According to the global cancer statistics, the
incidence of lung cancer is second only to breast cancer,
accounting for >21% of all cancers (1). Non-small cell lung cancer (NSCLC) is
the primary pathological subtype of lung cancer, accounting for
~85% of all cases lung cancer (2). In addition, NSCLC can be further
sub-divided into lung squamous cell carcinoma, lung adenocarcinoma
(LUAD) and large-cell lung cancer (3). Early diagnosis of NSCLC is of
importance to both the improved cure rates and superior prognosis
(4,5). Although novel targeted drug
therapies have made considerable progress, both the overall
survival rates and early diagnosis rates of patients remain <20%
(6,7). Therefore, it is necessary to
discover novel predictive biomarkers and therapeutic targets for
NSCLC.
Until recently, long non-coding (lncRNAs) have been
considered to be 'junk' material on the genome that serves little
purpose. However, as genomic research improves, roles of lncRNAs
were progressively revealed in numerous diseases (8,9).
LncRNAs are RNA sequences that consist of >200 nucleotides and
serve important roles in transcriptional regulation and epigenetic
gene regulation (10). In
addition, lncRNAs confer obvious advantages in epigenetic
regulation (11). A number of
lncRNAs have high tissue expression specificity and are
evolutionary conserved (12).
Previous pan-cancer transcriptome analysis showed that the
expression of lncRNAs were frequently dysregulated and in manner
that was specific to multitude of tumors, including lung cancer
(13), breast cancer (14), and glioblastoma (8,15-18).
LncRNAs are closely associated with the occurrence
and progression of NSCLC, notably by regulating the development of
drug resistance and radiation sensitivity in patients with NSCLC.
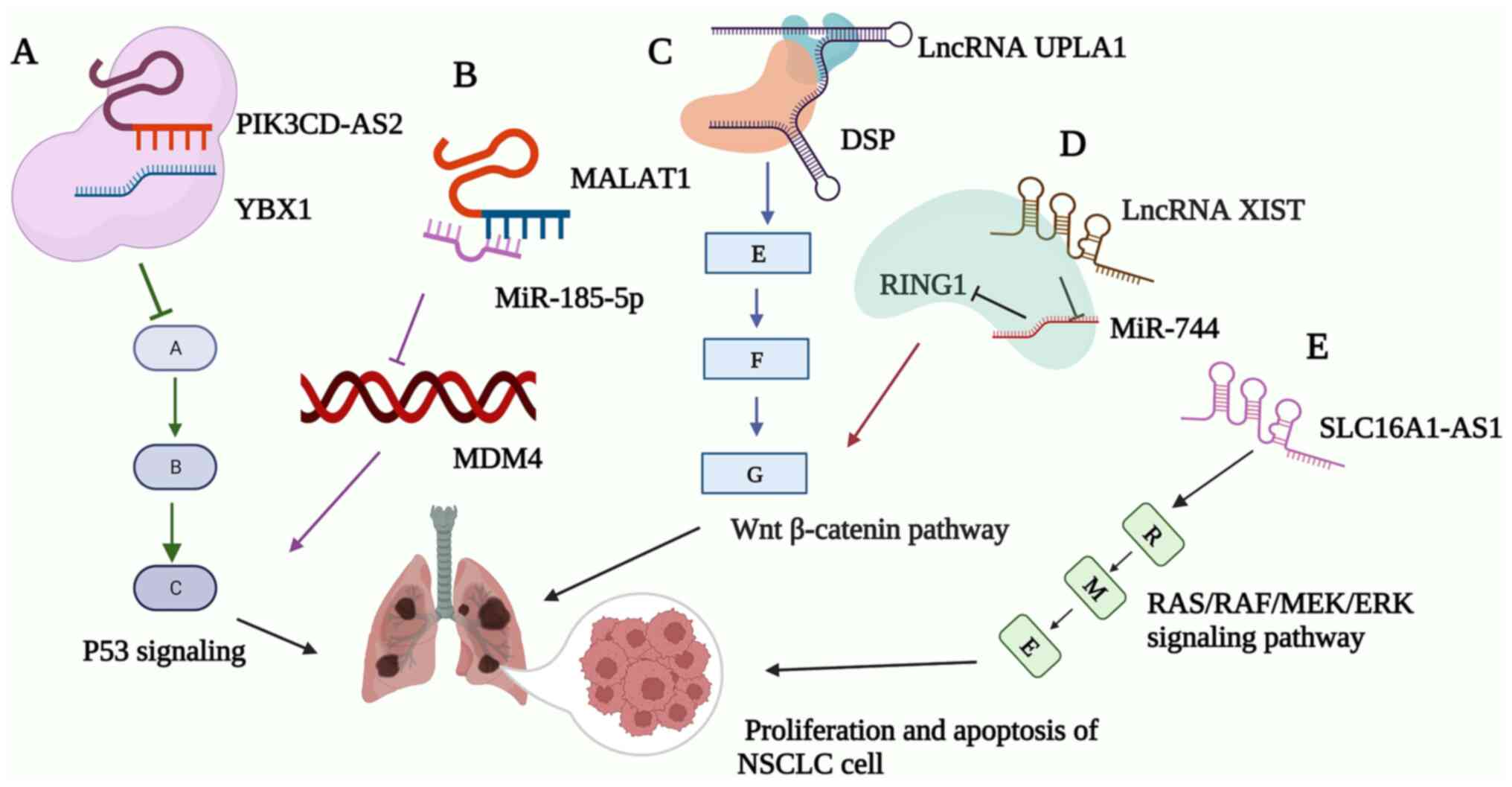
Overexpression of PIK3CD antisense RNA 2 (PIK3CD-AS2) was found to
promote NSCLC cell proliferation, apoptosis and progression through
the PIK3CD-AS2/Y-box binding protein 1 (YBX1)/p53 signaling axis
(19). Elucidating the mechanism
of lncRNAs on NSCLC would be beneficial for the development of
therapeutic strategies against its tumorigenesis. However, the
detailed mechanisms remain to be fully elucidated. The present
review therefore summarized the recent progress on lncRNA research
and their potential underlying mechanisms revealed in NSCLC, to
provide reference for the potential implications of lncRNAs in
NSCLC.
The majority of lncRNAs are similar to mRNAs, in
that they are transcribed by RNA polymerase II from the genomic
loci in chromatin (20). LncRNAs
can be classified according to their positions relative to the
encoding genes (8), namely long
intergenic RNAs, intron lncRNAs, antisense lncRNAs, bidirectional
lncRNAs and enhancer lncRNAs. LncRNAs can be classified into
oncogenes and tumor suppressor genes in accordance with whether
their expression can promote tumor development. In general, lncRNAs
that are overexpressed to promote tumor development are classified
as oncogenes, whilst lncRNAs that function in the opposite way
manner would be deemed to be tumor suppressor genes (21,22). In addition, lncRNAs can be
classified into cis-acting lncRNAs and trans-acting lncRNAs
according to whether it serves a cis-regulatory or trans-regulatory
role in cancers (23).
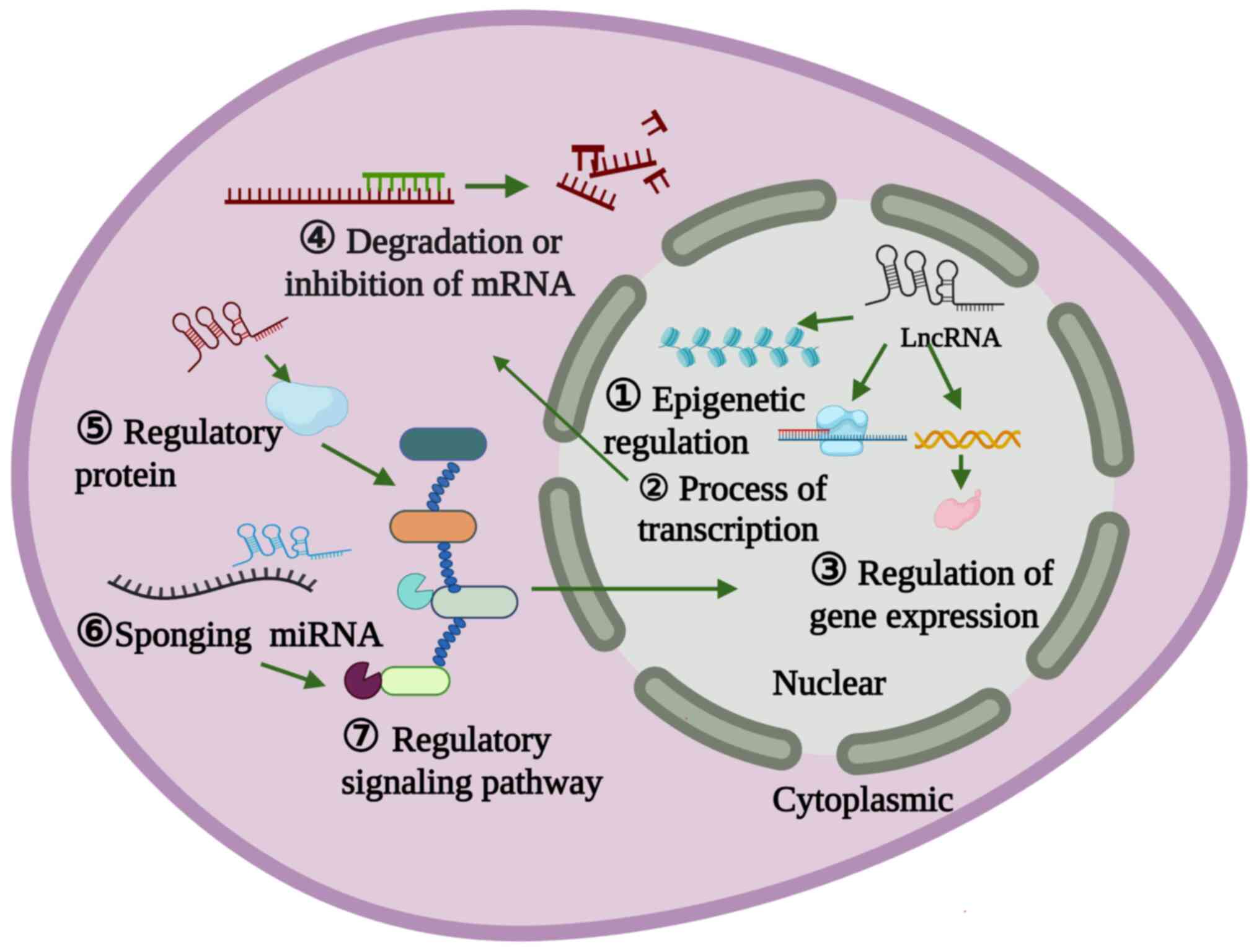
LncRNAs show a diverse array of characteristic
functions, in addition to having high tissue and sex specificity
(24). The functions of lncRNAs
are largely reflected by their subcellular localization (Fig. 1). Nuclear lncRNAs typically
regulate chromatin organization, transcriptional and
post-transcriptional gene expression, where they can also serve as
structural scaffolds anchoring nuclear domains to regulate
biological processes (25). By
contrast, cytoplasmic lncRNAs generally regulate various functions,
including mRNA conversion, translation, protein stability, cytokine
sponging and cell signaling (26). LncRNAs can interact with different
types of biomolecules, which would be of great significance in the
proliferation and apoptosis, invasion and migration,
epithelial-mesenchymal transition (EMT) and metastasis, in addition
to drug resistance of NSCLC cells. Therefore, monitoring changes in
lncRNA expression and elucidating its functional mechanisms are
likely to have clinical implications for the diagnosis, treatment
and prognosis of NSCLC.
The occurrence and development of malignancies are
frequently accompanied with changes in cell cycle and apoptosis
signaling. As summarized in the present review, lncRNAs can
regulate the activity of signaling cascades by binding to proteins
and affecting their stability. In addition, lncRNAs can serve as a
competitive endogenous RNA by interacting with miRNAs to regulate
downstream target gene expression. Conversely, miRNAs can regulate
the expression of lncRNAs, since certain lncRNAs share structural
similarities with certain mRNAs. Several lncRNAs associated with
NSCLC proliferation and apoptosis are summarized in this
section.
P53 is an important tumor suppressor, that can
regulate apoptosis, autophagy and senescence (27). In particular, splice factor YBX1
is a negative p53 regulator that serves an essential role in
autophagy in NSCLC (28,29). PIK3CD-AS2 was found to inhibit p53
signaling by binding with YBX1, protecting YBX1 from ubiquitination
and degradation (Fig. 2A)
(19). In addition,
metastasis-associated LUAD transcript 1 (MALAT1) was reported to be
associated with a number of cancers (30-33). Murine double minute 4 (MDM4), an
essential negative regulator of p53, was frequently found to be
overexpressed in cancer cells expressing wild-type p53. As shown in
Fig. 2B, overexpression of MALAT1
can upregulate miR-185-5p expression to reduce the expression of
MDM4, which inhibited the migration and invasion of NSCLC (34). In another study, MALAT1 was
demonstrated to promote the proliferation of NSCLC through the
MALAT1-FOXP3-GINS1 axis (35). In
conclusion, targeting PIK3CD-AS2 and MALAT1 may be a NSCLC
treatment strategy for restoring p53 function in tumors.
As a critical component of desmosomal plaque
proteins, desmoplakin (DSP) can also serve as a tumor suppressor by
inhibiting the Wnt/β-catenin signaling pathway in lung cancer
(36). This pathway is central to
the tumorigenesis, prognosis and therapeutic resistance of NSCLC
(37-41). As revealed in Fig. 2C, upregulation promoting
LUAD-associated transcript-1 (UPLA1) was found to be closely
associated with cell proliferation, migration and apoptosis in
NSCLC cells by regulating the DSP/Wnt/β-catenin pathway (42). LncRNA candidate gene for
X-inactivation center (XIST) inhibited the miR-744/really
interesting new gene 1 (RING1) pathway whilst activating that of
Wnt/β-catenin signaling (Fig.
2D), which inhibited the proliferation of NSCLC cells (43).
The RAS/RAF/MEK/ERK signaling pathway is an
extensively studied signaling pathway, particularly in cancer
(44,45). Hyperactivation of MAPK signaling
has been found to induce the occurrence of cancer (46). As demonstrated in Fig. 2E, lncRNA SLC16A1 antisense
transcript 1 (SLC16A1-AS1) affected the overall survival and
progression-free survival of patients with NSCLC by regulating the
RAS/RAF/MEK pathway (47).
SLC16A1-AS1 has also been reported in other cancers (48). In brief, SLC16A1-AS1 can
potentially serve a role in regulating the proliferation and
apoptosis of NSCLC.
In conclusion, PIK3CD-AS2, MALAT1, UPLA1, XIST and
SLC16A1-AS1 can all potentially serve different roles in the cell
proliferation, migration and apoptosis of NSCLC cells by
intervening in various regulatory pathways. They can be exploited
for the treatment of NSCLC. The role and mechanism of lncRNAs in
proliferation and apoptosis of NSCLC are listed in Table I.
Cancer metastasis increases the mortality rate of
NSCLC, which requires cell migration and the maintenance of
activity by altering the cell arrangement of EMT (82). A large number of lncRNAs have been
found to possibly regulate the migration and invasion of NSCLC.
Nuclear lncRNAs can not only induce methylation to regulate the
transcription of genes and binding of transcription factors to gene
promoters (83), but they can
also recruit other components to regulate mRNA (84). LncRNAs associated with migration,
invasion and EMT of NSCLC are summarized in Table II.
Elevated expression of the transcription factor
c-Myc has been frequently observed in human cancers, which is also
associated with increased tumor invasion and adverse clinical
outcomes (94,95). C-Myc promotes tumor cell
proliferation by amplifying the output of the existing gene
expression program (96). A
previous study identified a novel oncogenic axis involving long
intergenic non-coding RNA 01234 (linc01234), RNA-binding protein
heterogeneous nuclear ribonucleoprotein A2/B1, miR-106b-5p,
downregulating cryptochrome 2 and c-Myc (89). The upregulation of linc01234 in
NSCLC was positively associated with poorer prognosis. In addition,
linc01234 was found to facilitate the migration and invasion of
NSCLC cells through different pathway in cytoplasm and nucleus
(90). Specifically, linc01234
inhibited cell migration functioning as a competing endogenous RNA
for miR-340-5p and miR-27b-3p in the cytoplasm. In the nucleus,
linc01234 can interact with RNA-binding proteins lysine-specific
demethylase 1 and enhancer of zeste homolog 2 (EZH2), which led to
histone modification and the transcriptional suppression of B-cell
translocation gene 2, an anti-proliferative gene. Linc01123 also
promoted proliferation and aerobic glycolysis in NSCLC cell through
the miR-199a-5p/c-Myc axis, whilst inhibiting the malignancy of
LUAD through the miR-449b-5p/NOTCH1 axis (93). This suggests that linc01234 and
linc01123 can be used as potential biomarkers and therapeutic
targets for NSCLC.
Apart from c-Myc, lncRNAs have also been found to
regulate to activity of SRY-related HMG box 4 transcription factor
(SOX4), which is a master regulator of EMT. It can promote
tumorigenesis by endowing cells with migratory and invasive
properties, stemness and resistance to apoptosis (97,98). Cancer susceptibility candidate
(CASC) 15 is a hypoxia-sensitive lncRNA that appears to be
important for NSCLC cell migration and proliferation (75). CASC15 is transcriptionally
activated by hypoxia signaling in NSCLC cells, in a process that is
dependent hypoxia-inducible factor 1α (HIF-1α) and hypoxia response
elements (HREs). CASC15 served an essential role in the development
and progression of NSCLC through the HIF-1α/CASC15/SOX4/β-catenin
pathway. Accordingly, inhibiting the HIF-1α/CASC15/SOX4/β-catenin
axis may be a novel therapeutic strategy for treating patients with
NSCLC. The expression of long intergenic non-coding RNA 00301
(linc00301) was found to be upregulated in NSCLC and associated
with prognosis (99). The
linc00301 carcinogenic mechanism was found to involve the forkhead
box C1 (FOXC1)/linc00301/EZH2/EAF2/pVHL/HIF1α and
FOXC1/linc00301/miR-1276/HIF1α pathways, which offered novel ideas
and potential therapeutic targets.
In conclusion, linc01234, linc01123 and CASC15 are
potential therapeutic targets for improving NSCLC by inhibiting
migration, invasion and EMT. Additional mechanistic studies have
shown the signaling pathways that are involved downstream of c-Myc
and SOX4. In addition, as shown in Table II, lncCRYBG3, linc01426 and
HOTAIR were also found to be associated with migration, invasion
and EMT in NSCLC. Research on the relationship between lncRNAs and
NSCLC progression provided insight into the treatment of NSCLC.
NSCLC is not susceptible to immunotherapy or
chemotherapy, which reduces its overall survival (100,101). In addition to recruiting
epigenetic regulatory complexes, lncRNAs can also act as sponges of
miRNAs after gene transcription to regulate downstream signal
transduction cascades (102).
LncRNAs have been documented to exert an impact on therapeutic
resistance of NSCLC by regulating gene transcription (103). LncRNAs were found to be
associated with drug sensitivity in the treatment of NSCLC, such as
cisplatin and EGFR-tyrosine kinase inhibitors gefitinib and
afatinib (Table III).
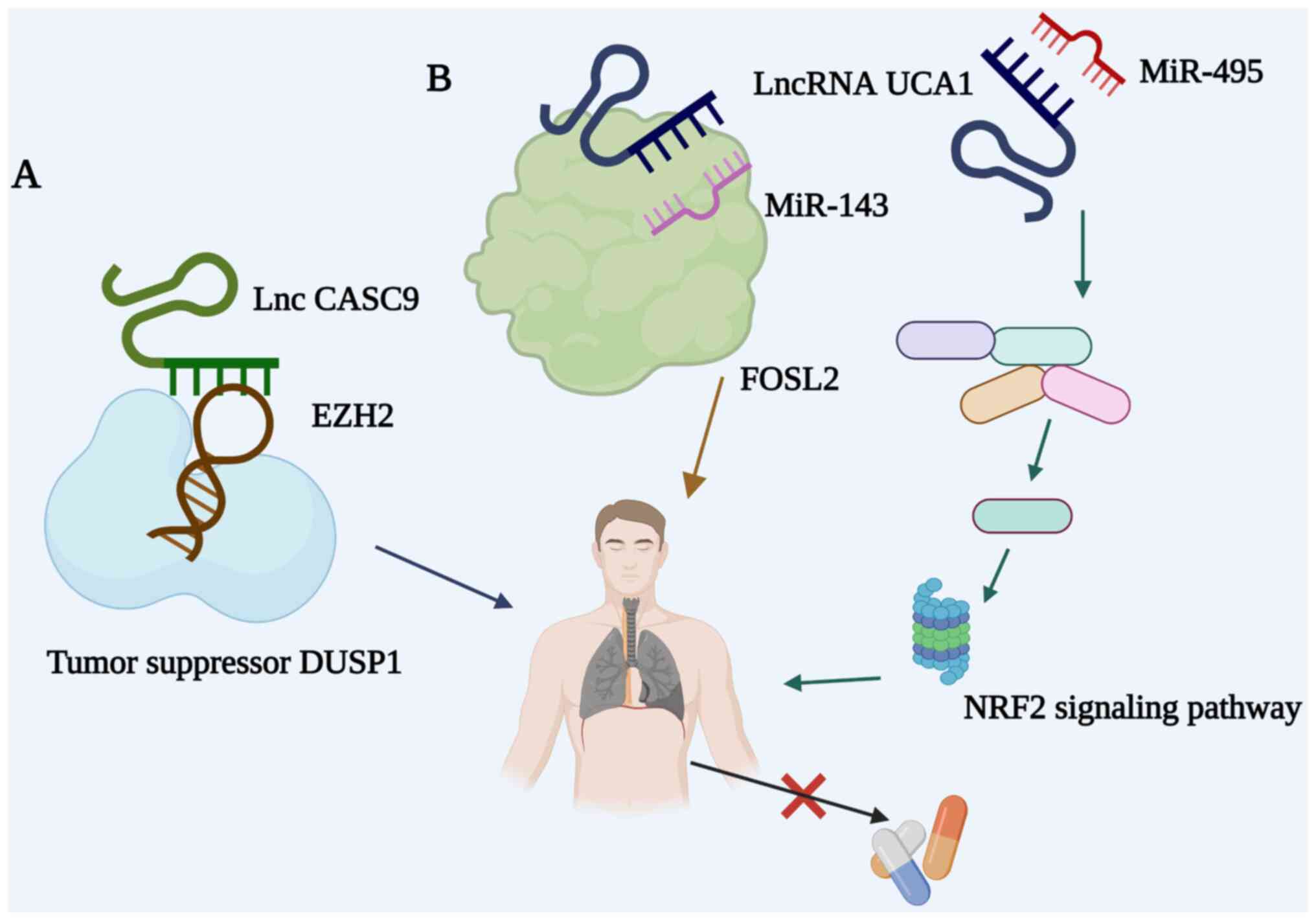
Histone methyl transferase EZH2 trimethylates
histone H3 (H3K27me3) at lysine 27 kept enzymatic activity in
cancer cells. The effect of candidate tumor susceptibility gene 9
(CASC9) on the sensitivity of NSCLC was associated with EZH2 and
dual specificity phosphatase 1 (DUSP1), reducing the sensitivity of
NSCLC to gefitinib (105).
Ectopic expression of DUSP1 was found to reduce NSCLC resistance to
gefitinib, suggesting that the CASC9/EZH2/DUSP1 axis can be a
target for overcoming EGFR resistance in NSCLC (Fig. 3A). In addition, linc00525 was
found to act on NSCLC through H3K27me3, rendering it another
potential therapeutic target for LUAD (51). Therefore, since both CASC9 and
linc00525 had an impact on drug resistance in NSCLC, they may
provide novel targets for drug resistance therapy in NSCLC.
LncRNAs can regulate drug sensitivity in NSCLC
through different pathways. Exosome-derived lncRNA urothelial
carcinoma-associated 1 (UCA1) was found to be overexpressed in
gefitinib-resistant NSCLC cells. In Fig. 3B, lncRNA UCA1 functioned as an
endogenous competitive RNA that can bind miR-143 to regulate the
expression of FOSL2 (119).
Overexpression of lncRNA UCA1 contributed to the development of
resistance to cisplatin through the UCA1/miR-495/NRF2 signaling
pathway (108). In addition,
lncRNA UCA1 induced resistance to gefitinib by epigenetically
silencing CDKN1A in NSCLC (109). Therefore, lncRNA UCA1 provides
another insight into the regulatory mechanisms of
gefitinib-resistant and cisplatin resistance in patients with
NSCLC.
A previous study identified the biological function
and mechanism of long intergenic non-protein coding RNA 1116
(linc01116) in the drug resistance of cancer cells (110). Linc01116 facilitated gefitinib
resistance in NSCLC cells by affecting interferon-induced protein
44 (IFI44) expression. IFI44 was involved in the IFN/STAT1 pathway
which could mediate resistance and radiotherapy in the tumor
microenvironment (120,121). Linc01116 was also associated
with cisplatin resistance in LUAD (122). Increasing the expression of
linc01116 was found to be associated with poorer outcomes in
patients with LUAD (123).
Conversely, downregulation of linc01116 expression inhibited cell
proliferation and blocked the cell cycle inhibition of EMT
(124). In addition, linc01116
could regulate iron-metabolism and AKT signaling in LUAD (125,126). In conclusion, linc01116 may be a
valuable prognostic biomarker and target to improve drug
sensitivity for patients with NSCLC.
In conclusion, the relationship between lncRNAs and
drug resistance in NSCLC was partially elucidated, which
represented a promising approach for predicting the chemotherapy
response of NSCLC. Studies on CASC9, lncRNA UCA1 and linc01116 in
drug resistance provided an insight into strategies for improving
therapeutic resistance in patients with NSCLC.
Radiotherapy serves an irreplaceable role in
improving local lesions and overall survival of patients with NSCLC
(127,128). As understanding into the
interaction between radiotherapy and cancer deepens, accumulating
studies have combined radiotherapy with novel drugs for NSCLC
treatment, such as immunotherapy and DNA damage response inhibitors
(129-131). LncRNAs could influence
radio-sensitivity by regulating the DNA damage response, stagnation
of autophagy, apoptosis and cell cycle progression (132,133). The relationship between lncRNAs
and NSCLC radio-sensitivity are listed in Table III.
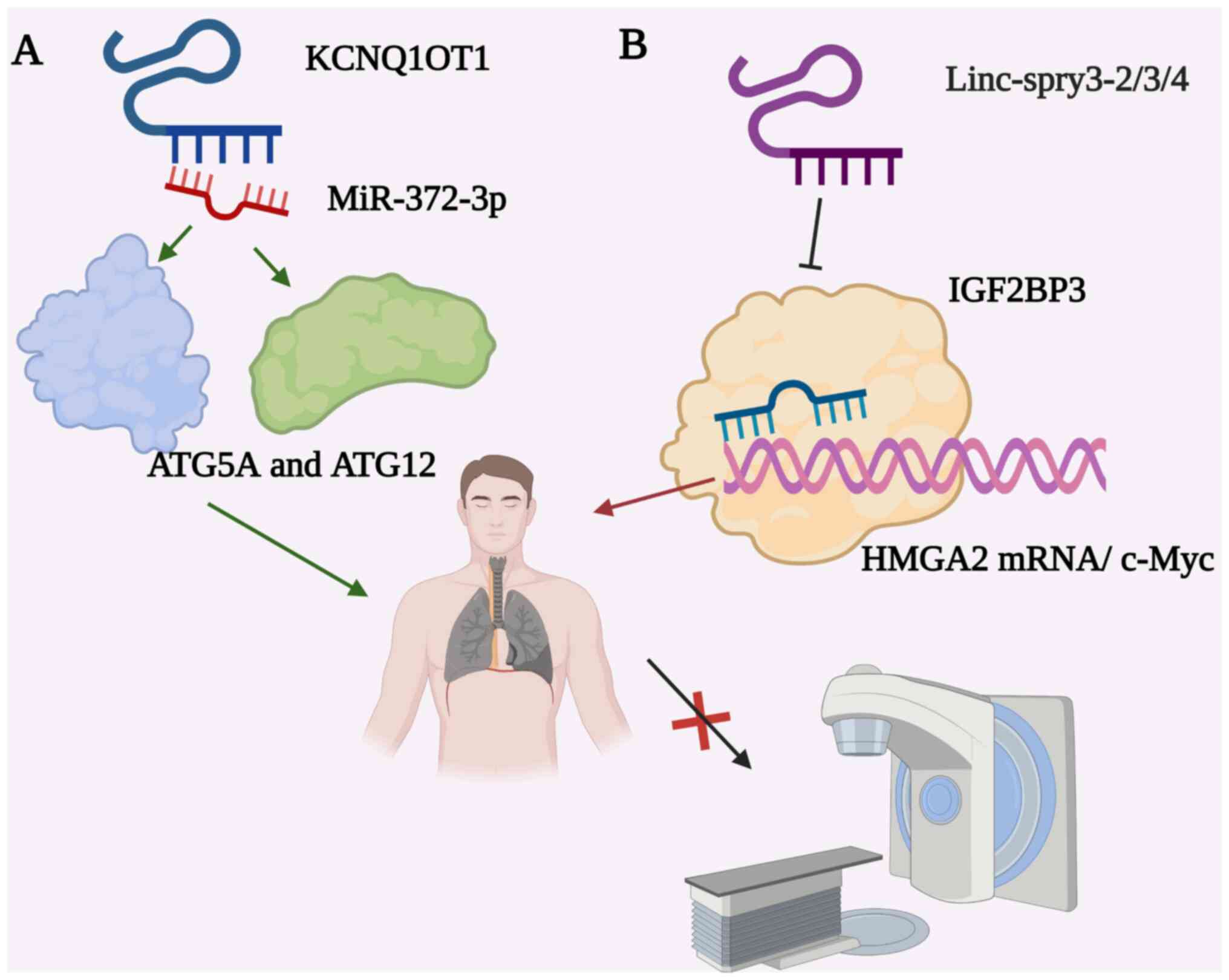
Knockdown of KCNQ1 opposite strand/antisense
transcript 1 (KCNQ1OT1) was found to improve the resistance of LUAD
to paclitaxel. KCNQ1OT1 promoted cell proliferation, migration and
invasion by regulating the miR-129-5p/JAG1 axis (134). As shown in Fig. 4A, KCNQ1OT1 affected cell
proliferation, autophagy and apoptosis by regulating the
miR-204-5p/autophagy-related (ATG) 3 axis (135). Higher expression levels of
KCNQ1OT1 were found to be associated with autophagy and decreased
sensitivity to radiation therapy (112). KCNQ1OT1 induced stereotactic
radiotherapy resistance in LUAD by stimulating miR-372-3p to induce
ATG5 and ATG12 dependent autophagy. This suggested that KCNQ1OT1 is
a potential target for enhancing the anti-tumor effect of
radiotherapy.
Human Y chromosome deletion and rearrangement were
shown to be associated with the occurrence and development of
certain malignancies (136);
however, on the possible association between NSCLC and lncRNAs on Y
chromosome has not been reported. Long chain non-coding
testicle-specific transcription Y-related gene 15 (TTTY15) was
previously found to be was associated with the progression of NSCLC
(137). LncRNAs in Y chromosome
DYZ1 regulated the radiation response. Linc-spry3-2/3/4 transcripts
were found to inhibit tumor growth, where their Y chromosome inlay
deletion (LOY) may lead to radiation resistance in NSCLC cells
(24). Further study revealed
that lncRNAs interfered with the stabilization of high mobility
group AT-Hook 2 (HMGA2) and c-Myc to reduce radio-sensitivity, by
binding to IGF2BP3 (Fig. 4B). It
revealed a negative correlation between the linc-SPRY3-2/3/4 or LOY
and overall survival. In summary, these findings suggested that
linc-spry3-2/3/4 is a promising marker of radiotherapy in patients
with NSCLC.
In brief, KCNQ1OT1, TTTY15, and linc-spry3-2/3/4
were associated with radio-sensitivity of NSCLC. As the
understanding into the mechanism of interaction between lncRNAs and
radiotherapy deepens, lncRNAs may prove to be a potential strategy
enhancing the antitumor effects of radiotherapy in patients.
The incidence of NSCLC has remained high, which is
coupled with the 5-year survival rate remaining low. Pathological
staging is particularly necessary for designating the treatment of
NSCLC (138,139). Therefore, in addition to the
current traditional imaging and pathological examination
techniques, it is necessary to identify novel characteristic
diagnostic biomarkers of NSCLC. LncRNAs can be classified according
to the location, function, mechanisms or its roles in the tumors.
LncRNAs are involved in proliferation and apoptosis, migration,
invasion and EMT, development of drug resistance and radiation
sensitivity in NSCLC. Therefore, they have the potential to serve
as molecular diagnostic biomarkers, therapeutic targets and
prognostic indicators for NSCLC. This is because they have a wide
array of characteristic functions, high tissue and sex
specificity.
Nevertheless, the application of lncRNAs in clinical
therapies patients still had several challenges. Although lncRNAs
are promising as an innovative tool, certain lncRNAs lack
specificity. It is therefore crucial to identify the most specific
lncRNAs associated with tumor staging. In addition, although
evidence has been accumulating about the utility of lncRNAs, the
structure and functional information on these lncRNAs remain to be
fully elucidated, which impedes the application of lncRNAs for
clinical diagnosis and treatment. In spite of lncRNAs having high
tissue specificity and evolutionary conservation, the conservation
among the various species is unsatisfactory. Accordingly, rigorous
preclinical studies were required prior to the application of
lncRNAs for NSCLC treatment.
All data generated or analyzed during this study are
included in this published article.
PYT conceived the idea and participated in preparing
the figures and tables of the manuscript. DJS participated in the
preparation and proofreading of the manuscript. WX, LXC and HL
supervised the project and guided the manuscript. All authors read
and approved the final manuscript.
Not applicable.
Not applicable.
The authors declare that they have no competing
interests.
The authors would like to acknowledge the support
from the Joint National Local Engineering Research Center of Fujian
and Taiwan Chinese Medicine Molecular Biotechnology, Fujian Key
Laboratory of Chinese Materia Medica, Fujian University Key
Laboratory for Research and Development of TCM Resources, at Fujian
University of Traditional Chinese Medicine.
The present study was supported by the National Natural Science
Foundation of China (NSFC) (grant no. 82204224), the Chunhui
Program-Cooperative Research Project of the Ministry of Education,
Liaoning Natural Science Foundation (grant no. 2022-MS-241), the
China Postdoctoral Science Foundation (grant no. 2021M693957), the
Shenyang Young and Middle-aged Innovative Talents Support Program
(grant no. RC210446), and the Project of the Educational Department
of Liaoning (grant no. LJKZ0919).
|
1
|
Sung H, Ferlay J, Siegel RL, Laversanne M,
Soerjomataram I, Jemal A and Bray F: Global cancer statistics 2020:
GLOBOCAN estimates of incidence and mortality Worldwide for 36
cancers in 185 countries. CA Cancer J Clin. 71:209–249. 2021.
View Article : Google Scholar
|
|
2
|
Xie S, Wu Z, Qi Y, Wu B and Zhu X: The
metastasizing mechanisms of lung cancer: Recent advances and
therapeutic challenges. Biomed Pharmacother. 138:1114502021.
View Article : Google Scholar
|
|
3
|
Campbell JD, Alexandrov A, Kim J, Wala J,
Berger AH, Pedamallu CS, Shukla SA, Guo G, Brooks AN, Murray BA, et
al: Distinct patterns of somatic genome alterations in lung
adenocarcinomas and squamous cell carcinomas. Nat Genet.
48:607–616. 2016. View
Article : Google Scholar : PubMed/NCBI
|
|
4
|
Chaft JE, Rimner A, Weder W, Azzoli CG,
Kris MG and Cascone T: Evolution of systemic therapy for stages
I-III non-metastatic non-small-cell lung cancer. Nat Rev Clin
Oncol. 18:547–557. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Howlader N, Forjaz G, Mooradian MJ, Meza
R, Kong CY, Cronin KA, Mariotto AB, Lowy DR and Feuer EJ: The
effect of advances in lung-cancer treatment on population
mortality. N Engl J Med. 383:640–649. 2020. View Article : Google Scholar
|
|
6
|
Siegel RL, Miller KD, Fuchs HE and Jemal
A: Cancer statistics, 2021. CA Cancer J Clin. 71:7–33. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Sheng GT, Duan HJ, Sun ZG and Chu HQ:
Advances in therapeutic nanodrug delivery systems for infectious
lung diseases: A review. Acta Materia Medica. 1:343–364. 2022.
View Article : Google Scholar
|
|
8
|
Ginn L, Shi L, Montagna M and Garofalo M:
LncRNAs in non-small-cell lung cancer. Noncoding RNA.
6:252020.PubMed/NCBI
|
|
9
|
Li Z, Meng X, Wu P, Zha C, Han B, Li L,
Sun N, Qi T, Qin J, Zhang Y, et al: Glioblastoma Cell-Derived
lncRNA-containing exosomes induce microglia to produce complement
C5, promoting chemotherapy resistance. Cancer Immunol Res.
9:1383–1399. 2021. View Article : Google Scholar
|
|
10
|
Rinn JL and Chang HY: Genome regulation by
long noncoding RNAs. Annu Rev Biochem. 81:145–166. 2012. View Article : Google Scholar
|
|
11
|
Bánfai B, Jia H, Khatun J, Wood E, Risk B,
Gundling WE Jr, Kundaje A, Gunawardena HP, Yu Y, Xie L, et al: Long
noncoding RNAs are rarely translated in two human cell lines.
Genome Res. 22:1646–1657. 2012. View Article : Google Scholar :
|
|
12
|
Guttman M, Amit I, Garber M, French C, Lin
MF, Feldser D, Huarte M, Zuk O, Carey BW, Cassady JP, et al:
Chromatin signature reveals over a thousand highly conserved large
non-coding RNAs in mammals. Nature. 458:223–227. 2009. View Article : Google Scholar :
|
|
13
|
Wang S, Zeng F, Liang S, Wang Q, Wen Y,
Wang Q, Zhang J, Li M, Fang S, Wei T, et al: lncRNA Linc00173
modulates glucose metabolism and multidrug chemoresistance in SCLC:
Potential molecular panel for targeted therapy. Mol Ther.
30:17872022. View Article : Google Scholar :
|
|
14
|
Qiao Y, Jin T, Guan S, Cheng S, Wen S,
Zeng H, Zhao M, Yang L, Wan X, Qiu Y, et al: Long non-coding RNA
Lnc-408 promotes invasion and metastasis of breast cancer cell by
regulating LIMK1. Oncogene. 40:4198–4213. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Du M, Zheng R, Ma G, Chu H, Lu J, Li S,
Xin J, Tong N, Zhang G, Wang W, et al: Remote modulation of lncRNA
GCLET by risk variant at 16p13 underlying genetic susceptibility to
gastric cancer. Sci Adv. 6:eaay55252020. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zhao W and Xie Q: Exosomal lncRNA-Mediated
intercellular communication promotes glioblastoma chemoresistance.
Cancer Immunol Res. 9:13722021. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zhang B, Zhang M, Yang Y, Li Q, Yu J, Zhu
S, Niu Y and Shang Z: Targeting KDM4A-AS1 represses AR/AR-Vs
deubiquitination and enhances enzalutamide response in CRPC.
Oncogene. 41:387–399. 2022. View Article : Google Scholar :
|
|
18
|
Meessen J, Bär C, di Dona FM, Staszewsky
LI, Di Giulio P, Di Tano G, Costa A, Leonardy J, Novelli D, Nicolis
EB, et al: LIPCAR Is increased in chronic symptomatic HF patients.
A Sub-Study of the GISSI-HF trial. Clin Chem. 67:1721–1731. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zheng X, Zhang J, Fang T, Wang X, Wang S,
Ma Z, Xu Y, Han C, Sun M, Xu L, et al: The long non-coding RNA
PIK3CD-AS2 promotes lung adenocarcinoma progression via
YBX1-mediated suppression of p53 pathway. Oncogenesis. 9:342020.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Eidem TM, Kugel JF and Goodrich JA:
Noncoding RNAs: Regulators of the mammalian transcription
machinery. J Mol Biol. 428:2652–2659. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Tao H, Zhang Y, Li J, Liu J, Yuan T, Wang
W, Liang H, Zhang E and Huang Z: Oncogenic lncRNA BBOX1-AS1
promotes PHF8-mediated autophagy and elicits sorafenib resistance
in hepatocellular carcinoma. Mol Ther Oncolytics. 28:88–103. 2023.
View Article : Google Scholar :
|
|
22
|
Liu B, Qu X, Wang J, Xu L, Zhang L, Xu B,
Su J and Bian X: LINC00365 functions as a tumor suppressor by
inhibiting HIF-1α-mediated glucose metabolism reprogramming in
breast cancer. Exp Cell Res. 425:1135142023. View Article : Google Scholar
|
|
23
|
Kopp F and Mendell JT: Functional
classification and experimental dissection of long noncoding RNAs.
Cell. 172:393–407. 2018. View Article : Google Scholar :
|
|
24
|
Brownmiller T, Juric JA, Ivey AD, Harvey
BM, Westemeier ES, Winters MT, Stevens AM, Stanley AN, Hayes KE,
Sprowls SA, et al: Y Chromosome LncRNA are involved in radiation
response of male non-small cell lung cancer cells. Cancer Res.
80:4046–4057. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Sun Q, Hao Q and Prasanth KV: Nuclear long
noncoding RNAs: Key regulators of gene expression. Trends Genet.
34:142–157. 2018. View Article : Google Scholar :
|
|
26
|
Noh JH, Kim KM, McClusky WG, Abdelmohsen K
and Gorospe M: Cytoplasmic functions of long noncoding RNAs. Wiley
Interdiscip Rev RNA. 9:e14712018. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Jing X, Yang F, Shao C, Wei K, Xie M, Shen
H and Shu Y: Role of hypoxia in cancer therapy by regulating the
tumor microenvironment. Mol Cancer. 18:1572019. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Kashihara M, Azuma K, Kawahara A, Basaki
Y, Hattori S, Yanagawa T, Terazaki Y, Takamori S, Shirouzu K,
Aizawa H, et al: Nuclear Y-box binding protein-1, a predictive
marker of prognosis, is correlated with expression of HER2/ErbB2
and HER3/ErbB3 in non-small cell lung cancer. J Thorac Oncol.
4:1066–1074. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Cui Y, Li F, Xie Q, Zhao S, Guo T, Guo P,
Hu S, Hao J, Tian C, Yu W, et al: YBX1 mediates autophagy by
targeting p110β and decreasing the sensitivity to cisplatin in
NSCLC. Cell Death Dis. 11:4762020. View Article : Google Scholar
|
|
30
|
Zhao Y, Zhou L, Li H, Sun T, Wen X, Li X,
Meng Y, Li Y, Liu M, Liu S, et al: Nuclear-Encoded lncRNA MALAT1
epigenetically controls metabolic reprogramming in HCC cells
through the mitophagy pathway. Mol Ther Nucleic Acids. 23:264–276.
2021. View Article : Google Scholar
|
|
31
|
Cao DW, Liu MM, Duan R, Tao YF, Zhou JS,
Fang WR, Zhu JR, Niu L and Sun JG: The lncRNA Malat1 functions as a
ceRNA to contribute to berberine-mediated inhibition of HMGB1 by
sponging miR-181c-5p in poststroke inflammation. Acta Pharmacol
Sin. 41:22–33. 2020. View Article : Google Scholar
|
|
32
|
Barik GK, Sahay O, Behera A, Naik D and
Kalita B: Keep your eyes peeled for long noncoding RNAs: Explaining
their boundless role in cancer metastasis, drug resistance, and
clinical application. Biochim Biophys Acta Rev Cancer.
1876:1886122021. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Knutsen E, Harris AL and Perander M:
Expression and functions of long non-coding RNA NEAT1 and isoforms
in breast cancer. Br J Cancer. 126:551–561. 2022. View Article : Google Scholar
|
|
34
|
Wang D, Zhang S, Zhao M and Chen F: LncRNA
MALAT1 accelerates non-small cell lung cancer progression via
regulating miR-185-5p/MDM4 axis. Cancer Med. 9:9138–9149. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Li M, Shi M, Hu C, Chen B and Li S: MALAT1
modulated FOXP3 ubiquitination then affected GINS1 transcription
and drived NSCLC proliferation. Oncogene. 40:3870–3884. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Yang L, Chen Y, Cui T, Knösel T, Zhang Q,
Albring KF, Huber O and Petersen I: Desmoplakin acts as a tumor
suppressor by inhibition of the Wnt/β-catenin signaling pathway in
human lung cancer. Carcinogenesis. 33:1863–1870. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Wang R, Liu J, Li K, Yang G, Chen S, Wu J,
Xie X, Ren H and Pang Y: An SETD1A/Wnt/β-catenin feedback loop
promotes NSCLC development. J Exp Clin Cancer Res. 40:3182021.
View Article : Google Scholar
|
|
38
|
Liao Y, Feng J, Sun W, Wu C, Li J, Jing T,
Liang Y, Qian Y, Liu W, Wang H, et al: CIRP promotes the
progression of non-small cell lung cancer through activation of
Wnt/β-catenin signaling via CTNNB1. J Exp Clin Cancer Res.
40:2752021. View Article : Google Scholar
|
|
39
|
Shapiro M, Akiri G, Chin C, Wisnivesky JP,
Beasley MB, Weiser TS, Swanson SJ and Aaronson SA: Wnt pathway
activation predicts increased risk of tumor recurrence in patients
with stage I nonsmall cell lung cancer. Ann Surg. 257:548–554.
2013. View Article : Google Scholar
|
|
40
|
Pang B, Wang Y and Chang X: A Novel Tumor
suppressor gene, ZNF24, inhibits the development of NSCLC by
inhibiting the WNT signaling pathway to induce cell senescence.
Front Oncol. 11:6643692021. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Zhang K, Wang J, Yang L, Yuan YC, Tong TR,
Wu J, Yun X, Bonner M, Pangeni R, Liu Z, et al: Targeting histone
methyltransferase G9a inhibits growth and Wnt signaling pathway by
epigenetically regulating HP1α and APC2 gene expression in
non-small cell lung cancer. Mol Cancer. 17:1532018. View Article : Google Scholar
|
|
42
|
Han X, Jiang H, Qi J, Li J, Yang J, Tian
Y, Li W, Jing Q and Wang C: Novel lncRNA UPLA1 mediates
tumorigenesis and prognosis in lung adenocarcinoma. Cell Death Dis.
11:9992020. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Wang J, Cai H, Dai Z and Wang G:
Down-regulation of lncRNA XIST inhibits cell proliferation via
regulating miR-744/RING1 axis in non-small cell lung cancer. Clin
Sci (Lond). 133:1567–1579. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Kun E, Tsang YTM, Ng CW, Gershenson DM and
Wong KK: MEK inhibitor resistance mechanisms and recent
developments in combination trials. Cancer Treat Rev.
92:1021372021. View Article : Google Scholar
|
|
45
|
Weissman R, Diamond EL, Haroche J, Durham
BH, Cohen F, Buthorn J, Amoura Z, Emile JF, Mazor RD, Shomron N, et
al: MicroRNA-15a-5p acts as a tumor suppressor in histiocytosis by
mediating CXCL10-ERK-LIN28a-let-7 axis. Leukemia. 36:1139–1149.
2022. View Article : Google Scholar :
|
|
46
|
Han J, Liu Y, Yang S, Wu X, Li H and Wang
Q: MEK inhibitors for the treatment of non-small cell lung cancer.
J Hematol Oncol. 14:12021. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Liu HY, Lu SR, Guo ZH, Zhang ZS, Ye X, Du
Q, Li H, Wu Q, Yu B, Zhai Q, et al: lncRNA SLC16A1-AS1 as a novel
prognostic biomarker in non-small cell lung cancer. J Investig Med.
68:52–59. 2020. View Article : Google Scholar :
|
|
48
|
Logotheti S, Marquardt S, Gupta SK,
Richter C, Edelhäuser BAH, Engelmann D, Brenmoehl J, Söhnchen C,
Murr N, Alpers M, et al: LncRNA-SLC16A1-AS1 induces metabolic
reprogramming during bladder cancer progression as target and
co-activator of E2F1. Theranostics. 10:9620–9643. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Zang X, Gu J, Zhang J, Shi H, Hou S, Xu X,
Chen Y, Zhang Y, Mao F, Qian H, et al: Exosome-transmitted lncRNA
UFC1 promotes non-small-cell lung cancer progression by
EZH2-mediated epigenetic silencing of PTEN expression. Cell Death
Dis. 11:2152020. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Yang Z, Lin X, Zhang P, Liu Y, Liu Z, Qian
B, Liu X and Shao G: Long non-coding RNA LINC00525 promotes the
non-small cell lung cancer progression by targeting miR-338-3p/IRS2
axis. Biomed Pharmacother. 124:1098582020. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Fang P, Chen H, Ma Z, Han C, Yin W, Wang
S, Zhu H, Xia W, Wang J, Xu L, et al: LncRNA LINC00525 suppresses
p21 expression via mRNA decay and triplex-mediated changes in
chromatin structure in lung adenocarcinoma. Cancer Commun (Lond).
41:596–614. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Yang M, He X, Huang X, Wang J, He Y and
Wei L: LncRNA MIR4435-2HG-mediated upregulation of TGF-β1 promotes
migration and proliferation of nonsmall cell lung cancer cells.
Environ Toxicol. 35:582–590. 2020. View Article : Google Scholar
|
|
53
|
Xu F, Hua Q, Zhang A, Di Z, Wang Y, Zhao
L, Yang H, Liu J and Huang G: LncRNA AC020978 facilitates non-small
cell lung cancer progression by interacting with malate
dehydrogenase 2 and activating the AKT pathway. Cancer Sci.
112:4501–4514. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Cai Y, Wu Q, Liu Y and Wang J: AZIN1-AS1,
A novel oncogenic LncRNA, promotes the progression of non-small
cell lung cancer by regulating MiR-513b-5p and DUSP11. Onco Targets
Ther. 13:9667–9678. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Wu S, Liu B, Zhang Y, Hong R, Liu S, Xiang
T, Tao T, Cai J, Wu J, Li M and Guan H: Long non-coding RNA LEISA
promotes progression of lung adenocarcinoma via enhancing
interaction between STAT3 and IL-6 promoter. Oncogene.
40:3449–3459. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Wang RQ, Long XR, Zhou NN, Chen DN, Zhang
MY, Wen ZS, Zhang LJ, He FZ, Zhou ZL, Mai SJ and Wang HY: Lnc-GAN1
expression is associated with good survival and suppresses tumor
progression by sponging mir-26a-5p to activate PTEN signaling in
non-small cell lung cancer. J Exp Clin Cancer Res. 40:92021.
View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Li C, Zhao W, Pan X, Li X, Yan F, Liu S,
Feng J and Lu J: LncRNA KTN1-AS1 promotes the progression of
non-small cell lung cancer via sponging of miR-130a-5p and
activation of PDPK1. Oncogene. 39:6157–6171. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Liu C, Li X, Hao Y, Wang F, Cheng Z, Geng
H and Geng D: STAT1-induced upregulation of lncRNA KTN1-AS1
predicts poor prognosis and facilitates non-small cell lung cancer
progression via miR-23b/DEPDC1 axis. Aging (Albany NY).
12:8680–8701. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Zhu Y, Li J, Bo H, He D, Xiao M, Xiang L,
Gong L, Hu Y, Zhang Y, Cheng Y, et al: LINC00467 is up-regulated by
TDG-mediated acetylation in non-small cell lung cancer and promotes
tumor progression. Oncogene. 39:6071–6084. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Yang J, Liu Y, Mai X, Lu S, Jin L and Tai
X: STAT1-induced upregulation of LINC00467 promotes the
proliferation migration of lung adenocarcinoma cells by
epigenetically silencing DKK1 to activate Wnt/β-catenin signaling
pathway. Biochem Biophys Res Commun. 514:118–126. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Chang Y and Yang L: LINC00467 promotes
cell proliferation and stemness in lung adenocarcinoma by sponging
miR-4779 and miR-7978. J Cell Biochem. 121:3691–3699. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Xue F, Yang C, Yun K, Jiang C, Cai R,
Liang M, Wang Q, Bian W, Zhou H, Liu Z and Zhu L: RETRACTED
ARTICLE: Reduced LINC00467 elevates microRNA-125a-3p to suppress
cisplatin resistance in non-small cell lung cancer through
inhibiting sirtuin 6 and inactivating the ERK1/2 signaling pathway.
Cell Biol Toxicol. 39:3652023. View Article : Google Scholar
|
|
63
|
Yin H, Chen L, Piao S, Wang Y, Li Z, Lin
Y, Tang X, Zhang H, Zhang H and Wang X: M6A RNA
methylation-mediated RMRP stability renders proliferation and
progression of non-small cell lung cancer through regulating
TGFBR1/SMAD2/SMAD3 pathway. Cell Death Differ. 30:605–617. 2023.
View Article : Google Scholar
|
|
64
|
Sun CC, Zhu W, Li SJ, Hu W, Zhang J, Zhuo
Y, Zhang H, Wang J, Zhang Y, Huang SX, et al: Correction to:
FOXC1-mediated LINC00301 facilitates tumor progression and triggers
an immune-suppressing microenvironment in non-small cell lung
cancer by regulating the HIF1α pathway. Genome Med. 13:252021.
View Article : Google Scholar
|
|
65
|
Shi L, Magee P, Fassan M, Sahoo S, Leong
HS, Lee D, Sellers R, Brullé-Soumaré L, Cairo S, Monteverde T, et
al: A KRAS-responsive long non-coding RNA controls microRNA
processing. Nat Commun. 12:20382021. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
La Montagna M, Shi L, Magee P, Sahoo S,
Fassan M and Garofalo M: AMPKα loss promotes KRAS-mediated lung
tumorigenesis. Cell Death Differ. 28:2673–2689. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Shi J, Yang C, An J, Hao D, Liu C, Liu J,
Sun J and Jiang J: KLF5-induced BBOX1-AS1 contributes to cell
malignant phenotypes in non-small cell lung cancer via sponging
miR-27a-5p to up-regulate MELK and activate FAK signaling pathway.
J Exp Clin Cancer Res. 40:1482021. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Ni J, Zhang X, Li J, Zheng Z, Zhang J,
Zhao W and Liu L: Tumour-derived exosomal lncRNA-SOX2OT promotes
bone metastasis of non-small cell lung cancer by targeting the
miRNA-194-5p/RAC1 signalling axis in osteoclasts. Cell Death Dis.
12:6622021. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Lv X, Lian Y, Liu Z, Xiao J, Zhang D and
Yin X: Exosomal long non-coding RNA LINC00662 promotes non-small
cell lung cancer progression by miR-320d/E2F1 axis. Aging (Albany
NY). 13:6010–6024. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Liu S, Zhan N, Gao C, Xu P, Wang H, Wang
S, Piao S and Jing S: Long noncoding RNA CBR3-AS1 mediates
tumorigenesis and radiosensitivity of non-small cell lung cancer
through redox and DNA repair by CBR3-AS1/miR-409-3p/SOD1 axis.
Cancer Lett. 526:1–11. 2022. View Article : Google Scholar
|
|
71
|
Wang C, Meng X, Zhou Y, Yu J, Li Q, Liao
Z, Gu Y, Han J, Linghu S, Jiao Z, et al: Long Noncoding RNA
CTD-2245E15.3 Promotes Anabolic Enzymes ACC1 and PC to support
non-small cell lung cancer growth. Cancer Res. 81:3509–3524. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Wang S, Wang T, Liu D and Kong H: LncRNA
MALAT1 aggravates the progression of non-small cell lung cancer by
stimulating the expression of COMMD8 via targeting miR-613. Cancer
Manag Res. 12:10735–10747. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Jin D, Guo J, Wu Y, Du J, Yang L, Wang X,
Di W, Hu B, An J, Kong L, et al: m6A mRNA methylation
initiated by METTL3 directly promotes YAP translation and increases
YAP activity by regulating the MALAT1-miR-1914-3p-YAP axis to
induce NSCLC drug resistance and metastasis. J Hematol Oncol.
12:1352019. View Article : Google Scholar
|
|
74
|
Jin S, He J, Zhou Y, Wu D, Li J and Gao W:
LncRNA FTX activates FOXA2 expression to inhibit non-small-cell
lung cancer proliferation and metastasis. J Cell Mol Med.
24:4839–4849. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Sun J, Xiong Y, Jiang K, Xin B, Jiang T,
Wei R, Zou Y, Tan H, Jiang T, Yang A, et al: Hypoxia-sensitive long
noncoding RNA CASC15 promotes lung tumorigenesis by regulating the
SOX4/β-catenin axis. J Exp Clin Cancer Res. 40:122021. View Article : Google Scholar
|
|
76
|
Fan H, Yuan J, Li Y, Jia Y, Li J, Wang X
and Li X: MKL1-induced lncRNA SNHG18 drives the growth and
metastasis of non-small cell lung cancer via the miR-211-5p/BRD4
axis. Cell Death Dis. 12:1282021. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Chen J, Liu A, Wang Z, Wang B, Chai X, Lu
W, Cao T, Li R, Wu M, Lu Z, et al: LINC00173.v1 promotes
angiogenesis and progression of lung squamous cell carcinoma by
sponging miR-511-5p to regulate VEGFA expression. Mol Cancer.
19:982020. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Xiao L, Li Y, Zeng X, Zhou Z, Hu S, Zhang
S, Zhou Y, Zhang Z, Zhao H, Zhao H, et al: Silencing of LOC389641
impairs cell proliferation and induces autophagy via EGFR/MET
signaling in lung adenocarcinoma. Aging (Albany NY). 13:2539–2552.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Cao G, Tan B, Wei S, Shen W, Wang X, Chu
Y, Rong T and Gao C: Down-regulation of MBNL1-AS1 contributes to
tumorigenesis of NSCLC via sponging miR-135a-5p. Biomed
Pharmacother. 125:1098562020. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Hua Q, Mi B, Xu F, Wen J, Zhao L, Liu J
and Huang G: Hypoxia-induced lncRNA-AC020978 promotes proliferation
and glycolytic metabolism of non-small cell lung cancer by
regulating PKM2/HIF-1α axis. Theranostics. 10:4762–4778. 2020.
View Article : Google Scholar :
|
|
81
|
Chen Q, Guo SM, Huang HQ, Huang GP, Li Y,
Li ZH, Huang R, Xiao L, Fan CR, Yuan Q and Zheng SL: Long noncoding
RNA SBF2-AS1 contributes to the growth and metastatic phenotypes of
NSCLC via regulating miR-338-3p/ADAM17 axis. Aging (Albany NY).
12:17902–17920. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Folkman J: What is the evidence that
tumors are angiogenesis dependent? J Natl Cancer Inst. 82:4–6.
1990. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Quinodoz SA, Jachowicz JW, Bhat P,
Ollikainen N, Banerjee AK, Goronzy IN, Blanco MR, Chovanec P, Chow
A, Markaki Y, et al: RNA promotes the formation of spatial
compartments in the nucleus. Cell. 184:5775–5790.e5730. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Markaki Y, Gan Chong J, Wang Y, Jacobson
EC, Luong C, Tan SYX, Jachowicz JW, Strehle M, Maestrini D,
Banerjee AK, et al: Xist nucleates local protein gradients to
propagate silencing across the X chromosome. Cell.
184:6174–6192.e6132. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Guo Z, Dai Y, Hu W, Zhang Y, Cao Z, Pei W,
Liu N, Nie J, Wu A, Mao W, et al: The long noncoding RNA CRYBG3
induces aneuploidy by interfering with spindle assembly checkpoint
via direct binding with Bub3. Oncogene. 40:1821–1835. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Tian B, Han X, Li G, Jiang H, Qi J, Li J,
Tian Y and Wang C: A Long intergenic non-coding RNA, LINC01426,
promotes cancer progression via AZGP1 and predicts poor prognosis
in patients with LUAD. Mol Ther Methods Clin Dev. 18:765–780. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Liu X, Yin Z, Xu L, Liu H, Jiang L, Liu S
and Sun X: Upregulation of LINC01426 promotes the progression and
stemness in lung adenocarcinoma by enhancing the level of SHH
protein to activate the hedgehog pathway. Cell Death Dis.
12:1732021. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Jia D, Xing Y, Zhan Y, Cao M, Tian F, Fan
W, Huang J, Cui Y, Gu R, Cui Y, et al: LINC02678 as a novel
prognostic marker promotes aggressive non-small-cell lung cancer.
Front Cell Dev Biol. 9:6869752021. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Chen Z, Chen X, Lei T, Gu Y, Gu J, Huang
J, Lu B, Yuan L, Sun M and Wang Z: Integrative analysis of NSCLC
identifies LINC01234 as an oncogenic lncRNA that interacts with
HNRNPA2B1 and regulates miR-106b biogenesis. Mol Ther.
28:1479–1493. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Chen Z, Chen X, Lu B, Gu Y, Chen Q, Lei T,
Nie F, Gu J, Huang J, Wei C, et al: Up-regulated LINC01234 promotes
non-small-cell lung cancer cell metastasis by activating VAV3 and
repressing BTG2 expression. J Hematol Oncol. 13:72020. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Zheng F, Li J, Ma C, Tang X, Tang Q, Wu J,
Chai X, Xie J, Yang XB and Hann SS: Novel regulation of miR-34a-5p
and HOTAIR by the combination of berberine and gefitinib leading to
inhibition of EMT in human lung cancer. J Cell Mol Med.
24:5578–5592. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Hua Q, Jin M, Mi B, Xu F, Li T, Zhao L,
Liu J and Huang G: LINC01123, a c-Myc-activated long non-coding
RNA, promotes proliferation and aerobic glycolysis of non-small
cell lung cancer through miR-199a-5p/c-Myc axis. J Hematol Oncol.
12:912019. View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Zhang M, Han Y, Zheng Y, Zhang Y, Zhao X,
Gao Z and Liu X: ZEB1-activated LINC01123 accelerates the
malignancy in lung adenocarcinoma through NOTCH signaling pathway.
Cell Death Dis. 11:9812020. View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Chen H, Liu H and Qing G: Targeting
oncogenic Myc as a strategy for cancer treatment. Signal Transduct
Target Ther. 3:52018. View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Gabay M, Li Y and Felsher DW: MYC
activation is a hallmark of cancer initiation and maintenance. Cold
Spring Harb Perspect Med. 4:a0142412014. View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Scafuro M, Capasso L, Carafa V, Altucci L
and Nebbioso A: Gene Transactivation and Transrepression in
MYC-Driven Cancers. Int J Mol Sci. 22:34582021. View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Vervoort SJ, van Boxtel R and Coffer PJ:
The role of SRY-related HMG box transcription factor 4 (SOX4) in
tumorigenesis and metastasis: friend or foe? Oncogene.
32:3397–3409. 2013. View Article : Google Scholar
|
|
98
|
Zhang J, Liang Q, Lei Y, Yao M, Li L, Gao
X, Feng J, Zhang Y, Gao H, Liu DX, et al: SOX4 induces
epithelial-mesenchymal transition and contributes to breast cancer
progression. Cancer Res. 72:4597–4608. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Sun CC, Zhu W, Li SJ, Hu W, Zhang J, Zhuo
Y, Zhang H, Wang J, Zhang Y, Huang SX, et al: FOXC1-mediated
LINC00301 facilitates tumor progression and triggers an
immune-suppressing microenvironment in non-small cell lung cancer
by regulating the HIF1α pathway. Genome Med. 12:772020. View Article : Google Scholar
|
|
100
|
Lu Z, Fang Z, Guo Y, Liu X and Chen S:
Cisplatin resistance of NSCLC cells involves upregulation of
visfatin through activation of its transcription and stabilization
of mRNA. Chem Biol Interact. 351:1097052022. View Article : Google Scholar
|
|
101
|
Błach J, Wojas-Krawczyk K, Nicoś M and
Krawczyk P: Failure of immunotherapy-the molecular and
immunological origin of immunotherapy resistance in lung cancer.
Int J Mol Sci. 22:90302021. View Article : Google Scholar
|
|
102
|
Wang CJ, Zhu CC, Xu J, Wang M, Zhao WY,
Liu Q, Zhao G and Zhang ZZ: The lncRNA UCA1 promotes proliferation,
migration, immune escape and inhibits apoptosis in gastric cancer
by sponging anti-tumor miRNAs. Mol Cancer. 18:1152019. View Article : Google Scholar : PubMed/NCBI
|
|
103
|
Wei MM and Zhou GB: Long Non-coding RNAs
and their roles in non-small-cell lung cancer. Genomics Proteomics
Bioinformatics. 14:280–288. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
104
|
Huang J, Pan B, Xia G, Zhu J, Li C and
Feng J: LncRNA SNHG15 regulates EGFR-TKI acquired resistance in
lung adenocarcinoma through sponging miR-451 to upregulate MDR-1.
Cell Death Dis. 11:5252020. View Article : Google Scholar : PubMed/NCBI
|
|
105
|
Chen Z, Chen Q, Cheng Z, Gu J, Feng W, Lei
T, Huang J, Pu J, Chen X and Wang Z: Long non-coding RNA CASC9
promotes gefitinib resistance in NSCLC by epigenetic repression of
DUSP1. Cell Death Dis. 11:8582020. View Article : Google Scholar : PubMed/NCBI
|
|
106
|
Bing Z, Han J, Zheng Z and Liang N:
FOXO3-induced oncogenic lncRNA CASC9 enhances gefitinib resistance
of non-small-cell lung cancer through feedback loop. Life Sci.
287:1200122021. View Article : Google Scholar : PubMed/NCBI
|
|
107
|
Li Z, Niu H, Qin Q, Yang S, Wang Q, Yu C,
Wei Z, Jin Z, Wang X, Yang A and Chen X: lncRNA UCA1 mediates
resistance to cisplatin by regulating the miR-143/FOSL2-Signaling
pathway in ovarian cancer. Mol Ther Nucleic Acids. 17:92–101. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
108
|
Li C, Fan K, Qu Y, Zhai W, Huang A, Sun X
and Xing S: Deregulation of UCA1 expression may be involved in the
development of chemoresistance to cisplatin in the treatment of
non-small-cell lung cancer via regulating the signaling pathway of
microRNA-495/NRF2. J Cell Physiol. 235:3721–3730. 2020. View Article : Google Scholar
|
|
109
|
Xu T, Yan S, Wang M, Jiang L, Ma P, Lu B,
Chen Q, Wei C and Wang Z: LncRNA UCA1 induces acquired resistance
to gefitinib by epigenetically silencing cdkn1a expression in
non-small-cell lung cancer. Front Oncol. 10:6562020. View Article : Google Scholar : PubMed/NCBI
|
|
110
|
Wang H, Lu B, Ren S, Wu F, Wang X, Yan C
and Wang Z: Long Noncoding RNA LINC01116 contributes to gefitinib
resistance in non-small cell lung cancer through regulating IFI44.
Mol Ther Nucleic Acids. 19:218–227. 2020. View Article : Google Scholar
|
|
111
|
Fu J, Cai H, Wu Y, Fang S and Wang D:
Elevation of FGD5-AS1 contributes to cell progression by improving
cisplatin resistance against non-small cell lung cancer cells
through regulating miR-140-5p/WEE1 axis. Gene. 755:1448862020.
View Article : Google Scholar : PubMed/NCBI
|
|
112
|
He H, Song X, Yang Z, Mao Y, Zhang K, Wang
Y, Su B, Li Q, Chen H and Li Y: Upregulation of KCNQ1OT1 promotes
resistance to stereotactic body radiotherapy in lung adenocarcinoma
by inducing ATG5/ATG12-mediated autophagy via miR-372-3p. Cell
Death Dis. 11:8832020. View Article : Google Scholar : PubMed/NCBI
|
|
113
|
Dong Z, Yang P, Qiu X, Liang S, Guan B,
Yang H, Li F, Sun L, Liu H, Zou G and Zhao K: KCNQ1OT1 facilitates
progression of non-small-cell lung carcinoma via modulating
miRNA-27b-3p/HSP90AA1 axis. J Cell Physiol. 234:11304–11314. 2019.
View Article : Google Scholar
|
|
114
|
Shu D, Xu Y and Chen W: Knockdown of
lncRNA BLACAT1 reverses the resistance of afatinib to non-small
cell lung cancer via modulating STAT3 signalling. J Drug Target.
28:300–306. 2020. View Article : Google Scholar
|
|
115
|
Ju ZS, Sun B, Bao D and Zhang XF: Effect
of lncRNA-BLACAT1 on drug resistance of non-small cell lung cancer
cells in DDP chemotherapy by regulating cyclin D1 expression. Eur
Rev Med Pharmacol Sci. 24:9465–9472. 2020.PubMed/NCBI
|
|
116
|
Zeng Z, Zhao G, Zhu H, Nie L, He L, Liu J,
Li R, Xiao S and Hua G: LncRNA FOXD3-AS1 promoted chemo-resistance
of NSCLC cells via directly acting on miR-127-3p/MDM2 axis. Cancer
Cell Int. 20:3502020. View Article : Google Scholar : PubMed/NCBI
|
|
117
|
Yang D, Feng W, Zhuang Y, Liu J, Feng Z,
Xu T, Wang W, Zhu Y and Wang Z: Long non-coding RNA linc00665
inhibits CDKN1C expression by binding to EZH2 and affects cisplatin
sensitivity of NSCLC cells. Mol Ther Nucleic Acids. 23:1053–1065.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
118
|
Yu Z, Wang G, Zhang C, Liu Y, Chen W, Wang
H and Liu H: LncRNA SBF2-AS1 affects the radiosensitivity of
non-small cell lung cancer via modulating microRNA-302a/MBNL3 axis.
Cell Cycle. 19:300–316. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
119
|
Boras B, Jones RM, Anson BJ, Arenson D,
Aschenbrenner L, Bakowski MA, Beutler N, Binder J, Chen E, Eng H,
et al: Discovery of a Novel Inhibitor of Coronavirus 3CL Protease
for the Potential Treatment of COVID-19. bioRxiv. 2021.
|
|
120
|
Gaston J, Cheradame L, Yvonnet V, Deas O,
Poupon MF, Judde JG, Cairo S and Goffin V: Intracellular STING
inactivation sensitizes breast cancer cells to genotoxic agents.
Oncotarget. 7:77205–77224. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
121
|
Duarte CW, Willey CD, Zhi D, Cui X, Harris
JJ, Vaughan LK, Mehta T, McCubrey RO, Khodarev NN, Weichselbaum RR
and Gillespie GY: Expression signature of IFN/STAT1 signaling genes
predicts poor survival outcome in glioblastoma multiforme in a
subtype-specific manner. PLoS One. 7:e296532012. View Article : Google Scholar : PubMed/NCBI
|
|
122
|
Wang J, Gao J, Chen Q, Zou W, Yang F, Wei
C and Wang Z: LncRNA LINC01116 contributes to cisplatin resistance
in lung adenocarcinoma. Onco Targets Ther. 13:9333–9348. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
123
|
Geng W, Lv Z, Fan J, Xu J, Mao K, Yin Z,
Qing W and Jin Y: Identification of the prognostic significance of
somatic Mutation-Derived LncRNA signatures of genomic instability
in lung adenocarcinoma. Front Cell Dev Biol. 9:6576672021.
View Article : Google Scholar : PubMed/NCBI
|
|
124
|
Zeng L, Lyu X, Yuan J, Wang W, Zhao N, Liu
B, Sun R, Meng X and Yang S: Long non-coding RNA LINC01116 is
overexpressed in lung adenocarcinoma and promotes tumor
proliferation and metastasis. Am J Transl Res. 12:4302–4313.
2020.PubMed/NCBI
|
|
125
|
Shang B, Li Z, Li M, Jiang S, Feng Z, Cao
Z and Wang H: Silencing LINC01116 suppresses the development of
lung adenocarcinoma via the AKT signaling pathway. Thorac Cancer.
12:2093–2103. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
126
|
Yao J, Chen X, Liu X, Li R, Zhou X and Qu
Y: Characterization of a ferroptosis and iron-metabolism related
lncRNA signature in lung adenocarcinoma. Cancer Cell Int.
21:3402021. View Article : Google Scholar : PubMed/NCBI
|
|
127
|
Faivre-Finn C, Vicente D, Kurata T,
Planchard D, Paz-Ares L, Vansteenkiste JF, Spigel DR, Garassino MC,
Reck M, Senan S, et al: Four-year survival with durvalumab after
chemoradiotherapy in stage III NSCLC-an update from the PACIFIC
trial. J Thorac Oncol. 16:860–867. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
128
|
Chen Y, Gao M, Huang Z, Yu J and Meng X:
SBRT combined with PD-1/PD-L1 inhibitors in NSCLC treatment: A
focus on the mechanisms, advances, and future challenges. J Hematol
Oncol. 13:1052020. View Article : Google Scholar : PubMed/NCBI
|
|
129
|
Nickoloff JA: Toward greater precision in
cancer radiotherapy. Cancer Res. 81:3156–3157. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
130
|
Tu X, Qin B, Zhang Y, Zhang C, Kahila M,
Nowsheen S, Yin P, Yuan J, Pei H, Li H, et al: PD-L1 (B7-H1)
Competes with the RNA exosome to regulate the DNA damage response
and can be targeted to sensitize to radiation or chemotherapy. Mol
Cell. 74:1215–1226.e4. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
131
|
Tchelebi LT, Eng C, Messick CA, Hong TS,
Ludmir EB, Kachnic LA and Zaorsky NG: Current treatment and future
directions in the management of anal cancer. CA Cancer J Clin.
72:183–195. 2022. View Article : Google Scholar
|
|
132
|
Barcena-Varela M and Lujambio A: A novel
long noncoding RNA finetunes the DNA damage response in
hepatocellular carcinoma. Cancer Res. 81:4899–4900. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
133
|
Liu JY, Chen YJ, Feng HH, Chen ZL, Wang
YL, Yang JE and Zhuang SM: LncRNA SNHG17 interacts with LRPPRC to
stabilize c-Myc protein and promote G1/S transition and cell
proliferation. Cell Death Dis. 12:9702021. View Article : Google Scholar : PubMed/NCBI
|
|
134
|
Wang Y, Zhang L, Yang J and Sun R: LncRNA
KCNQ1OT1 promotes cell proliferation, migration and invasion via
regulating miR-129-5p/JAG1 axis in non-small cell lung cancer.
Cancer Cell Int. 20:1442020. View Article : Google Scholar : PubMed/NCBI
|
|
135
|
Kang Y, Jia Y, Wang Q, Zhao Q, Song M, Ni
R and Wang J: Long Noncoding RNA KCNQ1OT1 promotes the progression
of non-small cell lung cancer via regulating miR-204-5p/ATG3 Axis.
Onco Targets Ther. 12:10787–10797. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
136
|
Dumanski JP, Halvardson J, Davies H,
Rychlicka-Buniowska E, Mattisson J, Moghadam BT, Nagy N, Węglarczyk
K, Bukowska-Strakova K, Danielsson M, et al: Immune cells lacking Y
chromosome show dysregulation of autosomal gene expression. Cell
Mol Life Sci. 78:4019–4033. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
137
|
Lai IL, Chang YS, Chan WL, Lee YT, Yen JC,
Yang CA, Hung SY and Chang JG: Male-specific long noncoding RNA
TTTY15 inhibits non-small cell lung cancer proliferation and
metastasis via TBX4. Int J Mol Sci. 20:34732019. View Article : Google Scholar : PubMed/NCBI
|
|
138
|
Yang CJ, Kumar A, Deng JZ, Raman V, Lui
NS, D'Amico TA and Berry MF: A National analysis of short-term
outcomes and long-term survival following thoracoscopic versus open
lobectomy for clinical Stage II Non-Small-Cell lung cancer. Ann
Surg. 273:595–605. 2021. View Article : Google Scholar
|
|
139
|
Zhang SB, Hong M, Sun XY, Huang D, He DH,
Chen YF, Yuan Y and Liu YQ: Silybin has therapeutic efficacy
against non-small cell lung cancer through targeting of Skp2. Acta
Materia Medica. 1:302–313. 2022. View Article : Google Scholar
|
|
140
|
Jiang N, Meng X, Mi H, Chi Y, Li S, Jin Z,
Tian H, He J, Shen W, Tian H, et al: Circulating lncRNA XLOC_009167
serves as a diagnostic biomarker to predict lung cancer. Clin Chim
Acta. 486:26–33. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
141
|
Zhu Z, Wang H, Pang Y, Hu H, Zhang H and
Wang W: Exosomal long non-coding RNA UCA1 functions as growth
inhibitor in esophageal cancer. Aging (Albany NY). 12:20523–20539.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
142
|
Yu AM and Tu MJ: Deliver the promise: RNAs
as a new class of molecular entities for therapy and vaccination.
Pharmacol Ther. 230:1079672022. View Article : Google Scholar :
|