1. Introduction
Alzheimer's disease (AD) is a complex disorder
characterized by the gradual loss of memory and the
self-sufficiency of patients, the deterioration of thinking, and of
the usage and understanding of written and spoken language, social
isolation. It is also associated with behavioral changes, due to a
confused state accompanied by apathy, depression and aggression
(1-6). This disease occurs mainly in
individuals >60 years of age; however, there is an increasing
prevalence of the disorder in ~40% of the population <65 years
of age (7). Considering this
increasing prevalence of the disease, as well as the considerable
socio-economic burden and the absence of any specialized treatment,
it is important to make efforts to enhance the understanding of the
pathophysiological mechanisms that lead to the development of AD
(2,5,8).
AD occurs mainly sporadically without being due to a
specific genetic background, with age being the main risk factor
(1). The progressive atrophy of
the cerebral regions of the hippocampus and cortex are
representative macroscopic features of the disease and are clearly
visible on a neuroimaging examination, while extracellular deposits
of the amyloid-β peptide (Aβ1-42) and intraneuronal tangles of
hyperphosphorylated forms of microtubule-associated protein tau are
some of the microscopic features of the disease. The activation of
microglia associated with β-amyloid behavior, as well as the
inflammatory response have been the focus of several studies on the
contribution of β-amyloid cataracts to the development of AD
(9-13).
Duing the early onset of AD, causal mutations in
specific genes have been identified. The main genes are amyloid
precursor protein (APP), presenilin-1 (PSEN1), and presenilin-2
(PSEN2). PSEN1 and PSEN2 are proteases involved in the conversion
of APP into Aβ42, and are related neurotoxic products. The
production of Aβ42 does not necessarily increase due to the
abnormal variants of presenilin, but causes the production of other
forms with a high tendency to cause agglomeration (14-16). On the other hand, the
apolipoprotein E (ApoE) gene is the most well-known and important
risk factor for the development of AD. This gene has three
isoforms; however, 25% of AD cases carry the ε4 allele, while
generally, the presence of two ApoE4 alleles increases the risk
10-fold compared to the presence of one allele (6,17).
Although the mechanisms through which ApoE4 increases the risk of
developing AD are not yet known, when this protein is poorly
lipidated it binds to Aβ42 and is associated with its greater
accumulation and oligomerization in the brain, as well as with
reduced extracellular, microglial and the blood-brain
barrier-mediated clearance of Aβ42. Thus, the destructive effect of
Aβ42 on the function of synapses is aggravated (18-20).
A number of recent studies have focused on the
search and detection of genetic loci or genes that are risk factors
for AD, through the study and analysis of the genome (21). Genetic loci or genes, such as
NME8, FERMT2, PICALM, PTK2B, CD2AP, CD33, CELF1SLC24A4/RIN3,
FERMT2, CASS4 and DGS2 appear to be associated with the development
of late-onset AD due to the single nucleotide polymorphisms (SNPs)
that they contain (22). Aside
from genetic factors, an array of environmental and lifestyle
factors are known as risk factors for the development of AD,
ranging from exposure to toxins to a high-cholesterol diet, while
non-coding RNAs (ncRNAs) and other epigenetic mechanisms play a key
role in their harmful effects (19,23,24).
Currently, various ncRNAs have been detected and
studied in accordance with their involvement in AD. The main
categories of these molecules are microRNAs (miRNAs/miRs), circular
RNAs (circRNAs), Piwi-interacting RNAs (piRNAs) and long non-coding
RNAs (lncRNAs). The functions of these molecules, and their ability
to interact with each other, as well with DNA and proteins result
in the regulation of gene expression, since they either promote or
inhibit the expression of genes. In addition, their expression is
altered due to a pathological condition, rendering them effective
biomarkers for the early diagnosis of diseases, including cancer
and neurodegenerative diseases.
The emergence of 'omics' technologies has
revolutionized the study of complex pathologies and diseases,
including AD. The applications of omics platforms involve the
recognition and the study of genes (genomics), messenger RNAs
(mRNAs, transcriptomics), epigenomic factors (epigenomics),
proteins (proteomics), metabolites (metabolomics) and lipids
(lipidomics). In addition, the interest of the gut microbiota (the
microbiome/microbiomics) is increasing due to its association with
various diseases (25). The
analysis and combination of data derived from different omics
technologies is crucial for complex pathologies, such as AD in
order to acquire a complete knowledge of the disease. Cerebrospinal
fluid (CSF) and blood were the main samples used in omics studies
on patients with AD, with the former being in close contact with
neurons, containing several soluble biomarkers of the brain, and
thus reflecting the changes that occur during the disease (26,27). More specifically, the increase in
total tau protein (t-tau) and phosphorylated tau protein (p-tau),
as well as the decrease in Aβ42 in the CSF, reflect the formation
of amyloid plaques and neurofibrillary tangles in the brain tissue,
which are characteristics of AD. However, the etiology of AD
depends on, and is due to multiple factors, including genetic
alterations, proteins and ncRNAs (28).
Omics technologies are a promising tool for studying
the multifaceted pathology of AD. With the advancement of
technology, the era of omics enables the collection of diverse
data, as well as the analysis and filtering of data, which is
carried out through cutting-edge computational tools. The
importance and value of omics and ncRNAs are highlighted through
the process of the development of personalized diagnostic and
therapeutic tools. For this purpose, various studies have focused
on novel pathways and networks, demonstrating novel pathological
mechanisms related to AD and linked to other diseases (29-31). The present review aimed to
summarize the current evidence on the role and utilization of
ncRNAs as biomarkers in AD, as well as to describe the application
of omics technologies and large-scale data in the efficient
prediction, diagnosis and treatment of AD.
2. Role of ncRNAs in AD
ncRNAs have several distinct classes. The most
well-studied category is that of miRNAs, whose function is now
well-understood. The role of miRNA epigenetic and genetic defects
has been shown in a variety of diseases, including cardiovascular
diseases (32), metabolic
syndrome (33) and cancer
(34) and pathologies of the
nervous system, such as AD (35).
However, in addition to miRNAs, there are several other classes of
ncRNAs, such as small nucleolar RNAs (snoRNAs), circRNAs, piRNAs, Y
RNAs and the large heterogeneous group of lncRNAs, key factors in
the development of various human disorders, including AD, and with
potential use as biomarkers (36)
(Table I).
 | Table IncRNAs in AD. |
Table I
ncRNAs in AD.
| miRNAs | (Refs.) | circRNAs | (Refs.) | piRNAs | (Refs.) | lncRNAs | (Refs.) |
|---|
| miR-106a | (42) | ciRS-7 | (71,81-83) | piR-38240 | (92) | BACE1-AS | (100-103) |
| miR-520c | (42) | circHOMER1 | (63,84) | piR-34393 | (92) | NAT-Rad18 | (104,105) |
| miR-20a | (43,44) | circCORO1C | (63,84) | piR-40666 | (92) | 51A | (105,106) |
| miR-19 | (43,44) | circRNA
KIAA1586 | (63,85) | piR-51810 | (92) | 17A | (107) |
| miR-106b | (43,44) | circHDAC9 | (63,86) | piR-hsa-1282 | (92) | BCYRN1 | (105,109) |
| miR-20a family | (43,44) |
circRNA_0000950 | (63,87) | piR-hsa-23538 | (92) | AD-linc2 | (111) |
| miR-101 | (45) | circNF1-419 | (63,88,89) | piR-hsa-23566 | (92) | HAO2-AS | (111) |
| miR-147 | (46,47) | | | piR-hsa-27400 | (92) | EBF3-AS | (111) |
| miR-124 | (48-50) | | | piR-hsa-27725 | (92) | AD-linc1 | (111) |
| miR-339-5p | (36) | | | piR-hsa-28116 | (92) | MAGI2-AS3 | (108) |
| miR-195 | (61) | | | piR-hsa-28189 | (92) | | |
| miR-107 | (62) | | | piR-hsa-28390 | (92) | | |
| miR-9 | (64) | | | piR-hsa-29114 | (92) | | |
|
miR-144/miR-451 | (66) | | | piR-hsa-7193 | (92) | | |
| miR-181c | (71,72) | | | | | | |
| miR-146a | (73-75) | | | | | | |
| miR-298/328 | (57) | | | | | | |
| miR-135a | (58) | | | | | | |
| miR-135b | (59) | | | | | | |
| miR-455-3p | (76) | | | | | | |
| miR-485-3p | (77) | | | | | | |
miRNAs
miRNAs are a class of small ncRNAs ~20-24
nucleotides in length, which bind to the 3′untranslated region
(3′UTR) of target mRNAs, leading to post-transcriptional silencing
either by transcription, degradation, or translational repression.
To date, >2,000 miRNAs have been identified in the human genome
that play a key role in critical biological processes (37), such as development, apoptosis,
signal transduction and proliferation. With respect to the brain,
they are expressed in neurons and are involved in processes of
neuronal differentiation, synaptogenesis and plasticity (38). According to the literature, miRNAs
have a significant impact on the development of several
neurological diseases and disorders, such as AD, Parkinson's
disease, amyotrophic lateral sclerosis and Huntington's disease
(39,40).
As regards AD, miRNAs have been shown to be involved
in Aβ pathology by regulating APP expression and other enzymes
involved in Aβ processing, such as β-secretase (BACE1). The first
study to record the regulatory role of miRNAs in APP mRNA involved
the homologous APP gene in C. elegans, APL-1, which showed
its developmental regulation by miRNA let-7 (41). Subsequent studies have
demonstrated that APP is regulated by miRNA in humans, where the
overexpression of miR-106a and miR-520c has been shown to lead to
the translational suppression of APP mRNA, thereby significantly
reducing APP levels (42). In
addition, miR-20a, miR-19, miR-106b, the miR-20a family (43,44) and miR-101 (45) have been shown to directly regulate
APP mRNA in human cells in vitro. In addition, the effect of
SNPs on miRNA binding sites in the 3′-UTR of APP mRNA in AD
pathology and the risk of AD are demonstrated, where more
specifically, miR-147 and miR-20a were shown to be the affected
SNPs variants associated with AD in the 3′-UTR of APP mRNA
(46,47). Finally, miRNAs, such as miR-124,
which regulates the expression of the polypyrimidine tract-binding
protein 1 (PTB1) in neuronal cell lines, have also been implicated
in the regulation of the alternative splicing of APP (48-50). In general, there is significant
evidence of the increased levels of exon 7 and 8 isoforms of APP in
brains of patients with AD, while abnormal APP splicing has been
shown to be associated with an increased Aβ production (51,52).
The importance of BACE1 activity in AD lies in the
fact that this factor cleaves APP as the first and rate-limiting
step in the formation of Aβ (53). In this case, miRNAs belonging to
the miR-29 family have been well-studied in vitro and in
vivo. The three major mature miRNAs in this family are miR-29a,
miR-29b and miR-29c, the latter of which has been shown to regulate
BACE1 expression by targeting the 3′-UTR in both human and mouse
cell lines (54-56). The aforementioned miR-29, as well
as other miRNAs that directly target BACE1 in vitro, such as
miR-298/328 (57), miR-135a,
miR-135b (59), miR-9 (60), miR-298, miR-339-5p, miR-195
(61) and miR-107 (62), are deregulated in brains affected
by AD, mainly exhibited a reduced expression (36,63).
miR-9 is another miRNA involved in Aβ regulation,
targeting calcium/calmodulin-dependent protein kinase kinase 2
(CAMKK2), thereby attenuating Aβ-induced synaptic toxicity
(64). In addition, this miRNA
appears to be involved in insulin signaling, which may be
associated with an increased risk of developing diabetes in
patients with AD (65). The
miR-144/miR-451, finally, has been shown to regulate α-secretase
ADAM10, which protects the brain from the production of Aβ
(66).
The search and discovery of specialized and
effective biomarkers for the prediction and early detection of AD
is of utmost importance for the better management of symptoms and
timely intervention (67,68). For this purpose, miRNAs have been
proposed as promising candidate biomarkers due to their high
stability under storage and handling conditions (69). Through qPCR and RNA-seq studies,
it has now become possible to identify circulating miRNAs in plasma
and CSF that serve as biomarkers for AD and to construct miRNA
catalogs that are differentially expressed between AD and control
groups to identify new biomarkers (70). Two miRNAs that have been
identified as suitable biomarkers for AD are miR-181c (71,72) and miR-146a (73-75). The former was found to be
downregulated in the serum and CSF of patients with AD, while the
latter was found to be upregulated in brains affected by AD, as
well as in the CSF of patients with AD. miR-455-3p is another
potential biomarker for AD, as its level is higher in the serum of
patients with AD (76), as well
as miR-485-3p, whose expression was found to be upregulated in
patients with AD and cell models (77).
circRNAs
circRNAs are a class of non-coding RNAs that
originate primarily from the exonic regions of the genes encoding
proteins. Their length is variable, while they display significant
stability. These ncRNAs act as regulators of miRNAs, to which they
bind through specific binding sites. circRNAs are expressed in
central nervous system (CNS) tissue and tend to accumulate during
the normal aging process of the brain, exhibiting susceptibility to
age-related neurodegenerative diseases, such as AD. This renders
them potential therapeutic targets and biomarkers for the diagnosis
and treatment of AD (78-80).
A well-studied circRNA that has been linked to AD is
ciRS-7. This RNA binds to the well-preserved miRNA-7, which is
abundant in the human brain. More specifically, ciRS-7 contains
several binding sites specific for miRNA-7 and acts as a 'sponge',
thus inhibiting the functions of miRNA-7 (81). In the hippocampus of patients with
AD, there is a downregulation of ciRS-7 and, consequently, of its
activity as a miRNA-7 sponge, causing the latter to exhibit
increased endogenous levels in AD (71,82). The upregulation of miRNA-7 has the
ability to target and downregulate ubiquitin protein ligase, UBE2A,
which is involved in the autophagic clearance of amyloid peptides
in the brain affected by AD (83).
In addition, additional studies have reported two
other circRNAs that are dysregulated in cortical areas in AD,
namely circHOMER1 and circCORO1C. These ncRNAs are significantly
associated with the neuropathological status of AD, as they bind
two miRNAs, miR-651 and miR-105, respectively, which target both
APP and SNCA42 and are associated with the pathology of AD
(63,84). circRNA KIAA1586 is another circRNA
that functions as a miRNA sponge, which specifically binds several
miRNAs, including miR-29b, miR-101 and miR-15a, that regulate
different AD-associated genes (63,85). Moreover, circHDAC9 binds miR-138
and its expression is decreased in AD, resulting in the
downregulation of ADAM10 by miR-138, thus promoting Aβ production
(63,86).
In this context, circRNA_0000950 functions as a
miR-103 sponge, leading to the upregulation of the
prostaglandin-endoperoxide synthase 2 and interleukin (IL)-6 and
IL-1β, as well as tumor necrosis factor (TNF), and results in an
increase in neuronal cell apoptosis (63,87). CircNF1-419 is related to early
neuropathological changes and interacts with dynamin-1 and adaptor
protein 2 B1. Its overexpression reduces the levels of AD marker
proteins, such as tau, p-tau, Aβ1-42 and ApoE, and inflammatory
factors, including TNF and the nuclear factor kappa B subunit 1,
resulting in delayed senile dementia and AD progression (63,88,89). It is therefore clear that circRNAs
may play a critical role in AD, mainly as miRNA sponges (63), where the inhibition of 'sponging
miRNA activity', which translates to the upregulation of specific
miRNAs, is a possible reason for the downregulation of important
genes associated with the brain in AD (83).
piRNAs
piRNAs are also a class of ncRNAs that are ~24-34
nucleotides in length and are associated with AD, as they are
involved in CNS stress and physical damage response. These ncRNAs
interact with a specific family of Argonaute-associated 'MILI/MIWI'
RNA-binding proteins and can affect the cytoplasmic translation of
mRNAs into proteins, as well as the transcription of genes by
influencing histones and the methylation of DNA (17).
In general, piRNAs appear to be overexpressed in
neurodegenerative diseases (90).
In the study by Qiu et al (91), 9,453 piRNAs were detected in the
brains of patients with AD, and 103 piRNAs were associated with the
risk of developing AD, of which 81 were upregulated and 22 were
downregulated. piRNAs are considered a potential biomarker for AD
due to their association with SNPs of genome-wide significant risk,
such as ApoE (91). In addition,
in the study by Roy et al (92), 146 piRNAs were found to be
upregulated in patients with AD, while they were associated with
five critical AD-related pathway targets. More specifically, the
enrichment of the CYCS, LIN7C, KPNA6 and RAB11A genes was observed,
regulated by four piRNAs, piR-38240, piR-34393, piR-40666 and
piR-51810 (92). Finally, the
analysis of two different AD datasets led to the identification of
10 overlapping, differentially expressed piRNAs, with potential as
biomarkers for AD. These piRNAs include piR-hsa-1282,
piR-hsa-23538, piR-hsa-23566, piR-hsa-27400, piR-hsa-27725,
piR-hsa-28116, piR-hsa-28189, piR-hsa-28390, piR-hsa-29114 and
piR-hsa-7193 (92).
lncRNAs
lncRNAs are another class of ncRNAs that has
attracted scientific interest in the battle against
neurodegenerative diseases. They are >200 nucleotides in length
and can be derived from different regions of the genome, such as
promoters, enhancers, introns, UTRs, overlapping or non-coding
isoforms of coding genes, antisense (AS) to other transcripts and
pseudogenes (93,94). Through technological advances and
new laboratory techniques, a vast amount of information has been
collected on the role of lncRNAs in a variety of vital biological
processes (95), including
transcription (96), alternative
splicing (97), translation,
apoptosis (98) and the cell
cycle (99).
RNA sequencing methods have enabled the study of
lncRNAs and their role in various diseases, including AD. In
general, the majority of examples of lncRNAs whose activity has
been studied belong to the subcategory of AS lncRNAs. A
well-studied lncRNA with an elucidated involvement in AD is lncRNA
BACE1-AS (100), which is
transcribed from the complementary strand of the BACE1 gene. This
lncRNA is in direct involvement with elevated levels of Aβ 1-42 in
AD, as it drives the feed-forward regulation of β-secretase
(63,101,102), and it can bind to miR-214-3p,
promoting autophagy-mediated neuronal damage through the
miR-214-3p/ATG5 signaling axis in AD (103). In addition to BACE1-AS lncRNA,
NAT-Rad18 (104,105) and 51A (63,105,106) are two other lncRNAs involved in
AD, as the former has been shown to be upregulated in rat neurons
in response to the Aβ peptide, and the latter affects the formation
of Aβ and is upregulated in AD by overlapping it with SORL1. 17A is
another AS lncRNA, which is complementary to an endogenous region
of the GABA receptor gene. The expression of this lncRNA leads to
the production of alternative splicing transcripts of this receptor
which, in combination with its upregulation in AD, leads to
increased Aβ secretion in neuroblastoma cells (107). According to the study by Zhang
and Wang (108), the expression
of MAGI2-AS3 lncRNA was increased in AD cell models. This lncRNA is
a regulator of cell viability in diverse diseases. Its
overexpression enhances the effects of Aβ25-35 on neuronal
viability and neuroinflammation, and its knockdown reduces
neurotoxicity and neuroinflammation, highlighting the potential
role of MAGI2-AS3 in AD progression and treatment. Moreover, this
lncRNA functions as an miR-374b-5p sponge, a miRNA that targets
BACE1 mRNA and interacts with AKT1, RECK, WNT16, VEGFA, TACC1 and
SRSF7 mRNAs, and with compounds including cisplatin, gemcitabine,
tamoxifen and 5-fluorouracil (108). Long intergenic ncRNAs (lincRNAs)
are another subcategory of lncRNAs that are abundant in the human
genome. The primate-specific BC200 RNA (BCYRN1) is a lincRNA that
may be involved in AD. This RNA was detected in the dendritic
domains of neurons and its downregulation was observed during aging
(63,105,109).
The critical role of lncRNAs in AD was first
evidenced through the study of Zhou and Xu (110), which used an algorithm to
analyze microarray data from the brain and identified ~100 lncRNAs
that were altered in AD. Notably, a number of these lncRNAs were
specific to the brain and could be used as biomarkers for AD, since
altered expression signatures of lncRNAs provided the ability to
predict AD with the same accuracy as the altered protein-coding
genes, while requiring fewer lncRNAs for optimal prediction in
comparison to protein-coding genes (110). In addition, in a study by
Magistri et al (111),
significant alterations in the lncRNA expression profile were
observed in brains affected by AD, with the majority of the altered
lncRNAs found to be intergenic. According to the results of that
study, the AS lncRNAs, AD-linc2, HAO2-AS and EBF3-AS were dependent
on neuronal activity, while AD-linc1 was upregulated in response to
Aβ (111). These efforts are
only the beginning of a long road toward the complete elucidation
of the involvement of lncRNAs in AD, with further studies required
to explore their potential as novel biomarkers and pharmacological
targets.
In summary, the main ncRNAs that are involved in AD
are classified into four categories, miRNAs, piRNAs, circRNAs and
lncRNAs. These non-coding molecules function via several
mechanisms, such as inhibiting or promoting the expression of genes
that are associated with AD, or they can be used as effective
biomarkers as their expression is altered in AD-affected brains.
These two types of classification of those ncRNAs, based on their
category and based on their function, are illustrated in Fig. 1.
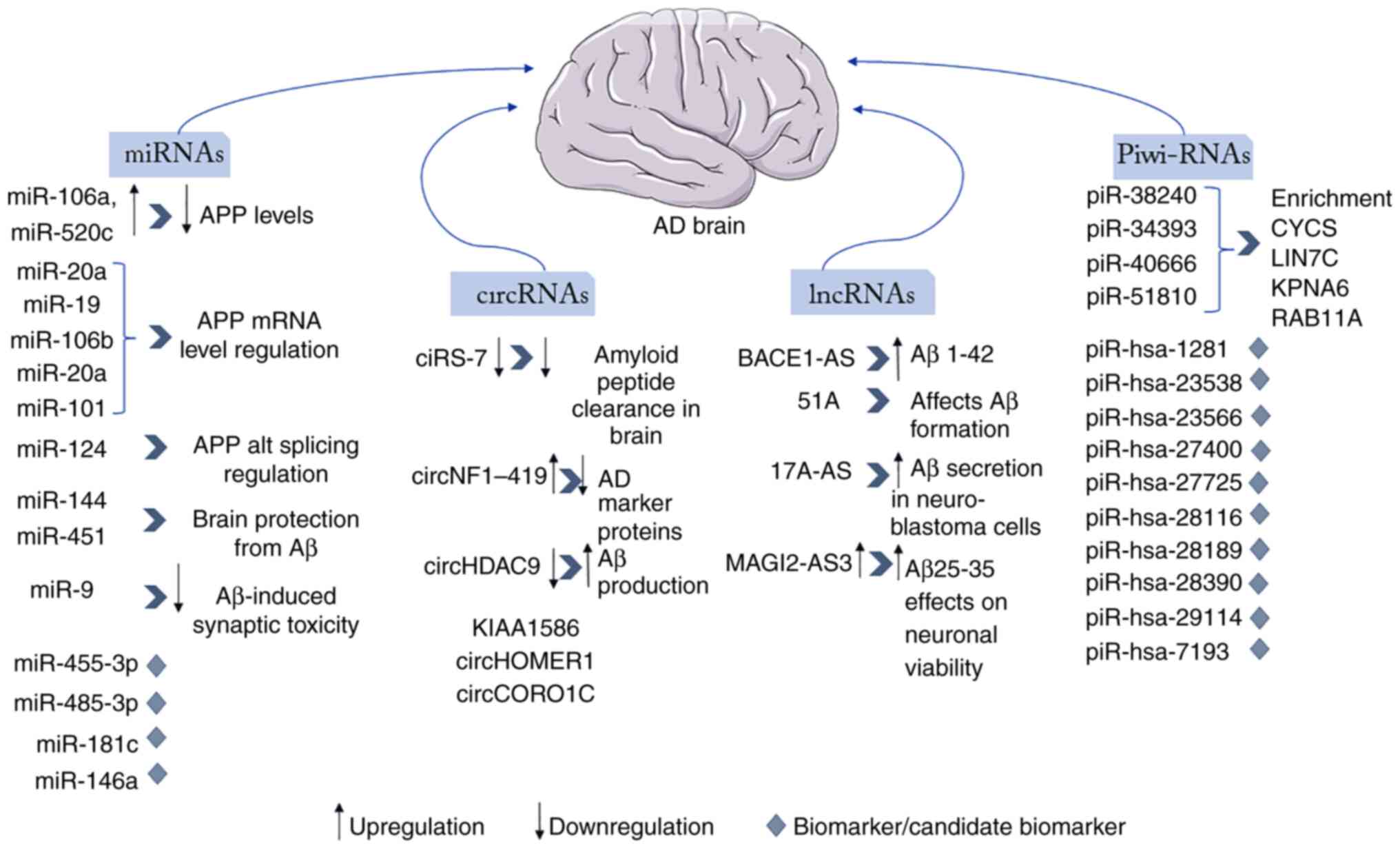 | Figure 1Illustration of the main ncRNAs of
each category (miRNAs, circRNAs, piRNAs and lncRNAs) that are
involved in AD and their mode of function and regulation in AD,
including their upregulation or downregulation and the results of
this alteration in their expression levels, as well their usage as
biomarkers. ncRNAs, non-coding RNAs; miRNAs/miRs, microRNAs;
circRNAs, circular RNAs; piRNAs, Piwi-interacting RNAs; lncRNAs,
long non-coding RNAs; AD, Alzheimer's disease. |
3. Application of omics technologies in
AD
The wave of 'omics' has been taking the scientific
world by storm, encompassing genomics, transcriptomics,
epigenomics, proteomics, metabolomics and lipidomics as part of a
rounded approach (112). When
faced with complex diseases, such as AD, the efficient analysis and
integration of data that omics technologies yield is critical for
the development of diagnostics and therapeutics (28,113,114).
Genomics
Genetics studies, genome-wide association studies
(GWAS) and next-generation sequencing (NGS) technologies have
helped to gain knowledge about the genes which are associated with
a risk of developing AD. Genetic studies have enabled the detection
of rare mutations in the genes of APP, PSEN1 and PSEN2, which are
associated with the dominantly inherited early-onset AD, as well as
the identification of genetic components that affect the
development of the sporadic cases of the disease. In addition,
genomics analysis has shown that gene-gene interaction can be a
significant risk factor for the development of AD (115,116).
GWAS can lead to the identification of genes with
common variants involved in various diseases. By comparing the
whole genome set of genetic variants in different individuals, GWAS
shed light on SNP characteristics of different diseases,
highlighting the possibility of associations between the detected
variants. Several genomic loci of interest have been identified
that may increase the risk of an individual for developing
late-onset AD, including genes involved in β-amyloid processing and
clearance, immune response and inflammation, such as CR1, CD33,
MS4A, ABCA7 EPHA1 and MEF2C, in the metabolism of cholesterol, such
as ApoE, SORL1, ABCA7 and CLU, and in the regulation of
endocytosis, such as BIN1, CD2AP, PICALM, EPHA1, SORL1 (117,118).
In addition to GWAS, NGS technologies, including
whole genome sequencing and whole exome sequencing, enable the
detection of rare mutations that affect complex diseases.
Significant are the findings of NGS studies, which have identified
new risk genes with low-frequency variants, including TREM2, which
encodes the ADAM10 activation receptor expressed in myeloid cells
and leads to defective α-secretory activity, phospholipase D3,
UNC5C and AKAP9 (119,120).
Epigenomics
Non-hereditary epigenetic changes have the potential
to equally affect the risk of developing AD. Epigenetic
modifications mainly involve DNA methylation and histone
modification, thereby regulating gene expression. In the case of
AD, the reduced methylation of DNA is described, while at the same
time, a number of genes that have been characterized in AD exhibit
a high level of methylation in their promoters and in the cytosines
that precede guanines (CpG islands) (121). Two large-scale epigenome-wide
association studies have identified four new genetic loci,
including RHBDF2, RPL13, C10orf54-CDH23 and ANK1, with differential
methylation, suggesting an association with the risk of developing
AD (122,123). However, histone modification
studies have yielded conflicting results regarding histone
acetylation levels in AD (124-127). Thus, the heterogeneity of
results from epigenomics studies indicates that the further
evaluation of epigenomics alterations is necessary, reflecting both
changes in cell composition and cell-specific changes associated
with AD pathology.
Transcriptomics
Transcriptome analysis provides the ability to
evaluate the number of transcripts that result through alternative
splicing, novel transcript identification and long and small
ncRNAs. Several studies have highlighted the critical role of
ncRNAs, mostly focusing on the role of miRNAs in AD (68,128-130). As aforementioned, miRNA profiles
can be investigated in several biological fluids, including blood
and CSF (131), as several
AD-related miRNAs have been identified following an analysis of
brain tissue in patients with AD (132). The analysis of circulating
miRNAs is very promising for the study of the pathogenesis of AD;
however, the heterogeneity of the research results requires the
participation of a greater number of patients for the study of the
transcriptome.
Proteomics
Proteomics studies provide the ability to discover
and record potential biomarkers and validate potential candidate
proteins in various diseases. Furthermore, mass spectrometry (MS)
offers effective capabilities in the analysis and determination of
proteins in combination with chromatographic or other separation
techniques (133).
Through a standard proteomics platform, which
includes two-dimensional gel electrophoresis in combination with
MS, novel candidate biomarkers were identified in the biological
fluids of patients with AD, primarily involved in the processing
pathway of the Aβ peptide (134,135). Additionally, proteomics studies
have identified eight protein biomarkers among 100 candidates, the
levels of which tend to decrease in cases of AD (136).
A notable finding of extensive target proteomics
studies is the presence of different isoforms of Aβ peptides, which
arise through alternative pathways of APP degradation, identifying
the isoforms Aβ42, Aβ40 and Aβ38, as well as APPα and APPβ in CSF
and brain tissue samples from patients with AD (137-139). In addition, a significant
increase in neurogranin levels in CSF was evidenced in patients
with AD (140), in contrast to a
significant decrease in ApoE levels in the serum of patients with
AD (141).
Through a proteomics study, the interaction of brain
transglutaminase interaction with APP, huntingtin and α-synuclein
was observed, thus demonstrating the role of brain transglutaminase
in the formation of protein aggregates in various neurodegenerative
diseases (142). In addition,
oxidatively modified proteins associated with tau and Aβ pathology
have been identified in the brains of patients with AD through
redox proteomics studies (143).
Lastly, the study by Chiasserini et al (144) yielded information on 1,315
proteins, including neurodegenerative disease biomarkers such as
APP, prion protein and DJ-1.
Metabolomics
Metabolomics studies examine and focus on
metabolites, which are small molecules (<1,500 Da) that are
involved in numerous biological functions and vary as a result of
genetic, transcriptional and protein changes, as well as
environmental influences. The main techniques used in metabolomics
studies are MS and nuclear magnetic resonance spectroscopy
(28).
Significant alterations in the abundance of
metabolites in the biological fluids of patients with AD have been
observed. According to a previous study, a change in eight
metabolites was detected, including acylcarnitine, sphingomyelins
and glycerophospholipids, which were significantly increased in the
CSF of patients (145).
Furthermore, the quantitative analysis of 17 metabolites led to the
observation of a significant increase in glycine and
S-adenosylhomocysteine levels, with a concomitant decrease in the
levels of S-adenosylmethionine in the CSF of patients diagnosed
with AD (146). Similarly,
shifts of 13 key metabolites were recorded at different stages of
AD by researchers at the Alzheimer Disease Metabolomics Consortium,
exhibiting associations with the CSF Aβ42 and t-tau/Aβ42 ratio,
with cognitive function or brain atrophy (147). Finally, through two previous
studies, 10 and 24 plasma lipids were detected to predict the
conversion of healthy individuals to patients with AD (148,149).
Big data analysis
The development and analysis of a wealth of big data
have been accompanied by advances in bioinformatics and
computational programming (150,151). An example of a large-scale omics
platform for AD is the Dominantly Inherited Alzheimer Network
(DIAN) Central Archive (https://dian.wustl.edu/), which provides all the
cognitive information, biomarkers and brain imaging information for
AD, while allowing the data analysis of different domains and wide
access to them (28).
The analysis and interpretation of big data are one
of the most important issues of concern to the scientific
community, with the main goal of analyzing the cross-platform
association between data from different omics technologies.
Furthermore, computational biology pipelines can aid the
development of antibody-interacting drugs against neurodegenerative
diseases, such as AD, a rapidly emerging field of increasing
medicinal interest (152). Big
data allow for rapid advances in personalized medicine; however,
the heterogeneity and variability of omics data hinder the
application of omics science (28).
Alterations in genomics, proteomics, epigenomics,
transcriptomics, metabolomics and lipidomic levels may be
associated with the development of neurodegenerative diseases, such
as AD, worldwide. The development of computational tools for big
data analysis based on phenotypic analytical prediction models
remains a challenge for de novo drug design or effective
drug repurposing (28). Deep
learning methods can provide a potential solution to this
challenge, allowing the exploitation of multi-omics data and
helping to form an accurate representation of AD patients. This
approach can allow researchers to develop effective personalized
treatments and early diagnostic tools, as well as guide the design
or repurposing of drugs in complex neurodegenerative pathologies
(153). An integrative
multi-omics approach, as previously described by Clark et al
(154), yielded promising
results, identifying novel molecular and pathway alterations which
are related to the pathophysiological processes of AD. A notable
strength of this approach is the identification of the main axes of
inter-individual heterogeneity, critical for the development of
tailored therapeutic interventions (154).
4. Conclusions and future perspectives
AD is a multifactorial disease, which in addition to
the genetic and hereditary background, also occurs sporadically,
due to epigenetic factors, which include ncRNAs. These molecules
are implicated in a wide range of cellular processes and human
diseases, including neurodegeneration. Several studies at various
levels, including the molecular, cellular, physiological and
epidemiological ones, have detected a rising number of ncRNAs
involved in AD. As aforementioned, they participate in the three
major pathogenic traits of AD that include the formation of Aβ
plaques, the phosphorylation of tau, and the establishments of an
inflammatory zone. In order to identify these ncRNAs, various
studies have examined both cultured cell models and biological
samples, such as brain, CSF and serum. In this context, their
identification can be managed using sensitive methods of RNA
analysis, including reverse transcription followed by conventional
PCR or qPCR analyses for the survey of individual RNAs, or
RNA-sequencing and analysis by microarray for the survey of larger
panels of RNAs. These methods would be particularly useful if the
diagnostic and prognostic ncRNAs were tested in tissues and fluids
easily accessible, such as blood, urine and some epithelia
(129).
In conjunction with these non-coding biomarkers,
omics technologies are a promising tool for the study of AD
pathology and patient variation, providing the ability to combine
and correlate different types of data, the analysis of which could
lead to personalized therapy and de novo development of more
effective drugs. The integration of omics and clinical data and the
development of novel experimental and computational strategies are
essential in multifactorial diseases, such as AD. In these cases,
high-throughput omics technologies can lead to the better
understanding of the pathological changes in the brain, to the
development of more accurate tools for the early diagnosis and
prediction of the disease, to the development of new drugs, as well
to the selection of the most beneficial and personalized
therapies.
Availability of data and materials
Not applicable.
Authors' contributions
All authors (KP, EP, LP, ID, TM, KD, DAS, FB, GPC,
GNG, EE and DV) contributed to the conceptualization, design,
writing, drafting, revising, editing and reviewing of the
manuscript. All authors have read and approved the final
manuscript. Data authentication is not applicable.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
DAS is the Editor-in-Chief for the journal, but had
no personal involvement in the reviewing process, or any influence
in terms of adjudicating on the final decision, for this article.
The other authors declare that they have no competing
interests.
Acknowledgments
Not applicable.
Funding
The authors would like to acknowledge funding from the following
organizations: i) AdjustEBOVGP-Dx (RIA2018EF-2081): Biochemical
Adjustments of native EBOV Glycoprotein in Patient Sample to Unmask
target Epitopes for Rapid Diagnostic Testing. A European and
Developing Countries Clinical Trials Partnership (EDCTP2) under the
Horizon 2020 'Research and Innovation Actions' DESCA; and ii)
'MilkSafe: A novel pipeline to enrich formula milk using omics
technologies', a research co-financed by the European Regional
Development Fund of the European Union and Greek national funds
through the Operational Program Competitiveness, Entrepreneurship
and Innovation, under the call RESEARCH-CREATE-INNOVATE (project
code: T2EDK-02222).
References
|
1
|
Cuyvers E and Sleegers K: Genetic
variations underlying Alzheimer's disease: Evidence from
genome-wide association studies and beyond. Lancet Neurol.
15:857–868. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ballard C, Gauthier S, Corbett A, Brayne
C, Aarsland D and Jones E: Alzheimer's disease. Lancet.
377:1019–1031. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Braak H, Thal DR, Ghebremedhin E and Del
Tredici K: Stages of the pathologic process in Alzheimer disease:
Age categories from 1 to 100 years. J Neuropathol Exp Neurol.
70:960–969. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Fliss R, Le Gall D, Etcharry-Bouyx F,
Chauviré V, Desgranges B and Allain P: Theory of Mind and social
reserve: Alternative hypothesis of progressive Theory of Mind decay
during different stages of Alzheimer's disease. Soc Neurosci.
11:409–423. 2016. View Article : Google Scholar
|
|
5
|
Scheltens P, Blennow K, Breteler MM, de
Strooper B, Frisoni GB, Salloway S and Van der Flier WM:
Alzheimer's disease. Lancet. 388:505–517. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Krokidis MG, Exarchos TP and Vlamos P:
Data-driven biomarker analysis using computational omics approaches
to assess neurodegenerative disease progression. Math Biosci Eng.
18:1813–1832. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Verheijen J and Sleegers K: Understanding
Alzheimer disease at the interface between genetics and
transcriptomics. Trends Genetics. 34:434–447. 2018. View Article : Google Scholar
|
|
8
|
Prince M, Bryce R, Albanese E, Wimo A,
Ribeiro W and Ferri CP: The global prevalence of dementia: A
systematic review and metaanalysis. Alzheimers Dement. 9:63–75.e62.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Heppner FL, Ransohoff RM and Becher B:
Immune attack: The role of inflammation in Alzheimer disease. Nat
Rev Neurosci. 16:358–372. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Medeiros R, Kitazawa M, Passos GF,
Baglietto-Vargas D, Cheng D, Cribbs DH and LaFerla FM:
Aspirin-triggered lipoxin A4 stimulates alternative activation of
microglia and reduces Alzheimer disease-like pathology in mice. Am
J Pathol. 182:1780–1789. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Prokop S, Miller KR and Heppner FL:
Microglia actions in Alzheimer's disease. Acta Neuropathol.
126:461–477. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Heneka MT, Kummer MP and Latz E: Innate
immune activation in neurodegenerative disease. Nat Rev Immunol.
14:463–477. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Labzin LI, Heneka MT and Latz E: Innate
Immunity and Neurodegeneration. Annu Rev Med. 69:437–449. 2018.
View Article : Google Scholar
|
|
14
|
Aubry S, Shin W, Crary JF, Lefort R,
Qureshi YH, Lefebvre C, Califano A and Shelanski ML: Assembly and
interrogation of Alzheimer's disease genetic networks reveal novel
regulators of progression. PLoS One. 10:e01203522015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Chouraki V and Seshadri S: Genetics of
Alzheimer's disease. Adv Genet. 87:245–294. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Greenough MA: The Role of presenilin in
protein trafficking and degradation-implications for metal
homeostasis. J Mol Neurosci. 60:289–297. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Millan MJ: Linking deregulation of
non-coding RNA to the core pathophysiology of Alzheimer's disease:
An integrative review. Prog Neurobiol. 156:1–68. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Huang YA, Zhou B, Wernig M and Südhof TC:
ApoE2, ApoE3, and ApoE4 differentially stimulate APP transcription
and Aβ secretion. Cell. 168:427–441.e21. 2017. View Article : Google Scholar
|
|
19
|
Jiang T, Yu JT, Tian Y and Tan L:
Epidemiology and etiology of Alzheimer's disease: From genetic to
non-genetic factors. Curr Alzheimer Res. 10:852–867. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Kanekiyo T, Xu H and Bu G: ApoE and Aβ in
Alzheimer's disease: Accidental encounters or partners? Neuron.
81:740–754. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Vlachakis D, Papakonstantinou E, Sagar R,
Bacopoulou F, Exarchos T, Kourouthanassis P, Karyotis V, Vlamos P,
Lyketsos C, Avramopoulos D and Mahairaki V: Improving the utility
of polygenic risk scores as a biomarker for Alzheimer's disease.
Cells. 10:2021. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Yan Y, Zhao A, Qui Y, Li Y, Yan R, Wang Y,
Xu W and Deng Y: Genetic Association of FERMT2, HLA-DRB1, CD2AP,
and PTK2B Polymorphisms with Alzheimer's disease risk in the
southern Chinese population. Front Aging Neurosci. 12:162020.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Chin-Chan M, Navarro-Yepes J and
Quintanilla-Vega B: Environmental pollutants as risk factors for
neurodegenerative disorders: Alzheimer and Parkinson diseases.
Front Cell Neurosci. 9:1242015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Reitz C, Brayne C and Mayeux R:
Epidemiology of Alzheimer disease. Nat Rev Neurol. 7:137–152. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Tremlett H, Bauer KC, Appel-Cresswell S,
Finlay BB and Waubant E: The gut microbiome in human neurological
disease: A review. Ann Neurol. 81:369–382. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Blennow K, Dubois B, Fagan AM, Lewczuk P,
de Leon MJ and Hampel H: Clinical utility of cerebrospinal fluid
biomarkers in the diagnosis of early Alzheimer's disease.
Alzheimers Dement. 11:58–69. 2015. View Article : Google Scholar
|
|
27
|
Sancesario GM and Bernardini S: How many
biomarkers to discriminate neurodegenerative dementia? Crit Rev
Clin Lab Sci. 52:314–326. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Sancesario GM and Bernardini S:
Alzheimer's disease in the omics era. Clin Biochem. 59:9–16. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Trushina E, Dutta T, Persson X-MT, Mielke
MM and Petersen RC: Identification of altered metabolic pathways in
plasma and CSF in mild cognitive impairment and Alzheimer's disease
using metabolomics. PLoS One. 8:e636442013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Nday CM, Eleftheriadou D and Jackson G:
Shared pathological pathways of Alzheimer's disease with specific
comorbidities: Current perspectives and interventions. J Neurochem.
144:360–389. 2018. View Article : Google Scholar
|
|
31
|
Morgan SL, Naderi P, Koler K, Pita-Juarez
Y, Prokopenko D, Vlachos IS, Tanzi RE, Bertram L and Hide WA: Most
pathways can be related to the pathogenesis of Alzheimer's disease.
Front Aging Neurosci. 14:8469022022. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Colpaert RMW and Calore M: Epigenetics and
microRNAs in cardiovascular diseases. Genomics. 113:540–551. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Ramzan F, Vickers MH and Mithen RF:
Epigenetics, microRNA and metabolic syndrome: A comprehensive
review. Int J Mol Sci. 22:50472021. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Suzuki H, Maruyama R, Yamamoto E and Kai
M: Epigenetic alteration and microRNA dysregulation in cancer.
Front Genet. 4:2582013. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Lau P, Bossers K, Janky R, Salta E,
Frigerio CS, Barbash S, Rothman R, Sierksma AS, Thathiah A,
Greenberg D, et al: Alteration of the microRNA network during the
progression of Alzheimer's disease. EMBO Mol Med. 5:1613–1634.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Maoz R, Garfinkel BP and Soreq H:
Alzheimer's disease and ncRNAs. Adv Exp Med Biol. 978:337–361.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Sun AX, Crabtree GR and Yoo AS: MicroRNAs:
Regulators of neuronal fate. Curr Opin Cell Biol. 25:215–221. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Huang Y, Shen XJ, Zou Q, Wang SP, Tang SM
and Zhang GZ: Biological functions of microRNAs: A review. J
Physiol Biochem. 67:129–139. 2011. View Article : Google Scholar
|
|
39
|
Fiore R, Khudayberdiev S, Saba R and
Schratt G: MicroRNA function in the nervous system. Prog Mol Biol
Transl Sci. 102:47–100. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Goodall EF, Heath PR, Bandmann O, Kirby J
and Shaw PJ: Neuronal dark matter: The emerging role of microRNAs
in neurodegeneration. Front Cell Neurosci. 7:1782013. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Niwa R, Zhou F, Li C and Slack FJ: The
expression of the Alzheimer's amyloid precursor protein-like gene
is regulated by developmental timing microRNAs and their targets in
Caenorhabditis elegans. Dev Biol. 315:418–425. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Patel N, Hoang D, Miller N, Ansaloni S,
Huang Q, Rogers JT, Lee JC and Saunders AJ: MicroRNAs can regulate
human APP levels. Mol Neurodegener. 3:102008. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Fan X, Liu Y, Jiang J, Ma Z, Wu H, Liu T,
Liu M, Li X and Tang H: miR-20a promotes proliferation and invasion
by targeting APP in human ovarian cancer cells. Acta Biochim
Biophys Sin (Shanghai). 42:318–324. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Hébert SS, Horré K, Nicolaï L, Bergmans B,
Papadopoulou AS, Delacourte A and De Strooper B: MicroRNA
regulation of Alzheimer's Amyloid precursor protein expression.
Neurobiol Dis. 33:422–428. 2009. View Article : Google Scholar
|
|
45
|
Vilardo E, Barbato C, Ciotti M, Cogoni C
and Ruberti F: MicroRNA-101 regulates amyloid precursor protein
expression in hippocampal neurons. J Biol Chem. 285:18344–18351.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Glinsky GV: An SNP-guided microRNA map of
fifteen common human disorders identifies a consensus disease
phenocode aiming at principal components of the nuclear import
pathway. Cell Cycle. 7:2570–2583. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Delay C, Calon F, Mathews P and Hébert SS:
Alzheimer-specific variants in the 3′UTR of Amyloid precursor
protein affect microRNA function. Mol Neurodegener. 6:702011.
View Article : Google Scholar
|
|
48
|
Smith P, Al Hashimi A, Girard J, Delay C
and Hébert SS: In vivo regulation of amyloid precursor protein
neuronal splicing by microRNAs. J Neurochem. 116:240–247. 2011.
View Article : Google Scholar
|
|
49
|
Kong Y, Wu J, Zhang D, Wan C and Yuan L:
The role of miR-124 in drosophila Alzheimer's disease model by
targeting delta in notch signaling pathway. Curr Mol Med.
15:980–989. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Schonrock N, Matamales M, Ittner LM and
Götz J: MicroRNA networks surrounding APP and amyloid-β
metabolism-implications for Alzheimer's disease. Exp Neurol.
235:447–454. 2012. View Article : Google Scholar
|
|
51
|
Rockenstein EM, McConlogue L, Tan H, Power
M, Masliah E and Mucke L: Levels and alternative splicing of
amyloid beta protein precursor (APP) transcripts in brains of APP
transgenic mice and humans with Alzheimer's disease. J Biol Chem.
270:28257–28267. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Donev R, Newall A, Thome J and Sheer D: A
role for SC35 and hnRNPA1 in the determination of amyloid precursor
protein isoforms. Mol Psychiatry. 12:681–690. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Yang LB, Lindholm K, Yan R, Citron M, Xia
W, Yang XL, Beach T, Sue L, Wong P, Price D, et al: Elevated
beta-secretase expression and enzymatic activity detected in
sporadic Alzheimer disease. Nat Med. 9:3–4. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Yang G, Song Y, Zhou X, Deng Y, Liu T,
Weng G, Yu D and Pan S: MicroRNA-29c targets β-site amyloid
precursor protein-cleaving enzyme 1 and has a neuroprotective role
in vitro and in vivo. Mol Med Rep. 12:3081–3088. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Lei X, Lei L, Zhang Z, Zhang Z and Cheng
Y: Downregulated miR-29c correlates with increased BACE1 expression
in sporadic Alzheimer's disease. Int J Clin Exp Pathol.
8:1565–1574. 2015.PubMed/NCBI
|
|
56
|
Zong Y, Wang H, Dong W, Quan X, Zhu H, Xu
Y, Huang L, Ma C and Qin C: miR-29c regulates BACE1 protein
expression. Brain Res. 1395:108–115. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Boissonneault V, Plante I, Rivest S and
Provost P: MicroRNA-298 and microRNA-328 regulate expression of
mouse beta-amyloid precursor protein-converting enzyme 1. J Biol
Chem. 284:1971–1981. 2009. View Article : Google Scholar
|
|
58
|
Liu T, Huang Y, Chen J, Chi H, Yu Z, Wang
J and Chen C: Attenuated ability of BACE1 to cleave the amyloid
precursor protein via silencing long noncoding RNA BACE1-AS
expression. Mol Med Rep. 10:1275–1281. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Zhang Y, Xing H, Guo S, Zheng Z, Wang H
and Xu D: MicroRNA-135b has a neuroprotective role via targeting of
β-site APP-cleaving enzyme 1. Exp Ther Med. 12:809–814. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Xie H, Zhao Y, Zhou Y, Wang D, Zhang S and
Yang M: MiR-9 regulates the expression of BACE1 in dementia induced
by chronic brain hypoperfusion in rats. Cell Physiol Biochem.
42:1213–1226. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Zhu HC, Wang LM, Wang M, Song B, Tan S,
Teng JF and Duan DX: MicroRNA-195 downregulates Alzheimer's disease
amyloid-β production by targeting BACE1. Brain Res Bull.
88:596–601. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Wang WX, Rajeev BW, Stromberg AJ, Ren N,
Tang G, Huang Q, Rigoutsos I and Nelson PT: The expression of
microRNA miR-107 decreases early in Alzheimer's disease and may
accelerate disease progression through regulation of beta-site
amyloid precursor protein-cleaving enzyme 1. J Neurosci.
28:1213–1223. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Rybak-Wolf A and Plass M: RNA dynamics in
Alzheimer's disease. Molecules. 26:51132021. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Chang F, Zhang LH, Xu WP, Jing P and Zhan
PY: microRNA-9 attenuates amyloidβ-induced synaptotoxicity by
targeting calcium/calmodulin-dependent protein kinase kinase 2. Mol
Med Rep. 9:1917–1922. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Janson J, Laedtke T, Parisi JE, O'Brien P,
Petersen RC and Butler PC: Increased risk of type 2 diabetes in
Alzheimer disease. Diabetes. 53:474–481. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Cheng C, Li W, Zhang Z, Yoshimura S, Hao
Q, Zhang C and Wang Z: MicroRNA-144 is regulated by activator
protein-1 (AP-1) and decreases expression of Alzheimer
disease-related a disintegrin and metalloprotease 10 (ADAM10). J
Biol Chem. 288:13748–13761. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Dubois B, Padovani A, Scheltens P, Rossi A
and Dell'Agnello G: Timely diagnosis for Alzheimer's disease: A
literature review on benefits and challenges. J Alzheimers Dis.
49:617–631. 2016. View Article : Google Scholar
|
|
68
|
Wei W, Wang ZY, Ma LN, Zhang TT, Cao Y and
Li H: MicroRNAs in Alzheimer's disease: Function and potential
applications as diagnostic biomarkers. Front Mol Neurosci. 13:2020.
View Article : Google Scholar
|
|
69
|
Schwarzenbach H, Nishida N, Calin GA and
Pantel K: Clinical relevance of circulating cell-free microRNAs in
cancer. Nat Rev Clin Oncol. 11:145–156. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Bekris LM and Leverenz JB: The biomarker
and therapeutic potential of miRNA in Alzheimer's disease.
Neurodegener Dis Manag. 5:61–74. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Cogswell JP, Ward J, Taylor IA, Waters M,
Shi Y, Cannon B, Kelnar K, Kemppainen J, Brown D, Chen C, et al:
Identification of miRNA changes in Alzheimer's disease brain and
CSF yields putative biomarkers and insights into disease pathways.
J Alzheimers Dis. 14:27–41. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Siedlecki-Wullich D, Català-Solsona J,
Fábregas C, Hernández I, Clarimon J, Lleó A, Boada M, Saura CA,
Rodríguez-Álvarez J and Miñano-Molina AJ: Altered microRNAs related
to synaptic function as potential plasma biomarkers for Alzheimer's
disease. Alzheimers Res Ther. 11:462019. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Denk J, Boelmans K, Siegismund C, Lassner
D, Arlt S and Jahn H: MicroRNA profiling of CSF reveals potential
biomarkers to detect Alzheimer's disease. PLoS One.
10:e01264232015. View Article : Google Scholar
|
|
74
|
Alexandrov PN, Dua P and Lukiw WJ:
Up-regulation of miRNA-146a in progressive, Age-related
inflammatory neurodegenerative disorders of the human CNS. Front
Neurol. 5:1812014. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Arena A, Iyer A, Milenkovic I, Kovacs GG,
Ferrer I, Perluigi M and Aronica E: Developmental expression and
dysregulation of miR-146a and miR-155 in Down's syndrome and mouse
models of Down's syndrome and Alzheimer's disease. Curr Alzheimer
Res. 14:1305–1317. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Kumar S and Reddy PH: Elevated levels of
MicroRNA-455-3p in the cerebrospinal fluid of Alzheimer's patients:
A potential biomarker for Alzheimer's disease. Biochim Biophys Acta
Mol Basis Dis. 1867:1660522021. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Yu L, Li H, Liu W, Zhang L, Tian Q, Li H
and Li M: MiR-485-3p serves as a biomarker and therapeutic target
of Alzheimer's disease via regulating neuronal cell viability and
neuroinflammation by targeting AKT3. Mol Genet Genomic Med.
9:e15482021. View Article : Google Scholar
|
|
78
|
Andreeva K and Cooper NGF: Circular RNAs:
New players in gene regulation. Adv Bioscience Biotechnol. 06(06):
82015. View Article : Google Scholar
|
|
79
|
Gruner H, Cortés-López M, Cooper DA, Bauer
M and Miura P: CircRNA accumulation in the aging mouse brain. Sci
Rep. 6:389072016. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Akhter R: Circular RNA and Alzheimer's
disease. Adv Exp Med Biol. 1087:239–243. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Hansen TB, Jensen TI, Clausen BH, Bramsen
JB, Finsen B, Damgaard CK and Kjems J: Natural RNA circles function
as efficient microRNA sponges. Nature. 495:384–388. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Lukiw WJ: Circular RNA (circRNA) in
Alzheimer's disease (AD). Front Genet. 4:3072013. View Article : Google Scholar
|
|
83
|
Lonskaya I, Shekoyan AR, Hebron ML,
Desforges N, Algarzae NK and Moussa CE: Diminished parkin
solubility and Co-localization with intraneuronal amyloid-β are
associated with autophagic defects in Alzheimer's disease. J
Alzheimers Dis. 33:231–247. 2013. View Article : Google Scholar
|
|
84
|
Dube U, Del-Aguila JL, Li Z, Budde JP,
Jiang S, Hsu S, Ibanez L, Fernandez MV, Farias F, Norton J, et al:
An atlas of cortical circular RNA expression in Alzheimer disease
brains demonstrates clinical and pathological associations. Nat
Neurosci. 22:1903–1912. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Zhang Y, Yu F, Bao S and Sun J: Systematic
characterization of circular RNA-associated CeRNA network
identified novel circRNA biomarkers in Alzheimer's disease. Front
Bioeng Biotechnol. 7:2222019. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Lu Y, Tan L and Wang X: Circular
HDAC9/microRNA-138/Sirtuin-1 pathway mediates synaptic and amyloid
precursor protein processing deficits in Alzheimer's disease.
Neurosci Bull. 35:877–888. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Yang H, Wang H, Shang H, Chen X, Yang S,
Qu Y, Ding J and Li X: Circular RNA circ_0000950 promotes neuron
apoptosis, suppresses neurite outgrowth and elevates inflammatory
cytokines levels via directly sponging miR-103 in Alzheimer's
disease. Cell Cycle. 18:2197–2214. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Diling C, Yinrui G, Longkai Q, Xiaocui T,
Yadi L, Xin Y, Guoyan H, Ou S, Tianqiao Y, Dongdong W, et al:
Circular RNA NF1-419 enhances autophagy to ameliorate senile
dementia by binding Dynamin-1 and Adaptor protein 2 B1 in AD-like
mice. Aging (Albany NY). 11:12002–12031. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Zhang M and Bian Z: The emerging role of
circular RNAs in Alzheimer's disease and Parkinson's disease. Front
Aging Neurosci. 13:6915122021. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Huang X and Wong G: An old weapon with a
new function: PIWI-interacting RNAs in neurodegenerative diseases.
Transl Neurodegener. 10:92021. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Qiu W, Guo X, Lin X, Yang Q, Zhang W,
Zhang Y, Zuo L, Zhu Y, Li CR, Ma C and Luo X: Transcriptome-wide
piRNA profiling in human brains of Alzheimer's disease. Neurobiol
Aging. 57:170–177. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Roy J, Sarkar A, Parida S, Ghosh Z and
Mallick B: Small RNA sequencing revealed dysregulated piRNAs in
Alzheimer's disease and their probable role in pathogenesis. Mol
Biosyst. 13:565–576. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Mercer TR, Dinger ME and Mattick JS: Long
non-coding RNAs: Insights into functions. Nat Rev Genet.
10:155–159. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Wilusz JE, Sunwoo H and Spector DL: Long
noncoding RNAs: Functional surprises from the RNA world. Genes Dev.
23:1494–1504. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Clark MB and Mattick JS: Long noncoding
RNAs in cell biology. Semin Cell Dev Biol. 22:366–376. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Martianov I, Ramadass A, Serra Barros A,
Chow N and Akoulitchev A: Repression of the human dihydrofolate
reductase gene by a non-coding interfering transcript. Nature.
445:666–670. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Tripathi V, Ellis JD, Shen Z, Song DY, Pan
Q, Watt AT, Freier SM, Bennett CF, Sharma A, Bubulya PA, et al: The
nuclear-retained noncoding RNA MALAT1 regulates alternative
splicing by modulating SR splicing factor phosphorylation. Mol
Cell. 39:925–938. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Huarte M, Guttman M, Feldser D, Garber M,
Koziol MJ, Kenzelmann-Broz D, Khalil AM, Zuk O, Amit I, Rabani M,
et al: A large intergenic noncoding RNA induced by p53 mediates
global gene repression in the p53 response. Cell. 142:409–419.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Mourtada-Maarabouni M, Hedge VL, Kirkham
L, Farzaneh F and Williams GT: Growth arrest in human T-cells is
controlled by the non-coding RNA growth-arrest-specific transcript
5 (GAS5). J Cell Sci. 121:939–946. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Li F, Wang Y, Yang H, Xu Y, Zhou X, Zhang
X, Xie Z and Bi J: The effect of BACE1-AS on β-amyloid generation
by regulating BACE1 mRNA expression. BMC Mol Biol. 20:232019.
View Article : Google Scholar
|
|
101
|
Faghihi MA, Modarresi F, Khalil AM, Wood
DE, Sahagan BG, Morgan TE, Finch CE, St Laurent G III, Kenny PJ and
Wahlestedt C: Expression of a noncoding RNA is elevated in
Alzheimer's disease and drives rapid feed-forward regulation of
beta-secretase. Nat Med. 14:723–730. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
102
|
Zeng T, Ni H, Yu Y, Zhang M, Wu M, Wang Q,
Wang L, Xu S, Xu Z, Xu C, et al: BACE1-AS prevents BACE1 mRNA
degradation through the sequestration of BACE1-targeting miRNAs. J
Chem Neuroanat. 98:87–96. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
103
|
Zhou Y, Ge Y, Liu Q, Li YX, Chao X, Guan
JJ, Diwu YC and Zhang Q: LncRNA BACE1-AS promotes
autophagy-mediated neuronal damage through the miR-214-3p/ATG5
signalling axis in Alzheimer's disease. Neuroscience. 455:52–64.
2021. View Article : Google Scholar
|
|
104
|
Parenti R, Paratore S, Torrisi A and
Cavallaro S: A natural antisense transcript against Rad18,
specifically expressed in neurons and upregulated during
beta-amyloid-induced apoptosis. Eur J Neurosci. 26:2444–2457. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
105
|
Li D, Zhang J, Li X, Chen Y, Yu F and Liu
Q: Insights into lncRNAs in Alzheimer's disease mechanisms. RNA
Biol. 18:1037–1047. 2021. View Article : Google Scholar
|
|
106
|
Ciarlo E, Massone S, Penna I, Nizzari M,
Gigoni A, Dieci G, Russo C, Florio T, Cancedda R and Pagano A: An
intronic ncRNA-dependent regulation of SORL1 expression affecting
Aβ formation is upregulated in post-mortem Alzheimer's disease
brain samples. Dis Model Mech. 6:424–433. 2013.
|
|
107
|
Massone S, Vassallo I, Fiorino G,
Castelnuovo M, Barbieri F, Borghi R, Tabaton M, Robello M, Gatta E,
Russo C, et al: 17A, a novel non-coding RNA, regulates GABA B
alternative splicing and signaling in response to inflammatory
stimuli and in Alzheimer disease. Neurobiol Dis. 41:308–317. 2011.
View Article : Google Scholar
|
|
108
|
Zhang J and Wang R: Deregulated lncRNA
MAGI2-AS3 in Alzheimer's disease attenuates amyloid-β induced
neurotoxicity and neuroinflammation by sponging miR-374b-5p. Exp
Gerontol. 144:1111802021. View Article : Google Scholar
|
|
109
|
Mus E, Hof PR and Tiedge H: Dendritic
BC200 RNA in aging and in Alzheimer's disease. Proc Natl Acad Sci
USA. 104:10679–10684. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
110
|
Zhou X and Xu J: Identification of
Alzheimer's disease-associated long noncoding RNAs. Neurobiol
Aging. 36:2925–2931. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
111
|
Magistri M, Velmeshev D, Makhmutova M and
Faghihi MA: Transcriptomics profiling of Alzheimer's disease reveal
neurovascular defects, altered Amyloid-β homeostasis, and
deregulated expression of long noncoding RNAs. J Alzheimers Dis.
48:647–665. 2015. View Article : Google Scholar
|
|
112
|
Subramanian I, Verma S, Kumar S, Jere A
and Anamika K: Multi-omics data integration, interpretation, and
its application. Bioinformatics Biol Insights.
14:11779322198990512020. View Article : Google Scholar
|
|
113
|
Peña-Bautista C, Baquero M, Vento M and
Cháfer-Pericás C: Omics-based Biomarkers for the Early Alzheimer
disease diagnosis and reliable therapeutic targets development.
Curr Neuropharmacol. 17:630–647. 2019. View Article : Google Scholar :
|
|
114
|
Tan MS, Cheah PL, Chin AV, Looi LM and
Chang SW: A review on omics-based biomarkers discovery for
Alzheimer's disease from the bioinformatics perspectives:
Statistical approach vs machine learning approach. Comput Biol Med.
139:1049472021. View Article : Google Scholar : PubMed/NCBI
|
|
115
|
Giri M, Zhang M and Lü Y: Genes associated
with Alzheimer's disease: An overview and current status. Clin
Interv Aging. 11:665–681. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
116
|
Ridge PG, Mukherjee S, Crane PK and Kauwe
JSK; Alzheimer's Disease Genetics Consortium: Alzheimer's disease:
Analyzing the missing heritability. PLoS One. 8:e797712013.
View Article : Google Scholar : PubMed/NCBI
|
|
117
|
Lambert JC, Ibrahim-Verbaas CA, Harold D,
Naj AC, Sims R, Bellenguez C, DeStafano AL, Bis JC, Beecham GW,
Grenier-Boley B, et al: Meta-analysis of 74,046 individuals
identifies 11 new susceptibility loci for Alzheimer's disease. Nat
Genet. 45:1452–1458. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
118
|
Naj AC, Jun G, Beecham GW, Wang LS,
Vardarajan BN, Buros J, Gallins PJ, Buxbaum JD, Jarvik GP and Crane
PK: Common variants at MS4A4/MS4A6E, CD2AP, CD33 and EPHA1 are
associated with late-onset Alzheimer's disease. Nat Genet.
43:436–441. 2011. View
Article : Google Scholar : PubMed/NCBI
|
|
119
|
Jonsson T, Stefansson H, Steinberg S,
Jonsdottir I, Jonsson PV, Snaedal J, Bjornsson S, Huttenlocher J,
Levey AI, Lah JJ, et al: Variant of TREM2 associated with the Risk
of Alzheimer's disease. N Eng J Med. 368:107–116. 2012. View Article : Google Scholar
|
|
120
|
Cruchaga C, Karch CM, Jin SC, Benitez BA,
Cai Y, Guerreiro R, Harari O, Norton J, Budde J, Bertelsen S, et
al: Rare coding variants in the phospholipase D3 gene confer risk
for Alzheimer's disease. Nature. 505:550–554. 2014. View Article : Google Scholar
|
|
121
|
Bennett DA, Yu L, Yang J, Srivastava GP,
Aubin C and De Jager PL: Epigenomics of Alzheimer's disease. Transl
Res. 165:200–220. 2015. View Article : Google Scholar
|
|
122
|
Lunnon K, Smith R, Hannon E, De Jager PL,
Srivastava G, Volta M, Troakes C, Al-Sarraj S, Burrage J, Macdonald
R, et al: Methylomic profiling implicates cortical deregulation of
ANK1 in Alzheimer's disease. Nat Neurosci. 17:1164–1170. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
123
|
De Jager PL, Srivastava G, Lunnon K,
Burgess J, Schalkwyk LC, Yu L, Eaton ML, Keenan BT, Ernst J, McCabe
C, et al: Alzheimer's disease: Early alterations in brain DNA
methylation at ANK1, BIN1, RHBDF2 and other loci. Nat Neurosci.
17:1156–1163. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
124
|
Zhang K, Schrag M, Crofton A, Trivedi R,
Vinters H and Kirsch W: Targeted proteomics for quantification of
histone acetylation in Alzheimer's disease. Proteomics.
12:1261–1268. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
125
|
Lu X, Deng Y, Yu D, Cao H, Wang L, Liu L,
Yu C, Zhang Y, Guo X and Yu G: Histone acetyltransferase p300
mediates histone acetylation of PS1 and BACE1 in a cellular model
of Alzheimer's disease. PLoS One. 9:e1030672014. View Article : Google Scholar : PubMed/NCBI
|
|
126
|
Rao JS, Keleshian VL, Klein S and Rapoport
SI: Epigenetic modifications in frontal cortex from Alzheimer's
disease and bipolar disorder patients. Transl Psychiatry.
2:e1322012. View Article : Google Scholar : PubMed/NCBI
|
|
127
|
Narayan PJ, Lill C, Faull R, Curtis MA and
Dragunow M: Increased acetyl and total histone levels in
post-mortem Alzheimer's disease brain. Neurobiol Dis. 74:281–294.
2015. View Article : Google Scholar
|
|
128
|
Zhang Y, Zhao Y, Ao X, Yu W, Zhang L, Wang
Y and Chang W: The role of Non-coding RNAs in Alzheimer's disease:
From regulated mechanism to therapeutic targets and diagnostic
biomarkers. Front Aging Neurosci. 13:6549782021. View Article : Google Scholar : PubMed/NCBI
|
|
129
|
Idda ML, Munk R, Abdelmohsen K and Gorospe
M: Noncoding RNAs in Alzheimer's disease. Wiley Interdiscip Rev
RNA. Jan 12–2018.Epub ahead of print. View Article : Google Scholar
|
|
130
|
Wang M, Qin L and Tang B: MicroRNAs in
Alzheimer's disease. Front Genet. 10:1532019. View Article : Google Scholar : PubMed/NCBI
|
|
131
|
Formosa A, Piro MC, Docimo R, Maturo P,
Sollecito DR, Kalimutho M, Sancesario G, Barlattani A, Melino G,
Candi E and Bernardini S: Salivary miRNAome profiling uncovers
epithelial and proliferative miRNAs with differential expression
across dentition stages. Cell Cycle. 10:3359–3368. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
132
|
Dehghani R, Rahmani F and Rezaei N:
MicroRNA in Alzheimer's disease revisited: Implications for major
neuropathological mechanisms. Rev Neurosci. 29:161–182. 2018.
View Article : Google Scholar
|
|
133
|
Shevchenko G, Konzer A, Musunuri S and
Bergquist J: Neuroproteomics tools in clinical practice. Biochim
Biophys Acta. 1854:705–717. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
134
|
Henkel AW, Müller K, Lewczuk P, Müller T,
Marcus K, Kornhuber J and Wiltfang J: Multidimensional plasma
protein separation technique for identification of potential
Alzheimer's disease plasma biomarkers: A pilot study. J Neural
Transm (Vienna). 119:779–788. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
135
|
Thambisetty M, Simmons A, Velayudhan L,
Hye A, Campbell J, Zhang Y, Wahlund LO, Westman E, Kinsey A,
Güntert A, et al: Association of plasma clusterin concentration
with severity, pathology, and progression in Alzheimer disease.
Arch Gen Psychiatry. 67:739–748. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
136
|
Korolainen MA, Nyman TA, Aittokallio T and
Pirttilä T: An update on clinical proteomics in Alzheimer's
research. J Neurochem. 112:1386–1414. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
137
|
Pannee J, Portelius E, Oppermann M, Atkins
A, Hornshaw M, Zegers I, Höjrup P, Minthon L, Hansson O, Zetterberg
H, et al: A selected reaction monitoring (SRM)-based method for
absolute quantification of Aβ 38, Aβ 40, and Aβ 42 in cerebrospinal
fluid of Alzheimer's disease patients and healthy controls. J
Alzheimers Dis. 33:1021–1032. 2013. View Article : Google Scholar
|
|
138
|
Erik P, Niklas M, Ulf A, Kaj B and Henrik
Z: Novel AβIsoforms in Alzheimer's disease-their role in diagnosis
and treatment. Curr Pharmaceutical Design. 17:2594–2602. 2011.
View Article : Google Scholar
|
|
139
|
Brinkmalm G, Brinkmalm A, Bourgeois P,
Persson R, Hansson O, Portelius E, Mercken M, Andreasson U, Parent
S, Lipari F, et al: Soluble amyloid precursor protein α and β in
CSF in Alzheimer's disease. Brain Res. 1513:117–126. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
140
|
Thorsell A, Bjerke M, Gobom J, Brunhage E,
Vanmechelen E, Andreasen N, Hansson O, Minthon L, Zetterberg H and
Blennow K: Neurogranin in cerebrospinal fluid as a marker of
synaptic degeneration in Alzheimer's disease. Brain Res.
1362:13–22. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
141
|
Han SH, Kim JS, Lee Y, Choi H, Kim JW, Na
DL, Yang EG, Yu MH, Hwang D, Lee C and Mook-Jung I: Both targeted
mass spectrometry and flow sorting analysis methods detected the
decreased serum apolipoprotein E level in Alzheimer's disease
patients. Mol Cell Proteomics. 13:407–419. 2014. View Article : Google Scholar :
|
|
142
|
André W, Nondier I, Valensi M, Guillonneau
F, Federici C, Hoffner G and Djian P: Identification of brain
substrates of transglutaminase by functional proteomics supports
its role in neurodegenerative diseases. Neurobiol Dis. 101:40–58.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
143
|
Sultana R, Perluigi M and Butterfield DA:
Lipid peroxidation triggers neurodegeneration: A redox proteomics
view into the Alzheimer disease brain. Free Radic Biol Med.
62:157–169. 2013. View Article : Google Scholar :
|
|
144
|
Chiasserini D, van Weering JRT, Piersma
SR, Pham TV, Malekzadeh A, Teunissen CE, de Wit H and Jiménez CR:
Proteomic analysis of cerebrospinal fluid extracellular vesicles: A
comprehensive dataset. J Proteomics. 106:191–204. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
145
|
Koal T, Klavins K, Seppi D, Kemmler G and
Humpel C: Sphingomyelin SM(d18:1/18:0) is significantly enhanced in
cerebrospinal fluid samples dichotomized by pathological
amyloid-β42, tau, and phospho-tau-181 levels. J Alzheimers Dis.
44:1193–1201. 2015. View Article : Google Scholar
|
|
146
|
Guiraud SP, Montoliu I, Da Silva L, Dayon
L, Galindo AN, Corthésy J, Kussmann M and Martin FP:
High-throughput and simultaneous quantitative analysis of
homocysteine-methionine cycle metabolites and co-factors in blood
plasma and cerebrospinal fluid by isotope dilution LC-MS/MS. Anal
Bioanal Chem. 409:295–305. 2017. View Article : Google Scholar
|
|
147
|
Toledo JB, Arnold M, Kastenmüller G, Chang
R, Baillie RA, Han X, Thambisetty M, Tenenbaum JD, Suhre K,
Thompson JW, et al: Metabolic network failures in Alzheimer's
disease: A biochemical road map. Alzheimers Dement. 13:965–984.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
148
|
Mapstone M, Cheema AK, Fiandaca MS, Zhong
X, Mhyre TR, MacArthur LH, Hall WJ, Fisher SG, Peterson DR, Haley
JM, et al: Plasma phospholipids identify antecedent memory
impairment in older adults. Nat Med. 20:415–418. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
149
|
Fiandaca MS, Zhong X, Cheema AK, Orquiza
MH, Chidambaram S, Tan MT, Gresenz CR, FitzGerald KT, Nalls MA,
Singleton AB, et al: Plasma 24-metabolite panel predicts
preclinical transition to clinical stages of Alzheimer's disease.
Front Neurol. 6:2372015. View Article : Google Scholar : PubMed/NCBI
|
|
150
|
Pimplikar SW: Multi-omics and Alzheimer's
disease: A slower but surer path to an efficacious therapy? Am J
Physiol Cell Physiol. 313:C1–C2. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
151
|
Moreno-Indias I, Lahti L, Nedyalkova M,
Elbere I, Roshchupkin G, Adilovic M, Aydemir O, Bakir-Gungor B,
Santa Pau EC, D'Elia D, et al: Statistical and machine learning
techniques in human microbiome studies: Contemporary challenges and
solutions. Front Microbiol. 12:6357812021. View Article : Google Scholar : PubMed/NCBI
|
|
152
|
Papageorgiou L, Papakonstantinou E, Salis
C, Polychronidou E, Hagidimitriou M, Maroulis D, Eliopoulos E and
Vlachakis D: Drugena: A fully automated immunoinformatics platform
for the design of antibody-drug conjugates against
neurodegenerative diseases. Adv Exp Med Biol. 1194:203–215. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
153
|
Termine A, Fabrizio C, Strafella C, Caputo
V, Petrosini L, Caltagirone C, Giardina E and Cascella R:
Multi-Layer picture of neurodegenerative diseases: Lessons from the
use of big data through artificial intelligence. J Pers Med.
11:2802021. View Article : Google Scholar : PubMed/NCBI
|
|
154
|
Clark C, Dayon L, Masoodi M, Bowman GL and
Popp J: An integrative multi-omics approach reveals new central
nervous system pathway alterations in Alzheimer's disease.
Alzheimers Res Ther. 13:712021. View Article : Google Scholar : PubMed/NCBI
|















