Introduction
Follicles form the basic reproductive unit of female
mammalians and are important not only for ovulation, but also for
the production of hormones that maintain the secondary sexual
characteristics and early pregnancy (1). According to the previously reported
morphological and functional changes that occur during follicular
development, follicles can be divided into primordial, primary,
secondary and mature follicles (2-4).
In particular, formation and activation of primordial follicles is
key for determining reproductive ability (5). In the majority of mammalian species,
the formation of primordial follicles typically occurs during the
embryonic stages or around birth (6-8).
By contrast, in adult ovaries, primordial follicles cannot be
renewed and regenerated (6-8).
Therefore, the size of the primordial follicle pool is mainly used
to determine the reproductive capacity of female mammals throughout
their lifetime (6-8).
The primordial follicle is comprised of an immature
oocyte and several flattened precursor granulosa cells wrapped
around it. After the primordial follicle becomes activated to form
a primary follicle, it then consists of an oocyte along with one or
several layers of cubic granulosa cells wrapped around it. At
present, a number of studies have been performed on the formation
and activation of primordial follicles, which mainly reported the
involvement of various signaling pathways, including Notch, PI3K,
Janus kinase, TGF-β and KIT (9-13).
However, the mechanism underling primordial follicle formation and
activation remains poorly understood.
Calmodulin-dependent protein kinase (CaMK) serves an
important role in reproductive regulation. CaMK belongs to the
serine/threonine kinase family and consists of four members, namely
CaMKI, II, IV and K, where CaMKII is the most widely studied
(14-16). CaMKII is a 8-12 polymer and is
comprised of four homologous genes CaMKIIA, CaMKIIB, CaMKIIG and
CaMKIID, along with their corresponding expression products
CaMKIIα, CaMKIIβ, CaMKIIγ and CaMKIIδ (17). Previous studies have demonstrated
that CaMKII is closely associated with the activation of oocytes,
where ~80% of MII-phase mouse oocytes with CaMKIIγ expression
knocked down can be activated after the overexpression of CaMKIIγ
or CaMKIIδ (18). CaMKII also
serves an important role in the development of early mouse embryos.
It has been previously revealed that the expression of CaMKIIγ can
be detected in mouse embryos in the two-celled stage, where it can
regulate the development of these embryos by regulating the
activation levels of cAMP response element-binding protein and
cAMP-dependent transcription factor (19). Although these findings suggest
that CaMKII can serve an important role in reproductive regulation,
the effect of CaMKII on follicular development remains unknown.
KN93 is a CaM-binding specific antagonist that can
reversibly and competitively inhibit CaMKII activity (20,21). Several studies have previously
reported that KN93 can affect physiological and pathological
processes in the body by inhibiting CaMKII, including the brain,
nervous system, cardiovascular system and cancer (22-25). Combined with previous reports, it
is likely that CaMKII serves an important role in reproductive
regulation. However, to the best of our knowledge, the effects of
KN93 as a specific inhibitor of CaMKII on follicular development
remain to be reported.
In mice, the embryonic females from 17.5 days
post-coitus (dpc) is the initial stage of primordial follicular
formation, whereas the neonatal females from 5 days post-partum
(dpp) is the stage of primordial follicular bank formation. For
this reason, in the present study, the expression levels of CaMKII
in the ovaries of mice at different development stages (17.5, 1, 3
and 5 dpp) were measured using immunofluorescence. It was revealed
that the expression of CaMKII was obviously changed in ovary of
mice at the various developmental stages. Therefore, it was
hypothesized that CaMKII has a potential role in regulating
follicular development. Based on this, KN93 was selected to inhibit
CaMKII to study the possible role and potential mechanism of KN93
in regulating follicular development after inhibiting CaMKII.
Materials and methods
Animal and ovary collection
ICR mice were selected for the present study because
of their high fecundity characteristics. Adult ICR mice, 10 males
and 60 females (8-10 weeks old; male, 34-38 g; female, 24-28 g),
were purchased from Beijing Vital River Laboratory Animal
Technology Co., Ltd. All mice were housed in an environment with
50±10% humidity, lighting (12-h light/dark cycle) and temperature
(24-26°C) conditions with free access to food and water. Animal
experiments were approved by the Animal Ethical and Welfare
Committee of Ningxia University (approval no. IACUC-N
DLAC-2020019). Mice health and behavior were monitored daily. No
mouse death occurred during the experiment. The total cumulative
duration of the experiment was 8 months, excluding some time
gaps.
All female mice were randomly divided into four
groups as follows: 17.5 dpc Embryonic ovaries group (45 mice), 1
dpp neonatal ovaries group (five mice), 3 dpp neonatal ovaries
group (five mice) and 5 dpp neonatal ovaries group (five mice).
These female mice were randomly assigned in batches to mate with
adult males at a ratio of 1:1 overnight. Mice with a vaginal plug
in the next morning were defined at 0.5 dpc. Whereas, mice without
a vaginal plug were scheduled for another round of mating. The day
after partum was considered to be 1 dpp. The mice at 17.5 days of
pregnancy (45 mice) were euthanized by cervical dislocation.
Following sacrifice, the abdomen was immediately cut open to obtain
the fetuses in utero. These fetuses were also euthanized by
cervical dislocation to collect embryonic ovaries (17.5 dpc).
Additionally, female mice at 1, 3 and 5 days after birth (10 each)
were randomly selected and euthanized by cervical dislocation to
collect neonatal ovaries (1, 3 and 5 dpp). Mouse ovaries were
separated in cold PBS under a stereo-microscope in sterile
conditions. During this procedure, care was taken to ensure that
the structure of the whole ovaries was not damaged. After the
experiment, all remaining animals were euthanized by cervical
dislocation. In the present study, all animal's mortalities were
confirmed by determining the lack of heartbeat and respiration.
Ovary culture
The 17.5 dpc ovaries were cultured in six-well
culture plates in 1,500 µl DMEM/Ham F12 nutrient mixture
(Gibco; Thermo Fisher Scientific, Inc.) supplemented with
insulin-transferrin-sodium selenite (1:100; Sigma-Aldrich; Merck
KGaA) and 1% penicillin-streptomycin solution at 37°C, 5%
CO2 and saturated humidity. The mouse ovaries were
randomly assigned so that there were the same number of ovaries in
each group. The culture medium was exchanged once every 2 days. The
different concentration (5, 10 and 10 µM) of KN93 (cat. no.
S6787; Selleck Chemicals) was added in the treatment group. The
control group was instead treated with an equivalent volume of
DMSO. The ovaries were then cultured for 4 days to assess the role
of KN93.
Hematoxylin staining
Ovaries were fixed in 4% paraformaldehyde at 4°C
overnight, embedded in paraffin and sectioned serially at 5
µm. Sections were then adhered onto the slides and stained
with hematoxylin at room temperature (12 sec) for detecting the
presence of oocytes and follicles with the Nikon 80i digital
fluorescence microscope (Nikon Corporation).
Immunofluorescence staining
Ovaries were fixed in 4% paraformaldehyde at 4°C
overnight, embedded in paraffin and sectioned serially at 5
µm. The sections were deparaffinized at 60°C for 20 min and
washed for 5 min with xylene (cat. no. 33535; Yantai Shuangshuang
Chemical Co., Ltd.) twice. Then the sections were rehydrated in
descending alcohol (cat. no. 64-17-5; Tianjin Damao Chemical
Reagent Factory) series and subjected to high temperature (95-98°C)
antigen retrieval in 0.01% sodium citrate buffer (pH 6.0). The
sections were then rinsed thoroughly with PBS, blocked with normal
donkey serum (cat. no. ZX108; Beijing Zoman Biotechnology Co.,
Ltd.) in PBS for 1 h at room temperature and incubated with primary
antibodies for 12-16 h at 4°C. The antibodies used were as follows:
Anti-CaMKII antibody (1:200; cat. no. ab52476; Abcam), DEAD-box
helicase 4 (DDX4; 1:50; cat. no. ab27591; Abcam) and anti-forkhead
box L2 (FOXL2) antibody (1:100; cat. no. NB100-1277; Novus
Biologicals, LLC). Subsequently, the ovarian sections were rinsed
thoroughly with PBS and incubated with Alexa Fluor 488 AffiniPure
donkey anti-mouse IgG (H+L; 1:200; cat. no. 34106ES60; Shanghai
Yeasen Biotechnology Co., Ltd.) or Donkey anti-goat IgG (H+L)
highly cross-adsorbed secondary antibody, Alexa Fluor™ Plus 555
(1:200; cat. no. A32816; Invitrogen; Thermo Fisher Scientific,
Inc.) for 1 h at 37°C. The slides were then rinsed in PBS, stained
with Hoechst 33342 at 37°C (1:1,000; cat. no. B2261; Sigma-Aldrich;
Merck KGaA) for 5 min and sealed in the anti-fade fluorescence
mounting medium (cat. no. 20180116; Applygen Technologies, Inc.)
with coverslips. Sections were examined and images were captured
using the Nikon 80i digital fluorescence microscope (Nikon
Corporation).
Protein isolation, quantification
digestion and desalting
The proteins were extracted from the ovary samples
of control group and KN93 treatment group, and the Bradford method
was used to determine protein concentration according to the
previously described method (26). Proteins were reduced with 1 M
DL-Dithiothreitol (DTT; cat. no. 3483-12-3; Beijing Solarbio
Science & Technology Co., Ltd.) at 56°C for 30 min, cooled to
room temperature, alkylated with 0.55 M iodoacetamide (cat. no.
144-48-9; Shanghai Aladdin Bio-chem Technology Co., Ltd.) in a
darkroom for 30 min at room temperature and 10 mM DTT was added and
precipitated at -20°C for 2 h. The sample was centrifuged at 4°C,
13,000 × g for 20 min and the supernatant discarded. Subsequently,
1 ml cold acetone (cat. no. 144-48-9; Sinopharm Chemical Reagent
Co., Ltd.) was added to the precipitate to give a final
concentration of 10 mM DTT. The precipitate was thoroughly
mashed-up, vortexed and left to stand for 30 min at −20°C, then
centrifuged at 4°C, 13,000 × g for 20 min and the supernatant
discarded. The precipitate was air-dried, with 8 M Urea (cat. no.
U5378; Sigma-Aldrich; Merck KGaA) lysis buffer was added and
sonicated at 4°C for 5 min (working 1 sec, stopping 2 sec), and
then centrifuged at 4°C, 13,000 × g for 20 min. The supernatant was
collected and the Bradford method was used to determine protein
concentration. The reduced and alkylated proteins were digested
using trypsin (cat. no. V5111; Promega Corporation) in a volume
ratio of 1:100 (enzyme/protein) at 37°C for >8 h. A total of 10
mg of C18 column material was weighed, corresponding to every 100
µg of peptide sample. The column material was activated
using 1 ml methanol (cat. no. 10014118; Sinopharm Chemical Reagent
Co., Ltd.), centrifuged instantaneously with shaking at room
temperature and the supernatant was discarded. Added 1 ml 0.1%
formic acid (FA; cat. no. 80065518; Sinopharm Chemical Reagent Co.,
Ltd.) to acidify at room temperature for 30 sec, centrifuged
instantaneously with shaking at room temperature and discarded the
supernatant. Peptide samples were acidified with an equal volume of
0.1% FA, shaken, vortexed into a centrifuge tube, mixed at room
temperature for 30 min by muter mixer and centrifuged
instantaneously with shaking at room temperature to discard the
supernatant. The sample was then washed twice with 0.1% FA + 3%
acetonitrile (ACN; cat. no. 40064193; Sinopharm Chemical Reagent
Co., Ltd.) for desalting and eluted with 1 ml 0.1% FA + 80% ACN.
The eluted peptide was dried with a vacuum concentrator.
HPLC-MS/MS analysis
The analysis was performed using QEXactive HF-X
(Thermo Fisher Scientific, Inc) liquid mass spectrometry system.
The samples were separated by a liquid phase UltiMate 3000 RSLCnano
system (Thermo Fisher Scientific, Inc.) at a nanoliter flow rate.
The peptide samples were dissolved by loading buffer, inhaled by
automatic sampler and bound to C18 capture column (3 µm, 120
Å, 100 µm x 20 mm; Thermo Fisher Scientific, Inc), and then
eluted to an analysis column (2 µm, 120 Å, 750 µm x
250 mm) for separation. An analytical gradient was established
using two mobile phases (mobile phase A: 98% water, 2% ACN, 0.1%
FA; and mobile phase B: 98% ACN, 2% water, 0.1% FA). The flow rate
of the liquid phase was set at 300 nl/min. In the Data Dependent
Acquisition (DDA) mode analysis of MS, each scan cycle contains a
full MS scan (R=60 K; AGC=3e6; Max IT=20 MS; Scan range=350-1,800
m/z). And then 20 MS/MS scans (R=15 K; AGC=2e5; Max IT=100 MS).
Electrospray ionization ion source ion type was positive ion,
atomizer voltage was set to 2.2 kV, impact gas pressure was
automatically optimized with sample type, nitrogen temperature (ion
transfer tube temperature) was 320°C, HCD impact energy was set to
28. The filter window for the quad pole was set to 1.6 DA. The
dynamic exclusion time of ion repeated collection was set to 35 sec
and raw data for mass detection (.raw) was generated.
Data analysis and bioinformatics
analysis
The raw MS data obtained from three biological
replicates were combined and imported into MaxQuant software
(version 1.6.17.0; Max Planck Institute of Biochemistry) to
identify and quantify the proteins. For protein identification, the
MS data were aligned to the Uniport Mus_musculus protein database
(Proteome ID: UP000000589; https://www.uniprot.org/proteomes/UP000000589)
represented by the file uniport
(PR1-21010013-PR1-21010015-uniprot-Mus_
musculus-10090-v20210123.fasta). The MS/MS tolerance of first
search with an error window of 20 ppm and then with a main search
error of 4.5 ppm. The proteins were cleaved using trypsin, and the
two missed cleavages were accepted. Peptide identifications with
false discovery rates >1% were discarded. Proteins with an
adjusted [log2 (fold-change)]>2 and P<0.05 or [log2
(fold-change)] <0.5 and P<0.05 were identified differentially
expressed proteins. Gene Ontology (GO; http://www.geneontology.org/) annotations and Kyoto
Encyclopedia of Genes and Genomes (KEGG; http://www.genome.jp/kegg/) metabolic pathway analyses
were performed to identify possible enrichment of the differential
expressed proteins with particular biological characteristics.
Taking P-value ≤0.01 as the threshold, the GO terms or KEGG terms
that meet this condition in the DEPs were called the enrichment GO
terms or KEGG terms. Furthermore, all interactions and network
construction were performed in the STRING 11.0 database (http://string-db.org/).
Western blotting
Each protein sample was obtained from ≥ eight
ovaries after extraction in the WIP Tissue and cell lysis solution
containing 1 mM PMSF (cat. no. 8553S; Cell Signaling Technology,
Inc.) according to the manufacturer's protocols. Protein
concentration was measured using a BCA assay (cat. no. P0012;
Beyotime Institute of Biotechnology). The samples (10 µg)
were separated on 10% SDS-PAGE and then transferred onto PVDF
membranes. The membranes were blocked with 5% skimmed milk powder
for 1 h at room temperature. The membranes were then incubated
overnight at 4°C with the appropriate primary antibodies. CaMKII
(1:300; cat. no. ab52476; Abcam), DDX4 (1:500; cat. no. ab27591;
Abcam), FOXL2 (1:500; cat. no. NB100-1277; Novus Biologicals, LLC),
cytochrome P450 family 51 family A member 1 (CYP51A1; 1:2,000; cat.
no. 13431-1-AP; ProteinTech Group, Inc.), fas-associated death
domain (FADD; 1:5,000; cat. no. ab124812; Abcam), neural cell
adhesion molecule 1 (NCAM1; 1:1,000; cat. no. ab220360; Abcam),
cytochrome c, testis (Cyct; 1:5,000; cat. no. ab133504;
Abcam), insulin-like growth factor binding protein 3 (IGFBP3;
1:1,000; cat. no. ab220429; Abcam), plastin 1 (1:1,000; cat. no.
A15303; ABclonal Biotech Co., Ltd.), PDZ domain-containing 1
(PDZK1; 1:5,000; cat. no. ab92491; Abcam), zona pellicida
sperm-binding protein (ZP2; 1:2,000; cat. no. A10126; ABclonal
Biotech Co., Ltd.), Y-box binding protein 2 (YBX2; 1:5000; cat. no.
ab154829; Abcam) and Bcl-2-interacting protein 3-like (BNIP3L;
1:5,000; cat. no. ab109414; Abcam). After rinsing thoroughly with
TBST, the membranes were incubated with secondary antibodies
(1:5,000; cat. no. ZB-2301; ZSGB-BIO). The membranes were then
visualized using SuperSignal West Pico chemiluminescent detection
system (cat. no. 34080; Thermo Fisher Scientific, Inc.). GAPDH
(1:1,000; cat. no. AF7021; Affinity Biosciences) was used as an
intrinsic control. An ImageJ software (version 1.8.0; National
Institutes of Health) was used to quantify the relative expression
of each protein.
Statistical analysis
All experiments were repeated ≥3 times. The data
were analyzed using unpaired Student's t-test by GraphPad Prism 8
(GraphPad Software, Inc.) and presented as the mean ± standard
deviation (SD). P<0.05 was considered to indicate a
statistically significant difference.
Results
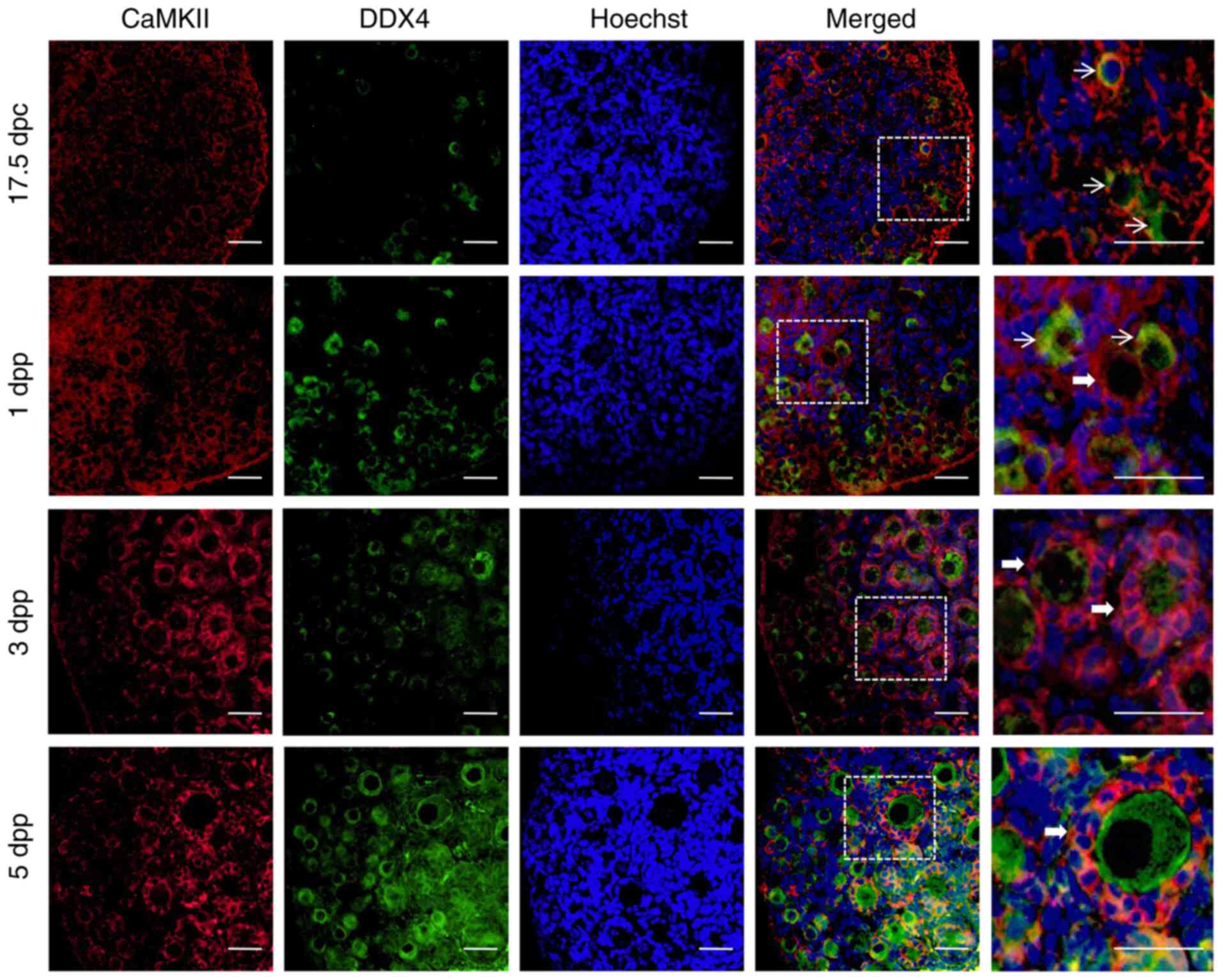
Expression of CaMKII in the embryonic and
neonatal mouse ovaries
To explore the potential role of CaMKII in
follicular development, ovaries from 17.5 dpc embryonic mice and
ovaries from 1, 3 and 5 dpp newborn mice were collected to prepare
tissue sections. Immunofluorescence was first performed in the
follicles to detect the localization and expression pattern of
CaMKII at different developmental stages by co-staining CaMKII and
the oocyte specific marker DDX4. The expression of CaMKII was
detected in ovaries at different developmental stages. In
particular, CaMKII was revealed to be mainly localized to the
cytoplasm of ovarian granulosa cells in embryonic and newborn mice
(Fig. 1). It has been previously
reported that the initial stage of primordial follicle formation in
mouse ovaries is embryonic 17.5 dpc (27). In the present study, CaMKII was
mainly expressed in the ovarian cortex of 17.5 dpc embryos, where
only a small number of primordial follicles were revealed (Fig. 1). In the ovaries of neonatal 1 dpp
mice, CaMKII expression extended from the cortex into the medulla
region (Fig. 1), where more
primordial follicles were revealed. In the ovaries of 3 and 5 dpp
neonatal mice, primordial follicles were more extensively activated
and developed into primary follicles. During these two stages of
development, CaMKII was mainly expressed in the medulla region of
the ovary and in the granulosa cell cytoplasm within the primordial
and primary follicle (Fig. 1).
These results suggested that CaMKII was involved in follicular
development by regulating the function of granulosa cells,
especially the formation or activation of primordial follicles.
KN93 inhibits CaMKII and delays
follicular development
To study the effects of CaMKII on follicular
development further, KN93, an inhibitor of CaMKII, was used in the
present study. Different doses of KN93 (5, 10 and 15 µM)
were selected to treat 17.5 dpc embryonic ovaries for 4 days in
vitro before H&E staining to detect the progress of ovarian
development. It was revealed that 5 µM exerted no notable
effects on ovarian development, whilst 15 µM KN93 mediated
marked influence on the development of the ovary but seriously
damaged its structure. By contrast, the effects of 10 µM
were relatively moderate, which not only exerted notable impact on
the development of ovarian follicles but also induced relatively
little damage to the ovarian tissue structure (Fig. S1). Therefore, 10 µM was
selected as the final dose for subsequent experiments in the
present study.
H&E staining also indicated that the development
of follicles in the control group was markedly faster compared with
that in the 10 and 15 µM KN93 treatment group. In addition,
it was revealed that a large number of primordial follicles in the
control group had migrated to the ovarian medulla region, which
were activated and developed into primary follicles. However, no
similar phenomenon was observed in the 10 and 15 µM KN93
treatment groups (Fig. S1).
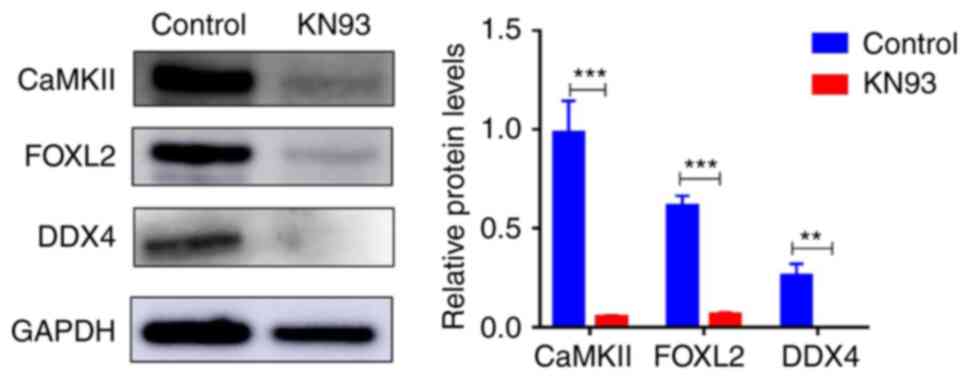
Subsequently, western blotting revealed that KN93
could significantly inhibit CaMKII expression in addition to
significantly reducing the expression of the oocyte specific marker
DDX4 and the granulosa cell specific marker FOXL2 in follicles
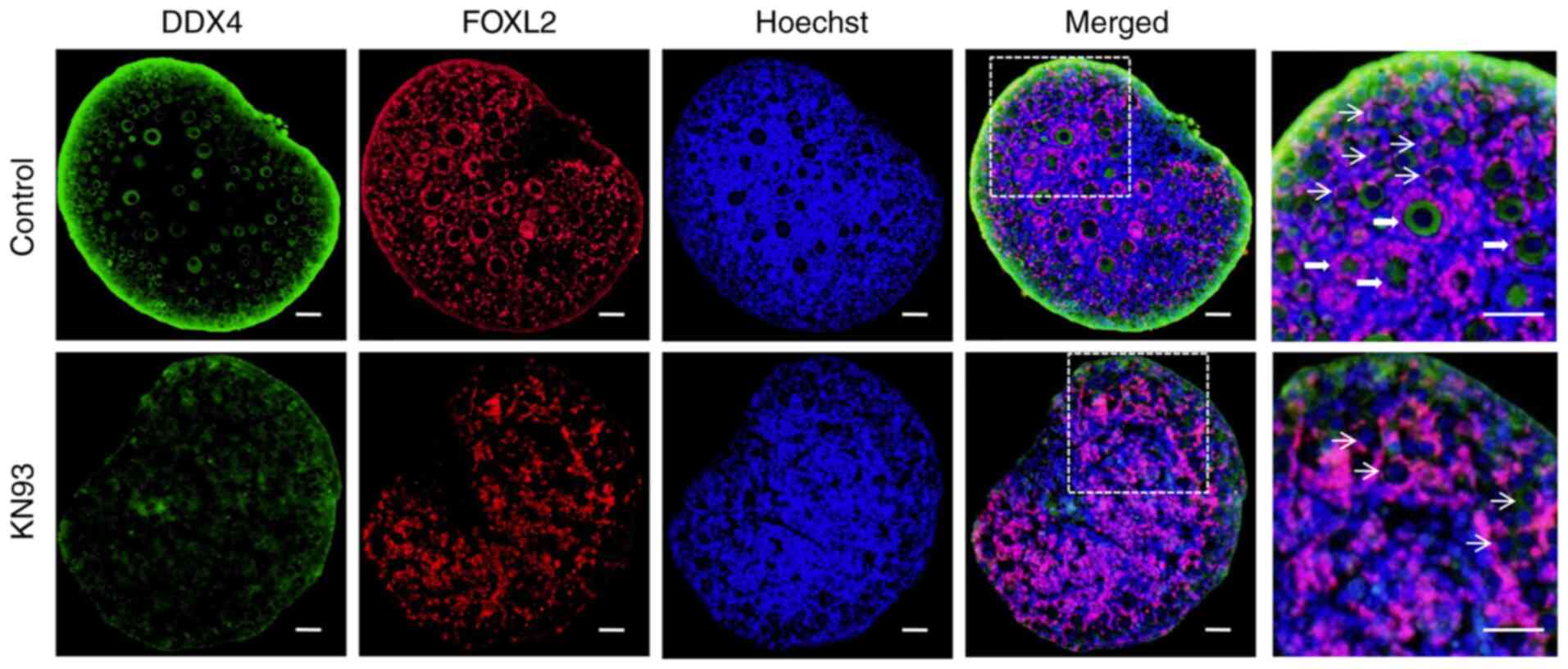
(Fig. 2). Immunofluorescence
detection results validated the western blotting data, in that the
fluorescence intensity of the oocyte marker DDX4 and the granulosa
cell marker FOXL2 in the ovarian follicles from the KN93 treatment
group was weaker compared with that in the control group (Fig. 3). In the enlarged view of the
ovaries in the control group, a large number of primordial and
primary follicles could be observed, whilst there are only a small
number of primordial follicles in the KN93 group. This suggested
that the development of follicles in the KN93 treatment group was
inhibited, consistent with the results of hematoxylin staining
(Fig. 3). These results suggested
that KN93 downregulated the expression of the oocyte marker DDX4
and the granulosa cell marker FOXL2 in follicles by inhibiting
CaMKII, thereby delaying follicular development.
Differentially expressed proteins
affected by KN93 as revealed by proteomic techniques
To investigate the proteins affected by KN93
inhibition of CaMKII during ovarian development further, the
ovaries of 17.5 dpc embryonic mice treated with KN93 and cultured
for 4 days in vitro were collected. The protein samples were
then detected by proteomics techniques. After detection, a total of
5,843 proteins were identified. Principal component analysis of the
identified proteins indicated that the three samples of the KN93
treatment group and the three samples of the control group were
clustered into different regions. This suggested that the genetic
background of the three samples of the KN93 treatment group and the
three samples of the control group showed good consistency. In
addition, this suggested that the experimentally identified
proteins could be used for further analysis (Fig. 4A). Among the 5,843 identified
proteins, a total of 262 differentially expressed proteins were
identified in the KN93 treatment group compared with the control
group, including 168 proteins that were upregulated
[log2 (fold-change)≥0.5 and P<0.01] and 94 proteins
that were downregulated [log2 (fold-change) ≤0.5 and
P<0.01] (Fig. 4B).
Subsequently, 10 proteins from this list of 262 differentially
expressed proteins were randomly selected for western blotting
verification. It was revealed that two of the 10 proteins (Cyct and
IGFBP3) did not concur with the proteomic results, which showed
that there may be false positive results in proteomics, whilst the
other eight proteins were consistent with the proteomic results and
supported the high accuracy of this proteomic detection method
(Fig. 4C and D). Taken together,
these results suggested that KN93 inhibition of CaMKII could induce
expression changes of various proteins during ovarian
development.
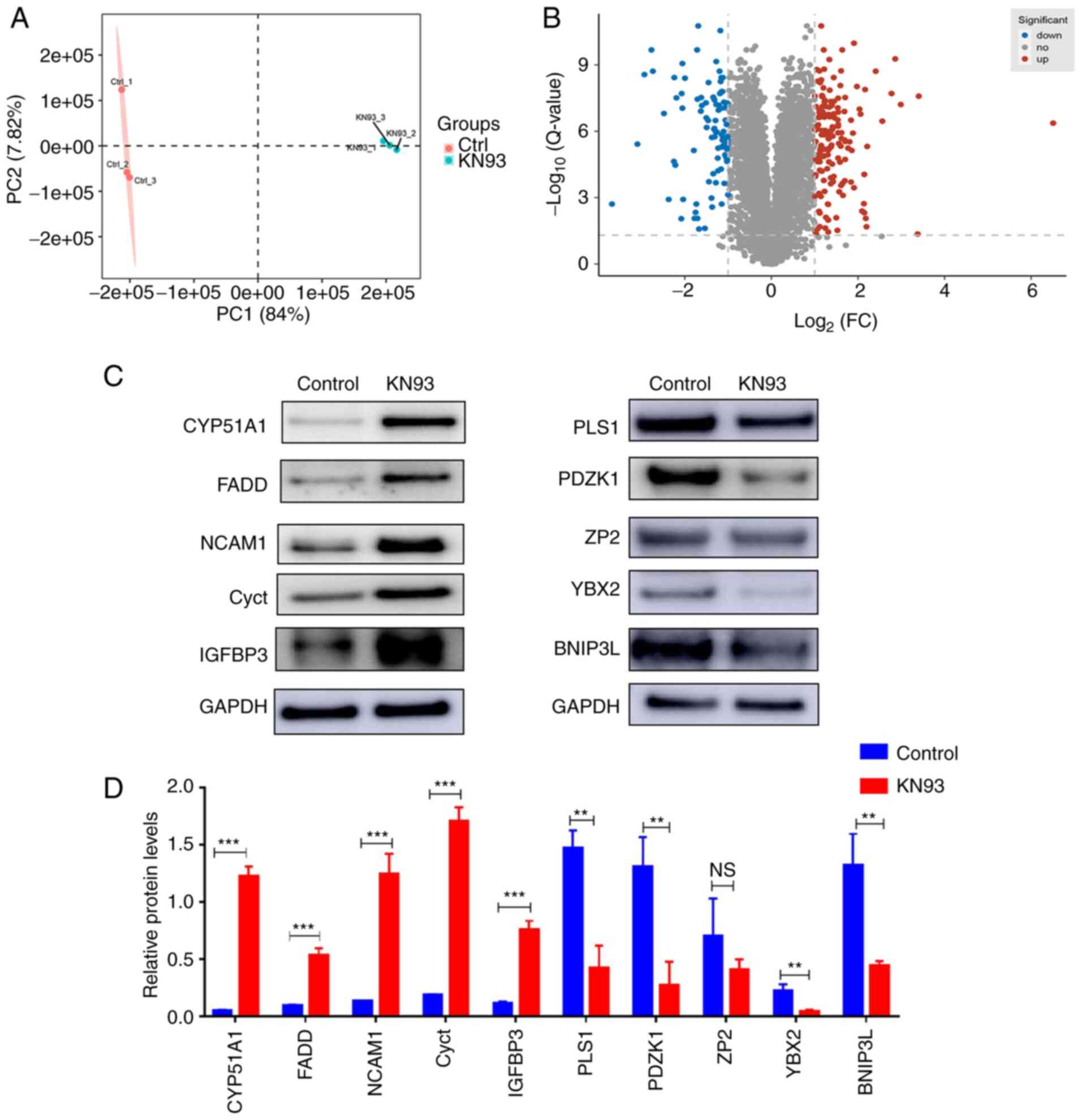 | Figure 4Proteomic techniques revealed
differential proteins influenced by KN93. (A) Principal component
analysis of the identified proteins in the KN93 treatment group and
control group. Red and blue dots represent the KN93 treatment group
and control group, respectively. (B) Volcano plot indicated the
differential proteins between KN93 treatment group and control
group. Red dots indicated significantly upregulated proteins with
adjusted P-values <0.01, blue dots indicated significantly
down-regulated proteins with adjusted P-values <0.01 and gray
dots indicated non-significantly expressed proteins. (C)
Differential proteins were verified by western blotting, which was
then (D) quantified. GAPDH was used as a loading control. The data
are presented as the mean ± SD (n=3).
**P<0.01,***P<0.001. NS, no significant
difference; CYP51A1, cytochrome P450 family 51 family A member 1;
FADD, fas-associated death domain; NCAM1, neural cell adhesion
molecule 1; Cyct, cytochrome c, testis; IGFBP3, insulin-like
growth factor binding protein 3; PLS1, plastin-1; PDZK1, PDZ
domain-containing 1; ZP2, zona pellicida sperm-binding protein;
YBX2, Y-box binding protein 2; BNIP3L, Bcl-2-interacting protein
3-like. |
Function analysis of differentially
expressed proteins that are affected by KN93
To further investigate the biological functions and
enrichment pathways of the differentially expressed proteins
induced by KN93 during ovarian development, GO and KEGG enrichment
analyses were performed for the obtained proteins in the present
study. It was revealed that a number of cell molecules, cell
components, biological processes or pathways associated with
reproductive development, cell proliferation and differentiation or
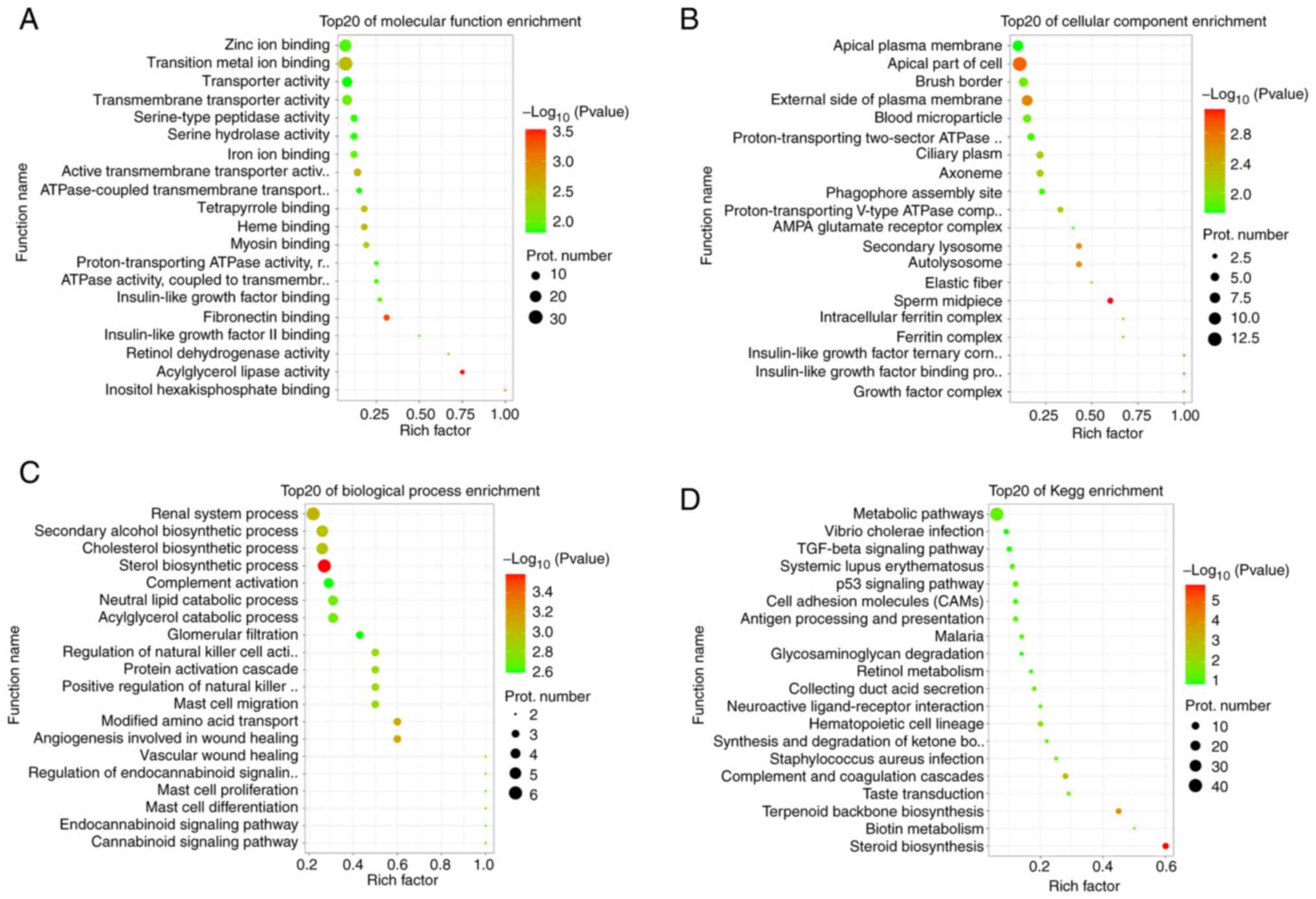
migration were enriched. In the GO terms of molecular function,
'retinol dehydrogenase activity', 'insulin-like growth factor
binding', 'insulin-like growth factor II binding', 'iron ion
binding' and 'zinc ion binding' were enriched (Fig. 5A). In the GO terms of cell
components, 'insulin-like growth factor binding protein complex',
'growth factor complex', 'insulin-like growth factor ternary
complex', 'autolysosome', 'intracellular ferritin complex' and
'ferritin complex' were enriched (Fig. 5B). In the GO terms of biological
process, 'sterol biosynthetic process', 'renal system process',
'cholesterol biosynthetic process', 'secondary alcohol biosynthetic
process', 'mast cell differentiation', 'mast cell proliferation'
and 'mast cell migration' were enriched (Fig. 5C). Furthermore, important
biological pathways that were enriched in terms of biological
pathway mainly included 'steroid biosynthesis', 'p53 signaling
pathway', 'metabolic pathways', 'retinol metabolism', 'cell
adhesion molecules (CAMs)', and 'TGF-β signaling' pathway (Fig. 5D). Comprehensive analysis results
revealed that KN93 inhibited CaMKII, which may serve an important
role in reproductive regulation through these differentially
expressed proteins. In particular, the differential proteins
enriched in the biological pathways closely associated with
reproductive development and metabolism may serve a key role in
follicular development.
Potential mechanism of KN93 retarding
follicular development
To further explore the potential mechanism of
delaying follicular development after KN93 inhibits CaMKII, the
present study extracted the main differentially expressed proteins
enriched in the four biological pathways closely associated with
reproductive development and metabolism obtained in the present
study (Table I). It was revealed
that 19 important differentially expressed proteins were enriched
in the four biological pathways, among which all four
differentially expressed proteins were downregulated whereas 15
differentially expressed proteins were upregulated (Table I). There are 15 differentially
expressed proteins enriched in 'metabolic pathways', which included
guanine deaminase, V-type proton ATPase subunit a, NADP-dependent
steroid dehydrogenase-like (Nsdhl), Atp6v1c1, alcohol dehydrogenase
1 (Adh1), hydroxymethylglutaryl-CoA synthase 1 (Hmgcs1), fatty acid
synthase (Fasn), α-L-iduronidase, dimethylallyltranstransferase
(Fdps), DNA-directed RNA polymerase subunit, lanosterol synthase
(Lss), N-acetylgalactosaminyltransfer ase 7, farnesyl-diphosphate
farnesyltransferase 1 (Fdft1), NADH-ubiquinone oxidoreductase chain
5 and Lanosterol 14-alpha demethylase (Table I). Subsequently, four
differentially expressed proteins were enriched in the 'p53
signaling pathway', including Igfbp3, Cyct, insulin-like growth
factor-binding protein 5 (Igfbp5) and neuroserpin. In addition,
four differential proteins were enriched in the 'steroid
biosynthesis', including Nsdhl, Lss, Fdft1 and Cyp51a1. Only one
differential protein was revealed to be enriched in 'retinol
metabolism', which was Adh1. It was also revealed that Nsdhl, Lss,
Fdft1 and Cyp51a1 were simultaneously enriched in 'steroid
biosynthesis' and 'metabolic pathways'. Adh1 was enriched in both
'retinol metabolism' and 'metabolic pathways' (Table I). These results suggested that
Nsdhl, Lss, Fdft1, Cyp51a1 and Adh1 were the key proteins affected
by KN93.
 | Table IDifferential proteins enriched in the
four key KEGG pathways closely related to reproductive development
and metabolism. |
Table I
Differential proteins enriched in the
four key KEGG pathways closely related to reproductive development
and metabolism.
| Protein name | Accession | Description | KN93: Control
state | Fold-change | Enrichment
pathway |
|---|
| Igfbp3 | P47878 | Insulin-like growth
factor-binding protein 3 | Down | 0.147 | p53 signaling
pathway |
| Cyct | P00015 | Cytochrome c | Down | 0.219 | p53 signaling
pathway |
| Gda | Q9R111 | Guanine
deaminase | Down | 0.237 | Metabolic
pathways |
| Igfbp5 | Q07079 | Insulin-like growth
factor-binding protein 5 | Down | 0.306 | p53 signaling
pathway |
| Tcirg1 | Q9JHF5 | V-type proton
ATPase subunit a | Up | 2.823 | Metabolic
pathways |
| Nsdhl | Q9R1J0 |
Sterol-4-alpha-carboxylate
3-dehydrogenase, decarboxylating | Up | 2.915 | Steroid
biosynthesis, metabolic pathways |
| Atp6v1c1 | Q9Z1G3 | V-type proton
ATPase subunit C 1 | Up | 2.916 | Metabolic
pathways |
| Adh1 | P00329 | Alcohol
dehydrogenase 1 | Up | 3.002 | Retinol metabolism,
metabolic pathways |
| Serpini1 | O35684 | Neuroserpin | Up | 3.026 | p53 signaling
pathway |
| Hmgcs1 | Q8JZK9 |
Hydroxymethylglutaryl-CoA synthase,
cytoplasmic | Up | 3.048 | Metabolic
pathways |
| Fasn | P19096 | Fatty acid
synthase | Up | 3.113 | Metabolic
pathways |
| Idua | Q8BLF6 |
α-L-iduronidase | Up | 3.264 | Metabolic
pathways |
| Fdps | Q920E5 | Farnesyl
pyrophosphate synthase | Up | 3.745 | Metabolic
pathways |
| Znrd1 | G3UXD3 | DNA-directed RNA
polymerase subunit | Up | 3.748 | Metabolic
pathways |
| Lss | Q8BLN5 | Lanosterol
synthase | Up | 4.634 | Steroid
biosynthesis, Metabolic pathways |
| Galnt7 | Q80VA0 |
N-acetylgalactosaminyltransferase 7 | Up | 5.906 | Metabolic
pathways |
| Fdft1 | P53798 | Squalene
synthase | Up | 7.25 | Metabolic pathways,
Steroid biosynthesis |
| Mtnd5 | P03921 | NADH-ubiquinone
oxidoreductase chain 5 | Up | 10.587 | Metabolic
pathways |
| Cyp51a1 | Q8K0C4 | Lanosterol 14-alpha
demethylase | Up | 91.003 | Steroid
biosynthesis, Metabolic pathways |
Furthermore, the 19 important differentially
expressed proteins in four biological pathways were incorporated
into the interaction analysis. In total, seven of the 19
differentially expressed proteins were revealed to have direct
interactions, including Nsdhl, Lss, Fdft1, Cyp51, Hmgcs1, Fasn and
Fdps (Fig. 6A). Combined with the
analysis results aforementioned, it was revealed that among the
seven differentially expressed proteins, Nsdhl, Lss, Fdft1 and
Cyp51a1 were enriched in 'steroid biosynthesis', whilst Hmgcs1,
Fasn and Fdps were enriched in 'metabolic pathways' (Table I; Fig. 6A). This result indicated that
after KN93 inhibited CaMKII, it may be involved in the regulation
of follicular development by affecting the expression levels of
Nsdhl, Lss, Fdft1, Cyp51a1, Hmgcs1, Fasn and Fdps in 'steroid
biosynthesis' and 'metabolic pathways'.
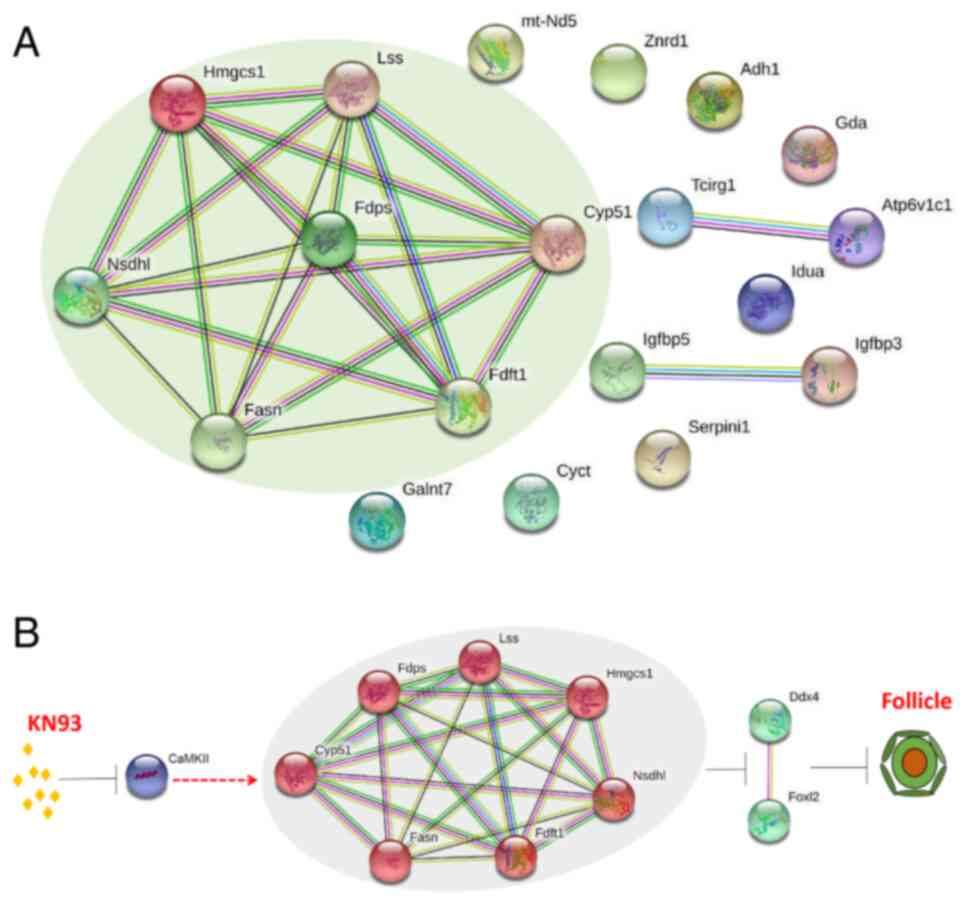 | Figure 6Analysis of potential mechanism for
KN93 retarding ovarian follicle development. (A) Interaction
analysis of differential proteins in four key KEGG enriched
pathways. Nsdhl, Lss, Fdft1, Cyp51a1, Hmgcs1, Fasn and Fdps have
direct interactions, which was marked through the pale green
elliptic region. (B) Model of KN93 retarding follicular
development. After CaMKII was inhibited by the inhibitor KN93,
seven interacting proteins were upregulated (Nsdhl, Lss, Fdft1,
Cyp51a1, Hmgcs1, Fasn and Fdps), then downregulated oocyte marker
DDX4 and granulosa cell marker FOXL2 in follicles, ultimately
slowing down primordial follicles formation and activation, thereby
leaded to the delay of the ovarian follicle development. Nsdhl,
NADP-dependent steroid dehydrogenase-like; Lss, lanosterol
synthase; Fdft1, farnesyl-diphosphate farnesyltransferase; Cyp51a1,
cytochrome P450 family 51 family A member 1; Hmgcs1,
hydroxymethylglutaryl-CoA synthase 1; Fasn, fatty acid synthase;
Fdps, dimethylallyltranstransferase; CaMKII, calmodulin-dependent
protein kinase II; DDX4, DEAD-box helicase 4; FOXL2, forkhead box
L2. |
Based on these studies, the present study mapped the
potential mechanism of KN93 inhibiting CaMKII and delaying
follicular development further, where the possible regulatory
mechanism was as follows: KN93 Inhibits CaMKII, which upregulates
the expression levels of Nsdhl, Lss, Fdft1, Cyp51a1, Hmgcs1, Fasn
and Fdps. This then downregulates the expression of the oocyte
marker DDX4 and the granulosa cell marker FOXL2 in follicles, which
ultimately slowed down the formation and activation of primordia
follicles, leading to the delay in the follicle development
(Fig. 6B).
Discussion
The ovary is the reproductive gland of female
mammals, where the follicle forms the basic structural and
functional unit of the ovary (28). Follicles provide stable and
controllable environments for oocyte development in addition to
precisely controlling the mobilization of stored follicles, which
can provide healthy oocytes for fertilization in a sustainable
manner (29,30). By contrast, the somatic cells in
the follicle can form a closely coordinated neuroendocrine system
with the hypothalamus and the pituitary gland through endocrine and
paracrine regulation, which regulates almost all physiological
activities that occur during sexual reproduction (31). Follicular development begins with
the formation of primordial follicles in the ovary. Formation of
primordial follicles not only determines whether there are oocytes
available for fertilization, establishment and maintenance of the
primordial follicle reserve pool also directly determines the
reproductive life span of female animals (32,33). It has been reported that in the
ovaries of mice at 17.5 days of embryonic stage, only part of the
cysts rupture to form a small number of primordial follicles
(27). By contrast, in the
ovaries of mice after birth, a large number of cysts rupture and
primordial follicles are formed continuously, before formalizing
the primordial follicle pool during the first 4-5 days after birth
(34). Research has demonstrated
that there are two different types of primordial follicles in the
ovary. The first type is those in the medulla of the ovary, which
are activated immediately after formation, whereas the second type
is those in the cortex, which are gradually activated during the
subsequent reproductive years (35). Once the primordial follicle is
activated, it begins further development into primary follicles and
more advanced follicles (36).
The formation and activation of primordial follicles
is a complex and delicate process that involves the co-ordination
of molecular signaling pathways in the ovary. CaMKII is one such
important regulatory molecule that has been demonstrated to serve a
role in reproductive processes, such as oocyte and early embryonic
development (18,19). However, the effects of CaMKII on
follicular development remains unknown. In the present study,
CaMKII was expressed in the ovaries of mice at embryonic stage 17.5
days and postnatal stage 1, 3 and 5 days, suggesting the
possibility of CaMKII regulating follicular development. It was
then revealed that CaMKII was mainly expressed in the cortical
regions of the ovaries in mice at 17.5 days embryonic stage and
those at 1-day postnatal stage. By contrast, it was mainly
expressed in the primordial and primary follicles in the medulla
region of the ovaries of neonatal mice at 3 and 5 days postnatal.
These results suggested that CaMKII could activate primordial
follicles. In addition, in the ovaries of newborn mice that are 3
and 5 days old, CaMKII was mainly expressed in the cytoplasm of
granulosa cells in the primordial and primary follicles in the
ovarian medulla region. Therefore, it was hypothesized that CaMKII
could transform the flattened granulosa cells in the primordial
follicles into cuboid granulosa cells, which could activate and
develop into primary follicles. However, this hypothesis requires
further experimental confirmation.
After the formation of the final primordial
follicles pool, the periodic activation of primordial follicles
will continuously deplete the number of primordial follicles,
shortening the reproductive lifespan. Therefore, adequately
regulating the speed of primordial follicle activation will prolong
the reproductive life of the ovaries (5). A number of studies have previously
reported that KN93 is a specific inhibitor of CaMKII that can
regulate various physiological and pathological processes by
specifically inhibiting CaMKII activity (20,21). However, to the best of our
knowledge, it has not been previously reported whether KN93 can
delay follicular development by inhibiting CaMKII.
In the present study, the ovaries of embryonic mice
(17.5 dpc) treated with KN93 was observed following hematoxylin
staining. It was revealed that a large proportion of primordial and
primary follicles were observed in the deep cortex and medulla of
the ovaries in the control group, but no such phenomenon was
observed in the follicles in the KN93 group. These results
suggested that KN93 could inhibit follicular development.
Immunofluorescence detection also verified that the majority of the
primordial and primary follicles were localized to the deeper
layers of the cortex and medulla in the ovaries in the control
group. However, again no such phenomenon was observed in the KN93
treatment group, further confirming that KN93 exerted an inhibitory
effect on follicular development. It has been previously reported
that DDX4 and FOXL2 are markers of oocyte and granulosa cells in
the follicles that can be used for determining the fate of
follicular development (37,38). In the present study,
immunofluorescence revealed that the fluorescence intensity of the
oocyte marker DDX4 in follicles treated with KN93 was significantly
weaker compared with that in the control group. By contrast, the
fluorescence intensity of the granulosa cell marker FOXL2 in
follicles was not altered compared with that in the control group.
However, western blotting demonstrated that the expression of DDX4,
CaMKII and FOXL2 in the ovaries after KN93 treatment group was
significantly decreased. These results suggested that KN93 could
reduce the expression levels of DDX4 and FOXL2 by inhibiting CaMKII
to delay the development of follicles and activation of primordial
follicles.
Although the results aforementioned reported that
KN93 can inhibit CaMKII, reduce DDX4 and FOXL2 expression and delay
follicular development, a more detailed mechanistic model
underlying the effects of KN93 on follicular development remains to
be fully elucidated. This is because after KN93 inhibits CaMKII
activity, its downstream signaling target proteins remain unknown.
To reveal the downstream target proteins that were altered after
KN93 treatment, proteomics techniques were applied to measure these
proteins in vitro in cultured ovaries treated with KN93. The
expression levels of 262 proteins were demonstrated to be
significantly altered between the KN93 treatment group and the
control group. Furthermore, the expression of 10 differentially
expressed proteins were measured by western blotting to validate
the accuracy of the proteomics technique. Previous studies reported
that a wide range of important signaling pathways are involved in
the regulation of follicular development, including Notch, PI3K,
KIT, TGF-β and JNK (9-13). In the present study, GO and KEGG
enrichment analysis was performed on the 262 differentially
expressed proteins obtained, which were mainly enriched in
molecular and biological pathways associated with reproductive
development, cell proliferation and differentiation or migration.
In addition, four pathways were revealed to be closely associated
reproductive development, namely 'steroid biosynthesis', 'p53
signaling pathway', 'retinol metabolism' and 'metabolic
pathways'.
Discovery of the biological pathways aforementioned
provided potentially important targets for studying the mechanism
underlying the KN93-mediated delayed in follicular development.
Therefore, the differentially expressed proteins were enriched in
the four biological pathways of 'steroid biosynthesis', 'p53
signaling pathway', 'retinol metabolism and 'metabolic pathways'.
Finally, 19 enriched differentially expressed proteins were
obtained from these four biological pathways. Nevertheless, western
blotting detection of these 19 differentially expressed proteins
could not be carried out in the current study. This was because 10
differentially expressed proteins were randomly detected by
proteomics using western blotting, which confirmed the reliability
of the proteomic detection results in the study. Another important
reason was that the ovarian materials of embryonic mice were
limited. The ovaries required in the study were from the female
embryos of 17.5 d pregnant mice and collected after in vitro
culture for 4 days. The amount of proteins extracted from every
8-10 ovaries could only complete the detection of two proteins at a
time. Therefore, to complete the western blotting analysis of the
19 differentially expressed proteins, enough ovaries must be
collected, which would take several months or longer. Although
these 19 differentially expressed proteins cannot be detected one
by one in the present study, these 19 differentially expressed
proteins will be studied one by one in the follow-up study.
In spite of this, for these 19 differential
proteins, an interaction analysis was performed further, where
among the 19 differentially expressed proteins, seven were
upregulated, including Nsdhl, Lss, Fdft1, Cyp51a1, Hmgcs1, Fasn and
Fdps, all of which interact which each other. The current results
suggested that these seven differentially expressed proteins were
the key downstream targets after CaMKII inhibition by KN93. A
number of studies have revealed that Nsdhl, Lss, Fdft1, Cyp51a1,
Hmgcs1, Fasn and Fdps can serve important roles in cholesterol
synthesis by promoting this process (39-42). In addition, another study has also
revealed that cholesterol synthesis is closely associated with
ovarian reserve function, whereby patients with diminished ovarian
reserves tended to exhibit significantly lower cholesterol
metabolic levels compared with those with normal ovarian reserve
(43). Based on these reports, it
was hypothesized that these seven upregulated proteins may be
involved in cholesterol synthesis during follicular development,
thereby improving ovarian reserve function. In addition, the
potential mechanism underlying the effects of KN93 inhibiting
CaMKII on delaying follicular development can be summarized as
follows: KN93 Inhibits CaMKII, which upregulates the expression of
Nsdhl, Lss, Fdft1, Cyp51a1, Hmgcs1, Fasn and Fdps to promote
cholesterol synthesis. This then downregulates the expression of
the oocyte marker DDX4 and the granulosa cell marker FOXL2 in
follicles, leading to the delay in the formation and activation of
primordial follicles. Although the potential mechanism of KN93
inhibiting CaMKII in delaying follicular development has been
suggested by data from the present study, its authenticity require
validation in further studies.
To the best of our knowledge, the present study
demonstrated for the first time that KN93, an inhibitor of CaMKII,
can directly delay follicular development in mouse ovaries and
reveal its underlying regulatory mechanism. Therefore, these
results suggest that KN93 may be a promising therapeutic agent for
delaying follicular development, thereby prolonging the
reproductive life of female individuals in the future.
Supplementary Data
Availability of data and materials
The mass spectrometry proteomics data have been
deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the
iProX partner repository with the dataset identifier PXD035136. The
datasets used and/or analyzed during the current study are
available from the corresponding author on reasonable request.
Authors' contributions
JY, XX and YM performed the experiments. YY and CW
analyzed the data. GX conducted literature search and analyzed the
data. XD analyzed and interpreted the immunofluorescence images. XL
and XD can confirm the authenticity of all the raw data. XL
designed the study. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
All experiments were reviewed and approved by the
Animal Ethical and Welfare Committee of Ningxia University
(approval no. IACUC-NDLAC-2020019).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgments
Not applicable.
Funding
The present study was supported by the Natural Science
Foundation of Ningxia Province (grant no. 2020AAC03115), the
Cultivation Foundation for Outstanding Young Teachers of Ningxia
Higher Education Institutions (grant no. 15-1464) and National Key
Research and Development Program of China (grant no.
2018YFC1003701).
References
|
1
|
Shah JS, Sabouni R, Vaught KCC, Owen CM,
Albertini DF and Segars JH: Biomechanics and mechanical signaling
in the ovary: A systematic review. J Assist Reprod Genet.
35:1135–1148. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Green LJ and Shikanov A: In vitro culture
methods of preantral follicles. Theriogenology. 86:229–238. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
West ER, Shea LD and Woodruff TK:
Engineering the follicle microenvironment. Semin Reprod Med.
25:287–299. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Simon LE, Kumar TR and Duncan FE: In vitro
ovarian follicle growth: A comprehensive analysis of key protocol
variables. Biol Reprod. 103:455–470. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Zhang T, He M, Zhao L, Qin S, Zhu Z, Du X,
Zhou B, Yang Y, Liu X, Xia G, et al: HDAC6 regulates primordial
follicle activation through mTOR signaling pathway. Cell Death Dis.
12:5592021. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Guo R and Pankhurst MW: Accelerated
ovarian reserve depletion in female anti Müllerian hormone knockout
mice has no effect on lifetime fertility†. Biol Reprod.
102:915–922. 2020. View Article : Google Scholar
|
|
7
|
Chen J, Liu W, Lee KF, Liu K, Wong BPC and
Yeung WSB: Overexpression of Lin28a induces a primary ovarian
insufficiency phenotype via facilitation of primordial follicle
activation in mice. Mole Cell Endocrinol. 539:1114602022.
View Article : Google Scholar
|
|
8
|
Zhang J, Yan L, Wang Y, Zhang S, Xu X, Dai
Y, Zhao S, Li Z, Zhang Y, Xia G, et al: In vivo and in vitro
activation of dormant primordial follicles by EGF treatment in
mouse and human. Clin Transl Med. 10:e1822020. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Xu J and Gridley T: Notch2 is required in
somatic cells for break-down of ovarian germ-cell nests and
formation of primordial follicles. BMC Biol. 11:132013. View Article : Google Scholar
|
|
10
|
Zhang H and Liu K: Cellular and molecular
regulation of the activation of mammalian primordial follicles:
Somatic cells initiate follicle activation in adulthood. Hum Reprod
Update. 21:779–786. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Huang K, Wang Y, Zhang T, He M, Sun G, Wen
J, Yan H, Cai H, Yong C, Xia G and Wang C: JAK signaling regulates
germline cyst breakdown and primordial follicle formation in mice.
Biol Open. 7:bio0294702018.
|
|
12
|
Wang ZP, Mu XY, Guo M, Wang YJ, Teng Z,
Mao GP, Niu WB, Feng LZ, Zhao LH and Xia GL: Transforming growth
factor-β signaling participates in the maintenance of the
primordial follicle pool in the mouse ovary. J Biol Chem.
289:8299–8311. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Komatsu K and Masubuchi S: Increased
supply from blood vessels promotes the activation of dormant
primordial follicles in mouse ovaries. J Reprod Dev. 66:105–113.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Hea Q, Chengb J and Wang Y: Chronic CaMKII
inhibition reverses cardiac function and cardiac reserve in HF
mice. Life Sci. 219:122–128. 2019. View Article : Google Scholar
|
|
15
|
Zhu Q, Hao L, Shen Q, Pan J, Liu W, Gong
W, Hu L, Xiao W, Wang M, Liu X, et al: CaMK II Inhibition
attenuates ROS dependent necroptosis in acinar cells and protects
against acute pancreatitis in mice. Oxid Med Cell Longev.
17:41873982021.
|
|
16
|
Altobelli GG, Van Noorden S, Cimini D,
Illario M, Sorriento1 D and Cimini V: Calcium/calmodulin-dependent
kinases can regulate the TSH expression in the rat pituitary. J
Endocrinol Invest. 44:2387–2394. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Hoch B, Haase H, Schulze W, Haqemann D,
Morano I, Krause EG and Karczewski P: Differentiation-dependent
expression of cardiac delta-CaMKII isoforms. J Cell Biochem.
68:259–268. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Medvedev S, Stein P and Schultz RM:
Specificity of calcium/calmodulin-dependent protein kinases in
mouse egg activation. Cell Cycle. 13:1482–1488. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Jin XL and O'Neill C: The presence and
activation of two essential transcription factors (cAMP response
element-binding protein and cAMP-dependent transcription factor
ATF1) in the two-cell mouse embryo. Biol Reprod. 82:459–468. 2010.
View Article : Google Scholar
|
|
20
|
Wang J, Xu X, Jia W, Zhao D, Boczek T, Gao
Q, Wang Q, Fu Y, He M, Shi R, et al: Calcium-/calmodulin-dependent
protein kinase II (CaMKII) inhibition induces learning and memory
impairment and apoptosis. Oxid Med Cell Longev. 2021:46350542021.
View Article : Google Scholar :
|
|
21
|
Johnson CN, Pattanayek R, Potet F, Rebbeck
RT, Blackwell DJ, Nikolaienko R, Sequeira V, Meur RL, Radwański PB,
Davis JP, et al: The CaMKII inhibitor KN93-calmodulin interaction
and implications for calmodulin tuning of NaV1.5 and RyR2 function.
Cell Calcium. 82:1020632019. View Article : Google Scholar :
|
|
22
|
Yang X, Wu N, Song L and Liu Z:
Intrastriatal injections of KN-93 ameliorates levodopa-induced
dyskinesia in a rat model of Parkinson's disease. Neuropsychiatr
Dis Treat. 9:1213–1220. 2013.PubMed/NCBI
|
|
23
|
Liu Y, Liang Y, Hou B, Liu M, Yang X, Liu
C, Zhang J, Zhang W, Ma Z and Gu X: The inhibitor of
calcium/calmodulin-dependent protein kinase II KN93 attenuates bone
cancer pain via inhibition of KIF17/NR2B traffiffifficking in mice.
Pharmacol Biochem Behav. 124:19–26. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Edvinsson L, Povlsen GK, Ahnstedt H and
Waldsee R: CaMKII inhibition with KN93 attenuates endothelin and
serotonin receptor-mediated vasoconstriction and prevents
subarachnoid hemorrhage-induced deficits in sensorimotor function.
J Neuroinflammation. 11:2072014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Chen W, An P, Quan XZ, Zhang J, Zhou ZY,
Zou LP and Luo HS: Ca2+/calmodulin-dependent protein
kinase II regulates colon cancer proliferation and migration via
ERK1/2 and p38 pathways. World J Gastroenterol. 23:6111–6118. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Shin H, Han C, Labuz ML, Kim J, Kim J, Cho
S, Gho YS, Takayama S and Parka J: High-yield isolation of
extracellular vesicles using aqueous two-phase system. Sci Rep.
5:131032015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
He Y, Chen Q, Dai J, Cui Y, Zhang C, Wen
X, Li J, Xiao Y, Peng X, Liu M, et al: Single-cell RNA-Seq reveals
a highly coordinated transcriptional program in mouse germ cells
during primordial follicle formation. Aging Cell. 20:e134242021.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Bazot M, Robert Y, Mestdagh P, Boudghène F
and Rocourt N: Ovarian functional disorders. J Radiol.
81:1801–1818. 2000.In French.
|
|
29
|
Fortune JE, Rivera GM and Yang MY:
Follicular development: The role of the follicular microenvironment
in selection of the dominant follicle. Anim Reprod Sci. 82:109–126.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Sun L, Chen L, Jiang Y, Zhao Y, Wang F,
Zheng X, Li C and Zhou X: Metabolomic profifiling of ovary in mice
treated with FSH using ultra performance liquid chromatography/mass
spectrometry. Biosci Rep. 38:BSR201809652018. View Article : Google Scholar
|
|
31
|
Hirshfifield AN: Development of follicles
in the mammalian ovary. Int Rev Cytol. 124:43–101. 1991. View Article : Google Scholar
|
|
32
|
Li T, Liu X, Gong X, Qiukai E and Zhang X
and Zhang X: microRNA 92b-3p regulate primordial follicle assembly
by targeting TSC1 in neonatal mouse ovaries. Cell Cycle.
18:824–833. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Wang J, Ge W, Zhai QY, Liu JC, Sun XW, Liu
WX, Li L, Lei CZ, Dyce PW, De Felici M and Shen W: Single-cell
transcriptome landscape of ovarian cells during primordial follicle
assembly in mice. PLoS Biol. 18:e30010252020. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Bristol-Gould SK, Kreeger PK, Selkirk CG,
Kilen SM, Cook RW, Kipp JL, Shea LD, Mayo KE and Woodruff TE:
Postnatal regulation of germ cells by activin: The establishment of
the initial follicle pool. Dev Biol. 298:132–148. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Yin H, Kristensen SG, Jiang H, Rasmussen A
and Andersen CY: Survival and growth of isolated pre-antral
follicles from human ovarian medulla tissue during long-term 3D
culture. Hum Reprod. 31:1531–1539. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Zhai J, Zhang J, Zhang L, Liu X, Deng W,
Wang H, Zhang Z, Liu W, Chen B, Wu C, et al: Autotransplantation of
the ovarian cortex after in vitro activation for infertility
treatment: A shortened procedure. Hum Reprod. 36:2134–2147. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Song K, Ma W, Huang C, Ding J, Cui D and
Zhang M: expression pattern of mouse vasa homologue (MVH) in the
ovaries of C57BL/6 female mice. Med Sci Monit. 22:2656–2663. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Georges A, Auguste A, Bessière L, Vanet A,
Todeschini AL and Veitia RA: FOXL2: A central transcription factor
of the ovary. J Mol Endocrinol. 52:R17–R33. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Su Y, Sugiura K, Wigglesworth K, O'Brien
MJ, Affourtit JP, Pangas SA, Matzuk MM and Eppig JJ: Oocyte
regulation of metabolic cooperativity between mouse cumulus cells
and oocytes: BMP15 and GDF9 control cholesterol biosynthesis in
cumulus cells. Development. 135:111–121. 2008. View Article : Google Scholar
|
|
40
|
Ershov P, Kaluzhskiy L, Mezentsev Y,
Yablokov E, Gnedenko O and Ivanov A: Enzymes in the cholesterol
synthesis pathway: Interactomics in the cancer context.
Biomedicines. 9:8952021. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Nakamura T, Iwase A, Bayasula B, Nagatomo
Y, Kondo M, Nakahara T, Takikawa S, Goto M, Kotani T, Kiyono T and
Kikkawa F: CYP51A1 induced by growth differentiation factor 9 and
follicle-stimulating hormone in granulosa cells is a possible
predictor for unfertilization. Reprod Sci. 22:377–384. 2015.
View Article : Google Scholar :
|
|
42
|
Jin Y, Chen Z, Dong J, Wang B, Fan S, Yang
X and Cui M: SREBP1/FASN/cholesterol axis facilitates
radioresistance in colorectal cancer. FEBS Open Bio. 11:1343–1352.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Yang X, Zhao Z, Fan Q, Li H, Liu LZC and
Liang X: Cholesterol metabolism is decreased in patients with
diminished ovarian reserve. Reprod Biomed Online. 44:185–192. 2022.
View Article : Google Scholar
|