Introduction
Experimental models have been used to study the
molecular changes associated with cardiac diseases. However, these
models have significant drawbacks and do not always reflect the
clinical setting. For example, the commonly used nontransgenic
models of coronary artery disease involve acute ischemia rather
than progressive atherosclerotic coronary disease which triggers
slow progressive cardiac remodeling (1). It is highly informative to study
human hearts directly; however, the collection of samples is
challenging. Studies using human cardiac ventricular tissues for
molecular investigations are few and have largely focused on global
gene expression in failing and non-failing donor hearts and
comparing atrial with ventricular tissue (2–5).
For example, a study comparing gene expression in right atrial
appendage and left ventricle of patients undergoing two types of
cardiac surgery (coronary artery bypass graft and aortic valve
replacement) identified ~2% of the detected genes as differentially
expressed (6). Although gene
expression profiling is informative, it is not conclusive as it may
not always reflect changes in protein levels associated with
disease progression (7). For
this reason, we have carried out comparative studies investigating
disease-induced changes in protein expression in congenital hearts
(8–10) and in adult patients with either
aortic valve stenosis (AVS) or coronary artery disease (CAD)
(11). In the latter, we have
shown that different cardiac diseases trigger different proteomic
remodeling in the diseased left (LV) and relatively less diseased
right ventricles (RV) and that the extent of remodeling varies with
disease type.
Investigating the effect of disease on cardiac
molecular changes provides critical information about signaling
pathways to improve our understanding of disease-induced cardiac
remodeling (11). However,
information is also required concerning the acute molecular changes
associated with cardiopulmonary bypass and cardioplegic ischemic
arrest during open-heart surgery which are likely to have
implications for postoperative morbidity. Distinguishing such
changes from molecular changes due to chronic disease requires
information gathered from pre- and post-operative levels. In the
present study, we collected pre- and post-cardioplegic arrest
biopsies from diseased LVs and relatively normal RVs of patients
undergoing surgery for CAD or AVS in order to investigate the acute
changes in protein expression during open heart surgery. We also
compared differences in protein expression between CAD and AVS
patients at each timepoint. This part of the study provides an
update to our previous work (11); the data are improved due to the
use of the latest proteomic technology which allows detection of a
greater number of proteins as well as the analysis of relative
phosphoprotein expression, a larger n number of available samples
and the inclusion of a comparison of post-reperfusion samples. The
information collected will help in the design of cardioprotective
interventions taking into consideration the involvement of
pathology and ventricular chambers.
Materials and methods
Patients and tissue collection
The data presented in this work are a sub-study of a
two-centre randomized controlled trial investigating the effect of
the upper limb remote ischemic preconditioning (RIPC) in patients
undergoing isolated coronary artery bypass grafting and/or aortic
valve replacement on cardiac injury, metabolic stress and
inflammatory response (12,13). The trial was conducted in
accordance with the Declaration of Helsinki at the Hammersmith
Hospital and the Bristol Royal Infirmary. It was sponsored by the
Imperial College of London, approved by the London-Harrow Research
Ethics Committee (REC number 12/LO/1361) and registered to the
International Standard Randomized Controlled Trial Number (ISRCTN)
registry (ID 33084113).
Inclusion, exclusion criteria and trial conduct were
previously published (12,13). A group of patients who had either
CAD (n=6) or AVS (n=6) were randomly selected from the trial and
used for this sub-study. All 6 CAD patients had 3 grafts for
diseases in the following vessels: left anterior descending, obtuse
marginal branch and the right coronary artery. One patient had an
additional left main disease. Both CAD and AVS patients were
control patients; no patients who received RIPC intervention were
included. Frozen biopsies were obtained using a Trucut needle from
the left and the right ventricles pre and Post cardioplegic arrest
during coronary artery bypass graft or aortic valve surgery.
However, a few biopsies did not yield enough protein for the
proteomic analysis and were therefore not included. In total 40
biopsies were processed (n=20 from CAD and n=20 from AVS
patients).
Anesthesia, surgery and cardioplegia
management
Anesthetic management, cardiopulmonary bypass (CPB),
cardioplegia, surgical techniques and any other aspect of pre- and
post-operative management were in accordance with existing
protocols at both centers. Surgery proceeded as per routine
practice in each center. Cardioplegic ischemic arrest was induced
using cold cardioplegia (4 parts blood:1 part cardioplegia ratio)
given at a temperature of approximately 4°C as described in detail
elsewhere (12,13). In agreement with reported studies
and clinical practice, the cross-clamp time was significantly
longer in AVS patients (Table
I).
 | Table IKey patient characteristics and
intraoperative data. |
Table I
Key patient characteristics and
intraoperative data.
|
Characteristics | CAD (n=6) | AVS (n=6) |
|---|
| Mean age, years
(range) | 72±3 (60–80) | 72±2 (65–77) |
| Sex (M/F) | 4/2 | 4/2 |
| NYHA | I: 3 | II: 3 |
| II: 3 | III: 3 |
| Diabetes | 4 | 2 |
| Hypertension | 5 | 4 |
| Cross clamp time
(min) | 47.5±5.3 | 75.5±7.1a |
| Bypass time
(min) | 86.8±7.1 | 112.3±9.2 |
| | (P=0.053) |
Sample preparation
Proteins were extracted in radioimmunoprecipitation
assay (RIPA) buffer (1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS,
in PBS) containing phosphatase and protease inhibitors, and
quantified using the Bradford assay. Aliquots of 100 µg of
10 samples (including a reference sample prepared as a mixture from
all samples) per experiment were digested with trypsin (2.5
µg trypsin per 100 µg protein; 37°C, overnight),
labelled with Tandem Mass Tag (TMT) 10Plex reagents according to
the manufacturer's protocol (Thermo Fisher Scientific, Inc.), and
all the labelled samples were pooled. For the total proteome
analysis, aliquots of 50 µg of the pooled sample were
evaporated to dryness and re-suspended in buffer A (20 mM ammonium
hydroxide, pH 10.0) prior to fractionation by high pH
reversed-phase chromatography using an Ultimate 3000 liquid
chromatography system (Thermo Fisher Scientific, Inc.). The samples
were processed in randomly selected groups of 9, with the same
pooled sample included in each run to allow comparison between
runs. The samples were loaded onto an XBridge BEH C18 Column (130
Å, 3.5 µm, 2.1×150 mm, Waters) in buffer A and peptides
eluted with an increasing gradient of buffer B (20 mM ammonium
hydroxide in acetonitrile, pH 10.0) from 0–95% over 60 min. The
resulting fractions were evaporated to dryness and re-suspended in
1% formic acid prior to analysis by nano-LC MSMS using an Orbitrap
Fusion Tribrid mass spectrometer (Thermo Fisher Scientific,
Inc.).
For the phosphoproteome analysis, the remainder of
the TMT-labelled pooled sample was evaporated to dryness and
subjected to TiO2-based phosphopeptide enrichment
according to the manufacturer's instructions (Pierce/Thermo Fisher
Scientific, Inc.). The phospho-enriched sample was evaporated to
dryness and then re-suspended in 1% formic acid prior to analysis
by nano-LC MSMS using an Orbitrap Fusion Tribrid mass spectrometer
(Thermo Fisher Scientific, Inc.).
Nano-LC mass spectrometry
High pH RP fractions (total proteome analysis) or
the phospho-enriched fraction (phospho-proteome analysis) were
further fractionated using an Ultimate 3000 nano-HPLC system in
line with an Orbitrap Fusion Tribrid mass spectrometer (Thermo
Fisher Scientific, Inc.). In brief, peptides in 1% (vol/vol) formic
acid were injected onto an Acclaim PepMap C18 nano-trap column
(Thermo Fisher Scientific, Inc.). After washing with 0.5% (vol/vol)
acetonitrile 0.1% (vol/vol) formic acid, peptides were resolved on
a 250 mm × 75 µm Acclaim PepMap C18 reverse phase analytical
column (Thermo Fisher Scientific, Inc.) over a 150 min organic
gradient, using seven gradient segments (1–6% solvent B over 1 min,
6–15% B over 58 min, 15–32% B over 58 min, 32–40% B over 5 min,
40–90% B over 1 min, held at 90% B for 6 min and then reduced to 1%
B over 1 min) with a flow rate of 300 nl·min−1. Solvent
A was 0.1% formic acid and Solvent B was aqueous 80% acetonitrile
in 0.1% formic acid. Peptides were ionized by nano-electrospray
ionization at 2.0 kV using a stainless-steel emitter with an
internal diameter of 30 µm (Thermo Fisher Scientific, Inc.)
and a capillary temperature of 275°C.
All spectra were acquired using an Orbitrap Fusion
Tribrid mass spectrometer controlled by Xcalibur 3.0 software
(Thermo Fisher Scientific, Inc.) and operated in data-dependent
acquisition mode using an SPS-MS3 workflow. FTMS1 spectra were
collected at a resolution of 120,000, with an automatic gain
control (AGC) target of 200,000 and a maximum injection time of 50
msec. The Top N most intense ions were selected for MS/MS.
Precursors were filtered according to charge state (to include
charge states 2–7) and with mono-isotopic precursor selection.
Previously interrogated precursors were excluded using a dynamic
window (40 sec +/-10 ppm). The MS2 precursors were isolated with a
quadrupole mass filter set to a width of 1.2 m/z. ITMS2 spectra
were collected with an AGC target of 5,000, max injection time of
120 msec and CID collision energy of 35%.
For FTMS3 analysis, the Orbitrap was operated at
60,000 resolution with an AGC target of 50,000 and a max injection
time of 120 msec. Precursors were fragmented by high-energy
collision dissociation (HCD) at normalized collision energy of 55%
to ensure maximal TMT reporter ion yield. Synchronous Precursor
Selection (SPS) was enabled to include up to five MS2 fragment ions
in the FTMS3 scan.
Data processing and analysis
The raw data files were processed and quantified
using Proteome Discoverer software version 1.4 (Thermo Fisher
Scientific, Inc.) and peptide sequences searched against the
Uniprot Human database (134,169 sequences) using the SEQUEST
algorithm (https://www.uniprot.org/)Peptide
precursor mass tolerance was set at 10 ppm, and MS/MS tolerance was
set at 0.6 Da. Search criteria included oxidation of methionine
(+15.9949) as a variable modification and carbamido-methylation of
cysteine (+57.0214) and the addition of the TMT mass tag (+229.163)
to peptide N-termini and lysine as fixed modifications. For the
phosphoproteome analysis, phosphorylation of serine, threonine and
tyrosine (+79.966) were also included as variable modifications.
Searches were performed with full tryptic digestion and a maximum
of one missed cleavage was allowed. The reverse database search
option was enabled and all peptide data were filtered to satisfy a
false discovery rate (FDR) of 5%.
Only proteins or phosphoproteins that were detected
in all biopsies for each comparison were included in the analysis
(14). Values are presented as a
ratio to the internal standard (a pool of all samples) and
represent the median of the measured peptide(s) for each protein.
Values for phosphoproteins are expressed relative to the total
protein expression-this allows determination of genuine changes in
levels of phosphorylation rather than changes in the overall
abundance of a protein whose phosphorylation level remains
unchanged. Fold changes (Post reperfusion/pre ischemic cardioplegic
arrest and AVS/CAD) in biopsies were calculated, and
log2(fold change) was plotted
against-log10(P-value) on a volcano plot. A change in
protein expression greater or less than 1.3× and with P<0.05
(Student's unpaired, two-tailed, t-test) was considered
significant. These cut-offs were chosen in accordance with previous
studies (9,11,15). No further correction for multiple
testing was made; several features of proteomics studies such as
this one (e.g. small n numbers and ratio compression caused by TMT
tagging) are known to make it difficult to apply multiple
correction techniques effectively, often leading to a significant
number of false-positive results (16). Proteins or phosphoproteins with a
significant difference in expression between pre and post samples
or between AVS and CAD samples (P<0.05) were loaded into
Ingenuity Pathway Analysis (IPA) software (QIAGEN Inc., https://www.qiagenbioinformatics.com/products/ingenuitypathway-analysis)
to determine the significantly (P<0.05) enriched canonical
pathways (P-value of overlap calculated by Fisher's exact test
right tailed). For the comparison between pathologies, all proteins
were included in the IPA analysis, not only those that were present
in every biopsy. The IPA analysis is routinely used, recommended,
and was purchased by our Proteomics Centre and our bioinformatics
specialist. IPA is a well-known and extensively used software
package for proteomics data analysis as confirmed by the large
number of peer-reviewed publications reporting its use. Quick Go
software (https://www.ebi.ac.uk/QuickGO/) was used to analyze
gene ontology (GO) enrichment and heatmaps were plotted using R
software (https://www.r-project.org/; version
3.5.1). In an earlier study, we carried out western blotting
analysis on selected proteins extracted from human cardiac biopsies
and showed strong correlation with our proteomic analysis thus
providing some support for the validity of our analytical methods
(11).
Results
Effect of ischemic cardioplegic arrest on
cardiac proteome/phosphoproteome in LV and RV of patients with
coronary artery disease (CAD)
Approximately 7,000 proteins were detected over all
the samples in this study. However, only 3,371 proteins were
detected in all pre and post ischemic biopsies collected from the
LV of CAD patients (n=6 and n=5 for pre and post samples
respectively) and 3,120 proteins were detected in all pre and post
biopsies collected from the RV of CAD patients (n=6 and n=3). Only
these proteins were used in our analysis (see exclusions in
Materials and methods).
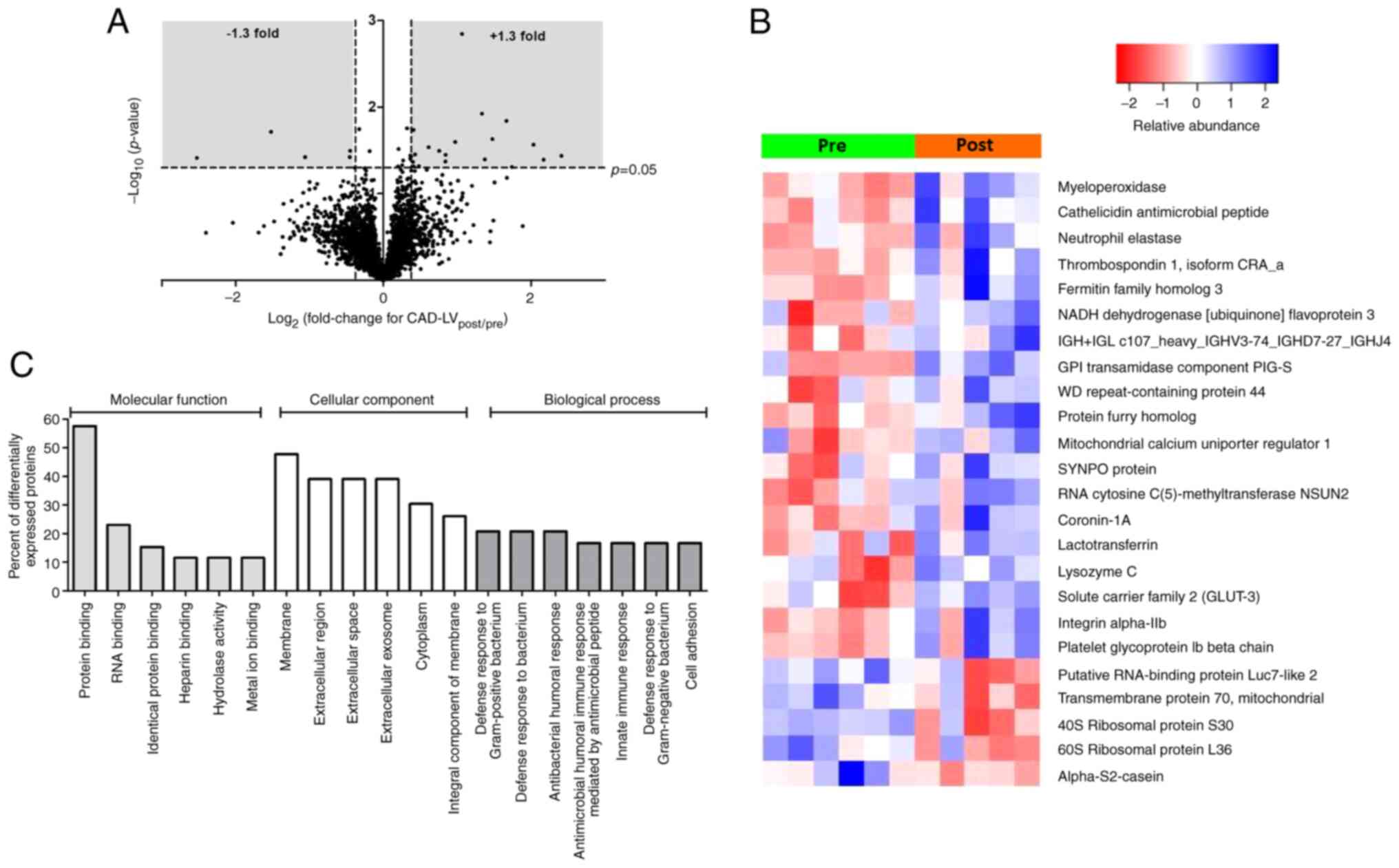
Fig. 1A shows a
volcano plot for the changes (post/pre ischemia) in protein
expression found in the LV of CAD patients as a result of ischemic
cardioplegic arrest and reperfusion. The expression of 24 proteins
were significantly altered, of which 19 demonstrated increased
expression and 5 decreased expression (Table SI). A heat map (Fig. 1B) shows the changes in individual
biopsies. Several of the proteins with the greatest increase in
expression are likely to have emanated from components of the
systemic circulation (e.g. platelet glycoprotein Iβ, integrin
α-IIb, neutrophil elastase and myeloperoxidase). Other proteins
which were differentially expressed are of cardiac origin, such as
the mitochondrial calcium uniporter regulator 1, NADH dehydrogenase
flavoprotein 3 and GLUT3. Gene Ontology (GO) analysis of these
proteins indicates involvement of biological processes that are
associated with changes in immune responses (Fig. 1C) while IPA canonical pathway
analysis highlighted significant enrichment of pathways involved in
protein translation (eukaryotic initiation factors 2 and 4,
Table II).
 | Table IIEnriched canonical pathways for the
total protein analysis of the LV and RV of CAD patients. |
Table II
Enriched canonical pathways for the
total protein analysis of the LV and RV of CAD patients.
| Ventricle | Ingenuity canonical
pathway | P-value of
overlap | Molecules |
|---|
| LV | Airway pathology in
chronic obstructive pulmonary disease | 0.004 | ELANE, MPO |
| Melatonin
degradation III | 0.005 | MPO |
| Regulation of eIF4
and p70S6K signalling | 0.026 | AGO1, FAU,
ITGA2B |
| Airway inflammation
in asthma | 0.030 | ELANE |
| EIF2
signalling | 0.045 | AGO1, FAU,
RPL36 |
| RV | Mitochondrial
dysfunction |
1.0×10−11 | ATP5PD, COX7A2L,
CYB5A, CYC1, MAOA, MAOB, MAPK9, NDUFA10, NDUFA5, NDUFA7, NDUFB1,
NDUFB10, NDUFB2, NDUFB6, NDUFB7, NDUFS8, NDUFV1, NDUFV2, NDUFV3,
UQCRQ |
| Oxidative
phosphorylation |
1.3×10−11 | ATP5PD, COX7A2L,
CYB5A, CYC1, NDUFA10, NDUFA5, NDUFA7, NDUFB1, NDUFB10, NDUFB2,
NDUFB6, NDUFB7, NDUFS8, NDUFV1, NDUFV2, NDUFV3, UQCRQ |
| Sirtuin signalling
pathway |
1.4×10−6 | CYC1, NDUFA10,
NDUFA5, NDUFA7, NDUFB1, NDUFB10, NDUFB2, NDUFB6, NDUFB7, NDUFS8,
NDUFV1, NDUFV2, NDUFV3, PARP1, PPIF, SIRT2 |
| Estrogen receptor
signalling |
4.4×10−6 | CYC1, MYL3, MYL4,
MYL7, NDUFA10, NDUFA5, NDUFA7, NDUFB1, NDUFB10, NDUFB2, NDUFB6,
NDUFB7, NDUFS8, NDUFV1, NDUFV2, NDUFV3, RALA |
| TCA cycle II
(eukaryotic) |
4.9×10−6 | CS, DLST, FH,
IDH3A, IDH3G, SUCLG1 |
| Serotonin receptor
signalling |
1.6×10−5 | MAOA, MAOB, QDPR,
SPR |
| Glucocorticoid
receptor signalling |
2.2×10−5 | CYC1, FGG, HSPA5,
MAPK9, NDUFA10, NDUFA5, NDUFA7, NDUFB1, NDUFB10, NDUFB2, NDUFB6,
NDUFB7, NDUFS8, NDUFV1, NDUFV2, NDUFV3, RALA |
| Agranulocyte
adhesion and diapedesis |
4.7×10−5 | MYH3, MYH6, MYH7,
MYH9, MYL3, MYL4, MYL7, PODXL |
| Calcium
signalling |
1.6×10−4 | MYH3, MYH6, MYH7,
MYH9, MYL3, MYL4, MYL7, TNNI1, TRDN |
| Melatonin
degradation II |
5.1×10−4 | MAOA, MAOB |
| Hepatic
fibrosis/hepatic stellate cell activation |
9.5×10−4 | MYH3, MYH6, MYH7,
MYH9, MYL3, MYL4, MYL7 |
| Dilated
cardiomyopathy signalling pathway |
1.0×10−3 | MYH3, MYH6, MYH7,
MYH9, MYL3, MYL4, MYL7, TNNI1 |
| Cellular effects of
sildenafil (Viagra) |
1.4×10−3 | MYH3, MYH6, MYH7,
MYH9, MYL3, MYL4, MYL7 |
| ILK signalling |
1.5×10−3 | MAPK9, MYH3, MYH6,
MYH7, MYH9, MYL3, MYL4, MYL7, RHOT1 |
| Dopamine receptor
signalling |
1.9×10−3 | MAOA, MAOB,
PPP1R14C, QDPR, SPR |
| Tight junction
signalling |
2.0×10−3 | MYH3, MYH6, MYH7,
MYH9, MYL3, MYL4, MYL7, VAPA |
| Gα12/13
signalling |
2.6×10−3 | F2, MAPK9, MYL3,
MYL4, MYL7, RALA |
| Actin cytoskeleton
signalling |
6.3×10−3 | F2, MYH3, MYH6,
MYH7, MYH9, MYL3, MYL4, MYL7, RALA |
| RHOGDI
signalling |
6.8×10−3 | MYH3, MYH6, MYH7,
MYH9, MYL3, MYL4, MYL7, RHOT1 |
| Phenylalanine
degradation IV (mammalian, via side chain) |
1.3×10−2 | MAOA, MAOB |
Phosphoprotein analysis for the LV of CAD patients
revealed 28 phosphoproteins differentially expressed in pre vs.
post samples when adjusted for total protein content (Table SII). A total of 23
phosphoproteins showed increased expression while 5 showed
decreased expression. A volcano plot can be found in Fig. S1A. IPA canonical pathway
analysis (Table III)
highlighted enrichment of signalling pathways relating to several
proinflammatory cytokines [interleukin (IL)-6 and IL-22].
 | Table IIIEnriched canonical pathways for the
relative phosphoprotein analysis of the LV and RV of CAD
patients. |
Table III
Enriched canonical pathways for the
relative phosphoprotein analysis of the LV and RV of CAD
patients.
| Ventricle | Ingenuity canonical
pathway | P-value of
overlap | Molecules |
|---|
| LV | Endocannabinoid
cancer inhibition pathway | 7.08E-06 | MAPK14, MAPK3,
PRKAR1A, VIM |
| Amyloid
processing | 1.23E-05 | MAPK14, MAPK3,
PRKAR1A |
| Sertoli
cell-sertoli cell junction signalling | 3.02E-05 | MAPK14, MAPK3,
PRKAR1A, SPTBN1 |
| Melatonin
signalling | 3.47E-05 | MAPK3, PRKAR1A,
SLC2A4 |
| Apelin adipocyte
signalling pathway | 5.89E-05 | MAPK14, MAPK3,
PRKAR1A |
| BMP signalling
pathway | 6.17E-05 | MAPK14, MAPK3,
PRKAR1A |
| Cardiac hypertrophy
signalling | 7.24E-05 | HSPB1, MAPK14,
MAPK3, PRKAR1A |
| Insulin secretion
signalling pathway | 8.32E-05 | MAPK14, MAPK3,
PRKAR1A, SLC2A4 |
| Antioxidant action
of vitamin C | 1.26E-04 | MAPK14, MAPK3,
SLC2A4 |
| CDK5
signalling | 1.29E-04 | MAPK14, MAPK3,
PRKAR1A |
| Renin-angiotensin
signalling | 1.58E-04 | MAPK14, MAPK3,
PRKAR1A |
| Gαs signalling | 1.62E-04 | ADD1, MAPK3,
PRKAR1A |
| Endocannabinoid
developing neuron pathway | 1.70E-04 | MAPK14, MAPK3,
PRKAR1A |
| IL-6
signalling | 1.95E-04 | HSPB1, MAPK14,
MAPK3 |
| IL-22
signalling | 2.04E-04 | MAPK14, MAPK3 |
| Role of JAK family
kinases in IL-6-type cyto kine signalling | 2.24E-04 | MAPK14, MAPK3 |
| IL-17A signalling
in gastric cells | 2.40E-04 | MAPK14, MAPK3 |
| Insulin receptor
signalling | 2.51E-04 | MAPK3, PRKAR1A,
SLC2A4 |
| Dilated
cardiomyopathy signalling pathway | 2.82E-04 | MAPK14, MAPK3,
PRKAR1A |
| Endocannabinoid
neuronal synapse pathway | 2.88E-04 | MAPK14, MAPK3,
PRKAR1A |
| RV | p38 MAPK
signalling | 0.001 | HSPB1, MAPK14 |
| IL-6
signalling | 0.001 | HSPB1, MAPK14 |
| Acute phase
response signalling | 0.003 | HNRNPK, MAPK14 |
| Cardiac hypertrophy
signalling | 0.005 | HSPB1, MAPK14 |
| Parkinson's
signalling | 0.007 | MAPK14 |
| IL-22
signalling | 0.010 | MAPK14 |
| Role of JAK family
kinases in IL-6-type cytokine signalling | 0.010 | MAPK14 |
| IL-17A signalling
in gastric cells | 0.011 | MAPK14 |
| 4-1BB signalling in
T lymphocytes | 0.014 | MAPK14 |
| Inhibition of
angiogenesis by TSP1 | 0.014 | MAPK14 |
| IL-17A signalling
in fibroblasts | 0.016 | MAPK14 |
| April mediated
signalling | 0.018 | MAPK14 |
| B cell activating
factor signalling | 0.018 | MAPK14 |
| nNOS signalling in
skeletal muscle cells | 0.020 | SNTA1 |
| iNOS
signalling | 0.020 | MAPK14 |
| Cardiac hypertrophy
signalling (enhanced) | 0.021 | HSPB1, MAPK14 |
| Amyloid
processing | 0.021 | MAPK14 |
| UVC-induced MAPK
signalling | 0.021 | MAPK14 |
| UVB-induced MAPK
signalling | 0.022 | MAPK14 |
| EGF signalling | 0.023 | MAPK14 |
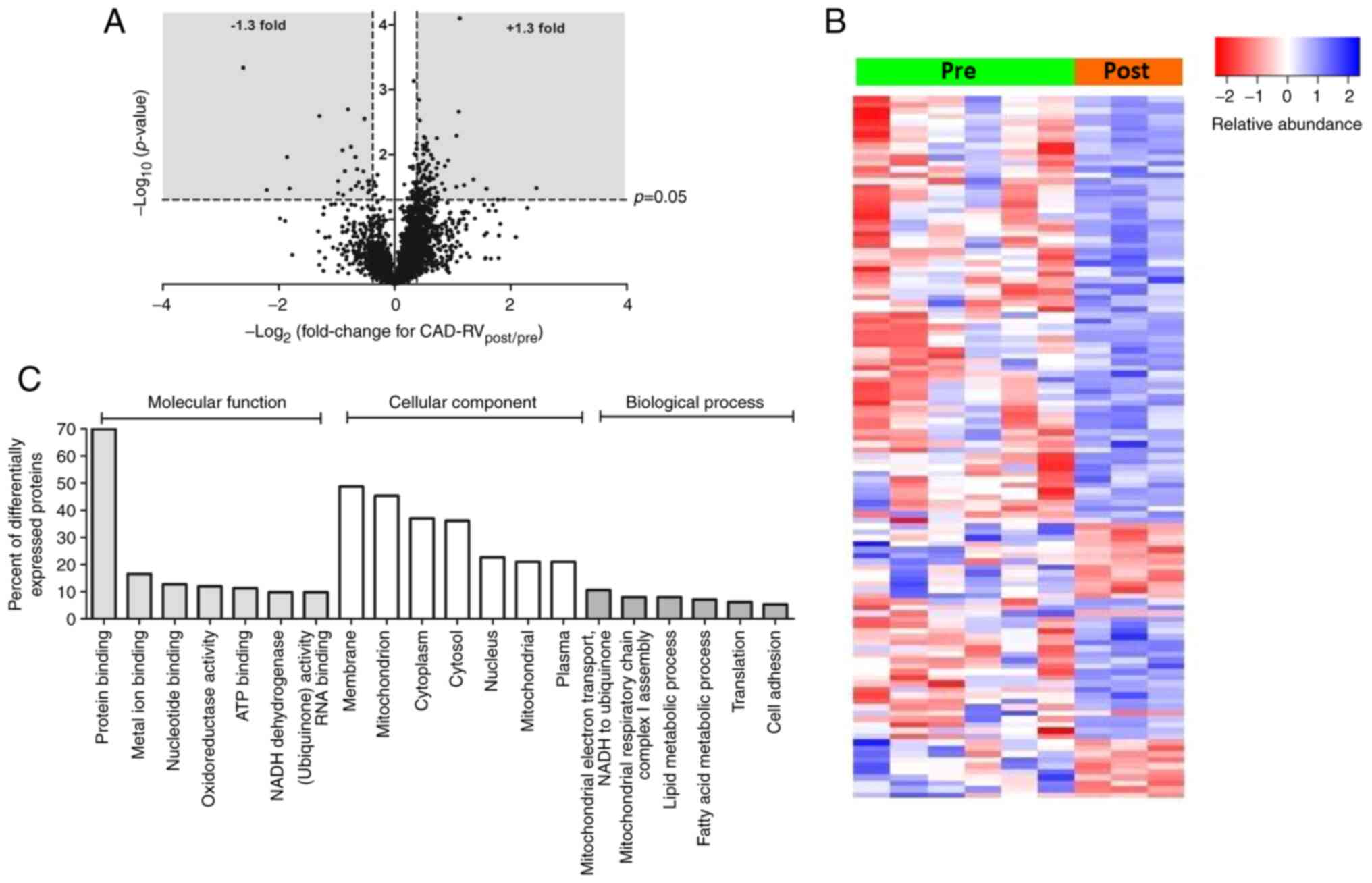
In the RV of CAD patients, 120 proteins showed a
significant change in expression between pre and post samples, with
the majority (95 proteins) showing an increase in expression
(Fig. 2A and Table SIII). Many of these are
mitochondrial proteins, including several proteins associated with
NADH dehydrogenase or the TCA cycle. As with the LV, there was also
elevation of several proteins known to originate in the circulation
(e.g. blood components). Several of the proteins that decreased in
expression as a result of surgery were related to the contractile
myofilaments. A heat map analysis shows the fold change (FC) levels
for individual biopsies and clearly indicate that despite the
smaller number of post biopsies, the effects were marked (Fig. 2B). Further ontology analysis of
these proteins indicated involvement of biological processes that
are associated with changes in metabolic/energetic processes,
several of which relate to mitochondria (Fig. 2C). Similarly, IPA canonical
pathway analysis (Table II)
revealed mitochondrial dysfunction to be a highly enriched pathway
(P-value of overlap 1.0×10−11).
Phosphoprotein analysis for the RV of CAD patients
revealed 13 phosphoproteins differentially expressed in pre vs.
post samples when adjusted for total protein content (9 with
increased expression and 4 with decreased expression; Table SIV and Fig. S1B). Similarily for
the LV, phosphoprotein analysis for this group of patients, IPA
canonical pathway analysis of the RV phosphoproteome highlighted
several inflammatory pathways as being significantly enriched,
including IL-6 signalling, IL-22 signalling and acute phase
response signalling (Table
III).
Effect of ischemic cardioplegic arrest on
cardiac proteome/phosphoproteome in LV and RV of patients with
aortic valve stenosis (AVS)
For AVS patients, 3,220 proteins were detected in
all pre and post ischemic biopsies collected from the LV (n=6 and
n=5 for pre and post samples respectively) and 3,208proteins were
detected in all pre and post biopsies collected from the RV (n=5
and n=4). Only these proteins were used in our analysis (see
exclusions in Materials and methods).
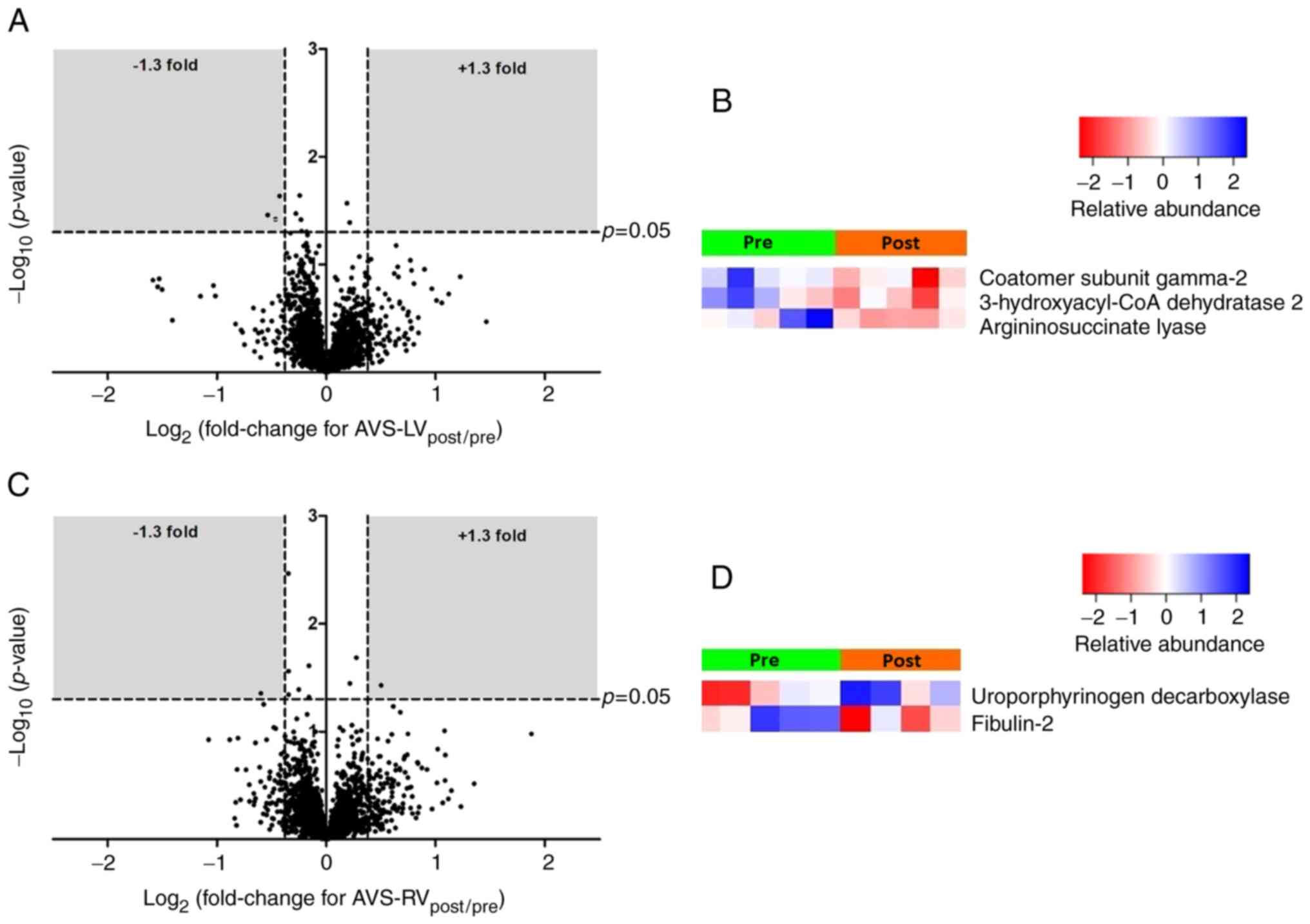
Fig. 3 shows
volcano plots and heatmaps for the LV (Fig. 3A and B) and RV (Fig. 3C and D) of AVS patients. The
differentially expressed proteins for both ventricles are listed in
Table SV. In the LV, the
expression of three proteins [coatomer subunit gamma-2,
very-long-chain (3R)-3-hydroxyacyl-CoA dehydratase 2 and
argininosuccinate lyase] was significantly decreased. In the RV,
one protein showed increased expression (uroporphyrin
decarboxylase) and one protein showed decreased expression
(fibulin-2). IPA canonical pathway analysis (Table SVI) was uninformative given the
small number of proteins involved.
Analysis of phosphoprotein expression (relative to
total protein expression) in AVS patients (Table SVII) found 14 phosphoproteins
with altered expression in the LV (11 increased and 3 decreased,
Fig. S1C) and 4 proteins with
altered expression in the RV (all with increased expression,
Fig. S1D). Canonical pathway
analysis (Table IV) found
enrichment of several pro-inflammatory pathways, including IL-6 in
both ventricles and IL-17 in the LV.
 | Table IVEnriched canonical pathways for the
relative phosphoprotein analysis of the LV and RV of AVS
patients. |
Table IV
Enriched canonical pathways for the
relative phosphoprotein analysis of the LV and RV of AVS
patients.
| Ventricle | Ingenuity canonical
pathway | P-value of
overlap | Molecules |
|---|
| LV | Dilated
cardiomyopathy signalling pathway | 4.90E-05 | GSK3B, MAPK14,
TTN |
| Adrenomedullin
signalling pathway | 1.23E-04 | GSK3B, MAPK14,
TTN |
| IL-17A signalling
in fibroblasts | 1.66E-04 | GSK3B, MAPK14 |
| Cardiac hypertrophy
signalling | 2.63E-04 | GSK3B, HSPB1,
MAPK14 |
| Amyloid
processing | 2.95E-04 | GSK3B, MAPK14 |
| IL-17A signalling
in airway cells | 5.13E-04 | GSK3B, MAPK14 |
| IL-7 signalling
pathway | 6.92E-04 | GSK3B, MAPK14 |
| ERBB
signalling | 1.00E-03 | GSK3B, MAPK14 |
| ATM signalling | 1.07E-03 | HP1BP3, MAPK14 |
| p53 signalling | 1.10E-03 | GSK3B, MAPK14 |
| Hepatic fibrosis
signalling pathway | 1.10E-03 | GSK3B, MAPK14,
TTN |
| Mouse embryonic
stem cell pluripotency | 1.23E-03 | GSK3B, MAPK14 |
| p38 MAPK
signalling | 1.58E-03 | HSPB1, MAPK14 |
| Endocannabinoid
developing neuron pathway | 1.74E-03 | GSK3B, MAPK14 |
| IL-6
signalling | 1.86E-03 | HSPB1, MAPK14 |
| Cardiac hypertrophy
signalling (enhanced) | 2.24E-03 | GSK3B, HSPB1,
MAPK14 |
| Endocannabinoid
cancer inhibition pathway | 2.29E-03 | GSK3B, MAPK14 |
| Factors promoting
cardiogenesis in vertebrates | 2.57E-03 | GSK3B, MAPK14 |
| WNT/β-catenin
signalling | 3.39E-03 | APPL1, GSK3B |
| IL-17
signalling | 3.89E-03 | GSK3B, MAPK14 |
| RV | Aldosterone
signalling in epithelial cells | 0.000 | CRYAB, HSPB1 |
| Protein
ubiquitination pathway | 0.001 | CRYAB, HSPB1 |
| Death receptor
signalling | 0.020 | HSPB1 |
| p38 MAPK
signalling | 0.025 | HSPB1 |
| Ferroptosis
signalling pathway | 0.026 | HSPB1 |
| IL-6
signalling | 0.027 | HSPB1 |
| Aryl hydrocarbon
receptor signalling | 0.033 | HSPB1 |
| ERK/MAPK
signalling | 0.045 | HSPB1 |
Comparision of protein/phosphoprotein
expression in AVS patients compared to CAD patients pre-ischaemic
cardioplegic arrest
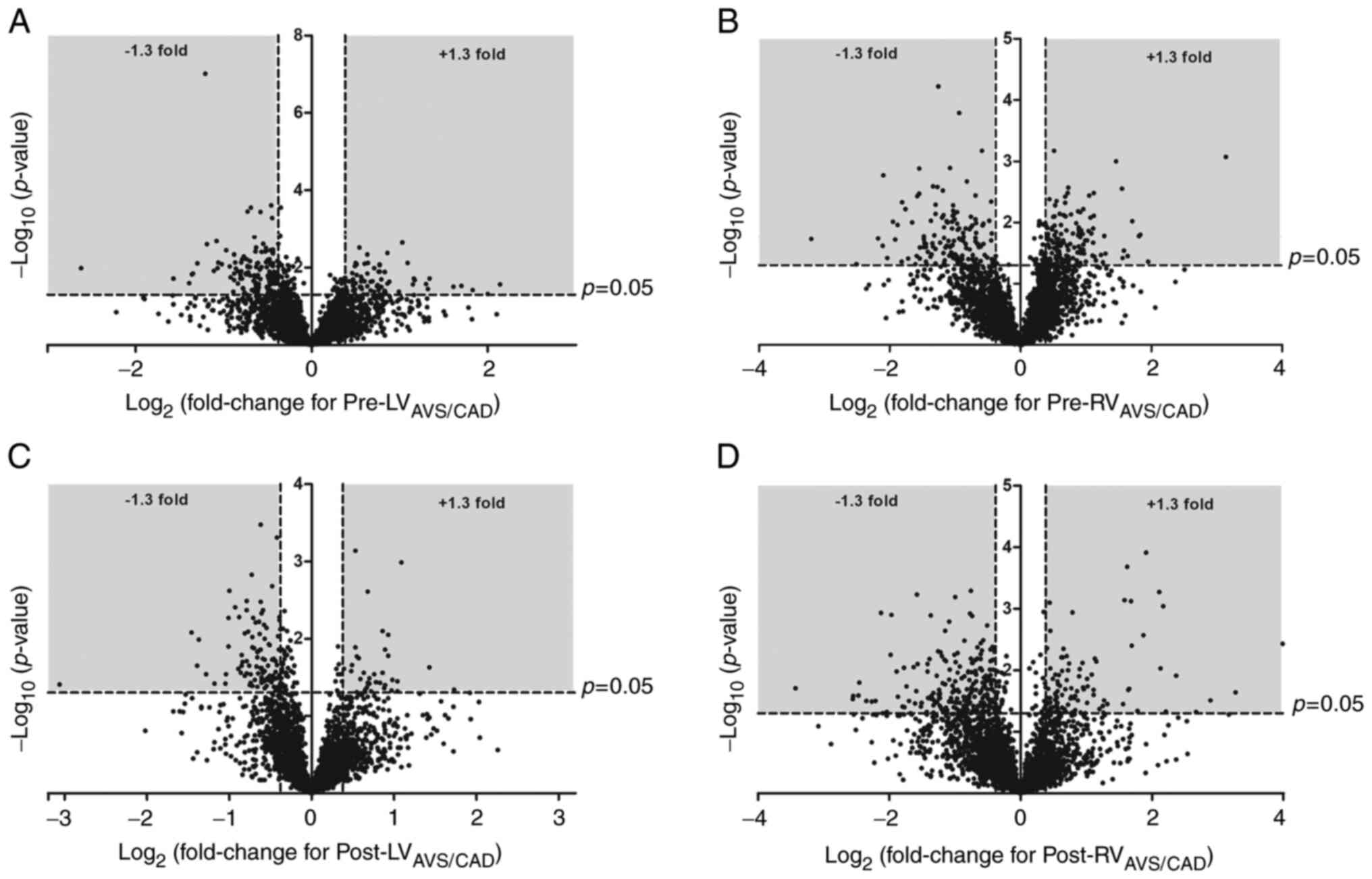
A total of 3,009 proteins were detected in all LV
biopsies from AVS (n=6) and CAD (n=6) patients pre-ischaemic
cardioplegic arrest while 3,000 were found in all RV biopsies from
AVS (n=5) and CAD (n=6) patients pre-ischaemic cardioplegic arrest.
In the LV, the expression of 206 proteins were significantly
altered (135 with decreased expression in AVS patients relative to
CAD patients and 71 with increased expression). The differentially
expressed proteins are listed in Table SVIII, and a volcano plot can be
found in Fig. 4A. In the RV, the
expression of 273 proteins were significantly altered (140 with
decreased expression in AVS patients relative to CAD patients and
133 with increased expression; Table SIX and Fig. 4B). This included a significant
underexpression of MAP2K1 and MAP2K2 in AVS patients compared to
CAD patients. IPA canonical pathway analysis (Table SX) highlighted phospholipase C
signalling as the most significantly enriched pathway in the LV
(P-value of overlap=4.57×10−5); other significantly
enriched pathways included Gαs signalling and
α-adrenergic signalling. In the RV, several canonical pathways
relating to inflammation were enriched, including acute phase
response signalling, IL-2 signalling and complement system
pathways.
Phosphoproteomic analysis found 24 significantly
enriched phosphoproteins in the LV pre-ischaemic cardioplegic
arrest comparision (7 with increased expression in AVS patients
relative to CAD patients and 17 with decreased expression; Table SXI and Fig. S2A) and 11
significantly enriched phosphoproteins in the RV pre-ischaemic
cardioplegic arrest comparision (7 with increased expression in AVS
patients relative to CAD patients and 4 with decreased expression;
Table SXII and Fig. S2B).
Notable differences in the LV include a significant decrease in
phosphorylated myosin heavy chains-11 and -9 in the LV of AVS
patients compared to CAD patients. IPA canonical signalling pathway
analysis (Table SXIII)
highlighted several enriched pathways including PKA signalling in
both ventricles and cardiac β-adrenergic signalling and calcium
signalling in the LV.
Comparision of protein/phosphoprotein
expression in AVS patients compared to CAD patients
post-reperfusion
A total of 2,981 proteins were detected in all LV
biopsies from AVS (n=5) and CAD (n=5) patients post-reperfusion
while 3,218 were found in all RV biopsies from AVS (n=4) and CAD
(n=3) patients post-reperfusion. In the LV, the expression of 135
proteins were significantly different between groups (99 with
decreased expression in AVS patients relative to CAD patients and
36 with increased expression; Table
SXIV and Fig. 4C). In the
RV, the expression of 314 proteins were significantly altered (217
with decreased expression in AVS patients relative to CAD patients
and 97 with increased expression; Table SXV and Fig. 4D). IPA canonical pathway analysis
(Table SXVI) highlighted the
significant enrichment of several pathways involved in inflammation
(IL-6 and IL-2 signalling in the LV and IL-8 signalling and
complement system pathways in the RV) as well as calcium signalling
pathways in the RV.
Phosphoproteomic analysis found 19 significantly
differentially expressed phosphoproteins in the LV post-reperfusion
comparision (4 with increased expression in AVS patients relative
to CAD patients and 15 with decreased expression; Table SXVII and Fig. S2C) and 20
significantly differentially expressed phosphoproteins in the RV
post-reperfusion comparision (9 with increased expression in AVS
patients relative to CAD patients and 11 with decreased expression;
Table SXVIII and Fig. S2D). IPA
canonical signalling pathway analysis (Table SXIX) highlighted several
enriched pathways including PKA signalling, cAMP-mediated
signalling and calcium signalling in the LV.
Discussion
In this study, proteomic analysis involving tandem
mass tagging followed by reverse phase nano-liquid chromatography
mass spectrometry/mass spectrometry (LC-MS/MS) was utilized to
investigate the effect of open heart surgery using cold blood
cardioplegic arrest and reperfusion on the changes in protein
levels in biopsies collected from left ventricles (LVs) and right
venticles (RVs) of patients with either corony artery disease (CAD)
or aortic valve stenosis (AVS).
The length of time spent on cross-clamp (ischemic
cardioplegic arrest) was significantly longer in AVS patients
compared to CAD patients. Surprisingly, despite this difference,
CAD patients demonstrated far greater changes in protein expression
than AVS patients. This study shows for the first time that the
diseased LV of AVS patients shows little change in its proteome
during surgery compared to LV of CAD patients. Similarly, the
relatively normal RV of AVS patients show little change in its
proteome compared to RV proteome of CAD patients which demonstrated
the greatest number of changes in protein expression including
altered expression of a significant number of mitochondrial
proteins.
Cardioplegic arrest and reperfusion
increase proteins of systemic origin in both LV and RV of CAD
patients
It is widely assumed that proteins measured in
cardiac biopsies are related to the main cell types of the
myocardium (e.g. myocytes, smooth muscle cells, fibroblasts,
endothelia cells). However, the possibility that some of the
proteins have come from blood components (e.g. fragmented
platelets, neutrophils) cannot be discounted. This is particularly
relevant during reperfusion where neutrophils are known to
penetrate the stressed/injured myocardium. Additionally, platelets
fragmented by cardiopulmonary bypass would penetrate the
vasculature and become attached to the myocardium. We found a
significant increase in platelet proteins including integrin α-IIb
[which is an important mediator of platelet aggregation via the
integrin α(IIb)β(3) complex] and
platelet membrane glycoprotein Ib β in the LV of CAD patients post
surgery.
It is important to point out that integrins, which
are fast turnover focal adhesion proteins, are also found in
cardiomyocytes and act to connect the Z-disc with the sarcolemma
(17). Mechanical forces
including stretching of cardiomyocytes have been shown to markedly
increase integrin expression (18). Furthermore β3 integrin has been
implicated in cardiac hypertrophy and associated signaling
(19–21) and cardiac repair including
fibrosis after myocardial infarction (22–24). Unlike β3 integrin, little is
known about the role/significance of α2 integrin in cardiomyocytes
but it has an important role in cell adhesion and motility, and
immune/blood cell regulation when bound to β1 integrin (25).
Other proteins that increase in LV of CAD patients
during surgery which are likely to originate outside the myocardium
include neutrophil elastase [likely due to activation of
neutrophils triggered by cardiopulmonary bypass (26)], coronin-1A [predominantly
expressed in leukocytes and important for integrin-mediated
leukocyte adhesion (27)] and
myeloperoxidase [likely due to its release by neutrophils, and
linked to adverse left ventricular remodeling following myocardial
ischemia (28)]. Finally,
cathelicidin antimicrobial peptide was also increased;
cathelicidins are peptides which are expressed at high levels in
neutrophils and some epithelia and can act as a natural antibiotic
(29). Cathelicidin
antimicrobial peptide has been shown to protect cardiomyocytes
against ischemia/reperfusion (I/R) injury via activation of
survival signaling (30,31).
Similarly, there was also an increase in proteins
associated with systemic blood components in the RV of CAD patients
during surgery. These included platelet glycoprotein 4, protein
disulfide-isomerase A6 (which plays a role in platelet aggregation
and activation), and Ras-related protein Ral-A which is abundantly
present in human platelets.
Cardioplegic arrest and reperfusion
trigger changes in expression of proteins involved in translation
and inflammation in LV and RV of CAD patients
Exposure to CPB and cardioplegic arrest is known to
induce inflammation (32), and
indeed canonical pathway analysis of the phosphoprotein datasets
found a significant enrichment of inflammatory canonical signalling
pathways including IL-6, IL-22, IL-17 and acute phase response
signalling in the RV and IL-6 and IL-22 in the LV. These changes
were due in part to a significant increase in the relative
expression of phosphorylated MAPK14 and phosphorylated heat shock
protein β-1 (HSPβ1) in both ventricles, and phosphorylated MAPK3 in
the LV.
40S ribosomal protein S30 and 60S ribosomal protein
L36 were 2 of only 5 proteins to demonstrate decreased expression
in the LV of CAD patients following surgery. These are
ribonucleoproteins involved in RNA processing and DNA repair
(32). Canonical pathway
analysis demonstrated that changes in the expression of these
ribosomal proteins contributed to significant enrichment of
eukaryotic initiation factor (eIF) signalling pathways, which are
involved in translation. Protein translation was also affected in
the RV, which demonstrated significant increases in the expression
of eIF3E, phospho-eIF4B and 60S acidic ribosomal protein P0.
Cardioplegic arrest and reperfusion
trigger changes in mitochondrial proteome consistent with
mitochondrial impairment, especially in the RV of CAD patients
The signifcant alteration in mitochondrial proteins
in the RV of CAD patients is evident in both the canonical pathway
analysis, which lists mitochondrial dysfunction as a highly
enriched pathway (Table II),
and the Gene Ontology analysis (Fig.
2C). In contrast, the LV of CAD patients shows relatively few
changes in the expression of mitochondrial proteins. These
differences are unlikley to be due to the disease status as all 6
CAD patients had 3 similar vessel disease (see Materials and
methods). Significant dynamic changes occur to the mitochondria
during I/R which include swelling and fission which are likely to
require additional mitochondrial protein synthesis. Recent research
involving paired atrial biopsies collected before and after
cardiopulmonary bypass (CPB) with cold cardioplegia for CAD and/or
valve surgery has demonstrated that these procedures are associated
with mitochondrial DNA damage and biogenesis (33).
A key finding in the present study is the increase
in expression of several proteins (NADH dehydrogenase flavoprotein
1, flavoprotein 2, flavoprotein 3, 1 α subcomplex subunit 5, 1 β
subcomplex subunit 6, 1 β subcomplex subunit 7) associated with
mitochondrial complex I in the RV of CAD patients. In the LV, only
flavoprotein 3 demonstrated increased expression. Complex I is the
largest and most complicated enzyme of the respiratory chain, known
to release deleterious oxygen radicals and whose dysfunction has
been linked to a number of hereditary and degenerative diseases
(34).
Several proteins increased in the mitochondria in
the RV were related to lipid metabolism and the tricarboxylic acid
(TCA) cycle. Both delta(3,5)-Delta(2,4)-dienoyl-CoA isomerase and
methylmalonyl-CoA epimerase are involved in fatty acid β-oxidation
and short-chain fatty acid catabolic process. Proteins associated
with the TCA cycle include malate dehydrogenase, isocitrate
dehydrogenase and fumarylacetoacetate hydrolase. The latter
catalyzes the hydrolysis of 4-fumarylacetoacetate into acetoacetate
and fumarate which end up in the TCA cycle, or are used for
biosynthetic purposes (35).
Signalling relating to the TCA cycle was also highlighted as a
significant canonical pathway for the RV (P-value of
overlap=4.9×10−6).
Other mitochondrial proteins that increased during
surgery in the RV include proteins involved in protein biosynthesis
(mitochondrial Isoleucine--tRNA ligase), mitochondrial
dicarboxylate carrier (solute carrier family 25 member 10) and the
optic atrophy 3 protein (OPA3) which is in the inner mitochondrial
membrane (36) and implicated in
dilated cardiomyopathy (37).
The expression of adenylate kinase 2 and ATP synthase protein 8
were also increased in the RV; these proteins play important roles
in cellular energy homeostasis and ATP synthesis. In the LV, there
was an increase in the expression of mitochondrial calcium
uniporter regulator 1 [an activator of the mitochondria
Ca2+ uniporter that is responsible for mitochondrial
calcium uptake (38), a process
that is augmented during I/R]. Interestingly, there was also a
decrease in the expression of the mitochondrial transmembrane
protein 70 (TMEM70) in the LV of CAD patients; TMEM70 facilitates
the biogenesis of mammalian F1Fo ATP synthase and knockout of this
protein is lethal as it impairs mitochondrial energy production
(39).
Cardioplegic arrest and reperfusion
trigger changes in contractile machinary proteins in RV of CAD
patients following cardioplegic arrest and reperfusion
There was a decrease in motor proteins (mostly
myosin-related) in the RV of CAD patients. These were MYH3
(developmental), MYH7, MYH6 (hypertrophic and dilated
cardiomyopathy), myosin light chain 3, unconventional myosin-Ib,
F-actin-capping protein subunit β (which plays a role in the
regulation of cell morphology and cytoskeletal organization),
kinesin-like protein KIF1C (involved in cytoskeleton-dependent
intracellular transport) and 39S ribosomal protein L17 (involved in
mitochondrial translational elongation and termination).
Interestingly, the contractile machinary of the heart appears to be
more affected by surgery in the RV compared to the LV. It is
difficult to address this issue without having detailed pre- and
post-operative functional/structural data using a large number of
patients. There have been suggestions that the RV of Coronary
artery bypass grafting (CABG) patients selectively sustains more
disruptions following CABG surgery (40–42). It is likely that the adaptation
to the disease is different between LV and RV and therefore could
alter vulnerability to I/R.
Small number of changes in protein and
phosphoprotein expression during cardioplegic arrest and
reperfusion in both LV and RV of AVS patients
A major finding in this study was that surgery
caused very few changes in the proteome of either ventricle of AVS
patients. There was a decrease in the expression of only 3 proteins
in the LV (involved in protein cellular transport, fatty acid
metabolism and urea synthesis) and altered expression of only 2
proteins in the RV (an extracellular matrix glycoprotein and a
protein involved in heme synthesis). The surprising observation
that there are far fewer changes in the total protein expression of
AVS patients compared to CAD patients suggests that cold ischemic
cardioplegic arrest and reperfusion of a heart with hypertrophic LV
does not impose significant molecular stress. This is in contrast
to the coronary diseased LV and is consistent with proposed
strategies to protect the hypertrophic heart (43).
The phosphoproteome showed slightly more
significance, with 14 differentially expressed phosphoproteins in
the LV and 4 in the RV. Here there are some similarities to the
phosphoproteome analysis in CAD patients, with a significant
increase in the relative expression of HSPβ1 in both ventricles and
a significant increase in the relative expression of MAPK14 in the
LV. As with the CAD patients, these findings cause significant
enrichment of pro-inflammatory signalling pathways (IL-6 signalling
in both ventricles and also IL-17 signialling in the LV).
Comparison between the
proteome/phosphoproteome of the LV and RV of AVS patients compared
to CAD patients pre-ischemia and at post reperfusion
The data obtained in this study have been used to
update a previous study authored by Littlejohns et al
(11) in which the pre-ischemic
proteomes of the LV and RV of AVS patients were compared with the
corresponding ventricles of CAD. In this previous study,
approximately 500 proteins were identified in all biopsies. By
contrast, in the present study given the advancement of proteomic
technology and a larger sample size, we were able to identify
approximately 3,000 proteins present in all biopsies for each
comparision. Of these detectable proteins, 206 were differentially
expressed between AVS and CAD patients in the LV biopsies, and 273
in the RV biopsies taken pre-ischaemic cardioplegic arrest. In the
early study only 9 and 73 differentially expressed proteins were
detected in the LV and RV biopsies, respectively. Thus, the present
study allows us to expore the differences between pathologies in
greater detail.
Differences in adrenergic and calcium
signalling pathways between LV and RV of AVS and corresponding LV
and RV of CAD patients before ischemia and after reperfusion
IPA canonical pathway analysis of the LV
pre-ischaemic cardioplegic arrest samples identified phospholipase
C (PLC) signalling as the most significantly enriched canonical
pathway. Other enriched pathways included Gαs signalling
and α-adrenergic signalling. Similarly, the phosphoprotein analysis
also identified several enriched pathways relating to adrenergic
signalling (e.g. protein kinase A signalling in both ventricles,
and cardiac β-adrenergic signalling in the LV). Calcium signalling
was also identified as a significantly enriched canonical pathway
in the comparision of the LV phosphoproteome of samples taken
pre-ischaemic cardioplegic arrest, driven largely by changes in
expression of isoforms of myosin heavy chain such as MYH9 and
MYH11, both of which are significantly underexpressed in AVS
patients compared to CAD patients. PLC and α- and β-adrenergic
signalling pathways are all known to be affected by or implicated
in cardiac hypertrophy (44–46), while the expression of myosin
heavy chains is also known to be altered in hypertrophic or failing
hearts (47,48). It is likely therefore that
differences in adrenergic and calcium signalling pathways
pre-ischaemic cardioplegic arrest are the result of the different
effects of the two pathology types on the basal physiological state
of the hearts.
Post-reperfusion, there were also differences in
adrenergic and calcium signalling pathways; PKA signalling and
cAMP-mediated signalling were identified as significantly enriched
phosphoprotein canonical pathways in the post-reperfusion LV
samples and calcium signalling was a significantly enriched
canonical pathway in both the RV total proteome and the LV
phosphoproteome post-reperfusion analysis. These differences may
represent a continuation of the basal differences already noted,
but may also be affected by different responses of the two types of
heart to exposure to cardiopulmonary bypass and cardioplegic
arrest.
Differences in inflammatory pathways
between LV and RV of AVS and corresponding LV and RV of CAD
patients before ischemia and after reperfusion
Differences in inflammatory pathways also exist
between the two pathologies. In the RV pre-ischaemic cardioplegic
arrest biopsies the most significantly enriched canonical pathway
was acute phase response signalling (a group of proteins which
respond to inflamation); IL-2 signalling (a pro-inflammatory
cytokine) and complement system pathways were also significantly
enriched. These effects were in part due to increased expression of
MAP2K1 and MAP2K2 in the RV pre-ischaemic cardioplegic arrest
samples from CAD patients compared to AVS patients. Differences in
basal levels of inflammation are likely to exist between CAD and
AVS hearts-for example one study demonstrated increased systemic
levels of pro-inflammatory IL-3 in patients with CAD (49).
There are also differences in inflammatory pathways
between AVS and CAD hearts post-reperfusion. In the LV, there was
significant enrichment of IL-6 and IL-2 canonical signalling
pathways while in the RV there was significant enrichment of IL-8
signalling as well as complement system pathways. As previously
mentioned, exposure to CPB and cardioplegic arrest is known to
induce inflammation (32); our
results suggest that the degree of inflammation was different
between patients with different pathologies. This may relate to the
fact that the cross-clamp time was longer in the AVS group, but may
also reflect an influence of the underlying pathology on the
inflammatory response to surgery.
In conclusion, the study presented in this
manuscript demonstrates for the first time that LVs and RVs of
patients with either AVS or CAD respond differently at the
molecular level to cold ischemic cardioplegic arrest and
reperfusion. These molecular changes are disease-dependent, with
CAD patients showing a larger number of proteins changing during
the surgery despite having shorter cardioplegic arrest; notable
changes include a higher number of differentially expressed
mitochondrial proteins and lower number of contractile proteins in
the RV of CAD patients. Enrichment analysis indicates involvement
of canonical pathways associated with inflammation, mitochondria
and contractility. In addition to intra-disease changes, each
ventriclular chamber showed inter-disease differences measured
before and after cardioplegic arrest. It is important to note that
this is largely a descriptive study designed to report the changes
in the proteome/phosphoproteome in different ventricular chambers
of two groups of patients with different pathologies. Further
research is needed to identify the mechanisms involved and to
investigate the significance of these changes. In particular,
future research could investigate the relevant functional
impairment and differences between sub-groups of patients, like
those with differing degrees of CAD, sex differences and the
presence of co-morbidities.
Supplementary Data
Availability of data and materials
The datasets used and/or analysed during the current
study are publically available at Open Science Framework
(https://doi.org/10.17605/OSF.IO/BPJGX).
Authors' contributions
Conceptualization of the study was achieved by MSS,
GDA and BCR. Formal analysis was conducted by SAG, KLS, MJK, PAL
and KH. Funding acquisition was acquired by CE, BCR, GDA, PPP and
MSS. Investigation (preparing the samples and performing the
proteomic analysis) was conducted by SAG, KLS, MJK, PAL and KH. The
research methodology was carried out by MM, FF, BCR, GDA and MSS.
Writing of the original draft was performed by SAG and KLS. Review
and editing was conducted by BCR, GDA and MSS. All authors read and
approved the manuscript and agree to be accountable for all aspects
of the research in ensuring that the accuracy or integrity
(particularly the data) of any part of the work are appropriately
investigated and resolved.
Ethics approval and consent to
participate
The study was conducted according to the guidelines
of the Declaration of Helsinki and approved by the London-Harrow
Research Ethics Committee (reference number REC number 12/LO/1361).
The study was registered to the International Standard Randomized
Controlled Trial Number (ISRCTN) registry with the ID 33084113
(doi: 10.1186/ISRCTN33084113) and to the UK controlled randomized
trial number (UKCRN) registry ID 13672 and sponsored by the
Imperial College of London. Informed, written consent was obtained
from all subjects involved in the study.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgements
We would like to acknowledge the help of the Cardiac
Surgery teams at both institutions, the Bristol Trials Centre
(BRI-Hub), Bristol and Mrs. Hua Lin for her technical
assistance.
Funding
This study was funded/supported by the British Heart Foundation
(RG/15/5/31446) and the NIHR Biomedical Research Centre at
University Hospitals Bristol NHS Foundation Trust and the
University of Bristol (UK).
Abbreviations:
|
AVS
|
aortic valve stenosis
|
|
CAD
|
coronary artery disease
|
|
FC
|
fold change
|
|
GO
|
Gene Ontology
|
|
IPA
|
Ingenuity Pathway Analysis
|
|
LC-MS/MS
|
liquid chromatography mass
spectrometry/mass spectrometry
|
|
LV
|
left ventricle
|
|
RV
|
right ventricle
|
|
TMT
|
tandem mass tags
|
|
TBS
|
Tris-buffered saline
|
|
SEM
|
standard error of the mean
|
References
|
1
|
Chase A, Jackson CL, Angelini GL and
Suleiman MS: Coronary artery disease progression is associated with
increased resistance of hearts and myocytes to cardiac insults.
Crit Care Med. 35:2344–2351. 2007. View Article : Google Scholar
|
|
2
|
Barth AS, Merk S, Arnoldi E, Zwermann L,
Kloos P, Gebauer M, Steinmeyer K, Bleich M, Kääb S, Hinterseer M,
et al: Reprogramming of the human atrial transcriptome in permanent
atrial fibrillation: Expression of a ventricular-like genomic
signature. Circ Res. 96:1022–1029. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Barth AS, Merk S, Arnoldi E, Zwermann L,
Kloos P, Gebauer M, Steinmeyer K, Bleich M, Kääb S, Pfeufer A, et
al: Functional profiling of human atrial and ventricular gene
expression. Pflugers Arch. 450:201–208. 2005. View Article : Google Scholar
|
|
4
|
Ellinghaus P, Scheubel RJ, Dobrev D,
Ravens U, Holtz J, Huetter J, Nielsch U and Morawietz H: Comparing
the global mRNA expression profile of human atrial and ventricular
myocardium with high-density oligonucleotide arrays. J Thorac
Cardiovasc Surg. 129:1383–1390. 2005. View Article : Google Scholar
|
|
5
|
Kääb S, Barth AS, Margerie D, Dugas M,
Gebauer M, Zwermann L, Merk S, Pfeufer A, Steinmeyer K, Bleich M,
et al: Global gene expression in human myocardium-oligonucleotide
microarray analysis of regional diversity and transcriptional
regulation in heart failure. J Mol Med (Berl). 82:308–316. 2004.
View Article : Google Scholar
|
|
6
|
Asp J, Synnergren J, Jonsson M, Dellgren G
and Jeppsson A: Comparison of human cardiac gene expression
profiles in paired samples of right atrium and left ventricle
collected in vivo. Physiol Genomics. 44:89–98. 2012. View Article : Google Scholar
|
|
7
|
Borchert B, Tripathi S, Francino A,
Navarro-Lopez F and Kraft T: The left and right ventricle of a
patient with a R723G mutation of the beta-myosin heavy chain and
severe hypertrophic cardiomyopathy show no differences in the
expression of myosin mRNA. Cardiol J. 17:518–522. 2010.
|
|
8
|
Bond AR, Iacobazzi D, Abdul-Ghani S,
Ghorbel M, Heesom K, Wilson M, Gillett C, George SJ, Caputo M,
Suleiman S and Tulloh RMR: Changes in contractile protein
expression are linked to ventricular stiffness in infants with
pulmonary hypertension or right ventricular hypertrophy due to
congenital heart disease. Open Heart. 5:e0007162018. View Article : Google Scholar
|
|
9
|
Bond AR, Iacobazzi D, Abdul-Ghani S,
Ghorbel MT, Heesom KJ, George SJ, Caputo M, Suleiman MS and Tulloh
RM: The cardiac proteome in patients with congenital ventricular
septal defect: A comparative study between right atria and right
ventricles. J Proteomics. 191:107–113. 2019. View Article : Google Scholar
|
|
10
|
Iacobazzi D, Suleiman MS, Ghorbel M,
George SJ, Caputo M and Tulloh RM: Cellular and molecular basis of
RV hypertrophy in congenital heart disease. Heart. 102:12–17. 2016.
View Article : Google Scholar
|
|
11
|
Littlejohns B, Heesom K, Angelini GD and
Suleiman MS: The effect of disease on human cardiac protein
expression profiles in paired samples from right and left
ventricles. Clin Proteomics. 11:342014. View Article : Google Scholar
|
|
12
|
Fiorentino F, Angelini GD, Suleiman MS,
Rahman A, Anderson J, Bryan AJ, Culliford LA, Moscarelli M, Punjabi
PP and Reeves BC: Investigating the effect of remote ischaemic
preconditioning on biomarkers of stress and injury-related
signalling in patients having isolated coronary artery bypass
grafting or aortic valve replacement using cardiopulmonary bypass:
Study protocol for a randomized controlled trial. Trials.
16:1812015. View Article : Google Scholar
|
|
13
|
Moscarelli M, Fiorentino F, Suleiman MS,
Emanueli C, Reeves BC, Punjabi PP and Angelini GD: Remote ischaemic
preconditioning in isolated aortic valve and coronary artery bypass
surgery: A randomized trial†. Eur J Cardiothorac Surg. 55:905–912.
2019. View Article : Google Scholar
|
|
14
|
Välikangas T, Suomi T and Elo LL: A
comprehensive evaluation of popular proteomics software workflows
for label-free proteome quantification and imputation. Brief
Bioinform. 19:1344–1355. 2018.
|
|
15
|
She S, Jiang L, Zhang Z, Yang M, Hu H, Hu
P, Liao Y, Yang Y and Ren H: Identification of the C-reactive
protein interaction network using a bioinformatics approach
provides insights into the molecular pathogenesis of hepatocellular
carcinoma. Cell Physiol Biochem. 48:741–752. 2018. View Article : Google Scholar
|
|
16
|
Pascovici D, Handler DC, Wu JX and Haynes
PA: Multiple testing corrections in quantitative proteomics: A
useful but blunt tool. Proteomics. 16:2448–2453. 2016. View Article : Google Scholar
|
|
17
|
Israeli-Rosenberg S, Manso AM, Okada H and
Ross RS: Integrins and integrin-associated proteins in the cardiac
myocyte. Circ Res. 114:572–586. 2014. View Article : Google Scholar :
|
|
18
|
Sharp WW, Simpson DG, Borg TK, Samarel AM
and Terracio L: Mechanical forces regulate focal adhesion and
costamere assembly in cardiac myocytes. Am J Physiol.
273:H546–H556. 1997.
|
|
19
|
Balasubramanian S, Quinones L, Kasiganesan
H, Zhang Y, Pleasant DL, Sundararaj KP, Zile MR, Bradshaw AD and
Kuppuswamy D: β3 integrin in cardiac fibroblast is critical for
extracellular matrix accumulation during pressure overload
hypertrophy in mouse. PLoS One. 7:e450762012. View Article : Google Scholar
|
|
20
|
Johnston RK, Balasubramanian S,
Kasiganesan H, Baicu CF, Zile MR and Kuppuswamy D: Beta3
integrin-mediated ubiquitination activates survival signaling
during myocardial hypertrophy. FASEB J. 23:2759–2771. 2009.
View Article : Google Scholar
|
|
21
|
Suryakumar G, Kasiganesan H,
Balasubramanian S and Kuppuswamy D: Lack of beta3 integrin
signaling contributes to calpain-mediated myocardial cell loss in
pressure-overloaded myocardium. J Cardiovasc Pharmacol. 55:567–573.
2010. View Article : Google Scholar
|
|
22
|
Chen C, Li R, Ross RS and Manso AM:
Integrins and integrin-related proteins in cardiac fibrosis. J Mol
Cell Cardiol. 93:162–174. 2016. View Article : Google Scholar :
|
|
23
|
Jenkins WS, Vesey AT, Stirrat C, Connell
M, Lucatelli C, Neale A, Moles C, Vickers A, Fletcher A, Pawade T,
et al: Cardiac αV β3 integrin expression
following acute myocardial infarction in humans. Heart.
103:607–615. 2017. View Article : Google Scholar
|
|
24
|
Sun M, Opavsky MA, Stewart DJ, Rabinovitch
M, Dawood F, Wen WH and Liu PP: Temporal response and localization
of integrins beta1 and beta3 in the heart after myocardial
infarction: Regulation by cytokines. Circulation. 107:1046–1052.
2003. View Article : Google Scholar
|
|
25
|
Adorno-Cruz V and Liu H: Regulation and
functions of integrin α2 in cell adhesion and disease. Genes Dis.
6:16–24. 2018. View Article : Google Scholar
|
|
26
|
Wada T: Coagulofibrinolytic changes in
patients with post-cardiac arrest syndrome. Front Med (Lausanne).
4:1562017. View Article : Google Scholar
|
|
27
|
Pick R, Begandt D, Stocker TJ, Salvermoser
M, Thome S, Böttcher RT, Montanez E, Harrison U, Forné I, Khandoga
AG, et al: Coronin 1A, a novel player in integrin biology, controls
neutrophil trafficking in innate immunity. Blood. 130:847–858.
2017. View Article : Google Scholar
|
|
28
|
Mollenhauer M, Friedrichs K, Lange M,
Gesenberg J, Remane L, Kerkenpaß C, Krause J, Schneider J, Ravekes
T, Maass M, et al: Myeloperoxidase mediates postischemic
arrhythmogenic ventricular remodeling. Circ Res. 121:56–70. 2017.
View Article : Google Scholar
|
|
29
|
Di Nardo A, Vitiello A and Gallo RL:
Cutting edge: Mast cell antimicrobial activity is mediated by
expression of cathelicidin antimicrobial peptide. J Immunol.
170:2274–2278. 2003. View Article : Google Scholar
|
|
30
|
Bei Y, Pan LL, Zhou Q, Zhao C, Xie Y, Wu
C, Meng X, Gu H, Xu J, Zhou L, et al: Cathelicidin-related
antimicrobial peptide protects against myocardial
ischemia/reperfusion injury. BMC Med. 17:422019. View Article : Google Scholar
|
|
31
|
Zheng X, Peng M, Li Y, Wang X, Lu W, Wang
X, Shan Y, Li R, Gao L and Qiu C: Cathelicidin-related
antimicrobial peptide protects against cardiac fibrosis in diabetic
mice heart by regulating endothelialmesenchymal transition. Int J
Biol Sci. 15:2393–2407. 2019. View Article : Google Scholar
|
|
32
|
Paparella D, Yau TM and Young E:
Cardiopulmonary bypass induced inflammation: Pathophysiology and
treatment. An update Eur J Cardiothorac Surg. 21:232–244. 2002.
View Article : Google Scholar
|
|
33
|
Andres AM, Tucker KC, Thomas A, Taylor DJ,
Sengstock D, Jahania SM, Dabir R, Pourpirali S, Brown JA, Westbrook
DG, et al: Mitophagy and mitochondrial biogenesis in atrial tissue
of patients undergoing heart surgery with cardiopulmonary bypass.
JCI Insight. 2:e893032017. View Article : Google Scholar
|
|
34
|
Wirth C, Brandt U, Hunte C and Zickermann
V: Structure and function of mitochondrial complex I. Biochim
Biophys Acta. 1857:902–914. 2016. View Article : Google Scholar
|
|
35
|
Weiss AKH, Loeffler JR, Liedl KR, Gstach H
and Jansen-Dürr P: The fumarylacetoacetate hydrolase (FAH)
superfamily of enzymes: Multifunctional enzymes from microbes to
mitochondria. Biochem Soc Trans. 46:295–309. 2018. View Article : Google Scholar
|
|
36
|
Da Cruz S and Martinou JC: Purification
and proteomic analysis of the mouse liver mitochondrial inner
membrane. Methods Mol Biol. 432:101–116. 2008. View Article : Google Scholar
|
|
37
|
Davies VJ, Powell KA, White KE, Yip W,
Hogan V, Hollins AJ, Davies JR, Piechota M, Brownstein DG, Moat SJ,
et al: A missense mutation in the murine Opa3 gene models human
costeff syndrome. Brain. 131:368–380. 2008. View Article : Google Scholar
|
|
38
|
Stoll S, Xi J, Ma B, Leimena C, Behringer
EJ, Qin G and Qiu H: The valosin-containing protein protects the
heart against pathological Ca2+ overload by modulating
Ca2+ uptake proteins. Toxicol Sci. 171:473–484. 2019.
View Article : Google Scholar
|
|
39
|
Vrbacký M, Kovalčíková J, Chawengsaksophak
K, Beck IM, Mráček T, Nůsková H, Sedmera D, Papoušek F, Kolář F,
Sobol M, et al: Knockout of Tmem70 alters biogenesis of ATP
synthase and leads to embryonal lethality in mice. Hum Mol Genet.
25:4674–4685. 2016.
|
|
40
|
Yadav H, Unsworth B, Fontana M, Diller GP,
Kyriacou A, Baruah R, Mayet J and Francis DP: Selective right
ventricular impairment following coronary artery bypass graft
surgery. Eur J Cardiothorac Surg. 37:393–398. 2010.
|
|
41
|
Singh A, Huang X, Dai L, Wyler D,
Alfirevic A, Blackstone EH, Pettersson GB and Duncan AE: Right
ventricular function is reduced during cardiac surgery independent
of procedural characteristics, reoperative status, or
pericardiotomy. J Thorac Cardiovasc Surg. 159:1430–1438.e4. 2020.
View Article : Google Scholar
|
|
42
|
Rösner A, Avenarius D, Malm S, Iqbal A,
Schirmer H, Bijnens B and Myrmel T: Changes in right ventricular
shape and deformation following coronary artery bypass
surgery-insights from echocardiography with strain rate and
magnetic resonance imaging. Echocardiography. 32:1809–1820. 2015.
View Article : Google Scholar
|
|
43
|
Suleiman MS, Hancock M, Shukla R,
Rajakaruna C and Angelini GD: Cardioplegic strategies to protect
the hypertrophic heart during cardiac surgery. Perfusion. 26(Suppl
1): S48–S56. 2011. View Article : Google Scholar
|
|
44
|
Cotecchia S, Del Vescovo CD, Colella M,
Caso S and Diviani D: The alpha1-adrenergic receptors in cardiac
hypertrophy: Signaling mechanisms and functional implications. Cell
Signal. 27:1984–1993. 2015. View Article : Google Scholar
|
|
45
|
Dent MR, Dhalla NS and Tappia PS:
Phospholipase C gene expression, protein content, and activities in
cardiac hypertrophy and heart failure due to volume overload. Am J
Physiol Heart Circ Physiol. 287:H719–H727. 2004. View Article : Google Scholar
|
|
46
|
Khalilimeybodi A, Daneshmehr A and
Sharif-Kashani B: Investigating β-adrenergic-induced cardiac
hypertrophy through computational approach: Classical and
non-classical pathways. J Physiol Sci. 68:503–520. 2018. View Article : Google Scholar
|
|
47
|
Nakao K, Minobe W, Roden R, Bristow MR and
Leinwand LA: Myosin heavy chain gene expression in human heart
failure. J Clin Invest. 100:2362–2370. 1997. View Article : Google Scholar
|
|
48
|
Woo A, Rakowski H, Liew JC, Zhao MS, Liew
CC, Parker TG, Zeller M, Wigle ED and Sole MJ: Mutations of the
beta myosin heavy chain gene in hypertrophic cardiomyopathy:
Critical functional sites determine prognosis. Heart. 89:1179–1185.
2003. View Article : Google Scholar
|
|
49
|
Rudolph T, Schaps KP, Steven D, Koester R,
Rudolph V, Berger J, Terres W, Meinertz T and Kaehler J:
Interleukin-3 is elevated in patients with coronary artery disease
and predicts restenosis after percutaneous coronary intervention.
Int J Cardiol. 132:392–397. 2009. View Article : Google Scholar
|