1. Introduction
Dark DNA, the non-coding portion of the genome,
constitutes ~98% of human DNA, while genomes of other large
multicellular eukaryotes also appear to be mostly comprised of DNA
that does not encode for proteins as well (1,2).
New sequencing technologies made it possible to investigate this
dark side of the genome, previously referred to as 'junk' (1). It appears that the extended DNA
sequences of higher eukaryotic organisms that do not encode
proteins are eventually transcribed in RNA to a very large
percentage. Up to 80-90% of the genome of eukaryotic organisms is
estimated to being transcribed, suggesting a potential role for
such molecules (3). Furthermore,
as the complexity of an organism increases, so does the percentage
of dark DNA, while non-coding RNAs (ncRNAs) appear to dominate the
regulation of its genome, in contrast to protein-coding genes
(4).
Although the functions of all these transcripts were
initially unknown, RNA has been shown to have a very wide range of
biological functions. Specifically, RNA transcripts are being used
as a means of gene regulation in higher eukaryotes, both by
cis and trans mechanisms (5). Research has indicated that up to 20%
of these dark DNA regions play a vital role in controlling gene
expression by regulating when and where a gene is activated or
deactivated (6). ncRNAs appear to
use a number of different mechanisms in order to regulate gene
expression. RNA transcripts have the ability to function as
transcriptional or post-transcriptional regulators or as regulators
of epigenetic modifications (7,8).
RNA transcripts also appear to have tissue-, time- and
cell-specific expression. One such example is microRNA
(miRNA/miR)-18a, a ncRNA involved in the site-specific regulation
of glucocorticoid receptor (GR) expression and the stress response.
This expression of this ncRNA appears to be regulated throughout
the lifetime of an organism, with miR-18a levels declining from the
embryonic stage to adulthood (9).
Finally, given that the non-coding part of the genome is extremely
larger than the protein-coding one, the genetic cause of numerous
diseases may be related to mutations within ncRNAs, including
neurological and psychiatric disorders, among several others
(7,10).
Moreover, the expression of these transcripts is
dependent on the specific developmental stage of the organism in
order to optimally regulate its internal dynamic equilibrium.
ncRNAs are also essential for the function of organisms, since they
allow them to respond to environmental signals, with a prominent
position among them being to maintain their homeostasis in response
to stress, a state where homeostasis is threatened (11). Differentiation, development, the
maintenance of homeostasis, stress responses and plasticity are,
therefore, mediated via epigenetic mechanisms, such as ncRNA
expression (7).
ncRNAs are thus ideal molecules to mediate stress
responses, and research has suggested that ncRNAs are key players
of epigenetic responses in the brain (12). Other studies have demonstrated
that exposure to stress induces brain-specific alterations in both
long ncRNA and small ncRNA expression levels (13,14). The present review aimed to collect
the information available thus far concerning ncRNA alterations
under stress conditions in order to highlight their mediating role
in the stress response.
2. Classification of non-coding RNAs
ncRNAs can be divided into two main categories,
structural and regulatory (11).
The former group includes well-known classes of ncRNAs, such as
ribosomal, transporter and small nuclear RNAs. Regulatory ncRNAs,
although a slightly more difficult to classify, can be divided into
small ncRNAs (sncRNAs) and long ncRNAs (lncRNAs) depending on their
length. sncRNAs include miRNAs, small interfering RNAs (siRNAs),
P-element induced wimpy testis (PIWI)-interacting RNAs (piRNAs) and
small nucleolar RNAs (snoRNAs). There are also ncRNAs derived from
enhancer RNAs (eRNAs), super-enhancers RNAs (seRNAs) and ncRNAs
derived from introns and telomeric sequences (TERRA) (15-20).
miRNAs are single-stranded RNA molecules that
suppress translation in a number of eukaryotic organisms through
incomplete mating, of six to eight nucleotides in length, with
their target mRNAs. In the case of perfect complementarity with
their mRNA targets, they cause their degradation via the
RNA-induced silencing complex, a phenomenon known as RNA
interference (16,21). miRNAs are derived from introns and
exons of both protein-coding and non-coding transcripts synthesized
by RNA polymerase II (21); thus,
a particular set of miRNAs can be synthesized under specific
conditions and in certain cell types only (22). By controlling various biological
processes and strictly regulating their expression, miRNAs are
central players in a wide range of developmental processes, such as
cell proliferation or stress responses (23).
siRNAs, produced from longer double-stranded RNAs or
long hairpins (often of exogenous origin), are considered to be
more specific than miRNAs and usually target homologous sequences
for gene silencing (17).
However, both miRNAs and siRNAs are produced by similar pathways
and have similar mechanisms of action; therefore the distinction
between them becomes blurred (17). While siRNAs were originally
considered to be primarily an antiviral mechanism, other findings
suggest that they ultimately play a much broader role in genome
regulation (16,17).
piRNAs are integrated into the PIWI subfamily of
Argonaute proteins. piRNAs in mammals appear to function primarily
in the reproductive cell line, where they target and suppress the
expression of transposable elements in order to maintain genomic
stability (15). The reason
transposons, although comprising up to 70% of the genome, have not
led organisms to extinction is that they are controlled by a number
of mechanisms in the cell, including piRNAs (24). It is worth mentioning that
transposons themselves can regulate gene expression through their
ncRNAs. The expression of transposons responds to environmental
signals and a number of them are activated under various forms of
cellular stress, sometimes resulting in inherited mutations of
certain genes (25).
lncRNAs are a heterogeneous class of ncRNA
regulators, >200 nucleotides in size. lncRNAs have minimal
conservation, unlike other classes of ncRNAs, although their
promoters are highly conserved (18,26,27). It has been suggested that their
conserved secondary structure is the key to their interaction with
protein molecules (11). A number
of nuclear lncRNAs appear to be involved in gene-regulating
processes, where they can regulate both their neighboring
environment and act in distant genomic loci. In other words, they
may be involved in the specific repression of a promoter or
transcription activation (8,28).
They also apppear to play a critical role in genomic imprinting,
the process through which a gene expresses only one of its two
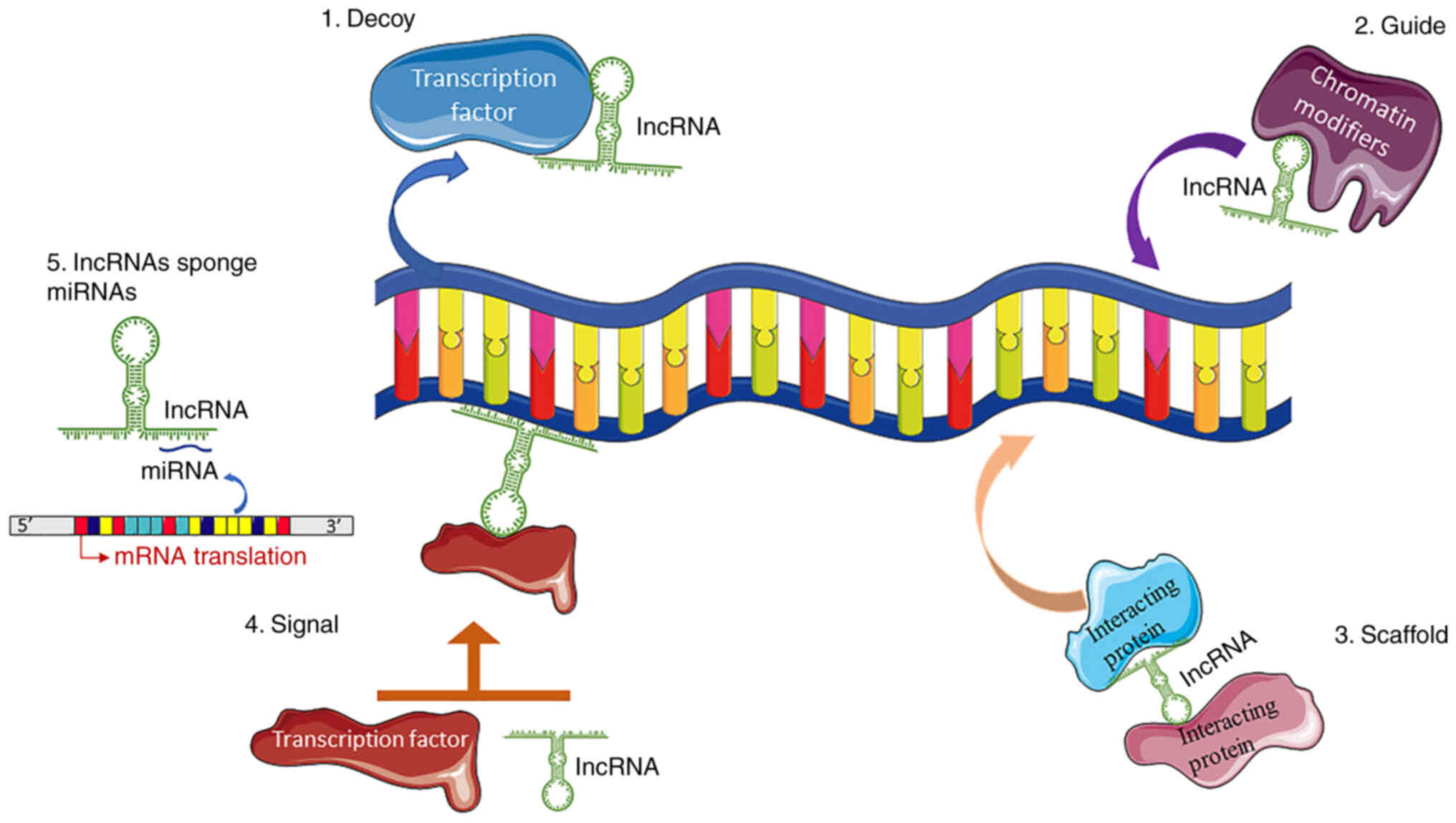
alleles (29). Based on their
modes of action, they can be classified as 'signals' for the
incorporation of temporal, spatial and developmental information,
as baits with the ability to isolate RNA and protein molecules in
order to suspend their functions (decoys), as 'guides' that lead
molecules to specific genomic sites, or finally, as scaffolds for
the creation of macromolecular complexes with various functions
(Fig. 1) (26). By modulating transcription,
lncRNAs can be considered as sensors of environmental signals, such
as stress, playing a key role in regulating transcriptional
responses to external, as well as developmental stimuli through
their interaction with transcription factors (15).
Sense and antisense transcripts are also derived
from certain enhancers and are therefore termed eRNAs (19). The majority of these eRNAs are
transcribed at lower levels than other ncRNAs and are rapidly
degraded by protein complexes, such as the human PM/Scl complex,
while they can also undergo methylation (30). They are divided into two
categories: Small, bidirectional, non-polyadenylated eRNAs and
long, unidirectional, polyadenylated eRNAs (31). Enhancer transcripts may play a
structural role in creating or stabilizing enhancer-promoter loops.
The transcription of eRNAs has been found to be associated with
mRNA synthesis in neighboring genes, suggesting their involvement
in transcriptional regulation, while subsequent studies have
demonstrated their ability to orchestrate time and tissue-specific
gene expression (19,32). Super-enhancers (SEs) are areas
where multiple enhancers are assembled together. They also have the
ability to transcribe, producing seRNAs, which exert their action
through cis and trans mechanisms (31).
A large number of functional ncRNAs are produced in
introns, including snoRNAs, piRNAs and lncRNAs (20,33). Although introns are believed to
degrade immediately following cleavage by primary transcripts,
there is strong evidence to indicate that intron RNAs can be
processed into smaller RNAs with significant half-lives and
specific subcellular locations, while the splicing of introns
appears to provide plasticity to the type of RNA produced from a
genetic locus (34).
Additionally, ncRNAs also appear to be produced by pseudogenes,
offering another mechanism to control gene function (35).
lncRNA molecules may also be transcribed from the
sub-telomere sequences. These lncRNAs carry telomere repeats in
their sequences, and are involved in the maintenance and regulation
of telomere homeostasis. The heterogeneous lncRNA produced by these
regions is termed TERRA (36),
and it has been shown that it may be subject to developmental
regulation and, in turn, may play a key role in orchestrating
certain aspects of the complex chromosomal transactions that occur
during cell differentiation (37).
3. Stress and ncRNAs
ncRNAs have been linked to a variety of disorders of
the stress system, such as anxiety, and major depressive and
bipolar disorders (38) (Table I). These RNA molecules appear to
be a conserved mechanism of how genes are being regulated in
response to a stressor among numerous animals (39).
 | Table IStress and ncRNAs. |
Table I
Stress and ncRNAs.
ncRNAs and their
stress-related targets
|
|---|
| ncRNAs | Stress-related
targets of ncRNAs |
Effect/function | (Refs.) |
|---|
| miR-34c | CS-R1 mRNA | Downregulation | (40) |
| miR-132 and
miR-128 | BDNF gene | Downregulation | (48) |
| miR-34b and
miR-27a | CRHR1 mRNA | Downregulation | (49) |
| miR-15a | FKBP51 gene | Downregulation | (9,50) |
| lncRNA GAS5 | GR | GRE decoy in
regulating glucocorticoid feedback | (51-53) |
| Lethe lncRNA | NF-κB | Inhibition of
NF-κB/DNA binding and inhibition of activation of NF-κB target
genes | (57,58) |
| IL1β-eRNA | IL1β and CXCL8 | Positive
correlation | (65) |
|
| ncRNAs regulated by
stress induction |
|
| ncRNAs | | Effect after stress
induction | (Refs.) |
|
| miR-186 and
miR-381 | | Overexpression | (47) |
| miR-709 | | Downregulation | (47) |
| lncRNA Gomafu | | Downregulation | (14) |
A number of studies have demonstrated that that the
expression of specific miRNAs changes in response to stress. For
example, a clear association between the manifestation of anxiety
behavior and the differential expression of specific miRNAs has
been observed in mice (40),
while exposure to environmental stressors regulates the expression
not only of miRNAs, but also of factors involved in their
biogenesis (41). lncRNAs have
also been shown to regulate gene expression in various mental
illnesses (42,43). In addition, research on the
effects of early-life stress in rodents has revealed long-term
effects that vary, depending on both the genetic background and
exposure to stressors, which may lead to epigenetic alterations in
genes involved in the stress circuit (14). Furthermore, researchers have
demonstrated that acquired changes due to traumatic stress, as well
as regulated fear responses, can be inherited for up to two
generations in mice (44).
Research on C. elegans and mice has demonstrated that
sncRNAs mediate a non-Mendelian inheritance of traits or phenotypes
acquired during life. sncRNAs are abundant in germ cells and are
influenced by environmental factors, such as early traumatic
stress, contributing to the possible onset of pathological features
later in life (45).
As regards the roles of miRNAs, the latter appear to
be responsible for the regulation of genes associated with the
activity of the hypothalamic-pituitary-adrenal (HPA) axis (38). Their levels are altered by stress,
glucocorticoids and mood stabilizers, suggesting that miRNAs may be
vital to the aetiology of anxiety disorders (40), in which stress is a critical
factor both in influencing their onset and maintenance (46). miRNAs target and regulate
stress-related proteins and play a role in the specific regulation
of genes associated with susceptibility to anxiety disorders. An
example of a protein targeting miRNA, is miR-34c, which targets the
corticosteroid type 1 receptor and facilitates the recovery process
following stressful situations (40). Moreover, in a previous study, the
exposure of rats to mild restraint stress for 2 weeks altered the
expression of certain miRNAs in the cerebellum; miR-186 and miR-381
were overexpressed, while miR-709 was underexpressed; these changes
may be involved in the long-term adaptation of organisms to stress
(47). An additional two miRNAs,
miRNA-132 and miRNA-128, have also been found to affect stress by
targeting the brain-derived neurotrophic factor gene (48), while the expression of numerous
miRNAs has been found to be increased in primary hypothalamic
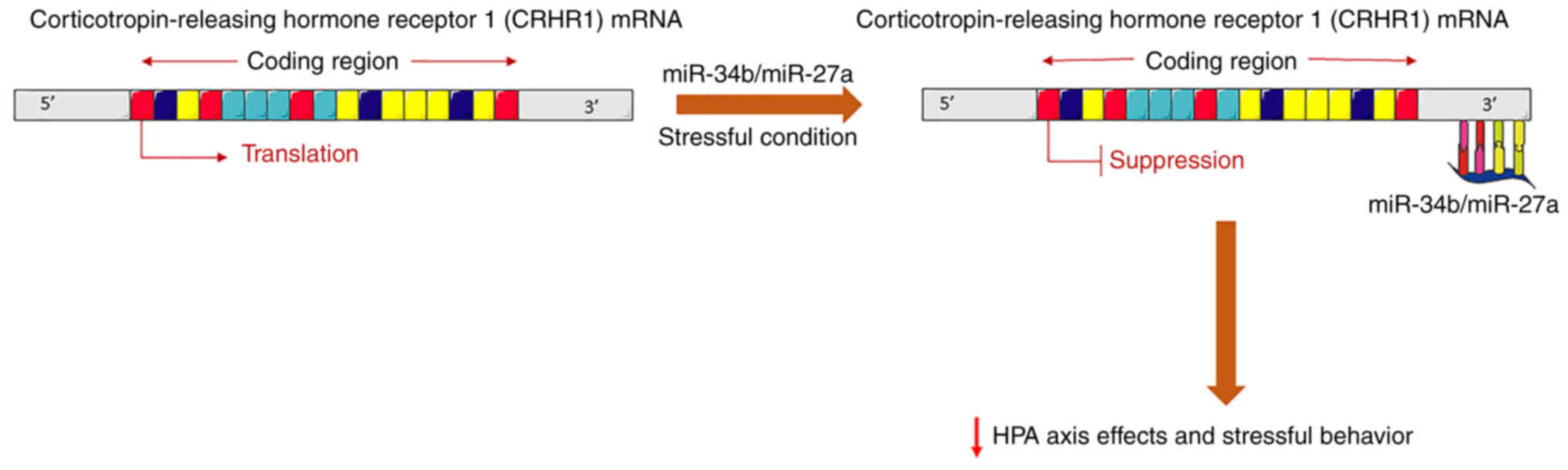
neurons following stress. miR-34b and miR-27a, for example, have
been found to be negatively associated with corticotropin releasing
hormone receptor 1 (CRHR1) mRNA levels. In particular, the
overexpression of miR-34b reduces both the CRHR1 mRNA and protein
levels, thereby reducing the effects of the HPA axis and stressful
behavior (49) (Fig. 2). In the amygdala, miR-15a has
been found to play an essential role in regulating behavioral
responses to chronic stress. As a target of miR-15a, the FKBP51
gene, is known to play a role in the transcriptional activation of
glucocorticoid receptor following an increase in cortisol levels,
and has been found to be involved in a number of stress-related
psychiatric disorders (50). Mice
expressing decreased levels of miR-15a in the amygdala following
exposure to chronic stress also exhibit severe anxiety behavior. In
humans, exposure to a traumatic event in childhood has also been
found to be associated with elevated levels of miR-15a in
peripheral blood (9).
It is now known that lncRNAs regulate genes in
mental illnesses, such as post-traumatic stress disorder (PTSD),
major depressive disorder, schizophrenia and autism spectrum
disorders, and have been associated with >200 illnesses
(42,43). lncRNAs are also involved in the
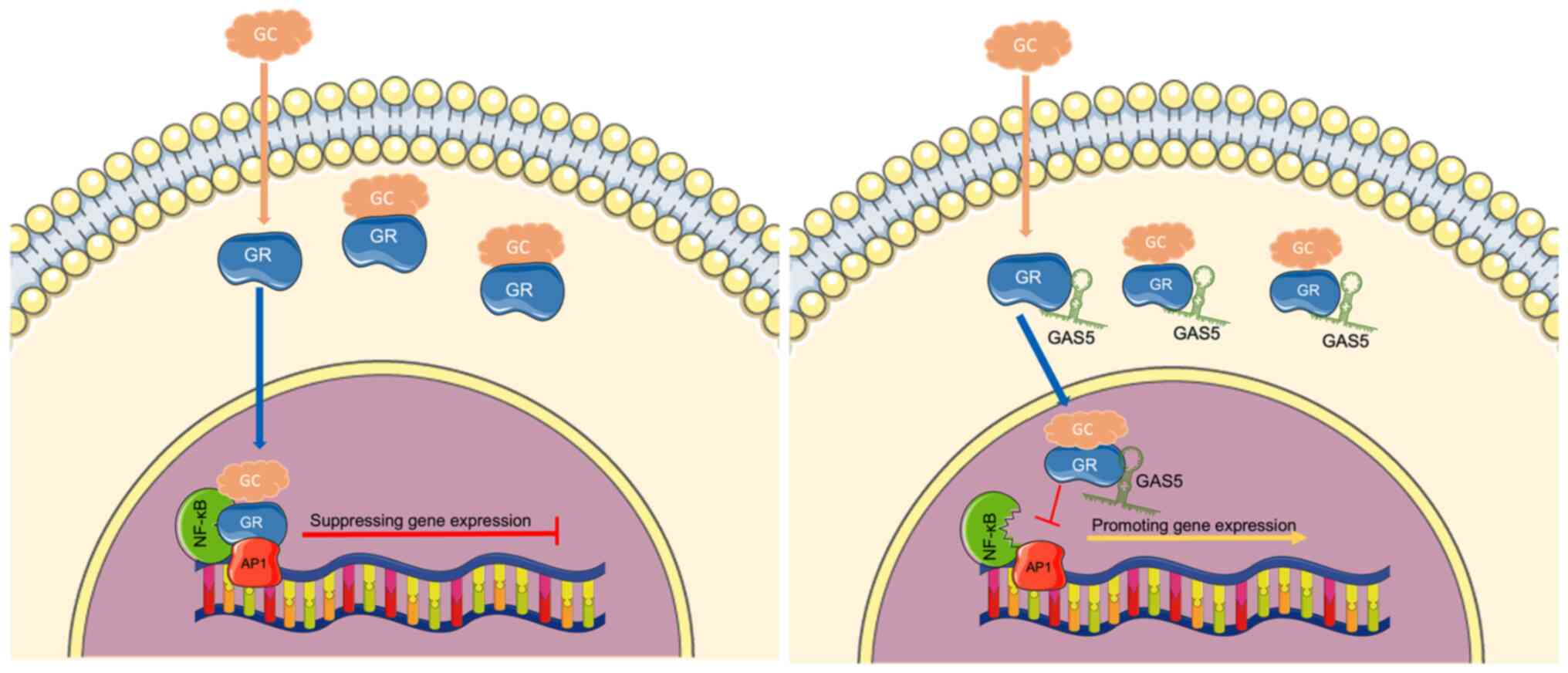
regulation of the HPA axis negative feedback. Growth
arrest-specific transcript 5 (Gas5), a lncRNA that interacts with
glucocorticoid receptors, interferes with the binding of the
glucocorticoid response element (GRE) to DNA and, thus inhibits its
transcriptional activity (51).
Thus, Gas5 can act as bait and GRE decoy in regulating
glucocorticoid feedback. Gas5 can alter the fate of cells by making
them more or less sensitive to apoptotic or other growth-related
stimuli by regulating the activity of glucocorticoids and, perhaps,
other steroid hormones. This lncRNA is involved in regulating
certain immune functions, as well as the pathogenesis of
autoimmune, inflammatory and infectious diseases, such as multiple
sclerosis, partly through modulating GR transcriptional activity
(52,53) (Fig.
3). It is important to note that Gas5 can be used as a
biomarker for personalized therapies and a novel therapeutic target
(54). In another study, the
expression profiles of lncRNAs in the medial prefrontal cortex
revealed that the lncRNA Gomafu was significantly downregulated in
adult mice following the induction of fear. On the other hand,
stress reactivity and anxiety behaviors have also been reported
following mutations in this lncRNA, while the reduction of Gomafu
expression in the brain cortex has also been associated with
schizophrenia (14). The lncRNA,
Neat1, has also been shown to be involved in stress signaling in
the brain, and enhances adaptive behavior in response to stress,
while its loss in mice leads to hyperactivity, deficits in social
interaction, and a panic escape response (55).
The pro-inflammatory cytokine, TNFα, which has been
found to regulate hundreds of lncRNAs, functions through the
transcription factor, NF-κB, thus playing a role in a variety of
processes, such as innate and adaptive immunity, inflammation,
apoptosis and aging (56). Lethe
is a lncRNA derived from a pseudogene that is selectively induced
by proinflammatory cytokines and interacts with an NF-κB subunit to
inhibit its binding to DNA and the subsequent activation of NF-κB
target genes. The loss of Lethe ncRNA expression is age-related and
may be one of the causes of increased NF-κB activity in aging.
Notably, Lethe is also selectively induced by dexamethasone,
suggesting that a potential anti-inflammatory therapeutic effect
may be due in part to the activation of Lethe's negative feedback
system (57). Finally, Lethe also
exerts a protective effect on sepsis-induced brain injury in mice,
by regulating autophagy, which is generally known to be controlled
by glucocorticoids in mouse cortical neurons (58).
A ground-breaking discovery in nematodes has
provided information on the mechanisms through which the nervous
system communicates through sncRNAs within the gamete line in order
to influence animal behavior across generations (59). That research demonstrated that
small RNA molecules in the nervous system can regulate genes in the
reproductive line, allowing specific behaviors to be modified for a
number of generations. These findings suggest that small RNAs,
particularly piRNAs, play a critical role in the epigenetic
inheritance of learned behaviors, enhancing an organism's chances
of survival. One of the proposed mechanisms for transmitting
learned behaviors to offspring is the natural transfer of small
RNAs from neurons to gametes resulting in inheritable changes
(44). It has also been
documented that piRNAs mediate the suppression of retrotransposon
mobilization, a mechanism that contributes to different transfer
rates in brain regions involved in learning and memory (44). It is also important to state that
the mobility of retrotransposons increases with age, which can
contribute to the observed neuronal decline associated with age.
Thus, activating the expression of certain piRNAs, which show
age-onset rhythmicity, may be a novel strategy for maintaining the
genomic integrity, which is threatened by stress as an organism
grows (60).
According to several studies, changes in eRNA levels
are also observed under conditions of stress, as their up- or
downregulation appears to lead to pathology. In models of mice with
myocardial infarction and transaortic constriction, the expression
of various eRNAs was induced, suggesting that these molecules are
differentially expressed in response to stress and may promote
abnormal transcription (61).
Another study revealed that hypoxia-inducible factor 1α-activated
eRNA (HERNA1) was a defining factor in heart disease, and it was
shown that the inactivation of HERNA1 by antisense oligonucleotides
in vivo prevented cardiac pathogenesis and improved the
overall survival of ill mice (62,63). Other studies have shown that in
the human BEAS-2B cell line, there are certain eRNAs responsible
for controlling anti-inflammatory genes, whose expression is
regulated by glucocorticoid receptor in association with NF-κB
(64). eRNAs also have been found
to regulate the expression of their target genes, such as
IL1β-eRNA, a molecule that regulates the expression of genes
involved in inflammation (65).
4. Possible applications
As miRNAs can cross the blood-brain barrier, they
could potentially be quantified by routine examinations as markers
for various neurodegenerative and neurodevelopmental disorders
(66). The experimental up- or
downregulation of certain miRNA expression which is altered
following stress have been shown to influence stress-associated
behavior in animal models of anxiety disorders (67). So far, several studies have linked
circulating miRNAs to perceived stress and anxiety. For example,
before their final examinations, the stress levels of medical
students have been shown to be significantly associated with blood
miR-16 levels (68). Furthermore,
miR-125a expression has been shown to be reduced in individuals
with PTSD, while blood levels of miR-22, -138-2, -148a, -339, -488
and -491 have been found to be associated with panic and phobic
disorders (69). It should be
noted that extracellular miRNAs are quite stable in the
circulation. This makes them attractive candidates for monitoring
the progression of a disease and its treatment. Another advantage
of using ncRNAs as biomarkers is that changes in their expression
in body fluids are also predicted to occur earlier than changes
observed in conventional biomarkers (70). Similar to miRNAs, lncRNAs can be
released into the extracellular space and detected in body fluids
or as circulating lncRNAs. Despite a large number of publications
on the use of circulating ncRNAs as potential clinical biomarkers,
to date, only one ncRNA has been converted to an FDA-approved
diagnostic marker (71).
Recent developments in basic and clinical research
have demonstrated that RNA molecules are a valuable tool in the
therapy of neurodegenerative diseases (72). ncRNAs could, therefore, become
novel targets for therapeutic interventions. For example, miR-223
may be a neuroprotective agent after a stroke, as it has been
observed to minimize the death of neuronal cells following an
ischemic attack (73). Moreover,
several different artificial oligonucleotides have been used to
inhibit the activity of miRNAs in cell lines or in vivo. For
example, 2'-O-methyl-oligonucleotides inhibit let-7b and miR-124,
thereby inducing nerve stem cell proliferation (73). The main issue with oligonucleotide
therapy appears to be the way in which the active oligonucleotide
would be 'delivered' to its site of action in the cytosol or in the
cell nucleus within tissues (74). Nevertheless, efforts have been
made to overcome these obstacles, as novel lipid nanoparticles are
being developed to shield miRNAs for tumor-targeted delivery to
combat metastatic cancers (75).
There are also evolved regions in the dark genome
that are related to stress and that have been associated with
schizophrenia and bipolar disorders or even with psychosis and
suicide. Those dark genome parts are specific to humans as they
have not been found in the genomes of other vertebrates. It is
possible that these regions evolved rapidly in humans as our
intelligence and cognitive abilities developed. However, it appears
they are easily disrupted, leading to the manifestation of
schizophrenia and bipolar disorders. This is a breakthrough, as it
applies to numerous individuals in modern societies and as many of
the available drugs are designed to target gene-encoded proteins
and not the dark part of the genome. Thus, there is hope and
potential in the future treatment of schizophrenia and bipolar
disorder as novel pharmacological targets may be identified.
5. Conclusions and future perspectives
At the beginning of the 21st century, it was
demonstrated that, although only ~2% of the human genome encodes
proteins, ~80% of it is transcribed to RNA. Thus, the idea of the
existence of 'junk DNA' was debunked, and many of the transcribed
non-coding RNAs were shown to be involved in almost every level of
gene expression regulation, with their expression being quickly
adjusted to environmental changes. The latter property renders them
ideal for regulating the stress response. In fact, research has
indicated that the various classes of non-coding RNAs, such as
miRNAs, lncRNAs, piRNAs, etc., and the factors involved in their
biogenesis, show variations in their levels in response to
stressful stimuli. A prime example is that of the glucocorticoid
receptors, which are targeted by a variety of miRNA molecules, as
evidenced by decreased expression of these receptors by miR-18α or
by miR-124. It is known that the regulation of gene expression is
mainly aimed at helping organisms adapt to their environment and to
ensure the optimal changes for their survival. The use of ncRNA
molecules as mediators of these responses has ultimately proven to
be a truly intelligent mechanism of adaptation. Thus, ncRNAs may be
used both as biomarkers and as therapeutic molecules; their
potential clinical uses are a very promising field of research.
Availability of data and materials
Not applicable.
Authors' contributions
All authors (KM, EP, TM, LP, KP, KID, KD, DAS, FB,
GPC, EE and DV) contributed to the conceptualization, design,
writing, drafting, revising, editing and reviewing of the
manuscript. All authors have read and approved the final
manuscript. Data authentication is not applicable.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
DAS is the Editor-in-Chief for the journal, but had
no personal involvement in the reviewing process, or any influence
in terms of adjudicating on the final decision, for this article.
The other authors declare that they have no competing
interests.
Acknowledgments
Not applicable.
Funding
The authors would like to acknowledge funding from the following
organizations: i) AdjustEBOVGP-Dx (RIA2018EF-2081): Biochemical
Adjustments of native EBOV Glycoprotein in Patient Sample to Unmask
target Epitopes for Rapid Diagnostic Testing. A European and
Developing Countries Clinical Trials Partnership (EDCTP2) under the
Horizon 2020 'Research and Innovation Actions' DESCA; and ii)
'MilkSafe: A novel pipeline to enrich formula milk using omics
technologies', a research co-financed by the European Regional
Development Fund of the European Union and Greek national funds
through the Operational Program Competitiveness, Entrepreneurship
and Innovation, under the call RESEARCH-CREATE-INNOVATE (project
code: T2EDK-02222).
References
|
1
|
Trayhurn P: Of genes and genomes- and dark
matter. Br J Nutr. 91:1–2. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Palazzo AF and Lee ES: Non-coding RNA:
What is functional and what is junk? Front Genet. 6:22015.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Pennisi E: Shining a light on the genome's
'Dark Matter.'. Science. 330:16142010. View Article : Google Scholar
|
|
4
|
Amaral PP and Mattick JS: Noncoding RNA in
development. Mamm Genome. 19:454–492. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Jacob F and Monod J: Genetic regulatory
mechanisms in the synthesis of proteins. J Mol Biol. 3:318–356.
1961. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Nerenz RD and Lefferts J: Our genome's
'Dark Matter' is the next frontier in molecular diagnostics. Clin
Chem. 63:792–793. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Qureshi IA, Mattick JS and Mehler MF: Long
non-coding RNAs in nervous system function and disease. Brain Res.
1338:20–35. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Salviano-Silva A, Lobo-Alves SC, Almeida
RC, Malheiros D and Petzl-Erler ML: Besides pathology: Long
Non-Coding RNA in cell and tissue homeostasis. Noncoding RNA.
4:32018. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
de Kloet ER, Fitzsimons CP, Datson NA,
Meijer OC and Vreugdenhil E: Glucocorticoid signaling and
stress-related limbic susceptibility pathway: About receptors,
transcription machinery and microRNA. Brain Res. 1293:129–141.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
de Almeida RA, Fraczek MG, Parker S,
Delneri D and O'Keefe RT: Non-coding RNAs and disease: The
classical ncRNAs make a comeback. Biochem Soc Trans. 44:1073–1078.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Kaikkonen MU, Lam MT and Glass CK:
Non-coding RNAs as regulators of gene expression and epigenetics.
Cardiovasc Res. 90:430–440. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Babenko O, Golubov A, Ilnytskyy Y,
Kovalchuk I and Metz GA: Genomic and epigenomic responses to
chronic stress involve miRNA-mediated programming. PLoS One.
7:e294412012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Rinaldi A, Vincenti S, De Vito F, Bozzoni
I, Oliverio A, Presutti C, Fragapane P and Mele A: Stress induces
region specific alterations in microRNAs expression in mice. Behav
Brain Res. 208:265–269. 2010. View Article : Google Scholar
|
|
14
|
Daskalakis NP, Provost AC, Hunter RG and
Guffanti G: Noncoding RNAs: Stress, glucocorticoids, and
posttraumatic stress disorder. Biol Psychiatry. 83:849–865. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Hombach S and Kretz M: Non-coding RNAs:
Classification, biology and functioning. Adv Exp Med Biol.
937:3–17. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Mattick JS and Makunin IV: Small
regulatory RNAs in mammals. Hum Mol Genet. 14(Spec No 1):
R121–R132. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Lam JK, Chow MY, Zhang Y and Leung SW:
siRNA Versus miRNA as therapeutics for gene silencing. Mol Ther
Nucleic Acids. 4:e2522015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Harrow J, Frankish A, Gonzalez JM,
Tapanari E, Diekhans M, Kokocinski F, Aken BL, Barrell D, Zadissa
A, Searle S, et al: GENCODE: The reference human genome annotation
for The ENCODE Project. Genome Res. 22:1760–1774. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Pennacchio LA, Bickmore W, Dean A, Nobrega
MA and Bejerano G: Enhancers: Five essential questions. Nat Rev
Genet. 14:288–295. 2013. View
Article : Google Scholar : PubMed/NCBI
|
|
20
|
Jo BS and Choi SS: Introns: The functional
benefits of introns in genomes. Genomics Inform. 13:112–118. 2015.
View Article : Google Scholar
|
|
21
|
Mattick JS and Makunin IV: Non-coding RNA.
Hum Mol Genet. 15(Spec No 1): R17–R29. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wahid F, Shehzad A, Khan T and Kim YY:
MicroRNAs: Synthesis, mechanism, function, and recent clinical
trials. Biochim Biophys Acta. 1803:1231–1243. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Croce CM and Calin GA: miRNAs, cancer, and
stem cell division. Cell. 122:6–7. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Maggert KA: Stress: An evolutionary
mutagen. Proc Natl Acad Sci USA. 116:17616–17618. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wheeler BS: Small RNAs, big impact: Small
RNA pathways in transposon control and their effect on the host
stress response. Chromosome Res. 21:587–600. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Wang KC and Chang HY: Molecular mechanisms
of long noncoding RNAs. Mol Cell. 43:904–914. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Amaral PP, Dinger ME and Mattick JS:
Non-coding RNAs in homeostasis, disease and stress responses: An
evolutionary perspective. Brief Funct Genomics. 12:254–278. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Cipolla GA, de Oliveira JC, Salviano-Silva
A, Lobo-Alves SC, Lemos DS, Oliveira LC, Jucoski TS, Mathias C,
Pedroso GA, Zambalde EP and Gradia DF: Long Non-Coding RNAs in
multifactorial diseases: Another layer of complexity. Noncoding
RNA. 4:132018. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Kung JT, Colognori D and Lee JT: Long
noncoding RNAs: Past, present, and future. Genetics. 193:651–669.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
de Lara JC, Arzate-Mejía RG and
Recillas-Targa F: Enhancer RNAs: Insights into their biological
role. Epigenet Insights. 12:25168657198460932019. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Wu M and Shen J: From super-enhancer
Non-coding RNA to immune checkpoint: Frameworks to functions. Front
Oncol. 9:13072019. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Chen H, Du G, Song X and Li L: Non-coding
transcripts from enhancers: New insights into enhancer activity and
gene expression regulation. Genomics Proteomics Bioinformatics.
15:201–207. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Parenteau J and Abou Elela S: Introns:
Good day junk is bad day treasure. Trends Genet. 35:923–934. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Hubé F and Francastel C: Mammalian
introns: When the junk generates molecular diversity. Int J Mol
Sci. 16:4429–4452. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Pink RC, Wicks K, Caley DP, Punch EK,
Jacobs L and Carter DR: Pseudogenes: Pseudo-functional or key
regulators in health and disease? RNA. 17:792–798. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Oliva-Rico D and Herrera LA: Regulated
expression of the lncRNA TERRA and its impact on telomere biology.
Mech Ageing Dev. 167:16–23. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Luke B and Lingner J: TERRA: Telomeric
repeat-containing RNA. EMBO J. 28:2503–2510. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Schmidt U, Keck ME and Buell DR: miRNAs
and other non-coding RNAs in posttraumatic stress disorder: A
systematic review of clinical and animal studies. J Psychiatr Res.
65:1–8. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Storey KB and Wu CW: Stress response and
adaptation: A new molecular toolkit for the 21st entury. Comp
Biochem Physiol. Part A Mol Integr Physiol. 165:417–428. 2013.
View Article : Google Scholar
|
|
40
|
Malan-Müller S, Hemmings SM and Seedat S:
Big effects of small RNAs: A review of MicroRNAs in Anxiety. Mol
Neurobiol. 47:726–739. 2013. View Article : Google Scholar :
|
|
41
|
Hollins SL and Cairns MJ: MicroRNA: Small
RNA mediators of the brains genomic response to environmental
stress. Prog Neurobiol. 143:61–81. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Nemoto T and Kakinuma Y: Involvement of
Noncoding RNAs in stress-related neuropsychiatric diseases caused
by DOHaD Theory: ncRNAs and DOHaD-Induced neuropsychiatric
diseases. Adv Exp Med Biol. 1012:49–59. 2018. View Article : Google Scholar
|
|
43
|
Levran O, Correa da Rosa J, Randesi M,
Rotrosen J, Adelson M and Kreek MJ: A non-coding CRHR2 SNP
rs255105, a cis-eQTL for a downstream lincRNA AC005154.6, is
associated with heroin addiction. PLoS One. 13:e01999512018.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Kim KW: PIWI Proteins and piRNAs in the
Nervous System. Mol Cells. 42:828–835. 2019.PubMed/NCBI
|
|
45
|
Gapp K, Jawaid A, Sarkies P, Bohacek J,
Pelczar P, Prados J, Farinelli L, Miska E and Mansuy IM:
Implication of sperm RNAs in transgenerational inheritance of the
effects of early trauma in mice. Nat Neurosci. 17:667–669. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Bystritsky A, Khalsa SS, Cameron ME and
Schiffman J: Current diagnosis and treatment of anxiety disorders.
P T. 38:30–57. 2013.PubMed/NCBI
|
|
47
|
Schouten M, Aschrafi A, Bielefeld P,
Doxakis E and Fitzsimons CP: microRNAs and the regulation of
neuronal plasticity under stress conditions. Neuroscience.
241:188–205. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Li YJ, Xu M, Gao ZH, Wang YQ, Yue Z, Zhang
YX, Li XX, Zhang C, Xie SY and Wang PY: Alterations of serum levels
of BDNF-related miRNAs in patients with depression. PLoS One.
8:e636482013. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Zhu J, Chen Z, Tian J, Meng Z, Ju M, Wu G
and Tian Z: miR-34b attenuates trauma-induced anxiety-like behavior
by targeting CRHR1. Int J Mol Med. 40:90–100. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Volk N, Pape JC, Engel M, Zannas AS,
Cattane N, Cattaneo A, Binder EB and Chen A: Amygdalar MicroRNA-15a
is essential for coping with chronic stress. Cell Rep.
17:1882–1891. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Kino T, Hurt DE, Ichijo T, Nader N and
Chrousos GP: Noncoding RNA Gas5 is a growth arrest and
starvation-associated repressor of the glucocorticoid receptor. Sci
Signal. 3:ra82010. View Article : Google Scholar :
|
|
52
|
Mayama T, Marr AK and Kino T: Differential
expression of glucocorticoid receptor Noncoding RNA repressor Gas5
in autoimmune and inflammatory diseases. Horm Metab Res.
48:550–557. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Moradi M, Gharesouran J, Ghafouri-Fard S,
Noroozi R, Talebian S, Taheri M and Rezazadeh M: Role of NR3C1 and
GAS5 genes polymorphisms in multiple sclerosis. Int J Neurosci.
130:407–412. 2020. View Article : Google Scholar
|
|
54
|
Lucafò M, Bravin V, Tommasini A,
Martelossi S, Rabach I, Ventura A, Decorti G and De Iudicibus S:
Differential expression of GAS5 in rapamycin-induced reversion of
glucocorticoid resistance. Clin Exp Pharmacol Physiol. 43:602–605.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Kukharsky MS, Ninkina NN, An H, Telezhkin
V, Wei W, Meritens CR, Cooper-Knock J, Nakagawa S, Hirose T,
Buchman VL and Shelkovnikova TA: Long non-coding RNA Neat1
regulates adaptive behavioural response to stress in mice. Transl
Psychiatry. 10:1712020. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Tilstra JS, Clauson CL, Niedernhofer LJ
and Robbins PD: NF-κB in aging and disease. Aging Dis. 2:449–465.
2011.
|
|
57
|
Rapicavoli NA, Qu K, Zhang J, Mikhail M,
Laberge RM and Chang HY: A mammalian pseudogene lncRNA at the
interface of inflammation and anti-inflammatory therapeutics.
ELife. 2:e007622013. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Mai C, Qiu L, Zeng Y and Jian HG: LncRNA
Lethe protects sepsis-induced brain injury via regulating autophagy
of cortical neurons. Eur Rev Med Pharmacol Sci. 23:4858–4864.
2019.PubMed/NCBI
|
|
59
|
Posner R, Toker IA, Antonova O, Star E,
Anava S, Azmon E, Hendricks M, Bracha S, Gingold H and Rechavi O:
Neuronal small RNAs control behavior transgenerationally. Cell.
177:1814–1826.e15. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Kuintzle RC, Chow ES, Westby TN, Gvakharia
BO, Giebultowicz JM and Hendrix DA: Circadian deep sequencing
reveals stress-response genes that adopt robust rhythmic expression
during aging. Nat Commun. 8:145292017. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Minerath RA, Hall DD and Grueter CE:
Targeting transcriptional machinery to inhibit enhancer-driven gene
expression in heart failure. Heart Fail Rev. 24:725–741. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Mirtschink P, Bischof C, Pham MD, Sharma
R, Khadayate S, Rossi G, Fankhauser N, Traub S, Sossalla S, Hagag
E, et al: Inhibition of the hypoxia-inducible factor 1α-induced
cardio-specific HERNA1 enhance-templated RNA protects from heart
disease. Circulation. 139:2778–2792. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Aguilo F, Li S, Balasubramaniyan N, Sancho
A, Benko S, Zhang F, Vashisht A, Rengasamy M, Andino B, Chen CH, et
al: Deposition of 5-Methylcytosine on enhancer RNAs enables the
coactivator function of PGC-1α. Cell Rep. 14:479–492. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Sasse SK, Gruca M, Allen MA, Kadiyala V,
Song T, Gally F, Gupta A, Pufall MA, Dowell RD and Gerber AN:
Nascent transcript analysis of glucocorticoid crosstalk with TNF
defines primary and cooperative inflammatory repression. Genome
Res. 29:1753–1765. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
IIott NE, Heward JA, Roux B, Tsitsiou E,
Fenwick PS, Lenzi L, Goodhead I, Hertz-Fowler C, Heger A, Hall N,
et al: Long non-coding RNAs and enhancer RNAs regulate the
lipopolysaccharide-induced inflammatory response in human
monocytes. Nat Commun. 5:39792014. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Hanna J, Hossain GS and Kocerha J: The
potential for microRNA therapeutics and clinical research. Front
Genet. 10:4782019. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Peedicayil J: The potential role of
epigenetic drugs in the treatment of anxiety disorders.
Neuropsychiatr Dis Treat. 16:597–606. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Katsuura S, Kuwano Y, Yamagishi N,
Kurokawa K, Kajita K, Akaike Y, Nishida K, Masuda K, Tanahashi T
and Rokutan K: MicroRNAs miR-144/144* and miR-16 in peripheral
blood are potential biomarkers for naturalistic stress in healthy
Japanese medical students. Neurosci Lett. 516:79–84. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Scott KA, Hoban AE, Clarke G, Moloney GM,
Dinan TG and Cryan JF: Thinking small: Towards microRNA-based
therapeutics for anxiety disorders. Expert Opin Investig Drugs.
24:529–542. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Moldovan L, Batte KE, Trgovcich J, Wisler
J, Marsh CB and Piper M: Methodological challenges in utilizing
miRNAs as circulating biomarkers. J Cell Mol Med. 18:371–390. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Anfossi S, Babayan A, Pantel K and Calin
GA: Clinical utility of circulating non-coding RNAs-an update. Nat
Rev Clin Oncol. 15:541–563. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Pascale E, Divisato G, Palladino R,
Auriemma M, Ngalya EF and Caiazzo M: Noncoding RNAs and midbrain DA
neurons: Novel molecular mechanisms and therapeutic targets in
health and disease. Biomolecules. 10:12692020. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Varela MA, Roberts TC and Wood MJA:
Epigenetics and ncRNAs in brain function and disease: Mechanisms
and prospects for therapy. Neurotherapeutics. 10:621–631. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Juliano RL: The delivery of therapeutic
oligonucleotides. Nucleic Acids Res. 44:6518–6548. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
O'Neill CP and Dwyer RM:
Nanoparticle-Based delivery of tumor suppressor microRNA for cancer
therapy. Cells. 9:5212020. View Article : Google Scholar : PubMed/NCBI
|