolic pressure; Su, SU5416; Hx, hypoxia; CH,
chrysin; dP/dt, differential of pressure with time. |
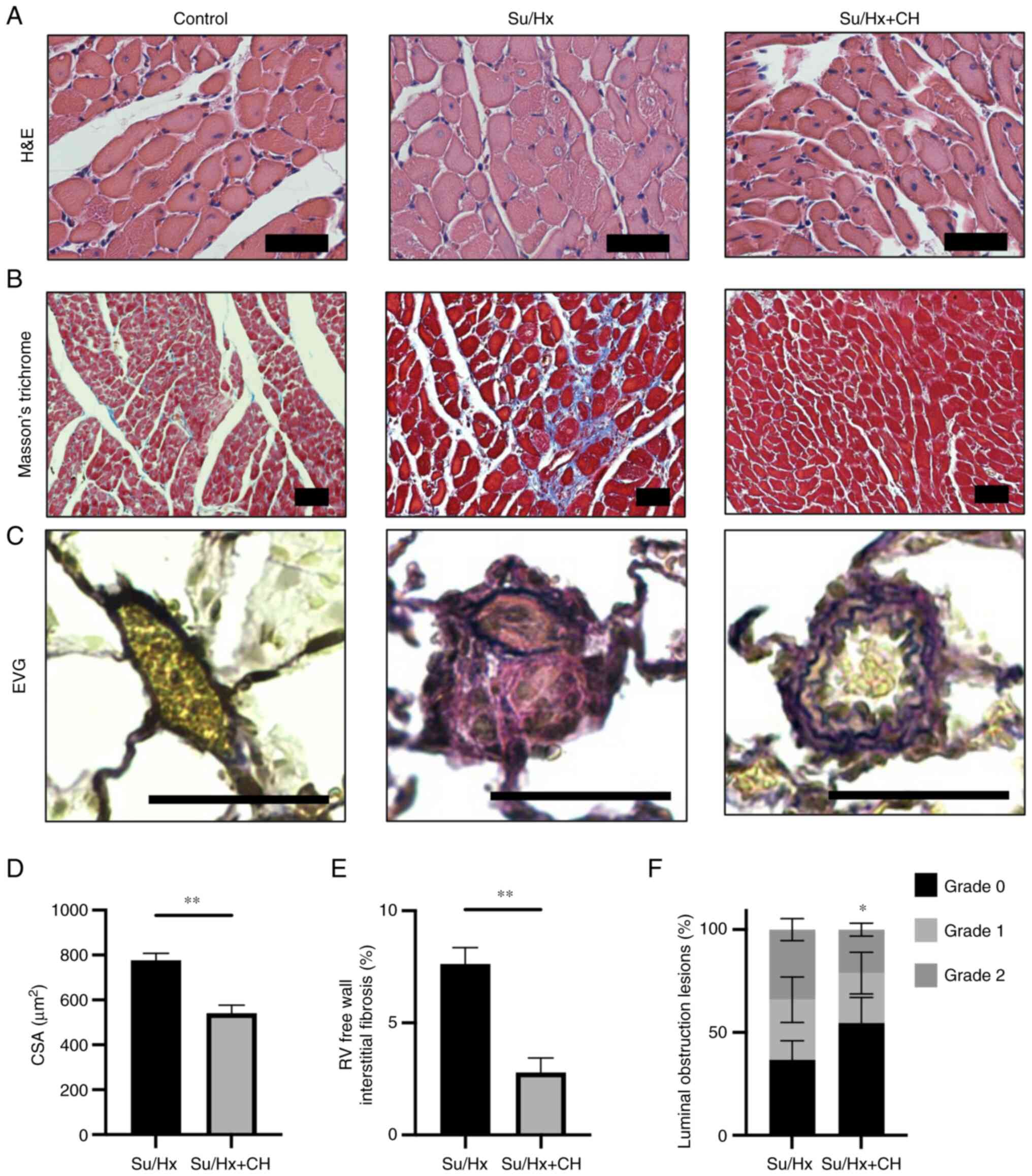
Histological analysis of right heart and
pulmonary artery
Myocyte CSA was visualized using H&E staining
(Fig. 7A). The fibrotic area was
stained blue using Masson's trichrome stain (Fig. 7B). RV free wall myocyte size was
significantly smaller in Su/Hx + CH than Su/Hx (Fig. 7D), suggesting CH treatment may
have ameliorated RV hypertrophy. RV free wall fibrosis was
significantly higher in Su/Hx than Su/Hx + CH (Fig. 7E), suggesting CH may have
decreased RV wall fibrosis. Pulmonary artery remodeling was
pathologically quantified using EVG staining (Fig. 7C). The extent of pulmonary vessel
luminal obstruction in grade 2 was significantly lower in Su/Hx +
CH compared with Su/Hx (Fig.
7F), suggesting CH alleviated pulmonary artery remodeling.
Discussion
The present study investigated the molecular
mechanisms underlying the effects of CH on RV remodeling and
dysfunction in the development of PH. Gene expression changes in RV
of Su/Hx rats in the presence or absence of CH were analyzed using
RNA-seq. Furthermore, metabolomic profiling of RVs was used to
determine metabolic regulation by CH and pathophysiological changes
were observed in Su/Hx + CH rats. Transcriptome analysis suggested
that pathways associated with mitochondrial energy metabolism were
downregulated in Su/Hx and restored in Su/Hx + CH, such as
generation of precursor metabolites and energy, mitochondrial ATP
synthesis, oxidative phosphorylation and TCA cycle. CH protected
against downregulation of these genes. Metabolome analysis revealed
that CH increased the total concentration of adenylate, which
suggested an improvement in whole energy production. CH decreased
accumulation of LCFA. RT-qPCR revealed that expression of PPARγ, a
master regulator of FA metabolism and mitochondrial biogenesis, was
increased in RV of Su/Hx + CH rats. Genes associated with FA
transport and mitochondrial biogenesis were also increased in the
RV of Su/Hx + CH rats.
Regulation of mitochondrial biogenesis in the
pulmonary vascular wall and RV is a therapeutic target in PH
(24,25). RT-qPCR revealed that expression
of PPARγ was increased in RV of Su/Hx + CH rats. PPARγ, a member of
the nuclear hormone receptor superfamily, is ubiquitously expressed
in myocardium, pulmonary vascular smooth muscle and endothelial
cells and serves a key role in lipid metabolism and mitochondrial
dynamics, resulting in inhibited ventricular hypertrophy (26-28). PPARγ-associated signaling is
involved in multiple processes, including regulation of
mitochondrial biogenesis, induction of antioxidant genes,
transrepression of inflammatory signaling and regulation of Nitric
oxide bioavailability in PH (29-33).
CH is a phytochemical categorized as a flavonoid
based on its chemical structure. It is present in propolis, honey,
passion fruit, mushrooms and other plant sources; it has been used
to treat numerous types of degenerative disorder and exhibits
anti-inflammatory activity (34-36). In addition, it has been used in
the treatment of numerous types of metabolic malfunction, such as
metabolic syndrome (37,38).
In rats exposed to chronic Hx, CH exhibits
cardioprotective effects, such as antitumor and antioxidant
effects, leading to prevention of vascular remodeling (9,39). To the best of our knowledge,
however, its precise molecular effects on RV have not been defined.
In a previous report, CH was shown to serve as a PPARγ agonist and
directly upregulate transcriptional activity of PPARγ (40). Here, transcriptome analysis
indicated that CH partially improved Su/Hx-induced downregulation
of genes associated with mitochondrial function, such as aerobic
electron transport chain, mitochondrial ATP synthesis-coupled
electron transport and mitochondrial respiratory chain complex
assembly. Likewise, mitochondrial oxidative phosphorylation and TCA
cycle downregulation in RV of Su/Hx rats was partly reversed by CH.
CH may upregulate mitochondrial energy production, which is
associated with an increase in total adenylate production.
Improvement of energy production in mitochondria by upregulation of
PPARγ may be a key treatment target of CH for RV dysfunction in
PAH.
FA metabolic regulation in PH may be cell- and
etiology-specific, thus complicating therapeutic targeting
(41). In the Randle cycle, FA
or glucose exhibit a negative regulatory association. FA and
glucose oxidation share a reciprocal mechanism (42). Although FA oxidation is
considered to account for a large part of cellular energy
production in the RV, it has not been completely defined.
RV-specific accumulation of increased LCFAs may be a hallmark of
lipotoxicity (43,44). Here, CH upregulated gene
expression of Cd36 and Cpt1b, which transport Fas across plasma and
mitochondrial membranes (45,46). Moreover, metabolome analysis
indicated that CH decreased accumulation of LCFAs in the RV,
particularly palmitic and stearic acid. It was hypothesized that
decreasing LCFA accumulation may be another mechanism by which CH
improves RV remodeling.
The present study has certain limitations. The
cardiovascular unit is composed of two primary functional units: RV
and pulmonary vasculature. As right heart failure in PH is the
consequence of increased afterload, a full understanding of
physiological and pathobiological aspects in the cardiopulmonary
unit is required to understand the molecular mechanisms underlying
PH (47). Decreased oxidative
phosphorylation and increased glycolysis are considered primary
metabolic pathways disrupted in PH (48). Mitochondria in RV cardiomyocytes
and pulmonary vascular cells may exhibit similar respiratory
suppression followed by increased glucose uptake and glycolysis in
PH. The complexities of metabolic reprogramming and mitochondrial
dysfunction may promote cell survival and proliferation, while
metabolic switching in pulmonary vascular cells may drive extensive
remodeling, which has been proposed as a metabolic theory (48,49). The aforementioned studies suggest
CH may act on both RV and pulmonary vasculature. The present study
indicated that CH altered gene expression and mitochondrial
metabolite levels in cardiomyocytes. However, whether these
transcriptomic changes were solely due to the direct effects of CH
on RV remains unclear as transcriptome analysis of resistant
pulmonary blood vessels using laser microdissection (50) was not performed in this
study.
The present study did not fully elucidate the
mitochondrial dynamics. CH upregulated numerous mitochondrial
fusion genes. Expression of DRP1, a representative fission marker,
was higher in Su/Hx +CH than Su/Hx, although this was not
statistically significant. For functional efficiency of DRP1,
phosphorylation at the serine 616 residue is key in addition to the
gene expression (51). However,
protein level of phosphorylated DRP1 was not evaluated in the
present study. Further investigation is needed to elucidate the
effect of CH on mitochondrial dynamics.
CH has multiple effects, such as anti-oxidation and
anti-inflammatory, in addition to aforementioned metabolic effects
(9-11,34-38). The present transcriptome analysis
did not reveal the anti-inflammatory effect of CH, which has been
previously reported (10,11).
The mild effects of CH as a flavonoid (including anti-oxidation,
anti-inflammatory and anti-diabetic effects) (52) may make it superior to selective
synthetic agents; however, the entire potential mechanism and side
effects of CH in PAH need to be clarified before approval as a
therapeutic drug (9,10).
In conclusion, the present study suggested that CH
altered gene expression and mitochondrial metabolite levels in
cardiomyocytes, resulting in improved RV remodeling and dysfunction
in a Su/Hx PAH rat model. CH may be a potential candidate for
therapeutic options in PAH.
Supplementary Data
Availability of data and materials
The datasets generated and/or analyzed during the
current study are available in the NCBI Gene Expression Omnibus
database repository, ncbi.nlm.nih.gov/geo (accession no. GSE186989). Other
datasets used and/or analyzed during the current study are
available from the corresponding author on reasonable request.
Authors' contributions
KT, SS, YK, JDK and YN were involved in study design
and conceptualization. TK, AY, AN and TJS performed animal
experiments. KT and JDK confirm the authenticity of all the raw
data. TK and AN wrote the manuscript. KT, YK and JDK critically
revised the manuscript. YK, YN and TS were involved in the
interpretation of data and supervision of the study. All authors
have read and approved the final manuscript.
Ethics approval and consent to
participate
All animal procedures were approved by the Review
Board for Animal Experiments of Chiba University (approval no.
30-126) and performed in accordance with the guidelines of the
Animal Research Committee of Laboratory Animal Center, Graduate
School of Medicine, Chiba University.
Patient consent for publication
Not applicable.
Competing interests
The authors declare they have no competing
interests.
Acknowledgments
The authors would like to thank Ms. Sumina Atarashi
and Ms. Tomoko Misawa for technical advice and Ms. Chieko Handa for
administrative support (all Department of Respirology, Graduate
School of Medicine, Chiba University).
Funding
This study was supported by Japan. Agency for Medical Research
and Development-Core Research for Evolution Science and Technology
(grant no. 21gm1410010s0101), KAKENHI (grant no. 19H03664) and
Intractable Respiratory Diseases and Pulmonary Hypertension
Research Group, Ministry of Health, Labor and Welfare, Japan (grant
no. 20FC1027).
Abbreviations:
|
Cpt1b
|
carnitine palmitoyl transferase 1B
|
|
Drp1
|
dynamin-related protein 1
|
|
Ea
|
arterial elastance
|
|
ECM
|
extracellular matrix
|
|
Ees
|
end-systolic elastance
|
|
LC-TOFMS
|
liquid chromatography time-of-flight
mass spectrometry
|
|
Mfn
|
mitofusin
|
|
Opa1
|
optic atrophy 1
|
|
PAH
|
pulmonary arterial hypertension
|
|
PPARγ
|
peroxisome proliferator-activated
receptor γ
|
|
RVSP
|
right ventricle systolic pressure
|
|
Su/Hx
|
SU5416 followed by chronic hypoxia
|
|
Su/Hx + CH
|
SU5416 followed by chronic hypoxia and
chrysin administration
|
|
Tfam
|
mitochondrial transcription factor
A
|
References
|
1
|
Voelkel NF, Quaife RA, Leinwand LA, Barst
RJ, McGoon MD, Meldrum DR, Dupuis J, Long CS, Rubin LJ, Smart FW,
et al: Right ventricular function and failure: Report of a national
heart, lung, and blood institute working group on cellular and
molecular mechanisms of right heart failure. Circulation.
114:1883–1891. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Tonelli AR, Arelli V, Minai OA, Newman J,
Bair N, Heresi GA and Dweik RA: Causes and circumstances of death
in pulmonary arterial hypertension. Am J Respir Crit Care Med.
188:365–369. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Frieler RA and Mortensen RM: Immune cell
and other noncardiomyocyte regulation of cardiac hypertrophy and
remodeling. Circulation. 131:1019–1030. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Bishop SP and Altschuld RA: Increased
glycolytic metabolism in cardiac hypertrophy and congestive
failure. Am J Physiol. 218:153–159. 1970. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Partovian C, Adnot S, Eddahibi S, Teiger
E, Levame M, Dreyfus P, Raffestin B and Frelin C: Heart and lung
VEGF mRNA expression in rats with monocrotaline- or hypoxia-induced
pulmonary hypertension. Am J Physiol. 275:H1948–H1956.
1998.PubMed/NCBI
|
|
6
|
Oikawa M, Kagaya Y, Otani H, Sakuma M,
Demachi J, Suzuki J, Takahashi T, Nawata J, Ido T, Watanabe J and
Shirato K: Increased [18F]fluorodeoxyglucose accumulation in right
ventricular free wall in patients with pulmonary hypertension and
the effect of epoprostenol. J Am Coll Cardiol. 45:1849–1855. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Panche AN, Diwan AD and Chandra SR:
Flavonoids: An overview. J Nutr Sci. 5:e472016. View Article : Google Scholar
|
|
8
|
García-Lafuente A, Guillamón E, Villares
A, Rostagno MA and Martínez JA: Flavonoids as anti-inflammatory
agents: Implications in cancer and cardiovascular disease. Inflamm
Res. 58:537–552. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Li XW, Wang XM, Li S and Yang JR: Effects
of chrysin (5,7-dihydroxyflavone) on vascular remodeling in
hypoxia-induced pulmonary hypertension in rats. Chin Med. 10:42015.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Yang M, Xiong J, Zou Q, Wang DD and Huang
CX: Chrysin attenuates interstitial fibrosis and improves cardiac
function in a rat model of acute myocardial infarction. J Mol
Histol. 49:555–565. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Kseibati MO, Sharawy MH and Salem HA:
Chrysin mitigates bleomycin-induced pulmonary fibrosis in rats
through regulating inflammation, oxidative stress, and hypoxia. Int
Immunopharmacol. 89:1070112020. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Abe K, Toba M, Alzoubi A, Ito M, Fagan KA,
Cool CD, Voelkel NF, McMurtry IF and Oka M: Formation of plexiform
lesions in experimental severe pulmonary arterial hypertension.
Circulation. 121:2747–2754. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Oka M, Homma N, Taraseviciene-Stewart L,
Morris KG, Kraskauskas D, Burns N, Voelkel NF and McMurtry IF: Rho
kinase-mediated vasoconstriction is important in severe occlusive
pulmonary arterial hypertension in rats. Circ Res. 100:923–929.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Toba M, Alzoubi A, O'Neill KD, Gairhe S,
Matsumoto Y, Oshima K, Abe K and Oka M: Temporal hemodynamic and
histological progression in Sugen5416/hypoxia/normoxia-exposed
pulmonary arterial hypertensive rats. Am J Physiol Heart Circ
Physiol. 306:H243–H250. 2014. View Article : Google Scholar :
|
|
15
|
Taraseviciene-Stewart L, Kasahara Y, Alger
L, Hirth P, Mc Mahon G, Waltenberger J, Voelkel NF and Tuder RM:
Inhibition of the VEGF receptor 2 combined with chronic hypoxia
causes cell death-dependent pulmonary endothelial cell
proliferation and severe pulmonary hypertension. FASEB J.
15:427–438. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Sanada TJ, Hosomi K, Shoji H, Park J,
Naito A, Ikubo Y, Yanagisawa A, Kobayashi T, Miwa H, Suda R, et al:
Gut microbiota modification suppresses the development of pulmonary
arterial hypertension in an SU5416/hypoxia rat model. Pulm Circ.
10:20458940209291472020. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Guidelines of the Animal Research
Committee of Laboratory Animal Center, Graduate School of Medicine,
Chiba University. https://www.chiba-u.ac.jp/general/JoureiV5HTMLContents/act/frame/frame110000180.htm.
Accessed March 3, 2022.
|
|
18
|
Kato F, Sakao S, Takeuchi T, Suzuki T,
Nishimura R, Yasuda T, Tanabe N and Tatsumi K: Endothelial
cell-related autophagic pathways in Sugen/hypoxia-exposed pulmonary
arterial hypertensive rats. Am J Physiol Lung Cell Mol Physiol.
313:L899–L915. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Takeuchi T, Sakao S, Kato F, Naito A, Jujo
T, Yasuda T, Tanabe N and Tatsumi K: Pulmonary haemodynamics are
correlated with intimal lesions in a rat model of severe PAH:
Attenuation of pulmonary vascular remodelling with ambrisentan.
Histol Histopathol. 31:1357–1365. 2016.PubMed/NCBI
|
|
20
|
Ge SX, Son EW and Yao R: iDEP: An
integrated web application for differential expression and pathway
analysis of RNA-Seq data. BMC Bioinformatics. 19:5342018.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Kuleshov MV, Jones MR, Rouillard AD,
Fernandez NF, Duan Q, Wang Z, Koplev S, Jenkins SL, Jagodnik KM,
Lachmann A, et al: Enrichr: A comprehensive gene set enrichment
analysis web server 2016 update. Nucleic Acids Res. 44:W90–W97.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Sakao S, Kawakami E, Shoji H, Naito A,
Miwa H, Suda R, Sanada TJ, Tanabe N and Tatsumi K: Metabolic
remodeling in the right ventricle of rats with severe pulmonary
arterial hypertension. Mol Med Rep. 23:2272021. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
24
|
Yeligar SM, Kang BY, Bijli KM, Kleinhenz
JM, Murphy TC, Torres G, San Martin A, Sutliff RL and Hart CM:
PPARγ regulates mitochondrial structure and function and human
pulmonary artery smooth muscle cell proliferation. Am J Respir Cell
Mol Biol. 58:648–657. 2018. View Article : Google Scholar :
|
|
25
|
Gomez-Arroyo J, Mizuno S, Szczepanek K,
Van Tassell B, Natarajan R, dos Remedios CG, Drake JI, Farkas L,
Kraskauskas D, Wijesinghe DS, et al: Metabolic gene remodeling and
mitochondrial dysfunction in failing right ventricular hypertrophy
secondary to pulmonary arterial hypertension. Circ Heart Fail.
6:136–144. 2013. View Article : Google Scholar
|
|
26
|
Vidal-Puig AJ, Considine RV, Jimenez-Liñan
M, Werman A, Pories WJ, Caro JF and Flier JS: Peroxisome
proliferator-activated receptor gene expression in human tissues.
Effects of obesity, weight loss, and regulation by insulin and
glucocorticoids. J Clin Invest. 99:2416–2422. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Qi HP, Wang Y, Zhang QH, Guo J, Li L, Cao
YG, Li SZ, Li XL, Shi MM, Xu W, et al: Activation of peroxisome
proliferator-activated receptor γ (PPARγ) through NF-κB/Brg1 and
TGF-β1 pathways attenuates cardiac remodeling in
pressure-overloaded rat hearts. Cell Physiol Biochem. 35:899–912.
2015. View Article : Google Scholar
|
|
28
|
Lee TW, Bai KJ, Lee TI, Chao TF, Kao YH
and Chen YJ: PPARs modulate cardiac metabolism and mitochondrial
function in diabetes. J Biomed Sci. 24:52017. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Dominy JE and Puigserver P: Mitochondrial
biogenesis through activation of nuclear signaling proteins. Cold
Spring Harb Perspect Biol. 5:a0150082013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Fan W and Evans R: PPARs and ERRs:
Molecular mediators of mitochondrial metabolism. Curr Opin Cell
Biol. 33:49–54. 2015. View Article : Google Scholar :
|
|
31
|
Ricote M and Glass CK: PPARs and molecular
mechanisms of transrepression. Biochim Biophys Acta. 1771:926–935.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Calnek DS, Mazzella L, Roser S, Roman J
and Hart CM: Peroxisome proliferator-activated receptor gamma
ligands increase release of nitric oxide from endothelial cells.
Arterioscler Thromb Vasc Biol. 23:52–57. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Pascual G, Fong AL, Ogawa S, Gamliel A, Li
AC, Perissi V, Rose DW, Willson TM, Rosenfeld MG and Glass CK: A
SUMOylation-dependent pathway mediates transrepression of
inflammatory response genes by PPAR-gamma. Nature. 437:759–763.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Del Fabbro L, Rossito Goes A, Jesse CR, de
Gomes MG, Cattelan Souza L, Lobo Ladd FV, Lobo Ladd AAB, Nunes
Arantes RV, Reis Simionato A, Oliveira MS, et al: Chrysin protects
against behavioral, cognitive and neurochemical alterations in a
6-hydroxydopamine model of Parkinson's disease. Neurosci Lett.
706:158–163. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Jiang Y, Gong FL, Zhao GB and Li J:
Chrysin suppressed inflammatory responses and the inducible nitric
oxide synthase pathway after spinal cord injury in rats. Int J Mol
Sci. 15:12270–12279. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Tang H, Tao A, Song J, Liu Q, Wang H and
Rui T: Doxorubicin-induced cardiomyocyte apoptosis: Role of
mitofusin 2. Int J Biochem Cell Biol. 88:55–59. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Yamamoto Y: Effects of dietary chrysin
supplementation on blood pressure and oxidative status of rats fed
a high-fat high-sucrose diet. Food Sci Technol Res. 20:295–300.
2014. View Article : Google Scholar
|
|
38
|
Andrade N, Andrade S, Silva C, Rodrigues
I, Guardão L, Guimarães JT, Keating E and Martel F: Chronic
consumption of the dietary polyphenol chrysin attenuates metabolic
disease in fructose-fed rats. Eur J Nutr. 59:151–165. 2020.
View Article : Google Scholar
|
|
39
|
Fu B, Xue J, Li Z, Shi X, Jiang BH and
Fang J: Chrysin inhibits expression of hypoxia-inducible
factor-1alpha through reducing hypoxia-inducible factor-1alpha
stability and inhibiting its protein synthesis. Mol Cancer Ther.
6:220–226. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Liang YC, Tsai SH, Tsai DC, Lin-Shiau SY
and Lin JK: Suppression of inducible cyclooxygenase and nitric
oxide synthase through activation of peroxisome
proliferator-activated receptor-gamma by flavonoids in mouse
macrophages. FEBS Lett. 496:12–18. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Xu W, Janocha AJ and Erzurum SC:
Metabolism in pulmonary hypertension. Annu Rev Physiol. 83:551–576.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Randle PJ, Garland PB, Hales CN and
Newsholme EA: The glucose fatty-acid cycle. Its role in insulin
sensitivity and the metabolic disturbances of diabetes mellitus.
Lancet. 1:785–789. 1963. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Brittain EL, Talati M, Fessel JP, Zhu H,
Penner N, Calcutt MW, West JD, Funke M, Lewis GD, Gerszten RE, et
al: Fatty acid metabolic defects and right ventricular lipotoxicity
in human pulmonary arterial hypertension. Circulation.
133:1936–1944. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Hemnes AR, Brittain EL, Trammell AW,
Fessel JP, Austin ED, Penner N, Maynard KB, Gleaves L, Talati M,
Absi T, et al: Evidence for right ventricular lipotoxicity in
heritable pulmonary arterial hypertension. Am J Respir Crit Care
Med. 189:325–334. 2014. View Article : Google Scholar :
|
|
45
|
Kampf JP and Kleinfeld AM: Is membrane
transport of FFA mediated by lipid, protein, or both? An unknown
protein mediates free fatty acid transport across the adipocyte
plasma membrane. Physiology (Bethesda). 22:7–14. 2007.
|
|
46
|
McGarry JD and Brown NF: The mitochondrial
carnitine palmitoyltransferase system. From concept to molecular
analysis. Eur J Biochem. 244:1–14. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Vonk Noordegraaf A, Chin KM, Haddad F,
Hassoun PM, Hemnes AR, Hopkins SR, Kawut SM, Langleben D, Lumens J
and Naeije R: Pathophysiology of the right ventricle and of the
pulmonary circulation in pulmonary hypertension: An update. Eur
Respir J. 53:18019002019. View Article : Google Scholar
|
|
48
|
Ryan JJ and Archer SL: Emerging concepts
in the molecular basis of pulmonary arterial hypertension: Part I:
Metabolic plasticity and mitochondrial dynamics in the pulmonary
circulation and right ventricle in pulmonary arterial hypertension.
Circulation. 131:1691–1702. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Han S and Chandel NS: Lessons from cancer
metabolism for pulmonary arterial hypertension and fibrosis. Am J
Respir Cell Mol Biol. 65:134–145. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Laumanns IP, Fink L, Wilhelm J, Wolff JC,
Mitnacht-Kraus R, Graef-Hoechst S, Stein MM, Bohle RM, Klepetko W,
Hoda MA, et al: The noncanonical WNT pathway is operative in
idiopathic pulmonary arterial hypertension. Am J Respir Cell Mol
Biol. 40:683–691. 2009. View Article : Google Scholar
|
|
51
|
Kar UP, Dey H and Rahaman A: Regulation of
dynamin family proteins by post-translational modifications. J
Biosci. 42:333–344. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Naz S, Imran M, Rauf A, Orhan IE, Shariati
MA, Iahtisham-Ul-H, Yasmin Iqra, Shahbaz M, Qaisrani TB, Shah ZA,
et al: Chrysin: Pharmacological and therapeutic properties. Life
Sci. 235:1167972019. View Article : Google Scholar : PubMed/NCBI
|